Abstract
Chicken infectious anaemia (CIA) is an important viral disease of chicken causing significant immunosuppression and severe anaemia worldwide. Occurrence of severe disease and mortality is noticed in young chicks (2–3 weeks). Vertical mode of transmission increases chance of infection and persistence of virus among the infected flocks. The current study was conducted in Punjab state for confirmation and genetic characterization of CAV among chicken flocks of various poultry farms. DNA was extracted from the tissue samples and subjected to polymerase chain reaction (PCR) of VP1 gene and whole genome. PCR products were further sequenced for confirmation of chicken infectious anaemia virus (CIAV) genome in the clinical samples. PCR amplification of DNA from the tissue samples yielded expected product size of 1350 bases of VP1 gene and 2.3 kb of whole genome. Out of 16 commercial poultry farms, 11 were confirmed with presence of CIAV, and out of 65 birds, 39 were found positive (60%) for CIAV genes. Among the various organs, the presence of viral gene was detected at highest level in thymus when compared with other organs. It is concluded that chicken infectious anaemia virus detected from Punjab state is closely related to other Indian isolates and neighbouring countries which necessitates need of more intensive studies with a greater number of samples for implementing effective control measures.
Electronic supplementary material
The online version of this article (10.1007/s42770-019-00160-9) contains supplementary material, which is available to authorized users.
Keywords: Chicken infectious anaemia virus, Molecular, Sequencing, Characterization, Phylogeny
Introduction
Chicken infectious anaemia virus (CIAV) is an economically important viral pathogen causing severe immunosuppression and anaemia among chicken worldwide. Since its first isolation from chicken during twentieth century [1], the virus has been reported from various poultry producing countries throughout the world including India [2–5]. Previous studies have identified chicken as common natural host of CIAV. But recently, it has been demonstrated in faecal samples collected from cat and dog and it is closely related Gyrovirus in blood samples from human [6, 7]. Based on these findings and close relation to other Anellovirus, it has been shifted from the family Circoviridae to Anelloviridae under the genus Gyrovirus [8, 9] as per the latest ICTV classification. It has features in common with viruses such as Torque Teno virus (TTV) and Torque Teno mini virus (TTMV), which are members of the genus Anellovirus.
It is one of the smallest viruses having an average size of 25–26 nm with non-enveloped icosahedral symmetry. The genome consists of negative-sense, circular, single-stranded DNA (2.3 kb) with three important overlapping viral genes encodes VP1, VP2 and VP3 proteins. VP1 (51.6 kDa) is capsid protein (structural protein) with the highest variable region and highly immunogenic. VP2 (28 kDa) is scaffold protein with phosphatase activity, whereas VP3 (13.6 kDa) is apoptin, a non-structural protein responsible for induction of apoptosis of infected cells [10–13]. It causes severe clinical form of disease in young chicks of 10–12 days with severe morbidity and mortality. Anaemia in young chicks is the only specific sign of infection with CIAV. Transmission of CIAV is either vertical through fertilized eggs or horizontal through oral and/or faecal route (direct or indirect contact with virus contaminated litter). The affected birds may show depression, more or less pale, depressed weight gain and retarded growth. Multiple focal haemorrhages on muscles and necrosis of the wing and skin are the signs usually observed in infected birds. Thymic and bone marrow atrophy can be seen as gross lesions [14–16], and morbidity can be up to 10–30% [17]. The birds may recover, but retarded growth along with secondary bacterial/virus infections were frequently seen in the field condition [18, 19]. Subclinical nature of the CIAV gained much attention recently as the virus has been reported along with many other major immunosuppressive viral diseases like infectious bursal disease (IBD), Marek’s disease (MD), reticuloendotheliosis (RE), adenovirus infection and Newcastle disease (ND) [17, 19, 20]. Co-infection of CIAV with other viruses and bacteria can enhance the pathogenicity of later, thereby negative impact on bird performance [21]. Study on interaction of CIAV and many poultry viral vaccine shows aggravating influence of CIAV on residual pathogenicity of vaccine strain and vaccination failure [11, 21, 22]. CIA is of great concern, as the fast pace in growth of poultry sector worldwide and changing preference for animal protein (red meat to white meat) [23]. Therefore, given their subclinical nature, high prevalence and aggravating influence on other vaccine strains, CIAV infection is of considerable economic importance. Seroprevalence (86%) and molecular epidemiological study (73.3%) from various regions of India show high prevalence of CIAV [24–26]. CIA has been reported to be included in the list of emerging avian viral diseases which is having severe threatening impact on the fast-growing poultry sector.
Since there is no routine vaccination of birds against CIAV in India, the virus may persist in the areas with high density of poultry population without any containment. As per the latest (19th livestock census) census in India, the total poultry population has increased by 12.39% and the total poultry in the country is 729.2 million numbers [27]. Punjab is one of the states where poultry sector has grown to the tune of 57.17% in last 5 years [27]. So far, no systematic molecular study for genetic characterization of CIAV has been done in the Punjab state and, considering the chance of its occurrence in other species, the current study was projected to detect and confirm the presence of chicken infectious anaemia virus among commercial poultry farms by PCR method which is highly sensitive and specific for CIAV. Further, it was characterized by gene/genome sequencing and phylogenetic analysis using bioinformatic tools. The study will be expected to provide awareness about current status of CIAV in the state and further its presence in other mammals can be explored.
Methods
Tissue specimen collection
The tissue samples viz. the thymus, liver, bone marrow, spleen and bursa were collected randomly from 65 dead birds of 16 flocks in various regions of Punjab between January 2016 and December 2017. All tissue samples were collected in sterile phosphate-buffered saline (PBS—pH 7.2 to 7.4) and processed as per standard procedure. Each tissue was washed twice with sterile PBS. Triturated and homogenized with sterile sand, mortar and pestle to prepare 10% (w/v) suspension. Centrifuged at 2500×g at 4 °C for 20 min, supernatant was collected (repeated centrifugation twice to remove all tissue debris). Incubated was the inoculum with 1% antimicrobial (strepto-penicillin, Sigma) solution at 37 °C for 30 min and stored at − 80 °C until further use.
DNA extraction
DNA was extracted from each tissue inoculums (thymus, liver, bone marrow, spleen and bursa) using DNA extraction kit (DNeasy Blood and Tissue Kit, QIAGEN, Germany) as per the manufacture’s protocol. The DNA extracted (ng/μl) was measured and quantified (NanoDrop, Thermo Fisher Scientific) spectrophotometrically and stored at − 20 °C for further use.
Detection of CIAV by PCR
To detect CIAV-specific gene in the total DNA extracted, PCR method was performed using published primers [26] specific for VP1 gene. Oligonucleotides of VP1 forward primer with the 18 bases, i.e. 5′ ATG GCA AGA CGA GCT CGC 3′, and VP1 reverse primer with 18 bases, i.e. 5′ TCA GGG CTG CGT CCC CCA 3′, were employed for amplification of VP1 gene. The primers amplify the CIAV DNA to produce 1350 base product. The PCR protocol was optimized using PCR master mix (GoTaq green, Promega). A 25-μl reaction volume was prepared in a 200-μl PCR tube contains 12.5 μl of PCR master mix (2×), 1 μl of forward and reverse primer (10 pmol) each, 3 μl of template DNA (0.45 μg) and reaction volume was adjusted to 25 μl using nuclease free water (NFW). All the components were mixed properly (all work was done on cooler box), and amplification was carried out in the thermal cycler (Bio-Rad, CA). The amplification included various steps, that is, initial denaturation at 94 °C—5 min one cycle, 35 cycles of primary amplification (denaturation at 94 °C for 1 min, annealing 59 °C—1 min and extension 72 °C-1.5 min) and a final extension at 72 °C—10 min. The amplified PCR products were visualized with ethidium bromide stained 1.5% agarose gel prepared in 1× TBE buffer. The gel was electrophoresed at 90 V for 1 h, visualized on a gel documentation system (Syngene, USA) and analysed.
Sequencing of VP1-specific DNA of CIAV
PCR product was purified using a gel extraction kit (Wizard SV Gel and PCR Clean-Up System, Promega) as per the manufacture’s protocol. Purity of extracted DNA was checked and sent to Eurofins genomic India Pvt. Limited for bidirectional sequencing by Sangers’ method (BigDye V3.1 terminator technology). The data obtained were analysed by DNASTAR and MEGA7.0 software. The DNA sequences were aligned with other published reference sequences (retrieved from GenBank, NCBI) using ClustalW method of MegaAlign programme (DNASTAR, USA) and MEGA7 [28]. Molecular phylogenetic analysis was done using maximum likelihood method (MEGA7, USA). The evolutionary history was inferred by using the maximum likelihood method based on the Kimura 2-parameter model [29]. The bootstrap consensus tree inferred from 1000 replicates [30] was taken to represent the evolutionary history of the taxa analysed. The sequence of CIAV VP1 genes was submitted in GenBank and accession number obtained.
Whole genome amplification of CIAV DNA
PCR amplifying the entire length of CIAV genome was optimized using published primers [19]. DNA from samples positive for VP1 gene was selected for whole genome amplification. The forward primer includes 27 oligonucleotides, i.e. 5′ GAA TTC CGA GTG GTT ACT ATT CCA TCA 3′, and reverse primer includes 28 oligonucleotides, i.e. 5′ GAT AGT GCG ATA AAT CTA TTT TCT GCG T 3′. The PCR protocol was optimized using various components (Q5 HiFi reaction buffer (5×, New England Biolabs, USA), dNTPs mix (10 mM each, New England Biolabs, USA), forward primer (10 pmol), reverse primer (10 pmol), Q5 high fidelity DNA polymerase (2 U/μl, New England Biolabs, USA), template DNA (0.5 μg)), and total volume was adjusted to 50 μl using nuclease free water. For amplifying the whole genome of CIAV, a temperature gradient ranging from 50 to 60 °C was used to optimize the annealing temperature. The various steps included are initial denaturation at 94 °C—4 min, 35 cycles of primary amplification (denaturation at 94 °C for 1 min, annealing 54 °C—1 min and extension 72 °C—2 min) and a final extension at 72 °C—10 min. The amplified PCR products were visualized as explained earlier (used 0.75% agarose gel). Further, gel-purified PCR amplicon (Wizard SV Gel and PCR Clean-Up System, Promega) was sent for sequencing from Eurofins genomic India Pvt. Limited.
Genetic and phylogenetic analysis of CIAV genome
The sequence data generated from DNA of CIAV were analysed by using DNASTAR (Lasergene) software to obtain the full-length nucleotide sequence of the CIAV genome. The nucleotide sequences of CIAV isolate generated in the current study and the complete genomes available in the public database (NCBI/GenBank) were analysed phylogenetically to find out the identity/divergence of each isolates. Multiple sequence alignments were performed using ClustalW algorithm, and phylogenetic tree was generated by using MEGA7 software [28]. The evolutionary history was inferred using the maximum likelihood (ML) method, and the bootstrap consensus tree was inferred from 1000 replicates to represent the evolutionary history of the taxa analysed.
Results
History and clinical findings
Birds included in the present study were based on history of one or more CIA-specific clinical signs in young dead birds and/or based on necropsy findings. Clinical signs reported were droopiness, sudden death and high mortality rate among chicken flocks. Lesions observed in the necropsy finding include paleness of the carcass; haemorrhage in the subcutaneous tissues; muscle serosa and proventriculus; bursa of fabricius, pale bone marrow and liver; and gangrenous dermatitis of the wing. For molecular detection of field CIAV, a total of 295 tissue samples (thymus, bone marrow, liver, spleen and bursa of fabricius) from 65 dead birds were screened by PCR.
Detection of CIAV by PCR
DNA was extracted from each tissue samples and quantified by Nanodrop spectrophotometry. The concentration of DNA eluted was ranged from 0.15 μg to 0.85 μg/μl. The concentration of DNA was optimized to 0.45–0.5 μg/μl in PCR protocol run in the present study. The DNA of CIAV was detected by VP1-specific PCR from the various tissue samples. VP1 gene–specific primer yielded expected amplified products of 1350 bases in both positive control and suspected clinical samples. No band was observed in the non-template (NTC) control (Fig. 1). Out of 65 birds, 39 (60%) were found positive for CIAV. A total of 16 farms were screened for the presence of CIAV; out of it, 11 (69%) were found positive for CIAV. Among the various lymphoid tissues screened, viral presence was observed highest in the thymus tissues. Out of 50 thymus tissues screened, 33 (66%) were found positive for CIAV and least was seen in the liver and the spleen with 28 each (56%). All young chicks of < 20 days except one were found highly positive for CIAV.
Fig. 1.
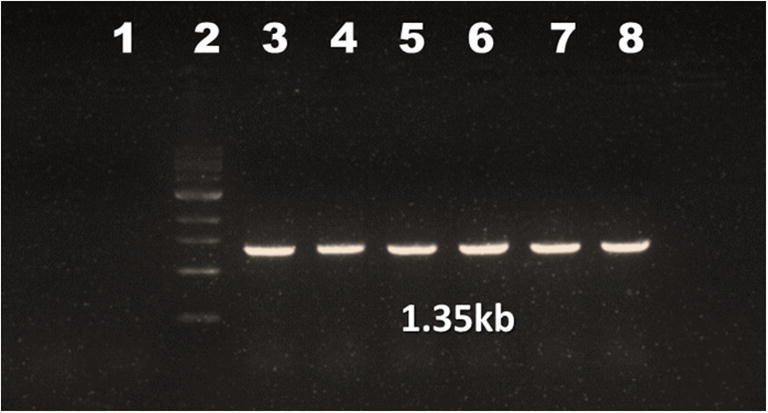
Agarose gel electrophoresis of PCR amplifying VP1 gene of CIAV field isolates. Lane 1: non-template control (NTC); lane 2: marker (1Kb); lane 3: positive control; lane 4–8: samples (1350 bases)
Genetic characterization and phylogenetic analysis of VP1 gene
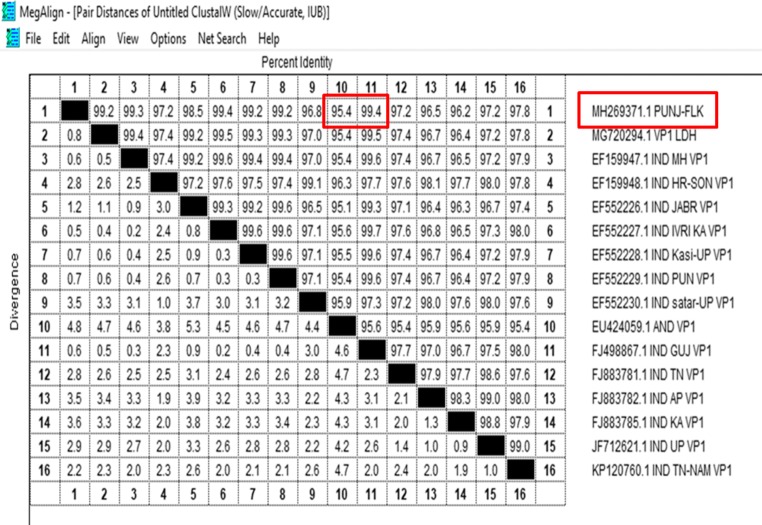
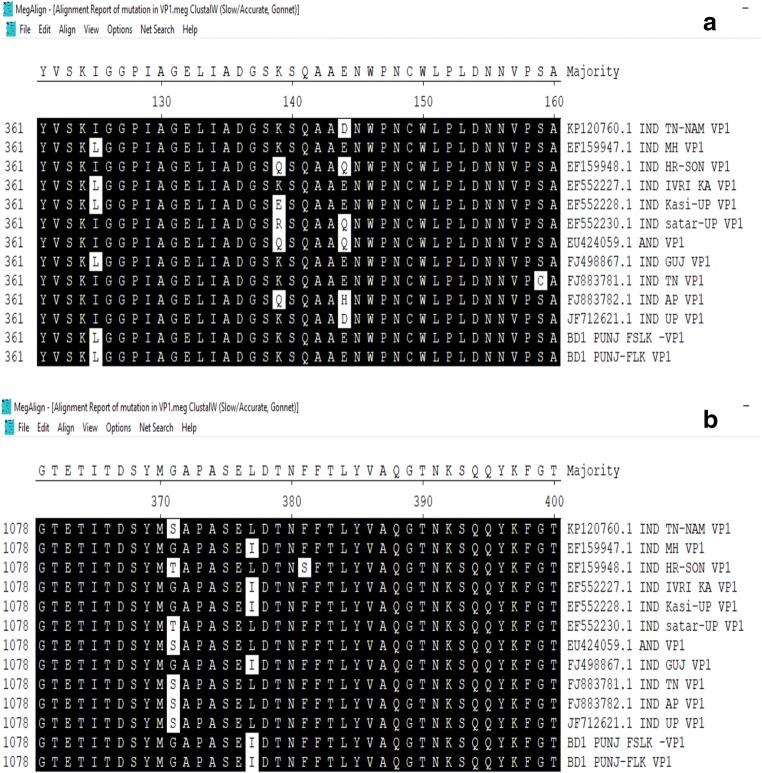
From the PCR positive tissue samples, highly positive (by DNA concentration) purified DNA samples were sent for Sanger’s sequencing. Initial BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) analysis of the VP1 gene sequences showed high similarity (99%) with the sequences available in GenBank (NCBI). Further, multiple sequence alignment of VP1 gene sequences using MegAlign programme (DNASTAR) showed 99.4% identity with isolate from other part of India (Fig. 2). Many point mutations could be observed in VP1 gene, and deduced alignment of the amino acid sequence showed some conservative mutations (L to I and S to A) in viral protein 1 (Fig. 3).
Fig. 2.
Distance matrix of VP1 gene nucleotide sequences of CIAV. Percentage identity/divergence matrix generated by multiple alignments using ClustalW algorithm (MegAlign, DNASTAR) of VP1 gene nucleic acid sequences. Upper triangle represents the percentage of identity between two viruses. Lower triangle represents the percentage of divergence between two viruses
Fig.3.
a, b Amino acid alignment of coding region of VP1 protein. Multiple alignments using ClustalW algorithm (MegAlign, DNASTAR) of VP1 protein amino acid sequences deduced from PUNJ/FSLK isolate. a K-139, E-144, and b Q-394
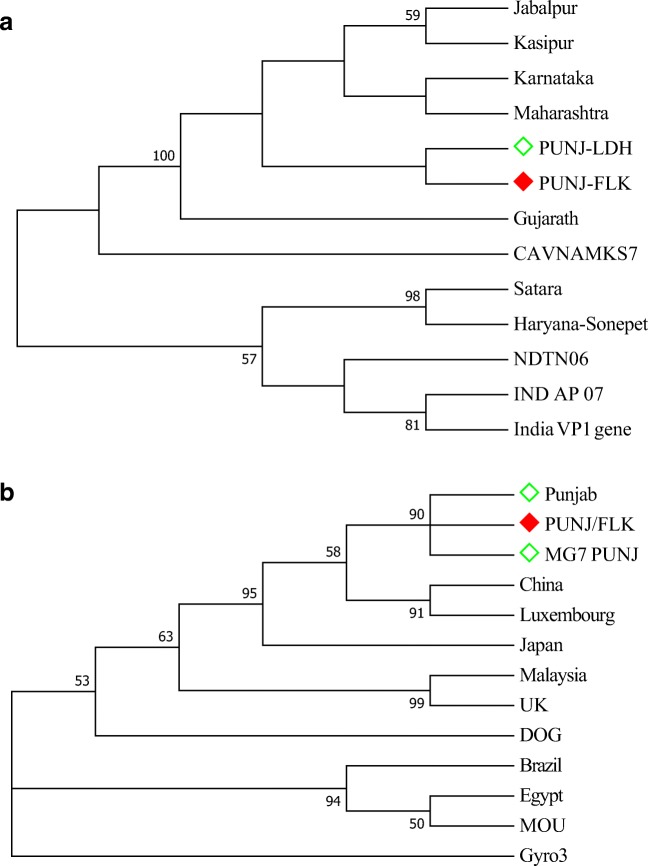
On the basis of VP1 gene sequence, the isolate from the present study did not show distinct Clad formation with any of the reference sequences but it was clustered with many of the Indian isolates and they were branched at close proximity to the isolate from present study (MH269371.1 PUNJ-FLK). Exception of one isolate from Tamil Nadu which branched distantly from Punjab isolate. Further analysis with various sequences from different countries indicated that the Indian isolates are closely branched to form a cluster. Also, they were distantly related to avian Gyro virus and other Chicken anaemia virus isolates from dog and cat (Fig. 4).
Fig. 4.
Molecular phylogenetic analysis of VP1 gene from Indian (a) and global (b) isolates. The evolutionary history was inferred by using the maximum likelihood method based on the Hasegawa-Kishino-Yano model. The bootstrap consensus tree inferred from 1000 replicates is taken to represent the evolutionary history of the taxa analysed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. Red box—isolate from present study
Whole genome amplification of CIAV DNA
PCR amplifying the entire length of CIAV genome was optimized and an amplicon size approximately 2.3 kb was observed at temperature range between 52 and 56 °C after resolving in agar gel electrophoresis (Fig. 5). In the present study, a temperature of 54 °C was selected (as annealing temperature) for amplification of full-length CIAV genome.
Fig. 5.
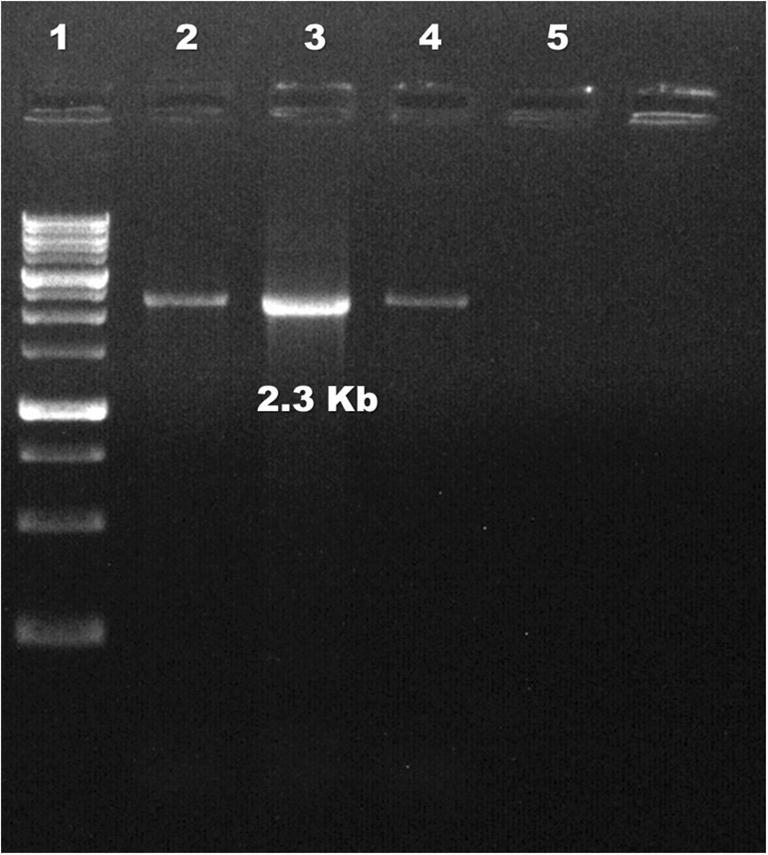
Agarose gel electrophoresis of PCR amplifying whole genome of CIAV. Lane 1: marker (1 kbp); lane 2: positive control; lanes 3, 4: samples; lane 5: NTC
Genetic and phylogenetic analyses of CIAV genome
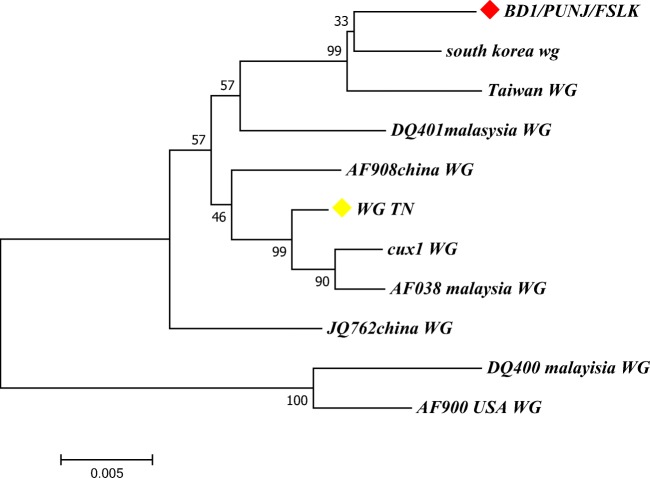
The nucleotide sequences obtained were assembled, and BLAST analysis (https://blast.ncbi.nlm.nih.gov/Blast.cgi) showed high percentage of identity (> 99%) with other reference sequences of CIAV isolates available in the GenBank. After sequencing, we obtained a partial sequence of the isolate (named BD1/PUNJ/FSLK) which is composed of 1.753 kb of CIAV genome. The sequence was consisting of three overlapping ORF of VP1, VP2 and VP3 genes. VP2 and VP3 genes have complete coding sequences consisting of 652 and 364 bases respectively, whereas VP1 gene was incomplete at 3′ end. Multiple sequence alignment of BD1/PUNJ/FSLK sequence with reference sequences retrieved from NCBI revealed that highest percentage identity with sequence from South Korea (98.9%) and Taiwan (98.8%). BD1/PUNJ/FSLK showed 98.2% identity with Indian isolate from Tamil Nadu. There were many point mutations/substitution that could be observed in the various region of the genome sequence which consequently led to amino acid changes in the coding region like isoleucine to leucine (I to L), serine to glycine (S to G) and alanine to serine (A to S). ML tree was constructed based on reference nucleotide sequences using Tamura-Nei model. The phylogenetic tree constructed shows three distinct clusters and BD1/PUNJ/FASLK closely branched with isolates from South Korea and Taiwan (Fig. 6). Other isolates, one from India (Tamil Nadu), Malaysia and China, were found in two other clusters. USA isolate was found distantly related to BD1/PUNJ/FASLK.
Fig. 6.
Molecular phylogenetic analysis of CIAV genome. The evolutionary history was inferred by using the maximum likelihood method based on the Tamura-Nei model. The bootstrap consensus tree inferred from 1000 replicates is taken to represent the evolutionary history of the taxa analysed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. BD1/PUNJ/FSLK grouped with South Korea and Taiwan with bootstrap value of 99
Discussion
For detection of CIAV from the chicken flocks, various lymphoid tissues were collected from dead birds suspected of CIAV infection and screened by PCR. DNA was isolated from the samples, and molecular characterization of CIAV has been done. The serological and antigenic prevalence of chicken infectious anaemia has been reported from all parts of the world, and reports suggest the alarming situation of this disease in poultry sector [15, 31]. In the present study, it was also observed high incidence of chicken anaemia among poultry flocks in Punjab region. Out of the 16 farms screened, 11 (68.75%) were found positive for the presence of CIAV in the unvaccinated flocks. The presence of CIAV was high in thymus but all tissues were showed its presence. Therefore, the detected virus may be a wild-type/mutated variant circulating among bird population. Various strains and genotypes were reported earlier from the various region of the world, but only one serotype of CIAV was reported till date [5, 23, 32–34]. CIAV can infect all age group, sex, breed, layers, broilers and breeders, and it is ubiquitous in nature. Various age groups ranging from less < 7 days to > 60 days were also included in the present study. Among these groups, chicks with age group of 0–20 days (92.86%) had highest incidence of infection and disease. It is primarily a disease of young chicks of < 3 weeks of age, but virus can persistently infect older birds also. Similar findings were reported by various researchers [1, 26]. Vertical mode of transmission and viral persistence makes difficult to control.
Molecular detection and characterization of the CIAV genome have been carried out by targeting VP1 gene and partial genome (comprising of all three ORF) of CIAV. The genome is highly conserved throughout the length of DNA except VP1 gene which has maximum nucleotide variation. Among the three genes of CIAV, hyper variable region is residing in the VP1 gene and it transcribes into highly immunogenic protein [35] that forms the base of capsid in matured virus particle. The immunogenicity and virulence of the virus are determined by the presence of few amino acids in VP1 gene. Therefore, monitoring of CIAV circulating in the field at various interval of time is necessary to determine the genotype and pathotype present in the field. There were many points mutations could be observed in VP1 gene, and deduced alignment of the amino acid sequence shows some conservative mutation (L to I and S-A) in VP1 viral protein. Various researchers targeted VP1gene and genetically characterized CIAV, and they suggested that amino acid Q (glutamine) at position 394 in VP1 may be a major determinant of pathogenicity and amino acids at 139 and 144 are also major concerns [24, 32, 33, 36]. The isolate from the present study had amino acid K at 139, E at 144 and Q at 394 positions (Fig. 5). Some other variations in amino acid observed are L at 135, S at 287, I at 376 and S at 413. Also, few reports suggest that nucleotide at 75I, 89T, 125L, 141L and 144E associated with lower pathogenicity [37, 38]. In the present study, the nucleotides at these positions observed were 75V, 89T, 125L, 141Q and 144E. There was no nucleotide substitution, or mutations were observed in VP2 and VP3 coding region of CIAV genome of BD1/PUNJ/FSLK isolate.
Percentage identity/divergence matrix generated by multiple sequence alignments of VP1 gene showed 99.8% identity with isolate from Gujarat, and highest divergence is from isolates of Maharashtra. On the basis of VP1 gene sequence, phylogenetic tree constructed with the isolate from the present study did not show any distinct Clad formation along with various other Indian isolates. The isolates from Karnataka, Gujarat, Maharashtra, UP and Punjab were branched at close proximity to isolate from present study. Isolate from Tamil Nadu branched distantly from Punjab isolates. Sequence analysis of the isolates from the present study is closely related to other reported CIAV isolates from various parts of India. The genome of CIAV has been analysed by various researchers and isolates from different region of the world show limited sequence variability. Present study also agrees with the previous reports. Phylogenetic analysis shows close relatedness to South Korea and Taiwan, but there is no clear geographical distribution of different isolates as they are interspersed in the tree generated (data given in supplementary document). All isolates, perhaps, belong to single serotype but different genotypes. The whole genome analysis of various isolates shows that there could be intergenotypic recombination event in the VP1 gene that may decide the virulence of virus [5, 32, 33, 39]. Highly conservative nature of the VP2 and VP3 viral protein and various point mutations in hypervariable region of VP1 may allow this ubiquitous virus to escape the vaccine induced immunity. Since VP1 is highly immunogenic and variable, it can be used for molecular epidemiology and at the same time highly conserved VP2 and VP3 can be used to explore the route of origin of CIAV. Few reports suggest the possible occurrence of CIAV in mammals like dog and cat, but it can only cause severe infection and diseases in its natural host [1, 40, 41]. One report from Italy showed the presence of Gyrovirus genome (closely related to CIAV) in the blood of human patients, and reports from China suggest the CIAV presence in dog and mouse [6, 7]. Therefore, we have compared our sequence with these isolates also (AY846844.1 China, AM407857.1 Luxemburg, KP780288.1 Japan, MG827098.1 Egypt, MG846491.1 Brazil, AF390038.1 Malaysia, AJ133508.1 UK, KU645525.1 MOUSE, KU645524.1 DOG, MG366592.1 Gyro3, EF552229.1 Punjab, MG720294.1 MG7 Punj, MH269371.1 PUNJ-FLK). Phylogenetic tree constructed with these isolates appeared to be distantly related to chicken anaemia virus. It formed two clusters—one with all chicken anaemia isolates including isolate from the present study. Another cluster formed with CIAV isolates from other mammals and isolates from Brazil and Egypt.
Conclusion
In this study, molecular characterization of Chicken infectious anaemia virus circulating among various flocks in Punjab region has been done. Sixty percent of the birds screened were found positive for genome of CIAV by PCR method. Among the various age groups, chicks of < 20 days were found highly positive for CIAV. The presence of virus was observed in almost all the lymphoid organs with highest was in thymus tissues. Molecular study and phylogenetic analysis indicated that isolates of CIAV circulating in Punjab might have originated from the various CIAV isolates reported from different parts of the India and/or from the neighbouring countries. The genetic map of the genome especially VP1 gene from the present study suggests that the isolate currently circulating among chicken flocks is highly pathogenic, and it may have direct/indirect role in higher mortality observed in chicks (Author’s personal view—the farms with high mortality among chicks showed specific signs of CIA at the time of sample collection). However, the presence of other immunosuppressive viruses along with CIAV needs to be studied further to find out the role of each virus in morbidity and mortality among chicks. This study may give the limelight on the genetic feature of CIAV circulating among poultry flocks, and the information generated could be used for the development of diagnostics and preventive measures. However, as per the recent reports, the role of CIAV in other mammals including human being needs to be further studied.
Electronic supplementary material
(DOCX 82.1 kb)
Acknowledgements
Authors are thankful to the Director of Research, Guru Angad Dev Veterinary and Animal Sciences University, Ludhiana for providing necessary facilities.
Funding information
The Department of Science and Technology (DST) granted the project and financial assistance was received from DST in the form of WOS-A fellowship to the first author.
Compliance with ethical standards
Conflict of interests
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Yuasa N, Taniguchi, Yoshida I. Isolation and some characteristics of an agent inducing anemia in chicks. Avian Dis. 1979;23:366–385. doi: 10.2307/1589567. [DOI] [Google Scholar]
- 2.Yuasa N, Imai K. Pathogenicity and antigenicity of eleven isolates of chicken anaemia agent (CAA) Avian Pathol. 1986;15:639–645. doi: 10.1080/03079458608436327. [DOI] [PubMed] [Google Scholar]
- 3.Gowthaman V, Singh SD, Dhama K, Barathidasan R, Srinivasan P, Mahajan NK, Ramakrishnan MA. Molecular characterization of chicken anemia virus isolated from commercial poultry with respiratory disease complex in India. Adv Ani Vet Sci. 2014;2(3):171–176. doi: 10.14737/journal.aavs/2014/2.3.171.176. [DOI] [Google Scholar]
- 4.Li Y, Wang Y, Fang L, Fu J, Cui S, Zhao Y, Cui Z, Chang S, Zhao P (2016) Genomic analysis of the chicken infectious anemia virus in a specific pathogen-free chicken population in China. Biomed Res Int 2016:4275718. 10.1155/2016/4275718 [DOI] [PMC free article] [PubMed]
- 5.Ou SC, Lin HL, Liu PC (2018) Epidemiology and molecular characterization of chicken anaemia virus from commercial and native chickens in Taiwan. Transbound Emerg Dis 65:1–9 [DOI] [PubMed]
- 6.Maggi F, Macera L, Focosi D, Vatteroni LM, Boggi U, Antonelli G, Eloit M, Pistello M. Human gyrovirus DNA in human blood, Italy. Emerg Infect Dis. 2012;18:6. doi: 10.3201/eid1811.121226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang L, Li Y, Wang Y, Fu J, Cui S, Li X, Chang S, Zhao P (2017) Genetic analysis of two chicken infectious anemia virus variants-related Gyrovirus in stray mice and dogs: the first report in China 2015. Biomed Res Int 2017:6707868. 10.1155/2017/6707868 [DOI] [PMC free article] [PubMed]
- 8.ICTV (2011) Virus taxonomy (classification and nomenclature of viruses). The online (9th) report of the International Committee on Taxonomy of Viruses. https://talk.ictvonline.org/taxonomy/
- 9.ICTV (2017) Virus taxonomy (classification and nomenclature of viruses). The online (10th) report of the International Committee on Taxonomy of Viruses. https://talk.ictvonline.org/taxonomy/
- 10.Noteborn MHM, De Boer GF, Van Roozelaar DJ, Karreman C, Kranenburg O, Jeurissen SHM, Hoeben RC, Zantema A, Koch G, Van Ormondt H, Van Der Eb AJ. Characterization of cloned chicken anemia virus DNA that contains all elements for the infectious replication cycle. J Virol. 1991;65:3131–3139. doi: 10.1128/JVI.65.6.3131-3139.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noteborn MHM, Koch G. Chicken anaemia virus infection: molecular basis of pathogenicity. Avian Pathol. 1995;24:11–31. doi: 10.1080/03079459508419046. [DOI] [PubMed] [Google Scholar]
- 12.Renshaw RW, Soine C, Weinkle T, O’Connell PH, Ohashi K, Watson S, Lucio B, Harrington S, Schat KA. A hypervariable region in VP1 of chicken infectious anemia virus mediates rate of spread and cell tropism in tissue culture. J Virol. 1996;70:8872–8878. doi: 10.1128/JVI.70.12.8872-8878.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nayabian H, Karim M. Molecular characterization of the chicken anaemia viruses isolated from broiler farms of west Azerbaijan, Iran. Avian Pathol. 2013;42(2):108–113. doi: 10.1080/03079457.2013.766668. [DOI] [PubMed] [Google Scholar]
- 14.Jeurissen SHM, Wagenaar F, Pol JMA, Van Der Eb AJ, Noteborn MHM (1992) Chicken anemia virus causes apoptosis of thymocytes after in vivo infection and of cell lines after in vitro infection. J Virol 66(12):7383–7388 [DOI] [PMC free article] [PubMed]
- 15.Todd D. Circoviruses: immunosuppressive threats to avian species: a review. Avian Pathol. 2000;29(5):373–394. doi: 10.1080/030794500750047126. [DOI] [PubMed] [Google Scholar]
- 16.Smyth JA, Moffett DA, Connor TJ, McNulty MS. Chicken anaemia virus inoculated by the oral route causes lymphocyte depletion in the thymus in 3-weekold and 6-week-old chickens. Avian Pathol. 2006;35(3):254–259. doi: 10.1080/03079450600717349. [DOI] [PubMed] [Google Scholar]
- 17.Adair BM. Immunopathogenesis of chicken anemia virus infection. Dev Comp Immunol. 2000;24:247–255. doi: 10.1016/S0145-305X(99)00076-2. [DOI] [PubMed] [Google Scholar]
- 18.Bulow VV, Schat KA (1997) Chicken infectious anemia. In: Calnek BW, Barens HJ, Beard CW, McDougald LR, Saif YM (eds) Disease of poultry. 10th ed. Iowa State University Press, pp 739–756
- 19.Krishan G, Shukla SK, Bhatt P, Wani MY, Dhama K, Malik YPS, Kumar R. Clinical association of chicken anemia virus with other infectious poultry diseases in north India and Nepal: its pathological studies, molecular epidemiology and RFLP pattern of PCR amplified full length viral genome. Adv Anim Vet Sci. 2015;3(7):395–405. doi: 10.14737/journal.aavs/2015/3.7.395.405. [DOI] [Google Scholar]
- 20.Hegazy AM, Abdallah FM, Abd-El Samie LK, Nazim AA. Chicken infectious anemia virus (CIAV) in broilers and laying hens in Sharkia Province, Egypt. J Am Sci. 2010;6:9. [Google Scholar]
- 21.Lebdah MA, Megahed MM, Hassanin OAA, Ali AMI. The negative impact of chicken infectious anemia virus infection on immune responses to different Newcastle disease virus vaccination programs. Zagazig Vet J. 2016;44(2):138–148. doi: 10.21608/zvjz.2016.7856. [DOI] [Google Scholar]
- 22.De Boer GF, Van Roozelaar DJ, Moormann RJ, Jeurissen SHM, Van Den Wijngaard JC, Hilbink F, Koch G. Interaction between chicken anaemia virus and live Newcastle disease vaccine. Avian Pathol. 1994;23:263–275. doi: 10.1080/03079459408418994. [DOI] [PubMed] [Google Scholar]
- 23.Bennett CE, Thomas R, Williams M, Zalasiewicz J, Edgeworth M, Miller H, Coles B, Foster A, Burton EJ, Marume U. The broiler chicken as a signal of a human reconfigured biosphere. R Soc Open Sci. 2018;5:180325. doi: 10.1098/rsos.180325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Natesan S, Kataria JM, Dhama K, Rahul S, Baradhwaj N. Biological and molecular characterization of chicken anaemia virus isolates of Indian origin. Virus Res. 2006;118(1–2):78–86. doi: 10.1016/j.virusres.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 25.Bhatt P, Shukla SK, Mahendran M, Dhama K, Chawak MM, Kataria JM. Prevalence of chicken infectious anemia virus (CIAV) in commercial poultry flocks of northern India: a serological survey. Transbound Emerg Dis. 2011;58:458–460. doi: 10.1111/j.1865-1682.2011.01215.x. [DOI] [PubMed] [Google Scholar]
- 26.Wani MY, Dhama K, Barathidasan R, Gowthaman V, Tiwari R, Bhatt P, Mahajan NK, Chawak MM, Singh SD, Kataria JM. Molecular detection and epidemiology of chicken infectious anemia virus in India. South Asian J Exp Biol. 2013;3(4):145–151. [Google Scholar]
- 27.Livestock census (2012) 19th Livestock census-2012 all India report. Ministry of Agriculture, Department of Animal Husbandry, Dairying and Fisheries, Krishi Bhawan http://dahd.nic.in/sites/default/filess/Livestock 5_0.pdf. Accessed 26 March 2015
- 28.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 30.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 31.Schat KA, van Santen VL. Chicken infectious anemia. In: Swayne DE, Glisson JR, McDougald LR, Nolan LK, Suarez DL, Nair V, editors. Disease of poultry. 13. Ames: Wiley; 2013. pp. 248–264. [Google Scholar]
- 32.Kim HR, Kwon YK, Bae YC, Oem JK, Lee OS. Molecular characterization of chicken infectious anemia viruses detected from breeder and broiler chickens in South Korea. Poult Sci. 2010;89:2426–2431. doi: 10.3382/ps.2010-00911. [DOI] [PubMed] [Google Scholar]
- 33.Olszewska-Tomczyk M, Świętoń E, Minta Z, Śmietanka K. Occurrence and phylogenetic studies of chicken anemia virus from polish broiler flocks. Avian Dis. 2016;60(1):70–74. doi: 10.1637/11277-091415-ResNote.1. [DOI] [PubMed] [Google Scholar]
- 34.Li Y, Fang L, Cui S, Fu J, Li X, Zhang H, Cui Z, Chang S, Shi W, Zhao P. Genomic characterization of recent chicken anemia virus isolates in China. Front Microbiol. 2017;8:401. doi: 10.3389/fmicb.2017.00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Todd D, Creelan JL, Mackie DP, Rixon F, McNulty MS. Purification and biochemical characterization of chicken anaemia agent. J Gen Virol. 1990;71:819–823. doi: 10.1099/0022-1317-71-4-819. [DOI] [PubMed] [Google Scholar]
- 36.Erfan AM, Abdullah AS, Naguib MM (2018) Characterization of full genome sequences of chicken anemia viruses circulating in Egypt reveals distinct genetic diversity and evidence of recombination. Virus Res. 10.1016/j.virusres.2018.05.008 [DOI] [PubMed]
- 37.Todd D, Alistair NJ, Scott NW, Ball BJ, Borghmans, Brian MA. Molecular basis of the attenuation exhibited by molecularly cloned highly passaged chicken anemia virus isolates. J Virol. 2002;76:8472–8474. doi: 10.1128/JVI.76.16.8472-8474.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kye SJ, Kim JY, Seul HJ, Kim S, Kim SE, Lee HS, Sorn S, Choi KS. Phylogenetic analysis and genetic characterization of chicken anemia virus isolates from Cambodia. Poult Sci. 2013;92:2681–2686. doi: 10.3382/ps.2013-03204. [DOI] [PubMed] [Google Scholar]
- 39.AboElkhair M, El-Razak AGA, Metwally AEY (2014) Molecular characterization of chicken anemia virus circulating in chicken flocks in Egypt. Adv Virol 2014:797151. 10.1155/2014/79715 [DOI] [PMC free article] [PubMed]
- 40.Rosenberger JK, Cloud SS. Chicken anemia virus. Poult Sci. 1998;77:1190–1192. doi: 10.1093/ps/77.8.1190. [DOI] [PubMed] [Google Scholar]
- 41.Oluwayelu DO (2010) Diagnosis and epidemiology of chicken infectious anemia in Africa. Afr J Biotechnol 9(14):2043–2049
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 82.1 kb)