Abstract
Single-particle tracking experiments have measured escape times of DNA-binding species diffusing in living cells: CRISPR-Cas9, TetR, and LacI. The observed distribution is a truncated power law. Working backward from the experimental results, the observed distribution appears inconsistent with a Gaussian distribution of binding energies. Working forward, the observed distribution leads to transient anomalous subdiffusion, in which diffusion is anomalous at short times and normal at long times, here only mildly anomalous. Monte Carlo simulations are used to characterize the time-dependent diffusion coefficient D(t) in terms of the anomalous exponent α, the crossover time tcross, and the limits D(0) and D(∞) and to relate these quantities to the escape time distribution. The simplest interpretations identify the escape time as the actual binding time to DNA or the period of one-dimensional diffusion on DNA in the standard model combining one-dimensional and three-dimensional search, but a more complicated interpretation may be required. The model has several implications for cell biophysics. 1) The initial anomalous regime represents the search of the DNA-binding species for its target DNA sequence. 2) Non-target DNA sites have a significant effect on search kinetics. False positives in bioinformatic searches of the genome are potentially rate-determining in vivo. For simple binding, the search would be speeded if false-positive sequences were eliminated from the genome. 3) Both binding and obstruction affect diffusion. Obstruction ought to be measured directly, using as the primary probe the DNA-binding species with the binding site inactivated and eGFP as a calibration standard among laboratories and cell types. 4) Overexpression of the DNA-binding species reduces anomalous subdiffusion because the deepest binding sites are occupied and unavailable. 5) The model provides a coarse-grained phenomenological description of diffusion of a DNA-binding species, useful in larger-scale modeling of kinetics, FCS, and FRAP.
Significance
DNA-binding biomolecules such as transcription factors diffuse in the nucleus until they find their biological target and bind to it. A biomolecule may bind to many false-positive sites before it reaches its target, and the search process is a research topic of considerable interest. Experimental results from other laboratories show a truncated power-law distribution of escape times from the binding sites. I show by Monte Carlo simulations that this escape time distribution implies that the protein shows transient anomalous subdiffusion, defined as anomalous subdiffusion at short times and normal diffusion at long times. Implications of the model for controls, experiments, and interpretation of experiments are discussed.
Introduction
Many biomolecules in a cell, whether in the nucleus, cytoplasm, or plasma membrane, must find their biological targets in a crowded sticky environment. I consider here the “sticky” aspect: a biomolecule diffuses in the cell and encounters a variety of binding sites that are more or less similar to its biological target. The more similar the site is to the target, the stronger the binding is, and the longer the delay. The false-positive binding sites thus act as traps, delaying the diffusing species. Eventually, the diffusing species reaches its target site, assumed to be the deepest binding site. The specific situation considered is a DNA-binding biomolecule in the nucleus, because distributions of escape times have been measured in single-particle tracking (SPT) experiments. The corresponding model is a point tracer carrying out a random walk on a cubic lattice containing a set of point traps with a prescribed distribution of mean escape times.
Long ago, I examined transient anomalous subdiffusion (TASD) due to traps (1) and the interpretation of TASD in terms of the search of diffusing particles for their biological target (2). Experimental advances make it timely to revisit this topic. SPT experiments have caught up with modeling. Specifically, three SPT studies reported the escape times (equivalently, the binding, residence, or dwell times) for biomolecules binding to DNA in living mammalian cells and found a truncated power-law (TPL) distribution: repressor TetR (3), repressor LacI (4), and CRISPR-Cas9 (5).
The change in the biological context of the model is important. My earlier work was phrased in terms of cell membranes and assumed a two-dimensional (2D) triangular lattice in which the sites had various distributions of binding energies from the physics literature (1) or a discrete hierarchy of traps (2) with escape times satisfying Shlesinger’s definition of fractal time (6). The work here is phrased in terms of binding of biomolecules to DNA and assumes a three-dimensional (3D) cubic lattice with escape time distributions based on experimental distributions in vivo. Qualitatively, the behavior here is similar to the discrete case, but the dependence on the escape time distribution is more complicated than one would expect from the discrete case. Furthermore, I begin to examine whether distributions of binding energies to DNA from the literature are sufficient to explain the observed distribution of escape times.
As in (2), I discuss TASD in terms of the search of a biomolecule for its biological target in the nucleus. The basic picture is that the observed TPL distribution of escape times from traps leads to TASD on length scales much greater than the trap spacing, the deepest trap is the biological target of the diffusing molecule, and the anomalous regime represents the search for that target in a background of competing binding sites.
I consider only diffusion here, not kinetics. Woringer and Darzacq (7) briefly reviewed the physics literature on fractal kinetics to assess its applicability to binding to DNA. Work by Slutsky et al. (8) and by Bénichou et al. (9) examined anomalous diffusion in the nucleus in terms of the first passage time, so their results are directly linked to search kinetics. The question addressed here is different: what do SPT measurements reveal about anomalous subdiffusion in the nucleus? Much work has been done on modeling chromatin structure, ranging from bottom-up physics starting with simple polymer models to top-down genomics starting with Hi-C results, reviewed in (10, 11, 12, 13), for example. Examination of these models is beyond the scope of this work, except for some discussion of the general topic of obstructed diffusion.
The problem addressed here is multiscale, organized in order of increasing lengths as 1) molecular scale: binding of the mobile species to particular DNA base pair sequences and a combination of one-dimensional (1D) and 3D diffusion among individual binding sites; 2) SPT localization scale: binding and diffusion at the resolution of the SPT experiments, around 25 nm; and 3) μm scale: TASD over a longer length scale, as measured by FRAP, FCS, and long SPT trajectories. In this problem, cause and effect flow from shorter to longer lengths, but the SPT binding measurements are at the intermediate scale, so the modeling has to start in the middle.
In this article, I first describe the TPL and summarize the experimental results in terms of the TPL parameters. Then, I work backward from the observed TPL form to begin to examine its molecular origins. Next, I work forward from the observed TPL form to show that it implies TASD over longer length scales, and I discuss the relationship of the TPL parameters to the TASD parameters. Then, I examine the connections of the TPL model to mechanisms of anomalous subdiffusion from the physics literature, and models of search in the nucleus from the biophysics literature. Finally, in Conclusions, I consider the implications of the TPL model for experiment, both the experiments the model suggests and the interpretation of experimental results.
Methods
Monte Carlo runs in Fortran were used to find the mean-square displacement as a function of time for tracers carrying out a random walk among TPL traps. Methods are considered in (1,14). The Methods section of the Supporting Materials and Methods discusses these calculations in detail (Supporting Materials and Methods, Section S1.1), as well as the analysis of TASD curves (Supporting Materials and Methods, Section S1.2) and Monte Carlo calculations in Mathematica (Supporting Materials and Methods, Section S1.3). In the Monte Carlo results, is in units of lattice constants and t is in units of Monte Carlo steps, one attempt to move by one tracer. The relation between Monte Carlo units and physical units is discussed in the Conversion to Physical Units.
Results
TPL distribution
Here, I describe the TPL distribution and summarize the experimental results. I then describe two important properties of the experimental distributions: the physical reasons for truncation and the delay between preparation of the system and observation.
Definition
In this section and in Supporting Materials and Methods, Section S2, T is simply a random variable, either the mean escape time or the observed escape time. In later sections, these escape times must be distinguished. The published distributions of observed escape times (3, 4, 5) are given in terms of the survival function
| (1) |
where PDF is the probability density function and CDF is the cumulative density function (15). The PDF is thus the negative derivative of the survival function, and S(t) ∝ 1/tm − 1 yields PDF(t) ∝ 1/tm. The experimental results give a TPL distribution of escape times T, which is defined by the PDF
| (2) |
| (3) |
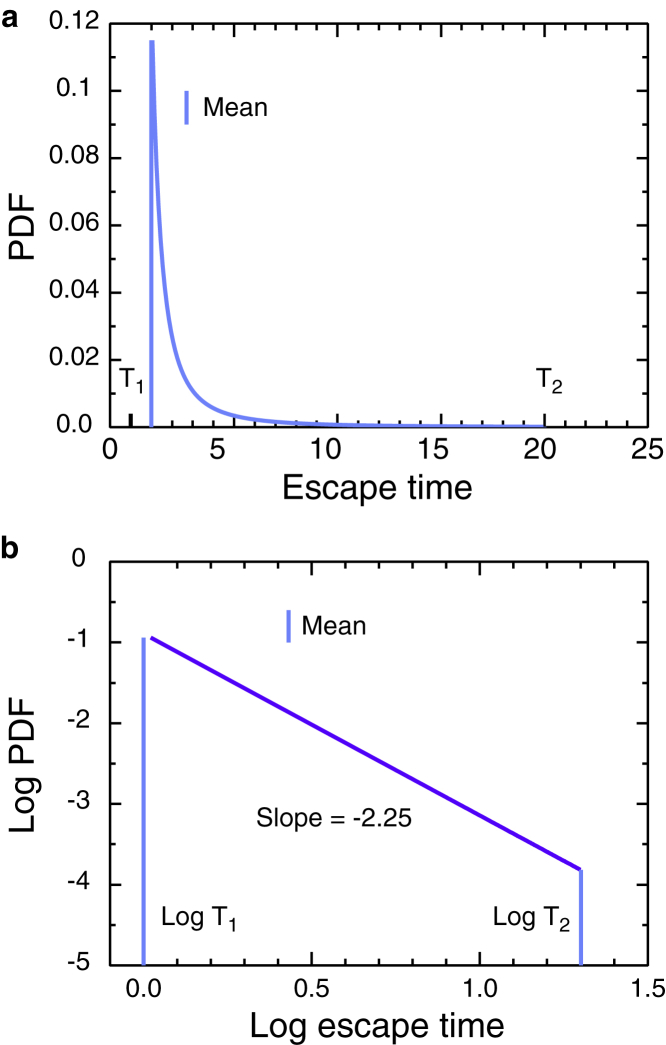
specified by a power m > 0, a minimum escape time T1, and a maximum T2. Power-law distributions are reviewed by Newman (16). If the PDF is truncated at T1 and T2, the CDF and survival function are also truncated there. Fig. 1 shows the shape of the distribution in linear and logarithmic plots, the linear plot to show that most of the traps are shallow, and the logarithmic plot to show the presence of the deeper traps, which have a major influence on diffusion. Fig. 2 shows the dependence on m and on T2 as logarithmic plots. The cumulative density function of the TPL distribution is
| (4) |
and the mean is
| (5) |
Figure 1.
TPL distribution for m = 2.25, T1 = 1, and T2 = 200 as (a) linear and (b) log-log plots. Vertical lines are means. To see this figure in color, go online.
Figure 2.
Log-log plots of TPL distributions for (a) the specified values of the exponent m at fixed values of T1 = 1 and T2 = 200, and (b) the specified values of T2 at constant T1 = 1 and m = 2.25. Vertical lines are means. To see this figure in color, go online.
Further properties are given in Supporting Materials and Methods, Section S2, and the case m = 1.
Experimental results
Three SPT studies measured the escape times (equivalently, the binding, residence, or dwell times) for biomolecules interacting with DNA in living mammalian cells and found a TPL distribution: 1) repressor TetR in the nucleus of U2OS human osteosarcoma cells (3). A bacterial transcription factor was used with an artificially introduced bacterial target site; 2) repressor LacI in the nucleus of U2OS cells (4); and 3) CRISPR-Cas9 in the nucleus of 3T3 cells (5). Nonsense sgRNA (single-guide RNA) was used to examine the search phase rather than the bound phase. In all three studies, the final power-law histograms were constructed by combining several experimental histograms taken at different frame rates, as shown in the original figures. All three final distributions have a width of 2.5 decades, suggesting that the range might result from experimental constraints. Table 1 lists the TPL parameters of the experimental TPL curves, which were presented in the original publications as plots of log S(t) vs. log t, where S(t) is the survival function (Eq. 1) and m is the corresponding exponent in Eq. 2.
Table 1.
Experimental Results
In the Monte Carlo calculations here, the typical value of m is taken to be 2.25, the average of the values from (4,5). A wide range of m is used to include the value from (3) and the error bars of (5).
A related study examined CRISPR-Cas9 binding to DNA curtains (17). Here, DNA strands are anchored at one end in a microfluidic flow cell and are extended by fluid flow to form an array of parallel strands. In some experiments, downstream anchoring groups are then attached to a support so that the parallel geometry can be maintained without fluid flow. Fluorescent-labeled DNA-binding biomolecules are observed by single-molecule imaging using total internal reflection microscopy (18). An interesting result is that at the spatial resolution of the experiments, CRISPR-Cas9 used predominantly a 3D search, both in the DNA curtain and in cells (5,17).
Physical basis for truncation
Why is the distribution truncated at short and long times? The truncation at short times is somewhat subjective: how short a delay is scored as binding? Experimentalists must define a threshold to distinguish binding events from the episodes of localization that occur by chance in a pure random walk (19). The truncation at long times may be absolute or experimental. Absolute corresponds to the escape time from the target; experimental, to the longest escape time observable given experimental constraints, including the rarity of extreme escape times.
Experimental delay
The experimental measurements are incomplete. The histogram for TetR (3) covers a range of escape times of roughly 0.01–600 s, but the authors report that practical reasons lead to a 1 min delay between mixing and the start of the SPT measurements. The effect of this delay on TASD is discussed in Supporting Materials and Methods, Section S4.4, and photoactivation experiments to reduce the delay are discussed in Supporting Materials and Methods, Section S4.4.4.
The measurements on CRISPR-Cas9 interacting with a DNA curtain (17) would have a similar limitation if the measurement is of a tracer that is already in the DNA curtain because one does not know how long it has already been there equilibrating. To avoid this problem, observations ought to be limited to tracers seen to enter the curtain region.
From binding energies to observed escape times
Proceeding backward, I start with the observed escape times on the length scale of SPT localization and examine the underlying molecular binding. I consider the simplest explanation, that the distribution of binding energies leads directly to the distribution of escape times, and find the explanation questionable. I also examine the relation between the distribution of observed escape times and the underlying distribution of mean escape times.
The connection between observed escape times and binding energies is made in two steps to better match the literature. First, I find the distribution of mean escape times corresponding to the distribution of binding energies. In the physics literature, this is the usual derivation of a power-law distribution of mean escape times. In the statistics literature, this is the derivation of the distribution of a function of a stochastic variable. Second, I find the distribution of observed escape times τ from the distribution of mean escape times T. In the physics literature, this is a very common procedure to express the escape times from a statistical distribution of traps in terms of a Laplace transform. In the statistics literature, this is a compound distribution, in which a parameter of the primary distribution is a stochastic quantity distributed according to the parameter distribution. (There is no consensus on nomenclature, but “compound distribution” seems reasonable.) Both steps involve Boltzmann factors, a deterministic one in the first step and a stochastic one in the second.
I summarize some basic properties of the mean escape times T. Attempts to escape occur at the Arrhenius rate
| (6) |
where ν is the frequency of escape attempts, ΔE is the absolute value of the free energy of binding, and boldface kT is the thermal energy, so the mean escape time is
| (7) |
The escape probability per Monte Carlo time step is
| (8) |
This probability generates the geometrical distribution of observed escape times in the Monte Carlo simulations. The Boltzmann factor for a tracer to be in the ith trap is
| (9) |
Step 1. From binding energies to mean escape times
What is the relation between the distribution of binding energies and the distribution of mean escape times, and what is the temperature dependence? The basic physical picture is that the system is at thermal equilibrium at the ambient temperature, except that the tracer is not necessarily equilibrated with the trap distribution. The distribution of binding energies is ϕ(ΔE, parameters), where ΔE has dimensions of energy and ϕ has dimensions of 1/energy, so that ϕ(ΔE)dΔE is dimensionless. The parameters are constant values for the ambient temperature. The distribution of mean escape times depends on temperature only through the expression for the escape time T as a function of binding energy.
In this picture, the relation between the distributions is given by a standard transformation from statistics (20). The starting point is Eq. 7 with ν = 1. Then
| (10) |
| (11) |
The distribution of mean escape times g(T) is
| (12) |
obtained by a mapping of CDFs in some probability textbooks, or more formally as the Jacobian of the transformation. Then
| (13) |
Here, the mean escape time T is dimensionless, equal to the number of time steps δt, either Monte Carlo time steps (see Conversion to Physical Units) or the duration of one frame in an SPT experiment.
What causes a power-law distribution of mean escape times? The simplest mechanism is Arrhenius escape from an exponential distribution of binding energies (3,16,21, 22, 23). Suppose that the binding energies are exponentially distributed on [0, ∞), so that the PDF is
| (14) |
where is the mean binding energy. Then, from Eq. 13,
| (15) |
A pure exponential distribution of binding energies thus leads to a pure power-law distribution of mean escape times. A truncated exponential distribution leads to a TPL distribution, as shown in detail in Supporting Materials and Methods, Section S3.
Gaussian distributions of binding energies are often used in the DNA literature (24,25). These distributions are justified on the grounds that transcription factors typically bind around 10 nucleotides, range 5–31 (26), providing enough averaging to give an approximate Gaussian, whether the protein is reading hydrogen bonding or elastic interactions. As an aside, this is roughly enough averaging. One algorithm to generate Gaussian random numbers is to generate 12 random numbers uniformly distributed between 0 and 1, sum them, and subtract 6, though this method gives such poor-quality results that it is highly unrecommended (14).
Gaussian distributions of binding energies have a very limited ability to produce a power-law distribution of mean escape times. The Gaussian distribution is
| (16) |
where μ is the mean and σ is the standard deviation. Proceeding as in the exponential case yields a log-normal distribution of escape times
| (17) |
Here kT, μ, and σ have dimensions of energy, and the mean escape time T is dimensionless. In the numerical example of Fig. S3-2, g(T) from the Gaussian case is clearly far from a power law for the parameters chosen. Log-normal distributions are known to have tails resembling power-law distributions over a limited range of the variable (27, 28, 29), but not under the conditions here. Take the logarithm of g(T) and rearrange to give
| (18) |
If σ is increased to decrease the quadratic term, the coefficient of the linear term is driven toward −1, which is fine if one is trying to reproduce Zipf’s law or 1/f noise, but not useful here. Simultaneously matching the experimental slope and the range of linearity is difficult or impossible. Or, to put a more positive spin on it, the experimental signature of Arrhenius escape from Gaussian traps is a TPL with a slope near −1, potentially with a range of a few decades depending on the standard deviation of ΔE.
Some labs have used modified distributions such as an asymmetric Gaussian (for example, a saddle-point expansion plus first-order correction (30)), or Gaussian-like curves with tails that are defined numerically from experiment or bioinformatics (31) or defined analytically on the basis of an analogy between protein-protein binding and protein folding (32). But it is hard to see how these modifications could yield a TPL distribution over 2.5 decades in three different experimental systems.
Supporting Materials and Methods, Section S3 examines in detail the distributions of mean escape times from three truncated binding energy distributions: exponential, Gaussian, and lognormal. Log-log plots of these distributions illustrate the limits on power-law dependence from Gaussian and log-normal binding energies.
It seems difficult to get appropriate TPL distributions of escape times from plausible binding energy distributions. Presumably, a distinct mechanism is responsible. One possibility is obstruction, say trapping in dead ends, but it is not obvious how to make dead ends affecting diffusion on a scale <25 nm out of DNA with a persistence length of 50 nm. Tightly bent DNA structures would be required; these structures were reviewed by Garcia et al. (33). Similarly, the fractal model of the nucleus (see, for example, Bancaud et al. (34)) is a coarse-grained model with beads of diameter 20–40 nm (35). Another possibility is the combined effects of multiple traps, recapture, and trap geometry, potentially important because a 25-nm segment of DNA includes 75 base pairs. Several groups have examined this effect ((36, 37, 38) and Note S7 of (3)), but further work remains to be done. Other possibilities include viscoelasticity with crowding (39), and a nonequilibrium state produced by active cellular processes such as chromatin remodeling. Possible mechanisms will be examined in future work, beginning with (40).
Step 2. From mean escape times to observed escape times
There are two distinct escape times in the binding site problem, the observed escape time τ for each escape event and the underlying mean escape time T. The distribution of τ is the experimental histogram of escape times f(τ). The distribution of T is g(T), determined by the distribution of binding energies ϕ(ΔE), so it is in principle predictable from genomics in terms of frequencies of DNA sequences, free energies of binding to DNA sequences, and the accessibility of particular sequences to the binding species. The distribution of τ is broader than the distribution of T because of the scatter in τ for a fixed value of T. For Arrhenius escape, f(τ) is readily calculated for a given g(T), but finding g(T) from an observed f(τ) is problematic, as discussed in Supporting Materials and Methods, Section S3.2.
For escape from confinement, one representation of the escape process would be in terms of a mean escape time that depends on the size and shape of the confining region, the size of the opening(s), and the initial position of the tracer. The observed escape time then reflects the statistical fluctuations in escape time for that specific geometry. One would then average over initial positions and at least some of the shape parameters of the confining region.
TASD
Here, I proceed forward, starting with the experimental TPL on the length scale of SPT localization and ending with TASD, observed over μm lengths by FCS, FRAP, and long SPT measurements. I first review the method of analysis of TASD data: Monte Carlo or experimental. The effects may be small on the scale of the usual linear plot of mean-square displacement versus time, but the underlying structure can be seen by appropriate data analysis. Then, I show how the TASD varies with TPL parameters, as families of curves in the text and as plots of TASD parameters versus TPL parameters in the Supporting Materials and Methods. These Monte Carlo results show first, that the effects, though sometimes small, vary systematically with the TPL inputs; and second, that the dependence on TPL parameters here is more complex than earlier work on TASD resulting from a discrete distribution of escape times (2) suggests. Finally, I consider two important topics for the interpretation of these results: the conversion from Monte Carlo units to physical units and the requirement for a nonequilibrium state.
I argue that this structure is worth examining as a signature of the biological search process. Unfortunately one cannot in general uniquely invert an observed TASD plot of log D(t) vs. log t to give the distribution of escape times responsible for it, particularly when the functional form of the escape time distribution is not known a priori. But, one can readily use Monte Carlo simulations to obtain TASD curves from an assumed escape time distribution and potentially exclude the distribution.
Analysis of TASD
I review the method of analyzing mean-square displacement data, either SPT or Monte Carlo, to reveal the TASD substructure. In TASD, diffusion is subdiffusive at short times, that is, the mean-square displacement is proportional to a fractional power of time, and diffusion is normal at long times, that is, the mean-square displacement is proportional to time, giving
| (19) |
where is the mean-square displacement, t is time, α is the anomalous diffusion exponent, and tcross is the crossover time. In other words, the diffusion coefficient D(t) ∝ /t is time-dependent but reaches a constant value at large times. In subdiffusion, diffusion is hindered, and α < 1.
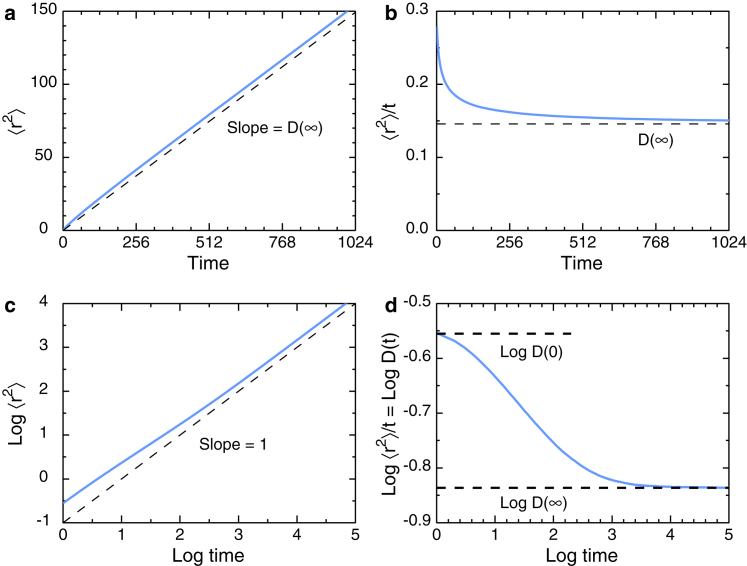
The simplest plot of the data is the standard plot of vs. time, Fig. 3 a, which gives for the most part an apparently straight line of slope D(∞) = constant. In this example, curvature is evident for t ≤ 128. Taking out the asymptotic time dependence reveals the structure more clearly. A plot of /t vs. t (Fig. 3 b) shows that the time range of the nonlinear regime is actually larger than Fig. 3 a suggests. The power-law form in Eq. 19 implies that a log-log plot is needed, and the simple plot of log vs. log t in Fig. 3 c shows the change in slope due to the change in exponent. The plots in Fig. 3, b and c each emphasize the initial structure, and combining them provides the clearest depiction of the structure, as shown in Fig. 3 d, log/t vs. log t. The plot of Fig. 3 d shows the initial structure by removing the asymptotic time dependence and can also be interpreted as the time dependence of log D(t), suitably normalized (41).
Figure 3.
How to find the TASD structure in SPT or Monte Carlo data. Various plots of the same Monte Carlo data for a 3D random walk on a simple cubic lattice are given. Linear plots: (a) vs. t and (b) /t vs. t are shown. Log-log plots: (c) log vs. log t and (d) log/t vs. log t are shown. Here, is in units of lattice constants, and t is in units of Monte Carlo steps. Conversion of Monte Carlo units to physical units is discussed in Conversion to Physical Units. The mean-square displacement is found by ensemble averaging, with no averaging over time segments within a trajectory. This choice of averaging is essential, as discussed in Nonequilibrium Requirement. To see this figure in color, go online.
The analysis of this plot (41) is sketched in Fig. 4. The initial behavior depends on the dynamics used (Brownian versus Newtonian) and on the nature of the trap (point traps here versus multisite dead ends in a percolating cluster). The anomalous exponent α is given by the slope of the anomalous region around the vertical midpoint, and the horizontal region represents normal diffusion with coefficient D(∞). The intersection of the slope line and the horizontal D(∞) line is one reasonable choice for the crossover time tcross, as discussed in Supporting Materials and Methods, Section S1.2.
Figure 4.
Analysis of transient anomalous subdiffusion in a plot of log/t vs. log t. To see this figure in color, go online.
Monte Carlo results for TASD
In this section, I present the results of Monte Carlo simulations of subdiffusion (Supporting Materials and Methods, Section S1.1). Here, T is taken to be the mean escape time, assumed to follow a TPL distribution as in Fig. 2. The actual escape time for each visit to a trap site is a random variate generated in the Monte Carlo program.
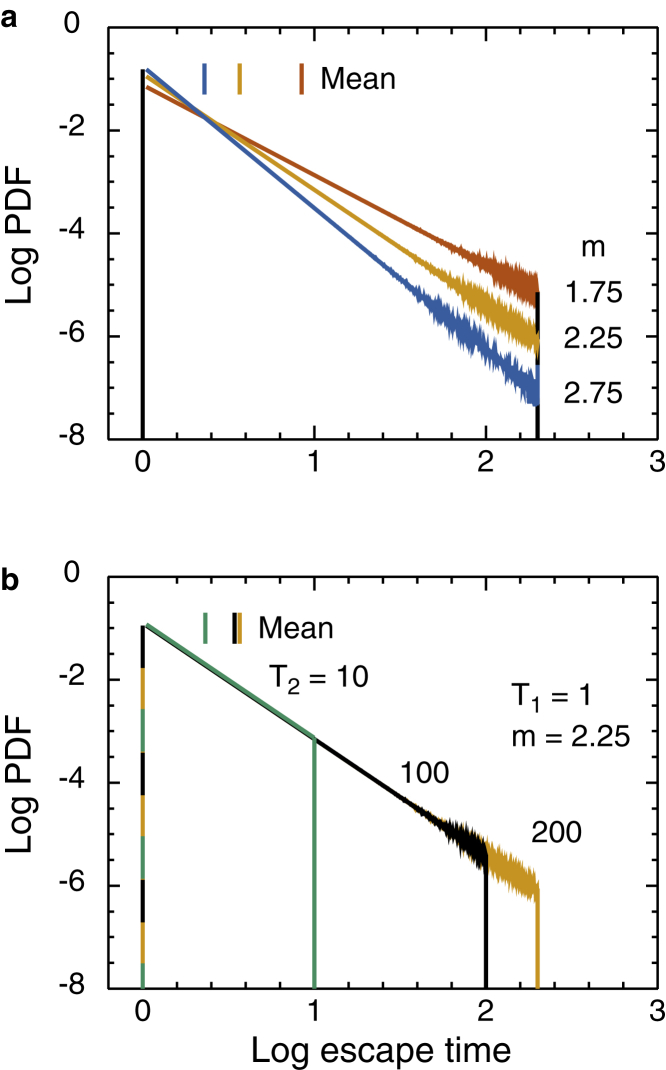
Fig. 5 a shows the effect of varying the exponent m at constant minimum and maximum times T1 = 1 and T2 = 200. The curves are of similar shape, but the value of log tcross changes with m as discussed in Supporting Materials and Methods, Section S4.1. Fig. 5 b shows the effect of increasing T1 at constant T2 and m. As T1 is increased, the curves flatten and D(t) decreases. In the limit T1 = T2, the curve is flat with D(t) = 1/T2. Fig. 5 c shows the effect of increasing T2 at constant T1 and m. As T2 increases, diffusion becomes more anomalous—the slope gets steeper—for a longer time. The top curve, T2 = T1 = 1, is the control; uniform traps yield normal diffusion at all times. Fig. 5 d shows that the trap concentration has a major effect on the anomalous exponent α at constant T1, T2, and m. As the fraction of traps increases, the curves grow steeper in the anomalous regime. The crossover time does not change much, so the change in slope is almost entirely due to the changes in D(0) and D(∞), Eqs. 21 and 25, with each nontrap site contributing T = 1 to the averages. Much, but not all, of the concentration dependence in the log D(t) curves can be removed by rescaling log D(t) as shown in Supporting Materials and Methods, Section S4.1. In simulations, the limiting values of D are trivial to calculate from the TPL parameters. In experiments, one ought to make good measurements of D(0) and D(∞), using fast and slow frame rates if necessary, to get good values. The amount of averaging needed to show the TASD structure is discussed in Supporting Materials and Methods, Section S4.3.
Figure 5.
Log-log plots of D(t) vs. t. Note the difference in the vertical scales for the first two panels and the last two. For comparison, the curve for the standard case T1 = 1, T2 = 200, m = 2.25, trap fraction 1, is shown in magenta in all panels. Blue lines indicate m = 1, 2, 3 in (a) and T2 a power of 10 in panel (c). (a) Vary m at constant T1 = 1, T2 = 200, trap fraction 1. Values of m are 1.00–3.00 in steps of 0.25. (b) Vary T1 at constant T2 = 200, m = 2.25, fraction of traps 1, for the indicated values of T1. The T2 = 1 curve is actually for T1 = 1, T2 = 1.01, so that the standard program could be used without modification. (c) Vary T2 at constant T1 = 1, m = 2.25, trap fraction 1. Values of T2 are from 1 to 5000 in a 1, 2, 5 series. (d) Vary fraction of traps at constant T1 = 1, T2 = 200, m = 2.25. Trap fractions are 0.1–1.0 in steps of 0.1. To see this figure in color, go online.
Four parameters characterize D(t): D(0) and D(∞) describe the initial and final values, the slope α describes how anomalous the diffusion is, and tcross locates the transition from anomalous to normal diffusion. Here, D(0) and D(∞) are obtained from simple formulas based on the TPL, but α and tcross are obtained from Monte Carlo calculations. Prediction of α and tcross will be discussed in future work.
The initial and final diffusion coefficients in a system with binding are given by a simple argument. The normalized diffusion coefficient is the product of the squared displacement for a move and the probability of that move. For a single step, the squared displacement is 1 in units of the lattice constant . For the first move, the probability is
| (20) |
where the subscript indicates an average over the TPL distribution, so
| (21) |
and, from Eq. S2–7,
| (22) |
Here, D(0) is normalized to 1 when all sites are nonbinding. For a move at long times, the probability is
| (23) |
where the subscript th indicates a thermal average over the TPL distribution, so from Eq. S2–14,
| (24) |
and
| (25) |
From Eq. S2–7,
| (26) |
More formal arguments for Eq. 26 are given in the literature (42, 43, 44).
The results here for the 3D case are qualitatively similar to earlier results (1) for the 2D case. The main effect of dimensionality is through the probability that a diffusing particle revisits a site in a lattice model. For normal diffusion in an infinite system, the number of distinct sites visited (DSV) can be written as a function of number of steps n, as DSV(1D) ∝ , DSV(2D) ∝ n/ln n, DSV(3D) ∝ n. These are the leading terms; there are higher-order correction terms (45). For anomalous subdiffusion on an infinite fractal, the DSV depends on the so-called spectral or fracton dimension, which is not necessarily an integer. This dimension is one of the standard quantities used to characterize fractals (45).
Conversion to physical units
A key question is how to translate Monte Carlo units into physical units, particularly how to translate tcross into clock time. The conversion is based on one of the fundamental equations of diffusion, in three dimensions
| (27) |
where is the lattice constant, D0 the diffusion coefficient in the absence of hindrances, and δt the unit of time. Choose any two and calculate the third.
In earlier work on pure obstructed diffusion in 2D membranes (46), the bilayer was taken to be on average a triangular lattice of lipids, was chosen to be the lipid diameter, and D0 was chosen as the value for the tracer in a pure lipid bilayer. The resulting is the characteristic time to diffuse the length of the bond joining adjacent lattice points.
The case of the nucleus is more complicated. Ultimately one would like to model all four hindrances to diffusion—binding, obstruction, viscoelasticity, and crowding—but at this stage, it is appropriate to model binding in isolation, and take the other hindrances into account approximately through D0. I choose D0 to be the value from FRAP experiments on green fluorescent protein (GFP) in the nucleus of H1299 human large cell lung carcinoma cells (47). This is a long-range diffusion coefficent for a protein that is presumably nonbinding. The value is D0 = 41.6 μm2/s, here rounded off to 40 μm2/s. What value to use for ? Analogy with the membrane case suggests choosing to be the size of a transcription factor binding site, roughly 10 base pairs (26), so 3 nm. Then δt = /6D0 = 37.5 ns, which is very poorly matched to the time resolution of the SPT experiments. A single SPT time step of 5 ms would require over 133,000 Monte Carlo steps. A better choice is to take to be the SPT spatial resolution of 25 nm, so that δt = 2.6 μs. This is still not well matched to the SPT experiment; a single SPT time step corresponds to 1920 Monte Carlo time steps, and an escape time of 5 s requires 1.8 million Monte Carlo time steps. But any further increase in would make the Monte Carlo resolution less than the experimental resolution. Better to spend computer time on longer simulations than to throw away hard-earned experimental spatial resolution. With this choice of , the Monte Carlo calculations are done on a simple cubic lattice of resels (resolution elements), which seems appropriate.
Nonequilibrium requirement
The TASD mechanism discussed here requires the system to be out of thermodynamic equilibrium, at least partially. Here, I assume that the tracer is in local thermodynamic equilibrium, so its probability of being in a trap is given by a Boltzmann factor, as in Eq. 9. But the tracer is not in global equilibrium with the population of traps, and TASD reveals this global equilibration process, a process that requires the tracer to sample the population of traps adequately, as will be discussed in future work.
A fundamental property of diffusion in the presence of time-independent (quenched) energy traps is that it is highly sensitive to the initial conditions, specifically the time since the interaction of the tracer with the traps is turned on. In the nonequilibrium case, fresh tracer diffuses with a time-dependent diffusion coefficient D(t) as shown in Fig. 3 d. In the equilibrium case, stale tracer diffuses with a small constant diffusion coefficient D(∞), determined largely by the escape time from the deepest traps. In the physics literature, this sensitivity is called “aging,” the dependence of diffusion on the time elapsed since the initial preparation of the system (22). Ordinary diffusion is said to be stationary; that is, the diffusive behavior for a given time interval is statistically the same as the behavior for any another time interval of the same duration. There is no absolute clock. For diffusion with binding to nonuniform static traps, there is an absolute clock. The search process is that clock, and its zero time is defined as the laboratory time at which the interaction is turned on.
In the nucleus, the interaction is turned on by entry of the tracer into the nucleus, or by biochemical activation. Normanno et al. (3) reported a time of 1 min between mixing and the start of observations, a significant delay. The modeling of delay effects is discussed in Supporting Materials and Methods, Section S4.4, but the main implication of the modeling is that experimental data is needed. Photoactivation of the DNA-binding species would be very useful experimentally to reduce the delay and to define “t = 0” precisely for ensemble averaging of measured trajectories. Photoactivated probes are discussed briefly in Supporting Materials and Methods, Section S4.4.4.
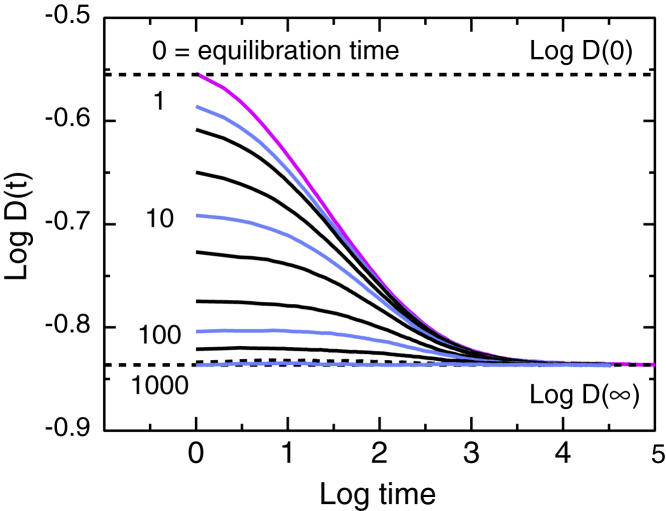
The effect of delay can be illustrated by Monte Carlo calculations in which the tracer takes a prescribed number of equilibration steps before log D(t) is recorded. (See (2) for the 2D case.) For an equilibration time of 0, the usual TASD results are recovered For the largest equilibration time, diffusion is normal at all times but slow, with D = D(∞). Intermediate equilibration times flatten the initial part of the D(t) curve, as shown in Fig. 6. The theoretical importance of this result is that one can go smoothly from pure TASD to slow pure normal diffusion by tuning a single well-defined parameter. The experimental importance is that the changes in shape at small times are the signature of partial equilibration of the tracer with the binding sites, for example, during the delay between mixing and the start of SPT observations. Importantly, no such changes occur for pure obstructed diffusion. The plot of log D vs. log t is independent of initial delay time (1).
Figure 6.
Annealing. Change in shape of log D(t) vs. log t for various equilibration times 0, 1, 2, 5, 10, 20, 50, 100, 200, 500, 1000″, with T1 = 2, T2 = 200, m = 2.25, trap fraction 1. In general, significant equilibration effects occur for equilibration times < tcross. Powers of 10 are shown in blue; the standard unequilibrated case, in magenta. To see this figure in color, go online.
Discussion
How does the model presented here connect to results in the physics and biophysics literature?
Connections to models of anomalous subdiffusion in the physics literature
Several different physical mechanisms can lead to anomalous subdiffusion, pure or transient. See, for example, the book by ben-Avraham and Havlin (48) and recent reviews (22,49, 50, 51, 52). Supporting Materials and Methods, Section S5 further discusses experiments to distinguish these mechanisms and some types of tracers that might be useful in these experiments.
Nonequilibrium trapping in energy traps
As already discussed, the tracer must not be in equilibrium with the system of traps. If it is equilibrated, it diffuses normally, at a rate dominated by the escape time from the deeper traps.
Obstruction
Obstruction can lead to anomalous subdiffusion, either pure or transient (41). There is no dependence on equilibration time, but obstructed diffusion is highly sensitive to tracer size, as discussed in the next section.
Anticorrelated motion
In the physics literature, this category has often been taken to be fractional Brownian motion as defined by Mandelbrot and Van Ness (53) in terms of a correlation function of extremely macroscopic inspiration, Hurst’s classic studies of flooding on the Nile. A more microscopic starting point for cell biophysics is viscoelasticity (50,54, 55, 56).
All of the above
These processes are nonexclusive and can operate in parallel. In a cell, it would be much more surprising to have one of the processes dominant than to have all three act simultaneously. The obvious example is that chromatin can act as an obstacle and as a set of binding sites. For examples of how to sort out anomalous subdiffusion mechanisms, see Golan and Sherman (57) for the T-cell plasma membrane, Kepten et al. (58) for diffusion of telomeres, Szymanski and Weiss (59) for a model crowded system, and Thiel et al. (60) for modeling.
A series of experiments and simulations by Izeddin et al. (61) addressed the question of physics models. The experiments used PALM SPT to examine motion of two nuclear factors in U2OS human osteosarcoma cells. Trajectories were analyzed by plots of mean-square displacement versus time, histograms of step sizes, and histograms of the angles between consecutive displacements at various time lags as a test for anticorrelation on various length scales. Four proteins were used, all with the photoconvertible label Dendra2: 1) free Dendra2. It is structurally similar to GFP and is assumed to be freely diffusing because GFP has no known binding partners in mammalian cells; 2) Dendra2 fused to the proto-oncogene c-Myc, which binds directly to DNA. Fast, slow, and immobile populations were observed, and the angular distribution of moves was approximately uniform; 3) Dendra2 fused to the cyclin T1 subunit of P-TEFb, which binds to the transcription machinery. Motion was subdiffusive with α ∼ 0.6, and the angular distribution of moves favored reversal; and 4) Dendra2 fused to histone H2B was bound, and the angular distribution favored reversal. Results were compared to a variety of models based on the physics literature and interpreted in terms of target-searching strategies. For c-Myc, the search involved global exploration of space, which leaves sites unvisited. For P-TEPb, the search involved local (compact) exploration of space, thus a redundant search.
Connection to models of the nucleus in the biophysics literature
Several interpretations are possible of the diffusion model presented here.
No interpretation is needed
A TPL distribution of mean escape times necessarily implies transient anomalous subdiffusion, independent of the microscopic mechanism producing the escape times. But it is useful to go beyond this to ask what happens during the escape time.
Binding
The diffusing particle is bound to an immobile site on the DNA, escapes, and then diffuses freely or at the rate set by pure obstruction. This model is the one discussed in the most detail here.
Facilitated diffusion
To reduce a vast literature to a single paragraph, the key problem in DNA binding is searching. How does a DNA-binding species find its biological target fast enough among the huge number of potential binding sites? The problem is solved by the classic Berg-von Hippel facilitated diffusion model (62,63), in which the search is speeded by switching between 3D and 1D diffusion. A major issue within the facilitated diffusion model is the speed-stability paradox, involving the energy landscape of the protein-DNA interaction. A smooth landscape is required for rapid motion, but a rugged landscape is required for specific binding. A solution to this problem is the two-state protein model, in which the protein can exist in recognition and search states (24). Recent reviews include the very succinct (64), more detailed discussions in (25,65,66), and a physics perspective in (67), but see (68) for an argument that the speed-selectivity paradox is not real.
The simplest mapping between facilitated diffusion and the model presented here identifies the escape time from a trap as a period of 1D diffusion, and diffusion between traps as a period of 3D diffusion, as suggested by Normanno et al. (3). This mapping is consistent with the resolution of the experiments. Estimates of the 1D diffusion length vary among systems; Barbi et al. (69) used a value of 170 base pairs in the model summarized in the next paragraph. The SPT lateral resolution is 25 nm or 75 base pairs (1 base pair = 0.332 nm), so a period of 1D diffusion of 170 base pairs is below the 1D detection limit of three points for a straight line. The persistence length of DNA is 50 nm (13), so the DNA in one resolution element could be slightly curved, but not coiled unless bound in a nucleosome. Recent SPT measurements on RNA polymerase (RNAP) in Escherichia coli cells found three distinct diffusive modes, which were interpreted as DNA-bound RNAP, RNAP rapidly associating and dissociating with DNA, and freely diffusing RNAP (Bettridge et al. (70,71)).
Modeling by Barbi et al. (69,72) showed that the local energy landscape for protein binding to DNA can lead to 1D anomalous subdiffusion of the protein. Combining their 1D results with the 3D results presented here leads to what might be called nested TASD. The first level is 1D TASD because of the detailed DNA sequence, resulting from variations in either the H-bond binding energy (69,72) or the elastic energy of DNA bent by protein (8). This 1D diffusion crosses over to normal when the DNA-binding species has sampled the DNA energy landscape sufficiently. The second level is 3D TASD because of the distribution of escape times from the 1D diffusion state. The nested model must include unbinding and binding events, and thus some modeling of DNA geometry, at least at the level of the approximate 3D separation of binding sites. Nested TASD can be considered a multilevel equilibration process.
Obstruction and geometrical traps
Diffusion in the nucleus is likely to be affected by obstruction, especially by geometrical trapping in dead ends (see Supporting Materials and Methods, Section S5). Obviously, the geometrical traps must be large enough for the tracer to enter, as in size exclusion chromatography. If the trap size is below the resolution of the SPT measurement, the tracer will appear to be stationary. If motion in the trap can be resolved, the well-known form of for confined motion (73) will be observed, with eventual escape to free diffusion. This form is distinct from the form for anomalous subdiffusion, as shown in cartoon form in Fig. 7. It is informative to make two plots, vs. t and log D vs. log t. In their work on TetR diffusion in the nucleus, Normanno et al. (3) observed some trajectories consistent with pure confinement, with timescales <1 ms and confinement radii in the range of 0.5–1 μm. (See their Fig. S2, Confined diffusion analysis, and their Note S4, Single-particle tracking analysis).
Figure 7.
Cartoon of vs. time for geometrical trapping and for pure anomalous subdiffusion. (a) Pure confinement and transient confinement with eventual escape and free diffusion. The plateau is a measure of the corral size. The escape time is random, so in an actual average of SPT trajectories, the transition between confined motion inside the dead end and free motion outside would be smeared out. (b) Pure anomalous subdiffusion. There is no plateau. To see this figure in color, go online.
For rigid immobile obstacles, there is a well-defined percolation threshold. The experimental signature of this case is that diffusion is highly sensitive to the size of the tracer. The factor controlling diffusion is not the area fraction of obstacles but the excluded area fraction, which is highly sensitive to tracer size (for 2D immobile obstacles (74)). Bénichou et al. (9) modeled chromatin as a 3D percolating cluster at the threshold. They argued that on a large scale, chromatin is effectively branched as a result of intersegmental transfer of the diffusing species.
For soft or mobile obstacles, there is no permanent obstruction. The escape times depend in part on obstacle dynamics, such as mobility, dissociation rates, or rates of conformational fluctuations. Chow and Skolnick (75) present a coarse-grained model of gated diffusion of LacI in the E. coli nucleoid, in which LacI moves from cage to cage when the DNA wall moves. Diffusion may still be highly sensitive to tracer size as a result of the lower probability of large fluctuations. High concentrations of mobile obstacles lead to a glassy state. For a model of the nucleus based on glassy dynamics, see Kang et al. (76).
Conclusions
The model has several implications for experiments and the interpretation of experiments.
Search
Experimental examination of the TASD substructure of SPT measurements is useful, even though the effect is sometimes a small one that might be overlooked in the usual linear plot of mean-square displacement versus time, especially in noisy data. The phenomenon of aging is important in a static trap model. That is, the time in a log-log plot of D(t) is the clock time since the start of the interaction of the tracer with the DNA, so trajectories must be ensemble averaged, not averaged over time segments within an individual trajectory. Using a photoactivated tracer (Supporting Materials and Methods, Section S4.4.4) would be worth the effort because photoactivation improves the initial time resolution (Supporting Materials and Methods, Section S4.4) and defines zero time for ensemble averaging.
Escape time distributions
I have shown the relation between the observed escape times and the mean escape times for a TPL distribution of mean escape times. The distribution of observed escape times is broadened by the statistics of escape. The mean escape time distribution is static; the observed distribution is static plus dynamic. For binding, the mean escape time distribution will eventually be predictable from genomics, though this distribution of binding energies must be corrected for the physical accessibility of binding sites to a tracer of given size and shape. For obstruction, constructing the escape time distribution requires a geometric model of chromatin.
Limiting measurements
The values of D(0) and D(∞) capture much of the dependence of D(t) on the TPL parameters. These values are trivial to calculate from the parameters and essential to measure experimentally, using fast and slow frame rates if necessary. See Supporting Materials and Methods, Section S4.1.
Experimental controls
As claimed in the section on models of anomalous subdiffusion from the physics literature, binding, obstruction, viscoelasticity, and crowding may all affect diffusion in a cell simultaneously. Sorting out these mechanisms requires a variety of experimental controls even more extensive than usual. As a baseline, D in buffer ought to be measured for each probe. For comparisons among cell lines and laboratories, D of eGFP ought to be measured. Two types of controls are needed for nonspecific binding by the DNA-binding species of interest: “specific nonspecific” binding, in which the recognition site binds to DNA sequences that only partially match the true target site; and “nonspecific nonspecific” binding, in which nonrecognition sites bind to DNA, say through electrostatic interactions. Natural selection may have reduced nonspecific binding, as discussed by Qian and Kussell (77), and Buchanan (78).
The probe for “specific nonspecific” binding ought to be the DNA-binding species of interest with the binding site inactivated as unobtrusively as possible. For example, the pioneering single-particle measurements of Elf et al. (79) on transcription factor dynamics used LacI (360 amino acids) with the DNA-binding domain deleted (41 amino acids); the fusion to Venus fluorescent protein (238 amino acids) was most likely a much larger perturbation. Elf and Barkefors (80) recently reviewed single-molecule kinetic measurements in live cells, with an extensive discussion of labels and perturbations from labeling.
The comparison of nonbinding and binding tracers is essential to be able to distinguish TASD due to obstruction from TASD due to binding. A general problem in diffusion measurements in complex systems is that a measurement of D or D(t) is not uniquely invertible. By an appropriate choice of parameters, the three anomalous subdiffusion mechanisms mentioned above can presumably be tuned to produce very similar curves, and the mechanisms may well operate in parallel. Despite this limitation, diffusion measurements are important to biophysicists because diffusion is important to cells. Diffusion is a means of transport that uses thermal energy, not metabolic energy.
Histograms of escape times for nonbinding tracers would help to characterize confinement events due to obstruction, and apparent confinement events due to fluctuations in pure random walks, in the context of the experimental conditions and the criteria used to define confinement. These criteria ought to be described in detail, as in Gorman et al. (81), for example. Nonspecific binding can be characterized by comparing the escape time histograms for the DNA-binding tracer and the corresponding nonbinding control.
The controls for obstruction ought to be done experimentally. Modeling obstruction requires a model of chromatin organization in the nucleus, a complicated and unsettled issue at best, requiring multiscale modeling. See, for example, (10, 11, 12, 13,82).
Subresel dynamics
In the three sets of SPT experiments (3, 4, 5), the resolution element was significantly larger than a transcription factor binding site, so it is necessary to consider the dynamics of the DNA-binding species within a resel. Experimental data would be a useful constraint, particularly a histogram of the escape time from a single resel for a sample of distinct resels. These histograms ought to be reported for the DNA-binding species, the nonbinding analogs, and eGFP or another general calibration protein.
Effect of deep traps
Eliminating deep traps may affect diffusion and kinetics. First, false-positive sites affect diffusion, and the effect is greater the more similar they are to the target sequence. As a result, in bioinformatics searches for binding sites, the occurrence and frequency of false-positive sites are important results, not annoying artifacts. (For a general discussion of weak binding sites on DNA, see Tanay (83).) The search process in vivo might be speeded by (re)designing the genome to eliminate false-positive sites, that is, by separating the target sequences in affinity space. Hahn et al. (84) found a mild form of this effect in prokaryotic genomes. Sheinman et al. (25) used extreme value statistics to examine the gap in binding energy between the target site and the deepest false-positive sites and distinguished the gapped, marginally gapped, and nongapped cases.
Second, overexpression of the diffusing species affects diffusion. The level of expression is potentially an uncontrolled variable affecting TASD or a means of modulating and studying TASD. In their work on TetR in U2OS cells, Normanno et al. (3) did a key experiment to address (and refute for their system) this point. They tracked labeled TetR with and without 1000-fold excess of unlabeled TetR. They saw no effect. The traps were not saturable, at least at this level of excess. Perhaps the traps were highly abundant geometric traps. This sort of test ought to be standard practice. As these authors point out, quantifying the number of binding sites in the nucleus would be very useful.
This effect of eliminating deep binding sites can be approximated very simply if the concentrations of binding sites are known. If N unlabeled tracers are added, assume that they fill the N deepest sites, in effect reducing the upper limit T2 on the mean escape time and making diffusion less anomalous for a shorter time as shown in Fig. 5. This description is somewhat fictitious in that it assumes that N unlabeled tracers at zero temperature fill the deepest traps permanently, and then a single labeled tracer at ambient temperature diffuses, entering into and escaping from the remaining traps. But the picture captures enough of the physics simply enough to be a named model, the step approximation. A more complete treatment includes the chemical potential, to allow thermal excitation of all bound tracers. Such a treatment is a standard result in the field of diffusion in inhomogeneous catalyst pellets (85, 86, 87). Models of DNA binding that include the chemical potential are called “biophysical” or “thermodynamic” or “statistical thermodynamic.” See, for example, (83,88, 89, 90). A rigorous approach takes into account the binding equilibria of multiple species to multiple sites, as in the program of Wasson and Hartemink (91).
Applications of model
The TASD model provides a simple Monte Carlo prediction of D(t) based on SPT measurements, suitable for use in larger-scale modeling of FRAP, FCS, and kinetics. It would be interesting to do FRAP or FCS on the same system as this SPT work, with the corresponding Monte Carlo simulations.
In summary, the Monte Carlo simulations presented here show that a TPL distribution of escape times leads to transient anomalous subdiffusion, as expected. I have described the parameters characterizing each and begun to examine the connection between the two sets of parameters. I have discussed the biological interpretation and the experiments and controls suggested by modeling. Further work will examine the detailed connection of the TPL and TASD parameters and the basis for a TPL distribution of escape times.
Acknowledgments
The early parts of this work were supported by the National Institute of General Medical Sciences of the U.S. National Institutes of Health under award R01-GM038133. The content is solely the responsibility of the author and does not necessarily represent the official views of the National Institutes of Health.
Editor: Andrew Spakowitz.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2020.03.015.
Supporting Citations
References (92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129, 130, 131, 132, 133, 134, 135, 136, 137, 138, 139, 140, 141, 142, 143) appear in the Supporting Material.
Supporting Material
References
- 1.Saxton M.J. Anomalous diffusion due to binding: a Monte Carlo study. Biophys. J. 1996;70:1250–1262. doi: 10.1016/S0006-3495(96)79682-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saxton M.J. A biological interpretation of transient anomalous subdiffusion. I. Qualitative model. Biophys. J. 2007;92:1178–1191. doi: 10.1529/biophysj.106.092619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Normanno D., Boudarène L., Dahan M. Probing the target search of DNA-binding proteins in mammalian cells using TetR as model searcher. Nat. Commun. 2015;6:7357. doi: 10.1038/ncomms8357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caccianini L., Normanno D., Dahan M. Single molecule study of non-specific binding kinetics of LacI in mammalian cells. Faraday Discuss. 2015;184:393–400. doi: 10.1039/c5fd00112a. [DOI] [PubMed] [Google Scholar]
- 5.Knight S.C., Xie L., Tjian R. Dynamics of CRISPR-Cas9 genome interrogation in living cells. Science. 2015;350:823–826. doi: 10.1126/science.aac6572. [DOI] [PubMed] [Google Scholar]
- 6.Shlesinger M.F. Fractal time in condensed matter. Annu. Rev. Phys. Chem. 1988;39:269–290. [Google Scholar]
- 7.Woringer M., Darzacq X. Protein motion in the nucleus: from anomalous diffusion to weak interactions. Biochem. Soc. Trans. 2018;46:945–956. doi: 10.1042/BST20170310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slutsky M., Kardar M., Mirny L.A. Diffusion in correlated random potentials, with applications to DNA. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2004;69:061903. doi: 10.1103/PhysRevE.69.061903. [DOI] [PubMed] [Google Scholar]
- 9.Bénichou O., Chevalier C., Voituriez R. Facilitated diffusion of proteins on chromatin. Phys. Rev. Lett. 2011;106:038102. doi: 10.1103/PhysRevLett.106.038102. [DOI] [PubMed] [Google Scholar]
- 10.Imakaev M.V., Fudenberg G., Mirny L.A. Modeling chromosomes: beyond pretty pictures. FEBS Lett. 2015;589:3031–3036. doi: 10.1016/j.febslet.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosa A., Zimmer C. Computational models of large-scale genome architecture. Int. Rev. Cell Mol. Biol. 2014;307:275–349. doi: 10.1016/B978-0-12-800046-5.00009-6. [DOI] [PubMed] [Google Scholar]
- 12.Huet S., Lavelle C., Bancaud A. Relevance and limitations of crowding, fractal, and polymer models to describe nuclear architecture. Int. Rev. Cell Mol. Biol. 2014;307:443–479. doi: 10.1016/B978-0-12-800046-5.00013-8. [DOI] [PubMed] [Google Scholar]
- 13.Amitai A., Holcman D. Polymer physics of nuclear organization and function. Phys. Rep. 2017;678:1–83. [Google Scholar]
- 14.Saxton M.J. Modeling 2D and 3D diffusion. Methods Mol. Biol. 2007;400:295–321. doi: 10.1007/978-1-59745-519-0_20. [DOI] [PubMed] [Google Scholar]
- 15.Forbes C.S., Evans M., Peacock J.B. Fourth Edition. Wiley; Hoboken, NJ: 2011. Statistical Distributions. [Google Scholar]
- 16.Newman M.E.J. Power laws, Pareto distributions and Zipf’s law. Contemp. Phys. 2005;46:323–351. [Google Scholar]
- 17.Sternberg S.H., Redding S., Doudna J.A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorman J., Fazio T., Greene E.C. Nanofabricated racks of aligned and anchored DNA substrates for single-molecule imaging. Langmuir. 2010;26:1372–1379. doi: 10.1021/la902443e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saxton M.J. Lateral diffusion in an archipelago. Single-particle diffusion. Biophys. J. 1993;64:1766–1780. doi: 10.1016/S0006-3495(93)81548-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rose C., Smith M.D. Springer; New York: 2002. Mathematical Statistics with Mathematica. [Google Scholar]
- 21.Bernasconi J., Beyeler H.U., Strassler S. Anomalous frequency-dependent conductivity in disordered one-dimensional systems. Phys. Rev. Lett. 1979;42:819–822. [Google Scholar]
- 22.Metzler R., Jeon J.-H., Barkai E. Anomalous diffusion models and their properties: non-stationarity, non-ergodicity, and ageing at the centenary of single particle tracking. Phys. Chem. Chem. Phys. 2014;16:24128–24164. doi: 10.1039/c4cp03465a. [DOI] [PubMed] [Google Scholar]
- 23.Nelson J. Continuous-time random-walk model of electron transport in nanocrystalline TiO2 electrodes. Phys. Rev. B Condens. Matter Mater. Phys. 1999;59:15374–15380. [Google Scholar]
- 24.Slutsky M., Mirny L.A. Kinetics of protein-DNA interaction: facilitated target location in sequence-dependent potential. Biophys. J. 2004;87:4021–4035. doi: 10.1529/biophysj.104.050765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheinman M., Bénichou O., Voituriez R. Classes of fast and specific search mechanisms for proteins on DNA. Rep. Prog. Phys. 2012;75:026601. doi: 10.1088/0034-4885/75/2/026601. [DOI] [PubMed] [Google Scholar]
- 26.Stewart A.J., Hannenhalli S., Plotkin J.B. Why transcription factor binding sites are ten nucleotides long. Genetics. 2012;192:973–985. doi: 10.1534/genetics.112.143370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montroll E.W., Shlesinger M.F. On 1/f noise and other distributions with long tails. Proc. Natl. Acad. Sci. USA. 1982;79:3380–3383. doi: 10.1073/pnas.79.10.3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitzenmacher M. A brief history of generative models for power law and lognormal distributions. Internet Math. 2004;1:226–251. [Google Scholar]
- 29.Malevergne Y., Pisarenko V., Sornette D. Testing the Pareto against the lognormal distributions with the uniformly most powerful unbiased test applied to the distribution of cities. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2011;83:036111. doi: 10.1103/PhysRevE.83.036111. [DOI] [PubMed] [Google Scholar]
- 30.Aurell E., d’Hérouël A.F., Vergassola M. Transcription factor concentrations versus binding site affinities in the yeast S. cerevisiae. Phys. Biol. 2007;4:134–143. doi: 10.1088/1478-3975/4/2/006. [DOI] [PubMed] [Google Scholar]
- 31.Djordjevic M., Sengupta A.M., Shraiman B.I. A biophysical approach to transcription factor binding site discovery. Genome Res. 2003;13:2381–2390. doi: 10.1101/gr.1271603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng X., Wang J. The universal statistical distributions of the affinity, equilibrium constants, kinetics and specificity in biomolecular recognition. PLoS Comput. Biol. 2015;11:e1004212. doi: 10.1371/journal.pcbi.1004212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia H.G., Grayson P., Wiggins P.A. Biological consequences of tightly bent DNA: the other life of a macromolecular celebrity. Biopolymers. 2007;85:115–130. doi: 10.1002/bip.20627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bancaud A., Huet S., Ellenberg J. Molecular crowding affects diffusion and binding of nuclear proteins in heterochromatin and reveals the fractal organization of chromatin. EMBO J. 2009;28:3785–3798. doi: 10.1038/emboj.2009.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mirny L.A. The fractal globule as a model of chromatin architecture in the cell. Chromosome Res. 2011;19:37–51. doi: 10.1007/s10577-010-9177-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loverdo C., Bénichou O., Desbiolles P. Quantifying hopping and jumping in facilitated diffusion of DNA-binding proteins. Phys. Rev. Lett. 2009;102:188101. doi: 10.1103/PhysRevLett.102.188101. [DOI] [PubMed] [Google Scholar]
- 37.Parsaeian A., de la Cruz M.O., Marko J.F. Binding-rebinding dynamics of proteins interacting nonspecifically with a long DNA molecule. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2013;88:040703. doi: 10.1103/PhysRevE.88.040703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amitai A. Chromatin configuration affects the dynamics and distribution of a transiently interacting protein. Biophys. J. 2018;114:766–771. doi: 10.1016/j.bpj.2017.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guigas G., Kalla C., Weiss M. Probing the nanoscale viscoelasticity of intracellular fluids in living cells. Biophys. J. 2007;93:316–323. doi: 10.1529/biophysj.106.099267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saxton M.J. Single-particle tracking of DNA-binding biomolecules in the nucleus: why a power-law distribution of dwell times? Biophys. J. 2020;118:458a. [Google Scholar]
- 41.Saxton M.J. Anomalous diffusion due to obstacles: a Monte Carlo study. Biophys. J. 1994;66:394–401. doi: 10.1016/s0006-3495(94)80789-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haus J.W., Kehr K.W., Lyklema J.W. Diffusion in a disordered medium. Phys. Rev. B Condens. Matter. 1982;25:2905–2907. [Google Scholar]
- 43.Bouchaud J.P., Georges A. Anomalous diffusion in disordered media: statistical mechanisms, models and physical applications. Phys. Rep. 1990;195:127–293. [Google Scholar]
- 44.Haus J.W., Kehr K.W. Diffusion in regular and disordered lattices. Phys. Rep. 1987;150:263–406. [Google Scholar]
- 45.Klafter J., Sokolov I.M. Oxford University Press; Oxford, UK: 2011. First Steps in Random Walks: From Tools to Applications. [Google Scholar]
- 46.Saxton M.J. Lateral diffusion in a mixture of mobile and immobile particles. A Monte Carlo study. Biophys. J. 1990;58:1303–1306. doi: 10.1016/S0006-3495(90)82470-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hinow P., Rogers C.E., DiBenedetto E. The DNA binding activity of p53 displays reaction-diffusion kinetics. Biophys. J. 2006;91:330–342. doi: 10.1529/biophysj.105.078303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.ben-Avraham D., Havlin S. Cambridge University Press; Cambridge, UK: 2000. Diffusion and Reactions in Fractals and Disordered Systems. [Google Scholar]
- 49.Barkai E., Garini Y., Metzler R. Strange kinetics of single molecules in living cells. Phys. Today. 2012;65:29–35. [Google Scholar]
- 50.Höfling F., Franosch T. Anomalous transport in the crowded world of biological cells. Rep. Prog. Phys. 2013;76:046602. doi: 10.1088/0034-4885/76/4/046602. [DOI] [PubMed] [Google Scholar]
- 51.Krapf D. Mechanisms underlying anomalous diffusion in the plasma membrane. Curr. Top. Membr. 2015;75:167–207. doi: 10.1016/bs.ctm.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 52.Sokolov I.M. Models of anomalous diffusion in crowded environments. Soft Matter. 2012;8:9043–9052. [Google Scholar]
- 53.Mandelbrot B.B., Van Ness J.W. Fractional Brownian motions, fractional noises, and applications. SIAM Rev. 1968;10:422–437. [Google Scholar]
- 54.Weber S.C., Theriot J.A., Spakowitz A.J. Subdiffusive motion of a polymer composed of subdiffusive monomers. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2010;82:011913. doi: 10.1103/PhysRevE.82.011913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weiss M. Single-particle tracking data reveal anticorrelated fractional Brownian motion in crowded fluids. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2013;88:010101. doi: 10.1103/PhysRevE.88.010101. [DOI] [PubMed] [Google Scholar]
- 56.Goychuk I. Viscoelastic subdiffusion in a random Gaussian environment. Phys. Chem. Chem. Phys. 2018;20:24140–24155. doi: 10.1039/c8cp05238g. [DOI] [PubMed] [Google Scholar]
- 57.Golan Y., Sherman E. Resolving mixed mechanisms of protein subdiffusion at the T cell plasma membrane. Nat. Commun. 2017;8:15851. doi: 10.1038/ncomms15851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kepten E., Bronshtein I., Garini Y. Ergodicity convergence test suggests telomere motion obeys fractional dynamics. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2011;83:041919. doi: 10.1103/PhysRevE.83.041919. [DOI] [PubMed] [Google Scholar]
- 59.Szymanski J., Weiss M. Elucidating the origin of anomalous diffusion in crowded fluids. Phys. Rev. Lett. 2009;103:038102. doi: 10.1103/PhysRevLett.103.038102. [DOI] [PubMed] [Google Scholar]
- 60.Thiel F., Flegel F., Sokolov I.M. Disentangling sources of anomalous diffusion. Phys. Rev. Lett. 2013;111:010601. doi: 10.1103/PhysRevLett.111.010601. [DOI] [PubMed] [Google Scholar]
- 61.Izeddin I., Récamier V., Darzacq X. Single-molecule tracking in live cells reveals distinct target-search strategies of transcription factors in the nucleus. eLife. 2014;3:e02230. doi: 10.7554/eLife.02230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Berg O.G., von Hippel P.H. Diffusion-controlled macromolecular interactions. Annu. Rev. Biophys. Biophys. Chem. 1985;14:131–160. doi: 10.1146/annurev.bb.14.060185.001023. [DOI] [PubMed] [Google Scholar]
- 63.von Hippel P.H., Berg O.G. Facilitated target location in biological systems. J. Biol. Chem. 1989;264:675–678. [PubMed] [Google Scholar]
- 64.Tafvizi A., Mirny L.A., van Oijen A.M. Dancing on DNA: kinetic aspects of search processes on DNA. ChemPhysChem. 2011;12:1481–1489. doi: 10.1002/cphc.201100112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kong M., Van Houten B. Rad4 recognition-at-a-distance: physical basis of conformation-specific anomalous diffusion of DNA repair proteins. Prog. Biophys. Mol. Biol. 2017;127:93–104. doi: 10.1016/j.pbiomolbio.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Normanno D., Dahan M., Darzacq X. Intra-nuclear mobility and target search mechanisms of transcription factors: a single-molecule perspective on gene expression. Biochim. Biophys. Acta. 2012;1819:482–493. doi: 10.1016/j.bbagrm.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 67.Bauer M., Rasmussen E.S., Metzler R. Real sequence effects on the search dynamics of transcription factors on DNA. Sci. Rep. 2015;5:10072. doi: 10.1038/srep10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Veksler A., Kolomeisky A.B. Speed-selectivity paradox in the protein search for targets on DNA: is it real or not? J. Phys. Chem. B. 2013;117:12695–12701. doi: 10.1021/jp311466f. [DOI] [PubMed] [Google Scholar]
- 69.Barbi M., Place C., Salerno M. Base-sequence-dependent sliding of proteins on DNA. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2004;70:041901. doi: 10.1103/PhysRevE.70.041901. [DOI] [PubMed] [Google Scholar]
- 70.Bettridge K., Bohrer C., Xiao J. Investigating RNAP search dynamics in live E. coli cells using single molecule and statistical methods. Biophys. J. 2017;112:312A. doi: 10.1016/j.bpj.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bettridge K., Verma S., Xiao J. Single molecule tracking reveals the role of transitory dynamics of nucleoid-associated protein HU in organizing the bacterial chromosome. bioRxiv. 2020 doi: 10.1101/2019.12.31.725226. [DOI] [Google Scholar]
- 72.Barbi M., Place C., Salerno M. A model of sequence-dependent protein diffusion along DNA. J. Biol. Phys. 2004;30:203–226. doi: 10.1023/B:JOBP.0000046728.51620.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Saxton M.J. Single-particle tracking: effects of corrals. Biophys. J. 1995;69:389–398. doi: 10.1016/S0006-3495(95)79911-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Saxton M.J. Two-dimensional continuum percolation threshold for diffusing particles of nonzero radius. Biophys. J. 2010;99:1490–1499. doi: 10.1016/j.bpj.2010.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chow E., Skolnick J. DNA internal motion likely accelerates protein target search in a packed nucleoid. Biophys. J. 2017;112:2261–2270. doi: 10.1016/j.bpj.2017.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kang H., Yoon Y.-G., Hyeon C. Confinement-induced glassy dynamics in a model for chromosome organization. Phys. Rev. Lett. 2015;115:198102. doi: 10.1103/PhysRevLett.115.198102. [DOI] [PubMed] [Google Scholar]
- 77.Qian L., Kussell E. Genome-wide motif statistics are shaped by DNA binding proteins over evolutionary time scales. Phys. Rev. X. 2016;6:041009. [Google Scholar]
- 78.Buchanan M. Focus: evolution thins out distracting DNA. Physics. 2016;9:119. [Google Scholar]
- 79.Elf J., Li G.-W., Xie X.S. Probing transcription factor dynamics at the single-molecule level in a living cell. Science. 2007;316:1191–1194. doi: 10.1126/science.1141967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Elf J., Barkefors I. Single-molecule kinetics in living cells. Annu. Rev. Biochem. 2019;88:635–659. doi: 10.1146/annurev-biochem-013118-110801. [DOI] [PubMed] [Google Scholar]
- 81.Gorman J., Wang F., Greene E.C. Single-molecule imaging reveals target-search mechanisms during DNA mismatch repair. Proc. Natl. Acad. Sci. USA. 2012;109:E3074–E3083. doi: 10.1073/pnas.1211364109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hacker W.C., Li S., Elcock A.H. Features of genomic organization in a nucleotide-resolution molecular model of the Escherichia coli chromosome. Nucleic Acids Res. 2017;45:7541–7554. doi: 10.1093/nar/gkx541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tanay A. Extensive low-affinity transcriptional interactions in the yeast genome. Genome Res. 2006;16:962–972. doi: 10.1101/gr.5113606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hahn M.W., Stajich J.E., Wray G.A. The effects of selection against spurious transcription factor binding sites. Mol. Biol. Evol. 2003;20:901–906. doi: 10.1093/molbev/msg096. [DOI] [PubMed] [Google Scholar]
- 85.Kirchheim R. Hydrogen solubility and diffusivity in defective and amorphous metals. Prog. Mater. Sci. 1988;32:261–325. [Google Scholar]
- 86.Cameron L.M., Sholl C.A. The average jump rate and diffusion in disordered systems. J. Phys. Condens. Matter. 1999;11:4491–4497. [Google Scholar]
- 87.Dvoyashkin M., Khokhlov A., Valiullin R. Pulsed field gradient NMR study of surface diffusion in mesoporous adsorbents. Microporous Mesoporous Mater. 2009;125:58–62. [Google Scholar]
- 88.Berg O.G., von Hippel P.H. Selection of DNA binding sites by regulatory proteins. Statistical-mechanical theory and application to operators and promoters. J. Mol. Biol. 1987;193:723–750. doi: 10.1016/0022-2836(87)90354-8. [DOI] [PubMed] [Google Scholar]
- 89.Roider H.G., Kanhere A., Vingron M. Predicting transcription factor affinities to DNA from a biophysical model. Bioinformatics. 2007;23:134–141. doi: 10.1093/bioinformatics/btl565. [DOI] [PubMed] [Google Scholar]
- 90.Ruan S., Stormo G.D. Inherent limitations of probabilistic models for protein-DNA binding specificity. PLoS Comput. Biol. 2017;13:e1005638. doi: 10.1371/journal.pcbi.1005638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wasson T., Hartemink A.J. An ensemble model of competitive multi-factor binding of the genome. Genome Res. 2009;19:2101–2112. doi: 10.1101/gr.093450.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Saxton M.J. Anomalous subdiffusion in fluorescence photobleaching recovery: a Monte Carlo study. Biophys. J. 2001;81:2226–2240. doi: 10.1016/S0006-3495(01)75870-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bulnes F.M., Pereyra V.D., Riccardo J.L. Collective surface diffusion: n-fold way kinetic Monte Carlo simulation. Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Topics. 1998;58:86–92. [Google Scholar]
- 94.Press W.H., Teukolsky S.A., Flannery B.P. Second Edition. Cambridge University Press; Cambridge, UK: 1992. Numerical Recipes in FORTRAN. The Art of Scientific Computing. [Google Scholar]
- 95.Ellery A.J., Baker R.E., Simpson M.J. Communication: distinguishing between short-time non-Fickian diffusion and long-time Fickian diffusion for a random walk on a crowded lattice. J. Chem. Phys. 2016;144:171104. doi: 10.1063/1.4948782. [DOI] [PubMed] [Google Scholar]
- 96.Wolfram Language & System Documentation Center Random number generation. 2019. https://reference.wolfram.com/language/tutorial/RandomNumberGeneration.html Accessed 30 Jul 2019.
- 97.Wolfram S. Wolfram Media, Inc; Champaign, IL: 2016. Idea Makers: Personal Perspectives on the Lives & Ideas of Some Notable People. [Google Scholar]
- 98.Mathematica Stack Exchange Quality of random numbers. 2012. https://mathematica.stackexchange.com/questions/3208/quality-of-random-numbers Accessed 30 Jul 2019.
- 99.Gentle J.E. Springer; New York: 1998. Random Number Generation and Monte Carlo Methods. [Google Scholar]
- 100.Wikipedia Generalized mean. 2019. https://en.wikipedia.org/wiki/Generalized_mean Accessed 30 Jul 2019.
- 101.Cantrell D.W., Weisstein E.W. Power mean. MathWorld–A Wolfram Web Resource. http://mathworld.wolfram.com/PowerMean.html Accessed 30 Jul 2019.
- 102.Zaninetti L. A left and right truncated lognormal distribution for the stars. Adv. Astrophys. 2017;2:197–213. [Google Scholar]
- 103.Pottier N. Relaxation time distributions for an anomalously diffusing particle. Physica A. 2011;390:2863–2879. [Google Scholar]
- 104.Liebovitch L.S., Toth T.I. Distributions of activation energy barriers that produce stretched exponential probability distributions for the time spent in each state of the two state reaction AB. Bull. Math. Biol. 1991;53:443–455. [Google Scholar]
- 105.Metzler R., Klafter J., Volk M. Multiple time scales for dispersive kinetics in early events of peptide folding. Chem. Phys. Lett. 1998;293:477–484. [Google Scholar]
- 106.Ostrowsky N., Sornette D., Pike E.R. Exponential sampling method for light-scattering polydispersity analysis. Opt. Acta (Lond.) 1981;28:1059–1070. [Google Scholar]
- 107.Berberan-Santos M.N., Bodunov E.N., Valeur B. Mathematical functions for the analysis of luminescence decays with underlying distributions 1. Kohlrausch decay function (stretched exponential) Chem. Phys. 2005;315:171–182. [Google Scholar]
- 108.Barone P., Ramponi A., Sebastiani G. On the numerical inversion of the Laplace transform for nuclear magnetic resonance relaxometry. Inverse Probl. 2001;17:77–94. [Google Scholar]
- 109.Plonka A., Kroh J., Berlin Y.A. Photodissociation of carbon monoxy myoglobin: kinetics of carbon-monoxide rebinding. Chem. Phys. Lett. 1988;153:433–435. [Google Scholar]
- 110.Istratov A.A., Vyvenko O.F. Exponential analysis in physical phenomena. Rev. Sci. Instrum. 1999;70:1233–1257. [Google Scholar]
- 111.Epstein C.L., Schotland J. The bad truth about Laplace’s transform. SIAM Rev. 2008;50:504–520. [Google Scholar]
- 112.Lee S.B. Correction-to-scaling of random walks in disordered media. Int. J. Mod. Phys. B. 2003;17:4867–4881. [Google Scholar]
- 113.Saxton M.J. Single-particle tracking: the distribution of diffusion coefficients. Biophys. J. 1997;72:1744–1753. doi: 10.1016/S0006-3495(97)78820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Berg O.G., Winter R.B., von Hippel P.H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981;20:6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 115.Halford S.E., Marko J.F. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nemirovsky A.M., Mártin H.O., Coutinho-Filho M.D. Universality in the lattice-covering time problem. Phys. Rev. A. 1990;41:761–767. doi: 10.1103/physreva.41.761. [DOI] [PubMed] [Google Scholar]
- 117.Brummelhuis M.J.A.M., Hilhorst H.J. Covering of a finite lattice by a random walk. Physica A. 1991;176:387–408. [Google Scholar]
- 118.Ellis-Davies G.C.R. Caged compounds: photorelease technology for control of cellular chemistry and physiology. Nat. Methods. 2007;4:619–628. doi: 10.1038/nmeth1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lee H.-M., Larson D.R., Lawrence D.S. Illuminating the chemistry of life: design, synthesis, and applications of “caged” and related photoresponsive compounds. ACS Chem. Biol. 2009;4:409–427. doi: 10.1021/cb900036s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jain P.K., Ramanan V., Bhatia S.N. Development of light-activated CRISPR using guide RNAs with photocleavable protectors. Angew. Chem. Int. Ed. Engl. 2016;55:12440–12444. doi: 10.1002/anie.201606123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gautier A., Gauron C., Vriz S. How to control proteins with light in living systems. Nat. Chem. Biol. 2014;10:533–541. doi: 10.1038/nchembio.1534. [DOI] [PubMed] [Google Scholar]
- 122.Zhang K., Cui B. Optogenetic control of intracellular signaling pathways. Trends Biotechnol. 2015;33:92–100. doi: 10.1016/j.tibtech.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kolar K., Weber W. Synthetic biological approaches to optogenetically control cell signaling. Curr. Opin. Biotechnol. 2017;47:112–119. doi: 10.1016/j.copbio.2017.06.010. [DOI] [PubMed] [Google Scholar]
- 124.Motta-Mena L.B., Reade A., Gardner K.H. An optogenetic gene expression system with rapid activation and deactivation kinetics. Nat. Chem. Biol. 2014;10:196–202. doi: 10.1038/nchembio.1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hughes R.M. A compendium of chemical and genetic approaches to light-regulated gene transcription. Crit. Rev. Biochem. Mol. Biol. 2018;53:453–474. doi: 10.1080/10409238.2018.1487382. [DOI] [PubMed] [Google Scholar]
- 126.Saxton M.J. Wanted: scalable tracers for diffusion measurements. J. Phys. Chem. B. 2014;118:12805–12817. doi: 10.1021/jp5059885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hughes B.D. Clarendon Press; Oxford, UK: 1996. Random Walks and Random Environments. Random Environments, Volume 2. [Google Scholar]
- 128.Tischer C., Altmann S., Florin E.-L. Three-dimensional thermal noise imaging. Appl. Phys. Lett. 2001;79:3878–3880. [Google Scholar]
- 129.Bartsch T.F., Kochanczyk M.D., Florin E.-L. Nanoscopic imaging of thick heterogeneous soft-matter structures in aqueous solution. Nat. Commun. 2016;7:12729. doi: 10.1038/ncomms12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Reisch A., Heimburger D., Klymchenko A.S. Protein-sized dye-loaded polymer nanoparticles for free particle diffusion in cytosol. Adv. Funct. Mater. 2018;28:1805157. [Google Scholar]
- 131.Howarth M., Liu W., Ting A.Y. Monovalent, reduced-size quantum dots for imaging receptors on living cells. Nat. Methods. 2008;5:397–399. doi: 10.1038/nmeth.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lees E.E., Gunzburg M.J., Mulvaney P. Experimental determination of quantum dot size distributions, ligand packing densities, and bioconjugation using analytical ultracentrifugation. Nano Lett. 2008;8:2883–2890. doi: 10.1021/nl801629f. [DOI] [PubMed] [Google Scholar]
- 133.Sperling R.A., Pellegrino T., Parak W.J. Electrophoretic separation of nanoparticles with a discrete number of functional groups. Adv. Funct. Mater. 2006;16:943–948. [Google Scholar]
- 134.Lin C.-A.J., Sperling R.A., Parak W.J. Design of an amphiphilic polymer for nanoparticle coating and functionalization. Small. 2008;4:334–341. doi: 10.1002/smll.200700654. [DOI] [PubMed] [Google Scholar]
- 135.Cutler P.J., Malik M.D., Lidke K.A. Multi-color quantum dot tracking using a high-speed hyperspectral line-scanning microscope. PLoS One. 2013;8:e64320. doi: 10.1371/journal.pone.0064320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dross N., Spriet C., Langowski J. Mapping eGFP oligomer mobility in living cell nuclei. PLoS One. 2009;4:e5041. doi: 10.1371/journal.pone.0005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Baum M., Erdel F., Rippe K. Retrieving the intracellular topology from multi-scale protein mobility mapping in living cells. Nat. Commun. 2014;5:4494. doi: 10.1038/ncomms5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Erdel F., Baum M., Rippe K. The viscoelastic properties of chromatin and the nucleoplasm revealed by scale-dependent protein mobility. J. Phys. Condens. Matter. 2015;27:064115. doi: 10.1088/0953-8984/27/6/064115. [DOI] [PubMed] [Google Scholar]
- 139.Brittingham G., Delarue M., Holt L.J. mTORC1 controls rheology and phase separation by tuning ribosome concentration. Mol. Biol. Cell. 2017;28:3727. (P2593) [Google Scholar]
- 140.Pinglay S., Brittingham G., Holt L.J. Genetically Encoded Multimeric nanoparticles (GEMs) to visualize the biophysical properties of the nucleus. Mol. Biol. Cell. 2017;28:3727. (P3116) [Google Scholar]
- 141.Delarue M., Brittingham G.P., Holt L.J. mTORC1 controls phase separation and the biophysical properties of the cytoplasm by tuning crowding. Cell. 2018;174:338–349. doi: 10.1016/j.cell.2018.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Giessen T.W. Encapsulins: microbial nanocompartments with applications in biomedicine, nanobiotechnology and materials science. Curr. Opin. Chem. Biol. 2016;34:1–10. doi: 10.1016/j.cbpa.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 143.King N.P., Bale J.B., Baker D. Accurate design of co-assembling multi-component protein nanomaterials. Nature. 2014;510:103–108. doi: 10.1038/nature13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.