Abstract
This study reports the optimization of milk-clotting protease production from Aspergillus oryzae DRDFS13 under solid-state fermentation (SSF) in both one-variable-at-a-time and response surface methodology (RSM). The production and optimization of milk-clotting protease obtained from Aspergillus oryzae DRDFS13 under solid-state fermentation (SSF) using different agro-industrial wastes as solid substrates were studied. The agro-industrial wastes used included wheat bran, rice bran, pea bran, and grass pea bran. The chemical composition of the best solid substrate was tested using standard methods. Others cultivation parameters were studied, and the results showed that the optimum fermentation medium composed of wheat bran, casein (1% w/w), and glucose (0.5% w/w) and the conditions for maximum milk-clotting protease production were at the moisture content of 55.0%, inoculum of 0.5*106 spores/mL, incubation temperature of 30 °C, pH of 6.0, and fermentation time of 5 days. The highest milk-clotting activity was obtained from the crude enzyme extracted using 0.1 M NaCl and partial purification of the crude enzyme using chilled acetone, and 80% (NH4)2SO4 increased the ratio of MCA/PA from 0.56 to 1.30 and 0.65, respectively. Moreover, the highest MCA (137.58 U/mL) was obtained at a casein concentration of 0.5%, pH 4.0, and 25 °C, using RSM. Thus, results from the present study showed that the optimization of milk-clotting protease production from A. oryzae DRDFS 13 under SSF by both one-variable-at-a-time and RSM significantly increased the milk-clotting activity. This is the first report from a fungus in the Ethiopian setting and a modest contribution to highlight the potential of harnessing microbial protease enzymes for industrial applications.
Electronic supplementary material
The online version of this article (10.1007/s42770-020-00243-y) contains supplementary material, which is available to authorized users.
Keywords: Aspergillus oryzae, Milk-clotting activity, Optimization, Milk-clotting protease solid-state fermentation
Introduction
Rennet (EC 3.4.23.4), is a complex enzyme traditionally extracted from slaughtered young calf and used for milk-clotting. It contains a mixture of proteolytic enzymes such as acid proteases (aspartic proteases) that are employed in cheese maturation and production with its appropriate flavor and texture [1–3]. However, an increase in demand for global cheese production coupled with a shortage in calf rennet production and ethical issues necessitated a search for alternative rennet substitutes from microorganisms [1].
These enzymes are extracted from mass cultured fungal and bacterial sources that are widely used for cheese production, and for not only cheese making but also for use as digestive aids, beer clarifiers, protein modifiers, and microbial milk-clotting enzymes constitute 33% of the total protease utilization and largely replaced animal rennet for milk-clotting [1, 4].
Microbial substitutes for rennet production earlier focused on fungal substitutes such as Mucor and Endothia [5, 6]. However, the search for an ideal rennet continues for the production of milk-clotting enzymes from a number of strains of fungi mainly from the genera Aspergillus, Penicillium, Rhizopus, Mucor, Humicola, Thermoascus, and Thermomyces [7, 8].
Aspergillus oryzae (A. oryzae) is one of the species known for its capacity to secrete high levels of enzymes in their growth environment [7]. It is a non-toxigenic strain, for it lacks the genes responsible for aflatoxin production. Consequently, it is recognized as a “Generally Recognized as Safe (GRAS)” organism by the US Food and Drug Administration. It has a long history of use in the production of traditional fermented foods, and in the modern food industry, due to its high-level production of proteolytic active enzymes [4, 9–11]. The fungus possesses more secretory proteinase genes that function in acidic pH. It grows on cheap agricultural by-products such as wheat bran, rice bran, and bagasse. The enzyme secretion is initiated when the fungus grows on substrates with low amino acids and sugar contents since transport-related gene families are expressed to utilize external nutrient resources for its growth [9, 12].
The production of enzymes by fungi, in general, is undertaken using a time-tested, cost-effective method known as solid-state fermentation (SSF). It is suitable for fungi growth using agricultural by-products as substrates and requirement of the less sterile environment and skill and recovery of concentrated and more stable enzymes than those obtained in submerged fermentation [11].
It is established that the production of extracellular proteases is affected by the composition of media and other factors, such as temperature, pH, incubation time, and inoculum density [13]. About 30–40% of the production cost of a protease enzyme is accounted for the cost of growth medium [13]. Therefore, the search for local and low-cost substrates, together with optimization of cultivation conditions, is necessary to significantly minimize the cost of enzyme production in the context of traditional “one-variable-at-a-time” strategic operation in biotechnology.
Ethiopia is endowed with the highest number of cattle herd in Africa [14]. Although the hitherto cheese production is mainly contributed by household and cottage level production, the ever-expanding dairy industries around the peri-urban centers are importing rennet enzymes for cheese production [15]. This necessitates the search for rennet substitutes from fungi at a local level.
In this study, the production of extracellular milk-clotting protease by locally isolated fungal species, Aspergillus oryzae DRDFS 13, under solid-state fermentation (SSF) using cheap agro-industrial by-product (wheat bran) was optimized. The effects of several physicochemical and environmental factors were investigated to select the optimal conditions that ensure the best milk-clotting activity by application of “one-factor-at-a-time” method. This is the first report of screening and optimization for milk-clotting activity from a local isolate, Aspergillus oryzae DRDFS 13, under solid-state fermentation with potential for industrial application after subsequent studies.
Materials and methods
Microorganism and growth conditions
The fungus was isolated from soil samples in Ethiopia and identified as Aspergillus oryzae DRDFS13 using ITS primers(ITS86F (F): 5′- GTG AAT CAT CGA ATC TTT GA-3′ and ITS4 (R): 5′- TCC TCC GCT TAT TGA TAT GC -3′) at Eurofins Genomics, Germany, after cultural and morphological studies. The fungus was deposited in the fungus culture collection at Addis Ababa University and Jacobs University (Bremen, Germany).
Raw materials
Four different agro-industrial substrates, wheat bran, rice bran, pea bran, and grass pea bran, were purchased from the local market and properly dried at 60 °C.
Inoculum preparation
Aspergillus oryzae DRDFS13 was grown into 50-mL Falcon tubes containing 20 mL of potato dextrose agar (PDA) and incubated at 30 °C for 5 days. Sporulated culture lawn was mixed well in sterilized distilled water. The spore suspension was diluted and adjusted to a concentration of 106 spores/mL using a hemocytometer.
Experimental setup for preliminary screening for production of MCE under SSF
The experimental setup for solid-state fermentation was according to [16] with slight modification. The inoculum (1 mL of 106 spores/mL spores suspension) was transferred into 250-mL Erlenmeyer flasks containing 10 g of wheat bran, 12.43% crude protein, 3.77% crude fat, 47.23% carbohydrate, 18.77% fiber, 7.4% ash, and 10.40% moisture content, moistened with 12 mL of (0.2 M) HCl. The flasks were incubated at 30 °C for 6 days.
The solid-state fermentation media were also prepared in 250-mL Erlenmeyer flasks containing wheat bran (10 g), casein or skim milk (2.0 g), and 10 mL of salt solution (g/L, 2.0, KNO3; 0.5, MgSO4·7H2O; 1.0, K2HPO4; 0.439, ZnSO4·7H2O; 1.116, FeSO4·7H2O; 0.203, MnSO4·7H2O; and pH 7.0). The medium was sterilized at 121 °C for 30 min, and after sterilization, the flasks were inoculated with 10% of spore suspension (1 mL of 106 spores/mL) and incubated at 30 °C for 5 days under static conditions [17].
Enzyme extraction
Enzyme extraction was made according to [16, 18]. The enzyme was extracted from solid medium using 100 mL (1:10 ratio of bran solvent w/v) of distilled water, 0.1 M NaCl solution, and a solution of Tween-80 (0.1%) by shaking on a rotary shaker (MaxQ 2000 Open-Air Platform Shaker, Thermo Fisher Scientific, USA) (240 rpm, 40 min, at room temperature) and filtered using cotton cloth. The filtrate was centrifuged (Heraeus Pico17/21 centrifuge, Thermo Electron Led, Germany) at 10,000g for 10 min at 4 °C. The supernatant was used as crude enzyme source for milk-clotting and protease activities.
Assay for milk-clotting activity (MCA)
The milk-clotting activity of the enzyme extract was assayed according to [6]; 0.5 mL of the crude enzyme was added to 5 mL of reconstituted skim milk (Nestle TM) in 12-mL test tube pre-incubated at 35 °C for 10 min. Reconstituted skim milk (Nestle TM) solution is consisted of 10-g dry skim milk/100 mL and 0.01 M CaCl2 (AppliChemTM). The clotting time was quantified in Soxhlet unit (SU) according to the following formula:
where T is the clotting time (s) and D is dilution of crude enzyme.
One SU is expressed as the quantity of enzyme required to clot 1 mL of a solution comprising 0.1-g skim milk powder and 0.01 M calcium chlorides at 35 °C within 40 min.
Assay for protease activity
For assaying the proteolytic activity, 0.5 mL of the enzyme extract was mixed with 2.5 mL of 1% (w/v) alkali-soluble casein in 20-mM potassium phosphate buffer at pH 6.5 [6], and the mixture was incubated in a water bath at 35 °C for 10 min. The mixture was then mixed with 2.5 mL of 0.44 M trichloroacetic acid and filtered through Whatman no.1 filter paper. Then, 1 mL of the filtrate was mixed with 1 mL of 2 N Folin’s phenol reagent (three times diluted) and 2.5 mL of 0.55 M sodium carbonate solution and incubated at 35 °C for 20 min to observe color development and determine the optical density (OD) at 660 nm. One unit (1 U) of enzyme activity was defined as the amount of enzyme that liberated 1 μg of tyrosine per 1 mL in 1 min.
where PA, protease activity; μTry, μg of tyrosine equivalent released; Vt, total volume of assay in mL (5 mL of substrate plus 1 mL of enzyme plus 5 mL of TCA); Vs, sample volume (i.e., the volume of protease used for assay in mL); T, reaction time (i.e., time of incubation in minutes, 10 min); and Va, volume of assayed (i.e., the final volume of the product used in calorimetric determination).
Effect of media on milk-clotting protease (MCP) production
Initial screening of the media composition for maximum milk-clotting protease (MCP) production was performed by a one-variable-at-a-time approach [4, 13]. Thus, 1 mL of 1*106 spore/mL from Aspergillus oryzae DRDFS13 was inoculated into 10 g of each substrate (SS media) in 250-mL Erlenmeyer flasks hydrated with 12-mL HCl (0.2 M) except 6-mL HCl (0.2 M) for rice bran and incubated at 30 °C for 144 h. The extraction was made as before, and the crude enzyme was assayed for milk-clotting and protease activity according to [6]. After having tested the effect of substrates on enzyme production, the highest enzyme-producing substrate was selected and tested for further optimization.
Effect of incubation time
After inoculation, the flasks were incubated at 30 °C for different time periods ranging from 24 to 144 h, and enzyme activity was monitored according to [6].
Biomass determination
The biomass was indirectly detected by the glucosamine (GlcN) released from the cell wall according to [19].
Where
ΔA650 = A650 (sample)-A650 (sample blank)
e^[(ΔA650-b)/m]] = mgGlcN
Standard curve values (logarithmic fit) = b&m
Sample’s amount = 0.1gdfs
Sample’s dilution = DFsample
Effect of incubation temperature
The fungal spores were inoculated into SSF medium in 250-mL Erlenmeyer flask and incubated at 25, 30, 35, and 40 °C for 120 h to obtain the optimum temperature for MCP production, and the milk-clotting and protease activities were assayed according to [6].
Effect of inoculum size
The effect of inoculum size on MCP production was studied by inoculating 0.1 (1*105 spores/mL), 0.5 (5*105 spores/mL), 1 (1*106 spores/mL), 2 (2*106 spores/mL), 3 (3*106 spores/mL), and 4 mL (4*106 spores/mL) spore suspension in to SSF media and assayed for milk-clotting and protease activities according to [6].The following optimization study was undertaken by inoculating 0.5 mL of 1*106 spore/mL using 10 g of wheat bran on SSF minimal medium in 250-mL Erlenmeyer flask and incubated at 30 °C for 120 h unless stated otherwise.
Effect of moisture content
The effect of initial moisture content on enzyme production was tested by moistening substrate using distilled water in different percentages of moisture content, 45, 50, 55, 60, 65, and 70%, to find out the best percentage for enzyme production [20]. The milk-clotting and protease activities were assayed according to [6].
Effect initial media pH
The effect of initial media pH on milk-clotting protease production was studied by adjusting the SSF medium to pH 4.0, 4.5, 5.0, 5.5, 6.0, 6.5, and 7.0 using HCl or diluted NaOH. The milk-clotting and protease activities were assayed according to [6].
Effect of supplementary carbon and nitrogen sources
The SSF production medium was supplemented with different C sources (glucose, galactose, fructose, sucrose, maltose, lactose, and starch) and N sources (casein, skim milk, yeast extract, and urea) and inorganic nitrogen source (NH4Cl, (NH4)2SO4 and NH4NO3) at a level of 1% w/w of the SSF medium. Further, the amount of best nitrogen and carbon source (0.5, 1, 2, 3, 4, and 5%) was optimized [21]. The milk-clotting and protease activities were assayed according to [6].
The extraction efficiency of some solvents
The efficiency of some solvents was checked by extracting the milk-clotting protease using distilled water, 0.1 M NaCl, and 0.1% Tween 80 at 1:10 bran to solvent ratio. The crude enzyme extract was then assayed for milk-clotting activity and protease activity according to [6].
Optimization of major factors affecting milk-clotting protease production by response surface methodology (RSM)
Experimental design
The effects of factors that have shown significant outcome on milk-clotting protease production during optimization by one-variable-at-a-time were further determined by response surface methodology (RSM). The RSM was used for studying the effects of interaction among pH, temperature, and casein concentration on milk-clotting protease production. The optimized ranges for the selected variables were pH (4–7), incubation temperature (25–40 °C), and casein concentration (0.5–5%). RSM design was adopted to optimize the levels of the three factors, with three center points yielding a set of 20 experiments. The factors at three different levels (− 1, 0, + 1) with minimum and maximum range of values were presented in Table 1. The replicates (treatments 13–20 in Table 4) at the center of the design were used for estimation of the pure error sum of squares. The experiments were randomized to maximize the effects of unknown variability due to irrelevant factors in the observed responses [4].
Table 1.
Experimental range and levels of the three independent variables used in RSM in terms of actual and coded factors
| S.No | Variables | Range and levels | ||
|---|---|---|---|---|
| − 1 | 0 | + 1 | ||
| 1. | pH | 4 | 5.5 | 7 |
| 2. | Temp | 25 | 32.5 | 40 |
| 3. | Casein conc. | 0.5 | 2.75 | 5 |
– 1, minimum value; 0, average value; + 1: maximum value
Partial purification of milk-clotting protease
Acetone precipitation
The crude enzyme extract was precipitated with chilled acetone (acetone or 75% acetone). Two volumes of chilled acetone were slowly added to the extract, and the precipitate was then allowed to settle for 1 h at – 18 °C to permit complete precipitation. The precipitated protein was separated by centrifuging (Heraeus Pico17/21 centrifuge, Thermo Electron Led, Germany) at 10000g for 10 min at 4 °C. The pellet was then dried in the open air for 30 min and dissolved in 0.02 M phosphate buffer, pH 6.0 to remove trace amounts of acetone [22].
Ammonium sulfate precipitation
The crude enzyme extract was precipitated with ammonium sulfate according to [23]. Accordingly, 300 mL of the crude extract was added to 1200-mL (100%) saturated ammonium sulfate to precipitate. Then, the enzymatic solution was decanted for one night at 4 °C and then centrifuged (Heraeus Pico17/21 centrifuge, Thermo Electron Led, Germany) at 10000 g for 10 min at 4 °C, and the pellet was suspended in the phosphate buffer (0.02 M; pH 6).
Inhibition study
The effect of protease inhibitors to inhibit protease activity was studied according to the methods of [24] after the crude enzyme was produced using optimized parameters (one-variable-at-a-time). The inhibitors used include cysteine protease inhibitor, iodoacetamide (1 and 10 mM); aspartic protease inhibitor, pepstatin A (0.02, 0.04, 0.6, 0.08, and 0.1 mM); metalloprotease inhibitor, EDTA (5 and 10 mM); and serine protease inhibitor, phenyl-methane sulphonyl fluoride (PMSF) (1 and 10 mM). After incubating the enzyme with inhibitor at 35 °C for 30 min, residual activity was assayed under optimal conditions. The control enzyme activity, without the inhibitor, was taken as 100%.
Data analysis
Data analyses were performed using SAS software version 9 (Inc. Cary NC USA). The experiments were carried out in triplicate. Analysis of variance (ANOVA) and means comparisons was done by Duncan’s multiple range tests.
Results and discussion
Selection of solid substrate for production of milk-clotting protease (MCP) by Aspergillus oryzae DRDFS13
In this study, several agro-industrial substrates (wheat bran, rice bran, pea bran, and grass pea bran) were screened for production of MCP from A. oryzae DRDFS13 under SSF. Although the fungus grew on all substrates (wheat bran, pea bran, grass pea, and rice bran), it did not show milk-clotting activity, except on wheat bran with milk-clotting activity (MCA) of 77.74 U/mL (Fig. 1). Although the particle size of each substrate was not determined, the difference may be due to differences in particle size between wheat bran and the other three substrates which is partly related to porosity [25]. Wheat bran is considered as the best substrate for the production of acid protease from A. oryzae MTCC 5341 [4], other Aspergillus species [20, 26], and Mucor sp. [27].In the present study, both wheat bran moistened with a mineral solution and wheat bran moistened with HCL were used for milk-clotting protease production; however, the result showed that wheat bran moistened with HCL was better for MCP production (Fig. 1).
Fig. 1.
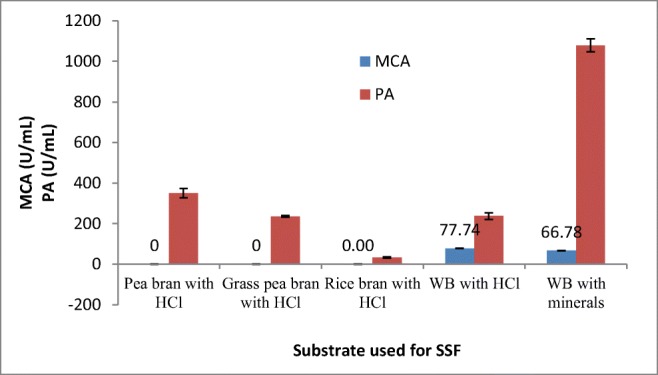
Screening of various agro-industrial residues for MCP production by A. oryzae DRDFS13 in SSF. MCA, milk-clotting activity; PA, protease activity; WB, wheat bran; initial media pH 4.2; no C and N supplement; temp 30 °C
The effect fermentation time on biomass and milk-clotting protease production by Aspergillus oryzae DRDFS13
The growth and milk-clotting protease production in SSF were studied for 6 days under previously established optimal conditions (Table 1). Chymosin (commercial enzyme) was used as a positive control during the milk-clotting activity test. The fungus showed a steady growth starting from 10.15 mg GLcN/gdfs to 19.77 ± 8.20 mg GLcN/gdfs implying a twofold increase in biomass after 2 days of incubation. Similarly, the highest biomass levels of M. mucedo DSM 809 was achieved between 2 and 3 days [24]. This could be interpreted as microbe that undergoes lag phase before active growth and starts to decline in biomass when the nutrients depleted. Biomass production was supported by carbohydrates consumption, which was almost exhausted at the time of maximum enzyme production.
The milk-clotting activity was detected after 96 h of incubation, and the maximum value of MCA (79.6 U/mL) was recorded at a 120 h incubation period. Whereas the protease activity was noticed from the second day (100.24 U/mL) and the highest PA (231.24 U/mL) was recorded on the 6th day of the incubation period (Table 2), this might be due to the gene encoding aspartic protease enzyme that could be induced later than other types of protease. The maximum MCA recorded from M. circinelloides, A. oryzae MTCC, and Aspergillus sp. on the 5th day of fermentation time under SSF is comparable with the present study [4, 21, 28]. But, further incubation showed a reduction in enzyme production. The reduction in enzyme yield after the optimum period was probably due to the depletion of nutrients available to microbial growth [21]. On the other hand, the highest milk-clotting activity was obtained from A. niger FFB1 on the 3rd day [20], and maximum acid protease activity was noticed from A. oryzae HG76 at 80 h incubation time [29] which was different from the present study.
Table 2.
Biomass, MCA, and PA of the crude enzyme from A. oryzae DRDFS13
| Sample | Day | pH mean ± SD |
Biomass in mg GlcN/gdfs | MCA(U/mL) mean ± SD |
PA(U/mL) mean ± SD |
Ratio MCA/PA |
|---|---|---|---|---|---|---|
| Blank | _ | 3.93 ± 0.01a | – | NDc | 0.00 ± 0.00f | ND |
| Aspergillus oryzae DRDFS13 | 0 | 3.91 ± 0.01a | 10.15 ± 0.11c | NDc | 0.00 ± 0.00f | ND |
| 1 | 3.78 ± 0.00a | 12.43 ± 2.02c | NDc | 0.00 ± 0.00f | ND | |
| 2 | 3.90 ± 0.09a | 19.77 ± 8.20a | NDc | 100.24 ± 6.88e | ND | |
| 3 | 3.85 ± 0.01a | 16.64 ± 6.48b | NDc | 129.07 ± 1.64d | ND | |
| 4 | 3.90 ± 0.02a | 11.43 ± 0.18c | 63.76 ± 6.62b | 196.39 ± 3.12b | 0.32 | |
| 5 | 3.94 ± 0.01a | 11.30 ± 0.04c | 79.60 ± 3.16a | 172.26 ± 10.53c | 0.46 | |
| 6 | 4.02 ± 0.20a | 10.96 ± 0.71c | 70.20 ± 1.20b | 231.24 ± 1.01a | 0.30 | |
| Chymosin | _ | – | – | 2181.81 ± 0.00 | 281.75 ± 24.79 | 7.74 |
Chymosin, commercially produced pure enzyme used as positive control; ND, not determined (if milk did not clot within 40 min); MCA, milk-clotting activity (U/mL); PA, protease activity (U/mL); ratio, the ratio of MCA/PA; SD, standard deviation; mean, average of two measurements; different letters (a, b, c, d) designate significantly different means as determined by Duncan multiple mean comparison test (P < 0.05)
The effect of incubation temperature on milk-clotting protease (MCP) production by Aspergillus oryzae DRDFS13
The study indicated that the MCA activity started to steadily increase to 60–70 U/mL between 25 and 30 °C and abruptly declined to 30–40 U/mL at 35 °C (Fig. 2). Other studies also showed slight variations in the temperature optima for the production protease enzymes by Aspergillus spp. from above 30 °C (32 °C) [28] and a decline at 35 °C from A. oryzae MTCC 5341 [4]. Although the enzymes showed slight variation, no significant difference showed; in their temperature optimal activities, they were within the optimum temperature of 30 °C for the production of these enzymes with solid-state fermentation by other fungi.
Fig. 2.
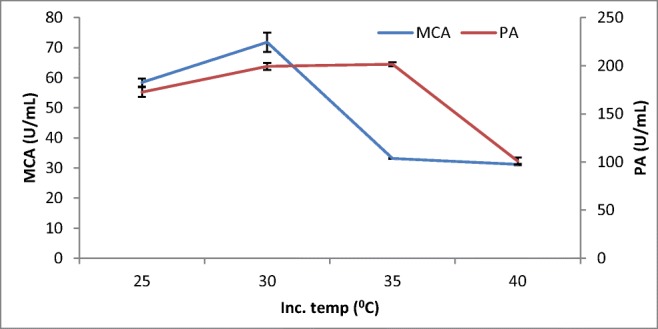
Effect of incubation temperature on MCP production by A. oryzaeDRDFS13in SSF. MCA, milk-clotting activity; PA, protease activity; WB moistened with HCL; initial media pH 4.2; no C and N supplement
The effect of inoculum size on milk-clotting protease production by Aspergillus oryzae DRDFS13
The highest milk-clotting activity was recorded from the crude enzyme extract produced by A. oryzae DRDFS13 using an inoculum size of 0.5*106 spores/mL followed by 0.1*106 spores/mL (Fig. 3). This was comparable to the highest enzyme activity displayed by A. niger under SSF using an inoculum size of 1*106 spores/mL [20]. However, the acid protease activity observed from A. oryzae HG76 at an inoculum concentration of 0.75*106 mL−1 under SSF is similar to the present study [29].
Fig. 3.
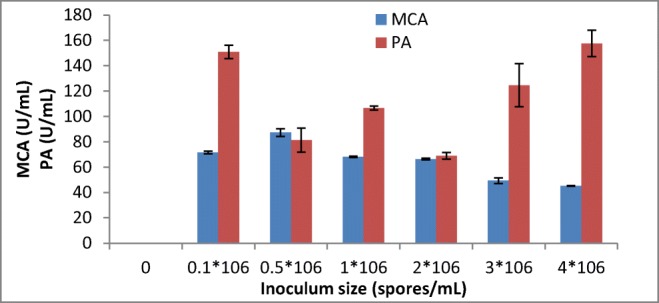
Effect of inoculum size on MCP production by A. oryzae DRDFS13 under SSF. MCA, milk-clotting activity; PA, protease activity; substrate, WB moistened with HCL; initial media pH 4.2; no C and N supplement; temp, 30 °C
As the level of inoculum size increased to 4*106 spores/mL, the milk-clotting activity of the protease was decreased to lower activity (45.16 U/mL). Similarly, a milk-clotting activity of the protease lowered when an inoculum concentration of 4.5 × 106 spores/mL was used in SSF [20]. The decrease in MCA that has been observed with higher inoculum size could be due to the shortage of the nutrients available for the larger biomass and faster growth of the culture. Hence, a balance between the proliferating biomass and available material is vital for maximum enzyme production [21].
The effect moisture content on milk-clotting protease production by Aspergillus oryzae DRDFS13
In the present study, the maximum milk-clotting activity of 100.43 ± 0.84 was observed with 55% moisture content (Fig. 4). Different studies showed that different SSF systems gave optimum protease activities at various moisture contents ranging from 40 to 60% with various fungi: A. oryzae HG76, Rhizopus stolonifer, Rhizomucor miehei, and A. niger FFB1 [20, 29–31]. [21] reported maximum enzyme production from M. circinelloides at 20% moisture content in SSF. This may be related to the different types and porosity of substrates used for SSF.
Fig. 4.
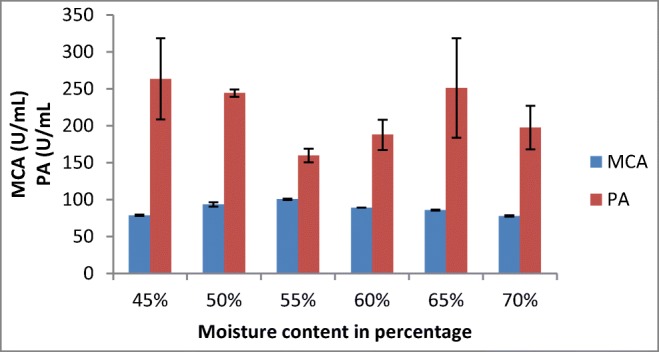
Effect of moisture content on MCP production by A. oryzae DRDFS13 in SSF. MCA, milk-clotting activity; PA, protease activity; substrate, WB moistened with HCL; initial media pH 4.2; no C and N supplement; temp, 30 °C
The effect initial media pH on milk-clotting protease production by Aspergillus oryzae DRDFS13
The pH of the medium strongly affects many enzyme processes and transport of various compounds across the cell membrane. Thus, the effect of initial media pH on the production of MCP is illustrated in Fig. 5. The MCA gradually increased from pH 4.0 to pH 6 where the maximum milk-clotting activity was recorded at pH 6. This was similar to maximum MCA production recorded from A. oryzae MTCC 5341 and Rhizopus stolonifer at pH 6.3 and pH 6, respectively [4, 31]. Other studies indicated optimum MCA production by A. oryzae and Mucor circinelloides at pH 7.0 [21, 26]. The protease production by A. niger FFB1 and A. oryzae MTCC 5341 and other Aspergillus sp. was maximum between pH 4 and pH 5 [4, 20, 28]indicating the pH optima difference between acidophilic and neutrophilic fungi irrespective of their taxa.
Fig. 5.
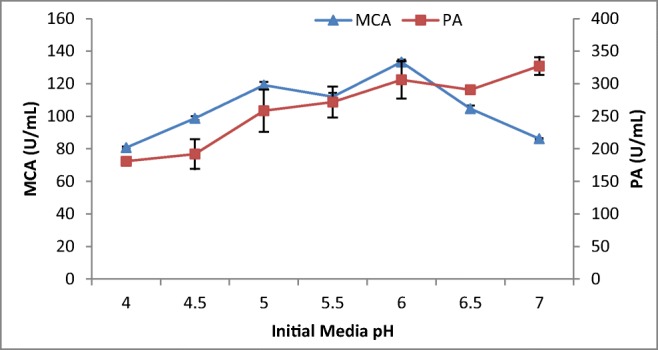
Effect of initial media pH on MCP production by A. oryzae DRDFS13 under SSF. MCA, milk-clotting activity; PA, protease activity; substrate, WB moistened with HCL; no C and N supplement; temp, 30 °C; moist. content, 55%
The effect supplementary nitrogen source on milk-clotting protease production by Aspergillus oryzae DRDFS13
Different nitrogen sources, i.e., skim milk, casein, yeast extract, urea, NH4NO3, NH4SO4, and NH4Cl, were used in the SSF medium for the production of milk-clotting protease from A. oryzae DRDFS1 (Fig. 6). Supplementation of casein (1%) as a nitrogen source resulted in an increase in enzyme production, which is comparable to the highest MCA acquired from A. oryzae MTCC 5341 [4] and Rhizomucor nainitalensis [32] on the same substrate. Although the effect of specific nitrogen supplement on milk-clotting enzyme production differs from organism to organism, the complex nitrogen sources such as casein and peptone are usually used for milk-clotting enzyme production [26].
Fig. 6.
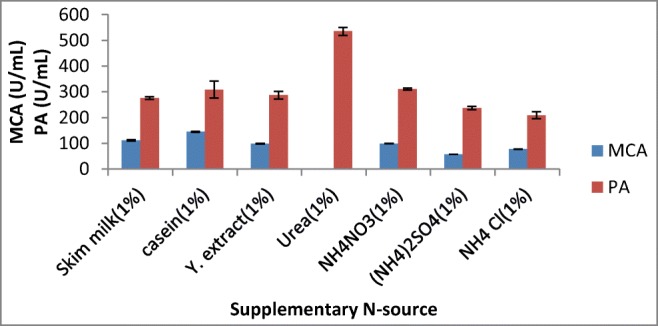
Effect of supplementary N source on MCP production by A. oryzae DRDFS1 3. MCA, milk-clotting activity; PA, protease activity; substrate, WB moistened with HCL; int. media pH 6.0; temp, 30 °C; moist. content, 55%
The effect of casein concentration on milk-clotting protease production by Aspergillus oryzae DRDFS13
The maximum MCA was observed at 1% casein concentration (Supplementary Data Fig. 1). Similar to the present study, the highest MCA was recorded from Rhizopus stolonifer [31] and Rhizomucor nainitalensis [32] at 1% and 1.5% casein concentration, respectively. Similarly, the addition of 2 g of casein into the solid substrate showed a significant increase in the production of microbial rennet from M. miehei [33].
The effect supplementary carbon source on milk-clotting protease production by Aspergillus oryzae DRDFS13
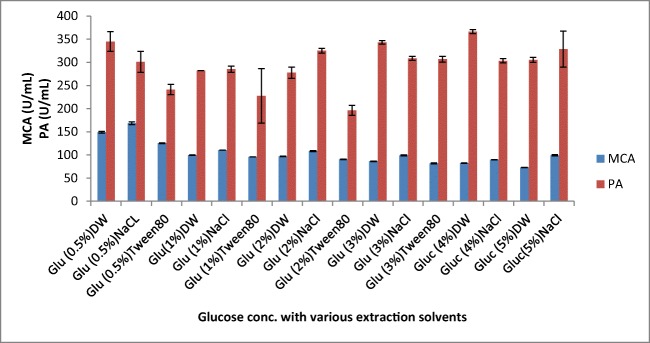
Among the various carbon sources tested, glucose at 0.5% was found to be the best source for milk-clotting enzyme production followed by galactose and starch (Figs. 7 and 8). Although it is argued that the simplest carbon sources like glucose in the media enhance protease production by A. oryzae [34], other studies also showed supplementation with maltose [35], and lactose [31] showed maximum milk-clotting activity in the SSF media.
Fig. 7.
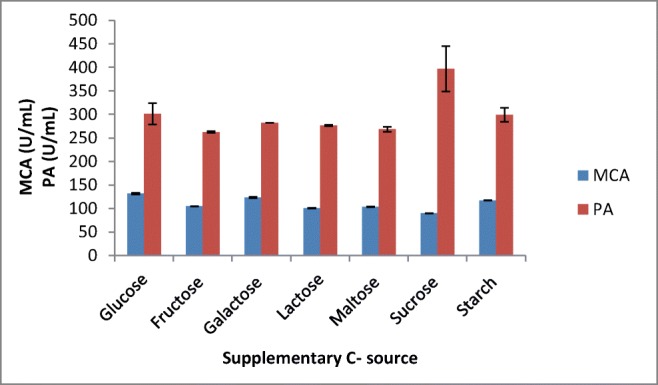
Effect of supplementary C source on MCP production by A. oryzae DRDFS13 in SSF. MCA, milk-clotting activity; PA, protease activity; substrate, WB moistened with HCL; int. media pH 6.0; temp, 30 °C; moist. content, 55%; suppl. N source, 1% casein
Fig. 8.
Effect glucose concentration and different extraction solvents on MCP production by A. oryzae DRDFS13. MCA, milk-clotting activity; PA, protease activity; DW, distilled water; NaCl, sodium chloride; Glu, glucose; substrate, WB moistened with HCL; int. media pH 6.0; temp, 30 °C; moist. content, 55%; suppl. N source, 1% casein; suppl. C source, 0.5% glu
The effect extraction solvents on the activity of MCP produced by A. oryzae DRDFS13
The extraction efficiency is critical to the recovery of the enzyme from the fermented biomass; hence, the selection of a suitable solvent is necessary [36]. The highest milk-clotting activity was recorded using 0.1 M NaCl followed by distilled water as an extraction solvent for enzyme leaching in SSF (Fig. 8). The acid protease enzyme extracted from A. oryzae MTCC 5341 using 0.1 M NaCl solution that showed significant MCA was comparable with the present study [1]. However, on the economic feasibility of fermentation, distilled water was chosen as a suitable solvent for further extraction of crude protease from fermented solids [28].
Partial purification of milk-clotting protease from A. oryzae DRDFS13
Partial purification of the crude enzyme extract obtained from A. oryzae DRDFS13 using chilled acetone and 80% (NH4)2SO4 increased the ratio of MCA/PA while reducing the MCA and PA. Chymosin (commercial enzyme) was used as a positive control during the milk-clotting activity test. The ratio of MCA/PA of the crude enzyme increased by 2.3-fold (from 0.56 to 1.3) by acetone and by 1.2-fold (from 0.56 to 0.65) via 80% (NH4)2SO4 (Table 3). Similarly, partial purification of protease from A. oryzae MTCC 5341, A. terreus, and P. aeruginosa using chilled acetone increased the purification by 5-fold, 2.6-fold, and 1.6-fold, respectively [9, 22, 37].
Table 3.
The effect of acetone and ammonium sulfate precipitation on MCA/PA ratio of MCP from A. oryzae DRDFS13
| Sample | Extraction solvent | Precipitating solvents | MCA (U/mL) Mean ± SD |
PA (U/mL) Mean ± SD |
Ratio (MCA/PA) |
|---|---|---|---|---|---|
| Crude enzyme (0.5% glucose) | 1 M NaCl | – | 168.47 ± 2.96a | 300.80 ± 22.52a | 0.56 |
| Crude enzyme (0.5% glucose) | 1 M NaCl | Acetone | 84.96 ± 1.06c | 65.28 ± 3.07c | 1.30 |
| Crude enzyme (0.5% glucose) | 1 M NaCl | 80% (NH4)2SO4 | 120.19 ± 6.80b | 185.01 ± 0.32b | 0.65 |
| Chymosin | _ | _ | 2533.33 ± 188.56 | 99.76 ± 5.36 | 25.39 |
Chymosin, commercially produced pure enzyme used as positive control; MCA, milk-clotting activity (U/mL); PA, protease activity (U/mL); ratio, the ratio of MCA/PA; SD, standard deviation; mean, average of two measurements; different letters (a, b, c, d) designate significantly different means as determined by Duncan multiple mean comparison test (P < 0.05)
Production of the milk-clotting protease (MCP) under SSF using response surface methodology
Three factors, i.e., pH, casein concentrations, and temperature, were found to be the most significant factors affecting the production of MCP under SSF conditions, but their possible interactions were not evaluated. Thus, a statistical design response surface methodology was employed here to study their possible interactions for its effect on milk-clotting protease production. Consequently, the data were fitted with the following regression equation which is an empirical relationship between the enzyme yields and test variables:
MCA (U/mL) = + 323.85921 – (14.75783* pH) – (4.90801 *Caesin conc.) – (4.99157 *Temp.)
The regression equation obtained from the ANOVA showed that the multiple correlation coefficient (R2) was 0.8402. The R2 value > 0.75 indicates the fitness of the model [38]. The “adjusted R2” is 0.8102, and the predicted R2 is 0.7161, indicating that the model is good as for a good statistical model, the R2 should be in the range of 0–1.0, and the nearer to 1.0 the value is the more fit the model is deemed to be [38]. The “adequate precision value” of the present model was 17.467suggesting that the model can be used to navigate the design space. The “precision value” is an index of the signal-to-noise ratio, and values of higher than 4 are prerequisites for a model to be a good fit [38]. The model showed standard deviation, mean, and predicted R2 values of 18.08, 66.97, and 0.7161, respectively (Table 4).
Table 4.
ANOVA table for milk-clotting activity in response surface linear model
| Model terms | Value | Model terms | Value |
|---|---|---|---|
| Std. Dev. | 18.08 | R2 | 0.8402 |
| Mean | 66.97 | Adj R2 | 0.8102 |
| C.V. % | 27.00 | Pred R2 | 0.7161 |
| PRESS | 9293.13 | Adeq Precision | 17.467 |
| Model F-value | 28.04 | Lack of fit | 4799.77 |
The results from the RSM experiments were presented in Table 5 as both predicted and experimental values. The highest MCA (137.58 U/mL) from Aspergillus oryzae DRDFS13 was recorded at a casein concentration of 0.5%, pH 4.0, and 25 °C. Generally, the optimization of the physicochemical and nutritional factors for the production of the MCE by RSM increased the MCA by ≈2.0-fold. Similar to the present study, [39] reported the optimization of protease production from Paecilomyces marquandii in central composite design (CCD) increased the enzyme activity by 2.57-fold. In the same way, the MCA of MCE from B. subtilis B1 was improved by 5.57-fold after optimization by response surface methodology [40]. In another study, optimizing the production of MCE enzyme from Mucor mucedo KP736529 by the CCD led to a 6.12-fold increase in MCA compared with initial activity [41]. According to [42], the activity of acid protease from a mixed culture of A. oryzae AS3042 and A. niger SL-09 was increased by five fold using response surface methodology (RSM) optimization.
Table 5.
Experimental design used in the RSM studies of three independent variables with 3 center points for MCP production by A. oryzae DRDFS13 under SSF
| Run order | A pH |
B Casein conc. |
C Temp. |
MCA (U/mL) | |
|---|---|---|---|---|---|
| Actual value | Predicted value | ||||
| 1. | 4.00 | 0.50 | 25.00 | 134.85 | 137.58 |
| 2. | 7.00 | 0.50 | 25.00 | 107.99 | 93.31 |
| 3. | 4.00 | 5.00 | 25.00 | 131.18 | 115.50 |
| 4. | 7.00 | 5.00 | 25.00 | 79.34 | 71.23 |
| 5. | 4.00 | 0.50 | 40.00 | 83.05 | 62.72 |
| 6. | 7.00 | 0.50 | 40.00 | 0.00 | 18.44 |
| 7. | 4.00 | 5.00 | 40.00 | 15.95 | 40.62 |
| 8. | 7.00 | 5.00 | 40.00 | 14.55 | −3.65 |
| 9. | 2.98 | 2.75 | 32.50 | 132.24 | 104.20 |
| 10. | 8.02 | 2.75 | 32.50 | 49.49 | 29.74 |
| 11. | 5.5 | −1.03 | 32.50 | 69.78 | 85.54 |
| 12. | 5.5 | 6.53 | 32.50 | 30.57 | 48.40 |
| 13. | 5.5 | 2.75 | 19.89 | 101.95 | 129.93 |
| 14. | 5.5 | 2.75 | 45.11 | 0.00 | 4.01 |
| 15. | 5.5 | 2.75 | 32.5 | 56.65 | 66.97 |
| 16. | 5.5 | 2.75 | 32.5 | 72.89 | 66.97 |
| 17. | 5.5 | 2.75 | 32.5 | 59.54 | 66.97 |
| 18. | 5.5 | 2.75 | 32.5 | 56.94 | 66.97 |
| 19. | 5.5 | 2.75 | 32.5 | 79.18 | 66.97 |
| 20. | 5.5 | 2.75 | 32.5 | 63.22 | 66.97 |
MCA, milk-clotting activity (U/mL); casein conc., casein concentration (%); and temp., temperature in °C
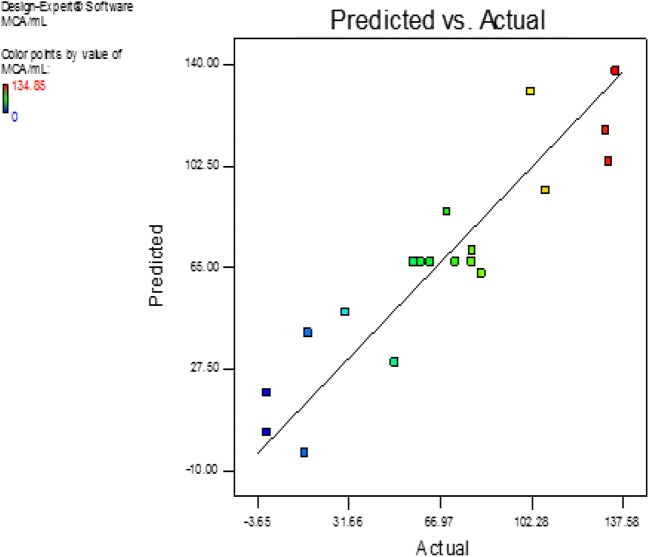
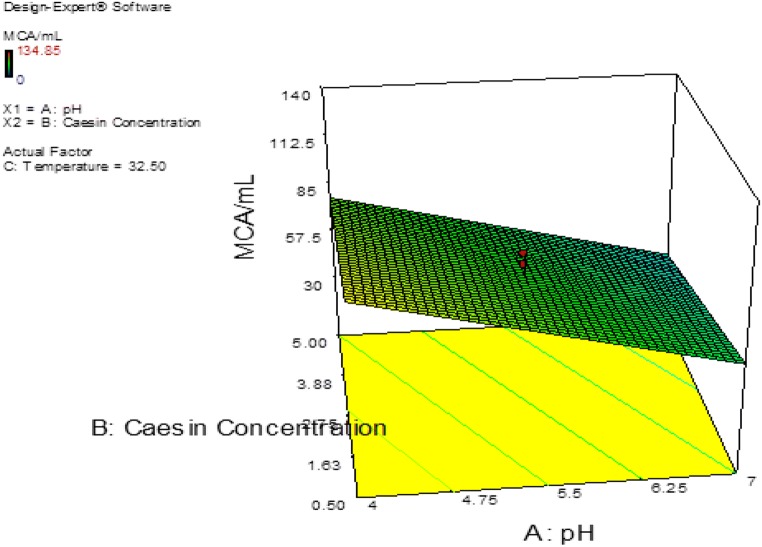
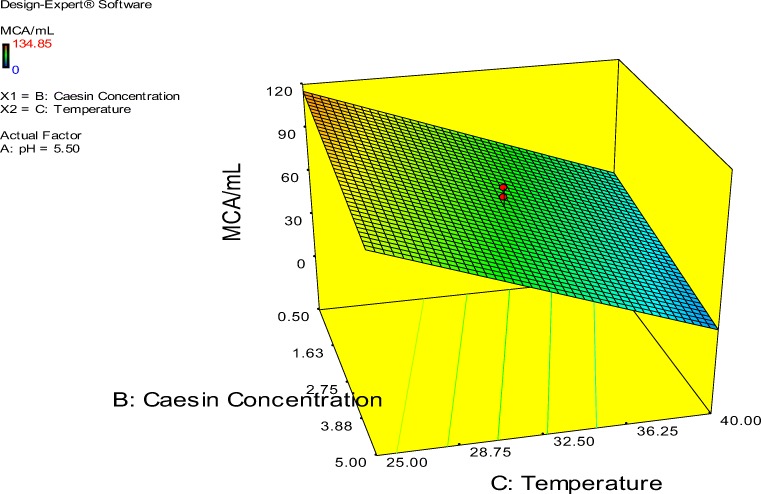
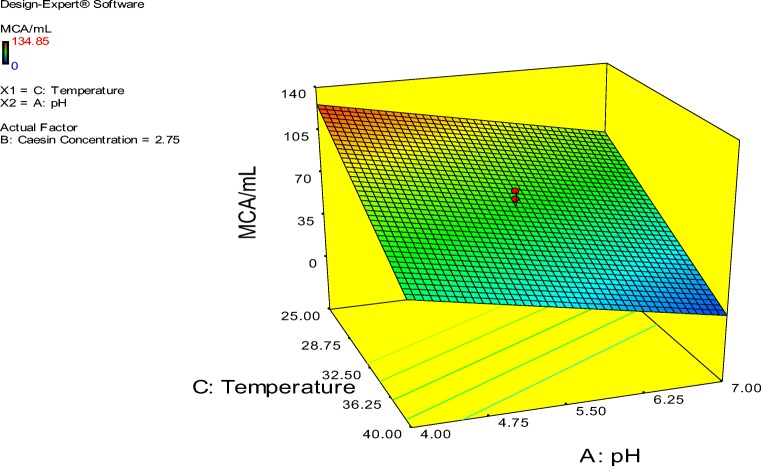
The probability plot showed a satisfactory correlation between the experimental and predictive values (Fig. 9). In order to determine the optimal levels of each factor for maximum milk-clotting protease production, three-dimensional response surface plots were constructed. Figure 10 shows the response for the interactive factors, casein and pH, when the temperature was at 32.5. The maximum milk-clotting activity in this condition was predicted to be 132.24 U/mL. The production of milk-clotting protease production varied considerably over the range tested from 30.57 to 132.24 U/mL. Figure 11. indicates the response for the interactive factors, casein and temperature, when the pH was 5.5. The maximum milk-clotting activity in this condition was predicted to be 101.95 U/mL. The production of milk-clotting protease production varied considerably over the range tested from 0.00 to 101.95 U/mL.
Fig. 9.
Comparative plot showing predicted vs. actual values of milk-clotting protease from Aspergillus oryzae DRDFS13
Fig. 10.
Response surface curves of milk-clotting protease production from Aspergillus oryzae DRDFS 13 showing the interaction between casein and pH
Fig. 11.
Response surface curves of milk-clotting protease production from Aspergillus oryzae DRDFS 13 showing the interaction between casein and temperature
Figure 12 shows the response for the interactive factors, pH and temperature, when the casein concentration was 2.75. The maximum milk-clotting activity in this condition was predicted to be 45.11 U/mL. The production of milk-clotting protease production varied considerably over the range tested from 32.5 to 101.95 U/mL.
Fig. 12.
Response surface curves of milk-clotting protease production from Aspergillus oryzae DRDFS 13 showing the interaction between temperature and pH
Inhibition study
Incubation of crude enzyme with pepstatin A completely inhibited the milk-clotting activity of the enzyme. On the other hand, more than 68% of residual MCA was recorded using iodoacetamide, EDTA, and PMSF as protease inhibitors (Table 6). Similar results were reported by [1, 24, 43–48].
Table 6.
Inhibition study for the crude enzyme from A. oryzae DRDFS13
| Sample | Fermentation Time (days) |
MCA(U/mL) Mean ± SD |
Inhibition study Residual MCA (%) |
|||
|---|---|---|---|---|---|---|
| Pep A (1 mM) |
I.A (10 mM) |
EDTA (10 mM) |
PMSF (10 mM) |
|||
| Aspergillus oryzae DRDFS13 | 0 | NDc | NDd | NDd | NDd | NDd |
| 1 | NDc | NDd | NDd | NDd | NDd | |
| 2 | NDc | NDd | NDd | NDd | NDd | |
| 3 | NDc | NDd | NDd | NDd | NDd | |
| 4 | 63.76 ± 6.62b | 0.00d | 97.14c | 94.56c | 95.78c | |
| 5 | 174.61 ± 3.18a | 0.00d | 72.94b | 69.82b | 76.22b | |
| 6 | 170.22 ± 1.21a | 0.00d | 84.19ab | 89.52a | 74.80b | |
D, not determined; MCA, milk-clotting activity (U/mL); Pep A, pepstatin A; I.A, iodoacetamide; EDTA, ethylenediaminetetraacetic acid; PMSF, phenyl-methane sulphonyl fluoride; SD, standard deviation; mean is average of two measurements; different letters (a, b, c, for MCA (U/mL) designate significantly different means as determined by Duncan multiple mean comparison test (P < 0.05); different letters (a, b, c,d for PepA, I.A, EDTA, and PMSF) designate significantly different means as determined by Duncan multiple mean comparison test (P < 0.05)
Comparative analysis of the efficiency of A. oryzae DRDFS 13 on MCA under SSF in comparison with other fungi
In general, the milk-clotting activity of protease enzyme produced from Aspergillus oryzae DRDFS 13 was comparable with the milk-clotting activity of protease reported by [20]. However, the milk-clotting activity recorded for milk-clotting protease in this study was higher than the milk-clotting activity of proteases produced from Aspergillus oryzae [26], Aspergillus flavo furcates DPUA 1461 [49], Aspergillus spp. [28], and Rhizopus stolonifer [31]. On the other hand, the activity of milk-clotting protease from Aspergillus oryzae DRDFS 13 was lower than the milk-clotting activity of proteases produced from Aspergillus oryzae HG76 [29] and Mucor spp. [50]. The variation recorded in milk-clotting activity could be due to the difference of the substrate combination, mineral supplements, effectiveness of the fungus species, incubation time, and/or moisture content used in solid-state fermentation.
Conclusion
Optimization of the milk-clotting protease enzyme production from A. oryzae DRDFS 13 by both one-variable-at-a-time and response surface methodology (RSM) significantly improved the milk-clotting activity by twofold.
The milk-clotting activity was increased by twofold in one-variable-at-a-time optimization (from 77.74 U/mL to 168.47) when A. oryzae DRDFS13 was cultivated at temperature (30 °C), pH (6.0), moisture content (55%), and inoculum size (5*105 spores/mL) for 5 days using wheat bran as solid substrate under solid-state fermentation. The milk-clotting activity was also increased by about twofold using RSM optimization (from 77.74 U/mL to 137.85 U/mL) when A. oryzae DRDFS13 was cultivated at a casein concentration of 0.5%, pH 4.0, and 25 °C.
Electronic supplementary material
(DOCX 152 kb)
Acknowledgments
Moreover, we would like to acknowledge the Department of Microbial, Cellular, and Molecular Biology of Addis Ababa University and Downstream Processing Laboratory, Department of Life Sciences and Chemistry, Jacobs University, Bremen Germany for the facilitation of laboratory space and provision of basic facilities.
Funding
This work was supported by Microbial, Cellular, and Molecular Biology Department, Addis Ababa University.
Compliance with ethical standards
Conflict of interests
The authors declare that there is no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Vishwanatha SA, KS. Rao AAG, Singh Production and characterization of a milk-clotting enzyme from Aspergillus oryzae MTCC 5341. Appl Microbiol Biotechnol. 2010;85:1849–1859. doi: 10.1007/s00253-009-2197-z. [DOI] [PubMed] [Google Scholar]
- 2.Rao MB, Tanksale AM, Ghatge MS, Deshpande VV. Molecular and biotechnological aspects of microbial proteases †. MIicrobiology Mol Biol Rev. 1998;62(3):597–635. doi: 10.1128/mmbr.62.3.597-635.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Talib MA, Abubakar MA, Jideani IA. Comparative study of bacteriological and organoleptic properties of white pickled cheese produced using calf rennet. ACT-Biotechnology Res Commun. 2011;1(1):36–39. [Google Scholar]
- 4.Vishwanatha KS, Appu Rao AG, Singh SA. Acid protease production by solid-state fermentation using Aspergillus oryzae MTCC 5341: optimization of process parameters. J Ind Microbiol Biotechnol. 2010;37(2):129–138. doi: 10.1007/s10295-009-0654-4. [DOI] [PubMed] [Google Scholar]
- 5.Sardinas JL. Rennin enzyme of Endothia parasitica. Appl Microbiol. 1968;16(2):248–255. doi: 10.1128/am.16.2.248-255.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arima K, Yu J, Iwasaki S. Milk-clotting enzyme from Mucor pusillus var. Lindt. Methods Enzymol. 1970;19:446–459. [Google Scholar]
- 7.de Souza PM, Bittencourt ML d A. A biotechnology perspective of fungal proteases. Braz J Microbiol. 2015;46(2):337–346. doi: 10.1590/S1517-838246220140359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.V. M. S. * and J. V. R. Kalaskar Vishalkirti Vijay, Narayanan Kasinathan, “Optimization of extracellular acid protease production from Aspergillus Niger by factorial design,” J Microbiol Biotechnol Food Sci, vol. 4, no. 2, pp. 132–136, 2014
- 9.Vishwanatha KS, Rao AGA, Singh SA. Characterisation of acid protease expressed from Aspergillus oryzae MTCC 5341. Food Chem. 2009;114(2):402–407. [Google Scholar]
- 10.de Castro RJS, Nishide TG, Sato HH. Production and biochemical properties of proteases secreted by Aspergillus niger under solid state fermentation in response to different agroindustrial substrates. Biocatal Agric Biotechnol. 2014;3(4):236–245. [Google Scholar]
- 11.Soares O, De Oliveira RL, Souza-motta CM, Lúcia A, Porto F, Porto TS. Novel Protease from Aspergillus tamarii URM4634 : production and characterization using inexpensive agroindustrial substrates by solid-state fermentation. Adv Enzym Res. 2016;4:125–143. [Google Scholar]
- 12.Ferea TL, Botstein D, Brown PO, Rosenzweig RF. Systematic changes in gene expression patterns following adaptive evolution in yeast. Proc Natl Acad Sci. 1999;96(17):9721–9726. doi: 10.1073/pnas.96.17.9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siala R, Frikha F, Mhamdi S, Nasri M, Kamoun AS. Optimization of acid protease production by Aspergillus Niger I1 on shrimp peptone using statistical experimental design. Sci World J. 2012;2012:1–11. doi: 10.1100/2012/564932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tadesse G, Mihret, Fentahun M, Tadesse Dairy farming and its economic importance in Ethiopia : a review. World J Dairy Food Sci. 2017;12(1):42–51. [Google Scholar]
- 15.Zelalem S. Yilma; Emmannuelle. Guernebleich and Ameha: A Review of the Ethiopian Dairy Sector Edited by; 2011. [Google Scholar]
- 16.Fernandez-Lahore HM, Fraile E, Cascone O. Acid protease recovery from a solid-state fermentation system. J Biotechnol. 1998;62:83–93. [Google Scholar]
- 17.Garcia G, De Oliveira GM, Ribeiro EJ, Monti R, Contiero J (2005) Microbial rennet produced by Mucor miehei in solid-state and submerged fermentation. 48(November):931–937
- 18.da Silveira GG, de Oliveira GM, Ribeiro EJ, Monti R, Contiero J. Microbial rennet produced by Mucor miehei in solid-state and submerged fermentation. Brazilian Arch Biol Biotechnol. 2005;48(6):931–937. [Google Scholar]
- 19.Zamani A, Jeihanipour A, Edebo L, Niklasson C, Taherzadeh AMJ. Determination of glucosamine and N-acetyl glucosamine in fungal cell walls. J Agric Food Chem. 2008;56:8314–8318. doi: 10.1021/jf801478j. [DOI] [PubMed] [Google Scholar]
- 20.Bensmail S, Aicha M, Fazouane F. Optimization of milk-clotting protease production by a local isolate of Aspergillus niger ffb1 in solid-state fermentation. J Microbiol Biotechnol Food Sci. 2015;4(5):467–472. [Google Scholar]
- 21.Sathya R, Pradeep BV, Angayarkanni J, Palaniswamy M. Production of milk clotting protease by a local isolate of Mucor circinelloides under SSF using agro-industrial wastes. Biotechnol Bioprocess Eng. 2009;14(6):788–794. [Google Scholar]
- 22.Palpperumal S, Sankaralingam S, Kathiresan D, Harinathan B, Shankar T, Prabhu D. Partial purification and characterization of a haloalkaline protease from Pseudomonas aeruginosa. Br Microbiol Res J. 2016;15(3):1–7. [Google Scholar]
- 23.Nouani A, Mati FM, Belbraouet S, Bellal M. Purification and characterization of a milk-clotting protease from Mucor pusillus: method comparison. Afr J Biotechnol. 2011;10(9):1655–1665. [Google Scholar]
- 24.Yegin S, Goksungur Y, Fernandez-Lahore M. Purification, structural characterization , and technological properties of an aspartyl proteinase from submerged cultures of Mucor mucedo DSM 809. Food Chem. 2012;133(4):1312–1319. [Google Scholar]
- 25.Janser R, De Castro S, Sato HH. Production and biochemical characterization of protease from Aspergillus oryzae : an evaluation of the physical – chemical parameters using agroindustrial wastes as supports. Biocatal Agric Biotechnol. 2014;3(3):20–25. [Google Scholar]
- 26.Patil P, Avanti K, Pallavi K. Production of milk clotting enzyme from Aspergillus oryzae under solid-state fermentation using mixture of. Int J Sci Res Publ. 2012;2(10):1–12. [Google Scholar]
- 27.Khademi F, Abachi S, Mortazavi A, Ehsani MA, Tabatabaei MR, Malekzadeh F. Optimization of fungal rennet production by local isolate of Rhizomucor nainitalensis under solid substrate fermentation system. IOSR J Pharm Biol Sci. 2013;5(2):115–121. [Google Scholar]
- 28.Radha S, Sridevi A, HimakiranBabu R, Nithya VJ, Prasad NBL, Narasimha G. Medium optimization for acid protease production from Aspergillus sps under solid state fermentation and mathematical modelling of protease activity. J Microbiol Biotech Res. 2012;2(1):6–16. [Google Scholar]
- 29.Li C, Xu D, Zhao M, Sun L, Wang Y. Production optimization, purification, and characterization of a novel acid protease from a fusant by Aspergillus oryzae and Aspergillus niger. Eur Food Res Technol. 2014;238(6):905–917. [Google Scholar]
- 30.Aljammas HA, Al Fathi H, Alkhalaf W. Study the influence of culture conditions on rennin production by Rhizomucor miehei using solid-state fermentations. J Genet Eng Biotechnol. 2018;16(1):213–216. doi: 10.1016/j.jgeb.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gais S, Fazouane F, Mechakra A. Production of milk clotting protease by Rhizopus Stolonifer through optimization of culture conditions. Int J Biol Biomol Agric Food Biotechnol Eng. 2009;3(6):340–344. [Google Scholar]
- 32.F. Khademi, S. Abachi, A. Mortazavi, M. A. Ehsani, M. R. Tabatabaei, and F. A. Malekzadeh, “Screening and isolation of substitute-rennet producing thermophilic phycomycetes, by modified Warcup method and improved selective medium,” IOSR J Agric Vet Sci, vol. 2, no. 2, pp. 30–37, 2013
- 33.B. F. Araújo, E. L. P. Ramos, J. Contiero, G. L. S. Ferreira, and G. G. Silveira, “The role of the type of substrate, particle size, and coagulations analytical method on microbial rennet synthesis by Mucor miehei Cooney & R . Emers ., 1964 ( Fungi : Zygomycota ) via solid-state fermentation,” Brazilian J Biol Sci, vol. 2, no. 4, pp. 245–251, 2015
- 34.M. S. and Y. M. Dhurway KS. Malakar R, Tiwari A. Production, purification and characterization of Protease from ‘Aspergillus oryzae. Discov Biotechnol. 2012;1(1):33–38. [Google Scholar]
- 35.Abdel-Fattah A, Saleh SA. Production and isolation of milk-clotting enzyme from Aspergillus versicolor. Zentralbl Bakteriol Naturwiss. 1979;134(6):547–550. doi: 10.1016/s0323-6056(79)80079-8. [DOI] [PubMed] [Google Scholar]
- 36.Ahmed Samia A. Optimization of production and extraction parameters of Bacillus megaterium Levansucrase using solid-state fermentation. J Appl Sci Res. 2008;4(10):1199–1204. [Google Scholar]
- 37.Niyonzima FN, More SS. Purification and characterization of detergent-compatible protease from Aspergillus terreus gr. Biotech. 2015;5:61–70. doi: 10.1007/s13205-014-0200-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dutt K, Gupta P, Saran S, Misra S, Rajendra KS. Production of Milk-clotting protease from Bacillus subtilis. Appl Biochem Biotechnol. 2009;158:761–772. doi: 10.1007/s12010-008-8504-9. [DOI] [PubMed] [Google Scholar]
- 39.Soares V, FilippeFreitas E, Braga FR, Leonardo HGA, De Araújo V, Ferreira SR, Araujo JM, De Oliveira A, Longo, De Vilela JH, Ribeiro and Queiróz Optimization of medium composition for protease production by Paecilomyces marquandii in solid-state- fermentation using response surface methodology. Afr J Microbiol Res. 2010;4(24):2699–2703. [Google Scholar]
- 40.Ding Z, Liu S, Gu Z, Zhang L, Zhang K. Production of milk-clotting enzyme by Bacillus subtilis B1 from wheat bran. Afr J Biotechnol. 2011;10(46):9370–9378. [Google Scholar]
- 41.Ayana IAAA, Ibrahim AE, Saber WIA. Statistical optimization of Milk clotting enzyme biosynthesis by Mucor mucedo KP736529 and its further application in cheese production. Int J Dairy Sci. 2015;10(2):61–76. [Google Scholar]
- 42.Leng Y, Xu Y. Improvement of acid protease production by a mixed culture of Aspergillus Niger and Aspergillus oryzae using solid-state fermentation technique. Afr J Biotechnol. 2011;10(35):6824–6829. [Google Scholar]
- 43.N. Hsiao, Y. Chen, Y. Kuan, Y. Lee, S. Lee, H. Chan, and C. Kao, “Purification and characterization of an aspartic protease from the Rhizopus oryzae protease extract , Peptidase R,” Electron J Biotechnol, vol. 17, pp. 89–94, 2014
- 44.Fernandez-Lahore HM, Auday RM, Fraile ER, de Jimenez MB, Bonino, Pirpignani L, Machalinski C, Cascone O. Purification and characterization of an acid proteinase from mesophilic Mucor sp. solid-state cultures. J Pept Res. 1999;53:599–605. doi: 10.1034/j.1399-3011.1999.00043.x. [DOI] [PubMed] [Google Scholar]
- 45.Yin L-J, Chou Y-H, Jiang S-T. Purification and characterization of acidic protease from Aspergillus oryzae BCRC 30118. J Mar Sci Technol. 2013;21(1):105–110. [Google Scholar]
- 46.Merheb-Dini C, Gomes E, Boscolo M, da Silva R. Production and characterization of a milk-clotting protease in the crude enzymatic extract from the newly isolated Thermomucor indicae seudaticae N31 (milk-clotting protease from the newly isolated) Food Chem. 2010;120(1):87–93. [Google Scholar]
- 47.P. M. Souza, G. Werneck, B. Aliakbarian, F. Siqueira, E. X. F. Filho, P. Perego, A. Converti, P. O. Magalhaes, and A. P. Junior, “Production , purification and characterization of an aspartic protease from Aspergillus foetidus,” Food Chem Toxicol, vol. 109, pp. 1103–1110, 2017 [DOI] [PubMed]
- 48.Rodrigues R, Caroline L, De Oliveira G, Aparecida M, Juliano L, De Oliveira AHC, Rosa JC, Cabral H. Biochemical and milk-clotting properties and mapping of catalytic subsites of an extracellular aspartic peptidase from basidiomycete fungus Phanerochaete chrysosporium. Food Chem. 2017;225:45–54. doi: 10.1016/j.foodchem.2017.01.009. [DOI] [PubMed] [Google Scholar]
- 49.Alecrim MM, Palheta RA, Teixeira MFS, Oliveira IMDA. Milk-clotting enzymes produced by Aspergillus flavo furcatis strains on Amazonic fruit waste. Int J Food Sci Technol. 2015;50:151–157. [Google Scholar]
- 50.H. M. Fernandz-Lahore, R. M. Auday, E. R. Fraile, B. J. M. Bonino, L. Pirpignani, C. Machalinski, and O. Cascone, “Purification and characterization of an acid proteinase from mesophilic Mucor sp. solid-state cultures,” J Pept Res, vol. 53, pp. 599–605, 1999 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 152 kb)