Abstract
Protein detection techniques such as western blotting and ELISA rely on housekeeping proteins as standards for sample normalization. However, clinical or animal tissue specimens are heterogeneous due to presence of contaminating cell types and tissues (e.g., blood vessels and muscle) or cellular decay during tissue storage and isolation which may compromise protein integrity. This biological heterogeneity may invalidate the assumption that housekeeping proteins are invariable across various specimens. This study provides data that advocate for protein standardization based on total protein staining in rabbit posterior capsular tissues. We compared the classical normalization markers glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and β-tubulin (TUBB) with other proteins that have low variation in expression (i.e., FTL, FTH1, EEF1A1, TPT1) based on RNAseq data for human posterior capsular tissues. Histological examination revealed a high degree of qualitative variation in microscopic images of capsular tissue specimens. This variation is reflected by significant differences in specific protein signals for all housekeeping proteins as detected by western blot analysis. However, total protein staining, which combines the intensity of multiple gel electrophoretic bands, normalizes natural biological variation observed for individual housekeeping proteins and permits assessment of protein integrity. Therefore, we propose that normalization based on total protein staining increases accuracy of protein quantification of heterogeneous tissue specimen samples.
Keywords: protein quantification, tissue samples, housekeeping, total protein
1. INTRODUCTION
Quantitative proteomics has led to advancements in our understanding of the physiological differences between biological samples in different disease states. The techniques for protein quantification have continued to evolve over the last half century, all of which require appropriate standards for accurate protein measurement. Housekeeping proteins that are typically highly abundant have been utilized as primary standards of normalization for various protein quantification techniques (e.g., western blotting, ELISA). Normalization using housekeeping proteins is based on the assumption that such proteins are invariable regardless of the sample quality or experimental condition. Since this assumption requires experimental validation for each set of protein samples, it is desirable to consider additional normalization methods to mitigate the variability seen with such samples.
Housekeeping proteins that are relatively invariable in one cell type or tissue may be more variable in other cell types or tissues. Tissue specimens collected from patient biopsies or animal models may exhibit a high degree of heterogeneity, which results from contaminating cell types and tissues (e.g., blood vessels and muscle). For example, because GAPDH is abundant in tissue with high metabolic rates (e.g., muscle), GAPDH levels may differ between specimens with different tissues that differ in glycolytic activity. Also, because GAPDH is a cytoplasmic protein, it is expected to differ in cell types with skewed ratios between cytoplasmic and nuclear compartments. Furthermore, cellular decay during tissue storage and isolation may compromise protein integrity and prevent reliable detection.
Previous studies and our own experience indicate that single housekeeping proteins may not suffice for normalization techniques in heterogeneous tissue samples1, 2. Therefore, we considered total protein stains to serve as a loading control for our samples3-11. Total protein staining has many benefits over housekeeping proteins for protein normalization4. For example, the collection of bands detected by protein stains are typically abundant and proportional to pre-estimated total amounts of protein quantified by spectrophotometric methods. In addition, western blots rely on the detection of proteins with defined molecular weights. However, single or multiple cleavage events of the same housekeeping protein would prevent reliable detection of single proteins at the expected molecular weight either due to a change in migration or loss of the epitope. We hypothesized that total protein staining is a viable and more reliable alternative for normalization of heterogeneous tissues samples compared to commonly used housekeeping proteins.
2. MATERIALS AND METHODS
2.1. Histologic Processing.
Rabbit posterior capsule tissue specimens were derived from waste tissue from rabbits that were sacrificed for studies on treatment of arthrofibrosis12. Samples were harvested by dissection and immediately placed into 5 mL of 10% neutral buffered formalin (NBF), stored at room temperature (22°C) for 48 – 72 hours, and subsequently processed routinely into paraffin. For paraffin embedding tissue was transferred into a 95% ethanol solution and placed in an Isotemp® Vacuum Oven at 65°C and 12 mm Hg for 1.5 hours. Tissue was then removed from the oven and left at standard temperature and pressure (STP) for 1 hour. The previous step was repeated with 100% ethanol after which the samples were left at STP for 3 hours. Next, the samples were suspended in xylenes and placed into the oven for 1 hour and then left at STP overnight. The samples were then transferred into 50% xylenes/50% paraffin solution and placed in the oven for 2 hours. Finally, the samples were infiltrated with 100% paraffin, embedded in paraffin, and sectioned serially at 5 microns (μM). Sections were then stained with hematoxylin and eosin (H&E). Staining was performed on all samples at our institution’s Pathology Research Core.
2.2. Protein Analysis and Western Blot Analysis.
Frozen posterior capsule rabbit tissues were placed in liquid nitrogen and ground to a powder with mortar and pestle. Samples were then processed in a standard fashion in order to quantify EEF1A1, FTL, FTH1, TPT1, GAPDH, and β-Tubulin levels via western blotting. Subsequent to freezing and grinding, the powdered posterior capsule samples were transferred to an eppendorf tube and 700μL of buffer solution (RIPA, 5mM ethylene diamine tetraacetic acid [EDTA] & 1X protease inhibitor cocktail [ThermoFisher Scientific] was added. The powdered tissue and buffer solution was then vortexed for 1 minute, set on dry ice for 10 seconds, and then placed at room temperature (RT) for 50 seconds; this cycle was then repeated additional 4 times. Solutions were then spun down at 12,000 x g for 15 minutes at 4°C. Supernatants were cleared of any cell debris and fatty layers, placed into Pierce™ Protein Concentrators PES (ThermoFisher Scientific), 3K molecular weight cutoff (MWCO) and centrifuged at 15,000 x g for 35 minutes at 4°C to achieve ~300mL concentrate. Ten aliquots were made, and concentrations were determined using DC Protein Assay Kit II (Bio-Rad) prior to storage at −80 °C.
Each protein sample for western blotting was composed of 1x Laemmli buffer such that 20μg/lane was loaded (12.5 μL in each well). Each sample was then heated for 10 minutes at 95°C and resolved with hand-cast midi 12% protein gels, 10-well (Bio-Rad). Each polyacrylamide gel was then laid into a Bio-Rad transfer cassette with a nitrocellulose membrane and transferred utilizing a Bio-Rad Mini Trans-Blot® Cell at 4 °C for 90 minutes at 95V. The membrane was rinsed 3 times in PBS for 1 min, stained with 15mL Ponceau S (total protein) stain (Sigma-Aldrich) for 15min, and subsequently washed 3 times in 5% acetic acid for 3 min each, with subsequent imaging for total protein quantification. The membrane was then washed 4 times in TBST (0.1% Tween20) for 5 min each, to fully remove total protein stain. The membrane was then blocked with StartingBlock™ T20 (ThermoFisher Scientific) blocking buffer for one hour at RT, rocking. Primary antibodies were added in same blocking buffer, rocking overnight at 4°C (Figure 2B.). The membrane was then rinsed 4 times with TBST (0.1% Tween20) for 5 minutes each. Secondary antibody (Figure 2C.) was then added for 1 hour rocking at RT. The membrane was rinsed 4 times with TBST (0.1% Tween20) for 5 minutes and finally in PBS for 1 minute. The membrane was subsequently developed with SuperSignal™ West Pico PLUS Chemiluminescent Substrate, and final images were captured. All images were taken utilizing the Bio-Rad ChemiDocTM Touch Imaging System.
Figure 2.
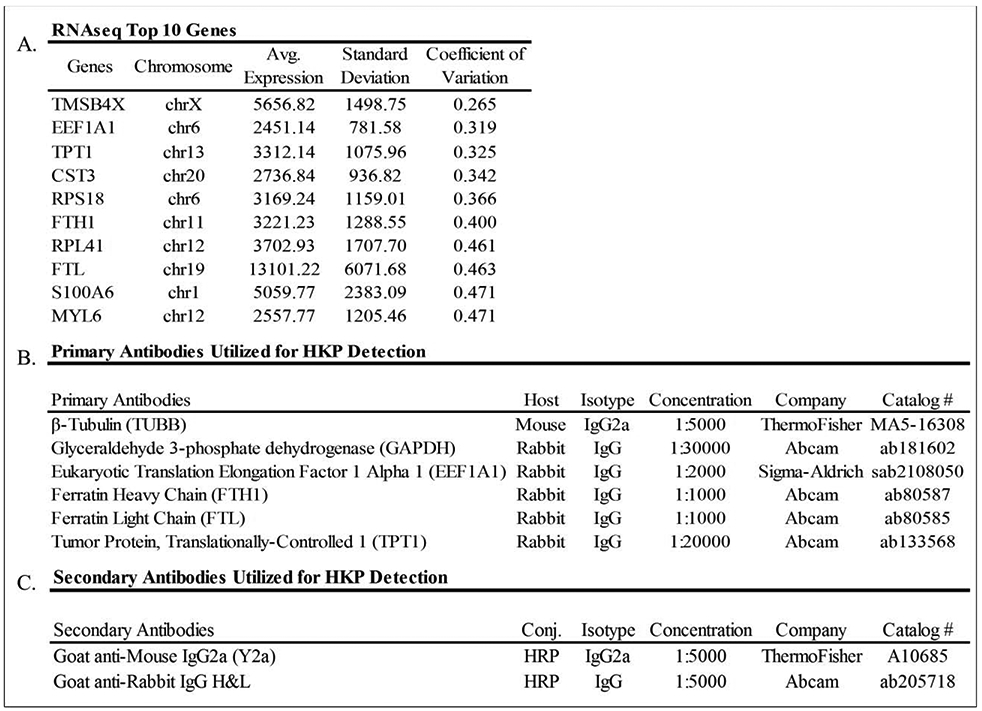
Selection of novel protein normalization markers based on RNA-seq data. The data are derived from mRNAs (in reads per million mapped reads, RPKMs) derived from human posterior capsular tissues obtained from patients undergoing primary knee arthroplasty (A). Description of all primary (B) and secondary (C) antibodies utilized.
2.3. Protein Quantitation and Statistical Analysis.
Protein bands detected on western blots by Ponceau S total protein staining or immunodetection were quantified utilizing Image Lab™ Software (Bio-Rad). Densitometric quantifications were reported in reference to average protein levels across all samples11. Statistical analysis was performed using a Welch’s ANOVA test due to unequal sample variation for housekeeping proteins while a Brown-Forsythe ANOVA test was utilized for total protein as similar sample variation was observed. Protein integrity was estimated by quantifying the combined band intensities of all proteins above 50kDa divided by the total signal of proteins below 50kDa.
2.4. Bioinformatic Analysis of Protein and mRNA Expression Data.
The human proteomic database (ProteomicsDB;https://www.proteomicsdb.org)permits housekeeping protein selection based on protein expression from 27 different human tissues13, 14 However, this database does not contain posterior capsule tissues. Therefore, we consulted RNA-seq data from a large musculoskeletal RNA-seq data within our institute which contains mRNA expression values in knee capsular tissues from primary total knee arthroplasty (pTKA) patients and a variety of other joint tissues. We retrieved data from pTKA samples (GEO accession number: 135854) with mRNA levels expressed as reads per kilobasepair per million mapped reads (RPKM).
3. RESULTS
3.1. Histological Analysis.
Histological examination of 6 rabbit posterior capsule samples revealed that samples were primarily composed of fibrous capsular tissue and associated adipose tissues interspersed with blood vessels. Occasionally, these samples contain small amounts of extraneous tissue (e.g., muscle) (data not shown). Qualitative histologic annotation for adipose tissue, fibrous tissue, and blood vessels revealed 2 samples with high adipose and low fibrous tissue content while 2 samples were about 50/50 and 2 samples with low adipose and high fibrous tissue content. Primary qualitative analysis revealed significantly different tissue compositions (Figure 1).
Figure 1.
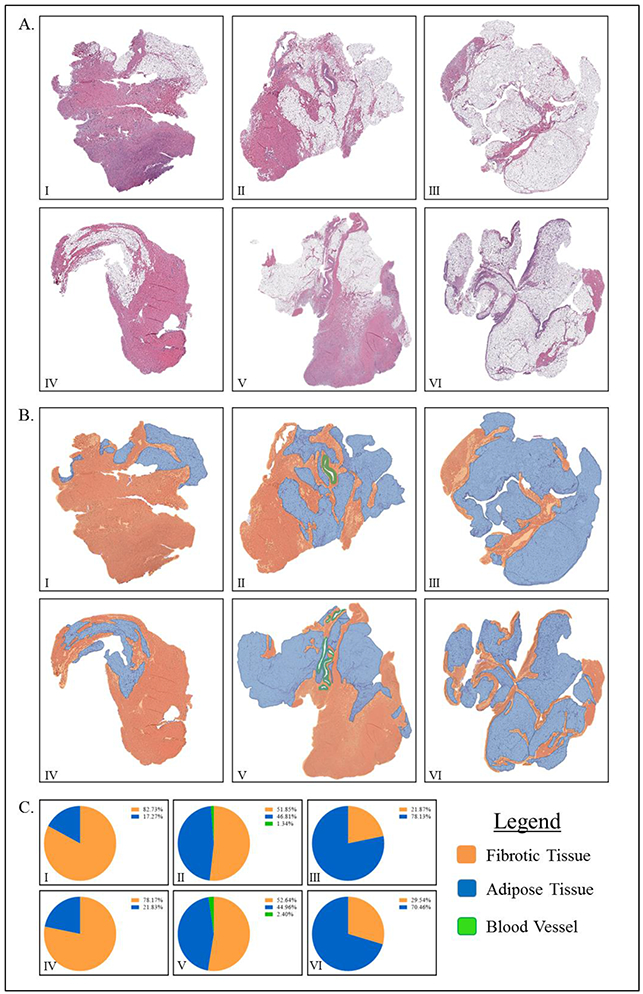
H&E staining of rabbit posterior capsule specimens qualitatively illustrates the high degree of histological variation in tissue samples.
3.2. Selection of Housekeeping Proteins for Sample Normalization.
Following selection of two classical housekeeping proteins (e.g., GAPDH and β-tubulin), the utilization and manipulation of RNA sequencing data from human posterior capsule tissue allowed for the selection of four possible alternative housekeeping proteins. Transcripts from the RNA-seq data were filtered for highest expression (RPKM>10) and lowest coefficient of variation (CV<0.5), as well as further prioritized based on antibody availability15. This filtering procedure yielded a number of proteins, and four that met our criteria were selected for further study: ferratin light chain (FTL), ferratin heavy chain (FTH1), eukaryotic translation elongation factor 1 alpha 1 (EEF1A1), and translationally-controlled tumor protein (TPT1)15. Thus, for this study, two commonly utilized and four sample specific housekeeping proteins were selected to be trialed for their effectiveness of normalization when compared to that of a total protein stain.
3.3. Housekeeping and Total Protein Analysis:
The protein levels of GAPDH, β-tubulin, FTL, FTH1, EEF1A1, and TPT1 in 4 posterior capsule samples were measured via western blot analysis (Figure 3). Via chemiluminescent substrate signal detection and normalized to average protein across all samples, the quantified volume of all housekeeping proteins varied significantly between all rabbit knee samples (p<0.05). It should be noted that when GAPDH levels were measured, blot development revealed a molecular weight of 50kD, different from the usual 37kD band. Furthermore, to ensure that inadequate loading was not the cause for the observed protein level differences, each protein was utilized in an attempt to normalize the other 5 proteins. This process was repeated for each protein individually, without success (data not shown).
Figure 3.
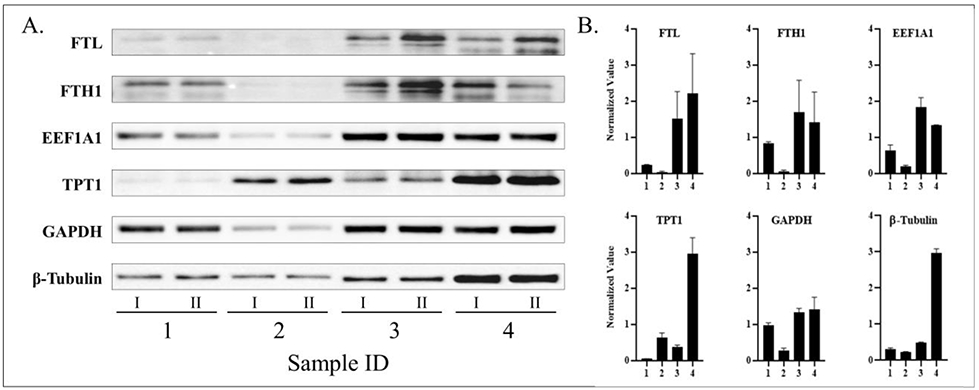
Western blot analysis (A) and quantification (B) of housekeeping proteins.
Likewise, total protein was measured via Ponceau S Stain, as per normal SOP this staining serves to confirm completion of a successful protein transfer to NC membrane. Via colormetric pixel density analysis and normalized to average total protein across all samples, the quantified total protein volumes did not different significantly from each other (p>0.05).
In conclusion, western blot analysis of 4 equally loaded samples, confirmed via total protein staining, showed there to be statistically significant differences between all samples for all common and selected housekeeping proteins.
3.4. Protein Integrity Score Analysis.
Ponceau S staining of electrophoretically separated proteins not only provides a quantitative measure of total protein, but also generates information about protein integrity. Therefore, we developed a scoring system that is based on the natural tendency of proteins to degrade over time. We selected serum albumin (67kDa) as a proxy for overall protein quality of each sample, because albumin is a very abundant protein that is found in all samples and its identity was confirmed by a proteomics approach (data not shown). We estimated total protein integrity primarily based on the degradation of albumin by quantifying the total amount of protein above and below 50kDa using standard imaging software (Biorad Image Lab). Densitometric volume analysis of total protein staining revealed that samples 1, 3, and 4 had integrity scores of 3.54, 2.89, and 4.30, respectively, while analysis of sample 2 revealed a score of 0.80 (Figure 4). Thus, quantitation of protein integrity permits rational selection of suitable protein samples for downstream analyses based on numerical parameters.
Figure 4.
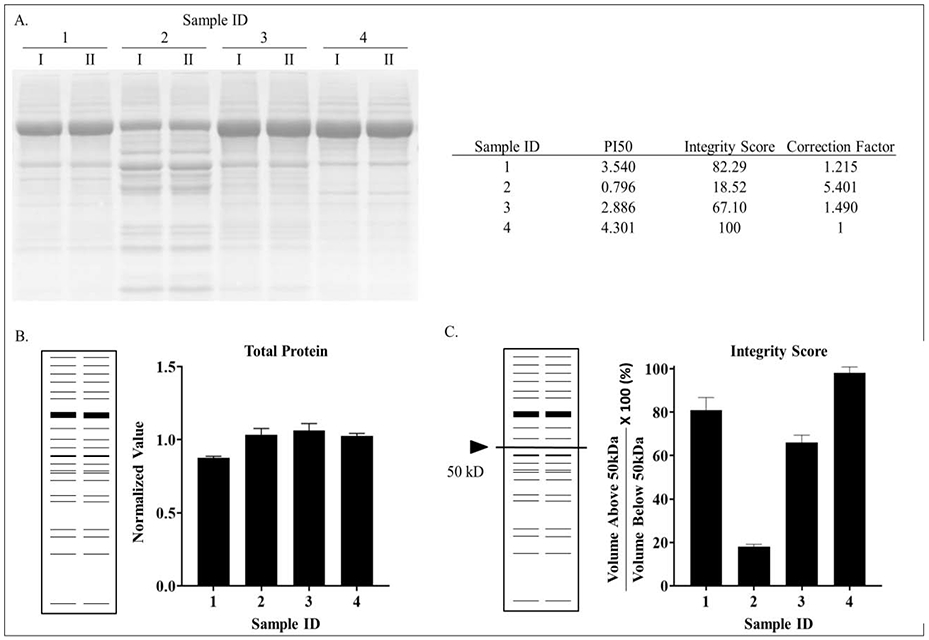
Staining of electrophoretically separated proteins (A), quantitation of total protein staining (B) and protein integrity scores (C). Integrity scores were calculated based on the total protein volume above 50kDa divided by the total protein below 50kDa. This value was then divided by the highest score of all samples and then multiplied by 100% to yield a percent protein integrity based off of the least degraded sample.
4. DISCUSSION
In this study, immunoblot analysis of rabbit knee posterior capsular tissue capsular tissues revealed that common housekeeping proteins, such as GAPDH or β-tubulin, varied significantly in these heterogeneous tissue samples, thus precluding their use in normalization. Rather, normalization based on total protein quantification appeared to be more reliable. The finding that protein quantification based on housekeeping proteins was not reliable in these tissue samples is consistent with many other studies that have assessed their use and discussed caveats and assumptions that remain to be validated2, 4-7, 16-18. Normalization techniques using housekeeping proteins are critically dependent on the assumption that these proteins have equal expression across all samples under all experimental conditions and do not vary over a developmental or treatment time course. It is clear from our studies that these conditions are not met for housekeeping proteins in capsular joint tissues that differ from one another based on the experimental conditions and possibly other factors.
Total protein staining not only allows for accurate protein quantification but also provides other significant investigative advantages. Commonly, specific housekeeping proteins are chosen based on the expression profile of the protein of interest. For example, if a select protein of interest has a relative low expression it must be matched to a housekeeping protein with similar expression to avoid signal saturation when blotting. This has the potential to be a substantial limitation based on the fact that signal saturation is achieved for many of these housekeeping proteins at as low as 5μg, yet commonly loaded quantities are between 30-140μg, well beyond the saturation point of many4, 10. Interestingly, signal saturation has yet to be reported as an issue when similar volumes of protein are loaded (30-140μg) and total protein stains are utilized4. Beyond reductions in technical and biologic variation, the use total protein stains represents a cheaper and faster normalization method that correspondingly permits evaluation of protein integrity and effective transfer prior to immunodetection.
This study not only identified a reliable loading control for these samples, but also provided a set of characteristics, through qualitative histologic analysis, that may be useful in the future for the identification of samples that could potentially benefit from the use of total protein as a loading control. Though various studies have suggested that total protein may be a loading control that is suitable for all samples, further experimentation is needed to confirm and validate this hypothesis. It remains necessary for each study to determine which internal control is selected for normalization and to validate assumptions about its numerical consistency relative to proteins of interest that are modulated. This becomes critically important to understanding the physiological differences between biological samples of different disease states and their response to treatment.
4. CONCLUSION
In summary, our reported data demonstrate that the use of commonly utilized (GAPDH and β-tubulin) and tissue specific housekeeping proteins (FTL, FTH1, EEF1A1, and TPT1) are not invariably expressed between heterogeneous samples of capsular tissue. Therefore, total protein levels appear to represent the most reliable loading control for western blot analysis of rabbit posterior capsule tissue.
Tissue samples are heterogeneous and may contain other cell types
Use of housekeeping proteins for normalization relies on sample homogeneity
Standard housekeeping proteins significantly vary in different tissues
Total protein stains may represent a more robust method for normalization
ACKNOWLEDGMENTS
The authors would like to acknowledge lab members of the Abdel and van Wijnen laboratories for their critical review of this work and their insightful discussions and/or assistance with reagents and procedures. Research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) of the National Institutes of Health (NIH) under Award Number AR072597-01A1 (Abdel) and the Anna-Maria and Stephen Kellen Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. We would like to thank the Pathology Research Core at the Mayo Clinic for their work on the immunohistochemistry.
Abbreviations
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- TUBB
β-tubulin
- NBF
neutral buffered formalin
- STP
standard temperature and pressure
- H&E
hematoxylin and eosin
- RT
room temperature
- MWCO
molecular weight cutoff
- RPKM
reads per kilobasepair per million mapped reads
- FTL
ferratin light chain
- FTH1
ferratin heavy chain
- EEF1A1
eukaryotic translation elongation factor 1 alpha 1
- TPT1
translationally-controlled tumor protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: No benefits in any form have been received or will be received by any authors from a commercial party related directly or indirectly to the subject of this article.
REFERENCES
- 1.Petrak J, Ivanek R, Toman O, Cmejla R, Cmejlova J, Vyoral D, et al. Deja vu in proteomics. A hit parade of repeatedly identified differentially expressed proteins. Proteomics. 2008. May;8(9):1744–9. Epub 2008/04/30. [DOI] [PubMed] [Google Scholar]
- 2.Ferguson RE, Carroll HP, Harris A, Maher ER, Selby PJ, Banks RE. Housekeeping proteins: a preliminary study illustrating some limitations as useful references in protein expression studies. Proteomics. 2005. February;5(2):566–71. Epub 2005/01/04. [DOI] [PubMed] [Google Scholar]
- 3.Thacker JS, Yeung DH, Staines WR, Mielke JG. Total protein or high-abundance protein: Which offers the best loading control for Western blotting? Analytical biochemistry. 2016. March 1;496:76–8. Epub 2015/12/27. [DOI] [PubMed] [Google Scholar]
- 4.Moritz CP. Tubulin or Not Tubulin: Heading Toward Total Protein Staining as Loading Control in Western Blots. Proteomics. 2017. October;17(20). Epub 2017/09/25. [DOI] [PubMed] [Google Scholar]
- 5.Lee HG, Jo J, Hong HH, Kim KK, Park JK, Cho SJ, et al. State-of-the-art housekeeping proteins for quantitative western blotting: Revisiting the first draft of the human proteome. Proteomics. 2016. July;16(13):1863–7. Epub 2016/04/30. [DOI] [PubMed] [Google Scholar]
- 6.Hu X, Du S, Yu J, Yang X, Yang C, Zhou D, et al. Common housekeeping proteins are upregulated in colorectal adenocarcinoma and hepatocellular carcinoma, making the total protein a better "housekeeper". Oncotarget. 2016. October 11;7(41):66679–88. Epub 2016/08/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilda JE, Gomes AV. Stain-Free total protein staining is a superior loading control to beta-actin for Western blots. Analytical biochemistry. 2013. September 15;440(2):186–8. Epub 2013/06/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eaton SL, Roche SL, Llavero Hurtado M, Oldknow KJ, Farquharson C, Gillingwater TH, et al. Total protein analysis as a reliable loading control for quantitative fluorescent Western blotting. PloS one. 2013;8(8):e72457. Epub 2013/09/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dittmer A, Dittmer J. Beta-actin is not a reliable loading control in Western blot analysis. Electrophoresis. 2006. July;27(14):2844–5. Epub 2006/05/12. [DOI] [PubMed] [Google Scholar]
- 10.Chen W, Xu WH. beta-Actin as a loading control: Less than 2 mug of total protein should be loaded. Electrophoresis. 2015. September;36(17):2046–9. Epub 2015/06/03. [DOI] [PubMed] [Google Scholar]
- 11.Aldridge GM, Podrebarac DM, Greenough WT, Weiler IJ. The use of total protein stains as loading controls: an alternative to high-abundance single-protein controls in semi-quantitative immunoblotting. Journal of neuroscience methods. 2008. July 30;172(2):250–4. Epub 2008/06/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trousdale WH, Salib CG, Reina N, Lewallen EA, Viste A, Berry DJ, et al. A Drug Eluting Scaffold for the Treatment of Arthrofibrosis. Tissue engineering Part C, Methods. 2018. September;24(9):514–23. Epub 2018/08/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilhelm M, Schlegl J, Hahne H, Gholami AM, Lieberenz M, Savitski MM, et al. Mass-spectrometry-based draft of the human proteome. Nature. 2014. May 29;509(7502):582–7. Epub 2014/05/30. [DOI] [PubMed] [Google Scholar]
- 14.Activities at the Universal Protein Resource (UniProt). Nucleic acids research. 2014. January;42(Database issue):D191–8. Epub 2013/11/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salib CG, Lewallen EA, Paradise CR, Tibbo ME, Robin JX, Trousdale WH, et al. Molecular pathology of adverse local tissue reaction caused by metal-on-metal implants defined by RNA-seq. Genomics. 2018. September 21. Epub 2018/09/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rivero-Gutierrez B, Anzola A, Martinez-Augustin O, de Medina FS. Stain-free detection as loading control alternative to Ponceau and housekeeping protein immunodetection in Western blotting. Analytical biochemistry. 2014. December 15;467:1–3. Epub 2014/09/07. [DOI] [PubMed] [Google Scholar]
- 17.Barber RD, Harmer DW, Coleman RA, Clark BJ. GAPDH as a housekeeping gene: analysis of GAPDH mRNA expression in a panel of 72 human tissues. Physiological genomics. 2005. May 11;21(3)389–95. Epub 2005/03/17. [DOI] [PubMed] [Google Scholar]
- 18.Welinder C, Ekblad L. Coomassie staining as loading control in Western blot analysis. J Proteome Res. 2011. March 4;10(3):1416–9. Epub 2010/12/29. [DOI] [PubMed] [Google Scholar]


