Abstract
Chromatin can be viewed as a hierarchically structured fiber that regulates gene expression. It consists of a complex network of DNA and proteins whose characteristic dynamical modes facilitate compaction and rearrangement in the cell nucleus. These modes stem from chromatin’s fundamental unit, the nucleosome, and their effects are propagated across length scales. Understanding the effects of nucleosome dynamics on the chromatin fiber, primarily through post-translational modifications that occur on the histones, is of central importance to epigenetics. Within the last decade, imaging and chromosome conformation capture techniques have revealed a number of structural and statistical features of the packaged chromatin fiber at a hitherto unavailable level of resolution. Such experiments have led to increased efforts to develop polymer models that aim to reproduce, explain, and predict the contact probability scaling and density heterogeneity. At nanometer scales, available models have focused on the role of the nucleosome and epigenetic marks on local chromatin structure. At micrometer scales, existing models have sought to explain scaling laws and density heterogeneity. Less work, however, has been done to reconcile these two approaches: bottom-up and top-down models of chromatin. In this perspective, we highlight the multiscale simulation models that are driving toward an understanding of chromatin structure and function, from the nanometer to the micron scale, and we highlight areas of opportunity and some of the prospects for new frameworks that bridge these two scales. Taken together, experimental and modeling advances over the last few years have established a robust platform for the study of chromatin fiber structure and dynamics, which will be of considerable use to the chromatin community in developing an understanding of the interplay between epigenomic regulation and molecular structure.
Main Text
Genome packaging poses intriguing questions that are relevant not only to biology but also to polymer physics and chemistry. The human genome consists of billions of basepairs of DNA that are densely packed in the cell nucleus, well below the theoretical packing limit dictated by the persistence length of double-stranded DNA (∼50 nm). Such DNA packaging occurs over multiple length scales, eventually leading to a dense nuclear environment. Despite the dense packaging, DNA must be rendered accessible at the gene scale (∼kbp) for necessary processes such as replication (1), transcription (2), and DNA repair (3). The chromatin fiber provides an avenue for such functionality through its hierarchical and dynamic structure, overcoming strict DNA-packaging constraints to facilitate organization and epigenomic regulation across length scales (4,5). In particular, epigenomic regulation influences chromatin structure through covalent modifications of the fiber, such as post-translational modifications (PTMs) and DNA methylation, and substitutions of structural proteins.
There are well-documented links between chromatin structure, gene regulation, and epigenetics, but less is known about how these phenomena directly influence one another. Moreover, these features or processes are manifested at all relevant chromosomal length scales, thereby requiring multiscale methodologies for their study. Because the structure of chromatin is inherently dynamic and falls within the “dark region” (6) between conventional microscopy and x-ray crystallography, multiscale modeling is particularly useful in “filling the gaps.” Thus far, modeling efforts to describe chromatin have adopted two perspectives. Bottom-up approaches, including detailed descriptions of nucleosomes, have been primarily focused on the role of the nucleosome in chromatin; how does the nucleosome facilitate regulation of the underlying DNA, and how does the epigenome, in the form of PTMs and histone variants, affect nucleosome structure and dynamics? Bottom-up methods make use of atomistic and coarse-grained molecular models. Top-down approaches rely on chromosome conformation capture and immunoprecipitation techniques such as Hi-C (7), ChIA-PET (8), ChIP-seq (9), and Micro-C (10,11) to understand the three-dimensional (3D) genome—the physical organization of the genome in the nucleus. Such methods rely on polymer physics and statistical approaches to develop models that replicate specific behavior.
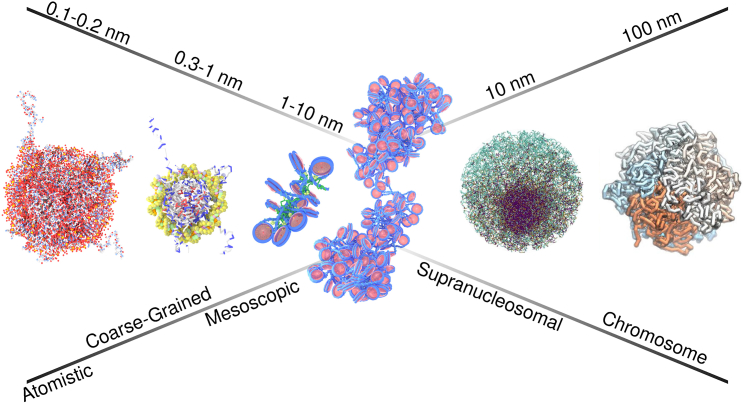
Bottom-up models of increasing sophistication have been able to capture nucleosome physics up to 10–100 kbp, and top-down methods parameterized with vast amounts of experimental data are broaching the 3D genome at a <1-kbp resolution. At this point in time, chromatin models are reaching a “convergence” of sorts, in which top-down and bottom-up methodologies can both resolve the intermediate length scales that hold many of the secrets that encode epigenetic information. Despite the wide disparity between these modeling philosophies, there is now a crossover and tremendous potential for cross-fertilization. At intermediate length scales, these two types of models are stretched to their limits; by combining the two, it should be possible to arrive at more complete and content-rich representations of chromatin. (Fig. 1).
Figure 1.
Current modeling approaches to chromatin. From the left, bottom-up modeling approaches begin with all-atom resolution, with each bead around the scale of 1–2 Å. The coarse-grained scale representation here is that of the three sites per nucleotide DNA model and the atomic-interaction-based coarse-grained protein model (22,97). Particles at this coarse-grained scale are in the range of 3–6 Å. The colors of the proteins here represent the net charge of the residue: white for no charge, blue for +1 charge, and red for −1 charge. The mesoscopic model is the 1CPN model, which represents each nucleosome as a cylinder and DNA as a sphere on the scale of 1 nm (53). Here, the 1CPN configuration is an oligomer with linker histones bound to the nucleosome. From the right, top-down models use coarse polymer representations of chromatin. The chromosomal model is the minimal chromatin model at a 50 kbp per bead resolution (81). The supranucleosomal model is the mean-field model representing chromatin at a nucleosome per bead resolution (90). This image has been published with approval of the Proceedings of the National Academy of Sciences of the United States of America. These models converge at intermediate, mesoscopic length scales and offer nucleosome-scale resolution and capture the relevant, underlying physics. To see this figure in color, go online.
A complete toolset is needed to address the following questions: what physics from the nucleosome scale influence the 3D genome? What physics do not? And how can they be manipulated? In this perspective, we discuss recent modeling advances, the potential to bridge length scales and timescales and, ultimately, the impact that forming such bridges will have in chromatin research.
Bottom-up design philosophy
In bottom-up models, significant efforts must be devoted to achieve representability. In contrast, however, they can achieve higher levels of transferability than very coarse, top-down models. For example, if a nucleosome is the system of choice, a bottom-up model that is able to reproduce the effects of histone modification on DNA affinity is likely to be capable of reproducing how nucleosomes interact and the thermodynamics that emerge from those interactions in a wide variety of contexts. Here, we briefly highlight various modeling and experimental efforts that, when coupled, can start with chromatin at the atomistic scale and take that information to the mesoscale. We also discuss several relevant questions that arise at each length scale.
Atomistic simulations: nucleosome structure and the epigenome
Experiments that rely on x-ray crystallography have provided an atomistically detailed view of the nucleosome, in which a stable complex is formed between an octamer of histones wrapped ∼1.7 times by a 147-bp, left-handed DNA superhelix (12,13). That view led directly to the first fully atomistic simulations of the nucleosome (14). Initial work at this scale focused on nucleosome stability and dynamics (15). More recent, ambitious efforts have considered nucleosome structure and function influenced by nucleosome-binding proteins (16), histone variants (17,18), or PTMs (19,20). These efforts are starting to elucidate major pieces of the role of the nucleosome in the epigenetic landscape. For example, Bowerman et al. revealed that H2A histone variants alter nucleosome dynamics, whereas Melters et al. explained that the enhanced elasticity of Cenp-A nuclesomes is related to enhanced levels of transcription (17,18).
Unfortunately, encompassing every permutation of histone modifications, histone variants, and nucleosome-binding proteins with atomistic resolution is beyond the currently available computational resources. And, although bottom-up methods are more likely to be transferable, transferability is not necessarily guaranteed. In general, atomistic models or force fields tend to not be as transferable as it is often presumed. The onset of machine learning for force field development could increase the transferability of atomistic simulations and address histone modifications with higher fidelity. More importantly, the question of how histone interactions translate into chromatin changes at longer scales is not accessible at this level of resolution (21), and careful coarse graining is therefore necessary to address it.
Coarse-grained nucleosome models feasibly resolve longer timescale and length-scale nucleosome dynamics while drawing information from the atomistic scale. Through coarse graining, nucleosome models can be better compared to experimental data, but extensive validation is needed to establish their limits of applicability. As a result, representability at this length scale often becomes a time-consuming endeavor. Nevertheless, significant progress in chromatin research has been performed at the coarse-grained nucleosome scale. Recent efforts have resolved several key questions raised by experiments (6,15,22). Freeman et al., for example, explained that the affinity of particular sequences of DNA to the histone core is encoded in the curvature of such sequences (22). Such models have thermodynamically assessed experimental force-induced unwrapping (6,23), sequence-dependent nucleosome stability (15,22), and nucleosome sliding (24,25).
Although single-nucleosome studies have been the primary focus of atomistic and coarse-grained models, multinucleosome studies have been carried out with coarse-grained models. With multiple nucleosomes, additional information is necessary. Internucleosome interactions and the length of linker DNA must also be considered—both are known to greatly influence the structure of chromatin (26,27). The internucleosome interaction has been difficult to quantify. Experiments pulling isolated chromatin fibers have attempted to extract the energy of association between nucleosomes within that context (28). Three significant studies have reported values of nucleosome interaction energies, with variable results (29, 30, 31). Support for these results in the form of atomistic simulations cite the histone tails as the primary contributor for these interactions (20). In particular, the positively-charged residues of the H4 tail are reported to attract nearby nucleosomes through interactions with an acidic patch located on the H2A histone of the nucleosome core (20). Acetylation of these residues disrupts these interactions and, to some degree, lowers the attractive interactions between nucleosomes (32, 33, 34). This phenomenon serves to underscore that epigenetic modifications directly influence structural changes in chromatin. One could conclude that the ubiquitous H4K16ac modification is associated with transcription because the chromatin becomes unraveled after this change in attractions between nucleosomes (35,36). The picture outlined above has now been put on firmer grounds through extensive simulations with coarse-grained models, in which internucleosome interactions were derived from more-detailed representations, leading to excellent agreement with experiments with and without H4K16ac (37). Importantly, such models provide a generalized description of nucleosome interactions as a function of both relative orientation and separation distance. Such a description is invaluable from a chromatin modeling perspective because interactions within the chromatin fiber have thus far been poorly characterized.
With such detailed characterization of nucleosome physics now available, current modeling efforts have taken different directions. Drawing inspiration from recent atomistic studies, one such direction has sought to investigate the influence of histone variants, nucleosome-binding proteins, etc., on nucleosome thermodynamics and structure. Another direction has sought to use the information now available at this scale to build models capable of describing longer molecules and scales. Examples are provided by the work of Watanabe et al., who have investigated small complexes with these models, such as dinucleosomes with the HP1 protein and trinucleosomes, to represent their effect on chromatin structure (38,39). Because of the limitations in sampling, however, such efforts can currently provide reliable descriptions of one or a few nucleosomes; scaling up to even a few kilobasepairs requires that a different, more coarse-grained class of models be developed.
Nucleosome-based coarse graining: mesoscale modeling
Within the constellation of nucleosome-based coarse-grained models, the “mesoscale” currently represents the largest scale. At this scale, current models can describe oligomers of nucleosomes connected by linker DNA. A prominent example of the questions that arise at such length scales is provided by the structure of nanoscopic chromatin fibers or aggregates. Early experiments with isolated chromatin oligomers in vitro originally proposed the now-controversial concept of a 30-nm fiber (27,32,40,41). Although recent in vivo imaging has contradicted the notion of a static 30-nm chromatin fiber, in vitro evidence is strong (42, 43, 44). To ultimately understand how epigenetic phenomena influence the structure of chromatin at this level, the structure of unmodified chromatin and what drives its formation must be assessed. Put simply, the basis for chromatin fiber structure derives from DNA deformation penalties and the mechanical modes that limit them. Generally, these are balanced by energetic processes that are favorable. These include internucleosome contacts, nucleosome-positioning energy, and DNA-protein interactions. This balance is modulated by the amount of linker DNA connecting each nucleosome. From experiments and simulation, it is found that linker DNA lengths that are in integer amounts of DNA pitch (∼10 bp) lead to compact structures, whereas deviations frustrate the fiber, leading to larger structures. This property influences the eukaryotic genome, and high-throughput sequencing of linker DNA has revealed peaks of intensity at integer lengths of DNA pitch (45, 46, 47).
In general, models at this scale coarse-grain DNA as a few basepairs per bead, and the nucleosome is represented as a single entity. The DNA is treated as a worm-like chain (WLC) model (48), and different variants treat the nucleosome and internucleosome interactions in their own way. Pioneering work by Arya et al. introduced a coarse-grained scheme that reproduces the electronic field of the nucleosome with a reduced representation, and it has been successful in predicting chromatin fiber structure (49,50). This method, known as the discrete surface charge optimization (DISCO), reduces the nucleosome to ∼300 pseudo-charges distributed on the nucleosome surface. Building on that work, in recent years, additional modeling approaches at this length scale have emerged. A different approach to nucleosome coarse-graining draws from experimental values of nucleosome-interaction strengths (51). Additional efforts have sought to develop a nucleosome-resolution model with specified topological interactions to inform the assembly of nucleosomes in the chromatin fiber (52). Recently, we have introduced the one cylinder per nucleosome (1CPN) model, built off of previous nucleosome-centric results and other, well-founded models at this length scale (53,54). Another model at this scale incorporates nonhistone proteins that bend DNA, resulting in heterogeneous packing in oligonucleosome fibers (55).
In addition to unaltered chromatin, these models incorporate some of the more-well-characterized epigenetic phenomena. For example, the work started by Arya and Schlick incorporated the H1 linker histone, which was further extended to achieve better resolution and more variants in recent work (50,56,57). These efforts influenced the linker histone model described in the 1CPN model of Lequieu et al. (53) Recent work by Bascom et al. has pushed modeling efforts at this scale to understand how packaging of the HOXC gene hub is influenced by the amount of linker histones, H4 acetylation, and variable linker lengths (58).
Models at the mesoscale are increasing their reach onto larger length scales as a part of broad efforts to understand how nucleosomes influence the supranucleosomal scale. This work is only beginning to knock at the door of higher-order structures such as chromatin topologically associating domains (TADs), compartments, and territories. Although the recent in silico advancements are exciting, larger phenomena that occur with 100-kbp to 1-Mbp structures are still out of reach. The multiscale modeling approach highlighted above demonstrates the importance of nucleosome physics on the overall structure of chromatin, and moving forward, it will be important to further characterize the effects of individual linker lengths and linker histones on chromatin structure and their impact on chromosome packaging. Note that several experimental studies have provided information about the distribution of linker lengths (45,47,59), which have only recently been integrated into in silico work (58,60). An additional key study for nucleosome positioning is Micro-C, which has been integrated into the model introduced by Wiese et al. to predict the structure of chromatin with experimentally informed heterogeneous linker spacing (61). To capture larger-scale chromatin packaging, top-down modeling approaches are still necessary.
Top-down models
Top-down models have aimed to explain chromatin conformation and deep-sequencing experiments at coarse levels of resolution. In general, top-down models offer an easy avenue for representability that is accompanied by a low chance of transferability. As mentioned earlier, a vast amount of information pertaining to the 3D genome has emerged from high-throughput sequencing and chromosome conformation capture methods (3C, 5C, and Hi-C) (62, 63, 64, 65). In particular, Hi-C uses high-throughput sequencing to measure the contact probability of genomic segments as a function of genomic distance (62,64,66). These methods are helping elucidate some of the higher length-scale organization of the genome. Processes such as chromatin looping, TADs, and chromatin compartmentalization have been gradually elucidated by relying on Hi-C studies (62,64,66). The advent of these methods provides a direct link between the epigenome and the structural organization of chromatin. This is further underscored by the fact that chromatin compartments strongly correlate with associated epigenetic marks (64). Recently, single-cell Hi-C and novel sequential fluorescence in situ hybridization have revealed that population-average features such as TADs are in fact present in single nuclei (67), thereby providing an additional incentive to rely on modeling approaches that resolve the 3D genome.
These chromatin structures play vital roles in transcription and repair as well as cell development. Disruption of TAD boundaries is linked to genetic diseases, including cancer (68). Arising from these higher-order structures is a heterogeneous DNA-packaging density. Current label-free imaging technologies, like partial-wave spectroscopy (PWS), have quantified the density heterogeneity of nuclear chromatin (69,70). By capturing fluctuations in DNA density with up to 20-nm resolution, these methods also reveal that the degree of heterogeneity differentiates cancerous from healthy cells and helps connect physical changes in chromatin to genetic diseases at small, molecular scales (71).
The resolution of Hi-C data is relatively coarse, and polymer models of chromatin, informed by these experiments, have been useful in interpreting such data (72). Initial work attempted to use established polymer physics to describe chromatin packaging. However, this framework was abandoned because no ideal polymer model is able to predict the anomalous contact-scaling probability of chromatin as a function of genomic separation (73). From Hi-C, this corresponding exponent is in the range of 0.75–1.33 and falls out of the theory of the scaling of an ideal chain (1.5) and that of a poor-solvent chain (1.0) (7,63,74). Accounting for this anomaly, polymer models incorporate chromatin looping in the form of the strings and binders switch model (75), slip springs (52), or active extrusion (76). The work of Fudenberg et al. has been important in describing the active extrusion process and its influence on the genome (77,78).
Because chromatin compartments depending on epigenetic mark have been uncovered through Hi-C, they have been the focus of some modeling approaches. One such approach is a static definition of epigenetic state, in which monomers of a polymer are “colored” according to their state, similar to block copolymer models. The model of Jost et al. incorporates four such colors, representing different epigenetic states to predict the structure of Drosophila chromosomes from relevant Hi-C data (79). However, epigenetic state is capable of “spreading” through a feedback loop mechanism. The block copolymer type model has also been extended to incorporate a change of epigenetic state through a “two-state” kinetic model between active and inactive states of chromatin. This model of Michieletto et al. is able to recapitulate the stability of epigenetic domains subject to perturbations, thereby providing some insight into the presence of TADs (80). These modeling approaches are seminal in providing insight into the dynamic interplay between epigenomic regulation and chromatin structure at a coarse scale.
A different approach to unraveling the structure of chromatin has come from maximal-entropy minimization between models and experimental data (81), which can reproduce chromosome conformation contact maps and are able to describe the underlying structures of chromatin. One such approach of Georgetti et al. informed the structure of the mouse X-inactivation center region with a simple polymer model from 5C data with high fidelity (82). The maximal-entropy model was later extended to cover entire chromsomes through Hi-C data in the minimal chromatin model of di Pierro et al. (81) Expanding upon this work, di Pierro et al. drew information from ChIP-seq data and, through machine-learning methods, discovered that the same Hi-C contact maps could be attained. This leads to the surprising conclusion that the structure of chromatin is encoded in the epigenetic marks (83). This model has also been physically characterized and demonstrates varying chromatin diffusion rates of chromosome compartments (84).
In addition to incorporating Hi-C data, the prevalence of single-particle tracking through optical microscopy leads to easy quantification of chromatin diffusion and the tendency to form loops between distal elements. Rouse polymer models of chromatin that incorporate such quantities have been especially helpful in elucidating chromatin packaging and dynamics (85). Recent work of Socol et al. demonstrates that Rouse models require extension in the form of transient chromatin contacts, known as the RouseTIC model, to predict both single-particle tracking of in vitro chromatin and Hi-C of the yeast genome (86). Rouse models and looping implementations have been the subject of multiple reviews that are recommended for further interest (76,85,87,88).
A key setback of current top-down approaches is a lack of transferability. A recent novel approach to budding yeast nucleus simulations by Arbona et al. uses Bayesian inference to learn a fixed class of polymer and microtubule parameters using Hi-C, imaging, simulations, and Micro-C XL data (89). We find approaches that link experiments and simulations through data-driven methods to be an exciting avenue to determine fundamental physical parameters. Approaches that draw from multiple experiments such as those of Arbona et al. and Socol et al. could even reconcile the differences in single-nucleus versus population-average effects that have been the source of discrepancies in results (88).
Where bottom-up meets top-down
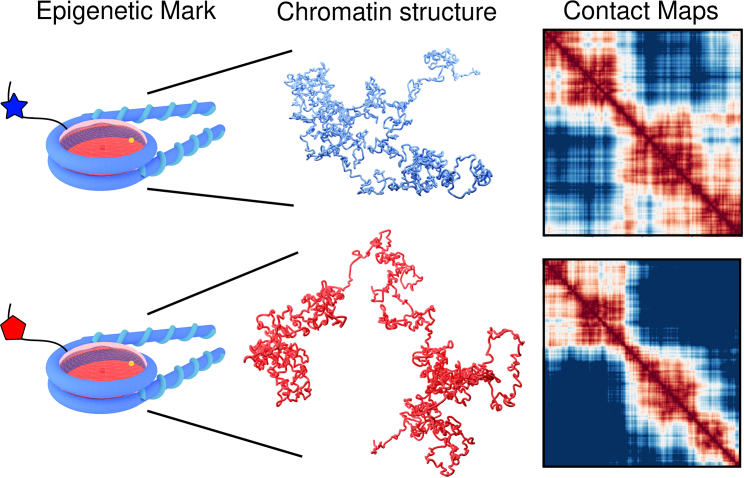
Although prior modeling has laid the groundwork for understanding the physics that underlie each length scale, we now come back to the initial question of how the nucleosome influences chromosomal organization. To incorporate higher-order effects and preserve a nucleosome-first approach, additional features must be included in available models. The outlook is optimistic, with advances from both approaches reaching similar scales. Recently, MacPherson et al. introduced a bottom-up mean-field chromatin model that is also discretized at the nucleosome level (90,91). This model incorporates chromatin compartmentalization through a Hamiltonian that accounts for constitutive heterochromatin formation. We find this model to be a promising candidate for bridging the gap between the two scales. Additionally, the recent self-returning random walk model, at a similar level of resolution, has been promising at capturing both the packaging heterogeneity predicted by PWS and ChromSTEM with the contact probability scaling of Hi-C (74). With advances at the mesoscale, chromatin can be studied in considerable detail, with resolution ranging from nanometers and the histone level to the gene or kbp scales of DNA. With careful consideration, the nucleosome scale physics from mesoscale models could be incorporated in these models. Similarly, the addition of the maximal-entropy methodology of di Pierro et al. of mapping ChIP-seq or Hi-C to such a model provides an exciting prospect (Fig. 2).
Figure 2.
This figure illustrates some of the prospects for chromatin models. Ideally, modeling could incorporate single epigenetic marks, shown here as a red and blue mark to a histone tail. Such marks influence the chromatin fiber structure and can be predicted by modeling efforts. Here, the blue mark results in a condensed fiber and the red in an extended fiber. The subsequent contact maps are provided for these schematic fibers to highlight the comparison to top-down methods and the potential to be compared with Hi-C data. To see this figure in color, go online.
Another promising avenue for crossover is the potential for mesoscale models to uncover larger building blocks of chromatin. Although the nucleosome is the fundamental unit, recent experimental evidence points toward larger potential candidates. Evidence of small fibers comprising the genome has already been the subject of recent super-resolution imaging techniques, such as stochastic optical reconstruction super-resolution microscopy (STORM), and recent chromosome conformation capture methods. One STORM experiment links pluripotency and chromatin structure to the size of clutch-like motifs, which are on the order of 4–16 nucleosomes (92). These clutches are also linked to the DNA-packaging density through acetylations (93). Advances in chromosome conformation capture methods can also map the orientations of all entering and exiting DNA, called Hi-CO (94). These methods have uncovered two recurrent structural tetranucleosome motif. Perhaps nucleosome-resolution models may not be necessary after all, and simpler, less-demanding alternatives could be considered.
The current grand challenge for chromatin modeling is to explain epigenetic phenomena in terms of mechanistic, molecular processes. To effectively understand the structure of chromatin, even at these small scales, one must consider the effect of variable DNA linker length, PTMs, linker histones, histone variants, etc. Perhaps first on that list is to identify the modifications that have direct impact on the structural properties of chromatin. In this vein, atomistic simulations have identified the effects of a few such modifications. In addition, we emphasize that chromatin represents a far-from-equilibrium system, largely as a result of ATP-dependent chromatin remodelers and elongation by RNA polymerase II. Through unique inference about these molecular processes, a few models are beginning to introduce these effects with success, providing great promise for their use in future studies (52,95,96). This wide parameter space is still in need of significant computational, experimental, and theoretical efforts from the chromatin community; multiscale approaches, such as those outlined here, are likely to play an important role in that endeavor. Bridging the length scales of chromatin modeling will allow for a hitherto unprecedented fundamental understanding of chromatin. With exciting new advances in experimental techniques, simulations could provide the critical insights into chromatin structural functionality that are needed to make progress.
Author Contributions
J.M. and J.d.P. wrote the article.
Acknowledgments
The authors thank Joshua Lequieu and Andrés Córdoba for their insights into chromatin multiscale modeling. We also thank Marc Morgan, Vadim Backman, Ali Shilatifard, and Hao Zhang for their helpful discussions with chromatin imaging and experiments. Additionally, we thank Soren Kyhl for conversations on the potential of future models. Lastly, we thank Nicholas Jackson, Viviana Palacio-Betancur, and Cody Bezik with editing this work.
This work was supported by the National Science Foundation under grant EFRI CEE 1830969.
Editor: Tamar Schlick.
References
- 1.Groth A., Rocha W., Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007;128:721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 2.Li B., Carey M., Workman J.L. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 3.Adkins N.L., Niu H., Peterson C.L. Nucleosome dynamics regulates DNA processing. Nat. Struct. Mol. Biol. 2013;20:836–842. doi: 10.1038/nsmb.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowman G.D., Poirier M.G. Post-translational modifications of histones that influence nucleosome dynamics. Chem. Rev. 2015;115:2274–2295. doi: 10.1021/cr500350x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kurdistani S.K., Grunstein M. Histone acetylation and deacetylation in yeast. Nat. Rev. Mol. Cell Biol. 2003;4:276–284. doi: 10.1038/nrm1075. [DOI] [PubMed] [Google Scholar]
- 6.Zhang B., Zheng W., Wolynes P.G. Exploring the free energy landscape of nucleosomes. J. Am. Chem. Soc. 2016;138:8126–8133. doi: 10.1021/jacs.6b02893. [DOI] [PubMed] [Google Scholar]
- 7.Lieberman-Aiden E., van Berkum N.L., Dekker J. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li G., Fullwood M.J., Sung W.K. ChIA-PET tool for comprehensive chromatin interaction analysis with paired-end tag sequencing. Genome Biol. 2010;11:R22. doi: 10.1186/gb-2010-11-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson D.S., Mortazavi A., Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 10.Hsieh T.H., Weiner A., Rando O.J. Mapping nucleosome resolution chromosome folding in yeast by Micro-C. Cell. 2015;162:108–119. doi: 10.1016/j.cell.2015.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsieh T.H., Fudenberg G., Rando O.J. Micro-C XL: assaying chromosome conformation from the nucleosome to the entire genome. Nat. Methods. 2016;13:1009–1011. doi: 10.1038/nmeth.4025. [DOI] [PubMed] [Google Scholar]
- 12.Luger K., Mäder A.W., Richmond T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 13.Davey C.A., Sargent D.F., Richmond T.J. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J. Mol. Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 14.Bishop T.C. Molecular dynamics simulations of a nucleosome and free DNA. J. Biomol. Struct. Dyn. 2005;22:673–686. doi: 10.1080/07391102.2005.10507034. [DOI] [PubMed] [Google Scholar]
- 15.Eslami-Mossallam B., Schiessel H., van Noort J. Nucleosome dynamics: sequence matters. Adv. Colloid Interface Sci. 2016;232:101–113. doi: 10.1016/j.cis.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 16.Öztürk M.A., Cojocaru V., Wade R.C. Dependence of chromatosome structure on linker histone sequence and posttranslational modification. Biophys. J. 2018;114:2363–2375. doi: 10.1016/j.bpj.2018.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowerman S., Wereszczynski J. Effects of MacroH2A and H2A.Z on nucleosome dynamics as elucidated by molecular dynamics simulations. Biophys. J. 2016;110:327–337. doi: 10.1016/j.bpj.2015.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Melters D.P., Pitman M., Dalal Y. Intrinsic elasticity of nucleosomes is encoded by histone variants and calibrated by their binding partners. Proc. Natl. Acad. Sci. USA. 2019;116:24066–24074. doi: 10.1073/pnas.1911880116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z., Kono H. Distinct roles of histone H3 and H2A tails in nucleosome stability. Sci. Rep. 2016;6:31437. doi: 10.1038/srep31437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang R., Erler J., Langowski J. Histone acetylation regulates chromatin accessibility: role of H4K16 in inter-nucleosome interaction. Biophys. J. 2017;112:450–459. doi: 10.1016/j.bpj.2016.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jung J., Nishima W., Sanbonmatsu K.Y. Scaling molecular dynamics beyond 100,000 processor cores for large-scale biophysical simulations. J. Comput. Chem. 2019;40:1919–1930. doi: 10.1002/jcc.25840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freeman G.S., Lequieu J.P., De Pablo J.J. DNA shape dominates sequence affinity in nucleosome formation. Phys. Rev. Lett. 2014;113:168101. doi: 10.1103/PhysRevLett.113.168101. [DOI] [PubMed] [Google Scholar]
- 23.Lequieu J., Córdoba A., de Pablo J.J. Tension-dependent free energies of nucleosome unwrapping. ACS Cent. Sci. 2016;2:660–666. doi: 10.1021/acscentsci.6b00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lequieu J., Schwartz D.C., de Pablo J.J. In silico evidence for sequence-dependent nucleosome sliding. Proc. Natl. Acad. Sci. USA. 2017;114:E9197–E9205. doi: 10.1073/pnas.1705685114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brandani G.B., Takada S. Chromatin remodelers couple inchworm motion with twist-defect formation to slide nucleosomal DNA. PLoS Comput. Biol. 2018;14:e1006512. doi: 10.1371/journal.pcbi.1006512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Routh A., Sandin S., Rhodes D. Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proc. Natl. Acad. Sci. USA. 2008;105:8872–8877. doi: 10.1073/pnas.0802336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song F., Chen P., Li G. Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science. 2014;344:376–380. doi: 10.1126/science.1251413. [DOI] [PubMed] [Google Scholar]
- 28.Meng H., Andresen K., van Noort J. Quantitative analysis of single-molecule force spectroscopy on folded chromatin fibers. Nucleic Acids Res. 2015;43:3578–3590. doi: 10.1093/nar/gkv215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kruithof M., Chien F.T., van Noort J. Single-molecule force spectroscopy reveals a highly compliant helical folding for the 30-nm chromatin fiber. Nat. Struct. Mol. Biol. 2009;16:534–540. doi: 10.1038/nsmb.1590. [DOI] [PubMed] [Google Scholar]
- 30.Cui Y., Bustamante C. Pulling a single chromatin fiber reveals the forces that maintain its higher-order structure. Proc. Natl. Acad. Sci. USA. 2000;97:127–132. doi: 10.1073/pnas.97.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Funke J.J., Ketterer P., Dietz H. Uncovering the forces between nucleosomes using DNA origami. Sci. Adv. 2016;2:e1600974. doi: 10.1126/sciadv.1600974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collepardo-Guevara R., Portella G., Orozco M. Chromatin unfolding by epigenetic modifications explained by dramatic impairment of internucleosome interactions: a multiscale computational study. J. Am. Chem. Soc. 2015;137:10205–10215. doi: 10.1021/jacs.5b04086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Potoyan D.A., Papoian G.A. Regulation of the H4 tail binding and folding landscapes via Lys-16 acetylation. Proc. Natl. Acad. Sci. USA. 2012;109:17857–17862. doi: 10.1073/pnas.1201805109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winogradoff D., Echeverria I., Papoian G.A. The acetylation landscape of the H4 histone tail: disentangling the interplay between the specific and cumulative effects. J. Am. Chem. Soc. 2015;137:6245–6253. doi: 10.1021/jacs.5b00235. [DOI] [PubMed] [Google Scholar]
- 35.Vettese-Dadey M., Grant P.A., Workman J.L. Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J. 1996;15:2508–2518. [PMC free article] [PubMed] [Google Scholar]
- 36.Brownell J.E., Zhou J., Allis C.D. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 37.Moller J., Lequieu J., de Pablo J.J. The free energy landscape of internucleosome interactions and its relation to chromatin fiber structure. ACS Cent. Sci. 2019;5:341–348. doi: 10.1021/acscentsci.8b00836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang L., Takada S. Histone acetylation dependent energy landscapes in tri-nucleosome revealed by residue-resolved molecular simulations. Sci. Rep. 2016;6:34441. doi: 10.1038/srep34441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watanabe S., Mishima Y., Takada S. Interactions of HP1 bound to H3K9me3 dinucleosome by molecular simulations and biochemical assays. Biophys. J. 2018;114:2336–2351. doi: 10.1016/j.bpj.2018.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olins A.L., Olins D.E. Spheroid chromatin units (v bodies) Science. 1974;183:330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- 41.Robinson P.J., Rhodes D. Structure of the ‘30 nm’ chromatin fibre: a key role for the linker histone. Curr. Opin. Struct. Biol. 2006;16:336–343. doi: 10.1016/j.sbi.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 42.Maeshima K., Hihara S., Eltsov M. Chromatin structure: does the 30-nm fibre exist in vivo? Curr. Opin. Cell Biol. 2010;22:291–297. doi: 10.1016/j.ceb.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 43.Ou H.D., Phan S., O’Shea C.C. ChromEMT: visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science. 2017;357:eaag0025. doi: 10.1126/science.aag0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tremethick D.J. Higher-order structures of chromatin: the elusive 30 nm fiber. Cell. 2007;128:651–654. doi: 10.1016/j.cell.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 45.Brogaard K., Xi L., Widom J. A map of nucleosome positions in yeast at base-pair resolution. Nature. 2012;486:496–501. doi: 10.1038/nature11142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Widom J. A relationship between the helical twist of DNA and the ordered positioning of nucleosomes in all eukaryotic cells. Proc. Natl. Acad. Sci. USA. 1992;89:1095–1099. doi: 10.1073/pnas.89.3.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chereji R.V., Ramachandran S., Henikoff S. Precise genome-wide mapping of single nucleosomes and linkers in vivo. Genome Biol. 2018;19:19. doi: 10.1186/s13059-018-1398-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marko J.F., Siggia E.D. Stretching DNA. Macromolecules. 1995;28:8759–8770. [Google Scholar]
- 49.Arya G., Zhang Q., Schlick T. Flexible histone tails in a new mesoscopic oligonucleosome model. Biophys. J. 2006;91:133–150. doi: 10.1529/biophysj.106.083006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arya G., Schlick T. A tale of tails: how histone tails mediate chromatin compaction in different salt and linker histone environments. J. Phys. Chem. A. 2009;113:4045–4059. doi: 10.1021/jp810375d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Norouzi D., Zhurkin V.B. Topological polymorphism of the two-start chromatin fiber. Biophys. J. 2015;108:2591–2600. doi: 10.1016/j.bpj.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brackley C.A., Johnson J., Marenduzzo D. Nonequilibrium chromosome looping via molecular slip links. Phys. Rev. Lett. 2017;119:138101. doi: 10.1103/PhysRevLett.119.138101. [DOI] [PubMed] [Google Scholar]
- 53.Lequieu J., Córdoba A., de Pablo J.J. 1CPN: a coarse-grained multi-scale model of chromatin. J. Chem. Phys. 2019;150:215102. doi: 10.1063/1.5092976. [DOI] [PubMed] [Google Scholar]
- 54.Wedemann G., Langowski J. Computer simulation of the 30-nanometer chromatin fiber. Biophys. J. 2002;82:2847–2859. doi: 10.1016/S0006-3495(02)75627-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bajpai G., Padinhateeri R. Irregular chromatin: packing density, fiber width, and occurrence of heterogeneous clusters. Biophys. J. 2020;118:207–218. doi: 10.1016/j.bpj.2019.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luque A., Collepardo-Guevara R., Schlick T. Dynamic condensation of linker histone C-terminal domain regulates chromatin structure. Nucleic Acids Res. 2014;42:7553–7560. doi: 10.1093/nar/gku491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perišic O., Portillo-Ledesma S., Schlick T. Sensitive effect of linker histone binding mode and subtype on chromatin condensation. Nucleic Acids Res. 2019;47:4948–4957. doi: 10.1093/nar/gkz234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bascom G.D., Myers C.G., Schlick T. Mesoscale modeling reveals formation of an epigenetically driven HOXC gene hub. Proc. Natl. Acad. Sci. USA. 2019;116:4955–4962. doi: 10.1073/pnas.1816424116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Voong L.N., Xi L., Wang X. Insights into nucleosome organization in mouse embryonic stem cells through chemical mapping. Cell. 2016;167:1555–1570.e15. doi: 10.1016/j.cell.2016.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bascom G.D., Kim T., Schlick T. Kilobase pair chromatin fiber contacts promoted by living-system-like DNA linker length distributions and nucleosome depletion. J. Phys. Chem. B. 2017;121:3882–3894. doi: 10.1021/acs.jpcb.7b00998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wiese O., Marenduzzo D., Brackley C.A. Nucleosome positions alone can be used to predict domains in yeast chromosomes. Proc. Natl. Acad. Sci. USA. 2019;116:17307–17315. doi: 10.1073/pnas.1817829116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dixon J.R., Selvaraj S., Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rao S.S., Huntley M.H., Aiden E.L. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rao S.S.P., Huang S.C., Aiden E.L. Cohesin loss eliminates all loop domains. Cell. 2017;171:305–320.e24. doi: 10.1016/j.cell.2017.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sanborn A.L., Rao S.S.P., Aiden E.L. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl. Acad. Sci. USA. 2015;112:E6456–E6465. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schwarzer W., Abdennur N., Spitz F. Two independent modes of chromatin organization revealed by cohesin removal. Nature. 2017;551:51–56. doi: 10.1038/nature24281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bintu B., Mateo L.J., Zhuang X. Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science. 2018;362:eaau1783. doi: 10.1126/science.aau1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hnisz D., Weintraub A.S., Young R.A. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science. 2016;351:1454–1458. doi: 10.1126/science.aad9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dong B., Almassalha L.M., Backman V. Superresolution intrinsic fluorescence imaging of chromatin utilizing native, unmodified nucleic acids for contrast. Proc. Natl. Acad. Sci. USA. 2016;113:9716–9721. doi: 10.1073/pnas.1602202113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Almassalha L.M., Tiwari A., Backman V. The global relationship between chromatin physical topology, fractal structure, and gene expression. Sci. Rep. 2017;7:41061. doi: 10.1038/srep41061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Almassalha L.M., Bauer G.M., Backman V. Label-free imaging of the native, living cellular nanoarchitecture using partial-wave spectroscopic microscopy. Proc. Natl. Acad. Sci. USA. 2016;113:E6372–E6381. doi: 10.1073/pnas.1608198113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rosa A., Everaers R. Structure and dynamics of interphase chromosomes. PLoS Comput. Biol. 2008;4:e1000153. doi: 10.1371/journal.pcbi.1000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mirny L.A. The fractal globule as a model of chromatin architecture in the cell. Chromosome Res. 2011;19:37–51. doi: 10.1007/s10577-010-9177-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang K., Li Y., Szleifer I. Physical and data structure of 3D genome. Science Advances. 2020;6 doi: 10.1126/sciadv.aay4055. eaay4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Barbieri M., Chotalia M., Nicodemi M. Complexity of chromatin folding is captured by the strings and binders switch model. Proc. Natl. Acad. Sci. USA. 2012;109:16173–16178. doi: 10.1073/pnas.1204799109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fiorillo L., Bianco S., Chiariello A.M. A modern challenge of polymer physics: novel ways to study, interpret, and reconstruct chromatin structure. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2019 doi: 10.1002/wcms.1454. Published online November 16, 2020. [DOI] [Google Scholar]
- 77.Fudenberg G., Imakaev M., Mirny L.A. Formation of chromosomal domains by loop extrusion. Cell Rep. 2016;15:2038–2049. doi: 10.1016/j.celrep.2016.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nuebler J., Fudenberg G., Mirny L. Chromatin organization by an interplay of loop extrusion and compartmental segregation. Proc. Natl. Acad. Sci. USA. 2018;115:E6697–E6706. doi: 10.1073/pnas.1717730115. Published online October 3, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jost D., Carrivain P., Vaillant C. Modeling epigenome folding: formation and dynamics of topologically associated chromatin domains. Nucleic Acids Res. 2014;42:9553–9561. doi: 10.1093/nar/gku698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Michieletto D., Orlandini E., Marenduzzo D. Polymer model with epigenetic recoloring reveals a pathway for the de novo establishment and 3D organization of chromatin domains. Phys. Rev. X. 2016;6:041047. [Google Scholar]
- 81.Di Pierro M., Zhang B., Onuchic J.N. Transferable model for chromosome architecture. Proc. Natl. Acad. Sci. USA. 2016;113:12168–12173. doi: 10.1073/pnas.1613607113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Giorgetti L., Galupa R., Heard E. Predictive polymer modeling reveals coupled fluctuations in chromosome conformation and transcription. Cell. 2014;157:950–963. doi: 10.1016/j.cell.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Di Pierro M., Cheng R.R., Onuchic J.N. De novo prediction of human chromosome structures: epigenetic marking patterns encode genome architecture. Proc. Natl. Acad. Sci. USA. 2017;114:12126–12131. doi: 10.1073/pnas.1714980114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Di Pierro M., Potoyan D.A., Onuchic J.N. Anomalous diffusion, spatial coherence, and viscoelasticity from the energy landscape of human chromosomes. Proc. Natl. Acad. Sci. USA. 2018;115:7753–7758. doi: 10.1073/pnas.1806297115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Amitai A., Holcman D. Polymer physics of nuclear organization and function. bioRxiv. 2017 doi: 10.1101/076661. [DOI] [Google Scholar]
- 86.Socol M., Wang R., Bancaud A. Rouse model with transient intramolecular contacts on a timescale of seconds recapitulates folding and fluctuation of yeast chromosomes. Nucleic Acids Res. 2019;47:6195–6207. doi: 10.1093/nar/gkz374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tiana G., Giorgetti L. Integrating experiment, theory and simulation to determine the structure and dynamics of mammalian chromosomes. Curr. Opin. Struct. Biol. 2018;49:11–17. doi: 10.1016/j.sbi.2017.10.016. [DOI] [PubMed] [Google Scholar]
- 88.Le Dily F., Serra F., Marti-Renom M.A. 3D modeling of chromatin structure: is there a way to integrate and reconcile single cell and population experimental data? Wiley Interdiscip. Rev. Comput. Mol. Sci. 2017;7:e1308. [Google Scholar]
- 89.Arbona J.M., Herbert S., Zimmer C. Inferring the physical properties of yeast chromatin through Bayesian analysis of whole nucleus simulations. Genome Biol. 2017;18:81. doi: 10.1186/s13059-017-1199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.MacPherson Q., Beltran B., Spakowitz A.J. Bottom-up modeling of chromatin segregation due to epigenetic modifications. Proc. Natl. Acad. Sci. USA. 2018;115:12739–12744. doi: 10.1073/pnas.1812268115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Macpherson Q., Beltran B., Spakowitz A.J. Chromatin compaction leads to a preference for peripheral heterochromatin. Biophys. J. 2020;118:1479–1488. doi: 10.1016/j.bpj.2020.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ricci M.A., Manzo C., Cosma M.P. Chromatin fibers are formed by heterogeneous groups of nucleosomes in vivo. Cell. 2015;160:1145–1158. doi: 10.1016/j.cell.2015.01.054. [DOI] [PubMed] [Google Scholar]
- 93.Otterstrom J., Castells-Garcia A., Lakadamyali M. Super-resolution microscopy reveals how histone tail acetylation affects DNA compaction within nucleosomes in vivo. Nucleic Acids Res. 2019;47 doi: 10.1093/nar/gkz593. 8740–8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ohno M., Ando T., Taniguchi Y. Sub-nucleosomal genome structure reveals distinct nucleosome folding motifs. Cell. 2019;176:520–534.e25. doi: 10.1016/j.cell.2018.12.014. [DOI] [PubMed] [Google Scholar]
- 95.Laghmach R., Di Pierro M., Potoyan D.A. Mesoscale liquid model of chromatin recapitulates nuclear order of eukaryotes. Biophys. J. 2020;118:2130–2140. doi: 10.1016/j.bpj.2019.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Michieletto D., Colì D., Orlandini E. Nonequilibrium theory of epigenomic microphase separation in the cell nucleus. Phys. Rev. Lett. 2019;123:228101. doi: 10.1103/PhysRevLett.123.228101. [DOI] [PubMed] [Google Scholar]
- 97.Li W., Wolynes P.G., Takada S. Frustration, specific sequence dependence, and nonlinearity in large-amplitude fluctuations of allosteric proteins. Proc. Natl. Acad. Sci. USA. 2011;108:3504–3509. doi: 10.1073/pnas.1018983108. [DOI] [PMC free article] [PubMed] [Google Scholar]