Abstract
This study aimed to assess the safety aspects of 15 lactic acid bacteria (LAB) strains previously isolated from a dairy environment with relation to their beneficial features. LAB strains were assessed using phenotypic methods according to their production of virulence factors at 25 °C and 37 °C, as well as by examining their potential resistance to 15 antibiotics. Polymerase chain reaction (PCR) was also used to identify the presence of 50 genes associated with virulence factors and antibiotic resistance in the strains. None of the strains presented hemolytic activity or the production of gelatinase, lipase, deoxyribonuclease, or the tested biogenic amines. Based on the disk diffusion assay, all strains were resistant to oxacillin and sulfa/trimethoprim. Further, some were resistant to gentamicin (14), clindamycin (11), vancomycin (9), rifampicin (8), erythromycin (5), tetracycline (4), ampicillin (2), and chloramphenicol (1); no strain was resistant to imipenem. Regarding virulence- and antibiotic-resistance-related genes, 19 out of 50 tested genes were present in some strains; there was a variable association of expression. Based on the obtained data, the isolates presented relatively safe characteristics and behavior, findings that should lead to further studies to assess their potential usage as beneficial cultures in the food industry.
Keywords: Lactic acid bacteria, Virulence factors, Antibiotic resistance
Introduction
Lactic acid bacteria (LAB) is a heterogeneous group of bacteria with a known history about their use in fermented products, fermentation processes, and for production of antimicrobial substances, including organic acids and antimicrobial proteins, such as bacteriocins [1]. In addition, LAB are known for their ability in providing beneficial effects on consumers, being several strains described as probiotics [2]. Among LAB, Lactobacillus strains are the most described as probiotics, followed by Streptococcus, Leuconostoc, Pediococcus, and Enterococcus [3].
Scientific knowledge regarding beneficial LAB has advanced significantly in terms of selection and characterization of new useful cultures, with a particular focus on benefits to consumer health [4–6]. The available studies in this area are also focused on the safety features of the selected LAB strains. Beneficial and safety aspects must be considered in order to assess the potential use of the selected LAB strains for human consumption [7]. Despite being characterized as beneficial, LAB can express a variety of pathogenic mechanisms and cause disease in human and other animal hosts [8]. Further, several factors are reported as hazardous, especially those related to antibiotic-resistance genes, genes that facilitate genetic material exchange, possible complications in the gastrointestinal tract (GIT), and indiscriminate use of antibiotics in human and veterinary medicine practice [9–11]. These factors can pose significant risks to public health, and they highlight the relevance of characterizing them in all bacteria that will be introduced into the food chain or recommended to be applied as probiotics [11].
A probiotic strain must resist and persist in the GIT and provide benefits to the host. It must also be safe and hazard-free for consumers [6, 7]. Based on a number of phenotypic and molecular assays, probiotic strains are generally recognized as safe (GRAS) if they possess only a minimal possibility for antibiotic-resistance gene transfer. They should be safe for human and animal food consumption and demonstrate proven health-promoting effects (e.g., non-invasive in epithelial cell line models, produce anti-inflammatory rather than pro-inflammatory cytokines) [7]. However, a GRAS status is not enough to indicate that a strain is safe, because other virulence factors are not considered in this evaluation [12, 13]. Deep research evaluation for the safety aspects of each specific strain needs to be performed in order to confirm the safety of that strain to be applied in food fermentation processes or as a beneficial culture for human or animal applications. Additionally, for these beneficial cultures to be considered safe for human health, they cannot cause disease (such as bacteremia) or be related to any toxic or metabolic effects, and they should not be able to transfer antibiotic-resistance genes [13, 14].
Thus, studies that involve bacteria with beneficial potential must characterize their virulence potential to exclude potential hazards. Here, we aimed to characterize the safety of a panel of LAB strains previously isolated from a dairy environment and characterized as possessing beneficial features [15, 16]. The examined parameters were related to virulence activity, biogenic amine production, and antibiotic resistance.
Material and methods
Bacterial strains and growth conditions
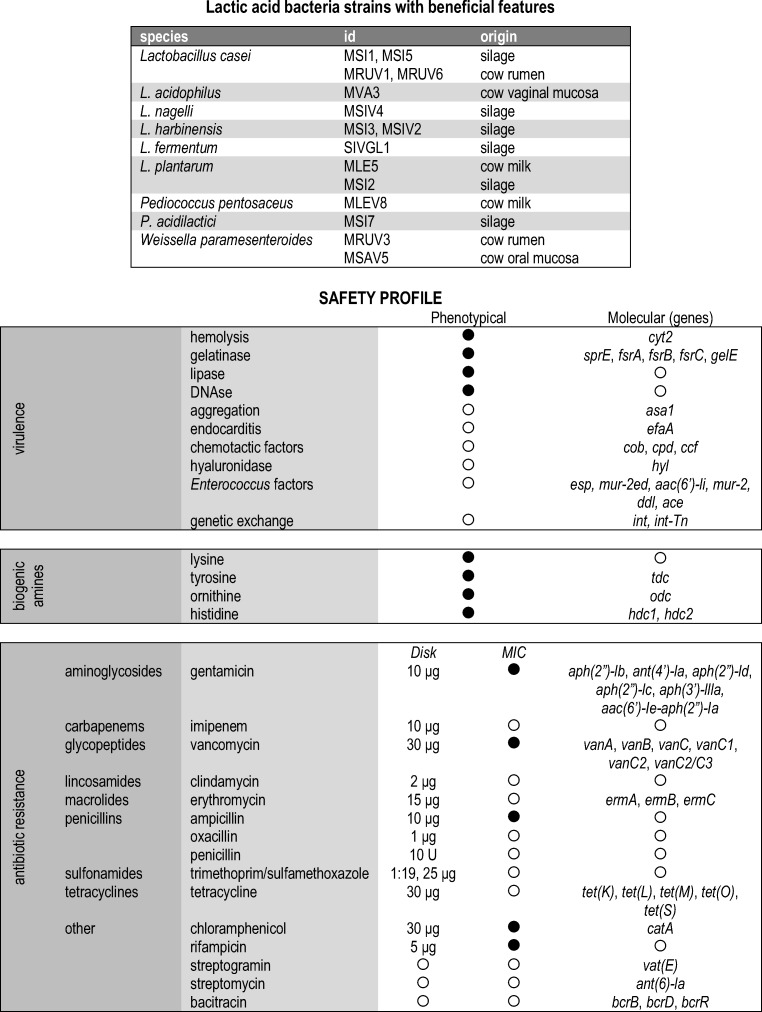
In a previous study, 15 strains were selected from an LAB collection isolated from a dairy environment and characterized as beneficial by phenotypic and molecular methods [15, 16]. These strains were characterized based on their genetic profiles by repetitive palindromic sequence polymerase chain reaction (rep-PCR) and random amplified polymorphic DNA (RAPD), and identified based on physiological and biochemical features and 16S rRNA as Lactobacillus spp. (n = 11), Pediococcus spp. (n = 2), and Weissella spp. (n = 2) [15, 16]. The selected strains were named as MSI1, MSI5, MRUV1, and MRUV6 (Lactobacillus casei), MVA3 (L. acidophilus), MSIV4 (L. nagelli), MSI3 and MSIV2 (L. harbinensis), SIVGL1 (L. fermentum), MLE5 and MSI2 (L. plantarum), MLEV8 (Pediococcus pentosaceus), MSI7 (P. acidilactici), and MRUV3 and MSAV5 (Weissella paramesenteroides). The strains were stored in de Man, Rogosa, and Sharpe (MRS) broth (Oxoid Ltd., Basingstoke, England) supplemented with 25% (v/v) glycerol at − 80 °C. For use, stock cultures were streaked on MRS agar (Oxoid), incubated at 37 °C for 24 h, when isolated colonies were transferred to MRS broth and incubated at 37 °C for 24 h. The selected strains were subjected to a panel of phenotypical and molecular assays in order to characterize their safety profiles. Figure 1 summarizes the adopted analysis, and further details are briefly described below.
Fig. 1.
Identification and origins of 15 lactic acid bacteria strains isolated from a dairy environment [15, 16] and the schematic description of the phenotypical and molecular assays to characterize their safety profiles. Conducted assays are indicated by filled circle, disk concentrations, and genes; open circle indicates non-conducted assays
Phenotypical characterization of safety
The selected strains were subjected to phenotypical assays to characterize their safety according to Barbosa et al. [17]. For hemolytic activity, the cultures were streaked on trypticase soya agar (Oxoid) with 5% (v/v) defibrinated horse blood. After incubation at 25 °C and 37 °C for 24 h, the hemolytic activity of each isolate was measured and classified as total or β-hemolysis (clear halos around the colonies), partial or α-hemolysis (greenish halos around the colonies), or γ-hemolysis (absence of hemolysis). For gelatinase production, 1-μL aliquots of the examined cultures was spotted on Luria Bertani (LB) agar (BD, Franklin Lakes, NJ, USA) with 3% (w/v) gelatin (BD). After incubation at 25 °C and 37 °C for 48 h, the plates were maintained at 4 °C for 4 h, and gelatin hydrolysis was recorded by the formation of opaque halos around the colonies. Lipase production was assessed by spotting 1 μL of each culture on LB with 0.2% (w/v) CaCl2 (Sigma-Aldrich, Inc., St. Louis, MO, USA) and 1% (v/v) Tween 80 (Sigma-Aldrich). After incubation at 25 °C and 37 °C for 48 h, transparent halos around the colonies were recorded as lipase production. DNAse activity was identified by spotting 1-μL aliquots of the cultures on the surface of DNAse methyl green agar (BD). After incubation at 25 °C and 37 °C for 48 h, the formation of clear halos around the colonies was identified as a positive result. All assays were conducted in three independent repetitions.
The potential to produce biogenic amines was assessed according to Bover-Cid and Holzapfel [18] and Joosten and Northolt [19]. Aliquots (0.5 mL) from each culture were transferred five times consecutively to MRS broth supplemented with 0.005% (w/v) pyridoxal-5-phosphate (Sigma-Aldrich); 0.1% (w/v) biogenic amine precursors were added individually: lysine, tyrosine, ornithine, and histidine (Sigma-Aldrich). The incubation was performed at 25 °C and 37 °C for 24 h, and the last transfer performed was streaked in duplicate on a modified MRS agar that was supplemented with one of each biogenic amine precursor described as above (1% w/v). The plates were incubated at 25 °C and 37 °C for 4 days, and positive results were recorded when the color changed from yellow to purple.
Antibiotic resistance was characterized based on disk diffusion assay (Oxoid) and Etest® strips (bioMérieux SA, Marcy l’Etoile, France). For the disk diffusion assay, each strain was diluted using 0.85% NaCl (w/v) until the turbidity was equivalent to 0.5 McFarland standard and swabbed onto the surface of a MRS agar plate (where the antibiotic disks were placed). A panel of 12 antibiotics from 9 classes were considered (Fig. 1). The plates were incubated at 37 °C for 18 h, when the diameter of the inhibition halos were measured and the isolates were characterized as presenting resistance (R), intermediary resistance (IR), or sensitivity (S) [20–22]. In addition, LAB strains were subjected to the antimicrobial susceptibility test based on their minimum inhibitory concentrations (MIC) against five selected antibiotics (Fig. 1), by using Etest® strips, allowing their characterization as resistant (R) or susceptible (S) [20–22]. Breakpoint values for Streptococcus spp. indicated by the Clinical and Laboratory Standards Institute [22] were considered in the analysis.
Molecular characterization of safety
LAB strains were subjected to DNA extraction using the ZR Fungal/Bacterial DNA Kit (Zymo Research, Irvine, CA, USA). After measuring their concentration and quality (NanoDrop 2000, Thermo Scientific Inc., Waltham, MA, USA), DNA samples were subjected to PCR assays to detect the presence of 50 genes related to virulence, biogenic amines, and antibiotic resistance (Fig. 1). The primers and references for PCR conditions are described in Table 1. PCR products were separated on 0.8 to 2.0% (w/v) agarose gels in 0.5× Tris-borate-ethylene diamine tetra acetic acid (TBE) and stained with Tris-acetic acid-EDTA (TAE) buffer that contained GelRed (Biotium Inc., Hayward, CA, USA) at 0.5 μg/mL.
Table 1.
Primers and references for the PCR protocols conducted in the present study to assess the presence of virulence, biogenic amines, and antibiotic-resistance genes in 15 lactic acid bacteria strains with beneficial features obtained from a dairy processing environment [15, 16]. Functions of target genes are presented in Fig. 1
| Target gene | Primers | Reference |
|---|---|---|
| cyt2 | F: ACTCGGGGATTGATAGGC | Vankerckhoven et al. [23] |
| R: GCTGCTAAAGCTGCGCTT | ||
| sprE | F: TTGAGCTCCGTTCCTGCCGAAAGTCATTC | Nakayama et al. [24] |
| R: TTGGTACCGATTGGGGAACCAGATTGACC | ||
| fsrA | F: ATGAGTGAACAAATGGCTATTTA | Nakayama et al. [24] |
| R: CTAAGTAAGAAATAGTGCCTTGA | ||
| fsrB | F: GGGAGCTCTGGACAAAGTATTATCTAACCG | Nakayama et al. [24] |
| R: TTGGTACCCACACCATCACTGACTTTTGC | ||
| fsrC | F: ATGATTTTGTCGTTATTAGCTACT | Nakayama et al. [24] |
| R: CATCGTTAACAACTTTTTTACTG | ||
| gelE | F: TATGACAATGCTTTTTGGGAT | Vankerckhoven et al. [23] |
| R: AGATGCACCCGAAATAATATA | ||
| asa1 | F: GCACGCTATTACGAACTATGA | Vankerckhoven et al. [23] |
| R: TAAGAAAGAACATCACCACGA | ||
| efaA | F: GCCAATTGGGACAGACCCTC | Martin-Platero et al. [25] |
| R: CGCCTTCTGTTCCTTCTTTGGC | ||
| cob | F: AACATTCAGCAAACAAAGC | Eaton and Gasson [26] |
| R: TTGTCATAAAGAGTGGTCAT | ||
| cpd | F: TGGTGGGTTATTTTTCAATTC | Eaton and Gasson [26] |
| R: TACGGCTCTGGCTTACTA | ||
| ccf | F: GGGAATTGAGTAGTGAAGAAG | Eaton and Gasson [26] |
| R: AGCCGCTAAAATCGGTAAAAT | ||
| hyl | F: ACAGAAGAGCTGCAGGAAATG | Vankerckhoven et al. [23] |
| R: GACTGACGTCCAAGTTTCCAA | ||
| esp | F: AGATTTCATCTTTGATTCTTG | Vankerckhoven et al. [23] |
| R: AATTGATTCTTTAGCATCTGG | ||
| mur-2ed | F: AACAGCTTACTTGACTGGACGC | Robredo et al. [27] |
| R: GTATTGGCGCTACTACCCGTATC | ||
| aac(6′)-Ii | F: GCGGTAGCAGCGGTAGACCAAG | Costa et al. [28] |
| R: GCATTTGGTAAGACACCTACG | ||
| mur-2 | F: CGTCAGTACCCTTCTTTTGCAGAGTC | Chu et al. [29] |
| R: GCATTATTACCAGTGTTAGTGGTTG | ||
| ddl | F: ATCAAGTACAGTTAGTCT | Dutka-Malen et al. [30] |
| R: ACGATTCAAAGCTAACTG | ||
| ace | F: GAATTGAGCAAAAGTTCAATCG | Martin-Platero et al. [25] |
| R: GTCTGTCTTTTCACTTGTTTC | ||
| int | F: GCGTGATTGTATCTCACT | Gevers et al. [31] |
| R: GACGCTCCTGTTGCTTCT | ||
| int-Tn | F: TGACACTCTGCCAGCTTTAC | Barbeyrac et al. [32] |
| R: CCATAGGAACTTGACGTTCG | ||
| tdc | F: GAYATNATNGGNATNGGNYTNGAYCARG | Rivas et al. [33] |
| R: CCRTARTCNGGNATAGCRAARTCNGTRTG | ||
| odc | F: GTNTTYAAYGCNGAYAARCANTAYTTYGT | Rivas et al. [33] |
| R: ATNGARTTNAGTTCRCAYTTYTCNGG | ||
| hdc1 | F: AGATGGTATTGTTTCTTATG | Favaro et al. [34] |
| R: AGACCATACACCATAACCTT | ||
| hdc2 | F: AAYTCNTTYGAYTTYGARAARGARG | Favaro et al. [34] |
| R: ATNGGNGANCCDATCATYTTRTGNCC | ||
| aph(2″)-Ib | F: TATGGATCCATGGTTAACTTGGACGCTGAGAT | Kao et al. [35] |
| R: TAAGCTTCCTGCTAAAATATAAACATCTCTGCT | ||
| ant(4′)-Ia | F: CAAACTGCTAAATCGGTAGAAGCC | Vakulenko and Mobashery [36] |
| R: GGAAAGTTGACCAGACATTACGAACT | ||
| aph(2″)-Id | F: GTGGTTTTTACAGGAATGCCATC | Fortina et al. [5] |
| R: CCCTCTTCATACCAATCCATATAACC | ||
| aph(2″)-Ic | F: CCACAATGATAATGACTCAGTTCCC | Fortina et al. [5] |
| R: CCACAGCTTCCGATAGCAAGAG | ||
| aph(3′)-llla | F: GCCGATGTGGATTGCGAAAA | Fortina et al. [5] |
| R: GCTTGATCCCCAGTAAGTCA | ||
| aac(6′)-Ie-aph(2″)-Ia | F: CCAAGAGCAATAAGGGCATA | Van de Klundert and Vliegenthart [37] |
| R: CACTATCATAACCACTACCG | ||
| vanA | F: TCTGCAATAGAGATAGCCGC | Martin-Platero et al. [25] |
| R: GGAGTAGCTATCCCAGCATT | ||
| vanB | F: GCTCCGCAGCCTGCATGGACA | Paulsen et al. [38] |
| R: ACGATGCCGCCATCCTCCTGC | ||
| vanC | F: GGTATCAAGGAAACCTC | Dutka-Malen et al. [30] |
| R: CTTCCGCCATCATAGCT | ||
| vanC1 | F: GCTGAAATATGAAGTAATGACCA | Miele et al. [39] |
| R: CGGCATGGTGTTGATTTCGTT | ||
| vanC2 | F: CTCCTACGATTCTCTTG | Dutka-Malen et al. [30] |
| R: CGAGCAAGACCTTTAAG | ||
| vanC2/C3 | F: CTCCTACGATTCTCTTG | Dutka-Malen et al. [30] |
| R: CGAGCAAGACCTTTAAG | ||
| ermA | F: TCTAAAAAGCATGTAAAAGAA | Sutcliffe et al. [40] |
| R: CTTCGATAGTTTATTAATATTAG | ||
| ermB | F: CATTTAACGACGAAACTGGC | Jensen et al. [41] |
| R: GGAACATCTGTGGTATGGCG | ||
| ermC | F: ATCTTTGAAATCGGCTCAGG | Jensen et al. [41] |
| R: CAAACCCGTATTCCACGATT | ||
| tet(K) | F: TTAGGTGAAGGGTTAGGTCC | Aarestrup et al. [42] |
| R: GCAAACTCATTCCAGAAGCA | ||
| tet(L) | F: CATTTGGTCTTATTGGATCG | Aarestrup et al. [42] |
| R: ATTACACTTCCGATTTCGG | ||
| tet(M) | F: GTTAAATAGTGTTCTTGGAG | Aarestrup et al. [42] |
| R: CTAAGATATGGCTCTAACAA | ||
| tet(O) | F: GATGGCATACAGGCACAGAC | Aarestrup et al. [42] |
| R: CAATATCACCAGAGCAGGCT | ||
| tet(S) | F: TGGAACGCCAGAGAGGTATT | Aarestrup et al. [42] |
| R: ACATAGACAAGCCGTTGACC | ||
| catA | F: GGATATGAAATTTATCCCTC | Aarestrup et al. [42] |
| R: CAATCATCTACCCTATGAAT | ||
| vat(E) | F: ACGTTACCCATCACTATG | Duh et al. [43] |
| R: GCTCCGATAATGGCACCGAC | ||
| ant(6)-Ia | F: ACTGGCTTAATCAATTTGGG | Fortina et al. [5] |
| R: GCCTTTCCGCCACCTCACCG | ||
| bcrB | F: AAAGAAACCGACTGCTGATA | Manson et al. [44] |
| R: GCTTACTTGTATAGCAGAGA | ||
| bcrD | F: AGGATTCGGCCGAATGGCACTTGATTTTAT | Manson et al. [44] |
| R: GTTTCTTCGCGAAATTGCCGTTATAAGTAA | ||
| bcrR | F: AACAAACAGGGAGCGGCCGCATGGAATTTA | Manson et al. [44] |
| R: TGATGTTCGCGATTTCATTCCCATCTGCTT |
Results and discussion
Here, we considered as much as we were able of different safety features usually assessed to characterize beneficial and probiotic strains before use for potential human consumption [45]. Based on the adopted panel (Fig. 1, Table 1), Table 2 summarizes the results for the detected safety features in the 15 LAB strains. None of the investigated strains presented hemolysis, gelatinase or lipase production, and DNase activity in the in vitro tests at either 25 °C or 37 °C. Similarly, there was no in vitro detection of biogenic amines production. All investigated strains showed negative results for the presence of genes related to the production of lysine, histidine, and ornithine biogenic amines (Table 2), as expected for safe cultures [46, 47]. Only some strains presented positive results for tdc (L. casei MSI1, from silage, MRUV6, from cow rumen, and L. acidophilus MVA3, from cow vaginal mucosa), related to production of tyrosine [18, 33].
Table 2.
Phenotypic and genotypic antibiotic resistance and resistance and virulence genes detected by PCR of lactic acid bacteria
| Species | Strain | Phenotypic antibiotic resistance | Resistance and virulence genes detected by PCR | |
|---|---|---|---|---|
| Disk diffusion assay | MIC (μg/mL) | |||
| L. casei | MSI1 | GEN, CLI, ERY, OXA, TRS, RIF | VAN (A), AMP (1.0) | vanC2, bcrB, mur-2ed, tdc |
| MSI5 | GEN, VAN, CLI, OXA, TRS | VAN (A), | vanC2, tet(S), bcrR, cpd | |
| MRUV1 | GEN, VAN, AMP, OXA, TRS, TET | VAN (A), AMP (1.0) | vanA, ant(4′)-Ia, int | |
| MRUV6 | GEN, VAN, OXA, TRS | VAN (A), AMP (1.0) | ermA, ant(4′)-Ia, tdc | |
| L. acidophilus | MVA3 | GEN, VAN, CLI, ERY, OXA, TRS, RIF | VAN (A), AMP (1.0), RIF (A) | tet(K), tet(S), ermA, ant(4′)-Ia, bcrR, asa1, ccf, tdc |
| L. nagelli | MSIV4 | GEN, CLI, OXA, TRS, RIF | VAN (A), AMP (50.0) | ccf, int |
| L. harbinensis | MSI3 | GEN, VAN, CLI, OXA, TRS, TET | GEN (A), VAN (A), AMP (1.0), CHL (A) | asa1 |
| MSIV2 | VAN, CLI, OXA, TRS | VAN (A), RIF (32.0) | vanC2, cpd | |
| L. fermentum | SIVGL1 | GEN, VAN, AMP, OXA, TRS | VAN (A), AMP (1.5) | -- |
| L. plantarum | MLE5 | GEN, CLI, ERY, OXA, TRS, RIF | VAN (A), AMP (0.64) | vanC1, aph(3′)-IIIa |
| MSI2 | GEN, VAN, CLI, OXA, TRS, CHL, RIF | VAN (A) | -- | |
| P. pentosaceus | MLEV8 | GEN, CLI, ERY, OXA, TRS, RIF | VAN (A), AMP (50) | ermB, aac(6′)-Ie-aph(2″)-Ia |
| P. acidilactici | MSI7 | GEN, VAN, OXA, TRS, TET | VAN (A) | tet(K), int |
| W. paramesenteroides | MRUV3 | GEN, VAN, CLI, OXA, TRS, TET, RIF | VAN (A), AMP (50), CHL (50.0), RIF (4.0) | vanA, ant(4′)-Ia, int |
| MSAV5 | GEN, CLI, ERY, OXA, TRS, RIF | -- | mur-2ed, cpd, hyl, int | |
GEN gentamicin, IMI imipenem, VAN vancomycin, CLI clindamycin, ERY erythromycin, AMP ampicillin, OXA oxacillin, PEN penicillin, TRS trimethoprim/sulfamethoxazole, TET tetracycline, CHL chloramphenicol, RIF rifampicin, STG streptogramin, STM streptomycin, BAC bacitracin, A absence of inhibition zone
Breaking points have been determined according to manufacturer instructions (bioMérieux, France), EUCAST, and CLSI standard [20–22]
The absence of hemolytic activity and biogenic amines production in Lactobacillus was already reported and it is to be expected. Indeed, this characteristic is essential for beneficial and probiotic candidate strains [45, 48, 49]. However, some Lactobacillus spp. are described as biogenic amine producers, and they may be present in microbiota related to a dairy environment, perhaps introduced through contamination at some steps in the dairy production processes [46]. The production of extracellular enzymes and hemolytic activity are not exhibited by Pediococcus spp., as shown by Borges et al. [50]. To the best of our knowledge, there is no study about hemolytic activity or extracellular enzymes for W. paramesenteroides. Jeong and Lee [51] described a W. paramesenteroides strain that produces histamine and tyramine. Only two strains, L. fermentum SIVGL1 (silage) and L. plantarum MSI2 (silage), did not present any positive result for any of the assayed virulence related genes; L. harbinensis MSI3 (silage) presented only positive result for asa1. Asa1 is responsible for aggregation substance capacity, and this protein increases bacterial adherence to renal tubular cells and heart endocardial cells, enhances internalization in intestinal epithelial cells, and increases the valvular vegetation mass in an animal model of endocarditis [23]. However, Asa1 can be an important adhering feature to help the persistence of a beneficial strain in the GIT [15]. The presence of Enterococcus spp.–related genes indicates the ability of the strains in acquiring mobile elements from other bacteria, what can explain the presence of cpd, ccf, int, and hyl in some of the strains (Table 2), and considered a special concern related to antibiotic-resistance spread in GIT [26].
Most of the evaluated strains were susceptible to the majority of the tested antibiotics. Based on the results obtained by the disk diffusion, all strains were resistant to oxacillin and sulfa/trimethoprim, and just one strain (L. harbinensis MSIV2, silage) was susceptible to gentamicin (all other 14 strains were resistant). Most of the strains were also resistant to clindamycin, vancomycin, and rifampicin (Table 2). No strain was resistant to more than 7 of the 12 evaluated antibiotics. L. acidophilus MVA3 (cow vaginal mucosa), L. plantarum MSI2 (silage), and W. paramesenteroides MRUV3 (cow rumen) were the only three strains that exhibited more variable resistance to the tested antibiotics; they showed resistance to seven antibiotics. As strains obtained from similar samples did not show equivalent resistance patterns (Fig. 1, Table 2), apparently, the origin of the strains is not related to this feature, as indicated by Duar et al. [52]. The results obtained in this study are in accordance with the results obtained by other authors who investigated antibiotic resistance in beneficial LAB [10, 53]. Based on MIC, L. harbinensis MSI3 and W. paramesenteroides MRUV3 were the only two strains that presented high levels of resistance, to four antibiotics and susceptible only to only rifampicin and gentamicin, respectively (Table 2). Ampicillin and vancomycin were the two antibiotics to which most strains showed resistance, a potential concern as vancomycin is considered a key indicator in the evaluation of LAB safety [10, 45]. However, several reports demonstrate that LAB resistance to vancomycin can be intrinsic, and more dedicated studies need to be performed in order to draw conclusions about safety related to sensitivity to this specific antibiotic. Gentamicin, chloramphenicol, and rifampicin were the antibiotics to which the 15 evaluated LAB strains were most susceptible (Table 2). The results obtained in this study agree with those reported by other authors related to the susceptibility of the lactobacilli strains to the selected antibiotics [10, 12]. In addition to the genus Lactobacillus, Munoz-Atienza et al. [54] described antibiotic resistance in the Weissella and Pediococcus genera. Although lactobacilli and other LAB are considered safe by several regulatory agencies, they should still be characterized on a strain-by-strain basis as safe in order to be applied as probiotic cultures [55]. Beneficial bacteria are considered alternatives for antibiotic therapy, once the spread of antibiotic resistance is a worldwide concern in a context of One Health [56], leading to a careful analysis of potential probiotic candidates as resistant or resistance-related gene carriers [57]. As many of the probiotic carriers are fermented foods, it is also important to verify the potential transfer of resistance genetic elements to starter cultures, what would spread this feature in a food industry and be potentially transferred to pathogenic and spoilage bacteria, jeopardizing the safety and quality of end products [58].
Based on the One Health approach, beneficial and probiotic strains must be checked for the presence of antibiotic-resistance genes, independently of presenting or not phenotypical resistance [57]. Based on the obtained results (Table 2), it was possible to characterize a diversity pattern of antibiotic-related genes in the assessed strains, and many of them can be located in plasmids, facilitating their transference to other bacteria [26]. Considering the molecular panel included in our study (Fig. 1), the most frequent antibiotic-resistance genes were related to vancomycin and gentamicin, as observed by the disk diffusion assay and MIC (Table 2). However, direct correspondence between phenotypical and molecular results for antibiotic resistance was not observed; this lack of correspondence can be considered expected in some situations, as the presence of a genetic element related to resistance is not necessarily an indicative of expression and thus resistance [59]. Bacteria usually adopt a number of resistance mechanisms for different antibiotics, resulting in alternative pathways to express this feature; because of this, it is currently well accepted the need for a wide approach, based on phenotypical and molecular assays, to characterize the antibiotic resistance in bacteria, mainly when they are supposed to be included in foods for human consumption, as considered for beneficial and probiotic strains [13, 57, 59].
None of the 15 tested strains were positive for the following genes: vanB, vanC-1, vanC2/C3, tet(L), tet(M), tet(O), int-Tn, ermC, catA, aph(2″)-Ib, aph(2″)-Id, aph(2″)-Ic, aph(3′)-IIIa, vat(E), bcrD, ant(6)-la, mur-2, ddl, ace, cyt2, esp, efaA, cob, sprE, fsrA, fsrB, fsrC, gelE, odc, hdc1, and hdc2. The results obtained in this study agree with those obtained by other authors who investigated LAB probiotic candidates [10, 53, 54] and highlight the safety profiles of the selected strains.
Based on the obtained results, the selected LAB strains can be considered relatively safe to be used as beneficial cultures in the food industry. None strain presented phenotypical production of the assayed virulence features and biogenic amines, and resistance was limited to a few antibiotics. In addition, only a few genetic markers related to virulence, biogenic amines, and antibiotic resistance were detected. Based on the characterized beneficial features [15, 16], and the safety profile identified in this study, the LAB strains can be considered candidates for specific studies to characterize their probiotic potential and industrial use.
Funding information
This study was supported by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, Brasília, DF, Brazil—financial code 001), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Brasília, DF, Brazil), and Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG, Belo Horizonte, MG, Brazil).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Highlights
• The safety aspects of LAB isolated from a dairy environment was evaluated.
• Presence of 49 virulence factors and antibiotic resistance genes was studied.
• Physiological expression of virulence factors, biogenic amine, and antibiotic resistance was tested.
Note
This manuscript was organized based on results obtained by the first author during her Doctorate training, and fully described in her thesis [16].
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Luis Augusto Nero, Email: nero@ufv.br.
Svetoslav Dimitrov Todorov, Email: slavi310570@abv.bg.
References
- 1.Leroy F, De Vuyst L. Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci Technol. 2004;15(2):67–78. doi: 10.1016/j.tifs.2003.09.004. [DOI] [Google Scholar]
- 2.Kechagia M, Basoulis D, Konstantopoulou S, Dimitriadi D, Gyftopoulou K, Skarmoutsou N, Fakiri EM. Health benefits of probiotics: a review. ISRN Nut. 2013;2013:481651. doi: 10.5402/2013/481651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fontana L, Bermudez-Brito M, Plaza-Diaz J, Munoz-Quezada S, Gil A. Sources, isolation, characterisation and evaluation of probiotics. Br J Nutr. 2013;109(Suppl 2):S35–S50. doi: 10.1017/S0007114512004011. [DOI] [PubMed] [Google Scholar]
- 4.FAO/WHO (2001) Expert consultation on evaluation of health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria, Córdoba
- 5.Fortina MG, Ricci G, Borgo F, Manachini PL, Arends K, Schiwon K, Abajy MY, Grohmann E. A survey on biotechnological potential and safety of the novel Enterococcus species of dairy origin E italicus. Int J Food Microbiol. 2008;123(3):204–211. doi: 10.1016/j.ijfoodmicro.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 6.Colombo M, Todorov SD, Eller M, Nero LA. The potential use of probiotic and beneficial bacteria in the Brazilian dairy industry. J Dairy Res. 2018;85(4):487–496. doi: 10.1017/S0022029918000845. [DOI] [PubMed] [Google Scholar]
- 7.FAO/WHO . Health and nutritional properties and guidelines for evaluation. Rome: FAO Food and Nutrition; 2006. [Google Scholar]
- 8.Wilson JW, Schurr MJ, LeBlanc CL, Ramamurthy R, Buchanan KL, Nickerson CA. Mechanisms of bacterial pathogenicity. Postgrad Med J. 2002;78(918):216–224. doi: 10.1136/pmj.78.918.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bautista-Gallego J, Arroyo-López FN, Rantsiou K, Jiménez-Díaz R, Garrido-Fernández A, Cocolin L. Screening of lactic acid bacteria isolated from fermented table olives with probiotic potential. Food Res Int. 2013;50(1):135–142. doi: 10.1016/j.foodres.2012.10.004. [DOI] [Google Scholar]
- 10.Casado Muñoz MC, Benomar N, Lerma LL, Gálvez A, Abriouel H. Antibiotic resistance of Lactobacillus pentosus and Leuconostoc pseudomesenteroides isolated from naturally-fermented Aloreña table olives throughout fermentation process. Int J Food Microbiol. 2014;172:110–118. doi: 10.1016/j.ijfoodmicro.2013.11.025. [DOI] [PubMed] [Google Scholar]
- 11.van Reenen CA, Dicks LMT. Horizontal gene transfer amongst probiotic lactic acid bacteria and other intestinal microbiota: what are the possibilities? A review. Arch Microbiol. 2011;193(3):157–168. doi: 10.1007/s00203-010-0668-3. [DOI] [PubMed] [Google Scholar]
- 12.Rubio R, Jofré A, Martín B, Aymerich T, Garriga M. Characterization of lactic acid bacteria isolated from infant faeces as potential probiotic starter cultures for fermented sausages. Food Microbiol. 2014;38:303–311. doi: 10.1016/j.fm.2013.07.015. [DOI] [PubMed] [Google Scholar]
- 13.Sharma P, Tomar SK, Goswami P, Sangwan V, Singh R. Antibiotic resistance among commercially available probiotics. Food Res Int. 2014;57:176–195. doi: 10.1016/j.foodres.2014.01.025. [DOI] [Google Scholar]
- 14.Snydman DR. The safety of probiotics. Clin Infect Dis. 2008;46(Supp 2):S104–S111. doi: 10.1086/523331. [DOI] [PubMed] [Google Scholar]
- 15.Colombo M, Castilho NPA, Todorov SD, Nero LA. Beneficial properties of lactic acid bacteria naturally present in dairy production. BMC Microbiol. 2018;18(1):219. doi: 10.1186/s12866-018-1356-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colombo M. Beneficial properties and safety of lactic acid bacteria isolated from the dairy production environment. Viçosa: Universidade Federal de Viçosa; 2017. [Google Scholar]
- 17.Barbosa J, Gibbs PA, Teixeira P. Virulence factors among enterococci isolated from traditional fermented meat products produced in the North of Portugal. Food Control. 2010;21(5):651–656. doi: 10.1016/j.foodcont.2009.10.002. [DOI] [Google Scholar]
- 18.Bover-Cid S, Holzapfel WH. Improved screening procedure for biogenic amine production by lactic acid bacteria. Int J Food Microbiol. 1999;53(1):33–41. doi: 10.1016/S0168-1605(99)00152-X. [DOI] [PubMed] [Google Scholar]
- 19.Joosten HMLJ, Northolt MD. Detection, growth, and amine-producing capacity of lactobacilli in cheese. Appl Environ Microbiol. 1989;55(9):2356–2359. doi: 10.1128/AEM.55.9.2356-2359.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.EUCAST (2019) Antimicrobial susceptibility testing-EUCAST disk diffusion method. EUCAST
- 21.EUCAST (2019) Clinical breakpoints and dosing of antibiotics
- 22.CLSI (2017) Performance standards for antimicrobial susceptibility testing, 27th edn. CLSI supplement M100, Clinical and Laboratory Standards Institute
- 23.Vankerckhoven V, Van Autgaerden T, Vael C, Lammens C, Chapelle S, Rossi R, Jabes D, Goossens H. Development of a multiplex PCR for the detection of asa1, gelE, cylA, esp, and hyl genes in enterococci and survey for virulence determinants among European hospital isolates of Enterococcus faecium. J Clin Microbiol. 2004;42(10):4473–4479. doi: 10.1128/JCM.42.10.4473-4479.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakayama J, Kariyama R, Kumon H. Description of a 23.9-kilobase chromosomal deletion containing a region encoding fsr genes which mainly determines the gelatinase-negative phenotype of clinical isolates of Enterococcus faecalis in urine. Appl Environ Microbiol. 2002;68(6):3152–3155. doi: 10.1128/AEM.68.6.3152-3155.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin-Platero AM, Valdivia E, Maqueda M, Martinez-Bueno M. Characterization and safety evaluation of enterococci isolated from Spanish goats’ milk cheeses. Int J Food Microbiol. 2009;132(1):24–32. doi: 10.1016/j.ijfoodmicro.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 26.Eaton TJ, Gasson MJ. Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl Environ Microbiol. 2001;67(4):1628–1635. doi: 10.1128/AEM.67.4.1628-1635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robredo B, Singh KV, Baquero F, Murray BE, Torres C. From vanA Enterococcus hirae to vanA Enterococcus faecium: a study of feed supplementation with avoparcin and tylosin in young chickens. Antimicrob Agents Chemother. 1999;43(5):1137–1143. doi: 10.1128/AAC.43.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Costa Y, Galimand M, Leclercq R, Duval J, Courvalin P. Characterization of the chromosomal aac(6′)-Ii gene specific for Enterococcus faecium. Antimicrob Agents Chemother. 1993;37(9):1896–1903. doi: 10.1128/aac.37.9.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chu C, Kariyama R, Daneo-Moore L, Shockman GD. Cloning and sequence analysis of the muramidase-2 gene from Enterococcus hirae. J Bacteriol. 1992;174(5):1619–1625. doi: 10.1128/JB.174.5.1619-1625.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995;33(1):24–27. doi: 10.1128/JCM.33.1.24-27.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gevers D, Danielsen M, Huys G, Swings J. Molecular characterization of tet(M) genes in Lactobacillus isolates from different types of fermented dry sausage. Appl Environ Microbiol. 2003;69(2):1270–1275. doi: 10.1128/AEM.69.2.1270-1275.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barbeyrac B, Dupon M, Rodriguez P, Renaudin H, Bébéar C. A Tn1545-like transposon carries the tet(M) gene in tetracycline resistant strains of Bacteroides ureolyticus as well as Ureaplasma urealyticum but not Neisseria gonorrhoeae. J Antimicrob Chemother. 1996;37(2):223–232. doi: 10.1093/jac/37.2.223. [DOI] [PubMed] [Google Scholar]
- 33.Rivas B, Marcobal A, Muñoz R. Improved multiplex-PCR method for the simultaneous detection of food bacteria producing biogenic amines. FEMS Microbiol Lett. 2005;244(2):367–372. doi: 10.1016/j.femsle.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 34.Favaro L, Basaglia M, Casella S, Hue I, Dousset X, Franco BDGM, Todorov SD. Bacteriocinogenic potential and safety evaluation of non-starter Enterococcus faecium strains isolated from home made white brine cheese. Food Microbiol. 2014;38:228–239. doi: 10.1016/j.fm.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 35.Kao SJ, Il Y, Clewell DB, Donabedian SM, Zervos MJ, Petrin J, Shaw KJ, Chow JW. Detection of the high-level aminoglycoside resistance gene aph(2″)-Ib in Enterococcus faecium. Antimicrob Agents Chemother. 2000;44(10):2876–2879. doi: 10.1128/AAC.44.10.2876-2879.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vakulenko SB, Mobashery S. Versatility of aminoglycosides and prospects for their future. Clin Microbiol Rev. 2003;16(3):430–450. doi: 10.1128/CMR.16.3.430-450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van de Klundert JAM, Vliegenthart JS. PCR detection of genes coding for aminoglycoside-modifying enzymes. In: Persing D, Smith T, Tenover T, White T, editors. Diagnostic molecular microbiology: principles and applications. Washington: American Society for Microbiology; 1993. pp. 547–552. [Google Scholar]
- 38.Paulsen IT, Banerjei L, Myers GSA, Nelson KE, Seshadri R, Read TD, Fouts DE, Eisen JA, Gill SR, Heidelberg JF. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science. 2003;299(5615):2071–2074. doi: 10.1126/science.1080613. [DOI] [PubMed] [Google Scholar]
- 39.Miele A, Bandera M, Goldstein BP. Use of primers selective for vancomycin resistance genes to determine van genotype in enterococci and to study gene organization in VanA isolates. Antimicrob Agents Chemother. 1995;39(8):1772–1778. doi: 10.1128/AAC.39.8.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sutcliffe J, Grebe T, Tait-Kamradt A, Wondrack L. Detection of erythromycin-resistant determinants by PCR. Antimicrob Agents Chemother. 1996;40(11):2562–2566. doi: 10.1128/AAC.40.11.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jensen LB, Frimodt-Moller N, Aarestrup FM. Presence of erm gene classes in Gram-positive bacteria of animal and human origin in Denmark. FEMS Microbiol Lett. 1999;170(1):151–158. doi: 10.1111/j.1574-6968.1999.tb13368.x. [DOI] [PubMed] [Google Scholar]
- 42.Aarestrup FM, Agerso Y, Gerner-Smidt P, Madsen M, Jensen LB. Comparison of antimicrobial resistance phenotypes and resistance genes in Enterococcus faecalis and Enterococcus faecium from humans in the community, broilers, and pigs in Denmark. Diagn Microbiol Infect Dis. 2000;37(2):127–137. doi: 10.1016/S0732-8893(00)00130-9. [DOI] [PubMed] [Google Scholar]
- 43.Duh R, Singh KV, Malathum K, Murray BE. In vitro activity of 19 antimicrobial agents against enterococci from healthy subjects and hospitalized patients and use of an ace gene probe from Enterococcus faecalis for species identification. Microb Drug Resist. 2001;7(1):39–46. doi: 10.1089/107662901750152765. [DOI] [PubMed] [Google Scholar]
- 44.Manson JM, Keis S, Smith JMB, Cook GM. Acquired bacitracin resistance in Enterococcus faecalis is mediated by an ABC transporter and a novel regulatory protein, BcrR. Antimicrob Agents Chemother. 2004;48(10):3743–3748. doi: 10.1128/AAC.48.10.3743-3748.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adams MR. Safety of industrial lactic acid bacteria. J Biotechnol. 1999;68(2):171–178. doi: 10.1016/S0168-1656(98)00198-9. [DOI] [PubMed] [Google Scholar]
- 46.Papageorgiou M, Lambropoulou D, Morrison C, Kłodzińska E, Namieśnik J, Płotka-Wasylka J. Literature update of analytical methods for biogenic amines determination in food and beverages. Trends Anal Chem. 2018;98:128–142. doi: 10.1016/j.trac.2017.11.001. [DOI] [Google Scholar]
- 47.Vankerckhoven V, Huys G, Vancanneyt M, Vael C, Klare I, Romond M, Entenza JM, Moreillon P, Wind RD, Knol J, Wiertz E, Pot B, Vaughan EE, Kahlmeter G, Goossens H. Biosafety assessment of probiotics used for human consumption: recommendations from the EU-PROSAFE project. Trends Food Sci Technol. 2008;19(2):102–114. doi: 10.1016/j.tifs.2007.07.013. [DOI] [Google Scholar]
- 48.Casarotti SN, Carneiro BM, Todorov SD, Nero LA, Rahal P, Penna ALB. In vitro assessment of safety and probiotic potential characteristics of Lactobacillus strains isolated from water buffalo mozzarella cheese. Ann Microbiol. 2017;67(4):289–301. doi: 10.1007/s13213-017-1258-2. [DOI] [Google Scholar]
- 49.Pisano MB, Viale S, Conti S, Fadda ME, Deplano M, Melis MP, Deiana M, Cosentino S. Preliminary evaluation of probiotic properties of Lactobacillus strains isolated from sardinian dairy products. Biomed Res Int. 2014;2014:9. doi: 10.1155/2014/286390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Borges S, Barbosa J, Silva J, Teixeira P. Evaluation of characteristics of Pediococcus spp. to be used as a vaginal probiotic. J Appl Microbiol. 2013;115(2):527–538. doi: 10.1111/jam.12232. [DOI] [PubMed] [Google Scholar]
- 51.Jeong D, Lee J. Antibiotic resistance, hemolysis and biogenic amine production assessments of Leuconostoc and Weissella isolates for kimchi starter development. LWT - Food Sci Technol. 2015;64(2):1078–1084. doi: 10.1016/j.lwt.2015.07.031. [DOI] [Google Scholar]
- 52.Duar RM, Lin XB, Zheng J, Martino ME, Grenier T, Pérez-Muñoz ME, Leulier F, Gänzle M, Walter J. Lifestyles in transition: evolution and natural history of the genus Lactobacillus. FEMS Microbiol Rev. 2017;41(Supp_1):S27–S48. doi: 10.1093/femsre/fux030. [DOI] [PubMed] [Google Scholar]
- 53.Santos KMO, Vieira ADS, Salles HO, Oliveira JS, Rocha CRC, Borges MF, Bruno LM, Franco BDGM, Todorov SD. Safety, beneficial and technological properties of Enterococcus faecium isolated from Brazilian cheeses. Braz J Microbiol. 2015;46(1):237–249. doi: 10.1590/s1517-838246120131245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Munoz-Atienza E, Gomez-Sala B, Araujo C, Campanero C, del Campo R, Hernandez P, Herranz C, Cintas L. Antimicrobial activity, antibiotic susceptibility and virulence factors of lactic acid bacteria of aquatic origin intended for use as probiotics in aquaculture. BMC Microbiol. 2013;13(1):15. doi: 10.1186/1471-2180-13-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morovic W, Roper JM, Smith AB, Mukerji P, Stahl B, Rae JC, Ouwehand AC. Safety evaluation of HOWARU® Restore (Lactobacillus acidophilus NCFM, Lactobacillus paracasei Lpc-37, Bifidobacterium animalis subsp. lactis Bl-04 and B. lactis Bi-07) for antibiotic resistance, genomic risk factors, and acute toxicity. Food Chem Toxicol. 2017;110:316–324. doi: 10.1016/j.fct.2017.10.037. [DOI] [PubMed] [Google Scholar]
- 56.Imperial ICVJ, Ibana JA. Addressing the antibiotic resistance problem with probiotics: reducing the risk of its double-edged sword effect. Front Microbiol. 2016;7:1983–1983. doi: 10.3389/fmicb.2016.01983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Courvalin P. Antibiotic resistance: the pros and cons of probiotics. Dig Liver Dis. 2006;38:S261–S265. doi: 10.1016/S1590-8658(07)60006-1. [DOI] [PubMed] [Google Scholar]
- 58.Verraes C, Van Boxstael S, Van Meervenne E, Van Coillie E, Butaye P, Catry B, de Schaetzen M-A, Van Huffel X, Imberechts H, Dierick K, Daube G, Saegerman C, De Block J, Dewulf J, Herman L. Antimicrobial resistance in the food chain: a review. Int J Environ Res Public Health. 2013;10(7):2643–2669. doi: 10.3390/ijerph10072643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Corona F, Martinez JL. Phenotypic resistance to antibiotics. Antibiotics (Basel) 2013;2(2):237–255. doi: 10.3390/antibiotics2020237. [DOI] [PMC free article] [PubMed] [Google Scholar]