Abstract
Growth factors (GFs) are critical components in governing cell fate during tissue regeneration. Their controlled delivery is challenging due to rapid turnover rates in vivo. Functionalized hydrogels, such as heparin-based hydrogels, have demonstrated great potential in regulating GF release. While the retention effects of various concentrations and molecular weights of heparin have been investigated, the role of geometry is unknown. In this work, 3D printing is used to fabricate GF-embedded heparin-based hydrogels with arbitrarily complex geometry (i.e., teabag, flower shapes). Simplified cylindrical core–shell structures with varied shell thickness are printed, and the rates of GF release are measured over the course of 28 days. Increasing the shell layers’ thickness decreases the rate of GF release. Additionally, a mathematical model is developed, which is found capable of accurately predicting GF release kinetics in hydrogels with shell layers greater than 0.5 mm thick (R2 > 0.96). Finally, the sequential release is demonstrated by printing two GFs in alternating radial layers. By switching the spatial order, the delivery sequence of the GFs can be modulated. This study demonstrates how 3D printing can be utilized to fabricate user-defined structures with unique geometry in order to control the rate of GF release in hydrogels.
Keywords: 3D printing, controlled drug release, heparin, hyaluronic acid, hydrogels, sequential release, vascular endothelial growth factors
1. Introduction
Growth factors (GFs) regulate proliferation and differentiation of cells in order to promote tissue regeneration.[1,2] GF turnover is rapid in vivo, resulting in short serum half-lives. In order to better stimulate tissue regeneration, tissue engineering strategies often seek to control the release of GFs.[3] Owing to their controllable degradability and capability to protect enveloped molecules from degradation, hydrogels are often employed to regulate GF release.[4,5] They are cross-linked hydrophilic polymers with water as the dispersion medium. The high water content—typically 70–99%—supports encapsulation of hydrophilic molecules, such as GFs, without denaturation and aggregation.[3] Their network structures are also believed to hamper penetration of various proteins, thus preventing premature degradation of encapsulated bioactive molecules from diffusing enzymes.[6] The rich water content also provides physical environments similar to native tissue, thus offering an intimate environment for cells to reside and grow. However, due to high water content, GFs tend to diffuse out quickly from hydrogels since there are no moieties for them to attach.
Heparin is a natural linear polysaccharide that is most noted for its anticoagulation effects. It consists of an alternating sequence of disaccharide units with 1→4 linked 2-O-sulfated iduronic acid and 6-O-sulfated, N-sulfated glucosamine. These building blocks contribute three sulfate moieties per repeating unit, making heparin have the highest negative charge density of any known biological molecule.[7] Due to its high negative charge density, heparin can trap positively charged common proteins, such as GFs, by electrostatic forces, which can be used to prolongate GF release from hydrogels that traditionally are released rapidly from hydrogels.[8–11] Previous studies have discovered that the kinetics of GF release can be modulated by varying the molecular weight and concentration of heparin in the hydrogel; increased heparin molecular weight and increased heparin concentration result in protracted GF release. This is likely due to a higher electrostatic retention force, which keeps GFs from exiting the hydrogels.[12] However, the low molecular weights of heparin (5–15 kDa) and the repulsion of negative charges limit their crosslinkability. Additional modifications such as acrylation also reduce heparins’ solubility in water to form a stable hydrogel. Thus, most heparin-based systems require another component to create a matrix for modified heparins to reside in.[9,12] Hyaluronic acid (HA) is a hydrogel which has been widely engineered for applications such as wound healing and atopic dermatitis due to its role in granulation and cell migration.[13–15] Previous studies have shown that the synthesis of glycidyl methacrylate HA (HA-GM) allows HA to be compatible with light-based 3D printing, and provides a mechanism to tune the physical properties and geometry of the hydrogel.[16] The combination of HA and heparin further allows for the ability to modify GF release kinetics over extended periods of time from hydrogels.[17]
To better understand the hydrogel-based GF delivery system, there is a need to theoretically model the drug release profiles from hydrogels. Mathematical models of mass transport in traditional polymeric controlled drug delivery systems have been developed, guiding the design and fabrication of such a delivery system, and enabling accurate prediction of drug release kinetics.[18] While the effect of physical shape on the drug release kinetics of conventional dosage forms (i.e., capsules) has been extensively investigated, the influence of hydrogel shape on long-term GF release kinetics remains to be quantified.[19] Fundamental studies of the diffusion of proteins in hydrogels have provided some insights on their basic mechanisms. However, due to the lack of control over the shape of hydrogels during fabrication, the role of spatial arrangement within the hydrogels on release kinetics is largely unknown.[20,21]
Recent developments in light-based 3D printing allow for the precise control of hydrogel geometry. As one of the most advanced additive manufacturing methods, digital light projection (DLP) based 3D printing techniques have become pivotal in the rapid fabrication of customized hydrogel systems due to its speed and printing resolution.[22–25] DLP-based 3D printing uses a light modulating device, such as a digital micromirror device (DMD), to reflect light in user-defined patterns onto a photosensitive resin. This printing technique has been used to fabricate hydrogel structures with tissue informed, complex geometry, as well as incorporate biomaterials such as GFs and cells directly into the scaffold structure.[26,27] The fast printing speed (typically within 1 min) is also crucial in printing GF-containing hydrogels where release occurs rapidly.[10] Furthermore, DLP-based 3D printing can be used to print functional biomaterials which have been modified to support photopolymerization—such as thiolated heparin (Hep-SH)—in order to tune the hydrogel properties like GF retention without affecting other qualities such as mechanical properties.[8,17,28,29] Therefore, as the influence of hydrogel geometry on GF release kinetics is unknown, the goal of this study is to investigate how geometry influences GF release kinetics in a hydrogel. Hep-SH and HA-GM were synthesized, combined with GFs, and 3D printed into arbitrarily complex shapes (Figure 1). A core + shell hydrogel laden with GFs was fabricated using a DLP-based 3D printer, then growth factor release kinetics were evaluated over the course of 28 days (Figure 2). We hypothesized that a barrier layer of HA-GM + Hep-SH could prolong GF release and this bilayer structure could additionally be used for the sequential release of multiple GFs. Finally, the GF release profile from this structure was analyzed, and a mathematical model of release kinetics of each 3D hydrogel design was developed in order to understand how geometry informs the ability to predict GF release kinetics for these structures.
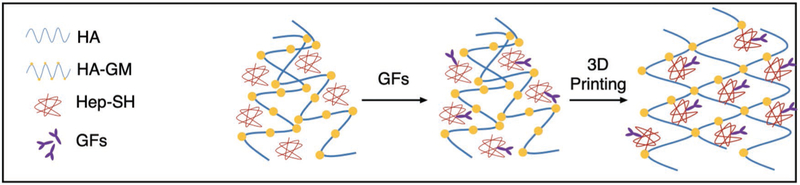
Figure 1.
Schematic of synthesis and polymerization. Thiolated heparin (Hep-SH) was synthesized and mixed with glycidyl methacrylated hyaluronic acid (HA-GM) and growth factors (GFs), followed by 3D printing into a stable hydrogel.
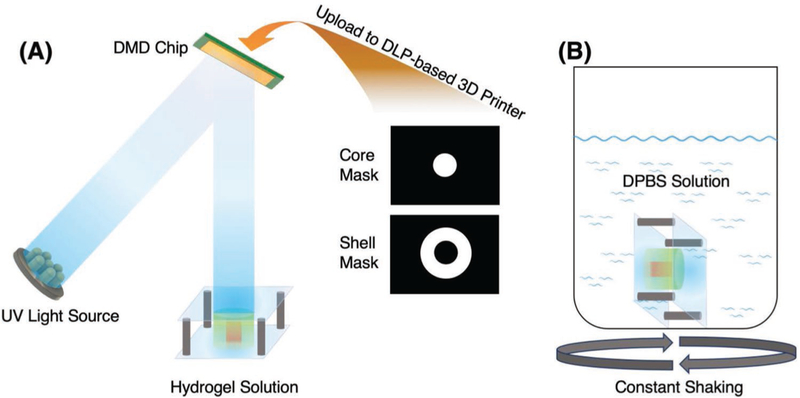
Figure 2.
A) Schematic of the DLP-based 3D printing process. The bilayer core–shell structure was fabricated by two digital masks. The first hydrogel was printed into a cylindrical shape and the remaining unprinted hydrogel was rinsed. Then the second hydrogel was printed into a ring shape around the core. B) The printed hydrogel structures were immersed into DPBS solution to study GF release kinetics.
2. Results and Discussion
2.1. Synthesis Confirmation and UV Crosslinking Confirmation from Fourier Transform Infrared (FTIR) Spectroscopy
To confirm the substitution of the thiol group on heparin, we conducted Ellman’s test on the heparin solution, before and after the reaction. The assay results indicated 33% substitution of the carboxylic group by the thiol group, similar to previous studies.[8,30] The substitution by thiol groups is less than half of the overall sulfate groups on the heparin backbone, leaving the majority of the negative charges untouched. The substitution was further confirmed by comparing the FTIR and 1H-NMR of the product and native heparin (Figure S1, Supporting Information). The broad absorption peak over 1600 cm−1 indicated the substitution by the secondary amine from cystamine. The heparin and Hep-SH were dissolved in deuterium oxide (D2O) and examined by nuclear magnetic resonance (NMR). The chemical shift at δ 2.6 and δ 2.85 ppm also confirmed the substitution of the thiol group in the Hep-SH. To further confirm the thiol–ene reaction, we mixed Hep-SH with PEGDA and photoinitiator. After UV exposure, the reduction of the absorption peak of alkene around 900 cm−1 confirmed the thiol–ene reaction of heparin.
2.2. Rapid 3D Printing of Multimaterial Structures with Complex Geometry
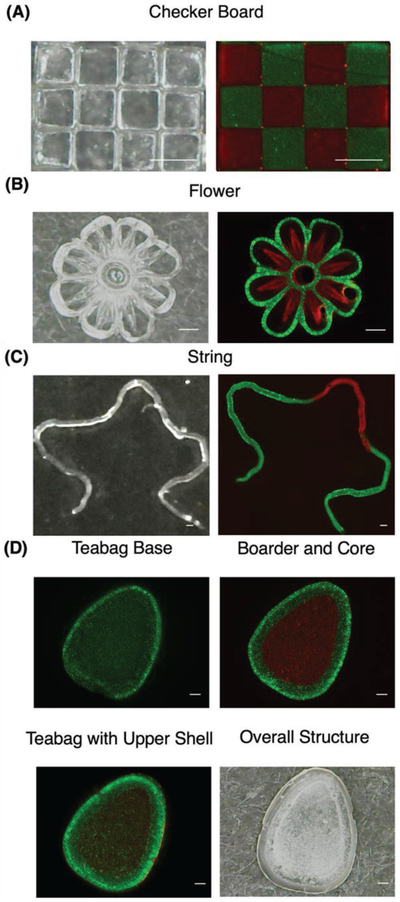
To demonstrate the capability of 3D printing of complex microstructures with multiple materials, checker board, flower, string, and teabag structures were printed with fluorescein isothiocyanate (FITC) and tetramethylrhodamine isothiocyanatedextran (TRITC-dextran) containing hydrogels (Figure 3). The checker board structure contains 200 µm × 200 µm squares in 3 × 4 arrays with distinct edges separating each other. The flower was designed with a 40 µm line width and 1 mm in overall diameter and the string was printed in three masks with 100 µm in line width and 8 mm in total length, demonstrating printing of multicomponent structures in both micrometer and millimeter scales. The teabag structure was constructed to be 1 mm thick. All structures except the teabag design were printed within 1 min. The total processing time for the teabag design was 3 min due to the multiple steps involved. Their fluorescent images indicate that DLP-based 3D printing can be used to rapidly print high-resolution, multicomponent structures, with arbitrarily complex geometries. Combining with the inherent retaining effects of heparins, it could be used for delivery of GF or other complex molecules.
Figure 3.
A–D) Optical and fluorescent images of checker board, flower, string, and teabag-inspired structure printed with FITC (green) and TRITC-dextran (red) containing hydrogels. Scale bar = 200 µm.
2.3. Delayed Release of GFs from HA-GM + Hep-SH Hydrogels
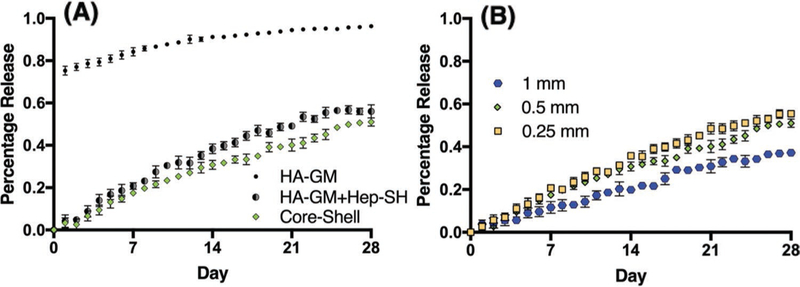
Previous studies have demonstrated the synthesis and polymerization of protein retention hydrogels and the difference in retention between Hep-SH and HA-GM.[17] The large pore sizes of HA allow encapsulated GFs to diffuse out of the hydrogels without obstruction. Thus, it provides an ideal platform to test the GF retention effect due to the addition of heparin. The initial concentration gradient of GFs between the HA-GM hydrogel and the Dulbecco’s phosphate buffered saline (DPBS) solution act to drive pierce bicichoninic acid (BCA) out of the hydrogel so that equilibrium is reached. Without any effective retaining agents, the release profile follows zero-order pulsatile release, with more than 70% of bovine serum albumin (BSA) diffused out of the hydrogel after day 1 (Figure 4A). The initial burst diffusion of GFs quickly depletes the reservoir of GFs in the hydrogel, thus reducing the chemical potential. The diffusion of BCA out of the HA-GM hydrogel plateaued after 15 days, reaching a local concentration equilibrium after 20 days where 95% of the initial amount of BCA was released from the hydrogel.
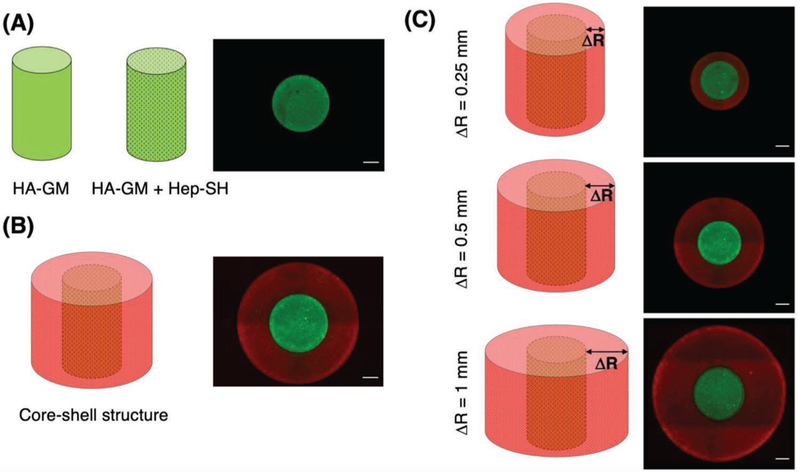
Figure 4.
Design for 3D printing and structural validation with FITC (green) and TRITC-dextran (red) containing hydrogels. A) Cylindrical core-only structures, B) cylindrical core–shell structures, and C) core–shell structures with varied outer shell thicknesses. Scale bar = 200 µm.
To evaluate the GF retention effect of heparin, we incorporated Hep-SH into the BSA containing HA-GM hydrogel and printed into the same cylindrical structure. The incorporation of Hep-SH into the HA-GM hydrogel was found to significantly decrease the amount of BSA released (p < 0.0001). A post hoc analysis showed a significant decrease in the amount of GF retained at all time points (p < 0.0001). The release profile of BSA from the Hep-SH + HA-GM hydrogel indicated that less than 10% of BSA had diffused out of the hydrogel on the first day, much less than the 70% BSA diffusion in the HA-GM-only hydrogel (Figure 4B). This is likely due to the increased electrostatic forces attributed to the sulfate groups in heparin. After 28 days, the Hep-SH + HA-GM hydrogel retained 44% of BSA, compared to 4% of the initial BSA in the HA-GM-only hydrogel. These experiments confirm HA-GM itself has poor retention of GFs, but the addition of Hep-SH drastically improves drug retention.
2.4. Effect of a GF-Laden Core with Hep-SH + HA-GM Shell on GF Release
The systematic release of GFs is integral to restoring native tissue function after injury. The ability to tailor the release of various GFs in a tissue engineered system allows for a more physiologically relevant replication of the natural cascade of GFs after injury. The DLP-based 3D printing system has previously been used to fabricate HA-GM hydrogels with various geometric designs.[16,31,32] In this study, we hypothesized that the core–shell design would further inhibit GF release, through the addition of a GF retention boundary with no initial concentration of GF. The DLP 3D printer was used to fabricate a multilayer and multimaterial structure, consisting of two layers: (1) a cylindrical HA-GM + Hep-SH core with BSA, and (2) a HA-GM + Hep-SH shell with no GF incorporated.
BSA release in core-only and core–shell capsules was examined over the course of 28 days (Figure 5A). The inclusion of the core layer resulted in a significant decrease in the amount of BSA released at all time points after 8 days (p = 0.036 to p < 0.0001) with the exception of day 15 where a trend was found (p = 0.0534). This confirms the hypothesis that the inclusion of an additional hydrogel layer not containing any GFs can result in prolonged GF release. For example at day 20, 49% of the BSA had diffused out of the core-only structure, compared to 39% of BSA that had diffused out of the cylindrical core– shell structure. These experiments demonstrate the GF release profile of a hydrogel can be easily modified by the inclusion of a heparin containing shell, which serves to further slowdown diffusion of GFs out of a hydrogel structure.
Figure 5.
Release kinetics of various structures with different compositions and shell thicknesses. A) Release profile of HA-GM-only versus HA-GM with Hep-SH core-only versus core–shell structure. Two-way repeated measures ANOVA with Sidak post hoc tests revealed that incorporating Hep-SH ( ) into the HA-GM-only (
) into the HA-GM-only ( ) hydrogel significantly decreased the amount of GF released retained the GF for all time points (p < 0.0001). Furthermore, an additional layer of Hep-SH not containing any GF (
) hydrogel significantly decreased the amount of GF released retained the GF for all time points (p < 0.0001). Furthermore, an additional layer of Hep-SH not containing any GF ( ) was found to further decrease total GF releases after day 8 (p = 0.036 to p < 0.0001). B) Release profile of core–shell structures with different shell thicknesses. Doubling the layer thickness reduced the release rates after day 3 (p = 0.0137 to p < 0.0001) whereas halving the layer increased the release after day 14 (p = 0.0009 to p < 0.0001). Data presented as mean ± SD, n = 4 for all data points
) was found to further decrease total GF releases after day 8 (p = 0.036 to p < 0.0001). B) Release profile of core–shell structures with different shell thicknesses. Doubling the layer thickness reduced the release rates after day 3 (p = 0.0137 to p < 0.0001) whereas halving the layer increased the release after day 14 (p = 0.0009 to p < 0.0001). Data presented as mean ± SD, n = 4 for all data points
In order to further control GF release kinematics, the effect of varying the thickness of the outer shell was examined. Core–shell scaffolds were printed with a shell size double (1 mm) and half (250 µm) of the original design (0.5 mm). All release profiles still follow first-order release. Doubling the shell size resulted in a significantly decreased amount of GF released at all time points after day 3 (p = 0.0137 to p < 0.0001; Figure 5B). At the conclusion of the experiment, only 37% of the BSA had diffused out of the cylindrical core–shell structure with doubled shell thickness, compared to 51% of the BSA in the baseline design. Similarly, reducing the shell size resulted in a significantly increased amount of drug released at all time points after day 14 (p = 0.0009 to p < 0.0001). After 28 days, 56% of the BSA had diffused out of the cylindrical core–shell structure with halved thickness. Overall, the change in shell thickness significantly altered GF release kinetics (p < 0.0001). These observations indicate the release kinetics of a GF in a HA-GM + Hep-SH structure can be further tuned by varying the geometrical design, which can be easily realized by 3D printing.
2.5. Mathematical Model Derivation
In order to understand the release mechanism of GFs from the hydrogel systems, we derived a mathematical model based on the Fick’s second law
| (1) |
where c denotes the concentration of the GFs within the hydrogel system, D is the diffusion coefficient of the GF, t and x are time and position, respectively. Since both caps of the cylindrical structure were covalently bonded to the coverslips, the GF can only diffuse out of the hydrogel radially. As hydrogel swelling reaches equilibrium after the first day under incubation, the effect of the geometric change and water penetration over time was considered fixed. Also, the initial distribution of GFs within the hydrogel was considered homogeneous at the beginning of the experiment, therefore the initial conditions were defined as
| (2) |
where ci is the initial GF concentration and R is the equilibrium radius of the cylinder after swelling. The aqueous solution is well stirred, contains a volume much greater than the volume of the printed structure, and was therefore considered a perfect sink. Thus, the concentration of GFs at far away was assumed to be zero. No accumulation of GFs is considered at the surface of the film. Therefore, the rate at which the GFs diffuse to the hydrogel/solution boundary through the shell was equivalent to the rate at which GFs were depleted from the hydrogel. This is proportional to the concentration gradient between the surface (cs) and the concentration required to maintain equilibrium with the surrounding environment (c∞). Therefore, this boundary condition can be represented mathematically as
| (3) |
where h is the mass transfer coefficient in the shell. Using Laplace transform,[19] Equation (1) can be solved as
For the cylindrical structure
| (4) |
For the cylindrical core–shell structure
| (5) |
where Mt and M∞ are the cumulative amounts of GF released at time t and equilibrium, respectively. They are results from concentration items (cs and c∞) from Equation (3) transformed over time. A represents the total surface area of the hydrogel, D denotes the diffusion coefficient of GF in the hydrogel. H is the height of the cylinder. The partition coefficient (K) is a measure of relative concentration between the interface shell and the solution at the equilibrium condition. It is related to the initial GF concentration and shell thickness. V represents the volume of the GF containing hydrogel, R is the radius of the cylinder, and ΔR represents the thickness of the shell.
2.6. Modeling Drug Release Profiles to Predict Release Kinetics
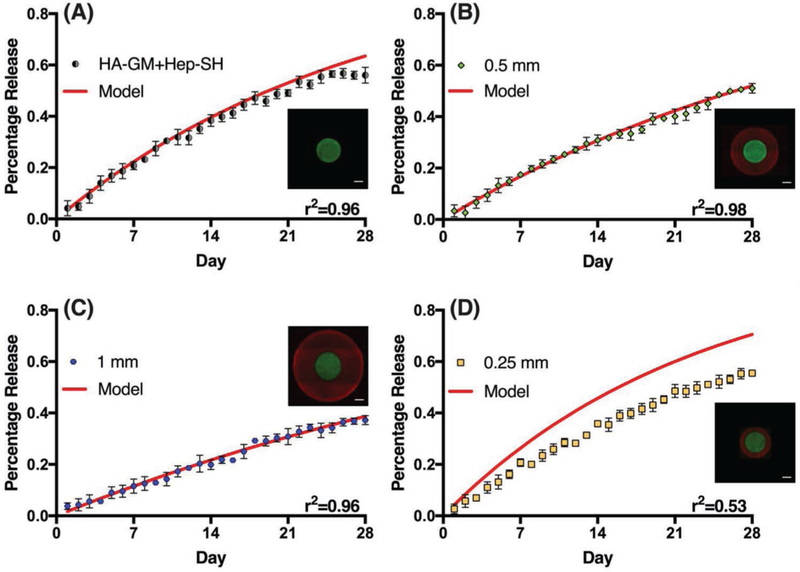
Understanding the relationship between the scaffold geometry and GF release kinetics allows for predictive design of GF release platforms. In order to predict the GF release profiles from Equations (4) and (5), we first had to measure the intrinsic diffusion coefficient (D) and partition coefficient (K) of BSA in a HA-GM + Hep-SH hydrogel. To estimate the intrinsic diffusion coefficient, we initially fit the release profile of BSA in a core-only hydrogel to Equation (4). The diffusion coefficient of BSA in a HA-GM + Hep-SH hydrogel was found to be 0.0045 mm2 d−1 with 96% of the variance explained by the model. Then, using the measured diffusion coefficient, the partition coefficient was obtained by fitting the release profile of BSA in a baseline core–shell design to Equation (5). The partition coefficient of BSA in a HA-GM + Hep-SH hydrogel was found to be 0.485 with 98% of the variance predicted by the model.
In order to evaluate how well the models predict BSA release in structures with different shell thicknesses, models with ΔR = 1 mm, where the shell thickness is twice of the core radius (2:1 shell-to-core thickness ratio) and ΔR = 250 µm where the shell thickness is half of the core radius (1:2 shell-to-core thickness ratio) were compared to experimentally measured data (Figure 6). By systematically changing the core-to-shell thickness ratio, we are able to carefully evaluate how the size of the core–shell design can influence GF release kinetics. Excellent agreement was found for the model with ΔR = 1 mm, with the model explaining 96% of the variance. However, poor agreement was found for the model with ΔR = 250 µm, with the model explaining 53% of the variance in the data. This may be due to a higher concentration gradient across the shell of the thinner shell structures, resulting in a larger driving force and less obstruction for BSA to diffuse out of the structure. This will affect the relative concentrations at equilibrium, which in turn would affect the partition coefficient (K). With a larger driving force and a shorter distance to travel before exiting into the aqueous solution, the concentration of GF inside the hydrogel will be lower in the sample with a thinner shell at equilibrium. Therefore, we hypothesize that the partition coefficient for the ΔR = 0.25 mm should be smaller than what was measured in the ΔR = 0.5 mm model.
Figure 6.
Evaluation of models predicting BSA release in structures with different designs and shell thicknesses. A) HA-GM + Hep-SH core-only structure, B) baseline core–shell structure with shell thickness = 0.5 mm, C) core–shell structure with shell thickness = 1 mm, D) core–shell structure with shell thickness = 0.25 mm. Scale bar = 200 µm. Data presented as mean ± SD with the coefficients of determination (r2) stated at the bottom right of each graph.
2.7. Sequential GF Release
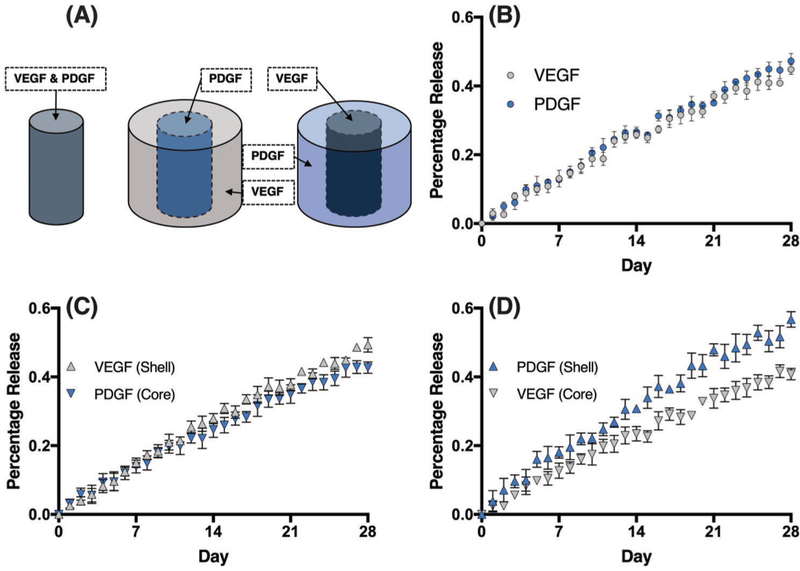
In vivo, GFs are sequentially released in order to initialize cell differentiation and tissue regeneration. For example, the sequential release of vascular endothelial growth factor (VEGF) followed by platelet derived growth factor (PDGF) has been shown to improve the regeneration of cardiomyocytes.[33,34] As the DLP-based 3D printed system is capable of printing cylindrical core–shell structures with spatial control to alter the release kinetics of GFs, we sought to simultaneously release multiple GFs over an extended period of time. Additionally, we sought to investigate how the spatial ordering of GFs within the cylindrical core–shell structure affects the sequence and rate of GF release (Figure 7A). As a control, VEGF and PDGF were mixed into a HA-GM + Hep-SH hydrogel, and printed into a cylindrical structure and the release of each GF was examined by ELISA. The release profile indicated that both GFs are released following first-order pulsatile release similar to BSA. Additionally, PDGF was found to diffuse slightly faster than VEGF (p = 0.0115; Figure 7B).
Figure 7.
Sequential release of VEGF and PDGF. A) Schematics of structural designs. B) Release profile of nonhierarchical structure. PDGF ( ) was found to have a higher diffusion rate than VEGF (
) was found to have a higher diffusion rate than VEGF ( ) with p = 0.0115. C) Release profile of structure with shell containing VEGF (
) with p = 0.0115. C) Release profile of structure with shell containing VEGF ( ) and core containing PDGF (
) and core containing PDGF ( ). The geometrical design reduced the diffusion of VEGF as compared to PDGF (p < 0.0001). D) Release profile of structure with reversed spatial order with PDGF in the shell (
). The geometrical design reduced the diffusion of VEGF as compared to PDGF (p < 0.0001). D) Release profile of structure with reversed spatial order with PDGF in the shell ( ) and VEGF in the core (
) and VEGF in the core ( ). The PDGF was found to diffuse faster than VEGF in this design (p < 0.0001). Data presented as mean ± SD, n = 4.
). The PDGF was found to diffuse faster than VEGF in this design (p < 0.0001). Data presented as mean ± SD, n = 4.
In order to deliver the VEGF earlier than PDGF, we adapted a similar core–shell approach by printing structures loaded with PDGF first in the core, followed by printing the shell loaded with VEGF. The release profile indicated that VEGF was released faster than PDGF (p < 0.0001) and reached 40% release almost 3 days before PDGF reached the same level (Figure 7C). These data support that the sequential release of different GFs from the same structure can be obtained by using the core–shell design. To further explore the flexibility of cylindrical core–shell structure designs, the spatial order of the GFs was reversed by printing a structure with VEGF in the core and PDGF in the shell to examine whether the system will work in the opposite order. As anticipated, the release profile indicated that PDGF was released faster than VEGF (p < 0.0001), reaching 40% released 5 days before VEGF (Figure 7D). Additionally, the amount of VEGF released decreased when printed into the core of the structure as compared to the homogeneously mixed structure (p = 0.0135), supporting that the core–shell structure decreases the GF release rate, even if there is another GF within the shell layer. As PDGF was released faster in the homogeneously mixed structure, it is unsurprising that the difference in the release rates of VEGF and PDGF was greatest when PDGF was located in the shell of the structure. This likely can be contributed to the higher native diffusion rate of PDGF in this hydrogel system. Finally, both GFs had a significant decrease in release rate when printed in the core of the structure compared to the shell, respectively (VEGF: p = 0.0002; PDGF: p < 0.0001). These observations further support that the core–shell design can be utilized to modify not only the GF release rates but also the sequence in which GFs are predominantly released from a hydrogel.
3. Conclusions
This study demonstrates the effect of geometry and spatial ordering of GF-laden hydrogels on GF release kinetics. Heparin was incorporated into a hydrogel to provide retention for GFs. Capsule-inspired, cylindrical core–shell hydrogel structures were 3D printed to study the effect of spatial arrangement on GF release kinetics. The DLP-based 3D printing system used in this study demonstrated excellent speed and flexibility for rapid fabrication of precision engineered multimaterial structures. Furthermore, an initial diffusion model was evaluated in order to predict GF release kinetics for structures with varying wall thickness. While the diffusion model was ineffective at predicting diffusion kinetics in structures with shell layers approaching 0 mm thickness, the model had excellent agreement with experimental data in structures with a shell layer greater than or equal to 0.5 mm. The failure of the model to predict release kinetics in structures with a smaller shell thickness may be the result from the change in geometric design, which will affect the relative concentration of GFs across the shell at equilibrium which in turn affects the partition coefficient. Finally, the core–shell model was utilized to demonstrate the simultaneous release of multiple growth factors, and how geometry can be manipulated in order to preferentially release different GFs in a particular sequence. In addition to geometry, future studies can further manipulate GF release kinetics by varying heparin molecular weight, which may increase control over GF release. Additionally, since hydrogel-based GF delivery systems yield high initial release of GF due to high water content, the ability to control initial delivery release is of interest. Overall, this study demonstrates that the geometry of 3D-printed, GF-laden hydrogels can be modified in order to release GF with predictable release kinetics over long periods of time.
4. Experimental Section
Materials:
HA with a molecular weight of 200 kDa was purchased from Lifecore Biomedical (Chaska, MN). 1-Ethyl-3-[3-(dimethylamino) propyl] carbodiimide (EDC), sodium chloride, Ellman’s test kit, and BCA kit were purchased from Thermo Fisher Scientific (Waltham, MA). 1-Hydroxybenzotriazole (HOBt) was purchased from Tokyo Chemical Industries America (Portland, OR). Hydrochloric acid (HCl) cystamine, l-cystine, tris(2-carboxyethyl) phosphine (TCEP), GM, dimethyl phenylphosphonite, 2,4,6-trimethylbenzoyl chloride, lithium bromide, 2-butanone, FITC-dextran, TRITC-dextran, D2O, and triethylamine (TEA) were purchased from Millipore Sigma (St. Louis, MO). Heparin was purchased from Celsus Laboratory (Cincinnati OH). Dialysis shells (10 000 MWCO) were purchased from Spectrum Laboratories (Rancho Dominguez, CA). All chemicals were used as received. 1× DPBS was purchased from Gibconco (Carlsbad, CA). Rat-VEGF and rat-PDGF were purchased from R&D Systems (Minneapolis, MN). The respective ELISA kits were purchased from RayBiotech Life (Norcross, GA). BSA was purchased from Gemini Bio-products (Woodlands, CA). Sodium hydroxide (NaOH) was purchased from Spectrum Chemical (Gardena, CA). Polydimethylsiloxane (PDMS) sheets with defined thickness were purchased from Specialty Manufacturing, Inc. (Saginaw, MI).
Synthesis of Hep-SH:
Hep-SH was synthesized by adapting a previous protocol.[17] 200 mg of heparin was dissolved into 40 mL of MilliQ water and reacted with 91.04 mg of EDC and 65.05 mg HOBt. The pH was adjusted to 6.8 before 78.06 mg of cystamine was added. The pH of the reaction solution was readjusted to 6.8 and left at room temperature for reaction over 5 h. The reaction solution was then dialyzed against deionized (DI) water for 24 h before lyophilization. The lyophilized powder was then dissolved in 40 mL of water before 966.7 mg of TCEP was added. The pH of the solution was readjusted to 7.5. The neutralization process was continued for 1 h at room temperature. The final product was dialyzed against 5 M NaCl solution with pH adjusted to 5 by HCl for 24 h before changing into the solution of the same pH without the salt. The final product was then lyophilized and stored in −20 °C before use. The reagent and product were examined by FTIR spectroscopy (PerkinElmer, Waltham, MA) and NMR (Bruker, Billerica, MA) to confirm the substitution. The degree of substitution was determined by Ellman’s test.
Synthesis of HA-GM and Photoinitiator:
HA-GM was synthesized following a previously reported method.[16] Briefly, HA was dissolved in acetone/water solution with 1:1 volume ratio. After the HA powder fully dissolved, an equal amount of TEA and GM was added dropwisely. The reaction continued for 24 h before dialysis against distilled water. HA-GM foam was obtained after lyophilization, which was redissolved into DPBS at predetermined concentrations for printing.
The photoinitiator was synthesized by a previously reported protocol.[35] Dimethyl phenylphosphonate was mixed with 2,4,6-trimethylbenzoyl chloride under argon for 18 h before heated to 50 °C. Lithium bromide was mixed with 2-butanone and added into the reaction solution dropwisely. The reaction was cooled to room temperature while precipitates were collected and dried.
3D Printing of Hep-SH with HA-GM:
To investigate the effects of Hep-SH and spatial designs on the release of GFs, the hydrogels were printed into various shapes using the DLP-based 3D printing system.[36] The main components of the DLP-based 3D printing system include a computer for design and control, an ultraviolet (UV) light source (Ominicure 2000, Waltham, MA) for photopolymerization, lenses and DMD chip (Texas Instruments, Dallas, TX ) for light patterning, and precision three-axial stages. The DMD chip consists of 4 millions of micromirrors, each can be individually controlled using a binary, user-defined image. To print a structure, the UV light is projected onto the DMD chip, which can in turn project the light in a user-defined pattern onto a reservoir containing a photopolymerizable hydrogel (Figure 2A). The prepolymer solution consisted of 1% w/v Hep-SH, 4% w/v HA-GM, and photoinitiator in DPBS. To test their respective release kinetics, 200 mg of BSA, 0.1 mg of PDGF, and 0.01 mg of VEGF were dissolved in 1 mL of the prepolymer solution, respectively. During the 3D printing process, the mixed solution was loaded into the reservoir between two methacrylated glass slides as reported previously.[32] The glass slides were separated by a 1 mm tall PDMS sheet. Upon exposure of UV light, the hydrogel covalently bonded to the glass surface to prevent diffusion in the z-direction. After printing, the PDMS sheets were removed and UV glue was applied and cured at the four corners of the glass sides to prevent deformation of the hydrogel structure during postprinting handling. For GF release tests, the structure, together with the glass slides, was then immersed into DPBS reservoir to test the release kinetics (Figure 2B). The overall printing process takes 45 s. To demonstrate the versatility and complexity of the structures that DLP technologies can print, a checkerboard, flower, string, and teabag structure were printed with FITC and TRITC-dextran containing hydrogels (Figure 3). For the checkerboard, the first mask was designed with alternating squares. Hydrogel containing FITC-dextran was printed followed by brief wash. The second mask contains complimentary squares to the first mask, and used to print hydrogel containing TRTIC-dextran was printed (Figure 3A). The flower and string designs were printed similarly with multiple masks (Figure 3B,C). Additionally, a teabag-inspired structure, known for core–shell features with sustaining release of payloads, was printed in four steps (Figure 3D). First, the base was printed with FITC-dextran containing hydrogel, followed by the border and core segments printed with FITC and TRITC-dextran containing hydrogels, respectively. Finally, the top shell was printed to seal the payload inside.
Core–Shell Structure Designs for Drug Release Evaluation:
To quantify GF release kinetics, several cylindrical core–shell structures with variations of hydrogel composition and geometric shape were systematically tested (outlined in Table 1). First, in order to validate the ability of heparin to prolong GF release, HA-GM and HA-GM + Hep-SH hydrogels were mixed with BSA and printed into a single cylindrical core (Figure 4A). To further prolong GF release, a bilayer cylindrical core–shell structure, where a BSA containing HA-GM + Hep-SH hydrogel was surrounded by a BSA free HA-GM + Hep-SH hydrogel shell, was additionally printed around the core (Figure 4B). To further evaluate the effect of geometric design on the release profile, the thickness of the outer shell of the cylindrical core–shell structures was varied from 0.25 to 1 mm (Figure 4C). Finally, in order to determine whether the release kinetics of multiple GFs could be simultaneously controlled by the spatial order, VEGF and PDGF were encapsulated at either the core or the shell region.
Table 1.
Composition and geometric design of different 3D-printed scaffolds.
| Composition | Structure (growth factor) | Shell thickness [mm] |
|---|---|---|
| HA-GM | Core (BSA) | 0 |
| HA-GM, Hep-SH | Core (BSA) | 0 |
| HA-GM, Hep-SH | Core (BSA)/shell (none) | 0.5 |
| HA-GM, Hep-SH | Core (BSA)/shell (none) | 1 |
| HA-GM, Hep-SH | Core (BSA)/shell (none) | 0.25 |
| HA-GM, Hep-SH | Core (VEGF, PDGF) | 0 |
| HA-GM, Hep-SH | Core (VEGF)/shell (PDGF) | 0.5 |
| HA-GM, Hep-SH | Core (PDGF)/shell (VEGF) | 0.5 |
GF Release of the Hydrogel:
To assess the retention effect of HA-GM and BSA, VEGF and PDGF were added to HA-GM as payloads and printed into cylindrical and core–shell structures according to Table 1. Their release pattern was studied by either Pierce BCA assay or ELISA. GF release properties of the hydrogel structures were assessed every day for 28 d in sealed containment with 1 mL of DPBS. Every 24 h, 20 µL aliquots of DPBS were removed from the DPBS reservoir for analysis, and 20 µL of fresh DPBS was added to the aqueous solution in order to maintain the same volume of fluid in the solution. For each GF release profile, data points from four structures in separate DPBS reservoirs were collected. At the end of 28 days, the concentration of the GFs in each aliquot was determined by Pierce BCA assay for BSA release and ELISA for VEGF and PDGF releases. UV–vis spectrophotometer (Infinite 200 Pro, Tecan, Mannedorf, Switzerland) was used to examine the absorbance and fluorescence of each data point and compared to the standard curves to determine the concentration. These concentration values were then adjusted by the dilution factors from prior removals of DPBS to determine the release profile.
Statistics:
All concentration values used in this analysis were generated based on spectrophotometric comparison to standard curves to quantify the concentration of BSA, VEGF, and PDGF, respectively, as described before. For each geometric design, four identical samples were evaluated. To compare the difference in the drug release profile over time between the HA-GM and HA-GM + Hep-SH hydrogels, a two-way, repeated measures analysis of variance (ANOVA) (levels: time, Hep-SH ±) with a post hoc Sidak test was used. To compare the difference in the drug release profile over time between the core-only and cylindrical core–shell structures, a two-way ANOVA (levels: time, shell ±) with a post hoc Sidak test was used. The study of shell thickness on retention effect was conducted by a two-way ANOVA (levels: time, shell thickness) with a post hoc Sidak test with shell thickness of 0.5 mm as a control. The sequential release of multiple GFs by alternating spatial orders of multiple GFs was investigated by a two-way ANOVA with post hoc Sidak test (levels: time, spatial order). In order to evaluate the amount of variance explained by the mathematical model of GF release, the coefficient of determination (r2) was calculated between experimental data and the corresponding mathematical model for each geometric design. The threshold for significance (α) was set to 0.05 for all analysis. All statistics were performed using Prism (Version 7.0a, La Jolla, CA). All data are reported as mean ± standard deviation.
Supplementary Material
Acknowledgements
This work was supported in part by grants from the National Institutes of Health (R21AR074763, R01EB021857, R33HD090662) and National Science Foundation (1644967, 1937653).
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Conflict of Interest
The authors declare no conflict of interest.
Contributor Information
Pengrui Wang, Materials Science and Engineering Program, University of California San Diego, La Jolla, CA 92093, USA.
David Berry, Department of NanoEngineering, University of California San Diego, La Jolla, CA 92093, USA.
Amy Moran, Chemical Engineering Program, University of California San Diego, La Jolla, CA 92093, USA.
Frank He, Department of Bioengineering, University of California San Diego, La Jolla, CA 92093, USA.
Trevor Tam, Department of Bioengineering, University of California San Diego, La Jolla, CA 92093, USA.
Luwen Chen, Department of Chemistry and Biochemistry, University of California San Diego, La Jolla, CA 92093, USA.
Shaochen Chen, Materials Science and Engineering Program, University of California San Diego, La Jolla, CA 92093, USA; Department of NanoEngineering, University of California San Diego, La Jolla, CA 92093, USA; Chemical Engineering Program, University of California San Diego, La Jolla, CA 92093, USA; Department of Bioengineering, University of California San Diego, La Jolla, CA 92093, USA.
References
- [1].Lee K, Silva EA, Mooney DJ, Soc JR., Interface 2011, 8, 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Stocum DL, Science 1997, 276, 15. [DOI] [PubMed] [Google Scholar]
- [3].Li J, Mooney DJ, Nat. Rev. Mater 2016, 1, 16071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Yamamoto M, Ikada Y, Tabata Y, J. Biomater. Sci., Polym. Ed 2001, 12, 77. [DOI] [PubMed] [Google Scholar]
- [5].Whitaker MJ, Quirk RA, Howdle SM, Shakesheff KM, J. Pharm. Pharmacol 2001, 53, 1427. [DOI] [PubMed] [Google Scholar]
- [6].Su J, Hu B-H, Lowe WL, Kaufman DB, Messersmith PB, Biomaterials 2010, 31, 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Butterworth PJ, Cell Biochem. Funct 2005, 23, 293. [Google Scholar]
- [8].Fu A, Gwon K, Kim M, Tae G, Kornfield JA, Biomacromolecules 2015, 16, 497. [DOI] [PubMed] [Google Scholar]
- [9].Pike DB, Cai S, Pomraning KR, Firpo MA, Fisher RJ, Shu XZ, Prestwich GD, Peattie RA, Biomaterials 2006, 27, 5242. [DOI] [PubMed] [Google Scholar]
- [10].Wu J, Ye J, Zhu J, Xiao Z, He C, Shi H, Wang Y, Lin C, hang H, Zhao Y, Fu X, Chen H, Li X, Li L, Zheng J, Xiao J, Biomacromolecules 2016, 17, 2168. [DOI] [PubMed] [Google Scholar]
- [11].Tae G, Kim YJ, Choi WI, Kim M, Stayton PS, Hoffman AS, Biomacromolecules 2007, 8, 1979. [DOI] [PubMed] [Google Scholar]
- [12].Jha AK, Mathur A, Svedlund FL, Ye J, Yeghiazarians Y, Healy KE, J. Controlled Release 2015, 209, 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ballios BGG, Cooke MJJ, Donaldson L, Coles BLKLK, Morshead CMM, van der Kooy D, Shoichet MSS, van der Kooy D, Shoichet MSS, Stem Cell Rep 2015, 4, 1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Collins MN, Birkinshaw C, 2013, 92, 1262. [DOI] [PubMed] [Google Scholar]
- [15].Rape AD, Zibinsky M, Murthy N, Kumar S, Nat. Commun 2015, 6, 8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wang P, Li X, Zhu W, Zhong Z, Moran A, Wang W, Zhang K, Chen S, Bioprinting 2018, 12, e00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Jha AK, Tharp KM, Ye J, Santiago-Ortiz JL, Jackson WM, Stahl A, Schaffer DV, Yeghiazarians Y, Healy KE, Biomaterials 2015, 47, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Siepmann J, Siepmann F, J. Controlled Release 2012, 161, 351. [DOI] [PubMed] [Google Scholar]
- [19].Muschert S, Siepmann F, Leclercq B, Carlin B, Siepmann J, J. Controlled Release 2009, 135, 71. [DOI] [PubMed] [Google Scholar]
- [20].Zustiak SP, Boukari H, Leach JB, Soft Matter 2010, 6, 3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hettiaratchi MH, Schudel A, Rouse T, García AJ, Thomas SN, Guldberg RE, McDevitt TC, APL Bioeng 2018, 2, 026110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zhang AP, Qu X, Soman P, Hribar KC, Lee JW, Chen S, He S, Adv. Mater 2012, 24, 4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Soman P, Chen S, Fozdar D, U.S. Patent No. 9 631 171, 2017.
- [24].Zhu W, Ma X, Gou M, Mei D, Zhang K, Chen S, Curr. Opin. Biotechnol 2016, 40, 103. [DOI] [PubMed] [Google Scholar]
- [25].Cha C, Soman P, Zhu W, Nikkhah M, Camci-Unal G, Chen S, Khademhosseini A, Biomater. Sci 2014, 2, 703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Koffler J, Zhu W, Qu X, Platoshyn O, Dulin JN, Brock J, Graham L, Lu P, Sakamoto J, Marsala M, Chen S, Tuszynski MH, Nat. Med 2019, 25, 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Berry DB, You S, Warner J, Frank LR, Chen S, Ward SR, Tissue Eng., Part A 2017, 23, 980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Liang Y, Kiick KL, Acta Biomater 2014, 10, 1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Benoit DSW, Anseth KS, Acta Biomater 2005, 1, 461. [DOI] [PubMed] [Google Scholar]
- [30].Gwon K, Kim E, Tae G, Acta Biomater 2017, 49, 284. [DOI] [PubMed] [Google Scholar]
- [31].Zhu W, Qu X, Zhu J, Ma X, Patel S, Liu J, Wang P, Lai CSE, Gou M, Xu Y, Zhang K, Chen S, Biomaterials 2017, 124, 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ma X, Qu X, Zhu W, Li Y-S, Yuan S, Zhang H, Liu J, Wang P, Lai CSE, Zanella F, Feng G-S, Sheikh F, Chien S, Chen S, Proc. Natl. Acad. Sci 2016, 113, 2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bai Y, Bai L, Zhou J, Chen H, Zhang L, Cell. Immunol 2018, 323, 19. [DOI] [PubMed] [Google Scholar]
- [34].Awada HK, Johnson NR, Wang Y, J. Controlled Release 2015, 207, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Qu X, Zhu W, Huang S, Li JY-S, Chien S, Zhang K, Chen S, Biomaterials 2013, 34, 9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Pyo S-H, Wang P, Hwang HH, Zhu W, Warner J, Chen S, ACS Appl. Mater. Interfaces 2017, 9, 836. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.