The calcium-dependent protein kinase CPK1 regulates a pivotal developmental switch in senescence and leaf cell death through direct phosphorylation of transcription factor ORE1.
Abstract
Calcium-regulated protein kinases are key components of intracellular signaling in plants that mediate rapid stress-induced responses to changes in the environment. To identify in vivo phosphorylation substrates of CALCIUM-DEPENDENT PROTEIN KINASE1 (CPK1), we analyzed the conditional expression of constitutively active CPK1 in conjunction with in vivo phosphoproteomics. We identified Arabidopsis (Arabidopsis thaliana) ORESARA1 (ORE1), the developmental master regulator of senescence, as a direct CPK1 phosphorylation substrate. CPK1 phosphorylates ORE1 at a hotspot within an intrinsically disordered region. This augments transcriptional activation by ORE1 of its downstream target gene BIFUNCTIONAL NUCLEASE1 (BFN1). Plants that overexpress ORE1, but not an ORE1 variant lacking the CPK1 phosphorylation hotspot, promote early senescence. Furthermore, ORE1 is required for enhanced cell death induced by CPK1 signaling. Our data validate the use of conditional expression of an active enzyme combined with phosphoproteomics to decipher specific kinase target proteins of low abundance, of transient phosphorylation, or in yet-undescribed biological contexts. Here, we have identified that senescence is not just under molecular surveillance manifested by stringent gene regulatory control over ORE1. In addition, the decision to die is superimposed by an additional layer of control toward ORE1 via its posttranslational modification linked to the calcium-regulatory network through CPK1.
INTRODUCTION
Plant calcium-dependent protein kinases (CDPKs) have been characterized as enzymes in which, within the same protein molecule, the input (calcium binding) by a sensor domain controls the output (substrate phosphorylation) by a kinase effector domain. This dual sensor-effector role is evident in the conserved modular protein structure whereby the variable N-terminal domain precedes the protein kinase domain and the calcium-activation domain (CAD). The CAD encompasses an inhibitory pseudosubstrate region and the calmodulin-like domain, which in canonical CDPKs carries four consensus EF-hand calcium binding motifs. This modular structure is corroborated by biochemical studies with isolated and recombinant enzymes, which phosphorylate substrates only in the presence of calcium (Harmon et al., 2000; Harper et al., 2004; Liese and Romeis, 2013).
Although this overall biochemical mechanism of CDPK action is widely accepted, knowledge about biological processes that involve specific enzymes as well as in vivo substrates is rare. Associated with their role as calcium sensors, CDPKs have predominantly been investigated in signaling processes (Boudsocq and Sheen, 2013; Schulz et al., 2013; Simeunovic et al., 2016; Yip Delormel and Boudsocq, 2019). Plants respond to sudden changes in their environment and to abiotic or biotic stress factors with a rapid increase in the cytoplasmic calcium concentration (Kudla et al., 2010). CDPKs (known as CPKs in Arabidopsis [Arabidopsis thaliana]) perceive and translate these stimulus-induced calcium changes into the phosphorylation of specific target proteins, resulting in the activation of further downstream processes, including the onset of transcriptional reprogramming. According to this scheme, CPKs have been identified as crucial early signaling components that guarantee plant survival under drought conditions as well as upon pathogen attack. Arabidopsis guard-cell-expressed CPKs have been shown both to phosphorylate and to activate guard-cell S-type anion channels SLAH3 and SLAC1 (Geiger et al., 2010, 2011; Brandt et al., 2012), controlling stomatal ion conductance and closure. In innate immune signaling, Arabidopsis CPKs were identified in a functional genomics screen, and transiently expressed constitutive active variants of CPK family members mediate pathogen-related flg22 peptide-induced transcriptional reprogramming (Boudsocq et al., 2010; Gao et al., 2013). Targeted in vivo phosphoproteomics and biochemical studies with recombinant proteins identified plasma-membrane-localized NADPH-oxidase RESPIRATORY BURST OXIDASE HOMOLOGUE D as a direct in vivo substrate for CPKs (Dubiella et al., 2013; Kadota et al., 2014). In addition to membrane-localized target proteins, CPKs were shown to phosphorylate ABA-RESPONSIVE BINDING FACTOR (ABF) and WRKY transcription factors to mediate ABA and defense signaling, respectively (Choi et al., 2005; Zhu et al., 2007; Zhao et al., 2011; Gao et al., 2013).
Arabidopsis CPK1 represents the prototype of a canonical CDPK with four EF-hand motifs. CPK1 was among the first CDPKs to be biochemically characterized in detail, and the conceptual models for calcium binding and regulation derived from this characterization are still valid today (Harper et al., 1994, 2004). By contrast, the biological function of this isoform remains much less clear. Phylogenetically, CPK1 classifies into subgroup 1 of the CDPK gene family (Cheng et al., 2002), which contains several members for which functions in plant stress signaling have been shown. In Arabidopsis, CPK1-overexpressing plants were reported to exhibit enhanced resistance to the fungal pathogens Fusarium oxysporum and Botrytis cinerea and to bacterial infection by Pseudomonas syringae pv tomato DC 3000, whereas cpk1 mutant plants appeared to be more susceptible (Coca and San Segundo, 2010). Also, CPK1 and its closest homolog CPK2 contribute to Arabidopsis effector-triggered immune responses, and both enzymes were found to phosphorylate NADPH-oxidase homologues in vitro (Gao et al., 2013).
When analyzing the regulation of senescence, the identification of a CDPK phosphorylation-substrate relationship is more challenging because neither the nature of an inducing stimulus triggering changes in the intracellular calcium concentration nor the concrete biological function of a CDPK possibly involved in this developmental process is known. The plant’s competence to senesce is regulated by transcriptional networks. The transcription factor ORE1 (ORESARA1/ANAC092) is a master regulator of developmental leaf senescence, and it controls a senescence-associated gene regulatory network required for subsequent nutrient remobilization and chlorophyll degradation (Kim et al., 2009; Balazadeh et al., 2010; Qiu et al., 2015). ORE1 expression itself is strictly controlled by leaf age and abiotic stresses that are known to promote senescence, including N limitation, darkness, and salinity. Whereas in young leaves, high levels of microRNA miR164 posttranscriptionally suppress ORE1 expression, miR164 levels decline in older leaves, thereby relieving ORE1 suppression. In addition, ORE1 expression is directly activated in older leaves not only by upstream transcription factor EIN3 (Kim et al., 2014), a key regulator of ethylene signaling, and by ARABIDOPSIS THALIANA ACTIVATING FACTOR 1 (ATAF1), an ABA-induced transcription factor (Garapati et al., 2015) but also by circadian clock component PSEUDO-RESPONSE REGULATOR 9 (PRR9; Kim et al., 2018). Furthermore, regulation of ORE1 protein stability by ubiquitination and deubiquitination is known to play an important role in senescence induced by nitrogen deficiency. In this biological process, ORE1 levels are fine tuned by its polyubiquitination through NITROGEN LIMITATION ADAPTATION (NLA) and PHOSPHATE 2 (PHO2), leading to subsequent ORE1 degradation, as well as the counteracting deubiquitination by UBIQUITIN-SPECIFIC PROTEASE12 (UBP12) and UBP13, which stabilizes ORE1 and promotes senescence (Park et al., 2018, 2019). In addition to acting in leaf senescence, a partially redundant function of ORE1 has recently been documented in the stigma longevity by controlling the expression of programmed cell-death-associated genes (Gao et al., 2018).
To identify in vivo phosphorylation substrates of CPK1 and to address CPK1 function in plant development beyond immune signaling, we report here a novel approach in which the conditional expression of constitutively active CPK1-VK (consisting of the variable N-terminal domain and the adjacent protein kinase domain) is combined with subsequent in vivo phosphoproteomics analysis. The constitutively active CPK1-VK variant lacks its CAD domain and shows calcium-independent kinase activity. In planta, this enzyme variant is expected to phosphorylate its substrate proteins (in the absence of a yet-unknown external/endogenous biological stimulus), thereby allowing researchers to address developmental processes experimentally. To escape the potential lethality of transgenic plants harboring constitutively active enzyme variants, CPK1-VK was expressed from an ethanol-inducible promoter (Caddick et al., 1998). In a modification to the original protocol, the native promoter of CPK1 (CPK1pro) was used to drive the expression of the conditional transcriptional regulator alcR to mimic temporal and spatial activity of the native CPK1 gene in planta.
Our screen identified an ORE1 peptide that became transiently phosphorylated in vivo by CPK1. Phosphorylation occurred within an intrinsically disordered region of the ORE1 protein that is functionally required for ORE1-dependent target gene activation, but not DNA binding, and plant senescence promotion. ORE1 variants that carry mutations or lack this phosphorylation hotspot display low transactivation ability. Our data not only link CPK1 to the induction of senescence-related cell death but also show that senescence master regulator ORE1, known to be strictly controlled at the transcript level by gene regulatory networks, is subject to an additional layer of control, namely the posttranslational modification of ORE1 catalyzed by CPK1, a member of the calcium-regulatory kinase network.
RESULTS
Induced Expression of CPK1-VK in Its Native Plant Tissue Yields an Active, Calcium-Independent Enzyme That Triggers Cell Death
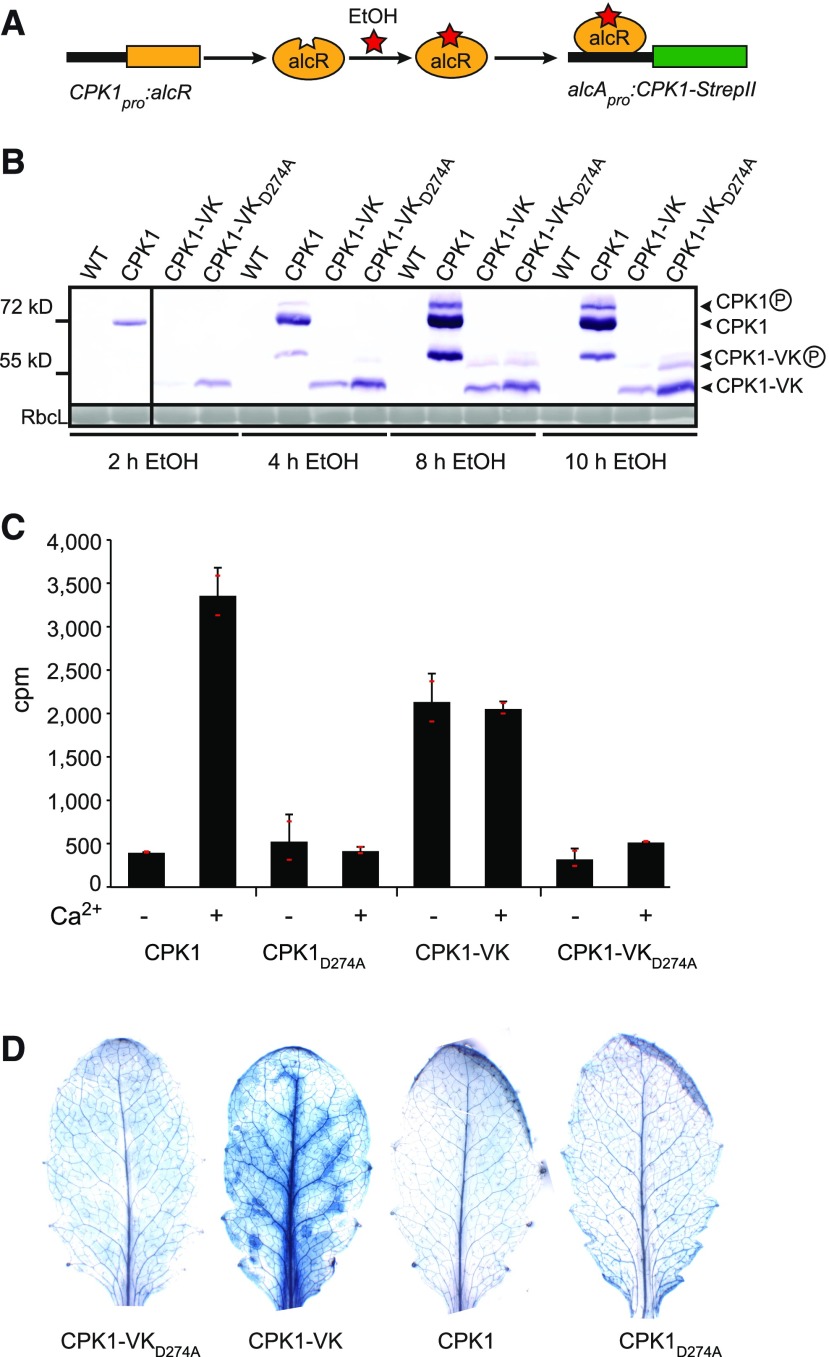
Ethanol-inducible CPK1-expressing lines encoding StrepII-tagged full-length CPK1, truncated CPK1-VK (amino acids 1 to 413), or kinase-deficient variants CPK1D274A and CPK1-VKD274A, were generated in cpk1-1 (SALK_096452) knockout plants (Figures 1A and 1B; Supplemental Figure 1). Gene expression is controlled by alcApro, the fungal alcA promoter (Caddick et al., 1998). In modification of the original protocol, expression of the corresponding ethanol binding transcriptional regulator alcR is driven by CPK1pro. Here, in contrast to previous studies that used the constitutive cauliflower mosaic virus (CaMV) 35S promoter (35Spro), upon exposure of plants to ethanol vapor, CPK1 variants are synthesized in cells, tissues, and organs where native CPK1 protein is typically produced. Expression of constructs, as analyzed by RT-PCR, was observed as early as 30 min after ethanol induction (Supplemental Figure 2). CPK1 proteins were detected at ∼72 kD (full length) and ∼50 kD (CPK1-VK) after 2 h by immunoblotting with standard alkaline phosphatase detection (Figure 1B) and after 1 h using sensitive horseradish peroxidase-derived luminescence detection (Supplemental Figure 3). Maximum protein levels were reached at 8 h (Figure 1B).
Figure 1.
Induced Protein Synthesis of Constitutively Active CPK1 in Its Native Plant Tissue Triggers Cell Death.
(A) Experimental design for ethanol-inducible synthesis of CPK1. In the presence of ethanol, the ethanol binding fungal transcriptional regulator alcR, which is expressed from the native CPK1 promoter, binds to the fungal alcA promoter, allowing ethanol inducible expression of CPK1-StrepII.
(B) Ethanol-induced protein accumulation. At the indicated times after exposure of transgenic and wild-type plants to ethanol vapor, CPK1, the truncated variant lacking the C-terminal calcium-activation domain CPK1-VK, and respective kinase-deficient variants carrying the D274A amino acid substitution in the kinase-active center were StrepII-affinity purified from 250 mg ground material of a pool of 4 to 5 Arabidopsis rosettes. Proteins were analyzed by immunoblot with Strep-Tactin alkaline phosphatase (top). An equal amount of initial protein per sample prior to purification is demonstrated by Ponceau staining of the large subunit of Rubisco (RbcL; bottom). The vertical line separates immunoblots from different gels.
(C) Kinase activity of affinity-purified CPK1 proteins as in (B), purified from a pool of three rosettes per line after 12 h exposure to ethanol vapor, on peptide substrate Syntide-2 in the presence of 50 μM CaCl2 (+Ca2+) or 2 mM EGTA (−Ca2+). The values and error bars represent the means and SDs of two independent measurements; Cpm, counts per minute. Individual values are depicted as small red horizontal lines. This is a qualitative comparison, showing which CPK1 variant is calcium dependent in its activity and which is catalytically inactive.
(D) Cell death symptoms in plants, in which the expression of CPK1, CPK1-VK, and of the kinase-deficient variants CPK1D274A and CPK1-VKD274A was induced by 12 h exposure to ethanol vapor, analyzed by trypan blue staining after further 27 h.
With increasing time, additional slower migrating bands for CPK1 and CPK1-VK appeared. These bands most likely represent differentially phosphorylated forms of CPK1, consistent with the literature, where 10 in vivo phosphosites of CPK1 have been listed (PhosPhAt 4.0 database; Durek et al., 2010) or where multiple differentially phosphorylated bands became evident in SDS gels for the CPK1 ortholog NtCDPK2 from Nicotiana tabacum (Witte et al., 2010). For the native CPK1 protein in wild-type Columbia (Col-0) plants, which also appears as multiple bands in SDS gels, it has been shown that slower migrating bands in the PhosTag gel system are indeed phosphorylated (Durian et al., 2020). In vitro protein kinase assays with purified, immobilized proteins, originating from leaf material after ethanol incubation, displayed a calcium-dependent increase in phosphorylating activity toward peptide substrate Syntide-2. This is not observed with kinase-deficient variants carrying the D274A amino acid substitution in the active center of the kinase domain (Figure 1C) In contrast, CPK1-VK lacking the CAD shows constitutive phosphorylating activity irrespective of the absence or presence of calcium in the assay as previously shown by Harper and colleagues (Figure 1C; Harper et al., 1994). When plants were treated with ethanol vapor for a longer period (12 h), cell death symptoms developed during subsequent 27 h of growth in the absence of ethanol only in CPK1-VK-synthesizing plants (Figure 1D).
Differential In Vivo Phosphoproteomics Identified Transient Phosphorylation of Transcription Factor ORE1
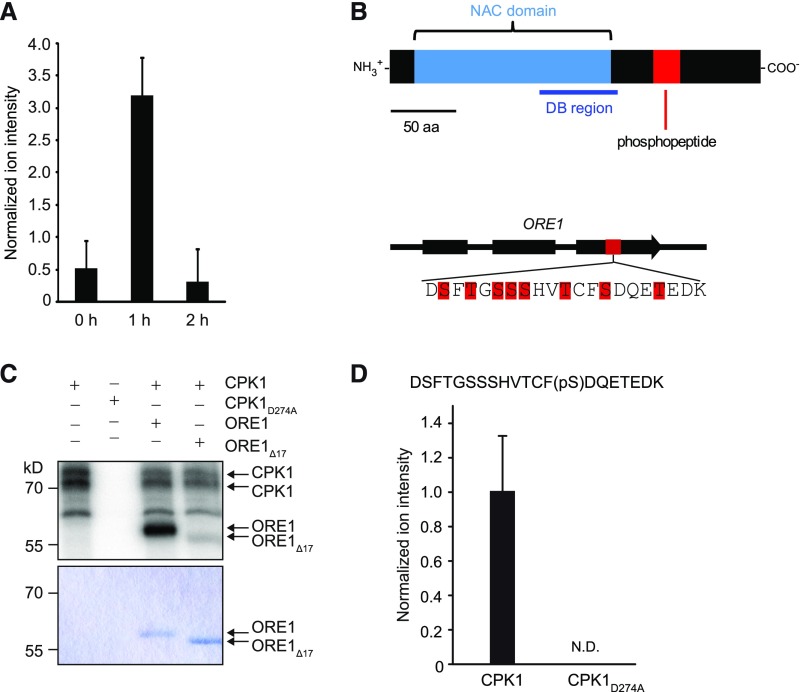
To identify in vivo substrates of CPK1, plants expressing CPK1-VK and CPK1-VKD274A were exposed to ethanol vapor, and leaf material was harvested before (0 h) and 1 h and 2 h after treatment. Protein extracts were subjected to differential phosphopeptide analysis, and several differentially phosphorylated peptides were detected by mass spectrometry. One of these peptides showed a transient 7-fold accumulation at the 1-h time point in the CPK1-VK-derived samples (Figure 2). This peptide (Figure 2A) represented a multiply phosphorylated 21-amino-acid peptide derived from the NAC transcription factor ORE1 (At5g39610; ORESARA1/ ANAC092 (Figure 2B; Supplemental Figures 4A to 4C; Kim et al., 2009; Balazadeh et al., 2010). The peptide encompasses amino acids 204 to 224 of the 285-amino-acid ORE1 protein and is located C-terminal to the NAC DNA binding region (Figure 2B).
Figure 2.
Differential In Vivo Phosphoproteomics Identifies CPK1-Mediated Phosphorylation of the Transcriptional Regulator ORE1.
(A) Transient in vivo accumulation of phosphorylated ORE1 peptide in plants expressing CPK1-VK. After exposure of alcApro:CPK1-VK transgenic plants to ethanol vapor for the indicated times, leaf material was harvested, and protein extracts were analyzed by mass spectrometry. Identification of peptides was performed by LC-MS/MS (LTQ Orbitrap), and data show normalized intensities, with bars representing the means and error bars representing the SEs of three biological replicates. For each biological replicate, proteins from five pooled Arabidopsis rosettes were extracted and equal amounts of precipitated total proteins per sample were used for tryptic digestion and subsequent phosphopeptide enrichment.
(B) Structures of ORE1 and ORE1. Top, Diagram of the protein structure of ORE1 showing the NAC domain, the DNA binding (DB) region and, in red, the phosphopeptide quantified in (A). Scale bar = 50 amino acid residues. Bottom, Diagram showing the exon (thick box) and intron (thin line) structure of ORE1. The region of the DNA sequence encoding the phosphorylated peptide that was identified is indicated in red. The amino acid sequence of the peptide is shown below and the Ser and Thr residues within it are marked in red.
(C) In vitro phosphorylation of ORE1 by CPK1. Recombinant ORE1-GST and ORE1Δ17-GST, lacking 17 amino acid residues comprising all phosphorylation sites shown in red in (B), were incubated with CPK1 or CPK1D274A. Both were StrepII-affinity purified from leaf material after transient expression in Nicotiana benthamiana leaves. Proteins were separated by SDS-PAGE, and phosphorylation in the presence of calcium and [γ-32P]-ATP was determined by autoradiography and phosphoimaging (top). Protein amount was confirmed by Coomassie Brilliant Blue staining (bottom).
(D) In vivo phosphorylation of ORE1. ORE1 was transiently coexpressed with either CPK1 or CPK1D274A in Col-0 Arabidopsis mesophyll protoplasts. Protein extracts were analyzed by LC-MS/MS, and normalized ion intensities of the phosphorylated ORE1-peptide DSFTGSSSHVTCF(pS)DQETEDK were determined. The mean of three independent transfection assays (n = 3) and SD are shown. N.D., not detectable.
To test whether CPK1 directly phosphorylates ORE1 protein, we conducted in vitro protein kinase assays with immobilized CPK1 and CPK1D274A, purified from leaf material. Recombinant ORE1-GST (glutathione S-transferase) or ORE1Δ17-GST, a variant lacking a stretch of 17 amino acids encompassing eight Ser and Thr residues was used as substrate. The deletion of this particular 17-amino-acid stretch (amino acids 205 to 221) was chosen because it has not been possible to resolve by mass spectrometry which amino acid residues of the eight potential Ser and Thr phosphorylation sites present within the identified 21-amino-acid peptide were phosphorylated in vivo. CPK1 showed efficient catalytic activity toward itself and to ORE1 (substrate phosphorylation band at ∼58 kD; Figure 2C, top). ORE1Δ17 became much less phosphorylated by CPK1, although comparable protein amounts were used (Figure 2C, bottom). In the absence of calcium, only residual weak kinase activity was observed (Supplemental Figures 5C and 5D), and no phosphorylation activity occurred with CPK1D274A.
To independently validate in vivo phosphorylation of ORE1 by CPK1, either full-length CPK1 or CPK1D274A was transiently coexpressed with ORE1 in Arabidopsis mesophyll cell protoplasts. Full-length CPK1 allows efficient protein kinase and ORE1 substrate protein expression sufficient for mass spectrometry detection, whereas expression of CPK1-VK induced early cell death (see below; Figure 3C). Targeted phosphopeptide analysis by mass spectrometry of protein extracts detected a single phosphorylated form of the identified 21-amino-acid ORE1-peptide (DSFTGSSSHVTCF(pS)DQETEDK) in the presence of active CPK1, but not of CPK1D274A (Figure 2D; Supplemental Figures 4D and 4E). Whereas this single phosphorylation verifies ORE1 as an in vivo substrate of CPK1, we cannot conclude from these data whether this single phosphosite is more relevant because it becomes phosphorylated first by CPK1-FL. In addition, by targeted analysis including known CPK1-phosphopeptides (PhosPhAt 4.0 database; Durek et al., 2010), we detected two phosphopeptides originating from CPK1, likely due to CPK1 autophosphorylation—a 7-amino-acid peptide VS(pS)AGLR (amino acids 128 to 134; Supplemental Figures 4D and 6) and a C-terminal 7-amino-acid peptide (SF(pS)IALK; amino acids 603 to 609). Phosphorylation at Ser 130 was shown to be an in vitro autophosphorylation site by Swatek et al. (2014).
Figure 3.
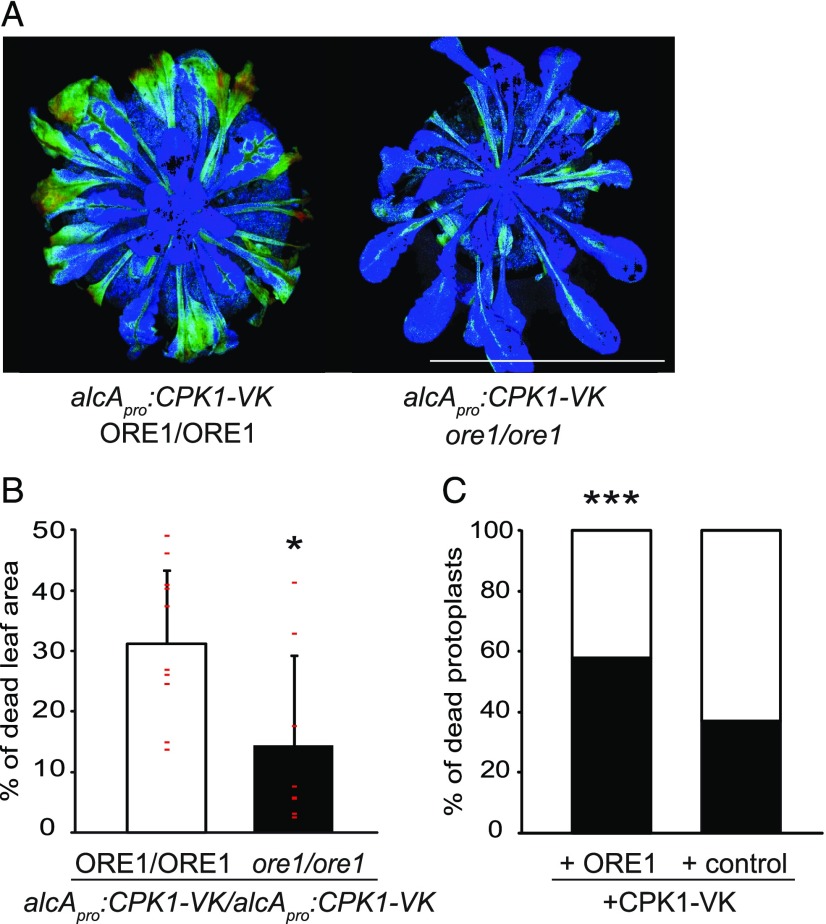
Induction of Cell Death by Constitutively Active CPK1-VK Requires ORE1.
(A) Five-week-old homozygous alcApro:CPK1-VK plants in either the Col-0 wild type (ORE1/ORE1) or the homozygous ore1-1/ore1-1 mutant background were exposed to ethanol vapor for 12 h. Cell death development was assessed after a further 22 h by chlorophyll fluorescence (PAM) measurements. Leaf areas in light blue, green, or red color (order indicates increasing severity, with red being the most severe) are of reduced photosynthetic activity indicative of cell death, whereas dark blue areas document normal activity. Bar = 3.4 cm.
(B) Quantification of leaf areas with cell death symptoms in (A) was calculated as the percentage of areas of reduced photosynthetic activity (light blue, green, and red) to the total photosynthetically active area. Data show means and SDs. At least eight independent plants were assessed per line. Individual values are depicted as small red horizontal lines. The asterisk indicates a significant difference from the other plant line using the Mann-Whitney U test; *P < 0.05 (Supplemental Data Set).
(C) CPK1-VK-mediated cell death in Arabidopsis leaf mesophyll protoplasts. CPK1-VK was transiently coexpressed with either ORE1 or an empty vector control in protoplasts derived from ore1-1 plants. Both coexpression assays were performed in parallel. The percentage of dying protoplasts (black part of the bars) between the interval of 3 h and 26 h after transfection and the corresponding percentage of surviving cells during the same interval (white part of the bars) were determined by staining with either propidium iodide or fluorescein diacetate for dead and living cells, respectively. Analysis was conducted with a fluorescence microscope. n (total number of cells counted per combination and time point, i.e., 3 h, 26 h) ≥ 165. Asterisks indicate a significant difference from the other coexpression assay. Statistical analysis was done by logistic regression; ***P < 0.001 (Supplemental Data Set).
To independently investigate specificity of CPK1 phosphorylation of ORE1, we conducted in vitro protein kinase assays with immobilized CPK1-VK and CPK6-VK proteins that were purified from plant extracts, and CPK autophosphorylation and substrate phosphorylation of ORE1 were compared (Supplemental Figures 5A and 5B). Strong ORE1 phosphorylation was evident by CPK1-VK but not by CPK6-VK, whereas both kinases were capable of autophosphorylation in vitro (Supplemental Figure 5B).
CPK1 Controls the Expression of ORE1 and of ORE1 Target Genes
ORE1 (ANAC092) is a NAC transcription factor consisting of an N-terminal NAC DNA binding domain and a C-terminal transactivating regulatory domain, and ORE1 plays a crucial role for positively regulating programmed cell death during senescence in Arabidopsis. ORE1 itself is transcriptionally activated during the onset of senescence (Kim et al., 2009). ORE1 activates the expression of several senescence-related target genes, and direct binding of ORE1 to the promoters of senescence-enhanced BFN1 (BIFUNCTIONAL NUCLEASE1) and NAC transcriptional regulator VNI2 (VND-INTERACTING2) has been demonstrated (Balazadeh et al., 2008, 2010; Matallana-Ramirez et al., 2013).
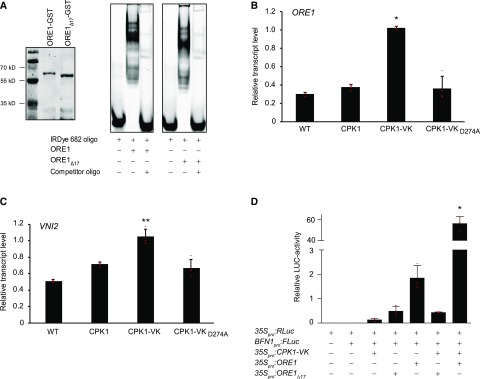
Because the identified peptide is located C-terminal to the DNA binding region of ORE1, we assessed the DNA binding ability to a 40-bp promoter region from BFN1, a target of ORE1 (Figure 4; Matallana-Ramirez et al., 2013). Recombinant purified ORE1-GST and ORE1Δ17-GST (Figure 4A, left) were used for in vitro DNA binding reactions and were analyzed by an electrophoretic mobility shift assay (Figure 4A, right). Both ORE1 and ORE1Δ17 showed a positive electrophoretic mobility shift assay (EMSA) shift, indicating that the deletion of the 17-amino-acid peptide did not compromise DNA binding.
Figure 4.
CPK1 Controls the Expression of ORE1 and of ORE1 Target Genes.
(A) Electrophoretic mobility shift assay. Recombinant ORE1-GST and ORE1Δ17-GST as in Figure 2C were affinity purified, expression was confirmed by immunoblot detection of ORE1-GST and ORE1Δ17-GST using the anti-GST-antibody (left) as described in “Methods.” Proteins were subjected to in vitro DNA binding assays using a labeled (5′-DY682) 40-bp region of the BFN1 promoter (right) in the absence or presence of a 200-fold excess of unlabeled oligomeric competitor fragment as indicated.
(B) and (C) RT-qPCR expression analysis, using the reference gene ACTIN2, of ORE1 (B) and ORE1-regulated target gene VNI2 (C) in wild-type and CPK1-expressing plants after a 10-h exposure to ethanol vapor. Relative transcript levels (= 2−ΔΔCt) are shown. Data show means and SDs of four biological replicates, each replicate consisting of cDNA from one individual Arabidopsis rosette. Individual 2−ΔΔCt values are depicted as small red horizontal lines. Asterisks denote statistically significant differences from the Col-0 wild type (WT; Kruskal-Wallis test and Dunn-Bonferoni post hoc test; *P < 0.05; **P < 0.005; Supplemental Data Set).
(D) ORE1-dependent BFN1-promoter activation is mediated by CPK1. Arabidopsis ore1-1 protoplasts were transfected with either 35Spro:RLuc (transfection control) or BFN1pro:FLuc (reporter) constructs in combination with the constitutively active kinase (35Spro:CPK1-VK) and either full-length (35Spro:ORE1) or mutated ORE1 transcription factor (35Spro:ORE1Δ17), which lacks 17 amino acids encompassing the CPK1-dependent phosphorylation sites. The relative luciferase (LUC) activity was determined as the ratio of the signals of target luciferase (FLUC) and control (RLUC), normalizing for transfection efficiency in different samples. Data show means of relative LUC activities of three biological replicates, with each replicate consisting of an independent transfection assay. Error bars represent the SDs. Individual values are depicted as small red horizontal lines. The asterisk denotes a statistically significant difference from all other coexpression combinations; one-way analysis of variance, Tukey post hoc test; *P < 0.05 (Supplemental Data Set).
We next investigated whether CPK1 activity not only results in ORE1 phosphorylation but also leads to further ORE1 gene expression and function. In leaves harvested 10 h after exposure to ethanol vapor, ORE1 transcripts accumulated to ∼3-fold higher levels in CPK1-VK-expressing plants than in wild-type plants (Figure 4B) and, in accordance with this, transcript abundance of ORE1 target gene VNI2 increased as well (Figure 4C).
To address whether ORE1-mediated transactivation is controlled by CPK1 and whether the identified CPK1-phosphorylated amino acids within ORE1 are required for this, we conducted a transient BFN1 promoter-firefly luciferase (Fluc) reporter assay using Arabidopsis protoplasts derived from the ore1-1 (anac092-1) mutant (Balazadeh et al., 2010). The ore1-1 mutant allele has been characterized as a true null mutant (Balazadeh et al., 2010; Trivellini et al., 2012) despite showing residual ORE1 transcript when primers upstream of the T-DNA insertion between coding sequence positions 743 and 744 (He et al., 2005) are used. Based on using 35Spro:RLuc as a control, BFN1pro:FLuc promoter activity strongly increased when ORE1, but not ORE1Δ17, was coexpressed (Figure 4D). In the presence of CPK1-VK, in addition to ORE1, a significant further increase in BFN1 promoter activation (∼30-fold) occurred. This strong CPK1-VK-dependent increase was not evident with ORE1Δ17 or when CPK1-VK was expressed without the ORE1 transcription factor.
To exclude the possibility that this diminished transactivation of BFN1pro:FLuc by ORE1Δ17 was solely due to potential structural changes caused by the deletion of 17 amino acids, we generated two ORE1-variants, ORE1A and ORE1D, that carry amino acid substitutions in six out of eight possible Ser/Thr phosphorylation sites within the 17-amino-acid stretch (amino acids 205 to 221). These two variants mimic a sixfold nonphosphorylated form (ORE1A) and a sixfold phosphorylated form (ORE1D) of ORE1, respectively. Both phosphosite variants showed diminished transactivation of BFN1pro:FLuc compared to wild-type ORE1 (Supplemental Figure 7A). The transactivation activity of the two phosphosite variants did not change much when CPK1-VK was coexpressed (Supplemental Figure 7B). Note that the 17-amino-acid stretch has in vivo been identified as a sixfold phosphorylated peptide, but the exact location of the six phosphorylation sites distributed on eight potential Ser and Thr residues in this stretch (Figures 2A and 2B) has not been resolved for technical reasons. Hence, the sixfold phosphomimic ORE1D can only serve as a proxy for the native multiple phosphorylation within the region of amino acids 205 to 221 of ORE1. Thus, CPK1 phosphorylates ORE1 within an intrinsically disordered region located at the C-terminal transcription regulatory domain of the ORE1 protein and controls transactivation.
ORE1 Is Required for Cell Death Induced by CPK1-VK
Because ORE1 mediates senescence-related programmed cell death, we next investigated whether ORE1 is required for the cell death induced by CPK1-VK (Figure 1D). To this end, we generated crosses between the well-described ore1-1 mutant allele and the alcApro:CPK1-VK expression line, and selected double homozygous lines (Figure 3). The onset of cell death symptoms induced by a 12-h exposure to ethanol vapor was assessed by chlorophyll fluorescence measurements after 22 h (Figure 3A). Interestingly, the leaf areas that had decreased photosynthetic activity, indicative of cell death, were significantly reduced in size when CPK1-VK was expressed in the ore1-1/ore1-1 mutant compared to the expression in the wild-type (ORE1/ORE1) background (Figure 3B). In addition, transfection of CPK1-VK into ore1-1/ore1-1 protoplasts resulted in significantly more dying cells when ORE1 was coexpressed, than in the presence of CPK1-VK alone (Figure 3C).
ORE1, but Not ORE1Δ17, Promotes Senescence
Our findings presented above indicate a functional link between CPK1 and ORE1 in the process of cell death induction, although the question remained in which biological process this interaction would play a role. The transcription factor ORE1 is a central positive regulator of plant senescence (Kim et al., 2009). Because CPK1 has not yet been characterized in senescence, we generated crosses between ore1 and cpk1 and compared phenotype and physiological and molecular markers of the resulting homozygous double-mutant lines ore1 cpk1 in senescence.
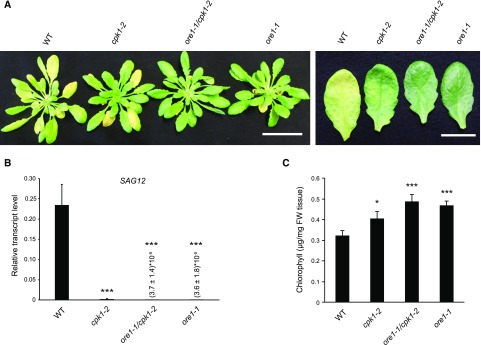
In dark-induced senescence assays with detached rosettes, both ore1-1 and cpk1-2 single mutants and the ore1-1 cpk1-2 double mutant showed less senescence-dependent chlorosis and SAG12 expression compared to the Col-0 wild type (Figure 5). Also, during natural developmental senescence induced by prolonged growth under long-day conditions, leaf yellowing and SAG12 transcript abundance were reduced in ore1 and cpk1 single mutants and even more prominently in ore1 cpk1 double mutant plants (Supplemental Figure 8).
Figure 5.
CPK1-Dependent ORE1 Function in Dark-Induced Senescence.
(A) Rosettes of 5-week-old plants of the Col-0 wild type (WT), cpk1-2, ore1-1, and the double mutant ore1-1/cpk1-2 were grown under short-day conditions and were then cut and stored in the dark for 6 d (left). Dark-induced senescence phenotypes of detached mature leaves (right). Bars = 4.5 cm (left), 1.5 cm (right).
(B) RT-qPCR expression analysis of SAG12, using the reference gene ACTIN2, in plants treated as in (A, left). Relative transcript levels are shown as 2−ΔCt values. Data show means and SDs of three biological replicates, each replicate consisting of cDNA from a pool of three rosettes. Asterisks indicate a significant difference from Col-0 (wild-type, WT) plants using one-way analysis of variance, Tukey post hoc test ***P < 0.0001 (Supplemental Data Set).
(C) Chlorophyll content of 5-week-old plants treated as in (A, left). Data show the mean values and SDs of three biological replicates, each replicate consisting of a pool of three rosettes. Asterisks indicate a significant difference from Col-0 (wild-type, WT) plants; one-way analysis of variance, Tukey post hoc test; *P < 0.05; ***P < 0.001 (Supplemental Data Set). FW, Fresh weight.
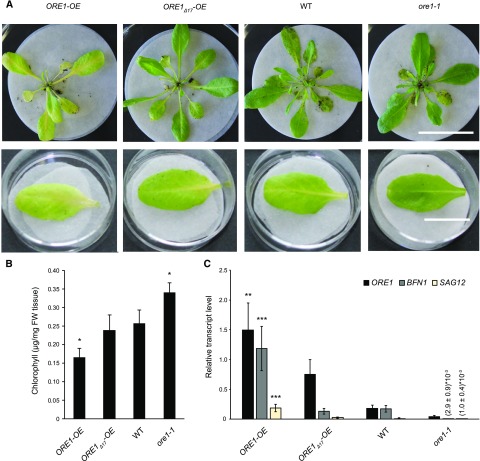
To investigate whether phosphorylation through CPK1 is required for ORE1 function in regulation of senescence, 4-week-old plants overexpressing either ORE1 or ORE1Δ17 were assessed for dark-induced senescence (Figure 6). Compared to the wild type, overexpression of ORE1 resulted in premature leaf senescence (Figure 6A) accompanied by a reduced chlorophyll content (Figure 6B) and increased expression of the senescence marker genes SAG12 and BFN1 (Figure 6C), consistent with previous observation (Balazadeh et al., 2010). ORE1Δ17-OE, which lacks the 17-amino-acid region encompassing the identified CPK1 phosphorylation sites, showed no early senescence, accumulation of senescence marker genes, or alteration in chlorophyll content. Similar results were observed when evaluating these lines in natural, developmental senescence (Supplemental Figure 9). Importantly, ORE1Δ17-OE mirrors Col-0 phenotypes rather than those of ore1-1 in both developmental and dark-induced senescence, excluding a dominant-negative effect of the ORE1Δ17 protein variant. Residual ORE1 transcripts in the ore1-1 mutant amplified in region 459 to 576 of the coding sequence in our RT-qPCR experiments is consistent with the literature and the T-DNA insertion between coding-sequence positions 743 and 744 (He et al., 2005). As noted earlier, ore1-1 has been characterized as a true null mutant (Balazadeh et al., 2010; Trivellini et al., 2012).
Figure 6.
ORE1 but Not ORE1Δ17 Promotes Dark-Induced Senescence.
(A) Senescence phenotypes of plants of the Col-0 wild-type (WT), ore1-1, and the overexpressing lines ORE1-OE and ORE1Δ17-OE. The ORE1Δ17-OE line expresses a mutated ORE1 transcription factor that lacks 17 amino acids encompassing the CPK1-dependent phosphorylation sites. Plants were grown under short-day conditions for 4 weeks. Rosettes were cut and stored in the dark for 4 d on filter papers wetted with distilled water (top). Dark-induced senescence phenotypes of detached mature leaves (bottom). Bars = 4.2 cm (top), 1.7 cm (bottom).
(B) Chlorophyll content of 4-week-old plants treated as in (A, top). Data show means and SDs of three biological replicates, each replicate consisting of a pool of three rosettes. Asterisks indicate a significant difference from Col-0 (wild-type, WT) plants; one-way analysis of variance, Tukey post hoc test; *P < 0.05 (Supplemental Data Set). FW, Fresh weight.
(C) RT-qPCR expression analysis of ORE1, BFN1, and senescence marker gene SAG12, using the reference gene ACTIN2, in plants treated as in (A, top). Relative transcript levels are shown as 2−ΔCt values. Data show means and SDs of three biological replicates, each replicate consisting of cDNA from a pool of three rosettes. Asterisks indicate a significant difference from Col-0 (wild-type, WT) plants; one-way analysis of variance, Tukey post hoc test **P < 0.01; ***P < 0.001 (Supplemental Data Set).
DISCUSSION
Senescence in plants is a highly coordinated process that evolved to ensure maximal recovery of nutrients from dying organs (aged leaves) in order to benefit newly forming organs (young leaves, seeds, and fruits). The competence of leaves to senesce is governed by interconnected transcriptional networks, in which NAC transcription factor ORE1 functions as a master regulator by activating the expression of various genes known to play critical roles in senescence (Kim et al., 2009; Balazadeh et al., 2010). Expression of ORE1 itself is subject to a sophisticated regulatory network that robustly prevents its precocious activation during leaf development or in the absence of abiotic stress. This regulation involves various upstream transcription factors including, e.g., EIN3 (ethylene signaling), ATAF1 (response to carbon starvation and elevated ABA), and PHYTOCHROME INTERACTING FACTOR (PIF) 4 and PIF5, which integrate plant development with light conditions (Kim et al., 2009, 2014; Garapati et al., 2015; Zhang et al., 2015). In addition, ORE1 transcript abundance is negatively controlled by miR164 during early leaf growth (Kim et al., 2009). Both EIN3 and the circadian clock transcription factor PRR9 directly activate ORE1 transcription and additionally directly repress the transcription of miR164, the posttranscriptional repressor of ORE1, hence creating two feed-forward pathways for ORE1 expression (Li et al., 2013; Kim et al., 2014, 2018). Our data presented here identify a key layer of regulation in developmental leaf senescence by which the function of ORE1 is regulated at the posttranslational level by calcium-regulated phosphorylation by CPK1.
In our in vivo phosphoproteomics screen with inducible CPK1-VK, we identified an ORE1 peptide as being a multiply phosphorylated peptide. The respective ORE1 peptide is located C-terminal to the DNA binding region and encompasses amino acids 204 to 224, which contains eight Ser and Thr residues as potential phosphorylation sites compatible with the interpretation as a phosphorylation hotspot (Christian et al., 2012). Interestingly, this stretch is located within an unstructured region of the ORE1 protein spanning amino acids 170 to 230, for which a high disorder probability is predicted by Protein DisOrder prediction System (PrDOS; Ishida and Kinoshita, 2007). Intrinsically disordered regions are known to be modified by (clustered) phosphorylation. They can be found in proteins that exert key regulatory functions (Iakoucheva et al., 2004) in which (multiple) phosphorylation, and thus the introduction of (multiple) negative charges causes changes in the protein structure that mediate the control of biological processes such as molecular recognition or transcription. NAC transcription factors are generally known to comprise intrinsically disordered regions (Jensen et al., 2010).
NAC transcription factors consist of an N-terminal DNA binding domain and a C-terminal transcription regulatory domain responsible for transactivation. The latter is characterized by group-specific sequence motifs with a high degree of intrinsic disorder (Jensen et al., 2010). Our EMSA analysis showed that ORE1 and ORE1Δ17 bound equally well to a 40-bp promoter fragment of the ORE1 target gene BFN1 (Figure 4A). In contrast, the transactivation assay reveals that the coexpression of CPK1-VK with ORE1, but neither with ORE1Δ17 nor with ORE1A or ORE1D carrying either multiple A or multiple D amino acid substitutions in that region, leads to a significant increase in the BFN1 promoter readout (Figure 4D; Supplemental Figure 7). These data are consistent with the concept of a phosphorylation hotspot targeted by CPK1 in the transcription regulatory domain, thereby directing protein folding of the disordered region and inducing transactivation activity. Furthermore, plants overexpressing ORE1, but not ORE1Δ17, displayed an early promotion of senescence accompanied by a reduced chlorophyll content and showed increased gene expression of ORE1 target gene BFN1 and senescence marker SAG12 in both dark-induced senescence and natural senescence (Figure 6; Supplemental Figure 9). Deduced from these data, in vivo activation of ORE1 by CPK1 may allow a rapid transient acceleration of transcriptional reprogramming and output.
Our gain-of-function results of CPK1-VK phosphorylating ORE1 are corroborated by reduced senescence-related cell death symptoms, when CPK1-VK is expressed in the ore1-1 background (Figure 3). Thus, the onset of senescence and its progression requires changes in the intracellular calcium concentration that are sufficient to activate calcium-dependent enzymes such as CPK1 in vivo. Currently, neither the source of the calcium, whether apoplastic or from internal stores, nor the molecular components or the nature of the stimulus triggering these calcium changes is known in the context of leaf senescence. Changes in the cytoplasmic calcium concentration undergo circadian oscillations, which are subject to modulation by red and blue light via respective photoreceptors, including phytochrome B. These oscillations may encode temporal information regulating the cellular physiology (Xu et al., 2007). Also, plant age-related changes in either the basal intracellular calcium concentration and/or changes in the circadian rhythm of calcium oscillations may exist (Li et al., 2016) that interconnect with the circadian transcriptional regulation of ORE1 and its posttranscriptional repressor miR164 at the onset of developmental leaf senescence (Kim et al., 2018). These changes in calcium levels and dynamics may become decoded by signaling mediator CPK1 and fine tune the integration of environmental and endogenous signals into senescence-executing processes.
Thus, ongoing transcriptional reprogramming results in ORE1 accumulation during the later stages of leaf development. CPK1-dependent phosphorylation of ORE1 combined with ORE1 transcription factor-mediated control of gene expression (including its own gene) subsequently constitutes a feed-forward loop that initiates senescence-related programmed cell death (Figure 7). This interpretation does not exclude additional functions of CPK1 to contribute in the early onset of immune signaling (Gao et al., 2013). Recruitment of CDPKs in various biological processes involving different protein targets has been reported for example for Arabidopsis CPK6 phosphorylating SLAC1 in abiotic, and RBOHD in biotic, stress signaling (Brandt et al., 2012; Kadota et al., 2014).
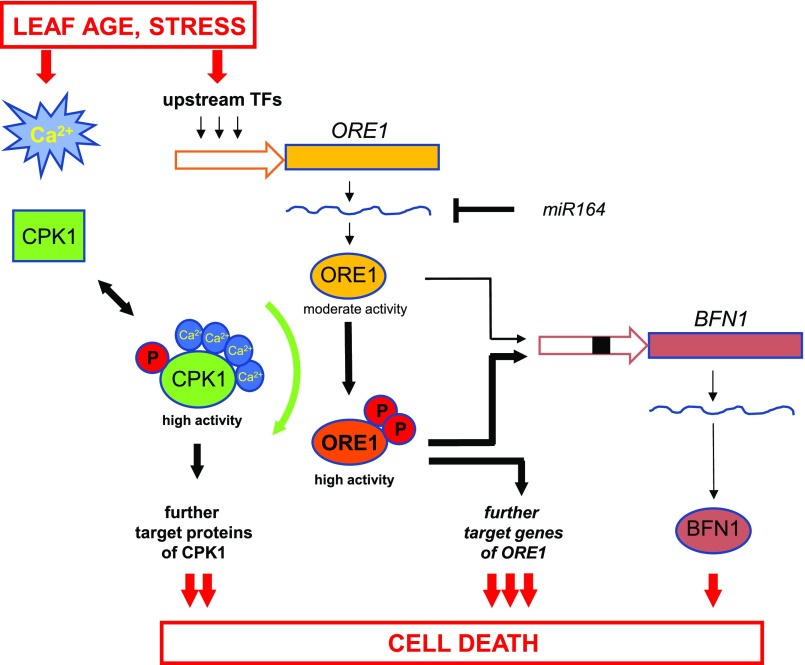
Figure 7.
Model for Cell Death Regulation Controlled by CPK1 and ORE1.
Senescence- and stress-related cell death in plants is governed by interconnected gene networks converging on ORE1, which acts as a master switch that is positively and negatively regulated by upstream transcription factors and miR164, respectively. The ORE1 protein undergoes an additional layer of control by posttranslational modification via phosphorylation by CPK1, a member of the calcium-regulatory network. This leads to high ORE1 activity and subsequent induction of expression of target gene(s) such as BFN1. The linkage between the reversible calcium-dependent signaling network via CPK1 and the irreversible gene-regulatory network via ORE1 guarantees a highly coordinated response, enabling, for example, a maximal retrieval of nutrients from the dying cells.
Although the mechanism of senescence-related calcium elevation is yet unknown, it is conceivable that members of the CYCLIC-NUCLEOTIDE-GATED CHANNEL (CNGC) gene family may play a role as Ca2+ channels responsible for either establishing or maintaining respective developmental cytoplasmic Ca2+ levels. In Arabidopsis, the expression of plasma-membrane-located CNGC2 increases during age-dependent leaf senescence, and the participation of CNGC2 in developmentally regulated cell death has been postulated (Köhler et al., 2001). Interestingly, calmodulin-gated CNGC2 and CNGC4 have recently also been characterized in pathogen-associated molecular pattern-triggered immunity, mediating calcium influx into the cytosol (Tian et al., 2019).
By taking advantage of recent progress in phosphoproteomic mass spectrometry in combination with the inducible expression of a constitutively active CPK1 in its native spatial and developmental environment, we identified an in vivo phosphorylation substrate for CPK1. Transcription factors are often of low abundance and are thus notoriously difficult to identify, even by targeted in vivo mass spectrometry. This is even more challenging when phosphorylation occurs transiently, as seen here for ORE1, despite expressing a constitutively active kinase. Alternative strategies, such as protein-protein interaction mapping, may also fail if distinct stable phosphorylation patterns are a prerequisite for the interaction. Furthermore, using the native CPK1 promoter to drive the expression of the ethanol binding transcriptional regulator alcR reduces off-target effects that may have been more common in case of the CaMV 35S promoter due to potential misexpression in time and space where and/or when native CPK1 would not be present. Thus, the investigation of a rapid response to a biochemically active enzyme allows a recording in time lapse of an otherwise long adaptive or developmental process, and in addition enables studies in the absence of (and the knowledge of) the primary endogenous or exogenous stimulus of a biological process.
As illustrated in our model in Figure 7, our discovery that CPK1 phosphorylates ORE1 uncovered a crucial direct link between two key regulatory concepts: the reversibility of a calcium-regulated protein kinase that controls via rapid transient phosphorylation and a transcriptional program that irreversibly leads to the ultimate possible response—death.
METHODS
Plant Material
Arabidopsis (Arabidopsis thaliana) ecotype Col-0 wild type and derived transgenic overexpressing and mutant plants were grown in a growth chamber at 20°C and 60% relative humidity with an 8-h photoperiod (light intensity 150 μmol/m2s) in compost soil (42.42% [w/w] Einheitserde P, 42.42% [w/w] Einheitserde T, and 15.15% [w/w] (Perligran [Kausek]). CPK1 full-length, truncated CPK1-VK, and kinase-deficient variants for ethanol-inducible overexpression were cloned and lines selected as described in the Supplemental Methods. Overexpression of CPK1 variants was induced by incubating 5-week-old plants in pots in a closed ethanol/water-vapor atmosphere (0.1% [v/v] ethanol and 99.9% [v/v] water in the liquid phase not in contact with the plants). Homozygous cpk1-1 (SALK_096452), cpk1-2 (SALK_080155c), and ore1-1 (SALK_090154) mutants were selected by PCR using primers designed according to the Salk Institute Genomic Analysis Laboratory website (http://signal.salk.edu/tdnaprimers.2.html), primers that were also used for the identification of double-homozygous mutants of these lines. For developmental senescence assays, a 35Spro:ORE1 line (Matallana-Ramirez et al., 2013) and a 35Spro:ORE1Δ17 line (Supplemental Methods) were grown together with ore1-1 and Col-0 in a growth chamber at 20°C and 60% relative humidity with a 16 h-photoperiod (light intensity, 240 μmol/m2s) for 6 weeks.
DNA Constructs and Transgenic Arabidopsis Overexpression Lines
The generation of DNA constructs and transgenic lines is described in detail in Supplemental Methods. In brief, the binary construct pI4-cpk1pro-alcR-alcApro-CPK1-VK-Strep and the respective CPK1-VKD274A, CPK1, and CPK1D274A variants, coding for different forms of StrepII-tagged (Witte et al., 2004) CPK1, were used to transform Arabidopsis cpk1-1 mutant plants for the ethanol-inducible expression system. The 35Spro:ORE1-construct using vector pGreen0229 is described in Balazadeh et al. (2010); the corresponding overexpression line has a genetic wild-type (Col-0) background. Transgenic Arabidopsis lines with the 35Spro:ORE1Δ17 in vector pGreen0229 were generated in the ore1-1 mutant background.
CPK1 Protein Extraction, Affinity Purification, and Detection
For protein purification, 0.5 g of leaf material was ground in liquid nitrogen and thawed in 1.5 mL extraction buffer (100 mM Tris, pH 8.0; 100 mM NaCl; 5 mM EDTA; 5 mM EGTA; 20 mM DTT; 0.5 mM 4-(2-aminoethyl)benzenesulfonylfluoride [AEBSF]; 10 mM NaF; 10 mM Na3VO4; 2 µL/mL plant protease inhibitor cocktail [Sigma P9599]; 2 µg/mL aprotinin; 2 µg/mL leupeptin; 10 mM β-glycerophosphate; 0.5% [v/v] Triton X-100; 100 µg/mL avidin). After centrifugation at 21,000g at 4°C for 20 min, 1 mL of the supernatant was incubated with 40 μL of Strep-Tactin MacroPrep (50% [v/v] slurry, IBA) and incubated in a rotation wheel for 20 min at 4°C. The Strep-Tactin-matrix was pelleted by centrifugation for 1 min at 700g, and the supernatant was discarded. The matrix with bound protein was washed four times with 1 mL washing buffer (100 mM Tris, pH 8.0; 100 mM NaCl; 0.5 mM EDTA; 2 mM DTT; 0.05% [v/v] Triton X-100). Purified StrepII-tagged CPK1 was either used for in vitro kinase assays bound to the matrix or eluted by incubating the matrix for 5 min at 90°C in SDS-loading buffer (60 mM Tris, pH 6.8; 100 mM DTT; 10% [v/v] glycerol; 2% [w/v] SDS; 0.004% [w/v] bromophenol blue) for subsequent gel electrophoresis and immunoblotting. StrepII-tagged CPK1 proteins were visualized using Strep-Tactin alkaline phosphatase or Strep-Tactin horseradish peroxidase conjugate (IBA). For the Strep-Tactin alkaline phosphatase conjugate, the nitrocellulose membranes were incubated for detection in alkaline phosphatase buffer (100 mM Tris-HCl, pH 9.5; 100 mM NaCl; 5 mM MgCl2) with 163 µg/mL 5-bromo-4-chloro-3-indolyl phosphate disodium salt and 327 µg/mL nitroblue tetrazolium chloride. For the Strep-Tactin horseradish peroxidase conjugate, the nitrocellulose membranes were incubated with SuperSignal West Femto ECL substrate (Thermo Fisher Scientific), and luminescence was detected with Image Station 440 CF (Kodak).
Transient Expression in Arabidopsis Leaf Mesophyll Protoplasts and Viability Assays
Protoplast isolation and transfection were performed as described by Yoo et al. (2007). In brief, leaf strips were incubated in 6 mL enzyme solution for 3 h. The same volume of W5 buffer was added and protoplasts were centrifuged. Cell densities were adjusted in W5 buffer to 2 × 105 cells/mL, and cells were rested on ice for 30 min. Transfections used 11,000 cells per transfection using a 30% (w/v) polyethylene glycol solution and a total amount of either 21 µg (for mass spectrometry [MS] analysis) or 25 μg (viability assays) plasmid DNA. Cells were then incubated in 6-well plates coated with 5% (v/v) calf-serum albumin until staining and counting (in the case of the viability assay). Protoplasts were stained 3 h and 26 h after transfection with 5 µg/mL propidium iodide and 5 µg/mL fluorescein diacetate. Dead and living protoplasts were counted in a counting chamber using a fluorescence microscope (Nikon Eclipse 90i). Logistic regression was used as statistical tool to analyze differences in cell death rates (Supplemental Data Set).
Trypan Blue Staining and Chlorophyll Fluorescence Measurements
The expression of CPK1 proteins was induced in 5-week-old plants by incubation in an ethanol/water-vapor atmosphere for 12 h. Plants were subsequently returned to normal growth conditions in the growth chamber for additional 27 h. Whole-leaf rosettes were harvested and immediately submerged in trypan blue staining solution (0.83 mg/mL trypan blue; 8% [v/v] lactic acid; 8% [v/v] glycerol; 8% [v/v] phenol; 67% [v/v] ethanol) and boiled for 4 min. Rosettes were kept in this solution at room temperature for additional 4.5 h and were subsequently destained in 15.1 M chloral hydrate for 13.5 d with four changes of the chloral hydrate solution. The rosettes were washed three times in 5% [v/v] ethanol, and cut leaves for photography were incubated in 25% [v/v] glycerol. Chlorophyll a fluorescence analyses were conducted in 5-week-old plants using a PAM fluorimeter (Walz), and the maximum quantum efficiency of PSII (FV/FM) was determined as previously described (Schreiber et al., 1995).
In Vitro Protein Kinase Assays
To assay CDPK kinase activity (Romeis et al., 2001), we analyzed Strep-Tactin-bound CPK proteins that were purified from either the rosettes of transgenic Arabidopsis plants (Figure 1C) or leaf material after transient expression in Nicotiana benthamiana. After affinity purification, equal aliquots of Strep-Tactin MacroPrep resin-bound CPK variants were used for a CPK expression control after gel electrophoresis and subsequent immunoblot detection with Strep-Tactin alkaline phosphatase conjugate. When using the same variant in different samples, an equal amount of resin per sample was used. Otherwise, resin amounts were adjusted to obtain equal protein expression strength based on the results of the immunoblots. In the case of the synthetic peptide substrate Syntide-2, Strep-Tactin MacroPrep resin-bound CPK1 was resuspended in 20 μL buffer E (50 mM Hepes, pH 7.4; 2 mM DTT; 0.1 mM EDTA), and 5 μL of the slurry was mixed with 20 μL buffer E and 5 μL reaction mix (60 mM MgCl2, 60 µM CaCl2; 60 µM syntide-2; 6 µM ATP; 18 µCi [γ-32P]-ATP). For the negative controls, the reaction mix contained 12 mM EGTA instead of CaCl2. After 10 min at room temperature, the kinase reaction was stopped by adding 3 μL 10% (v/v) phosphoric acid. Twenty microliters of the supernatant were spotted on P81 phosphocellulose paper squares (Merck Millipore), which were allowed to dry and subsequently washed four times with 1% (v/v) phosphoric acid. Radioactivity was determined after a 30-min incubation of paper squares in scintillation mixture ROTISZINT eco plus (0016.3; Carl Roth) using a scintillation counter (Hidex Plate Chameleon 425-104 multilabel counter). For recombinant protein substrates, 0.5 to 1.0 µg of each protein in 15 μL GST-elution buffer (100 mM Tris, pH 8; 20 mM reduced GSH) was incubated with 16 μL slurry of Strep-Tactin-matrix-bound CPK1 in buffer E and 6 μL reaction mix. After 30 min at 25°C in a shaker (650 rpm), samples were centrifuged for 1 min at 800g, and 15 μL of the supernatant were transferred to a test tube containing 4 μL of 5× SDS-loading buffer (300 mM Tris, pH 6.8; 500 mM DTT; 50% [v/v] glycerol; 10% [w/v] SDS; 0.02% [w/v] bromophenol blue) and 1.5 μL EDTA (50 mM) were added. The mixture was heated for 5 min at 95°C, and 17 μL of each sample were loaded on a SDS-polyacrylamide gel. Following electrophoresis, the gel was stained with colloidal Coomassie Brilliant Blue and dried. Protein bands of phosphorylated proteins were visualized by autoradiography on a PhosphoImager (BAS-MS, Fujifilm) via a FLA2000G scanner (Fujifilm) using the software Science Lab 99 (Fujifilm).
RT-PCR and RT-qPCR Analyses
RNA was extracted from Arabidopsis leaves using the Trizol method (Chomczynski and Sacchi, 2006). For RT-PCR-analysis, 1 µg of RNA treated with RNase-free DNase (Fermentas) was used in a reaction mixture containing 50 mM Tris, pH 8.3; 75 mM KCl; 3 mM MgCl2; 10 mM DTT; 0.5 µg oligo(dT)-oligonucleotides, and 200 units of moloney murine leukemia virus reverse transcriptase (Promega). The reaction mixture was incubated for 90 min at 42°C and the reverse transcriptase was subsequently inactivated for 15 min at 70°C. cDNA was used for PCR, and amplified fragments were separated on 1% (w/v) agarose gels and detected by ethidium bromide staining. As a control for consistent cDNA amounts, cDNA of ACTIN2 (At3g18780) was used.
For RT-qPCR, 2 µg of DNase-treated RNA were used for reverse transcription with SuperscriptIII First Strand Synthesis SuperMix (Invitrogen) according to the manufacturer’s protocol. Real-time qPCR was performed in a final volume of 10 μL according to the instructions of Power SYBR Green PCR master mix (Applied Biosystems) using the CFX96 system (Bio-Rad). Postamplification dissociation curves were analyzed for amplification specificity by identifying the occurrence of only one major peak ACTIN2 was used as an internal control for the quantification of gene expression. The relative transcript level 2− (ΔΔCt) (Livak and Schmittgen, 2001) was calculated, using the sample with the lowest Ct of target gene and reference gene as calibrator sample. Alternatively, the 2−ΔCt method was used. Sequences of primers used are presented in the Supplemental Table
Protein Expression in Escherichia coli and Purification of GST-Tagged Proteins
Induction of protein expression using vectors pDEST24 (Invitrogen; for ORE1+ORE1Δ17) and pGEX4T-3 (for GST-protein) in E. coli BL21(DE3)-cells was achieved by using autoinduction medium (Novagen overnight express instant TB medium, EMD Millipore Chemicals) according to the manufacturer’s instructions. Cells were harvested by centrifugation (15 min; 4°C; 4500g) and frozen. To a cell pellet from a 50-mL culture, 1 mL lysis buffer (50 mM Tris-HCl, pH 8; 250 mM NaCl; 1 mM EDTA; 0.2% [v/v] Triton X-100; 1 mM DTT; 1:200 protease inhibitor mix for E. coli cell extract [Sigma Aldrich]; 1 mM AEBSF; 20 mg lysozyme) was added and incubated for 15 min at room temperature. After sonication and centrifugation, 1 mL supernatant was added to 100 μL 50% (v/v) GST-Bind Resin-slurry (Merck Millipore) in lysis buffer, and the mixture was incubated for 1.5 h at 4°C in a rotation wheel at 10 rpm. The resin was pelleted (3 min, 4°C, 700g) and washed four times with 1 mL washing buffer (100 mM Tris-HCl, pH 8; 150 mM NaCl; 0.2% [v/v] Nonidet-P40; 1 mM AEBSF). Elution from the glutathione matrix was achieved by adding 50 μL elution buffer (100 mM Tris-HCl, pH 8; 20 mM reduced L-glutathione) to 50 μL matrix and incubating the slurry for 10 min at room temperature in a shaker (700 rpm). ORE1-GST and ORE1Δ17-GST proteins were confirmed by immunoblot analysis using 1:2500 dilution of monoclonal anti-GST antibody produced in mouse (Sigma-Aldrich G1160, product no. 71097).
Protein Preparation for MS and Phosphopeptide Enrichment
For each biological replicate (Figure 2A) five Arabidopsis rosettes were pooled and ground in liquid N2, and 500 mg of this powder was extracted with 1.5 mL extraction buffer. An amount of 250 µg acetone-precipitated total proteins from this extract was dissolved in 50 μL of a mixture of 6 M urea/2 M thiourea, pH 8. After addition of iodoacetamide to a final concentration of 2.5 mM and incubation for 20 min at room temperature, samples were predigested with 1.25 µg endoproteinase Lys-C (WAKO Chemicals) for 2.5 h at room temperature. Samples were diluted with four volumes Tris-HCl, pH 8, and digested with 0.5 µg/µg sequencing grade modified trypsin (Promega) for 16 h at room temperature. Samples were acidified with 2% (v/v) trifluoroacetic acid (TFA) to reach pH ≤ 3. Desalting was performed via C18 tips (Rappsilber et al., 2003). For enrichment of phosphopeptides, 2.5 mg of TiO2 beads (GL-Sciences) were equilibrated with 200 μL of solution C (300 mg/mL lactic acid; 80% [v/v] acetonitrile; 0.1% [v/v] TFA). The slurry was placed in a self-made microcolumn in a 200-μL pipette tip with an Empore C8 disk (3M) as a plug and centrifuged (2000g, 5 min). Desalted peptide samples were mixed (1:1) with solution C and loaded onto the TiO2-column (centrifugation 1000g, 5 min). Columns were washed first with 200 μL solution C and subsequently with 200 μL of a mixture of 0.1% (v/v) TFA and 5% (v/v) acetonitrile. Phosphopeptides were eluted from TiO2 beads using 5% (v/v) ammonium hydroxide and 5% (v/v) piperidine successively (Nakagami et al., 2010). Eluates were immediately acidified with 44 μL 10% (v/v) TFA to reach pH < 3. Prior to mass spectrometric analysis, enriched phosphopeptides were desalted over C18 tips.
Liquid Chromatography-MS/MS of Peptides and Phosphopeptides
Peptide mixtures after phosphopeptide enrichment were analyzed by liquid chromatography tandem MS (LC-MS/MS) using a nanoflow Easy-nLC (Thermo Fisher Scientific) for HPLC, and an Orbitrap hybrid mass spectrometer (LTQ-Orbitrap, Thermo Fisher Scientific) as a mass analyzer. Peptides were eluted from a 75-μm analytical column (Reprosil C18, Dr. Maisch) on a linear gradient running from 4 to 64% (v/v) acetonitrile in 90 min and sprayed directly into the LTQ-Orbitrap mass spectrometer. Proteins were identified by MS/MS by information-dependent acquisition of fragmentation spectra of multiply charged peptides. Up to five data-dependent MS/MS spectra were acquired in the linear ion trap for each full-scan spectrum acquired at 60,000 full-width half-maximum resolution in the Orbitrap. Overall cycle time was approximately 1 s. Multistage activation was chosen for fragmentation to achieve simultaneous fragmentation of parent ion and neutral loss peaks of phosphopeptides (Schroeder et al., 2004).
Protein identification and ion intensity quantitation were performed by MaxQuant version 1.3.0.5 (Cox and Mann, 2008). Spectra were matched against the Arabidopsis proteome (The Arabidopsis Information Resource 10, 35,386 entries) using Andromeda (Cox et al., 2011). Carbamidomethylation of Cys was set as a fixed modification; oxidation of Met as well as phosphorylation of Ser, Thr, and Tyr was set as variable modifications. Mass tolerance for the database search was set to 20 ppm on full scans and 0.5 daltons for fragment ions. Multiplicity was set to 1. For label-free quantitation, retention time matching between runs was chosen within a time window of two min. Peptide false discovery rate (FDR) and protein FDR were set to 0.01, while site FDR was set to 0.05. Hits to contaminants (e.g., keratins) and reverse hits identified by MaxQuant were excluded from further analysis.
Mass Spectrometric Data Analysis and Statistics
Ion intensity values were used for quantitative data analysis. cRacker (Zauber and Schulze, 2012) was used for label-free data analysis of phosphopeptide ion intensities based on the MaxQuant output (evidence.txt). All phosphopeptides and nonphosphopeptides were used for quantitation. Within each sample, ion intensities of each phosphopeptide ions species (each m/z) were normalized against the total ion intensities of all nonphosphopeptides in that sample (phosphopeptide ion intensity/total sum of ion intensities nonphosphopeptides). Subsequently, each phosphopeptide ion species (i.e., each m/z value) was scaled against the average normalized intensities of that ion across all treatments. For each phosphopeptide, ion intensity values from three biological replicates were then averaged after normalization and scaling.
Transactivation Assays
The ∼1.0-kb upstream promoter region of BFN1 (BFN1pro) was amplified by PCR from Arabidopsis genomic DNA and inserted into pENTR/D-TOPO vector (Invitrogen). The sequence-verified entry clones were then transferred to the p2GWL7.0 vector (Ghent University; http://gateway.psb.ugent.be/vector) harboring the firefly luciferase (FLuc) coding region by LR recombination to generate the BFN1pro:FLuc reporter vector. Luciferase activity was assayed with the Dual Luciferase Reporter Assay System (Promega). The effector, reporter, and control (35Spro:RLuc or UBIpro:GUS) plasmids were cotransfected using polyethylene glycol into mesophyll cell protoplasts prepared from ore1-1 (Balazadeh et al., 2010) rosette leaves (at 38 d after sowing) as reported by Yoo et al. (2007) using 6 µg DNA of each construct. Protoplasts were incubated at room temperature during 14 h in darkness. Luminescence was recorded using a GloMax 20∕20 Luminometer (Promega) and a TriStar LB 941 multimode microplate reader (Berthold). Normalization and relative promoter activity were calculated based on the activity of the internal control reporter.
EMSAs
Purified GST-ORE1 and GST-ORE1Δ17 proteins were detected using the anti-GST antibody described above (1:10,000 dilution) via immunoblot. EMSAs were performed as previously described by (Wu et al., 2012). Binding reactions were performed using the Odyssey infrared EMSA kit (LI-COR) following the manufacturer’s instructions. DNA-protein complexes were separated in a 6% (w/v) retardation gel (EC6365BOX, Invitrogen), and DY682 signal was detected using the Odyssey infrared imaging system from LI-COR.
Chlorophyll Concentration Measurements
Chlorophyll pigment was extracted by treating 50 mg plant tissue with 1 mL 80% (v/v) acetone in water (v/v) overnight at 4°C. Spectrophotometric absorbance readings were performed at both 633 and 647 nm in a spectrophotometer (NanoSpec 2, Nanolytik). The total chlorophyll content was calculated using the formula below and normalized to fresh weight. Chla + b = [12.25(A663) − 2.79(A647)] + [21.50(A647) − 5.10(A663)]
Statistics
For statistical analyses, we first tested for a normal distribution of the replicate values within a sample group. In the case of a normal distribution of the replicate values in all the multiple sample groups, a one-way ANOVA with Tukey post hoc test was performed. When not all replicate values were normally distributed, either a Mann-Whitney U test (in the case of only two sample groups, i.e., Figure 3B) or a Kruskal-Wallis test with Dunn-Bonferoni post hoc test was performed. For data comprising only two possible states (i.e., Figure 3C, “protoplasts dying within 23 h” and “protoplasts surviving these 23 h”) a logistic regression was used to analyze the differences in cell death rates between the two coexpression groups. The parameters of the statistical tests are given in the Supplemental Data Set. Sample sizes were selected to meet the standard in the field in dependence of the type of experiment (biochemistry, gene expression data, plant growth assay) and are stated in each figure for the respective experiment.
Data Availability
The authors declare that the data supporting the findings of this study are available within the article and its supplemental files or are available from the corresponding author on request.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: ACTIN2, At3g18780; BFN1, At1g11190; CPK1, At5g04870; ORE1, At5g39610; SAG12, At5g45890. Germplasms used in the Col-0 background are cpk1-1 (SALK_096452), cpk1-2 (SALK_080155c), and ore1-1 (SALK_090154).
Supplemental Data
Supplemental Figure 1. Ethanol-induced CPK1D274A protein accumulation
Supplemental Figure 2. Ethanol-inducible gene expression of CPK1 variants
Supplemental Figure 3. Ethanol-induced CPK1-VK and CPK1-VKD274A protein accumulation at 1 h
Supplemental Figure 4. Fragment spectra of identified phosphopeptides of ORE1
Supplemental Figure 5. Specificity of ORE1 phosphorylation by CPK1
Supplemental Figure 6. In vivo autophosphorylation of CPK1
Supplemental Figure 7. ORE1-dependent BFN1 promoter activation is dependent on intact phosphorylation sites on the CPK1 target peptide
Supplemental Figure 8. CPK1-dependent ORE1 function in developmental senescence
Supplemental Figure 9. ORE1 function in developmental senescence
Supplemental Table. Oligonucleotide sequences
Supplemental Methods. Generation of constructs and plant lines
Supplemental References. References for the Supplemental Table and Supplemental Methods
Supplemental Data Set. Statistics tables
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
This research was funded by the Deutsche Forschungsgemeinschaft (DFG; Priority Program SPP1212 to T.R.), the Collaborative Research Centre (grant SFB973 to B.M.-R., S.B., and T.R.), the Academy of Finland (postdoctoral project grant 289687 to G.D.), and the Academy of Finland Center of Excellence in Primary Producers (grant 307335 to G.D.).
AUTHOR CONTRIBUTIONS
G.D., C.-P.W., B.M.-R., S.B., and T.R. conceived and designed the research. G.D., M.S., L.P.M.-R., S.M.S., S.S., T.G., M.H., C.-P.W., W.S., and S.B. performed experiments. G.D., W.S., S.B., and T.R. analyzed the data. G.D. and T.R. wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Balazadeh S., Riaño-Pachón D.M., Mueller-Roeber B.(2008). Transcription factors regulating leaf senescence in Arabidopsis thaliana. Plant Biol (Stuttg) 10 (Suppl 1): 63–75. [DOI] [PubMed] [Google Scholar]
- Balazadeh S., Siddiqui H., Allu A.D., Matallana-Ramirez L.P., Caldana C., Mehrnia M., Zanor M.-I., Köhler B., Mueller-Roeber B.(2010). A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J. 62: 250–264. [DOI] [PubMed] [Google Scholar]
- Boudsocq M., Sheen J.(2013). CDPKs in immune and stress signaling. Trends Plant Sci. 18: 30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq M., Willmann M.R., McCormack M., Lee H., Shan L., He P., Bush J., Cheng S.-H., Sheen J.(2010). Differential innate immune signalling via Ca(2+) sensor protein kinases. Nature 464: 418–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt B., Brodsky D.E., Xue S., Negi J., Iba K., Kangasjärvi J., Ghassemian M., Stephan A.B., Hu H., Schroeder J.I.(2012). Reconstitution of abscisic acid activation of SLAC1 anion channel by CPK6 and OST1 kinases and branched ABI1 PP2C phosphatase action. Proc. Natl. Acad. Sci. USA 109: 10593–10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caddick M.X., Greenland A.J., Jepson I., Krause K.P., Qu N., Riddell K.V., Salter M.G., Schuch W., Sonnewald U., Tomsett A.B.(1998). An ethanol inducible gene switch for plants used to manipulate carbon metabolism. Nat. Biotechnol. 16: 177–180. [DOI] [PubMed] [Google Scholar]
- Cheng S.H., Willmann M.R., Chen H.C., Sheen J.(2002). Calcium signaling through protein kinases. The Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol. 129: 469–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H.I., Park H.-J., Park J.H., Kim S., Im M.-Y., Seo H.-H., Kim Y.-W., Hwang I., Kim S.Y.(2005). Arabidopsis calcium-dependent protein kinase AtCPK32 interacts with ABF4, a transcriptional regulator of abscisic acid-responsive gene expression, and modulates its activity. Plant Physiol. 139: 1750–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N.(2006). The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: Twenty-something years on. Nat. Protoc. 1: 581–585. [DOI] [PubMed] [Google Scholar]
- Christian J.-O., Braginets R., Schulze W.X., Walther D.(2012). Characterization and prediction of protein phosphorylation hotspots in Arabidopsis thaliana. Front Plant Sci 3: 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coca M., San Segundo B.(2010). AtCPK1 calcium-dependent protein kinase mediates pathogen resistance in Arabidopsis. Plant J. 63: 526–540. [DOI] [PubMed] [Google Scholar]
- Cox J., Mann M.(2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26: 1367–1372. [DOI] [PubMed] [Google Scholar]
- Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., Mann M.(2011). Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10: 1794–1805. [DOI] [PubMed] [Google Scholar]
- Dubiella U., Seybold H., Durian G., Komander E., Lassig R., Witte C.-P., Schulze W.X., Romeis T.(2013). Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc. Natl. Acad. Sci. USA 110: 8744–8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durek P., Schmidt R., Heazlewood J.L., Jones A., MacLean D., Nagel A., Kersten B., Schulze W.X.(2010). PhosPhAt: The Arabidopsis thaliana phosphorylation site database. An update. Nucleic Acids Res. 38: D828–D834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durian G., et al. (2020). PROTEIN PHOSPHATASE 2A-B′γ controls Botrytis cinerea resistance and developmental leaf senescence. Plant Physiol. 182: 1161–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Chen X., Lin W., Chen S., Lu D., Niu Y., Li L., Cheng C., McCormack M., Sheen J., Shan L., He P.(2013). Bifurcation of Arabidopsis NLR immune signaling via Ca2+-dependent protein kinases. PLoS Pathog. 9: e1003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z., et al. (2018). KIRA1 and ORESARA1 terminate flower receptivity by promoting cell death in the stigma of Arabidopsis. Nat. Plants 4: 365–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garapati P., Xue G.-P., Munné-Bosch S., Balazadeh S.(2015). Transcription factor ATAF1 in Arabidopsis promotes senescence by direct regulation of key chloroplast maintenance and senescence transcriptional cascades. Plant Physiol. 168: 1122–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger D., Maierhofer T., Al-Rasheid K.A.S., Scherzer S., Mumm P., Liese A., Ache P., Wellmann C., Marten I., Grill E., Romeis T., Hedrich R.(2011). Stomatal closure by fast abscisic acid signaling is mediated by the guard cell anion channel SLAH3 and the receptor RCAR1. Sci. Signal. 4: ra32. [DOI] [PubMed] [Google Scholar]
- Geiger D., Scherzer S., Mumm P., Marten I., Ache P., Matschi S., Liese A., Wellmann C., Al-Rasheid K.A.S., Grill E., Romeis T., Hedrich R.(2010). Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc. Natl. Acad. Sci. USA 107: 8023–8028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon A.C., Gribskov M., Harper J.F.(2000). CDPKs: a kinase for every Ca2+ signal? Trends Plant Sci. 5: 154–159. [DOI] [PubMed] [Google Scholar]
- Harper J.F., Breton G., Harmon A.(2004). Decoding Ca(2+) signals through plant protein kinases. Annu. Rev. Plant Biol. 55: 263–288. [DOI] [PubMed] [Google Scholar]
- Harper J.F., Huang J.-F., Lloyd S.J.(1994). Genetic identification of an autoinhibitor in CDPK, a protein kinase with a calmodulin-like domain. Biochemistry 33: 7267–7277. [DOI] [PubMed] [Google Scholar]
- He X.-J., Mu R.-L., Cao W.-H., Zhang Z.-G., Zhang J.-S., Chen S.-Y.(2005). AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 44: 903–916. [DOI] [PubMed] [Google Scholar]
- Iakoucheva L.M., Radivojac P., Brown C.J., O’Connor T.R., Sikes J.G., Obradovic Z., Dunker A.K.(2004). The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 32: 1037–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T., Kinoshita K.(2007). PrDOS: Prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 35: W460–W464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen M.K., Kjaersgaard T., Nielsen M.M., Galberg P., Petersen K., O’Shea C., Skriver K.(2010). The Arabidopsis thaliana NAC transcription factor family: Structure-function relationships and determinants of ANAC019 stress signalling. Biochem. J. 426: 183–196. [DOI] [PubMed] [Google Scholar]
- Kadota Y., Sklenar J., Derbyshire P., Stransfeld L., Asai S., Ntoukakis V., Jones J.D., Shirasu K., Menke F., Jones A., Zipfel C.(2014). Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol. Cell 54: 43–55. [DOI] [PubMed] [Google Scholar]
- Kim H., Kim H.J., Vu Q.T., Jung S., McClung C.R., Hong S., Nam H.G.(2018). Circadian control of ORE1 by PRR9 positively regulates leaf senescence in Arabidopsis. Proc. Natl. Acad. Sci. USA 115: 8448–8453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.J., et al. (2014). Gene regulatory cascade of senescence-associated NAC transcription factors activated by ETHYLENE-INSENSITIVE2-mediated leaf senescence signalling in Arabidopsis. J. Exp. Bot. 65: 4023–4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., Woo H.R., Kim J., Lim P.O., Lee I.C., Choi S.H., Hwang D., Nam H.G.(2009). Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 323: 1053–1057. [DOI] [PubMed] [Google Scholar]
- Köhler C., Merkle T., Roby D., Neuhaus G.(2001). Developmentally regulated expression of a cyclic nucleotide-gated ion channel from Arabidopsis indicates its involvement in programmed cell death. Planta 213: 327–332. [DOI] [PubMed] [Google Scholar]
- Kudla J., Batistic O., Hashimoto K.(2010). Calcium signals: The lead currency of plant information processing. Plant Cell 22: 541–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Zhang G., Gonzales N., Guo Y., Hu H., Park S., Zhao J.(2016). Ca(2+) -regulated and diurnal rhythm-regulated Na(+) /Ca(2+) exchanger AtNCL affects flowering time and auxin signalling in Arabidopsis. Plant Cell Environ. 39: 377–392. [DOI] [PubMed] [Google Scholar]
- Li Z., Peng J., Wen X., Guo H.(2013). Ethylene-insensitive3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant Cell 25: 3311–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liese A., Romeis T.(2013). Biochemical regulation of in vivo function of plant calcium-dependent protein kinases (CDPK). Biochim. Biophys. Acta 1833: 1582–1589. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D.(2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Matallana-Ramirez L.P., Rauf M., Farage-Barhom S., Dortay H., Xue G.-P., Dröge-Laser W., Lers A., Balazadeh S., Mueller-Roeber B.(2013). NAC transcription factor ORE1 and senescence-induced BIFUNCTIONAL NUCLEASE1 (BFN1) constitute a regulatory cascade in Arabidopsis. Mol. Plant 6: 1438–1452. [DOI] [PubMed] [Google Scholar]
- Nakagami H., Sugiyama N., Mochida K., Daudi A., Yoshida Y., Toyoda T., Tomita M., Ishihama Y., Shirasu K.(2010). Large-scale comparative phosphoproteomics identifies conserved phosphorylation sites in plants. Plant Physiol. 153: 1161–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B.S., Yao T., Seo J.S., Wong E.C.C., Mitsuda N., Huang C.-H., Chua N.-H.(2018). Arabidopsis NITROGEN LIMITATION ADAPTATION regulates ORE1 homeostasis during senescence induced by nitrogen deficiency. Nat. Plants 4: 898–903. [DOI] [PubMed] [Google Scholar]
- Park S.H., Jeong J.S., Seo J.S., Park B.S., Chua N.H.(2019). Arabidopsis ubiquitin-specific proteases UBP12 and UBP13 shape ORE1 levels during leaf senescence induced by nitrogen deficiency. New Phytol. 223: 1447–1460. [DOI] [PubMed] [Google Scholar]
- Qiu K., Li Z., Yang Z., Chen J., Wu S., Zhu X., Gao S., Gao J., Ren G., Kuai B., Zhou X.(2015). EIN3 and ORE1 accelerate degreening during ethylene-mediated leaf senescence by directly activating chlorophyll catabolic genes in Arabidopsis. PLoS Genet. 11: e1005399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J., Ishihama Y., Mann M.(2003). Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75: 663–670. [DOI] [PubMed] [Google Scholar]
- Romeis T., Ludwig A.A., Martin R., Jones J.D.G.(2001). Calcium-dependent protein kinases play an essential role in a plant defence response. EMBO J. 20: 5556–5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber U., Hormann H., Neubauer C., Klughammer C.(1995). Assessment of photosystem II photochemical quantum yield by chlorophyll fluorescence quenching analysis. Aust. J. Plant Physiol. 22: 209–220. [Google Scholar]
- Schroeder M.J., Shabanowitz J., Schwartz J.C., Hunt D.F., Coon J.J.(2004). A neutral loss activation method for improved phosphopeptide sequence analysis by quadrupole ion trap mass spectrometry. Anal. Chem. 76: 3590–3598. [DOI] [PubMed] [Google Scholar]
- Schulz P., Herde M., Romeis T.(2013). Calcium-dependent protein kinases: Hubs in plant stress signaling and development. Plant Physiol. 163: 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeunovic A., Mair A., Wurzinger B., Teige M.(2016). Know where your clients are: Subcellular localization and targets of calcium-dependent protein kinases. J. Exp. Bot. 67: 3855–3872. [DOI] [PubMed] [Google Scholar]
- Swatek K.N., Wilson R.S., Ahsan N., Tritz R.L., Thelen J.J.(2014). Multisite phosphorylation of 14-3-3 proteins by calcium-dependent protein kinases. Biochem. J. 459: 15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian W., Hou C., Ren Z., Wang C., Zhao F., Dahlbeck D., Hu S., Zhang L., Niu Q., Li L., Staskawicz B.J., Luan S.(2019). A calmodulin-gated calcium channel links pathogen patterns to plant immunity. Nature 572: 131–135. [DOI] [PubMed] [Google Scholar]
- Trivellini A., Jibran R., Watson L.M., O’Donoghue E.M., Ferrante A., Sullivan K.L., Dijkwel P.P., Hunter D.A.(2012). Carbon deprivation-driven transcriptome reprogramming in detached developmentally arresting Arabidopsis inflorescences. Plant Physiol. 160: 1357–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte C.-P., Noël L.D., Gielbert J., Parker J.E., Romeis T.(2004). Rapid one-step protein purification from plant material using the eight-amino acid StrepII epitope. Plant Mol. Biol. 55: 135–147. [DOI] [PubMed] [Google Scholar]
- Witte C.-P., Keinath N., Dubiella U., Demoulière R., Seal A., Romeis T.(2010). Tobacco calcium-dependent protein kinases are differentially phosphorylated in vivo as part of a kinase cascade that regulates stress response. J. Biol. Chem. 285: 9740–9748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A., et al. (2012). JUNGBRUNNEN1, a Reactive Oxygen Species–Responsive NAC Transcription Factor, Regulates Longevity in Arabidopsis. Plant Cell 24: 482–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Hotta C.T., Dodd A.N., Love J., Sharrock R., Lee Y.W., Xie Q., Johnson C.H., Webb A.A.R.(2007). Distinct light and clock modulation of cytosolic free Ca2+ oscillations and rhythmic CHLOROPHYLL A/B BINDING PROTEIN2 promoter activity in Arabidopsis. Plant Cell 19: 3474–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip Delormel T., Boudsocq M.(2019). Properties and functions of calcium-dependent protein kinases and their relatives in Arabidopsis thaliana. New Phytol. 224: 585–604. [DOI] [PubMed] [Google Scholar]
- Yoo S.-D., Cho Y.-H., Sheen J.(2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zauber H., Schulze W.X.(2012). Proteomics wants cRacker: Automated standardized data analysis of LC-MS derived proteomic data. J. Proteome Res. 11: 5548–5555. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu Z., Chen Y., He J.X., Bi Y.(2015). PHYTOCHROME-INTERACTING FACTOR 5 (PIF5) positively regulates dark-induced senescence and chlorophyll degradation in Arabidopsis. Plant Sci. 237: 57–68. [DOI] [PubMed] [Google Scholar]
- Zhao R., Sun H.-L., Mei C., Wang X.-J., Yan L., Liu R., Zhang X.-F., Wang X.-F., Zhang D.-P.(2011). The Arabidopsis Ca(2+) -dependent protein kinase CPK12 negatively regulates abscisic acid signaling in seed germination and post-germination growth. New Phytol. 192: 61–73. [DOI] [PubMed] [Google Scholar]
- Zhu S.Y., et al. (2007). Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell 19: 3019–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors declare that the data supporting the findings of this study are available within the article and its supplemental files or are available from the corresponding author on request.