Abstract
BackgroundGenetic determinants of susceptibility to severe acute respiratory syndrome coronavirus (SARS-CoV) infection remain unknown. We assessed whether mannose-binding lectin (MBL) gene polymorphisms were associated with susceptibility to SARS-CoV infection or disease severity in an ethnically homogeneous population born in northern China
MethodsThe frequencies of 1 mutation in codon 54 and 3 promoter polymorphisms at nt −550, −221, and 4 were ascertained in 352 patients with SARS and 392 control subjects, by means of polymerase chain reaction direct sequencing
ResultsOf 352 patients with SARS and 392 control subjects, 120 (34.4%) and 91 (23.2%) were carriers of the codon 54 variant, respectively (odds ratio [OR], 1.73 [95% confidence interval {CI}, 1.25–2.39]; P=.00086). A total of 123 (36.0%) of 352 patients with SARS and 100 (25.5%) of 392 control subjects had haplotype pairs associated with medium or low expression of MBL (OR, 1.67 [95% CI, 1.21–2.29]; P=.00187). The population-attributable fraction of patients with SARS that was associated with having the codon 54 variant was 20.1% (95% CI, 7.9%–32.3%)
Conclusions MBL gene polymorphisms were significantly associated with susceptibility to SARS-CoV infection; this might be explained by the reduced expression of functional MBL secondary to having the codon 54 variant
Severe acute respiratory syndrome (SARS) is a newly identified human infectious disease that is caused by a novel coronavirus (CoV), SARS-CoV [1–3]. By late July 2003, >8000 cases of SARS and >700 SARS-related deaths were reported from >25 countries worldwide. Its rapid transmission, high infectivity, unpredictable clinical progression, and high mortality make SARS a potential global threat [3], but the pathogenesis of SARS is still not fully understood. Mapping the genetic variants that account for disease risk can identify disease pathways in complex disorders. Thus, a hope to resolve the etiology of SARS has prompted a search for genetic variants in candidate susceptibility genes. Individual susceptibility to SARS-CoV infection seems to be variable. For example, subjects seropositive for SARS-CoV may develop SARS or have only mildly symptomatic infections [4]. Like that of other major infectious diseases [5, 6], the development of SARS may also be the result of a complex interaction between the microbe, host genetic factors, and the environment. Early reports have shown that higher age, diabetes mellitus, and heart disease are risk factors for the development of SARS [7, 8], but the extent to which genetic factors might influence susceptibility to SARS-CoV infection remains unknown
Variation in host immunity has long been regarded as an important factor in determining susceptibility to infectious diseases. Indeed, recent case-control studies have suggested an association between the development of SARS and HLA-B*4601, HLA-B*0703, and HLA-DRB1*0301 [9, 10]. Although the adaptive immune response is a highly effective defense mechanism against SARS-CoV, a potential weakness is the time delay between the recognition of a new pathogen and the production of sufficient antibodies to provide protection. Specific antibodies can be detected ⩾10 days after the onset of symptoms (mean time, 20 days) [2]. Thus, other factors, in addition to antibodies, are likely to determine the differences in susceptibility during this vulnerable period. In addition to the adaptive immune response, there exists an innate immune response that involves a series of soluble and membrane-bound proteins that bind and facilitate the killing and clearance of invading microbes [11, 12]. This innate immune response is present from birth and provides the first line of defense for the human body. The variability in the response to SARS-CoV suggests that the innate immune response might be involved in an individual’s susceptibility to SARS-CoV infection during the period before the production of specific antibodies
Mannose-binding lectin (MBL) is a serum protein belonging to the family of collectin and plays a critical role in the innate immune response [13–15]. MBL is an acute-phase reactant of hepatic origin that can bind through multiple lectin domains to repeating mannose and N-acetylglucosamine sugar motifs that are characteristically displayed at high densities on bacterial and fungal cells and on viruses and protozoa but not on mammalian cells [16, 17]. After binding to a pathogen, MBL initiates at least 2 protective functions that are well defined. First, through the lectin pathway, MBL can mediate the activation of the complement system without the participation of antibodies; second, MBL can promote opsonophagocytosis by collectin receptors directly [18–20]. Experiments in vitro and in vivo have shown that an MBL deficiency is likely to have a major effect on innate immune activation and appears to predispose individuals to serious infection [21, 22]
The amounts of MBL in human plasma are genetically determined. Three polymorphisms, encoding for structurally variant proteins, have been identified in codons 52, 54, and 57 (denoted as variants D, B, and C, respectively; the wild type is denoted as A) in the MBL gene. Individuals with any heterozygous variant have only ∼10% of the MBL plasma concentrations that those with the wild type have, and those with homozygous or compound heterozygous variants have almost undetectable MBL plasma concentrations [23, 24]. In addition, polymorphisms at nt −550, −221, and 4 (referred to as the H/L, Y/X, and P/Q variants, respectively) in the promoter region, which are reported to control MBL plasma concentrations of structurally normal proteins, have also been described [25, 26]. Moreover, the combination of structural polymorphisms and promoter polymorphisms in this gene results in well-described MBL expression groups (high, medium, and low) [25, 26]. Variant alleles causing low plasma concentrations of functional MBL have been shown to be associated in both children and adults with an increased risk of developing infections [19, 27–29]. The aim of the present study was to assess whether the variant MBL alleles, which cause defects in the activation of the innate immune system, are associated with susceptibility to SARS-CoV infection or disease severity
Patients, Materials, and Methods
Case patients and control subjectsIn total, 352 case patients who were serologically confirmed as having SARS and whose conditions fulfilled the World Health Organization case definition of SARS were recruited from the hospitalization wards at Xiaotangshan Hospital (Beijing, China) between May and July 2003 [30]. The clinical profiles and backgrounds of all subjects were extracted from medical records. Of the 352 case patients, 52 (patient group 1 [P1]) had comorbid conditions (including diabetes mellitus, hypertension, heart diseases, tuberculosis, asthma, and malignancy), which have been shown to be risk factors for the development of SARS [7, 8]. Of the 300 case patients without comorbid conditions, 20 were patients with severe SARS (patient group 2 [P2]), who were identified by their admissions to intensive care units or deaths, and the remaining 280 were patients with mildly symptomatic SARS (patient group 3 [P3]). By checking the medical records or by interviewing the case patients or their guardians, we determined that all case patients were genetically unrelated adults born in northern China. For the 352 patients, the male/female ratio was 1.2, and the mean age was 34.9 years (range, 15–77 years). A total of 392 age-, sex-, and ethnicity-matched healthy genetically unrelated adults born in northern China were recruited as control subjects; the male/female ratio was 1.5, and the mean age was 32.7 years (range, 17–68 years). Written, informed consent was obtained from all the participants or their guardians, and the study was performed with the approval of the Ethical Committee of the Chinese Human Genome Center. Genomic DNA was extracted from 5-mL peripheral blood samples using standard phenol-chloroform protocols. DNA samples were diluted to a concentration of 10 ng/μL and were distributed in 96-well plates
Validation of MBL variantsTwo pairs of primer sets covering the genomic sequence of exon 1 and the promoter region of the MBL gene, which spans ∼1.1 kb (from nt −730 to 375; GenBank accession no. AF080508.1), were designed on the basis of size and overlap of polymerase chain reaction (PCR) amplicons. The screening panel included DNA from 174 individuals randomly selected, without regard to disease status, from the total study population of 744 individuals. The primers for the target regions were designed using the Web-based software Primer3 (available at: http://www-genome.wi.mit.edu/). DNA samples from the 174 individuals were amplified and purified. Then the PCR products were sequenced using an ABI PRISM Dye Terminator Sequencing Kit with Amplitaq DNA polymerase (ABI) and loaded onto an ABI 3730 sequencer. Polymorphism candidates were identified by the PolyPhred program (available at: http://droog.mbt.washington.edu/PolyPhred.html) and were inspected by 2 observers. Polymorphism positions and individual genotypes were confirmed by reamplifying and resequencing the polymorphism sites from the opposite strand. The primers are available on request
GenotypingThe promoter polymorphisms at nt −550, −221, and 4 (i.e., the H/L, Y/X, and P/Q variants, respectively) and the structural polymorphism at nt 230 (i.e., the codon 54 A/B variant) were selected for genotyping by use of PCR direct sequencing in the case-control population. The primers for PCR and sequencing and the reaction parameters were identical with those used for the polymorphism validation procedure mentioned above. Genotyping of all 744 samples was performed in a blinded manner so that the case or control status was unknown. The accuracy of genotyping data for each polymorphism was validated by masking, choosing at random, and resequencing 15% of the samples from case patients and control subjects
Statistical analysisThe haplotypes of the MBL genes from the samples were assigned by the PHASE program [31]. PHASE is an implementation of the Stephens-Donnelly method of haplotype reconstruction, which uses a Bayesian approach that incorporates a priori expectations of haplotypic structure from population genetic and coalescent theory. The pairwise linkage disequilibrium (LD) calculation (Lewontin’s D′ and r2) was performed using Arlequin software (available at: http://lgb.unige.ch/arlequin/)
Genotype and allele frequencies for each polymorphism were determined by gene counting. The significance of deviations from the Hardy-Weinberg equilibrium was tested using the random-permutation procedure implemented in Arlequin. Unconditional logistic regression analysis was performed to evaluate whether there was an association between specific MBL alleles and susceptibility to SARS-CoV infection or disease severity after adjustment for age and sex, and the P values, odds ratios (ORs), and 95% confidence intervals (CIs) were calculated. In view of the multiple comparisons in the case-control study, the correction factor n(m-1) (in which there are n loci with m alleles each) was applied to correct the significance level. These analyses were performed using SPSS software (version 9.0; SPSS)
Results
Resequencing of the ∼1.1-kb genomic region in the MBL gene in 174 individuals revealed 12 polymorphisms (table 1). Consistent with the findings of studies published elsewhere [24, 32], the codon 52 and 57 mutations were not detected in our study population of individuals born in northern China. The frequencies of the variant alleles H/L, Y/X, and P/Q measured here—that is, 0.445, 0.141, and 0.144, respectively—are very similar to those reported in other Chinese subpopulations [24, 32]. Seven polymorphisms (i.e., −427A→C, −349A→G, −336A→G, 6-bp insertion or deletion at nt −329 to −324, −70C→T, P/Q, and 366G→A) were in absolute LD in the study population (D′=1 and r2=1), suggesting that any 1 of them can represent the other 6 polymorphisms as a marker, and, in the present study, P/Q was chosen for its influence on functional MBL serum concentration [25, 26]. We therefore selected the variants H/L, Y/X, P/Q, and A/B as markers for use in the subsequent genotyping analysis
Table 1.
Positions and frequencies of polymorphisms screened from the mannose-binding lectin (MBL) gene
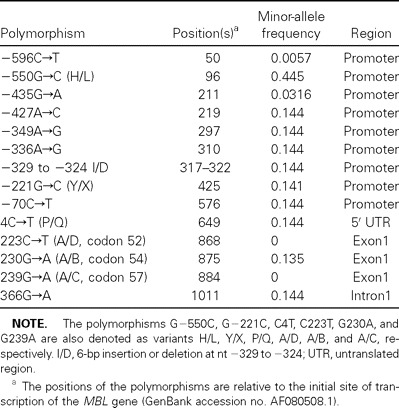
Table 2 indicates that the genotype distributions of the 4 mutations in any patient category or in control subjects were in Hardy-Weinberg equilibrium. On the basis of unconditional logistic regression analysis with adjustments for age and sex, a statistically significant association was observed between susceptibility to SARS-CoV infection and having the codon 54 variant. Subjects that were homozygous or heterozygous for the variant B allele (i.e., the B/B or the A/B genotype) had an increased susceptibility to SARS-CoV infection, compared with those homozygous for the wild-type A allele (P1, P2, and P3 vs. control subjects, OR, 1.73 [95% CI, 1.25–2.39]; P=.00086) (table 2). After stratification by comorbid conditions, the association was pronounced only in patients without comorbid conditions (P2 and P3 vs. control subjects, OR, 1.84 [95% CI, 1.32–2.57]; P=.00031) (table 2). The association remained significant even after correction for multiple comparisons. The nt −221 promoter variant showed only a marginally significant association with susceptibility to SARS-CoV infection, but this type of association was not observed after correction for multiple testing. Neither variant H/L nor variant P/Q was found to be significantly associated with susceptibility to SARS-CoV infection
Table 2.
The distribution of mannose-binding lectin (MBL) genotype and haplotype pairs associated with various levels of MBL expression in patients with severe acute respiratory syndrome (SARS) and control subjects
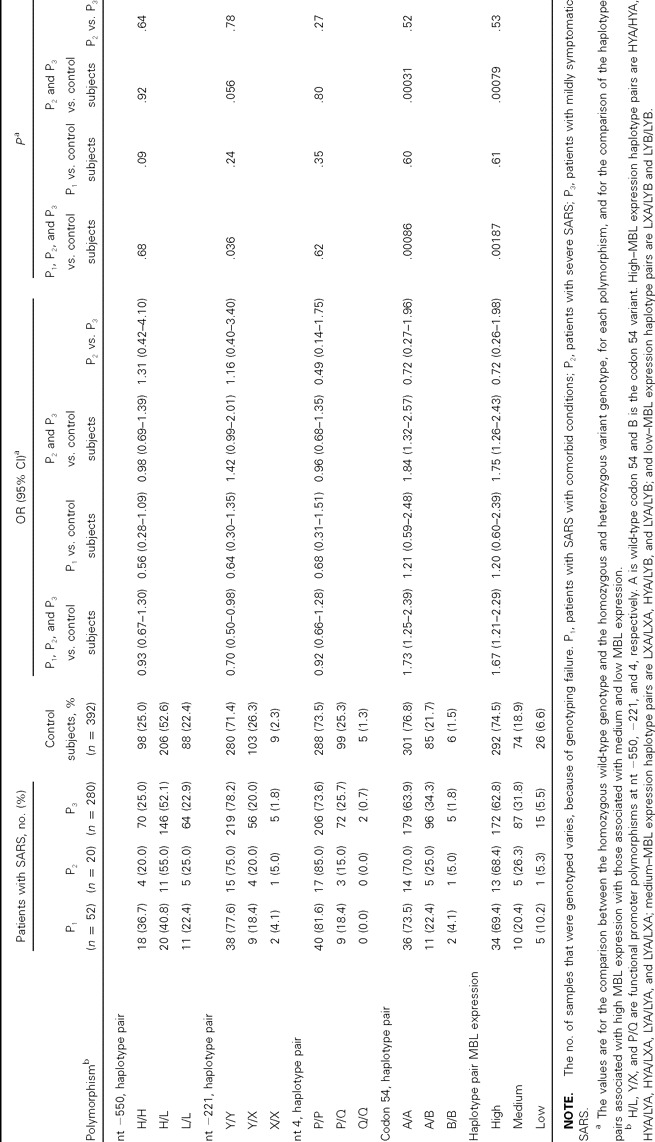
The combination of structural mutations and promoter polymorphisms in the MBL gene results in well-described MBL expression groups (high, medium, and low) [25]. We assigned all case patients and control subjects to groups on the basis of having haplotypes associated with high, medium, or low MBL expression, to assess whether haplotype pairs associated with various levels of MBL expression were associated with susceptibility to SARS-CoV infection. In 352 case patients, 219 (64.0%) were in the high–MBL expression group, 102 (29.8%) were in the medium–MBL expression group, and 21 (6.1%) were in the low–MBL expression group. In 392 control subjects, this distribution was 292 (74.5%), 74 (18.9%), and 26 (6.6%) control subjects, respectively (OR, 1.67 [95% CI, 1.21–2.29]; P = .00187, with adjustment for age and sex) (table 2). Similarly, the association remained significant only in patients without comorbid conditions (P2 and P3 vs. control subjects, OR, 1.75 [95% CI, 1.26–2.43]; P=.00079) (table 2)
The frequency distribution of the alleles and genotypes of the 4 polymorphisms among 20 patients with severe SARS (P2) and the remaining 280 case patients (P3) was also analyzed, but there was no significant difference. The distribution of haplotype pairs associated with various levels of MBL expression did not show any association with the severity of SARS when age and sex were accounted for (table 2)
Discussion
This study has shown that, in a population of individuals born in northern China, having the codon 54 variant in the MBL gene is significantly associated with susceptibility to SARS-CoV infection but not with disease severity. To our knowledge, this is the first report of an association between the MBL gene and susceptibility to SARS-CoV infection. This indicates that the MBL gene may play a role in the pathogenesis of this disease
Nongenetic factors can confound the contribution of genetic factors to the development of disease. Indeed, early reports have shown that some nongenetic factors, such as comorbid conditions, are risk factors for the development of SARS [7, 8]. However, after stratification by comorbid conditions, the significant association between having the codon 54 variant and susceptibility to SARS-CoV infection was pronounced only in patients without comorbid conditions, which indicates that these MBL polymorphisms exerted an independent effect on the risk of developing SARS
The MBL gene encodes a homotrimeric molecule with a carbohydrate recognition domain and a collagenous tail [33]. Formation of a triple helix in the collagenous tail is impaired by the variant B allele, and this impaired helix formation disrupts polymerization and leads to enzymatic degradation and functional deficiency of MBL [24]. The nt −221 promoter variant X allele has a smaller but detectable effect on serum concentrations of functional MBL, and its interplay with structural mutations results in a more substantial decrease in the expression of MBL [25]. The individuals bearing medium- or low-expression haplotype pairs had an increased risk of developing SARS (table 2), which suggests that MBL deficiency possibly plays a role in susceptibility to SARS-CoV infection
Our observed genetic associations are plausible from a biological perspective, although the mechanism by which MBL deficiency is associated with increased susceptibility to SARS-CoV infection requires investigation. Various studies have shown that MBL binds to clinically relevant pathogens, including HIV-1 and influenza virus [16, 17]. Recently, it has been reported that, in chickens, MBL can take part in the neutralization of infectious bronchitis virus (IBV), a member of the CoV family, before the humoral antibody response takes over [34]. The S protein of SARS-CoV, which plays an important role in infection, is also glycosylated with high mannose or hybrid oligosaccharides [35–37]. Thus, it is very likely that MBL can bind to the S protein of SARS-CoV and exert its role as an antivirus, although additional studies are needed to clarify this possibility
MBL can act directly as an opsonin, by binding to carbohydrates on pathogens and then interacting with MBL receptors on phagocytic cells. Additionally, it can also trigger the opsonic activity of complement that might lead to the deposition of C3b or iC3b on targets and stimulate phagocytic uptake via complement receptors [18–20]. One speculative interpretation of our results is that low serum concentrations of functional MBL caused by the variant B allele might result in reduced opsonization and favor the survival of SARS-CoV
In reviewing the results of the present study, however, one must keep a potential limit in mind. We selected the control subjects from the general population and not from those who spontaneously recovered from SARS-CoV infection (i.e., those who were exposed to SARS-CoV but did not develop SARS, who are very difficult to identify at present). This results in a type II error (a false-negative error). Because the unexposed individuals in the general population remain at risk for the development of SARS, the inclusion of such a control group limits the ability to obtain positive results when comparing contributions of polymorphisms in control subjects with those in case patients. Thus, from this point of view, it seems unlikely that the association between having the codon 54 variant and susceptibility to SARS-CoV infection is a false-positive error (i.e., it is unlikely that there was a type I error)
Several association studies have shown that, in both children and adults, the MBL gene is related to the susceptibility to various specific infections [19, 27–29]. Most of the results, however, could not be replicated in subsequent studies in other populations because of multiple factors, such as small sample size, marginal statistical significance, genotyping complexity, or ethnic heterogeneity in the study groups. Although the highly significant association between MBL deficiency and increased susceptibility to SARS-CoV infection derives from a biologically based a priori hypothesis, our initial finding should be independently verified in other subpopulations of ethnic Chinese origin (e.g., Guangdong, Taiwanese, or Hong Kong Chinese) or in those of different ancestry, such as whites
In the present study, we did not observe an association between MBL polymorphisms and SARS disease severity. This result may be due to the limited number of patients with severe SARS in the present study; it certainly warrants confirmation in future studies
The observation that MBL mutations are associated with increased susceptibility to SARS-CoV infection may have implications for transmission precautions and clinical practices for this disease. First, individuals who harbor the susceptibility genotype in the MBL gene are at high risk for developing SARS. The usefulness of general screenings to detect MBL mutations and MBL deficiency in a defense against future SARS outbreaks should be determined. Second, vaccines against SARS are expected to exert a greater influence in the future control of SARS. Our results may help in the selection of candidates for vaccination and in the confirmation of effective vaccines for high-risk groups. Third, replacement MBL therapy has been used in patients with repeated infections, and a possible beneficial effect was seen [38]. The usefulness of this intervention in SARS might merit assessment. Finally, and perhaps most importantly, this study helps to explain why, in most settings, the attack rate for SARS is much lower than that for most respiratory viruses. In fact, in the present study, the frequencies of the genotype and haplotype pairs conferring protection from SARS-CoV infection were very high. For example, the frequencies of the codon 54 A/A genotype and high–MBL expression haplotype pairs reached 76.8% and 74.5%, respectively (table 2)
If being homozygous or heterozygous for the codon 54 variant (i.e., having the B/B or A/B genotype) is regarded as a risk factor for the development of SARS, then the population-attributable fraction associated with this genetic risk factor—the parameter that combines the strength of the epidemiological influence (relative risk) and the frequency of the genotype (population exposure rate)—can be estimated by the formula AF = f(RR−1)/[1 + f(RR−1)], where AF is the population-attributable fraction, f is the population exposure rate, and RR is the relative risk [39]. The AF calculated by the RR (OR, 1.73 [95% CI, 1.25–2.39]) (table 2) combined with the frequency of the A/B and B/B genotypes (34.4%) (table 2) indicates that 20.1% (95% CI, 7.9%–32.3%) of the increase in the risk of developing SARS can be attributed to the variant B allele. If this association is real, then the variant B allele is associated with a relatively low fraction of SARS-CoV infection in the Chinese individuals studied here, thereby suggesting that other genes are likely to modify the susceptibility to such an infection. When many susceptibility genes are identified and gene-gene interactions among polymorphisms in these genes are taken into account, the prediction of the susceptibility to SARS-CoV infection may become more accurate
In summary, our results reveal, for the first time, an association, between MBL mutations and increased susceptibility to SARS-CoV infection and provide support for the importance of MBL in the pathogenesis of SARS. The knowledge of genetic factors contributing to the pathogenesis of SARS as presented here could lead to improvements in treatment and prevention of this disease
Acknowledgment
We thank Emma Snyder, for helpful comments on the manuscript
Footnotes
Potential conflicts of interest: none reported
Financial support: Chinese High-Tech Program (grants 2001AA224011 and 2002BA711A10); Medicine and Health Research Program (grant 01Z018); Chinese National Science Fund for Creative Research Groups (grant 30321003)
References
- 1.Rota PA, Oberste MS, Monroe SS, et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 300:1394–9. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 2.Peiris JS, Lai ST, Poon LL, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 361:1319–25. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee N, Hui D, Wu A, et al. A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med. 348:1986–94. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- 4.Ho KY, Singh KS, Habib AG, et al. Mild illness associated with severe acute respiratory syndrome coronavirus infection: lessons from a prospective seroepidemiologic study of health-care workers in a teaching hospital in Singapore. J Infect Dis. 189:642–7. doi: 10.1086/381558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu CJ, Lok AS. Clinical significance of hepatitis B virus genotypes. Hepatology. 35:1274–6. doi: 10.1053/jhep.2002.33161. [DOI] [PubMed] [Google Scholar]
- 6.Segal S, Hill AV. Genetic susceptibility to infectious disease. Trends Microbiol. 11:445–8. doi: 10.1016/s0966-842x(03)00207-5. [DOI] [PubMed] [Google Scholar]
- 7.Booth CM, Matukas LM, Tomlinson GA, et al. Clinical features and short-term outcomes of 144 patients with SARS in the greater Toronto area. JAMA. 289:2801–9. doi: 10.1001/jama.289.21.JOC30885. [DOI] [PubMed] [Google Scholar]
- 8.Chan JW, Ng CK, Chan YH, et al. Short term outcome and risk factors for adverse clinical outcomes in adults with severe acute respiratory syndrome (SARS) Thorax. 58:686–9. doi: 10.1136/thorax.58.8.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin M, Tseng HK, Trejaut JA, et al. Association of HLA class I with severe acute respiratory syndrome coronavirus infection. BMC Med Genet. 4:9–15. doi: 10.1186/1471-2350-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ng MH, Lau KM, Li L, et al. Association of human-leukocyte-antigen class I (B*0703) and class II (DRB1*0301) genotypes with susceptibility and resistance to the development of severe acute respiratory syndrome. J Infect Dis. 190:515–8. doi: 10.1086/421523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fearon DT, Locksley RM. The instructive role of innate immunity in the acquired immune response. Science. 272:50–3. doi: 10.1126/science.272.5258.50. [DOI] [PubMed] [Google Scholar]
- 12.Fearon DT. Seeking wisdom in innate immunity. Nature. 388:323–4. doi: 10.1038/40967. [DOI] [PubMed] [Google Scholar]
- 13.Thiel S, Holmskov U, Hviid L, Laursen SB, Jensenius JC. The concentration of the C-type lectin, mannan-binding protein, in human plasma increases during an acute phase response. Clin Exp Immunol. 90:31–5. doi: 10.1111/j.1365-2249.1992.tb05827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ezekowitz RA, Day LE, Herman GA. A human mannose-binding protein is an acute-phase reactant that shares sequence homology with other vertebrate lectins. J Exp Med. 167:1034–46. doi: 10.1084/jem.167.3.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holmskov U, Thiel S, Jensenius JC. Collections and ficolins: humoral lectins of the innate immune defense. Annu Rev Immunol. 21:547–78. doi: 10.1146/annurev.immunol.21.120601.140954. [DOI] [PubMed] [Google Scholar]
- 16.Neth O, Jack DL, Dodds AW, Holzel H, Klein NJ. Mannose-binding lectin binds to a range of clinically relevant microorganisms and promotes complement deposition. Infect Immun. 68:688–93. doi: 10.1128/iai.68.2.688-693.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Townsend R, Read RC, Turner MW, Klein NJ, Jack DL. Differential recognition of obligate anaerobic bacteria by human mannose-binding lectin. Clin Exp Immunol. 124:223–8. doi: 10.1046/j.1365-2249.2001.01549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuhlman M, Joiner K, Ezekowitz RA. The human mannose-binding protein functions as an opsonin. J Exp Med. 169:1733–45. doi: 10.1084/jem.169.5.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turner MW. The role of mannose-binding lectin in health and disease. Mol Immunol. 40:423–9. doi: 10.1016/s0161-5890(03)00155-x. [DOI] [PubMed] [Google Scholar]
- 20.Matsushita M, Endo Y, Fujita T. MASP1 (MBL-associated serine protease 1) Immunobiology. 199:340–7. doi: 10.1016/S0171-2985(98)80038-7. [DOI] [PubMed] [Google Scholar]
- 21.Turner MW. Mannose-binding lectin: the pluripotent molecule of the innate immune system. Immunol Today. 17:532–40. doi: 10.1016/0167-5699(96)10062-1. [DOI] [PubMed] [Google Scholar]
- 22.Tabona P, Mellor A, Summerfield JA. Mannose binding protein is involved in first-line host defence: evidence from transgenic mice. Immunology. 85:153–9. [PMC free article] [PubMed] [Google Scholar]
- 23.Garred P, Larsen F, Madsen HO, Koch C. Mannose-binding lectin deficiency—revisited. Mol Immunol. 40:73–84. doi: 10.1016/s0161-5890(03)00104-4. [DOI] [PubMed] [Google Scholar]
- 24.Lipscombe RJ, Sumiya M, Hill AV, et al. High frequencies in African and non-African populations of independent mutations in the mannose binding protein gene. Hum Mol Genet. 1:709–15. doi: 10.1093/hmg/1.9.709. [DOI] [PubMed] [Google Scholar]
- 25.Madsen HO, Garred P, Thiel S, et al. Interplay between promoter and structural gene variants control basal serum level of mannan-binding protein. J Immunol. 155:3013–20. [PubMed] [Google Scholar]
- 26.Madsen HO, Satz ML, Hogh B, Svejgaard A, Garred P. Different molecular events result in low protein levels of mannan-binding lectin in populations from southeast Africa and South America. J Immunol. 161:3169–75. [PubMed] [Google Scholar]
- 27.Eisen DP, Minchinton RM. Impact of mannose-binding lectin on susceptibility to infectious diseases. Clin Infect Dis. 37:1496–505. doi: 10.1086/379324. [DOI] [PubMed] [Google Scholar]
- 28.Biezeveld MH, Kuipers IM, Geissler J, et al. Association of mannose-binding lectin genotype with cardiovascular abnormalities in Kawasaki disease. Lancet. 361:1268–70. doi: 10.1016/S0140-6736(03)12985-6. [DOI] [PubMed] [Google Scholar]
- 29.Best LG, Davidson M, North KE, et al. Prospective analysis of mannose-binding lectin genotypes and coronary artery disease in American Indians: the Strong Heart Study. Circulation. 109:471–5. doi: 10.1161/01.CIR.0000109757.95461.10. [DOI] [PubMed] [Google Scholar]
- 30.World Health Organization
- 31.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 68:978–89. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ip WK, To YF, Cheng SK, Lau YL. Serum mannose-binding lectin levels and MBL2 gene polymorphisms in different age and gender groups of southern Chinese adults. Scand J Immunol. 59:310–4. doi: 10.1111/j.0300-9475.2004.01392.x. [DOI] [PubMed] [Google Scholar]
- 33.Sastry K, Herman GA, Day L, et al. The human mannose-binding protein gene: exon structure reveals its evolutionary relationship to a human pulmonary surfactant gene and localization to chromosome 10. J Exp Med. 170:1175–89. doi: 10.1084/jem.170.4.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Juul-Madsen HR, Munch M, Handberg KJ, et al. Serum levels of mannan-binding lectin in chickens prior to and during experimental infection with avian infectious bronchitis virus. Poult Sci. 82:235–41. doi: 10.1093/ps/82.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han DP, Kim HG, Kim YB, Poon LL, Cho MW. Development of a safe neutralization assay for SARS-CoV and characterization of S-glycoprotein. Virology. 326:140–9. doi: 10.1016/j.virol.2004.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krokhin O, Li Y, Andonov A, et al. Mass spectrometric characterization of proteins from the SARS virus: a preliminary report. Mol Cell Proteomics. 2:346–56. doi: 10.1074/mcp.M300048-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bisht H, Roberts A, Vogel L, et al. Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc Natl Acad Sci USA. 101:6641–6. doi: 10.1073/pnas.0401939101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valdimarsson H, Stefansson M, Vikingsdottir T, et al. Reconstitution of opsonizing activity by infusion of mannan-binding lectin (MBL) to MBL-deficient humans. Scand J Immunol. 48:116–23. doi: 10.1046/j.1365-3083.1998.00396.x. [DOI] [PubMed] [Google Scholar]
- 39.Adams MJ, Jr, Khoury MJ, James LM. The use of attributable fraction in the design and interpretation of epidemiologic studies. J Clin Epidemiol. 42:659–62. doi: 10.1016/0895-4356(89)90009-7. [DOI] [PubMed] [Google Scholar]


