Abstract
Plant viruses rely on insect vectors for transmission among plant hosts, but many of the specifics of virus-vector interactions are not fully understood. Thrips tabaci, which transmits Tomato spotted wilt virus (TSWV) in a persistent and propagative manner, varies greatly in its ability to transmit different isolates of TSWV. Similarly, TSWV isolates are transmitted at different efficiencies by different populations of T. tabaci. This study characterizes differences in virus titers in the vector among TSWV isolate-T. tabaci isoline pairings in relation to differences in transmission rates, and demonstrates that although transmission rates were higher for sympatric than allopatric TSWV isolate-T. tabaci isoline pairings, virus titers in the thrips vector were significantly lower in the sympatric pairings. Results further demonstrate that TSWV titers in the vector were unrelated to virus titers in the leaf tissue from which they acquired the virus and provide evidence for the importance of specific vector-virus interactions and local adaptation in determining transmission efficiency of TSWV by T. tabaci.
Subject terms: Agroecology, Ecological epidemiology, Entomology, Coevolution
Introduction
Modern agricultural practices and globalization of trade have contributed to a surge of emerging plant viruses that are responsible for billions of dollars in annual crop losses. The expansion of agricultural land alters stable relationships between viruses, insects, and their natural plant hosts; providing opportunities for viruses and vectors to exploit widely available cultivated hosts1. Most plant viruses are dependent on insect vectors for plant-to-plant transmission. Specific interactions with their insect vectors are required for the viruses to move to new hosts but little is known about how these interactions impact the efficiency of transmission. This study examines the relationship between virus titer in the vector and transmission efficiency across multiple isolates of Tomato spotted wilt virus (TSWV) (Bunyavirales: Tospoviridae) and isolines of its thrips vector, Thrips tabaci, originating from multiple geographical locations and host plants. Specifically, we examine differences in transmission rates among TSWV isolate-T. tabaci isoline pairings in relationship to differences in virus titers in the vector and whether these relationships differ between sympatric and allopatric pairings of TSWV isolates and T. tabaci isolines.
Vector competence reflects the success of the virus in overcoming intrinsic, physiological, microbiota, and immunity barriers, and can be governed by genetic interactions between vector and viral genotypes that are subject to influence by extrinsic factors, including host plants and environmental conditions2–4. For persistently transmitted viruses, successful transmission requires that the viruses traverse anatomical barriers in their vectors. After ingestion, the virus must move from the gut lumen, across other tissues or through hemolymph, and into the salivary glands from which the virus can be transferred into plants during insect feeding5. Transmission and transmission efficiency of viruses are ultimately determined by the number of virions that accumulate in the appropriate salivary compartments after circulation and/or replication within the vector.
Several studies on vector taxa exhibiting different modes of transmission have shown positive correlations between viral titer within the source plant or insect vector and the efficiency of transmission of plant viruses6–10. Circulative, non-propagative viruses in the families Luteoviridae, Geminiviridae, and Nanoviridae are transmitted by aphids, whiteflies or leafhoppers. Because these viruses do not replicate in their vectors, higher virus titers in plants8–10 and/or longer feeding periods11 have been shown to increase the amount of virus acquired by the vector and increase transmission12. In persistent-propagative viruses, less is known about how virus titers in source plants and in the insect vector relate to transmission efficiency. There is evidence in the genus Tenuivirus that virus titer in source plants positively correlates with transmission efficiency by planthopper vectors7, but this relationship was not consistent among isolates of Maize stripe virus (MStV)6. Virus titer is also believed to be a determinant for transmission of TSWV because higher titers of TSWV in adult thrips have been related to higher transmission frequencies in the efficient vector F. occidentalis; however, differences in transmission among thrips species varying in the efficiency with which they transmit different tospoviruses are not fully explained by virus titers in the vector13–16.
Although the aforementioned studies document a relationship between virus titer and transmission frequency for certain virus-vector pairings, they focused on thrips from established laboratory colonies. They do not address the relationship between virus titer in the vector and inter-population variation in transmission within a single vector species or variation among isolates of TSWV. A previous study by Jacobson and Kennedy17 examining transmission of 89 distinct pairings between TSWV isolates and Thrips tabaci isolines showed a significant effect of virus isolate, thrips isoline, and their interaction on transmission efficiency. Although transmission rates ranged from 0–55% across all isolate by isoline pairings, the ability of a single T. tabaci-isoline to transmit multiple virus isolates varied up to 18-fold, and the transmission rates of each TSWV isolate by multiple T. tabaci isolines varied up to 45-fold. In addition, significantly higher transmission rates were observed among sympatric (originate from the same location) TSWV isolate-T. tabaci isoline pairings than allopatric pairings (originate from different locations), suggesting local adaptation between virus and vector resulting from coevolution in which local virus has greater infectivity than foreign virus on local vectors18. The purpose of this study was to examine whether or not the observed variation in transmission frequency was influenced by differences in virus titer within the vector. Both the leaf tissue and thrips used in the studies of Jacobson and Kennedy17 were flash frozen in liquid nitrogen immediately following their acquisition and inoculation access periods, respectively, and stored at −80 °C until used in this study. We used a subset of the TSWV isolate and T. tabaci isoline pairings studied by Jacobson and Kennedy17 to determine if TSWV titers in adult T. tabaci varied among the TSWV isolate and T. tabaci isoline pairings. We also examined whether differences in transmission rates were related to variation in virus titers in the leaf tissue from which the thrips acquired the virus and in the thrips vector.
Results
Thrips tabaci and TSWV isolates included in our study were subsamples of those tested by Jacobson and Kennedy17 in their characterization of differences in transmission efficiency among different pairings of T. tabaci isolines and TSWV isolates, both of which were obtained from multiple locations and host plants in North Carolina. The T. tabaci individuals studied were from clonal isolines of T. tabaci that were established from thelytokous females (parthenogenetic reproduction - producing only female offspring) collected at each location to minimize genetic variation within each of the isolines. Transmission efficiency of each TSWV isolate by each T. tabaci isoline was then characterized. Details of collection, establishment of clonal thrips isolines, TSWV isolates, transmission experiments, and results from transmission assays to characterize each TSWV isolate-isoline pairing are described in Jacobson and Kennedy17. They classified individual thrips as transmitting or non-transmitting based on a DAS-ELISA test of the leaf discs on which viruliferous thrips were allowed to feed during the inoculation access period (these leaf discs were no longer available for the experiments reported here). Samples of transmitting thrips, non-transmitting thrips, and infected leaf tissue used for acquisition of TSWV from each isolate-isoline pairing were flash frozen in liquid nitrogen and stored at −80 °C until used in the experiments reported here. A subset consisting of 12 of the 89 TSWV isolate-T. tabaci isoline pairings for which transmission efficiencies were reported by Jacobson and Kennedy17 were chosen for the experiments reported here; they were representative of the range in transmission efficiencies observed among the 89 TSWV isolate-T. tabaci isoline pairings and included both sympatric and allopatric pairings17 (Table 1). For each pairing, we used RT-qPCR to quantify TSWV titers in individual transmitting and non-transmitting thrips, and in the TSWV-infected, Emilia sonchifolia leaf tissue from which the thrips acquired the virus. Up to five transmitting and non-transmitting thrips were selected per TSWV isolate-T. tabaci isoline pairing unless transmission rates were so low that five transmitting individuals were not observed in transmission experiments. The numbers of transmitting and non-transmitting thrips for each TSWV isolate-T. tabaci isoline pairing subjected to RT-qPCR are shown in Supplementary Table S1.
Table 1.
TSWV isolate and Thrips. tabaci isoline pairings: North Carolina locations and host plants from which TSWV isolates and adult thrips used to initiate each T. tabaci isoline were collected and mean proportion of T. tabaci transmitting TSWV for each isolate-isoline pairing as reported by Jacobson and Kennedy (2013). * indicates sympatric isolate-isoline pairing.
| TSWV isolate | T. tabaci isoline | Location isolate-isoline | Host plant Isolate-isoline | Proportion thrips transmitting (n) |
|---|---|---|---|---|
| AM1 | IPOC1* | Cove City – Cove City | Nicotiana tabacum –Allium spp. | 0.20 (n = 65) |
| AM1 | Kin1 | Cove City – Kinston | N. tabacum – Allium cepa | 0.21 (n = 66) |
| AM1 | SH2 | Cove City – Jackson Springs | N. tabacum - Secale cerealae | 0.21 (n = 28) |
| AM1 | SH72 | Cove City – Jackson Springs | Capsicum annuum – Raphanus sativus var niger | 0.10 (n = 63) |
| SH3 | IPOC1 | Jackson Springs - Cove City | N. tabacum – Allium spp. | 0.03 (n = 67) |
| SH3 | Kin1 | Jackson Springs – Kinston | N. tabacum – A. cepa | 0.16 (n = 68) |
| SH3 | SH2* | Jackson Springs – Jackson Springs | N. tabacum – S. cerealae | 0.16 (n = 131) |
| SR3–3 | IPOC1* | Cove City – Cove City | N. tabacum – Allium spp. | 0.07 (n = 54) |
| SR3-3 | Kin1 | Cove City – Kinston | N. tabacum – A. sepa | 0.08 (n = 42) |
| SR3-3 | SH2 | Cove City – Jackson Springs | N. tabacum – S. cerealae | 0.06 (n = 141) |
| SR3-3 | SH72 | Cove City – Jackson Springs | N. tabacum - Raphanus sativus var niger | 0.20 (n = 20) |
| SHP | SH2* | Jackson Springs – Jackson Springs | Capsicum annuum - S. cerealae | 0.55 (n = 52) |
Primer efficiency and validation
Most of the published RT-qPCR primers for thrips internal controls were designed for Frankliniella occidentalis and did not show the specificity and consistency required for reproducible RT-qPCR with T. tabaci samples (Supplementary Table S2). EF1A primers from F. occidentalis demonstrated the most promising results for a heterologous internal control. However, homologous T. tabaci EF1A primers (Supplementary Table S3) amplified more robustly and consistently (Supplementary Fig. S1). Primer pair EF1A_346F and EF1A_456R was chosen as the internal control with a primer efficiency of 86% (Table 2).
Table 2.
Primers used in RT-PCR reactions.
| Primer Name | Sequence (5′-3′) |
|---|---|
| EF1A_346F | CGTCAAGGAACTTCGTCGTG |
| EF1A_456R | CACAGGGGTGTATCCGTTG |
| TSWVL_4382F | GCATGAAYTGGTTRCAAGGC |
| TSWVL_4493R | CAGAGTGCACAATCCATCTAG |
| Emilia_5.8 S_rRNAF | GTGTGAATTGCAGAATCCCGT |
| Emilia_5.8 S_rRNAR | CATGTGACGCCCAGGCA |
The elongation factor 1 alpha (EF1A) primers were used as a reference sequence in Thrips tabaci. TSWVL primers were used to target the L RNA sequence for TSWV, and the Emilia_5.8S primers were based on the reference sequence for Emilia sonchifolia.
Because E. sonchifolia does not have published RT-qPCR primers or a complete genome sequence, and previously published RT-qPCR primers developed from the highly conserved common plant genes actin, tRNA and profilin19 failed to amplify from E. sonchifolia source tissue, an alignment of 3 partial E. sonchifolia sequences for the 5.8 S rRNA gene and internal transcribed spacer (ITS) were utilized to design additional primer pairs (Supplementary Table S4). The ITS primers had poor amplification but the 5.8 S rRNA primers amplified most efficiently and consistently (Supplementary Fig. S2). Primer pair Emilia_5.8S_rRNAF and Emilia 5.8S_rRNAR was chosen as the internal control for the source leaf tissue with a primer efficiency of 97%.
The N gene primers for TSWV quantification produced inconsistent results (Supplementary Table S5). The L RNA primers were more consistent and robust overall than any of the N primers. Primer pair TSWVL4832F and TSWVL 4493 R was used to measure TSWV transcript levels with a primer efficiency of 92%. The L RNA is also a better representation of the viral titer present in infected tissue because it measures mostly genome replication, while the N gene primers measure both genome replication (S RNA) and N gene expression (amplified S mRNAs).
Viral titers in the source leaf tissues
There were no statistically significant differences in virus titers in the source leaf tissue used for virus acquisition among any of the TSWV isoline and T. tabaci isolate pairings [F = 0.25, df = (11, 12), P = 0.9853] (Fig. 1; Table 3). This result indicates that titer in the source leaves does not account for differences in virus titer or transmission among thrips included in this study because all thrips had the potential to ingest similar amounts of TSWV. The Ct values of the TSWV L RNA are very similar to the Ct values of the 5.8 S rRNA of E. sonchifolia showing the virus replicates to the level of ribosomal RNA in the source leaf tissue.
Figure 1.
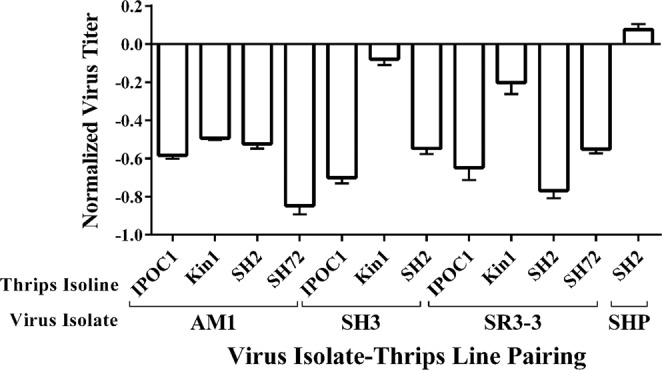
Log mean relative abundance ratio for TSWV LRNA in Emilia sonchifolia source leaf tissue for all TSWV isolate-Thrips tabaci isoline pairings. No significant differences in titer were found between any of the isolate-isoline pairings at P = 0.05. Error bars = standard error of mean.
Table 3.
Average Ct values (+ standard error) for the Emilia sonchifolia leaf tissue and Thrips tabaci individualsa.
| TSWV Isolate | Thrips Isoline | E. sonchifolia Ct Values | T. tabaci Ct Values (sd) | ||
|---|---|---|---|---|---|
| TSWV-L RNA | Emilia 5.8S rRNA | TSWV-L RNA | Thrips EF1A | ||
| AM1 | IPOC1* | 14.7 ± 0.1 | 12.2 ± 0.1 | 30.5 ± 0.5 | 21.3 ± 0.1 |
| AM1 | Kin1 | 14.5 ± 0.0 | 12.3 ± 0.1 | 25.3 ± 0.2 | 20.8 ± 0.1 |
| AM1 | SH2 | 14.9 ± 0.1 | 12.5 ± 0.1 | 29.6 ± 0.2 | 19.6 ± 0.1 |
| AM1 | SH72 | 16.1 ± 0.3 | 12.6 ± 0.1 | 31.2 ± 0.3 | 18.9 ± 0.2 |
| SH3 | IPOC1 | 14.6 ± 0.2 | 11.7 ± 0.1 | 27.3 ± 0.3 | 20.6 ± 0.1 |
| SH3 | Kin1 | 12.6 ± 0.1 | 11.9 ± 0.2 | 34.2 ± 0.2 | 21.7 ± 0.3 |
| SH3 | SH2* | 14.4 ± 0.2 | 12.0 ± 0.1 | 35.8 ± 0.7 | 18.1 ± 0.0 |
| SR3-3 | IPOC1* | 15.2 ± 0.4 | 12.4 ± 0.1 | 30.0 ± 0.2 | 22.7 ± 0.1 |
| SR3-3 | Kin1 | 13.0 ± 0.4 | 11.8 ± 0.1 | 31.4 ± 0.2 | 21.2 ± 0.1 |
| SR3-3 | SH2 | 14.1 ± 0.2 | 10.9 ± 0.2 | 27.0 ± 0.2 | 22.5 ± 0.2 |
| SR3-3 | SH72 | 15.0 ± 0.0 | 12.5 ± 0.1 | 27.1 ± 0.3 | 24.2 ± 0.1 |
| SHP | SH2* | 12.7 ± 0.1 | 12.4 ± 0.1 | 25.7 ± 0.3 | 20.9 ± 0.1 |
aValues within columns are not significantly different (P > 0.05).
*TSWV isolate and thrips isoline pairings that are designated as sympatric.
Effects of TSWV isolate and T. tabaci isoline on virus titer in thrips
The main effect of TSWV isolate on virus titer per thrips was highly significant [F = 8.6, df = (3, 93), P < 0.0001] (Fig. 2), whereas the main effect of T. tabaci isoline [F = 2.3, df = (3, 93), P = 0.0820] (Fig. 3) was not. However, the isolate by isoline interaction was highly significant [F = 6.12, df = (5, 93), P < 0.0001 (Fig. 4)], indicating that the effect of TSWV isolate on virus titer per thrips varied depending on the T. tabaci isoline. In Jacobson and Kennedy17 this significant interaction term was consistent with higher transmission efficiency among sympatric than allopatric isolate-isoline pairings. Therefore, another analysis was conducted in which collection location (allopatric/sympatric) replaced the interaction term in the ANOVA model. Titers were measured in 39 individual thrips representing sympatric TSWV isolate-T. tabaci isoline pairings and 66 individual thrips representing allopatric TSWV isolate-T. tabaci isoline pairings. In this analysis, virus isolate [F = 12.21, df = (3, 94), P < 0.0001] and T. tabaci isoline [F = 5.53, df = (3, 94), P = 0.0015] main effects on virus titers per thrips were both significant. In addition, the effect of location was significant; mean virus titers of adult thrips were significantly lower for sympatric than allopatric virus isolate-thrips isoline pairings [F = 15.13, df = (1, 97), P = 0.0002] (Fig. 5).
Figure 2.
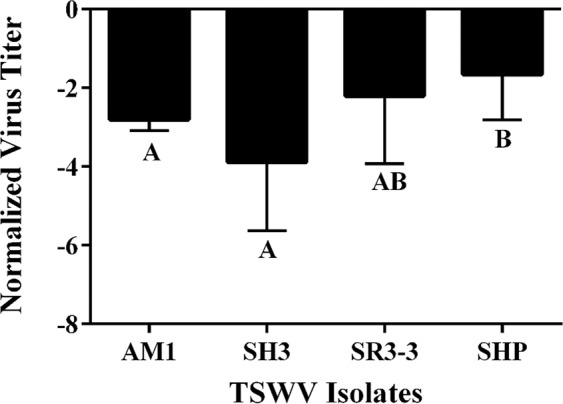
Mean normalized Log10 transformed virus titer of each Tomato spotted wilt virus (TSWV) isolate quantified from individual thrips. The main effect of virus isolate on titer is significant (P = 0.0011). Mean separation of TSWV isolates by LS Means at α = 0.05. Error bars = standard error of mean.
Figure 3.
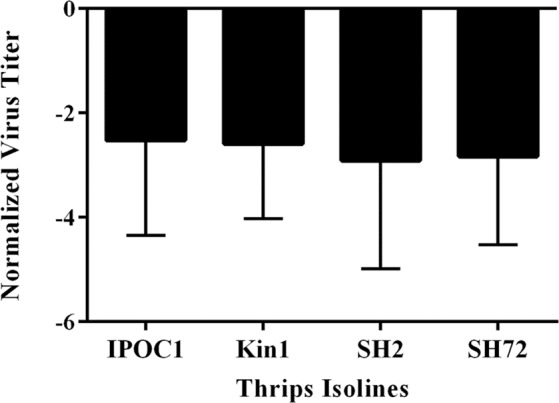
Mean normalized Log10 transformed titer of Tomato spotted wilt virus (TSWV) isolates averaged by Thrips tabaci isoline. The y-axis shows normalized virus titer of all thrips, both non-transmitting and transmitting. TSWV titers were not significantly different among isolines at P = 0.05. Error bars = standard error of mean.
Figure 4.
Mean normalized log10 transformed virus titer compared across all Tomato spotted wilt virus (TSWV) isolate-Thrips tabaci isoline pairings. Normalized TSWV titer means for all thrips, including transmitting and non-transmitting, shows titer is dependent on the interactions of the virus isolate and thrips isoline (P < 0.001). Mean separation of TSWV isolates by LS Means at α = 0.05. Error bars = standard error of mean.
Figure 5.
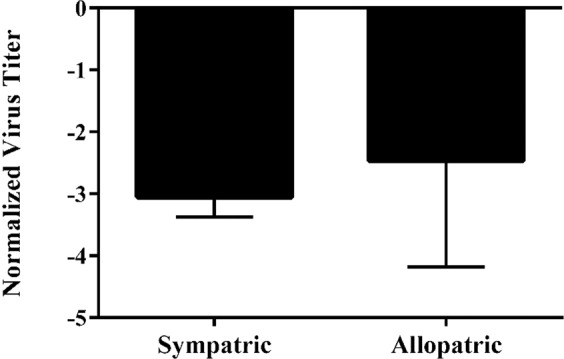
Mean normalized log10 transformed Tomato spotted wilt virus (TSWV) titer of sympatric and allopatric TSWV isolate-Thrips tabaci isoline pairings. The main effect of sympatry on titer is significant (P = 0.0002), meaning allopatric pairings have higher titers on average compared to sympatric pairings. Mean separation of TSWV isolates by LS Means at α = 0.05. Error bars = standard error of mean.
Relationship between TSWV transmission and virus titer in the vector
Of the 105 T. tabaci included in our study, 42 percent had transmitted TSWV to leaf discs and 58 percent had not transmitted. TSWV was detected in all 105 thrips regardless of whether they transmitted the virus, and the mean virus titer per thrips was significantly higher in transmitting than non-transmitting thrips [F = 11.23, df = (1, 100), P = 0.0011] (Fig. 6). However, even within individual TSWV isolate-T. tabaci isoline pairings virus titers were not always higher in transmitting than non-transmitting thrips, demonstrating that non-transmitters could support similar levels of viral replication (Supplementary Table S6; Supplementary Fig. S3). Because we measured virus titers in only a small sub-sample (n = 5–10) of the individual thrips used by Jacobson and Kennedy17 to characterize the transmission efficiency of each TSWV isolate-T. tabaci isoline pairing included in this study (Table 1), our subsamples could not be used to directly test associations between virus titers and transmission efficiencies of the individual isolate and isoline pairings.
Figure 6.
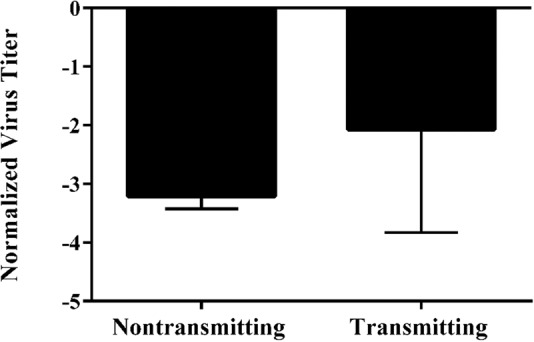
Mean normalized log10 transformed Tomato spotted wilt virus TSWV titer in transmitting thrips vs non-transmitting thrips. The main effect of transmission on titer is significant (P = 0.0011). Mean separation of TSWV isolates by LS Means at α = 0.05. Error bars = standard error of mean.
Discussion
The aims of this study were to characterize the relationship between virus titers in a thrips vector and transmission rates of TSWV, and to test whether these relationships differ between sympatric and allopatric pairings of the virus isolates and vector isolines. During these experiments, we developed a reliable method for quantifying L RNA, which is the only genome segment of TSWV that does not produce subgenomic RNA and, therefore, provides a measure of only genomic replication and not gene expression. In our study, virus titer in the leaf tissue from which thrips acquired virus did not account for differences in virus titers within thrips, suggesting vector-virus interactions are more important determinants of virus titers reached within thrips than viral load acquired during feeding.
Virus titers in adult T. tabaci did not fully explain the variation in transmission efficiency observed among the TSWV isolate-T. tabaci isoline pairings. Although mean virus titer in transmitting individuals was significantly higher than in non-transmitting individuals, there was extensive variation in virus titers among transmitting and non-transmitting individuals, both within and among TSWV isolate-T. tabaci isoline pairings. The effects of TSWV isolate and the interaction of TSWV isolate and T. tabaci isoline were consistently significant in all of our analyses, whereas the effect of T. tabaci isoline was significant only when collection location (sympatry/allopatry) was included as a main effect. Jacobson and Kennedy17, in their prior study involving a larger number of TSWV isolate-T. tabaci isoline pairings, found that transmission was higher in sympatric TSWV isolate-T. tabaci isoline pairs than allopatric pairs; a result indicative of local adaptation20. Within the subset of TSWV isolate-T. tabaci isoline pairings that were included in our study, the mean transmission rates for the sympatric and allopatric pairings were 25% and 12%, respectively, but the mean virus titers in thrips from the allopatric pairings were significantly higher than in the thrips from the sympatric pairings. This finding contrasts with our analysis comparing all transmitting and non-transmitting thrips, which showed that mean virus titers were higher in thrips that transmitted than in those that did not, and suggests that local adaptations between virus and vector leading to more efficient virus transmission involves more than simply higher virus replication or accumulation rates in the vector. In the collection locations for T. tabaci and TSWV isolates used in our study the most abundant vectors are F. fusca and F. occidentalis. The evidence for local adaptation between TSWV isolates and T. tabaci populations used in our study suggests that selection on the virus population for adaptation to specific vectors is sufficiently strong that even minor vector species such as T. tabaci can influence local vector-virus evolution17,21 (Table 1).
A genetic basis for differences in transmission efficiency of tospoviruses by T. tabaci and two other thrips species has been documented22–24 but the underlying mechanisms responsible for the differences have not been determined. Although TSWV titers in the vector F. occidentalis have been positively associated with vector competence (transmission yes or no) and frequency of transmission15,16, virus titers in the thrips are not necessarily indicative of titers in the salivary gland and salivary reservoir from which the virus is carried in saliva to the plant during feeding. There are numerous opportunities for virus replication and movement within the vector that may be compromised, resulting in reduced transmission efficiency25,26. Viral genes that encode nucleocapsid, glycoproteins, and NSs have been implicated in infection and movement in thrips vectors27–31. For TSWV to be transmitted by thrips, it must traverse the midgut following ingestion by first instars and infect the muscles surrounding the midgut where replication occurs. The virus must then move via the tubular salivary glands and the efferent duct that leads from the principal salivary glands to the salivary reservoir, and the level of salivary gland infection has been shown to be a critical determinant of transmission32–34. Viral receptor proteins putatively interact with the receptors of midgut cells in thrips, enabling cell-to-cell movement of TSWV via endocytosis to the salivary glands for transmission29,31,35–37. Even if replication occurs, infection may be limited to the midgut if virus cannot cross the basal lamina38, and midgut-limited infections can persist in adult thrips that cannot transmit the virus, as well as in non-vector species that have fed on virus infected plant tissue39,40. In aphids, the basal lamina of the accessory salivary gland acts as a selective barrier associated with differential transmission of Barley yellow dwarf virus41–43 and specific cells in the primary salivary gland control differential transmission of Tomato yellow leaf curl China virus and Tomato yellow leaf curl virus by different whiteflies in the Bemisia tabaci cryptic species complex44.
TSWV has been shown to elicit an immune response in F. occidentalis and F. fusca16,45–47 that limits virus movement, suggesting variation in vector competence may be affected by differences in immune responses of the thrips vectors. Similarly, the silencing suppressor (NSs) might also have a role, as it has also been shown to influence virus accumulation in F. occidentalis; although its exact role is unknown, it likely suppresses the immune response30.
Behavior and fitness-related effects were not examined in this study but have been documented to influence transmission of plant viruses by their vectors. Infection of TSWV in male thrips increased feeding and the number of non-ingestion probes48, which are believed to be largely responsible for transmission because they leave cells intact49. Both infections in thrips and host plants significantly alter survival and development times of F. occidentalis and F. fusca, and the magnitude of these effects is influenced by host plant, virus isolate and temperature3,4. Effects of plant and vector infections on fitness of T. tabaci have not been reported, but may contribute to the lower transmission rates and higher titers observed in allopatric T. tabaci-TSWV isolate pairings. Although not formally studied, poor survival of T. tabaci on TSWV-infected plants was observed in some of the transmission experiments (Jacobson and Kennedy personal observation). Plant-virus interactions leading to changes in plant attractiveness to vectors are documented for plant viruses and their vectors50 but have not been studied in this pathosystem.
Previous work demonstrated that transmission efficiency of TSWV by T. tabaci isolines likely depends on genotypic interactions between virus isolates and T. tabaci isolines17,21. Our results extend these findings by demonstrating that variation in virus titer in adult thrips is not entirely responsible for the observed variation in transmission efficiency, and provide additional support for the importance of specific vector-virus interactions and co-evolutionary dynamics determining transmission efficiency within a single vector species. A better understanding of virus replication, movement, and accumulation in the vector is needed to determine whether transmission outcomes are influenced by more efficient localization or replication in the salivary glands. Understanding epidemiologically important patterns of variation in vector competence also requires a better understanding of the mechanisms underlying transmission, similarities of these mechanisms across sympatric vector species, and the influence of vector-imposed selection pressure on viral populations.
Materials and Methods
T. tabaci and TSWV: Collecting, culturing and transmission assays
T. tabaci individuals and TSWV isolates were subsamples of those tested by Jacobson and Kennedy17 in their characterization of differences in transmission efficiency among different pairings of T. tabaci isolines and TSWV isolates obtained from multiple locations and host plants in North Carolina. The clonal isolines of T. tabaci were established from thelytokous females (parthenogenetic reproduction - producing only female offspring) collected at each location to minimize genetic variation within each of the isolines. Transmission efficiency was then characterized by each of the isolate by isoline pairings. Details of collection, establishment of clonal thrips isolines, TSWV isolate establishment, transmission experiments, and results from transmission assays to characterize each TSWV isolate-isoline pairing are described in Jacobson and Kennedy17. Individual thrips were classified as transmitting or non-transmitting based on a DAS-ELISA test of the leaf discs on which viruliferous thrips were allowed to feed during the inoculation access period. Samples of transmitting thrips, non-transmitting thrips, and infected leaf tissue used for acquisition of TSWV from each isolate-isoline pairing were flash frozen in liquid nitrogen and stored at −80 °C until used in the experiments reported here. Twelve of the 89 isolate-isoline pairings for which transmission efficiencies were reported by Jacobson and Kennedy17 were included in the experiments reported here; they were representative of the range in transmission efficiencies observed among the 89 isolate-isoline pairings and included both sympatric and allopatric isolate-isoline pairings17 (Table 1). Three biological replicates of source plant tissue from each isolate-isoline pairing was included in this study. Up to five transmitting and non-transmitting thrips were selected per isolate-isoline pairing unless transmission rates were so low that five transmitting individuals were not observed in transmission experiments. The numbers of transmitting and non-transmitting thrips for each isolate-isoline pairing subjected to RT- qPCR are shown in Supplementary Table S1.
Thrips and Leaf Disc RNA Extraction
Total RNA was extracted from 105 individual T. tabaci using TRIzol reagent (ThermoFisher Scientific, Waltham, Massachusetts) following the protocol used by Mason et al.51 with some modifications. For homogenization, each individual thrips was placed in a 1.5 ml microfuge tube, flash frozen in liquid N2, and then homogenized with a motorized micropestle (Kimble Chase, Vineland, NJ) followed by the addition of TRIzol. All incubation steps were at room temperature and all centrifugation steps were done at 16,000 x g. The pellets were air dried for 15 minutes, instead of vacuum concentrated, and resuspended in 8 µl (Diethyl pyrocarbonate; DEPC) treated water (Amresco, Solon, OH).
Total RNA was extracted from Emilia sonchifolia source leaf tissue (20 mg) with TRIzol using the manufacturers’ protocol. Homogenization was done using three Pyrex solid glass beads (3 mm; Corning, Corning, NY) in a 1.5 ml tube containing leaf tissue, flash frozen in liquid N2, and shaken for 20 seconds in a Silamat S6 mixer (Ivoclar Vivadent, Amherst, NY). Total RNA was resuspended in 50 µl of dH2O.
To ensure the integrity of flash frozen samples, nucleic acid concentrations of flash-frozen tissue were compared to fresh tissue samples for both E. sonchifolia and T. tabaci. Following total RNA extraction sample quality was confirmed by running the RNA on a gel to inspect quality as well as a NanoDrop 1000 (ThermoFisher Scientific, Waltham, MA) to confirm the quantity. Once the RNA extraction method was confirmed, it was used on the actual experiment samples and confirmed on the NanoDrop 1000.
cDNA Synthesis
cDNA synthesis was required to convert total RNA to the DNA template required for RT-qPCR. First strand cDNA was synthesized from total RNA using ProtoScript II reverse transcriptase (New England Biolabs, Ipswich, MA). Thrips and leaf disc cDNA was synthesized using 2 µl of total RNA in a 20 µl reaction primed with 2 µl of random hexamers (60 µM; Invitrogen, Carlsbad, CA). The primers, 1 µl dNTP mix (10 mM), total RNA and dH2O were combined to a final volume of 12 µl, heated at 70 °C for 5 minutes and chilled immediately on ice before the remaining reagents were added (ProtoScript reaction buffer, DTT (10 mM final), Murine RNase inhibitor (2U/µl final) and ProtoScript II reverse transcriptase (20U/µl final). The prep was then incubated at 25 °C for 5 minutes followed by incubation at 42 °C for 1 hour. The enzyme was inactivated at 80 °C for 5 minutes and stored at −20 °C until used for RT-qPCR.
Quantitative Real-Time PCR (RT-qPCR) Primer design
There are no published RT-qPCR primers nor was there a complete genome sequence for T. tabaci. Previously reported F. occidentalis actin primers as well as other internal control genes were assayed as possible internal control genes for T. tabaci19,52,53 (Supplementary Table S2). Primers were also designed and validated for the commonly used internal control, Elongation factor one alpha (EF1A), from an alignment of 7 published partial T. tabaci EF1A sequences (Genbank accession numbers: KM582809, AB894111, AB277263, AB277262, AB894109, AB894108, AB277575) (Supplementary Table S3).
Because E. sonchifolia does not have published RT-qPCR primers or a complete genome sequence, and previously published RT-qPCR primers developed from the highly conserved common plant genes actin, tRNA and profilin19. An alignment of 3 partial E. sonchifolia sequences for the 5.8 S rRNA gene and internal transcribed spacer (ITS) were also utilized to design additional primer pairs (accession numbers: JF733772, KU696022, MF440623) (Supplementary Table S4).
TSWV quantification was validated using a number of published RT-qPCR primers specific to the nucleocapsid (N) gene of the small (S) RNA segment16,19,54,55. Primers were also designed spanning the L RNA segment using an alignment of 5 published sequences (Genbank accession numbers: NC_002052, JN664254, HM581940, HM581934, JF960237) as another possible measurement of TSWV transcript levels in both thrips and infected source leaf tissue (Supplementary Table S5).
RT-qPCR
Viral titer was quantified using the primer efficiency method, in which the target gene expression is quantified relative to the expression of an internal control while accounting for primer efficiency56,57. Primer efficiency values (E) were calculated by 10(−1/slope) where the slope of the standard curve was obtained by plotting the concentration of five, five-fold dilutions of a single cDNA reaction made from thrips with a high TSWV titer against their Ct values. TSWV transcript levels were normalized to the internal controls by the inverse ratio of Pfaffl57: Einternal control Ct (internal control) /ELCt (L).
RT-qPCR was performed on a QuantStudio 6 Flex system (Applied Biosystems, Foster City, CA) with a 96-well fast block. iTaq Universal SYBR Green Supermix, SsoAdvanced SYBR Green Supermix, and SsoFast EvaGreen Supermix (all Bio-Rad, Hercules, CA) were compared for their ability to detect amplification of DNA. SsoAdvanced SYBR Green Supermix was chosen because it had the most reproducible results. All samples were run in triplicate for each primer pair to control for pipetting errors, along with no-template controls (Table 2). Reactions (20 µl) were performed in 0.1 ml 96-well plates using the manufacturer’s recommended protocol: initial denaturation at 95 °C for 30 seconds, 40 cycles of denaturation at 95 °C for 15 seconds and annealing/extension at 60 °C for 30 seconds, followed by a single melt curve stage of 95 °C for 15 seconds, 60 °C for 60 seconds, and 95 °C for 15 seconds. The number of denaturation/annealing/extension cycles was reduced to 30 for the source leaf tissue due to the higher initial RNA concentrations obtained.
Statistical analysis
Virus titers were expressed as the normalized values for the relative TSWV transcript levels in the leaf tissue used as the virus source for acquisition by the thrips or in individual thrips. Titers in the leaf discs used as virus sources were log transformed (base 10) and subjected to one-way ANOVA to test for differences among isolate-isoline pairings.
Further analyses examining the relationships between virus titer in individual thrips, virus isolate, and thrips isoline were conducted using the GLIMMIX procedure of the SAS system version 9.4 (SAS Institute, Cary, NC). Normalized values for the relative TSWV transcript levels were analyzed using a generalized linear mixed model with an assumed lognormal response distribution. An initial analysis tested a model in which the independent variables were virus isolate, isoline and their interaction. This relationship was examined further in a second model in which the interaction term was replaced with the variable “collection location,” in which virus isolate and isoline pairings were grouped as sympatric or allopatric. Similar analyses were conducted to test the association between virus titer in thrips and probability of transmission. In these analyses, transmission was treated as a binary variable with individual thrips in our subsamples classified as transmitting or non-transmitting. We could not directly test for associations between titers and transmission efficiencies of specific isolates-isoline pairings because the numbers of transmitting thrips were too low for some pairings to ensure inclusion of a sufficient number of transmitting thrips in a random sample of available thrips. Therefore, the thrips samples from each isolate-isoline pairing subjected to RT-qPCR were chosen to ensure adequate representation of transmitting and non-transmitting thrips.
Supplementary information
Acknowledgements
This work was supported by USDA NIFA Coordinated Agricultural Project Grant 2012-68004-20166. The authors would like to acknowledge Dorith Rotenberg for her help with RT-qPCR, and Thomas Chappell for his help with statistics.
Author contributions
Research conception, design of experiments was done by A.L.J. and G.G.K.; development of methodologies were done by J.A.L. and T.L.S.; J.A.L. and A.L.J. performed the experiments. Data curation, statistical analysis, and figure preparations were conducted by J.A.L., A.L.J. and G.G.K., The following authors: J.A.L., A.L.J., G.G.K. and T.L.S. contributed to writing, editing, and review of manuscript. Funding acquisition, supervision and project administration were provided by G.G.K. and T.L.S.
Data availability
The datasets generated or analyzed and not included in the manuscript (and its Supplemental Information files) are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-64507-1.
References
- 1.Gray SM, Banerjee N. Mechanisms of arthropod transmission of plant and animal viruses. MMBR. 1999;63:128–148. doi: 10.1128/mmbr.63.1.128-148.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agarwal A, Parida M, Dash PK. Impact of transmission cycles and vector competence on global expansion and emergence of arboviruses. Rev. Med. Virol. 2017;27:e1941. doi: 10.1002/rmv.1941. [DOI] [PubMed] [Google Scholar]
- 3.Stumpf CF, Kennedy GG. Effects of tomato spotted wilt virus (TSWV) isolates, host plants, and temperature on survival, size, and development time of Frankliniella fusca. Entomol. Exp. Appl. 2005;114:215–225. [Google Scholar]
- 4.Stumpf CF, Kennedy GG. Effects of tomato spotted wilt virus isolates, host plants, and temperature on survival, size, and development time of Frankliniella occidentalis. Entomol. Exp. Appl. 2007;123:139–147. [Google Scholar]
- 5.Hogenhout, S. A., Ammar, E. -D., Whitfield, A. E., & Redinbaugh, M. G. Insect Vector Interactions with Persistently Transmitted Viruses. Ann. Rev. Entomol.46, 327–359 (2008). [DOI] [PubMed]
- 6.Ammar E-D, Gingery RE, Madden L. Transmission efficiency of three isolates of maize stripe tenuivirus in relation to virus titre in the planthopper vector. Plant Pathol. 1995;44:239–243. [Google Scholar]
- 7.Ammar E-D. Effect of European wheat striate mosaic, acquired transovarially, on the biology of its planthopper vector Javesella pellucida. Ann. Appl. Biol. 1975;79:203–213. [Google Scholar]
- 8.Gill CC. Cyclical transmissibility of barley yellow dwarf virus from oats with increasing age of infection. Phytopathol. 1969;59:23–28. [Google Scholar]
- 9.Gray SM, Power AG, Smith DM, Seaman AJ, Altman NS. Aphid transmission of barley yellow dwarf virus: acquisition access periods and virus concentration requirements. Phytopathol. 1991;81:539–545. [Google Scholar]
- 10.Pereira A-M, Lister RM, Barbara PJ, Shaner GE. Relative transmissibility of barley yellow dwarf virus from sources with differing virus contents. Phytopathol. 1989;79:1353–1358. [Google Scholar]
- 11.Lett J-M, et al. Spatial and temporal distribution of geminiviruses in leafhoppers of the genus Cicadulina monitored by conventional and quantitative polymerase chain reaction. Phytopathol. 2002;92:65–74. doi: 10.1094/PHYTO.2002.92.1.65. [DOI] [PubMed] [Google Scholar]
- 12.Whitfield AE, Falk BW, Rotenberg D. Insect vector-mediated transmission of plant viruses. Virol. 2015;479:278–289. doi: 10.1016/j.virol.2015.03.026. [DOI] [PubMed] [Google Scholar]
- 13.Inoue T, Sakurai T, Murai T, Maeda T. Specificity of accumulation and transmission of tomato spotted wilt virus (TSWV) in two genera, Frankliniella and Thrips (Thysanoptera: Thripidae) Bull. of Entomol. Res. 2004;94:501–507. doi: 10.1079/ber2004326. [DOI] [PubMed] [Google Scholar]
- 14.Nagata T, Almeida ACL, Resende RO, DeÁvila AC. The competence of four thrips species to transmit and replicate four tospoviruses. Plant Pathol. 2004;53:136–140. [Google Scholar]
- 15.Okazaki O, et al. The effect of virus titre on acquisition efficiency of Tomato spotted wilt virus by Frankliniella occidentalis and the effect of temperature on detectable period of the virus in dead bodies. Australasian. Plant Pathol. 2011;40:120–125. [Google Scholar]
- 16.Rotenberg D, et al. Variation in Tomato spotted wilt virus titer in Frankliniella occidentalis and its association with frequency of transmission. Phytopathol. 2009;99:404–10. doi: 10.1094/PHYTO-99-4-0404. [DOI] [PubMed] [Google Scholar]
- 17.Jacobson AL, Kennedy GG. Specific insect-virus interactions are responsible for variation in competency of different Thrips tabaci isolines to transmit different tomato spotted wilt virus isolates. PLoS ONE. 2013;8:e54567. doi: 10.1371/journal.pone.0054567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morgan AD, Gandon S, Buckling A. The effect of migration on local adaptation in a coevolving host-parasite system. Nature. 2005;437:253–256. doi: 10.1038/nature03913. [DOI] [PubMed] [Google Scholar]
- 19.Boonham N, et al. The detection of Tomato spotted wilt virus (TSWV) in individual thrips using real time fluorescent RT-PCR (Taqman) J. Virol. Meth. 2002;101:37–48. doi: 10.1016/s0166-0934(01)00418-9. [DOI] [PubMed] [Google Scholar]
- 20.Gandon S, Michalakis Y. Local adaptation, evolutionary potential and host-parasite coevolution: interactions between migration, mutation, population size and generation time. J. Evol. Biol. 2002;15:451–462. [Google Scholar]
- 21.Jacobson AL, Booth W, Vargo EL, Kennedy GG. Thrips tabaci population genetic structure and polyploidy in relation to competency as a vector of tomato spotted wilt virus. PLoS ONE. 2013;8:e54484. doi: 10.1371/journal.pone.0054484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cabrera-La Rosa JC, Kennedy GG. Thrips tabaci and tomato spotted wilt virus: inheritance of vector competence. Entomol. Expt. et Appl. 2007;124:161–166. [Google Scholar]
- 23.Halaweh N, Poehling HM. Inheritance of vector competence by the thrips Ceratothripoides claratris (Shumsher) (Thysanoptera: Thripidae) J. Appl. Entomol. 2009;133:386–393. [Google Scholar]
- 24.Ogada PA, Debener T, Poehling HM. Inheritance genetics of the trait vector competence in Frankliniella occidentalis (western flower thrips) in the transmission of tomato spotted wilt virus. Ecol. & Evol. 2016;6:7911–7920. doi: 10.1002/ece3.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rotenberg D, Jacobson AL, Schneweis DJ, Whitfield AE. Thrips transmission of tospoviruses. Curr. Opin. Virol. 2015;15:80–89. doi: 10.1016/j.coviro.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 26.Oliver JE, Whitfield AE. The genus Tospovirus: Emerging Bunyaviruses that threaten food security. Annu. Rev. Virol. 2016;3:101–24. doi: 10.1146/annurev-virology-100114-055036. [DOI] [PubMed] [Google Scholar]
- 27.Ullman DE, German T, Sherwood JL, Wescot DM, Cantone FA. Immunocytochemical evidence that the nonstructural protein encoded by the S RNA of tomato spotted wilt tospovirus is present in thrips vector cells. Phytopathol. 1993;83:456–463. [Google Scholar]
- 28.Ullman DE, Wescot DM, Chenaut KD, Sherwood JL, German T. Compartmentalization, intracellular transport, and autophagy of tomato spotted wilt tospovirus proteins in infected thrips cells. Phytopathol. 1995;85:644–654. [Google Scholar]
- 29.Sin SH, McNulty BC, Kennedy GG, Moyer JW. Viral genetic determinants for thrips transmission of Tomato spotted wilt virus. PNAS. 2005;102:5168–5173. doi: 10.1073/pnas.0407354102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Margaria P, Bosco L, Vallino M. The NSs protein of Tomato spotted wilt virus is required for persistent infection and transmission by Frankliniella occidentalis. J. Virol. 2014;88:5788–5802. doi: 10.1128/JVI.00079-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montero-Astúa, M. et al. Disruption of vector transmission by a plant-expressed viral glycoprotein. MPMI. 27(3), 296–304 (2014). [DOI] [PubMed]
- 32.Montero-Astúa M, Ullman DE, Whitfield AE. Salivary gland morphology, tissue tropism and the progression of tospovirus infection in Frankliniella occidentalis. Virol. 2016;493:39–51. doi: 10.1016/j.virol.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 33.Nagata. T, Inoue-Nagata AK, Smid HM, Goldbach R, Peters D. Tissue tropism related to vector competence of Frankliniella occidentalis for tomato spotted wilt tospovirus. J. Gen. Virol. 1999;80:507–515. doi: 10.1099/0022-1317-80-2-507. [DOI] [PubMed] [Google Scholar]
- 34.Kritzman A, et al. The route of tomato spotted wilt virus inside the thrips body in relation to transmission efficiency. Archives Virol. 2002;147:2143–2156. doi: 10.1007/s00705-002-0871-x. [DOI] [PubMed] [Google Scholar]
- 35.Whitfield AE, et al. A soluble form of the tomato spotted wilt virus (TSWV) glycoprotein GN (GN -S) inhibits transmission of TSWV by Frankliniella occidentalis. Phyopathol. 2008;98:45–50. doi: 10.1094/PHYTO-98-1-0045. [DOI] [PubMed] [Google Scholar]
- 36.Bandla MD, Campbell LR, Ullman DE, Sherwood JL. Interaction of tomato spotted wilt tospovirus (TSWV) glycoproteins with a thrips midgut protein, a potential cellular receptor for TSWV. Phytopathol. 1998;88(2):98–104. doi: 10.1094/PHYTO.1998.88.2.98. [DOI] [PubMed] [Google Scholar]
- 37.Kikkert M, et al. Binding of tomato Spotted wilt virus to a 94-kDa thrips protein. Phytopathol. 1998;88:63–69. doi: 10.1094/PHYTO.1998.88.1.63. [DOI] [PubMed] [Google Scholar]
- 38.Thomas RE, Wu WK, Verleye D, Rai KS. Midgut basal lamina thickness and dengue-1 virus dissemination rates in laboratory strains of Aedes albopictus (Diptera: Culicidae) J. Med. Entomol. 1993;30:326–331. doi: 10.1093/jmedent/30.2.326. [DOI] [PubMed] [Google Scholar]
- 39.Ullman DE, Cho JJ, Mau RFL, Westcot DM, Custer DM. A midgut barrier to Tomato spotted wilt virus acquisition by adult western flower thrips. Phytopathol. 1992;82:1333–1342. [Google Scholar]
- 40.de Assis Filho FM, Stavisky J, Reitz SR, Deom CM, Sherwood JL. Midgut infection by tomato spotted wilt virus and vector incompetence of Frankliniella tritici. J. Appl. Entomol. 2005;129:548–550. [Google Scholar]
- 41.Gildow FE, Grey SM. The aphid salivary-gland basal lamina as a selective barrier associated with vector-specific transmission of barley yellow dwarf luteoviruses. Phytopathol. 1993;83:1293–1302. [Google Scholar]
- 42.Peiffer ML, Gildow FE, Gray SM. Two distinct mechanisms regulate luteovirus transmission efficiency and specificity at the aphid salivary gland. J. Gen. Virol. 1997;78:595–503. doi: 10.1099/0022-1317-78-3-495. [DOI] [PubMed] [Google Scholar]
- 43.Gray S, Gildow FE. Luteovirus-aphid interactions. Annu. Rev. Phytapathol. 2003;41:539–566. doi: 10.1146/annurev.phyto.41.012203.105815. [DOI] [PubMed] [Google Scholar]
- 44.Wei J, et al. Specific cells in the primary salivary glands of the whitefly Bemisia tabaci control retention and transmission of Begomoviruses. J. Virol. 2014;88:13460–13468. doi: 10.1128/JVI.02179-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Medeiros RB, Resende RDO, Ávila CD. The plant virus tomato spotted wilt tospovirus activates the immune system of its main vector, Frankliniella occidentalis. J. Virol. 2004;78:4976–4982. doi: 10.1128/JVI.78.10.4976-4982.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schneweis DJ, Whitfield AE, Rotenberg D. Thrips developmental stage-specific transcriptome response to tomato spotted wilt virus during the virus infection cycle in Frankliniella occidentalis, the primary vector. Virol. 2017;500:226–237. doi: 10.1016/j.virol.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 47.Shrestha A, et al. Transcriptome changes associated with Tomato spotted wilt virus infection in various life stages of its thrips vector, Frankliniella fusca (Hinds) J. Gen. Virol. 2017;98:2156–2170. doi: 10.1099/jgv.0.000874. [DOI] [PubMed] [Google Scholar]
- 48.Stafford CA, Walker GP, Ullman DE. Infection with a plant virus modifies vector feeding behavior. PNAS. 2011;108:9350–9355. doi: 10.1073/pnas.1100773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kindt F, Joosten NN, Peters D, Tjallingii WF. Characterization of the feeding behavior of western flower thrips in terms of electrical penetration graph (EPG) waveforms. J. Insect Physiol. 2003;49:183–191. doi: 10.1016/s0022-1910(02)00255-x. [DOI] [PubMed] [Google Scholar]
- 50.Eigenbrode SD, Bosque-Pérez NA, Davis TS. Insect-borne pathogens and their vectors: ecology, evolution, and complex interactions. Annu. Rev. Entomol. 2018;63:169–191. doi: 10.1146/annurev-ento-020117-043119. [DOI] [PubMed] [Google Scholar]
- 51.Mason G, Roggero P, Tavella L. Detection of tomato spotted wilt virus in its vector Frankliniella occidentalis by reverse transcription-polymerase chain reaction. J. Virol. Meth. 2003;109:69–73. doi: 10.1016/s0166-0934(03)00048-x. [DOI] [PubMed] [Google Scholar]
- 52.Yang CC, et al. Validation of reference genes for gene expression studies in nonviruliferous and viruliferous Frankliniella occidentalis (Thysanoptera: Thripidae) PeerJ PrePrints. 2014;2:e662v1. [Google Scholar]
- 53.Zheng. YT, Li HB, Lu MX, Du YZ. Evaluation and validation of reference genes for qRT-PCR normalization in Frankliniella occidentalis (Thysanoptera: Thripidae) PLoS ONE. 2014;9:e111369. doi: 10.1371/journal.pone.0111369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roberts CA, Dietzgen RG, Heelan LA, Maclean DJ. Real-time RT-PCR fluorescent detection of tomato spotted wilt virus. J. Virol. Meth. 2000;88:1–8. doi: 10.1016/s0166-0934(00)00156-7. [DOI] [PubMed] [Google Scholar]
- 55.Mortimer-Jones SM, Jones MGK, Jones RAC, Thomson G, Dwyer GI. A single tube, quantitative real-time RT-PCR assay that detects four potato viruses simultaneously. J. Virol. Meth. 2009;161:289–296. doi: 10.1016/j.jviromet.2009.06.027. [DOI] [PubMed] [Google Scholar]
- 56.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nature Protocols. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 57.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated or analyzed and not included in the manuscript (and its Supplemental Information files) are available from the corresponding author on reasonable request.



