Abstract
Purpose
Escherichia coli O157:H7 is one of the major foodborne pathogens of global public concern. Bacteriophages (phages) have emerged as a promising alternative to antibiotics for controlling pathogenic bacteria. Here, a lytic E. coli O157:H7-specific phage (KFS-EC) was isolated, identified, and characterized to evaluate its potential as a biocontrol agent for E. coli O157:H7.
Methods
KFS-EC was isolated from slaughterhouse in Korea. Morphological analysis, genomic analysis and several physiological tests were performed to identify and characterize the KFS-EC.
Results
A specificity test indicated KFS-EC was strictly specific to E. coli O157:H7 strains among 60 bacterial strains tested. Morphological and phylogenetic analyses confirmed that KFS-EC belongs to the Rb49virus genus, Tevenvirinae subfamily, and the Myoviridae family of phages. KFS-EC genome consists of 164,725 bp and a total of 270 coding sequence features, of which 114 open reading frames (ORFs) were identified as phage functional genes. KFS-EC does not contain genes encoding lysogenic property and pathogenicity, which ensure its safe application. KFS-EC was relatively stable (~1 log decrease) under stressed conditions such as temperatures (20 °C–50 °C), pHs (3–11), organic solvents (ethanol and chloroform), and biocides (0.1% citric acid, 1% citric acid, and 0.1% peracetic acid). KFS-EC was able to inhibit E. coli O157:H7 efficiently at a multiplicity of infection (MOI) of 0.01 for 8 h with greater inhibitory effect and durability and was stable at 4 °C and 22 °C over a 12-week storage period.
Conclusions
Our results suggest that KFS-EC could be used as a biocontrol agent to E. coli O157:H7.
Electronic supplementary material
The online version of this article (10.1007/s40201-020-00452-5) contains supplementary material, which is available to authorized users.
Keywords: Bacteriophage, Escherichia coli O157:H7, Biocontrol agent, Lytic activity, Stability
Introduction
Escherichia coli is a gram-negative, facultative anaerobic bacterium that is commonly found in the gastrointestinal tract of mammals [1]. Severe clinical illness can be caused by various strains of E. coli including enteroaggregative E. coli (EAEC), enteropathogenic E. coli (EPEC), enterotoxigenic E. coli (ETEC), enteroinvasive E. coli (EIEC), and enterohemorrhagic E. coli (EHEC) [2]. EHEC with the specific serotype O157:H7 can lead to the widespread incidence of Shiga-like toxin, and hence is known as a Shiga toxin-producing E. coli (STEC) serotype [3], after E. coli O157:H7 has been first recognized as one of foodborne pathogens in 1982 [4]. E. coli O157:H7 causes diarrhea, abdominal pain, vomiting, and mild fever. Moreover, some infections could be developed into a severe life-threatening complication known as hemolytic uremic syndrome [2]. Furthermore, some unique characteristics of E. coli O157:H7 such as low infectious dose (< 100 cells) [1] and long survival in the environment render it as a major pathogen in the public health aspect [5]. It was estimated that STEC causes ~2,801,000 acute illnesses globally and leads to ~230 deaths annually [6].
E. coli O157:H7 exist in environmental sources (water and soil) as well as various foods such as undercooked beef, poultry, raw milk, vegetables, and fruits [7]. Antibiotic-based control strategy has been used in the food industry to reduce and inhibit E. coli O157:H7. However, antibiotic treatment may no longer be the most effective method for the control of E. coli O157:H7 due to the sharp increase in antibiotic-resistant infections and strains as a result of the acquisition of genetic capacity [8] and alteration of the microbiome composition of host by antibiotic exposure [9]. Recently, bacteriophage (phage) treatment has emerged as a promising alternative to antibiotics [10, 11]. Phages are viruses that have the ability to infect and kill target bacteria [12]. For the consideration as an effective biocontrol agent, phages are required to have the capacity to lyse bacterial cells (strictly lytic phage) [13], be stable under stressed conditions such as extreme temperatures and pHs [14], and facilitate targeted elimination of a specific strain without disruption of other microbiota [15]. In addition, phage must be free of the pathogenic genes that encode toxins and allergens. The purposes of this study were not only to isolate and purify a novel E. coli O157:H7-specific phage but also to characterize its physical and genetic properties for the evaluation of its potential for use as a biocontrol agent for E. coli O157:H7.
Materials and methods
Bacterial strains
The bacterial strains used in this study (Table 1) were obtained from the American Type of Culture Collection (ATCC) and a laboratory in the Department of Plant and Food Sciences at Sangmyung University in Chungnam, Korea. Bacterial strains were cultivated in a 25 mL tryptic soy broth (TSB, Difco Laboratories Inc., Sparks, MD, USA) for 16 h at 37 °C with shaking. After washing three times with sterilized phosphate-buffered saline buffer (PBS, pH 7.4; Life Technologies Ltd., Paisley, UK) by centrifugation at 7000×g for 4 min, bacterial pellets were suspended in PBS. The concentration of each suspension was adjusted to 108 colony forming units (CFU)/mL by using standard curves constructed by measuring the optical density at 640 nm (OD640).
Table 1.
Specificity of KFS-EC against each bacterial strain
| Bacterial strains | Plaque formationa | EOPb | Bacterial strains | Plaque formation | EOP |
|---|---|---|---|---|---|
| Aeromonas hydrophila SNUFPC A3 | – | NT | E. coli K12 VSM 1692 | – | – |
| A. hydrophila SNUFPC A5 | – | NT | E. coli O157:H7 ATCC 43895 | + | 1.00 ± 0.00 |
| A. hydrophila SNUFPC A7 | – | NT | E. coli O157:H7 700,599 | + | 0.95 ± 0.03 |
| A. hydrophila SNUFPC A9 | – | NT | E. coli O157:H7 rif R | + | 0.94 ± 0.01 |
| A. hydrophila SNUFPC A10 | – | NT | E. coli O157:H7 204p | + | 0.88 ± 0.04 |
| A. hydrophila SNUFPC A11 | – | NT | E. coli O157:H7 204 | + | 0.90 ± 0.02 |
| A. hydrophila JUNAH | – | NT | E. coli O157:H7 10,536 | + | 0.54 ± 0.05 |
| A. salmonicida 1,608,061 | – | NT | Klebsiella pneumoniae ATCC 13883 | – | NT |
| A. salmonicida 1,608,062 | – | NT | Listeria innocua ATCC 33090 | – | NT |
| Bacillus cereus ATCC 13061 | – | NT | L. monocytogenes 1911 | – | NT |
| B. cereus ATCC 1611 | – | NT | L. monocytogenes G3982 4b | – | NT |
| B. cereus ATCC 21768 | – | NT | L. monocytogenes G6055 | – | NT |
| B. cereus NCCP 14795 | – | NT | L. monocytogenes H7738 | – | NT |
| B. cereus RB002 | – | NT | L. monocytogenes H7757 | – | NT |
| B. cereus RB003 | – | NT | Pseudomonas aeruginosa ATCC 9027 | – | NT |
| B. cereus RB005 | – | NT | Salmonella Enteritidis ATCC 13076 | – | NT |
| B. cereus RB011 | – | NT | S. Heidelberg | – | NT |
| B. cereus RB013 | – | NT | S. Hartford | – | NT |
| B. cereus RB015 | – | NT | S. Mission | – | NT |
| B. cereus RB020 | – | NT | S. Montevideo | – | NT |
| B. cereus RB037 | – | NT | S. Newport | – | NT |
| B. pumilus SR067 | – | NT | S. Salama | – | NT |
| B. safensis SR138 | – | NT | S. Senftenberg | – | NT |
| B. stratosphericus SR095 | – | NT | S. Typhimurium ATCC 13311 | – | NT |
| B. subtilis KACC 12680 | – | NT | Shigella boydii NCCP 11190 | – | NT |
| B. toyonensis SR070 | – | NT | S. flexneri 2457 | – | NT |
| Escherichia coli ATCC BAA-2192 | – | NT | S. sonnei ATCC 9290 | – | NT |
| E. coli ATCC BAA-2196 | – | NT | Staphylococcus aureus ATCC 25923 | – | NT |
| E. coli BW 25113 | – | NT | Vibrio parahaemolyticus ATCC 17802 | – | NT |
| E. coli K12 ER 2738 | – | NT | Yersinia enterocolitica ATCC 23715 | – | NT |
a+, plaque formation; −, no plaque formation
bEOP: efficiency of plating. EOP values were calculated by dividing the number of plaques on each tested strain by the number of plaques on the indicator strain (E. coli O157:H7 ATCC 43895). NT means not tested. EOP values are shown as the mean ± SD (n = 3)
Phage isolation
A wastewater sample was collected from a slaughterhouse in Daegu, Korea. Twenty-five milliliters of wastewater was mixed with 225 mL of TSB containing 1 mL of E. coli O157:H7 ATCC 43895 (108 CFU/mL). After incubation for 16 h at 37 °C with 160 rpm agitation, the mixture was centrifuged at 4000×g for 10 min at 4 °C. The supernatant was collected and filtered using a 0.20-μm cellulose acetate filter (Advantec Toyo Kaisha, Ltd., Tokyo, Japan). To perform a dot assay, 10 μL of the filtrate was dotted on the surface of pre-solidified TA soft agar (4 g agar, 8 g nutrient broth, 5 g NaCl, 0.2 g MgSO4, 0.05 g MnSO4, and 0.15 g CaCl2 per 1 L) containing 200 μL of overnight culture of E. coli O157:H7 ATCC 43895. After incubation for 16 h at 37 °C, the formation of plaques was observed.
A plaque assay was performed to isolate a single phage against E. coli O157:H7. An aliquot of 100 μL of serial dilutions of the filtrate and 200 μL of overnight culture of E. coli O157:H7 ATCC 43895 were placed into 4 mL of TA soft agar prior to pouring onto tryptic soy agar (TSA, Difco Laboratories Inc.) plate. After incubation for 16 h at 37 °C, a single plaque was picked and placed into a sodium chloride-magnesium sulfate (SM) buffer (50 mM Tris-HCl, 100 mM NaCl, 10 mM MgSO4, pH 7.5) with vigorous agitation for 1 h at 22 °C. The plaque assay was repeated 6 times to elute one single phage. The eluted single phage was named KFS-EC, where EC indicates the host genus and species name (E. coli) [16].
Propagation and purification of phage
To increase the concentration of the isolated KFS-EC, the propagation procedures described below were performed several times with increasing volumes of TA broth. One percent of the overnight culture of E. coli O157:H7 ATCC 43895 was inoculated into 3 mL of TA broth (8 g nutrient broth, 5 g NaCl, 0.2 g MgSO4, 0.05 g MnSO4, and 0.15 g CaCl2 per 1 L) and incubated for 2.5 h at 37 °C with shaking at 190 rpm prior to the addition of the eluted KFS-EC at a multiplicity of infection (MOI) of 1. The mixture was incubated at 37 °C with shaking at 190 rpm and centrifuged at 2400×g for 10 min at 4 °C. The supernatant was filtered through a 0.20-μm cellulose acetate filter (Advantec Toyo Kaisha, Ltd.) and the concentration of KFS-EC was measured at every step using the plaque assay. The final filtrate was mixed with 10% (w/v) polyethylene glycol 6000 (Sigma-Aldrich Co., St. Louis, MO, USA) and 10 mL of 1 M NaCl, followed by overnight precipitation at 4 °C. After centrifugation at 7200×g for 20 min at 4 °C, the supernatant was discarded, and the remaining pellet was suspended in 6 mL SM buffer and subjected to five-layer CsCl gradient ultracentrifugation at 39,000×g for 2 h at 4 °C. The bluish opalescent band was extracted, dialyzed and stored in SM buffer at 4 °C and the concentration of KFS-EC in SM buffer was determined by plaque assay before use.
Morphological characteristics of KFS-EC
KFS-EC (5 × 109 plaque forming units [PFU]) was adsorbed on a carbon-coated copper grid and negatively stained with 4% phosphotungstic acid (Sigma-Aldrich Co.). The morphological characteristics of KFS-EC were observed using transmission electron microscopy (TEM, H-7100, Hitachi Ltd., Tokyo, Japan) at an accelerating voltage of 100 kV with 40,000 to 60,000 × magnification.
Specificity and efficiency of plating (EOP) analysis
A dot assay was conducted to measure the specificity of KFS-EC by using various bacterial strains including E. coli O157:H7 strains (Table 1). An aliquot of 200 μL of each bacterial culture was mixed with 4 mL of 0.4% TA soft agar and overlaid on a TSA plate. Then, 10 μL of diluted KFS-EC was dotted on the plate and incubated for 16 h at 37 °C. KFS-EC susceptible bacterial strains were confirmed by the formation of a clear zone. Next, plaque assay was carried out to analyze the efficiency of plating (EOP) against phage susceptible bacterial strains. The number of plaques was determined after a 12-h incubation and the EOP was calculated by dividing the number of plaques on each tested strain by the number of plaques on the indicator strain (E. coli O157:H7 ATCC 43895). EOP value >0.5, >0.1 - <0.5, and 0.001–0.1 is viewed as high, medium and low lytic activity of the phage, respectively, while the EOP value of ≤0.001 denote the insufficient lytic activity of the phage against the bacterial strain [17].
Genome sequencing and annotation
Genomic DNA of KFS-EC was extracted using the Phage DNA Isolation Kit (Norgen Biotek Co., Thorold, ON, Canada) according to the manufacturer’s instructions. The concentration of genomic DNA was measured using a PicoDrop spectrophotometer (Picodrop Ltd., Hinxton, Cambridge, UK). Genome sequencing was performed by LabGenomics (Seongnam-si, Korea) using the paired-end Miseq sequencing platform (Illumina, Inc., San Diego, CA, USA). Totally, 2 × 1,558,510 reads were generated, and raw reads were trimmed to eliminate low quality reads and adaptor sequences using BBduk trimmer (version 1.0) [18]. Trimmed reads were assembled into contigs using Geneious v.11.1.4. (Biomatters Ltd., Auckland, New Zealand). Briefly, 2,605,946 of 3,115,931 reads were assembled and produced 17,152 contigs. They were used to reconstruct a 164,725 bp consensus sequence. The genome sequence of KFS-EC was initially compared with those available in the GenBank database using the PubMed NCBI BLAST program. Then, the genome sequence was annotated by Rapid Annotations using Subsystem Technology (RAST) [19] and confirmed with Genemark.hmm [20] and PHASTER [21]. All identified open reading frames (ORFs) were compared against the virulence factor database [22], ResFinder 2.1 [23], and Comprehensive Antibiotic Resistance Database (CARD) [24]. The predicted protein sequences were compared with the allergen database (http://www.allergenonline.com) from Food Allergy Research to confirm potential allergens in phage proteins. Potential tRNA genes in the genome sequence were predicted using ARAGORN and RNAmmer (version 1.2) in rapid prokaryotic genome annotation pipeline (Prokka) [25]. A genome map was generated and annotated according to the RAST subsystem.
Nucleotide sequence accession number
The complete genome sequence of KFS-EC is available in the GenBank database under nucleotide sequence accession number MH560358.
Phylogenetic analysis of KFS-EC
Eleven phage genome sequences from the GenBank database were included for phylogenetic analysis. These phages were selected as the representatives of genera within the subfamily Tevenvirinae, family Myoviridae [26]. The KFS-EC genome sequence with 11 phage sequences was subjected to genome-based phylogenetic analysis using the Virus Classification and Tree building Online Resource (VICTOR) [27]. All pairwise comparisons of the nucleotide sequences were conducted using the Genome-BLAST Distance Phylogeny (GBDP) method [28] under settings recommended for prokaryotic viruses [27]. The resulting intergenomic distances were used to infer a balanced minimum evolution tree with branch support via FASTME including SPR post-processing [29]. Branch support was inferred from 100 pseudo-bootstrap replicates each. Trees were rooted at the midpoint [30] and visualized with FigTree [31]. Taxon boundaries at the species, genus, and family levels were estimated with the OPTSIL program [32], the recommended clustering thresholds [27], and an F value (fraction of links required for cluster fusion) of 0.5 [33].
Effects of temperature, pH, and biocides on the lytic activity of KFS-EC
Lytic activity of KFS-EC was investigated by exposing the phage to various temperatures, pHs, and organic solvents and biocides. To analyze the effect of temperature on the lytic activity of KFS-EC, 100 μL of KFS-EC (106 PFU/mL) was mixed with 900 μL TSB (pH 7) and incubated at various temperatures (−70, −20, 4, 10, 20, 30, 40, 50, 60, and 70 °C) for 1 h. To measure the effect of pH, 100 μL (106 PFU/mL) of KFS-EC was mixed with 900 μL of TSB at various pHs (1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, and 12), adjusted with either 1 M HCl or 1 M NaOH, and incubated for 1 h at 22 °C. To observe the effect of organic solvents and biocides, 100 μL of KFS-EC was mixed with 900 μL of 100% organic solvent (chloroform, ethanol, or isopropanol) or biocide (0.1% peracetic acid, 1% peracetic acid, 100 ppm sodium hypochlorite, 200 ppm sodium hypochlorite, 0.1% citric acid, or 1% citric acid) and incubated for 1 h and 5 min, respectively, at 22 °C. Finally, the plaque assay described above was performed and the results were expressed as phage concentration (log PFU/mL).
Bacterial challenge assay and one-step growth curve
For the bacterial challenge assay, an overnight culture of E. coli O157:H7 ATCC 43895 was inoculated into 100 mL of fresh TSB medium (2% inoculum) and incubated for 3 h at 37 °C with shaking at 190 rpm. When the OD640 reached ~0.2 (108 CFU/mL), 1 mL of KFS-EC was added at a multiplicity of infection (MOI) of 0.01, 0.1, 1.0, 10, and 100 and incubated at 37 °C with shaking at 110 rpm for 26 h. Culture without the addition of KFS-EC was used as a control. The bacterial growth was monitored at 2-h intervals by measuring OD640 [13].
The mixture of the overnight culture of E. coli O157:H7 ATCC 43895 (30 mL; OD640 = 1.0) and KFS-EC at an MOI of 0.01 was incubated for 30 min at 37 °C for bacterial adsorption. To remove unabsorbed KFS-EC, the mixture was centrifuged at 11,400×g for 10 min at 4 °C. The pellet was resuspended with the same volume of TSB and incubated at 37 °C. Every 5 min, 1 mL of the incubated mixture was collected, and each collected suspension was evenly divided into 1.5-mL tubes. To determine the eclipse period, chloroform (f.c. 1%, v/v) was added to each 1.5-mL tube followed by vigorous vortexing for 30 s in order to release the internal KFS-EC. Chloroform-treated KFS-EC and non-chloroform-treated KFS-EC suspensions were diluted serially and plaque assay was performed to determine the latent period and burst size.
Storage study
KFS-EC (108 PFU/mL) in TSB was stored at various temperatures (−70, −20, 4, and 22 °C) to investigate its viability after storage. To minimize freezing damage on KFS-EC, 15% of glycerol was added and stored at −70 °C and − 20 °C. At 2-week intervals, each stored sample was collected and a plaque assay was performed to measure the lytic activity of KFS-EC at each collection time.
Statistical analysis
All data were expressed as the mean ± standard deviation (SD). GraphPad and InStatV.3 (GraphPad, San Diego, CA, USA) were used to perform the statistical analyses. Student’s paired t test for two groups and one-way analysis of variance (ANOVA) among more than two groups were used for the comparison. The significance level was determined at p < 0.05.
Results
Isolation and morphological analysis of KFS-EC
One phage (KFS-EC) was isolated from a wastewater sample taken from a slaughterhouse in Korea. Fig. S1 in Supplementary material shows the plaque formation of KFS-EC after isolation using a dot assay (Fig. S1[a], Supplementary material) and a plaque assay (Fig. S1[b], Supplementary material). Finally, KFS-EC was purified at a concentration of (5.32 ± 0.23) × 1011 PFU/mL. TEM images (Fig.1) show that the KFS-EC phage has an icosahedral shape (100.25 ± 7.26 nm in length 70.89 ± 8.90 in width) with a hexagonal head and a tail (113.88 ± 10.19 nm in length, 21.91 ± 2.59 nm in width). The tailed phages are normally classified into either Myoviridae (long, rigid, contractile tail), Podoviridae (very short, non-contractile tail), or Siphoviridae (long, flexible, non-contractile tail), which are families in the Caudovirales order [34]. Therefore, KFS-EC was presumed to belong to the Myoviridae family based on a comparison with the findings of previous studies (Table S1, Supplementary material).
Fig. 1.
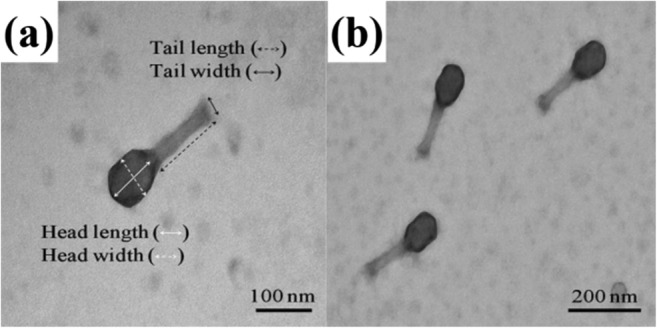
TEM images of KFS-EC at a magnification of (a) × 60.0 k and (b) × 40.0 k
Specificity and EOP analysis
A specificity test indicated that KFS-EC had the capability to form completely cleared zones on all tested E. coli O157:H7 strains among 60 bacterial strains including 11 E. coli strains (Table 1). Although EOP values was varied among E. coli O157:H7 strains, all EOP values were categorized into “high production” (EOP value >0.5)” against all E. coli O157:H7 strains tested [17].
Genome analysis
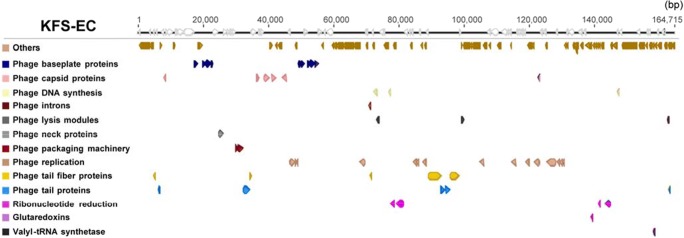
The KFS-EC genome was sequenced with 4746 × coverage. The final consensus sequence showed that it was 98% identical to the complete genomes of Enterobacteria phage RB49 and 99% identical to the complete genomes of Shigella phage Sf20 (GenBank accession no. AY343333 and MF327006, respectively) in a nucleotide BLAST comparison. The assembled KFS-EC genome consists of 164,725 bp with 40.5% G + C content and has terminally redundant ends of 812 bp. Annotation revealed a total of 270 coding sequence features in the genome, of which 114 ORFs were assigned the phage functional genes (Fig. 2; Table S2, Supplementary material). The phage structural genes consisted of genes for phage DNA replication (15 genes), phage baseplate proteins (10 genes), phage capsid proteins (7 genes), phage tail fiber proteins (5 genes), phage tail proteins (5 genes), phage lysis module (4 genes), phage DNA synthesis (3 genes), phage packaging machinery (2 genes), phage neck protein (1 gene), and a phage intron (1 gene). KFS-EC encoded thymidine kinase, thymidylate synthase, dNMP kinase, dCMP deaminase, RNA endonuclease, dihydrofolate reductase, and ribonucleotide reductase. Enzyme sequences were highly conserved (> 90% identity) with the enzyme sequences of Enterobacteria phage RB49. In addition, a tRNA synthetase (valyl-tRNA synthetase) gene, a glutaredoxin gene, and 4 ribonucleotide reduction genes were identified in the KFS-EC genome. The remaining 156 ORFs are hypothetical proteins. Potential tRNA sequences were not found in the KFS-EC genome. Notably, genes coding for lysogenic activity, virulence, antibiotic resistance, and potential allergens were not found in the KFS-EC genome.
Fig. 2.
Genome organization of KFS-EC. Predicted ORFs are indicated as arrows. Heads of arrows designate the direction of transcription. The colors were assigned according to subsystem classification (or functional categories) of the ORFs in the RAST pipeline. White arrows indicate ORFs encoding putative or hypothetical proteins. More detailed information is listed in Table S2
Phylogenetic analysis
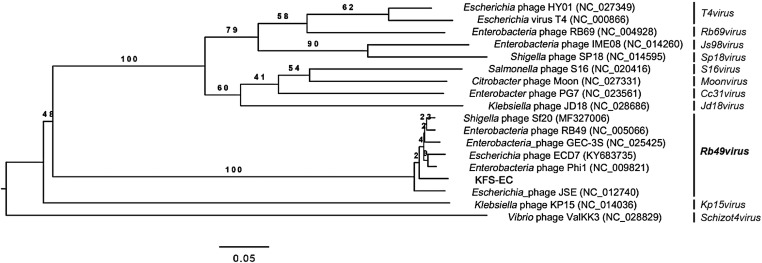
Enterobacteria phage RB49 and Shigella phage Sf20, whose genomes are highly similar (> 98%) to that of KFS-EC as described above, are classified into the Tevenvirinae subfamily and Myoviridae family by the International Committee on Taxonomy of Viruses (ICTV) taxonomy [26]. To obtain a more accurate classification of KFS-EC, a phylogenetic analysis with genome sequences of representative phages within Tevenvirinae was performed, revealing that KFS-EC is closely related to strains within the genus Rb49virus, as proposed by ICTV [26] (Fig. 3).
Fig. 3.
Phylogenetic tree of KFS-EC with closely related phage strains. The analysis was conducted using VICTOR [27] and the tree was constructed using FigTree [31]. KFS-EC is shown in the phylogenomic GBDP trees inferred using the formulas D0 and yielding an average support of 48%. The numbers above the branches are GBDP pseudo-bootstrap support values from 100 replications. The branch lengths of the resulting VICTOR trees are scaled in terms of the respective distance formula used. The numbers in parentheses are GenBank sequence accession numbers. Genus classification by ICTV taxonomy [26] is listed on the right
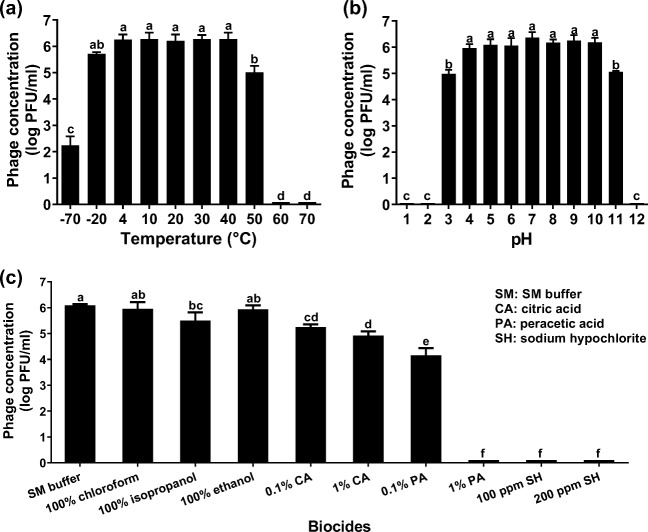
Effects of temperature, pH, and biocides on the lytic activity of KFS-EC
The lytic activity of KFS-EC was stable at temperatures between 4 and 40 °C (Fig. 4a). Slight decrease (~0.5 log) was observed at −20 °C, whereas the lytic activity of KFS-EC decreased at 50 °C (~1.2 log decrease) and at −70 °C (~4 log decrease) (p < 0.05). The lytic activity of KFS-EC phage was almost lost at 60 °C and 70 °C. With regard to the effect of pH, the lytic activity of KFS-EC phage was relatively stable (~l.0 log decrease) at pH values of 3–11, although significant reductions of KFS-EC concentration at pH 3 and 11 were observed (p < 0.05) (Fig. 4b). However, the lytic activity of KFS-EC was almost lost under extremely strong acidic and basic conditions (pH 1, 2, and 12). The lytic activity of KFS-EC was stable when exposed to 100% chloroform and 100% ethanol. However, the lytic activity of KFS-EC exposed to 100% isopropanol was significantly decreased (~0.6 log) compared with that of chloroform and ethanol (p < 0.05) (Fig. 4c). Moreover, sodium hypochlorite and 1% peracetic acid were the most effective phagicidal (virucidal) biocide, indicated by the almost complete inactivation of the phage, while KFS-EC showed relatively strong resistance against citric acid and 0.1% peracetic acid (Fig. 4c).
Fig. 4.
Stability of KFS-EC at various (a) temperatures, (b) pHs, and (c) biocides. KFS-EC (106 PFU/mL) in TSB was incubated at various temperatures, pHs and biocides and the phage concentration was measured by plaque assay as described in the materials and methods. The letters (a-d) indicate significant differences between treatment condition within groups p < 0.05. The data represents the mean ± SD (n = 3)
Bacterial challenge assay and one-step growth curve
E. coli O157:H7 ATCC 43895 was infected with KFS-EC at different MOIs at 37 °C (Fig. 5). Infection with KFS-EC began to considerably suppress host growth at the entire range of MOIs. When compared with the control (without phage exposure), phage treatment with even MOI of 0.01 decreased the turbidity of the cultures significantly (p < 0.05). Furthermore, growth inhibition of the host was then maintained until 8-h post-infection. In addition, there were no significant differences in the growth inhibition of E. coli O157:H7 ATCC 43895 among the different MOIs (0.01, 0.1, 1.0, 10, and 100) treatment during 8 h. Although re-growth of bacteria was observed after 10 h, the bacterial growth after exposure to KFS-EC remained lower than that of the control up to 20-h post infection, even at a very low MOI of 0.01.
Fig. 5.
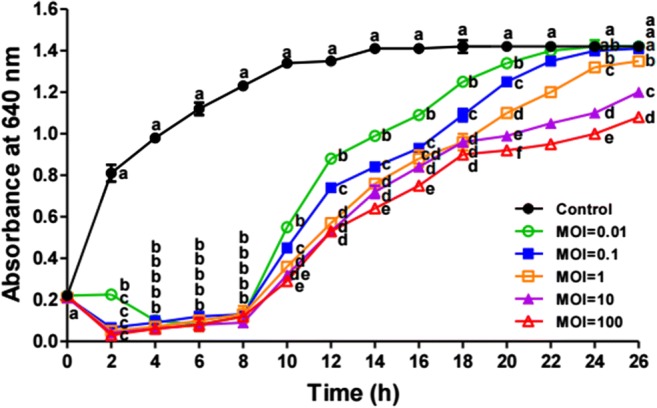
Bacterial challenge assay of KFS-EC with E. coli O157:H7. E. coli O157:H7 was incubated at 37 °C with KFS-EC at MOI of 0.01, 0.1, 1.0, 10, and 100, respectively and bacterial growth was determined by measuring absorbance. ●, control group without KFS-EC infection; ○, ■, □, ▲ and △, experimental groups with KFS-EC infection at a MOI of 0.01, 0.1, 1.0, 10, and 100, respectively. The letters (a-f) indicate significant differences between various MOI values at the same incubation time at p < 0.05. The data represents the mean ± SD (n = 3)
To determine the eclipse period, latent period, and burst size of KFS-EC, a one-step growth curve analysis was conducted with E. coli O157:H7 ATCC 43895 (Fig. 6). It indicated 5 min and 30 min for the eclipse period and latent period, respectively. The burst size was about ~150 PFU per infected cell.
Fig. 6.
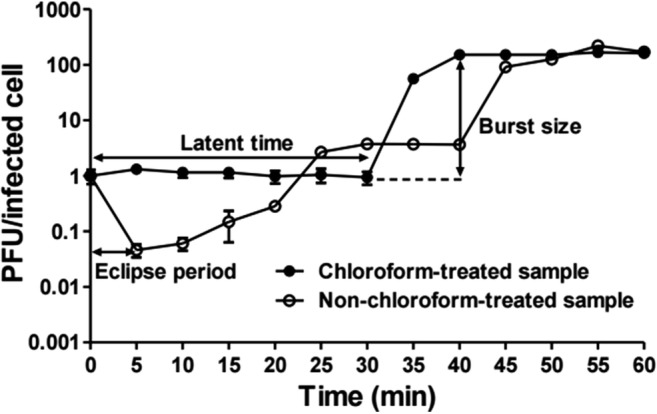
One step growth curve analysis of KFS-EC. The mixture of E. coli O157:H7 ATCC 43895 and KFS-EC at an MOI of 0.01 was incubated for 1 h at 37 °C under agitation. The mixture was collected every 5 min and divided into two groups as a non-chloroform treatment (○) and 1% chloroform treatment (●) for the determination of eclipse period, latent time, and burst size. The data represents the mean ± SD (n = 3)
Storage study
KFS-EC was exposed to deep freezing temperatures (−70 °C and − 20 °C), cold temperature (4 °C), and room temperature (22 °C) over a 12-week storage period to evaluate the potential effects of different temperatures on the stability of KFS-EC. Storage at 22 °C and 4 °C did not significantly inactivate KFS-EC for the duration of the storage period (Fig. 7). Although storage at −20 °C and − 70 °C significantly inactivated KFS-EC (p < 0.05), the overall titers were reduced only by ~1.0 log and ~1.7 log over the 12-week storage period, respectively.
Fig. 7.
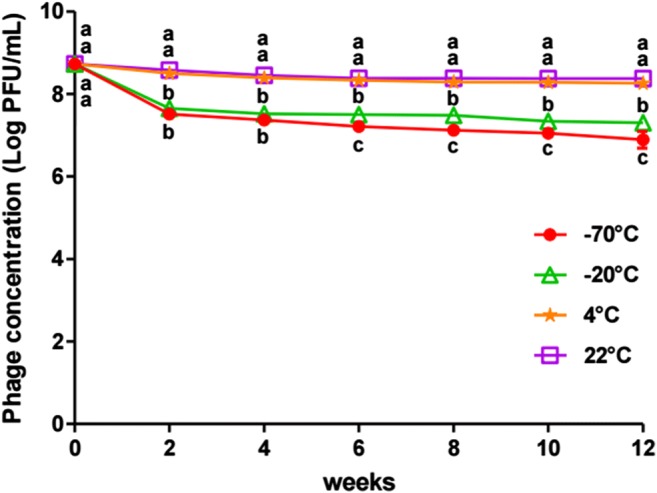
Stability of KFS-EC stored at various temperatures. KFS-EC in TSB was stored at various temperatures and collected every 2 weeks to investigate its lytic activity using plaque assay. The letters (a-c) indicate significant differences between various temperatures at the same storage periods at p < 0.05
Discussion
Our study presents a novel E. coli O157:H7-specific phage with morphological analysis, physiological tests, and genome analysis. To our knowledge, a total of 16 phages infecting E. coli O157:H7 have been reported since 2000 and 16 of these phages were classified into the families Myoviridae (9 strains), Siphoviridae (6 strains), and Podoviridae (1 strains) within the order Caudovirales (Table S1, Supplementary material). Our findings were consistent with these previous results because KFS-EC was found to belong to the Myoviridae family by TEM analysis (Fig. 1). Comparison of the morphological features of 9 Myoviridae phage strains revealed that its size was only similar with HY01 [13], FAHEc1 [35], and CEV1 [36] despite of different tail sizes with them (Table S1, Supplementary material). KFS-EC was strictly specific to E. coli O157:H7 only among 60 bacterial strains (Table 1), indicating that KFS-EC phage showed specific recognition only in E. coli O157:H7 strains. When compared with other previously reported phages, HY01 was specific to both E. coli O157:H7 and Shigella flexneri among 23 tested foodborne pathogens [13]. SFP10 phage [37] was specific to 11 strains of Salmonella (among 13 tested Salmonella) and 4 strains of E. coli O157:H7 from whole 32 tested foodborne pathogens.
The KFS-EC genome analysis showed highly similar features compared to closely related phages (Fig. 3). Their genome size (~164 kbp), G + C content (40.4–40.6%), and genomic sequence (>98% identity) were similar to those of other phage strains. KFS-EC has a G + C content of 40.06%, which is significantly lower than that of E. coli O157:H7 (average 50%) [11, 38]. Generally, the G + C content of a phage is lower than that of the host strains [39]. Previous studies showed that Enterobacteria phage RB49, Enterobacteria phage JSE, and Enterobacteria phage phi1 recognize E. coli B/5, E. coli K12 and E. coli K12F+ as a host, respectively [40, 41]. On the other hand, the KFS-EC phage showed specific recognition only to E. coli O157:H7 strains but not in other strains as mentioned above. Although there is more than 98% identity on a genomic basis, it has distinctive features among relative strains. On a molecular level, the specificity of host strains was decided by phage proteins. KFS-EC has a unique phage tail fiber protein (gp37 long tail fiber distal subunit; Fig. 2) in its genome. The nucleotide sequence of gp37 long tail fiber distal subunit in the genome sequence of KFS-EC shows less than 30% coverage when compared to those in the genome sequences of other relative phages (Table S3, Supplementary material). The long tail fibers are known to act as a reversible anchor to specific sites on the bacterial surface. It was reported that the gp37 protein of the isolated phage which infect E. coli O157:H7 show higher similarity to those of E. coli O157:H7-typing phages (AR1 and wV7) [42]. Thus, this specific recognition plays an important role in the selection of the host-phage [43, 44].
EOP data (Table 1) confirmed that KFS-EC could infect E. coli O157:H7 specifically with minimal disruption of other microorganisms [15, 35, 45], supporting phage application over antibiotic use [46]. From the result of the one-step growth curve (Fig. 6), the latent period and burst size of KFS-EC were determined to be 30 min and 150 PFU/cell, respectively, which indicates KFS-EC has strong lytic activity. Specifically, the latent period of KFS-EC was relatively longer than that of HY01 [13] (15 min) and SFP10 [37] (25 min) whereas the burst size of KFS-EC was larger than that of HY01 (100 PFU/cell) and SFP10 (25 PFU/cell). Conversely, CBA phage (Table S1, Supplementary material) showed a long latent period (40 min) and larger burst size (440 PFU/cell) than KFS-EC. Moreover, genomic analysis further showed that there are no coding genes for lysogenic property, virulence factor, antibiotic resistance, and potential allergen in the KFS-EC genome (Fig. 2). These results clearly indicate the potential of KFS-EC as a biocontrol agent with great effectiveness and safety.
The ability of phages to facilitate horizontal gene transfer through transduction is an important consideration in their use as biocontrol agents [47, 48]. Terminal redundancy (Fig. 2) and phylogenetic analysis (Fig. 3) suggest that KFS-EC is predicted to be pac-type phage-like, using a headful packaging mechanism [47]. Virulent pac-type phages do not display generalized transduction due to a tendency to degrade the host genome enzymatically [48]. Restriction–modification is one of the systems that bacteria utilize to guard against phage infection [49]. E. coli O157:H7 has type I restriction-modification system [50, 51]. Most restriction enzymes do not digest T4 DNA due to the modification of cytosine [49, 52, 53]. As a result, KFS-EC, which belongs to the genus T4virus, may be more infective to E. coli O157:H7.
Since E. coli O157:H7 can survive a relatively broad range of temperatures (7–46 °C) and pHs (4.4–9.0), the lytic activity of KFS-EC phage needs to cover these ranges for robust phage application. Temperature and pH stability studies demonstrated that KFS-EC sustained its lytic property through a broad range of temperatures (−20–50 °C) and pHs (3–11) (Fig. 4a and b), indicating greater stability of this phage compared with those in previous studies [13, 54]. Biocides are commonly used in the food industry for reducing and controlling microbial contamination in food [55]. Our results (Fig. 4c) also showed that KFS-EC was stable in the presence of biocides such as chloroform, ethanol, citric acid, and 0.1% peracetic acid but not sodium hypochlorite and 1% peracetic acid. When compared with DT1, DT2, DT3, DT4, DT5, and DT6 phages infecting E. coli O157:H7, our phage showed better stability in the presence of 100% ethanol and similar stability in the presence of sodium hypochlorite [56]. This means that KFS-EC would be stable and active against E. coli O157:H7 when exposed to various environmental conditions during food processing and storage [37].
In the bacterial challenge assay (Fig. 5), KFS-EC inhibited the growth of E. coli O157:H7 ATCC 43895 for 8 h at various MOIs. Mounting studies addressed that HY01 and SFP10 inhibited the growth of E. coli only for 1-h post infection [13, 37]. Vb_EcoS-B2phage and vB EcoS HSE also inhibited the growth of E. coli at an MOI of 10 [57, 58]. Our study revealed that KFS-EC showed excellent bacterial inhibition for longer periods even with a very low MOI of 0.01. Although re-growth of E. coli O157:H7 ATCC 43895 was observed, like in other studies [13, 37, 59], it could be controlled to some extent by storing phage-treated foods at cold temperatures or eliminating all host bacteria completely. However, complete elimination of host bacteria is not achievable due to re-growth of E. coli O157:H7. The reason for the re-growth could be explained by the appearance of bacteriophage insensitive mutants (BIMs) bacteria. Due to the BIMs, the surviving E. coli O157:H7 can continue to grow [13]. So, further practical approaches are required. Since KFS-EC showed excellent stability even at cold temperatures (Fig. 7), the combination of phage treatment with temperature will be considered in a future study. Due to the limited studies [60, 61] regarding the storage study of phages, it was difficult to compare our results with other studies objectively. Nonetheless, the fact that KFS-EC showed significantly better stability when stored at cold (4 °C) and room temperatures (22 °C) (Fig. 7) will enhance its practicability in use. Overall, the bacterial challenge and storage studies confirmed that KFS-EC might provide excellent practicability and cost efficiency, which will enhance the suitability and applicability of KFS-EC phage as a potential biocontrol agent.
Conclusions
The present study revealed that KFS-EC isolated from a wastewater sample taken from a slaughterhouse in Korea is specific to E. coli O157:H7 only. TEM analysis and phylogenetic analysis confirmed that it belongs to the Rb49virus genus within the Tevenvirinae subfamily within the Myoviridae family. Genomic analysis suggests that KFS-EC may be safe for use as a biocontrol agent because its genome does not have virulence factor, toxins, antibiotic resistance, and potential allergen-coding genes. Moreover, KFS-EC lytic activity was highly stable at various pHs and temperatures as well as in the presence of organic solvents and biocides. Additionally, KFS-EC can inhibit E. coli O157:H7 growth even at an MOI of 0.01 for 8 h and can be stable at cold and room temperatures for a relatively long period of time. Collectively, our results clearly speculate that KFS-EC would be useful for application as a biocontrol agent against E. coli O157:H7 in the food industry.
Electronic supplementary material
(DOCX 157 kb)
Author’s contributions
CL, IYC, DHP, and M-KP conducted this study, and drafted the manuscript. M-KP supervised the study in all steps. CL, IYC, and M-KP edited the manuscript. All authors read and approved the final manuscript.
Funding
This research was funded by the Basic Research Program through the National Research Foundation (NRF) of Korea founded by the Ministry of Education (NRF-2017R1D1A1B03035195).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Cheonghoon Lee and In Young Choi contributed equally to this work.
References
- 1.Adamu MT, Shamsul BMT, Desa MN, Khairani-Bejo S. A review on Escherichia coli O157:H7-the super pathogen. Health Environ J. 2014;5:78–93. [Google Scholar]
- 2.Saxena T, Kaushik P, Mohan MK. Prevalence of E. coli O157:H7 in water sources: an overview on associated diseases, outbreaks and detection methods. Diagn Microbiol Infect Dis. 2015;82:249–264. doi: 10.1016/j.diagmicrobio.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 3.Saeedi P, Yazdanparast M, Behzadi E, Salmanian AH, Mousavi SL, Nazarian S, et al. A review on strategies for decreasing E. coli O157:H7 risk in animals. Microb Pathog. 2017;103:186–195. doi: 10.1016/j.micpath.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 4.Riley LW, Remis RS, Helgerson SD, McGee HB, Wells JG, Davis BR et al. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N Engl J Med. 1983;308:681–5. [DOI] [PubMed]
- 5.Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–40. [DOI] [PubMed]
- 6.Majowicz SE, Scallan E, Jones-Bitton A, Sargeant JM, Stapleton J, Angulo FJ, et al. Global incidence of human Shiga toxin–producing Escherichia coli infections and deaths: a systematic review and knowledge synthesis. Foodborne Pathog Dis. 2014;11:447–55. [DOI] [PMC free article] [PubMed]
- 7.Holvoet K, Sampers I, Callens B, Dewulf J, Uyttendaele M. Moderate prevalence of antimicrobial resistance in Escherichia coli isolates from lettuce, irrigation water, and soil. Appl Environ Microbiol. 2013;79:6677–6683. doi: 10.1128/AEM.01995-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schroeder CM, Zhao C, DebRoy C, Torcolini J, Zhao S, White DG, et al. Antimicrobial resistance of Escherichia coli O157 isolated from humans, cattle, swine, and food. Appl Environ Microbiol. 2002;68:576–581. doi: 10.1128/AEM.68.2.576-581.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Francino MP. Antibiotics and the human gut microbiome: dysbioses and accumulation of resistances. Front Microbiol. 2016;6:1543. doi: 10.3389/fmicb.2015.01543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi IY, Lee J-H, Kim H-J, Park M-K. Isolation and characterization of a novel broad-host-range bacteriophage infecting Salmonella enterica subsp. enterica for biocontrol and rapid detection. J Microbiol Biotechnol. 2017;27:2151–2155. doi: 10.4014/jmb.1711.11017. [DOI] [PubMed] [Google Scholar]
- 11.Amarillas L, Chaidez C, González-Robles A, Lugo-Melchor Y, León-Félix J. Characterization of novel bacteriophage phiC119 capable of lysing multidrug-resistant Shiga toxin-producing Escherichia coli O157:H7. PeerJ. 2016;4:e2423. doi: 10.7717/peerj.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oh J-H, Park M-K. Recent trends in Salmonella outbreaks and emerging technology for biocontrol of Salmonella using phages in foods: a review. J Microbiol Biotechnol. 2017;27:2075–2088. doi: 10.4014/jmb.1710.10049. [DOI] [PubMed] [Google Scholar]
- 13.Lee H, Ku H-J, Lee D-H, Kim Y-T, Shin H, Ryu S, et al. Characterization and genomic study of the novel bacteriophage HY01 infecting both Escherichia coli O157:H7 and Shigella flexneri: potential as a biocontrol agent in food. PLoS One. 2016;11:e0168985. doi: 10.1371/journal.pone.0168985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hudson JA, Billington C, Carey-Smith G, Greening G. Bacteriophages as biocontrol agents in food. J Food Prot. 2005;68:426–437. doi: 10.4315/0362-028X-68.2.426. [DOI] [PubMed] [Google Scholar]
- 15.Carlton RM. Phage therapy: past history and future prospects. Arch Immunol Ther Exp. 1999;47:267–274. [PubMed] [Google Scholar]
- 16.Ackermann H-W, DuBow M, Jarvis A, Jones L, Krylov V, Maniloff J, et al. The species concept and its application to tailed phages. Arch Virol. 1992;124:69–82. doi: 10.1007/BF01314626. [DOI] [PubMed] [Google Scholar]
- 17.Mirzaei MK, Nilsson AS. Isolation of phages for phage therapy: a comparison of spot tests and efficiency of plating analyses for determination of host range and efficacy. PLoS One. 2015;10:e0118557. doi: 10.1371/journal.pone.0118557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bushnell, B. BB Map 2014. https://sourceforge.net/projects/bbmap/ Accessed 14 Feb 2019.
- 19.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, et al. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu W, Lomsadze A, Borodovsky M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 2010;38:e132. doi: 10.1093/nar/gkq275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen L, Zheng D, Liu B, Yang J, Jin Q. VFDB 2016: hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res. 2016;44:D694–D697. doi: 10.1093/nar/gkv1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, et al. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother. 2013;57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 26.Lefkowitz EJ, Dempsey DM, Hendrickson RC, Orton RJ, Siddell SG, Smith DB. Virus taxonomy: the database of the international committee on taxonomy of viruses (ICTV) Nucleic Acids Res. 2017;46:D708–D717. doi: 10.1093/nar/gkx932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meier-Kolthoff JP, Göker M. VICTOR: genome-based phylogeny and classification of prokaryotic viruses. Bioinformatics. 2017;33:3396–3404. doi: 10.1093/bioinformatics/btx440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meier-Kolthoff JP, Auch AF, Klenk H-P, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lefort V, Desper R, Gascuel O. FastME 2.0: a comprehensive, accurate, and fast distance-based phylogeny inference program. Mol Biol Evol. 2015;32:2798–2800. doi: 10.1093/molbev/msv150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farris JS. Formal definitions of paraphyly and polyphyly. Syst Zool. 1974;23:548–554. doi: 10.2307/2412474. [DOI] [Google Scholar]
- 31.Rambaut A. FigTree-version 1.4.3, a graphical viewer of phylogenetic trees; 2017. http://tree.bio.ed.ac.uk/software/figtree/ Accessed 14 Feb 2019.
- 32.Göker M, García-Blázquez G, Voglmayr H, Tellería MT, Martín MP. Molecular taxonomy of phytopathogenic fungi: a case study in Peronospora. PLoS One. 2009;4:e6319. doi: 10.1371/journal.pone.0006319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meier-Kolthoff JP, Hahnke RL, Petersen J, Scheuner C, Michael V, Fiebig A, et al. Complete genome sequence of DSM 30083T, the type strain (U5/41T) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand Genomic Sci. 2014;9:2. doi: 10.1186/1944-3277-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ackermann H-W. 5500 phages examined in the electron microscope. Arch Virol. 2007;152:227–243. doi: 10.1007/s00705-006-0849-1. [DOI] [PubMed] [Google Scholar]
- 35.Hudson JA, Billington C, Cornelius AJ, Wilson T, On SLW, Premaratne A, et al. Use of a bacteriophage to inactivate Escherichia coli O157:H7 on beef. Food Microbiol. 2013;36:14–21. doi: 10.1016/j.fm.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 36.Raya RR, Varey P, Oot RA, Dyen MR, Callaway TR, Edrington TS, et al. Isolation and characterization of a new T-even bacteriophage, CEV1, and determination of its potential to reduce Escherichia coli O157:H7 levels in sheep. Appl Environ Microbiol. 2006;72:6405–6410. doi: 10.1128/AEM.03011-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park M, Lee J-H, Shin H, Kim M, Choi J, Kang D-H, et al. Characterization and comparative genomic analysis of a novel bacteriophage, SFP10, simultaneously inhibiting both Salmonella enterica and Escherichia coli O157: H7. Appl Environ Microbiol. 2012;78:58–69. doi: 10.1128/AEM.06231-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Denou E, Bruttin A, Barretto C, Ngom-Bru C, Brüssow H, Zuber S. T4 phages against Escherichia coli diarrhea: potential and problems. Virology. 2009;388:21–30. doi: 10.1016/j.virol.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 39.Eppinger M, Mammel MK, Leclerc JE, Ravel J, Cebula TA. Genomic anatomy of Escherichia coli O157:H7 outbreaks. Proc Natl Acad Sci U S A. 2011;108:20142–20147. doi: 10.1073/pnas.1107176108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petrov VM, Ratnayaka S, Nolan JM, Miller ES, Karam JD. Genomes of the T4-related bacteriophages as windows on microbial genome evolution. Virol J. 2010;7:292. doi: 10.1186/1743-422X-7-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riede I, Drexler K, Schwarz H, Henning U. T-even-type bacteriophages use an adhesin for recognition of cellular receptors. J Mol Biol. 1987;194:23–30. doi: 10.1016/0022-2836(87)90712-1. [DOI] [PubMed] [Google Scholar]
- 42.Hamdi S, Rousseau GM, Labrie SJ, Tremblay DM, Kourda RS, Slama KB, et al. Characterization of two polyvalent phages infecting Enterobacteriaceae. Sci Rep. 2017;7:40349. doi: 10.1038/srep40349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song JY, Yoo RH, Jang SY, Seong W-K, Kim S-Y, Jeong H, et al. Genome sequence of enterohemorrhagic Escherichia coli NCCP15658. J Bacteriol. 2012;194:3749–3750. doi: 10.1128/JB.00653-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trojet SN, Caumont-Sarcos A, Perrody E, Comeau AM, Krisch HM. The gp38 adhesins of the T4 superfamily: a complex modular determinant of the phage’s host specificity. Genome Biol Evol. 2011;3:674–686. doi: 10.1093/gbe/evr059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adam E, Groenenboom AE, Kurm V, Rajewska M, Schmidt R, Tyc O, et al. Controlling the microbiome: microhabitat adjustments for successful biocontrol strategies in soil and human gut. Front Microbiol. 2016;7:1079. doi: 10.3389/fmicb.2016.01079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loc-Carrillo C, Abedon ST. Pros and cons of phage therapy. Bacteriophage. 2011;1:111–114. doi: 10.4161/bact.1.2.14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amarillas L, Rubí-Rangel L, Chaidez C, González-Robles A, Lightbourn-Rojas L, León-Félix J. Isolation and characterization of phiLLS, a novel phage with potential biocontrol agent against multidrug-resistant Escherichia coli. Front Microbiol. 2017;8:1355. doi: 10.3389/fmicb.2017.01355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meaden S, Koskella B. Exploring the risks of phage application in the environment. Front Microbiol. 2013;4:358. doi: 10.3389/fmicb.2013.00358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang J, Niu YD, Chen J, Anany H, Ackermann H-W, Johnson RP, et al. Feces of feedlot cattle contain a diversity of bacteriophages that lyse non-O157 Shiga toxin-producing Escherichia coli. Can J Microbiol. 2015;61:467–475. doi: 10.1139/cjm-2015-0163. [DOI] [PubMed] [Google Scholar]
- 50.Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, et al. Complete genome sequence of enterohemorrhagic Eschelichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 2001;8:11–22. doi: 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- 51.Weiserová M, Ryu J. Characterization of a restriction modification system from the commensal Escherichia coli strain A0 34/86 (O83:K24:H31) BMC Microbiol. 2008;8:106. doi: 10.1186/1471-2180-8-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miller ES, Kutter E, Mosig G, Arisaka F, Kunisawa T, Ruger W. Bacteriophage T4 genome. Microbiol Mol Biol Rev. 2003;67:86–156. doi: 10.1128/MMBR.67.1.86-156.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8:317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 54.Verma V, Harjai K, Chhibber S. Characterization of a T7-like lytic bacteriophage of Klebsiella pneumoniae B5055: a potential therapeutic agent. Curr Microbiol. 2009;59:274–281. doi: 10.1007/s00284-009-9430-y. [DOI] [PubMed] [Google Scholar]
- 55.Suarez VB, Reinheimer JA. Effectiveness of thermal treatments and biocides in the inactivation of Argentinian Lactococcus lactis phages. J Food Prot. 2002;65:1756–1759. doi: 10.4315/0362-028X-65.11.1756. [DOI] [PubMed] [Google Scholar]
- 56.Tomat D, Balagué C, Aquili V, Verdini R, Quiberoni A. Resistance of phages lytic to pathogenic Escherichia coli to sanitisers used by the food industry and in home settings. Int J Food Sci Technol. 2018;53:533–540. doi: 10.1111/ijfs.13626. [DOI] [Google Scholar]
- 57.Xu Y, Yu X, Gu Y, Huang X, Liu G, Liu X. Characterization and genomic study of phage Vb_EcoS-B2 infecting multidrug-resistant Escherichia coli. Front Microbial. 2018;9:793. doi: 10.3389/fmicb.2018.00793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peng Q, Yuan Y. Characterization of a newly isolated phage infecting pathogenic Escherichia coli and analysis of its mosaic structural genes. Sci Rep. 2018;8:8086. doi: 10.1038/s41598-018-26004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim M, Ryu S. Characterization of a T5-like coliphage, SPC35, and differential development of resistance to SPC35 in Salmonella enterica serovar Typhimurium and Escherichia coli. Appl Environ Microbiol. 2011;77:2042–2050. doi: 10.1128/AEM.02504-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Clark WA. Comparison of several methods for preserving bacteriophages. Appl Microbiol. 1962;10:466–471. doi: 10.1128/AEM.10.5.466-471.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jończyk E, Kłak M, Międzybrodzki R, Górski A. The influence of external factors on bacteriophages-review. Folia Microbiol. 2011;56:191–200. doi: 10.1007/s12223-011-0039-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 157 kb)