Abstract
Bacillus utilize preferred sugars such as glucose over other carbon sources due to carbon catabolite repression (CCR). Surfactin is a small signal molecule to regulate the quorum-sensing (QS) response such as biofilm formation and sporulation in B. subtilis. Here, the srfA operon for synthesis of surfactin was mutated for disrupting the production of surfactin in B. amyloliquefaciens. The srfA-mutant strain showed a defective biofilm and sporulation but could be restored by addition with surfactin, indicating that surfactin is a QS signal molecule in B. amyloliquefaciens. Unexpectedly, mutation of srfA also led to the cells’ death although nutrients were still enough to support the bacterial growth during this period. Analysis of transcriptomes found that the srfA-mutant strain could not relieve CCR to use non-preferred carbon sources after glucose exhaustion due to deficiency of surfactin. This was further verified by the fact that addition with glucose could dramatically restore the growth, and addition with surfactin could improve the enzymes’ activity (e.g., glucanase and α-amylase) to use non-preferred carbon sources in the srfA-mutant strain. After glucose exhaustion, the cells produce surfactin to relieve CCR for utilizing non-preferred sugars. As a signal molecule to regulate QS, surfactin also directly or indirectly relieves the CcpA-mediated CCR to utilize non-preferred carbon sources countering nutrient limitation (e.g., glucose deprivation) in the environment. In conclusion, our findings provide the first evidence that the QS signal molecule of surfactin is also involved in relieving the CcpA-mediated CCR in B. amyloliquefaciens.
Keywords: Bacillus amyloliquefaciens, surfactin, quorum-sensing, carbon catabolite repression, catabolite control protein A
Introduction
Bacillus are generally soil-dwelling Gram-positive bacteria ubiquitously distributed in the natural environment. Many Bacillus species, such as B. subtilis, B. licheniformis, and B. amyloliquefaciens, are used as hosts for making fermentation products and microbial pesticides. B. subtilis is also used as a model system for Gram-positive bacteria. To survive in the complex environment, Bacillus metabolize glucose as a primary carbon source. Upon deprivation, many genes and proteins will specifically respond to glucose starvation, including the genes required for transport and utilization of alternative carbon sources, as well as the genes that allow employment of survival strategies like quorum sensing (QS) (de Jong et al., 2012). However, how to link utilization of alternative carbon sources and QS is still unknown in Bacillus.
Bacillus utilize a large number of carbon sources, but a well-known phenomenon is that the most favored sugars (e.g., glucose, fructose, or malate) are used preferentially over other carbon sources. The mechanism of preferentially utilizing carbohydrates is complex, containing multiple specific and global regulatory strategies strictly controlled by carbon catabolite repression (CCR). By CCR, Bacillus first utilize preferred carbohydrates in the presence of non-preferred sources with significant quantities that is beneficial for a successful competition with other microorganisms in the natural environment (Marciniak et al., 2012; Kim and Burne, 2017). In B. subtilis, CCR is mainly exerted by a global transcription factor of CcpA (catabolite control protein A) (Yang et al., 2017). In the presence of preferable carbon sources such as glucose, the histidine-containing phosphotransfer protein (HPr) is phosphorylated by HPr kinase (HPrK) at Ser46 by the bifunctional HPr kinase/phosphorylase HPrK/P at high fructose-1,6-bisphosphate and ATP or pyrophosphate concentrations and then forms a complex with CcpA that can bind to a conserved cis-acting sequence called catabolite responsive elements (cre) to repress the gene transcription involved in catabolism of non-preferred carbon sources (Schumacher et al., 2011; Ishii et al., 2013; Charbonnier et al., 2017; Kim and Burne, 2017). After glucose exhaustion, HPrK/P dephosphorylates HPrSer46P in the presence of high inorganic phosphate (Pi) concentrations by phospho-phosphorylation of the Pi. The CcpA–HPrSer46P complex has a weak binding constant KA. Therefore, free HPrSer46P is present in the cell in excess to CcpA to keep CcpA saturated with HPrSer46P. In the presence of a high Pi concentration, HPrK/P dephosphorylates free HPrSer46P until the HPrSer46P concentration is so low that HPrSer46P dissociates from CcpA, and in consequence, CcpA dissociates from cre (Mijakovic et al., 2002; Poncet et al., 2004; Fujita, 2009).
With nutrient limitation (e.g., glucose deprivation), Bacillus will initiate a survival strategy called QS, by which the cells cooperate in a density-dependent manner through secretion and detection of specific autoinducer molecules such as ComX (Even-Tov et al., 2016; Pollak et al., 2016). In B. subtilis, ComX is a component of QS system encoded by the comQXPA operon (Tran et al., 2000; Stefanic and Mandic-Mulec, 2009), which contains an isoprenyl transferase of ComQ, a signal peptide of ComX, and the two-component system of ComP (histidine kinase) and ComA (response regulator) (Ansaldi et al., 2002; Ansaldi and Dubnau, 2004; Oslizlo et al., 2014). Pre-ComX is initially synthesized and then processed and modified by ComQ. Upon reaching a threshold concentration, ComX binds following leads to ComP autophosphorylation. ComP∼P transfers the phosphoryl group to ComA (Schneider et al., 2002), which controls the expression of srfA encoding a very large protein complex for non-ribosomal synthesis of the lipopeptide surfactin (Hamoen et al., 2003; Stefanic and Mandic-Mulec, 2009; Oslizlo et al., 2014). Surfactin is one of the most effective biosurfactants discovered so far (Stefanic and Mandic-Mulec, 2009; Oslizlo et al., 2014), as well as a signal molecule to trigger QS in B. subtilis (López et al., 2009a). Surfactin can cause pores on cell membrane leading the leakage of K+ from cytoplasm that acts as a signal to activate the membrane histidine kinase KinC and eventually phosphorylate Spo0A, a global regulator of QS in B. subtilis (López et al., 2009b; Banse et al., 2011; Bendori et al., 2015). High levels of Spo0A∼P are required for sporulation while low levels of Spo0A∼P are essential for biofilm formation (López et al., 2009b, 2010; Vlamakis et al., 2013).
In the natural environment, available carbon sources are a mixture of diverse carbohydrates including both of hexose and pentose sugars like glucose, xylose, arabinose, etc. (Li et al., 2014). CCR enables competitive advantages in carbon catabolism. By CCR, Bacillus utilize, out of a mixture of compounds, preferred carbon source (e.g., glucose) to allow a fast growth (Fujita, 2009). Accordingly, glucose content might be a restrictive nutrient factor for cell density in a given environment. Consequently, we hypothesized that after glucose exhaustion, CCR will be relieved to utilize non-preferred sugars supporting the initiated QS response such as biofilm formation and sporulation in Bacillus.
In addition to surfactin, Bacillus also produce other lipopeptides such as iturin and fengycin, which can interact with the biological membrane of pathogenic fungi to induce cell leakage and death (Koumoutsi et al., 2004; Zeriouh et al., 2014). Thereby, Bacillus are popularly used as biological control agents by virtue of their lipopeptide products (Xu et al., 2013; Zeriouh et al., 2014; Yang et al., 2015). Previously, we isolated a strain of B. amyloliquefaciens WH1 with production of several lipopeptides such as surfactin, iturin, and fengycin (Qi et al., 2010). In this study, we mutated the gene cluster for biosynthesis of surfactin (srfA), fengycin (fenA), and iturin (ituB), respectively. Unexpectedly, mutation of srfA caused a seriously defective growth in WH1. Further analysis found that surfactin, a signal peptide to regulate QS, is also involved in relieving CCR in B. amyloliquefaciens, by which QS is tightly linked with CCR to counter carbon sources limitation in the natural environment.
Materials and Methods
Bacterial Strains, Plasmids, Primers, and Reagents
Experiments were performed with the strains and plasmids in Supplementary Table S1. Oligonucleotide primers, listed in Supplementary Table S2, were designed on the genome of B. amyloliquefaciens. Materials for DNA manipulation including T4 DNA ligase, DNA marker, Pfu DNA Polymerase, and Plasmid Miniprep Kit were from Takara Bio (China). Nucleotide sequences were determined by Beijing Genomics institution (China). Surfactin was chemically synthesized by Chinese Peptide Company (China) (Zhang et al., 2017). Bacitracin was from Lifecome Biochemistry Co., Ltd., China. Amphotericin B and Nystatin were purchased from Biosharp, China. All other chemicals were of analytical grade supplied by Sinopharm Chemical Reagent (China).
Construction of Mutant Strains
Genes including srfA, ituB, fenA, comA, and ccpA were mutated by double crossover homologous recombination method (Qi et al., 2014). Firstly, two approximately 500-bp arms homologous to the 5′ and 3′ coding region of the above genes were amplified by PCR from B. amyloliquefaciens WH1 by primers LF and LR for L arm and RF and RR for R arm (Supplementary Table S2) and then ligated by splicing with Overlapping Extension PCR (SOE-PCR) with primers LF and RR, respectively. After digestion by BamHI and XbaI, the DNA fragments were subcloned into the vector T2(2) with a temperature-sensitive replicon from B. subtilis (Qi et al., 2014).
The resulting plasmids were transformed into B. amyloliquefaciens WH1 by our previous methods (Qi et al., 2014). Transformants were selected by kanamycin resistance (20 μg/ml) and then verified by PCR using LF and RR primers (Supplementary Table S2). The selected transformants were cultured in LB medium containing kanamycin at 45°C for 8 h to promote the first crossover and then the mutants with first crossover were selected by PCR with single-crossover LF and RR primers (Supplementary Table S2). Thereafter, the selected mutants with single crossover were cultured in LB medium at 37°C for 8 h and then the cultures were serially diluted following spread on LB agar plates for 100 μl/plate. After being cultured at 37°C for 24 h, the colonies were picked up and replicated on kanamycin plates for selecting sensitive ones. Knockout strains were screened out and had looped out of the kanamycin-resistant gene by the second crossover and confirmed by PCR with LF and RR primers (Supplementary Table S2) followed by nucleotide sequencing of the PCR products.
Analysis of Phenotype
The wild-type and mutant strains were cultured on LB agar plates and then the morphology of colonies was observed by microscope. Robust pellicle (floating biofilm) was determined in 24-well plates with 2 ml of LB medium each well. The multiwell plates were inoculated with different strains and then cultured at 28°C for 48 h to allow float biofilm formation.
The growth of different strains was detected in LB medium. The strains were incubated at 28°C and 180 rpm and then the OD600 value of culture was determined by a spectrophotometer, and the cells collected at several time points (18, 20, 26, and 29 h) were stained by crystal violet following observation by microscope. WH1 and ΔsrfA (srfA-mutant strain) were also respectively incubated at 28°C and 180 rpm with a ventilation of 3 L/min in a 5-L bioreactor (National center of Bio-Engineering and Technology, China) and then the dissolved O2 of culture was recorded by a sensor.
Analysis of Lipopeptides
Lipopeptides were crudely purified from the culture of different strains. Briefly, the pH of culture supernatant was adjusted to 4.0 for precipitation of proteins and then to 2.0 for precipitation of lipopeptides. After centrifugation, the pellets were collected and dissolved in water and then extracted by the same volume of n-butanol for determination of the lipopeptides by MALDI-TOF (Qi et al., 2010). Additionally, the agar plates containing sheep blood cells were used for culturing the wild-type and mutant strains and then the hemolytic activity was determined to evaluate the surfactin production in different strains (Shaligram and Singhal, 2010).
Effect of Surfactin and Antibiotics on Bacterial Growth and Biofilm Formation
ΔsrfA was cultured in LB medium supplemented with surfactin or other antibiotics including bacitracin, amphotericin B, and nystatin (80 μg/ml) at 28°C and 180 rpm for 24 h and then the bacterial growth was recorded. ΔsrfA was also cultured in 24-well plates with 2 ml of LB medium added with surfactin, bacitracin, amphotericin B, or nystatin at different concentrations (0, 20, 40, 60, and 80 μg/ml) in each well at 28°C for 48 h to allow biofilm formation. The comA-null mutant strain (ΔcomA) was also cultured in LB medium supplemented with surfactin (80 μg/ml) as above and then the bacterial growth was observed.
Comparison of Transcriptomes Between WH1 and ΔsrfA
Transcriptome analysis was performed by SHBIO Technology Co., Ltd. (China). WH1 and ΔsrfA were cultured in LB medium at 28°C and 180 rpm for 18 h (the highest biomass of ΔsrfA at this time point). Culture (1.5 ml) of three test tubes per replicate was centrifuged for collecting cell pellets and then used for isolating RNA by RNeasy Mini Kit (Qiagen, Germany). Enrichment of mRNA was done with the RiboZero rRNA Removal Kit for Bacteria according to the manufacturer’s directions. Library preparations for paired-end sequencing on the Illumina HiSeq 1500 platform were performed according to the protocols of Illumina, and the raw Illumina pair-end read data for all samples were deposited in the Short Read Archive under the Bioproject: PRJNA560470.
Obtained sequences were mapped onto the reference genome of B. amyloliquefaciens (NCBI Accession no: CP000560.1). To quantify gene transcription from the obtained sequence reads, RPKM values (Reads Per Kilobase per Million mapped reads) were calculated using the number of reads mapped to a gene, the total amount of mapped reads in the experiment, and the length in base pairs for a gene, and then used for comparison of normalized gene transcription rates. Differential gene transcriptions between ΔsrfA and WH1 were evaluated using the software platform ReadXplorer. The thresholds for significant changes in gene transcription were fold-changes of greater than 2 or less than 0.5 with an adjusted p-value of 0.05 (Kröber et al., 2016). By KEGG analysis, the metabolic pathways were constructed to describe the differential gene expression (DGE) involved in carbon metabolism between these two strains.
Some key gene transcriptions involved in carbon metabolism were quantified by quantitative real-time PCR (qRT-PCR). RNA was extracted as above and then cDNA was produced by reverse transcription with 1 μg RNA, iScript Select cDNA Synthesis Kit, and random oligonucleotide primers (Bio-Rad). qRT-PCR was performed with cDNA, SsoAdvanced Universal SYBR Green Supermix (Bio-Rad), and target-specific primers (Supplementary Table S3) in CF96 Real-Time System (Bio-Rad) as follows: 1 cycle of 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, and 46°C for 20 s. All expression data were normalized to the copy number of 16S rRNA in each sample.
Effect of Glucose on Bacterial Growth
The glucose content in LB medium was enzymatically detected using SBA-40D Bio-analyzer (Shandong Academy of Sciences, China). ΔsrfA was cultured at 28°C and 180 rpm in LB medium, or LB medium supplemented with glucose (4 g/L), and then the bacterial growth was observed after 24 h. ΔsrfA and WH1 were also cultured in M9 medium (5.92 mg MgSO4⋅7H2O, 0.11 mg CaCl2, 17.0 g Na2HPO4⋅12H2O, 3.0 g KH2PO4, 0.5 g NaCl, and 1.0 g NH4Cl in 1 L) with mixed sugars (0.10 g/L glucose and 3.90 g/L xylose) at 28°C and 180 rpm and then the bacterial growth was observed after 24 h.
Detecting Activity of Amylase and Glucanase
The activity of α-amylase and glucanase in WH1 and ΔsrfA was respectively determined by measuring the amount of reducing sugars released from soluble starch or glucan using alkaline DNS oxidant reagent (Kanchi et al., 2018). WH1 and ΔsrfA were cultured in LB or LB added with surfactin (80 μg/ml) at 28°C for 15 h, and then the culture was centrifuged at 12,000 rpm and 4°C for collecting the supernatant as crude enzyme solution. Crude enzyme solution (0.5 ml) was mixed with 1 ml of 2% (w/v) soluble starch or glucan (20,000 Da), incubated at 37°C for 5 min, and then the reaction was stopped by addition of 1 ml of DNS reagent. The solution was incubated in boiling water bath for 5 min to form color and then the OD540 value was detected by a spectrophotometer after dilution with water. Measurements were also performed on a blank without substrate but with crude enzyme solution, and a control without enzyme solution but with substrate at the same time. Each measurement procedure was performed three times, with measurements taken in triplicate. The enzyme activity was defined as the amount of reducing sugar produced by enzymes in 1 ml of crude enzyme solution per minute (μg/ml.min).
The α-amylase activity was also determined on LB agar plates added with starch. Briefly, the wild-type WH1 and mutants of ΔsrfA, ΔccpA (ccpA-null mutant strain), and ΔsrfAΔccpA (srfA and ccpA mutant strain) were cultured on LB agar plates containing 4 g/L of starch at 28°C for 24 h, and then the plates were stained by iodine to determine the bacterial α-amylase activity. The digested starch could not be stained by iodine showing a clear zone around bacterial colony on the plates.
Statistical Analysis
All experiments are repeated in triplicate. Data between two groups were compared by a Student t test with a significance level of ∗p < 0.05 and ∗∗p < 0.01.
Results
Lipopeptide Production in Different Strains
The genes for biosynthesis of surfactin (srfA), iturin (ituB), and fengycin (fenA) were mutated in B. amyloliquefaciens WH1, respectively (Supplementary Figure S1). We determined the lipopeptide production in these strains by MALDI-TOF (Deng, 2009). The results showed that WH1 produced surfactin (1044), iturin (1081, 1095), and fengycin (1464, 1502); ΔsrfA produced iturin (1043, 1081, 1095) and fengycin (1435, 1449, 1463, 1501); ΔituB produced surfactin (1044) and fengycin (1435, 1464, 1502); and ΔfenA produced surfactin (1044) and iturin (1081) (Supplementary Figure S2A). Thereby, the mutant strains of ΔsrfA, ΔituB, and ΔfenA lose the ability to produce surfactin, iturin, and fengycin, respectively. However, it is surfactin rather than iturin and fengycin, with a strong hemolytic activity (Shaligram and Singhal, 2010). Here, WH1 showed an obvious hemolytic activity around colony as well as ΔituB and ΔfenA, but the hemolytic activity disappeared in ΔsrfA (Supplementary Figure S2B). The result further indicated that ΔsrfA loses the ability to produce surfactin.
Mutation of srfA Led to a Defective Biofilm Formation and Growth in B. amyloliquefaciens
The colony morphology of ΔsrfA was flat and unwrinkled, very different from WH1, ΔituB, and ΔfenA (Supplementary Figure S2C). We further investigated the biofilm formation of mutant strains in LB medium, and found that both ΔituB and ΔfenA could form a robust pellicle like WH1, but ΔsrfA only produced a thin and fragile pellicle (Supplementary Figure S2D). The result suggested that surfactin is required for the biofilm formation in B. amyloliquefaciens. Unexpectedly, mutation of srfA also resulted in a significantly different growth profile from WH1, ΔituB, and ΔfenA. In 18 h, ΔsrfA showed a similar growth profile to WH1. Thereafter, it fell fast into death while WH1, ΔituB, and ΔfenA all grew well in this period (Figure 1A). WH1 could form spores as well as ΔituB and ΔfenA, but ΔsrfA could not sporulate in LB medium (Figure 1B).
FIGURE 1.
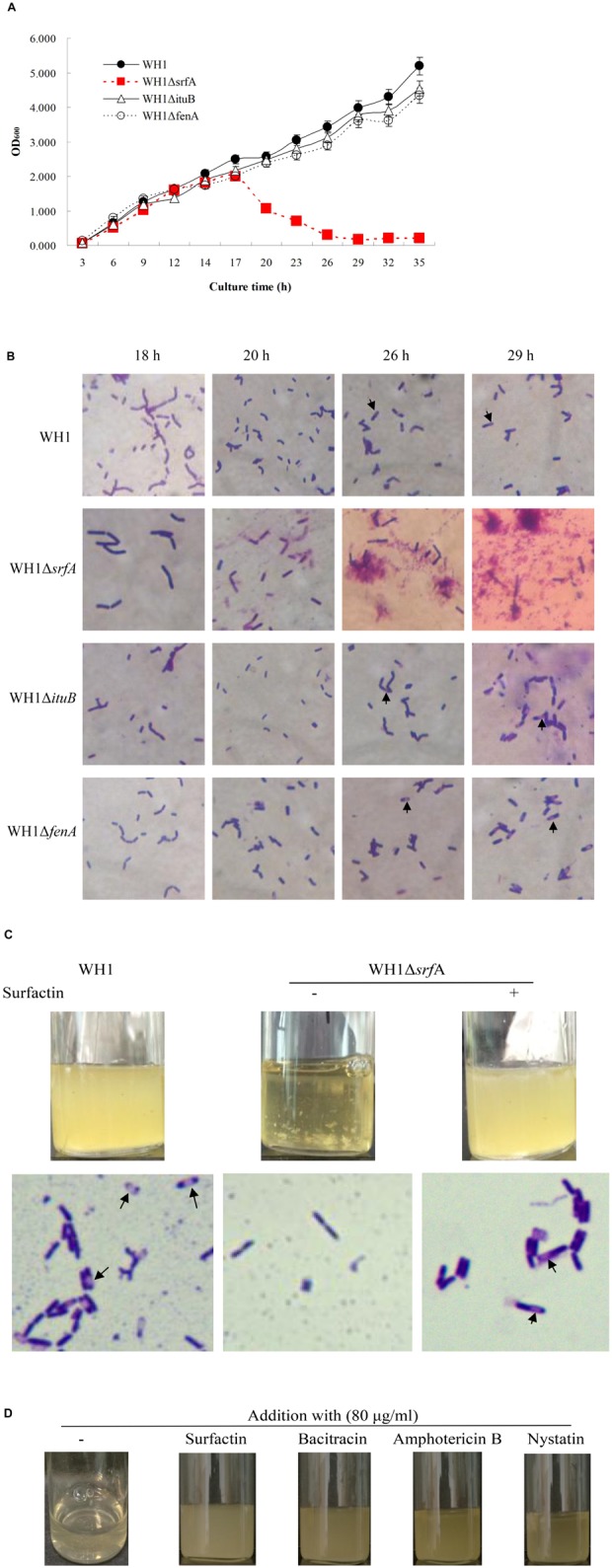
Mutation of srfA resulted in a defective bacterial growth. (A) Growth curves of WH1 and lipopeptide gene mutant strains. Mutation of srfA led to a defective bacterial growth. (B) Cells of WH1 and lipopeptide gene mutant strains (magnification 1000×). ΔsrfA died after 18 h without sporulation while other strains grew well with sporulation. Arrows indicate the spores. (C) Addition with surfactin dramatically restored the growth and sporulation in ΔsrfA (magnification 1000×). Spores are indicated by arrows. (D) Addition with bacitracin, nystatin, or amphotericin B partially restored the growth in ΔsrfA.
The defective growth of ΔsrfA could be restored by addition with surfactin in LB medium (Figure 1C). Addition with surfactin could also restore the sporulation (Figure 1C) and biofilm formation (Supplementary Figure S3) in ΔsrfA. Besides, for surfactin, other antibiotics such as bacitracin, amphotericin, and nystatin could also effectively alleviate the death of ΔsrfA due to lack of surfactin (Figure 1D). Except for Amphotericin B, both Bacitracin and Nystatin could also partially restore the biofilm formation in ΔsrfA, but were weaker than surfactin (Supplementary Figure S3).
Knockout of comA Resulted in a Similar Phenotype to ΔsrfA
ComA∼P is required for the transcription of srfA operon in B. subtilis (Gonźalez-Pastor, 2011). Here, we deleted comA in B. amyloliquefaciens. As expected, knockout of comA resulted in a similarly defective growth, biofilm formation, and colony morphology to ΔsrfA (Supplementary Figure S4). Nevertheless, the defective growth of ΔcomA could also be restored by addition with surfactin, which further verified that surfactin is very important for the growth of B. amyloliquefaciens.
Disordered Carbon Metabolism in ΔsrfA
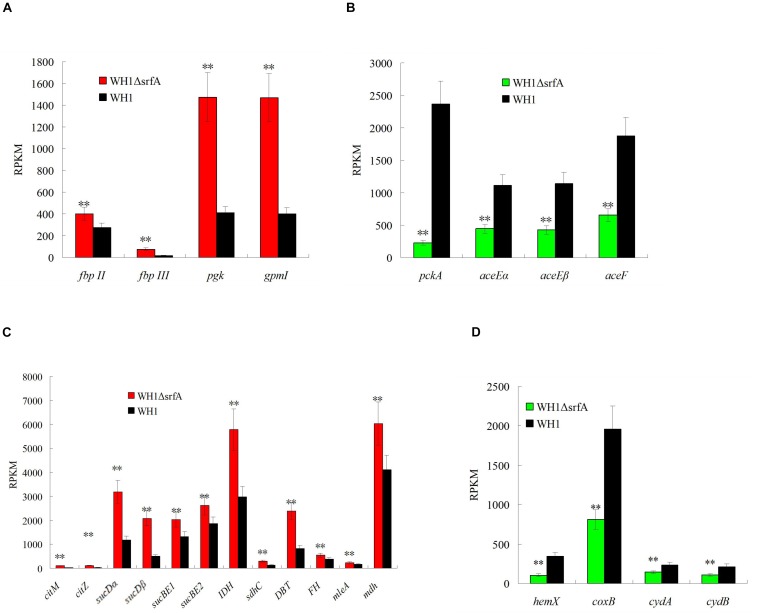
Mutation of srfA led to the cell death in B. amyloliquefaciens. For clues to explain this phenomenon, we compared the transcriptomes between ΔsrfA and WH1, and found that ΔsrfA showed a significant difference of genes transcription about cellular process, metabolic process, single organism process, and catalytic activity from WH1 (Supplementary Figure S5). Further analysis revealed that carbon metabolism was very different between WH1 and ΔsrfA. The carbon metabolism was seriously damaged in ΔsrfA. Compared to WH1, the Embden–Meyerhof pathway (EMP) that catalyzes glucose to pyruvate was enhanced in ΔsrfA. The key gene transcriptions involved in EMP such as fbp (fructose-1,6-bisphosphatase), pgk (phosphoglycerate kinase), and gpmI (2,3-bisphosphoglycerate-independent phosphoglycerate mutase) were all significantly up-regulated in ΔsrfA (Figure 2A). Pyruvate is catalyzed to acetyl-CoA and then enters into the citrate cycle (TCA). In this progress, several key gene transcriptions such as pckA (phosphoenolpyruvate carboxykinase), aceE (pyruvate dehydrogenase E1 component), and aceF (pyruvate dehydrogenase E2 component) were significantly down-regulated (Figure 2B), and the key gene transcriptions involved in TCA such as citM (citrate transporter), citZ (citrate synthase), sucD (succinyl-CoA synthetase), sucB (2-oxoglutarate dehydrogenase), IDH (isocitrate dehydrogenase), sdhC (succinate dehydrogenase cytochrome b-556 subunit), DBT (branched-chain alpha-keto acid dehydrogenase subunit E2), FH (fumarate hydratase), mleA (malate dehydrogenase), and mdh (malate dehydrogenase) were all significantly up-regulated in ΔsrfA (Figure 2C). In the TCA cycle, NADH is produced and then enters into the oxidative phosphorylation for production of ATP. In this progress, several key gene transcriptions such as hemX (cytochrome c assembly protein), coxB (cytochrome c oxidase), cydA (cytochrome d ubiquinol oxidase subunit I), and cydB (cytochrome d ubiquinol oxidase subunit II) were all significantly down-regulated in ΔsrfA (Figure 2D). Thereby, the basic carbon metabolism is disordered in ΔsrfA due to deficiency of surfactin.
FIGURE 2.
Comparison of the gene transcription involved in EMP, pyruvate dehydrogenation, TCA, and oxidative phosphorylation between ΔsrfA and WH1. (A) EMP. (B) Pyruvate dehydrogenation. (C) TCA. (D) Oxidative phosphorylation. Data were analyzed on the basis of transcriptomes. Double stars indicate significant difference (p < 0.01) between ΔsrfA and WH1.
Dissolved O2 in Culture Was Not Influenced by Surfactin
Surfactin is a powerful biosurfactant, thus possibly influences the dissolved O2 content in culture following with an effect on the cellular carbon and energy metabolism. However, the dissolved O2 content in ΔsrfA was not lower than WH1 (Supplementary Figure S6), implying there is no limitation of dissolved O2 in the culture of ΔsrfA.
Obstacle to Use Non-preferred Carbon Sources Due to Deficiency of Surfactin
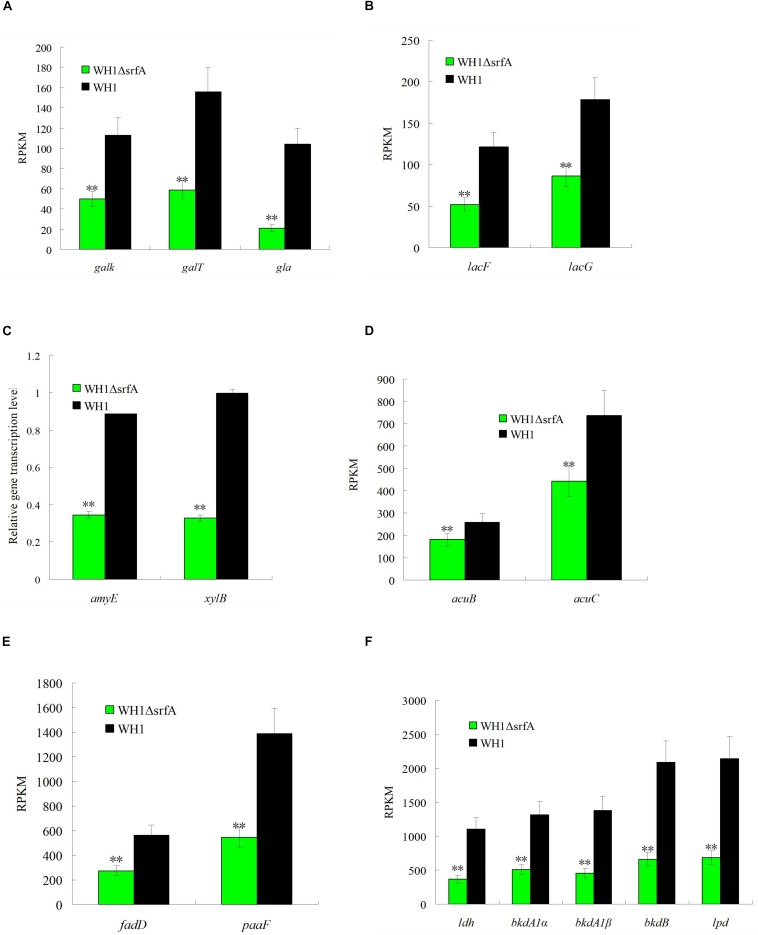
It was paradoxical that the death of ΔsrfA began at ∼18 h because the residual nutrients were still enough for growth. Thereby, we further analyzed the ability to utilize non-preferred carbon sources such as galactose, lactose, xylose, fatty acids, amino acids, acetoin, etc. in ΔsrfA. Compared to WH1, the genes transcription involved in utilization of galactose such as galK (galactokinase), galT (UDP-glucose-hexose-1-phosphate uridylyltransferase) and gla (alpha-galactosidase) were all significantly down-regulated in ΔsrfA (Figure 3A). The key genes transcription involved in lactose metabolism such as lacF (PTS system, lactose-specific IIA component) and lacG (6-phospho-beta-galactosidase) were also significantly down-regulated in ΔsrfA (Figure 3B). Other key genes involved in utilization of non-preferred carbon sources such as amyE (α-amylase) for metabolizing starch and xylB (Xylulose kinase) for utilizing xylose were both significantly down-regulated in ΔsrfA (Figure 3C). Acetoin is a product from carbon overflow metabolism, and can be used as a non-preferred carbon source in B. subtilis. The results showed the genes transcription involved in utilization of acetoin such as acetoin utilization proteins (acuB and acuC) were significantly down-regulated in ΔsrfA (Figure 3D).
FIGURE 3.
Comparison of the gene transcription involved in utilization of non-preferred carbon sources between ΔsrfA and WH1. (A) Galactose. (B) Lactose. (C) Starch and xylose. (D) Acetoin. (E) Fatty acids. (F) Branched-chain amino acids. (A), (B), (D), (E), and (F) were analyzed on the basis of transcriptomes, and (C) was analyzed by qRT-PCR. Double stars indicate significant difference (p < 0.01) between ΔsrfA and WH1.
In addition to extracellular carbon sources, the key genes transcription involved in utilizing intracellular carbon sources such as fatty acids and branched-chain amino acids were also significantly down-regulated in ΔsrfA. Compared to WH1, the transcription of genes involved in degradation of fatty acids such as long-chain acyl-CoA synthetase (ACSL/fadD) and enoyl-CoA hydratase (paaF/echA) were both significantly down-regulated in ΔsrfA (Figure 3E). The branched-chain amino acids including Ala, Val, Leu and Ile can be utilized as carbon sources in B. subtilis. The transcription of several key genes involved in degradation of branched-chain amino acids such as leucine dehydrogenase (ldh), 2-oxoisovalerate dehydrogenase E1 component alpha subunit (bkdA1) and E2 component (dihydrolipoyl transacylase) (bkdB), and dihydrolipoamide dehydrogenase (lpd/pdhD), were all significantly down-regulated in ΔsrfA (Figure 3F). All of these results indicated the ability to use non-preferred carbon sources is weakened in ΔsrfA.
High Expression of ccpA in ΔsrfA
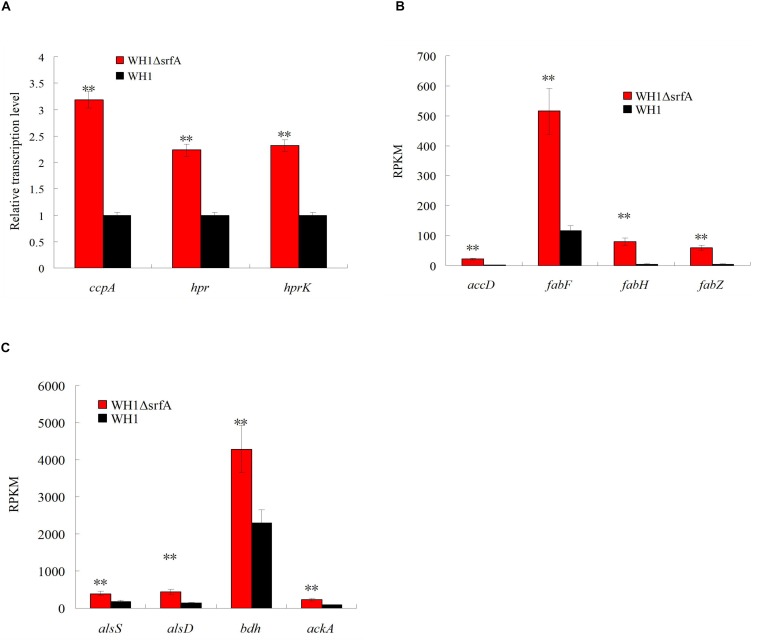
CcpA inhibits utilization of non-preferred carbon source by forming a complex with HPr∼P in B. subtilis. Analysis by qRT-PCR, we found the genes transcription of ccpA, hpr and hprK (HPr kinase) were all significantly up-regulated in ΔsrfA when compared to WH1 (Figure 4A), indicated the CcpA-mediated CCR is enhanced in this strain. CcpA also promotes the genes transcription in many carbon metabolism such as EMP, biosynthesis of fatty acids, carbon overflow. The results showed the genes transcription involved in biosynthesis of fatty acids such as accD (acetyl-CoA carboxylase carboxyl transferase), fabF (3-oxoacyl-[acyl-carrier-protein] synthase II), fabH (3-oxoacyl-[acyl-carrier-protein] synthase III), and fabZ (3-hydroxyacyl-[acyl- carrier-protein] dehydratase), were all significantly up-regulated in ΔsrfA (Figure 4B). The genes transcription involved in carbon overflow metabolism such as alsS (acetolactate synthase), alsD (acetolactate decarboxylase), bdh (2,3-butanediol dehydrogenase) and ackA (acetate kinase), were all significantly up-regulated in ΔsrfA (Figure 4C). These results all suggested that the enhanced ccpA transcription improves the genes transcription involved in bio-synthesis of fatty acids, carbon overflow metabolism and EMP (Figure 2A) in ΔsrfA.
FIGURE 4.
Comparison of the transcription of ccpA and the genes controlled by CcpA between ΔsrfA and WH1. (A) Transcription of ccpA, hpr, and hprK. (B) Transcription of genes involved in biosynthesis of fatty acids. (C) Transcription of genes involved in carbon overflow metabolism. (A) was analyzed by qRT-PCR, and (B) and (C) were analyzed on the basis of transcriptomes. Double stars indicate significant difference (p < 0.01) between ΔsrfA and WH1.
High Expression of Genes Involved in Chemotaxis and Motility in ΔsrfA
Because of the disability to produce surfactin, ΔsrfA could not use non-preferred carbon sources after glucose exhaustion. Thereby, the cells searched for a new environment with enough glucose for survival by up-regulation of the genes transcription involved in chemotaxis such as methyl-accepting chemotaxis protein (mcpA, mcpB, mcpC, tlpA, tlpB, yfmS), chemotaxis protein (cheC, cheV, and cheW), chemotaxis protein methyltransferase (cheR), sensor histidine kinase (cheA), chemotaxis response regulator (cheY) and chemoreceptor glutamine deamidase (cheD) in ΔsrfA (Supplementary Figure S7A). Also, the transcription of genes involved in motility such as swarming motility protein (swrAA, swrB, and swrC), flagellar motor protein (motA, motB, motP, and motS), flagellar capping protein (fliD), flagellar assembly protein (fliS, fliT, and fliW), flagellar motor switch protein (fliM, fliN), flagellar biosynthesis protein (fliP, fliQ, fliR, and fliZ), flagellar hook-basal body protein (fliE), flagellar basal body-associated protein (fliL), and flagellar basal-body rod protein (flhO), were all significantly up-regulated in ΔsrfA (Supplementary Figure S7B).
Weakened Enzyme Activities to Utilize Non-preferred Carbon Sources in ΔsrfA
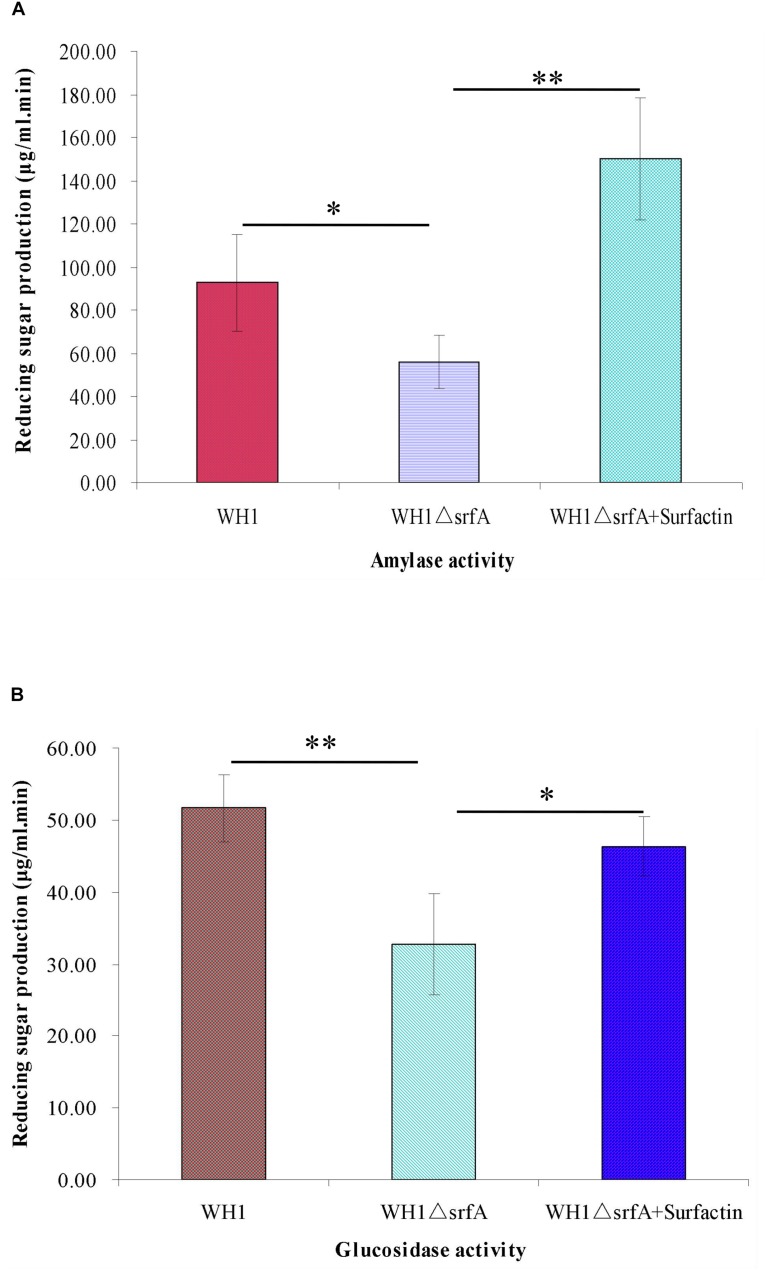
Many enzymes for utilizing non-preferred carbon sources such as amylase for utilizing starch and glucosidase for utilizing glucan, are controlled by the CcpA-mediated CCR in B. subtilis. We determined the activity of α-amylase and glucosidase in WH1 and ΔsrfA respectively, and found the α-amylase activity was significantly weakened in ΔsrfA when compared to WH1 (Figure 5A). Similarly, the glucosidase activity of ΔsrfA was also significantly lower than WH1 (Figure 5B). However, addition with surfactin could dramatically increase the activity both of α-amylase (Figure 5A) and glucosidase (Figure 5B) in ΔsrfA. The results further verified that ΔsrfA could not effectively utilize the non-preferred carbon sources such as starch and glucan after glucose exhaustion due to deficiency of surfactin.
FIGURE 5.
Comparison of the activity of α-amylase and glucanase between WH1 and ΔsrfA. (A) Amylase activity. (B) Glucanase activity. The activity of α-amylase and glucosidase was significantly weakened in ΔsrfA but could be dramatically improved by addition with surfactin. Single star and double stars indicate significant (p < 0.05) and very significant (p < 0.01) difference between two groups, respectively.
Addition With Glucose Dramatically Restoring Defective Growth in ΔsrfA
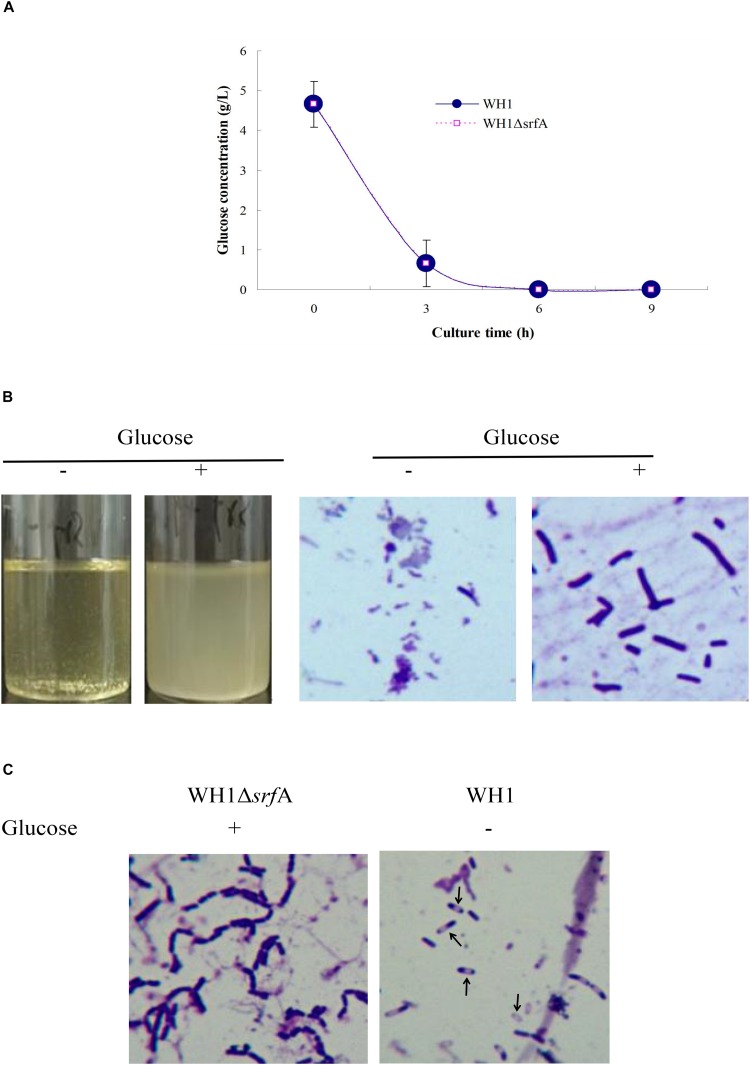
The glucose concentration was about 4.66 g/L in the fresh LB medium. ΔsrfA showed a similar profile to utilize the glucose in LB medium like WH1 in 6 h (Figure 6A). We added glucose (4 g/L) into the fresh LB medium, and found addition with glucose could dramatically alleviate the cell death in ΔsrfA due to deficiency of surfactin. Under microscope, the bacterial cells grew well in the LB medium supplemented with glucose (Figure 6B). However, addition with glucose could not restore the sporulation in ΔsrfA (Figure 6C). We further used the modified M9 medium containing a mixed carbon sources (glucose and xylose) to study whether ΔsrfA could utilize xylose after glucose exhaustion. The result showed ΔsrfA grew weakly in the M9 medium with mixed sugars when compared to WH1. After glucose exhaustion, WH1 could use xylose for growth while ΔsrfA could not due to lack of surfactin (Supplementary Figure S8).
FIGURE 6.
Addition with glucose restored the growth of ΔsrfA in LB medium. (A) Residual glucose in LB medium. Both WH1 and ΔsrfA showed a similar profile to utilize the glucose in LB medium. (B) Addition with glucose (4 g/L) dramatically restored the growth of ΔsrfA in LB medium (magnification 1000×). (C) Addition with glucose could not restore the sporulation in ΔsrfA (magnification 1000×). Spores are indicated by arrows.
Mutation of srfA Had Obviously an Effect on CcpA Dependent CCR
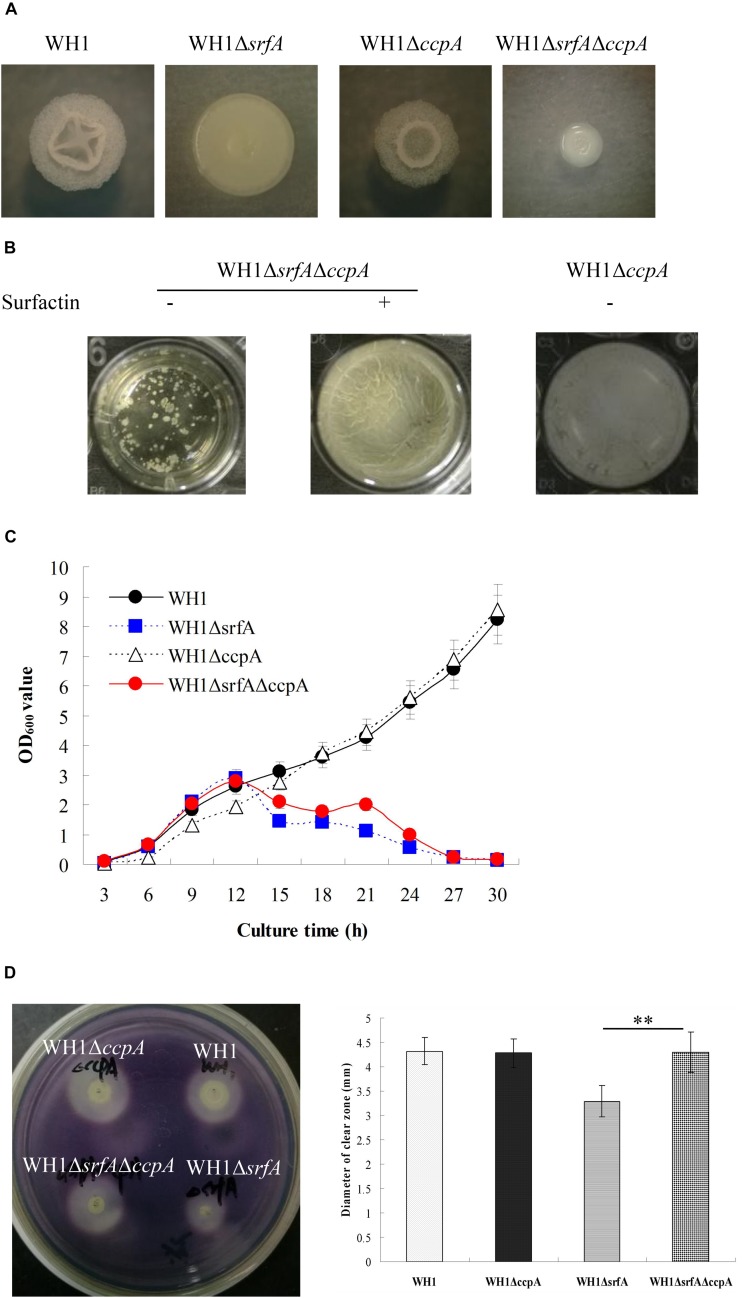
CcpA is known to repress the genes transcription involved in utilization of non-preferred carbon sources. To know whether CcpA was required for surfactin to relieve CCR, we deleted the ccpA gene in WH1 and ΔsrfA respectively (Supplementary Figures S9A,B). The colony morphology of ΔccpA was slightly different from WH1, but ΔsrfAΔccpA was very different from WH1, ΔsrfA and ΔccpA (Figure 7A). ΔsrfAΔccpA formed a dispersed and fragile float biofilm that was also very different from WH1, ΔsrfA and ΔccpA. After addition with surfactin, the defective biofilm could also be restored in ΔsrfAΔccpA (Figure 7B), suggested CcpA is not required for the role of surfactin in regulating biofilm formation.
FIGURE 7.
Mutation of srfA had obviously an effect on CcpA-dependent CCR. (A) Colony morphology of ccpA-null mutant strains. (B) Addition with surfactin restored the biofilm formation in ΔsrfAΔccpA. (C) Growth curves of ccpA-null mutant strains in LB medium. Knockout of ccpA could partially restore the growth of ΔsrfA in LB medium. (D) The α-amylase activity of wild type and mutants on LB agar plates supplemented with starch. The α-amylase activity was weakened in ΔsrfA compared to WH1, but could be restored by further knockout of ccpA in ΔsrfA. Double stars indicate significant difference (p < 0.01) between two groups.
Growth curves showed that knockout of ccpA alone did not influence the bacterial growth in LB medium, but ΔsrfAΔccpA grew better than ΔsrfA after 12 h (Figure 7C), suggesting that further knockout of ccpA is favorable for restoring the growth of ΔsrfA in LB medium. The amyE gene encoding extracellular α-amylase is subject to the CcpA-mediated CCR in B. subtilis (Fujita, 2009). Here, the α-amylase activity was weakened in ΔsrfA compared to WH1 (Figure 7D), consistent with the results in Figure 5A. However, the weakened enzyme activity could be restored by knockout of ccpA in ΔsrfA (Figure 7D). These results suggested that mutation of srfA might obviously affect the CcpA-mediated CCR in B. amyloliquefaciens.
Discussion
B. amyloliquefaciens is a close relative to and shares many characteristics with B. subtilis. Previously, we isolated a strain of B. amyloliquefaciens WH1 with production of surfactin, iturin and fengycin that are potential to be used as anti-fungal pesticides (Qi et al., 2010), and delivery system for oral delivery of protein/peptide drugs (Xing et al., 2018). For high production of single lipopeptide, we mutated the lipopeptides gene including srfA, ituB and fenA in WH1, respectively. As expected, mutation of these genes disrupted the lipopeptides production. Differently, mutation of srfA led to a defective biofilm formation and sporulation, suggesting surfactin regulates the QS response in B. amyloliquefaciens. This is agreement with the previous report that surfactin is a QS signal molecule in B. subtilis (López et al., 2009b). Unexpectedly, mutation of srfA also led to a severely defective growth that is not reported previously. Apparently, the death of ΔsrfA was not attributed to nutrients limitation because WH1 grew well during this period. In fact, the cells death was due to the deficiency of surfactin because compensation with surfactin could restore the bacterial growth in ΔsrfA. This was further verified by knockout of comA, which is essential for the srfA transcription in B. subtilis (Gonźalez-Pastor, 2011). ΔcomA showed a defective growth like ΔsrfA that could also be partially alleviated by addition with surfactin.
Surfactin has a structure consisting of a cyclic heptapeptide headgroup (Glu-Leu-D-Leu-Val-Asp-D-Leu-Leu) linked to a C13-15 β-hydroxy fatty acid by lactone bond (Zhang et al., 2017). This structure endows surfactin with a powerful biosurfactant activity by its amphiphilic nature, a polar amino acid head and a hydrocarbon chain (Carrillo et al., 2003). Thereby, surfactin is considered as a powerful biosurfactant that can cause pores on membrane by its biosurfactant activity (Shaligram and Singhal, 2010). Besides for surfactin, other antibiotics such as bacitracin, amphotericin B and nystatin could also partially restore the growth and biofilm formation in ΔsrfA. Possibly, these antibiotics seem to have some common properties with surfactin, which can substitute surfactin signaling even they are with different structures.
Addition with surfactin could also restore the biofilm formation and sporulation in ΔsrfA, consistent with the previous reports (López et al., 2009a, b, 2010; Gonźalez-Pastor, 2011). This raises another question that the death of cells is due to the defective biofilm formation and sporulation in ΔsrfA? Our data showed that besides for surfactin, glucose could also restore the growth of ΔsrfA; however, addition with glucose could not restore the sporulation in this strain. Therefore, it seems that the disability to sporulate is not the reason for the cell death of ΔsrfA. Actually, it likes that the cells had a defective carbon metabolism in ΔsrfA because the nutrients in medium were enough for growth but they still needed additional glucose for survival. This conception was further strengthened by the data of transcriptomes that revealed a severely defective carbon metabolism in ΔsrfA. As a feedback of glucose limitation, the EMP pathway was significantly enhanced in ΔsrfA (Fujita, 2009). In the progress of catalyzing pyruvate to acetyl-CoA, several key genes transcription were significantly down-regulated due to lack of pyruvate. Thereby, there was enough acetyl-CoA in cells. As a feedback to acetyl-CoA limitation, several key genes transcription in TCA were up-regulated in ΔsrfA. In the TCA cycle, NADH is produced, then enters into the oxidative phosphorylation for producing ATP (Fujita, 2009). Due to lack of NADH, several key genes transcription involved in this progress were significantly down-regulated in ΔsrfA. All of these results suggested that the basic carbon metabolism is disordered in ΔsrfA.
What is the reason for the disordered carbon metabolism in ΔsrfA? Because surfactin is one of the most effective biosurfactants discovered so far (Shaligram and Singhal, 2010), we deduced that surfactin might influence the dissolved O2 content in culture. Without enough O2, the carbon and energy metabolism will be limited in cells (Zhu et al., 2015). However, the dissolved O2 content in ΔsrfA was not lower than WH1, suggested the disordered carbon metabolism was not attributed to the limitation of dissolved O2 in ΔsrfA. In fact, the ability to utilize non-preferred carbon sources were defective in ΔsrfA because the transcription of genes involved in utilizing galactose, lactose, xylose, starch, acetoin, fatty acids and branched-chain amino acids were all significantly down-regulated in this strain. This was also consistent with the activity of α-amylase and glucosidase which were both significantly decreased in ΔsrfA but could be restored by addition with surfactin. Thereby, ΔsrfA could not utilize non-preferred carbon sources after glucose exhaustion due to deficiency of surfactin. This was further verified by the result that addition with glucose could dramatically restore the growth of ΔsrfA. Additionally, the dissolved O2 content also supported that ΔsrfA is defective to use non-preferred carbon sources. After glucose was rapidly exhausted, ΔsrfA could not use non-preferred carbon sources hence there was no carbon source to be used for consuming O2 (Zhu et al., 2015). As a result, the dissolved O2 content was increased in ΔsrfA.
Generally, the ability to use non-preferred carbon sources are controlled by CcpA in Bacillus spp. such as B. subtilis (Gorke and Stulke, 2008; Fujita, 2009). Thereby, the disability to utilize non-preferred carbon sources might be due to the enhanced repression mediated by CcpA in ΔsrfA. Indeed, the genes transcription of ccpA, hpr and hprK (Müller et al., 2006; Schumacher et al., 2011), were all significantly up-regulated in ΔsrfA due to deficiency of surfactin. Addition to repression, CcpA also promotes many genes transcription involved in carbon metabolism (Fujita, 2009; Yang et al., 2017). In ΔsrfA, several genes transcription involved in EMP, carbon overflow metabolism and biosynthesis of fatty acids were all significantly up-regulated. These results also indirectly confirmed that the CcpA-mediated CCR is enhanced in ΔsrfA. Nevertheless, the growth of ΔsrfA could be partially restored by further knockout of ccpA. Consistently, the extracellular α-amylase activity was weakened in ΔsrfA but could be restored by further knockout of ccpA. Thereby, the srfA deletion has obviously an effect on the CcpA dependent CCR in B. amyloliquefaciens.
It seems that the lack of surfactin causes not only an inability to use non-preferred carbon sources but additionally lysis of the cells. It might be expected that a deficit of nutrients would first only stop growth for a while before lysis occurs due to starvation. The srfA mutant does not stop only growth but starts to lyse immediately after 12 h. Thereby, in order to survival the cells of ΔsrfA would search for a new environment with enough glucose. Indeed, many genes transcription involved in chemotaxis and motility (Mirouze and Dubnau, 2013; van Gestel et al., 2015), were significantly up-regulated in ΔsrfA. The result is consistent with the previous report that after nutrients limitation, B. subtilis could synthesize a complex motility and chemotaxis system searching for nutrients in the environment (Hamoen et al., 2003).
Although both of surfactin and glucose could restore the growth in ΔsrfA, it was only surfactin rather than glucose with an ability to restore the sporulation in this strain. Addition with glucose only supplemented a preferred carbon source for ΔsrfA while addition with surfactin could trigger a signaling pathway to relieve CCR in this mutant strain. The signaling triggered by surfactin could not only relieve CCR to utilize non-preferred carbon sources, but also regulate the QS response such as biofilm formation and sporulation in B. amyloliquefaciens (López et al., 2009a, 2010; Vlamakis et al., 2013; Xu et al., 2013; Zeriouh et al., 2014).
Surfactin is a QS molecule in B. subtilis (López et al., 2010), by which the cells undergo several adaptive responses to nutrients depletion and high cell density in the environment (Ohsawa et al., 2006). In B. subtilis, prolonged nutritional stress can result in the development of QS response such as biofilm formation and sporulation (Hamoen et al., 2003). Here, the preferred carbon sources such as glucose were exhausted that acts as a nutritional stress to induce production of surfactin triggering the QS response in B. amyloliquefaciens. Possibly, the signaling triggered by surfactin has either a direct or an indirect effect on HPrSer46P dephosphorylation by HPrK/P (Mijakovic et al., 2002; Poncet et al., 2004; Fujita, 2009). For example, it can cause low intracellular Pi levels or it reduces somehow HPrK/P phospho-phosphorylase activity, but further studies are needed for verifying this conception.
By surfactin, QS tightly links with carbon metabolism to counter nutrients limitation in the natural environment. Generally, the available carbon sources are composed of diverse sugars in the complex environment (Li et al., 2014), and the preferred carbon sources (e.g., glucose) are preferentially utilized by cells for a fast growth (Müller et al., 2006). After exhaustion, the cells produce surfactin to relieve CCR utilizing the non-preferred sugars. Thereby, as a signal molecule to regulate QS, surfactin also directly or indirectly relieves CCR to utilize non-preferred carbon sources countering nutrients limitation (e.g., glucose deprivation) in the environment. In conclusion, our findings provide the first evidence that the QS signal molecule of surfactin is also involved in reliving the CcpA-mediated CCR in B. amyloliquefaciens.
Data Availability Statement
The raw data were uploaded to SRA database and the BioProject ID is PRJNA560470.
Author Contributions
GQ and XZ conceived, designed, and coordinated the study, and drafted the manuscript. BC, JW, and JD constructed mutant strains, determined phenotype, and performed biochemical studies. BC determined lipopeptides. JD and JW analyzed transcriptomes. All authors analyzed the data, provided critical input into the final manuscript, and approved the final version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. Project 31870030 supported by the National Natural Science Foundation of China.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00631/full#supplementary-material
References
- Ansaldi M., Dubnau D. (2004). Diversifying selection at the Bacillus quorum-sensing locus and determinants of modification specificity during synthesis of the ComX pheromone. J. Bacteriol. 186 15–21. 10.1128/jb.186.1.15-21.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansaldi M., Marolt D., Stebe T., Mandic-Mulec I., Dubnau D. (2002). Specific activation of the Bacillus quorum-sensing systems by isoprenylated pheromone variants. Mol. Microbiol. 44 1561–1573. 10.1046/j.1365-2958.2002.02977.x [DOI] [PubMed] [Google Scholar]
- Banse A. V., Hobbs E. C., Losick R. (2011). Phosphorylation of Spo0A by the histidine kinase KinD requires the lipoprotein Med in Bacillus subtilis. J. Bacteriol. 193 3949–3955. 10.1128/JB.05199-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendori S. O., Pollak S., Hizi D., Eldar A. (2015). The RapP-PhrP quorum-sensing system of Bacillus subtilis strain NCIB3610 affects biofilm formation through multiple targets, due to an atypical signal-insensitive allele of RapP. J. Bacteriol. 197 592–602. 10.1128/JB.02382-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo C., Teruel J. A., Aranda F. J., Ortiz A. (2003). Molecular mechanism of membrane permeabilization by the peptide antibiotic surfactin. Biochim. Biophys. Acta 1611 91–97. 10.1016/s0005-2736(03)00029-4 [DOI] [PubMed] [Google Scholar]
- Charbonnier T., Le Coq D., McGovern S., Calabre M., Delumeau O., Aymerich S., et al. (2017). Molecular and physiological logics of the pyruvate-induced response of a novel transporter in Bacillus subtilis. mBio 8:e00976-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong I. G., Veening J. W., Kuipers O. P. (2012). Single cell analysis of gene expression patterns during carbon starvation in Bacillus subtilis reveals large phenotypic variation. Environ. Microbiol. 14 3110–3121. 10.1111/j.1462-2920.2012.02892.x [DOI] [PubMed] [Google Scholar]
- Deng J. (2009). Characterization of the Antifungal Substances Produced by Bacillus Amyloliquefaciencs Strain YN-1. dissertation/master’s thesis in Chinese, Huazhong Agricultural University, Wuhan. [Google Scholar]
- Even-Tov E., Omer Bendori S., Valastyan J., Ke X., Pollak S., Bareia T., et al. (2016). Social evolution selects for redundancy in bacterial quorum sensing. PLoS Biol. 14:e1002386. 10.1371/journal.pbio.1002386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y. (2009). Carbon catabolite control of the metabolic network in Bacillus subtilis. Biosci. Biotechnol. Biochem. 73 245–259. 10.1271/bbb.80479 [DOI] [PubMed] [Google Scholar]
- Gonźalez-Pastor J. E. (2011). Cannibalism: a social behavior in sporulating Bacillus subtilis. FEMS Microbiol. Rev. 35 415–424. 10.1111/j.1574-6976.2010.00253.x [DOI] [PubMed] [Google Scholar]
- Gorke B., Stulke J. (2008). Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 6 613–624. 10.1038/nrmicro1932 [DOI] [PubMed] [Google Scholar]
- Hamoen L. W., Venema G., Kuipers O. P. (2003). Controlling competence in Bacillus subtilis: shared use of regulators. Microbiology 149 9–17. 10.1099/mic.0.26003-0 [DOI] [PubMed] [Google Scholar]
- Ishii H., Tanaka T., Ogura M. (2013). The Bacillus subtilis response regulator gene degU is positively regulated by CcpA and by catabolite-repressed synthesis of ClpC. J. Bacteriol. 195 193–201. 10.1128/JB.01881-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanchi S., Sabela M. I., Mdluli P. S., Bisetty K. (2018). Smartphone based bioanalytical and diagnosis applications: a review. Biosens. Bioelectron. 102 136–149. 10.1016/j.bios.2017.11.021 [DOI] [PubMed] [Google Scholar]
- Kim J. N., Burne R. A. (2017). CcpA and CodY Coordinate acetate metabolism in Streptococcus mutans. Appl. Environ. Microbiol. 83:e03274-16. 10.1128/AEM.03274-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koumoutsi A., Chen X. H., Henne A., Liesegang H., Hitzeroth G., Franke P., et al. (2004). Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens Strain FZB42. J. Bacteriol. 186 1084–1096. 10.1128/jb.186.4.1084-1096.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kröber M., Verwaaijen B., Wibberg D., Winkler A., Pühler A., Schlüter A. (2016). Comparative transcriptome analysis of the biocontrol strain Bacillus amyloliquefaciens FZB42 as response to biofilm formation analyzed by RNA sequencing. J. Biotechnol. 231 212–223. 10.1016/j.jbiotec.2016.06.013 [DOI] [PubMed] [Google Scholar]
- Li L., Li K., Wang K., Chen C., Gao C., Ma C., et al. (2014). Efficient production of 2,3-butanediol from corn stover hydrolysate by using a thermophilic Bacillus licheniformis strain. Bioresour. Technol. 170 256–261. 10.1016/j.biortech.2014.07.101 [DOI] [PubMed] [Google Scholar]
- López D., Vlamakis H., Kolter R. (2010). Biofilms. Cold Spring Harb. Perspect. Biol. 2:a000398. 10.1101/cshperspect.a000398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López D., Fischbach M. A., Chu F., Losick R., Kolter R. (2009a). Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 106 280–285. 10.1073/pnas.0810940106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López D., Vlamakis H., Losick R., Kolter R. (2009b). Paracrine signaling in a bacterium. Gene Dev. 23 1631–1638. 10.1101/gad.1813709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak B. C., Pabijaniak M., de Jong A., Dûhring R., Seidel G., Hillen W., et al. (2012). High- and low-affinity cre boxes for CcpA binding in Bacillus subtilis revealed by genome-wide analysis. BMC Genomics 13:401. 10.1186/1471-2164-13-401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijakovic I., Poncet S., Galinier A., Monedero V., Fieulaine S., Janin J., et al. (2002). Pyrophosphate-producing protein dephosphorylation by HPr kinase/phosphorylase: a relic of early life? Proc. Natl. Acad. Sci. U.S.A. 99 13442–13447. 10.1073/pnas.212410399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirouze N., Dubnau D. (2013). Chance and necessity in Bacillus subtilis development. Microbiol. Spectrum 1:TBS-0004-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller W., Horstmann N., Hillen W., Sticht H. (2006). The transcription regulator RbsR represents a novel interaction partner of the phosphoprotein HPr-Ser46-P in Bacillus subtilis. FEBS J. 273 1251–1261. 10.1111/j.1742-4658.2006.05148.x [DOI] [PubMed] [Google Scholar]
- Ohsawa T., Tsukahara K., Sato T., Ogura M. (2006). Superoxide stress decreases expression of srfA through inhibition of transcription of the comQXP quorum-sensing locus in Bacillus subtilis. J. Biochem. 139 203–211. 10.1093/jb/mvj023 [DOI] [PubMed] [Google Scholar]
- Oslizlo A., Stefanic P., Dogsa I., Mandic-Mulec I. (2014). Private link between signal and response in Bacillus subtilis quorum sensing. Proc. Natl. Acad. Sci. U.S.A. 111 1586–1591. 10.1073/pnas.1316283111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak S., Omer-Bendoria S., Even-Tova E., Lipsmana V., Bareiaa T., Ben-Ziona I., et al. (2016). Facultative cheating supports the coexistence of diverse quorum-sensing alleles. Proc. Natl Acad. Sci. U.S.A. 113 2152–2157. 10.1073/pnas.1520615113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poncet S., Mijakovic I., Nessler S., Gueguen-Chaignon V., Chaptal V., Galinier A., et al. (2004). HPr kinase/phosphorylase, a Walker motif A-containing bifunctional sensor enzyme controlling catabolite repression in Gram-positive bacteria. Biochim. Biophys. Acta 1697 123–135. 10.1016/j.bbapap.2003.11.018 [DOI] [PubMed] [Google Scholar]
- Qi G., Kang Y., Li L., Xiao A., Zhang S., Wen Z., et al. (2014). Deletion of meso-2,3-butanediol dehydrogenase gene budC for enhanced D-2,3-butanediol production in Bacillus licheniformis. Biotechnol. Biofuels 7:16. 10.1186/1754-6834-7-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi G., Zhu F., Du P., Yang X., Qiu D., Yu Z., et al. (2010). Lipopeptide induces apoptosis in fungal cells by a mitochondria-dependent pathway. Peptides 31 1978–1986. 10.1016/j.peptides.2010.08.003 [DOI] [PubMed] [Google Scholar]
- Schneider K. B., Palmer T. M., Grossman A. D. (2002). Characterization of comQ and comX, two genes required for production of ComX pheromone in Bacillus subtilis. J. Bacteriol. 184 410–419. 10.1128/jb.184.2.410-419.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher M. A., Sprehe M., Bartholomae M., Hillen W., Brennan R. G. (2011). Structures of carbon catabolite protein A – (HPr-Ser46-P) bound to diverse catabolite response element sites reveal the basis for high-affinity binding to degenerate DNA operators. Nucleic Acids Res. 39 2931–2942. 10.1093/nar/gkq1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaligram N. S., Singhal R. S. (2010). Surfactin-A review on biosynthesis, fermentation, purification and applications. Food Technol. Biotechnol. 48 119–134. [Google Scholar]
- Stefanic P., Mandic-Mulec I. (2009). Social interactions and distribution of Bacillus subtilis pherotypes at microscale. J. Bacteriol. 191 1756–1764. 10.1128/JB.01290-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran L. S., Nagai T., Itoh Y. (2000). Divergent structure of the ComQXPA quorum-sensing components: molecular basis of strain-specific communication mechanism in Bacillus subtilis. Mol. Microbiol. 37 1159–1171. 10.1046/j.1365-2958.2000.02069.x [DOI] [PubMed] [Google Scholar]
- van Gestel J., Vlamakis H., Kolter R. (2015). From cell differentiation to cell collectives: Bacillus subtilis uses division of labor to migrate. PLoS Biol. 13:e1002141. 10.1371/journal.pbio.1002141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlamakis H., Chai Y., Beauregard P., Losick R., Kolter R. (2013). Sticking together: building a biofilm the Bacillus subtilis way. Nat. Rev. Microbiol. 11 157–168. 10.1038/nrmicro2960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing X., Zhao X., Ding J., Liu D., Qi G. (2018). Enteric-coated insulin microparticles delivered by lipopeptides of iturin and surfactin. Drug Deliv. 25 23–34. 10.1080/10717544.2017.1413443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Shao J., Li B., Yan X., Shen Q., Zhang R. (2013). Contribution of Bacillomycin D in Bacillus amyloliquefaciens SQR9 to antifungal activity and biofilm formation. Appl. Environ. Microbiol. 79 808–815. 10.1128/AEM.02645-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Wu H. J., Lin L., Zhu Q. Q., Borriss R., Gao X. W. (2015). A plasmid-born Rap-Phr system regulates surfactin production, sporulation and genetic competence in the heterologous host, Bacillus subtilis OKB105. Appl. Microbiol. Biotechnol. 99 7241–7252. 10.1007/s00253-015-6604-3 [DOI] [PubMed] [Google Scholar]
- Yang Y., Zhang L., Huang H., Yang C., Yang S., Gu Y., et al. (2017). A flexible binding site architecture provides new insights into CcpA global regulation in Gram-positive bacteria. mBio 8:e02004-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeriouh H., de Vicente A., Pérez-García A., Romero D. (2014). Surfactin triggers biofilm formation of Bacillus subtilis in melon phylloplane and contributes to the biocontrol activity. Environ. Microbiol. 16 2196–2211. 10.1111/1462-2920.12271 [DOI] [PubMed] [Google Scholar]
- Zhang L., Xing X., Ding J., Zhao X., Qi G. (2017). Surfactin variants for intra-intestinal delivery of insulin. Eur. J. Pharm. Biopharm. 115 218–228. 10.1016/j.ejpb.2017.03.005 [DOI] [PubMed] [Google Scholar]
- Zhu Y., Liu Y., Li J., Shin H. D., Du G., Liu L., et al. (2015). An optimal glucose feeding strategy integrated with step-wise regulation of the dissolved oxygen level improves N-acetylglucosamine production in recombinant Bacillus subtilis. Bioresour. Technol. 177 387–392. 10.1016/j.biortech.2014.11.055 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data were uploaded to SRA database and the BioProject ID is PRJNA560470.