Abstract
The development of native mass spectrometry (MS) has provided structural biologists an additional tool to probe the structures of large macromolecular systems. Surface-induced dissociation (SID) is one activation method used within tandem MS experiments that has proven useful in interrogating the connectivity and topology of biologically-relevant protein complexes. We present here the use of a tilted surface and ion carpet array within a new SID device design, enabling decreased dimensions along the ion path and fewer lenses to tune. This device works well in fragmenting ions of both low (peptides) and high (protein complexes) m/z. Results show that the ion carpet array, while enabling simplification of the back-end of the device, has deficiencies in product collection and subsequently signal at higher SID energies when fragmenting protein complexes. However, the use of the tilted surface was advantageous as it was an effective way to shorten the device and reduce the number of independent voltages.
Graphical abstract
INTRODUCTION
Evaluating the architecture of macromolecular complexes is key to understanding how they function and malfunction in biological processes.1 While there are many tools such as NMR, cryo-EM, and X-ray crystallography that structural biologists can utilize to gain information about the structures of these molecules, native mass spectrometry (MS) has evolved as a complementary technique. Native MS utilizes soft ionization techniques to enable the transfer of these macromolecules into the gas phase while retaining their noncovalent interactions and preserving the folded, native-like structure (e.g., kinetically trapping a solution-like structure with interfaces intact). Once the macromolecular complex is in the gas phase, MS is capable of providing details about molecular weight, stoichiometry, and ligand binding.2,3 A wide range of activation methods have been utilized within tandem MS to probe the substructures of protein complexes, the most common activation method being collision-induced dissociation (CID). CID involves accelerating ions through a neutral background gas; the ions undergo a stepwise build-up of internal energy, which typically culminates in restructuring of the complex and subsequent ejection of an unfolded, highly-charged monomer, leaving behind its complementary (n-1)mer.4 Despite the utility of CID (and its variant collision-induced unfolding) in elucidating stoichiometric and gas-phase stability information,5 this restructuring can lead to a loss in information about the connectivity between subunits.
Surface-induced dissociation (SID) has demonstrated utility in elucidating the substructure of noncovalent protein complexes.3,6 SID involves accelerating ions into a rigid surface. Although computational modeling of the SID process has been performed for molecules up to the size of small peptides and C60, computational models of SID do not exist for large protein complexes.7–9 It has been shown experimentally that the interaction time at the surface is short (picoseconds for ions below m/z 300),10 that energy deposition depends on the softness/hardness of the surface,8,9,11 and that higher-mass projectiles require higher acceleration energies to induce dissociation.12 Additionally, SID of protein complexes cleaves weaker interfaces at lower energies producing subcomplexes that are indicative of the connectivity and topology of the intact protein complex,13,14 that charge is distributed symmetrically upon SID of homo-oligomers,6 and that ligands can be retained in binding pockets or at surfaces by SID,3 in contrast to the restructuring and asymmetrical charging that is typical for multi-step CID.4 Because SID is an invaluable tool in native MS, we strive to develop improved/alternative devices so that SID can be easily utilized by a broader audience.
The ability to efficiently manipulate ions within a mass spectrometer is key to developing tools such as SID. A challenge in the case of in-line SID is the requirement to radially focus and bring ions back on-axis after the surface collision. Currently, this is accomplished using a series of independently-controlled DC lenses without the use of RF for radial confinement. The Jarrold group has shown that an “ion carpet” array can be utilized in both DC-only and DC+RF configurations for radial confinement of an ion beam following a drift region.15 An ion carpet array consists of a printed circuit board with concentric ring electrodes that are resistively linked to one another in series to create an effective “planar funnel,” guiding ions towards the center aperture opening. Here, we present the use of a DC-only ion carpet array to collect product fragments following surface collisions of small peptides and protein complexes within an SID device. Our current devices were optimized for native mass spectrometry applications. A goal in development of this new device was to ensure simple operation/similar tuning across a wide mass range to accommodate those groups that cannot fully dedicate an instrument to native MS.
EXPERIMENTAL
Sample preparation & ionization
Peptides and protein complexes that have been studied with SID on multiple platforms were chosen to demonstrate the efficacy of the tilted-surface/ion carpet SID design.3,6,14,16 Details regarding analyte sources and preparation are provided in the Supplementary Information. Ions were generated via nanoelectrospray ionization using borosilicate capillaries pulled in-house using a micropipette tip puller (Sutter Instruments model P-97, Novato, CA, USA).
SID device design & simulations
The tilted-surface/ion carpet SID device (Figure 1) was designed using Autodesk Inventor and the performance optimized using SIMION 8.1. Additional parameters and the corresponding Lua user program are outlined in the Supplementary Information. Collisions with background gas in the SID device were not modeled.
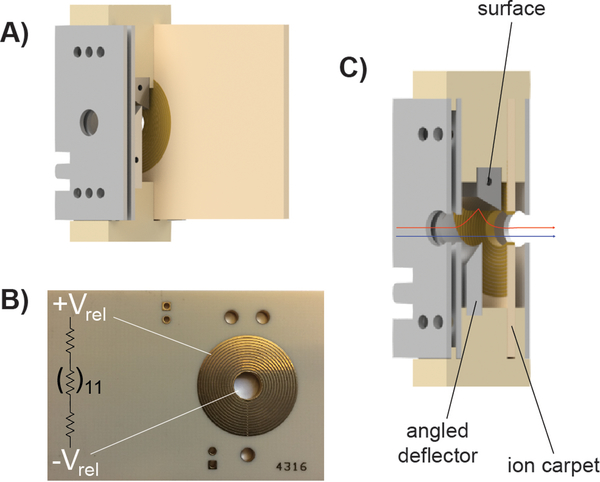
Figure 1.
A) Computer-aided drawing (CAD) of the tilted-surface/ion carpet SID device. B) Image of the ion carpet printed circuit board with a representation of the resistively linked (in series) concentric ring electrodes. C) Technical drawing of the device with a cutaway in which the blue arrow shows the ion path when the device is tuned for transmission/no ion activation. The red arrow indicates a stylized view of the ion path when the device is tuned for surface collision.
The ion carpet consists of a printed circuit board containing 14 concentric ring electrodes with 0.13 mm spacing between each ring and a 5 mm exit aperture. These electrodes are resistively linked to one another in series (Figure S1) allowing for a voltage gradient to form when the outermost and innermost ring are supplied with external DC. The ion carpet was fabricated by Advanced Circuits (Aurora, CO, USA) using a ceramic base (Rogers 4003)15 and gold-plated copper electrodes. The remaining portion of the SID device was fabricated by the OSU Arts & Sciences machine shop. The top and bottom brackets, which hold all electrodes and the ion carpet in place, were constructed from polyether ether ketone (PEEK). The surface electrode was made from polished stainless steel and the remaining electrodes were fabricated from aluminum.
Comparison experiments utilized a 10-lens SID device design described previously.14,17 A visual comparison between the two SID devices is shown in Figure S2. Because the goal of this work was to compare device designs but not evaluate surface material effects, experiments directly comparing the two device geometries with peptides and protein complexes utilized a polished stainless-steel surface in each device. While previous studies have utilized a fluorinated (FC12) self-assembled monolayer on a gold surface on a glass substrate,13 the size of the surface within the tilted-surface/ion carpet device is challenging to modify in this manner without components that could introduce fringe field effects (while maintaining electrical connectivity with the gold/glass surface plate); an alternative would be gold plating but this fell outside of the scope of the goals of the current work. While we still prefer the use of the fluorinated SAM surface material, its effect on large multiply-charged ions such as the peptides and proteins within this work is less significant. Thus, all experiments displayed here utilized a stainless-steel surface to test device geometry alone. DC voltages were applied to all electrodes using an external power supply controlled by Tempus software (Ardara Technologies, Ardara, PA, USA).
Instrumentation
All experiments were performed on a Synapt G2 Q-IM-TOF mass spectrometer (Waters Corporation, Wilmslow, UK). The stacked ring ion guide trap region was modified for each device to allow the SID device to be inserted between the trap and ion mobility (IM) region of the instrument (Figure S3). Each device can be tuned to allow ions to pass through without activation, enabling the original commercial functionality of the instrument, or tuned to guide ions towards the surface for collision. Tuning of both devices was accomplished by beginning with SIMION simulation-determined tune settings and then further adjusted experimentally to optimize for ion signal while minimizing harsh acceleration potentials that could cause unnecessary activation from ion-neutral(gas) collisions (Figure S4).
RESULTS/DISCUSSION
SID device simplification
SID has been previously implemented on Q-TOF, ICR, and Orbitrap platforms.14,16–18 The Q-TOF SID design has been installed in several laboratories.19,20 It is clear to us and other users that a device with fewer lenses to tune may improve ease-of-use and adaptation to additional laboratories. Additionally, it may be necessary to decrease the size of the SID device to maintain original functionality of the mass spectrometer when implementing SID on additional commercial instruments.18 Therefore, our main objectives within this work were to design a shorter SID device with fewer electrodes to enable simple operation and, eventually, automation. Both objectives have been accomplished by utilizing an ion carpet array to effectively replace the back half of the original design and shortening the front-end optics by using an angled surface. In making these changes, the number of independently-controlled voltages was decreased from 10 to 7 and the dimension of the device along the ion path was reduced from 3.0 cm to 1.6 cm.
Performance of the tilted-surface/ion carpet device
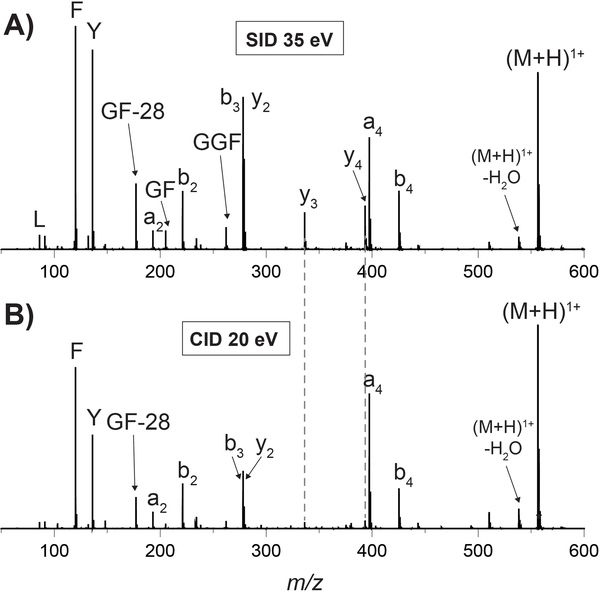
To evaluate the performance of these two major changes to the SID device design, the new device was compared with the previous 10-lens device in fragmenting multiple model peptides and protein complexes. SID energy (ΔV) is defined as the difference in potential between the trap exit and SID surface (see details in the Supplementary Information). Previous papers have studied the efficiency of energy deposition with SID, reporting that 7 to >20% of translational energy in the precursor ion is converted into internal energy with SID, though this depends on both surface material and ion identity.8–10,12,17,21,22 Leucine enkephalin was fragmented with the tilted-surface/ion carpet SID device (Figure 2). The higher abundance y-ions in SID (better b/y complementary ion ratios) are attributed to CID being a multiple-collision technique that causes secondary (and higher) fragmentation of primary fragment ions. These results align well with previously-published work.17 Fragmentation of peptides (Fibrinopeptide A shown in Figure S5) using this new device was successful (straightforward tuning and sufficient signal) across a broad energy range.
Figure 2.
Spectra of leucine enkephalin (YGGFL) monomer fragmented with A) 35 eV SID using the tilted-surface/ion carpet SID device and B) 20 eV CID in the trap cell. Because of the different effective target masses, collision energies were chosen to ~match precursor reduction rather than showing the same collision energy. SID experiments required analyte concentrations equal to and acquisition times comparable to CID. Additional energies and a comparison to the original 10-lens device are shown in Figures S6 and S7.
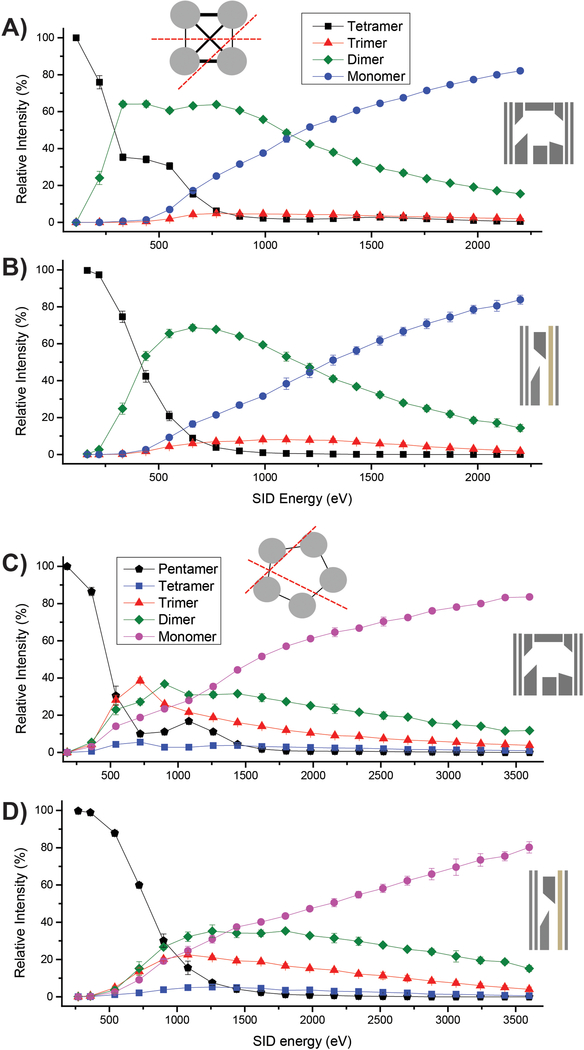
When fragmenting protein complexes, the combination of IM following SID enables determination of the relative intensities of each subcomplex or subunit at a specific energy without the requirement of isotopic resolution. The relative abundance of each subcomplex can be shown as a function of SID energy in energy-resolved mass spectrometry (ERMS) plots, as displayed for streptavidin (53 kDa homotetramer) and C-reactive protein (CRP; 115 kDa homopentamer) in Figure 3. The definition of ΔV for SID energy remained constant when comparing the two devices, but the surface collision angle, voltage of the angled deflector lens used to push ions to the surface, efficacy of product ion collection, and especially pressure in the region post-surface collision contribute to slightly different effective SID energies for each device. Despite identical placement of both devices within the instrument, the shorter dimension of the tilted-surface/ion carpet device results in a surface and post-surface region much closer to the entrance of the helium cell, and subsequent effects from gas flow and increased pressure. From these plots, the tilted-surface/ion carpet SID device shows a shallower onset between precursor and products in the low-energy regime. Analysis of the ion mobiligrams observed from these experiments on streptavidin and CRP indicate folded, native-like products with symmetric charge partitioning, aligning with previously-published data on SID of these proteins.14 An attractive feature of this device is the fact that, for the energy range of ΔV=10 V to 200 V, the tune settings used within the tilted-surface/ion carpet device remained consistent between experiments with peptides and protein complexes. The ion current resulting from higher-energy (ΔV>60 V) collisions of protein complexes in the tilted-surface/ion carpet SID device shows decreased signal compared to the original 10-lens device at the same energies regardless of the voltage gradient across the ion carpet, though characteristic SID products are still clearly observed with high S/N. This suggests that at higher-energy collisions, the ion carpet is not as effective in collecting large products. Thus, the inclusion of a more appropriate collection device (e.g. RF-confining trap) following the angled surface is likely necessary. The success with the angled surface implementation from this device was used as inspiration for another SID device implemented in an ICR platform that utilizes an RF-trapping cell immediately following the surface collision to more effectively collect the SID products.23
Figure 3.
SID-ERMS plots of A) and B) streptavidin 11+ and C) and D) the CRP 18+ precursor. Protein complexes were prepared under charge-reducing conditions using TEAA. A) and C) utilized the original 10-lens SID device and B) and D) utilized the new tilted-surface/ion carpet device. Grey protein cartoons show the expected SID cleavage interfaces. In all plots, error bars are indicative of the standard deviation from the mean of triplicate measurements. Representative mass spectra are shown in Figure S8.
CONCLUSIONS
We report here a new device that utilizes an ion carpet array and angled surface to enable a reduction in the number of independent voltages (from 10 to 7) and overall size along the ion path (from 3.0 cm to 1.6 cm) required to accomplish SID. Reducing the SID device size should enable this technology to be utilized in a broader range of mass spectrometers with reduced redesign of existing components. The ability to fragment a wide range of precursors utilizing the same tune settings along with decreasing the number of independent voltages allows for more straightforward tuning for the non-expert user and an improved potential for automated tuning (work in progress). Moving forward, it will be informative to investigate surface material and collision angle effects and reduce the countercurrent gas flow at the surface scattering region (e.g. by placing the short SID device in front of the trap SRIG) with the goal of enhancing product ion signal in the higher SID energy regime.
Supplementary Material
Acknowledgements
The authors thank Cathy McIntyre (Jonathan Amy Facility for Chemical Instrumentation, Purdue University) for assistance with the ion carpet design, Dalton T. Snyder for assistance with SIMION modeling, and Larry Antal (OSU Arts & Sciences Machine Shop) for assistance with SID device manufacturing. The authors gratefully acknowledge financial support from the National Science Foundation (NSF 1455654 to V.H.W.) and the National Institutes of Health (NIH P41GM128577 to V.H.W.).
Footnotes
COI
The author(s) declare no competing financial interest.
References:
- (1).Robinson CV; Sali A; Baumeister W The Molecular Sociology of the Cell. Nature 2007, 450 (7172), 973–982. 10.1038/nature06523. [DOI] [PubMed] [Google Scholar]
- (2).Sharon M How Far Can We Go with Structural Mass Spectrometry of Protein Complexes? J Am Soc Mass Spectrom 2010, 21 (4), 487–500. 10.1016/j.jasms.2009.12.017. [DOI] [PubMed] [Google Scholar]
- (3).Busch F; VanAernum ZL; Ju Y; Yan J; Gilbert JD; Quintyn RS; Bern M; Wysocki, Vicki H. Localization of Protein Complex Bound Ligands by Surface-Induced Dissociation High-Resolution Mass Spectrometry. Analytical Chemistry 2018, 90, 12796–12801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Benesch JLP; Robinson CV Mass Spectrometry of Macromolecular Assemblies: Preservation and Dissociation. Curr. Opin. Struct. Biol. 2006, 16, 245–251. [DOI] [PubMed] [Google Scholar]
- (5).Niu S; Ruotolo BT Collisional Unfolding of Multiprotein Complexes Reveals Cooperative Stabilization upon Ligand Binding. Protein Sci 2015, 24 (8), 1272–1281. 10.1002/pro.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Quintyn RS; Yan J; Wysocki VH Surface-Induced Dissociation of Homotetramers with D2 Symmetry Yields Their Assembly Pathways and Characterizes the Effect of Ligand Binding. Chem. Biol. 2015, 22, 583–592. [DOI] [PubMed] [Google Scholar]
- (7).Postawa Z; Czerwinski B; Szewczyk M; Smiley EJ; Winograd N; Garrison BJ Enhancement of Sputtering Yields Due to C60 versus Ga Bombardment of Ag{111} As Explored by Molecular Dynamics Simulations. Anal. Chem. 2003, 75 (17), 4402–4407. 10.1021/ac034387a. [DOI] [PubMed] [Google Scholar]
- (8).Meroueh O; Hase WL Dynamics of Energy Transfer in Peptide−Surface Collisions. J. Am. Chem. Soc. 2002, 124 (7), 1524–1531. 10.1021/ja011987n. [DOI] [PubMed] [Google Scholar]
- (9).Pratihar S; Barnes GL; Laskin J; Hase WL Dynamics of Protonated Peptide Ion Collisions with Organic Surfaces: Consonance of Simulation and Experiment. J. Phys. Chem. Lett. 2016, 7 (16), 3142–3150. 10.1021/acs.jpclett.6b00978. [DOI] [PubMed] [Google Scholar]
- (10).Williams ER; Fang L; Zare RN Surface Induced Dissociation for Tandem Time-of-Flight Mass Spectrometry. International Journal of Mass Spectrometry and Ion Processes 1993, 123, 233–241. [Google Scholar]
- (11).Laskin J; Futrell JH Energy Transfer in Collisions of Peptide Ions with Surfaces. J. Chem. Phys. 2003, 119 (6), 3413–3420. 10.1063/1.1589739. [DOI] [Google Scholar]
- (12).Dongre AR; Jones JL; Somogyi A; Wysocki, Vicki H. Influence of Peptide Composition, Gas-Phase Basicity, and Chemical Modification on Fragmentation Efficiency: Evidence for the Mobile Proton Model. J. Am. Chem. Soc. 1996, 118 (35), 8365–8374. [Google Scholar]
- (13).Harvey SR; Seffernick JT; Quintyn RS; Song Y; Ju Y; Yan J; Sahasrabuddhe AN; Norris A; Zhou M; Behrman EJ; et al. Relative Interfacial Cleavage Energetics of Protein Complexes Revealed by Surface Collisions. Proc Natl Acad Sci USA 2019, 116 (17), 8143–8148. 10.1073/pnas.1817632116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Zhou M; Dagan S; Wysocki VH Protein Subunits Released by Surface Collisions of Noncovalent Complexes: Nativelike Compact Structures Revealed by Ion Mobility Mass Spectrometry. Angew. Chem. 2012, 124 (18), 4412–4415. 10.1002/ange.201108700. [DOI] [PubMed] [Google Scholar]
- (15).Anthony SN; Shinholt DL; Jarrold MF A Simple Electrospray Interface Based on a DC Ion Carpet. International Journal of Mass Spectrometry 2014, 371, 1–7. 10.1016/j.ijms.2014.06.007. [DOI] [Google Scholar]
- (16).VanAernum ZL; Gilbert JD; Belov ME; Makarov AA; Horning SR; Wysocki, Vicki H. Surface-Induced Dissociation of Noncovalent Protein Complexes in an Extended Mass Range Orbitrap Mass Spectrometer. Analytical Chemistry 2019, 91, 3611–3618. 10.1021/acs.analchem.8b05605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Galhena AS; Dagan S; Jones CM; Beardsley RL; Wysocki VH Surface-Induced Dissociation of Peptides and Protein Complexes in a Quadrupole/Time-of-Flight Mass Spectrometer. Anal. Chem. 2008, 80, 1425–1436. [DOI] [PubMed] [Google Scholar]
- (18).Yan J; Zhou M; Gilbert JD; Wolff JJ; Somogyi A; Pedder RE; Quintyn RS; Morrison LJ; Easterling ML; Pasa-Tolić L; et al. Surface-Induced Dissociation of Protein Complexes in a Hybrid Fourier Transform Ion Cyclotron Resonance Mass Spectrometer. Anal. Chem. 2017, 89, 895–901. [DOI] [PubMed] [Google Scholar]
- (19).Shirzadeh M; Boone CD; Laganowsky A; Russell DH Topological Analysis of Transthyretin Disassembly Mechanism: Surface-Induced Dissociation Reveals Hidden Reaction Pathways. Anal. Chem. 2019, 91 (3), 2345–2351. 10.1021/acs.analchem.8b05066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Vimer S; Ben-Nissan G; Morgenstern D; Quintyn RS; Wysocki VH; Sharon M Multilevel Analysis of Ortholog Protein Complexes by Native Mass Spectrometry. Manuscript submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Wang Y; Hase WL; Song K Direct Dynamics Study of N-Protonated Diglycine Surface-Induced Dissociation. Influence of Collision Energy. J Am Soc Mass Spectrom 2003, 14 (12), 1402–1412. 10.1016/j.jasms.2003.08.014. [DOI] [PubMed] [Google Scholar]
- (22).Jo S-C; Cooks RG Translational to Vibrational Energy Conversion during Surface-Induced Dissociation of n-Butylbenzene Molecular Ions Colliding at Self-Assembled Monolayer Surfaces. Eur J Mass Spectrom (Chichester) 2003, 9 (4), 237–234. 10.1255/ejms.554. [DOI] [PubMed] [Google Scholar]
- (23).Snyder DT; Panczyk E; Stiving AQ; Gilbert JD; Somogyi A; Kaplan D; Wysocki VH Design and Performance of a Second-Generation Surface-Induced Dissociation Cell for Fourier Transform Ion Cyclotron Resonance Mass Spectrometry of Native Protein Complexes. Analytical Chemistry. 10.1021/acs.analchem.9b03746. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.