The metabolite transporter GPT1 is important for NADPH provision in both plastid and peroxisomes, mostly during pollen tube growth towards ovules, and is thus essential for fertilization in Arabidopsis.
Abstract
Studies on Glucose-6-phosphate (G6P)/phosphate translocator isoforms GPT1 and GPT2 reported the viability of Arabidopsis (Arabidopsis thaliana) gpt2 mutants, whereas heterozygous gpt1 mutants exhibited a variety of defects during fertilization/seed set, indicating that GPT1 is essential for this process. Among other functions, GPT1 was shown to be important for pollen and embryo-sac development. Because our previous work on the irreversible part of the oxidative pentose phosphate pathway (OPPP) revealed comparable effects, we investigated whether GPT1 may dually localize to plastids and peroxisomes. In reporter fusions, GPT2 localized to plastids, but GPT1 also localized to the endoplasmic reticulum (ER) and around peroxisomes. GPT1 contacted two oxidoreductases and also peroxins that mediate import of peroxisomal membrane proteins from the ER, hinting at dual localization. Reconstitution in yeast (Saccharomyces cerevisiae) proteoliposomes revealed that GPT1 preferentially exchanges G6P for ribulose-5-phosphate (Ru5P). Complementation analyses of heterozygous +/gpt1 plants demonstrated that GPT2 is unable to compensate for GPT1 in plastids, whereas GPT1 without the transit peptide (enforcing ER/peroxisomal localization) increased gpt1 transmission significantly. Because OPPP activity in peroxisomes is essential for fertilization, and immunoblot analyses hinted at the presence of unprocessed GPT1-specific bands, our findings suggest that GPT1 is indispensable in both plastids and peroxisomes. Together with its G6P-Ru5P exchange preference, GPT1 appears to play a role distinct from that of GPT2 due to dual targeting.
INTRODUCTION
In plant cells, the oxidative pentose phosphate pathway (OPPP) is found in plastids and the cytosol (reviewed in Kruger and von Schaewen, 2003), but it is also transiently present in peroxisomes (Meyer et al., 2011; Hölscher et al., 2014; 2016). The OPPP has distinct functions in each subcellular compartment and thus requires subcellular distribution of the corresponding enzymes and their metabolites.
In chloroplasts, NADPH is provided by photosynthetic electron flow to ferredoxin-NADP+ oxidoreductase (Palatnik et al., 2003). However at night, the OPPP is the main source of NADPH, and also in heterotrophic plastids of non-green tissues, upon Glu-6-phosphate (G6P) import (Dennis et al., 1997). The oxidation of 1 mole of G6P to ribulose-5-phosphate (Ru5P) produces 2 moles of NADPH (at the expense of CO2 release) via three enzymatic steps involving glucose-6-phosphate dehydrogenase (G6PD), 6-phosphogluconolactonase, and 6-phosphogluconate dehydrogenase. These irreversible OPPP reactions in the stroma are followed by reversible steps involving transketolase and transaldolase that create a broad range of phosphorylated intermediates. Because the reversible OPPP reactions are shared with the Calvin–Benson–Bassham cycle, they are essential for plant metabolism (reviewed in Kruger and von Schaewen, 2003). It is likely that only the irreversible OPPP reactions occur in the cytosol of plant cells (Schnarrenberger et al., 1995), which are linked to the full cycle in plastids via epimerization of Ru5P to Xylulose-5-phosphate (Xu5P) and subsequent import by the Xu5P/phosphate translocator (XPT) in the inner envelope membrane (Eicks et al., 2002).
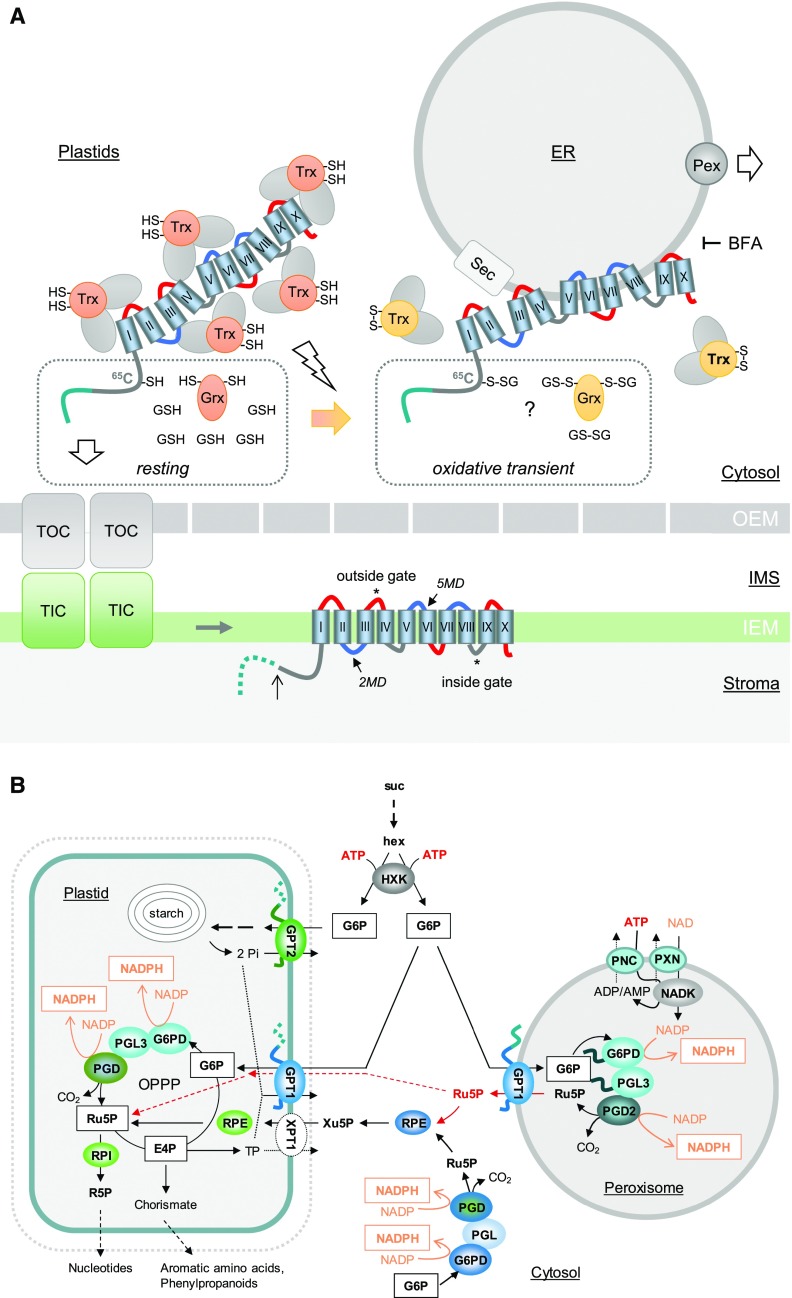
NADPH is mainly used in anabolic reactions in both plastids and the cytosol and primarily required for the biosynthesis of amino acids, fatty acids, and nucleotides (Geigenberger et al., 2005; Hutchings et al., 2005). Furthermore, NADPH is important for redox homeostasis of the glutathione pool (GSH/GSSG) via NADPH-dependent glutathione-disulfide reductases in the cytosol, plastids, mitochondria, and peroxisomes (Marty et al., 2009, 2019; Kataya and Reumann, 2010; Mhamdi et al., 2010). Hence, OPPP reactions play an important role in plant cells (Kruger and von Schaewen, 2003), particularly with the onset of stress or developmental change. Such conditions are often linked to physiological sink states, which are also induced by pathogen infection of leaves and related signaling. Sugar back-up in the cytosol stimulates G6PDH activity and NADPH production via the OPPP (Hauschild and von Schaewen, 2003; Scharte et al., 2009; Stampfl et al., 2016). Concomitantly activated NADPH oxidases at the plasma membrane (in plants, respiratory burst oxidase homologues called Rboh; Torres et al., 2002) use cytosolic NADPH for the extrusion of reactive oxygen species into the apoplast. Superoxide is converted to hydrogen peroxide (H2O2), which may react with macromolecules in the apoplast or enter the cell via aquaporins, leading to redox signaling in the cytosol. Peroxiredoxins dissipate H2O2, thereby retrieving electrons from glutaredoxins (Grxs) and thioredoxins (Trxs), which results in dithiol-disulfide changes in cognate Grx/Trx target enzymes (reviewed in Dietz, 2011; Noctor and Foyer, 2016; Liebthal et al., 2018; Waszczak et al., 2018).
OPPP enzymes have also been detected in purified plant peroxisomes (Corpas et al., 1998; del Río et al., 2002; Reumann et al., 2007; Hölscher et al., 2016), where they may serve as NADPH sources to establish redox homeostasis via dual cytosolic/peroxisomal GLUTATHIONE REDUCTASE1 (Kataya and Reumann, 2010). However, NADPH is also needed for metabolic reactions that occur mostly in peroxisomes, such as the removal of double bonds from unsaturated fatty acid/acyl chains before β-oxidation, including the final steps of auxin or jasmonic acid biosynthesis (Reumann et al., 2004).
We previously reported that dual targeting of Arabidopsis (Arabidopsis thaliana) enzymes G6PD1 (OPPP step 1) and PGL3 (OPPP step 2) to plastids and peroxisomes depends on the cytosolic redox state (Meyer et al., 2011; Hölscher et al., 2014). Furthermore, plants heterozygous for the only peroxisomal isoform, PGD2 (OPPP step 3), failed to produce homozygous offspring due to mutual sterility of the pgd2 gametophytes. These findings indicated that the OPPP plays essential roles in plant peroxisomes (Hölscher et al., 2016).
OPPP activity in organelles requires the flux of intermediates across the corresponding membranes. In Arabidopsis, G6P import into plastids involves G6P/phosphate translocators (GPT1 and GPT2) in the inner envelope membrane (Kammerer et al., 1998; Eicks et al., 2002; Knappe et al., 2003; Niewiadomski et al., 2005). By contrast, no peroxisomal transport protein for phosphorylated metabolites has thus far been identified. These substances are unable to pass through the porin-like channel used by malate and oxaloacetate (134 D and 130 D), as first described for spinach (Spinacia oleracea) peroxisomes (Reumann et al., 1996). Rokka et al. (2009) demonstrated that only molecules <200 D are able to pass through a corresponding porin-like channel in mammalian cells. Because G6P and Ru5P are larger than 200 D (258 and 230 D) and phosphorylated, it is unlikely that they are shuttled via peroxisomal porins. Thus, how OPPP substrates and products are transported across peroxisomal membranes (PerMs) has been unclear.
To provide the peroxisomal OPPP reactions with substrate, we reasoned that one of the two Arabidopsis GPT proteins may dually localize to plastids and peroxisomes in a manner similar to originally plastid-annotated OPPP isoforms G6PD1 (Meyer et al., 2011) and PGL3 (Kruger and von Schaewen, 2003; Reumann et al., 2004; Hölscher et al., 2014). GPT1 and GPT2 share 81% identity at the amino acid level and catalyze the import of G6P into heterotrophic plastids for NADPH provision via the stromal OPPP reactions and starch synthesis (Kammerer et al., 1998). GPT2 expression is most abundant in heterotrophic tissues (senescing leaves, sepals, seeds) and can be induced by high light in leaves (Athanasiou et al., 2010; Weise et al., 2019), whereas GPT1 is expressed ubiquitously, with the highest levels in reproductive tissues (Niewiadomski et al., 2005; Kunz et al., 2010). Interestingly, the loss of GPT2 function in Arabidopsis yielded vital plants (Niewiadomski et al., 2005; Athanasiou et al., 2010; Kunz et al., 2010; Dyson et al., 2014, 2015). By contrast, the lack of GPT1 was detrimental, leading to an early arrest of pollen and ovule development. The resulting gametophyte and embryo lethality was evidenced by pollen deformation and incompletely filled siliques (Niewiadomski et al., 2005; Andriotis et al., 2010; Flügge et al., 2011). Considering that the loss of PGD2 likewise did not yield homozygous offspring, GPT1 was a strong candidate for a peroxisomal substrate transporter.
Peroxisomal membrane proteins (PMPs) exhibit independent mPTS motifs of varying sequences (Rottensteiner et al., 2004). To date, two classes of PMPs have been identified. Class-I PMPs are directly inserted into PerMs from the cytosol, which involves the peroxins Pex3 and Pex19 (in some organisms also Pex16; Platta and Erdmann, 2007). Class-II PMPs are first inserted into the membrane of the endoplasmic reticulum (ER, via the Sec import complex) and then directed to the peroxisomal ER (perER), where peroxisomes are formed de novo (Theodoulou et al., 2013; Reumann and Bartel, 2016; Kao et al., 2018). The exact mechanism remains to be resolved, but Pex16 and Pex3 are most likely involved in ER recruitment and sorting to peroxisomes (Aranovich et al., 2014). Interestingly, the mutation of Arabidopsis PEX16 resulted in plants with a shrunken seed phenotype and impaired fatty acid biosynthesis (Lin et al., 1999, 2004) reminiscent of some gpt1 defects (Niewiadomski et al., 2005), but no defects in pollen germination.
Here we report that both GPT1 and GPT2 may insert into the ER, but only the N-terminal part of GPT1 is able to initiate ER targeting, a prerequisite shared with class-II PMPs. We co-expressed various reporter fusions to analyze the subcellular localization and test protein–protein interactions of GPT1 in plant cells. GPT1 formed homodimers at plastids, but not readily at the ER, and interacted with two cytosolic oxidoreductases listed by the Membrane-based Interactome Network Database (MIND; https://associomics.dpb.carnegiescience.edu) as Arabidopsis proteins with high interaction scores (38% confidence; Lalonde et al., 2010; Chen et al., 2012; Jones et al., 2014). In addition, we obtained evidence that GPT1 also contacts early peroxins involved in PMP delivery via the ER. GPT1-reporter fusions were rarely detected in membrane structures surrounding peroxisomes, indicating only transient presence at this location. Our main questions were as follows: Which protein part(s) confer dual targeting? How is this process regulated? Which OPPP metabolite exits the peroxisomes? And moreover, are some defects of heterozygous +/gpt1 plants (Niewiadomski et al., 2005) related to missing metabolite transport across PerMs during fertilization?
RESULTS
GPT1 Dually Targets Plastids and the ER
The alignment of GPT1 and GPT2 protein sequences from different Brassicaceae species (Supplemental Figure 1; Supplemental File 1) revealed that the isoforms mostly diverge at their N-terminal ends, whereas the central transmembrane regions (for substrate binding/transport) are highly conserved. We investigated subcellular targeting using various N- and C-terminal reporter fusions of the two Arabidopsis GPT isoforms and examined transfected protoplasts (of Arabidopsis or tobacco [Nicotiana tabacum]) by confocal laser-scanning microscopy (CLSM).
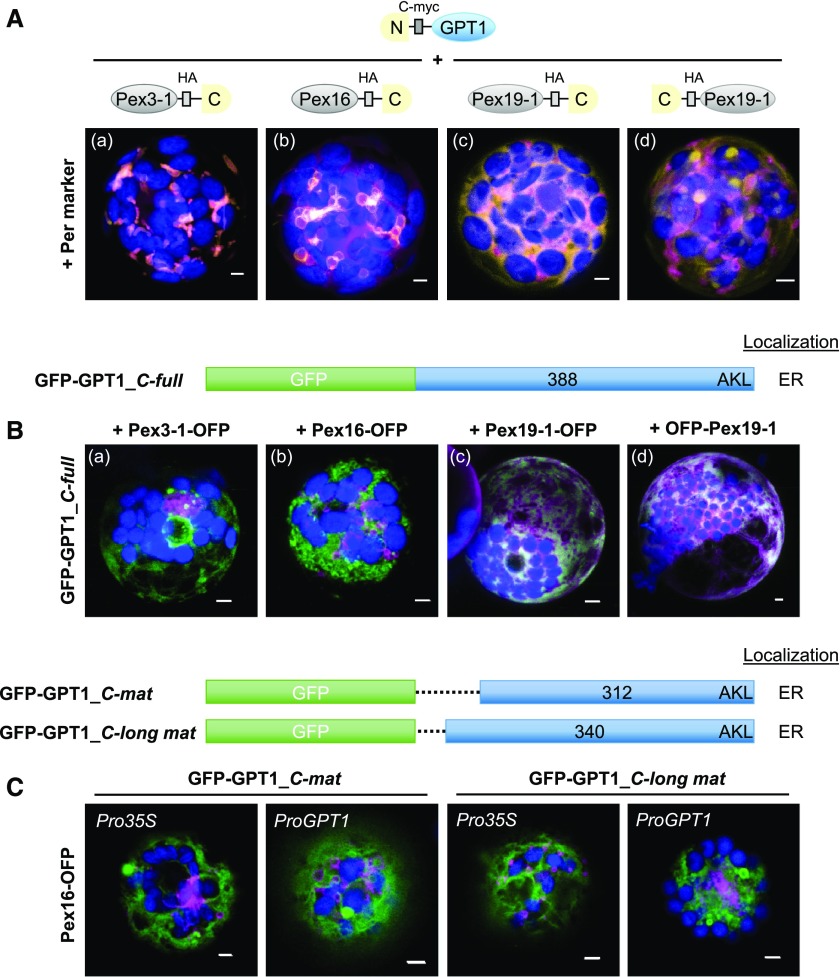
All N-terminally masked/truncated reporter-GPT variants (Supplemental Figure 2A) localized to the ER (Supplemental Figure 2B, green signals), as determined by co-expression with organelle markers (magenta signals), i.e., G/OFP-ER (OFP, orange fluorescent protein; Rips et al., 2014) or peroxisome marker G/OFP-PGL3_C-short (formerly named G/OFP-PGL3[∼50 amino acids]-SKL; Meyer et al., 2011). Note that the co-localization of green and magenta signals appears white. Both GPT fusions occasionally formed Z-membranes (Supplemental Figure 2B, white patches), a term coined for overexpressed, integral membrane proteins (Gong et al., 1996). The full-length fusion protein GFP-GPT1_C-full labeled ring-like substructures of the ER ∼3 µm in diameter (Supplemental Figure 2C, b) and interfered with import of the peroxisome marker (Supplemental Figure 2B, n). This was never observed for GFP-GPT2_C-full (Supplemental Figure 2B, p). Mutagenesis of the C-terminal GPT1 motif Ala-Lys-Leu (AKL) to Ala-Lys-Gln (AKQ) of GPT2 (or vice versa GPT2-AKQ to GPT2-AKL) had no effect on the localization of the fusion proteins.
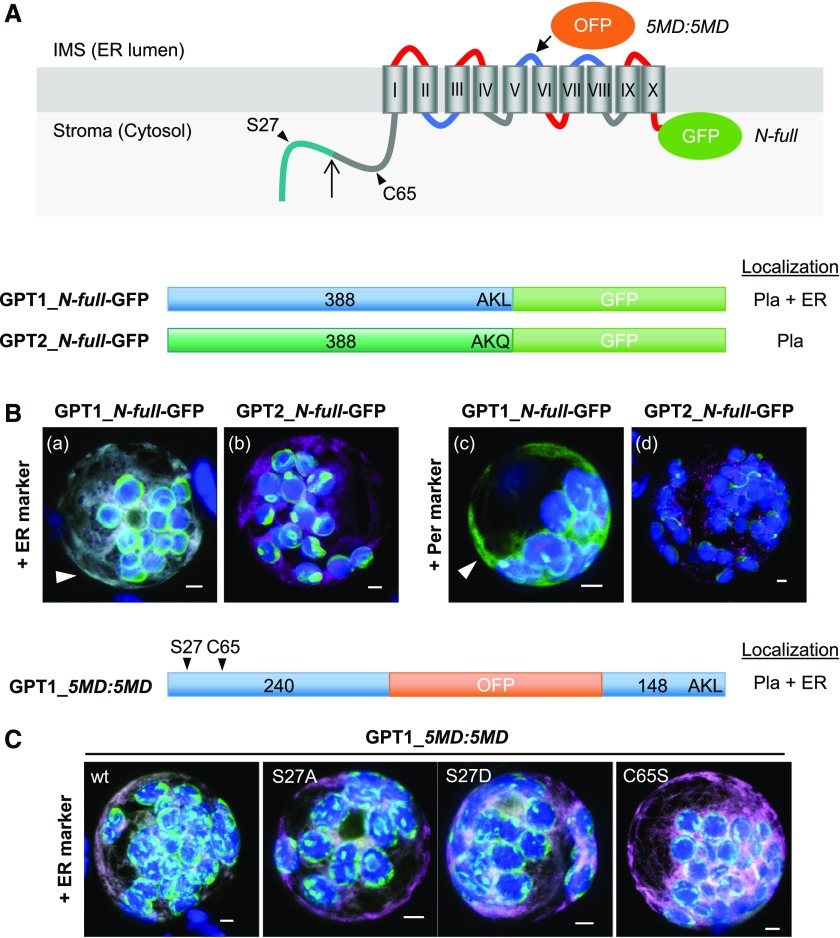
Among the GPT-reporter fusions (with opposite reporter orientation), the localization of GPT1 also differed from that of GPT2 (Figure 1). As GPT-GFP fusions that enable plastid import (Figure 1A), the full-length GPT1 version was detected at both plastids and the ER (Figure 1B, a and c, arrowheads), but GPT2 was only detected at plastids (Figure 1B, b and d, green signals; for single-channel images, see Supplemental Figure 3). The C-terminally truncated GPT-reporter versions are shown in Supplemental Figure 4. When the N terminus plus five membrane domains (N-5MD, 1 to 240 amino acids) was used, and OFP pointed to the intermembrane space (IMS, or ER lumen), the plastid surface was labeled (Supplemental Figure 4B, a to d; green signals). When the N terminus plus the first two membrane domains (N-2MD, 1 to 155 amino acids) was used, and GFP pointed to the stroma (or cytosol), patchy plastid labeling was mostly observed, indicating that the reporter was partially cleaved in the stroma (Supplemental Figure 4B, e to h). Importantly, when GPT1 was used, ER labeling was also detected (Supplemental Figure 4B, e and f, arrowheads), albeit to varying extents (Supplemental Figure 4C, a to e). Again, small ring-like structures of peroxisomal size were labeled, but none surrounded the peroxisomal marker (Supplemental Figure 4C, e, single sections). When the N terminus (N-term, 1 to 91/92 amino acids) fused to the reporter was used, only stroma labeling was observed for both GPT isoforms (Supplemental Figure 4B, i to l). These results indicate that the region comprising the GPT1 N terminus plus the first two membrane domains (N-2MD) is important for alternative targeting to the ER.
Figure 1.
GPT1 Reporter Fusions Dually Localize to Plastids and the ER.
(A) Topology model of Arabidopsis G6P/phosphate translocator (GPT) isoforms with 10 membrane domains (MD) depicted as barrels (Roman numerals), connected by hinge regions (red, positive; blue, negative; and gray, neutral net charge), and both N-/C-terminal ends facing the stroma (Lee et al. 2017). Relevant positions are indicated: Plastidic TP (green), TP processing site (upward arrow), N-terminal amino acids potentially modified/regulatory in GPT1 (arrowheads), medial OFP insertion (5MD:5MD), and C-terminal GFP fusion (N-full). Pla, plastids.
(B) and (C) Localization of the depicted GPT-reporter fusions upon transient expression in Arabidopsis protoplasts (24- to 48-h post transfection).
(B) With free N terminus, GPT1 targets both plastids and the ER (a and c, arrowheads), but GPT2 only targets plastids (b and d). Scale bars = 3 μm.
(C) The medial GPT1_5MD:5MD construct (wt, wild type) was used to analyze potential effects of single amino acid changes in the N terminus: Ser27-to-Ala (S27A, abolishing phosphorylation), Ser27-to-Asp (S27D, phospho-mimic), and Cys65-to-Ser (C65S, precluding Ser modification). All images show maximal projections of ∼30 optical sections (shown as merged; for single-channel images, see Supplemental Figure 5). Candidate fusions are shown in green, ER marker (B, OFP-ER; C, GFP-ER) or peroxisome marker (OFP-PGL3_C-short) in magenta, and chlorophyll fluorescence in blue. Co-localization of green and magenta (or very close signals <200 nm) appear white in the merge of all channels. Scale bars = 3 μm.
The First 155 Amino Acids of GPT1 Are Crucial for ER Targeting
To exclude localization artifacts caused by the masking of N- or C-terminal targeting signals, we also produced GPT fusion proteins with an internal reporter at two different positions (Supplemental Figure 5A). The GPT1 versions labeled both plastids and the ER (GPT1_2MD:8MD and GPT1_5MD:5MD; Supplemental Figure 5B, a, b, e, and f; arrowheads), whereas the corresponding GPT2 versions labeled only plastids (GPT2_2MD:8MD and GPT2_5MD:5MD; Supplemental Figure 5B, c, d, g, and h). We treated protoplasts expressing the GPT_2MD:8MD fusions with Brefeldin A (BFA), which interferes with the delivery of peroxisomal ascorbate peroxidase via the ER (Mullen et al., 1999). BFA treatment abolished GPT1 signals at the ER, but not at plastids (neither of GPT2; Supplemental Figure 6). These results confirm the notion that GPT proteins directly target plastids and that only GPT1 may insert into the ER.
Because the alternative localization of GPT1 appeared to involve the soluble N-terminal part of this protein, which strongly differs from that of GPT2 (Supplemental Figure 1), we changed amino acid positions in the medial GPT1_5MD:5MD fusion protein thought to be subject to post-translational modification by site-directed mutagenesis (Figure 1C; for single-channel images, see Supplemental Figure 3). However, neither changing Ser S27 (listed by PhosPhAt 4.0; Heazlewood et al., 2008; Durek et al., 2010; Zulawski et al., 2013) to Ala (A, abolishing phosphorylation) or Asp (D, to mimic phosphorylation; Ackerley et al., 2003) nor replacing the single Cys C65 by Ser (S, precluding redox modification) interfered with ER targeting.
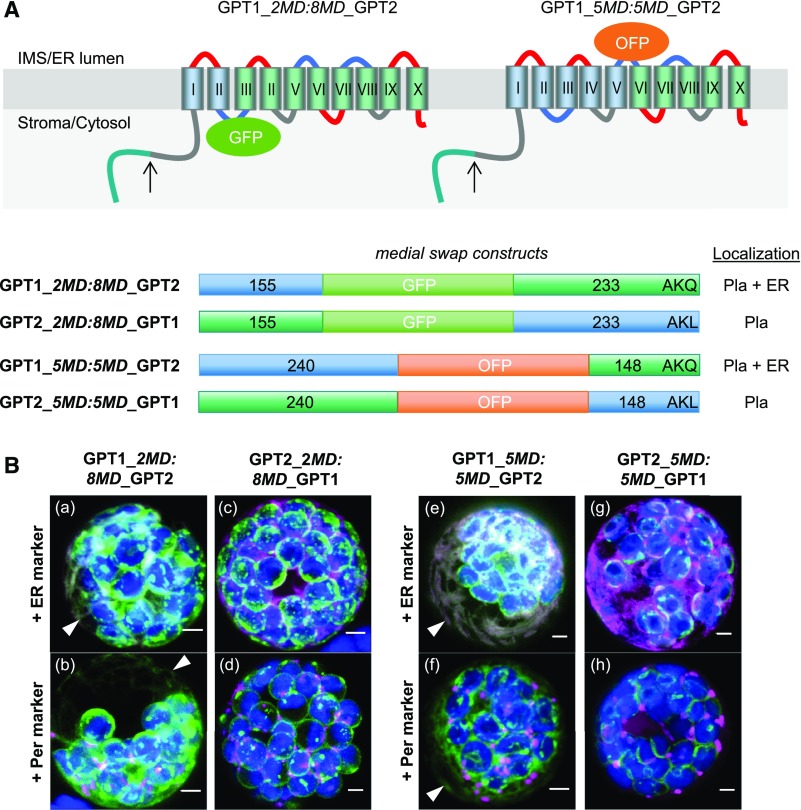
Domain swapping among the corresponding unmodified medial reporter constructs (Figure 2A) resulted in the dual localization of GPT1_2MD:8MD_GPT2 and GPT1_5MD:5MD_GPT2 to plastids and the ER (Figure 2B, a, b, e, and f; arrowheads), but GPT2_2MD:8MD_GPT1 and GPT2_5MD:5MD_GPT1 were solely detected at plastids (Figure 2B, e, d, g, and h; for single-channel images, see Supplemental Figure 7). These results demonstrate that the N terminus (plus the first two membrane domains) of GPT1 is crucial for initiating alternative ER targeting.
Figure 2.
Domain Swaps Demonstrate that the N Terminus of GPT1 Confers ER Targeting.
(A) Topology models of the GPT medial swap constructs showing the orientation of the inserted reporters: GFP facing the stroma/cytosol and OFP facing the IMS/lumen of the ER. Membrane domains (depicted as barrels, Roman numerals) of GPT1 in blue and of GPT2 in green. The upward arrows indicate TP cleavage sites (in the plastid stroma).
(B) Localization of the indicated medial swap constructs in Arabidopsis protoplasts (24- to 48-h post transfection). When headed by GPT1 (GPT1_2MD:8MD_GPT2 or GPT1_5MD:5MD_GPT2), both plastids and the ER (arrowheads) are labeled (a, b, e, and f); when headed by GPT2 (GPT2_2MD:8MD_GPT1 or GPT2_5MD:5MD_GPT1), only plastids are labeled (c, d, g, and h). All images show maximal projections of ∼30 optical sections (merged; for single-channel images, see Supplemental Figure 7). Candidate fusions are shown in green, ER marker (G/OFP-ER) or peroxisome marker (Per; G/OFP-PGL3_C-short) in magenta, and chlorophyll fluorescence in blue. Co-localization of green and magenta (and very close signals <200 nm) appear white in the merge of all channels. Scale bars = 3 μm.
GPT1 Dimer Formation Occurs at Plastids and Substructures of the ER
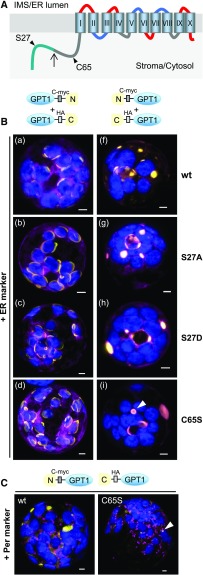
In their functional form, the plastidial phosphate translocators are dimers composed of two identical subunits (Knappe et al., 2003). We reasoned that if they are not necessary for ER targeting, amino acids S27 and/or C65 may be important for preventing GPT1 dimerization before reaching the final location(s). Therefore, we generated N- and C-terminal split yellow fluorescent protein (YFP) constructs of GPT1 and introduced the amino acid changes described above. We co-transfected Arabidopsis protoplasts with these constructs—plus organelle markers—and analyzed GPT1-dimer formation (Figure 3) by bimolecular fluorescence complementation (BiFC; Walter et al., 2004). Reconstitution of the GPT1-split YFP combinations was detected only at plastids (Figure 3B, a to d), with no effect of the indicated amino acid changes. When we used split YFP-GPT1 fusions (enforcing ER insertion), we observed large areas of signal accumulation in the ER (including perinuclear structures). This signal did not resemble the ER pattern that we usually observed (see Figures 1 and 2), and even the distribution of the ER marker was affected. Among the amino acid changes we analyzed, only C65S had an effect, resulting in the formation of hollow spherical structures surrounding single peroxisomes (Figure 3C, arrowhead) compared with the wild-type situation or S27 changes (Figure 3B, compare f and g to i, arrowhead; for single-channel images, see Supplemental Figure 8). Thus, ER insertion seems not to involve post-translational modification of the GPT1 N terminus, but the modification of C65 might negatively regulate sorting to PerMs.
Figure 3.
GPT1 Dimer Formation Occurs at Plastids and ER Substructures.
(A) Topology model of GPT1 with the N-terminal TP (blue-green) and cleavage site (upward arrow) plus positions of amino acids Ser (S27) and Cys (C65, arrowheads). The membrane domains are depicted as barrels (Roman numerals) connected by hinge regions of different net charge (red, positive; blue, negative; and gray, neutral).
(B) Localization of yellow BiFC signals (reconstituted split YFP, N+C halves) due to interaction of the GPT1 parts in Arabidopsis protoplasts (24- to 48-h post transfection). With unmasked N terminus, GPT1-labeled plastids and the ER (a to d), but with masked N terminus, it only labeled the ER (f to i). In addition to unmodified wild type (wt) GPT1, mutant combinations S27A (non-phosphorylated), S27D (phospho-mimic), and C65S (precluding Ser modification) were analyzed. GPT1 dimer formation occurred at plastid rims (a to d) or ER substructures (f to i), with the S27 changes having little impact, whereas C65S had visible effects (hollow sphere in i; surrounding a peroxisome in C, arrowheads). Note that structures with BiFC signals on the right (f to i) are also labeled by the ER marker (most obvious in g). Scale bars = 3 μm.
(C) Localization of the indicated split YFP combinations co-expressed with the peroxisome marker (Per). Note that in the case of C65S, the ring-like BiFC signal surrounds a peroxisome (arrowhead). All images show maximal projections of ∼30 optical sections (merged; for single-channel images, see Supplemental Figure 8). Organelle markers (OFP-ER or OFP-PGL3_C-short) are shown in magenta, chlorophyll fluorescence is shown in blue. Co-localization of yellow and magenta (or very close signals <200 nm) appear whitish in the merge of all channels. Scale bars = 3 μm.
Recruitment of GPT1 to the ER Involves Redox Transmitters
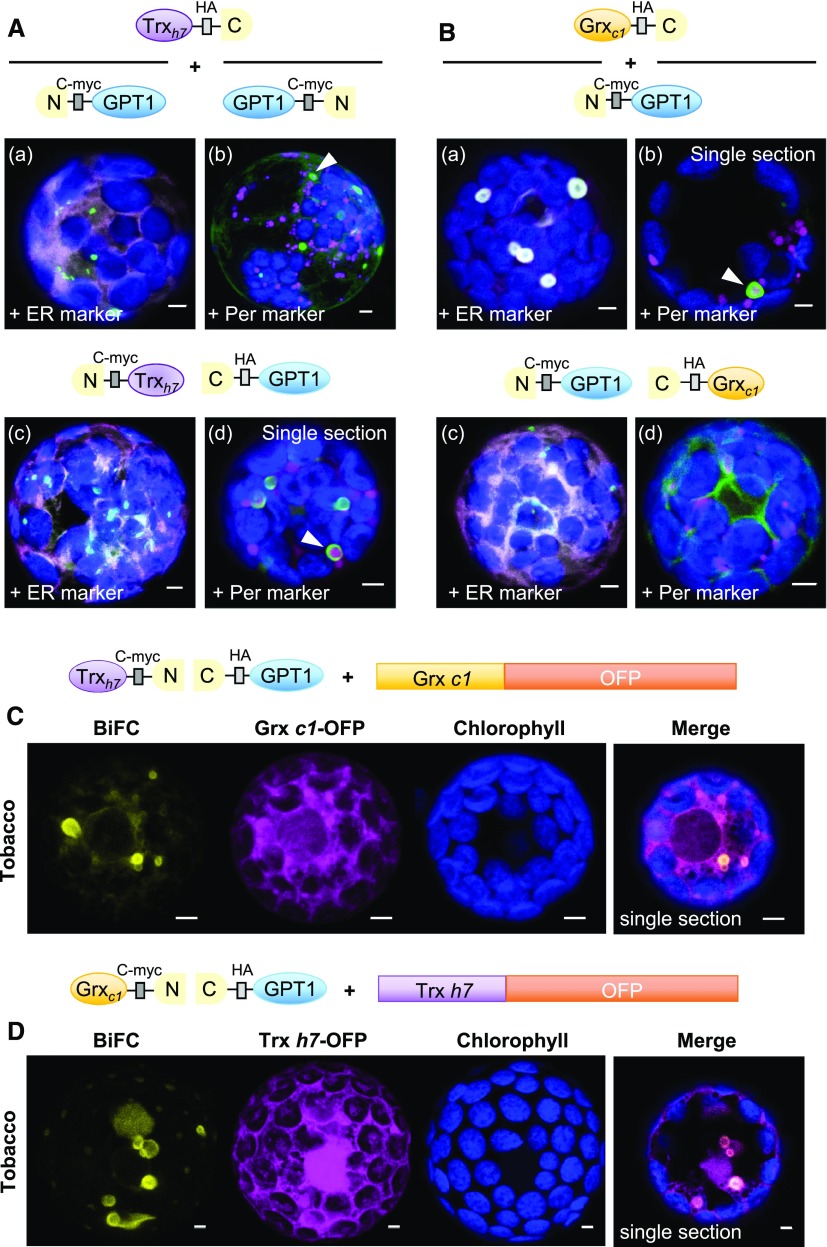
To identify potential interaction partners of GPT1, we searched the MIND of Arabidopsis proteins (based on split ubiquitin reconstitution in yeast [Saccharomyces cerevisiae]; Lalonde et al., 2010). Two cytosolic oxidoreductases, Thioredoxin h7 (Trxh7) and Glutaredoxin c1 (Grxc1), were among the 21 candidates listed with the highest scores (Supplemental Table 1). BiFC analyses in Arabidopsis protoplasts confirmed that GPT1 interacts with Trxh7 (Figure 4A) and Grxc1 (Figure 4B) at the ER and its substructures, but not at plastids (Figure 4A, b), and more clearly when the GPT1 N terminus was masked (enforcing ER insertion). Occasionally, ER-derived membranes around peroxisomes were labeled (Figure 4A, b and d, arrowheads; Figure 4B, b, arrowhead), which was less obvious when the Grxc1 N terminus was masked by split YFP (Figure 4B, c and d).
Figure 4.
GPT1 Interacts with Cytosolic Oxidoreductases Trxh7 and Grxc1 at the ER.
(A) and (B) Localization of GPT1 upon interaction with Trxh7 or Grxc1 in Arabidopsis protoplasts (24- to 48-h post transfection). The schemes illustrate different orientation of the candidate proteins with respect to free N- and C-terminal ends. GPT1 interacts with both oxidoreductases (green signals) at the ER and its spherical substructures (arrowheads), except when the N terminus of Grxc1 is masked (B, c and d). Note that these substructures differ from those labeled in Figure 3B. Merge of BiFC signals (green) with the ER marker (OFP-ER) or peroxisome marker (Per, OFP-PGL3_C-short) is shown in magenta, and chlorophyll fluorescence is shown in blue. Scale bars = 3 μm.
(C) and (D) Localization of split YFP reconstitution (BiFC, yellow signals) in heterologous tobacco protoplasts (24- to 48-h post transfection), testing a potential effect of the other oxidoreductase (co-expressed as OFP fusion, magenta). Note that similar ER substructures are labeled (merged, single sections). All other images show maximal projections of ∼30 optical sections. Chlorophyll fluorescence is shown in blue. Co-localization and very close signals (<200 nm) appear white in the merge of all channels. Scale bars = 3 μm.
To enhance interactions among the Arabidopsis proteins, we co-expressed selected BiFC combinations with the other oxidoreductases as OFP fusion proteins in a heterologous system (i.e., tobacco protoplasts). Similar results were obtained (Figures 4C and 4D), but smaller spherical structures (<3 µm) were also detected. Notably, in simple co-expression studies, Trxh7-OFP and Grxc1-OFP partially overlapped with the ER marker (Supplemental Figure 9B, white signals) and co-localized with GPT1_N-2MD-GFP at the ER (Supplemental Figure 9C). These results are consistent with the hypothesis that N-myristoylation (predicted for both oxidoreductases) might facilitate the insertion of GPT1 into the ER and/or its sorting to peroxisomes.
GPT1 Contacts Pex3 and Pex16 at the ER
While class-I PMPs are inserted into PerMs directly from the cytosol (involving Pex3 and Pex19), class-II PMPs are first inserted into the ER (Platta and Erdmann, 2007). Because Pex3, Pex16, and Pex19 also play central roles in ER insertion, PMP sorting, and peroxisome biogenesis (Reumann and Bartel, 2016; Kao et al., 2018), we analyzed their potential interactions with GPT1. Arabidopsis contains two PEX3 genes, PEX3-1 and PEX3-2 (Hunt and Trelease, 2004), one PEX16 gene (Karnik and Trelease, 2005), and two PEX19 genes, PEX19-1 and PEX19-2 (Hadden et al., 2006). Analysis of N- and C-terminal reporter fusions in protoplasts revealed mostly PerM labeling for the two Pex3 isoforms, ER and PerM labeling for Pex16 (see also Lansing et al., 2019), and mostly cytosolic distribution for the two Pex19 isoforms (Supplemental Figure 10, shown for one of the two Pex3 and Pex19 isoforms). OFP-Pex3-1 displayed weak signals in the cytosol. We conducted BiFC analyses with Pex3-1, Pex16, and Pex19-1. GPT1 interaction with Pex3-1 and Pex16 was detected at PerMs, partially contiguous with the ER (Figure 5A, a and b). By contrast, the interaction of GPT1 with Pex19 was mostly distributed across the cytosol, but also labeled spherical structures when the C-terminal farnesylation motif of Pex19 (McDonnell et al., 2016) was accessible (Figure 5A, d). Of note, the Pex16-GPT1 interaction interfered with import of the peroxisome marker (Figure 5A, b, magenta signals remained largely cytosolic), as already observed for GFP-GPT1_C-full (Supplemental Figure 2, n).
Figure 5.
Interaction versus Co-localization of GPT1 with Peroxin Factors at the ER.
(A) Localization of the indicated split YFP combinations (yellow BiFC signals) in Arabidopsis protoplasts (24- to 48-h post transfection). The peroxins Pex3, Pex16, and Pex19 are important for sorting class-II PMPs via the ER to peroxisomes. The soluble peroxisome marker (Per; OFP-PGL3_C-short) is shown in magenta and chlorophyll fluorescence in blue. Scale bars = 3 μm.
(B) Co-expression of GFP-GPT1 (green) and the corresponding Pex-OFP fusions (magenta) indicates that interaction with the Pex factors is transient (isoforms Pex3-2 = At1g48635 and Pex19-2 = At5g17550 gave comparable results; chlorophyll fluorescence in blue). Note that Pex16 co-expression has a vesiculating effect on GPT1 at the ER (merged; for single-channel images, see Supplemental Figure 10C). Scale bars = 3 μm.
(A) and (B) Maximal projections of ∼30 optical sections.
(C) Co-expression of the indicated GFP-GPT1 fusions with Pex16-OFP in Arabidopsis protoplasts (72-h post transfection). The C_mat version lacks the entire N-terminal part (including C65), whereas the C_long mat version lacks only the TP (Supplemental Figure 1). Besides the 35S promoter (Pro35S), these GFP fusions were also expressed from the GPT1 promoter (ProGPT1), with similar results. Images show single optical sections (merged; for single-channel images, see Supplemental Figure 11). GFP fusions are shown in green, Pex16-OFP is shown in magenta and chlorophyll fluorescence in blue. Co-localization of green and magenta (or very close signals <200 nm) appear white in the merge of all channels. Scale bars = 3 μm.
Co-expression of GFP-GPT1_C-full with the OFP-based Pex fusions resulted in different patterns (Figure 5B), suggesting that the interactions are merely transient. Pex3-1-OFP co-expression led to partial perinuclear localization of GFP-GPT1_C-full, reminiscent of the BiFC data obtained for GPT1 homomerization (Figure 5B, a compared with Figure 3, f to i). Interestingly, Pex16 had visible effects on GPT1 localization, promoting concentration/vesiculation at the ER (Figure 5B, b), a pattern similar to (but distinct from) that obtained using Pex16-OFP alone (Supplemental Figure 10, compare B to C). When co-expressed, Pex19-1 appeared to have little impact on GPT1 localization (Figure 5B, c and d).
To ensure that the co-expression patterns obtained with Pex16 were not artifacts due to use of the strong constitutive cauliflower mosaic virus (CaMV) 35S promoter (Pro35S), we also expressed two N-terminally truncated GPT1 versions (designed for stable plant transformation) from their own promoter (ProGPT1), which gave comparable results (Figure 5C, for single-channel images, see Supplemental Figure 11). Together with the BiFC analyses (Figure 5A), this demonstrated that ER-inserted GPT1 can be dragged to PerMs, and thus behaves like a class-II PMP, which requires a special trigger to contact interaction partners (including Pex3 and Pex16) to reach mature peroxisomes.
GPT1 May Be Recruited to Peroxisomes and Preferentially Exchanges G6P for Ru5P
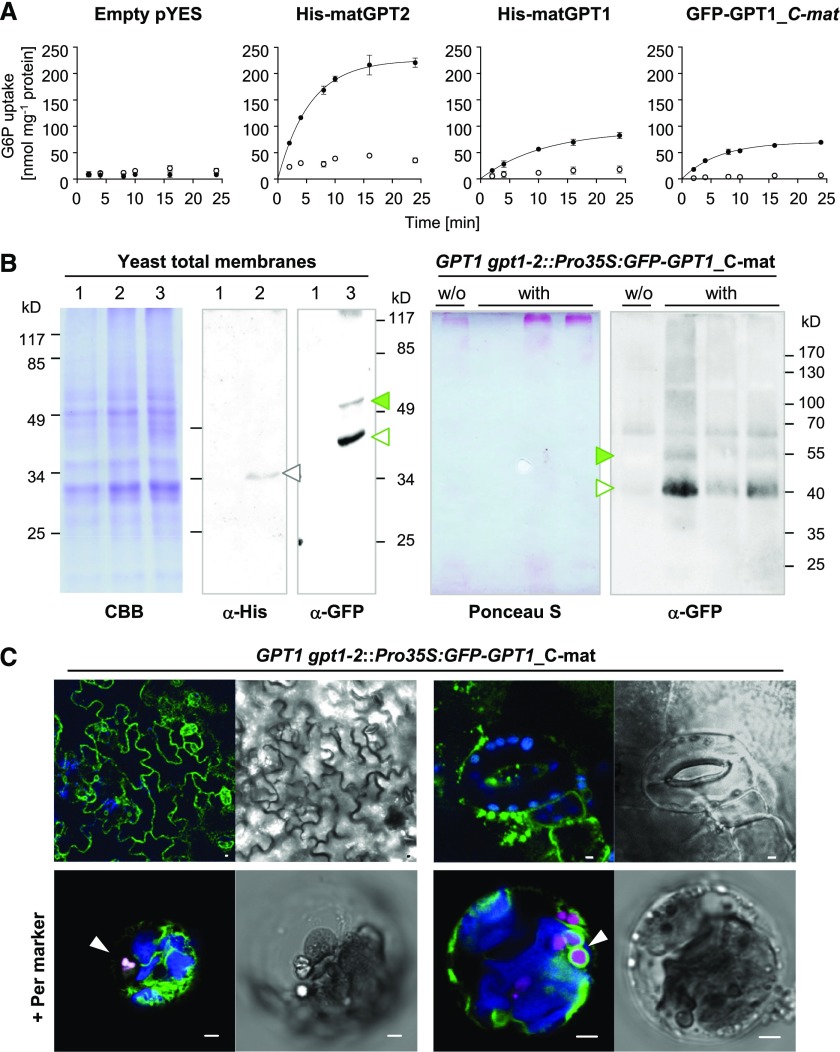
After plastid import, the N-terminal transit peptide (TP) of the precursor proteins is usually cleaved off (Chua and Schmidt, 1979; Schmidt et al., 1979). According to the recently described 3-dimensional structure of the Arabidopsis triose-phosphate/phosphate translocator (Lee et al., 2017), both the N- and C-terminal ends of GPT face the stroma. Thus when GPT1 is inserted into the ER, both the unprocessed N terminus and C-terminal end should point to the cytosol, which was confirmed by topology analyses using roGFP (redox-sensitive GFP; Supplemental Figure 12). To test whether N-terminal modification or the lack of transit-peptide processing might affect transport activity, we fused an N-terminal His tag (or GFP reporter) to the full-length and mature versions of GPT1 (with mature GPT2 used as a control) and measured metabolite exchange of the recombinant proteins in reconstituted yeast proteoliposomes (Linka et al., 2008). For the physiological exchange of G6P versus Pi using the mature GPT versions (Figure 6A), the transport rate of His-matGPT1 reached approximately one-third of that of His-matGPT2 (with comparable expression levels in yeast cells). N-terminal modification by GFP did not affect the transport rates of GPT1, but the presence of the TP (equivalent to localization at the ER/PerMs) reduced the transport rates by approximately half.
Figure 6.
Transport Activity and Localization of Mature GPT1 in Yeast and Plant Cells.
(A) Time-dependent uptake of radioactively labeled [14C]-G6P (0.2 mM) into reconstituted proteoliposomes preloaded with 10 mM of Pi (closed symbols), or without exchange substrate (open symbols) prepared from yeast cells harboring the empty vector (pYES) or the indicated GPT constructs. Note that transport rates of GPT1 are not influenced by the N-terminal tag (compare His-matGPT1 to GFP-matGPT1). In all graphs, the arithmetic mean of three technical replicates (±sd) was plotted against time (see Table 1 for substrate specificities).
(B) Immunoblot analysis upon expression in yeast and plant cells. Left representations, SDS gel of total yeast membrane fractions, stained with Coomassie Brilliant Blue (CBB) or blot detection by anti-His (α-His) or anti-GFP (α-GFP) antibodies: 1, empty vector; 2, His-matGPT1 (gray open triangle); and 3, GFP-matGPT1 (green closed and open triangles). Right representations, Blotted pellet fractions of leaf extracts (prepared without detergent) from Arabidopsis GPT1 gpt1-2::Pro35S:GFP-GPT1_C-mat plants (T2 progeny without [w/o] or with the transgene) developed with anti-GFP (α-GFP) antibodies. The Ponceau S-stained blot serves as a loading reference. Note that GFP-GPT1 (closed green and open triangles) extracted from yeast or plant membranes migrates similarly. Molecular masses of the bands are indicated (kD).
(C) Localization of GFP-GPT1_C-mat in heterozygous GPT1 gpt1-2 plants. Top, Green net-like structures (ER) in leaf epidermal cells (left), and spherical structures in seedlings (right); scale bars = 10 μm. Bottom, Pattern upon protoplast preparation and transfection with the peroxisome marker (Per; OFP-PGL3_C-short, magenta) in membranes surrounding peroxisomes (arrowheads). Chlorophyll fluorescence is shown in blue. All images show single optical sections. Co-localization (and very close signals <200 nm) appear white in the merge of all channels (bright field images are shown as reference). Scale bars = 3 μm.
We stably introduced the Pro35S:GFP-GPT1_C-mat construct into heterozygous +/gpt1-2 plants by floral dip transformation (Clough and Bent, 1998). Similar immunoblot patterns were obtained using GFP-GPT1 proteins extracted from yeast versus plant cells (Figure 6B, green arrowheads). In leaf cells of soil-grown plants, ER labeling dominated, but also spherical structures (≤3 µm) were detected (Figure 6C, top representations). The ER insertion of mature GPT1 occurs by default, but sorting to PerMs requires a stimulus. When mesophyll protoplasts were prepared from transgenic leaf material and transfected with the peroxisome marker (OFP-PGL3_C-short), GFP-labeled structures resembling newly forming peroxisomes appeared (Figure 6C, bottom representations; arrowheads).
If GPT1 imports G6P into peroxisomes, we wondered what might happen to Ru5P, the product of the three irreversible OPPP reactions, especially because bioinformatic and experimental analyses of the three Arabidopsis ribulose-phosphate 3-epimerase (RPE) isoforms (two cytosolic and one plastidic; Kruger and von Schaewen, 2003) did not provide hints about peroxisomal localization. We therefore analyzed whether the mature GPT versions (with N-terminal His tag) could exchange G6P for Ru5P. Indeed, matGPT1 and matGPT2 were able to catalyze the in vitro import of G6P versus Ru5P (Table 1). The relative velocity of matGPT1 was higher for G6P-Ru5P (116%) compared with Pi-Ru5P exchange (59%) and differed from that of matGPT2 (87% for G6P-Ru5P, 75% for Pi-Ru5P). Importantly, the exchange rates for 6-phosphogluconate (6PG < 10%) were negligible.
Table 1. Initial Velocities of Pi or G6P Import for Various Exchange Substrates.
| His-matGPT1 | His-matGPT2 | ||
|---|---|---|---|
| Pi versus | G6P | 9.9 (100%) | 19.3 (100%) |
| Ru5P | 5.8 (59%) | 14.4 (75%) | |
| 6PG | 0.8 (8%) | 1.2 (6%) | |
| G6P versus | Pi | 10.5 (100%) | 32.6 (100%) |
| Ru5P | 12.2 (116%) | 28.3 (87%) | |
| 6PG | 0.9 (9%) | 3.1 (10%) | |
Time-dependent uptake of [32P]-Pi or [14C]-G6P (0.2 mM) into liposomes reconstituted with total yeast membranes of cells expressing the indicated mature GPT versions (nmol mg−1 total protein). Proteoliposomes were preloaded with 10 mM of G6P, Ru5P, 6PG, or Pi. Relative velocities (given in parentheses) were compared to the counter-exchange experiment Pi/G6P or G6P/Pi, which was set to 100%. Values (bold) are given in percent.
Stress and Developmental Stimuli Enhance ER Targeting of GPT1
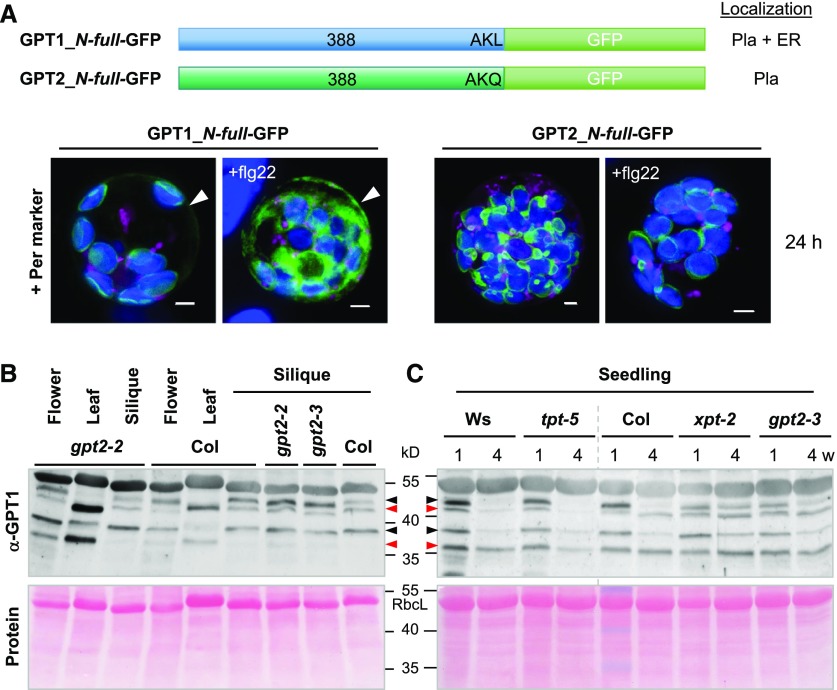
Because protoplast preparation (which is achieved by treating leaves with fungal enzymes) of stably transformed leaves led to the recruitment of GFP-GPT1_C-mat to peroxisomes, we tested whether treatment with a bacterial elicitor (flagellin, flg22) would also affect GPT localization. We co-transfected both the GPT1_ and GPT2_N-full-GFP constructs with the peroxisome marker OFP-PGL3_C-short into Arabidopsis protoplasts, divided the samples in half, and analyzed them after 24 h of mock or flg22 treatment. The latter led to enhanced GPT1 recruitment to the ER (Figure 7A, arrowheads) without having major effects on plastid localization (GPT2 was also unaffected; for single-channel images, see Supplemental Figure 13).
Figure 7.
GPT1 Levels at the ER Increased in Response to Stress Treatment and in Reproductive Arabidopsis Tissues.
(A) Arabidopsis protoplasts were co-transfected with the indicated GPT-GFP fusions and the peroxisome marker (Per, OFP-PGL3_C-short); Pla, plastids. The samples were split in half: one was treated with 0.2 μM of flagellin peptide (+flg22) and the other mock-incubated for 24 h. Note that flg22 treatment did not change GPT localization to plastids, but it increased the amount of the ER fraction of GPT1-GFP (arrowheads). All images show maximal projections of ∼30 single sections (merged; for single-channel images, see Supplemental Figure 13). GFP fusions are shown in green, peroxisome marker in magenta, and chlorophyll fluorescence in blue. Co-localization of magenta and green or very close signals (<200 nm) appear white in the merge of all channels. Scale bars = 3 μm.
(B) and (C) Protein extracts (without detergent) of flower, leaf, and green silique tissue were prepared from wild-type plants (Col, Ws) and the indicated homozygous mutant lines. Supernatant fractions were separated on 10% SDS gels and blotted onto nitrocellulose. After Ponceau-S staining, the blots were developed with GPT1-specific antibodies (α-GPT1) raised against the N terminus with His-tag (Supplemental Figure 14). Arrowheads mark double bands of full-length GPT1 (predicted size: 42.3 kD) and mature GPT1 (∼37 to 39 kD, depending on TP processing). Red arrowheads point to bands thought to represent a largely off-situation and black arrowheads the corresponding on-situation at either location (as deduced from comparison of leaf to silique tissue), likely due to protein modification.
(C) Immunoblot of seedlings harvested from germination plates (1% Suc) after 1- or 4-week (w) growth in short-day regime. Included mutant alleles: gpt2-2 (GK-950D09, T-DNA intron 2/exon 3), gpt2-3 (GK-780F12, T-DNA in exon 4), tpt-5 (FLAG_124C02, T-DNA in exon 9), and xpt-2 (SAIL_378C01, single exon; Hilgers et al., 2018). Note that the band pattern differs in OPPP-relevant gpt2 and xpt transporter mutants compared with wild-type Col and tpt-5 (wild-type Ws corresponds to tpt-5, vertical gray dashed line). Ponceau S-stained blots (protein) are shown as a loading reference; RbcL, large subunit of RubisCO. Molecular masses are indicated in kD.
In addition, His-tag versions of the GPT1 and GPT2 N termini were cloned. After overexpression in Escherichia coli, affinity-purified His-N1 and His-N2 were used to raise polyclonal antisera in rabbits. The resulting α-GPT1 antiserum specifically recognized the N terminus of GPT1 but not GPT2 (Supplemental Figure 14). Immunoblot analyses of different Arabidopsis tissues detected prominent high-molecular-weight bands in soluble fractions of flower, silique, and seedling tissue, but not leaf extracts (Figure 7B), with stronger labeling in gpt2 (Niewiadomski et al., 2005) and xpt-2 (Hilgers et al., 2018), but not triose-phosphate/phosphate translocator tpt-5 mutants (Figure 7C). In total, four bands were found in reproductive tissues/seedlings and three bands in leaves. The latter resembled those reported for 35S-labeled GPT upon import into isolated plastids, namely: precursor, weak intermediate, and N-terminally processed mature forms (Kammerer et al., 1998). Intermediates are unlikely to persist in planta. Thus, we reasoned that weak ∼39 kD bands in leaf extracts represent a minor share of active mature GPT1 in chloroplasts (as deduced from the stronger labeled top bands in gpt2 mutants compared with Col-0 wild type, Figure 7B, lower black arrowhead), migrating between less active mature and full-length versions (36.8 kD and 42.3 kD, red arrowheads). Conversely, top bands in reproductive flower and silique tissue (black arrowheads) would represent GPT1 in the perER/peroxisomes (Figure 7B, compare Col to gpt2-2 and gpt2-3). This was also observed in seedling extracts (Figure 7C). Interestingly, the pattern of tpt-5 mutants resembled that of the wild type (Wasilewski [Ws], Col), whereas unprocessed (top) bands persisted in extracts of 4-week–old seedlings harvested from OPPP-relevant xpt-2 and gpt2-3 mutants. However, additional treatments before SDS-PAGE/immuno-detection (−/+Lambda Protein Phosphatase, extraction −/+ phosphatase inhibitors; Supplemental Figure 14, F and G) or the use of 200 mM of the redox reagent dithiothreitol (DTTred) for tissue extraction and sample boiling did not result in visible differences.
GPT1 Is Required at Both Plastids and Peroxisomes during Fertilization
The loss of the last step of the OPPP in peroxisomes prevented the formation of homozygous offspring due to the mutual sterility of the pgd2 gametophytes (Hölscher et al., 2016). Based on this observation, we set out to rescue plastidial versus ER/peroxisomal defects by ectopic GPT expression in heterozygous +/gpt1 lines. First, the coding sequence of GPT2 was placed under the control of the constitutive mannopine synthase (MAS) promoter (Guevara-García et al., 1993) or the GPT1 promoter (position −1,958 to −1) and introduced into heterozygous +/gpt1 plants by floral dip transformation. The CaMV-35S promoter-driven GFP-GPT1_C-mat construct (targeting the ER/peroxisomes, Figure 6C) was included for comparison (Supplemental Figure 15A). Ectopic GPT2 expression merely rescued the gpt1 defect of incompletely filled siliques (Supplemental Figure 15B, a, b, and f). When driven by the GPT1 promoter, some siliques of the ProGPT1:GPT2 transformed plants were completely filled with seeds (Supplemental Figure 15B, d), whereas most siliques of the same plant/line showed erratic seed maturation (Supplemental Figure 15B, c) or seed abortion (Supplemental Figure 15B, e). The frequencies of unfertilized, aborted ovules are listed in Table 2. Compared with the untransformed heterozygous +/gpt1-1 or +/gpt1-2 lines (∼30%), there appeared to be a slight reduction for plants transformed with ProMAS:GPT2 (∼27%) compared with those transformed with ProGPT1:GPT2 (∼21%) or Ws wild type (∼7%), indicating some compensation by GPT2 on the female side. Attempted ER/peroxisomal rescue by Pro35S:GFP-GPT1_C-mat scored the highest values, with ∼34% aborted ovules.
Table 2. Seeds and Aborted Ovules without and upon Ectopic GPT Expression.
| Genotype | Normal seeds | Aborted ovules | Frequency (% ±sd) |
|---|---|---|---|
| GPT1 (Ws-2) | 439 | 39 | 8.3 ± 4.3 |
| GPT1 GPT1a | 755 | 53 | 6.6 |
| GPT1 gpt1-1 | 86 | 26 | 30.2 (mean) |
| GPT1 gpt1-1a | 507 | 236 | 32.0 |
| GPT1 gpt1-1::ProMAS:GPT2 (line 3) | 1,195 | 495 | 28.8 ± 7.2 |
| GPT1 gpt1-1::ProMAS:GPT2 (line 7) | 1,587 | 585 | 27.2 ± 8.8 |
| GPT1 gpt1-2 | 371 | 164 | 29.4 ± 6.9 |
| GPT1 gpt1-2a | 1,357 | 530 | 28.0 |
| GPT1 gpt1-2::ProGPT1:GPT2 (line 3) | 2,082 | 529 | 20.6 ± 8.9 |
| GPT1 gpt1-2::Pro35S:GFP-GPT1_C-mat (line 14.5) | 1,412 | 690 | 33.8 ± 9.8 |
| gpt1-2 gpt1-2::gGPT1-3.10a | 1,461 | 104 | 6.6 |
Arabidopsis ecotype Ws-2 and heterozygous +/gpt1-1 and +/gpt1-2 T-DNA lines compared to plastid-compensated GPT1 gpt1-2::ProMAS:GPT2 or::ProGPT1:GPT2 lines (T2 generation) and ER/peroxisomal-compensated line::Pro35S:GFP-GPT1_C-mat (T3 generation). Transformed progenies were initially selected on Hygromycin B. Values (bold) are given in percent.
Data from Niewiadomski et al. (2005) shown for comparison
Despite occasionally producing filled siliques, analyses of the ProGPT1:GPT2-compensated lines revealed no gpt1 homozygous plants (Table 3). Therefore, GPT1 gpt1-2::ProGPT1:GPT2 was reciprocally crossed with ER/peroxisomal GPT1 gpt1-2::Pro35S:GFP-GPT1_C-mat, forming seeds only with GPT1 gpt1-2::ProGPT1:GPT2 as the mother plant (Table 3). Because again no homozygous gpt1-2 alleles were found in the F2, several T2 plants of GPT1 gpt1-2::ProGPT1:GPT2 (line 3 #6 with ∼73% filled siliques; Supplemental Figure 16A) were super-transformed with ProGPT1:GPT1_N-long mat (ER/peroxisomal construct driven by the GPT1 promoter; Supplemental Figure 16B). This construct is based on OFP-Pex16 co-expression (Figure 5C) and GPT1-roGFP analyses (Supplemental Figure 12), but lacks the reporter. Surprisingly, siliques of heterozygous +/gpt1 plants carrying ProGPT1:GPT1_N-long mat (T1) were almost completely filled with seeds in both the presence and absence of ProGPT1:GPT2 (Supplemental Figure 16C, compare with bottom representations). These results highlight the major contribution of GPT1 in the ER/peroxisomes to fertilization and seed formation, which was also corroborated by the gpt1 transmission rates (Table 3).
Table 3. Transmission of the gpt1 Alleles with and without Ectopic GPT Expression.
| Genotype | + | +/gpt1 | gpt1 |
|---|---|---|---|
| GPT1 gpt1-1 | 79.3 (wild type = 184) | 20.7 (he = 48) | 0 (n = 232) |
| GPT1 gpt1-1::ProMAS:GPT2 (lines 3 and 7, T2) | 67.8 (wild type = 214) | 32.2 (he = 102) | 0 (n = 316) |
| GPT1 gpt1-2 | 74.8 (wild type = 95) | 25.2 (he = 32) | 0 (n = 127) |
| GPT1 gpt1-2::ProGPT1:GPT2 (line 3, T2) | 71.0 (wild type = 115) | 29.0 (he = 47) | 0 (n = 162) |
| GPT1 gpt1-2::Pro35S:GFP-GPT1_C-mat (T3) | 65.8 (wild type = 100) | 34.2 (he = 51) | 0 (n = 151) |
| GPT1 gpt1-2::ProGPT1:GPT2 (♀) x GPT1 gpt1-2::Pro35S:GFP-GPT1_C-mat (F2)a | 80.0 (wild type = 152) | 20.0 (he = 38) | 0 (n = 190) |
| GPT1 gpt1-2::ProGPT1:GPT2 (line 3, T3)::ProGPT1:GPT1_N-long mat (T2)a | 56.1 (wild type = 184) | 43.9 (he = 144) | 0 (n = 328) |
| GPT1 gpt1-2::ProGPT1:GPT1_N-long mat (T2)b | 54.4 (wild type = 68) | 45.6 (he = 57) | 0 (n = 125) |
Segregation analysis of heterozygous +/gpt1-1 and +/gpt1-2 lines upon selfing or transformation with the indicated GPT rescue constructs: GPT2 cDNA was driven by the constitutive MAS promoter (T2 generation) or the GPT1 promoter (T2 and T3 generation). ER/peroxisomal Pro35S:GFP-GPT1_C-mat was analyzed in parallel (transformed plants were selected on Hygromycin B). No homozygous gpt1 plants were found. Therefore, a plastid-compensated GPT1 gpt1-2::ProGPT1:GPT2 plant was reciprocally crossed with an ER/peroxisomal-compensated GPT1 gpt1-2::Pro35S:GFP-GPT1_C-mat plant. Only one combination set seeds, indicating that GPT2 is unable to rescue GPT1 function during pollen maturation. Still, no homozygous gpt1 plants were found. Thus, GPT1 gpt1-2::ProGPT1:GPT2 was super-transformed with ER/peroxisomal rescue construct ProGPT1:GPT1_N-long mat (lacking the TP region) and selected on Kanamycin. Among the progeny of individuals carrying all three T-DNA alleles, gpt1-2 transmission markedly improved, although no homozygous plants were found. Notably, this was also true for lines devoid of ProGPT1:GPT2. Values (bold) are given in percent. he, heterozygous; n, number analyzed.
Progeny of plants containing all three T-DNAs.
Based on two independent lines with similar T-DNA transmission.
In summary, compared with the untransformed +/gpt1 lines (21% to 25%), the number of heterozygous progeny appeared to increase only slightly in the presence of ProGPT1:GPT2 (29-32%), with the highest scores obtained using a GPT1 construct lacking the TP region (45.6%), independent of the presence of ProGPT1:GPT2. Thus, substantial recovery by GPT1 (solely targeting the ER/peroxisomes) was obtained without further contribution by GPT2 (solely targeting plastids), which was expressed from the same promoter.
DISCUSSION
GPT1 and GPT2 Differ in Several Aspects
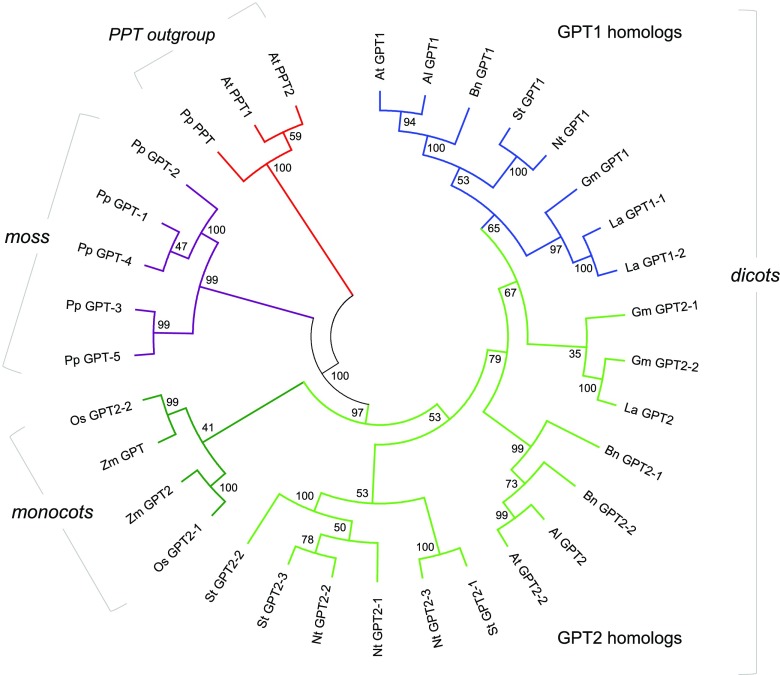
Based on the concept that peroxisomes developed from the proto-endomembrane system of the Archaebacterial host in an early pre-eukaryote (Tabak et al., 2006; Cavalier-Smith, 2009; van der Zand et al., 2010), and metabolite transporters were recruited to endosymbiont membranes (Tyra et al., 2007), it is conceivable that GPT had a pre-existing role in the secretory system. During land plant evolution, GPT1 developed a special role related to NADPH provision in plastids, whereas GPT2 mainly contributes to starch biosynthesis (Niewiadomski et al., 2005; Andriotis et al., 2010; Athanasiou et al., 2010; Kunz et al., 2010; Dyson et al., 2015). The functional specialization of GPT proteins is also supported by the late split of GPT1 from GPT2 in dicots (Figure 8; Supplemental Files 2 and 3) and the dichotomy of orthologous sequences in the monocot species rice (Oryza sativa) and maize (Zea mays). In rice, ADP-Glc and not G6P is imported by heterotrophic plastids as a precursor of starch biosynthesis (Cakir et al., 2016), except in pollen tissue, which imports G6P (Lee et al., 2016). Furthermore, the GPT1-interacting oxidoreductase Grxc1 (Supplemental Table 1, also listed as an interaction partner of GPT2, albeit with a lower score) is dicot-specific, while Grxc2 is present in all seed plants (Riondet et al., 2012; Li, 2014). In Arabidopsis, GPT2 mRNA is predominately found in heterotrophic tissues, whereas GPT1 is expressed ubiquitously (Niewiadomski et al., 2005), including rosette leaves (Supplemental Figure 17). Thus, basal G6P exchange, which is needed to stabilize the Calvin–Benson–Bassham cycle in chloroplasts (Sharkey and Weise, 2016), should involve GPT1 rather than GPT2. In addition, GPT2 may be induced under stress, e.g., by high light (Athanasiou et al., 2010; Preiser et al., 2019).
Figure 8.
Phylogenetic Analysis of GPT Sequences from Different Plants.
Selected GPT isoforms of the Brassicaceae, Fabaceae, Solanaceae, and Poaceae compared with the moss Phycomitrella patens. The Arabidopsis phosphoenolpyruvate/phosphate translocator (PPT) sequences served as the outgroup (red). The GPT accessions of P. patens (violet) form the base of the phylogenetic tree. GPT2 accessions (green) of monocotyledonous plants split off early (monocots, dark green), whereas the GPT1 accessions (blue) split much later from the GPT2 accessions (bright green) in the dicotyledonous branch (dicots). The tree with the highest log likelihood (−12,357.08) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches as bootstrap values (Felsenstein, 1985). This analysis involved 33 amino acid sequences (for sequence identifications, see Supplemental Table 2). There was a total of 642 positions in the final data set. Abbreviations: Al: Arabidopsis lyrata spp. lyrata; At, A. thaliana; Bn, Brassica napus; Gm, Glycine max; La, Lupinus angustifolius; Nt, N. tabacum; Os, O. sativa; Pp, P. patens; St, Solanum tuberosum; Zm, Z. mays.
The N Terminus of GPT1 Mediates the Dual Targeting of This Protein
Our analyses showed that the C-terminal peroxisomal targeting signal type 1 of GPT1 (PTS1 motif AKL; Gould et al., 1989; Reumann, 2004; Platta and Erdmann, 2007) is inactive, although reporter-GPT1 fusions interfered with import of the PTS1-based peroxisome marker. As expected for PMPs (Rottensteiner et al., 2004), GPT1 targeting was driven by other parts of the protein. Although the exact motif mediating ER import of GPT1 was not determined, domain swapping with GPT2 showed that the sequence lies within the first 155 amino acids (N terminus plus the first two membrane domains). Because the GPT1_N-long mat version (without TP) was also inserted into the ER, the region between K48 and the first membrane domain (A92) is probably crucial, partly lacking from GPT2, and strongly different from that of GPT2 (Supplemental Figure 1).
To exclude the possibility that GPT1 and GPT2 are inserted into the ER before plastid import (Baslam et al., 2016), we tested the effects of BFA, a fungal toxin that inhibits the formation of ER-derived coated vesicles (Orci et al., 1991; Klausner et al., 1992). Although BFA-induced compartments of merged ER/Golgi vesicles formed, GPT1 and GPT2 still localized to plastids. Furthermore, all medial swap constructs headed by GPT2 targeted plastids. Hence, in the case of dually targeted GPT1, threading into the plastidial TOC/TIC complex may prevent the binding of a factor (e.g., TPR7/OEP61; von Loeffelholz et al., 2011) that mediates post-translational ER import by the Sec pore complex (Figure 9A; TPR7 is an interaction partner of Sec62p; Mitterreiter et al., 2019). Alternatively, an ER-targeting suppressor region could be exposed to prevent signal-recognition particle binding, as shown for human PMP70 (Sakaue et al., 2016).
Figure 9.
Dual Targeting Model of GPT1 to Plastids and Peroxisomes for Proper OPPP Functioning.
(A) GPT1 precursors in the cytosol are covered with chaperons (gray ovals) and co-chaperons Trxh7 and Grxc1 as putative redox sensors/transmitters (orange, reduced state, -SH; yellow, oxidized state, -S-S-). The hydrophobic membrane domains (barrels) of GPT1 are labeled with Roman numerals. Hinge regions of negative net charge (blue) may facilitate ER insertion. Left, In the largely reduced state of the cytosolic glutathion pool (GSH), the N terminus of GPT1 (green) enters the TOC/TIC complex (translocon of the outer/inner chloroplast envelope), the membrane domains (MDs) integrate into the inner envelope membrane (IEM), and the TP is processed (open arrow)/degraded in the stroma (dotted line). Local oxidation (flash sign, open arrow) of the cytosolic glutathion pool (GSSG) likely retains GPT1 in the cytosol via a functional change in the bound redox transmitters (Grxc1 and Trxh7). Whether this involves glutathionylation of 65C in the GPT1 N terminus is unclear (question mark). ER insertion involves the Sec complex and sorting to PerMs via specific peroxins (Pex). BFA blocked the ER import of GPT1. OEM, outer envelope membrane.
(B) Scheme of sugar metabolism in a physiological sink state. Suc is cleaved by cytosolic invertase, yielding two hexoses (hex) that are activated by hexokinase (HXK), consuming ATP provided by glycolysis and mitochondrial respiration. In contrast to GPT2, GPT1 imports G6P into both plastids (in exchange for Pi released by GPT2-driven starch synthesis) and peroxisomes (in exchange for Ru5P, which may also enter plastids via GPT1, dashed red arrows), yielding 2 moles of NADPH in the oxidative part of the OPPP. NADP inside peroxisomes is formed by NAD kinase (NADK), which relies on ATP and NAD imported into peroxisomes via PNC (At3g05290; At5g27520) and PXN (At2g39970). The cytosolic OPPP reactions are usually linked via RPE and XPT to the complete pathway in the plastid stroma. Abbreviations: PGL, 6-Phosphogluconolactonase; PGD, 6-phosphogluconate dehydrogenase; RPE, ribulose-phosphate 3-epimerase; RPI, ribose-5-phosphate isomerase.
How dual targeting to secretory versus endosymbiont compartments may be regulated has been discussed by Porter et al. (2015). N-terminal phosphorylation might influence competition between chloroplast import and signal recognition particle binding (in the case of protein-disulfide isomerase RB60 from Chlamydomonas reinhardtii). However, phosphomimic/preclusion of phosphorylation had no influence on dual GPT1 targeting, nor did exchanging the single Cys (C65) in this protein (Figure 9). On the other hand, the C65S change in GPT1 enabled us to detect this protein version at PerMs, albeit a rare event (Figures 3B, i and 3C). Although conserved in distantly related species, C65 is not present in all Brassicaceae (Supplemental Figure 1) nor in GFP-GPT1_C-mature, which was detected around peroxisomes upon elicitation (Figure 6C). Thus, C65 is not required for GPT1 to reach peroxisomes, but it might play a role in the negative regulation of GPT1 transfer from the ER to peroxisomes.
Evidence That Redox Transmitters Help Recruit GPT1 to the ER/Peroxisomes
The release of GPT1 to peroxisomes might require its interaction with Grxc1 (and Trxh7). Grx is known to engage in monothiol-dithiol mechanisms, including glutathionylation (Riondet et al., 2012; Ukuwela et al., 2017). Triggered by oxidative transients (H2O2) that accompany stress signaling and developmental changes (2GSH → GSSG), redox-sensitive Cys residues may become sulfenylated or glutathionylated (reviewed in Zaffagnini et al., 2019). Reversion by GSH alone occurs slowly, but it occurs rapidly together with Grx and Trx (as shown for plastidial Amy3; Gurrieri et al., 2019). Perhaps this mechanism regulates the interaction of GPT1 with Pex16 and/or Pex3. In any case, GPT1 transport within the ER in monomeric form makes sense, because a potentially active translocator—en route to its final destination—is likely not tolerated. This idea is supported by our observation that the ER structure appeared aberrant when GPT1-dimer formation was enforced (Figure 3).
For indirect delivery of PMPs via the ER, it is still unclear how the processes of ER targeting and sorting to newly forming peroxisomes are regulated. It was suggested that cytosolic chaperones may guide Pex3 to the Sec translocon (Kim and Hettema, 2015) and that Pex16 may recruit Pex3 and other PMPs to the ER (Hua et al., 2015). We previously demonstrated that Trxs function as redox-dependent targeting regulators for OPPP enzymes. The co-chaperone function of Trx (holdase versus foldase) depends on the local redox state, and dual targeting of Arabidopsis G6PD1 and PGL3 is regulated by preventing folding, allowing plastid import; or supporting folding, as a prerequisite for peroxisome import (Meyer et al., 2011; Hölscher et al., 2014). Here we showed that treatment with flagellin/fungal enzymes enhances the ER localization/detection of GPT1 around peroxisomes. Moreover, the interaction of GPT1 with both oxidoreductases was detected at structures reminiscent of PerMs.
Trxs and Grxs can promote protein folding directly (Berndt et al., 2008), in addition to enhancing the activities of co-chaperones in a redox state-dependent manner (Park et al., 2009; Sanz-Barrio et al., 2012). However, the oligomerization state of Grxc1 is also influenced by the surrounding redox medium. Grxc1 is activated under oxidizing conditions, implying that it functions as a cytosolic redox sensor (Riondet et al., 2012; Ströher, 2012). Furthermore, Trxh7 and Grxc1 were found to be n-myristoylated in planta (Meng et al., 2010; Riondet et al., 2012; Traverso et al., 2013; Majeran et al., 2018). However, without the n-myristoylation motif (G2A), Grxc1 still appeared to promote the ER targeting of GPT1 in grxc1 mutant protoplasts (unpublished observations of H.L.), indicating that Grxc1 functions redundantly with one or more members of the Grx/Trx superfamily.
Interestingly, GPT1 is listed as a palmitoylation candidate (http://aramemnon.uni-koeln.de). Protein s-acylation (via Cys residues) is still a poorly understood process (usually preceded by n-myristoylation) that promotes membrane association, targeting, and partitioning into membrane subdomains (Aicart-Ramos et al., 2011; Hemsley, 2015). Analyzing the role of Grx/Trx n-myristoylation for putative s-palmitoylation of GPT1 will be a difficult task, considering the partial redundancy among Trx h2, h7, h8, h9, the Grx c1, c2 isoforms, and the described regulation of h-type Trx via Grxc1 (Meng et al., 2010; Rouhier, 2010; Riondet et al., 2012; Traverso et al., 2013; Majeran et al., 2018). Clearly, GPT1 is inserted into the ER in monomeric form and may be modified at C65 for ER retention (Figure 9A). Thus, dimer formation beyond the perER might occur after de-protection triggered by the cytosolic redox signaling that accompanies stress responses (Vandenabeele et al., 2004; Foyer et al., 2009) or certain developmental stages, such as pollen tube growth/navigation to ovules (Considine and Foyer, 2014; Hölscher et al., 2016).
GPT1 Behaves Like a Class-II PMP
Our BiFC data suggest that GPT1 contacts at least two of the three early peroxins (Kim and Mullen, 2013). Interaction with Pex3 and/or Pex16 was detected at the ER and PerMs, whereas interaction with Pex19 was mostly distributed across the cytosol, reflecting its function as a cytosolic cargo receptor (Hadden et al., 2006). Because simple co-expression with Pex19-reporter fusions did not appear to alter GPT1 localization, the dot-like structures labeled by GPT1-Pex19 detected by BiFC analyses might be a false-positive result. This would be in line with Pex19 being mainly involved in targeting class-I, but not class-II PMPs. Alternatively, Pex19 may aid in posttranslational ER targeting/insertion, as recently shown for a class of membrane proteins in human HeLa cells (Yamamoto, 2018). The focal localization of GPT1 at the ER, which was previously described for peroxisomal ascorbate peroxidase (pxAPX) in cottonseed and APX3 in Arabidopsis (Lisenbee et al., 2003; Narendra, 2006 ), was primarily detected upon BiFC. GPT1 dimers thus represent a forced interaction at the ER, which does not appear to occur under physiological conditions. GPT1 is usually distributed evenly across the ER, unless co-expressed with Pex16 that coexists at both the ER and PerMs (Lin et al., 2004; Karnik and Trelease, 2005). Interestingly, the presence of Pex16 influenced GPT1 localization at the ER, resulting in a similar but distinct pattern, which also occurred when GPT1 was driven by its own promoter (dark incubation in the presence of sugars activates GPT1 mRNA expression, Supplemental Figure 18). Considering that BiFC is not dynamic, and fluorescent signals persist once the split YFP halves are reconstituted (Robida and Kerppola, 2009), GPT1 was likely dragged to the peroxisomes upon (otherwise transient) interactions with the peroxins. This finding demonstrates that GPT1 can reach the PerMs (although not detected there, unless triggered); hence the transporter may first interact with Pex16 (to reach the perER; Hua et al., 2015) and then with Pex3 (during sorting to PerMs). In contrast to APX3, GPT1 is only needed at peroxisomes when the OPPP is required (Meyer et al., 2011; Hölscher et al., 2014; Lansing et al., 2019). This is in line with the observation that no other OPPP enzymes were identified by peroxisomal proteomics, aside from PGD2 (see Lansing et al., 2019 and references cited therein).
The Transport Preference of GPT1 Differs from That of GPT2
Plastidial TP sequences are cleaved off by the essential stromal processing peptidase, which is usually important for maturation, stabilization, and activation of the imported proteins (van Wijk, 2015). Here we showed that unprocessed GPT1 is also an active transporter. Furthermore, topology analyses using roGFP fusions indicated that upon ER insertion, both the N- and C termini of GPT1 face the cytosol (Supplemental Figure 12), similar to Arabidopsis PMP22 (Murphy et al., 2003) and the human Glc transporter (Mueckler and Lodish, 1986). These findings support the theory of Shao and Hegde (2011) that during post-translational ER import, type-I topology (N terminus facing the lumen) is strongly disfavored. This leads to obligate type-II topology (N terminus facing the cytosol), with membrane domain integration for the cytosolic hinge regions occurring according to the “positive inside rule” (von Heijne, 1986; Goder et al., 2004). The latter is not entirely true for the GPT proteins (color-coded in Supplemental Figure 1 and the topology models), which may facilitate post-translational ER insertion (Figure 9A).
The phosphate translocator family is known to mediate strict counter-exchange of various phosphorylated metabolites with inorganic phosphate (Pi). The ability to transport other intermediates (e.g., triose-phosphates ∼50% by GPT1 and 100% by GPT2; Niewiadomski et al., 2005) is usually disfavored due to the prevailing metabolite concentrations or competition with the preferred substrate (Flügge, 1999; Eicks et al., 2002). Here we showed that GPT1 and GPT2 exchange G6P for Ru5P, but GPT1 has a stronger preference for Ru5P. Thus, import of the OPPP substrate and export of its product across PerMs is warranted (Figure 9B). Moreover, the poor rates obtained with 6PG suggest that sugar-derived NADPH production usually involves all three OPPP steps (Meyer et al., 2011; Hölscher et al., 2014; Lansing et al., 2019), rendering a short-cut solely by Arabidopsis PGD2 in peroxisomes unlikely (Fernández-Fernández and Corpas, 2016; Hölscher et al., 2016).
This transport preference may also explain why Arabidopsis tpt xpt double mutants are viable (although strongly growth-compromised; Hilgers et al., 2018), and why rpi2 mutants, lacking one of the two cytosolic ribose-5-phosphate isomerase isoforms, form less starch in leaves than the wild type (Xiong et al., 2009). If Ru5P accumulates in the cytosol of the rpi mutants, minute amounts of GPT1 could drain some G6P from chloroplasts due to preferred exchange with Ru5P (Supplemental Figure 19). This scenario also supports a role for ubiquitously expressed GPT1 in stabilizing the Calvin–Benson–Bassham cycle (Sharkey and Weise, 2016), considering that GPT2 is usually absent from leaves (Supplemental Figure 14F). On the other hand, the lower transport capacity of GPT1 compared to GPT2 indicates that for GPT1 flux rates are not a limiting factor, in contrast to substrate preferences. This is in line with our complementation data, demonstrating that GPT2 cannot compensate for the absence of GPT1.
Dual Targeting of GPT1 Is Essential during Fertilization
Niewiadomski et al. (2005) found that the loss of GPT1 function strongly affects pollen maturation and embryo-sac development. In plants with reduced GPT1 levels, embryo development is normal up to the globular stage, but then embryos fail to differentiate further and accumulate starch (Andriotis et al., 2010; Andriotis and Smith, 2019). According to the Arabidopsis eFP Browser (Winter et al., 2007), mRNA expression is ∼3.5-fold higher for GPT2 versus GPT1 in this stage (Supplemental Figure 17), which explains the observed starch accumulation upon the loss of GPT1 activity. Accordingly, we suspect that ectopic GPT2 expression may rescue some plastidial functions, but not all phenotypes of the mutant gpt1 alleles, because swap constructs headed by GPT2 were never detected at the ER.
For heterozygous gpt1-2 plants transformed with ProGPT1:GPT2, filled siliques with green, non-aborted embryos and fertilized but later aborted brownish embryos were observed. Homozygous gpt1-2 T-DNA was absent from the progeny of this line and from ER/peroxisomal compensated Pro35S:GFP-GPT1_C-mat. Upon reciprocal crosses of these two lines, only one direction worked (Table 3), indicating that despite partial rescue of the female gpt1 defects (observed as filled siliques), plastid-confined GPT2 was unable to fully rescue GPT1’s functions during pollen maturation/tube growth. Pollen grains appeared normal, but again no homozygous gpt1-2 plants were found. These findings suggest that the remaining defects resulted from a unique function that GPT2 could not fulfill.
Notably, Pro35S:GFP-GPT1_C-mat (transport-competent ER/PerM control) did not rescue ovule abortion (Table 2), but led to a substantial increase in the number of heterozygous offspring compared with the parental line (Table 3). This may be an underestimation, because the CaMV-35S promoter is not well expressed in pollen, and its expression generally fluctuates in floral tissues (Wilkinson et al., 1997). By contrast, ProGPT1:GPT1_N-long mat (without TP) rescued seed set and increased gpt1-2 transmission up to 45.6%, independent of the presence of additional GPT2 in plastids. Together with the pollination defect (mentioned above) and successful complementation by a genomic GPT1 construct (Niewiadomski et al., 2005), our results indicate that for full rescue, GPT1 is also needed in plastids where the OPPP is mainly required for Ru5P provision to nucleotide biosynthesis (Figure 9B), as recently shown by Andriotis and Smith (2019). This nicely supports our previous finding that the loss of Ru5P formation in peroxisomes (due to missing PGD2 activity; Hölscher et al., 2016) prevents the formation of homozygous offspring due to the mutual sterility of male and female pgd2 gametophytes. Moreover, the low GPT transport rates for 6PG (and isoform redundancy at the PGL step; Lansing et al., 2019) suggests that usually no other OPPP intermediate is transported across PerMs. Transport preference for Ru5P may partially explain why GPT1 is indispensable in heterotrophic plastids (Figure 9B), there probably also accepting Pi as a counter-exchange substrate when it accumulates in the stroma due to GPT2-driven starch synthesis.
Finally, the dual targeting of GPT1 is supported by our immuno-detection of unprocessed (ER/peroxisomal) GPT1 in flower/silique and seedling tissues. In seedlings, the changing GPT1 pattern might reflect gradual adaptation to the photoautotrophic state. Moreover, the relative mobility and band intensities in wild type versus gpt2 (and other transporter mutants) indicate that GPT1 activity is regulated by post-translational modifications at both locations, perhaps phosphorylation of the mature protein part (up to five sites; Supplemental Figure 1, blue frames). Glutathionylation (300 D) of the single Cys in the GPT1 N terminus (C65; Figure 9A) cannot explain the observed size shifts, but palmitoylation might explain this observation (Greaves et al., 2008), which may add to the recently discovered role of palmitoylation in male and female gametogenesis (Li et al., 2019). In any case, Grx isoforms are important during fertilization, because Arabidopsis grxc1 grxc2 double mutants exhibited a lethal phenotype early after pollination (Riondet et al., 2012). Finding a potential link of these aspects to dual targeting of GPT1 will require more detailed studies.
In summary, our data present compelling evidence for dual targeting of GPT1 to both plastids and peroxisomes. Imported G6P is converted by the irreversible part of the OPPP to NADPH and Ru5P, the preferred exchange substrate of the GPT1 transporter, thus contributing to gametophyte and embryo development as well as pollen-tube guidance to ovules. Because the latter plays a dominant role in reproductive success, further analyses are required to determine the exact physiological context of GPT1’s presence at the ER/peroxisomes.
METHODS
Bioinformatics
The Arabidopsis Information Resource (TAIR) website (www.arabidopsis.org), Araport (www.araport.org/), PhosPhAt 4.0 (http://phosphat.uni-hohenheim.de/), and the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov) were used to obtain general information about Arabidopsis (Arabidopsis thaliana). Routine analyses were performed with programs on the ExPASy proteomics server (www.expasy.ch) and with Clustal W or Clustal Omega for multiple sequence alignments (www.ebi.ac.uk). Potential interaction candidates of GPT1 were searched in the MIND of Arabidopsis proteins (https://associomics.dpb.carnegiescience.edu; Lalonde et al., 2010). GPT mRNA expression levels were retrieved from the Arabidopsis eFP browser (Winter et al., 2007), and further information on plant membrane proteins were obtained from the ARAMEMNON database (http://aramemnon.uni-koeln.de).
Phylogenetic Analysis
For the phylogenetic tree, sequence information on different plants was retrieved from the National Center for Biotechnology Information, and for the moss Physcomitrella patens from www.cosmoss.org (Supplemental Table 2). Sequence alignments and evolutionary analyses were performed with the software MEGA X (Kumar et al., 2018), using the Maximum Likelihood method based on the Jones-Taylor-Thornton (JTT) matrix-based model (Jones et al., 1992). Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms (in MEGA X) to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value.
Cloning of Fluorescent Reporter Fusions
Open reading frames of candidate genes were obtained by RT-PCR using total RNA from Arabidopsis leaves as described in Hölscher et al. (2016), except for Trxh7, which was amplified from genomic DNA. The oligonucleotide primers are listed in Supplemental Table 3. Reporter constructs were cloned in plant expression vectors as described by Meyer et al. (2011) and Hölscher et al. (2016) and indicated in Supplemental Table 4.
Ratiometric Topology Analysis of roGFP Fusion Proteins
To clone the roGFP-GPT1_C-full construct, the GPT1 fragment was released from GFP-GPT1_C-full by SpeI/BamHI and inserted into proGFP-SDM-∆NcoI via the same sites (for roGFP and roGFP-ER controls, see Jeong et al., 2018). GFP signals were recorded at 405-nm and 488-nm excitation and analyzed ratiometrically using the software ImageJ (https://imagej.nih.gov/ij/index.html), as described by Marschall et al. (2016). Highly oxidized values are displayed in orange-to-yellow (405 nm/488 nm ratio > 1) and highly reduced values in pink-to-blue (405 nm/488 nm ratio < 1).
Site-Directed Mutagenesis
Single base changes designed to destroy restriction sites or alter amino acids were introduced with the Quik-Change PCR Mutagenesis Kit protocol (Stratagene), using the primer combinations listed in Supplemental Table 3 and Phusion High-Fidelity DNA Polymerase (Finnzymes). All base changes were confirmed by sequencing.
Heterologous Protein Expression in Yeast Cells
For in vitro-uptake studies, full-length or mature versions of GPT1 and GPT2 were amplified from cDNA using the corresponding primers and inserted into yeast vectors pYES2 or pYES-NTa via Acc65I (KpnI)/BamHI sites (Thermo Fisher Scientific). For full-length GPT1, the primer combinations were GPT1_Acc65I_s with GPT1+S_BamHI_as; for mature GPT1, GPT1_C-mat_Acc65I_s with GPT1+S_BamHI_as; and for mature GPT2, GPT2_C-mat_Acc65I_s with GPT2+S_BamHI_as (Supplemental Table 3). For the GFP-GPT1_C-mat version, PCR fragments (obtained with primers GPT1_C-mat_SpeI_s and GPT1+S_BamHI_as) were inserted into pGFP2-SDM via SpeI/BamHI sites, released with KpnI/BamHI, and cloned in pYES2. The resulting constructs were transformed into yeast strain INVSc1 (MATa, his3∆1, leu2, trp1-289, ura3-52/MATα,his3∆1, leu2, trp1-289, and ura3-52) using the lithium-acetate/PEG method (Gietz and Schiestl, 2007). Yeast clones were selected on synthetic complete medium (SC-Ura; 0.67% [w/v] YNB supplemented with appropriate amino acids and bases for uracil auxotrophy and 2% [w/v] Glu as a carbon source). Because protein expression was under the control of the Gal-inducible promoter pGAL1, the yeast cells were grown aerobically in SC-Ura supplemented with 2% (w/v) Gal for 6 h at 30°C. Harvest and enrichment of total yeast membranes without and with recombinant GPT proteins was performed according to Linka et al. (2008).
Uptake Studies Using Proteoliposomes
Yeast membranes were reconstituted into 3% (w/v) L-α-phosphatidylcholine using a freeze-thaw-sonication procedure for in vitro-uptake studies as described by Nozawa et al. (2007) and Linka et al. (2008). The proteoliposomes were preloaded with 10 mm) of KPi, G6P, Ru5P, or 6PG and also produced without preloading (negative control). Counter-exchange substrate that was not incorporated into proteoliposomes was removed via gel filtration on Sephadex G-25M columns (GE Healthcare). Transport assays were started by adding 0.2 mM [α-32P]-phosphoric acid (6,000 Ci/mmol) or 0.2 mM [14C]-G6P (290 mCi/mmol). The uptake reaction was terminated by passing the proteoliposomes over AG1-X8 anion-exchange columns (Dowex). The incorporated radiolabeled compounds were analyzed by liquid-scintillation counting. Time-dependent uptake data were fitted using nonlinear regression analysis based on one-phase exponential association using the software Prism 5.0 (GraphPad, www.graphpad.com). The initial uptake velocities were calculated using the equation , where Y0 was set to 0. The values for the plateau and k were extracted from nonlinear regression analyses using a global fit from three technical replicates, i.e., for the same protein batch, three experiments were conducted with the same yeast protein expression round. In detail, 6 × 50 mL Gal-induction cultures were grown. The combined culture volume (300 mL) was used to isolate yeast membranes, split into six proteoliposome samples (three without and three with preloading), and measured in the transport assay (kinetic with six time points), resulting in three uptake values per time point.
Arabidopsis Mutants
Heterozygous Arabidopsis gpt1-1 and gpt1-2 lines (Arabidopsis ecotype Wasilewski [Ws-2]) were kindly provided by Anja Schneider (Ludwig Maximilian University of Munich) and analyzed via PCR amplification from genomic DNA as suggested for the two T-DNA alleles (Niewiadomski et al., 2005). All oligonucleotide primers are listed in Supplemental Table 3 and all plant expression vectors in Supplemental Table 4. For the Feldman line, primers GPT1_EcoRI_s/GPT1-R5 were used for the wild-type allele, and F-RB/GPT1-R5 (Niewiadomski et al., 2005) for the gpt1-1 T-DNA allele. For the Arabidopsis Knockout Facility line, primers GPT1-F3/GPT1-R3 were used for the wild-type allele, and GPT1-F3/JL-202 (Niewiadomski et al., 2005) for the gpt1-2 T-DNA allele. To improve PCR analyses, GPT1-F3 was later replaced by primer gpt1-2_WT_s. Additional mutants used included gpt2-2 (GK-950D09), gpt2-3 (GK-780F12), and xpt-2 (SAIL_378C01) in the Columbia (Col) background, and tpt-5 (FLAG_124C02) in the Ws background. Mutant plants were identified by genomic PCR using the suggested gene-specific and T-DNA-specific primer combinations (Supplemental Table 3).
Plant Growth
Arabidopsis seeds were surface-sterilized in ethanol (vortexed for 5 s each time in 70% [v/v] ethanol, absolute ethanol, and 70% [v/v] ethanol), dried on sterile filter paper, spread on sterile germination medium (0.5× Murashige and Skoog salt mixture with vitamins, pH 5.7 to 5.8, 0.8% (w/v) agar; Duchefa) supplemented with 1% (w/v) Suc, and stratified for 2 to 3 d at 4°C. After propagation in a Percival growth cabinet for one week (short-day regime: 8 h light at 120 μE to 130 μE, combined OSRAM L 18W/640 cool white, top, and OSRAM Dulux L 55W/21-840, on two sides, 21°C, 16 h dark 19°C), the seedlings were transferred to sterile Magenta vessels (Sigma-Aldrich) and grown for 4 to 5 weeks in a tissue culture room (short-day regime: 9 h light at 120 μE to 130 μE, combined MASTER TL-D 58W/820 [Phillips] and Polylux XL FT8 58W/840 [General Electric] in vertical twin bulb set-ups, on one side, 21°C, 15 h dark 19°C) before aseptic rosette leaves were harvested for protoplast isolation. Alternatively, the seedlings were transferred to fertilized soil mix at the 4-leaf stage and grown in growth chambers, first under a short-day regime (8 h light at 130 μE to 150 μE, OSRAM Lumilux L36W/840 cool white, top, 21°C, 16 h dark 19°C) for ∼4 weeks before transfer to an equivalent growth chamber under a long-day regime (16 h light 21°C, 8 h dark 19°C) to promote flowering. For tobacco (Nicotiana tabacum var ‘Xanthi’) growth, sterile apical cuttings were cultivated on Murashige and Skoog agar supplemented with 2% (w/v) Suc in the tissue culture room (see above). The top leaves of 4-week–old plants were used for protoplast isolation.
Protoplast Transfection and Microscopy
The localization of fluorescent reporter fusions (all constructs driven by the CaMV-35S promoter, if not indicated otherwise) was determined by CLSM of freshly prepared mesophyll protoplasts transfected with plasmid DNA (Meyer et al., 2011). For co-expression analyses, 25 μg of test DNA (BiFC: 20 μg of each plasmid) was premixed with 5 μg of a subcellular marker construct (20 μg in the case of Pex16-OFP) before PEG transfection. After cultivation for 12 to 48 h at 21°C to 25°C in the dark (without or with the indicated drug/elicitor), fluorescent signals were recorded using a TCS SP2 or SP5 microscope (Leica) with excitation/emission wavelengths of 405 nm versus 488 nm/490 nm to 520 nm for roGFP, 488 nm/490 nm to 520 nm for GFP, 514 nm/520 nm to 550 nm for YFP, and 561 nm/590 nm to 620 nm for OFP (orange-shifted mRFP, monomeric red fluorescent protein, DsRed). All experiments were conducted at least three times. Transfection efficiency varied between 5 and 20%. For each sample, serial images of four to eight cells were recorded of which representative images are shown. For BiFC analyses, all four possible N and C split YFP orientations were cloned and analyzed. Other isoforms served as negative controls.
Production of GPT-Specific Antisera
The N-terminal GPT cDNA sequences (N1, 91 amino acids of GPT1; N2, 92 amino acids of GPT2) were amplified with primers suited for in-frame insertion behind the His-tag region in pET16b via NdeI/BamHI (Supplemental Table 2) and transformed into Escherichia coli XL-1 blue. Positive clones were verified by sequencing and retransformed into E. coli overexpression strain BL21. Two colonies each were inoculated in YT medium with antibiotics (Ampicillin and Chloramphenicol), transferred to Erlenmeyer flasks (4 × 80 mL each) and incubated on a shaker at 37°C until reaching 0.3 OD600. After further growth for 3 h at 37°C (>1.2 OD600), cells were harvested by centrifugation and resuspended in 8-mL extraction/binding buffer (pH 8) according to the QIAGEN Manual. After repeated sonication on ice (cell lysis), the suspension was centrifuged. The pellet (containing mainly inclusion bodies) was resuspended in 40-mL extraction/binding buffer, sonicated again and centrifuged (wash step). The resulting pellet was resuspended in 10-mL extraction/binding buffer with 6 mm of urea and tumbled overnight in the cold room at 4°C (solubilization step).
Recombinant protein purification started with centrifugation for 20 min at 39,000g and 4°C. The supernatant was pressed through a syringe-based sterile-filter device (0.45 μm), and an aliquot was withdrawn (before column). The filtered solution was mixed with Ni-NTA slurry (equilibrated in 10-Vol extraction/binding buffer with urea), tumbled at room temperature for 2 to 3 h, and filled into a plastic column, collecting the flow-through. Ni-NTA bound protein was rinsed with wash buffer (pH 6.3), resulting in 3 × 5 mL wash fractions (W1, W2, and W3). Weak interactions to the Ni-NTA resin were removed by washing with elution buffer containing 50 mm of imidazole, resulting in 4 × 500-μL elution fractions (E1 to E4). Bound His-N1 (or His-N2) was eluted with 250 mm of imidazole, resulting in final 6 × 500 μL-elution fractions (E1 to E6). SDS-PAGE (15% gels, followed by Coomassie Brilliant Blue staining or immunoblotting) was performed with 20-μL aliquots of all fractions after addition of 4 × SDS-loading buffer and warming for 5 min at 37°C. The purified proteins were diluted as requested before shipping for rabbit immunization (Eurogentec). Obtained successive bleeds were tested for labeling of specific bands on immunoblots (Supplemental Figure 14).
Immunoblot Analyses
Immunoblot analyses were usually conducted as previously described by Meyer et al. (2011), Hölscher et al. (2016), and Lansing et al. (2019) using 10, 12, or 15% separating gels with 10% (v/v) glycerol. Total proteins were separated by SDS-PAGE and transferred to nitrocellulose (90 min at 350 mA). Blots were blocked in TBS plus 0.1% (v/v) Tween-20 (TBST) with 2% (w/v) milk powder at room temperature for 2 h or in the cold overnight. Further incubation was at room temperature, first in GPT antiserum (diluted 1:5,000 to 1:10,000 in fresh blocking solution) for 2 h, followed by 3 × 10 min washes in TBST. Immuno-detection based on 2 h of incubation with GAR-HRP conjugate (1:15,000; Bio-Rad) and ECL reagents (GE Healthcare) employed a sensitive light recording device (MicroChemi; DNR Bio Imaging Systems) for 10 to 30 min. Other commercial antibodies (α-His, Miltenyi-Biotec; α-GFP, from either Invitrogen/Thermo Fisher Scientific or MoBiTec) were used according to the recommendations of the supplier.
Arabidopsis tissues were harvested from plants grown in soil or aseptic seedlings grown on germination plates (1% [w/v] Suc) at different time points. Protein extraction was conducted in 50 mm of HEPES-NaOH at pH 7.5, 2 mm of sodium pyrosulfite (Na2S2O5), 1 mm of Pefabloc SC, Protease Inhibitor Cocktail (1:100) for use with plant extracts (Sigma-Aldrich), and 280 mm of β-mercaptoethanol, if not stated otherwise. PageRuler Prestained Protein Ladder (Fermentas) served as molecular mass standard.
GPT Constructs for Rescue Analyses
For one of the plastidial rescue lines, expression from the MAS promoter was used (pBSK-pMAS-T35S, Supplemental Figure 20). The open reading frame of GPT2 was amplified from cDNA with primer combination GPT2_s_EcoRI/GPT2_as_PstI (all primers are listed in Supplemental Table 3) and inserted into pBSK-pMAS-T35S via EcoRI/PstI sites (pBSK-pMAS:GPT2). The entire expression cassette (pMAS:GPT2-T35S) was released with SalI/XbaI and inserted into binary vector pGSC1704-HygR (ProMAS:GPT2). For GPT1 promoter-driven GPT2, the 5′ upstream sequence of GPT1 (position −1 to −1,958) was amplified from genomic DNA using Phusion High-Fidelity DNA Polymerase (Finnzymes) and inserted blunt-end into pBSK via EcoRV (orientation was confirmed by sequencing). The GPT2-T35S part was amplified with primers GPT2_NdeI_s/T35S_SalI_as from pBSK-pMAS:GPT2 and inserted downstream of the GPT1 promoter via NdeI/SalI in pBSK. The final expression cassette (ProGPT1:GPT2-T35S), amplified with primers pGPT1_s/T35S_SalI_as, was digested with SalI and inserted into pGSC1704-HygR via SnaBI/SalI.
For the CaMV promoter-driven 35S:GFP-GPT1_C-mat construct, the expression cassette was released from vector pGFP2-SDM with PstI/EcoRI, the EcoRI site filled (using Klenow Fragment; Thermo Fisher Scientific) and inserted into binary vector pGSC1704-HygR via SdaI/SnaBI. For GPT1_N-long mat (also driven by the GPT1 promoter), fragments were amplified with primers GPT1_long mat-s and G6P_peroxi_Trans_full_BamHI from existing cDNA clones (upon insertion into the pGFP-NX backbone via XbaI and BamHI, removing GFP). The GPT1 promoter was amplified with primers P_GPT1_s and P_GPT1_as and inserted via PstI/SpeI into PstI/XbaI in the target plasmid, replacing the CaMV-35S promoter. The resulting cassette (GPT1 promoter, GPT1_N-long mat and NOS terminator) was amplified with primers P_GPT1_s and NosT_as, and after SalI digestion inserted into SalI/SnaBI-opened binary vector pDE1001 (both binary vectors were from Ghent University).
All binary constructs were transformed into Agrobacterium strain GV2260 (Scharte et al., 2009). Floral dip transformation of heterozygous +/gpt1 plants was conducted as described by Clough and Bent (1998). Seeds were selected on germination medium containing 15 μg mL−1 of Hygromycin B (Roche) or 50 μg mL−1 of Kanamycin (ProGPT1:GPT1_N-long mat) including 125 μg mL−1 of β-bactyl (SmithKline Beecham), and transferred to soil at the 4-leaf stage. After three weeks, wild-type and T-DNA alleles were genotyped as described above. ProMAS:GPT2 and ProGPT1:GPT2 constructs were amplified from genomic DNA using primers GPT2_C-4MD_SpeI_s and T35S_SalI_as. To test the presence of Pro35S:GFP-GPT1_C-mat, primer combinations P35S_s and GPT1_EcoRI_as or NosT_as were used. The presence of ProGPT1:GPT2 was detected with primers GPT2_XbaI_s and GPT2-Stop_BHI_as (discrimination between the cDNA-based complementation construct and wild-type sequence is based on size, i.e., absence or presence of introns), while GPT1_N-long mat was detected with primers GPT1_long mat_s and NosT_as.
Determination of Ovule-Abortion Frequencies
Siliques numbers 10 to 12 of the main inflorescence (counted from the top) were harvested and incubated in 8 mm of NaOH overnight. Images of bleached and unbleached siliques were recorded with transmitting light using a model no. MZ16 F stereo microscope (Leica) connected to a model no. DFC420 C camera (Leica). Aborted ovules were counted, and frequencies were calculated.
Accession Numbers
Sequence data from this article can be found in the GenBank/European Molecular Biology Laboratory libraries under the following accession numbers: At5g35790 (G6PD1); At5g24400 (PGL3); At3g02360 (PGD2); At1g63290, At3g01850, At5g61410 (RPE isoforms), At5g54800 (GPT1); At1g61800 (GPT2); At5g46110 (TPT); At5g17630 (XPT); At1g59730 (TRX-h7); At5g63030 (GRX-c1); At3g18160 (PEX3-1); At1g48635 (PEX3-2); At2g45690 (PEX16); At3g03490 (PEX19-1); and At5g17550 (PEX19-2).
Supplemental Data
Supplemental Figure 1. Alignment of GPT1 and GPT2 polypeptide sequences from six Brassicaceae.
Supplemental Figure 2. Localization of the N-terminally truncated and full-length reporter-GPT fusions.
Supplemental Figure 3. Single-channel images of Figures 1B and 1C.
Supplemental Figure 4. Localization of the C-terminally truncated GPT-reporter fusions.
Supplemental Figure 5. Localization of the two different GPT1 and GPT2 medial-reporter fusions.
Supplemental Figure 6. Brefeldin-A treatment of the medial GPT_2MD:8MD fusions.
Supplemental Figure 7. Single-channel images of Figure 2B.
Supplemental Figure 8. Single-channel images of Figures 3B and 3C.
Supplemental Figure 9. Trxh7 and Grxc1 partially localize at the ER, together with GPT1.
Supplemental Figure 10. OFP fusions of Pex3-1, Pex16, Pex19-1, and co-expression with GFP-GPT1.
Supplemental Figure 11. Single-channel images of Figure 5C.
Supplemental Figure 12. Ratiometric topology analysis of GPT1 at the ER using roGFP fusions.
Supplemental Figure 13. Single-channel images of Figure 7A.
Supplemental Figure 14. Generation and test of the polyclonal rabbit GPT1 antiserum.
Supplemental Figure 15. Ectopic GPT2 expression for plastidial rescue in heterozygous +/gpt1 lines.
Supplemental Figure 16. ER/peroxisomal rescue of GPT1 function in heterozygous +/gpt1-2 lines.
Supplemental Figure 17. mRNA-expression levels of Arabidopsis GPT1 and GPT2.
Supplemental Figure 18. GPT1 mRNA expression is induced by Suc in low light or darkness.
Supplemental Figure 19. Possible consequences of G6P-Ru5P exchange by GPT1 at chloroplasts.
Supplemental Figure 20. Vector map of pBSK-pMAS-T35S.
Supplemental Table 1. GPT1 search results of the MIND.
Supplemental Table 2. Protein sequences used to construct the phylogenetic tree.
Supplemental Table 3. Oligonucleotide primers used in this study.
Supplemental Table 4. Plant expression vectors used in this study.
Supplemental File 1. Text file of alignment data (Supplemental Figure 1).
Supplemental File 2. Phylogenetic tree with bootstrap values.
Supplemental File 3. Text file of alignment data (phylogenetic tree).
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
The authors thank Anja Schneider (Ludwig Maximilian University of Munich) for providing the heterozygous gpt1 lines, and the Arabidopsis Stock Centers (Nottingham Arabidopsis Stock Centre and Institut National de la Recherche Agronomique) for the other mutant lines. Andreas Meyer (Institute for Crop Science and Resource Conservation, University of Bonn) donated the roGFP2 plasmid, and Robert Marschall (Institut für Biologie und Biotechnologie der Pflanzen, IBBP) helped with ratiometric analyses in the software ImageJ. Stephan Rips gave advice on CLSM imaging, Olessia Becker was involved in GPT antigen production/antibody testing, Sebastian Hassa cloned expression vector pBSK:pMAS-T35S, Lennart Doering finished the GPT1-C65S split YFP constructs, and Wiltrud Krüger helped with routine lab work (members of the A.v.S work group, IBBP). Several Bachelor Degree students in the A.v.S. work group contributed by their thesis projects: Jan Wiese by RPE-isoform analyses, Margareta Westphalen to BiFC-interaction analyses, and Hinrik Plaggenborg to roGFP-topology analyses. This work was in part funded by the German Research Foundation (DFG grants SCHA 541/12 to A.v.S. and LI 1781/1-3 to N.L.).
AUTHOR CONTRIBUTIONS
M.-C.B., H.L., K.F., T.M., L.C., and N.L. performed the experiments; A.v.S., M.-C.B., H.L., and N.L. designed experiments and analyzed the data; M.-C.B., H.L.., and A.v.S. wrote the article; all authors read and approved the final version of the article.
Footnotes
Articles can be viewed without a subscription.
References
- Ackerley S., Thornhill P., Grierson A.J., Brownlees J., Anderton B.H., Leigh P.N., Shaw C.E., Miller C.C.J.(2003). Neurofilament heavy chain side arm phosphorylation regulates axonal transport of neurofilaments. J. Cell Biol. 161: 489–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aicart-Ramos C., Valero R.A., Rodriguez-Crespo I.(2011). Protein palmitoylation and subcellular trafficking. Biochim. Biophys. Acta 1808: 2981–2994. [DOI] [PubMed] [Google Scholar]
- Andriotis V.M.E., Pike M.J., Bunnewell S., Hills M.J., Smith A.M.(2010). The plastidial glucose-6-phosphate/phosphate antiporter GPT1 is essential for morphogenesis in Arabidopsis embryos. Plant J. 64: 128–139. [DOI] [PubMed] [Google Scholar]
- Andriotis V.M.E., Smith A.M.(2019). The plastidial pentose phosphate pathway is essential for postglobular embryo development in Arabidopsis. Proc. Natl. Acad. Sci. USA 116: 15297–15306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranovich A., Hua R., Rutenberg A.D., Kim P.K.(2014). PEX16 contributes to peroxisome maintenance by constantly trafficking PEX3 via the ER. J. Cell Sci. 127: 3675–3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiou K., Dyson B.C., Webster R.E., Johnson G.N.(2010). Dynamic acclimation of photosynthesis increases plant fitness in changing environments. Plant Physiol. 152: 366–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baslam M., Oikawa K., Kitajima-Koga A., Kaneko K., Mitsui T.(2016). Golgi-to-plastid trafficking of proteins through secretory pathway: Insights into vesicle-mediated import toward the plastids. Plant Signal. Behav. 11: e1221558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt C., Lillig C.H., Holmgren A.(2008). Thioredoxins and glutaredoxins as facilitators of protein folding. Biochim. Biophys. Acta 1783: 641–650. [DOI] [PubMed] [Google Scholar]
- Cakir B., et al. (2016). Analysis of the rice ADP-Glucose Transporter (OsBT1) indicates the presence of regulatory processes in the amyloplast stroma that control ADP-Glucose flux into starch. Plant Physiol. 170: 1271–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T.(2009). Predation and eukaryote cell origins: A coevolutionary perspective. Int. J. Biochem. Cell Biol. 41: 307–322. [DOI] [PubMed] [Google Scholar]
- Chen J., Lalonde S., Obrdlik P., Noorani Vatani A., Parsa S.A., Vilarino C., Revuelta J.L., Frommer W.B., Rhee S.Y.(2012). Uncovering Arabidopsis membrane protein interactome enriched in transporters using mating-based split ubiquitin assays and classification models. Front. Plant Sci. 3: 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua N.-H., Schmidt G.W.(1979). Transport of proteins into mitochondria and chloroplasts. J. Cell Biol. 81: 461–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F.(1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Considine M.J., Foyer C.H.(2014). Redox regulation of plant development. Antioxid. Redox Signal. 21: 1305–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpas F.J., Barroso J.B., Sandalio L.M., Distefano S., Palma J.M., Lupiáñez J.A., Del Río L.A.(1998). A dehydrogenase-mediated recycling system of NADPH in plant peroxisomes. Biochem. J. 330: 777–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Río L.A., Corpas F.J., Sandalio L.M., Palma J.M., Gómez M., Barroso J.B.(2002). Reactive oxygen species, antioxidant systems and nitric oxide in peroxisomes. J. Exp. Bot. 53: 1255–1272. [PubMed] [Google Scholar]
- Dennis D.T., Layzell D.B., Lefebvre D.D., Turpin D.H.(1997). Plant Metabolism, Dennis D.T., Layzell D.B., Lefebvre D.D., and Turpin D.H., eds, 2nd Edition (New Jersey: Prentice Hall; ). [Google Scholar]
- Dietz K.-J.(2011). Peroxiredoxins in plants and cyanobacteria. Antioxid. Redox Signal. 15: 1129–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durek P., Schmidt R., Heazlewood J.L., Jones A., MacLean D., Nagel A., Kersten B., Schulze W.X.(2010). PhosPhAt: The Arabidopsis thaliana phosphorylation site database. An update. Nucleic Acids Res. 38: 828–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson B.C., Allwood J.W., Feil R., Xu Y., Miller M., Bowsher C.G., Goodacre R., Lunn J.E., Johnson G.N.(2015). Acclimation of metabolism to light in Arabidopsis thaliana: The glucose 6-phosphate/phosphate translocator GPT2 directs metabolic acclimation. Plant Cell Environ. 38: 1404–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson B.C., Webster R.E., Johnson G.N.(2014). GPT2: A glucose 6-phosphate/phosphate translocator with a novel role in the regulation of sugar signalling during seedling development. Ann. Bot. 113: 643–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eicks M., Maurino V., Knappe S., Flügge U.-I., Fischer K.(2002). The plastidic pentose phosphate translocator represents a link between the cytosolic and the plastidic pentose phosphate pathways in plants. Plant Physiol. 128: 512–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J.(1985). Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- Fernández-Fernández Á., Corpas F.J.(2016). In silico analysis of Arabidopsis thaliana peroxisomal 6-phosphogluconate dehydrogenase. Scientifica (Cairo) 2016: 3482760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flügge U.-I.(1999). Phosphate translocators in plastids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 27–45. [DOI] [PubMed] [Google Scholar]
- Flügge U.-I., Häusler R.E., Ludewig F., Gierth M.(2011). The role of transporters in supplying energy to plant plastids. J. Exp. Bot. 62: 2381–2392. [DOI] [PubMed] [Google Scholar]
- Foyer C.H., Bloom A.J., Queval G., Noctor G.(2009). Photorespiratory metabolism: Genes, mutants, energetics, and redox signaling. Annu. Rev. Plant Biol. 60: 455–484. [DOI] [PubMed] [Google Scholar]
- Geigenberger P., Kolbe A., Tiessen A.(2005). Redox regulation of carbon storage and partitioning in response to light and sugars. J. Exp. Bot. 56: 1469–1479. [DOI] [PubMed] [Google Scholar]
- Gietz R.D., Schiestl R.H.(2007). High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2: 31–34. [DOI] [PubMed] [Google Scholar]
- Goder V., Junne T., Spiess M.(2004). Sec61p contributes to signal sequence orientation according to the positive-inside rule. Mol. Biol. Cell 15: 1470–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong F.-C., Giddings T.H., Meehl J.B., Staehelin L.A., Galbraith D.W.(1996). Z-membranes: Artificial organelles for overexpressing recombinant integral membrane proteins. Proc. Natl. Acad. Sci. USA 93: 2219–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S.J., Keller G.-A., Hosken N., Wilkinson J., Subramani S.(1989). A conserved tripeptide sorts proteins to peroxisomes. J. Cell Biol. 108: 1657–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves J., Salaun C., Fukata Y., Fukata M., Chamberlain L.H.(2008). Palmitoylation and membrane interactions of the neuroprotective chaperone cysteine-string protein. J. Biol. Chem. 283: 25014–25026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guevara-García A., Mosqueda-Cano G., Argüello-Astorga G., Simpson J., Herrera-Estrella L.(1993). Tissue-specific and wound-inducible pattern of expression of the mannopine synthase promoter is determined by the interaction between positive and negative cis-regulatory elements. Plant J. 4: 495–505. [DOI] [PubMed] [Google Scholar]
- Gurrieri L., Distefano L., Pirone C., Horrer D., Seung D., Zaffagnini M., Rouhier N., Trost P., Santelia D., Sparla F.(2019). The thioredoxin-regulated α-amylase 3 of Arabidopsis thaliana is a target of S-glutathionylation. Front. Plant Sci. 10: 993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadden D.A., Phillipson B.A., Johnston K.A., Brown L.A., Manfield I.W., El-Shami M., Sparkes I.A., Baker A.(2006). Arabidopsis PEX19 is a dimeric protein that binds the peroxin PEX10. Mol. Membr. Biol. 23: 325–336. [DOI] [PubMed] [Google Scholar]
- Hauschild R., von Schaewen A.(2003). Differential regulation of glucose-6-phosphate dehydrogenase isoenzyme activities in potato. Plant Physiol. 133: 47–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood J.L., Durek P., Hummel J., Selbig J., Weckwerth W., Walther D., Schulze W.X.(2008). PhosPhAt: A database of phosphorylation sites in Arabidopsis thaliana and a plant-specific phosphorylation site predictor. Nucleic Acids Res. 36: D1015–D1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemsley P.A.(2015). The importance of lipid modified proteins in plants. New Phytol. 205: 476–489. [DOI] [PubMed] [Google Scholar]
- Hilgers E.J.A., Schöttler M.A., Mettler-Altmann T., Krueger S., Dörmann P., Eicks M., Flügge U.I., Häusler R.E.(2018). The combined loss of triose phosphate and xylulose 5-phosphate/phosphate translocators leads to severe growth retardation and impaired photosynthesis in Arabidopsis thaliana tpt/xpt double mutants. Front. Plant Sci. 9: 1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hölscher C., Lutterbey M.-C., Lansing H., Meyer T., Fischer K., von Schaewen A.(2016). Defects in peroxisomal 6-phosphogluconate dehydrogenase isoform PGD2 prevent gametophytic interaction in Arabidopsis thaliana. Plant Physiol. 171: 192–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hölscher C., Meyer T., von Schaewen A.(2014). Dual-targeting of Arabidopsis 6-phosphogluconolactonase 3 (PGL3) to chloroplasts and peroxisomes involves interaction with Trx m2 in the cytosol. Mol. Plant 7: 252–255. [DOI] [PubMed] [Google Scholar]
- Hua R., Gidda S.K., Aranovich A., Mullen R.T., Kim P.K.(2015). Multiple domains in PEX16 mediate its trafficking and recruitment of peroxisomal proteins to the ER. Traffic 16: 832–852. [DOI] [PubMed] [Google Scholar]
- Hunt J.E., Trelease R.N.(2004). Sorting pathway and molecular targeting signals for the Arabidopsis peroxin 3. Biochem. Biophys. Res. Commun. 314: 586–596. [DOI] [PubMed] [Google Scholar]
- Hutchings D., Rawsthorne S., Emes M.J.(2005). Fatty acid synthesis and the oxidative pentose phosphate pathway in developing embryos of oilseed rape (Brassica napus L.). J. Exp. Bot. 56: 577–585. [DOI] [PubMed] [Google Scholar]
- Jeong I.S., et al. (2018). Purification and characterization of Arabidopsis thaliana oligosaccharyltransferase complexes from the native host: A protein super-expression system for structural studies. Plant J. 94: 131–145. [DOI] [PubMed] [Google Scholar]
- Jones A.M., et al. (2014). Border control—a membrane-linked interactome of Arabidopsis. Science 344: 711–716. [DOI] [PubMed] [Google Scholar]
- Jones D.T., Taylor W.R., Thornton J.M.(1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8: 275–282. [DOI] [PubMed] [Google Scholar]
- Kammerer B., Fischer K., Hilpert B., Schubert S., Gutensohn M., Weber A., Flügge U.I.(1998). Molecular characterization of a carbon transporter in plastids from heterotrophic tissues: The glucose 6-phosphate/phosphate antiporter. Plant Cell 10: 105–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao Y.T., Gonzalez K.L., Bartel B.(2018). Peroxisome function, biogenesis, and dynamics in plants. Plant Physiol. 176: 162–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnik S.K., Trelease R.N.(2005). Arabidopsis peroxin 16 coexists at steady state in peroxisomes and endoplasmic reticulum. Plant Physiol. 138: 1967–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataya A.R., Reumann S.(2010). Arabidopsis glutathione reductase 1 is dually targeted to peroxisomes and the cytosol. Plant Signal. Behav. 5: 171–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim P.K., Hettema E.H.(2015). Multiple pathways for protein transport to peroxisomes. J. Mol. Biol. 427: 1176–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim P.K., Mullen R.T.(2013). PEX16: A multifaceted regulator of peroxisome biogenesis. Front. Physiol. 4: 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausner R.D., Donaldson J.G., Lippincott-Schwartz J.(1992). Brefeldin A: Insights into the control of membrane traffic and organelle structure. J. Cell Biol. 116: 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knappe S., Flügge U.-I., Fischer K.(2003). Analysis of the plastidic phosphate translocator gene family in Arabidopsis and identification of new phosphate translocator-homologous transporters, classified by their putative substrate-binding site. Plant Physiol. 131: 1178–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger N.J., von Schaewen A.(2003). The oxidative pentose phosphate pathway: Structure and organisation. Curr. Opin. Plant Biol. 6: 236–246. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K.(2018). MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 35: 1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz H.-H., Häusler R.E., Fettke J., Herbst K., Niewiadomski P., Gierth M., Bell K., Steup M., Flügge U.-I., Schneider A.(2010). The role of plastidial glucose-6-phosphate/phosphate translocators in vegetative tissues of Arabidopsis thaliana mutants impaired in starch biosynthesis. Plant Biol (Stuttg) 12 (Suppl 1): 115–128. [DOI] [PubMed] [Google Scholar]
- Lalonde S., et al. (2010). A membrane protein/signaling protein interaction network for Arabidopsis version AMPv2. Front. Physiol. 1: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansing H., Doering L., Fischer K., Baune M.-C., von Schaewen A.(2019). Analysis of potential redundancy among Arabidopsis 6-phosphogluconolactonase (PGL) isoforms in peroxisomes. J. Exp. Bot. 71: 823–836. [DOI] [PubMed] [Google Scholar]
- Lee S.K., Eom J.S., Hwang S.K., Shin D., An G., Okita T.W., Jeon J.S.(2016). Plastidic phosphoglucomutase and ADP-glucose pyrophosphorylase mutants impair starch synthesis in rice pollen grains and cause male sterility. J. Exp. Bot. 67: 5557–5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Nishizawa T., Takemoto M., Kumazaki K., Yamashita K., Hirata K., Minoda A., Nagatoishi S., Tsumoto K., Ishitani R., Nureki O.(2017). Structure of the triose-phosphate/phosphate translocator reveals the basis of substrate specificity. Nat. Plants 3: 825–832. [DOI] [PubMed] [Google Scholar]
- Li S.(2014). Redox modulation matters: Emerging functions for glutaredoxins in plant development and stress responses. Plants (Basel) 3: 559–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Li H.J., Morgan C., Bomblies K., Yang W., Qi B.(2019). Both male and female gametogenesis require a fully functional protein S-acyl transferase 21 in Arabidopsis thaliana. Plant J. 100: 754–767. [DOI] [PubMed] [Google Scholar]
- Liebthal M., Maynard D., Dietz K.-J.(2018). Peroxiredoxins and redox signaling in plants. Antioxid. Redox Signal. 28: 609–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Cluette-Brown J.E., Goodman H.M.(2004). The peroxisome deficient Arabidopsis mutant sse1 exhibits impaired fatty acid synthesis. Plant Phys. 135: 814–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Sun L., Nguyen L.V., Rachubinski R.A., Goodman H.M.(1999). The Pex16p homolog SSE1 and storage organelle formation in Arabidopsis seeds. Science 284: 328–330. [DOI] [PubMed] [Google Scholar]
- Linka N., Theodoulou F.L., Haslam R.P., Linka M., Napier J.A., Neuhaus H.E., Weber A.P.(2008). Peroxisomal ATP import is essential for seedling development in Arabidopsis thaliana. Plant Cell 20: 3241–3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisenbee C.S., Karnik S.K., Trelease R.N.(2003). Overexpression and mislocalization of a tail-anchored GFP redefines the identity of peroxisomal ER. Traffic 4: 491–501. [DOI] [PubMed] [Google Scholar]
- Majeran W., Le Caer J.P., Ponnala L., Meinnel T., Giglione C.(2018). Targeted profiling of Arabidopsis thaliana subproteomes illuminates co- and posttranslationally N-terminal myristoylated proteins. Plant Cell 30: 543–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschall R., Schumacher J., Siegmund U., Tudzynski P.(2016). Chasing stress signals—exposure to extracellular stimuli differentially affects the redox state of cell compartments in the wild type and signaling mutants of Botrytis cinerea. Fungal Genet. Biol. 90: 12–22. [DOI] [PubMed] [Google Scholar]
- Marty L., et al. (2019). Arabidopsis glutathione reductase 2 is indispensable in plastids, while mitochondrial glutathione is safeguarded by additional reduction and transport systems. New Phytol. 224: 1569–1584. [DOI] [PubMed] [Google Scholar]
- Marty L., Siala W., Schwarzländer M., Fricker M.D., Wirtz M., Sweetlove L.J., Meyer Y., Meyer A.J., Reichheld J.-P., Hell R.(2009). The NADPH-dependent thioredoxin system constitutes a functional backup for cytosolic glutathione reductase in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 9109–9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell M.M., Burkhart S.E., Stoddard J.M., Wright Z.J., Strader L.C., Bartel B.(2016). The early-acting peroxin PEX19 is redundantly encoded, farnesylated, and essential for viability in Arabidopsis thaliana. PLoS One 11: e0148335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L., Wong J.H., Feldman L.J., Lemaux P.G., Buchanan B.B.(2010). A membrane-associated thioredoxin required for plant growth moves from cell to cell, suggestive of a role in intercellular communication. Proc. Natl. Acad. Sci. USA 107: 3900–3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer T., Hölscher C., Schwöppe C., von Schaewen A.(2011). Alternative targeting of Arabidopsis plastidic glucose-6-phosphate dehydrogenase G6PD1 involves cysteine-dependent interaction with G6PD4 in the cytosol. Plant J. 66: 745–758. [DOI] [PubMed] [Google Scholar]
- Mhamdi A., Mauve C., Gouia H., Saindrenan P., Hodges M., Noctor G.(2010). Cytosolic NADP-dependent isocitrate dehydrogenase contributes to redox homeostasis and the regulation of pathogen responses in Arabidopsis leaves. Plant Cell Environ. 33: 1112–1123. [DOI] [PubMed] [Google Scholar]
- Mitterreiter M.J., Bosch F.A., Brylok T., Schwenkert S.(2019). The ER luminal C-terminus of AtSec62 is critical for male fertility and plant growth in Arabidopsis thaliana. Plant J. 101: 5–17. [DOI] [PubMed] [Google Scholar]
- Mueckler M., Lodish H.F.(1986). The human glucose transporter can insert posttranslationally into microsomes. Cell 44: 629–637. [DOI] [PubMed] [Google Scholar]
- Mullen R.T., Lisenbee C.S., Miernyk J.A., Trelease R.N.(1999). Peroxisomal membrane ascorbate peroxidase is sorted to a membranous network that resembles a subdomain of the endoplasmic reticulum. Plant Cell 11: 2167–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy M.A., Phillipson B.A., Baker A., Mullen R.T.(2003). Characterization of the targeting signal of the Arabidopsis 22-kD integral peroxisomal membrane protein. Plant Physiol. 133: 813–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra S., et al. (2006). The Arabidopsis ascorbate peroxidase 3 is a peroxisomal membrane-bound antioxidant enzyme and is dispensable for Arabidopsis growth and development. Journal of Experimental Botany 57: 3033–3042. [DOI] [PubMed] [Google Scholar]
- Niewiadomski P., Knappe S., Geimer S., Fischer K., Schulz B., Unte U.S., Rosso M.G., Ache P., Flügge U.-I., Schneider A.(2005). The Arabidopsis plastidic glucose 6-phosphate/phosphate translocator GPT1 is essential for pollen maturation and embryo sac development. Plant Cell 17: 760–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G., Foyer C.H.(2016). Intracellular redox compartmentation and ROS-related communication in regulation and signaling. Plant Physiol. 171: 1581–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa A., Nanamiya H., Miyata T., Linka N., Endo Y., Weber A.P.M., Tozawa Y.(2007). A cell-free translation and proteoliposome reconstitution system for functional analysis of plant solute transporters. Plant Cell Physiol. 48: 1815–1820. [DOI] [PubMed] [Google Scholar]
- Orci L., Tagaya M., Amherdt M., Perrelet A., Donaldson J.G., Lippincott-Schwartz J., Klausner R.D., Rothman J.E.(1991). Brefeldin A, a drug that blocks secretion, prevents the assembly of non-clathrin-coated buds on Golgi cisternae. Cell 64: 1183–1195. [DOI] [PubMed] [Google Scholar]
- Palatnik J.F., Tognetti V.B., Poli H.O., Rodríguez R.E., Blanco N., Gattuso M., Hajirezaei M.R., Sonnewald U., Valle E.M., Carrillo N.(2003). Transgenic tobacco plants expressing antisense ferredoxin-NADP(H) reductase transcripts display increased susceptibility to photo-oxidative damage. Plant J. 35: 332–341. [DOI] [PubMed] [Google Scholar]
- Park S.K., et al. (2009). Heat-shock and redox-dependent functional switching of an h-type Arabidopsis thioredoxin from a disulfide reductase to a molecular chaperone. Plant Physiol. 150: 552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platta H.W., Erdmann R.(2007). Peroxisomal dynamics. Trends Cell Biol. 17: 474–484. [DOI] [PubMed] [Google Scholar]
- Porter B.W., Yuen C.Y.L., Christopher D.A.(2015). Dual protein trafficking to secretory and non-secretory cell compartments: Clear or double vision? Plant Sci. 234: 174–179. [DOI] [PubMed] [Google Scholar]
- Preiser A.L., Fisher N., Banerjee A., Sharkey T.D.(2019). Plastidic glucose-6-phosphate dehydrogenases are regulated to maintain activity in the light. Biochem. J. 476: 1539–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S.(2004). Specification of the peroxisome targeting signals type 1 and type 2 of plant peroxisomes by bioinformatics analyses. Plant Physiol. 135: 783–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S., Babujee L., Ma C., Wienkoop S., Siemsen T., Antonicelli G.E., Rasche N., Lüder F., Weckwerth W., Jahn O.(2007). Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. Plant Cell 19: 3170–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S., Bartel B.(2016). Plant peroxisomes: Recent discoveries in functional complexity, organelle homeostasis, and morphological dynamics. Curr. Opin. Plant Biol. 34: 17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S., Ma C., Lemke S., Babujee L.(2004). AraPerox. A database of putative Arabidopsis proteins from plant peroxisomes. Plant Physiol. 136: 2587–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S., Maier E., Benz R., Heldt H.W.(1996). A specific porin is involved in the malate shuttle of leaf peroxisomes. Biochem. Soc. Trans. 24: 754–757. [DOI] [PubMed] [Google Scholar]
- Riondet C., Desouris J.P., Montoya J.G., Chartier Y., Meyer Y., Reichheld J.P.(2012). A dicotyledon-specific glutaredoxin GRXc1 family with dimer-dependent redox regulation is functionally redundant with GRXC2. Plant Cell Environ. 35: 360–373. [DOI] [PubMed] [Google Scholar]
- Rips S., Bentley N., Jeong I.S., Welch J.L., von Schaewen A., Koiwa H.(2014). Multiple N-glycans cooperate in the subcellular targeting and functioning of Arabidopsis KORRIGAN1. Plant Cell 26: 3792–3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robida A.M., Kerppola T.K.(2009). Bimolecular fluorescence complementation analysis of inducible protein interactions: Effects of factors affecting protein folding on fluorescent protein fragment association. J. Mol. Biol. 394: 391–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokka A., Antonenkov V.D., Soininen R., Immonen H.L., Pirilä P.L., Bergmann U., Sormunen R.T., Weckström M., Benz R., Hiltunen J.K.(2009). Pxmp2 is a channel-forming protein in mammalian peroxisomal membrane. PLoS One 4: e5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottensteiner H., Kramer A., Lorenzen S., Stein K., Landgraf C., Volkmer-Engert R., Erdmann R.(2004). Peroxisomal membrane proteins contain common Pex19p-binding sites that are an integral part of their targeting signals. Mol. Biol. Cell 15: 3406–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhier N.(2010). Plant glutaredoxins: Pivotal players in redox biology and iron-sulphur centre assembly. New Phytol. 186: 365–372. [DOI] [PubMed] [Google Scholar]
- Sakaue H., Iwashita S., Yamashita Y., Kida Y., Sakaguchi M.(2016). The N-terminal motif of PMP70 suppresses cotranslational targeting to the endoplasmic reticulum. J. Biochem. 159: 539–551. [DOI] [PubMed] [Google Scholar]
- Sanz-Barrio R., Fernández-San Millán A., Carballeda J., Corral-Martínez P., Seguí-Simarro J.M., Farran I.(2012). Chaperone-like properties of tobacco plastid thioredoxins f and m. J. Exp. Bot. 63: 365–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharte J., Schön H., Tjaden Z., Weis E., von Schaewen A.(2009). Isoenzyme replacement of glucose-6-phosphate dehydrogenase in the cytosol improves stress tolerance in plants. Proc. Natl. Acad. Sci. USA 106: 8061–8066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt G.W., Devillers-Thiery A., Desruisseaux H., Blobel G., Chua N.-H.(1979). NH2-terminal amino acid sequences of precursor and mature forms of the ribulose-1,5-bisphosphate carboxylase small subunit from Chlamydomonas reinhardtii. J. Cell Biol. 83: 615–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnarrenberger C., Flechner A., Martin W.(1995). Enzymatic evidence for a complete oxidative pentose phosphate pathway in chloroplasts and an incomplete pathway in the cytosol of spinach leaves. Plant Physiol. 108: 609–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao S., Hegde R.S.(2011). Membrane protein insertion at the endoplasmic reticulum. Annu. Rev. Cell Dev. Biol. 27: 25–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkey T.D., Weise S.E.(2016). The glucose 6-phosphate shunt around the Calvin–Benson cycle. J. Exp. Bot. 67: 4067–4077. [DOI] [PubMed] [Google Scholar]
- Stampfl H., Fritz M., Dal Santo S., Jonak C.(2016). The GSK3/Shaggy-like kinase ASKα contributes to pattern-triggered immunity in Arabidopsis thaliana. Plant Physiol. 171: pp.01741.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ströher E.; Millar AH. (2012). The biological roles of glutaredoxins. Biochemical Journal 446: 333–348. [DOI] [PubMed] [Google Scholar]
- Tabak H.F., Hoepfner D., Zand A., Geuze H.J., Braakman I., Huynen M.A.(2006). Formation of peroxisomes: Present and past. Biochim. Biophys. Acta 1763: 1647–1654. [DOI] [PubMed] [Google Scholar]
- Theodoulou F.L., Bernhardt K., Linka N., Baker A.(2013). Peroxisome membrane proteins: Multiple trafficking routes and multiple functions? Biochem. J. 451: 345–352. [DOI] [PubMed] [Google Scholar]
- Torres M.A., Dangl J.L., Jones J.D.G.(2002). Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA 99: 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traverso J.A., Pulido A., Rodríguez-García M.I., Alché J.D.(2013). Thiol-based redox regulation in sexual plant reproduction: New insights and perspectives. Front. Plant Sci. 4: 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyra H.M., Linka M., Weber A.P.M., Bhattacharya D.(2007). Host origin of plastid solute transporters in the first photosynthetic eukaryotes. Genome Biol. 8: R212.1-R212.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukuwela A.A., Bush A.I., Wedd A.G., Xiao Z.(2017). Glutaredoxins employ parallel monothiol–dithiol mechanisms to catalyze thiol-disulfide exchanges with protein disulfides. Chem. Sci. (Camb.) 9: 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenabeele S., Vanderauwera S., Vuylsteke M., Rombauts S., Langebartels C., Seidlitz H.K., Zabeau M., Van Montagu M., Inzé D., Van Breusegem F.(2004). Catalase deficiency drastically affects gene expression induced by high light in Arabidopsis thaliana. Plant J. 39: 45–58. [DOI] [PubMed] [Google Scholar]
- van der Zand A., Braakman I., Tabak H.F.(2010). Peroxisomal membrane proteins insert into the endoplasmic reticulum. Mol. Biol. Cell 21: 2057–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijk K.J.(2015). Protein maturation and proteolysis in plant plastids, mitochondria, and peroxisomes. Annu. Rev. Plant Biol. 66: 75–111. [DOI] [PubMed] [Google Scholar]
- von Heijne G.(1986). Net N-C charge imbalance may be important for signal sequence function in bacteria. J. Mol. Biol. 192: 287–290. [DOI] [PubMed] [Google Scholar]
- von Loeffelholz O., Kriechbaumer V., Ewan R.A., Jonczyk R., Lehmann S., Young J.C., Abell B.M.(2011). OEP61 is a chaperone receptor at the plastid outer envelope. Biochem. J. 438: 143–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M., et al. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438. [DOI] [PubMed] [Google Scholar]
- Waszczak C., Carmody M., Kangasjärvi J.(2018). Reactive oxygen species in plant signaling. Annu. Rev. Plant Biol. 69: 209–236. [DOI] [PubMed] [Google Scholar]
- Weise S.E., Liu T., Childs K.L., Preiser A.L., Katulski H.M., Perrin-Porzondek C., Sharkey T.D.(2019). Transcriptional regulation of the glucose-6-phosphate/phosphate translocator 2 is related to carbon exchange across the chloroplast envelope. Front. Plant Sci. 10: 827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson J.E., Twell D., Lindsey K.(1997). Activities of CaMV 35S and nos promoters in pollen: Implications for field release of transgenic plants. J. Exp. Bot. 48: 265–275. [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J.(2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., DeFraia C., Williams D., Zhang X., Mou Z.(2009). Deficiency in a cytosolic ribose-5-phosphate isomerase causes chloroplast dysfunction, late flowering and premature cell death in Arabidopsis. Physiol. Plant. 137: 249–263. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y.; Toshiaki Sakisaka. (2018). The peroxisome biogenesis factors posttranslationally target reticulon homology domain-containing proteins to the endoplasmic reticulum membrane. Scientific Reports 8: 1027–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffagnini M., Fermani S., Marchand C.H., Costa A., Sparla F., Rouhier N., Geigenberger P., Lemaire S.D., Trost P.(2019). Redox homeostasis in photosynthetic organisms: Novel and established thiol-based molecular mechanisms. Antioxid. Redox Signal. 31: 155–210. [DOI] [PubMed] [Google Scholar]
- Zulawski M., Braginets R., Schulze W.X.(2013). PhosPhAt goes kinases—searchable protein kinase target information in the plant phosphorylation site database PhosPhAt. Nucleic Acids Res. 41: D1176–D1184. [DOI] [PMC free article] [PubMed] [Google Scholar]