Abstract
Plant cell suspension culture systems are valuable for the study of complex biological systems such as inducible defence responses and aspects of plant innate immunity. Perturbations to the cellular metabolome can be investigated using metabolomic approaches in order to reveal the underlying metabolic mechanism of cellular responses. Lipopolysaccharides from the sorghum pathogen, Burkholderia andropogonis (LPSB.a.), were purified, chemically characterised and structurally elucidated. The lipid A moiety consists of tetra- and penta-acylated 1,4’-bis-phosphorylated disaccharide backbone decorated by aminoarabinose residues, while the O-polysaccharide chain consists of linear trisaccharide repeating units of [→2)-α-Rha3CMe-(1 → 3)-α-Rha-(1 → 3)-α-Rha-(1 → ]. The effect of LPSB.a. in triggering metabolic reprogramming in Sorghum bicolor cells were investigated using untargeted metabolomics with liquid chromatography coupled to mass spectrometry detection. Cells were treated with LPSB.a. and the metabolic changes monitored over a 30 h time period. Alterations in the levels of phytohormones (jasmonates, zeatins, traumatic-, azelaic- and abscisic acid), which marked the onset of defence responses and accumulation of defence-related metabolites, were observed. Phenylpropanoids and indole alkaloids as well as oxylipins that included di- and trihydroxyoctadecedienoic acids were identified as signatory biomarkers, with marked secretion into the extracellular milieu. The study demonstrated that sorghum cells recognise LPSB.a. as a ‘microbe-associated molecular pattern’, perturbing normal cellular homeostasis. The molecular features of the altered metabolome were associated with phytohormone-responsive metabolomic reconfiguration of primary and secondary metabolites originating from various metabolic pathways, in support of defence and immunity.
Subject terms: Metabolomics, Metabolomics, Plant immunity, Plant immunity
Introduction
Plants are continuously exposed to an array of pathogens which can either be host-specific or can affect a wide range of hosts. Existing crop protection strategies are vulnerable against novel pathogens or pathogens developing increasing resistance against chemical controls1. One of the major diseases affecting Sorghum bicolor, an economically important grain crop, is bacterial leaf stripe disease caused by Burkholderia andropogonis. Recent metabolomics insights revealed aspects of the molecular mechanisms of the S. bicolor–B. andropogonis pathophysiological interaction2. The results revealed a dynamic and intricate network of the sorghum defence resources towards B. andropogonis in launching a heightened defensive capability for disease suppression2.
Plants employ several layers of defence to counter potential pathogens3,4. These passive (preformed) and active (induced) responses are triggered upon detection of conserved immunogenic motifs, epitopes or ‘patterns’ such as bacterial lipopolysaccharides (LPS), flagellin and fungal ergosterol, β-glucans and chitin5. These ‘microbe/pathogen-associated molecular patterns, M/PAMPs’ are perceived via surface-located ‘pattern-recognition receptors, PRRs’ to initiate signal transduction events linked to MAMP-triggered immunity or MTI6 that can be acquired in a systemic manner, contributing to broad-spectrum and durable resistance1.
LPS, an amphiphilic lipoglycan located in the outermost membrane of Gram-negative bacteria, provides a protective barrier against environmental stresses and is crucial for survival of the bacteria7,8. LPSs lies at the interface of the interaction of host plants with potential pathogens, and although structurally conserved, LPSs may differ within and across species to such an extent that a bacterial cell may contain varying LPS chemotypes9.
Generally, LPS has a tripartite structure, consisting of functionally distinct components; the O-polysaccharide (OPS) chain and the core oligosaccharide (COS) which is covalently attached to the lipid A (LA) moiety. The OPS-chain is the hydrophilic outermost component of LPS and exposed to the environment10,11. It is built up of repeating units of oligosaccharides and displays variation across Gram-negative bacterial species. The OPS acts in protection of the bacterial cells towards hostile surroundings as found in planta and is involved in microbial adhesion to host cells12. Bacteria possessing an OPS chain are termed smooth-type LPS (S-LPS) whereas the ones lacking it are rough-type LPS (R-LPS). The COS is located between the OPS-chain and LA moieties, and connected to the LA via 3-deoxy-D-manno-oct-2-ulosonic acid (Kdo). The LA domain is generally a bis-phosphorylated glucosamine disaccharide, varyingly acylated by fatty acids of various lengths embedded in the bacterial outer membrane13,14. The LA domain is highly conserved and contributes to LPS stability. However, environmental and growth conditions may alter its structure, thus affecting biological activity of the whole LPS molecule9,15.
In the case of plant pathogenic bacteria, knowledge about the structural features of LPSs contributing to M/PAMP activity is minimal6,8,16,17. Factors such as adaptation to different habitats and lifestyles, may lead to dynamic remodulation of the LPS structural features, thus contributing to size- and compositional heterogeneity. Differences in LPS structures can also lead to varying results as they may possibly be sensed by different receptors4,6,8. While some information about sensing of LA in Arabidopsis thaliana has been reported, mechanistic aspects of sensing of the OPS and COS remains unknown8 (Supplementary file 1).
In the current study, LPS isolated from B. andropogonis (LPSB.a.), was purified and the structures of the LA and OPS moieties elucidated. As a putative M/PAMP, the effect of LPSB.a. on triggering metabolic reprogramming and the extent to which this occurs, were investigated using a cell suspension culture system18,19. Here, sorghum cells were used in combination with a liquid chromatography-mass spectrometry platform for metabolite analysis. The metabolome reflects the ultimate changes in gene expression and, considering that it is highly responsive to changes in enzyme activity and metabolic fluxes2,20–23, measurement of dynamic changes of metabolites would reflect on differential and functional features of defence metabolism triggered by LPS perception. Such results would point to metabolic pathways involved in the antimicrobial defences of sorghum, thus contributing to increasing attempts to unravel the biochemical and molecular mechanisms involved in plant-microbe interactions, insights into plant innate immune responses and host-responses to bacterial infection.
Results
Compositional and structural analysis of LPS from Burkholderia andropogonis
LPSB.a. consists of a hydrophilic hetero-polysaccharide OPS covalently linked through a COS to the LA of Mr approximately 1.5 kDa. An LPS-specific SDS-PAGE analysis of purified LPSB.a. is shown in Fig. 1, where the LPS displays a characteristic ladder-like pattern indicating the occurrence of a different number of oligosaccharide repeating units that forms the extended OPS.
Figure 1.
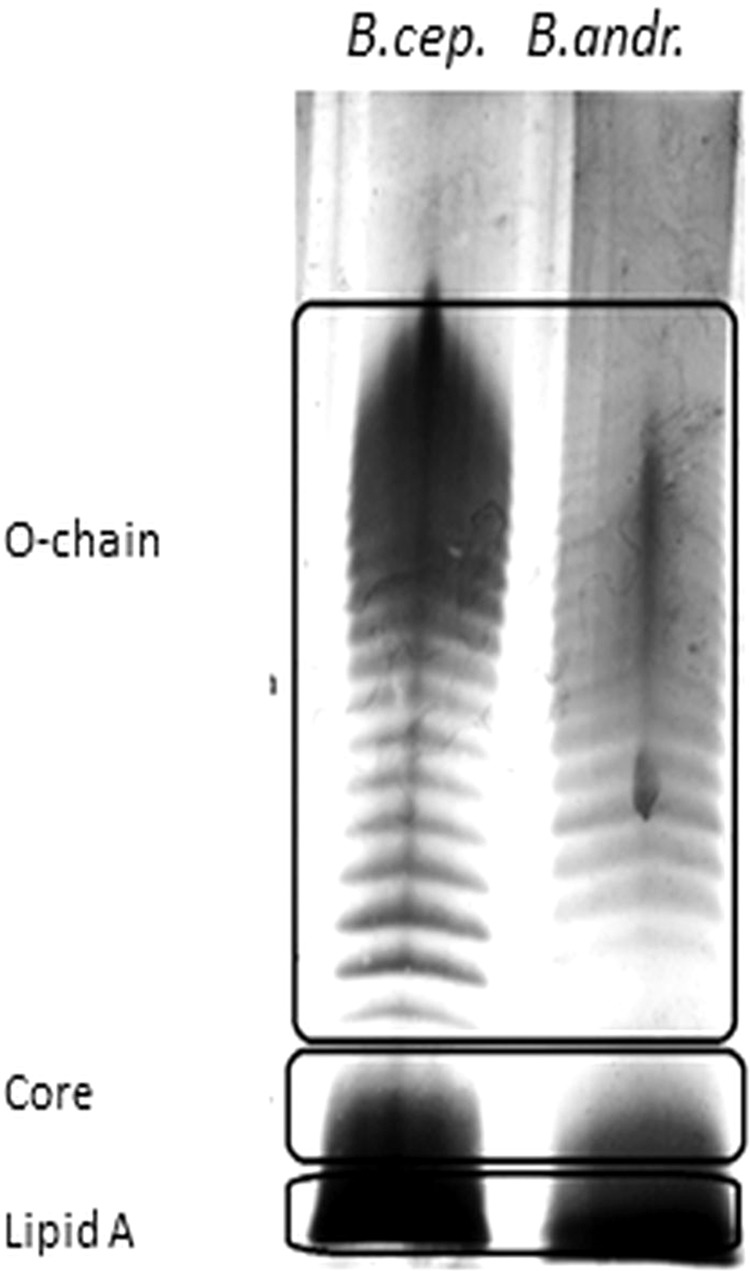
LPS-specific SDS-PAGE analysis of purified LPSB.a. LPS isolated from Burkholderia andropogonis (last lane) is compared to Burkholderia cepacia LPS (first lane). LPSB.cep. was used as a reference as it has been well-characterised11. The indicated regions show the three LPS components from the two Burkholderia species.
Compositional analysis of the sugars revealed mainly L-rhamnose (L-Rha) with minor amounts of 2,6-dideoxy-2-amino-D-glucose (D-QuiN), 4-amino-4-deoxy-L-arabinose (L-Ara4N), D-mannose (D-Man), D-galactose (D-Gal), D-glucose (D-Glc), 2-amino-2-deoxy-D-glucose (D-GlcN), L-glycero-D-manno-heptose (L,D-Hep), 3-deoxy-D-manno-oct-2-ulopyranosonic acid (D-Kdo) and D-glycero-D-talo-oct-2-ulopyranosonic acid (D-Ko). Linkage analysis highlighted the presence of 3-substituted L-Rha and the 2-substituted rhamnose derivative, 3-C-methyl-rhamnose (Rha3CMe). The absolute configuration of Rha3CMe remains to be determined.
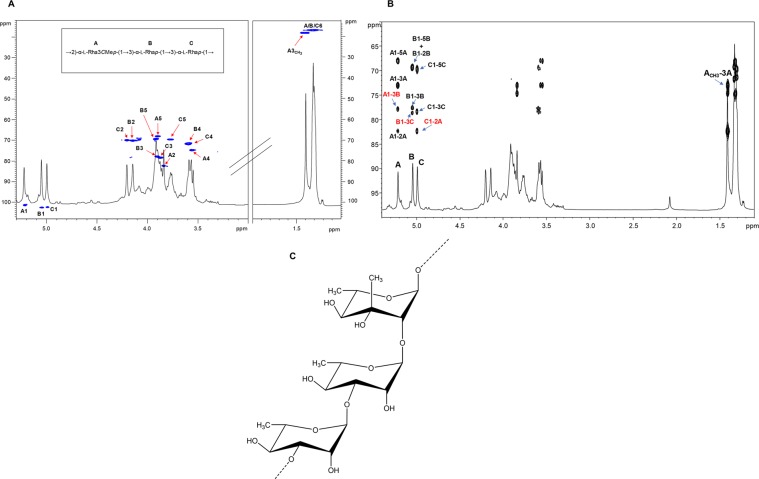
To define the OPS moiety of the LPSB.a., a complete set of 1D and 2D NMR spectroscopy experiments was executed (DQF-COSY, TOCSY, ROESY, NOESY, 1H,13C HSQC, 1H,13C HSQC-TOCSY, and 1H,13C HMBC), revealing a polysaccharide made up of the trisaccharide repeating unit [→2)-α-Rha3CMe-(1 → 3)-α-Rha-(1 → 3)-α-Rha-(1 → ]. The 1H NMR spectrum for the OPS moiety revealed three main anomeric proton signals detected at δH 5.21, 5.04, and 4.99 ppm relative to three different spin systems and referred to here as A, B, and C (Table S1). The intense methyl signals at δH 1.30, 1.31 ppm and the correlations in the total correlation spectroscopy (TOCSY) spectrum (Fig. S1) agreed with the presence of rhamnose units, as described above, in the sugar compositional analysis on intact LPSB.a. All sugar units were present as pyranose rings, according to their 13C chemical shift values (Table S1, Fig. 2A) and the long-range correlations between C-1/H-1 and H-5/C-5 in the 13C,1H HMBC spectrum (Fig. 2B, Table S2). The observation of a further signal a δH 1.40 (δC 18.3) corroborated the linkage analysis data relative to the presence of a sugar unit carrying an additional methyl group. Residues A (H-1 at 5.21 ppm), B (H-1 at 5.04 ppm), and C (H-1 at 4.99 ppm) were all identified as α-rhamnose units as demonstrated by the correlations in the TOCSY spectrum, with the methyl group signal resonating at δH 1.30/1.31 ppm, whereas the α-anomeric configuration was defined on the basis of the 1JC,H values and the intra-residual NOE contacts of H-1 with H-2 of each spin system. Finally, the manno configuration was confirmed by evaluation of 3JH,H coupling constant values10. Residue A, attributed to the Rha3CMe (evalose), was distinguished from the Rha residues B and C by the observation of HMBC correlations of the CH3-O group with C-3 at 73.0 ppm (Fig. 2B) as well as by the NOE correlations observed between the methyl group at 1.40 ppm with H-2 and H-5 of the same residue A, but not with H-4. This also proved that the methyl group was axial, and consequently that the -OH group at C-3 was equatorial (Fig. 2C).
Figure 2.
Structural characterisation of the O-polysaccharide (OPS) chain moiety from LPSB.a. (A) Zoom of the overlapped 1H and 1H, 13C HSQC spectra of the OPS from LPSB.a. after mild acid treatment. The heteronuclear correlations are indicated. Numbering of sugar residues is as reported in Table S1. (B) Zoom of the overlapped 1H, 1H, 13C HMBC spectra. The key inter-residual long-range correlations involving sugar moieties (A–C, Table S2) are indicated in red, whereas the intra-residual long-range contacts are reported in black; letters are as in Table S1. The one-bond heteronuclear correlation involving the C-methyl group of residue A is also reported in the spectrum. (C) The structure of the elucidated trisaccharide repeating unit of the LPSB.a. OPS moiety.
The down-field shifts of carbon signals identified glycosylated positions at O-2 of A, and O-3 of B and C, in full accordance with the linkage analysis data (Table S1, Fig. 2). The primary sugar sequence composing the OPS repeating unit was determined by analysis of NOESY, ROESY (not shown) and 1H, 13C HMBC (Fig. 2B) spectra. Briefly, the anomeric signal of residue A showed an NOE contact with H-3 of unit B, thus indicating a (1 → 3) linkage between A and B, also confirmed by long-range correlation of H-1 A with C-3 of B (Fig. 2B). An inter-residue NOE correlation was detected between H-1 B and H-3 of C, whose anomeric proton signal, in turn, presented an NOE contact with H-2 of the Rha3CMe A. The long-range correlations (Fig. 2B) observed between the anomeric proton of B and C-3 of C and between H-1 C and C-2 of A further confirmed the linkages (B1 → 3 C) and (C1 → 2 A). In conclusion, by merging NMR and linkage analysis data it was possible to define the complete OPS structure of the LPSB.a. as a linear trisaccharide repeating unit built up of α−3-C-methyl rhamnose spaced by two α-rhamnoses (Fig. 2C).
Investigation of the fatty acids content of the LA revealed the presence of (R)−3-hydroxyhexadecanoic acid (C16:0 (3-OH)) with an amide linkage, and of (R)−3-hydroxytetradecanoic (C14:0 (3-OH)) and tetradecanoic acid (C14:0) with an ester linkage. The overall chemical composition matched those of the prototype of Burkholderia S-LPSs/R-LPSs. Accordingly, the MALDI MS investigation of the LA component, obtained after mild acid hydrolysis of the pure LPSB.a., showed the occurrence of a mixture of mono- and bis-phosphorylated LA species (tetra- and penta-acylated) bound by one or two L-Ara4N residues (Fig. S2, Table S3) which is consistent with previously elucidated LA structures from the genus Burkholderia14,24.
Non-targeted metabolic profiling of LPSB.a.-treated cultured sorghum cells
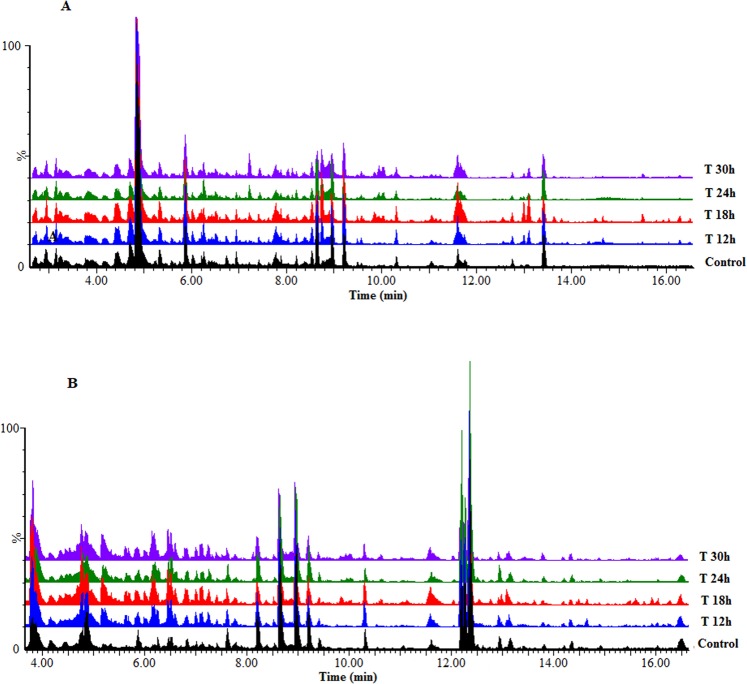
Metabolomics deals with the documentation and assessment of altered metabolome states and here ultra-high performance liquid chromatography – electrospray ionisation – quadrupole time-of-flight –high-definition mass spectrometry (UHPLC-ESI-QTOF-HDMS) was employed for the analysis of the endo- and exometabolomes of sorghum cells responding to LPSB.a. elicitation. Differences with regard to peak population and intensities across treated and non-treated cell extracts were visualised on MS chromatograms obtained from cell extracts (Fig. 3) and growth medium (Fig. S3). Thus, base peak intensity (BPI) chromatograms of these extracts displayed treatment- and time-related metabolic responses. The differences between control vs. treated samples, and those across the time points subsequently provided a visual picture of metabolic changes occurring due to the LPSB.a. treatment as a function of time.
Figure 3.
UHPLC-MS BPI chromatograms of methanolic intracellular extracts of sorghum cells treated with LPSB.a. (A): ESI(−) and (B): ESI( + ). Representative chromatograms of a control (non-treated 0 h) vs. treated samples (12–30 h) display variation related to treatment- and time-related metabolic changes occurring in the cells due to LPS treatment.
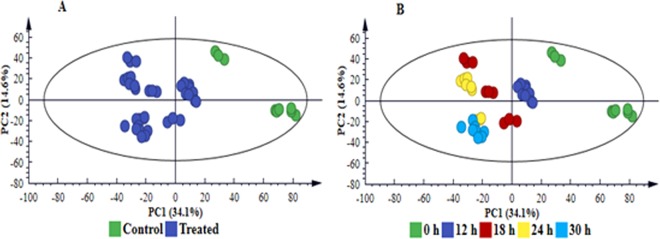
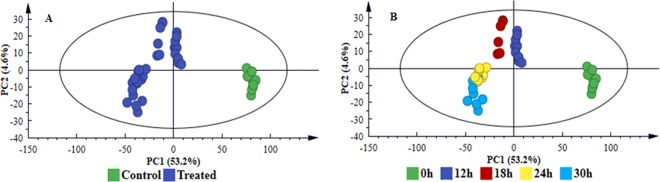
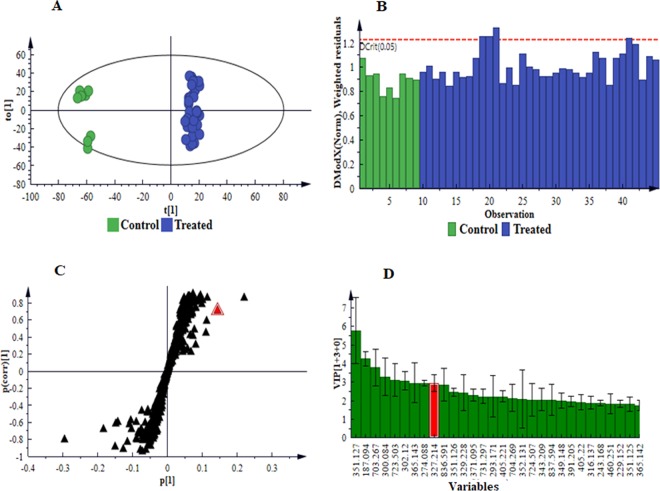
Multivariate statistical analyses (both unsupervised and supervised) were employed to mine the data and further investigate the observed treatment- and time-related differences. Thorough model validation was consistently applied as described in the experimental section; and only statistically valid models were retained and used. The unsupervised modelling, principal component analysis (PCA), permitted dimension reduction of the data and recognition of groupings, trends and outliers25,26. The computed PCA scores plot of cell extracts (Fig. 4 and S4) and medium extracts (Fig. 5 and S5), displayed a distinct separation between control and treated samples, revealing treatment-related sample clustering. Colour-coding based on time points showed a distinct sequential time clustering trend of samples, i.e. a time-related clustering for both extracts from cells (Fig. 4B and S4B) and medium (Fig. 5B and S5B). The clustering of samples highlighted in the PCA scores space, points to differential metabolic changes in cultured sorghum cells in response to LPSB.a. treatment.
Figure 4.
PC analyses of the LC-MS ESI(−) data for intracellular sorghum cell extracts. The 4-component model explains 61.9% variation in Pareto-scaled data, X, and the amount of predicted variation by the model, according to 7-fold cross-validation, is 51.4%. The 7-fold CV procedure is described in the experimental section. (A) Clusters coloured based on condition i.e. non-treated vs. treated shows clear separation between treated and control (non-treated, 0 h) samples. (B) is the same scores plot but coloured according to time and showing a clear sequential time trend clustering (from C0 h and T12–30 h). The related model for ESI( + ) data is supplied as Fig. S4, n = 9.
Figure 5.
PC analyses of the LC-MS ESI(−) data for extracellular sorghum cell extracts. The 3-component model explains 61.5% variation in Pareto-scaled data, X, and the amount of predicted variation by the model, according to 7-fold cross-validation, is 52.8%. The first 2 PCs were used to generate the above scores plot of all data. Clusters are coloured based on condition i.e. non-treated vs. treated and show clear separation between treated and control (non-treated, 0 h) samples (A) and according to time (B). The related model for ESI( + ) data is supplied as Fig. S5, n = 9.
For further characterisation and interpretation of the different clustering depicted in PC analyses, a supervised method, orthogonal projection to latent structures analysis (OPLS-DA), was used. The supervised modelling allowed identification of signatory biomarkers underlying the discrimination between the sample classes associated with the LPS treatment. Computed OPLS-DA scores plots showed clear discrimination between the control and treated samples (Fig. 6A and S6A-S8A). Evaluation of the goodness-of-fit (R2X(cum)), proportion of variance of the response variable explained by the model (R2Y(cum)) and predictive ability (Q2(cum)) parameters, indicated that the models were statistically reliable. Further validation also revealed the reliability of the models i.e. CV-ANOVA p-value of ≤ 0.0527,28. In addition, ‘distance to the model in space X’ (DModX) plots (Fig. 6B and S6B-S8B) for the generated OPLS-DA scores plots were used to assess moderate outliers.
Figure 6.
Supervised multivariate analyses of the LC-MS ESI(−) data for intracellular extracts. (A) Grouping of control (C 0 h) vs. treated (all time points combined) as indicated by an OPLS-DA score plot, n = 9 for each time point. This model comprises 1 predictive component and 3 orthogonal components (R2X = 60.0%, R2Y = 99.2% and Q2 = 95.1%). (B) A distance to the model in space X (DModX) plot to detect and evaluate moderate outliers (above the dashed red line, Dcrit). (C) An OPLS-DA loadings S-plot displaying the discriminating features (ions) that explain the clustering (sample grouping) observed in the OPLS-DA scores plot, with the features in the top right quadrant positively correlated to the treatment and those in the bottom left quadrant negatively correlated to the treatment. For instance, the selected m/z 327.2135 (annotated as trihydroxy-octadecadienoic acid II, Table 1) is positively correlated to the treatment. (D) A VIP plot summarising the importance of some of the variables in the projection of the model with the m/z values and jackknife confidence intervals reflecting the variable stability. A VIP value >1 indicates a significant variable in the complex analysis in comparing the difference between groups. For instance, the m/z 327.2135 (displayed as 327.214), selected from S-plot (C), shows a VIP score > 3, indicative of its importance to the model (contributing to the class separation). The equivalent set of graphs for ESI( + ) data is presented as Fig. S6, and in the case of extracellular extracts as Figs. S7: ESI(−) and S8: ESI( + ).
The OPLS-DA loadings S-plots (Fig. 6C and S6C-S8C) assisted in visualisation of the covariance and correlation between variables of modelled classes, and permitted the extraction of statistically significant features within the │p[1]│ ≥ 0.05 and │p(corr)│ ≥ 0.5 defined regions, responsible for discrimination between control and LPSB.a.-treated samples. The significance of the extracted variables towards discrimination of samples was assessed using the ‘variable importance in projection’ (VIP) plots (Fig. 6D and S6D-S8D). Only variables with a VIP score> 127,29 were considered significant and selected for further annotation.
Variables relating to the observed metabolic changes due to LPSB.a. elicitation, selected and validated with the aid of chemometrics tools mentioned above, were putatively identified (at Metabolomics Standards Initiative (MSI) level 2)30. Metabolites shown in Table 1 were annotated from both LC-MS ESI( − / + ) data, and had a VIP score > 1. Fold changes presented in Table 1 were obtained from the computed model of C 0 h vs. T 18 h, as this was the best time point representation of the overall metabolic changes. Equivalent tables expressing the fold changes and p-values generated from the computed OPLS-DA models of other time points (i.e. C 0 h vs. 12-, 24- and 30 h), for both intra- and extracellular data, are included in the supporting information data file Tables S4 and S5, respectively.
Table 1.
Annotated discriminatory metabolites from cell (intracellular) and medium (extracellular) extracts of Sorghum bicolor cultured cells treated with LPSB.a. All the metabolites had a VIP score> 1. Fold changes were obtained from OPLS-DA models of control (C0 h) vs. treated 18 h. (Similar data from 12-, 24- and 30 h time points is presented as Supplementary Information, Tables S4 and S5).
| Metabolites | m/z | Rt (min) | Adduct | Ion mode | Formula | Intracellular | Extracellular | Class | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p-value | Fold change | Trend | p-value | Fold change | Trend | |||||||
| L-Phenylalanine | 164.0686 | 1.84 | [M-H]- | neg | C9H11NO2 | 6.28E-06 | 1.4 | Increase | 3.07E-11 | 2.7 | Increase | Amino acid |
| L-Tryptophan | 203.0798 | 2.78 | [M-H]- | neg | C11H12N2O2 | 2.04E-06 | 1.4 | Increase | 0.001 | 1.4 | Increase | Amino acid |
| 16-Hydroxypalmitate | 273.2553 | 13.65 | [M + H]+ | pos | C16H32O3 | 0.490 | 1.3 | Increase | • | • | • | Fatty acid |
| 15-Hydroxylinoleic acid | 295.2253 | 14.29 | [M-H]- | neg | C18H32O3 | 0.001 | 2.1 | Increase | • | • | • | Fatty acid |
| Dihydroxy-octadecadienoic acid | 311.2242 | 11.79 | [M-H]- | neg | C18H32O4 | 1.28E-10 | 10.8 | Increase | 0.110 | 5.2 | Increase | Fatty acid |
| 9,10-Dihydroxy-12-octadecenoic acid | 313.2354 | 12.67 | [M-H]- | neg | C18H34O4 | 0.606 | 1.6 | Increase | • | • | • | Fatty acid |
| 9,10-Dihydroxystearic acid | 315.2511 | 13.51 | [M-H]- | neg | C18H36O4 | 2.37E-09 | 6.4 | Increase | • | • | • | Fatty acid |
| Trihydroxy-octadecadienoic acid І | 327.2149 | 9.72 | [M-H]- | neg | C18H32O5 | 0.377 | 113 | Increase | • | • | • | Fatty acid |
| Trihydroxy-octadecadienoic acid ІІ | 327.2135 | 11.05 | [M-H]- | neg | C18H32O5 | 3.97E-13 | 356.7 | Increase | 0.000 | 38.8 | Increase | Fatty acid |
| 9,12,13-Trihydroxy-10-octadecenoic acid | 329.2327 | 9.60 | [M-H]- | neg | C18H34O5 | 0.000 | 1.8 | Increase | 1.13E-08 | 9.2 | Increase | Fatty acid |
| Sophoraflavanone G | 423.1821 | 4.42 | [M-H]- | neg | C25H28O6 | 2.36E-05 | 0.8 | Decrease | 0.030 | 1.1 | Increase | Flavonoid |
| Apigenin-8-C-glucoside (vitexin) | 431.0974 | 5.58 | [M-H]- | neg | C21H20O10 | 0.702 | 1.1 | Increase | 0.537 | 1.2 | Increase | Flavonoid |
| Apigenin-6-C-xyloside-8-C-glucoside (vicenin-1) | 565.1545 | 4.94 | [M + H]+ | pos | C26H28O14 | 0.064 | 0.6 | Decrease | 0.004 | 1.4 | Increase | Flavonoid |
| Apigenin-6,8-di-C-glucoside(vicenin-2) | 595.1687 | 4.77 | [M + H]+ | pos | C27H30O15 | 0.082 | 0.4 | Decrease | 0.240 | 2.1 | Increase | Flavonoid |
| Apigenin 7,4’-dimethyl ether | 316.1157 | 8.29 | [M + H_NH3]+ | pos | C17H14O5 | 0.000 | 0.5 | Decrease | • | • | • | Flavonoid |
| 3’,4’5-Trihydroxy-3,7-dimethoxyflavone | 367.0221 | 3.90 | [M-H]- | neg | C17H20O9 | 0.032 | 0.8 | Decrease | • | • | • | Flavonoid |
| 4-Coumaroyl-3-hydroxyagmatine | 291.1471 | 5.72 | [M-H]- | neg | C14H20N4O3 | 0.013 | 0.7 | Decrease | 0.001 | 2.5 | Increase | HCA |
| 4-Coumaroylquinic acid | 337.1474 | 1.77 | [M-H]- | neg | C16H18O8 | 7.19E-09 | 0.6 | Decrease | 0.010 | 1.1 | Increase | HCA |
| Cinnamoylserotonin | 351.1251 | 2.43 | [M-H_HCOOH]- | neg | C19H18N2O2 | 7.77E-05 | 0.8 | Decrease | 0.000 | 1.2 | Increase | HCA |
| Feruloylserotonin | 351.1266 | 2.86 | [M-H]- | neg | C20H20N2O4 | 0.387 | 0.4 | Decrease | 0.771 | 1.2 | Increase | HCA |
| Sinapaldehyde glucoside | 369.1199 | 3.61 | [M-H]- | neg | C17H22O9 | 0.525 | 0.9 | Decrease | 1.68E-06 | 2.7 | Increase | HCA |
| 1-O-Coumaroyl-beta-D-glucose | 371.0957 | 4.94 | [M-H_HCOOH]- | neg | C15H18O8 | 5.26E-11 | 0.6 | Decrease | 7.70E-06 | 2.1 | Increase | HCA |
| Sinapoyl alcohol | 209.0764 | 6.72 | [M-H]- | neg | C11H14O4 | 1.55E-10 | 4.8 | Increase | 1.34E-05 | 1.8 | Increase | HCA |
| Dihydroconiferyl alcohol glucoside | 413.1422 | 3.27 | [M + H_HCOONa]+ | pos | C16H24O8 | 0.005 | 0.6 | Decrease | • | • | • | HCA |
| Indole-3-butyric acid | 272.0893 | 2.84 | [M + H_HCOONa]+ | pos | C12H13NO2 | 1.62E-05 | 0.5 | Decrease | • | • | • | Indole compound |
| N(6)-[(Indol-3-yl)acetyl]-L-lysine | 304.1667 | 4.20 | [M + H]+ | pos | C16H21N3O3 | 1.43E-07 | 5.8 | Increase | • | • | • | Indole compound |
| Indole-3-acetyl-myo-inositol | 353.1348 | 2.44 | [M-H_NH3]- | neg | C16H19NO7 | 0.479 | 1.8 | Increase | 0.000 | 2.9 | Increase | Indole compound |
| Indole-3-acetyl-beta-1-D-glucoside | 382.1121 | 3.93 | [M-H_HCOOH]- | neg | C16H19NO7 | 0.075 | 0.7 | Decrease | 0.925 | 1.0 | Increase | Indole compound |
| 6-Hydroxy-indole-3-acetyl-valine | 291.1294 | 3.89 | [M + H]+ | pos | C15H17N2O4 | 0.174 | 0.5 | Decrease | 0.098 | 2.1 | Increase | Indole compound |
| Traumatic acid | 297.1291 | 3.90 | [M + H_HCOONa]+ | pos | C12H20O4 | 0.027 | 4.4 | Increase | • | • | • | Phytohormone |
| (9 R,13 R)−1a,1b-Dihomo-jasmonic acid | 239.1638 | 12.19 | [M + H]+ | pos | C14H22O3 | 0.016 | 0.8 | Decrease | • | • | • | Phytohormone |
| Zeatin-7-beta-D-glucoside | 397.1826 | 6.73 | [M-H_NH3]- | neg | C16H23N5O6 | 0.216 | 0.8 | Decrease | • | • | • | Phytohormone |
| Zeatin | 220.1197 | 2.15 | [M + H]+ | pos | C10H13N5O | 2.99E-05 | 0.5 | Decrease | • | • | • | Phytohormone |
| Methyl jasmonate | 247.1298 | 2.52 | [M + H_Na]+ | pos | C13H20O3 | 1.21E-08 | 4.2 | Increase | • | • | • | Phytohormone |
| Dihydrozeatin riboside | 354.1769 | 6.20 | [M + H]+ | pos | C15H23N5O5 | 0.000 | 0.2 | Decrease | • | • | • | Phytohormone |
| Zeatin riboside | 374.1463 | 5.39 | [M + H_Na]+ | pos | C15H21N5O5 | 0.000 | 0.3 | Decrease | • | • | • | Phytohormone |
| Azelaic acid | 187.0935 | 6.74 | [M-H]- | neg | C9H16O4 | 7.24E-12 | 6.5 | Increase | 5.58E-07 | 1.9 | Increase | Phytohormone |
| Abscisic acid | 265.1552 | 3.35 | [M + H]+ | pos | C15H20O4 | 7.62E-08 | 3.5 | Increase | 4.39E-13 | 16.8 | Increase | Phytohormone |
| Agmatine | 173.0787 | 5.52 | [M-H_NaNa]- | neg | C5H14N4 | 0.000 | 1.4 | Increase | • | • | • | Polyamine |
| Riboflavin | 377.1476 | 4.49 | [M + H]+ | pos | C17H20N4O6 | 0.050 | 0.7 | Decrease | 7.65E-08 | 5.9 | Increase | Flavin |
• indicates the metabolite is absent in the particular extract. HCA = hydroxycinnamic acid. Metabolite annotation was achieved with the aid of the Taverna workbench (www.taverna.org.uk), databases such as Dictionary of Natural Products (DNP) (dnp.chemnetbase.com), ChemSpider (www.chemspider.com), PubChem (www.pubchem.ncbi.nlm.nih.gov), PlantCyc (www.plantcyc.org), SorgCyc (www.sorgcyc.org) and KNApSAcK (http://kanaya.naist.jp/knapsack_jsp/top.htm) and available literature2,23,85.
p-value refers to significance level of a metabolite. Fold change was calculated by dividing the average of the metabolite intensity in replicate samples of treated by the average of the metabolite intensity in replicate samples of control, a value ≥1 represents an increase (metabolite is higher in the treated samples than in the control) and value <1 represents a decrease (metabolite is higher in the control than in the treatment which led to decrease in levels).
Discussion
Structural features of the LPS from B. andropogonis
As complex amphiphilic lipoglycan macromolecules, LPSs can potentially contain MAMP structures within the OPS, COS and LA moieties7. Gram-negative bacteria have developed varied LPSs that can differ in composition. Not only the OPS, but also the bound COS and the LA region can exhibit structural variation, possibly as an adaptive mechanism to hostile environments9,11. The OPS chains, projecting from the cell surface of the bacteria are exposed to the environment and host defence systems. The structural variation of LPSs from pathogens and symbionts might be determining factors in the interactions of plants with microbes31. For example, the composition, size and structure of the OPS might be dependable indicators of virulence potential. As such, it appears to partake in the molecular communication between host plants and bacteria. With regards to the LA, various structural features were reported to play determining roles in the pathophysiological activity in animals. These include the quantity and position of acyl chains, the overall charge of the LA molecule as affected by the phosphorylation of the disaccharide backbone, and the presence/absence of additional polar headgroups9,11. Furthermore, the three-dimensional shape of LA, which in turn is a function of the structural features, has been linked to the differential biological activities of LPSs. Hexaacyl asymmetrical LA exhibits a conical conformation; pentaacyl LA has an intermediary form, and a cylindrical shape is associated with tetraacyl symmetrical LA32. In addition to the chemical composition, the structure and conformation of LA are thus central contributory factors during bacterial pathogenesis. Further interest in LA derives from the findings that bacteria are able to regulate the composition of their LA in response to environmental signals, thus modulating or even antagonizing the triggering of host defences9,11. Due to the restricted number of elucidated LA structures determined from plant-associated bacteria, no clear relationship between the structure and activity is currently known.
For the LPS from B. andropogonis, repeating component of the OPS was characterised as [→2)-α-Rha3CMe-(1 → 3)-α-Rha-(1 → 3)-α-Rha-(1 → ]. While rhamnose-containing OPSs in plant-associated bacteria is common, the occurrence of 3-C-methylrhamnose (evalose) is rare33. Both rhamnose and, particularly its methylated derivative provide highly lipophilic properties to the OPS of B. andropogonis which might play a role in the interaction between the bacterium and sorghum cells. Structural analysis on the LA component indicated a heterogeneous tetra- and penta-acylated, 1,4’-bis-phosphorylated glucosamine disaccharide backbone, further substituted by L-Ara4N. Analysis of the fatty acids revealed (R)−3-hydroxyhexadecanoic acid (16:0(3-OH)), (R)−3-hydroxytetradecanoic acid (14:0(3-OH)) and tetradecanoic acid (14:0), matching with the archetypal LA from Burkholderia genus. Potentially mimicking what happens in animals, such an under-acylation of the LA may be an important factor likely masking the bacterium from the plant immune system and thus favouring the spread of the infection34.
Previously, in a transcriptome investigation of the effect of LPS from B. cepacia on A. thaliana seedlings, we found that the LA and OPS-COS moieties were active in up-regulating subgroups of genes linked to defence over the same range of gene ontology categories as intact LPS11. However, the up-regulation observed in response to intact LPS was more wide-ranging. This suggests that although the molecular patterns of the LA and glycan moieties act as partial agonists, the intact LPS structure is necessary for full agonist activity11,14. Since the perception of individual MAMP-active moieties of LPSB.a. is not yet known, intact LPSB.a. was used for further investigation. The biological or immuno-modulatory activity of this LPS from the disease-causing B. andropogonis was studied by monitoring its ability to lead to reprogramming of the metabolome of sorghum cells.
Metabolic changes induced by LPSB.a. in Sorghum bicolor cells
The use of plant cell suspension culture systems as experimental models offers distinct advantages such as short growth cycles, rapid multiplication of a homogeneous population of cells, reduced complexity, decreased biological variability and improved experimental reproducibility. Plant cell suspensions are therefore highly suitable for metabolomics studies18,19, and depending on the cell line, ideal for studies on secondary metabolite biosynthetic pathways and inducible defence responses20–22.
To gain biological insights into the changes occurring in the sorghum cell culture system following LPSB.a. perception, the chemometrically selected variables were structurally annotated (Table 1). The endometabolome (intracellular/fingerprint) and exometabolome (extracellular/footprint) of the cells were characterised by metabolites associated with primary as well as secondary metabolism, and of diverse biochemical functions in plant defence. To have a comprehensive picture of metabolic reprogramming in sorghum cells induced by LPS treatment, relative quantification (expressed in fold changes) of the putatively identified metabolites was carried out, enabling a quantitative description of metabolic changes. Metabolomic analyses of the cultured cells system revealed the metabolic reprogramming in S. bicolor suspensions triggered by LPSB.a. to be similar to that in plant leaf tissue, triggered by the live pathogen (i.e. similar metabolite classes, that include amino acids, fatty acids, hydroxycinnamic acids, flavonoids, indoles and phytohormones)2. However, there were qualitative and quantitative differences in the levels of signatory metabolites identified from the plant vs. cell metabolomes. Nonetheless, the similarity in the induced metabolic profiles indicate that the cell suspension culture system is suitable for future research on this plant: microbe interaction.
LPSB.a. triggered an alteration of several plant hormones (jasmonates, zeatins, traumatic-, azelaic- and abscisic acid). The derivative of jasmonic acid (JA), methyl jasmonic acid (MeJA), accumulated in the intracellular milieu to significantly high levels (Table 1). As a well-known volatile plant stress hormone and one of the major players in induced systemic resistance (ISR) research, this molecule is involved in the activation of defence mechanisms such as programmed cell death (PCD), reactive oxygen species (ROS) production, lignin formation and stimulation of deposition of wax layers in plant tissues35–37. MeJA has been reported as a signalling molecule in elicitor-induced plant cell cultures and plant tissue, initiating secondary metabolite accumulation37,38. Studies on exogenous application of the hormone, revealed stimulation of phenylpropanoid, flavonoid, fatty acid and other secondary metabolic pathways28,35. A study on transcriptional profiling of genes induced by salicylic acid (SA) and MeJA in sorghum revealed that the hormones coordinately induced genes encoding various functionally important enzymes, such as phenylalanine ammonia-lyase and cinnamate-4-hydroxylase, amongst others ‒ catalysing the biosynthesis of anthocyanins, phytoalexins, lignin and other defence-related secondary metabolites of the phenylpropanoid pathway35,39.
Another fatty acid-derived phytohormone, traumatic acid, which accumulated exclusively in the intracellular milieu, exhibited significant accumulation (Table 1). The 12-oxo-trans-10-dodecenoic acid-derived hormone, which displayed very high levels during the early stages (12–18 h) of the treatment (Fig. 7), is generally known as a wound hormone, due to high accumulation around wounded areas40. Synthesis of traumatic acid is commonly associated with abiotic stress factors40, however, the hormone was also identified as a resistance-inducing metabolite in barley for conferring resistance to F. graminearum41.
Figure 7.
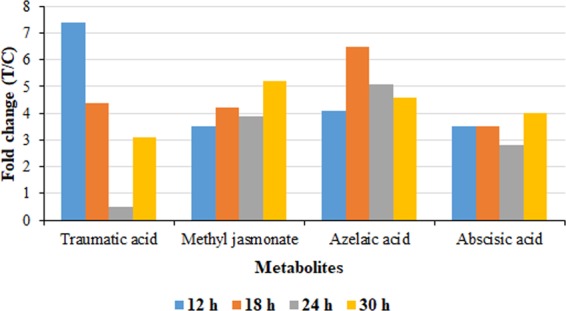
Relative quantification of significantly-accumulating plant hormones annotated in intracellular extracts and induced by LPS.a. treatment of sorghum cells. The graph shows the relative levels of each metabolite across different time points, expressed as fold changes, and computed from treated against control (C0 h) samples i.e. T/C, where fold change ≥1 represents significant accumulation, n = 9.
The hormones azelaic acid (AZA) and abscisic acid (ABA) were identified in high levels in both endo- and exometabolomes (Table 1). ROS accumulating in response to biotic stress (e.g. pathogen challenge42,43, can trigger the cleavage of fatty acids and yield products such as AZA, a signalling molecule also associated with systemic acquired resistance (SAR)19,44. Following pathogen infection, AZA ‒ as one of the signalling molecules - accumulates in petiole exudates and a small fraction translocates to distal tissue45. ABA is generally associated with defence responses to abiotic stress. However, the complex role of the hormone in plant immunity is continuously being uncovered46, with recently emerged insights into ABA’s role in plant–pathogen interactions as a positive or negative defence response regulator, depending on the phase/time of infection and nature of the pathogen47. Defence mechanisms such as stomatal closure, induced by ABA signalling in order to inhibit bacterial invasion, have been reported48. In some plant cell cultures, ABA has also been reported in the regulation of secondary metabolite biosynthesis38.
Metabolomic profiling of cultured cells revealed the intracellular induction of L-phenylalanine (phe) and L-tryptophan (trp), as well as the excretion thereof into the extracellular milieu (Table 1, S4 and S5). The role of these amino acids in plant defence includes functioning as regulators and precursors in various secondary metabolic pathways involved in MAMP-triggered immunity. Phe is a particularly important initiator/precursor molecule of the phenylpropanoid pathway and is also involved in SA biosynthesis, while trp is a major precursor in indolic secondary metabolite synthesis48. Moreover, the trp metabolic pathway is involved in defence responses in cereal crops, through the production of serotonin and conjugates49. The intracellular up-regulation of phe and trp (fold change>1) across the time points (Fig. S9) could be because of continuous requirement for precursors in the downstream synthesis of phe- and trp-derived metabolites, respectively. The presence of phenylpropanoids (and other related secondary metabolites) in the cell extracts indicates that these metabolic pathways were activated by the cells in response to ‘non-self’ perception4 of LPSB.a. as a MAMP. Similarly, the presence of trp and trp-derived metabolites such as indoles and serotonin conjugates, is also indicative of the active involvement of these pathways in LPSB.a.-triggered immune responses.
LPSB.a. elicitation also triggered significant changes in lipidome components as infographically shown in Fig. 8A-B. These hydroxy fatty acids accumulated in the intracellular milieu of treated cells to varying degrees, with levels fluctuating across the time points. Assessing the quality of the endo- and exolipidome, some fatty acids and derivatives were detected in both intra- and extracellular extracts, whereas others were exclusive to the endolipidome (15-hydroxylinoleic acid, 9,10-dihydroxy-12-octadecenoic acid, 9,10-dihydroxystearic acid, trihydroxy-octadecadienoic acid І and 16-hydroxypalmitate) (Table 1). Fatty acids and derivatives thereof are crucial in basal immunity and gene-mediated resistance and contribute to inducing SAR50,51. Fatty acids have been linked to plant defence responses as enhancers of structural defence (cell membrane and cell wall), antimicrobial compounds, key players in plant defence signalling pathways (e.g. oxylipins) and to act as precursor substrates in the production of JA, an important mediator of plant defence52,53.
Figure 8.
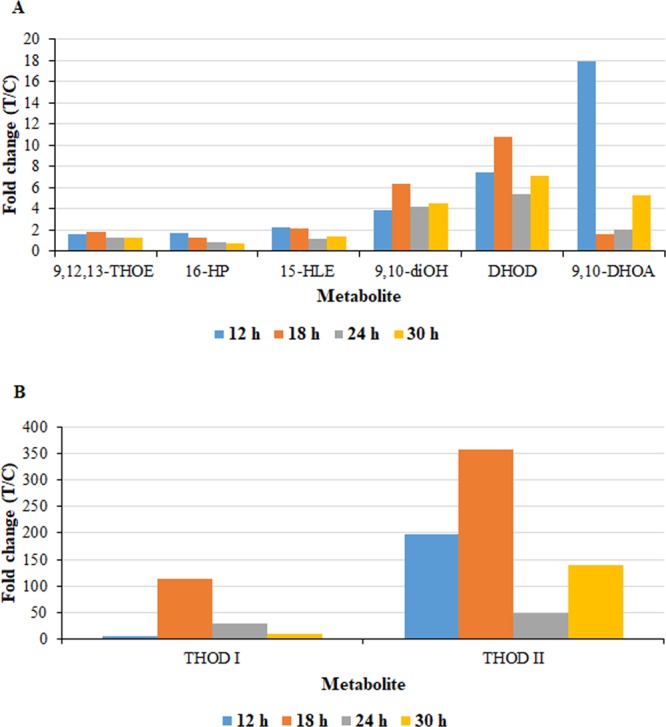
Relative quantification of fatty acids annotated in intracellular extracts and induced by LPSB.a. treatment of sorghum cells. The graph shows the relative levels of each metabolite across the time points, expressed as fold changes, and computed from treated against control (C0 h) i.e. T/C, where fold change ≥1 represents significant accumulation, n = 9. Graphical representation of the annotated fatty acids is divided into two graphs; (A) for reasonably high fold changes and (B) for very high fold changes. 9,12,13-THOE = 9,12,13-trihydroxy-10-octadecenoic acid; 15-HLE = 15-hydroxylinoleic acid; DHOD = dihydroxy-octadecadienoic acid; 16-HP = 16-hydroxypalmitate; 9,10-diOH = 9,10-dihydroxystearic acid; 9,10-DHOA = 9,10-dihydroxy-12-octadecenoic acid; THOD I = trihydroxy-octadecadienoic acid І; THOD II = trihydroxy-octadecadienoic acid ІІ.
Significant accumulation of oxylipins, trihydroxy-octadecadienoic acid І, trihydroxy-octadecadienoic acid ІІ and 9,12,13-trihydroxy-10-octadecenoic acid, and the dihydroxy-oxylipin, 9,10-dihydroxy-12-octadecenoic acid, was observed in LPSB.a.-treated cells. Based on correlative data and experimental work, several trihydroxy-oxylipins have been shown to exhibit antimicrobial activity and establish resistance towards fungal and some bacterial pathogens, and to orchestrate defence responses in cereal plants like barley against powdery mildew and rice against rice blast disease54,55. For such metabolites to be significantly effective in inhibiting pathogen growth in planta, they should be available in adequate concentrations54. An interesting observation was the massive accumulation of the oxylipin, trihydroxy-octadecadienoic acid ІІ, of 356.7-fold at the 18 h time point in the cells (Table 1). An unsupervised colour-coded PCA score plot (Fig. S10) revealed the presence of this metabolite only in treated cells (intracellular extracts) and absent in the control (non-treated) samples, implying a de novo biosynthesis of the metabolite as induced by LPS treatment. Trihydroxy-octadecadienoic acids such as 9,12,13-trihydroxy-10,15-octadecadienoic acid have been shown to possess antimicrobial activity towards both fungal and bacterial pathogens52,55.
The metabolomics results point to the importance of the oxylipin and fatty acid pathways in defence responses in sorghum cells, based on significant accumulation. However, the mechanisms by which oxylipins inhibit microbial growth through antimicrobial activity and to establish resistance in plants, is still largely undefined.
The pool size of induced metabolites may be determined by both the perceived stimulus and the metabolic activity occurring within the system22,56. Decrease in intracellular levels of secondary metabolites, accompanied by extracellular increases, may be due to the active translocation/secretion system of S. bicolor57 to the outside of the cell into the culture medium. This would functionally be similar to secretion into the apoplast/cell periphery in the tissue environment. Distribution of defence-related metabolites to the sites of early pathogen infection is crucial for the restriction of pathogen penetration and proliferation58, and a study on A. thaliana revealed the secretion of indolic glucosinolates upon LPS perception21.
Another possible explanation for the low intracellular levels of the phenylpropanoids and flavonoids in the cultured sorghum cells is the regulation of secondary metabolite levels by the cells, to avoid toxicity to the producing plant cell. In this regard, at certain levels some of the secondary metabolites become toxic to the producing cells, so the cells regulate levels in order to re-establish a form of homeostasis through interconversion, degradation, conjugation and secretion / translocation of potentially toxic metabolites22,58. Regulatory mechanisms by plants include transportation to the apoplast (via vesicles, simple diffusion, and transporter-mediated membrane transport) or specific organelles such as the vacuole, or other self-tolerance mechanisms59. Additionally, the decrease in intracellular phenolic levels could also be due to polymerisation of metabolites that act as monomers for polymers such as lignin in cell wall reinforcement, or due to transformation/conversion/conjugation into other defence-related metabolites60,61.
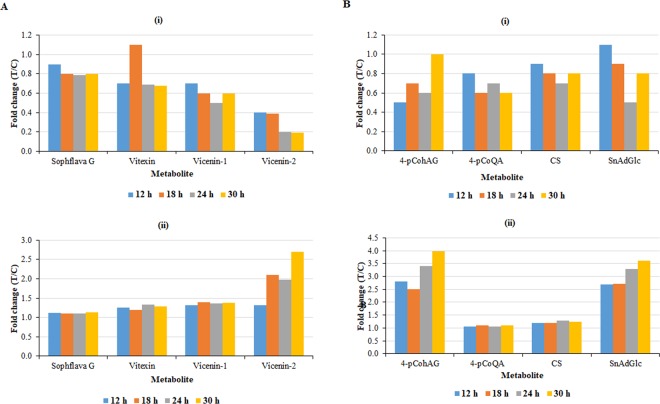
Elicitation of cultured cells with LPSB.a. induced alterations in the phenylpropanoid and flavonoid metabolic pathways (Table 1, S4, and S5). Phenylpropanoid-based metabolites from a number of sub-pathways are known to play central role(s) in plant defence62. Relative quantification revealed that most of the metabolites arising from these pathways were generally associated with a decrease in levels (fold change <1), at most of the time points in the intracellular milieu (except for sinapoyl alcohol, Fig. 9A (i) ‒ flavonoids and Fig. 9B (i) ‒ hydroxycinnamic acids). Out of the fourteen phenolic compounds annotated, only sinapoyl alcohol was positively correlated to the LPSB.a. treatment, i.e. the other thirteen compounds were located/extracted from the bottom left quadrant of the OPLS-DA S-plot (e.g. Figure 6A and S6C). In parallel, the flavonoids and hydroxycinnamic acids (HCAs) displayed a general increase in the extracellular milieu (fold change>1) as seen in Fig. 9A (ii) and 9B (ii), respectively.
Figure 9.
Relative quantification of some flavonoids (A) and hydroxycinnamic acids (B) annotated in (i) intracellular and (ii) extracellular extracts, and induced by LPSB.a. treatment of sorghum cells. The relative levels of each metabolite are expressed in fold changes, and computed from treated against control (C0 h) samples i.e. T/C, where fold change ≥1 represents significant accumulation, n = 9. Sophflava G = sophoraflavanone G; vicenin-1 = apigenin 6-C-xyloside-8-C-glucoside; vicenin-2 = apigenin-6,8-di-C-glucoside; vitexin = apigenin-8-C-glucoside 4-pCohAG = 4-coumaroyl-3-hydroxyagmatine; 4-pCoQA = 4-coumaroylquinic acid; CS = cinnamoylserotonin; SnAdGlc = sinapaldehyde glucoside.
A focus on the identified flavonoids indicates that flavones (apigenin derivatives) were the dominant subgroup (Table 1). The biological functions of metabolites belonging to this subgroup in plant defence, such as exhibiting antimicrobial properties towards various pathogens, have been described in several plants63. The tetrahydroxyflavanone, sophoraflavanone G, also identified amongst the flavonoids, has been reported to possess antibacterial properties, with the mechanism of action including alteration of bacterial membrane fluidity62,64. The roles of the identified HCAs in plant defence include cell wall strengthening, precursors to defence metabolites and as antimicrobial compounds22,65,66. Sinapoyl alcohol, a syringyl lignin precursor, is an important metabolite in structural/mechanical defence. This is through lignification i.e. polymerisation of monolignols (precursors of lignin), resulting in reinforcement of the cell wall to become more resistant to pathogen-derived degrading enzymes and penetration by mycotoxins. In general, increased accumulation of this precursor associated with lignin deposition has been reported following pathogen challenge. Treatment with LPSB.a. resulted in high accumulation of sinapoyl alcohol, suggesting the importance of the metabolite in activation of structural defences in cultured cells65–67.
Other metabolites identified in this study and reported to be deployed in cell wall strengthening, include 4-coumaroyl-3-hydroxyagmatine, feruloylserotonin, sinapaldehyde glucoside, cinnamoylserotonin and dihydroconiferyl alcohol glucoside (a guaiacyl lignin monomer). In other studies these metabolites have also been identified as resistance-related compounds41,60,67,68. Additionally, 4-coumaroylquinic acid has been shown to confer resistance against a range of pathogens. Agmatine, which can conjugate with HCAs to yield HCA-amides, and important metabolites in cell wall strengthening and as phytoalexins60,66,67, displayed an increase and was found exclusively in the endometabolome.
The metabolic reprograming in sorghum cells as a result of LPSB.a. treatment also involved some alterations in flavin and indole metabolism. The metabolite profiles of treated cells were characterised by down- and up-regulation of riboflavin in the intra- and extracellular milieu, respectively (Table 1, S4 and S5). This is indicative of the secretion of the metabolite into the latter. Riboflavin has been identified as a defence response/systemic resistance-inducing metabolite in various plant species against bacterial, fungal and viral pathogens23,69. In rice, the metabolite was shown to induce defence responses against Rhizoctonia solani and Pyricularia oryzae69. The major role of the octadecanoid pathway in riboflavin-induced resistance and basal resistance, together with the link between riboflavin accumulation and increased lignification, was also highlighted earlier36.
Indolic derivatives (mostly indole acetyl derivatives), accumulated to varying levels in both the intra- and extracellular milieus (Table 1). A number of plant species and plant cell cultures have been reported to accumulate indolic derivatives as phytoanticipins, phytoalexins, precursors or as signalling molecules in response to pathogen and abiotic stress21,47,49. Indole-3-acetyl-myo-inositol, significantly detected in both the intra- and extracellular milieus of the sorghum cells, has been linked to plant resistance in barley68. The data obtained in this study therefore suggests defence-related roles for indole derivatives in sorghum cells. Moreover, the detection of both trp and indole-containing metabolites as discriminant ions / signatory biomarkers provides an insight into the activation of defences related to trp metabolism in response to LPSB.a. treatment. Together with the metabolites associated with the phenylpropanoid pathway, this indicates activation of an integrated and complementary network of chemical-based plant defences.
Methods
Bacterial cell culturing, LPS isolation and purification
An overnight bacterial culture of B. andropogonis, strain BD 256, obtained from the Plant Protection Institute, Agricultural Research Council (Pretoria, South Africa), was initially prepared by inoculating 600 mL nutrient broth medium (Merck, Johannesburg, RSA) and incubated overnight on a shaker at 28 °C and speed of 130 rpm. This was followed by inoculating large scale cultures (3 L of nutrient broth), with 200 mL of the overnight bacterial culture, under strict sterile conditions, and incubating at 30 °C on a rotating shaker at 100 rpm for 14 d. Several large-scale culturing cycles were repeated for larger quantities of cell mass for LPS isolation and purification. Harvesting of bacterial cells was performed by centrifugation at 13,000 × g at 4 °C for 20 min, followed by lyophilisation for 48 h before the LPS isolation and purification steps. LPS was extracted from the bacterial cell biomass using the hot-phenol water extraction protocol as previously described70,71. This involved lysing bacterial cells and partitioning of the LPS into the aqueous phase for maximum yield. For LPS purification, enzymatic digestion of the RNA that possibly co-extracted into the water phase, was executed. Dried extracts were dissolved in 30 mL (1: 40 w/v) distilled water and sequentially treated with RNase and proteinase K (Sigma-Aldrich, Steinheim, Germany) as previously described14. Following incubation, an equal volume of phenol was added in order to denature and remove the enzymes. Following vortexing, the emulsion was centrifuged at 10,000 × g for 15 min to obtain the hydrophilic LPS in the water phase. This was dialysed for 3 d against distilled water at 5 °C and then lyophilised.
LPS-specific SDS-PAGE analysis and compositional analysis
The macromolecular nature of the purified LPS was evaluated by SDS-PAGE as previously described72. LPS from both B. andropogonis and B. cepacia70,72 were used for the analysis. Upon completion of electrophoresis, silver staining was performed for LPS visualisation72. For structural elucidation, an additional enzymatic treatment by RNase, DNase and Proteinase K (Roth, Germany) was performed in order to remove any contaminants. Subsequently a further step of purification by ultracentrifugation (100,000 ×g, 16 h, 4 °C) followed by size-exclusion chromatography on a Sephacryl S500 column was executed10,14. The LPS monosaccharide content was determined by analysis of the acetylated O-methyl glycoside derivatives obtained by treatment with HCl/MeOH (1.25 M, 85 °C, 24 h) followed by acetylation with acetic anhydride in pyridine (85 °C, 30 min). The absolute configuration was established through the evaluation of the O-octylglycoside derivatives71,73 of all sugars found in the LPS under investigation except for Rha3CMe. To define the sugar linkage pattern an aliquot of sample was methylated with CH3I, hydrolysed with trifluoroacetic acid (4 M, 100 °C, 4 h), carbonyl reduced with NaBD4 and acetylated with pyridine and acetic anhydride74. The total fatty acid content of intact LPS was determined by treating with HCl (4 M, 100 °C, 4 h) followed by a treatment with NaOH (5 M, 100 °C, 30 min). The pH was adjusted to reach slight acidity. After extraction in chloroform, fatty acids were methylated with diazomethane75. All chemical analyses were executed through the employment of gas-liquid chromatography (GLC-MS, Agilent Technologies 6850 A) equipped with a mass selective detector 5973 N and a Zebron ZB-5 capillary column.
Isolation of lipid A and the OPS chain from B. andropogonis LPS
The pure LPS (20 mg) underwent a mild hydrolysis with acetate buffer solution (pH 4.4, 100 °C, 2 h). After centrifugation, the supernatant containing the OPS product, was collected and lyophilised (15 mg). A further purification step by size-exclusion chromatography on Sephacryl S100 followed by a Superdex 30 (GE Healthcare Life Sciences) was also executed to purify the product10,11. The precipitate, containing the LA, was collected and washed several times with a freshly prepared Bligh/Dyer mixture (chloroform/methanol/water, 2:2:1.8, v/v/v)76. The organic phases were pooled, dried and analysed by matrix-assisted laser desorption ionisation mass spectrometry (MALDI MS).
NMR spectroscopy analysis
1D and 2D NMR spectra were executed on a Bruker 600 DRX instrument equipped with a cryo probe. The solvent was D2O. Spectra calibration was performed with internal acetone (δH 2.225 ppm, δC 31.45 ppm). The double-quantum filtered phase sensitive correlation spectroscopy (DQF-COSY) experiment was carried out by using data sets of 4096 × 256 points77,78. Total correlation spectroscopy (TOCSY) experiments were performed with spinlock times of 100 ms, using data sets (t1 × t2) of 4096 × 256 points. Rotating frame Overhauser enhancement spectroscopy (ROESY) and Nuclear Overhauser enhancement spectroscopy (NOESY) experiments were executed by using data sets (t1 × t2) of 4096 × 256 points and by using mixing times between 100 and 400 ms. In all homonuclear experiments the data matrix was zero-filled in both dimensions to give a matrix of 4 K × 2 K points and was resolution enhanced in both dimensions by a cosine-bell function before Fourier transformation. The determination of coupling constants was obtained by 2D phase sensitive DQF-COSY. Heteronuclear single quantum coherence (1H, 13C HSQC) and heteronuclear multiple bond correlation (1H, 13C HMBC) experiments were recorded in 1H-detection mode by single-quantum coherence with proton decoupling in the 13C domain using data sets of 2048 × 256 points. 1H, 13C HSQC was executed using sensitivity improvement and in the phase-sensitive mode using Echo/Antiecho gradient selection, with multiplicity editing during selection step79. The 1H, 13C HMBC experiment was optimised on long range coupling constants with low-pass J filter to suppress one-bond connectivity, using gradient pulses for selection. A delay of 60 ms was employed for the evolution of long range correlations. It was used at long range coupling constant value of 6 Hz. The data matrix in both heteronuclear experiments was extended to 2048 × 1024 points using forward linear prediction extrapolation80.
MALDI mass spectrometry analysis
Reflectron MALDI-TOF MS was performed on an ABSCIEX TOF/TOFTM 5800 Applied Biosystems mass spectrometer equipped with an Nd:YLF laser with a λ of 345 nm, a < 500-ps pulse length and a repetition rate of up to 1000 Hz. The LA fraction was dissolved in chloroform/methanol (1:1, v/v) as previously described12,81. The matrix was trihydroxyacetophenone dissolved in methanol/0.1% trifluoracetic acid/acetonitrile (7:2:1, v/v/v) at a concentration of 75 mg mL−1. Thereafter, 0.5 μL of the LA preparation and 0.5 μL of the matrix solution were deposited on the MALDI plate and dried at room temperature. Each spectrum was a result of the accumulation of 1500 laser shots82 and was recorded in negative polarity.
Preparation and elicitation of Sorghum bicolor cell suspensions
Seeds of white sorghum, Sorghum bicolor (L.) Moench, used for the generation of cell suspension cultures were obtained from Agricol (Pretoria, South Africa). Cell suspensions were generated from friable callus initiated and maintained on Murashige and Skoog medium83 containing [3% (w/v) sucrose, pH 5.8] including MS vitamins and phytohormones [3 mg L−1 2,4-dichlorophenoxyacetic acid (2,4-D) and 2.5 mg L−1 1-naphthaleneacetic acid (NAA)] as described previously84.
To minimise unplanned variation in experimental plant material three independent biological repeats were conducted and analysed. To achieve this, cell suspensions were sub-cultured before every biological repeat to generate sufficient cell material for subsequent metabolomics analysis. Four days after subculture, synchronised cells from replicate flasks were first combined to ensure a homogeneous culture. Equal aliquots (25 mL) of the cell suspensions were then redistributed into sterile 50 mL Falcon tubes with three replicates for each condition. Cells were treated with LPS dissolved in the culture medium and negative controls received the equivalent volume medium. The time study to monitor the dynamics of the changing metabolomes included intervals of 0, 12, 18, 24 and 30 h, post-inoculation (h.p.i.). Following extraction and sample preparation, each sample was analysed in triplicate by UHPLC-MS generating n = 9, (3 independent biological replicates x 3 analytical replicates) as required for downstream metabolomic data analysis.
LPSB.a. (isolated and purified as described above) was prepared in MS medium at room temperature to give a 10 mg mL−1 stock solution. The final concentration of LPSB.a. for elicitation of the cells was 100 µg mL−1. The treated and non-treated cell suspensions were placed horizontally on an orbital shaker and incubated at 130 rpm and 25 °C. A time study was conducted to monitor the response of the cells to treatment over time compared to untreated cells at 0 h. Cells were harvested at 12, 18, 24 and 30 h.p.i. by means of centrifugation using a swinging bucket bench top centrifuge at 5100 rpm and 4 °C for 25 min. Pelleted cells and medium supernatants were separated and immediately stored at –80 °C until both intracellular and extracellular metabolite extraction steps could be performed. The above procedures were performed under strict sterile conditions.
Metabolite extraction and sample preparation
Intracellular metabolites were extracted using 100% cold methanol. Initially the pelleted cells harvested by centrifugation were mixed with the extraction solvent in a ratio of 1:2 (w/v) and homogenised using an Ultra-Turrax homogeniser and sonication using a probe sonicator (Bandelin Sonopuls, Berlin, Germany) set at 55% power for 15 s, working at 4 °C. Samples were then prepared in a similar manner as previously described2, until reconstitution. For extracellular metabolites, the decanted supernatants (growth media) of each tube were first lyophilised and the obtained dried material weighed and kept at – 80 °C for reconstitution. The intra- and extracellular extracts were then reconstituted in 50% UHPLC-grade methanol (Romil Pure Chemistry, Cambridge, UK) in a 1:10 m/w ratio and filtered through 0.22 µm nylon filters into UHPLC glass vials fitted with 500 µL inserts.
Ultra-high-performance liquid chromatography-high definition mass spectrometry (UHPLC-HDMS) analyses
Cell (intracellular) and medium (extracellular) extracts were analysed on an UHPLC-HDMS system: Waters Acquity UHPLC coupled in tandem to a SYNAPT G1 Q-TOF high definition mass spectrometer (Waters Corporation, Milford, MA, USA) via an electrospray ionisation (ESI) interface. An optimised UHPLC-HDMS method previously reported by us[2]2 was adopted. In short, the samples were separated on a reverse phase C18 column (150 mm × 2.1 mm ×1.8 µm – HSS T3, Waters Corporation, Milford, MA, USA) at 60 °C. A binary mobile phase solvent system consisting of solvent A: 0.1% formic acid in MilliQ water and solvent B: 0.1% formic acid in acetonitrile (Romil, Cambridge, UK), was used. The flow rate was set to 0.4 mL min−1 with gradient elution of the following conditions: 2% B maintained for 1 min, 95% B at 15 min ‒ maintained for 2 min, and a change back to the initial conditions at 18 min, followed by a 2 min equilibration time of the column. The total chromatographic run time was 20 min and the injection volume was 4 µL. The MS detector acquired data in both ESI positive and negative modes. The conditions were set as follows: capillary voltage of 2.5 kV, sampling cone at 30 V, extraction cone at 4 V, cone gas flow 50 L h−1, desolvation gas flow 550 L h−1, source temperature at 120 °C, desolvation temperature at 450 °C, scan time of 0.1 s and mass range of 100–1000 Da. Each sample was analysed in triplicate. To ensure that high mass accuracy of analytes (1–3 mDa) was acquired, online mass correction was done using a lock spray source: leucine encephalin (50 pg mL−1), [M + H]+ = 55.2766 and [M – H]− = 554.2615. This reference calibrant was sampled every 15 s, producing an average intensity of 350 counts scan−1 in centroid mode. Sample analyses were randomised to provide stochastic stratification in sample acquisition to minimise measurement bias. To assess the reliability and reproducibility of the analyses, and for non-linear signal correction, quality control (QC) samples (prepared by combining equal volumes of all the individual samples) were used. The QC samples (6 injections) were analysed every 30–35 injections to monitor possible changes in the instrument response. QC runs were also performed at the beginning and end of each batch to ensure system equilibration. Blank samples (50% aqueous methanol) were randomly analysed to monitor background noise. To generate molecular fragment information for downstream structure elucidation and compound identification, a data-independent acquisition (DIA) method, namely MSE was applied: the MS analyses were set to carry out non-fragmented as well as five fragmenting experiments simultaneously by applying alternating collision energy of 0 eV (unfragmented) and from 10 to 50 eV (fragmented).
Data processing, multivariate data analyses and metabolite annotation
MassLynxTM XS software was used to extract raw data, obtained from UHPLC-HDMS, and the MarkerLynxTM software (Waters Corporation, Manchester, UK) for processing. Software parameters were the same for intra- and extracellular data, i.e. mass tolerance 0.01 Da, mass range 100–1000 Da, mass window 0.05 Da and a retention time (Rt) window of 0.20 min. The Rt range was 1.6–15 min for intracellular data and 1–13 min for extracellular data. The ion intensity threshold was set at 100 and 50 counts respectively for intracellular- and extracellular data. The data matrices obtained from MarkerLynx processing were exported into SIMCA 14 software (Umetrics, Umea, Sweden) for multivariate statistical analyses. The data were Pareto-scaled before principal component analysis (PCA) and orthogonal projection to latent structures discriminant analyses (OPLS-DA). Nonlinear iterative partial least squares (NIPALS) algorithm was a default SIMCA methodology used to handle the missing values. A seven-fold cross-validation (CV) was applied as a tuning and validating procedure in computing the models. This k-fold CV is the most common technique for model evaluation and selection in machine learning; where a dataset is iterated k times. In each round, the dataset is split into k parts: validation and training subsets. In other words, for a fixed number of components, the Y values of all individuals of each subset are predicted using a sub-model computed with the 6 other subsets, in this case of 7-fold CV. Results of this k-fold cross-validation procedure are summarised by the value of different quality parameters, such as R2 and Q2 metrics in this case. R2 indicates the goodness of fit (explained variation) and Q2 reflects the predictive ability of the model (predicted variation)25. Further metrics used to validate generated models are described in the results section. In addition to strong outliers detected through Hoteling’s T2-established normality (score space), moderate outliers were evaluated using the distance to the model in space X (DModX) plots permitted the assessment of moderate outliers. Furthermore, for variable selection, the extracted discriminating features were evaluated using variable importance in projection (VIP) scores. Metabolites were annotated as previously described2,23,85. The chemometrically selected variables were then structurally elucidated and putatively identified. Annotation was performed at level 2 of the Metabolomics Standards Initiative (MSI)30.
Availability of data and material
The study design information, LC-MS raw data, analyses and data processing information, and the meta-data were deposited to the EMBL-EBI metabolomics repository (MetaboLights50), with the identifier MTBLS1204 (http://www.ebi.ac.uk/metabolights/MTBLS.1204).
Supplementary information
Acknowledgements
We would like to thank Dr PA Steenkamp for the LC-MS instrumental analysis and Dr NE Madala who assisted with mass spectrometry data interpretation This work was supported by the South African National Research Foundation through grant support to IAD (grant number 95818). The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Author contributions
All authors contributed to the interpretation of results and approving the decision to submit the article for publication. I.A.D. and L.A.P. conceived the project and coordinated the research. C.R.M. performed the metabolomics experimental work, F.T. contributed to the analysis of the data and guided the chemometric analyses. F.D.L., A.S. and A.M. performed the structural analysis of the LPS. L.A.P. and F.D.L. assisted with writing, review and editing of the manuscript. All authors have read and approved the final version of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-64186-y.
References
- 1.Ranf S. Pattern recognition receptors – versatile genetic tools for engineering broad-spectrum disease resistance in crops. Agronomy. 2018;8:134. doi: 10.3390/agronomy8080134. [DOI] [Google Scholar]
- 2.Mareya, C. R. et al. Untargeted metabolomics reveal defensome-related metabolic reprogramming in Sorghum bicolor against infection by Burkholderia andropogonis. Metabolites, 9, 10.3390/metabo9010008 (2019). [DOI] [PMC free article] [PubMed]
- 3.Andolfo, G. & Ercolano, M.R. Plant innate immunity multicomponent model. Front. Plant Sci. 6, 10.3389/fpls.2015.00987 (2015). [DOI] [PMC free article] [PubMed]
- 4.Sanabria NM, Huang J-C, Dubery IA. Self/nonself perception in plants in innate immunity and defense. Self/ Nonself. 2010;1:40–54. doi: 10.4161/self.1.1.10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi, H.W. & Klessig, D.F. DAMPs, MAMPs and NAMPs in plant innate immunity. BMC Plant Biol. 16, 10.1186/s12870-016-0921-2. (2016). [DOI] [PMC free article] [PubMed]
- 6.Ranf S. Immune sensing of lipopolysaccharide in plants and animals: same but different. PLoS Pathog. 2016;12(6):e1005596. doi: 10.1371/journal.ppat.1005596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newman MA, von Roepenack E, Daniels M, Dow M. Lipopolysaccharides and plant responses to phytopathogenic bacteria. Mol. Plant Pathol. 2000;1:25–31. doi: 10.1046/j.1364-3703.2000.00004.x. [DOI] [PubMed] [Google Scholar]
- 8.Kutschera A, Ranf S. The multifaceted functions of lipopolysaccharide in plant-bacteria interactions. Biochimie. 2019;159:93–98. doi: 10.1016/j.biochi.2018.07.028. [DOI] [PubMed] [Google Scholar]
- 9.Di Lorenzo, F. et al. Lipopolysaccharides as microbe-associated molecular patterns: A structural perspective in Carbohydrates in drug design and discovery, (eds. Jimenez-Barbero, J., Canada, F.J. & Martín-Santamaría, S.) 38–63 (RSC Publishing, London, 2015).
- 10.De Castro. C, et al. Structural identification of the O-antigen fraction from the lipopolysaccharide of the Burkholderia ambifaria strain 19182. Carbohydr. Res. 2013;379:95–99. doi: 10.1016/j.carres.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 11.Madala NE, Molinaro A, Dubery IA. Distinct carbohydrate and lipid-based molecular patterns within lipopolysaccharides from Burkholderia cepacia contribute to defense-associated differential gene expression in Arabidopsis thaliana. Innate Immun. 2012;18:140–154. doi: 10.1177/1753425910392609. [DOI] [PubMed] [Google Scholar]
- 12.Di Lorenzo F. The lipopolysaccharide lipid A structure from the marine sponge-associated bacterium Pseudoalteromonas sp. 2A. Anton. van Leeuwenhoek. J. Microbiol. 2017;110:1401–1412. doi: 10.1007/s10482-017-0865-z. [DOI] [PubMed] [Google Scholar]
- 13.Sanabria NM, Dubery IA. Differential display profiling of the Nicotiana response to LPS reveals elements of plant basal resistance. Biochem. Biophys. Res. Commun. 2006;344:1001–1007. doi: 10.1016/j.bbrc.2006.03.216. [DOI] [PubMed] [Google Scholar]
- 14.Madala NE, Leone MR, Molinaro A, Dubery IA. Deciphering the structural and biological properties of the lipid A moiety of lipopolysaccharides from Burkholderia cepacia strain ASP B 2D, in Arabidopsis thaliana. Glycobiology. 2011;21:184–194. doi: 10.1093/glycob/cwq146. [DOI] [PubMed] [Google Scholar]
- 15.Di Lorenzo F, et al. The Lipid A from Rhodopseudomonas palustris strain BisA53 LPS possesses a unique structure and low immunostimulant properties. Chem. Euro. Journ. 2016;23:1–12. doi: 10.1002/chem.201604379. [DOI] [PubMed] [Google Scholar]
- 16.Newman, M.A., Sundelin, T., Nielsen, J.T. & Erbs, G. MAMP (microbe-associated molecular pattern) triggered immunity in plants. Front. Plant Sci. 4, 10.3389/fpls.2013.00139 PMID: 23720666 (2013). [DOI] [PMC free article] [PubMed]
- 17.Ranf, S. et al. A lectin S-domain receptor kinase mediates lipopolysaccharide sensing in Arabidopsis thaliana Nat. Immunol. 10, 1038/ni.3124 (2015). [DOI] [PubMed]
- 18.James JT, Tugizimana F, Steenkamp PA, Dubery IA. Metabolomic analysis of methyl jasmonate-induced triterpenoid production in the medicinal herb Centella asiatica (L.) urban. Molecules. 2013;18:4267–4281. doi: 10.3390/molecules18044267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Djami-Tchatchou, A.T., Ncube, E.N., Steenkamp, P.A. & Dubery I.A. Similar, but different: Structurally related azelaic acid and hexanoic acid trigger differential metabolomic and transcriptomic responses in tobacco cells. BMC Plant Biol. 17, 10.1186/s12870-017-1157-5 (2017). [DOI] [PMC free article] [PubMed]
- 20.Tugizimana F, Steenkamp PA, Piater LA, Dubery IA. Ergosterol-induced sesquiterpenoid synthesis in tobacco cells. Molecules. 2012;17:1698–1715. doi: 10.3390/molecules17021698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finnegan, T., Steenkamp, P.A., Piater, L.A. & Dubery, I.A. The lipopolysaccharide-induced metabolome signature in Arabidopsis thaliana reveals dynamic reprogramming of phytoalexin and phytoanticipin pathways. PLoS One, 11, 10.1371/journal.pone.0163572 (2016). [DOI] [PMC free article] [PubMed]
- 22.Mhlongo, M.I., Piater, L.A., Madala, N.E., Steenkamp, P.A & Dubery, I.A. Phenylpropanoid defences in Nicotiana tabacum cells: Overlapping metabolomes indicate common aspects to priming responses induced by lipopolysaccharides, chitosan and flagellin-22. PLoS One, 11, 10.1371/journal.pone.0151350 (2016). [DOI] [PMC free article] [PubMed]
- 23.Tugizimana, F., Djami-Tchatchou, A.T., Steenkamp, P.A., Piater, L.A. & Dubery, I.A. Metabolomic analysis of defense-related reprogramming in Sorghum bicolor in response to Colletotrichum sublineolum infection reveals a functional metabolic web of phenylpropanoid and flavonoid pathways. Front. Plant Sci. 9, 10.3389/fpls.2018.01840 (2019). [DOI] [PMC free article] [PubMed]
- 24.Di Lorenzo F, et al. Chemistry and biology of the potent endotoxin from a Burkholderia dolosa clinical isolate from a cystic fibrosis patient. ChemBioChem. 2013;14:1105–1115. doi: 10.1002/cbic.201300062. [DOI] [PubMed] [Google Scholar]
- 25.Tugizimana, F., Piater, L.A. & Dubery, I.A. Plant metabolomics: A new frontier in phytochemical analysis. S. Afr. J. Sci. 109, 10.1590/sajs.2013/20120005 (2013).
- 26.Jolliffe, I.T., & Cadima J. Principal component analysis: a review and recent developments. Philos Trans. Roy. Soc. A. 374, 10.1098/rsta.2015.0202 (2016). [DOI] [PMC free article] [PubMed]
- 27.Tugizimana, F., Steenkamp, P.A., Piater, L.A & Dubery, I.A. Multi-platform metabolomic analyses of ergosterol- induced dynamic changes in Nicotiana tabacum cells. PLoS One, 9, 10.1371/journal.pone.0087846 (2014). [DOI] [PMC free article] [PubMed]
- 28.Tugizimana F, Ncube EN, Steenkamp PA, Dubery IA. Metabolomics-derived insights into the manipulation of terpenoid synthesis in Centella asiatica cells by methyl jasmonate. Plant Biotechnol. Rep. 2015;9:125–136. [Google Scholar]
- 29.Tugizimana F, Steenkamp PA, Piater LA, Dubery IA. Mass spectrometry in untargeted liquid chromatography / mass spectrometry metabolomics: Electrospray ionisation parameters and global coverage of the metabolome. Rapid Commun. Mass Spectrom. 2018;32:121–132. doi: 10.1002/rcm.8010. [DOI] [PubMed] [Google Scholar]
- 30.Sumner LW, et al. Proposed minimum reporting standards for chemical analysis: Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI) Metabolomics. 2007;3:211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lerouge I, Vanderleyden J. O-Antigen structural variation: mechanisms and possible roles in animal/plant-microbe interactions. FEMS Microbiol. Rev. 2001;26:17–47. doi: 10.1111/j.1574-6976.2002.tb00597.x. [DOI] [PubMed] [Google Scholar]
- 32.Seydel U, Schromm AB, Blunck R, Brandenburg K. Chemical structure, molecular conformation and bioactivity of endotoxins. Chem Immunol. 2000;74:5–24. doi: 10.1159/000058754. [DOI] [PubMed] [Google Scholar]
- 33.Zdorovenko EL, et al. Structure of the O-polysaccharide of Azorhizobium caulinodans HAMBI 216; identification of 3-C-methyl-D-rhamnose as a component of bacterial polysaccharides. Carbohydr. Res. 2012;358:106–9. doi: 10.1016/j.carres.2012.06.012. [DOI] [PubMed] [Google Scholar]
- 34.Molinaro A, et al. Chemistry of Lipid A: At the heart of innate immunity. Chem. Eur. J. 2014;20:1–21. doi: 10.1002/chem.201403923. [DOI] [PubMed] [Google Scholar]
- 35.Salzman RA, et al. Transcriptional profiling of sorghum induced by methyl jasmonate, salicylic acid, and aminocyclopropane carboxylic acid reveals cooperative regulation and novel gene responses. Plant Physiol. 2005;138:352–368. doi: 10.1104/pp.104.058206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taheri P, Tarighi S. Riboflavin induces resistance in rice against Rhizoctonia solani via jasmonate-mediated priming of phenylpropanoid pathway. J Plant Physiol. 2010;167:201–208. doi: 10.1016/j.jplph.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 37.Petrussa E, et al. Plant flavonoids-biosynthesis, transport and involvement in stress responses. Int. J. Mol. Sci. 2013;14:14950–14973. doi: 10.3390/ijms140714950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao J, Davis LC, Verpoorte R. Elicitor signal transduction leading to production of plant secondary metabolites. Biotechnol. Adv. 2005;23:283–333. doi: 10.1016/j.biotechadv.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 39.Poloni A, Schirawski J. Red card for pathogens: Phytoalexins in sorghum and maize. Molecules. 2014;19:9114–9133. doi: 10.3390/molecules19079114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lukaszuk, E. & Ciereszko, I. Plant responses to wounding stress. In: Biological diversity – from cell to ecosystem (ed. Laska, G.) 73-85 (Polish Botanical Society, 2012). ISBN 978-83-62069-28-6.
- 41.Chamarthi SK, et al. Identification of fusarium head blight resistance related metabolites specific to doubled-haploid lines in barley. Eur. J. Plant Pathol. 2014;138:67–78. [Google Scholar]
- 42.Eckardt NA. Oxylipins signaling in plant stress responses. Plant Cell. 2008;20:495–497. doi: 10.1105/tpc.108.059485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao, Q.M., Zhu, S., Kachroo, P. & Kachroo, A. Signal regulators of systemic acquired resistance. Front. Plant Sci. 6, 10.3389/fpls.2015.00228 (2015). [DOI] [PMC free article] [PubMed]
- 44.Shah, J., Zeier, J. & Cameron, R.K. Long-distance communication and signal amplification in systemic acquired resistance. Front. Plant Sci. 4, 10.3389/fpls.2013.00030 (2013). [DOI] [PMC free article] [PubMed]
- 45.Shine MB, Xiao X, Kachroo P, Kachroo A. Signaling mechanisms underlying systemic acquired resistance to microbial pathogens. Plant Sci. 2018;279:81–86. doi: 10.1016/j.plantsci.2018.01.001. [DOI] [PubMed] [Google Scholar]
- 46.Huang J-C, Piater LA, Dubery IA. Identification and characterization of a differentially expressed NAC transcription factor gene in MAMP-treated. Arabidopsis thaliana. Physiol. Mol. Plant Pathol. 2012;80:19–27. [Google Scholar]
- 47.Denancé, N., Sánchez-Vallet, A., Goffner, D. & Molina, A. Disease resistance or growth: the role of plant hormones in balancing immune responses and fitness costs. Front. Plant Sci. 4, 10.3389/fpls.2013.00155 (2013). [DOI] [PMC free article] [PubMed]
- 48.Tzin, V. & Galili, G. The biosynthetic pathways for shikimate and aromatic amino acids in Arabidopsis thaliana. The Arabidopsis Book 8, e0132. 10.1199/tab.0132 (American Society of Plant Biologists, 2010). [DOI] [PMC free article] [PubMed]
- 49.Ishihara A, et al. The tryptophan pathway is involved in the defense responses of rice against pathogenic infection via serotonin production. Plant J. 2008;54:481–495. doi: 10.1111/j.1365-313X.2008.03441.x. [DOI] [PubMed] [Google Scholar]
- 50.Hou C, Forman R. Growth inhibition of plant pathogenic fungi by hydroxy fatty acids. J. Ind. Microbiol. Biotechnol. 2000;24:275–276. [Google Scholar]
- 51.Kachroo A, Kachroo P. Fatty acid–derived signals in plant defense. Annu. Rev. Phytopathol. 2009;47:153–176. doi: 10.1146/annurev-phyto-080508-081820. [DOI] [PubMed] [Google Scholar]
- 52.Prost I. Evaluation of the antimicrobial activities of plant oxylipins supports their involvement in defense against pathogens. Plant Physiol. 2005;139:1902–1913. doi: 10.1104/pp.105.066274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lim GH, Singhal R, Kachroo A, Kachroo P. Fatty acid– and lipid-mediated signaling in plant defense. Annu. Rev. Phytopathol. 2017;55:505–36. doi: 10.1146/annurev-phyto-080516-035406. [DOI] [PubMed] [Google Scholar]
- 54.Göbel C, Feussner I, Hamberg M, Rosahl S. Oxylipin profiling in pathogen-infected potato leaves. Biochim. Biophys. Acta (Mol. Cell. Biol. Lipids) 2002;1584:55–64. doi: 10.1016/s1388-1981(02)00268-8. [DOI] [PubMed] [Google Scholar]
- 55.Walters DR, Cowley T, Weber H. Rapid accumulation of trihydroxy oxylipins and resistance to the bean rust pathogen Uromyces fabae following wounding in Vicia faba. Ann. Bot. 2006;97:779–784. doi: 10.1093/aob/mcl034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gamir J, Pastor V, Kaever A, Cerezo M, Flors V. Targeting novel chemical and constitutive primed metabolites against Plectosphaerella cucumerina. Plant J. 2014;78:227–240. doi: 10.1111/tpj.12465. [DOI] [PubMed] [Google Scholar]
- 57.Ngara, R. et al. Identifying differentially expressed proteins in sorghum cell cultures exposed to osmotic stress. Sci. Rep. 8, 10.1038/s41598-018-27003-1 (2018). [DOI] [PMC free article] [PubMed]
- 58.Goossens A, Häkkinen S, Laakso I, Oksman-Caldentey K, Inzé D. Secretion of secondary metabolites by ATP-binding cassette transporters in plant cell suspension cultures. Plant Physiol. 2003;131:1161–1164. doi: 10.1104/pp.102.016329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shitan N. Secondary metabolites in plants: Transport and self-tolerance mechanisms. Biosci. Biotechnol. Biochem. 2016;80:1283–1293. doi: 10.1080/09168451.2016.1151344. [DOI] [PubMed] [Google Scholar]
- 60.Yogendra KN, et al. Quantitative resistance in potato leaves to late blight associated with induced hydroxycinnamic acid amides. Funct. Integr. Genom. 2014;14:285–298. doi: 10.1007/s10142-013-0358-8. [DOI] [PubMed] [Google Scholar]
- 61.Gauthier L, Atanasova-Penichon V, Chéreau S, Richard-Forget F. Metabolomics to decipher the chemical defense of cereals against Fusarium graminearum and deoxynivalenol accumulation. Int. J. Mol. Sci. 2009;16:24839–24872. doi: 10.3390/ijms161024839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Naoumkina. MA, et al. Genome-wide analysis of phenylpropanoid defence pathways. Mol. Plant Pathol. 2010;11:829–846. doi: 10.1111/j.1364-3703.2010.00648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Balmer D, De Papajewski DV, Planchamp C, Glauser G, Mauch-Mani B. Induced resistance in maize is based on organ-specific defence responses. Plant J. 2013;74:213–225. doi: 10.1111/tpj.12114. [DOI] [PubMed] [Google Scholar]
- 64.Kumar, S. & Pandey, A. Chemistry and biological activities of flavonoids: An overview. Sci. World J. Article ID 162750, 10.1155/2013/162750 (2013). [DOI] [PMC free article] [PubMed]
- 65.Wang Y, Chantreau M, Sibout R, Hawkins S. Plant cell wall lignification and monolignol metabolism. Front. Plant Sci. 2013;4:1–14. doi: 10.3389/fpls.2013.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pushpa D, Yogendra KN, Gunnaiah R, Kushalappa AC, Murphy A. Identification of late blight resistance-related metabolites and genes in potato through nontargeted metabolomics. Plant Mol. Biol. Report. 2014;32:584–595. [Google Scholar]
- 67.Gunnaiah R, Kushalappa AC. Metabolomics deciphers the host resistance mechanisms in wheat cultivar Sumai-3, against trichothecene producing and non-producing isolates of Fusarium graminearum. Plant Physiol. Biochem. 2014;83:40–50. doi: 10.1016/j.plaphy.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 68.Cuperlovic-Culf, M., Rajagopalan, N., Tulpan, D. & Loewen, M.C. Metabolomics and cheminformatics analysis of antifungal function of plant metabolites. Metabolites, 6, 10.3390/metabo6040031 (2016). [DOI] [PMC free article] [PubMed]
- 69.Taheri P, Tarighi S. A survey on basal resistance and riboflavin-induced defense responses of sugar beet against Rhizoctonia solani. J. Plant Physiol. 2011;168:1114–1122. doi: 10.1016/j.jplph.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 70.Gerber IB, Zeidler D, Durner J, Dubery IA. Early perception responses of Nicotiana tabacum cells in response to lipopolysaccharides from Burkholderia cepacia. Planta. 2004;218:647–657. doi: 10.1007/s00425-003-1142-0. [DOI] [PubMed] [Google Scholar]
- 71.De Castro C, Parrilli M, Holst O, Molinaro A. Microbe-associated molecular patterns in innate immunity. Extraction and chemical analysis of Gram-negative bacterial lipopolysaccharides. Methods Enzymol. 2010;480:89–115. doi: 10.1016/S0076-6879(10)80005-9. [DOI] [PubMed] [Google Scholar]
- 72.Coventry HS, Dubery IA. Lipopolysaccharides from Burkholderia cepacia contribute to an enhanced defensive capacity and the induction of pathogenesis-related proteins in Nicotiana tabacum. Physiol. Mol. Plant Pathol. 2001;58:149–158. [Google Scholar]
- 73.Leontein K. Assignment of absolute configuration of sugars by glc of their acetylated glycosides formed from chiral alcohols. Meth. Carbohydr. Chem. 1978;62:359–362. [Google Scholar]
- 74.Ciucanu I, Kerek F. A simple and rapid method for the permethylation of carbohydrates. Carbohydr Res. 1984;131:209–217. [Google Scholar]
- 75.Rietschel E. Absolute configuration of 3-hydroxy fatty acids present in lipopolysaccharides from various bacterial groups. Eur. J. Biochem. 1976;64:423–428. doi: 10.1111/j.1432-1033.1976.tb10318.x. [DOI] [PubMed] [Google Scholar]
- 76.Bligh EG, Dyer WJ. A rapid method for total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 77.Piantini U, Sorensen OW, Ernst RR. Multiple quantum filters for elucidating NMR coupling networks. J. Am. Chem. Soc. 1982;104:6800–6801. [Google Scholar]
- 78.Rance. M, et al. Improved spectral resolution in COSY (1)H NMR spectra of proteins via double quantum filtering. Biochem. Biophys. Res. Commun. 1983;425:527–533. doi: 10.1016/j.bbrc.2012.08.019. [DOI] [PubMed] [Google Scholar]
- 79.States DJ, Haberkorn RA, Ruben D. A two-dimensional nuclear Overhauser experiment with pure absorption phase in four quadrants. J. Magn. Res. 1982;8:286–292. [Google Scholar]
- 80.Stern AS, Li KB, Hoch JC. Modern spectrum analysis in multidimensional NMR spectroscopy: comparison of linear-prediction extrapolation and maximum-entropy reconstruction. J. Am. Chem. Soc. 2002;124:1982–1993. doi: 10.1021/ja011669o. [DOI] [PubMed] [Google Scholar]
- 81.Silipo A, et al. Structure elucidation of the highly heterogeneous lipid A from the lipopolysaccharide of the Gram-negative extremophile bacterium Halomonas magadiensis Strain 21 M1. Eur. J. Org. Chem. 2004;10:2263–2271. [Google Scholar]
- 82.Sturiale L, et al. Reflectron MALDI TOF and MALDI TOF/TOF mass spectrometry reveal novel structural details of native lipooligosaccharides. J. Mass Spectrom. 2011;46:1135–1142. doi: 10.1002/jms.2000. [DOI] [PubMed] [Google Scholar]
- 83.Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 1962;15:473–497. [Google Scholar]
- 84.Madala NE, Steenkamp PA, Piater LA, Dubery IA. Different metabolite distribution patterns in isonitrosoacetophenone elicited tobacco and sorghum cells as revealed by multivariate statistical models. Springer Plus Online. 2014;3:254. doi: 10.1186/2193-1801-3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kang J, Price WE, Ashton J, Tapsell LC, Johnson S. Identification and characterization of phenolic compounds in hydromethanolic extracts of sorghum wholegrains by LC-ESI-MSn. Food Chem. 2016;211:215–226. doi: 10.1016/j.foodchem.2016.05.052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The study design information, LC-MS raw data, analyses and data processing information, and the meta-data were deposited to the EMBL-EBI metabolomics repository (MetaboLights50), with the identifier MTBLS1204 (http://www.ebi.ac.uk/metabolights/MTBLS.1204).