Abstract
Women of Latin American origin in the United States (U.S.) are more likely to be diagnosed with advanced breast cancer and have a higher risk of mortality than non-Hispanic White women. Studies in U.S. Latinas and Latin American women have reported a high incidence of Human Epidermal Growth Factor Receptor 2 (HER2) positive (+) tumors; however, the factors contributing to this observation are unknown. Genome-wide genotype data for 1,312 patients from the Peruvian Genetics and Genomics of Breast Cancer Study (PEGEN-BC) was used to estimate genetic ancestry. We tested the association between HER2 status and genetic ancestry using logistic and multinomial logistic regression models. Findings were replicated in 616 samples from Mexico and Colombia. Average Indigenous American (IA) ancestry differed by subtype. In multivariate models, the odds of having an HER2+ tumor increased by a factor of 1.20 with every 10% increase in IA ancestry proportion (95%CI 1.07–1.35, p-value 0.001). The association between HER2 status and IA ancestry was independently replicated in samples from Mexico and Colombia. Results suggest that the high prevalence of HER2+ tumors in Latinas could be due in part to the presence of population-specific genetic variant(s) affecting HER2 expression in breast cancer.
Keywords: Genetic ancestry, breast cancer, subtypes, HER2+, Indigenous American
Introduction
Globally, more than two million women are diagnosed with breast cancer each year(1). The incidence of breast cancer varies by country, and within the United States risk of breast cancer differs among racial/ethnic subpopulations(2). The age-adjusted incidence rate for breast cancer in non-Hispanic White women in the United States is the highest at 126.1 per 100,000, followed by African American at 124 per 100,000, Hispanic/Latina at 93.9 per 100,000, Asian/Pacific Islander at 93.0 per 100,000, and American Indian/Alaska Native at 74.2 per 100,000(2). Mortality rates also differ by race/ethnicity, being highest in African American women at 28.1 per 100,000, followed by non-Hispanic Whites at 20.1 per 100,000, Hispanic/Latinas at 14.2, and with lowest rates among Asian/Pacific Islanders and American Indian/Alaska Natives at 11.2 and 11.4 respectively(2). Multiple studies have shown that while Hispanics/Latinas (Latinas) have lower breast cancer incidence and mortality rates than non-Hispanic White women, they have a higher risk of breast cancer specific mortality with hazard ratio estimates ranging from 1.1 to 1.3(3–7). This disparity could be in part explained by the fact that Latinas are more likely to be diagnosed with the more aggressive human epidermal growth factor receptor 2 positive (HER2+) and hormone receptor negative subtypes of the disease than non-Hispanic White women(8–15).
Previous studies suggest that reproductive factors or other behavior related exposures could explain some of the observed differences in the incidence of particular tumor subtypes between populations(16–18). In addition, genetic variants that are more strongly associated with particular tumor subtypes have been described(19). However, only few studies have reported specific factors associated with HER2+ subtypes and reports have been inconsistent(11, 19–21).
We have previously shown that a higher degree of Indigenous American (IA) genetic ancestry is associated with a lower incidence of breast cancer(22–24), and discovered a single nucleotide polymorphism (SNP) in the 6q25 chromosomal region near the estrogen receptor 1 (ESR1) gene that explained a large fraction of that lower risk with an allele that originated from IA ancestry and is highly protective for breast cancer, particularly estrogen receptor negative (ER-) tumors(25). In this study, we evaluated the association between continental genetic ancestry and breast cancer subtype among highly IA breast cancer patients from Peru, Mexico and Colombia, to gain insight into the possibility that differences in tumor subtype distribution between racial/ethnic groups might be in part driven by underlying population-specific genetic variants.
Methods
Study Participants
The Peruvian Genetics and Genomics of Breast Cancer Study (PEGEN-BC): As of October 2019, we have recruited 1,842 participants from the Instituto Nacional de Enfermedades Neoplásicas (INEN) in Lima, Peru. Women were invited to participate in the study if they had a diagnosis of invasive breast cancer in the year 2010 or later, and were between 21–79 years of age. A recruiter working locally at INEN, identified eligible breast cancer patients who had upcoming appointments. During the appointment check-in, the recruiter approached eligible patients, provided the patients with information about the study, and reviewed the written informed consent with them if they showed interest in participating in the study. At the end of their oncology clinic visit, blood was drawn by a certified phlebotomist at the INEN central laboratory.
Demographic and clinical data were abstracted from the electronic medical records and included place of birth, current residence, age at diagnosis, height, weight, and family history of breast cancer, as well as tumor characteristics (i.e. histological type, grade and stage, tumor size and ER, progesterone receptor (PR) and HER2 status). A short study survey complemented the information obtained from the medical records and included reproductive history (i.e. age at menarche, age at first pregnancy, number of pregnancies, breastfeeding history, and menopause status) and lifestyle factors (i.e. alcohol consumption and smoking status).
The present analysis includes 1,312 patients with available genome wide genotype data. This study was approved by the INEN and the University of California San Francisco institutional review boards and all individuals provided written informed consent to participate.
The Post Columbian Study of Environmental and Heritable Causes of Breast Cancer (COLUMBUS) is a case-control study of breast cancer with individuals recruited throughout large cancer hospitals in Colombia and Mexico. In Mexico, breast cancer patients were recruited at the Oncology Hospital, Centro Médico Nacional Siglo XXI, and Gynecology Hospital No. 4, Instituto Mexicano del Seguro Social, in Mexico City, Mexico, between 2008 and 2013. The study was approved by the ethics committee of each hospital and written informed consent was obtained from all patients prior to enrollment in the study. Cases were histologically confirmed by a local pathologist. Genomic DNA was isolated from whole blood obtained by a certified phlebotomist. In Colombia, breast cancer patients diagnosed between 2011 and 2016 were recruited through large cancer hospitals in five Colombian cities (Bogota, Ibague, Medellin, Neiva, and Pasto). After providing written informed consent, patients were interviewed in person by trained research nurses. The information collected includes sociodemographic characteristics, anthropometric measures, smoking and drinking habits, family history of cancer, and the complete pathology report. Genomic DNA was isolated from whole blood samples obtained by a certified phlebotomist. The study adhered to the Helsinki Declaration and received Institutional Review Board (IRB) approvals from University of Tolima and from all hospitals where recruitment was carried out.
Genotyping and Quality Control
In PEGEN-BC, DNA was extracted from whole blood following standard protocols and genotypic profiles were generated using the Affymetrix Precision Medicine Research Array(26). This array is designed to include a genome-wide imputation grid with about 800,000 markers and to cover five ancestral population groups, including African, admixed American, East Asian, European, and South Asian. Currently, 1,380 samples from 1,320 individuals have been genotyped. Quality control of the genotyped data was performed in PLINK(27). First, we removed 4 individuals with a genotype call rate less than 90%. After individual genotype call rate filtering, we removed markers on sex chromosomes and those with call rates below 98% (21,752 markers were removed). We used identity by descent to identify any related individuals. There were 60 sets of duplicates and only one sample per individual was kept. The concordance rate for these duplicate individuals was 0.99. There were four sets of first-degree relatives, of which only one sample per pair was included in downstream analyses. The final data set for association analyses included 1,312 samples. Markers with a minor allele frequency (MAF) < 1% were removed. The total number of markers included for imputation was 451,350. We imputed missing genotypes using the Michigan Imputation Server(28) (1000 Genomes Project (1000G) phase 3 samples as reference) and after imputation filtered out markers with r2 < 0.8 and MAF of <1%, which resulted in a total number of 6,915,815 markers.
The 447 Colombian samples included in the replication analysis were genotyped using Affymetrix Axiom UKBiobank Array and the 169 Mexican samples were genotyped on Affymetrix Precision Medicine Research Array. Quality control of the genotyped data has been previously described(29).
Continental and subcontinental structure characterization
After pruning 1000G reference samples (in Plink: window size =50, number of variants=5, Variance Inflation Factor (VIF) threshold=2), we merged them with the PEGEN-BC study imputed dataset which resulted in a total of 520,450 markers for downstream analyses. Individual genetic ancestry was determined using ADMIXTURE(30), unsupervised, with four populations to capture IA, European, African, and East Asian continental ancestry based on the known major continental influences to the population of Peru(31). Global genetic ancestry estimates and core analysis variables are available upon request. To further characterize the continental and subcontinental structure of the PEGEN-BC study participants we conducted Principal Component Analysis (PCA) using the program Plink 1.9(27) and T-Distributed Stochastic Neighbor Embedding (Rt-SNE) analyses with the Rtsne Package(32–34) using the first 50 principal components, in R 3.6.0(35). Rt-SNE was run multiple times (n=4) obtaining consistent clusters.
Individual genetic ancestry was also estimated with the program ADMIXTURE(30) in the replication samples. After pruning, 63,811 and 93,813 genotyped markers were used for the Colombian and Mexican samples respectively.
Breast Tumor Subtype Determination
Different approaches have been used to define breast cancer subtypes based on ER, PR and HER2 status by immunohistochemistry (IHC)(36). We used a cutoff of 1% to define ER/PR positivity(37). HER2 positivity was defined as cases with 3+ staining by IHC or with gene amplification by Fluorescence In Situ Hybridization (FISH) testing following a 2+ IHC result. For quality control, two independent pathologists from the University of California San Francisco reviewed the IHC slides at INEN for a subset of 52 patients. The concordance rate was high: 100% for ER, 87% for PR and 85% for HER2 (most of the discordant calls for HER2 were scored as ‘negative’ or 1+ at INEN and 2+ by the independent pathologists). Tumor subtypes were defined as: ER/PR+ HER2−, ER/PR+ HER2+, ER/PR- HER2+ and ER/PR- HER2−.
The IHC HER2 status for the Colombian and Mexican samples was obtained from pathology reports and medical records. The hospitals from where patients were recruited used the same HER2 positivity criteria as INEN.
Statistical Analysis
Patient characteristics were compared based on breast cancer subtype. T-tests were performed on normally distributed continuous variables, Kruskal-Wallis tests were performed on non-normally distributed continuous variables, and Chi-squared tests were used on categorical variables.
Logistic regression models were used to compare individuals with HER2− vs. HER2+ tumors. Multinomial logistic regression models defining the ER/PR+ HER2− group as the reference were used to test the association between tumor subtypes and IA genetic ancestry. Age at diagnosis, African ancestry and height were included as covariates. Height was included in the models due to its potential role as a confounder based on the comparison of patient characteristics between tumor subtypes. An additional model including stage (I-II, vs. III-IV) and region of residence (Coastal, Mountain or Amazonian) was run to assess the possible bias in the distribution of subtype by ancestry due to the potential correlation between IA ancestry and severity of the cases that are seen at INEN.
The replication analyses in the COLUMBUS consortium samples from Mexico and Colombia were conducted using logistic regression models comparing HER2+ vs. HER2− cases. Additional subtyping of HER2+ tumors in the replication analysis was not conducted due to the limited sample size.
All analyses were performed using Plink 1.9(27), R 3.6.0(35), or ADMIXTURE(30).
Results
Patient Characteristics
PEGEN-BC study patient characteristics by tumor subtype are presented in Table 1 (N=1,312). ER/PR+ HER2− tumors had a frequency of 49%, ER/PR+ HER2+ 18%, ER/PR- HER2+ 12% and ER/PR- HER2– 15%. Average age at diagnosis for all patients was 50 (SD = 11) and there was a non-significant trend towards younger age among women in the ER/PR- HER2− group. Average body mass index (BMI) for all subtypes was 28 (SD = 5), and was similar between groups. There was a statistically significant difference between the subtypes in measured height, where women with ER/PR- HER2+ tumors were shorter than those with other tumor subtypes (p-value= 0.033). Differences regarding reproductive variables by tumor subtype were not observed with the exception of menopausal status. There was a higher percent of pre-menopausal women among those with ER/PR- HER2− tumors (p-value = 0.023). We also observed statistically significant differences by histopathologic subtype (p-value <0.001), and clinical stage and grade (p-value <0.001) (Table 1). Lobular subtype had a frequency of 10% among women with ER/PR+ HER2− group compared to less than 3% in other groups. Stage and grade were higher in the ER/PR- subgroups (HER2+ and HER2−) compared to ER/PR+ tumors (Table 1).
Table 1.
Patient and tumor characteristics by ER, PR, HER2 based subtype in women with breast cancer from Lima, Peru (N=1,312).
| Variables | Overall | ER/PR+HER2− | ER/PR+HER2+ | ER/PR-HER2+ | ER/PR-HER2-010869Mc | P-value |
|---|---|---|---|---|---|---|
| 1,312* | 638(49) | 239(18) | 156(12) | 200(15) | ||
| Age at diagnosis, yrs. (mean (SD)) | 50(11) | 50(11) | 49(10) | 51(10) | 49(11) | 0.254 |
| First-degree family history (N (%)) | ||||||
| Yes | 98(8) | 49(8) | 16(7) | 7(4) | 15(8) | 0.561 |
| No | 1,214(92) | 589(92) | 223(93) | 149(96) | 185(92) | |
| rs140068132 genotype (N (%)) | ||||||
| AA | 970(74) | 446(70) | 182(76) | 124(80) | 156(78) | 0.006 |
| AG | 326(25) | 185(29) | 56(23) | 30(19) | 38(19) | |
| GG | 16(1) | 7(1) | 1(1) | 2(1) | 6(3) | |
| Anthropometric variables | ||||||
| Weight, Kg. (mean (SD)) | 64(12) | 65(13) | 65(12) | 62(11) | 64(13) | 0.146 |
| Height, cm. (mean (SD)) | 153(7) | 153(7) | 154(6) | 151(6) | 153(7) | 0.033 |
| Body Mass Index (BMI) (mean (SD)) | 28(5) | 28(5) | 28(5) | 27(5) | 27(5) | 0.731 |
| Reproductive variables | ||||||
| Age at menarche, yrs. (mean (SD)) | 13(2) | 13(2) | 13(2) | 13(2) | 13(2) | 0.678 |
| Age at first pregnancy, yrs. (mean (SD)) | 23(6) | 24(6) | 23(5) | 23(6) | 22(6) | 0.153 |
| Number of pregnancies (mean (SD)) | 3(2) | 3(2) | 3(2) | 3(2) | 3(2) | 0.968 |
| Post-menopausal (N (%)) | ||||||
| Yes | 1,071(82) | 530(83) | 194(81) | 128(82) | 160(80) | 0.023 |
| No | 149(11) | 55(9) | 33(14) | 15(10) | 32(16) | |
| Missing | 92(7) | 53(8) | 12(5) | 13(8) | 8(4) | |
| Breastfeeding history (N (%)) | ||||||
| Yes | 988(75) | 453(71) | 182(76) | 126(81) | 168(84) | 0.924 |
| No | 44(3) | 21(3) | 9(4) | 5(3) | 6(3) | |
| Missing | 280(21) | 164(26) | 48(20) | 25(16) | 26(13) | |
| Demographic variables and lifestyle | ||||||
| Genetic ancestry (mean (SD)) | ||||||
| Indigenous American | 76(16) | 75(18) | 77(16) | 80(14) | 77(16) | 0.023 |
| European | 17(12) | 17(12) | 17(12) | 15(10) | 16(11) | 0.12 |
| African | 4(7) | 4(8) | 5(8) | 3(5) | 5(7) | 0.319 |
| East Asian | 2(7) | 3(9) | 2(4) | 1(2) | 2(4) | 0.32 |
| Region of Residence (N (%)) | ||||||
| Amazonas | 81(6) | 39(6) | 13(5) | 7(4) | 17(8) | 0.237 |
| Coast | 1,090(83) | 535(84) | 203(85) | 125(80) | 157(78) | |
| Mountains | 137(10) | 62(10) | 23(10) | 24(15) | 25(12) | |
| Missing | 4(<1) | 2(<1) | 0(0) | 0(0.0) | 1(<1) | |
| Alcohol intake (N (%)) | ||||||
| Never | 389(30) | 186(29) | 77(32) | 42(27) | 69(34) | 0.755 |
| < one glass/day | 705(54) | 338(53) | 126(53) | 87(56) | 101(51) | |
| > one glass/day | 133(10) | 63(10) | 26(11) | 14(9) | 25(13) | |
| Missing | 85(6) | 51(8) | 10(4) | 13(8) | 5(2) | |
| Smoking history (N (%)) | ||||||
| Never | 894(68) | 422(66) | 171(72) | 107(69) | 146(73) | 0.281 |
| Past | 329(25) | 164(26) | 58(24) | 36(23) | 46(23) | |
| Current | 6(<1) | 3(<1) | 0(0) | 0(0) | 3(2) | |
| Missing | 83(6) | 49(8) | 10(4) | 13(8) | 5(2) | |
| Tumor characteristics | ||||||
| Histopathological variety (N (%)) | ||||||
| Ductal | 1,166(89) | 563(88) | 225(94) | 149(96) | 183(92) | <0.001 |
| Lobular | 83(6) | 63(10) | 8(3) | 4(3) | 5(2) | |
| Other | 22(2) | 10(2) | 3(1) | 2(1) | 7(4) | |
| Missing | 41(3) | 2(<1) | 3(1) | 1(<1) | 5(2) | |
| Grade (N (%)) | ||||||
| Low | 55(4) | 41(6) | 6(2) | 1(<1) | 5(2) | <0.001 |
| Intermediate | 515(39) | 363(57) | 75(31) | 21(14) | 32(16) | |
| High | 691(53) | 224(35) | 152(64) | 132(85) | 160(80) | |
| Missing | 51(4) | 10(2) | 6(2) | 2(1) | 3(2) | |
| Clinical stage (N (%)) | ||||||
| I | 67(5) | 36(6) | 11(5) | 5(3) | 13(7) | <0.001 |
| II | 442(34) | 256(40) | 75(31) | 36(23) | 53(26) | |
| III | 454(35) | 192(30) | 84(35) | 77(49) | 88(44) | |
| IV | 60(5) | 27(4) | 13(5) | 8(5) | 9(4) | |
| Missing | 289(22) | 127(20) | 56(23) | 30(19) | 37(19) | |
ER: estrogen receptor; PR: progesterone receptor; HER2: Human Epidermal Growth Factor Receptor 2. SD: Standard Deviation
ER, PR and HER2 status missing for 79 individuals (6%)
Distribution of Genetic Ancestry
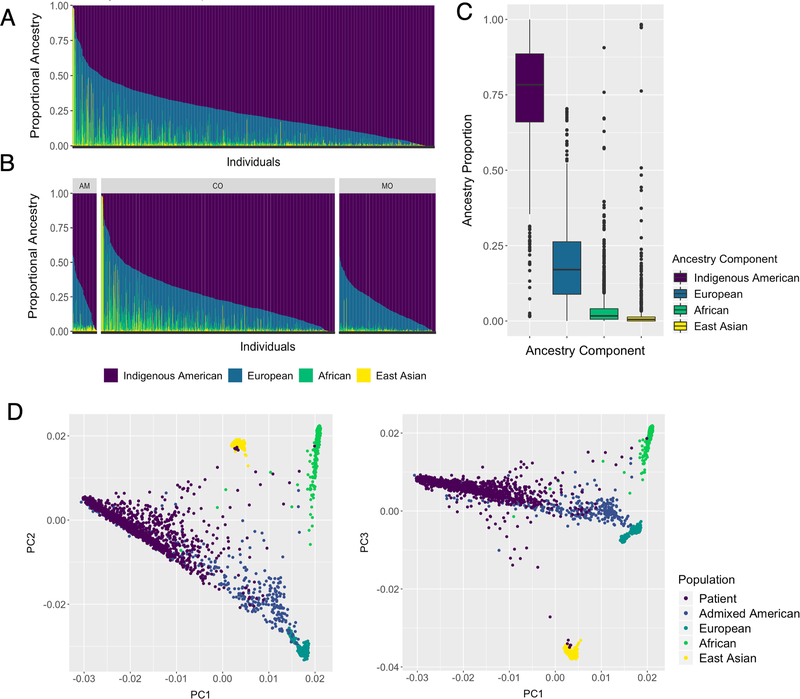
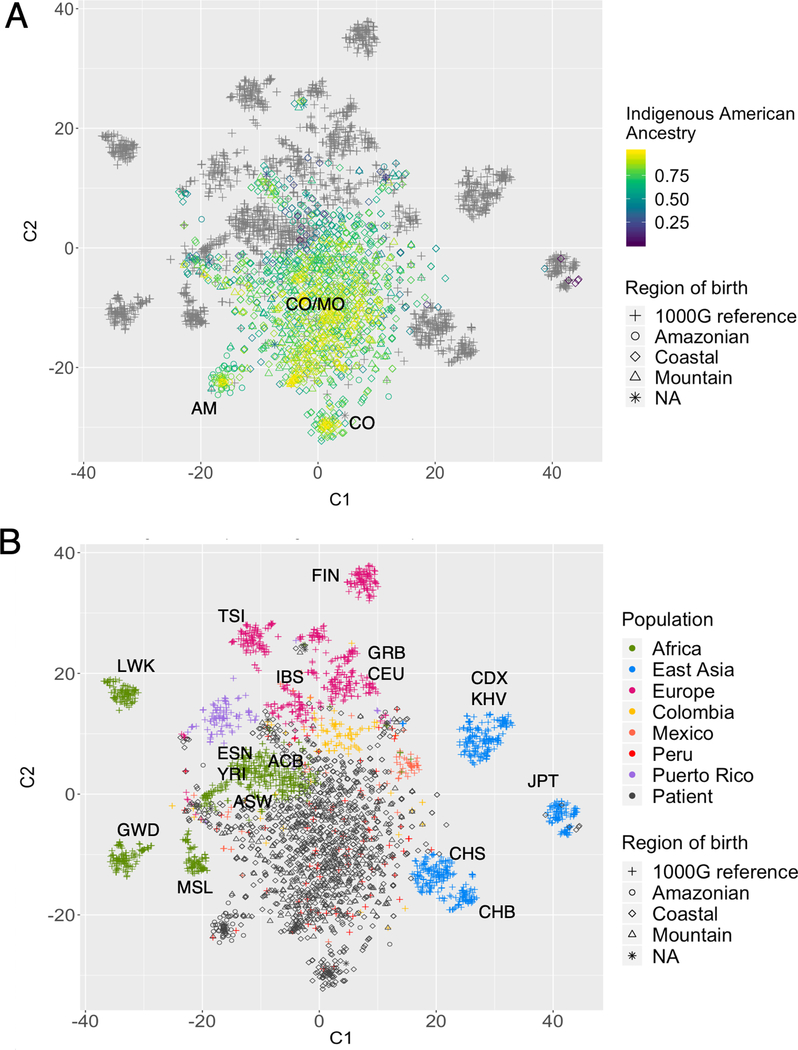
Results from the model-based continental genetic ancestry proportion estimation showed high IA ancestry proportions in the PEGEN-BC study patients (Table 1). The estimated average IA ancestry was 76%, the average European ancestry 18%, the average African ancestry 4%, and the average East Asian ancestry 2% (Table 1, Figure 1A, B and C). The PEGEN-BC study includes 128 (9.8%) participants with an estimated genetic IA ancestry of more than 95% and 75 (5.7%) had estimated genetic IA ancestry of more than 98%. PCA analysis shows that most of the INEN patients are concentrated in the highly IA range of genetic variation of the 1000G Admixed American populations, with few patients clustering with the Asian and African samples (Figure 1D). We were able to identify genetic structure at the subcontinental level using PCA and Rt-SNE approaches (Figure 2). The IA component of the study participants can be divided into three groups that correlate with the place of birth reported by the patient. We found two relatively small clusters with high IA ancestry cores, each of them composed mainly of patients from the Amazonian and the Coastal regions (Figure 2A, AM and CO clusters, respectively). A larger and more dispersed cluster, including patients mainly from the Coast and the Mountains (Figure 2A, CO/MO cluster), also contained patients with high IA ancestry. This observation is in line with previously described IA population migration patterns and genetic admixture in Peru(38). A small number of individuals in the study were identified who had high proportion of Asian or African ancestry (>50%), each group accounting for ~0.5% of the patients. Those with high East Asian ancestry clustered with the 1000G Japanese (Figure 2B), while patients with high degree of African ancestry clustered with West African populations (Figure 2B).
Figure 1:
Population structure of 1,312 Peruvian breast cancer patients. A and B) ADMIXTURE clustering analysis using unsupervised models assuming K = 4 for all patients and by region of birth, respectively. AM= Amazonian, CO= Coastal, MO= Mountain. C) Boxplots show the distribution of each ancestry component. Median ancestry value is represented as a solid line, interquartile range [IQR] as a box, and whiskers extend up to 1.5*IQR. Potential outliers are depicted as solid points. D) PCA including the PEGEN-BC study patients and 1000G samples. PC1, PCA2 and PC3: first three principal components.
Figure 2:
Rt-SNE analysis clusters continental subpopulations. A) IA ancestry values of Peruvian patients is shown. AM= Amazonian, CO= Coastal, MO= Mountain. B) Same plot showing continents and Latin American subpopulations in colors. Other continental subpopulations are annotated in the plot. ACB: African Caribbeans in Barbados; ASW: Americans of African Ancestry in SW USA; CDX: Chinese Dai in Xishuangbanna, China; CEU: Utah Residents (CEPH) with Northern and Western European Ancestry; CHB: Han Chinese in Beijing, China; CHS: Southern Han Chinese; ESN: Esan in Nigeria; FIN: Finnish in Finland; GBR: British in England and Scotland; GWD: Gambian in Western Divisions in the Gambia; IBS: Iberian Population in Spain; JPT: Japanese in Tokyo, Japan; KHV: Kinh in Ho Chi Minh City, Vietnam; LWK: Luhya in Webuye, Kenya; MSL: Mende in Sierra Leone; TSI: Toscani in Italy; YRI: Yoruba in Ibadan, Nigeria. C1 and C2: two t-SNE components.
Among women in the Amazonian cluster, the incidence of ER/PR-HER2− tumors was higher compared to other patients (19% vs. ~13%, respectively), concordant with what has been previously reported(39). However, likely due to the small size of the Amazonian cluster, this association is not statistically significant (p=0.89).
Subtypes and Continental Genetic Ancestry
As described above, univariate analysis showed a statistically significant difference in proportion of IA genetic ancestry between tumor subtypes (p-value = 0.023, Table 1). The average IA ancestry among participants with ER/PR- HER2+ tumors was 80% compared to 75% among participants with the ER/PR+ HER2− subtype. The results of a logistic regression model testing the association between HER2 status and IA ancestry showed a strong association with and without covariates (Table 2). The odds of having a HER2+ tumor increased by a factor of 1.19 per every 10% increase in IA ancestry (p-value=0.002). The results of the multinomial logistic regression analysis showed that the odds of ER/PR- HER2+ breast cancer increased by a factor of 1.22 per every 10% increase in IA ancestry (The ER/PR+ HER2− was defined as the reference group). This association remained statistically significant in the multivariable model (OR = 1.30, 95% CI = 1.10–1.55, p = 0.002) (Table 2). As expected given the high correlation between the IA and European components of ancestry, we observed a decrease in the odds of developing ER/PR- HER2+ disease with increasing European ancestry (Supplementary Table S1).
Table 2.
Association between ER/PR/HER2 defined groups and Indigenous American genetic ancestry (for every 10% increase)
| Covariates | Subtype | N | OR | 95% CI | P-value |
|---|---|---|---|---|---|
| By HER2 status | |||||
| Univariate | HER2− | 837 | Ref. | ||
| HER2+ | 392 | 1.10 | 1.02–1.19 | 0.012 | |
| Age dx + Afr + Height | HER2− | 688 | Ref. | ||
| HER2+ | 335 | 1.19 | 1.06–1.33 | 0.002 | |
| Age dx + Afr + Height + rs140068132 | HER2− | 688 | Ref. | ||
| HER2+ | 335 | 1.20 | 1.07–1.35 | 0.001 | |
| By ER/PR/HER2 status | |||||
| Univariate | ER/PR+ HER2− | 638 | Ref. | ||
| ER/PR- HER2+ | 156 | 1.22 | 1.08–1.37 | 0.001 | |
| ER/PR+ HER2+ | 239 | 1.06 | 0.97–1.16 | 0.175 | |
| ER/PR/HER2− | 200 | 1.07 | 0.97–1.18 | 0.165 | |
| Age dx + Afr + Height | ER/PR+ HER2− | 531 | Ref. | ||
| ER/PR- HER2+ | 129 | 1.30 | 1.08–1.55 | 0.004 | |
| ER/PR+ HER2+ | 209 | 1.17 | 1.02–1.33 | 0.024 | |
| ER/PR/HER2− | 158 | 1.10 | 0.95–1.27 | 0.196 | |
| Age dx + Afr + Height + rs140068132 | ER/PR+ HER2− | 531 | Ref. | ||
| ER/PR- HER2+ | 129 | 1.32 | 1.10–1.57 | 0.002 | |
| ER/PR+ HER2+ | 209 | 1.18 | 1.03–1.35 | 0.014 | |
| ER/PR/HER2− | 158 | 1.11 | 0.96–1.29 | 0.141 | |
Age dx: age at diagnosis; Afr: African genetic ancestry proportion
We also conducted the logistic regression analysis including the rs140068132-G protective SNP at 6q25 to test if the association between the HER2+ status and IA ancestry could be driven by this polymorphism. However, the association between HER2 status and IA ancestry was not explained by this protective variant (Table 2).
Given the Hospital-based nature of the study we evaluated the possibility that ancestry was associated with tumor subtype due to IA ancestry being higher among women from remote regions who would more likely be referred to INEN with aggressive tumors. However, in analyses that included stage at diagnosis and region of residence as covariates the association between genetic ancestry and tumor subtype was unchanged (Supplementary Table S2).
Replication of the association was conducted in samples from Mexico and Colombia. Given the relatively small sample size for replication we tested the association by HER2 status only (positive or negative), and analyzed samples from the two countries together including country as a covariate in the model (Table 3). The analysis showed a strong association between HER2 status and IA ancestry in the same direction as our analysis in the PEGEN-BC study patients (Table 3). For every 10% increase in IA ancestry among Mexican and Colombian breast cancer patients we observed a 28% increase in the odds of having a HER2+ tumor (Table 3). There was no evidence of heterogeneity by country (Colombia OR = 1.28, 95%CI 1.03–1.60; Mexico OR = 1.20, 95%CI 0.90–1.59).
Table 3:
Replication of Association between HER2* status and Indigenous American ancestry in Colombian and Mexican breast cancer patients, N=616
| Covariates | Subtype | N | OR | 95% CI | P-value |
|---|---|---|---|---|---|
| Country | HER2− | 460 | Ref. | ||
| HER2+ | 156 | 1.26 | 1.08–1.49 | 0.004 | |
| Age dx + Afr + Country | HER2− | 460 | Ref. | ||
| HER2+ | 156 | 1.27 | 1.07–1.52 | 0.006 |
Age dx: age at diagnosis; Afr: African genetic ancestry proportion.
Colombia ER/PR+ HER2– 231, ER/PR+ HER2+ 70, ER/PR− HER2+ 59 and ER/PR/HER2– 87; Mexico ER/PR+ HER2– 120, ER/PR+ HER2+ 18, ER/PR− HER2+ 9 and ER/PR/HER2– 22.
Discussion
Breast cancer subtype is a strong predictor of prognosis. Women diagnosed with ER/PR+ tumors tend to have better outcomes compared to women diagnosed with ER/PR- disease(36, 40). Multiple studies have been conducted evaluating factors (genetic, behavioral and environmental) that could explain subtype specific breast cancer risk(21, 41–45). Information about factors contributing to risk of developing HER2+ subtypes is limited but includes common germline variants(11, 19–21).
We evaluated the association between genetic ancestry and breast cancer subtype in a Latin America population with a high degree of IA genetic ancestry in order to assess the possibility that the previously reported higher relative frequency of HER2+ tumors among women of Latin American origin could be in part due to ancestry-specific genetic predisposition embedded within the IA ancestral component.
The clinical characteristics of the Peruvian breast cancer patients included in this study were similar to those described in other Latin American populations with young age and relatively high proportion of the most aggressive tumor subtypes (ER/PR-)(7, 46, 47). Furthermore, it reflects well the tumor characteristics of the overall patient population at INEN based on the similarities between our study patient characteristics and those included in a previously published review on INEN patient characteristics(48, 49). The average IA genetic ancestry in the Peruvian breast cancer patients was 76%, which was consistent with previous estimates of 70–80% IA ancestry in Peru(31, 50), including the 1000G Peruvians, who were also recruited in Lima. Taken together, these data suggest that our samples, even though they are clinic-based, are representative of the expected levels of IA ancestry in the general population of Peru.
Multiple studies have reported a higher incidence of HER2+ tumors in women of Latin American origin(8–12, 14). The results of the current analysis in breast cancer patients from Peru, and including independent replication in patients from Mexico and Colombia, strongly suggest there could be population-specific genetic variant(s) increasing the risk of HER2+ tumors among women with IA ancestry. These results are consistent with the previously reported suggestive association between IA ancestry and erbb2 gene expression (which codes for the HER2 protein) in breast tumors from Colombia(51).
There are various potential mechanisms behind the association between HER2 status and genetic ancestry, both environmental and/or genetic. Genetic risk factors could be acting in cis as well as in trans. Some examples of possible cis regulated mechanisms that could lead to differences in HER2 protein expression would be the presence of population-specific expression quantitative trait loci (eQTLs)(52), splice variants(53) or variants affecting the probability that erbb2 is amplified in tumor cells(54). Trans regulation of HER2 protein expression could result from eQTLs or splice variants for genes that regulate erbb2 expression. The reported associations between rare and common germline mutations in TP53 and HER2+ breast cancer (19, 55, 56) provide some support for this hypothesis. The exact mechanism for the association between TP53 mutations, SNPs, and HER2+ tumors is not well-understood, but is part of a larger body of subtype-specific common and rare variant associations with breast cancer(44, 57, 58). Large Admixture Mapping and genome wide association studies (GWAS) by tumor subtype in women of IA ancestry will be needed to identify the relevant germline variants and gain clarity regarding specific mechanisms for the observed association between IA ancestry and HER2 status.
The ER/PR- HER2+ tumor category had a larger OR than ER/PR+ HER2+ in the PEGEN-BC study, indicating that germline regulation of HER2 expression may not be the only mechanism at play in the relationship between HER2 status and IA ancestry. The difference between ER/PR+ vs. ER/PR- subtype could be random -both estimates are within each other’s 95% CI- or could be reflecting hormone receptor-context dependent expression and/or amplification of HER2(59).
In addition, environmental risk factors may also account for our observation. People of IA ancestry may have more exposure to risk factors that specifically increase the risk of HER2+ tumors. Some risk factors, such as body mass index, are known to differentially affect the risk of luminal vs. basal breast cancer subtypes(43). Although the HER2+ breast cancers are not known to have any specific environmental risk factors to date, additional studies including migration studies may help us understand whether this observation is related to environmental and/or genetic causes.
This is, to our knowledge, the first study evaluating the association between genetic ancestry and breast cancer subtypes in a sample of highly IA women. The IA genome is largely understudied. In 2016, Latinos made up 17.8% of the United States population(60). In that same year, however, that population made up only 0.54% of participants in GWAS(61). And just 0.05% of participants in GWAS were of non-admixed IA ancestry(61). Our study not only included admixed Latinas but a population in which 9.8% of the individuals had over 95% IA genetic ancestry. Additionally, our study collected detailed clinical data on tumor characteristics and other relevant patient information from electronic medical records, which allowed us to compare breast cancer subtypes. Discovering common variants of small effects contributing to breast cancer in non-White populations is challenging due to the relatively limited number of samples available for these groups. However, as was the case in the discovery of the 6q25 protective variant, leveraging genetic admixture could provide a shortcut for identifying specific areas that are likely to carry subtype specific risk variants (24, 25).
The PEGEN-BC study followed a hospital-based case-only design. As a result, the distribution of the subtypes in our study population might not represent the distribution of subtypes in the overall population of Peruvian breast cancer patients. It is possible that women with slow growing or easily treatable tumors either are never treated or receive care somewhere else. However, because this is the main cancer hospital in Peru and equally accessible (or inaccessible) to patients in remote areas such as the jungle lowlands or the mountains, our study participants are likely to be a good representation of patients from all over the country and of diverse breast cancer phenotypes, though likely from the lower end of the country’s income distribution. To account for this potential bias, we evaluated models that included stage and region of residence as covariates (Supplementary Table 1). Our results were unchanged.
This study reported a strong association between IA ancestry and HER2 status. The strongest association was observed for the ER/PR- HER2+ subtype, with individuals from this group having higher average IA ancestry compared to individuals with ER/PR+ HER2− tumors. Our results provide support for the hypothesis that there might be population-specific genetic risk factors predisposing women of IA ancestry to develop HER2+ tumors. Further studies are needed to confirm the genetic basis of this association and discover specific regions or variants within the IA genome. Ultimately, a better understanding of these and other etiological factors explaining breast cancer HER2 status will result in better subtype-specific polygenic risk prediction and improved targeted treatments, not only for women of Latin American origin, but in all women.
Supplementary Material
Statement of significance.
The positive association between Indigenous American genetic ancestry and HER2+ breast cancer suggests that the high incidence of HER2+ subtypes in Latinas might be due to population and subtype-specific genetic risk variants.
Acknowledgments
We want to thank the biobank at the Instituto Nacional de Enfermedades Neoplásicas, Lima, Peru, for their assistance managing and storing the material for the study. We also want to thank participants from the PEGEN-BC study and the COLUMBUS Consortium.
The PEGEN-BC study was supported by the National Cancer Institute [R01CA204797 (L.F.)] and the Instituto Nacional de Enfermedades Neoplásicas (Lima, Peru).
The COLUMBUS Consortium was supported by grants from School of Medicine (Dean’s Fellowship in Precision Health Equity to LC-C) and support from the Office of the Provost for LC-C’s Latinos United for Cancer Health Advancement, LUCHA, Initiative); GSK Oncology (Ethnic Research Initiative to LC-C and ME); The U.S. National Institutes of Health (Cancer Center Support Grant P30CA093372 from the National Cancer Institute). The contents of this article are solely the responsibility of the authors and do not reflect the official views of the National Institutes of Health. LC-C, MEB and ME are also grateful for support Colciencias (Graduate Studentship to Jennyfer Benavides, member of COLUMBUS, from Convocatoria para la Formación de Capital Humano de Alto Nivel para el Departamento de Tolima-COLCIENCIAS –755/2016), Universidad del Tolima (Grants to MEB and ME, project 10112), and Sistema Nacional de Regalías, Gobernación del Tolima (Grants to MEB and ME, projects 520120516 and 520115). JT was supported by Coordinacion Nacional de Investigación en Salud, IMSS, México, grant FIS/IMSS/PROT/PRIO/13/027 and by the Consejo Nacional de Ciencia y Tecnologia (Fronteras de la Ciencia grant 773), México.
The COLUMBUS Consortium (in alphabetical order) include: Jennyfer Benavides (Universidad del Tolima, Ibagué, Colombia), Mabel Bohorquez (Universidad del Tolima, Ibagué, Colombia), Fernando Bolaños (Hospital Hernando Moncaleano Perdomo, Neiva, Colombia), Luis G Carvajal-Carmona (University of California Comprehensive Cancer Center, Sacramento, Jenny Carmona (Dinámica IPS, Medellín, Colombia), Ángel Criollo (Universidad del Tolima, Ibagué, Colombia), Magdalena Echeverry (Universidad del Tolima, Ibagué, Colombia), Ana Estrada (Universidad del Tolima, Ibagué, Colombia),Gilbert Mateus (Hospital Federico Lleras Acosta, Ibagué, Colombia), Raúl Murillo (Pontificia Universidad Javeriana, Bogotá, Colombia), Justo Ramirez (Hospital Hernando Moncaleano Perdomo, Neiva, Colombia), Yesid Sánchez (Universidad del Tolima, Ibagué, Colombia), Carolina Sanabria (Instituto Nacional de Cancerología, Bogotá, Colombia), Martha Lucia Serrano (Instituto Nacional de Cancerología, Bogotá, Colombia), John Jairo Suarez (Universidad del Tolima, Ibagué, Colombia), Alejandro Vélez (Dinámica IPS, Medellín, Colombia, Hospital Pablo Tobón Uribe, Medellín, Colombia).
Footnotes
Disclosure of Potential Conflicts of Interest: The authors declare no potential conflicts of interest.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- 2.United States Department of Health and Human Services C for DC and P. National Program of Cancer Registries and Surveillance, Epidemiology, and End Results SEER*Stat Database: NPCR and SEER Incidence – U.S. Cancer Statistics 2001–2015 Public Use Research Database. 2018. [Google Scholar]
- 3.Hsu JL, Glaser SL, West DW. Racial/Ethnic Differences in Breast Cancer Survival Among San Francisco Bay Area Women. 1997;89:1311–1312. [DOI] [PubMed] [Google Scholar]
- 4.Li FP, Hankey BF, Clegg LX, Chu K, Edwards BK. Cancer Survival Among US Whites and Minorities. Arch Intern Med 2003;162:1985. [DOI] [PubMed] [Google Scholar]
- 5.Williams Pinheiro, Melanie Miller, Eric A, Easterday Stephanie, Moonie Sheniz, Trapido Edward J. PS. Cancer survival among Latinos and the Hispanic Paradox. Cancer Causes Control 2011;22:553–561. [DOI] [PubMed] [Google Scholar]
- 6.Ooi SL, Martinez ME, Li CI. Disparities in breast cancer characteristics and outcomes by race/ethnicity. Breast Cancer Res Treat 2011;127:729–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iqbal J, Ginsburg O, Rochon PA, Sun P, Narod SA. Differences in breast cancer stage at diagnosis and cancer-specific survival by race and ethnicity in the United States. JAMA 2015;313:165–73. [DOI] [PubMed] [Google Scholar]
- 8.Parise CA, Bauer KR, Caggiano V. Variation in breast cancer subtypes with age and race/ethnicity. Crit Rev Oncol Hematol 2010;76:44–52. [DOI] [PubMed] [Google Scholar]
- 9.Parise CA, Bauer KR, Brown MM, Caggiano V. Breast Cancer Subtypes as Defined by the Estrogen Receptor (ER), Progesterone Receptor (PR), and the Human Epidermal Growth Factor Receptor 2 (HER2) among Women with Invasive Breast Cancer in California, 1999–2004. Breast J 2009;15:593–602. [DOI] [PubMed] [Google Scholar]
- 10.Howlader N, Altekruse SF, Li CI, Chen VW, Clarke CA, Ries LAG, et al. US incidence of breast cancer subtypes defined by joint hormone receptor and HER2 status. J Natl Cancer Inst 2014;106:dju055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Banegas MP, Tao L, Altekruse S, Anderson WF, John EM, Clarke CA, et al. Heterogeneity of breast cancer subtypes and survival among Hispanic women with invasive breast cancer in California. Breast Cancer Res Treat 2014;144:625–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hines LM, Risendal B, Byers T, Mengshol S, Lowery J, Singh M. Ethnic Disparities in Breast Tumor Phenotypic Subtypes in Hispanic and Non-Hispanic White Women. J Women’s Heal 2011;20:1543–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Serrano-Gómez SJ, Fejerman L, Zabaleta J. Breast Cancer in Latinas: A focus on intrinsic subtypes distribution. Cancer Epidemiol Biomarkers Prev 2018;27:3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Serrano-Gomez SJ, Sanabria-Salas MC, Hernández-Suarez G, García O, Silva C, Romero A, et al. High prevalence of luminal B breast cancer intrinsic subtype in Colombian women. Carcinogenesis 2016;37:669–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martínez ME, Gomez SL, Tao L, Cress R, Rodriguez D, Unkart J, et al. Contribution of clinical and socioeconomic factors to differences in breast cancer subtype and mortality between Hispanic and non-Hispanic white women. Breast Cancer Res Treat 2017;166:185–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen L, Cook LS, Tang M-TC, Porter PL, Hill DA, Wiggins CL, et al. Body mass index and risk of luminal, HER2-overexpressing, and triple negative breast cancer. Breast Cancer Res Treat 2016;157:545–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ambrosone CB, Zirpoli G, Ruszczyk M, Shankar J, Hong CC, McIlwain D, et al. Parity and breastfeeding among African-American women: Differential effects on breast cancer risk by estrogen receptor status in the Women’s Circle of Health Study. Cancer Causes Control 2014;25:259–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bandera E V, Chandran U, Hong C-C, Troester MA, Bethea TN, Adams-Campbell LL, et al. Obesity, body fat distribution, and risk of breast cancer subtypes in African American women participating in the AMBER Consortium. Breast Cancer Res Treat 2015;150:655–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang H, Ahearn TU, Lecarpentier J, Barnes D, Beesley J, Jiang X, et al. Genome-wide association study identifies 32 novel breast cancer susceptibility loci from overall and subtype-specific analyses. bioRxiv 2019;778605.doi: 10.1101/778605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holm J, Eriksson L, Ploner A, Eriksson M, Rantalainen M, Li J, et al. Assessment of breast cancer risk factors reveals subtype heterogeneity. Cancer Res 2017;77:3708–3717. [DOI] [PubMed] [Google Scholar]
- 21.Gaudet MM, Gierach GL, Carter BD, Luo J, Milne RL, Weiderpass E, et al. Pooled Analysis of Nine Cohorts Reveals Breast Cancer Risk Factors by Tumor Molecular Subtype. Cancer Res 2018;78:6011–6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fejerman L, John EM, Huntsman S, Beckman K, Choudhry S, Perez-Stable E, et al. Genetic Ancestry and Risk of Breast Cancer among U.S. Latinas. 2008;doi: 10.1158/0008-5472.CAN-08-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fejerman L, Romieu I, John EM, Lazcano-ponce E, Beckman KB, Pérez-stable EJ, et al. European ancestry is positively associated with breast cancer risk in Mexican women. Natl Institutes Heath 2011;19:1074–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fejerman L, Chen GK, Eng C, Huntsman S, Hu D, Williams A, et al. Admixture mapping identifies a locus on 6q25 associated with breast cancer risk in US Latinas. Hum Mol Genet 2012;21:1907–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fejerman L, Ahmadiyeh N, Hu D, Huntsman S, Beckman KB, Caswell JL, et al. Genome-wide association study of breast cancer in Latinas identifies novel protective variants on 6q25. Nat Commun 2014;5:5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Affymextrix. Axiom ® Precision Medicine Research Array Data Sheet. [Google Scholar]
- 27.Purcell SM, Chang CC, Chow CC, Tellier LCAM, Lee JJ, Vattikuti S. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 2015;4:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Das S, Forer L, Schönherr S, Sidore C, Locke AE, Kwong A, et al. Next-generation genotype imputation service and methods. Nat Genet 2016;48:1284–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shieh Y, Fejerman L, Lott PC, Marker K, Sawyer SD, Hu D, et al. A polygenic risk score for breast cancer in U.S. Latinas and Latin-American women. J Natl Cancer Inst 2019;doi: 10.1093/jnci/djz174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alexander DH, Novembre J, Lange K Fast model-based estimation of ancestry in unrelated individuals. Genome Res 2009;19:1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sandoval JR, Salazar-Granara A, Acosta O, Castillo-Herrera W, Fujita R, Pena SDJ, et al. Tracing the genomic ancestry of Peruvians reveals a major legacy of pre-Columbian ancestors. J Hum Genet 2013;58:627–634. [DOI] [PubMed] [Google Scholar]
- 32.van der Maaten LJP, Hinton G Visualizing High-Dimensional Data Using t-SNE. J Mach Learn Res 2008;9(Nov):2579–2605. [Google Scholar]
- 33.van der Maaten LJP . Accelerating t-SNE using Tree-Based Algorithms. J Mach Learn Res 2014;15(Oct):3221–3245. [Google Scholar]
- 34.Krijthe JH. Rtsne: T-Distributed Stochastic Neighbor Embedding using a Barnes-Hut Implementation. available at <https://github.com/jkrijthe/Rtsne>. [Google Scholar]
- 35.Team RC. R: A language and environment for statistical computing. 2018;available at <https://www.r-project.org/>.
- 36.Blows FM, Driver KE, Schmidt MK, Broeks A, van Leeuwen FE, Wesseling J, et al. Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: A collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med 2010;7:e1000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang P, Tse GM. Immunohistochemical surrogates for molecular classification of breast carcinoma: A 2015 update. Arch Pathol Lab Med 2016;140:806–814. [DOI] [PubMed] [Google Scholar]
- 38.Harris DN, Song W, Shetty AC, Levano KS, Cáceres O, Padilla C, et al. Evolutionary genomic dynamics of Peruvians before, during, and after the Inca Empire. Proc Natl Acad Sci U S A 2018;115:E6526–E6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tamayo LI, Vidaurre T, Navarro Vásquez J, Casavilca S, Aramburu Palomino JI, Calderon M, et al. Breast cancer subtype and survival among Indigenous American women in Peru. PLoS One 2018;13:e0201287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A 2003;100:8418–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benefield HC, Zabor EC, Shan Y, Allott EH, Begg CB, Troester MA. Evidence for etiologic subtypes of breast cancer in the Carolina Breast Cancer Study. Cancer Epidemiol Biomarkers Prev 2019;cebp.0365.2019.doi: 10.1158/1055-9965.epi-19-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rey-Vargas L, Sanabria Salas MC, Fejerman L, Serrano-Gómez SJ. Risk factors for triple negative breast cancer among Latina women. Cancer Epidemiol Biomarkers Prev 2019;cebp.0035.2019.doi: 10.1158/1055-9965.epi-19-0035. [DOI] [PubMed] [Google Scholar]
- 43.Shieh Y, Scott CG, Jensen MR, Norman AD, Bertrand KA, Pankratz VS, et al. Body mass index, mammographic density, and breast cancer risk by estrogen receptor subtype. Breast Cancer Res 2019;21:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Milne RL, Kuchenbaecker KB, Michailidou K, Beesley J, Kar S, Lindström S, et al. Identification of ten variants associated with risk of estrogen-receptor-negative breast cancer. Nat Genet 2017;49:1767–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Purrington KS, Slager S, Eccles D, Yannoukakos D, Fasching PA, Miron P, et al. Genome-wide association study identifies 25 known breast cancer susceptibility loci as risk factors for triple-negative breast cancer. Carcinogenesis 2014;35:1012–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Srur-Rivero N, Cartin-Brenes M. Breast Cancer Characteristics and Survival in a Hispanic Population of Costa Rica. Breast Cancer (Auckl) 2014;5–7.doi: 10.4137/BCBCR.S15854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller KD, Goding Sauer A, Ortiz AP, Fedewa SA, Pinheiro PS, Tortolero-Luna G, et al. Cancer Statistics for Hispanics/Latinos, 2018. CA Cancer J Clin 2018;68:425–445. [DOI] [PubMed] [Google Scholar]
- 48.Vallejosa CS, Gómez HL, Cruza WR, Pintoa JA, Dyerb RR, Velardec R, et al. Breast Cancer Classification According to Immunohistochemistry Markers: Subtypes and Association With Clinicopathologic Variables in a Peruvian Hospital Database. Clin Breast Cancer 2010;10:294–300. [DOI] [PubMed] [Google Scholar]
- 49.Fejerman L, Ziv E. Population differences in breast cancer severity. Pharmacogenomics 2008;9:323–333. [DOI] [PubMed] [Google Scholar]
- 50.Homburger JR, Moreno-Estrada A, Gignoux CR, Nelson D, Sanchez E, Ortiz-Tello P, et al. Genomic Insights into the Ancestry and Demographic History of South America. In: Tarazona-Santos E, editor. PLOS Genet 2015;11:e1005602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Serrano-Gómez SJ, Sanabria-Salas MC, Garay J, Baddoo MC, Hernández-Suarez G, Mejía JC, et al. Ancestry as a potential modifier of gene expression in breast tumors from Colombian women. PLoS One 2017;12:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Su Y, Jiang Y, Sun S, Yin H, Shan M, Tao W, et al. Effects of HER2 genetic polymorphisms on its protein expression in breast cancer. Cancer Epidemiol 2015;39:1123–7. [DOI] [PubMed] [Google Scholar]
- 53.Caswell JL, Camarda R, Zhou AY, Huntsman S, Hu D, Brenner SE, et al. Multiple breast cancer risk variants are associated with differential transcript isoform expression in tumors. Hum Mol Genet 2015;24:7421–7431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.LaFramboise T, Weir BA, Zhao X, Beroukhim R, Li C, Harrington D, et al. Allele-Specific Amplification in Cancer Revealed by SNP Array Analysis. PLoS Comput Biol 2005;1:e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilson JRF, Bateman AC, Hanson H, An Q, Evans G, Rahman N, et al. A novel HER2-positive breast cancer phenotype arising from germline TP53 mutations. J Med Genet 2010;47:771–774. [DOI] [PubMed] [Google Scholar]
- 56.Melhem-Bertrandt A, Bojadzieva J, Ready KJ, Obeid E, Liu DD, Gutierrez-Barrera AM, et al. Early onset HER2-positive breast cancer is associated with germline TP53 mutations. Cancer 2012;118:908–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ha SM, Chae EY, Cha JH, Kim HH, Shin HJ, Choi WJ. Association of BRCA mutation types, imaging features, and pathologic findings in patients with breast cancer with BRCA1 and BRCA2 Mutations. Am J Roentgenol 2017;209:920–928. [DOI] [PubMed] [Google Scholar]
- 58.Chen H, Wu J, Zhang Z, Tang Y, Li X, Liu S, et al. Association between BRCA status and triple-negative breast cancer: A meta-analysis. Front Pharmacol 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Daemen A, Manning G. HER2 is not a cancer subtype but rather a pan-cancer event and is highly enriched in AR-driven breast tumors. Breast Cancer Res 2018;20:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.PEPASR6H Annual Estimates of the Resident Population by Sex, Age, Race, and Hispanic Origin for the United States and States : April 1, 2010 to July 1, 2014 2014 Population Estimates. 2016;available at <data.census.gov>. [Google Scholar]
- 61.Popejoy AB, Fullerton SM. Genomics is failing on diversity. Nature 2016;538:161–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.