Dear Editor,
Hospitals around the world are innovating their processes to separate COVID-19 patients from non-COVID-19 patients. Intensive care unit (ICU) capacity for COVID-19 patients has been expanded by converting post-anaesthesia care units (PACUs) into ICU, medical tents, deployment of care providers from OR to ICU, and numerous other innovations across geographies. Further capacity has been enabled by rescheduling non-urgent elective surgeries or, more generally, from learnings from prior reports following an influenza epidemic [1].
Epidemic models that describe how to ‘flatten the curve’ with public health initiatives are focused on the demand for care services. We have developed a simple patient simulation model (called icu-covid-sim) which explores the supply side: it supports hospital and regional planning decisions to ‘raise the bar’ with ICU capacity expansion activities and have deployed it on the web for others to use. The need for such a tool customized to support local and regional decision-making is highlighted by the high variability of critical care bed numbers across geographies [2].
The icu-covid-sim tool uses queueing theory and patient flow simulations [3]. It describes the maximum rate of COVID-19 patients which can be handled for a given number of ICU beds dedicated to COVID-19 patients, as a function of the arrival rate of COVID-19 patients, their length of stay (LOS) distribution, and the number of ICU beds dedicated to COVID-19 patients.
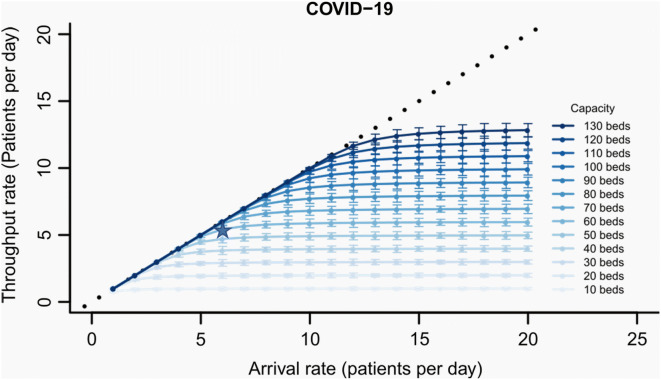
We used the tool in analysing the acceptable flow rates at the Amsterdam UMC (location AMC). Figure 1 shows the output for a range of ICU bed sizes for COVID-19 patients. The maximum average throughput of COVID-19 patients might be less than the average arrival rate due to bed blocking associated with randomness in the arrival times and LOS of each patient. From the figure, for example, we can see that with 60 COVID-19 ICU beds, approximately 5.5 COVID-19 patients/day can make it through if 6.0 such patients/day arrive. The other patients would need to be referred to another facility due to bed blocking, at that capacity.
Fig. 1.
Average throughput rate in long term of COVID patients per day as a function of the arrival rate and bed capacity. Throughput is the number of patients per day that can go through the system. Patient throughput rates would equal the patient arrival rates (the diagonal dotted line) if all patients were to be admitted to the ICU but is less than the arrival rate due to bed blocking. The star represents an example under 60 available beds where approximately 5.424 COVID-19 patients/day can go through if 6.0 such patients/day arrive. The bars represent stochastic variation over a simulated 3-month duration of a pandemic
Supplementary material [4] provides additional background for the conceptual model and analysis, and web links below provide source code, a user guide, and a publicly available web-based implementation of the model. Analysis also includes graphs with the percent utilization of those beds. Similar statistics are provided for non-COVID ICU bed capacity decisions. The graphs were computed assuming a LOS distributed for COVID-19 patients from Wuhan data [5], and with Amsterdam ICU data for non-COVIDs. The tool allows for hospitals to enter their own arrival rate and LOS distributions.
At a hospital level, the tool can inform decisions about how many COVID-19 patients can be handled. At a regional level, one might envisage using the tools for regional hospitals to compute how many referrals to a central hospital might be anticipated, for any given demand level for COVID-19 ICU resources. The icu-covid-sim tool is deployed at https://andres-alban.shinyapps.io/icu-covid-sim/ and source code and further modelling and usage details are available at https://github.com/sechick/icu-covid-sim/. See also [4].
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
AA, SEC, and WJW acknowledge funding from the European Union’s Horizon 2020 Research and Innovation Program under the Marie Skłodowska-Curie ESA-ITN Grant Agreement No 676129.
Study Group Collaborating Authors:
Alexander F. van der Sluijs, MD, Department of Intensive Care Medicine, Amsterdam UMC (location AMC), University of Amsterdam, Amsterdam, The Netherlands.
W. Joost Wiersinga, MD, PhD, MBA, Department of Intensive Care Medicine and Department of Medicine, Division of Infectious Diseases, Amsterdam UMC (location AMC), University of Amsterdam, Amsterdam, The Netherlands.
Author’s contribution
AA, SEC, and DS developed mathematical model. AA ran computer experiments. SEC was involved in the first draft of manuscript. WJW, DAD, AFvdS, WJW, APJ Vlaar provided medical expertise, anonymised statistical data. All were involved in final editing.
Funding
The study funders/sponsors had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Compliance with ethical standards
Conflicts of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Footnotes
The Study Group are listed in the acknowledgements section.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Andres Alban, Email: andres.alban@insead.edu.
Stephen E. Chick, Email: stephen.chick@insead.edu
Dave A. Dongelmans, Email: d.a.dongelmans@amsterdamumc.nl
Alexander P. J. Vlaar, Email: a.p.vlaar@amsterdamumc.nl
Danielle Sent, Email: d.sent@amsterdamumc.nl.
Study Group:
References
- 1.Sprung CL, Zimmerman JL, Christian MD, et al. Recommendations for intensive care unit and hospital preparations for an influenza epidemic or mass disaster: summary report of the European Society of Intensive Care Medicine’s Task Force for intensive care unit triage during an influenza epidemic or mass disaster. Intensive Care Med. 2010;36:428–443. doi: 10.1007/s00134-010-1759-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rhodes A, Ferdinande P, Flaatten H, et al. The variability of critical care bed numbers in Europe. Intensive Care Med. 2012;38:1647–1653. doi: 10.1007/s00134-012-2627-8. [DOI] [PubMed] [Google Scholar]
- 3.Law AM, Kelton WD. Simulation modeling and analysis. 4. New York: McGraw Hill; 2007. [Google Scholar]
- 4.Alban A, Chick SE, Dongelmans DA, Van der Sluijs AF, Wiersinga WJ, Vlaar APJ, Sent D (2020) ICU capacity management during the COVID-19 pandemic using a stochastic process simulation. https://ssrn.com/abstract=3570406, source code at https://github.com/sechick/icu-covid-sim/ [DOI] [PMC free article] [PubMed]
- 5.Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.