Abstract
Salmonella Dublin is a strongly adapted serovar that causes enteritis and/or systemic disease with high rates of mortality in cattle and occasionally infects humans. Despite the importance of this serovar, there is a lack of studies in Brazil. The aim of this study was to characterize the genetic diversity of 112 S. Dublin strains isolated from humans and animals in Brazil by CRISPR and CRISPR-MVLST and the relatedness among strains by MLST. In addition, the frequency of some important virulence genes was verified. The strains studied belonged to nine different sequence types, being all of them single- or double-locus variants of the ST10. CRISPR discriminated the strains into 69 subtypes with a similarity ≥ 84.4% and CRISPR-MVLST into 72 subtypes with a similarity ≥ 84.7%. The virulence genes ratB, lpfA, mgtC, avrA, sopB, sopE2, sifA, sseA, ssrA, csgA, fliC, and sinH were found in all the strains studied, while spvB, spvC, sodCl, rpoS, sipA, sipD, invA, and hilA were detected in ≥ 93.7% of the strains. In conclusion, the high similarity among the strains reinforces the clonal nature of the strains of this serovar that may have descended from a common ancestor that little differed over 33 years in Brazil. CRISPR and CRISPR-MVLST showed to be good alternatives to type S. Dublin strains. MLST suggested that S. Dublin strains from Brazil were phylogenetically related to strains from other parts of the globe. Moreover, the high frequency of virulence genes among the strains studied reinforces the capacity of S. Dublin to cause invasive diseases.
Electronic supplementary material
The online version of this article (10.1007/s42770-019-00156-5) contains supplementary material, which is available to authorized users.
Keywords: Salmonella Dublin, MLST, CRISPR, CRISPR-MVLST, Virulence genes
Introduction
Salmonellosis caused by non-typhoidal serovars is among the most common foodborne illnesses worldwide, accounting for 93.8 million cases of gastroenteritis and 155,000 deaths annually [1].
Salmonella enterica serovar Dublin (S. Dublin) is strongly adapted to cattle, responsible for causing enteritis and/or systemic disease with high rates of mortality. This fact becomes even more concerning due to the negative economic impact for many beef-producing countries, since S. Dublin infections in cattle may result in reduced milk production, abortion in pregnant cows, and eventually in deaths [2–4].
Occasionally, S. Dublin can also be isolated from serious and even fatal infections in humans, especially in patients with underlying immunosuppression conditions, and usually causing a serious disease that can even be indistinguishable from typhoid fever [2–4].
Similarly to most Salmonella serovars, the pathogenesis of S. Dublin strains is achieved, among others, by proteins coded by chromosomal genes responsible for multiple cellular functions as adhesion, acid and serum resistance, invasion of host cells, and survival within phagocytic cells. Such genes are mostly located in the Salmonella pathogenicity islands (SPIs), in special the most studied ones, SPI-1 and SPI-2. In addition, virulence plasmids such as the pSDL, that carries the spv operon, also play important roles in survival and growth of Salmonella Dublin into macrophages [5–9].
Some methodologies such as pulsed-field gel electrophoresis (PFGE), multiple-locus variable-number of tandem repeats analysis (MLVA), enterobacterial repetitive intergenic consensus PCR (ERIC-PCR), multilocus enzyme electrophoresis (MLEE), multilocus sequence typing (MLST), and clustered regularly interspaced short palindromic repeats (CRISPR) have been successfully used to subtype strains of many Salmonella serovars, including S. Dublin [9–13]. Although the great applicability, some methodologies, such as MLST and CRISPR, require excessive laboratory work and high costs that restricts the performing of these methodologies to small and representative sets of strains [14–17]. However, the evolution and costs reductions in whole-genome sequencing (WGS) have been providing a wider access to sequence larger sets of bacterial strains and consequently have been allowing different molecular analysis to be performed in a faster and easier way than the traditional typing [18, 19].
MLST is a method based on the analysis of a particular set of housekeeping genes for each bacterial species. The analysis of the alleles of seven housekeeping genes can be submitted to an online public database (https://enterobase.warwick.ac.uk/) that assigns the strains according to the specific alleles to a specific sequence type (ST), which allows the comparison of strains isolated in different parts of the globe [15]. This methodology has successfully contributed to the understanding of the epidemiology, evolution, and genotypic diversity of many Salmonella serovars and it has even been proposed as an alternative for traditional identification by serotyping [15, 19, 20].
CRISPR are short and highly conserved sequences of DNA direct repeats, which range from 21 to 48 base pairs (bp), usually specific for a determined CRISPR locus. These sequences are regularly interspaced by variable DNA sequences of constant and similar length, usually 20–58 bp, called spacers, which vary according to the species of the microorganisms or the CRISPR locus [21, 22]. Among Salmonella serovars, two non-coding CRISPR loci were identified in their genomes, and the analysis of the different spacers contained in their respective CRISPRs loci has been successfully used to subtype these serovars [18, 23–25].
Aiming to increase the discriminatory power of CRISPR technique, Liu et al. (2011) proposed the association of the two Salmonella CRISPR loci with the virulence genes fimH, responsible for bacterial binding to structures in the cell-host membrane, and sseL, responsible for inducing inflammation and killing macrophages [28]. This association originated the method known as CRISPR-multi-locus virulence sequence typing (CRISPR-MVLST) [26–28]. This methodology has also been successfully used to subtype serovars as Enteritidis, Newport, and Typhimurium [18, 23–25].
Few information is available about the molecular epidemiology of S. Dublin strains isolated worldwide, and most of the studies did not study sets of strains exclusively of this serovar, interfering on the understanding of the specific characteristics and traits of S. Dublin [9–12, 29]. Specifically in Brazil, only five studies molecularly typed strains of this serovar, among which, only two analyzed a large set exclusive of S. Dublin strains, making it difficult to evaluate the diversity of strains of this serovar circulating in this country [13, 30–33]. Furthermore, MLST, CRISPR, and CRISPR-MVLST have never been used for typing strains of this serovar in Brazil, according to the published literature.
Therefore, the aim of this study was to genotype S. Dublin strains isolated from humans and animals in Brazil between 1983 and 2016 by MLST and CRISPR and its variation CRISPR-MVLST. Moreover, the ability of CRISPR-based methodologies in subtyping S. Dublin strains was analyzed in addition to the pathogenic potential of these strains that was determined by searching for the frequency of 20 genes related to Salmonella virulence.
Material and methods
Bacterial strains
A total of 112 Salmonella Dublin strains isolated in Brazil from humans (82) between 1983 and 2016, and animals (30) between 1992 and 2015 were studied. These strains were previously described in Vilela et al. 2018 [13] and are representative isolates of the years, states, material, and source of isolation of the collections of two Salmonella reference laboratories in Brazil, the Adolfo Lutz Institute of São Paulo (IAL-SP), and Oswaldo Cruz Foundation of Rio de Janeiro (FIOCRUZ-RJ). Supporting Information Table S1 presents the year, source, and states of isolation of the 112 S. Dublin strains studied.
MLST
MLST was performed in silico for all the S. Dublin studied using the 112 whole-genome assembled sequences, previously obtained and described in Campioni et al. (2018) [34], following the Achtman scheme available at the Enterobase database (http://enterobase.warwick.ac.uk/species/senterica/allele_st_search) using the allele identification of seven specific housekeeping genes for S. enterica (aroC, dnaN, hemD, hisD, purE, sucA, and thrA). Allele identification was performed by uploading the assembled sequences of S. Dublin strains studied in the MLST web-based tool available in the Center for Genomic Epidemiology website (https://cge.cbs.dtu.dk/services/MLST/). The minimum spanning tree generated with STs and eBURST groups was generated with the software eBURSTv3.
CRISPR and fimH and sseL virulence genes analyses
The CRISPRs loci and the virulence genes fimH and sseL analysis were also performed for all the S. Dublin studied using the 112 whole-genome assembled sequences.
Analysis of CRISPR1 and CRISPR2 was performed, uploading the assembled sequences in the CRISPRFinder tool (available at http://crispr.i2bc.paris-saclay.fr/). This tool automatically analyzes the CRISPRs, as well as the length and location per contig of the direct repeats and spacers in each genome. To perform the analysis, only spacers were considered, as reported in previous studies [23–25, 28]. These spacers were manually listed in each of the S. Dublin strains studied, and a binary matrix with the presence or absence of every spacer in CRISPR1 and CRISPR2 was generated using Microsoft Excel.
The analysis of the sequences of the virulence genes fimH and sseL was performed using Basic Local Alignment Search Tool (BLAST) (available at blast.ncbi.nlm.nih.gov/Blast.cgi) by uploading the assembled sequences of all S. Dublin strains studied and aligning with sequences of the fimH and sseL genes, of 1005 bp and 954 bp, respectively, downloaded from GenBank (available at ncbi.nlm.gov/genbank/) as standard (accession numbers KF465864 and KJ095841 for the genes sseL and fimH, respectively). The sequences were analyzed using ChromasPro 2.33 (Technelysium Pty. Ltd.).
CRISPR-MVLST sequence types (CM-ST) were assigned based on the combination of CRISPR1 and CRISPR2 profiles and fimH and sseL alleles for all strains studied. A similarity dendrogram was generated based on the CRISPRs binary matrices and another one was generated based on CRISPRs binary matrices in addition to the allele types of fimH and sseL genes. The software BioNumerics 7.6 (Applied Maths) with the Unweighted Pair Group Method with Arithmetic Mean (UPGMA) algorithm was used to build both dendrograms. The discriminatory power of both CRISPR and CRISPR-MVLST was assessed by Simpson’s diversity index, as described by Hunter and Gaston (1988) [35].
Virulence gene detection
All the S. Dublin strains studied were tested for the presence of 21 virulence genes using the MyDbFinder tool, a web-based tool available in the Center for Genomic Epidemiology website (https://cge.cbs.dtu.dk/services/MyDbFinder/). Briefly, specific sequences of genes ratB, sodCl, lpfA, rpoS, mgtC, sipA, sipD, invA, avrA, hilA, sopB, sopE2, sifA, sseA, ssrA, spvB, spvC, csgA, fljB, fliC, and sinH were downloaded from GenBank (https://www.ncbi.nlm.nih.gov/genbank/) and combined in a single fasta format file to create a personalized database. This database was uploaded to MyDbFinder tool to perform the alignment between the assembled sequences of the S. Dublin strains studied with the sequences of the searched virulence genes and detecting if they were present or not among the strains studied. The parameters used were 90% of minimum identity and 60% of minimum length. The 21 virulence genes were searched; its respective functions and accession numbers are presented in Table 1.
Table 1.
Virulence genes searched in the 112 S. Dublin strains studied and its respective functions and accession numbers (GenBank)
| Virulence gene | Function | Accession number |
|---|---|---|
| ratB | Putative outer membrane protein | NP_461449 |
| sodCl | Gifsy-2 prophage: superoxide dismutase precursor (Cu-Zn) | NP_460019 |
| lpfA | Long polar fimbria protein LpfA | NP_462541 |
| rpoS | Sigma S (sigma 38) factor of RNA polymerase, the major sigma factor during stationary phase | NP_461845 |
| mgtC | Mg2+ transport protein | NP_462663 |
| sipA | Type III secretion system effector SipA, actin polymerizing activity | NP_461803 |
| sipD | Type III secretion system hydrophilic translocator, needle tip protein SipD | NP_461804 |
| invA | Type III secretion system major export apparatus protein InvA | NP_461817 |
| avrA | Putative inner membrane protein | NP_461786 |
| hilA | Invasion protein transcriptional activator | NP_461797 |
| sopB | Invasion gene D protein | NP_460064 |
| sopE2 | Type III secretion protein SopE2 | NP_460811 |
| sifA | Replication in macrophages; SIFA protein | NP_460194 |
| sseA | Chaperone for sseB and sseD | NP_460362 |
| ssrA | Hybrid sensor histidine kinase/response regulator | NP_460357 |
| spvB | Type III secretion system effector SpvB, ADP-ribosylation activity | NP_490529 |
| spvC | Type III secretion system effector SpvC, phosphothreonine lyase | NP_490528 |
| csgA | Curlin major subunit CsgA | NP_460115 |
| fljB | Phase 2 flagellin; flagellar synthesis | NP_461698 |
| fliC | Phase 1 flagellin; Filament structural protein | NP_460912 |
| sinH | Intimin-like protein | NP_461452 |
Results
MLST
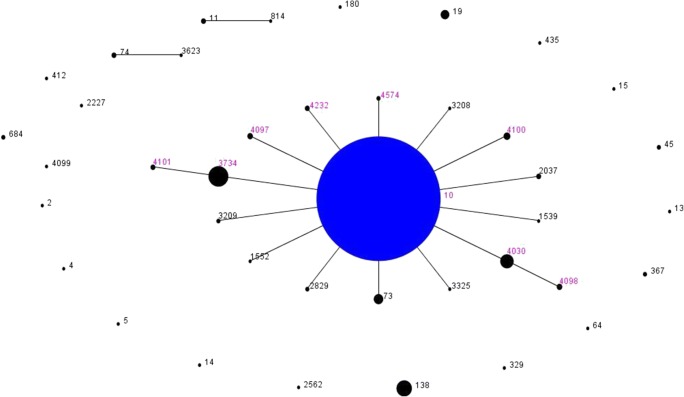
The STs detected, eBURST group, allelic profile of the housekeeping genes analyzed, and the percentage of detection among the strains studied are presented in Table 2. The minimum spanning tree generated with the STs detected is presented in Fig. 1. Among the S. Dublin strains studied, nine STs were detected and belonged to the same eBURST group eBG53 (Table 2). The most prevalent ST among the strains studied was ST10. The STs ST3734, ST4030, ST4097, ST4100, ST4232, and ST4574 were single-locus variants of the ST10, while ST4098 and ST4101 were double-locus variants of ST10 (Fig. 1, Table 2). Moreover, STs ST4097, ST4098, ST4100, ST4101, ST4232, and ST4574 were detected for the first time in the S. Dublin global database.
Table 2.
Sequence type (ST), eBURST group (eBG), number of strains, year of isolation, and allelic profile of 112 S. Dublin strains studied
| STs | eBG | Number of strains (%) | Year of isolation | aroC | dnaN | hemD | hisD | purE | sucA | thrA |
|---|---|---|---|---|---|---|---|---|---|---|
| 10 | 53 | 68 (60.7) | 1985–2016 | 5 | 2 | 3 | 6 | 5 | 5 | 10 |
| 3734 | 53 | 28 (25.0) | 1984–2013 | 5 | 2 | 3 | 6 | 5 | 671 | 10 |
| 4030 | 53 | 9 (8.0) | 1983–2005 | 5 | 2 | 612 | 6 | 5 | 5 | 10 |
| 4097 | 53 | 1 (0.9) | 2007 | 5 | 2 | 3 | 6 | 748 | 5 | 10 |
| 4098 | 53 | 1 (0.9) | 2005 | 5 | 2 | 612 | 6 | 749 | 5 | 10 |
| 4100 | 53 | 2 (1.8) | 1991 | 5 | 2 | 3 | 6 | 5 | 713 | 10 |
| 4101 | 53 | 1 (0.9) | 1998 | 5 | 2 | 3 | 6 | 750 | 671 | 10 |
| 4232 | 53 | 1 (0.9) | 1988 | 5 | 2 | 3 | 6 | 636 | 5 | 10 |
| 4574 | 53 | 1 (0.9) | 1990 | 5 | 2 | 3 | 6 | 800 | 5 | 10 |
Fig. 1.
Minimum spanning tree generated with the software eBURSTv3 for the 112 S. Dublin strains studied and the other strains of this serovar available in the Enterobase database. Each ST is represented by a dot. The pink numbers above black dots represent the STs detected in this study. The blue central dot represents the predicted primary founder ST of the clonal complex 53 (CC53), ST10. The diameter of each dot indicates the prevalence of the STs in the input data that generated the graphic. Black numbers above back dots represent other S. Dublin STs presented in the database
CRISPR and CRISPR-MVLST
Among the 112 S. Dublin strains studied, 59 CRISPR1, 12 CRISPR2, 2 fimH, and 4 sseL alleles were identified. When combined, these alleles generated 72 CM-STs (Table 3). CRISPR1 alleles ranged from one to nine spacers in size, while CRISPR2 alleles ranged from two to four spacers in size. In our analysis, we found 70 and 16 different spacers in CRISPR1 and CRISPR2 loci, respectively. The strain SD 721 presented a duplication in one of CRISPR1 spacers and was also included in the analysis. The complete absence of spacers was found in a single strain in CRISPR1 analysis and in 33 strains in CRISPR2 analysis.
Table 3.
CRISPR-MVLST sequence types (CM-ST) and respective frequencies identified in the 112 S. Dublin strains in this study
| CM-ST | Number of strains | Alleles | |||
|---|---|---|---|---|---|
| CRISPR1 | CRISPR2 | fimH | sseL | ||
| 1 | 9 | 53 | 2 | 1 | 1 |
| 2 | 7 | 5 | 11 | 1 | 1 |
| 3 | 7 | 43 | 2 | 1 | 1 |
| 4 | 6 | 5 | 7 | 1 | 1 |
| 5 | 5 | 5 | 1 | 1 | 1 |
| 6 | 5 | 29 | 2 | 1 | 1 |
| 7 | 4 | 35 | 1 | 1 | 1 |
| 8 | 2 | 5 | 8 | 1 | 1 |
| 9 | 2 | 5 | 10 | 1 | 1 |
| 10 | 2 | 10 | 1 | 1 | 1 |
| 11 | 2 | 36 | 1 | 1 | 1 |
| 12 | 2 | 49 | 2 | 1 | 1 |
| 13 | 1 | 1 | 4 | 1 | 1 |
| 14 | 1 | 2 | 8 | 1 | 1 |
| 15 | 1 | 3 | 4 | 1 | 1 |
| 16 | 1 | 4 | 1 | 2 | 1 |
| 17 | 1 | 4 | 4 | 1 | 1 |
| 18 | 1 | 4 | 11 | 1 | 2 |
| 19 | 1 | 5 | 12 | 1 | 1 |
| 20 | 1 | 5 | 7 | 1 | 3 |
| 21 | 1 | 6 | 8 | 1 | 1 |
| 22 | 1 | 6 | 1 | 1 | 1 |
| 23 | 1 | 7 | 1 | 1 | 1 |
| 24 | 1 | 8 | 11 | 1 | 1 |
| 25 | 1 | 9 | 8 | 1 | 1 |
| 26 | 1 | 11 | 4 | 1 | 1 |
| 27 | 1 | 12 | 1 | 1 | 1 |
| 28 | 1 | 13 | 1 | 1 | 1 |
| 29 | 1 | 14 | 4 | 1 | 1 |
| 30 | 1 | 15 | 7 | 1 | 1 |
| 31 | 1 | 16 | 7 | 1 | 1 |
| 32 | 1 | 17 | 11 | 1 | 1 |
| 33 | 1 | 18 | 1 | 1 | 1 |
| 34 | 1 | 19 | 9 | 1 | 1 |
| 35 | 1 | 20 | 6 | 1 | 1 |
| 36 | 1 | 21 | 5 | 1 | 1 |
| 37 | 1 | 22 | 7 | 1 | 1 |
| 38 | 1 | 23 | 1 | 1 | 1 |
| 39 | 1 | 24 | 1 | 1 | 1 |
| 40 | 1 | 25 | 7 | 1 | 1 |
| 41 | 1 | 26 | 1 | 1 | 1 |
| 42 | 1 | 27 | 2 | 1 | 1 |
| 43 | 1 | 28 | 1 | 1 | 1 |
| 44 | 1 | 30 | 2 | 1 | 1 |
| 45 | 1 | 31 | 2 | 1 | 4 |
| 46 | 1 | 32 | 2 | 1 | 1 |
| 47 | 1 | 33 | 1 | 1 | 4 |
| 48 | 1 | 34 | 1 | 1 | 1 |
| 49 | 1 | 37 | 1 | 1 | 1 |
| 50 | 1 | 38 | 1 | 1 | 1 |
| 51 | 1 | 38 | 1 | 1 | 1 |
| 52 | 1 | 40 | 1 | 1 | 1 |
| 53 | 1 | 41 | 1 | 1 | 1 |
| 54 | 1 | 42 | 1 | 1 | 1 |
| 55 | 1 | 43 | 2 | 2 | 1 |
| 56 | 1 | 44 | 2 | 1 | 1 |
| 57 | 1 | 45 | 2 | 1 | 1 |
| 58 | 1 | 46 | 2 | 1 | 1 |
| 59 | 1 | 47 | 2 | 1 | 1 |
| 60 | 1 | 48 | 2 | 1 | 1 |
| 61 | 1 | 50 | 2 | 1 | 1 |
| 62 | 1 | 51 | 2 | 1 | 1 |
| 63 | 1 | 52 | 2 | 1 | 1 |
| 64 | 1 | 52 | 3 | 1 | 1 |
| 65 | 1 | 53 | 1 | 1 | 1 |
| 66 | 1 | 53 | 2 | 1 | 2 |
| 67 | 1 | 54 | 2 | 1 | 1 |
| 68 | 1 | 55 | 2 | 1 | 1 |
| 69 | 1 | 56 | 2 | 1 | 1 |
| 70 | 1 | 57 | 2 | 1 | 1 |
| 71 | 1 | 58 | 2 | 1 | 1 |
| 72 | 1 | 59 | 1 | 1 | 1 |
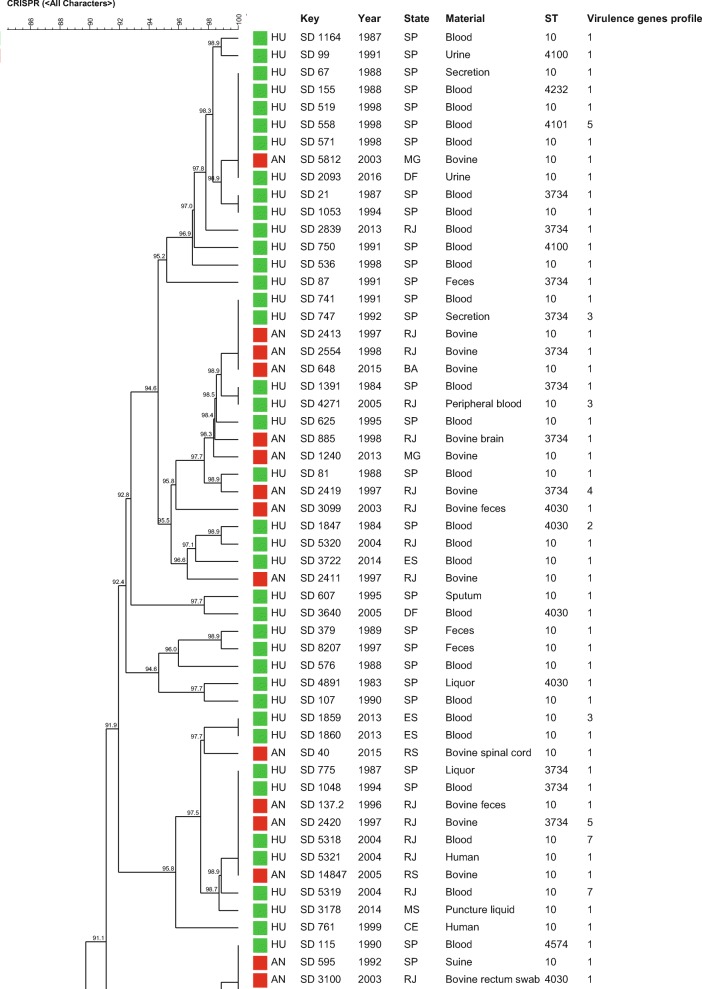
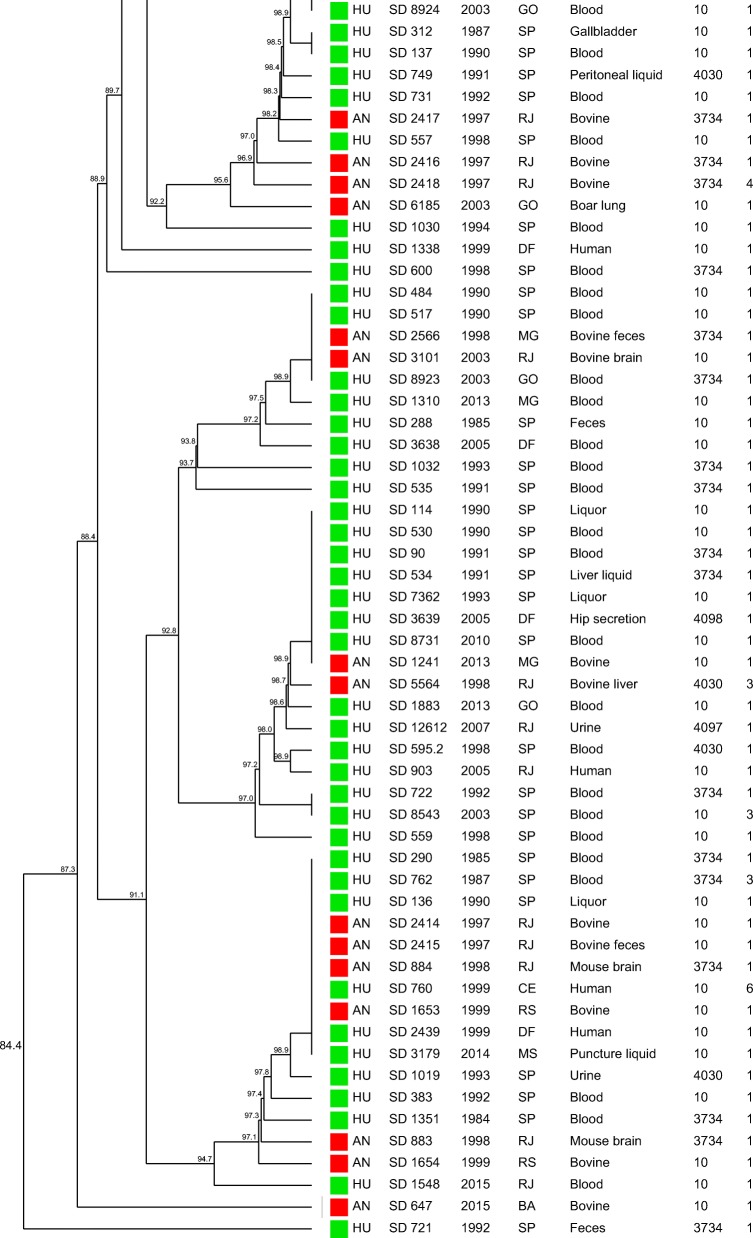
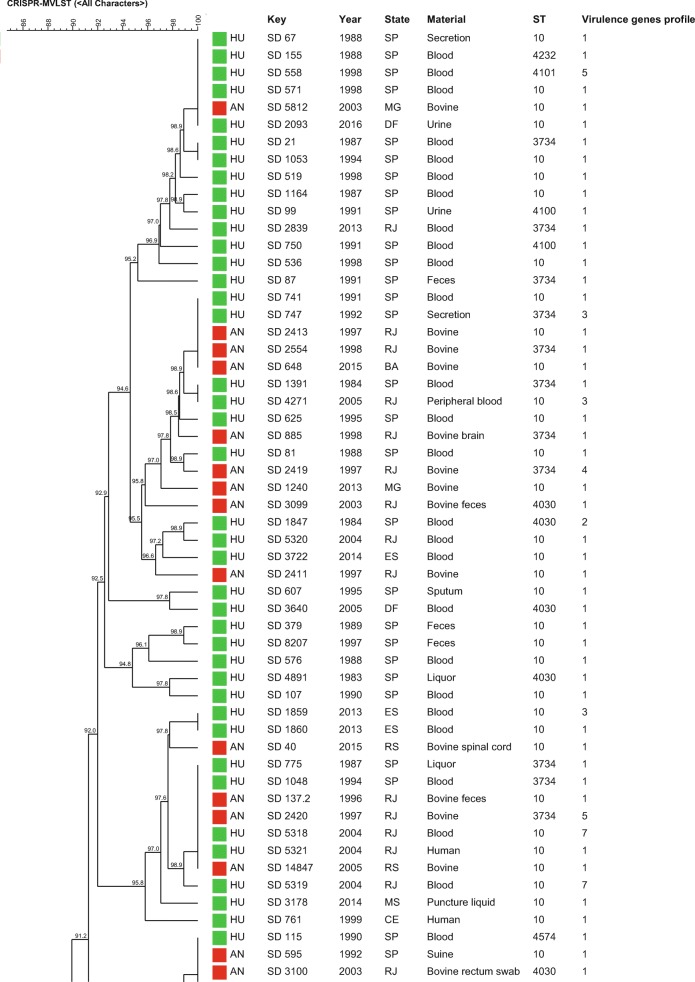
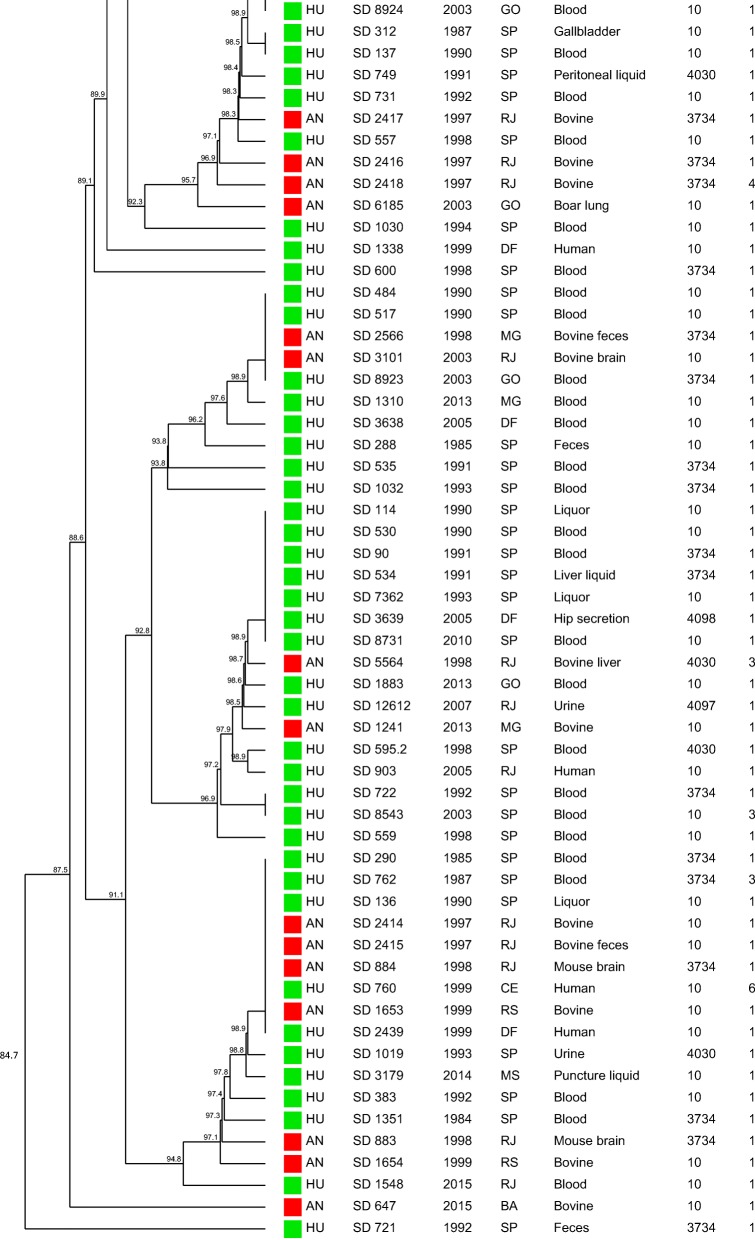
The similarity dendrogram generated with the binary matrix of CRISPR1 and CRISPR2 spacers grouped all the 112 S. Dublin strains in a single cluster (> 80% of similarity) presenting 69 CRISPR-types with a similarity ≥ 84.4% among the strains (Fig. 2). The association of the binary matrix of CRISPR1 and CRISPR2 spacers with the respective fimH and sseL gene loci also grouped all the 112 S. Dublin strains into a single cluster presenting 72 CRISPR-MVLST-types with a similarity ≥ 84.7% among the strains studied (Fig. 3). The discriminatory index (DI) for CRISPR and CRISPR-MVLST were 0.976 and 0.980, respectively.
Fig. 2.
Similarity dendrogram representing the genetic relationships among Salmonella Dublin strains based on the binary matrix of CRISPR1 and CRISPR2 spacers in the 112 strains studied. Green squares represent strains isolated from humans; red squares represent strains isolated from animals. Profile 1 (fljB negative), Profile 2 (fljB and spvB negative), Profile 3 (fljB, spvB, and spvC negative), Profile 4 (sipA, sipD, invA, hilA, and fljB negative), Profile 5 (sodCl and fljB negative), Profile 6 (rpoS and fljB negative), Profile 7 (sodCl, rpoS, and fljB negative)
Fig. 3.
Similarity dendrogram representing the genetic relationships among Salmonella Dublin strains based on the combination of the binary matrix of CRISPR1 and CRISPR2 spacers and the loci of fimH and sseL virulence genes in the 112 strains studied. Green squares represent strains isolated from humans; red squares represent strains isolated from animals. Profile 1 (fljB negative), Profile 2 (fljB and spvB negative), Profile 3 (fljB, spvB, and spvC negative), Profile 4 (sipA, sipD, invA, hilA, and fljB negative), Profile 5 (sodCl and fljB negative), Profile 6 (rpoS and fljB negative), Profile 7 (sodCl, rpoS, and fljB negative)
Detection of virulence genes
The frequency of 21 virulence genes showed that all the S. Dublin strains studied carried the genes ratB, lpfA, mgtC, avrA, sopB, sopE2, sifA, sseA, ssrA, csgA, fliC, and sinH. Moreover, spvB gene was present in 105 strains (93.7%), spvC in 106 strains (94.6%), sodCl in 108 strains 96.5%), rpoS in 109 strains (97.3%), sipA in 110 strains (98.2%), sipD in 110 strains (98.2%), invA in 110 strains (98.2%), and hilA was present in 110 strains (98.2%). On the other hand, fljB gene was not detected in any of the strains studied.
Discussion
Salmonella Dublin is a serovar strongly adapted to bovine hosts, but can be sporadically isolated from human clinical cases [1, 2]. Different molecular typing techniques have been used for epidemiological studies of S. Dublin strains worldwide [9–13]. The advancement in whole-genome sequencing allowed the sequencing of large sets of strains and the characterization by classic or newly developed methodologies, such as MLST and CRISPR [18, 19]. To our knowledge, no studies have been conducted to characterize the genotypic diversity in large sets composed exclusively of S. Dublin strains isolated in Brazil by MLST and/or CRISPR. In the present study, we used MLST, CRISPR, and CRISPR-MVLST to type 112 S. Dublin strains isolated from humans and animals between 1983 and 2016 in Brazil. In addition, we characterized the virulence potential of these strains searching for the frequency of 20 S. enterica virulence genes.
Due to laborious work and high costs to perform traditional MLST for large sets of strains, most studies only used this methodology to type small sets of representative strains [14–17]. However, WGS has been providing a faster and easier alternative to type in silico a large number of strains, contributing for a better characterization of many Salmonella serovars [18, 19].
In the present study, MLST revealed the presence of nine different STs among the strains studied (Table 2). The most prevalent ST was ST10, found in 68 strains (60.7%) out of 112 strains studied, that has also been the main ST reported for S. Dublin strains [14–17].
Moreover, other eight STs were detected in the strains studied. Among them, six (ST3734, ST4030, ST4097, ST4100, ST4232, and ST4574) were single-locus variants of ST10, while two (ST4098 and ST4101) were double-locus variants of ST10 (Fig. 1, Table 2). In addition, STs ST4097, ST4098, ST4100, ST4101, ST4232, and ST4574 were detected for the first time in S. Dublin strains.
The detection of only single- or double-locus variants of ST10 from clonal complex 10, as well as no prevalence of STs by source, material, or year of isolation among the strains studied, reinforces the proposed by Achtman et al. (2012) that different clonal complexes generally represent specific serovars, mainly due to the highly clonal characteristic of S. enterica [15].
Regarding CRISPR and CRISPR-MVLST analysis, the 112 S. Dublin strains studied showed 59 CRISPR1 and 12 CRISPR2 and 2 fimH and 4 sseL different alleles identified, which showed the high differentiation capacity of these methodologies, which were also confirmed by the high values of DI of 0.976 for CRISPR and of 0.980 for CRISPR-MVLST (Figs. 2 and 3, Table 1). However, despite the high discrimination power, the strains showed to be genetically related, with a similarity ≥ 84.4% among the strains for CRISPR and ≥ 84.7 for CRISPR-MVLST (Figs. 2 and 3).
The similar DI found in both methodologies mentioned above showed that the addition of fimH and sseL in the analysis did not increase the discriminatory power of the methodology on typing S. Dublin strains, which differed from previous studies with other Salmonella serovars [19, 23–25]. This fact reinforced that this was not due to a technique limitation but to the clonal characteristic of serovar Dublin strains. Similar to MLST, both methodologies grouped the strains independently of geographical, temporal, or isolation source characteristics, which reinforced the idea from previous studies of Salmonella Dublin strains from Brazil performed by our research group that suggest that these strains may have descended from a common ancestor that has little differentiated over the years [13].
In the previous study of our research group [13], the same 112 S. Dublin strains of this study were typed by pulsed-field gel electrophoresis (PFGE) and multilocus variable-number tandem repeat analysis (MLVA). Similar to the results found in the present study, PFGE also grouped the strains in a single cluster, with a similarity of ≥ 80.7%. However, that methodology showed a lower discriminatory power, differentiating the strains into 35 PFGE types and with a DI of 0.53. Regarding MLVA, the strains were classified in 89 types with a similarity of ≥23.3% and a DI of 0.95, closer to the results found in the present study. In addition, MLVA was able to group the strains into two different clusters that contained five and 106 strains, respectively, and also two strains showed to be single MLVA types [13].
The DIs observed for the four methodologies mentioned above showed that CRISPR and CRISPR-MVLST are good in silico techniques to type S. Dublin strains and alternatives to the non-WGS techniques, such as PFGE and MLVA, considered the gold standard methodologies to type Salmonella spp. Moreover, CRISPR methodologies are performed in silico, which minimizes reproducibility mistakes among different laboratories.
To our knowledge, until the writing of this manuscript, there were no studies performed that used CRISPR and CRISPR-MVLST methodologies to type exclusively S. Dublin strains, which makes the comparison of the results found in the present study difficult. However, other studies using these methodologies had been successfully conducted to subtype other Salmonella serovars, such as Typhimurium, Newport, Enteritidis, and Heidelberg [19, 23–25].
The presence of multiple virulence genes was verified in the strains studied (Table 1). The high prevalence of SPI-1 and SPI-2 genes detected in the S. Dublin strains studied reinforced the invasive potential and the capacity to cause serious disease of strains of this serovar [8, 36–38]. The absence of the genes spvB and spvC in seven and six strains studied, respectively, may be explained to a possible absence of pSDL2, a well-characterized S. enterica virulence plasmid encoded by genes of spv locus [39, 40].
Regarding the flagella-related genes, the fljB, responsible for phase-2 flagellin, was absent in all the strains studied, while fliC gene, responsible for phase-1 flagellin, was found in all the strains studied, which showed a prevalence of this type of flagella among the strains studies. It is interesting to mention that Yim et al. (2014) [16] showed differences in the expression of fliC gene in S. Dublin strains from Uruguay that may alter the flagella expression [16].
Although some genes such as hilA and rpoS are important transcriptional regulators and mainly detected in Salmonella, in this study, these genes were not detected in two and three of the strains studied, respectively. Previous studies have already reported the absence of hilA in Salmonella serovars [41, 42]. The absence of rpoS might have been due to a genome assembly drawback, which may have led to a non-detection of any gene fragment below the parameters established for the detection of this gene.
In conclusion, the high similarity among the strains reinforces the clonal nature of the strains of this serovar that may have descended from a common ancestor that little differed over 33 years in Brazil. CRISPR and CRISPR-MVLST showed to be good alternatives to type S. Dublin strains. MLST suggested that S. Dublin strains from Brazil were phylogenetically related to strains from other parts of the globe. Moreover, the high frequency of virulence genes among the strains studied reinforces the capacity of S. Dublin to cause invasive diseases.
Electronic supplementary material
(DOCX 13 kb)
Acknowledgments
The authors thank São Paulo Research Foundation - FAPESP for financial support and Dr. Marc Allard from FDA/CFSAN for providing whole-genome sequencing of the strains studied.
Funding information
This study was financed by São Paulo Research Foundation (FAPESP) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES) - Finance Code 001. During the course of the work, F.P. Vilela was supported by a scholarship from FAPESP (Proc. 2017/05756-7 and 2019/06947-6) and F. Campioni was supported by a postdoctoral fellowship from FAPESP (Proc. 2013/25191-3). Part of this work was also financed by a FAPESP grant (Proc. 2016/24716-3) under the supervision of J.P. Falcão that also received a productive fellowship (CNPq 303475/2015-3 and CNPq 304399/2018-3) from National Council for Scientific and Technological Development (CNPq).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Juliana Pfrimer Falcão, Email: jufalcao@fcfrp.usp.br.
Fábio Campioni, Email: campioni@fcfrp.usp.br.
References
- 1.Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O'Brien SJ, Jones TF, Fazil A, Hoekstra RM. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis. 2010;50:882–889. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- 2.Uzzau S, Brown DJ, Wallis T, Rubino S, Leori G, Bernard S, Casadesús J, Platt DJ, Olsen JE. Host adapted serotypes of Salmonella enterica. Epidemiol Infect. 2000;125:229–255. doi: 10.1017/S0950268899004379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nielsen LR. Review of pathogenesis and diagnostic methods of immediate relevance for epidemiology and control of Salmonella Dublin in cattle. Vet Microbiol. 2013;162:1–9. doi: 10.1016/j.vetmic.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 4.Pezoa D, Blondel CJ, Silva CA, Yang HJ, Andrews-Polymenis H, Santiviago CA, Contreras I. Only one of the two type VI secretion systems encoded in the Salmonella enterica serotype Dublin genome is involved in colonization of the avian and murine hosts. Vet Res. 2014;45:2–2. doi: 10.1186/1297-9716-45-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hur J, Choi YY, Park JH, Jeon BW, Lee HS, Kim AR, Lee JH. Antimicrobial resistance, virulence-associated genes, and pulsed-field gel electrophoresis profiles of Salmonella enterica subsp. enterica serovar Typhimurium isolated from piglets with diarrhea in Korea. Can J Vet Res. 2011;75:49–56. [PMC free article] [PubMed] [Google Scholar]
- 6.Shah DH, Zhou X, Addwebi T, Davis MA, Orfe L, Call DR, Guard J, Besser TE. Cell invasion of poultry-associated Salmonella enterica serovar Enteritidis isolates is associated with pathogenicity, motility and proteins secreted by the type III secretion system. Microbiol. 2011;157:1428–1445. doi: 10.1099/mic.0.044461-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zou M, Keelara S, Thakur S. Molecular characterization of Salmonella enterica serotype Enteritidis isolates from humans by antimicrobial resistance, virulence genes, and pulsed-field gel electrophoresis. Foodborne Pathog Dis. 2012;9:232–238. doi: 10.1089/fpd.2011.1012. [DOI] [PubMed] [Google Scholar]
- 8.Suez J, Porwollik S, Dagan A, Marzel A, Schorr YI, Desai PT, Agmon V, McClelland M, Rahav G, Gal-Mor O. Virulence gene profiling and pathogenicity characterization of non-typhoidal Salmonella accounted for invasive disease in humans. PLoS One. 2013;8:e58449–e58449. doi: 10.1371/journal.pone.0058449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liebana E, Garcia-Migura L, Clouting C, Cassar CA, Clifton-Hadley FA, Lindsay EA, Threlfall EJ, Chappell SA, Davies RH. Investigation of the genetic diversity among isolates of Salmonella enterica serovar Dublin from animals and humans from England, Wales and Ireland. J Appl Microbiol. 2002;93:732–744. doi: 10.1046/j.1365-2672.2002.01737.x. [DOI] [PubMed] [Google Scholar]
- 10.Zhao S, McDermott PF, White DG, Qaiyumi S, Friedman SL, Abbott JW, Glenn A, Ayers SL, Post KW, Fales WH, Wilson RB, Reggiardo C, Walker RD. Characterization of multidrug resistant Salmonella recovered from diseased animals. Vet Microbiol. 2007;123:122–132. doi: 10.1016/j.vetmic.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 11.Kjeldsen MK, Torpdahl M, Campos J, Pedersen K, Nielsen EM. Multiple-locus variable-number tandem repeat analysis of Salmonella enterica subsp. enterica serovar Dublin. J Appl Microbiol. 2014;116:1044–1054. doi: 10.1111/jam.12441. [DOI] [PubMed] [Google Scholar]
- 12.Vignaud ML, Cherchame E, Marault M, Chaing E, Le Hello S, Michel V, Jourdan-Da Silva N, Lailler R, Brisabois A, Cadel-Six S. MLVA for Salmonella enterica subsp. enterica Serovar Dublin: development of a method suitable for inter-laboratory surveillance and application in the context of a raw milk cheese outbreak in France in 2012. Front Microbiol. 2017;8:295. doi: 10.3389/fmicb.2017.00295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vilela FP, Frazão MR, Rodrigues DP, Costa RG, Casas MRT, Fernandes SA, Falcão JP, Campioni F. Genetic diversity, anti-microbial resistance, plasmid profile and frequency of the Vi antigen in Salmonella Dublin strains isolated in Brazil. Zoonoses Public Health. 2018;65:e34–e43. doi: 10.1111/zph.12407. [DOI] [PubMed] [Google Scholar]
- 14.Litrup E, Torpdahl M, Malorny B, Huehn S, Christensen H, Nielsen EM. Association between phylogeny, virulence potential and serovars of Salmonella enterica. Infect Genet Evol. 2010;10:1132–1139. doi: 10.1016/j.meegid.2010.07.015. [DOI] [PubMed] [Google Scholar]
- 15.Achtman M, Wain J, Weill FX, Nair S, Zhou Z, Sangal V, Krauland MG, Hale JL, Harbottle H, Uesbeck A, Dougan G, Harrison LH, Brisse S, the S.e.M.s.g Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathog. 2012;8:e1002776. doi: 10.1371/journal.ppat.1002776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yim L, Sasías S, Martínez A, Betancor L, Estevez V, Scavone P, Bielli A, Sirok A, Chabalgoity JA. Repression of flagella is a common trait in field isolates of Salmonella enterica Serovar Dublin and is associated with invasive human infections. Infect Immun. 2014;82:1465–1476. doi: 10.1128/IAI.01336-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silva C, Betancor L, García C, Astocondor L, Hinostroza N, Bisio J, Rivera J, Perezgasga L, Pérez Escanda V, Yim L, Jacobs J, García-Del Portillo F, SalmoIber CN, Chabalgoity JA, Puente JL. Characterization of Salmonella enterica isolates causing bacteremia in Lima, Peru, using multiple typing methods. PLoS One. 2017;12:e0189946–e0189946. doi: 10.1371/journal.pone.0189946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Almeida F, Medeiros MIC, Rodrigues DDP, Allard MW, Falcão JP. Molecular characterization of Salmonella typhimurium isolated in Brazil by CRISPR-MVLST. J Microbiol Methods. 2017;133:55–61. doi: 10.1016/j.mimet.2016.12.020. [DOI] [PubMed] [Google Scholar]
- 19.Almeida F, Seribelli AA, Silva P, Medeiros MIC, Rodrigues DDP, Moreira CG, Allard MW, Falcão JP. Multilocus sequence typing of Salmonella Typhimurium reveals the presence of the highly invasive ST313 in Brazil. Infect Genet Evol. 2017;51:41–44. doi: 10.1016/j.meegid.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 20.Campioni F, Pitondo-Silva A, Bergamini AM, Falcão JP. Comparison of four molecular methods to type Salmonella Enteritidis strains. APMIS. 2015;123:422–426. doi: 10.1111/apm.12367. [DOI] [PubMed] [Google Scholar]
- 21.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–1412. doi: 10.1128/JB.01415-07. [DOI] [PubMed] [Google Scholar]
- 22.Touchon M, Rocha EPC. The small, slow and specialized CRISPR and anti-CRISPR of Escherichia and Salmonella. PLoS One. 2010;5:e11126. doi: 10.1371/journal.pone.0011126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shariat N, DiMarzio MJ, Yin S, Dettinger L, Sandt CH, Lute JR, Barrangou R, Dudley EG. The combination of CRISPR-MVLST and PFGE provides increased discriminatory power for differentiating human clinical isolates of Salmonella enterica subsp. enterica serovar Enteritidis. Food Microbiol. 2013;34:164–173. doi: 10.1016/j.fm.2012.11.012. [DOI] [PubMed] [Google Scholar]
- 24.Shariat N, Kirchner MK, Sandt CH, Trees E, Barrangou R, Dudley EG. Subtyping of Salmonella enterica Serovar Newport outbreak isolates by CRISPR-MVLST and determination of the relationship between CRISPR-MVLST and PFGE results. J Clin Microbiol. 2013;51:2328–2336. doi: 10.1128/JCM.00608-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng X, Shariat N, Driebe EM, Roe CC, Tolar B, Trees E, Keim P, Zhang W, Dudley EG, Fields PI, Engelthaler DM. Comparative analysis of subtyping methods against a whole-genome-sequencing standard for Salmonella enterica serotype Enteritidis. J Clin Microbiol. 2015;53:212–218. doi: 10.1128/JCM.02332-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rytkönen A, Poh J, Garmendia J, Boyle C, Thompson A, Liu M, Freemont P, Hinton JCD, Holden DW. SseL, a Salmonella deubiquitinase required for macrophage killing and virulence. PNAS USA. 2007;104:3502–3507. doi: 10.1073/pnas.0610095104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kisiela D, Laskowska A, Sapeta A, Kuczkowski M, Wieliczko A, Ugorski M. Functional characterization of the FimH adhesin from Salmonella enterica serovar Enteritidis. Microbiol. 2006;152:1337–1346. doi: 10.1099/mic.0.28588-0. [DOI] [PubMed] [Google Scholar]
- 28.Liu F, Barrangou R, Gerner-Smidt P, Ribot EM, Knabel SJ, Dudley EG. Novel virulence gene and clustered regularly interspaced short palindromic repeat (CRISPR) multilocus sequence typing scheme for subtyping of the major Serovars of Salmonella enterica subsp. enterica. Appl Environ Microbiol. 2011;77:1946–1956. doi: 10.1128/AEM.02625-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olsen JE, Skov M. Genomic lineage of Salmonella enterica serovar Dublin. Vet Microbiol. 1994;40:271–282. doi: 10.1016/0378-1135(94)90116-3. [DOI] [PubMed] [Google Scholar]
- 30.FAd C, VRd S, Martins CHG, Fernandes SA, Zaia JE, Martinez R. Prevalence and antimicrobial susceptibility of Salmonella serotypes in patients from Ribeirão Preto, São Paulo, Brazil, between 1985 and 1999. Braz J Infect Dis. 2002;6:244–251. doi: 10.1590/S1413-86702002000500005. [DOI] [PubMed] [Google Scholar]
- 31.Tavechio AT, Ghilardi ÂCR, Peresi JTM, Fuzihara TO. Yonamine EK, Jakabi M, Fernandes SA. Salmonella serotypes isolated from nonhuman sources in São Paulo, Brazil, from 1996 through 2000. J Food Prot. 2002;65:1041–1044. doi: 10.4315/0362-028X-65.6.1041. [DOI] [PubMed] [Google Scholar]
- 32.Fernandes SA, Tavechio AT, Ghilardi ÂCR, Dias ÂMG, Almeida IAZC, LCVd M. Salmonella serovars isolated from humans in São Paulo state, Brazil, 1996-2003. Rev Inst Med Trop Sao Paulo. 2006;48:179–184. doi: 10.1590/S0036-46652006000400001. [DOI] [PubMed] [Google Scholar]
- 33.Vilela FP, Gomes CN, Passaglia J, Rodrigues DP, Costa RG, Tiba Casas MR, Fernandes SA, Falcão JP, Campioni F. Genotypic resistance to quinolone and tetracycline in Salmonella Dublin strains isolated from humans and animals in Brazil. Microb Drug Resist. 2018;25:143–151. doi: 10.1089/mdr.2017.0329. [DOI] [PubMed] [Google Scholar]
- 34.Campioni F, Vilela FP, Cao G, Kastanis G, Miller D, Sanchez Leon M, Tiba-Casas MR, Fernandes SA, Rodrigues DDP, Costa RG, Allard MW, Falcão JP (2018) Draft genome sequences of 112 Salmonella enterica Serovar Dublin strains isolated from humans and animals in Brazil. Genome Announc 6. 10.1128/genomeA.00405-18 [DOI] [PMC free article] [PubMed]
- 35.Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol. 1988;26:2465–2466. doi: 10.1128/JCM.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Norris FA, Wilson MP, Wallis TS, Galyov EE, Majerus PW. SopB, a protein required for virulence of Salmonella dublin, is an inositol phosphate phosphatase. PNAS USA. 1998;95:14057–14059. doi: 10.1073/pnas.95.24.14057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mirold S, Rabsch W, Tschäpe H, Hardt WD. Transfer of the Salmonella type III effector sopE between unrelated phage families. J Mol Biol. 2001;312:7–16. doi: 10.1006/jmbi.2001.4950. [DOI] [PubMed] [Google Scholar]
- 38.Mohammed M, Le Hello S, Leekitcharoenphon P, Hendriksen R. The invasome of Salmonella Dublin as revealed by whole genome sequencing. BMC Infect Dis. 2017;17:544–544. doi: 10.1186/s12879-017-2628-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mazurkiewicz P, Thomas J, Thompson JA, Liu M, Arbibe L, Sansonetti P, Holden DW. SpvC is a Salmonella effector with phosphothreonine lyase activity on host mitogen-activated protein kinases. Mol Microbiol. 2008;67:1371–1383. doi: 10.1111/j.1365-2958.2008.06134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guiney D, Fierer J (2011) The role of the spv genes in Salmonella pathogenesis. Front Microbiol 2(129). 10.3389/fmicb.2011.00129 [DOI] [PMC free article] [PubMed]
- 41.Ammar AM, Mohamed AA, El-Hamid MIA, El-Azzouny MM. Virulence genotypes of clinical Salmonella Serovars from broilers in Egypt. J Infect Dev Ctries. 2015;10(4):337–346. doi: 10.3855/jidc.7437. [DOI] [PubMed] [Google Scholar]
- 42.Allam SA, Mostafa NY, Kirrella GAK, Eleiwa NZ, El-Magd MA. Molecular detection of invA and hilA virulent genes in Salmonella serovars isolated from fresh water fish. Slov Vet Res. 2019;56(Suppl 22):693–698. doi: 10.26873/SVR-809-2019. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 13 kb)