Abstract
Background
Multifocal spherical nonstaining cavities and gram-positive, rod-shaped, and endospore-forming bacteria were found in the liver of a sow that died suddenly. Clostridium novyi type B was identified and isolated from the sudden death case, and the isolate was characterized by molecular analyses and bioassays in the current study.
Results
C. novyi was isolated from the liver of a sow that died suddenly and was confirmed as C. novyi type B by differential PCR. The C. novyi isolate fermented glucose and maltose and demonstrated lecithinase activity, and the cell-free culture supernatant of the C. novyi isolate exhibited cytotoxicity toward Vero cells, demonstrating that the isolate produces toxins. In addition, whole-genome sequencing of the C. novyi isolate was performed, and the complete sequences of the chromosome (2.29 Mbp) and two plasmids (134 and 68 kbp) were identified for the first time. Based on genome annotation, 7 genes were identified as glycosyltransferases, which are known as alpha toxins; 23 genes were found to be related to sporulation; 12 genes were found to be related to germination; and 20 genes were found to be related to chemotaxis.
Conclusion
C. novyi type B was isolated from a sow in a sudden death case and confirmed by biochemical and molecular characterization. Various virulence-associated genes were identified for the first time based on whole-genome sequencing.
Keywords: Anaerobe, Clostridium novyi, Isolation, Characterization, Whole-genome sequencing
Background
Clostridium novyi (C. novyi), originally named Bacillus oedematis maligni no. 2, was first isolated in 1894 from guinea pigs by Dr. Frederick Novy [1]. C. novyi is broadly distributed in soil, water and marine sediments and affects humans and animals worldwide [2–4]. C. novyi is a gram-positive, noncapsulated, motile obligatory anaerobe that produces endospores to resist unfavorable environments [1, 5].
Based on the toxins they produce, C. novyi are classified into four types: A, B, C and D. C. novyi type A produces alpha, gamma, delta and epsilon toxins. C. novyi type B produces alpha, beta, and zeta toxins, while type C produces gamma toxin [2, 6, 7]. C. novyi type D is considered to be a different species, Clostridium haemolyticum, because it does not produce alpha toxin and because the disease that it causes is different from those caused by types A and B [8]. C. haemolyticum produces beta, eta and theta toxins [9]. C. novyi type A is frequently involved in gas gangrene infections in humans and animals, while type B is the etiological agent of infectious necrotic hepatitis (black disease), which is typically observed in sheep, cattle and swine [10]. C. novyi type C is not known to induce illness in and is typically considered nonpathogenic toward laboratory animals [7]. C. novyi type D (C. haemolyticum) is responsible for hemoglobinuria in calves [7], while C. novyi types A and B (producing alpha toxin) cause sudden death in swine, and the carcasses exhibit gross distension and livers with gas bubble infiltration or sponge-like appearances [11, 12].
The 16S rRNA gene sequence has been used to detect genetic relatedness between different species of bacteria. Currently, next-generation sequencing is utilized as a rapid tool to perform whole-genome sequencing of clinical isolates. Indeed, this method has proved to be of great value for understanding bacterial evolution, outbreaks, toxigenicity, and antimicrobial resistance in a number of studies involving Vibrio cholera, Escherichia coli, Clostridium difficile, and Mycobacterium tuberculosis [12, 13]. In the present study, for the first time, C. novyi type B was isolated from a sudden death case of a sow in Korea, and the isolate was characterized by molecular analyses and bioassays. To the best of our knowledge, this is the first report of the complete genome sequence of C. novyi type B in Korea.
Results
Isolation of the C. novyi isolate
After 72 h of anaerobic incubation, colonies showing irregular shapes with unclear borders appeared on agar media, and gram-positive, rod-shaped, endospore-forming bacteria were identified in the colonies (Fig. 1a and b). Differential PCR was conducted on the DNA extracted from a single colony, which confirmed the isolate as C. novyi type B [14]. No other bacterial and viral pathogens were detected (data not shown).
Fig. 1.
Morphology of the Clostridium isolate based on staining. a Gram staining was performed on the cultured isolate, resulting in the detection of gram-positive rods. b Endospores of the cultured C. novyi isolate were observed after staining with 5% malachite green
Biochemical characterization of the C. novyi isolate
Biochemical analysis revealed that the C. novyi isolate generates gas when treated with hydrogen peroxide and exhibits beta hemolysis on blood agar. The biochemical characteristics of the C. novyi isolate are summarized in Table 1. The C. novyi isolate was positive for D-glucose, gelatin, D-maltose, salicin, L-rhamnose, D-cellobiose, and lecithinase and negative for L-tryptophan, urea, D-mannitol, D-lactose, D-saccharose, D-xylose, L-arabinose, esculin, glycerol, D-mannose, D-melezitose, D-sorbitol, D-trehalose, and catalase.
Table 1.
Biochemical test results for the C. novyi isolate
| Active ingredients | Results |
|---|---|
| D-Glucose | + |
| Gelatin | + |
| Lecithinase | + |
| L-Tryptophan | – |
| Urea | – |
| D-Mannitol | – |
| D-Lactose | – |
| D-Saccharose | + |
| D-Maltose | + |
| Salicin | + |
| D-Xylose | – |
| L-Arabinose | – |
| Esculin | – |
| Glycerol | – |
| D-Cellobiose | + |
| D-Mannose | – |
| D-Melezitose | – |
| D-Sorbitol | – |
| L-Rhamnose | + |
| D-Trehalose | – |
| Catalase | – |
| Endospore | + |
| Gram | + |
| Hemolysis | + |
+ positive; − negative
Phylogenetic analysis of the isolate based on the 16S rRNA gene sequence
Based on the 16S rRNA gene sequence analysis, the C. novyi isolate showed more than 99% similarity with C. novyi types B and C, C. haemolyticum, and C. botulinum types C and D. The C. novyi isolate exhibited 84 to 91% similarity with C. perfringens, C. sporogenes, and C. sordellii. A comparison of the 16S rRNA gene sequences of the C. novyi isolate with those of different Clostridium species is shown in Fig. 2.
Fig. 2.
Phylogenetic analysis of the 16S rRNA gene sequences of Clostridium spp. The tree was constructed using the neighbor-joining method with 1000 replicates for bootstrap values with MEGA 6. The scale bar represents 0.02 substitutions per nucleotide position. The circle indicates the isolate identified in this study
Histopathological examination of the liver
The liver tissues showed coagulative necrosis throughout the entire field. Multifocal spherical nonstaining cavities were found that were well demarcated, and the cavity margins were clear (Fig. 3a and b) Furthermore, gram-positive, rod-shaped bacteria were observed on the liver tissue (Fig. 3c).
Fig. 3.
Morphology of the Clostridium isolate based on staining. a and b Coagulation necrosis and multifocal spherical non-staining cavities were detected on tissue section. (c) Gram staining was performed on liver tissue (400×)
Cell culture assay of the C. novyi isolate on Vero cells
The cell-free supernatant of C. novyi reference strains, ATCC 17861 (C. novyi type A) and ATCC 25758 (C. novyi type B) were tested on Vero 76 cells that were observed for 5 days. Abnormal cells were detected in all of the strains at days 1 and 2 for the 1:4 and 1:8 dilutions, respectively. On day 3, rounding and retracting cells were observed up to dilutions of 1:32 from inoculation of cell-free supernatants of ATCC 17861 and ATCC 25758, while the cell-free supernatant of the C. novyi isolate (150775) showed abnormal cells up to dilutions of 1:16. After day 4, all tested wells that were inoculated with the cell-free supernatant of C. novyi strains exhibited rounding and retracting cells up to the 1:32 dilutions. No cytopathic effect was observed in the Vero cell controls. Vero 76 cells with or without cytopathic effect are shown in Fig. 4.
Fig. 4.
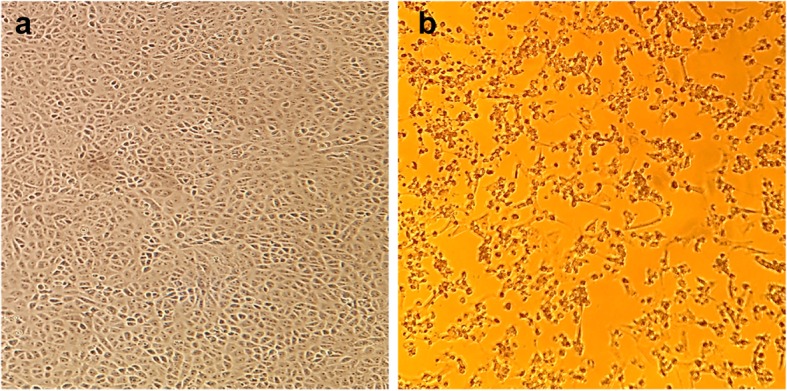
Cytopathic effect of the cell-free supernatant of the C. novyi isolate in Vero 76 cells. Morphological alterations of Vero 76 cells exposed to cell-free supernatant of the C. novyi isolate for 5 days. a Vero 76 cells without treatment; the cells show a fibroblast-like morphology. b Vero 76 cells inoculated with filtered (0.2 μm) C. novyi supernatant (1:32). A cytopathic effect (rounding and disintegration of cells) was observed in cells treated with C. novyi supernatant
Sequencing and genome features of the C. novyi isolate
The complete genome features of the C. novyi isolate are summarized in Table 2 and Fig. 5. The genome consisted of a single circular chromosome with two circular plasmids. The sizes of the C. novyi isolate chromosome and plasmids 1 and 2 were 2,296,219 bp, 134,627 bp, 68,232 bp, respectively. The GC content of the chromosomal DNA was 27.9%, and 33 rRNA genes, 84 tRNA genes, and 54 pseudogenes were identified. The GC content of plasmid 1 was 26.3%, and 13 pseudogenes were identified. The GC content of plasmid 2 was 25.5%, and 13 pseudogenes were identified. The assembled and annotated sequences of the C. novyi isolate chromosome and plasmids 1 and 2 were submitted to NCBI (accession numbers: CP029458.1, CP029459.1, and CP029460.1).
Table 2.
Genomic characteristic of the C. novyi isolate
| Chromosome | Plasmid 1 | Plasmid 2 | |
|---|---|---|---|
| Genome size (bp) | 2,296,219 | 134,627 | 68,232 |
| GC content (%) | 27.9 | 26.3 | 25.5 |
| Protein coding genes | 2009 | 141 | 54 |
| rRNAs | 33 | – | – |
| tRNAs | 84 | – | – |
| Pseudogenes | 54 | 13 | 13 |
Fig. 5.
Circular map of the chromosome and two plasmids of the C. novyi isolate. From the outside to the center: genes on the forward strand (colored by Clusters of Orthologous Genes (COG) categories), coding DNA sequence (CDS) on the forward strand, CDS on the reverse strand, genes on the reverse strand (colored by COG categories), GC content, and GC skew
Prediction of genes associated with pathogenicity
All of the identified genes of the C. novyi isolate are summarized in Additional file 1. A total of 35 genes related to sporulation and germination were detected in the whole genome of the C. novyi isolate. Among these genes, sporulation-related sigma factors (DFH04_RS02880, DFH04_RS08440, DFH04_RS08465, DFH04_RS08470, and DFH04_RS10365) and stage-specific sporulation genes were detected. Additionally, 7 glycosyltransferase genes associated with the monoglycosyltransferase activity of alpha toxin were identified (DFH04_RS05070, DFH04_RS05155, DFH04_RS10400, DFH04_RS10410, DFH04_RS10425, DFH04_RS10435, and DFH04_RS10440). Various genes encoding chemotaxis proteins were also identified in the C. novyi isolate genome, including a sensor kinase, the product of DFH04_RS10060; a deamidase, the product of DFH04_RS08070; a methyltransferase, the product of DFH04_RS10065; a docking protein, the product of DFH04_RS08070; and a phosphatase protein, the product of DFH04_RS10055. Additionally, 9 genes were identified as methyl-accepting chemotaxis proteins (MCPs).
Discussion
The isolation and identification of C. novyi is rarely successful because specimens must be delivered to laboratories very quickly under strict anaerobic conditions [12, 13, 15]. In addition, samples should be processed within 12 h of the death of pigs for accurate diagnosis and successful isolation of C. novyi before the proliferation of other bacteria in the carcass [11, 12, 16]. In this study, postmortem examination of a sow sudden death case was conducted, and a liver sample was collected within approximately 11 h. After extracting DNA from the sample, the presence of C. novyi in the sample was confirmed by differential PCR.
Biochemical assays of the isolated colonies showed the presence of lecithinase activity and glucose and maltose fermentation, which are the characteristic features of C. novyi. Previously, lecithinase activity and glucose and maltose fermentation have been associated with C. novyi types A and B [17]. On the basis of biochemical tests, our isolate could also be differentiated from other closely related Clostridia, such as C. haemolyticum, which lacks the ability to ferment maltose, while C. botulinum types C and D display variable maltose fermentation capabilities and maltose and lecithinase activity [17].
The 16S rRNA gene sequencing results for the C. novyi isolate showed that the isolate shared approximately 98–99% similarity with each type of C. novyi, C. haemolyticum and C. botulinum types C and D. In particular, the 16S rRNA gene sequence of the C. novyi isolate and that of C. haemolyticum showed the highest similarity (99.6%). Consistent with this finding, high similarity (> 99.9%) between the 16S rRNA gene sequence of C. novyi type B and that of C. haemolyticum has been reported in a previous study [18]. Therefore, different methods are needed to distinguish C. novyi type B from C. haemolyticum. The N- and C-terminal amino acid sequences of FliC are well preserved between C. novyi type B and C. haemolyticum, but the central region amino acid sequences are not [19]. Therefore, the C. novyi isolate was distinguished from C. haemolyticum using fliC gene primers.
In this study, virulence-associated genes were identified by whole-genome sequencing of the C. novyi isolate. The other identified genes are summarized in Additional file 1. Endospores are important contributors to pathogenesis before being the vegetative forms of C. novyi type B. Furthermore, the endospores of C. novyi are highly resistant to environmental conditions. Clostridium spp. initiate the sporulation process when unfavorable conditions are detected. The sporulation process is a carefully orchestrated cascade of events at both the transcriptional and posttranslational levels that involves a multitude of sigma factors, transcription factors, proteases, and phosphatases. Clostridium spp. genomes contain genes for all major sporulation-specific transcription and sigma factors. The sporulation process consists of several stages. Sporulation-specific sigma factors affect each sporulation stage [20]. Sporulation genes and sigma factor genes were detected in the C. novyi isolate; in addition, the C. novyi isolate was found to contain germination genes that initiate the germination process when a favorable environment is detected. Although the mechanism has not been completely elucidated, after C. novyi endospores are ingested, they are absorbed from the intestine and reach the liver via the portal circulation. Subsequently, the endospores are spread to other organs. The endospores germinate and produce toxins in organs when anaerobic conditions form [15].
The main pathogenic protein of C. novyi type B is alpha toxin. Alpha toxin is produced and released by the vegetative forms of C. novyi type B, and its monoglycosyltransferase activity inactivates several GTP-binding proteins in cells, resulting in modification and redistribution of the actin cytoskeleton. For these reasons, necrosis occurs in the liver, and the cut surface of the affected liver exhibits a sponge-like appearance [15, 21–25]. In this study, glycosyltransferase genes were detected in the genome of the C. novyi isolate. In addition, the effect of the toxins produced by the isolate was tested by observing the effect of the cell-free supernatant of the C. novyi isolate on Vero cells, although the alpha toxin of the isolate was not purified. The cytopathic effect produced was similar to the observations in a previous study in which the purified alpha toxin exhibited a strong effect on Vero cells, resulting in rounded cells in addition to lysed cells [26]. Histologically, multifocal spherical non-staining cavities were detected in tissue sections, and gram-positive rods were observed in the liver tissue by Gram staining. These findings were consistent with the results of previous studies [11, 12, 15, 16]. Based on above results, the C. novyi isolate was considered to have the capability of producing alpha toxin toward on Vero cells and liver tissue.
Chemotaxis enables bacteria to move according to chemical gradients. Chemotaxis affords key physiological benefits, including enhanced contact with growth substrates. Another important aspect of chemotaxis is that it plays a role in infection and disease, as chemotaxis signaling pathways are widely distributed among diverse pathogenic bacteria [27]. In this study, 15 genes related to chemotaxis, namely, DFH04_RS10060 (CheA), DFH04_RS10080 (CheW), DFH04_RS10075 (CheD), DFH04_RS08070 (CheV), DFH04_RS10055 (CheC), DFH04_RS10065 (CheR), and nine MCPs were detected in the genome of the C. novyi isolate. As reported previously [27], the products of these genes transfer signals by phosphorylation and activate the flagellum. The activated flagellum enables the bacterium to move toward an attractant. Chemotaxis-associated genes were also detected in a previous study based on Clostridium novyi-NT, which is an attenuated strain of C. novyi [28].
Conclusion
This C. novyi isolate was first isolated from a sudden death case of sow in Korea and confirmed by biochemical and molecular characterization. Furthermore, various virulence-associated genes were identified in the genome of the isolate, indicating that the isolate might have had a role in the sudden death of the sow. However, more research including animal experiments is needed to determine the pathogenicity of C. novyi. Nonetheless, the complete genomic sequence of C. novyi isolate will contribute to a better understanding of the biology of C. novyi and related species.
Methods
Sample collection and isolation of C. novyi
Diagnostic samples, primarily livers, collected from sows with sudden death within 12 h after death were submitted to the Jeonbuk National University Veterinary Diagnostic Center in 2015 (case no. 150557) [14]. Swab samples taken from affected sites on the livers were inoculated in Reinforced clostridial medium (RCM) (BD Biosciences, New Jersey, USA) and incubated anaerobically at 37 °C for 72 h in a gas jar. Then, the incubated broth medium was heated at 100 °C for 10 min, reinoculated in 5% sheep blood agar, and again incubated under the same conditions. Colonies suspected of being C. novyi that exhibited beta hemolysis on sheep blood agar plates were subsequently identified as C. novyi type B based on differential PCR, Gram staining and 16S rRNA gene sequencing [14].
The morphology of the bacteria in the identified colonies was further confirmed using endospore staining. Spores and vegetative cells were identified using a 5% malachite green staining solution (Thermo Fisher Scientific Inc., MA, USA) and a safranin staining solution (Gram stain kit solution), respectively, as described previously [29].
DNA extraction from isolates
DNA was extracted from colonies grown on 5% sheep blood agar for differential PCR and 16S rRNA gene sequencing by Patho Gene-spin™ (iNtRON Biotechnology, Seongnam, Korea), according to the manufacturer’s instructions.
Differential PCR for detection of the C. novyi isolate
The suspected colonies were confirmed using C. novyi specific flagellin gene (filC) primers, where fliC was amplified using differential primers for C. novyi types A and B, as well as C. haemolyticum, which is genetically close to C. noyvi [14, 19].
The PCR assays in this study were performed in a SimpliAmp Thermal Cycler (Applied Biosystems, Foster City, CA, USA) in a 20 μl reaction containing: 1× PCR buffer; 2 mM MgCl2; 250 μM each dNTP; 1 unit of DNA polymerase (AccuPower Multiplex PCR kit; Bioneer Inc., Alameda, CA, USA); and 10 pmol of each primer. The PCR conditions were as follows: initial denaturation at 94 °C for 5 min, followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 56.5 °C for 1 min and extension at 72 °C for 1 min, with a final extension at 72 °C for 5 min. The amplified PCR products were separated by electrophoresis in 2% (w/v) agarose gels and stained with Red Safe™ (iNtRON Biotechnology, Seongnam, Korea). The strains ATCC 17861 (C. novyi type A; 472 bp), ATCC 25758 (C. novyi type B; 551 bp), and KCTC 5570 (C. haemolyticum; 819 bp) were used as positive controls.
Histopathological examination of the liver
Approximately 2-cm3 liver tissue samples were fixed in 10% phosphate-buffered formalin, routinely processed, and embedded in paraffin. Tissue sections (4 μm) were prepared using a microtome (HM-340E, Thermo Fisher Scientific Inc., MA, USA), and the sections were placed onto glass slides. Hematoxylin and eosin (H&E) staining was performed according to standard techniques, and Gram staining was performed on liver tissue collected from the dead sow using a Gram stain kit according to the manufacturer’s instructions.
Identification and characterization of the C. novyi isolate
The biochemical characteristics of the C. novyi isolate were determined using an API 20A kit (BioMerieux, USA) according to the manufacturer’s instructions. The lecithinase activity of the C. novyi isolate was confirmed on an egg yolk agar plate (Kisan Biotech. Co., Seoul, Korea), where the isolate was inoculated onto the plate using a sterilized loop and incubated anaerobically for 72 h at 37 °C. The presence of opalescence around a colony was taken to indicate lecithinase activity. Additionally, a catalase test was performed using pure C. novyi isolates smeared on a microscope slide. A drop of 3% hydrogen peroxide (Wako, Osaka, Japan) was added, and the production of copious bubbles was taken to indicate that the bacteria were positive for catalase.
16S rRNA gene sequencing analysis
16S rRNA gene sequencing was performed by a sequencing facility (Biofact Co., Daejun, Korea) using a pure culture of the C. novyi isolate. The 16S rRNA gene sequence of the C. novyi isolate was identified by a BLAST search and compared with other Clostridium species sequences downloaded from the NCBI database. Multiple alignment of the 16S rRNA gene sequences was performed using MegAlign (DNASTAR, Madison, Wisconsin, USA), and a phylogenetic tree was constructed with the neighbor joining method using MEGA 6 [30].
Cell culture assay of the C. novyi isolate on Vero cells
Vero 76 cells (ATCC CRL-1587) were maintained in high-glucose Dulbecco’s modified Eagle’s medium (DMEM; Welgene, Korea) supplemented with heat-inactivated 5% fetal bovine serum (FBS; Invitrogen, USA), 2 mM L-glutamine, and a 100× antibiotic-antimycotic solution [Anti-Anti, Invitrogen; 1× solution contains 100 IU/ml penicillin, 100 μg/ml streptomycin, and 0.25 μg/ml Fungizone® (amphotericin B)] at 37 °C under a humidified 5% CO2 atmosphere.
The cell culture assay was performed in Vero 76 cells as described previously [26, 31]. Briefly, C. novyi cell-free supernatant obtained after centrifugation was prepared by filtrating the broth through a 0.2-μm cellulose acetate syringe filter (Corning, Germany). Two-fold serial dilutions of the filtered supernatant were prepared with cell medium for up to 8 dilutions. Cell culture assays were performed in 96-well tissue culture plates (Falcon, NY, USA). Subsequently, the supernatant of cultured Vero 76 cells was discarded, and the cells were washed twice with 1× PBS. Then, the diluted cell-free supernatant of the C. novyi isolate was added to the Vero 76 cells, which were then incubated at 37 °C under a humidified atmosphere with 5% CO2 for 5 days. The cell-free culture supernatants of reference strains of C. novyi type A (ATCC 17861) and C. novyi type B (ATCC25758) were used as positive controls.
Whole-genome sequencing
DNA was extracted from cell pellets of C. novyi isolate from 200-ml cultures using Patho Gene-spin™, according to the manufacturer’s instructions. A sample of high-quality, high-molecular-weight DNA was used to prepare size-selected SMRTbell templates of approximately 20 kb. A NanoDrop spectrophotometer (Thermo Fisher Scientific Inc., MA, USA) and a Qubit fluorometer (Thermo Fisher Scientific Inc., MA, USA) were used to measure the concentration of gDNA, and the sample passed quality control (QC) screening criteria (≥200 ng). For PacBio RS sequencing, 8 g of input gDNA was used for the 20-kb library preparation. For gDNA where the size range was less than 17 kb, we used a Bioanalyzer 2100 (Agilent Technologies, CA, USA) to determine the actual size distribution. If the apparent size of the gDNA was greater than 40 kb, the gDNA was sheared with a g-TUBE (Covaris Inc., Woburn, MA, USA) to produce library fragments in the optimal size range and purified using AMPure PB magnetic beads (Beckman Coulter Inc., CA, USA). Then, the concentration of the gDNA was measured using both a NanoDrop spectrophotometer and a Qubit fluorometer, and approximately 200 ng/μl gDNA was run on a field-inversion gel. A library was prepared in a total volume of 10 μl using a PacBio DNA Template Prep Kit 1.0 (for 3–10 kb), and SMRTbell templates were annealed using a PacBio DNA/Polymerase Binding Kit P6. A PacBio DNA Sequencing Kit 4.0 and 1 SMRT Cell was used for sequencing. SMRT Cells (Pacific Biosciences, CA, USA) with C4 chemistry were used, and 240-min movies were captured for each SMRT cell using the PacBio RS II (Pacific Biosciences, CA, USA) sequencing platform. The subsequent steps were based on the PacBio Sample Net-Shared Protocol, which is available at http://pacificbiosciences.com/.
Bioinformatics analysis
All runs were performed with diffusion-based loading and analyzed using standard primary data analysis methods by implementing HGAP and Quiver [32]. The coding DNA sequences were predicted with Prokaryotic Genome Annotation Pipeline version 4.5 on the NCBI website (https://www.ncbi.nlm.nih.gov/genome/annotation_prok/). Additional functional annotation was performed with the Rapid Annotation using Subsystem Technology server [33].
Supplementary information
Additional file 1. The identified genes of the C. novyi isolate.
Acknowledgements
We would like to thank the funding source and the Veterinary Diagnostic Center in Jeonbuk National University.
Abbreviations
- MCPs
Methyl-accepting chemotaxis proteins
- GTP
Guanosine 5′-triphosphate
- RCM
Reinforced clostridial medium
- NCBI
National center for biotechnology information
- CPE
Cytopathic effects
- HGAP
Hierarchical genome assembly process
- gDNA
Genomic DNA
Authors’ contributions
CGJ conducted the experiments and drafted the manuscript. SN and AK participated in the data analysis and helped draft the manuscript. BJS, BKJ and JHS contributed to the data analysis of the whole genome sequences. BK and MSY contributed to the histopathological examination. SHN and SCK contributed to the collection and processing of the samples. WIK designed the study and prepared the manuscript. All authors read and approved the final manuscript.
Funding
This research was financially supported by the Agriculture Science & Technology Development (PJ010530) in Rural Development Administration, Republic of Korea. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and in Additional file 1. Genome sequences obtained in the current study are deposited in GenBank under the accession numbers CP029458.1, CP029459.1, and CP029460.1. The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Ethics approval and consent to participate
Animal materials used in the present study were received as routine diagnostic submissions and were used under the agreement of Jeonbuk National University-Veterinary Diagnostic Center (JBNU-VDC). Furthermore, written consent for the use of sow samples were obtained from the owners. The authors also confirmed that ethical approval is not essential to use animal materials received for diagnostic purpose in accordance with the guidelines of the Jeonbuk National University-Institutional Animal Care and Use Committee (JBNU-IACUC).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Chang-Gi Jeong, Email: jcg0102@gmail.com.
Byoung-Joo Seo, Email: byoungjooseo@gamil.com.
Salik Nazki, Email: saliknazki@gmail.com.
Byung Kwon Jung, Email: bkjung@knu.ac.kr.
Amina Khatun, Email: amina.vet.sau.bd@gmail.com.
Myeon-Sik Yang, Email: 111@jbnu.ac.kr.
Seung-Chai Kim, Email: leesor2@jbnu.ac.kr.
Sang-Hyun Noh, Email: livensh@gmail.com.
Jae-Ho Shin, Email: jhshin@knu.ac.kr.
Bumseok Kim, Email: bskims@jbnu.ac.kr.
Won-Il Kim, Email: kwi0621@jbnu.ac.kr.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12917-020-02349-9.
References
- 1.Aronoff DM. Clostridium novyi, sordellii, and tetani: mechanisms of disease. Anaerobe. 2013;24:98–101. doi: 10.1016/j.anaerobe.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 2.Skarin H, Segerman B. Plasmidome interchange between Clostridium botulinum, Clostridium novyi and Clostridium haemolyticum converts strains of independent lineages into distinctly different pathogens. PLoS One. 2014;9(9):e107777. doi: 10.1371/journal.pone.0107777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nishida S, Nakagawara G. Isolation of toxigenic strains of Clostridium Novyi from soil. J Bacteriol. 1964;88:1636–1640. doi: 10.1128/JB.88.6.1636-1640.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith LD. The occurrence of Clostridium botulinum and Clostridium tetani in the soil of the United States. Health Lab Sci. 1978;15(2):74–80. [PubMed] [Google Scholar]
- 5.McGuigan CC, Penrice GM, Gruer L, Ahmed S, Goldberg D, Black M, et al. Lethal outbreak of infection with Clostridium novyi type a and other spore-forming organisms in Scottish injecting drug users. J Med Microbiol. 2002;51(11):971–977. doi: 10.1099/0022-1317-51-11-971. [DOI] [PubMed] [Google Scholar]
- 6.Oakley CL, Warrack GH, Clarke PH. The toxins of Clostridium oedematiens (cl. Novyi) J Gen Microbiol. 1947;1(1):91–107. doi: 10.1099/00221287-1-1-91. [DOI] [PubMed] [Google Scholar]
- 7.Eklund MW, Poysky FT, Peterson ME, Meyers JA. Relationship of bacteriophages to alpha toxin production in Clostridium novyi types a and B. Infect Immun. 1976;14(3):793–803. doi: 10.1128/IAI.14.3.793-803.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakamura S, Kimura I, Yamakwa K, Nishida S. Taxonomic relationships among Clostridium novyi types a and B, Clostridium haemolyticum and Clostridium botulinum type C. J Gen Microbiol. 1983;129:1473–1479. doi: 10.1099/00221287-129-5-1473. [DOI] [PubMed] [Google Scholar]
- 9.Hunter LC, Poxton IR. Clostridium botulinum types C and D and the closely related Clostridium novyi. Rev Med Microbiol. 2002;13(2):75–90. doi: 10.1097/00013542-200204000-00004. [DOI] [Google Scholar]
- 10.Brazier JS, Duerden BI, Hall V, Salmon JE, Hood J, Brett MM, et al. Isolation and identification of Clostridium spp. from infections associated with the injection of drugs: experiences of a microbiological investigation team. J Med Microbiol. 2002;51(11):985–989. doi: 10.1099/0022-1317-51-11-985. [DOI] [PubMed] [Google Scholar]
- 11.Akiyama N, Shibahara T, Ushiyama K, Shimizu H, Koizumi I, Osaki M. Fatal Clostridium novyi type B infection in a sow. Jarq-Jpn Agr Res Q. 2017;51(1):85–89. doi: 10.6090/jarq.51.85. [DOI] [Google Scholar]
- 12.Friendship CR, Bilkei G. Concurrent swine erysipelas and Clostridium novyi infections associated with sow mortality in outdoor sows in Kenya. Vet J. 2007;173(3):694–696. doi: 10.1016/j.tvjl.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 13.Karg H, Bilkei G. Causes of sow mortality in Hungarian indoor and outdoor pig production units. Berl Munch Tierarztl Wochenschr. 2002;115(9–10):366–368. [PubMed] [Google Scholar]
- 14.Chang-Gi J, Seo B-J, Kim W-I. Diagnosis on sudden death cases during summer season and isolated of Clostridium novyi. Korean J Vet Serv. 2016;39(2):131–136. doi: 10.7853/kjvs.2016.39.2.131. [DOI] [Google Scholar]
- 15.Nyaoke AC, Navarro MA, Beingesser J, Uzal FA. Infectious necrotic hepatitis caused by Clostridium novyi type B in a horse: case report and review of the literature. J Vet Diagn Investig. 2018;30(2):294–299. doi: 10.1177/1040638717737125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia A, Ayuso D, Benitez JM, Garcia WL, Martinez R, Sanchez S. Clostridium novyi infection causing sow mortality in an Iberian pig herd raised in an outdoor rearing system in Spain. J Swine Health Prod. 2009;17(5):264–268. [Google Scholar]
- 17.Quinn PJ, Carter ME, Markey B, Carter GR. Clinical veterinary microbiology. Edinburgh: MOSBY; 1999. p. 194.
- 18.Sasaki Y, Takikawa N, Kojima A, Norimatsu M, Suzuki S, Tamura Y. Phylogenetic positions of Clostridium novyi and Clostridium haemolyticum based on 16S rDNA sequences. Int J Syst Evol Microbiol. 2001;51(Pt 3):901–904. doi: 10.1099/00207713-51-3-901. [DOI] [PubMed] [Google Scholar]
- 19.Sasaki Y, Kojima A, Aoki H, Ogikubo Y, Takikawa N, Tamura Y. Phylogenetic analysis and PCR detection of Clostridium chauvoei, Clostridium haemolyticum, Clostridium novyi types a and B, and Clostridium septicum based on the flagellin gene. Vet Microbiol. 2002;86(3):257–267. doi: 10.1016/S0378-1135(02)00002-0. [DOI] [PubMed] [Google Scholar]
- 20.Al-Hinai MA, Jones SW, Papoutsakis ET. The Clostridium sporulation programs: diversity and preservation of endospore differentiation. Microbiol Mol Biol Rev. 2015;79(1):19–37. doi: 10.1128/MMBR.00025-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belyi Y, Aktories K. Bacterial toxin and effector glycosyltransferases. Biochim Biophys Acta. 2010;1800(2):134–143. doi: 10.1016/j.bbagen.2009.07.022. [DOI] [PubMed] [Google Scholar]
- 22.Aronoff DM, Kazanjian PH. Historical and contemporary features of infections due to Clostridium novyi. Anaerobe. 2018;50:80–84. doi: 10.1016/j.anaerobe.2017.12.012. [DOI] [PubMed] [Google Scholar]
- 23.Just I, Selzer J, Wilm M, von Eichel-Streiber C, Mann M, Aktories K. Glucosylation of rho proteins by Clostridium difficile toxin B. Nature. 1995;375(6531):500–503. doi: 10.1038/375500a0. [DOI] [PubMed] [Google Scholar]
- 24.Just I, Wilm M, Selzer J, Rex G, von Eichel-Streiber C, Mann M, et al. The enterotoxin from Clostridium difficile (ToxA) monoglucosylates the rho proteins. J Biol Chem. 1995;270(23):13932–13936. doi: 10.1074/jbc.270.23.13932. [DOI] [PubMed] [Google Scholar]
- 25.Just I, Selzer J, Hofmann F, Green GA, Aktories K. Inactivation of Ras by Clostridium sordellii lethal toxin-catalyzed glucosylation. J Biol Chem. 1996;271(17):10149–10153. doi: 10.1074/jbc.271.17.10149. [DOI] [PubMed] [Google Scholar]
- 26.Borrmann E, Schulze F. Detection of Clostridium novyi type B alpha toxin by cell culture systems. FEMS Immunol Med Microbiol. 1999;24(3):275–280. doi: 10.1111/j.1574-695X.1999.tb01294.x. [DOI] [PubMed] [Google Scholar]
- 27.Matilla MA, Krell T. The effect of bacterial chemotaxis on host infection and pathogenicity. FEMS Microbiol Rev. 2018;42(1):fux052. 10.1093/femsre/fux052. [DOI] [PubMed]
- 28.Bettegowda C, Huang X, Lin J, Cheong I, Kohli M, Szabo SA, et al. The genome and transcriptomes of the anti-tumor agent Clostridium novyi-NT. Nat Biotechnol. 2006;24(12):1573–1580. doi: 10.1038/nbt1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mormak DA, Casida LE. Study of Bacillus-Subtilis endospores in soil by use of a modified endospore stain. Appl Environ Microb. 1985;49(6):1356–1360. doi: 10.1128/AEM.49.6.1356-1360.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 31.Borrmann E, Schulze F, Cussler K, Hanel I, Diller R. Development of a cell culture assay for the quantitative determination of vaccination-induced antibodies in rabbit sera against Clostridium perfringens epsilon toxin and Clostridium novyi alpha toxin. Vet Microbiol. 2006;114(1–2):41–50. doi: 10.1016/j.vetmic.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 32.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10(6):563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 33.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, et al. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. The identified genes of the C. novyi isolate.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and in Additional file 1. Genome sequences obtained in the current study are deposited in GenBank under the accession numbers CP029458.1, CP029459.1, and CP029460.1. The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.