Abstract

Aspergillus fumigatus causes several nosocomial pulmonary infections and accounts for high morbidity and mortality rate globally. Among various virulence factors, 1,8-dihydroxynaphthalene-melanin plays an important role in the survival during unfavorable conditions both in vivo and in vitro, masks various molecular patterns associated with A. fumigatus, and protects it from the host immune system. In the present study, we aim to understand the potential of cis-9-hexadecenal as an antimelanogenic compound and its role in modulating other associated virulence factors in A. fumigatus. cis-9-Hexadecenal is a bioactive compound that belongs to C16 mono-unsaturated fatty-aldehyde groups. Minimum effective concentration of cis-9-hexadecenal affecting A. fumigatus melanin biosynthesis was determined using broth microdilution method. The spectrophotometric analysis revealed reduced melanin content (91%) and hydrophobicity (59%) at 0.293 mM of cis-9-hexadecenal. Cell surface organizational changes using electron microscopy showed altered demelanized smooth A. fumigatus conidial surface without any protrusions after cis-9-hexadecenal treatment. The transcript analysis of polyketide synthase (PKS) pksP/alb1 gene was quantified through qRT-PCR which revealed an upregulated expression. Total proteome profiling conducted through LC-MS-MS showed upregulated PKS enzyme but other downstream proteins involved in the 1,8-dihydroxynaphthalene-melanin biosynthesis pathway were absent. The homology modeling of PKS using Expasy’s web server predicted that PKS is stable at varied conditions and is hydrophilic in nature. The Ramachandran plot by PROCHECK confirmed the 3-D structure of PKS to be reliable. Docking analysis using AutoDock-4.2.6 predicted the binding of cis-9-hexadecenal and PKS at Thr-264 and Ser-171 residue via hydrogen bonding at a low binding energy of −4.95 kcal/mol.
1. Introduction
Rapid advancements in medical interventions in the past few years have revolutionized the treatment of life-threatening diseases such as cancer and autoimmune disorders. Ironically though, these modern advances are also leading to an alarming increase in the number of high risk patients getting susceptible to opportunistic pathogens.1Aspergillus fumigatus, ubiquitously present in the environment has emerged as a leading opportunistic fungal pathogen causing more than 90% of invasive fungal infections. This has in turn led to an alarmingly high rate of morbidity and mortality amongst immunocompromised patients.2A. fumigatus produces small spores (2–3 μm) that can easily surpass the human muco-cilliary clearance and it colonizes on the epithelial layer of respiratory airway, ultimately causing infections in patients with impaired immunity or those suffering from lung diseases.2
The limited spectrum of antifungal drugs available in the market target either ergosterol (polyenes and azoles) or fungal cell wall (echinocandins).3 These antifungal drugs are associated with severe side effects such as nephrotoxicity, hepatotoxicity, bronchospasm, haemoptysis, blurred vision, temporary blindness, and even skin cancer.4 The limitations of present antifungal drugs along with drug toxicity has enthused the search for identifying novel drug targets and new compound(s).5 It is more challenging to explore a unique target against A. fumigatus as the cellular mechanisms of mammalian cells and A. fumigatus are closely related.6
Multifactorial virulence traits contributing to the pathogenicity of A. fumigatus are related to its cell surface organization, adhesion molecules present on the conidial surface, and the secondary metabolites such as 1,8-dihydroxynaphthalene (DHN) melanin.7 DHN-melanin is present over the fungal cell wall and imparts greenish-gray color to A. fumigatus conidia. It plays a major adaptive role during harsh environmental conditions such as ultraviolet irradiation, reactive nitrogen species, and reactive oxygen species (ROS) and also in thermotolerance.8 It is closely associated with adhesion molecules such as hydrophobins forming rodlet layer that provides conidial hydrophobicity, physical resistance, and immunological inertness to A. fumigatus against the host immune system.9 DHN-melanin is also known to bind to the antimicrobial peptides and reduces the effectiveness of antifungal drugs.10
Unlike dihydroxyphenylalanine melanin that is present in mammalian cell,11 DHN-melanin biosynthesis in A. fumigatus is a polyketide-based pigment synthesis which consists of a cluster of six genes: pksP/alb1, ayg1, arp1, arp2, abr1, and abr2; expressed during conidiation.25 Polyketide synthase (PKS) protein, encoded by pksP/alb1gene, catalyzes the synthesis of heptaketide napthopyrone YWA1 using the endogenous precursor molecules malonyl-CoA and acetyl-CoA. The sequential reduction, dehydration, and polymerization lead to the formation of DHN-melanin.12 The microscopic and macroscopic studies have shown that pksP/alb1 mutant (ΔpksP) of A. fumigatus exclusively produces white, demelanized, and smooth conidial surface without any protrusions.12 However, mutation in other genes of this cluster, namely, ayg1, arp1, arp2, and abr1 produce yellowish, pinkish, brownish, and brownish-green colored colonies respectively, with protrusions similar to wild type (WT) A. fumigatus.13 The protrusions are important for cell–cell interaction and adherence to host epithelial cell surface which in turn enhances colonization and spreading of A. fumigatus linked infections.14 Furthermore, in vivo studies have shown that only ΔpksPA. fumigatus exhibits avirulence.9 The other colored mutants have unaltered virulence similar to the WT. ΔpksP conidia enhances phagolysosomal acidification, a critical step for killing of microbes inside host cell, making A. fumigatus highly susceptible to ROS and fast eradication by monocytes as compared to WT conidia.15
Exploration of bioactive compounds as potential antifungal agents is a prominent approach used in modern drug development due to their easy availability and minimal side effects.16 In the present study, the antimelanogenic antifungal effect of a natural compound cis-9-hexadecenal (C-9-H) was evaluated in A. fumigatus. C-9-H is present in many plants such as Myristica fragrans, Aegle marmelos, Thuja orientalis, Pentaclethra macrophylla Benth, Marchantia papillata, and Cuminum cyminum.17 It is a bioactive natural compound that belongs to group of organic C16 monounsaturated fatty aldehydes which has been reported to possess antimicrobial and anti-inflammatory properties.18 It has been observed that C-9-H inhibited biofilm formation in A. fumigatus and was not cytotoxic to human normal lung epithelial cell line L-132.17 ADME/Tox study has also revealed that C-9-H is a stable compound at varied physiological conditions and is safe for human consumption.17 It has been described as an aliphatic long-chain volatile compound that helps in fatty acid degradation and has characteristic strong fragrance.19 It has also been used in the synthesis of lipidized-soluble and long-acting insulin to enhance its hypoglycaemic effect.20 The in vitro combined effect of C-9-H and conventional antifungal drug Amphotericin B (AmpB) has been shown to display enhanced efficacy against A. fumigatus.17 However, the antimelanogenic activity of C-9-H against A. fumigatus is still unexplored, hence the compound needs to be explored further for its medicinal prospect which will broaden the available pharmacopoeias.
2. Results and Discussion
2.1. Determination of MEC of the Compound C-9-H for Antimelanogenic Activity in A. fumigatus
The compound C-9-H was dissolved in dimethylsulfoxide (DMSO) for all the experiments performed. DMSO-dissolved sample binds to the microbial cell membrane and increases the permeability of the membrane.21 In the present study, <5% concentration of DMSO was used. Radhika and Michael, 2013, recommended that the use of DMSO till 5% does not indicate any detectable effect during bioassays.22
Broth microdilution method was used for the determination of minimum effective concentration (MEC) which is referred as the lowest concentration of compound at which no pigmentation (demelanized conidia) was observed visually and phenotypically.23 The dilution method is considered as the best method for the determination of MIC/MEC value as it offers the possibility of estimation at varied concentrations of antimicrobial agents.24 The colony color after C-9-H treatment was compared with that of WT (positive control) and ΔpksP (mutant strain) in a 96-well plate. The result revealed that the phytocompound C-9-H inhibited melanin formation in A. fumigatus, leading to the appearance of pigmentless white conidia similar to ΔpksPA. fumigatus at a MEC of 0.293 mM (Figure 1).
Figure 1.

Estimation of MEC of cis-9-hexadecenal (A), AmpB (B), and DMSO (C). Demelanized colonies were observed in C-9-H-treated wells at 0.293 mM concentration, whereas no demelanization was observed in AmpB- and DMSO-treated wells.
The antimelanogenic effect of compound C-9-H was also analyzed against other melanized fungus viz., Magnaporthe oryzae, Aspergillus terreus, and Aspergillus flavus (Figure 2). The results revealed that only M. oryzae which produces melanin through DHN polyketide pathway,25 developed white demelanized colonies at 2.34 mM C-9-H concentration, whereas no demelanization was observed in A. terreus and A. flavus (Figure 2). A. terreus lacks pksP homolog in melanin biosynthesis pathway,26 whereas A. flavus produces melanin through non-DHN pathway.27
Figure 2.

MEC of the compound cis-9-hexadecenal for analyzing antimelanogenic efficacy in (A) M. oryzae, (B) A. terreus, and (C) A. flavus. Demelanized colonies were observed only in M. oryzae at 2.34 mM concentration of C-9-H whereas no demelanization was observed in A. terreus and A. flavus.
2.2. Phenotypic Characterization of C-9-H-Treated A. fumigatus in Comparison with DHN Melanin Inhibitors
Tricyclazoles (TC) and pyroquilones (PQ) are well-known DHN-inhibitors that inhibit the THN reductase enzyme encoded by arp2 gene of DHN-melanin biosynthesis gene cluster. Inhibition of arp2 gene or its product THN reductase as in case of TC and PQ leads to the formation of brown colored colonies. Inhibition of pksP/alb1 gene or its gene product PKS enzyme only leads to the formation of white demelanized colonies.12 Hence, the phenotypic effect of TC, PQ, C-9-H, and AmpB treatment was compared with that of WT and ΔpksPA. fumigatus. The culture plates were visually analyzed for change in colony color and radial growth on Czapek Dox agar (CzA) plate as a preliminary step in the identification of the target in DHN-melanin biosynthesis pathway. The results showed that WT A. fumigatus produced greenish-gray conidia on CzA plates, whereas TC- as well as PQ-treated plates were brown. Similar result was also reported by Pihet et al., 2009.12 This confirms that inhibition of either gene or gene product leads to similar alteration in conidial phenotype and produces same colored colonies. AmpB-treated colony formed regular greenish-gray conidia similar to WT A. fumigatus. White demelanized colonies were observed only in C-9-H-treated and ΔpksP plates (Figure 3), which may be attributed to the inhibition of DHN-melanin pathway at either pksP/alb1 gene or its gene product PKS protein level. Because demelanization leads to avirulence,28 further experiments were designed to understand the antimelanogenic associated virulence determinants related to surface organization after C-9-H treatment in A. fumigatus.
Figure 3.

Phenotype-based visual observation of A. fumigatus conidial color in the presence of DHN-melanin inhibitors (TC and PQ), phytocompound C-9-H, and antifungal drug AmpB. A. fumigatus was cultured on CzA supplemented with TC, PQ, AmpB, and C-9-H. The color of the colonies was observed visually and compared with that of WT and ΔpksP. WT—wild type, TC—tricyclazole, PQ—pyroquilon, C-9-H—cis-9-hexadecenal, AmpB—amphotericin B, ΔpksP—pksP mutant.
The radial growth assay results showed that WT A. fumigatus produced maximum diameter as compared to treated and ΔpksP colonies (Table 1). Because the radial diameter of antifungal agents has not been validated till date, it is expressed in terms of antifungal index percentage to better analyze the result of in vitro antifungal activity.29 The antifungal index value measures the ability of compounds to inhibit the fungal growth on the basis of radial diameter of fungal colonies on agar plate.29 The higher the antifungal index, the higher is the antifungal activity. The antifungal index percent of C-9-H was highest in comparison to other treatments in A. fumigatus. C-9-H has already been reported to possess antimicrobial and anti-inflammatory properties.17 It belongs to C16 long chain mono-unsaturated fatty aldehyde group that readily reacts with the nucleophilic groups of the cell membrane, resulting in alteration of membrane permeability and leakage of intracellular materials.19,30
Table 1. Comparative Results of Radial Growth Assaya.
| treatment | A. fumigatus | radial growth (mm) | antifungal index (%) |
|---|---|---|---|
| wild type | WT | 78 | |
| DHN-inhibitors | TC | 68 | 13 |
| PQ | 55 | 30 | |
| bioactive compound | C-9-H | 30 | 61 |
| antifungal drug | AmpB | 39 | 50 |
| pksP mutant | ΔpksP | 38 | 51 |
Wild-type (WT) A. fumigatus were cultured on CzA plates supplemented with tricyclazole (TC), pyroquilon (PQ), cis-9-hexadecenal (C-9-H), and amphotericin B (AmpB). The radial diameter of hyphal growth was measured after 4 days of incubation at 28 ± 2 °C and compared with that of WT (positive control) and ΔpksP (negative control).
2.3. Extraction, Physio-Chemical Characterization, and Spectrophotometric Analysis of Isolated Melanin
Melanin pigment is concentrated on the cell wall of A. fumigatus and gives a characteristic greenish-gray color to the conidia.12 To estimate reduction in melanin synthesis and its accumulation, melanin was extracted from WT, compound-treated, and ΔpksPA. fumigatus strain using 1 M KOH by autoclaving followed by acid hydrolysis. This method disrupts the linked proteins and other biological components associated with melanin, which can be further purified using acid treatment.31 The extracted pigment showed positive result for the physico-chemical tests used for fungal melanin diagnosis. The chemical tests were performed for the identification of isolated melanin pigment. The extracted melanin pigment was soluble at alkaline pH such as 1 M NaOH and 1 M KOH, insoluble at acidic pH such as 6 M HCl, organic solvents like chloroform, ethyl acetate, alcohol and acetone, and water.27 The melanin was decolorized by oxidizing agents such as H2O2 and KMnO4.27 The extracted pigment was precipitated in 1% FeCl3, which signified that it is a polyphenol.32
The spectrophotometric estimation showed 91% reduction in melanin content in A. fumigatus when treated with C-9-H at the concentration of 0.293 mM (Figure 4A; p ≤ 0.0087). The spectral study of melanin from both WT and treated cultures showed a characteristic peak in the UV region 200–260 nm (Figure 4B). Maximum light was absorbed in the UV region which gradually decreased with the increment in the wavelength. High absorbance in the UV range may be attributed to the presence of aromatic complex conjugated compounds in melanin.26 No melanin pigment was extracted from ΔpksP; therefore, the ΔpksP strain showed no characteristic peak depicting the absence of melanin. Because demelanization can also lead to avirulence,26 these results were further corroborated for other virulence factors in further analysis.
Figure 4.
Evaluation of the physical properties of the conidial surface. The WT A. fumigatus without any treatment (positive control), WT with C-9-H (0.293 mM), AmpB (0.012 mM; drug control) and ΔpksP strain (negative control) were grown on Czapek medium (n = 3). (A) Melanin estimation in treated and control group (p ≤ 0.0087), (B) UV–vis spectrum of melanin showing a characteristic peak at UV-region 200–260 with gradual decrease in absorption toward visible range where red, black, and blue curve shows the melanin spectra of WT, AmpB, and C-9-H respectively, and (C) altered membrane surface depicted by reduced cell surface hydrophobicity (CSH) (p ≤ 0.0071).
2.4. CSH in A. fumigatus
Cell Surface Hydrophobicity (CSH) contributes to the interaction between A. fumigatus and the host epithelial cell surface, which is an important factor for spreading infection. The presence of melanin increases hydrophobicity and adherence of the conidia to any surface.33A. fumigatus cell surface is charged due to ionization of free amine and carboxylic acid groups of proteins. The presence of melanin increases both negative charge and hydrophobicity of the conidia. It has been reported that the reduction in melanin formation is directly proportional to the increase/decrease in CSH and the adherence capacity of the conidia to any surface.33 Once adhered, the conidia proliferates into the hyphal structure to spread the infection inside host body. Therefore, reduction in hydrophobicity relates to reduction in adherence, thereby lowering the infection rate. The assessment of CSH was done by two-phase partitioning MATH assay using two solvent systems: water and hexadecane.34 Hexadecane provides a hydrophobic medium, whereas water provides the hydrophilic medium to the conidia. The higher amounts of conidia in hexadecane phase meant higher hydrophobicity. C-9-H treatment enhanced wettability of the conidia similar to that of ΔpksP. A decrease in the CSH for both C-9-H-treated and ΔpksP culture was observed as compared to the control wild type green conidia. The compound-treated and ΔpksP conidia showed CSH up to 59% and 74.5%, respectively, as compared to WT A. fumigatus (Figure 4C; p ≤ 0.0071). This decrease in hydrophobicity was also observed during the preparation of conidial suspensions. This suggested that restriction of the melanin biosynthetic pathway leads to reduction in CSH, contributing to loss of adherence property, colonization, and potential pathogenicity, thereby reducing the infection.12 Similar results were also reported on the ΔpksPA. fumigatus by Pihet et al., 2009.12
2.5. Cell Surface Morphology Study of A. fumigatus Using Electron Microscopy
2.5.1. Scanning Electron Microscopy
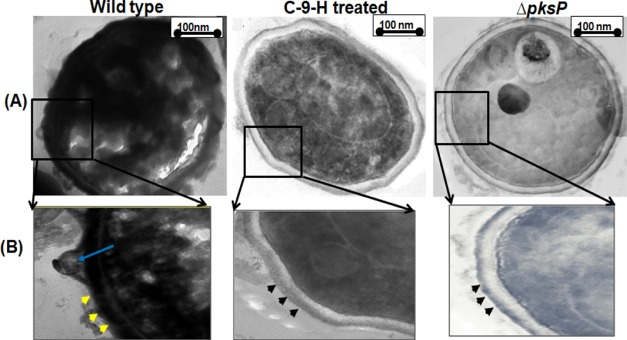
The electron microscopic results also ascertained a close resemblance in the cell surface morphology and melanin deposition in C-9-H-treated A. fumigatus. The scanning electron microscopy (SEM) micrograph revealed that both WT- and AmpB-treated A. fumigatus conidia had a rough echinulate surface with many protrusions (Figure 5). The conidial protrusions protect the fungus from phagocytosis and enhance its resistance to ROS produced by phagocytic cells in the host body.9 Both C-9-H-treated and ΔpksP-demelanized conidia revealed a smooth surface without any protrusions. Similar results were also observed by Pihet et al., 2009 in ΔpksPA. fumigatus.12 The protrusions are extremely important in adherence to human epithelial tissue and to spread infection.26 Conidia devoid of protrusions will not be able to adhere to the cell surface and hence would be less pathogenic.
Figure 5.
Visualization of the A. fumigatus conidial surface by SEM. Magnification is 50k× and scale corresponding to 200 nm. WT—wild type, C-9-H—cis-9-hexadecenal-treated, AmpB—amphotericin B–treated, and ΔpksP—pksP mutant A. fumigatus.
2.5.2. Transmission Electron Microscopy
The lateral conidial surface of the WT, C-9-H-treated and ΔpksPA. fumigatus conidia was visualized by transmission electron microscopy (TEM). The TEM micrograph of the WT conidia showed thick inner layer, indicating the presence of melanin concentrated on the membrane (Figure 6). TEM micrographs also revealed the presence of protrusions in WT conidia affirming the SEM observations. On the contrary, both C-9-H-treated and ΔpksPA. fumigatus conidial section revealed visibly clear inner surface without melanin deposition. There were no protrusions visualized in C-9-H-treated and ΔpksP conidial outer surface. Moreover, the accumulation of the cytoplasmic content was clearly seen in WT A. fumigatus conidia, whereas in C-9-H-treated and ΔpksP conidia cytoplasmic content was reduced which could be attributed to the alteration in membrane, leading to leakage of cellular contents.12 Similar observations have been reported in ΔpksPA. fumigatus.12 The membrane of C-9-H-treated conidia was observed to be partly aberrant in comparison to wild-type TEM micrograph. A similar TEM micrograph of disrupted membrane in Candida albicans was obtained upon treatment with aminopiperidine derived antifungal compound, suggesting the possibility of cellular leakage.35
Figure 6.
Ultrastructure of the lateral section of wild type (WT), cis-9-hexadecenal (C-9-H)–treated, and pksP mutant (ΔpksP) A. fumigatus conidia visualized by TEM. C-9-H and ΔpksP have a smooth surface (black arrowheads), whereas WT have a rough sough surface with melanin deposition (yellow arrowheads) and protrusions (blue arrow). Magnifications (A) 10k×, (B) 20k×.
2.6. Relative Expression Profiling of PKS pksP/alb1 Gene Using qRT-PCR
The mutant study of each genes involved in DHN-melanin biosynthesis, conducted elsewhere, revealed that only inhibition of pksP/alb1 gene or its gene product forms white avirulent colonies.12 Moreover, the present study showed that the characteristics of C-9-H-treated A. fumigatus was similar to that of ΔpksP strains. Hence, in order to understand the effect of C-9-H on the expression of pksP/alb1, the transcript profiling of pksP/alb1 gene was performed for WT- (positive control), C-9-H–treated, and ΔpksP (negative control) culture using real time-quantitative PCR (qRT-PCR). qRT-PCR is a sensitive and specific method for the analysis of gene expression on the basis of changes in the expression of a target gene relative to the reference gene expression.36 Reference genes are “house-keeping” genes whose expression remains constant at variant physiological conditions and are used for gene normalization.36 In the present study, β-actin was used as reference gene and expression stability was analyzed on the basis of threshold cycle (ΔCt) value.36 The relative expression of pksP/alb1 gene was 3.5-fold (p ≤ 0.0005) upregulated in C-9-H-treated sample as compared to WT sample, whereas there was no expression of pksP/alb1 gene in ΔpksPA. fumigatus (Figure 7). Upregulation of the gene expression is attributed in response to the development of any stress in the pathogen.37
Figure 7.
Relative expression fold of pksP/alb1 gene with wild type (WT), cis-9-hexadecenal (C-9-H) treated, and pksP mutant (ΔpksP) A. fumigatus (p ≤ 0.0005). RNA was extracted from 4 days old untreated WT, C-9-H treated, and (ΔpksP) A. fumigatus (n = 3). β-Actin expression was used as an internal control. mRNA expression corresponding to the pksp/alb1 gene was compared with that expressed in the untreated A. fumigatus.
2.7. Total Proteome Analysis
The total protein from 4 days old WT- and C-9-H-treated A. fumigatus culture was extracted and analyzed using nano-LC-MS-MS to correlate the transcriptional data and other enzymes of the DHN-melanin pathways. The result showed a combined total of 1805 proteins in both samples. On the basis of the cutoff value (>2 fold change), 309 proteins were found to be differentially expressed. There were 31 proteins that were present solely in control and 51 proteins were exclusively present in protein sample obtained from C-9-H-treated A. fumigatus.
These proteins were characterized into functional pathway gene ontology (GO) using cellular localizations and biological functions. GO analysis helps in characterization of proteins based on various functional pathways.38 On the basis of cellular functions, GO study showed that 36% and 7% of membrane and extracellular proteins, respectively, were differentially expressed in control and treated samples (Figure 8A). On the basis of biological functions, it was determined that 3% cell wall integrity proteins, 9% secondary metabolites, and 5% cell stress proteins were differentially expressed between proteins isolated from WT- and C-9-H-treated A. fumigatus, respectively (Figure 8B).
Figure 8.

GO study based on (A) cellular and (B) biological functions. On the basis of cellular localization 36% membrane proteins and 7% of extracellular proteins were differentially expressed in WT- and C-9-H-treated sample. Biological GO showed that 3% cell wall integrity proteins, 9% secondary metabolites, and 5% cell stress proteins were differentially expressed between proteins isolated from WT- and C-9-H-treated A. fumigatus respectively.
Amongst total proteome, the differentially expressed proteins related to the cell surface and toxins were further analyzed to find the upregulation /downregulation in WT- and C-9-H-treated sample which has been listed in Table 2. It was found that the PKS protein was also upregulated as observed in transcript analysis. The proteome profiling affirmed the transcript study similar to García-Martínez et al., 2007.39 The result also revealed that except PKS proteins, no other downstream enzymes involved in melanin synthesis pathway were present. The formation of each product in the DHN-melanin biosynthetic pathway is catalyzed by a specific enzyme.40 The upregulation of PKS protein and absence of other downstream proteins in the pathway, suggested that the identified compound C-9-H is inhibiting the initial step, conversion of malonyl-CoA, and acetyl-CoA to heptaketide, catalyzed by PKS.12 Because of the absence of any substrates for the subsequent step, the products are not observed in the present protein data set. This affirmed the inhibition of the downstream reactions in the DHN-melanin synthesis pathway, stopping the formation of the final product DHN-melanin. The analysis suggested that C-9-H is acting on stress responses in A. fumigatus and is also altering the expression of regulatory proteins responsible for maintaining cell membrane integrity.
Table 2. Fold Change in Proteins Previously Reported as Virulence Factors/Others in A. fumigatus.
| s. no. | protein Id | proteins | fold change | remarks |
|---|---|---|---|---|
| 1 | tr|A0A229Y072|A0A229Y072_ASPFM | integral membrane protein | 2.4 | downregulated |
| 2 | tr|A0A0J5SC73|A0A0J5SC73_ASPFM | protein transport protein sec13 | 2 | downregulated |
| 3 | tr|A0A0J5Q333|A0A0J5Q333_ASPFM | thioredoxin reductase GliT | 16 | downregulated |
| 4 | tr|B0Y816|B0Y816_ASPFC | methyltransferase GliN | 12 | downregulated |
| 5 | tr|A0A0J5PNZ4|A0A0J5PNZ4_ASPFM | secreted antimicrobial peptide | 2.4 | upregulated |
| 6 | tr|A0A0J5PG83|A0A0J5PG83_ASPFM | PKS | 2.5 | upregulated |
| 7 | tr|A0A0J5PDW0|A0A0J5PDW0_ASPFM | catalase | 2 | upregulated |
2.8. Structural Analysis of PKS Protein and In Silico Docking Analysis
Since both transcript and proteome study revealed upregulation in pksP/alb1 gene expression and PKS protein expression, respectively, the interaction between C-9-H and PKS protein was studied.
2.8.1. PKS Structure Prediction
The PKS protein sequence of A. fumigatus was retrieved from the NCBI, and homology modeling was performed using ExPASy’s web server tools. Homology modeling is considered as one of the best in silico approaches to predict and validate 3-D model. In this method, the alignment of known protein structures acts as templates and help in the prediction of the structure of unknown protein sequence. The higher the similarity percentage, the higher is the acceptability rate of the model. The Expasy’s Protparam computes the primary structure of protein on the basis of the physico-chemical parameters.41 PKS protein consists of 2146 amino acids and has a molecular weight of 234.4 kD. Protparam computes the extinction coefficient at various wavelengths including 276, 278, 279, 280, and 282 nm. However, the proteins show maximum absorbance at 280 nm with minimum interference. Therefore, on the basis of the concentrations of cystine, tyrosine, and tryptophan, the extinction coefficient is favored at 280 nm.42 In the present study, a high extinction coefficient of 236 960 M–1 cm–1 may be attributed to high concentration of tyrosine (2.5%).43 PKS has 239 negatively charged amino acid residues (aspartate and glutamate) and 203 positively charged (arginine and lysine) amino acid residues. High negative charge amino acid residue than positively charged residue signifies that the protein is extracellular. The pH at which the net charge of stable protein is zero is called the isoelectric point (pI), which predicts the alkaline or acidic nature of the protein.43 The pI of PKS protein is less than 7 (5.90), making the protein acidic.42 The computed pI helps in protein purification by the isoelectric point focusing on a polyacrylamide gel.43
The instability index and aliphatic index help in prediction of protein stability. The instability index of more than 40 depicts unstable protein.42 The instability index of the PKS protein was found to be <40, indicating it as a stable protein at varied conditions. The aliphatic index of a protein is the volume occupied by aliphatic amino acid side chains with respect to the total volume occupied by protein in a solution. The PKS protein of A. fumigatus has a high aliphatic index of 83.15 due to the high concentration of aliphatic amino acids. This predicts that the PKS protein is thermostable.42 The grand average of hydropathicity (GRAVY) index is the sum of hydropathy values of all the amino acids, divided by the number of residues in the given protein sequence.43 Low GRAVY index (−0.219) of PKS protein infers better aqueous interaction.
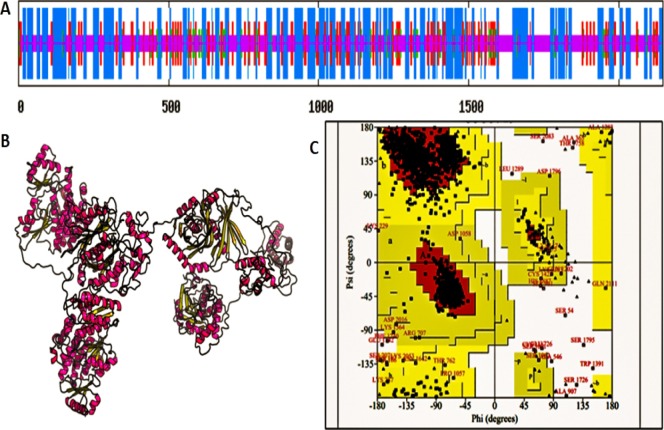
The secondary structure of the protein was predicted by Self-Optimized Prediction Method with Alignment (SOPMA) tool. SOPMA tool gives the structural information of protein sequence and helps to determine the percentage of α-helix, β-sheets, turns, and coils in query protein.42 The protein was found to have 37.09% α-helix (blue), 15.14% β-sheets (red), 6.01% turns (green), and 41.75% coils (purple) as seen in Figure 9A. These information have high importance in prediction of the tertiary protein structure.41
Figure 9.
(A) Secondary structure of PKS protein of A. fumigatus as predicted by SOPMA software. SOPMA: Self Optimized Prediction Method with Alignment. α-Helixes are depicted by blue lines, β-sheets by red lines, turns by green lines and coils by purple lines, (B) tertiary structure of PKS protein of A. fumigatus as predicted by RaptorX software, (C) Ramachandran plot of predicted 3-D model of PKS protein of A. fumigatus by PROCHECK software. The bottom left box indicates the presence of right-handed α-helix. The red region indicates the favored region where no steric clashes occur. The majority of amino acids are present in phi–psi distribution.
The tertiary structure was predicted by RaptorX server which computes the solvent accessibility, disordered regions, and binding sites of the PKS protein.44 The quality of the selected model is indicated on the basis of p-value, GDT (global distance test), and uGDT (un-normalized GDT). Smaller p-value meant a better quality of the 3-D model.45 The server selected the 6fijA model as the best model for PKS protein on the basis of low p-value 2.53 × 10–26 (Figure 9B). The server predicted that the PKS protein is multidomain protein with overall uGDT (GDT): 1364 (63%), 192 (8%) disordered positions which are good indicators of the predicted model. The solvent accessibility of any protein is divided into three categories based on cutoff values. If less than 10% surface is accessible to solvents, protein is under buried region. More than 42% solvent accessibility of protein is identified as exposed, and the values between 10 and 42% are medium.46 In the present study, 37% area was exposed followed by 33% buried region and 28% medium region for solvent accessibility. Appaiah and Vasu, 2016 reported that uGDT above 50 is a good indicator of any protein model with more than 100 residues.47 Because PKS protein has 2146 amino acid residues, the overall uGDT of 1364 infers the selected structure as of the high-quality model.
The structure was validated by the Ramachandran plot using PROCHECK server, which evaluates the stereochemistry of the protein model by determining residue-by-residue geometry and overall structure geometry.44 In the Ramachandran plot, the selected protein structure is considered a good-quality model on the basis of most favored (core), additional allowed, generously allowed and disallowed regions. A good quality model should have over 90% in the most favored regions.45 In the present study, 91.1% of the amino acid residues were present in the most favored region, whereas 6.9% residues were found in the additional allowed region. About 1.1% amino acid residues were seen to be in the generously allowed region and 0.9% in the disallowed region. The bottom left box indicated the presence of right-handed α-helix. The red region indicated favored region where no steric clashes occur. These results indicated that the majority of amino acids fall in phi–psi distribution. The Ramachandran plot revealed that the designed Protein Model for PKS protein of A. fumigatus was good and stable and was thus considered as reliable model.
2.8.2. In Silico Docking Analysis between C-9-H and PKS
After confirming the reliable 3-D model of PKS protein, the UCSF Chimera was used to visualize and analyze the interaction between C-9-H and PKS protein.46 The stability of the compound is an important parameter for their effectiveness. The computational study has reported C-9-H as a stable compound at varied conditions.17 In the docking analysis, low binding energy of −4.95 kcal/mol was observed which was stabilized by hydrophobic interaction and hydrogen bond between C-9-H and the amino acid residues of PKS protein. The hydrogen bonds were formed at Thr-264 and Ser 171 residue of PKS active domain (Figure 10). The binding features such as binding energy and hydrogen bonding between C-9-H to the PKS model was satisfactory and notable. The binding energy is based on the intermolecular interaction, entropic effects, and desolvation.48 The lowest binding energy predicts the strongest binding affinity. The interaction suggested the blockage of the active site of PKS by C-9-H, due to which further reaction in DHN-melanin biosynthesis might have stopped. This result was in sync with the proteome analysis where no protein of DHN-melanin pathway downstream to PKS protein was found.
Figure 10.

Docking study of the compounds with the active domain of A. fumigatus PKS protein. Protein–ligand interaction was visualized by UCSF Chimera. The bond between PKS and C-9-H is marked with blue arrow. The amino acid residues (SER-171 and THR 264) forming bonds are marked with blue circles. Green arrow shows the ligand C-9-H.
3. Conclusions
The study concludes that the compound cis-9-hexadecenal has potential antimelanogenic antifungal properties. It targets the cell wall organization, critical growth factor, and virulence in A. fumigatus as depicted by inhibition of DHN polyketide melanin synthesis, loss in cell surface protrusions, forming smooth cell wall and reduced CSH. Thus, it can decrease the pathogenicity of A. fumigatus and increase its susceptibility to available antifungal drugs. The efficacy of C-9-H can be further enhanced through structural modification. This study may provide a trail for elucidating a newer and safer therapeutics against A. fumigatus infections.
4. Materials and Methods
4.1. Procurement of Fungal Strains and the Natural Compound cis-9-Hexadecenal
A. fumigatus (ATCC-46645) and its pksP/alb1mutant (ΔpksP) strain9 were a kind gift from Prof. Axel Brakhage, Department of Molecular and Applied Microbiology, Leibniz Institute for Natural Product Research and Infection Biology-HKI, Germany. A. terreus (NCCPF-860035), A. flavus (MTCC 9367), and M. oryzae (ITCC 7019) were also used in the study. Aspergillus strains were maintained by subculturing on CzA (HiMedia, India) slants monthly. The fungus was grown on CzA at 28 ± 2 °C for 4 days to obtain conidial growth. M. oryzae was cultured on potato dextrose agar plates at 28 ± 2 °C for 7 days.
The compound C-9-H was procured from Carbosynth (Funakoshi Co, Japan) and was dissolved in 100% DMSO (HiMedia, India) to make the final concentration of 100 mg/mL. Stock solutions in DMSO were further diluted in media before performing any experiments so that the final concentration of DMSO did not exceed an amount that had any detectable effect in assays.49
4.2. Determination of MEC of the cis-9-Hexadecenal for Antimelanogenic Activity in A. fumigatus
The MEC is the concentration of cis-9-hexadecenal at which demelanized white A. fumigatus colonies can be visualized as compared to the greenish-gray WT (positive control) colonies and ΔpksP. This was determined by the broth microdilution method with some modification following the Clinical and Laboratory Standards Institute (CLSI).50 The experiment was conducted in triplicates. Variable concentrations of C-9-H (100 μL) in Czapek Dox Broth (CzB) were made in 96-well polystyrene plates (Tarson, India) to ascertain the concentrations from 20.92–0.016 mM. Further, 100 μL of A. fumigatus conidial suspension in CzB (approximately to 0.4 × 104 conidia/mL)24 were added to each well till 10th column of the 96-well plate. Column 11 was regarded as positive control having conidial suspension (100 μL) and CzB (100 μL). Column 12 was set as negative control comprising of CzB (200 μL) only. The MEC of the compound was visualized after 4 days of incubation at 28 ± 2 °C.
4.3. Phenotypic Comparison between cis-9-Hexadecenal and DHN-Melanin Inhibitors Treated A. fumigatus
Effects of DHN-melanin inhibitors TC (HiMedia, India) and PQ (HiMedia, India) were studied along with the compound C-9-H for asserting the phenotypic changes occurring after treatment with C-9-H. Sterilized CzA was supplemented with TC, PQ (20 μg/mL), and C-9-H at MEC and AmpB (HiMedia, India) in separate plates. After 30 min, 10 μL of prepared conidial suspension was added dropwise at the center of treated and control plates. All of the plates were incubated at 28 ± 2 °C for 4 days. Experiments were performed in triplicates, and percentage inhibition was interpreted on the basis of colony color and diameter on 4th day. The antifungal index percent was calculated to determine the antifungal activity using the following formula29
where Dt = diameter of the colony in treatment (t) and Dc = diameter of the colony on control (c).
4.4. Extraction, Physio-Chemical Characterization, and Spectrophotometric Analysis of Isolated Melanin
The isolation of cell–wall associated melanin from WT and treated A. fumigatus conidia was performed.27 Mycelial plug (1 cm diameter) was cut from the colonies grown on CzA, boiled in 5 mL of distilled water for 5 min, and then centrifuged for 5 min at 7000 rpm. The pellet was washed with sterile distilled water twice and melanin was extracted by autoclaving the pellet in the presence of 1 M KOH (3 mL; HiMedia, India). The extracted melanin was dried overnight at 20 °C in a dehumidified atmosphere. Further, acid hydrolysis was done to purify the extracted melanin by adding 7 M HCl (5 mL; HiMedia, India) in a sealed glass vial for 2 h at 100 °C. After cooling, the pigment was washed three times with distilled water, dried, and stored at 4 °C. Extracted and dried pigment from solid culture was dissolved in 1 M KOH and melanin was recorded as the absorption spectra at range of 200–800 nm using 1 M KOH as reference blank. The absorption spectra of the control and treated plates were analyzed.27
The pigments extracted from the isolates were confirmed as melanin on the basis of their physical and chemical properties. Characterization of melanin is based on the solubility in NaOH/KOH, insolubility in water or organic solvents, decolorization by the oxidizing agents (H2O2/KMnO4), and precipitation by 1% FeCl3.27
4.5. Cell Surface Hydrophobicity
Hydrophobicity of the microbial cell suspension was evaluated by two-phase partitioning using hexadecane as the hydrocarbon phase described by Kennedy et al., with minor modification.34 Briefly, conidia were harvested using phosphate-buffered saline (PBS), and their absorbance was set to 0.30 at 630 nm (A1). Hexadecane (500 μL; HiMedia, India) was then added and vortexed for 2 min. The suspension was incubated for 10 min at room temperature for phase separation. The absorbance of the aqueous phase was then measured at 630 nm (A2) and was compared to the initial absorbance. CSH percentage was calculated using following formulae
4.6. Cell Surface Morphology Studies of A. fumigatus Using Electron Microscopy
4.6.1. Scanning Electron Microscopy
Conidia from 4 days old A. fumigatus cultures grown on CzA medium with and without the compound (at MEC) were harvested. Conidia were fixed in 4% glutaraldehyde (HiMedia, India) in PBS for 24 h, under vacuum. Further conidia were washed, post-fixed with 1% osmium tetroxide (HiMedia, India) for 60 min, and dehydrated through series of ethanol washing with a gradual increase in concentration. The samples were then mounted on the aluminium sheet and coated with gold–palladium alloy. The observations were made using a Zeiss SEM, MA EVO—18 Special Edition.12
4.6.2. Transmission Electron Microscopy
A. fumigatus culture was grown in CzA medium with and/or without compound treatment for 4 days. Conidia were harvested, washed with distilled water, and fixed overnight at room temperature in 0.1 M sodium cacodylate buffer (pH 7.4; HiMedia, India) supplemented with 2.5% glutaraldehyde (HiMedia, India). After this, the conidia were incubated for 1.5 h at 20 °C in a solution of 4% formaldehyde–1% glutaraldehyde in 0.1% PBS and then further incubated in 2% osmium tetraoxide for 90 min. Dehydration was done by serially washing in ethanol solutions (50–95%) for 10 min, followed by two final washes in 100% ethanol for 15 min.51 The cells were embedded in Spurr’s resin, sectioned onto nickel grids, and examined on a JEOL 2100F transmission electron microscope to obtain micrographs.51
4.7. Relative Expression Study of pksP/alb1 Gene
To analyze the effect of the compound C-9-H on pksP/alb1 gene, transcript analysis was conducted using qRT-PCR technique.
4.7.1. Primer Designing
A. fumigatuspksp/alb1 and β-actin CDS gene sequences were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/pubmed) database for designing the primer for the expression analysis. The primers were designed by online available Primer 3 software (http://primer3.ut.ee/).
The primer sequences were then analyzed for potential hair pin formation and self-complementarity (http://www.basic.northwestern.edu/biotools/oligocalc.html). The primers used in the present study have been mentioned in Table 3.
Table 3. List of Primers Used in the Experiment.
| s. no. | gene name | accession no. | primer sequence (5′–3′) | amplicon size (bp) |
|---|---|---|---|---|
| 1 | pksP/alb1 | NC_007195.1 | F-GTCTACCTTCCTCACGACC | 167 |
| R-CAGCGTAGAGAGACGATGG | ||||
| 2 | β-actin | XM_742511.1 | F-ATCTGACGGACTACCTGATG | 179 |
| R-GTAGCAAAGCTTCTCCTTGA |
4.7.2. Total RNA Extraction, Purification, and cDNA Synthesis
Total RNA was extracted from A. fumigatus culture WT, compound treated, and ΔpksP using RNA-Xpress reagent (HiMedia, India) as per given manufacturer’s instruction. DNaseI treatment (Fermentas, USA) was given to the isolated RNA using the user guidelines provided by Fermentas to remove genomic DNA contamination. The purity of isolated RNA was measured by A260/280 and A260/230 ratios using nanodrop (Thermo Scientific Multiskan GO). Samples with <1.8 ratio for either of the absorbance ratio were not used for further analysis. Two micrograms of total RNA of each sample was used to synthesize first-strand cDNA by oligo (dT)18 primer using the high cDNA synthesis Kit (HiMedia, India).
4.7.3. Relative Expression Profiling of Genes by qRT-PCR
qRT-PCR amplification was carried out by using HiMedia LA-1012 Insta Q96-Real Time PCR, using a SYBR-green master mix (HiMedia, India), and the relative quantification of each individual expression of pksP/alb1 gene was performed using the comparative threshold cycle method.36 The amplification program used for real time was 95 °C for 3 min, 40 cycles at 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s. The melt curve analysis as per INSTA Q-96 programme RT-PCR was at 95 °C for 15 s, 60 °C for 60 s, and 72 °C for 30 s and holding time of 10 s. The results were analyzed using INSTA-Q96 software. At least two-fold up- or down-regulated gene expression was regarded as differentially expressed.
4.8. Total Proteome Analysis
4.8.1. Protein Extraction
A. fumigatus wet mat (1.5 g) with or without any treatment was ground in liquid nitrogen separately, and total protein was isolated at 4 °C in chilled 50 mM sodium phosphate buffer (pH 7.0) containing 2 mM EDTA (HiMedia, India), 0.2 mM dithiothreitol (HiMedia, India), and 1 mM PMSF (HiMedia, India), for 3 h under constant stirring. The extracted protein was then centrifuged at 12 000 rpm for 20 min at 4 °C. Trichloroacetic acid (5% of total volume) was added to the supernatant. The precipitate was washed with chilled acetone and dissolved in 6 M guanidium chloride (HiMedia, India).39
4.8.2. Liquid Chromatography Coupled with Tandem Mass Spectrometry (LC-MS-MS) and GO Analysis
Fifty microlitres of total protein sample was trypsinized for 16 h at 37 °C and then purified using column C-18 silica cartridge. The purified peptide mixture was dried by speed Vac and then resuspended in a buffer solution “A” comprising of 0.1% formic acid and 5% acetonitrile. The dried peptides (1 μg) were analyzed using a LC-MS-MS. An EASY-nLC 1000 system (Thermo Fisher Scientific, USA) coupled with QExactive MS (Thermo Fisher Scientific, USA) was employed to perform LC-MS-MS analysis.39 Nano-electro-spray was used as the ion source. The peptides were then loaded with buffer A and eluted at a flow rate of 300 nL/min using a 0–40% gradient of buffer “B” which consists of 0.1% formic acid and 95% acetonitrile. The total run time was for 90 min. The obtained raw files were analyzed with Proteome Discoverer against the Uniprot A. fumigatus. For SEQUEST/AMANDA search, the precursor and fragment mass tolerances were set at 10 ppm and 0.5 Da, respectively. Both peptide spectrum match and protein false discovery rate were set to 0.01 FDR.
The protein files obtained from LC-MS-MS were further screened for differentially expressed proteins on the basis of this cutoff value (>2-fold change) and GO analysis was performed for all groups.
4.9. Structural Analysis of PKS Protein and In Silico Docking Analysis
4.9.1. PKS Structure and Properties Prediction
The FASTA sequence of PKS protein of A. fumigatus was retrieved from NCBI. Because the three-dimensional structure of the protein was not available in Protein Data Bank (PDB), homology modeling was done using ExPASy web server.41
The primary sequence features were predicted by Protparam tool of Expasy server for the computation of various physical and chemical parameters for PKS sequence.41 The secondary structure of PKS sequence was predicted by Self-Optimized Prediction Method with Alignment (SOPMA) server.41 The tertiary structure of PKS protein was predicted using RaptorX server which predicts the protein structure and function.44 The best template having maximum identity with the target and modeling structure was then evaluated using PROCHECK.44
4.9.2. In Silico Docking Study
Molecular docking analysis was performed using AutoDock 4.2.6 for PKS protein. The Lamarckian genetic algorithm was used to perform the automated molecular dockings with the preset parameters. The total number of run was set to 50, and the lowest binding energy conformation was selected for UCSF Chimera analysis. The hydrogen bonds formed between C-9-H with the active site of PKS protein were analyzed.52
4.10. Statistical Analysis
For the statistical analysis, ANOVA was used, comparing the results of melanin estimation and CSH assay for compound-treated culture with wild type, drug-treated, and ΔpksP strain. All of the statistics was performed using GraphPad Prism software 8.0.2.263 version and Microsoft Excel. p ≤ 0.05 was considered statistically significant.
Acknowledgments
The authors thank Amity University Uttar Pradesh, India, for providing infrastructure for research. We thank Sonia Shishodia, Ph.D. Scholar, JUIT, Solan, HP, India, for her assistance in proteome result analysis.
Glossary
Abbreviations
- ΔpksP
pksP/alb1 mutant
- μL
microliter
- A. flavus
Aspergillus flavus
- A. fumigatus
Aspergillus fumigatus
- A. terreus
Aspergillus terreus
- AmpB
Amphotericin B
- ATCC
American Type Culture Collection
- bp
base pair
- cDNA
complementary deoxyribonucleic acid
- CLSI
Clinical and Laboratory Standards Institute
- CSH
cell surface hydrophobicity
- CzA
Czapek Dox Agar
- CzB
Czapek Dox Broth
- DHN
1,8-dihydroxynaphthalene
- DMSO
dimethylsulfoxide
- DNA
deoxyribonucleic acid
- dNTPs
deoxy nucleotide triphospahte
- EDTA
ethylenediamine tetra acetic acid
- FeCl3
ferric chloride
- GDT
global distance test
- GO
gene ontology
- GRAVY
grand average of hydropathicity
- H2O2
hydrogen peroxide
- HCl
hydrochloric acid
- kD
kilodalton
- KMnO4
potassium permanganate
- KOH
potassium hydroxide
- LC-MS-MS
liquid chromatography with tandem mass spectrometry
- mM
millimolar
- M. oryzae
Magnaporthe oryzae
- MATH
microbial adherence to hydrocarbon
- MEC
minimum effective concentration
- MTCC
Microbial Type Culture Collection
- NCCPF
National Culture Collection of Pathogenic Fungi
- nm
nanometer
- NaOH
sodium hydroxide
- pksP/alb1
polyketide synthase gene
- PBS
phosphate buffered saline
- PDA
potato dextrose agar
- PKS
polyketide synthase protein
- PMSF
phenyl methyl sulfonyl fluoride
- PQ
pyroquilone; qRT-PCR, real time quantitative polymerase chain reaction
- RNA
ribonucleic acid
- ROS
reactive oxygen species
- SEM
scanning electron microscopy
- SOPMA
Self-Optimized Prediction Method with Alignment
- TC
tricyclazoles
- TEM
transmission electron microscopy
- uGDT
un-normalized GDT
- WT
wild type
Author Contributions
S.H. carried out the experimental work, data interpretation, and prepared manuscript draft. L.G. helped in experimental work. J.S. supervised proteomics work. A.K.G. guided in qRT-PCR result analysis. P.V. designed the study and supervised the work. All authors have read and approved the final manuscript.
This work was financially supported by the DST-SERB in the form of fast track Young scientist (grant no: SB/YS/LS-72/2013), New Delhi, India.
The authors declare no competing financial interest.
References
- Bhetariya P. J.; Madan T.; Basir S. F.; Varma A.; Usha S. P. Allergens/antigens, toxins and polyketides of important Aspergillus species. Indian J. Clin. Biochem. 2011, 26, 104–119. 10.1007/s12291-011-0131-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Veerdonk F. L.; Gresnigt M. S.; Romani L.; Netea M. G.; Latgé J.-P. Aspergillus fumigatus morphology and dynamic host interactions. Nat. Rev. Microbiol. 2017, 15, 661–674. 10.1038/nrmicro.2017.90. [DOI] [PubMed] [Google Scholar]
- Scorzoni L.; de Paula e Silva A. C. A.; Marcos C. M.; Assato P. A.; de Melo W. C. M. A.; de Oliveira H. C.; Costa-Orlandi C. B.; Mendes-Giannini M. J. S.; Fusco-Almeida A. M. Antifungal therapy: new advances in the understanding and treatment of mycosis. Front. Microbiol. 2017, 8, 36. 10.3389/fmicb.2017.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly V.; Saint-Pierre-Chazalet M.; Saint-Julien L.; Bolard J.; Carbon C.; Yeni P. Inhibiting cholesterol synthesis reduces the binding and toxicity of amphotericin B against rabbit renal tubular cells in primary culture. J. Infect. Dis. 1992, 165, 337–343. 10.1093/infdis/165.2.337. [DOI] [PubMed] [Google Scholar]
- Boominathan M.; Ramamurthy V. Antimicrobial activity of Heliotropium indicum and Coldenia procumbens. J. Ecobiol. 2009, 24, 11–15. [Google Scholar]
- Sanglard D. Emerging threats in antifungal-resistant fungal pathogens. Front. Med. 2016, 3, 11. 10.3389/fmed.2016.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulussen C.; Hallsworth J. E.; Álvarez-Pérez S.; Nierman W. C.; Hamill P. G.; Blain D.; Rediers H.; Lievens B. Ecology of aspergillosis: Insights into the pathogenic potency of Aspergillus fumigatus and some other Aspergillus species. Microb. Biotechnol. 2016, 10, 296–322. 10.1111/1751-7915.12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankar J.; Tiwari S.; Shishodia S. K.; Gangwar M.; Hoda S.; Thakur R.; Vijayaraghavan P. Molecular insights into development and virulence determinants of Aspergilli: A proteomic perspective. Front. Cell. Infect. Microbiol. 2018, 8, 180. 10.3389/fcimb.2018.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayry J.; Beaussart A.; Dufrêne Y. F.; Sharma M.; Bansal K.; Kniemeyer O.; Aimanianda V.; Brakhage A. A.; Kaveri S. V.; Kwon-Chung K. J.; Latgé J.-P.; Beauvais A. Surface structure characterization of Aspergillus fumigatus conidia mutated in the melanin synthesis pathway and their human cellular immune response. Infect. Immun. 2014, 82, 3141–3153. 10.1128/iai.01726-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft C. A.; Culibrk L.; Moore M. M.; Tebbutt S. J. Interactions of Aspergillus fumigatus conidia with airway epithelial cells: a critical review. Front. Microbiol. 2016, 7, 472. 10.3389/fmicb.2016.00472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sponchiado S. R. P.; Sousa G. S.; Andrade J. C. R.; Lisboa H. F.; Gonçalves R. C. R.. Production of Melanin Pigment by Fungi and Its Biotechnological Applications, 1st ed.; Blumenberg M., Ed.; IntechOpen: London, UK, 2017; pp 47–79. [Google Scholar]
- Pihet M.; Vandeputte P.; Tronchin G.; Renier G.; Saulnier P.; Georgeault S.; Mallet R.; Chabasse D.; Symoens F.; Bouchara J.-P. Melanin is an essential component for the integrity of the cell wall of Aspergillus fumigatus conidia. BMC Microbiol. 2009, 9, 177. 10.1186/1471-2180-9-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Cuesta U.; Aparicio-Fernandez L.; Guruceaga X.; Martin-Souto L.; Abad-Diaz-de-Cerio L.; Antoran A.; Buldain I.; Hernando F. L.; Ramirez-Garcia A.; Rementeria A. Melanin and pyomelanin in Aspergillus fumigatus: from its genetics to host interaction. Int. Microbiol. 2020, 23, 55–63. 10.1007/s10123-019-00078-0. [DOI] [PubMed] [Google Scholar]
- Bayry J.; Aimanianda V.; Guijarro J. I.; Sunde M.; Latgé J.-P. Hydrophobins--unique fungal proteins. PLoS Pathog. 2012, 8, e1002700 10.1371/journal.ppat.1002700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin S.; Thywissen A.; Heinekamp T.; Saluz H. P.; Brakhage A. A. Melanin dependent survival of Aspergillus fumigatus conidia in lung epithelial cells. Int. J. Med. Microbiol. 2014, 304, 626–636. 10.1016/j.ijmm.2014.04.009. [DOI] [PubMed] [Google Scholar]
- Veeresham C. Natural products derived from plants as a source of drugs. J. Adv. Pharm. Technol. Res. 2012, 3, 200–201. 10.4103/2231-4040.104709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoda S.; Gupta L.; Agarwal H.; Raj G.; Vermani M.; Vijayaraghavan P. Inhibition of Aspergillus fumigatus biofilm and cytotoxicity study of natural compound cis-9-hexadecenal. J. Pure Appl. Microbiol. 2019, 13, 1207–1216. 10.22207/jpam.13.2.61. [DOI] [Google Scholar]
- Mujeeb F.; Bajpai P.; Pathak N. Phytochemical evaluation, antimicrobial activity, and determination of bioactive components from leaves of Aegle marmelos. BioMed Res. Int. 2014, 2014, 497606. 10.1155/2014/497606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisignano G.; Lagana M. G.; Trombetta D.; Arena S.; Nostro A.; Uccella N.; Mazzanti G.; Sajja A. In vitro antibacterial activity of some aliphatic aldehydes from Olea europaea L. FEMS Microbiol. Lett. 2006, 198, 9–13. 10.1111/j.1574-6968.2001.tb10611.x. [DOI] [PubMed] [Google Scholar]
- Mei H.; Yu X. C.; Chan K. K. NB1-C16-Insulin: Site-specific synthesis, purification and biological activity. Pharm. Res. 1999, 16, 1680–1686. 10.1023/a:1018993629553. [DOI] [PubMed] [Google Scholar]
- Hidayat J.; Margaret A. L.; Yolanda H.; Wibisono L. K. Hexane neem leaf extract more potent than ethanol extract against Aspergillus flavus. Universa Med. 2014, 3, 171–178. 10.18051/UnivMed.2014.v33.171-178. [DOI] [Google Scholar]
- Radhika S. M.; Michael A. In vitro antifungal activity of leaf extracts of Azadirachta indica. Int. J. Pharm. Pharm. Sci. 2013, 5, 723–725. [Google Scholar]
- Mircus G.; Hagag S.; Levdansky E.; Sharon H.; Shadkchan Y.; Shalit I.; Osherov N. Identification of novel cell wall destabilizing antifungal compounds using a conditional Aspergillus nidulans protein kinase C mutant. J. Antimicrob. Chemother. 2009, 64, 755–763. 10.1093/jac/dkp270. [DOI] [PubMed] [Google Scholar]
- Balouiri M.; Sadiki M.; Ibnsouda S. K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79. 10.1016/j.jpha.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob S.; Grötsch T.; Foster A. J.; Schüffler A.; Rieger P. H.; Sandjo L. P.; Liermann J. C.; Opatz T.; Thines E. Unravelling the biosynthesis of pyriculol in the rice blast fungus Magnaporthe oryzae. Microbiol 2017, 163, 541–553. 10.1099/mic.0.000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinekamp T.; Thywissen A.; Macheleidt J.; Keller S.; Valiante V.; Brakhage A. Aspergillus fumigatus melanins: interference with the host endocytosis pathway and impact on virulence. Front. Microbiol. 2013, 3, 440. 10.3389/fmicb.2012.00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal A. K.; Gajjar D. U.; Vasavada A. R. DOPA and DHN pathway orchestrate melanin synthesis in Aspergillus species. Med. Mycol. 2014, 52, 10–18. 10.3109/13693786.2013.826879. [DOI] [PubMed] [Google Scholar]
- Akoumianaki T.; Kyrmizi I.; Valsecchi I.; Gresnigt M. S.; Samonis G.; Drakos E.; Boumpas D.; Muszkieta L.; Prevost M.-C.; Kontoyiannis D. P.; Chavakis T.; Netea M. G.; van de Veerdonk F. L.; Brakhage A. A.; El-Benna J.; Beauvais A.; Latge J.-P.; Chamilos G. Aspergillus cell wall melanin blocks LC3-associated phagocytosis to promote pathogenicity. Cell Host Microbe 2016, 19, 79–90. 10.1016/j.chom.2015.12.002. [DOI] [PubMed] [Google Scholar]
- Rodianawati I.; Hastuti P.; Cahyanto M. N. Nutmeg’s (Myristica fragrans Houtt) oleoresin: effect of heating to chemical compositions and antifungal properties. Procedia Food Sci. 2015, 3, 244–254. 10.1016/j.profoo.2015.01.027. [DOI] [Google Scholar]
- Trombetta D.; Saija A.; Bisignano G.; Arena S.; Caruso S.; Mazzanti G.; Uccella N.; Castelli F. Study on the mechanisms of the antibacterial action, of some plant α,β-unsaturated aldehydes. Lett. Appl. Microbiol. 2002, 35, 285–290. 10.1046/j.1472-765x.2002.01190.x. [DOI] [PubMed] [Google Scholar]
- Pralea I.-E.; Moldovan R.-C.; Petrache A.-M.; Ilieş M.; Hegheş S.-C.; Ielciu I.; Nicoară R.; Moldovan M.; Ene M.; Radu M.; Uifălean A.; Iuga C.-A. From extraction to advanced analytical methods: The challenges of melanin analysis. Int. J. Mol. Sci. 2019, 20, 3943. 10.3390/ijms20163943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman N. M.; Ramasamy S. Genetic validation and spectroscopic detailing of DHN-melanin extracted from an environmental fungus. Biochem. Biophys. Rep. 2017, 12, 98–107. 10.1016/j.bbrep.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasowska A.; Sigler K. How microorganisms use hydrophobicity and what does this mean for human needs?. Front. Cell. Infect. Microbiol. 2019, 4, 112. 10.3389/fcimb.2014.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy M. J.; Rogers A. L.; Hanselmen L. R.; Soll D. R.; Yancey R. J. Variation in adhesion and cell surface hydrophobicity in Candida albicans white and opaque phenotypes. Mycopathologia 1988, 102, 149–156. 10.1007/bf00437397. [DOI] [PubMed] [Google Scholar]
- Hata M.; Ishii Y.; Watanabe E.; Uoto K.; Kobayashi S.; Yoshida K.-I.; Otani T.; Ando A. Inhibition of ergosterol synthesis by novel antifungal compounds targeting C-14 reductase. Med. Mycol. 2010, 48, 613–621. 10.3109/13693780903390208. [DOI] [PubMed] [Google Scholar]
- Gomes A. É. I.; Stuchi L. P.; Siqueira N. M. G.; Henrique J. B.; Vicentini R.; Ribeiro M. L.; Darrieux M.; Ferraz L. F. C. Selection and validation of reference genes for gene expression studies in Klebsiella pneumoniae using Reverse Transcription Quantitative real-time PCR. Sci. Rep. 2018, 8, 9001. 10.1038/s41598-018-27420-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafi A. S. A.; Santhanam J.; Khaithir T. M. N.; Musa N. F.; Huyop F. Determination of ergosterol as a potential biomarker in pathogenic medically important fungal isolates. Jurnal Sains Kesihatan Malaysia 2018, 16, 15–21. 10.17576/jskm-2018-1602-03. [DOI] [Google Scholar]
- Kurylo C. M.; Parks M. M.; Juette M. F.; Zinshteyn B.; Altman R. B.; Thibado J. K.; Vincent C. T.; Blanchard S. C. Endogenous rRNA sequence variation can regulate stress response gene expression and phenotype. Cell Rep. 2018, 25, 236–248. 10.1016/j.celrep.2018.08.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S.; Thakur R.; Goel G.; Shankar J. Nano-LC-Q-TOF analysis of proteome revealed germination of Aspergillus flavus conidia is accompanied by MAPK signalling and cell wall modulation. Mycopathologia 2016, 181, 769–786. 10.1007/s11046-016-0056-x. [DOI] [PubMed] [Google Scholar]
- García-Martínez J.; González-Candelas F.; Pérez-Ortín J. E. Common gene expression strategies revealed by genome-wide analysis in yeast. Genome Biol. 2007, 8, R222. 10.1186/gb-2007-8-10-r222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahrami A. A.; Bandhepour M.; Khalesi B.; Kazemi B. Computational Design and Analysis of a Poly-Epitope Fusion Protein: A New Vaccine Candidate for Hepatitis and Poliovirus. Int. J. Pept. Res. Ther. 2020, 26, 389–403. 10.1007/s10989-019-09845-z. [DOI] [Google Scholar]
- Lugani Y.; Sooch B. S. In silico Characterization of Cellulases from Genus Bacillus. Int. J. Curr. Res. Rev. 2017, 9, 30–37. 10.7324/ijcrr.2017.9136. [DOI] [Google Scholar]
- Satyanarayana S. D. V.; Krishna M. S. R.; Pavan Kumar P.; Jeereddy S. In silico structural homology modeling of nif A protein of rhizobial strains in selective legume plants. J. Genet. Eng. Biotechnol. 2018, 16, 731–737. 10.1016/j.jgeb.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Källberg M.; Wang H.; Wang S.; Peng J.; Wang Z.; Lu H.; Xu J. Template-based protein structure modeling using the RaptorX web server. Nat. Protoc. 2012, 7, 1511–1522. 10.1038/nprot.2012.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S. M.; Koringa P. G.; Reddy B. B.; Nathani N. M.; Joshi C. G. In silico analysis of consequences of non-synonymous SNPs of Slc11a2 gene in Indian bovines. Genom. Data 2015, 5, 72–79. 10.1016/j.gdata.2015.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C. C.; Meng E. C.; Morris J. H.; Pettersen E. F.; Ferrin T. E. Enhancing UCSF Chimera through web services. Nucleic Acids Res. 2014, 42, W478–W484. 10.1093/nar/gku377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appaiah P.; Vasu P. In silico Designing of Protein Rich in Large Neutral Amino Acids Using Bovine αs1 Casein for Treatment of Phenylketonuria. J. Proteonomics Bioinf. 2016, 9, 287–297. 10.4172/jpb.1000417. [DOI] [Google Scholar]
- Ferreira L.; dos Santos R.; Oliva G.; Andricopulo A. Molecular docking and structure-based drug design strategies. Molecules 2015, 20, 13384–13421. 10.3390/molecules200713384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szumilak M.; Galdyszynska M.; Dominska K.; Bak-Sypien I.; Merecz-Sadowska A.; Stanczak A.; Karwowski B.; Piastowska-Ciesielska A. Synthesis, biological activity and preliminary in silico ADMET screening of polyamine conjugates with bicyclic systems. Molecules 2017, 22, 794–816. 10.3390/molecules22050794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI) . Reference Method for Broth Dilution Testing of Filamentous Fungi, Approved Standard, 2nd ed.; CLSI Document M38-A2: Wayne, Pennsylvannia, USA, 2008.
- Graham L.; Orenstein J. M. Processing tissue and cells for transmission electron microscopy in diagnostic pathology and research. Nat. Protoc. 2007, 2, 2439–2450. 10.1038/nprot.2007.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaik A.; Thumma V.; Kotha A. K.; Kramadhati S.; Pochampally J.; Bandi S. Molecular docking analysis of UniProtKB nitrate reductase enzyme with known natural flavonoids. Bioinformation 2016, 12, 425–429. 10.6026/97320630012425. [DOI] [PMC free article] [PubMed] [Google Scholar]