Abstract
The transcription factor TCF7L2 is indispensable for intestinal tissue homeostasis where it transmits mitogenic Wnt/β-Catenin signals in stem and progenitor cells, from which intestinal tumors arise. Yet, TCF7L2 belongs to the most frequently mutated genes in colorectal cancer (CRC), and tumor-suppressive functions of TCF7L2 were proposed. This apparent paradox warrants to clarify the role of TCF7L2 in colorectal carcinogenesis. Here, we investigated TCF7L2 dependence/independence of CRC cells and the cellular and molecular consequences of TCF7L2 loss-of-function. By genome editing we achieved complete TCF7L2 inactivation in several CRC cell lines without loss of viability, showing that CRC cells have widely lost the strict requirement for TCF7L2. TCF7L2 deficiency impaired G1/S progression, reminiscent of the physiological role of TCF7L2. In addition, TCF7L2-negative cells exhibited morphological changes, enhanced migration, invasion, and collagen adhesion, albeit the severity of the phenotypic alterations manifested in a cell-line-specific fashion. To provide a molecular framework for the observed cellular changes, we performed global transcriptome profiling and identified gene-regulatory networks in which TCF7L2 positively regulates the proto-oncogene MYC, while repressing the cell cycle inhibitors CDKN2C/CDKN2D. Consistent with its function in curbing cell motility and invasion, TCF7L2 directly suppresses the pro-metastatic transcription factor RUNX2 and impinges on the expression of cell adhesion molecules. Altogether, we conclude that the proliferation-stimulating activity of TCF7L2 persists in CRC cells. In addition, TCF7L2 acts as invasion suppressor. Despite its negative impact on cell cycle progression, TCF7L2 loss-of-function may thereby increase malignancy, which could explain why TCF7L2 is mutated in a sizeable fraction of colorectal tumors.
Subject terms: Mitosis, Cancer genomics, Growth factor signalling, Extracellular matrix
Introduction
Aberrant Wnt/β-Catenin pathway activity plays a crucial role in virtually every aspect of colorectal carcinogenesis [1], and more than 90% of all colorectal tumors carry mutations in either tumor-suppressive or oncogenic Wnt/β-Catenin pathway components [2]. These tumor-promoting lesions interfere with the regulated activity of the Wnt/β-Catenin pathway and thereby affect proliferation, migration, invasion, and tumor initiation capacity of colorectal cancer (CRC) cells [1]. A mainstay of Wnt/β-Catenin signaling are changes in gene expression. These are predominantly elicited through the interaction of β-Catenin with members of the T-cell factor/lymphoid enhancer binding factor (TCF/LEF) family [3]. In mammals, this family consists of LEF1, TCF7, TCF7L1, and TCF7L2. All TCF/LEF family members possess as common structural features interaction domains for β-Catenin and Transducin-like Enhancer of Split/Groucho-related genes (TLE/GRG) corepressors, and an HMG-box DNA-binding motif [3]. Structural diversity outside these domains equips TCF/LEF family members with individual gene-regulatory capacity, and allows them to carry out both redundant and nonredundant functions as nuclear effectors of the Wnt/β-Catenin pathway [3–5].
Wnt/β-Catenin signaling governs multiple aspects of cellular dynamics and tissue architecture in the developing gastrointestinal tract and in adult intestinal tissue homeostasis [1, 6]. Particularly relevant for colorectal carcinogenesis are the stimulation of cell proliferation and the maintenance of intestinal stem cells [6] from which colorectal tumors appear to arise [7]. In the healthy gut, these Wnt/β-Catenin pathway functions are executed exclusively via TCF7L2 [8–11]. Accordingly, inactivation of the Tcf7l2 gene in mouse models and intestinal organoids is lethal due to diminished mitogenic activity and depletion of stem and progenitor cells [8–11]. There is also evidence that Tcf7l2 is indispensable for tumor initiation [11] which agrees well with the positive regulation of several oncogenes by TCF7L2 [12–16].
Its essential function in stimulating cell proliferation in the healthy murine intestine and its role in transmitting oncogenic Wnt/β-Catenin signals in mouse tumor models seemingly qualify TCF7L2 as a tumor-promoting factor also in human colorectal carcinogenesis. This view contrasts with the frequent occurrence of TCF7L2 loss-of-function mutations in CRC genomes [2, 17, 18], arguing that TCF7L2 activity may rather be tumor-suppressive. Indeed, TCF7L2 was claimed to function as haploinsufficient tumor suppressor in mice [9], and to restrict human CRC cell cycle progression [9, 19]. However, both findings were recently challenged [11]. Thus, the role of TCF7L2 in human CRC remains ambiguous. Specifically, it is unknown to which extent CRC cells tolerate complete absence of TCF7L2, and how this would affect cellular phenotypes. Likewise, a sustainable explanation for the high TCF7L2 mutation frequency is lacking. To address these issues, we systematically knocked-out TCF7L2 in CRC cell lines. Our results show that the vital necessity for TCF7L2 in healthy intestinal cells is broadly lost in the course of colorectal carcinogenesis. Even though TCF7L2-negative cells exhibit delayed G1/S transition, they are more migratory and invasive, and show enhanced collagen adhesion. Concomitantly, TCF7L2 deficiency disturbs gene-regulatory networks comprising cell cycle regulators, the pro-metastatic transcription factor RUNX2, and multiple cell adhesion molecules. Apparently, TCF7L2 has properties of a migration/invasion suppressor, which provides a biological rationale for the frequent mutation of TCF7L2 in CRC genomes.
Results
Human CRC cells survive without TCF7L2
We confirmed that murine intestinal organoids do not survive inactivation of Tcf7l2 (Supplementary Fig. S1). To test whether the essential function of TCF7L2 is preserved in human CRC cells, we utilized the CRISPR/Cas9 system to target TCF7L2 exon 6 (Fig. 1a) which is common to all known TCF7L2 RNA isoforms [20]. Expression patterns of TCF/LEF family members in colorectal tumors deviate from the healthy intestinal epithelium and are highly variable, as evident from CRC transcriptome data (Supplementary Fig. S2a, b), and immunohistochemical stainings of case-matched normal and CRC tissue specimens (Supplementary Figs. S3, S4). Consistent with this, we observed that CRC cell lines express diverse combinations of TCF/LEF factors (Supplementary Fig. S2c, d). To take into account the variability of TCF/LEF expression, we therefore selected the three CRC cell lines HT29, HCT116, and LoVo for genome editing. Among these, HT29 cells express TCF7 and TCF7L2 (Supplementary Fig. S2c, d), reflecting native TCF/LEF expression in the normal mouse and human colonic epithelium (Supplementary Figs. S3–S5). HCT116 cells additionally express TCF7L1 (Supplementary Fig. S2c, d). LoVo cells express all four TCF/LEF family members (Supplementary Fig. S2c, d). Furthermore, the cell lines chosen cover a range of different CRC-associated lesions in the Wnt/β-Catenin, MAP kinase, TP53, and TGFβ pathways (Supplementary Table S1) [2, 21–23]. Irrespective of their TCF/LEF status and the respective mutations in CRC driver genes, we obtained multiple clones with biallelic inactivation of TCF7L2 for all three cell lines (Fig. 1b, c; Supplementary Table S2). TCF7L2 knockout (KO) clones showed strongly reduced TCF7L2 transcription and complete absence of all TCF7L2 protein isoforms regardless of whether TCF7L2 was inactivated by intra-exon 6 mutations (HCT116 and HT29 TCF7L2KO cells) or exon 6 deletion (LoVo TCF7L2KOΔE6 cells). In contrast, a heterozygous (Het) HT29 cell clone carrying one TCF7L2 wildtype (WT) and one KO allele, expressed TCF7L2 at levels indistinguishable from TCF7L2WT cells. Notably, there was no compensatory upregulation of the other TCF/LEF family members in the absence of TCF7L2 (Supplementary Fig. S6). TCF7L2 deficiency did not affect the growth pattern and morphology of LoVo TCF7L2KOΔE6 cells (Fig. 1d), but HCT116 TCF7L2KO clones showed subtle morphological changes by forming more protrusions. HT29 TCF7L2KO cultures exhibited more pronounced alterations and grew as isolated, round cells instead of generating dense cell clusters like HT29 TCF7L2WT/Het clones. Furthermore, it appeared that HCT116 and HT29 TCF7L2KO cultures expanded slower than TCF7L2WT/Het cells (Fig. 1d). To ascertain that the phenotypic alterations observed with HCT116 and HT29 TCF7L2KO cells were not due to off target effects or differences in KO strategies, we created additional TCF7L2-deficient HCT116 and HT29 cell clones by inflicting the same exon 6 deletion as in LoVo cells. This yielded HCT116 and HT29 TCF7L2KOΔE6 cell clones exhibiting morphological changes highly similar, if not identical, to TCF7L2KO cells (Supplementary Fig. S7). As observed with the latter, no increase in expression of the other TCF/LEF family members was seen in HCT116 and HT29 TCF7L2KOΔE6 cell clones (Supplementary Fig. S8). Overall, we conclude that TCF7L2 is not essential for the survival of at least some human CRC cells. Nonetheless, TCF7L2 deficiency is accompanied by phenotypic changes in HCT116 and HT29 cells that manifest despite the presence of other TCF/LEF family members.
Fig. 1. CRC cell lines are viable without TCF7L2.
a Top: scheme of the TCF7L2 gene with its 17 exons (numbered boxes). Constitutively expressed exons are colored in black and red, alternatively spliced exons in gray. Asterisks mark start codons. Black arrowheads denote stop codons whose usage depends on the exon composition of alternatively spliced transcripts. The locations of gRNAs are indicated by red arrowheads. For HCT116 and HT29 cells, the gRNA g2 was used to disrupt the constitutive exon 6 (red box). For LoVo cells, gRNAs g1 and g3 were used to delete exon 6. Bottom: TCF7L2 protein structure. Functionally important domains are marked and connected to the exon(s) by which they are encoded. HMG-box DNA-binding domain. NLS nuclear localization sequence. BD binding domain. b The mRNA expression of TCF7L2 was analyzed by qRT-PCR in cell clones with biallelic wildtype (WT), heterozygous (Het), and biallelic knockout (KO) TCF7L2 genes with intra-exon 6 mutations or exon 6 deletion (KOΔE6). TCF7L2 transcript levels were normalized to GAPDH, and are displayed as relative expression (rel. expr.). Each dot represents an individual measurement, color of the dots identifies different cell clones. The box plots summarize and compare expression data from TCF7L2WT/Het, TCF7L2KO, and TCF7L2KOΔE6 cells. Linear mixed model (LMM) analysis was performed to assess significance (n = 3). c Nuclear extracts were generated from TCF7L2WT/Het, TCF7L2KO, and TCF7L2KOΔE6 clones and western blot analysis was performed to detect TCF7L2 expression. GSK3β was used as loading control. Molecular weights are given in kDa. Representative results from one of three independent biological replicates are shown. d Representative micrographs from one of three independent biological replicates showing the indicated TCF7L2WT, TCF7L2KO, and TCF7L2KOΔE6 cell clones 72 h after seeding the same starting numbers of cells. The scale bars represent 200 µm.
Comprehensive transcriptome analyses of TCF7L2KO clones reveal changes in Wnt signaling, proliferation, and adhesion
To capture the molecular effects of TCF7L2 ablation, we performed RNA-seq experiments. Principal component analyses showed that HCT116 and HT29 TCF7L2WT/Het and TCF7L2KO clones separate well along principal component 1 which explains most of the variance (Fig. 2a). Applying an adjusted p value < 0.05 and an absolute value of log2 fold change (log2FC) > 0.5 as thresholds, we identified 2148 differentially expressed genes (DEGs) in HCT116 TCF7L2KO versus TCF7L2WT cells (Supplementary Table S3) and 3084 DEGs in HT29 TCF7L2KO cells (Supplementary Table S4). Gene expression changes were mostly cell-line-specific. However, 630 DEGs were shared by HCT116 and HT29 TCF7L2KO cells (Fig. 2b, Supplementary Table S5). Among these, 280 DEGs were upregulated and 122 DEGs were downregulated in both cellular backgrounds. Expression of the remaining 228 DEGs changed in opposite directions (Fig. 2b). Altogether, TCF7L2 appears to drive both specific and common gene expression programs in CRC cell lines.
Fig. 2. Comprehensive transcriptome analyses of TCF7L2KO cells reveal commonly deregulated genes in HCT116 and HT29 cells.
a The number of raw counts was determined after alignment of RNA-seq data. This process was conducted with HCT116 and HT29 cell clones with biallelic WT, KO, and Het TCF7L2 genes (HCT116: WT #3, KO #18; HT29: WT #56, Het #62, KO #57, #83, #86) for each of two independent biological replicates. By applying principal component analysis (PCA) on the regularized-logarithm transformed values of the raw counts [69], the cell lines were depicted in two-dimensional PCA plots. The shape of the symbols identifies the replicates and their color indicates the TCF7L2 genotype. b Comparison of TCF7L2-dependent gene expression in HCT116 and HT29 cells. Numbers of DEGs in TCF7L2KO versus TCF7L2WT/Het cells, and their overlap between HCT116 and HT29 cells are shown in the Venn diagram on the left. The Venn diagram on the right differentiates between cell-line-specific and HCT116/HT29-common up- and downregulated genes. c Functional enrichment analysis of gene sets was performed on the TCF7L2-dependent DEGs common to HCT116 and HT29 cells using Fisher’s exact test. The top ten positively and negatively enriched Gene Ontology (GO) terms for “Biological function” are shown in the bar plot. The color of the bars indicates positive and negative enrichment scores as well as the corresponding log10 adjusted p values.
To identify cellular processes possibly controlled by TCF7L2 across cell lines, we performed functional enrichment analyses of gene sets using Fisher’s exact test for the overlapping DEGs in HCT116 and HT29 TCF7L2KO cells (Fig. 2c, Supplementary Table S6). According to these analyses, cell–cell and cell–matrix adhesion, migration, metabolic pathways, transcriptional regulation, and proliferation might be altered in TCF7L2KO cells (Fig. 2c), the latter being reminiscent of the reduced population dynamics exhibited by HCT116 and HT29 TCF7L2KO cells (see Fig. 1d), and the exhaustion of proliferating stem and progenitor cell compartments in Tcf7l2-deficient intestinal crypts [8, 10, 11]. Furthermore, Gene Ontology (GO) terms connected to Wnt signaling were among the features downregulated upon inactivation of TCF7L2 (Supplementary Table S6). Consistent with this, expression of the Wnt target genes MYC and TERT [12–14, 24] was strongly decreased in HCT116 and HT29 TCF7L2KO and TCF7L2KOΔE6 clones (Supplementary Fig. S9). Chromatin immunoprecipitation (ChIP) followed by quantitative PCR (qPCR) additionally demonstrated specific interactions of TCF7L2 with multiple regions at the MYC locus in TCF7L2WT/Het but not in TCF7L2KO cells (Supplementary Fig. S9e) [13, 14, 25–27]. Unfortunately, we could not detect TCF7L2 at a previously described TCF7L2-binding region at the TERT promoter (Supplementary Fig. S9f) [26, 27], albeit this does not rule out that TCF7L2 associates with other sites at the TERT locus. To further examine the impact of TCF7L2 deficiency on Wnt/β-Catenin target gene expression, we extended our analyses to AXIN2, ASCL2, EPHB3, and RNF43 [28–33]. However, these examinations were hampered by the fact that Wnt/β-Catenin targets are differentially expressed in CRC cells as reported by us and others [15, 34–37]. This likely explains why the comparatively low or undetectable expression of AXIN2, ASCL2, and EPHB3 in HCT116 cells was not further affected by TCF7L2 inactivation (Supplementary Fig. S10). Yet, their expression decreased in TCF7L2-deficient HT29 TCF7L2KO and TCF7L2KOΔE6 cells. Moreover, RNF43 was consistently downregulated in both HCT116 and HT29 TCF7L2KO and TCF7L2KOΔE6 cells (Supplementary Fig. S10). In agreement with the known function of TCF7L2, the results of the global and the targeted expression profiling in aggregate demonstrate that TCF7L2 deficiency vastly changes gene expression in HCT116 and HT29 cells and, as expected, impairs the transcriptional output of the Wnt/β-Catenin pathway.
TCF7L2 inactivation decreases proliferation
To experimentally address proliferative effects following loss of TCF7L2, we seeded the same numbers of TCF7L2WT/Het and TCF7L2KO and TCF7L2KOΔE6 cells and counted their progeny 96 h later. Both HCT116 and HT29 TCF7L2KO and TCF7L2KOΔE6 cells showed significantly lower cell numbers compared with TCF7L2WT/Het cells (Fig. 3a, Supplementary Fig. S11a, b). Flow cytometric analyses of DNA content further revealed increased G1 phase cell counts and decreased cell counts in S and G2/M phases in TCF7L2KO and TCF7L2KOΔE6 clones (Fig. 3b, Supplementary Fig. S11c, d) suggesting impaired G1/S phase progression. To provide a molecular explanation for the observed cell cycle changes, we extracted DEGs from the GO term REGULATION_OF_CELL_CYCLE (Fig. 3c, Supplementary Table S6). These DEGs include CDKN2C and CDKN2D which negatively control the G1/S transition. By quantitative reverse transcription polymerase chain reaction (qRT-PCR) and western blotting we verified upregulation of CDKN2C and CDKN2D in TCF7L2KO cells (Fig. 3d, e). Increased CDKN2C and CDKN2D transcript levels were also observed in HCT116 and HT29 TCF7L2KOΔE6 cells (Supplementary Fig. S11e, f). Altogether, TCF7L2 appears to promote proliferation of certain CRC cells like HCT116 and HT29 by suppressing cell cycle inhibitors while concomitantly stimulating the expression of mitogenic genes like MYC.
Fig. 3. Loss of TCF7L2 leads to reduced proliferation and delayed G1/S progression in CRC cells.
a 1 × 104 HCT116 cells and 5 × 104 HT29 cells with the genotypes indicated were seeded, and incubated for 96 h. Then, the resulting cell counts were determined and displayed as bar plots. For statistical analysis, LMM was applied (n = 3). b To analyze differences in cell cycle distribution of TCF7L2WT/Het and TCF7L2KO cells, cells were stained with propidium iodide and analyzed by flow cytometry. The proportions of cells in different cell cycle phases are depicted by the box plots. For statistical analysis LMM was performed (n = 3). ns not significant. c The GO term “REGULATION_OF_CELL_CYCLE” was used to extract genes differentially expressed in TCF7L2WT/Het and TCF7L2KO cell clones and connected to cell cycle control. The z-scores of the r-log normalized counts of the genes were plotted as heatmap. d, e Expression of the cell cycle inhibitors CDKN2C and CDKN2D was analyzed in HCT116 (d) and HT29 cells (e) by qRT-PCR and western blotting. Colored dots represent qRT-PCR results for individual cell clones. Box plots summarize qRT-PCR results from all clones according to TCF7L2 genotype. Data presented indicate TCF7L2 expression relative to that of GAPDH (rel. expr.). For statistical analysis, LMM was applied (n = 3). In western blotting experiments, α-TUBULIN (α-TUB) served as loading control. Molecular weights are given in kDa. Representative results from one of three independent biological replicates are shown. ns not significant.
Loss of TCF7L2 enhances migration and invasion of CRC cells
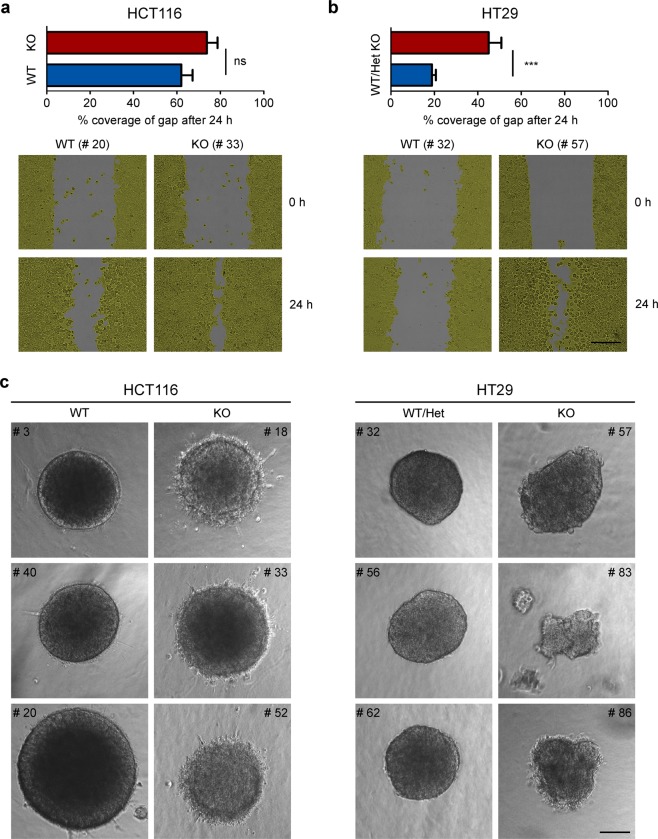
The RNA-seq results further indicated that TCF7L2 affects cell migration and adhesion. Indeed, two-dimensional wound-healing assays showed that HCT116 and HT29 TCF7L2KO cells are more migratory than TCF7L2WT/Het cells (Fig. 4a, b, Supplementary Fig. S12). Likewise, motility of TCF7L2KOΔE6 cells was enhanced (Supplementary Fig. S11g, h). We also compared invasive properties of TCF7L2KO and TCF7L2WT/Het cells in a three-dimensional collagen I invasion assay (Fig. 4c). HCT116 TCF7L2WT cells formed regularly shaped, round spheroids with smooth borders without evidence of invasion. In contrast, spheroids from HCT116 TCF7L2KO clones exhibited irregular, rough surfaces caused by the single cell emigration, and sprouting of short strands of cells into the surrounding collagen matrix. The invasive behavior of HT29 TCF7L2KO cells was less pronounced, but when compared with HT29 TCF7L2WT/Het cells, TCF7L2KO spheroids also presented with less regular and more raspberry-like shapes, and seemed to be less cohesive (Fig. 4c). We conclude that loss of TCF7L2 can promote migration and invasion of certain CRC cells.
Fig. 4. TCF7L2 deficiency enhances migration and invasion of CRC cells.
a, b The migratory capacity of HCT116 (a) and HT29 cells (b) with WT and mutant TCF7L2 genes was interrogated by wound-healing/gap closure assays. Bar graphs show the percent coverage of the gaps at 24 h after infliction. Shown are the combined mean values for all clones with the genotypes indicated. Error bars represent the SEM. Statistical analysis was performed using LMM (n = 4). Representative pictures of gaps at 0 and 24 h after wounding are shown in the lower panels. The scale bar represents 100 µm. c For three-dimensional invasion experiments, HCT116 and HT29 cells with WT and mutant TCF7L2 genes were allowed to form spheres in hanging drops prior to embedding in a collagen matrix. Seventy-two hours after embedding, pictures of the spheroids were taken. Representative pictures from one of four independent biological replicates are shown for each clone. The scale bar represents 100 µm.
Loss of TCF7L2 increases adhesion of CRC cells to collagen I
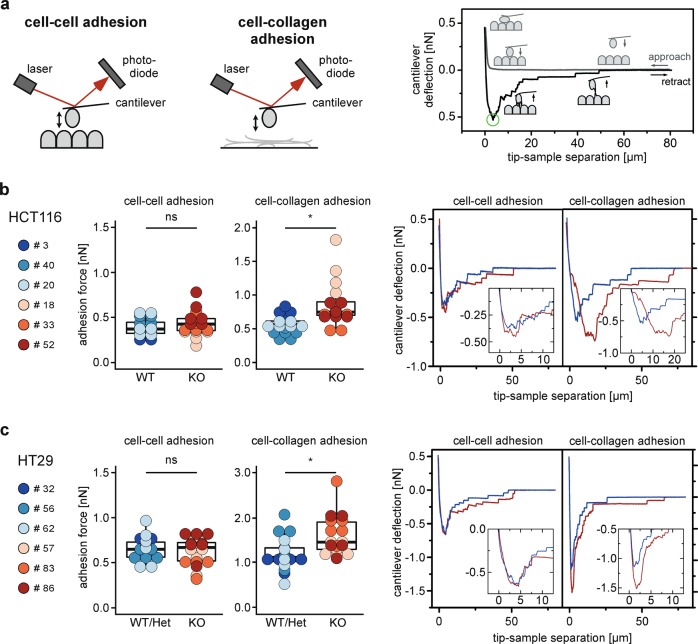
Next, we investigated whether cell–cell adhesion might be impaired by loss of TCF7L2. To address this, we used single-cell force spectroscopy (SCFS) which is based on atomic force microscopy and facilitates the quantification of cell–cell and cell–substrate interactions at a single cell level (Fig. 5a). Surprisingly, for both, HCT116 and HT29 cells, there was no difference in intercellular adhesion forces between TCF7L2WT/Het and TCF7L2KO cells (Fig. 5b, c). However, TCF7L2-deficient HCT116 and HT29 cell derivatives displayed enhanced adhesion to collagen I (Fig. 5b, c), which fits to the altered appearance and sprouting behavior of TCF7L2KO spheroids. Thus, TCF7L2 not only controls cell proliferation and migration, but also interactions of HCT116 and HT29 CRC cells with extracellular matrix components.
Fig. 5. Cells display increased adhesion to collagen I upon loss of TCF7L2.
a Left: scheme of the SCFS setup to measure cell–cell and cell–collagen adhesion. A cantilever with an immobilized cell is approached to substrates consisting of cell or collagen layers, and cantilever deflection is monitored via the reflection of a laser into a 4-quadrant photodiode. Cantilever bending is directly proportional to the force acting on the cantilever. Right: schematic force curves. During approach (gray curve) the cantilever deflection remains at 0 pN until contact with the sample is reached. Upon further approach the cantilever bends upward exerting force on the contact until the defined force set point is reached, and held for a defined timespan. Upon retraction (black curve) the contact force is continuously released until further retraction leads to downward bending as the cantilever starts pulling on the bonds that were formed during contact. The cell(s) and bonds are stretched until a critical value is reached, termed detachment force, Fdet (green circle), at which the bonds start to successively disengage until the cell is completely freed and the retraction curve arrives at the level of the approach curve. Statistical analyses of high numbers of retract curves from multiple cells yield information about the strength of cell–substrate adhesion. HCT116 (b) and HT29 cells (c) with WT und mutant TCF7L2 genes were measured by SCFS to quantify cell–cell and cell–collagen adhesion as indicated. The cell attached to the cantilever originated from the same clone as seeded on the coverslip. At least five cells were measured per cell clone and 30 force curves were recorded per cell. Dots represent the mean of the maximum adhesion force of each cell. Dot color identifies individual cell clones, and the box plot summarizes data according to TCF7L2 genotype. For statistical analysis, LMM was applied. ns not significant. Representative force curves for both experimental setups are shown on the right. WT curves blue, KO curves red. Inserts in graphs show blow ups of the force curves around Fdet.
Loss of TCF7L2 disturbs a gene-regulatory network related to cell adhesion and migration
To provide a mechanistic framework for the migration/invasion-related phenotypic alterations of TCF7L2-deficient cells we integrated our RNA-seq results with a query of the string database [38]. This identified a molecular network that linked TCF7L2 among others to the KLF4 and RUNX2 transcription factor genes, the laminin subunit LAMB3, and the integrin genes ITGA3, ITGB7, and ITGBL1 (Fig. 6a). KLF4 suppresses CRC cell migration and invasion [39–41], whereas RUNX2 and its target gene ITGBL1 are pro-migratory and pro-metastatic [42–45]. Similarly, overexpression of ITGA3 and ITGB7 augments migration, matrix adhesion, and metastasis formation [46, 47]. ITGA3 additionally mediates collagen adhesion [48]. Interestingly, ITGA3 marks numerous cell protrusions in HCT116 TCF7L2KO cells, which are not detected in TCF7L2WT cells (Supplementary Fig. S13). We additionally confirmed that KLF4 and RUNX2 were down- and upregulated, respectively, in HCT116 and HT29 TCF7L2KO and TCF7L2KOΔE6 cells (Fig. 6b, c, Supplementary Figs. S14–S17). Furthermore, compared with TCF7L2WT cells, expression of LAMB3, ITGBL1, ITGA3, and ITGB7 was elevated in TCF7L2-deficient cells (Fig. 6b, c, Supplementary Figs. S14, S17), except for ITGBL1, ITGA3, and ITGB7 whose transcript levels were largely unchanged in HT29 TCF7L2KOΔE6 cells (Supplementary Fig. S17). Possibly, TCF7L2 affects these genes only indirectly through intermediate regulatory steps which appear to be subject to clonal variation. We therefore focused on the robustly deregulated transcription factor genes KLF4 and RUNX2, and investigated whether TCF7L2 directly regulates them. To identify potential TCF7L2-binding sites at the KLF4 and RUNX2 loci, we explored existing TCF7L2 ChIP-seq datasets [26, 27], which suggested the presence of one promoter-proximal and two intragenic TCF7L2-binding regions at the KLF4 and RUNX2 loci, respectively (Supplementary Fig. S18). By ChIP-qPCR, however, we did not detect specific occupancy of the KLF4 promoter region by TCF7L2 (Supplementary Fig. S18a). In contrast, TCF7L2 associated with both RUNX2 intragenic elements in TCF7L2WT cells but not in TCF7L2KO cells (Supplementary Fig. S18b), thereby identifying RUNX2 as a direct target gene that is repressed by TCF7L2 in HCT116 and HT29 CRC cells.
Fig. 6. TCF7L2 deficiency disturbs a molecular network connected to migration and adhesion.
a The string database was used to detect a regulatory network comprising several transcription factors and cell adhesion molecules. Shading of the nodes represents the log2FC of gene expression in the respective cell line. Gray nodes denote genes that are not differentially expressed in the RNA-seq datasets. b, c Expression of components of the network shown in (a) was analyzed by qRT-PCR in HCT116 (b) and HT29 cells (c) with WT and mutant TCF7L2 genes. Expression was normalized to GAPDH and is shown as relative expression (rel. expr.). Colored dots represent results from independent biological replicates for the different cell clones. The box plots summarize data according to the TCF7L2 genotype. For statistical analysis, LMM was applied (n = 4). d Pairwise correlation analyses of the genes indicated based on the publicly available gene expression datasets for human CRC samples (GSE14333: 290 samples; TCGA_COADREAD: 212 samples). Red and blue color shading (scaling indicated by the colored bar) represents the Pearson correlation efficient for correlated and anticorrelated expression, respectively.
To assess whether the regulatory relationships among TCF7L2, KLF4, RUNX2 and the cell adhesion factors observed in CRC cell lines might be relevant for human CRC samples, we performed pairwise correlation analyses based on publicly available transcriptome data. In two different CRC cohorts, expression of RUNX2 and ITGBL1 was strongly positively correlated (Fig. 6d). This is consistent with the known direct control of ITGBL1 by RUNX2 and supports the significance of the analyses. Matching the results of our expression analyses in CRC cell lines, expression of TCF7L2 was anticorrelated to that of RUNX2 and ITGBL1 in CRC transcriptomes (Fig. 6d), whereas a positive correlation existed for expression of TCF7L2 and KLF4. The relationships between expression of TCF7L2 and RUNX2 and their potential targets ITGA3, ITGB7, and LAMB3 were more ambiguous, hinting at indirect and more complex connectivity, possibly depending on the precise cellular context. Nonetheless, the combined results of the expression analyses in cell lines and tumors suggest that loss of TCF7L2 can suppress a regulatory network that otherwise might mobilize CRC cells and thereby promote cancer cell dissemination.
The frequency of TCF7L2 mutations/copy number loss is positively correlated with increased tissue invasion and metastasis in colon adenomacarcinomas
The observed changes in CRC cell motility and invasiveness following inactivation of TCF7L2 prompted us to analyze potential relationships between the occurrence of TCF7L2 genomic alterations and colorectal tumor progression using publicly available data for colon adenocarcinoma (COAD) from The Cancer Genome Atlas (TCGA) collection. Upon stratification according to the TNM and AJCC staging systems, the percentages of patients with inactivating mutations/copy number loss in TCF7L2 turned out to be higher among tumors which had locally invaded to the submucosa and beyond (T rating > 1; stages I/II), as well as spread to the lymph nodes (N ≥ 1; stage III) or to distant organs (M = 1; stage IV) (Fig. 7, Supplementary Table S7). Thus, the frequency of TCF7L2 mutations/copy number loss are positively correlated with increased tissue invasion and metastasis which is consistent with a migration and invasion suppressor role for TCF7L2 in colorectal tumors.
Fig. 7. The frequency of TCF7L2 mutations/copy number loss increases during tumor progression.

The percentages of colon adenocarcinoma patients with TCF7L2 mutations/copy number loss in relation to different categories according to TNM and AJCC staging systems were computed and plotted as indicated. Spearman’s rank correlation analysis was used to examine whether the frequency of TCF7L2 mutations/copy number loss follows an increasing order in parallel with tumor progression. The individual Spearman’s rank correlation values (ρ) for all categories are shown above the bars. The mean of the correlation values across all groups (TNM and AJCC staging) is 0.925 (p value = 0.0003028 [95% CI 0.772–1.077]). Tis: carcinoma in situ. Increasing T ratings indicate tumor spread to the submucosa (T = 1) and beyond (T ≥ 2). N denotes absence (N = 0) or presence (N > 0) of lymph node metastases. M indicates absence (M = 0) or presence (M = 1) of distant organ metastases.
Discussion
Conflicting observations concerning the role of TCF7L2 in colorectal carcinogenesis prompted us to systematically investigate TCF7L2 dependence/independence of CRC cells and the phenotypic consequences of TCF7L2 inactivation. As key findings we present that TCF7L2-deficient CRC cells are viable and proliferate, thereby revealing a fundamental change in the functional importance of TCF7L2 compared with the healthy intestinal epithelium. In addition, TCF7L2 loss-of-function enhanced motility and invasiveness, two traits associated with increased malignancy. Thus, our data suggest that in certain CRC contexts TCF7L2 may function as invasion suppressor which provides an explanation for the observed frequency of TCF7L2 mutations.
The spectrum of mutations in TCF7L2 in CRC encompasses single nucleotide exchanges and small insertions/deletions leading to nonsense, missense, and frame-shift mutations throughout the entire coding region, as well as translocations, copy number changes and deep deletions, many of which are likely to result in alleles with partial or complete TCF7L2 loss-of-function [2, 17, 18]. As a caveat, the available genomic data and information about variant allele frequencies do not reveal whether a given tumor sample harbors one or two defective TCF7L2 alleles. In contrast, our study showcases the consequences of fully inactivated TCF7L2 and complete absence of TCF7L2 protein isoforms comparable to genetically engineered mouse models [8, 10, 11]. Thus, it is possible that colorectal tumors retaining one functional allele or expressing mutant TCF7L2 protein variants exhibit phenotypes different from TCF7L2KO and TCF7L2KOΔE6 cells.
By disrupting or deleting TCF7L2 exon 6 we readily achieved biallelic TCF7L2 inactivation in LoVo, HCT116, and HT29 cells. Likewise, SW480 cells tolerate KO of TCF7L2 [11]. These four CRC cell lines harbor various combinations of oncogenic lesions in the APC, CTNNB1, KRAS, BRAF, PIK3CA, TP53, SMAD4, and TGFBR2 genes [21], and represent different CRC subtypes [22, 23]. Thus, dispensability of TCF7L2 in CRC cells does not seem to be rare, but the mechanisms that allow CRC cells to acquire TCF7L2 independence do not become immediately apparent. According to one model, redundancy among TCF/LEF family members allows TCF7, TCF7L1, and LEF1 individually or collectively to substitute for TCF7L2 [11]. Indeed, inactivation of TCF7L2 in different cellular backgrounds came along with graded phenotypic changes, which inversely correlated with the range of TCF/LEF family members present. However, this could be a mere coincidence. For instance, TCF7 and TCF7L2 are coexpressed in the healthy intestinal epithelium, yet, TCF7 fails to rescue TCF7L2 inactivation. Likewise, the absence of the C-clamp domain from TCF7L1 and LEF1, as well as functional differences and even antagonism among TCF7L1, LEF1, and TCF7L2 [3–5, 20, 36, 49] suggest that other TCF/LEF family members possess only a limited potential to functionally replace TCF7L2. Alternative explanations for the survival of TCF7L2-deficient CRC cells, and the differential penetrance of the TCF7L2 loss-of-function phenotype could be that CRC cells in general appear to be variably dependent on TCF/LEF activity and even Wnt/β-Catenin pathway function [37, 50].
Deletion of TCF7L2 produced massive changes in gene expression and cellular behavior. Thus, unlike previously claimed [11], the absence of TCF7L2 in CRC cells is not phenotypically neutral. However, biallelic inactivation of TCF7L2 was necessary to bring out phenotypic changes, and no evidence for haploinsufficiency was obtained [9]. Our findings of decreased proliferation and G1/S cell cycle delay in the absence of TCF7L2 are completely concordant with substantial evidence connecting TCF7L2 to the control of cell division in the healthy intestinal epithelium and in CRC cells [8, 10–12, 37]. Fittingly, TCF7L2 stimulates the expression of several genes that promote cell cycle progression [12, 51, 52]. Based on our findings, TCF7L2 additionally suppresses cell cycle inhibitors. Thus, it appears that the role of TCF7L2 as positive regulator of proliferation in healthy intestinal cells is preserved in a CRC context. However, the exclusive reliance of intestinal stem and progenitor cells on mitogenic Wnt/β-Catenin signaling appears to be lost in the course of colorectal tumorigenesis.
The proliferation-stimulating function of TCF7L2 in CRC cells observed here disagrees with previous findings [19]. However, Tang et al. assessed proliferation only indirectly through a metabolic assay, which could be affected by TCF7L2 deficiency irrespective of any proliferative effects. Notably, TCF7L2 was linked to glycolysis [53, 54]. Likewise, our RNA-seq data imply that TCF7L2 influences cellular metabolism. Therefore, it is possible that the consequences of TCF7L2 knockdown in earlier work were largely misinterpreted.
Alongside with impaired proliferation, TCF7L2-deficient cells exhibited morphological changes, increased motility, invasion, and adhesion to collagen I. In line with this, loss of TCF7L2 was accompanied by a switch in transcription factor expression from migration and invasion-suppressing KLF4 to pro-migratory and pro-metastatic RUNX2. This was paralleled by the upregulation of LAMB3 and several integrins, although integrin deregulation was observed more robustly in HCT116 cells. This might be due to clonal variability in the expression of one or more intermediate regulators that connect TCF7L2 to the integrin genes, but could also hint that additional gene expression changes contribute to the more migratory and invasive phenotype following TCF7L2 ablation in a cell-type-specific manner. The cell guidance gene EPHB3 [32] which was downregulated specifically in HT29 TCF7L2KO and TCF7L2KOΔE6 cells, could be an example. Thus, the full range of genes related to cell–cell and cell–matrix adhesion under control of TCF7L2, KLF4, and RUNX2, and the precise regulatory relationships among these factors remain to be determined. In this regard, we already identified RUNX2 as a novel TCF7L2 target gene. Altogether, the observed gene expression changes provide a plausible molecular explanation for the increased migratory and invasive capacity of TCF7L2-deficient CRC cells, and mark TCF7L2 as an invasion suppressor gene.
Apparently, TCF7L2 controls highly divergent gene expression programs in CRC cells. Cell-line-specific TCF7L2-dependent gene expression patterns could represent different subprograms of the overall transcriptome whereby Wnt/β-Catenin signaling controls proliferation, lineage decisions, positioning, differentiation, and maturation of intestinal epithelial cells [1]. These subprograms might segregate in cancer cells due to epigenetic inactivation [55, 56] or because of variations in the activity of cooperating signal transduction pathways [35, 37, 57, 58].
In summary, the physiological functions of TCF7L2 appear to be largely retained during colorectal carcinogenesis: control of proliferation and motility of CRC cells precisely reflect the roles of Wnt/β-Catenin signaling and its nuclear effector TCF7L2 in the healthy intestinal epithelium where they stimulate proliferation of stem and progenitor cells, and control cell migration and allocation along the crypt axis [8, 10, 11, 32, 51]. However, whereas TCF7L2 is essential in noncancerous tissue, it is not required for CRC cell viability. The widespread dispensability of TCF7L2 combined with its traits of an invasion suppressor shed new light on the role of TCF7L2 in CRC cells, and are highly relevant for the assessment and potential selection of β-Catenin/TCF7L2 complexes as targets for therapeutic intervention in CRC.
Material and methods
Cell lines, oligonucleotides, and antibodies
Cell lines used and their culture conditions are presented in Supplementary Table S8. Oligonucleotides for ChIP-qPCR/qRT-PCR, and antibodies for immunodetection experiments are listed in Supplementary Tables S9 and S10, respectively.
Intestinal organoid culture and fractionation of the small intestine
The KO cassette was flipped out [59, 60] of the genome of C57BL/6NTac-Tcf7l2tm1a(EUCOMM)Wtsi/WtsiIeg mice (European Mouse Mutant Archive strain EM:07858), and offspring retaining the floxed Tcf7l2 exon 6 was bred to C57BL/6N-tgVillinCreERT2 animals [61]. Five females (18–26 weeks old) from the resulting C57BL/6N-Tcf7l2flox/flox; tgVillinCreERT2 mice were used to generate intestinal organoids as described [62], except that colon organoids additionally received 100 ng/ml murine Wnt3a (Peptrotech, Rocky Hill, NJ, USA). Mice were handled in accordance with legal regulations at the Center for Experimental Models and Transgenic Service of the University of Freiburg Medical Center (project registration number: X-17/07S). To delete Tcf7l2 exon 6, organoids were treated with 0.5 µM 4-hydroxytamoxifen for 24 h. The small intestinal epithelium was separated into crypt and villus fractions as previously described [35].
Genome editing
CRISPR/Cas9-mediated genome editing was performed as before [63], except that gRNAs were cloned into pMuLE_ENTR_U6_stuffer_sgRNA_scaffold_L1-R5 [64]. Target sequences of gRNAs were: 5′-GTTTTGGGGTCTACGTCGGC-3′ (TCF7L2 exon 6; GRCh38:10:113,141,184-113,141,316); 5′-GGCTGCTGGAACCGGCTTGA-3′ and 5′-ACGGAGCCAAGGTAGAGTGC-3′ (located in TCF7L2 introns 5 and 6). 1 × 106 cells were nucleofected with 600 ng of each gRNA construct, and 800 ng of Cas9-turboRFP or Cas9-GFP expression vectors. RFP+/GFP+ cells were single cell sorted into 96 well plates 72 h after nucleofection, expanded, and screened for TCF7L2 expression by western blotting (HCT116 and HT29 TCF7L2KO cells), or for exon 6 deletion by PCR (LoVo TCF7L2KOΔE6 cells, HCT116 and HT29 TCF7L2KOΔE6). Details about the genomic state of the TCF7L2 locus for all cell clones used in this study can be found in Supplementary Table S2.
Western blotting
Except for the experiments shown in Supplementary Fig. S7, for which whole cell lysates made with RIPA buffer were employed [63], we investigated TCF/LEF expression using nuclear extracts which were obtained following a published protocol [35], albeit without dounce homogenizing for disrupting the cells. All other antigens were analyzed in whole cell lysates which were prepared as before [35]. For antigen detection, cell lysates and nuclear extracts were separated by SDS-PAGE and further processed by western blotting as described [35].
Immunofluorescence and immunohistochemistry
Immunofluorescence staining of cultured cells was carried out as described [65] using 2 × 105 cells seeded on glass slides coated with 0.1% gelatin. For immunohistochemical analyses of mouse tissue, the gut of healthy C57BL/6N mice was isolated and washed with PBS prior to fixation in 4% PFA. The tissue was embedded in paraffin, cut (5–8 µm), deparaffinized, and antigens were retrieved by boiling in 0.01 M citrate buffer. After blocking endogenous peroxidase in 10% (v/v) methanol, 3% H2O2 (v/v) in H2O for 20 min at room temperature, the slides were washed once in PBS-Tween 20, and the sections were encircled by PAP pen. The subsequent staining procedure was performed using the Vectastain Elite ABC Peroxidase kit (#VEC-PK-6100, BIOZOL, Eching, Germany) following the manufacturer’s protocol with minor modifications: the blocking time was extended to 30 min, primary antibody incubation to overnight at 4 °C, and secondary antibody incubation to 1 h. DAB solution was used as peroxidase substrate solution. For counterstaining, the tissue sections were incubated for 5 min in filtrated Mayer’s hemalum solution (Merck, Darmstadt, Germany), rinsed with water for 15 min, and mounted. Formaldehyde-fixed and paraffin-embedded tissue specimens from patients with CRC who had undergone tumor surgery at the University of Regensburg between 2006 and 2018 for this diagnosis, were included in a retrospective histological analysis of tumor tissue and adjacent healthy colon tissue. Patients gave written informed consent and the study was performed according to the principles of Helsinki and approved by the Local Ethics Committee (No. 14-101-0014). Staining was performed as previously described [66]. A tissue microarray containing 90 adenocarcinoma tissue specimens with matched cancer-adjacent tissue or adjacent normal tissue (#CO1801, US Biomax Inc., Derwood, MD, USA) was processed and stained the same way.
Correlation analysis for TCF7L2 mutations/copy number loss and tumor staging
We extracted information about clinical parameters, somatic single nucleotide variations (SNV) and somatic copy number changes for a total of 351 patients from the TCGA COAD collection (https://portal.gdc.cancer.gov; date of download: November 5, 2019). In case of SNVs, we considered missense mutations, nonsense mutations, splice site alterations, and frame shift-causing insertions and deletions. The percentages of patients with TCF7L2 mutations/copy number loss for the different categories of the TNM and AJCC staging systems was computed, and plotted in the order of increasing ratings for each category. To examine whether the frequency of TCF7L2 mutations/copy number loss is positively correlated with increased TNM and AJCC staging, Spearman’s rank correlation analyses were performed as follows: for each group of TNM categories/AJCC stages, the correlation value was calculated between the percentages of the patients with TCF7L2 mutations/copy number loss from the different categories/stages and a predefined numerical vector. The numerical vectors are “1, 2, 3, 4, 5” for the T categories, “1, 2” for the N and M categories, and “1, 2, 3, 4” for the AJCC stages, thereby supporting strictly ascending orders. After computing the individual correlation values, a t test was used to calculate the average of the correlation values (alternative hypothesis: true = mean of the correlation values is not equal to 0).
Flow cytometry
3 × 105 cells/well were seeded in six-well plates and incubated for 48 h. Cells were then processed for flow cytometry as described [63]. BD FACSCalibur (BD Biosciences, Heidelberg, Germany) and CytoFLEX machines (Beckman Coulter, Krefeld, Germany) together with the FlowJo software were used for analysis.
Work with RNA and RNA-seq data analysis
RNA isolation, cDNA synthesis, and qRT-PCR were performed as described [63], except that a cDNA amount equivalent to 20 ng total RNA was used for qRT-PCR. For transcriptome analyses, extracted RNA was paired-end sequenced on an Illumina HiSeq4000 at the Genome and Proteome Core Facility of the German Cancer Research Center, Heidelberg, Germany. Paired-end reads were filtered using Trimmomatic [67]. Selected reads were aligned to the human reference genome GRCh37. Read counts per gene were quantified with STAR aligner [68]. After excluding genes with low expression levels, 23,209 and 23,030 genes were considered for further processing for HCT116 and HT29 cells, respectively. For HT29, the two replicates for each of the five clones were integrated by averaging the corresponding number of counts. The differential expression analysis is based on a generalized linear model with a negative binomial distribution (DESeq2) [69]. For each cell line, TCF7L2KO cells were compared with TCF7L2WT/Het cells. The 2148 and 3084 top DEGs for HCT116 and HT29 cells, respectively, were selected by applying a Benjamini–Hochberg false discovery rate-corrected p value < 0.05 and absolute values of log2FC > 0.5. From these sets of genes we determined the intersection of the significantly DEGs across the two cell lines. For functional enrichment analyses of gene sets, we applied a Fisher’s exact test [70] to the selected top DEGs from each cell line, and to the 402 overlapping genes that are deregulated in both cell lines. For this, the stats R package was employed using a related set of background genes and the GO term collection “Biological Processes” as annotation. For the cell line-specific analyses, the 23,209 (HCT116 cells) and the 23,030 (HT29 cells) genes identified by applying the DESeq2 package, were taken as the background. For the overlapping gene set, the union of the top DEGs in both cell lines (i.e., 4602 genes) was taken as background. We calculated enriched GO terms for up- and downregulated genes separately, and retained GO terms with a significant adjusted p value of < 0.05 for both the cell line-specific calculations and the overlapped genes. RNA-seq data were deposited at the GEO database under accession number GSE135328.
ChIP
Chromatin isolation and ChIP-qPCR were performed as before [35], except that nuclei were isolated by NEXSON [71]. For each ChIP, 100 µg of Chromatin and 20 µl of TCF7L2 antibody were used.
Cell migration and invasion
Cell migration assays were carried out as before [63] using 5 × 104 cells per chamber of ibidi® cell culture inserts (#81176 ibidi GmbH, Martinsried, Germany). Three-dimensional growth of cancer cell spheroids in collagen I and invasion of cells into the collagen matrix was analyzed as described [72]. Pictures of the spheroids were taken 72 h after embedding.
Single-cell force spectroscopy
SCFS was carried out on a JPK NanoWizard III/CellHesion200 device (JPK Instruments AG, Berlin, Germany) mounted on a Nikon inverted light microscope with a petri dish heater. Measurements were done in 35 mm petri dishes (TPP, Sigma-Aldrich, Taufkirchen, Germany) in CO2-independent medium (ThermoFisher Scientific, Dreieich, Germany) at 37 °C. Tipless cantilevers (MLCT-O10, Bruker, Bremen, Germany) were calibrated using the thermal noise method, and functionalized with mannose-binding concanavalin A using standard procedures [73]. Coverslips (9 mm Ø) were coated with cells or collagen I, and placed into 35 mm Ø petri dishes. For attachment of a single cell to the very front of the cantilever, a cell was “fished” from a droplet of cell suspension that was pipetted into the petri dish far away from the coated coverslips. Force curves were recorded using an approach/retract speed of 20 or 30 μm/s depending on cell type and substrate. Contact was held at a constant force of 0.50 nN for 1.5 s (HT29 cell–cell and cell–collagen contact, HCT116 cell–collagen contact) or 0.25 s (HCT116 cell–cell contact). Five to seven cells were probed per condition for each cell clone, and 30 approach-and-retraction cycles per cell were quantitatively evaluated using the JPK SPM data processing software.
Query of the STRING database and publicly available transcriptome data
A list comprising TCF7L2, RUNX2, KLF4, ITGA3, ITGBL1, and ITGB7 was submitted to the string database (state: January 2019) for network analysis using default settings and a single expansion step [38]. TCF/LEF expression data in human normal tissue und CRC samples were extracted from a publicly available microarray dataset [74], and the mean of the log2 expression values for genes-of-interest in the different samples was calculated using GraphPad Prism. The TCGA_COADREAD dataset was analyzed using cBioPortal (https://www.cbioportal.org/) [75, 76]. TCGA_COADREAD datasets containing gene expression information for human CRC samples (https://xenabrowser.net/) and the GSE14333 dataset were used for pairwise correlation analysis as described [35].
Statistical analysis
Statistical analysis was performed using a linear mixed model (LMM) to account for random and fixed effects. Normal distribution of data was assessed using the car R package [77]. For LMM analysis, the lme4 R package was applied [78]. The ggplot2 R package [79] was used to generate box plots depicting the median, the lower, and upper quartile. Whiskers represent 1.5 times the interquartile range. The p values for significant changes are represented as follows: *p < 0.05; **p < 0.01; ***p < 0.001. For each series of experiments the corresponding number (n) of independent biological replicates is given in the figure legends.
Supplementary information
Acknowledgements
The authors express their gratitude to K. Geiger, D. Herchenbach, J. Bodinek, and A. Schmitt for exquisite technical assistance, and to the team of the Genomics and Proteomics Core Facility, German Cancer Research Center/DKFZ, Heidelberg, Germany, for sequencing services. The studies reported herein were financially supported by the Deutsche Forschungsgemeinschaft (DFG) (DFG HE2004/11-1 to AH; CRC-850 subprojects B5 to AH, C11 to RK [KE2164/2-1] and Z1 to MB; 322977937/GRK2344 subprojects B02 to MB and B03 to AH), and by the German Federal Ministry of Education and Research (BMBF) within the framework of the e:Med research and funding concept CoNfirm (FKZ 01ZX1708F to MB). CL received a fellowship from the Carl-Zeiss-Foundation (Postdoc Program 2017, Az. 0563-2.8/685/4). JW was additionally supported by the Deutsche Forschungsgemeinschaft Research Training Group GRK2344 “MeInBio–BioInMe”. Open access funding provided by Projekt DEAL.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41388-020-1259-7) contains supplementary material, which is available to authorized users.
References
- 1.Lau W, de Barker N, Clevers H. WNT signaling in the normal intestine and colorectal cancer. Front Biosci. 2007;12:471–91. doi: 10.2741/2076. [DOI] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330. https://www.nature.com/articles/nature11252.pdf. [DOI] [PMC free article] [PubMed]
- 3.Cadigan KM, Waterman ML. TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harb Perspect Biol. 2012;4:a007906. http://cshperspectives.cshlp.org/content/4/11/a007906.full. [DOI] [PMC free article] [PubMed]
- 4.Atcha FA, Munguia JE, Li TWH, Hovanes K, Waterman ML. A new beta-catenin-dependent activation domain in T cell factor. J Biol Chem. 2003;278:16169–75. doi: 10.1074/jbc.M213218200. [DOI] [PubMed] [Google Scholar]
- 5.Hecht A, Stemmler MP. Identification of a promoter-specific transcriptional activation domain at the C terminus of the Wnt effector protein T-cell factor 4. J Biol Chem. 2003;278:3776–85. doi: 10.1074/jbc.M210081200. [DOI] [PubMed] [Google Scholar]
- 6.Clevers H. The intestinal crypt, a prototype stem cell compartment. Cell. 2013;154:274–84. http://www.sciencedirect.com/science/article/pii/S0092867413008386. [DOI] [PubMed]
- 7.Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608. https://www.nature.com/articles/nature07602.pdf. [DOI] [PubMed]
- 8.Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters PJ, et al. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet. 1998;19:379–83. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- 9.Angus-Hill ML, Elbert KM, Hidalgo J, Capecchi MR. T-cell factor 4 functions as a tumor suppressor whose disruption modulates colon cell proliferation and tumorigenesis. Proc Natl Acad Sci USA. 2011;108:4914–9. doi: 10.1073/pnas.1102300108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Es JH, Haegebarth A, Kujala P, Itzkovitz S, Koo B-K, Boj SF, et al. A critical role for the Wnt effector Tcf4 in adult intestinal homeostatic self-renewal. Mol Cell Biol. 2012;32:1918–27. doi: 10.1128/MCB.06288-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hrckulak D, Janeckova L, Lanikova L, Kriz V, Horazna M, Babosova O, et al. Wnt effector TCF4 is dispensable for Wnt signaling in human cancer cells. Genes (Basel) 2018;9:439.. doi: 10.3390/genes9090439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van de Wetering M, Sancho E, Verweij C, Lau W, de Oving I, Hurlstone A, et al. The β-Catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–50. doi: 10.1016/S0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 13.Yochum GS, Cleland R, Goodman RH. A genome-wide screen for beta-catenin binding sites identifies a downstream enhancer element that controls c-Myc gene expression. Mol Cell Biol. 2008;28:7368–79. doi: 10.1128/MCB.00744-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tuupanen S, Turunen M, Lehtonen R, Hallikas O, Vanharanta S, Kivioja T, et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet. 2009;41:885–90. doi: 10.1038/ng.406. [DOI] [PubMed] [Google Scholar]
- 15.Giakountis A, Moulos P, Zarkou V, Oikonomou C, Harokopos V, Hatzigeorgiou AG, et al. A positive regulatory loop between a Wnt-regulated non-coding RNA and ASCL2 controls intestinal stem cell fate. Cell Rep. 2016;15:2588–96. http://www.sciencedirect.com/science/article/pii/S2211124716306210. [DOI] [PubMed]
- 16.Beyes S, Andrieux G, Schrempp M, Aicher D, Wenzel J, Anton-Garcia P, et al. Genome-wide mapping of DNA-binding sites identifies stemness-related genes as directly repressed targets of SNAIL1 in colorectal cancer cells. Oncogene. 2019;38:647–6661. doi: 10.1038/s41388-019-0905-4. [DOI] [PubMed] [Google Scholar]
- 17.Sjöblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–74. https://science.sciencemag.org/content/sci/314/5797/268.full.pdf. [DOI] [PubMed]
- 18.Wood LD, Parsons DW, Jones S, Lin J, Sjöblom T, Leary RJ, et al. The genomic landscapes of human breast and colorectalcancers. Science. 2007;318:1108–13. https://science.sciencemag.org/content/sci/318/5853/1108.full.pdf. [DOI] [PubMed]
- 19.Tang W, Dodge M, Gundapaneni D, Michnoff C, Roth M, Lum L. A genome-wide RNAi screen for Wnt/beta-catenin pathway components identifies unexpected roles for TCF transcription factors in cancer. Proc Natl Acad Sci USA. 2008;105:9697–702. doi: 10.1073/pnas.0804709105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hrckulak D, Kolar M, Strnad H, Korinek V. TCF/LEF transcription factors: an update from the internet resources. Cancers (Basel) 2016;8:70.. doi: 10.3390/cancers8070070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mouradov D, Sloggett C, Jorissen RN, Love CG, Li S, Burgess AW, et al. Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer. Cancer Res. 2014;74:3238–47. doi: 10.1158/0008-5472.CAN-14-0013. [DOI] [PubMed] [Google Scholar]
- 22.Sadanandam A, Lyssiotis CA, Homicsko K, Collisson EA, Gibb WJ, Wullschleger S, et al. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med. 2013;19:619. https://www.nature.com/articles/nm.3175.pdf. [DOI] [PMC free article] [PubMed]
- 23.Berg KCG, Eide PW, Eilertsen IA, Johannessen B, Bruun J, Danielsen SA, et al. Multi-omics of 34 colorectal cancer cell lines - a resource for biomedical studies. Mol Cancer. 2017;16:116. doi: 10.1186/s12943-017-0691-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, Toh L, Lau P, Wang X. Human telomerase reverse transcriptase (hTERT) is a novel target of the Wnt/β-catenin pathway in human cancer. J Biol Chem. 2012;287:32494–511. doi: 10.1074/jbc.M112.368282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yochum GS. Multiple Wnt/ss-catenin responsive enhancers align with the MYC promoter through long-range chromatin loops. PLoS ONE. 2011;6:e18966. doi: 10.1371/journal.pone.0018966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mokry M, Hatzis P, Bruijn E, de Koster J, Versteeg R, Schuijers J, et al. Efficient double fragmentation ChIP-seq provides nucleotide resolution protein-DNA binding profiles. PLoS ONE. 2010;5:e15092. doi: 10.1371/journal.pone.0015092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frietze S, Wang R, Yao L, Tak YG, Ye Z, Gaddis M, et al. Cell type-specific binding patterns reveal that TCF7L2 can be tethered to the genome by association with GATA3. Genome Biol. 2012;13:R52. doi: 10.1186/gb-2012-13-9-r52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jho E-h, Zhang T, Domon C, et al. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol. 2002;22:1172–83. doi: 10.1128/MCB.22.4.1172-1183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leung JY, Kolligs FT, Wu R, Zhai Y, Kuick R, Hanash S, et al. Activation of AXIN2 expression by beta-catenin-T cell factor. A feedback repressor pathway regulating Wnt signaling. J Biol Chem. 2002;277:21657–65. doi: 10.1074/jbc.M200139200. [DOI] [PubMed] [Google Scholar]
- 30.Lustig B, Jerchow B, Sachs M, Weiler S, Pietsch T, Karsten U, et al. Negative feedback loop of Wnt signaling through upregulation of conductin/axin2 in colorectal and liver tumors. Mol Cell Biol. 2002;22:1184–93. doi: 10.1128/MCB.22.4.1184-1193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van der Flier LG, van Gijn ME, Hatzis P, Kujala P, Haegebarth A, Stange DE, et al. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell. 2009;136:903–12. http://www.sciencedirect.com/science/article/pii/S0092867409000798. [DOI] [PubMed]
- 32.Batlle E, Henderson JT, Beghtel H, van den Born, Maaike MW, Sancho E, et al. β-Catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/EphrinB. Cell. 2002;111:251–63. http://www.sciencedirect.com/science/article/pii/S0092867402010152. [DOI] [PubMed]
- 33.Koo B-K, Spit M, Jordens I, Low TY, Stange DE, van de Wetering M, et al. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature. 2012;488:665–9. doi: 10.1038/nature11308. [DOI] [PubMed] [Google Scholar]
- 34.Rönsch K, Jäger M, Schöpflin A, Danciu M, Lassmann S, Hecht A. Class I and III HDACs and loss of active chromatin features contribute to epigenetic silencing of CDX1 and EPHB tumor suppressor genes in colorectal cancer. Epigenetics. 2011;6:610–22. doi: 10.4161/epi.6.5.15300. [DOI] [PubMed] [Google Scholar]
- 35.Jägle S, Rönsch K, Timme S, Andrlová H, Bertrand M, Jäger M, et al. Silencing of the EPHB3 tumor-suppressor gene in human colorectal cancer through decommissioning of a transcriptional enhancer. Proc Natl Acad Sci USA. 2014;111:4886–91. doi: 10.1073/pnas.1314523111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schnappauf O, Beyes S, Dertmann A, Freihen V, Frey P, Jagle S, et al. Enhancer decommissioning by Snail1-induced competitive displacement of TCF7L2 and down-regulation of transcriptional activators results in EPHB2 silencing. Biochim Biophys Acta. 2016;1859:1353–67. doi: 10.1016/j.bbagrm.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 37.Rosenbluh J, Nijhawan D, Cox AG, Li X, Neal JT, Schafer EJ, et al. β-catenin driven cancers require a YAP1 transcriptional complex for survival and tumorigenesis. Cell. 2012;151:1457–73. doi: 10.1016/j.cell.2012.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–8. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dang DT, Chen X, Feng J, Torbenson M, Dang LH, Yang VW. Overexpression of Krüppel-like factor 4 in the human colon cancer cell line RKO leads to reduced tumorigenecity. Oncogene. 2003;22:3424–30. doi: 10.1038/sj.onc.1206413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang W, Zhu Y, Gao J, Fu J, Liu C, Liu Y, et al. MicroRNA-29a promotes colorectal cancer metastasis by regulating matrix metalloproteinase 2 and E-cadherin via KLF4. Br J Cancer. 2014;110:450. https://www.nature.com/articles/bjc2013724.pdf. [DOI] [PMC free article] [PubMed]
- 41.Lv H, Zhang Z, Wang Y, Li C, Gong W, Wang X. MicroRNA-92a Promotes Colorectal Cancer Cell Growth and Migration by Inhibiting KLF4. Oncol Res. 2016;23:283–90. doi: 10.3727/096504016X14562725373833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wen D, Li S, Jiang W, Zhu J, Liu J, Zhao S. miR-539 inhibits human colorectal cancer progression by targeting RUNX2. Biomed Pharmacother. 2017;95:1314–20.. doi: 10.1016/j.biopha.2017.09.044. [DOI] [PubMed] [Google Scholar]
- 43.Chai J, Guo D, Ma W, Han D, Dong W, Guo H, et al. A feedback loop consisting of RUNX2/LncRNA-PVT1/miR-455 is involved in the progression of colorectal cancer. Am J Cancer Res. 2018;8:538–50.. [PMC free article] [PubMed] [Google Scholar]
- 44.Guo Z-J, Yang L, Qian F, Wang Y-X, Yu X, Ji C-D, et al. Transcription factor RUNX2 up-regulates chemokine receptor CXCR4 to promote invasive and metastatic potentials of human gastric cancer. Oncotarget. 2016;7:20999–1012. doi: 10.18632/oncotarget.8236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li X-Q, Du X, Li D-M, Kong P-Z, Sun Y, Liu P-F, et al. ITGBL1 Is a Runx2 transcriptional target and promotes breast cancer bone metastasis by activating the TGFβ signaling pathway. Cancer Res. 2015;75:3302–13. doi: 10.1158/0008-5472.CAN-15-0240. [DOI] [PubMed] [Google Scholar]
- 46.Koshizuka K, Hanazawa T, Kikkawa N, Arai T, Okato A, Kurozumi A, et al. Regulation of ITGA3 by the anti-tumor miR-199 family inhibits cancer cell migration and invasion in head and neck cancer. Cancer Sci. 2017;108:1681–92.. doi: 10.1111/cas.13298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neri P, Ren L, Azab AK, Brentnall M, Gratton K, Klimowicz AC, et al. Integrin β7-mediated regulation of multiple myeloma cell adhesion, migration, and invasion. Blood. 2011;117:6202–13. doi: 10.1182/blood-2010-06-292243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramovs V, Secades P, Song J-Y, Thijssen B, Kreft M, Sonnenberg A. Absence of integrin α3β1 promotes the progression of HER2-driven breast cancer in vivo. Breast Cancer Res. 2019;21:63. doi: 10.1186/s13058-019-1146-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoverter NP, Zeller MD, McQuade MM, Garibaldi A, Busch A, Selwan EM, et al. The TCF C-clamp DNA binding domain expands the Wnt transcriptome via alternative target recognition. Nucleic Acids Res. 2014;42:13615–32. doi: 10.1093/nar/gku1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Varnat F, Siegl-Cachedenier I, Malerba M, Gervaz P. Ruiz i Altaba A. Loss of WNT-TCF addiction and enhancement of HH-GLI1 signalling define the metastatic transition of human colon carcinomas. EMBO Mol Med. 2010;2:440–57. doi: 10.1002/emmm.201000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holmberg J, Genander M, Halford MM, Anneren C, Sondell M, Chumley MJ, et al. EphB receptors coordinate migration and proliferation in the intestinal stem cell niche. Cell. 2006;125:1151–63. doi: 10.1016/j.cell.2006.04.030. [DOI] [PubMed] [Google Scholar]
- 52.Schuijers J, Junker JP, Mokry M, Hatzis P, Koo B-K, Sasselli V, et al. Ascl2 acts as an R-spondin/Wnt-responsive switch to control stemness in intestinal crypts. Cell Stem Cell. 2015;16:158–70. doi: 10.1016/j.stem.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 53.Geoghegan G, Simcox J, Seldin MM, Parnell TJ, Stubben C, Just S, et al. Targeted deletion of Tcf7l2 in adipocytes promotes adipocyte hypertrophy and impaired glucose metabolism. Mol Metab. 2019;24:44–63. doi: 10.1016/j.molmet.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiang J, Hu Q, Qin Y, Ji S, Xu W, Liu W, et al. TCF7L2 positively regulates aerobic glycolysis via the EGLN2/HIF-1α axis and indicates prognosis in pancreatic cancer. Cell Death Dis. 2018;9:321. doi: 10.1038/s41419-018-0367-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Sousa E, Melo F, Colak S, Buikhuisen J, Koster J, Cameron K.et al. Methylation of cancer-stem-cell-associated Wnt target genes predicts poor prognosis in colorectal cancer patients. Cell Stem Cell. 2011;9:476–85. http://www.sciencedirect.com/science/article/pii/S1934590911004887. [DOI] [PubMed]
- 56.Bormann F, Rodríguez-Paredes M, Lasitschka F, Edelmann D, Musch T, Benner A, et al. Cell-of-origin DNA methylation signatures are maintained during colorectal carcinogenesis. Cell Rep. 2018;23:3407–18. http://www.sciencedirect.com/science/article/pii/S2211124718307915. [DOI] [PubMed]
- 57.Rodilla V, Villanueva A, Obrador-Hevia A, Robert-Moreno À, Fernández-Majada V, Grilli A, et al. Jagged1 is the pathological link between Wnt and Notch pathways in colorectal cancer. Proc Natl Acad Sci USA. 2009;106:6315–20. doi: 10.1073/pnas.0813221106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beumer J, Clevers H. Regulation and plasticity of intestinal stem cells during homeostasis and regeneration. Development. 2016;143:3639–49.. doi: 10.1242/dev.133132. [DOI] [PubMed] [Google Scholar]
- 59.Farley FW, Soriano P, Steffen LS, Dymecki SM. Widespread recombinase expression using FLPeR (flipper) mice. Genesis. 2000;28:106–10. doi: 10.1002/1526-968X(200011/12)28:3/4<106::AID-GENE30>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 60.Kranz A, Fu J, Duerschke K, Weidlich S, Naumann R, Stewart AF, et al. An improved Flp deleter mouse in C57Bl/6 based on Flpo recombinase. Genesis. 2010;48:512–20. doi: 10.1002/dvg.20641. [DOI] [PubMed] [Google Scholar]
- 61.el Marjou F, Janssen K-P, Chang BH-J, Li M, Hindie V, Chan L, et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39:186–93. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- 62.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–5. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 63.Freihen V, Rönsch K, Mastroianni J, Frey P, Rose K, Boerries M, et al. SNAIL1 employs β-Catenin-LEF1 complexes to control colorectal cancer cell invasion and proliferation. Int J Cancer. 2020;146:2229–42. doi: 10.1002/ijc.32644. [DOI] [PubMed] [Google Scholar]
- 64.Albers J, Danzer C, Rechsteiner M, Lehmann H, Brandt LP, Hejhal T, et al. A versatile modular vector system for rapid combinatorial mammalian genetics. J Clin Investig. 2015;125:1603–19. doi: 10.1172/JCI79743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jägle S, Busch H, Freihen V, Beyes S, Schrempp M, Boerries M, et al. SNAIL1-mediated downregulation of FOXA proteins facilitates the inactivation of transcriptional enhancer elements at key epithelial genes in colorectal cancer cells. PLoS Genet. 2017;13:e1007109. doi: 10.1371/journal.pgen.1007109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Seebauer CT, Brunner S, Glockzin G, Piso P, Ruemmele P, Schlitt H-J, et al. Peritoneal carcinomatosis of colorectal cancer is characterized by structural and functional reorganization of the tumor microenvironment inducing senescence and proliferation arrest in cancer cells. Oncoimmunology. 2016;5. 10.1080/2162402X.2016.1242543. [DOI] [PMC free article] [PubMed]
- 67.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. https://genomebiology.biomedcentral.com/track/pdf/10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed]
- 70.Fisher RA. On the Interpretation of χ2 from contingency tables, and the calculation of P. J R Stat Soc. 1922;85:87. doi: 10.2307/2340521. [DOI] [Google Scholar]
- 71.Arrigoni L, Richter AS, Betancourt E, Bruder K, Diehl S, Manke T, et al. Standardizing chromatin research: a simple and universal method for ChIP-seq. Nucleic Acids Res. 2015;44:e67. doi: 10.1093/nar/gkv1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rönsch K, Jägle S, Rose K, Seidl M, Baumgartner F, Freihen V, et al. SNAIL1 combines competitive displacement of ASCL2 and epigenetic mechanisms to rapidly silence the EPHB3 tumor suppressor in colorectal cancer. Mol Oncol. 2015;9:335–54. doi: 10.1016/j.molonc.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kadem LF, Holz M, Suana KG, Li Q, Lamprecht C, Herges R, et al. Rapid reversible photoswitching of integrin-mediated adhesion at the single-cell level. Adv Mater. 2016;28:1799–802. doi: 10.1002/adma.201504394. [DOI] [PubMed] [Google Scholar]
- 74.Sabates-Bellver J, van der Flier LG, Palo M, de Cattaneo E, Maake C, Rehrauer H, et al. Transcriptome profile of human colorectal adenomas. Mol Cancer Res. 2007;5:1263–75. doi: 10.1158/1541-7786.MCR-07-0267. [DOI] [PubMed] [Google Scholar]
- 75.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fox J, Weisberg S. An R companion to applied regression. 3rd ed. Los Angeles: SAGE; 2019. [Google Scholar]
- 78.Bates D, Mächler M, Bolker B, Walker S. Fitting Linear Mixed-Effects Models Using lme4. J Stat Soft. 2015;67:1–48. doi: 10.18637/jss.v067.i01. [DOI] [Google Scholar]
- 79.Wickham H. ggplot2: elegant graphics for data analysis. New York, NY: Springer-Verlag; 2009. 10.1007/978-0-387-98141-3.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.