Abstract
Cells change their migration in response to chemical stimuli including the gradient of those stimuli. Cellular migration in the direction of a chemical gradient, known as chemotaxis, plays important roles in development, immune response, wound healing, and cancer metastasis. While chemotaxis modulate migrations of single cells as well as collectives of cells in-vivo, in-vitro researches mostly focus on single cell chemotaxis mainly due to the lack of experimental tools. To fill the gap, here we developed a unique experimental system which combines microfluidics and micropatternings to demonstrate the effect of chemical gradient on collective cell migration. Furthermore, we incorporated traction microscopy and monolayer stress microscopy into the system to characterize the changes in cellular force on substrates as well as between neighboring cells. As a proof-of-concept, we tested the migration of micropatterned circular islands of Madin-Darby canine kidney (MDCK) cells under the gradient of hepatocyte growth factor (HGF), a known scatter factor. We found that cells on the side of higher concentration of HGF migrated faster than cells on the other side within a cell island. Within the same island, we found that cellular traction was similar on both sides but intercellular stress was much less on the side of higher concentration of HGF. This novel experimental system would provide a new opportunity to study the mechanics of chemotactic migration of cellular collectives.
Keywords: microfluidics, traction microscopy, collective cell migration, chemotaxis, chemical gradient, micropatterning
SUMMARY
Collective cell migration in development, wound healing, and cancer metastasis is often guided by the gradients of growth factors or signaling molecules. Here we describe an experimental system combining traction microscopy with microfluidic system and demonstrate how to quantify the mechanics of collective migration under biochemical gradient.
INTRODUCTION
Cellular migration in biological system is a fundamental phenomenon for tissue formation, immune response, and wound healing. Cellular migration is also an important process in some diseases like cancer. Cells often migrate as a group rather than individuals, known as a collective cell migration. For the cells to move collectively as a group, sensing of microenvironment would be quintessential. For instance, cells perceive physicochemical stimuli and respond by changing the motility, cell-substrate interactions, and cell-cell interactions, resulting in directional migration along a chemical gradient. In this connection, rapid advancement has been made in lab-on-a-chip technologies that can create well controlled chemical microenvironments such as the gradient of a chemoattractant. However, their use in cell migration has been limited to single cell migration. How cellular collectives respond to a chemical gradient is not well understood. Thus, the development of a platform that enables the spatiotemporal control of soluble factor as well as in-situ observation of biophysical response of cells would help to unravel the mechanism of collective cell migration.
Here, we developed a multi-channeled microfluidic system that enables generation of a concentration gradient of soluble factors which would modulate migration of patterned cell clusters. In this study, we chose Hepatocyte growth factor (HGF) to regulate the migratory behavior of Madin-Darby canine kidney (MDCK). HGF is known to attenuate cell-cell integrity and enhance the motility of the cells. In the microfluidic system, we also incorporated the Fourier Transform Traction Microscopy and Monolayer Stress Microscopy and by doing so, we were able to analyze the motility, contractile force and intercellular tension induced by the constituent cells in response to HGF gradient. We found that within the same island, cells on the side of higher concentration of HGF migrated faster and showed reduced intercellular stress than cells on the other side. Our results suggest that this new experimental system would be suitable to explore many other questions in the field of collective cellular migration under chemical gradient of various soluble factors.
PROTOCOL
NOTE: Lithography of SU-8 molds for stencils (thickness = 250 μm) and microchannel parts (thickness = 150 μm), glass etching (depth = 100 μm) and cast fabrication were outsourced by sending designs using AUTO-CAD (Supplement File 1) to manufacturers.
1. Fabrication of Polydimethylsiloxane (PDMS) stencil and microchannel
1.1. Design the size of micropattern using a software (Auto-CAD, CA)
1.2. SU-8 molds on a silicon wafer (4” diameter) were custom-made (MicroFIT, KR)
1.2.1. For stencil, SU-8 thickness is ~ 250 μm
1.2.2. For microchannel, SU-8 thickness is ~ 150 μm
1.3. Prepare PDMS mixture by mixing the base elastomer and curing agent in a ratio of 10:1.
1.3.1. Place 15 ml of base elastomer in a 50-ml conical tube and add 1.5 ml of curing agent
1.3.1.1. Prepare 2 of this.
1.3.2. Vortex the PDMS mixture for 5 minutes.
1.3.3. Centrifuge the PDMS mixture at 1000 rpm for 1 min to remove bubbles
1.4. Fabricate PDMS stencil
1.4.1. Pour ~ 1 ml of the PDMS mixture on the wafer avoiding SU-8 patterned regions
1.4.2. Degas for 30 min in a vacuum chamber.
1.4.3. Place the wafer on a flat surface for over 30 min at room temperature.
1.4.4. Cure the PDMS in a dry oven at 80°C for over 2 hr.
1.4.5. Carefully peel off the PDMS from each SU-8 mold.
1.4.6. Trim the thin PDMS membrane using a 14 mm hollow punch.
1.4.7. Remove dust on the surface of the PDMS pieces and autoclave the PDMS stencils.
1.5. Fabricate PDMS microchannel
1.5.1. Pour ~30 ml of PDMS mixture over the SU-8 mold.
1.5.2. Degas for 30 min in a vacuum chamber.
1.5.3. Cure the PDMS in a dry oven at 80°C for over 2 hr.
1.5.4. Cut the PDMS to a size of 24 mm x 24 mm and carefully peel off PDMS block.
1.5.5. In each PDMS block, make one outlet and three inlets using a 1 mm biopsy punch.
2. Preparation of bottom glass with polyacrylamide (PA) gel
2.1. Rectangular slide glasses (24 mm x 24 mm x 1 mm) with rectangular micro-well (6 mm x 12 mm, 100 μm deep) were custom-made by cutting and etching glasses.
2.2. Surface treatment of bottom glass1
2.2.1. Remove dust and autoclave the glass.
2.2.2. Silanize the etched surface
2.2.2.1. Prepare a bind silane solution by mixing 200 ml of DIW, 80 μl of acetic acid, and 50 μl of 3-(Trimethoxysilyl)propyl methacrylate (TMSPMA, Sigma-Aldrich) for 1 hr.
CAUTION: TMSPMA is a combustible liquid. Follow the recommendation in material safety data sheets. Use only in a chemical fume hood.
2.2.2.2. Cover the etched surface of the bottom glass with 100 μl of the bind silane solution and leave the glass at room temperature for 1 hr.
2.2.2.3. Rinse the glass with DIW 3 times.
2.2.3. Let the glass dry at ambient air or by blowing air.
2.3. Cast PA gel of 6kPa in Young’s modulus2
2.3.1. Prepare a gel solution for PA gel
2.3.1.1. Prepare a fresh solution of 0.5 % (w/v) ammonium persulfate (APS) by dissolving 5 mg of APS (Bio-Rad, CA) in 1 ml of DIW.
2.3.1.2. Prepare a PA gel solution consisting of 138 μl of 40 % Acrylamide solution (Bio-Rad, CA), μl of 2 % Bis-acrylamide solution (Bio-Rad, CA), 5 μl of fluorescent particle solution (0.5 μm, Invitrogen, CA) and 655 μl of DIW.
CAUTION: Acrylamide and bisacrylamide solutions are toxic. Wear protective gloves, clothing, and eye protection. Protect the PA gel solution containing fluorescent particles from light.
2.3.2. After adding APS (Bio-Rad, CA), put 10 μl of mixed gel solution onto the rectangular micro-well and put a circular coverslip (18 mm, Marienfel, DE)
2.3.3. Flip the assembly of custom-glass, gel solution and coverslip and centrifuge for 10 min at 700 RPM to bring fluorescent particles on the top layer of PA gel
2.3.4. After 30 min, place the PA gel-filled bottom glass in a 35 mm cell culture dish, fill with 2 ml of DIW, and remove the coverslip on PA gel.
2.4. Coat collagen on PA gel
2.4.1. Activate PA gel surface using sulfosuccinimidyl 6-(4’-azido-2’-nitrophenylamino)hexanoate (Sulfo-SANPAH, ThermoFisher, US).
NOTE: Protect Sulfo-SANPAH from light.
2.4.1.1. Dissolve 1 mg/ml Sulfo-SANPAH in warm 50 mM HEPES buffer (Gibco, UK).
2.4.1.2. Drop 200 μl of Sulfo-SANPAH solution on gel surface and activate by ultraviolet (UV) light (365 nm wave length) for 10 min.
2.4.2. Rinse the gel with 0.1 M HEPES buffer twice and with PBS once.
2.4.3. Coat PA gel with collagen solution (100 μg/ml in PBS, rat tail collagen type I, Corning, US) at 4°C overnight.
2.4.4. On the following day, wash PA gel with PBS three times.
3. Micropatterning of cell islands
3.1. Pre-treatment of PDMS stencil with Pluronic F-127
3.1.1. Autoclave PDMS stencil.
3.1.2. Prepare F-127 solution (2 % (w/v) in PBS, Sigma-Aldrich, US).
3.1.3. Submerge PDMS stencil in F-127 solution and keep at 37 °C for 1 hr.
3.1.4. Wash PDMS stencil with PBS three times.
3.1.5. Remove liquid from both PDMS stencil and PA gel.
3.1.6. Put PDMS stencil on PA gel and add PBS on PDMS stencil.
3.1.6.1. Avoid bubbles in the holes of PDMS stencil.
3.1.7. Prepare cell solution (2 × 106 cell/ml) in cell culture media (Dulbecco’s modified Eagle’s medium (DMEM, Welgene, KR) with 10 % fetal bovine serum (FBS, Gibco, UK) and 1 % antibiotic antimycotic (AA, Gibco, UK)).
3.1.8. Remove PBS from PDMS stencil and put 200 μl of cell solution on PDMS stencil.
3.1.8.1. Keep the PA gel in incubator for 1hr so that cells can attach to the PA gel.
3.1.9. Gently wash off cell solution with cell culture media.
3.1.10. Remove PDMS stencil and add more cell culture media.
3.1.11. Check the formation of cell islands.
4. Assemble PA gel with PDMS microchannel
4.1. Remove dust on PDMS microchannel and autoclave them.
4.2. Do surface treatment with oxygen plasma (Femto Science, KR) for 30 sec.
4.3. Remove any fluid on the bottom glass which has PA gel in the rectangular micro-well
4.4. Put PDMS microchannel on top of the bottom glass and put on the custom glass holder.
4.4.1. Custom glass holder (Han-Gug Mechatronics, KR) was made to hold the assembly of
bottom glass and PDMS microchannel as well as to put on the stage of a conventional epi fluorescent microscope (JuLI stage, NanoEntek, KR).
4.5. Fill the microchannel with cell culture medium.
CAUTION: Make sure to remove all the bubbles trapped in the channels by gently flushing warm media with a pipette.
5. Integrated microfluidic system
5.1. Connect tubing
5.1.1. Prepare connectors by trimming the tip of needles (18 G, KOVAX, KR) and bending it 90 degrees.
5.1.2. Prepare tubing lines for 3 inlets and 1 outlet.
5.1.2.1. For inlet tubing, connect the trimmed needle and a 30 cm mini-volume line (Hyupsung, KR) with 3-way stopcock (Hyupsung, KR). Prepare 3 of this.
5.1.2.2. For outlet tubing, connect the trimmed needle and a 75 cm mini-volume line (Hyupsung, KR) with 3-way stopcock (Hyupsung, KR). Prepare 1 of this.
5.1.2.3. Fill the tubing lines with warm medium.
CAUTION: Make sure to remove all the bubbles trapped in the tubing lines by gently flushing warm media with a syringe.
5.1.3. Prepare reservoirs using syringe cylinders which connect with inlet tubing lines.
5.1.4. Plug the needle connectors of each tubing line into the 3 inlets and 1 outlet of the microfluidic device.
5.2. Fill the reservoirs with fresh medium or conditioned medium.
5.2.1. For the gradient test, fill the left inlet reservoir with 20 ng/ml of hepatocyte growth factor (HGF) in cell culture medium.
5.2.2. For the visualization of concentration gradient, add fluorescent dye (rhodamine B-dextran, 70kDa, Sigma-Aldrich, US) to the left inlet reservoir.
5.3. Connect the outlet tubing line to syringe pump (Chemyx, US).
NOTE: The operating mode of the syringe pump is ‘withdrawal’. Flow rate is changed according to the capacity of the syringe and the operating speed of the syringe pump.
5.4. Place the integrated microfluidic system on the stage of a conventional epi-fluorescent microscope.
6. Image acquisition
6.1. Time-lapse imaging
6.1.1. Take images every 10 min for 24 hr using a microscope with live-cell imaging set up (JuLI, NanoEnTek, KR).
6.1.1.1. In this experiment, the whole microscope system including samples was kept inside of an incubator
6.1.1.2. At each time point, a phase image was taken to visualize cell migration.
6.1.1.3. A green fluorescent image was taken to visualize fluorescent beads embedded in PA gel.
NOTE: These green fluorescent images will be used later for the calculation of traction.
6.1.1.4. A red fluorescent image was taken to visualize concentration gradient of a chemical
6.2. Reference image
6.2.1. After time-lapse imaging, cells were removed from PA gel using trypsin (Gibco, UK).
6.2.2. A green fluorescent image was taken as a reference image.
NOTE: This image will be used as a reference image for the traction recovery.
7. Data analysis
7.1. A custom-made code for data analysis was developed using Matlab (Mathworks, US) and the details have been described elsewhere3-5.
7.2. Phase-image analysis
7.2.1. Displacements in 2 consecutive phase images were calculated using particle image velocimetry.
7.3. Fourier Transform Traction Microscopy
7.3.1. Each green fluorescent image was compared with the reference image and the displacements in each image were calculated using particle image velocimetry.
7.3.2. From the displacements, traction made by cells on PA gel was recovered using Fourier Transform Traction Microscopy4-6
7.4. Monolayer Stress Microscopy
7.4.1. From the traction data, stresses within the monolayer of cell island were calculated using Finite Element Method3,5
REPRESENTATIVE RESULTS
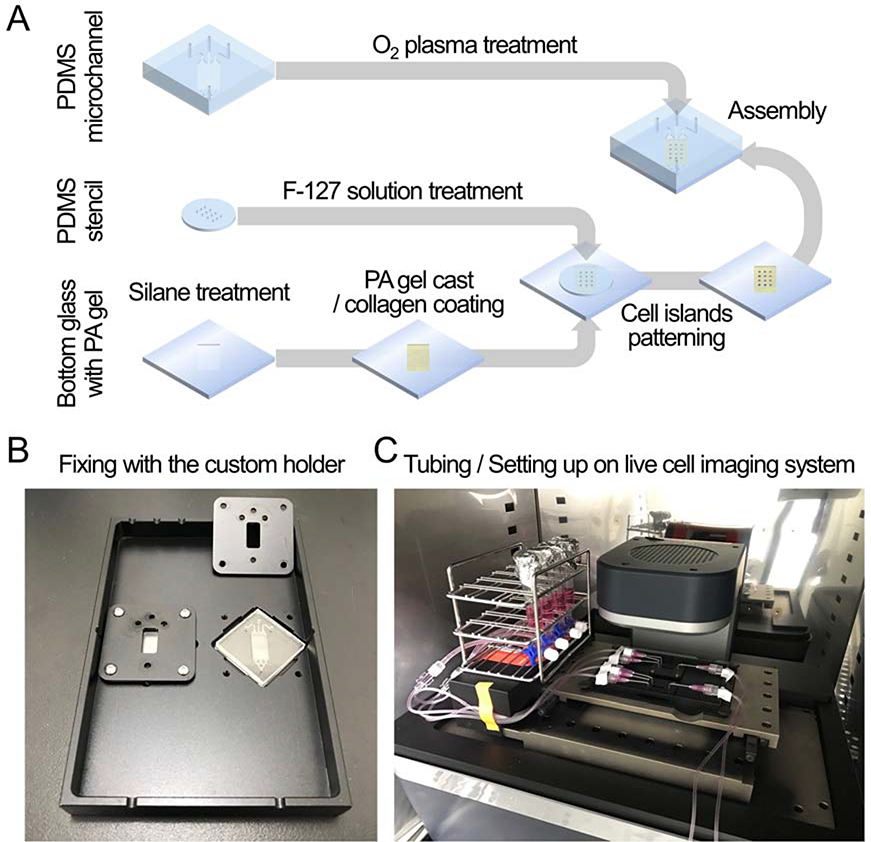
To explore the collective migration under a chemical gradient, we integrated a microfluidic system with traction microscopy (Fig 1). The microfluidic system was used to establish a stable chemical gradient over cellular collectives. The traction microscopy was used to measure mechanical characteristics of cellular collectives including migration dynamics, traction (contractile force per unit area) and intercellular stress within cellular collectives. To build the integrated system, we first cast polyacrylamide gel (PA gel) on custom-cut glass, and then we seeded MDCK cells within micropatterned islands made by PDMS stencil. For this experiment, we created twelve islands of MDCK cells (4 rows by 3 columns, diameter ~ 700 μm). After cells attached on PA gels, we removed the PDMS stencil to initiate collective migration. We put the pre-made microfluidic channel on top of custom-cut glass to build microfluidic channel above cellular islands (Fig 1A). We then inserted the sandwich of custom-cut glass and the microfluidic channel on the custom-built microscope stage. We kept the samples and the automated fluorescent microscope within an incubator for live cell imaging. Finally, we connected the microfluidic channels with external supply reservoirs and a syringe pump working as a drain (Fig 1B). Throughout the experiments, cells were kept in 5 % CO2 and at 37 °C.
Figure 1: Schematic of the integrated device preparation.
(A) To fabricate the integrated device, bottom glass with PA gel, on which cell islands were patterned using PDMS stencil, was covered with PDMS microchannel. (B) The integrated devices were fixed using a custom holder to prevent fluid leakage and component dissociation. (C) After the device filled with fluid without bubbles, reservoirs were connected to the inlets, and a syringe pump was connected to the outlet. All experimental set ups were put on a live cell imaging system.
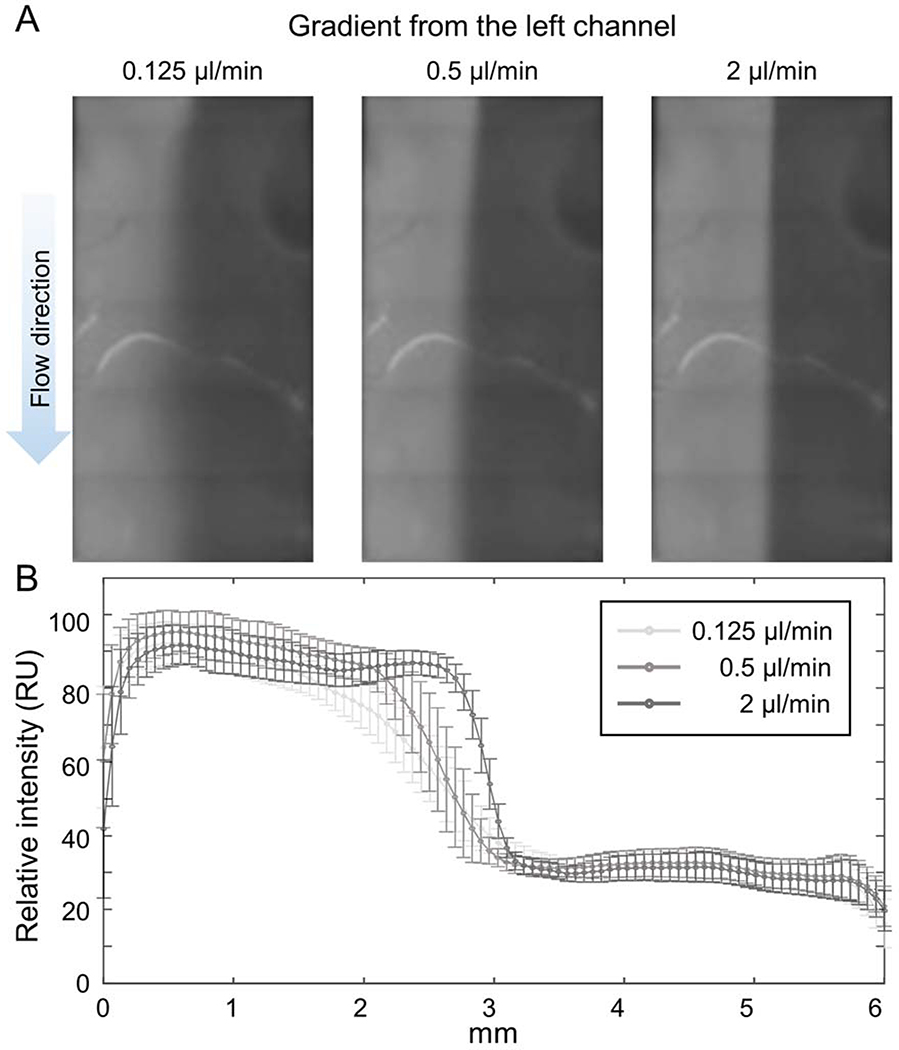
To establish a concentration gradient within microfluidic channel, we connected the left inlet to the supply reservoir containing fluorescent dye solution and the middle and the right inlets to the supply reservoirs containing water only. Then we connected the outlet to the syringe pump to suck the fluid out of the microfluidic channel. We found that the gradient of the fluorescent intensity of the fluorescent dye was quickly established at the flow rate of 0.125 ~ 2 μl/min (Fig 2A). The slope of the gradient depended on the flow rate and at the flow rate of 0.125 μl/min, the gradient was developed over 1.5 mm which is comparable size of cell islands (Fig 2B).
Figure 2: Establishing chemical gradient.
(A) Fluorescence images of rhodamine B-conjugated dextran (70 kDa) visualized the concentration gradient of the molecule under the fluid flow of 0.125, 0.5 and 2 μl/min. (B) The intensity profile of the fluorescence image showed that the concentration gradient in the device is adjustable by the flow rate. The error bars indicate standard deviations.
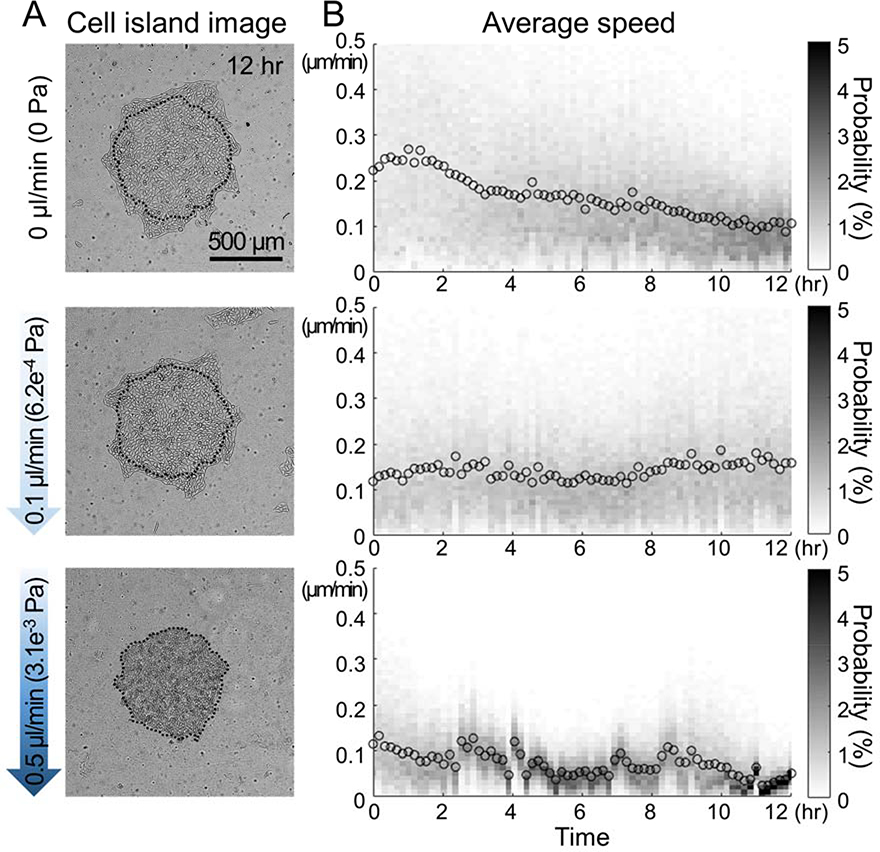
Next, we tested the integrated device with cells. We micropatterned the islands of MDCK cells and assembled the microfluidic channel as described above. First, without applying any flow in the microchannel, we checked that the cellular island spread well within the device and we found that over 12 hr period, the island expanded and the average migration speed was round 0.1 ~ 0.2 μm/min. While there were variations in the degree of island expansions but all cells looked healthy. Next, we applied flow over these islands and we found that at the flow rate of 0.1 μl/min, the cellular island spread well and they maintained average migration speed of 0.1 ~ 0.2μm/min for 12 hr. However, when we increased the flow rate to 0.5 μl/min, cells did not spread well and the average migration speed decreased below 0.1 μm/min. Furthermore, the morphologies of cells looked quite different and cells looked detaching from PA gels (Fig 3). Based on these data, we chose 0.1 μl/min as the flow rate for the following experiments.
Figure 3: Migration of MDCK cell islands under flow.
(A) The phase images at 12 hr of the MDCK cell island at each flow rate (0, 0.1, and 0.5 μl/min) showed that cell islands expanded well at a low flow rate but did not expand at a high flow rate. Dotted lines are the boundaries at 0 hr. (B) The mean and histogram of the cellular speed within each island every 10 min for 12 hr.
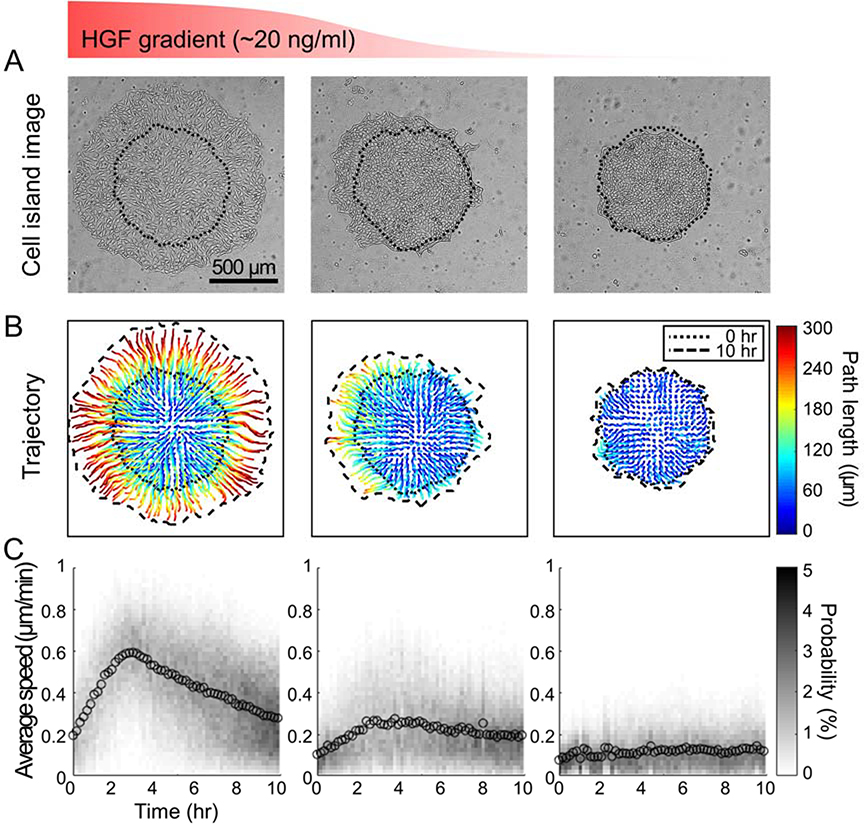
To test the effects of chemical gradient on collective cell migration, we chose HGF as a chemical which we have used in our previous study5,7. We connected the left inlet with the supply reservoir holding cell culture media with HGF solution (20 ng/mL) and the other 2 inlets with the reservoirs holding cell culture media. With the flow rate at 0.1 μl/min, we maintained the flow over the migrating cell islands (4 rows by 3 columns) for 10 hr (Fig 4). As shown in a separate experiment, the HGF concentration on the left column would be close to the concentration of suppling solution (20 ng/mL) and that on the right column would be closed to zero. The HGF concentration on the middle column would be gradually decreasing from the left half to the right half. When we compared the size of cell islands after 10 hr, the islands on the right column did not expand much but the islands on the left column became significantly bigger and expanded in all directions (Fig 4A). Such change in island size was supported by the trajectories of cells within each island (Fig 4B). Interestingly, the islands in the middle column did not expand toward right where the HGF concentration was low but expanded preferentially toward left where the HGF concentration was high. While there was little change in the average migration speed on the right column, on left and middle columns, the average migration speed increased gradually for the first 3 hr then gradually decreased (Fig 4C).
Figure 4: Migration of MDCK cell islands under flow and chemical gradient.
(A) The phase images at 10 hr (dotted line: cell island boundary at 0 hr) of the MDCK cell island under the HGF gradient (from left to right: 20 ~ 0 ng/ml) showed that island expanded more where HGF concentration was higher. (B) The trajectories of migration paths (color coding indicates the path length) of cells in the MDCK cell islands. Dotted lines and dashed lines are each cell island’s boundaries at 0 hr and 10 hr, respectively. (C) The mean and histogram of the cellular speed within each island every 10 min for 10 hr.
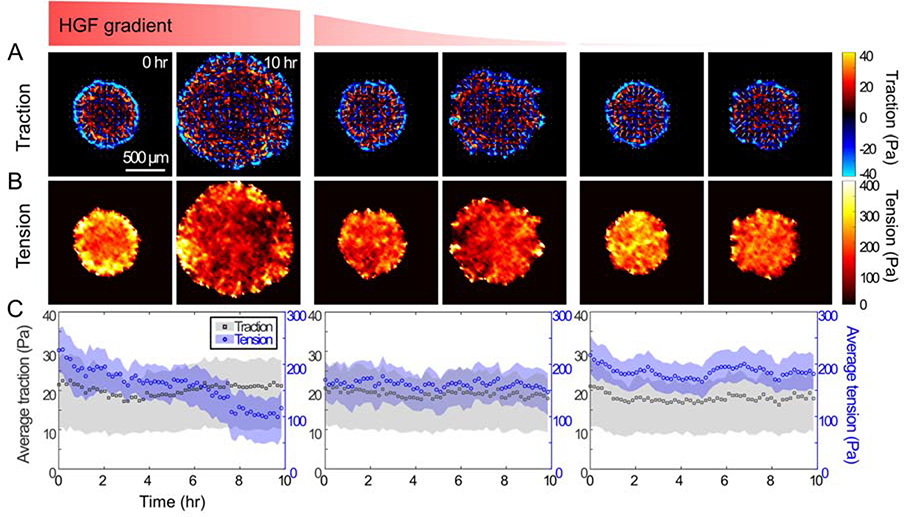
To measure the contractile force and inter-cellular stress, we combined the traction microscopy4,8,9 and the monolayer stress microscopy3,10 with the microfluidic channel. Over the course of 10 hr of applying chemical gradient, we took images of fluorescent particles in PA gels and analyzed the force (per unit area) applied by cells on the substrate using Fourier Transform Traction Microscopy4,6. We plotted traction in polar coordinate; blue color shows the force toward the center of island and red color shows the force away from the center. At time 0, all islands had similar traction distribution with strong inward traction on the edge and fluctuation within island. This fluctuation was similar to what we have previously shown4,5,11. After 10hr of applying HGF gradient, while the degree of island expansion was different in each column, the traction distributions were largely similar to the time 0 (Fig 5A). The average traction did not change for 10hr (Fig 5C). When we calculated the monolayer stress, however, each column showed different trends. On the right column where the HGF concentration was low, the average tension within islands maintained around 200 Pa throughout 10 hr. On the left column where the HGF concentration was high, the average tension within islands gradually decreased from 230 Pa to 100 Pa over 10 hr as previously shown5,7. On the middle column where the HGF concentration was high on the left half of the island and low on the right half, the average tension was maintained around 150 Pa.
Figure 5: Mechanical interactions of MDCK cell islands.
(A) The maps of traction (cell-substrate interaction) in the cell islands at 0 hr and 10 hr under HGF gradient. The maps were plotted in a radial coordination with outward traction using warm color and inward traction using cold color. (B) The maps of monolayer tension (cell-cell interactions) in the cell islands at 0 hr and 10 hr under HGF gradient. (C) The plot of average traction (gray square) and tension (blue circle) within the cell islands under HGF gradient over time every 10 min for 10 hr. The color bands represent the mid-quartile (25 ~ 75 %).
DISCUSSION
Collective migration of constituent cells is an important process during development and regeneration, and the migrating direction is often guided by the chemical gradient of growth factors12,13. During collective migration, cells keep interacting with neighboring cells and underlying substrates and such mechanical interactions give a rise to the emergent phenomemons such as durotaxis14, plithotaxis3 and kenotaxis15. However, all of these works were done in a single concentration of growth factors such as those in fetal bovine serum. To fill the gap, here we describe a new experimental platform that can measure mechanical interactions during collective cellular migration under a chemical gradient.
The new platform combines 3 experimental systems which have been used widely; micropatterning, traction microscopy and microfluidics. First we used micropatterning of cell islands for collective migration. Secondly, we used both traction microscopy and monolayer stress microscopy for mechanical measurements of migrating cells. Finally, we used microfluidics to establishing chemical gradient over migrating cells. With this experimental system, we demonstrated that cells within a single island migrated differently in response to different chemical gradients. Where the concentration of HGF (a known scatter factor) was higher, we found that cells migrated faster and lost most of cell-cell interactions.
To achieve high quality of data, there are a few key steps in the procedures that require particular attention. The PA gels in a custom-glass substrate have to be perfect in terms of the distribution of fluorescent particles and the surface coating of collagen because without high quality PA gels, one can’t acquire good data set for traction analysis and for monolayer stress analysis. Also, the microfluidic channels are very small and bubbles are easy to be trapped in them. Those bubbles not only disturb the flow but also can sweep the cells away if the trapped bubbles start to move along the flow.
This new platform would help us explore further the mechanics of collective cell migration under a chemical gradient, which is the key to understand the regeneration, cancer metastasis as well as development.
Supplementary Material
Table of Materials/Equipment
| Name of material / equipment | Company | Catalog number | Comments / Description |
|---|---|---|---|
| SU-8 master molds | MicroFIT | 4” diameter, custom-made | |
| 100 mm petri dishes | SPL | 10100 | 100 mm diameter, 15 mm height |
| Sylgard 184 Elastomer Kit | Dow Corning | PDMS | |
| 14 mm hollow punch | ILJIN | 124-0571 | |
| 1 mm Biopsy punch | Integra Miltex | 33-31AA-P/25 | |
| Bottom glass chip | MicroFIT | 24 × 24 × 1 mm, custom-made, rectangular groove (6 × 12 mm, depth : 100 μm) | |
| acetic acid | J.T. Baker | JT9508-03 | |
| 3-(Trimethoxysilyl)propyl methacrylate (TMSPMA) | Sigma-Aldrich | 440159-500ML | |
| Ammonium persulfate (APS) | Bio-Rad | 1610700 | |
| 40% Acrylamide Solution | Bio-Rad | 1610140 | Wear protective gloves, clothing, and eye protection. |
| 2% bis-acrylamide solution | Bio-Rad | 1610142 | Wear protective gloves, clothing, and eye protection. |
| tetramethylethylenedia mine (TEMED) | Bio-Rad | 1610800 | Wear protective gloves, clothing, and eye protection. |
| FluoSpheres amine-modified microspheres | Invitrogen | F8764 | , 0.2 μm, yellow-green fluorescent(505/515) |
| 18 mm Ø Coverslip | Marienfeld-Superior | 111580 | Circular 18 mm, thickness No. 1 (0.13 to 0.16 mm) |
| 35 mm cell culture dish | Corning | 430165 | |
| sulfosuccinimidyl 6-(4’-azido-2’-nitrophenylamino)hexanoate (Sulfo-SANPAH) | Thermo Scientific | 22589 | Store at −20°C. Store protected from moisture and light. |
| 1 M HEPES buffer solution | Gibco | 15630-056 | |
| Ultraviolet (UV) lamp | UVP LLC | 95-0248-02 | 365 nm wavelength |
| Collagen typeI, Rat tail | Corning | 354236 | |
| Dulbecco’s Phosphate Buffered Saline (PBS) | Biowest | L0615-500 | w/o Magnesium, Calcium |
| Pluronic F-127 | Sigma-Aldrich | P2443-250G | |
| Madin-Darby Canine Kidney (MDCK) cell | type II | ||
| Dulbecco’s Modified Eagle’s Medium (DMEM) | Welgene | LM 001-11 | |
| Fetal bovine serum (FBS) | Gibco | 26140-179 | |
| Antibiotic-Antimycotic | Gibco | 15240-062 | |
| 0.25% trypsin-EDTA (1X) | Gibco | 25200-056 | |
| Oxygen plasma system | Femto Science | CUTE-MPR | |
| Custom glass holder | Han-Gug Mechatronics | custom-made | |
| Steril hypodermic needle 18 G | KOVAX | Trim the tip of the needle and bend it 90 degrees for connecting in/out ports with volume line | |
| Syringes | KOVAX | 1, 3, 5, 10, or 50 cc for using inlet reservoir or outlet syringe pump | |
| 75 cm minimum volume line (for pediatric) | Hyupsung | HS-MV-75 | CAUTION: do not use if previously opened. do not resterlize or resuse |
| 30 cm minimum volume line (for pediatric) | Hyupsung | HS-MV-30 | CAUTION: do not use if previously opened. do not resterlize or resuse |
| 3-way stopcock | Hyupsung | HS-T-61N | CAUTION: do not use if previously opened. do not resterlize or resuse |
| Syringe pump | Chemyx Inc. | model fusion 720 | withdraw fluid |
| Hepatocyte Growth Factor (HGF) | Sigma-Aldrich | H1404-5UG | recombinant, human |
| Rhodamine B isothiocyanate–dextran | Sigma-Aldrich | R9379-100MG | 70 kDa, used to estimate spatiotemporal distribution of HGF in the microfluidic channel |
| JuLI stage live cell imaging system | NanoEnTek | Automated X-Y-Z stage and fluorsent imaging Incubator-compatible (37 °C and 5% CO2) |
ACKNOWLEDGMENTS
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) (No. NRF-2017R1E1A1A01075103), Korea University Grant, and the BK 21 Plus program.
Footnotes
DISCLOSURES:
The authors declare that they have no competing financial interests.
REFERENCES
- 1.Serra-Picamal X, Conte V, Sunyer R, Munoz JJ & Trepat X Mapping forces and kinematics during collective cell migration. Methods Cell Biol. 125 309–330, (2015). [DOI] [PubMed] [Google Scholar]
- 2.Denisin AK & Pruitt BL Tuning the Range of Polyacrylamide Gel Stiffness for Mechanobiology Applications. ACS Appl Mater Interfaces. 8 (34), 21893–21902, (2016). [DOI] [PubMed] [Google Scholar]
- 3.Tambe DT et al. Collective cell guidance by cooperative intercellular forces. Nat Mater. 10 (6), 469–475, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trepat X et al. Physical forces during collective cell migration. Nature physics. 5 (6), 426, (2009). [Google Scholar]
- 5.Jang H et al. Homogenizing cellular tension by hepatocyte growth factor in expanding epithelial monolayer. Sci Rep. 8 45844, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butler JP, Tolic-Norrelykke IM, Fabry B & Fredberg JJ Traction fields, moments, and strain energy that cells exert on their surroundings. Am J Physiol Cell Physiol. 282 (3), C595–605, (2002). [DOI] [PubMed] [Google Scholar]
- 7.Jang H et al. Traction microscopy with integrated microfluidics: responses of the multi-cellular island to gradients of HGF. Lab Chip. 19 (9), 1579–1588, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dembo M & Wang YL Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys J. 76 (4), 2307–2316, (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang N et al. Cell prestress. I. Stiffness and prestress are closely associated in adherent contractile cells. Am J Physiol Cell Physiol. 282 (3), C606–616, (2002). [DOI] [PubMed] [Google Scholar]
- 10.Tambe DT et al. Monolayer stress microscopy: limitations, artifacts, and accuracy of recovered intercellular stresses. PLoS One. 8 (2), e55172, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Notbohm J et al. Cellular Contraction and Polarization Drive Collective Cellular Motion. Biophys J. 110 (12), 2729–2738, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedl P & Gilmour D Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol. 10 (7), 445–457, (2009). [DOI] [PubMed] [Google Scholar]
- 13.Rorth P Collective guidance of collective cell migration. Trends Cell Biol. 17 (12), 575–579, (2007). [DOI] [PubMed] [Google Scholar]
- 14.Sunyer R et al. Collective cell durotaxis emerges from long-range intercellular force transmission. Science. 353 (6304), 1157–1161, (2016). [DOI] [PubMed] [Google Scholar]
- 15.Kim JH et al. Propulsion and navigation within the advancing monolayer sheet. Nat Mater. 12 (9), 856–863, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.