A genetic screen employed to identify genes that regulate flowering independently of environmental cues revealed a role for the chromatin remodeler CHR4 in promoting floral identity.
Abstract
Several pathways conferring environmental flowering responses in Arabidopsis (Arabidopsis thaliana) converge on developmental processes that mediate the floral transition in the shoot apical meristem. Many characterized mutations disrupt these environmental responses, but downstream developmental processes have been more refractory to mutagenesis. Here, we constructed a quintuple mutant impaired in several environmental pathways and showed that it possesses severely reduced flowering responses to changes in photoperiod and ambient temperature. RNA-sequencing (RNA-seq) analysis of the quintuple mutant showed that the expression of genes encoding gibberellin biosynthesis enzymes and transcription factors involved in the age pathway correlates with flowering. Mutagenesis of the quintuple mutant generated two late-flowering mutants, quintuple ems1 (qem1) and qem2. The mutated genes were identified by isogenic mapping and transgenic complementation. The qem1 mutant is an allele of the gibberellin 20-oxidase gene ga20ox2, confirming the importance of gibberellin for flowering in the absence of environmental responses. By contrast, qem2 is impaired in CHROMATIN REMODELING4 (CHR4), which has not been genetically implicated in floral induction. Using co-immunoprecipitation, RNA-seq, and chromatin immunoprecipitation sequencing, we show that CHR4 interacts with transcription factors involved in floral meristem identity and affects the expression of key floral regulators. Therefore, CHR4 mediates the response to endogenous flowering pathways in the inflorescence meristem to promote floral identity.
INTRODUCTION
Lateral shoot organs initiate from cells on the flanks of the shoot apical meristem (SAM), and the identity of the formed organs changes during development (Bowman and Eshed, 2000). In Arabidopsis (Arabidopsis thaliana), the transition from vegetative leaf initiation to flower production occurs in response to several environmental and endogenous cues. Important environmental signals that control flowering include seasonal fluctuations in temperature and daylength, which are mediated by the photoperiodic and vernalization pathways, whereas ambient changes in temperature also influence flowering time (Srikanth and Schmid, 2011; Andrés and Coupland, 2012). In addition, endogenous signals such as gibberellins (GAs) and the age of the plant contribute to the floral transition in the absence of inductive environmental cues (Wilson et al., 1992; Wang et al., 2009).
Three intersecting environmental pathways that promote flowering have been well characterized. The photoperiodic pathway promotes flowering under long days (LDs) but not under short days (SDs), in which plants flower much later. Exposure to LDs stabilizes the CONSTANS transcription factor (Valverde et al., 2004), which in turn activates transcription of FLOWERING LOCUS T (FT) and TWIN SISTER OF FT (TSF) in the leaf vascular tissue (Kardailsky et al., 1999; Kobayashi et al., 1999; Suárez-Lopez et al., 2001; An et al., 2004; Yamaguchi et al., 2005). The FT and TSF proteins, which are related to phosphatidyl-ethanolamine binding proteins, move to the SAM (Corbesier et al., 2007; Jaeger and Wigge, 2007; Mathieu et al., 2007), where they physically interact with the bZIP transcription factor FD (Abe et al., 2005, 2019; Wigge et al., 2005). In the SAM, the FT–FD protein complex promotes the transcription of genes encoding floral activators, such as SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1) and FRUITFULL (FUL), which induce the floral transition, as well as APETALA1 (AP1) and LEAFY (LFY), which promote floral meristem identity (Schmid et al., 2003; Wigge et al., 2005; Torti et al., 2012; Collani et al., 2019). Because they represent the mobile signal linking leaves and the SAM, FT and TSF are essential for the photoperiodic flowering response, and ft tsf double mutants are daylength-insensitive (Yamaguchi et al., 2005; Jang et al., 2009).
The seasonal cue of exposure to winter cold mediates flowering via the vernalization pathway, which represses transcription of the floral repressor FLOWERING LOCUS C (FLC; Michaels and Amasino, 1999; Sheldon et al., 1999). FLC is a MADS (MINICHROMOSOME MAINTENANCE FACTOR 1, AGAMOUS, DEFICIENS, SERUM RESPONSE FACTOR)-box transcription factor that forms regulatory complexes with other MADS-box floral repressors, such as SHORT VEGETATIVE PHASE (SVP; Li et al., 2008). Thus, vernalization reduces FLC transcription and promotes flowering via the endogenous and photoperiodic pathways, whereas mutants for FLC are essentially insensitive to vernalization. The genome-wide binding sites of FLC and SVP include those in several genes that promote flowering within the photoperiodic pathway, such as FT and SOC1 (Searle et al., 2006; Lee et al., 2007; Li et al., 2008; Deng et al., 2011; Mateos et al., 2015; Richter et al., 2019). Because FLC is stably repressed by exposure to cold, plants can flower through the photoperiodic pathway when they are exposed to LDs after cold exposure. Also, genes within the endogenous pathway that are repressed by FLC, such as SQUAMOSA PROMOTER BINDING PROTEIN-LIKE15 (SPL15), can promote flowering during vernalization (Deng et al., 2011; Hyun et al., 2019)
Arabidopsis also flowers rapidly when exposed to high temperatures, and this response can overcome the delay in flowering observed under SDs at lower growth temperatures (Balasubramanian et al., 2006). FT and TSF are transcribed at high temperature under SDs and promote early flowering; thus their transcriptional repression under SDs at lower temperatures is overcome at high temperatures (Kumar et al., 2012; Galvão et al., 2015; Fernández et al., 2016). Accordingly, MADS-box repressors of FT and TSF, particularly FLOWERING LOCUS M and SVP, do not accumulate under SDs at high temperature, and mutations in these genes reduce the flowering response to high temperature (Lee et al., 2007, 2013; Posé et al., 2013; Airoldi et al., 2015). The reduced activity of these repressors also enhances the response of the meristem to low levels of FT and TSF transcription in the leaves (Fernández et al., 2016). Triple mutants for FT, TSF, and SVP are insensitive to higher temperatures under SDs (Fernández et al., 2016).
In addition to these environmental pathways, there are several endogenous flowering pathways. A set of genes was ascribed to the autonomous flowering pathway, because they caused late-flowering under LDs and SDs and were therefore considered to promote flowering independently of photoperiodic cues (Koornneef et al., 1991). Mutations in all these genes caused elevated levels of FLC mRNA, and the encoded proteins contribute to FLC expression at the transcriptional and post-transcriptional levels (Whittaker and Dean, 2017). The late-flowering phenotype of autonomous pathway mutants can therefore be suppressed by mutations in FLC (Michaels and Amasino, 2001). In addition, GA is an important contributor to endogenous flowering regulation, because mutations or transgenes that strongly reduce GA levels almost abolished flowering under non-inductive SDs (Wilson et al., 1992; Galvão et al., 2012; Porri et al., 2012). Finally, microRNA156 (miR156) negatively regulates the floral transition and is developmentally regulated such that its abundance decreases progressively with increasing plant age (Wu and Poethig, 2006; Wang et al., 2009). This miRNA negatively regulates the accumulation of several SPL transcription factors, including SPL3, SPL9, and SPL15, which promote the floral transition, particularly under non-inductive SDs (Gandikota et al., 2007; Wang et al., 2009; Yamaguchi et al., 2009; Hyun et al., 2016; Xu et al., 2016). Thus, miR156/SPL modules have been associated with an endogenous flowering pathway related to plant age.
Here, we extend our understanding of the genetic basis of the floral transition by screening specifically for genes that regulate flowering independently of the environmental pathways. To this end, we constructed a high-order quintuple mutant, svp-41 flc-3 ft-10 tsf-1 soc1-2, which shows reduced sensitivity to environmental flowering signals because it is impaired in responses to photoperiod and high temperature. Using RNA sequencing (RNA-seq), we characterized the expression of flowering-related genes in this mutant, and we employed a forward genetics approach to identify genes controlling flowering time in this background. This allowed us to define a role for CHROMATIN REMODELING4 (CHR4) in promoting the floral transition.
RESULTS
Phenotypic and Molecular Characterization of a Quintuple Mutant Strongly Impaired in Responses to Environmental Cues
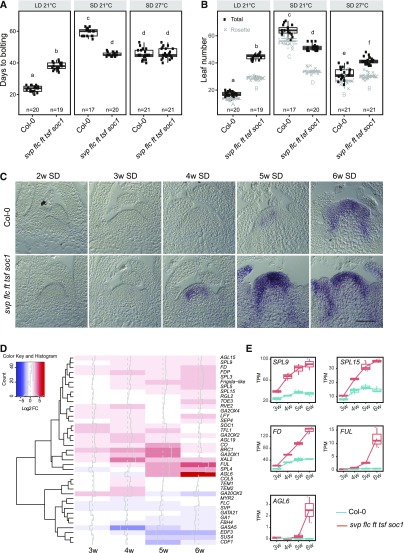
To assess the flowering time of Arabidopsis plants in which the major environmental pathways were inactivated, we constructed the quintuple mutant svp-41 flc-3 ft-10 tsf-1 soc1-2 (hereafter referred to as the quintuple mutant). The quintuple mutant showed a dramatically reduced flowering response to daylength compared with Columbia 0 (Col-0). Under LDs, the quintuple mutant bolted later and after forming more vegetative rosette leaves than the wild type (Col-0; Figures 1A and 1B). However, under SDs at 21°C, the mutant bolted much earlier than Col-0 in terms of days to flowering and rosette leaf number (Figures 1A and 1B). Bolting of the quintuple mutant was delayed by fewer than 10 d in SDs compared with LDs, whereas bolting in Col-0 was delayed by ∼50 d. Similarly, the quintuple mutant formed ∼5 more rosette leaves under SDs than LDs, whereas Col-0 formed over 40 more rosette leaves. The flowering time of the quintuple mutant was also insensitive to higher ambient temperatures under SDs when considering bolting time, but it displayed partial insensitivity in terms of rosette leaf number (Figures 1A and 1B). Finally, GA4 treatment accelerated flowering of Col-0 under SDs (Wilson et al., 1992) but had a smaller effect on the flowering time of the quintuple mutant (Supplemental Figures 1A and 1B). These results are consistent with the idea that the GA response and signaling are activated in the quintuple mutant, as shown for svp-41 mutants in Andrés et al. (2014). Overall, the quintuple mutant showed strongly impaired responses to environmental signals such as day-length and ambient temperatures, in terms of time to bolting and the number of rosette leaves formed. These data suggest that in the quintuple mutant, the floral transition occurs via endogenous mechanisms such as the GA or age pathway.
Figure 1.
Phenotypic and Molecular Characterization of the Quintuple Mutant svp flc ft tsf soc1.
(A) and (B) Days to bolting (A) and leaf number of plants grown (B) under LD-21°C, SD-21°C, and SD-27°C compared with Col-0. At least 17 plants were analyzed for each genotype. The data were analyzed with one-way ANOVA using Tukey’s honest significant difference as a post-hoc test. Different letters indicate significant differences (P ≤ 0.05). Whiskers represent a distance of 1.5 times the interquartile range.
(C) In situ hybridization analysis of FUL mRNA accumulation in SAMs of different genotypes grown in SDs. Plants were harvested each week between 2 weeks and 6 weeks after germination. Scale bar = 50 μm.
(D) Transcriptional profile comparisons in apices of svp flc ft tsf soc1. The analysis focuses on genes implicated in flowering time control. The data are represented as a heatmap to highlight upregulated (red) and downregulated genes (blue). Gene expression changes are represented as log2-fold changes.
(E) Box plots from RNA-seq data showing differential expression of SPL9, SPL15, FD, FUL, and AGL6 in the apices of svp flc ft tsf soc1 and Col-0 under SDs. The Y-axis shows transcripts per TPM. The X-axis shows time of sampling as weeks after sowing. Whiskers represent distance from the lowest to the largest data point.
In addition to effects on bolting time and vegetative rosette leaf number, the quintuple mutant produced more cauline leaves than Col-0 in all conditions tested (Supplemental Figure 1C). The quintuple mutant formed on average 4.5-fold more cauline leaves than Col-0 under LDs and 2.3-fold more under SDs. The increased cauline leaf number in the mutant compared with Col-0 suggests that the mutant is also impaired in the determination of floral meristem identity after floral induction and bolting, such that more phytomers contain cauline leaves and axillary shoots than in Col-0.
We then compared the developmental stage of the shoot apex of the quintuple mutant to that of Col-0 by performing in situ hybridizations for FUL transcript on apical cross sections of SD-grown plants of different ages (Figure 1C). FUL encodes a MADS-box floral activator that is partially genetically redundant with SOC1. FUL mRNA accumulates in the SAM during the early stages of the floral transition (Ferrándiz et al., 2000; Melzer et al., 2008; Torti et al., 2012). In the apices of SD-grown plants, FUL mRNA accumulated ∼1 week earlier in the quintuple mutant than in Col-0 (Figure 1C), which is consistent with the earlier flowering phenotype of the mutant.
Because the quintuple mutant flowers earlier under SDs and major regulators of flowering are inactivated, the transcriptional network associated with the floral transition is probably differentially expressed in the mutant compared with Col-0. To define these differences, we performed RNA-seq on apices of the quintuple mutant and Col-0 through a developmental time course under SDs. Apical samples were harvested from both genotypes 3, 4, 5, and 6 weeks after sowing. In vegetative apices of both genotypes at 3 weeks after sowing, only 46 genes were differentially expressed (adjusted P-value < 0.05) between the quintuple mutant and Col-0. At 4, 5, and 6 weeks, when the mutant flowered more rapidly than Col-0 (Figure 1C), 486, 736, and 568 genes, respectively, were differentially expressed in the mutant compared with Col-0 (Supplemental Data Set 1). At these time points, ∼45%, 14%, and 33% of the differentially expressed genes (DEGs), respectively, were more highly expressed in the quintuple mutant versus Col-0 (Supplemental Data Set 1). The mRNAs of SPL3, SPL4, SPL5, SPL9, SPL12, and SPL15, which are regulated by miR156 and contribute to the endogenous age-related flowering pathway, were more abundant in the quintuple mutant, which is consistent with promotion of flowering by the age pathway (Figures 1D and 1E). Moreover, the floral activators FD, FDP, and AGAMOUS-LIKE6 (AGL6) were more highly expressed in the mutant versus Col-0 (Figures 1D and 1E; Supplemental Figure 1D; Supplemental Data Set 1), and the expression of the floral repressors MADS-AFFECTING FLOWERING4 (MAF4) and MAF5 was attenuated in the quintuple mutant (Supplemental Data Set 1). Moreover, genes encoding enzymes involved in GA biosynthesis and catabolism were differentially expressed in the quintuple mutant (Figure 1D).
A Sensitized Mutant Screen in the Quintuple Mutant Background Identifies Two Loci that Promote Flowering
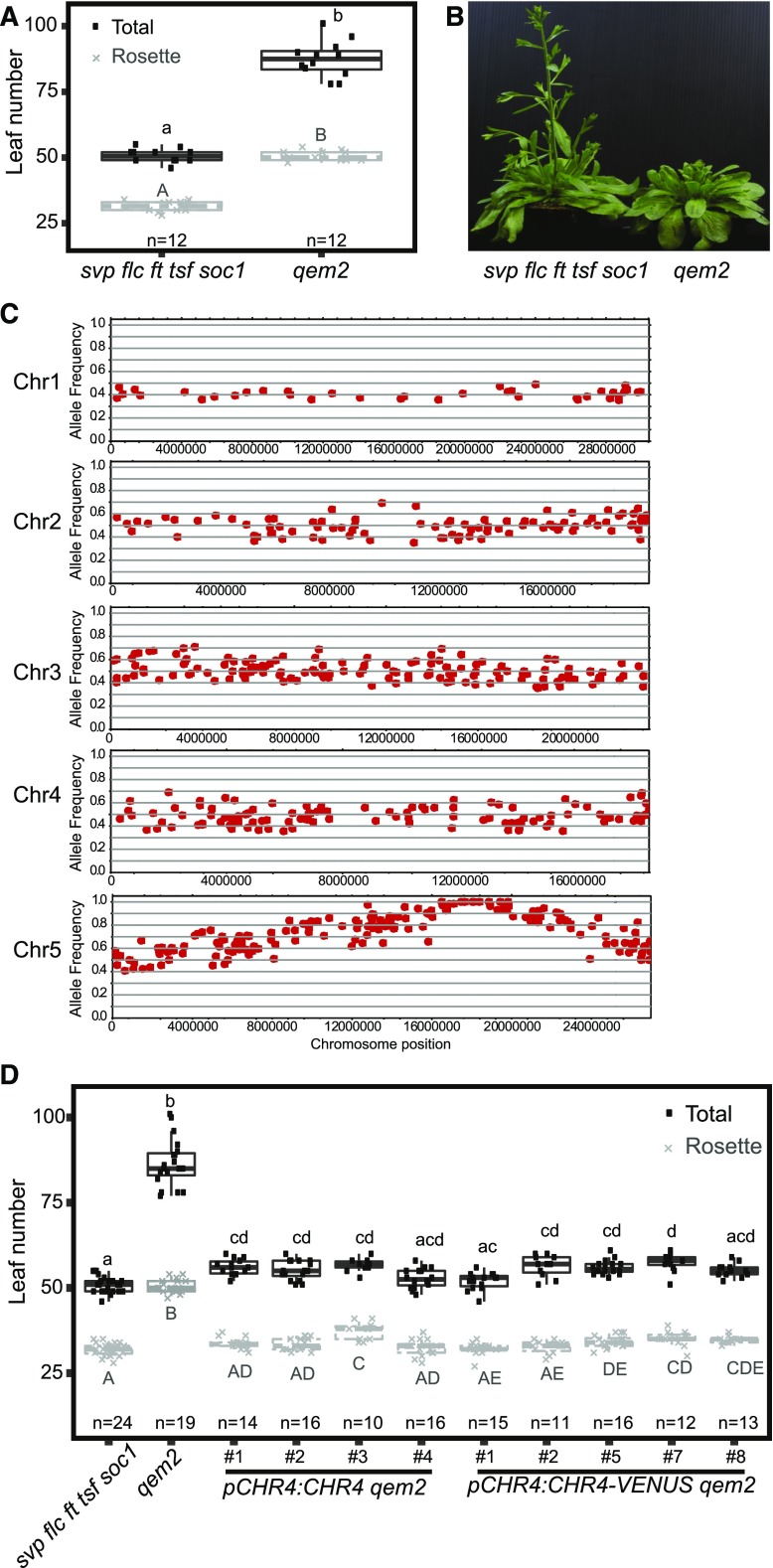
We then employed the quintuple mutant as a sensitized background for mutagenesis screening to identify genes that regulate flowering independently of environmental pathways. This approach was expected to identify mutations in endogenous components, because the major environmental floral response pathways are already impaired in the mutant, and mutations in the autonomous pathway should not be recovered, because FLC is inactive in the quintuple mutant. We screened the M2 generation for mutants with altered flowering behavior (see Methods). Two mutants showing delayed floral transition in the quintuple mutant background, quintuple ems1 (qem1) and qem2, were selected for detailed studies because they exhibited strong and reproducible phenotypes in the M3 generation. Both lines segregated the mutant phenotype in a 3:1 ratio in the BC1F2 generation (see Methods), suggesting that a single recessive mutation was responsible for the phenotypes of both mutants. Plants segregating the qem1 or qem2 phenotype in the respective BC1F2 populations were then bulk-harvested. Fast-isogenic mapping (see Methods; Hartwig et al., 2012) localized qem1 and qem2 with high confidence to different regions on chromosome 5 (Figure 2; Supplemental Figure 2).
Figure 2.
Molecular Genetic Analysis of qem2.
(A) Leaf number at flowering of plants grown under LDs. Twelve plants were analyzed per genotype. The data were compared with one-way ANOVA using Tukey’s honest significant difference as a post-hoc test. Different letters indicate significant differences (P ≤ 0.05). Whiskers represent the distance of 1.5 times the interquartile range.
(B) Images of qem2 and svp flc ft tsf soc1 plants ∼50 d after germination, showing that qem2 produces more leaves than svp flc ft tsf soc1 under LDs.
(C) Allele frequency estimates for EMS-induced mutations. Local allele frequencies indicate that the qem2 mutation localized to chromosome (Chr) 5.
(D) Leaf number for svp flc ft tsf soc1, qem2, gCHR4 qem2, and gCHR4-VENUS qem2 plants under LDs. At least 11 plants per genotype were analyzed. The data were compared with one-way ANOVA using Tukey’s honest significant difference as a post-hoc test. Different letters indicate significant differences (P ≤ 0.05). Whiskers represent a distance of 1.5 times the interquartile range.
The qem1 mutation localized to the same region of chromosome 5 as the GA 20-oxidase gene GA20ox2 (Supplemental Figure 2C; Supplemental Table). Mutation of GA20ox2 delays flowering and has a stronger effect in the svp-41 background (Rieu et al., 2008; Plackett et al., 2012; Andrés et al., 2014). In qem1, a single nucleotide polymorphism was identified in the first exon of GA20ox2 that was predicted to cause an amino acid substitution in the protein (ser137asn). To confirm that this mutation causes the late-flowering phenotype of qem1, we performed molecular complementation. Introducing the Col-0 genomic GA20ox2 locus into qem1 strongly reduced leaf number and flowering time, so that the transgenic lines flowered at a similar time or earlier than the quintuple mutant (Supplemental Figures 2D and 2E), confirming that the mutation in GA20ox2 was responsible for the delayed flowering of qem1. This result is consistent with the RNA-seq data showing that GA20ox2 mRNA is more highly expressed in the quintuple mutant background than in Col-0 (Supplemental Figure 1D and with the previous observation that svp-41 mutants contain higher levels of bioactive GAs than the wild type (Andrés et al., 2014). Therefore, the GA pathway likely plays a decisive role in promoting the floral transition in the quintuple mutant.
The qem2 mutant was later flowering and initiated more rosette and cauline leaves than the quintuple mutant (Figures 2A and 2B), indicating a delay in the floral transition and impaired floral meristem identity. The region of chromosome 5 to which qem2 mapped contained no previously described flowering-time genes (Figure 2C; Table 1). Three high-confidence polymorphisms predicted to cause non-synonymous mutations in the coding sequences At5g43450, At5g44690, and At5g44800 were identified (Table 1). At5g43450 encodes a protein with similarity to aminocyclopropane-1-carboxylate oxidase, At5g44690 encodes a protein of unknown function, and At5g44800 encodes the CHD3-like ATP-dependent chromatin-remodeling factor CHR4. In Arabidopsis, CHR4 is most closely related to PICKLE (PKL), which represses flowering via the GA pathway (Fu et al., 2016; Park et al., 2017) and promotes flowering via the photoperiodic pathway though FT activation (Jing et al., 2019a, 2019b). Both PKL and CHR4 are homologous to SWI/SWF nuclear-localized chromatin remodeling factors of the CHD3 family (Ogas et al., 1999), and CHR4 is also named PICKLE RELATED1 (Aichinger et al., 2009). The chr4 mutant shows no obvious mutant phenotype under standard growth conditions (Aichinger et al., 2009). However, CHR4 function has been implicated in floral organ development because it interacts with the MADS-domain transcription factors AGAMOUS (AG), AP3, PISTILLATA (PI), SEPALLATA3 (SEP3), and AP1, as revealed by immunoprecipitation of these factors (Smaczniak et al., 2012). Therefore, we hypothesized that the mutation in CHR4 caused the qem2 mutant phenotype. We tested this by introducing pCHR4:CHR4 and pCHR4:CHR4-VENUS constructs into qem2. The increased leaf number phenotype of qem2 was reduced to a similar number as in the progenitor quintuple mutant in all transformed lines (Figure 2D). Thus, we conclude that the later-flowering qem2 phenotype was caused by the mutation in CHR4.
Table 1. Candidate SNPs in qem2 Annotated in Genes.
| Chr | Pos | R | M | N | AF | Sh | Region | Gene ID | Type | AR | AM | Name |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 | 16,021,261 | C | T | 60 | 0.87 | 40 | CDS | At5g40010 | non-synonymous | G | S | ASD |
| 5 | 17,457,889 | C | T | 38 | 1 | 40 | CDS | At5g43450 | non-synonymous | D | N | — |
| 5 | 18,031,708 | G | A | 27 | 1 | 40 | CDS | At5g44690 | non-synonymous | R | STOP | — |
| 5 | 18,089,069 | G | A | 52 | 1 | 40 | CDS | At5g44800 | non-synonymous | A | V | CHR4 |
| 5 | 19,281,739 | G | A | 40 | 0.93 | 40 | CDS | At5g47530 | non-synonymous | G | E | — |
| 5 | 19,572,635 | G | A | 17 | 0.94 | 32 | 3′UTR | At5g48300 | — | — | — | ADG1 |
| 5 | 19,637,792 | G | A | 43 | 0.96 | 40 | CDS | At5g48460 | non-synonymous | A | V | ATFIM2 |
| 5 | 20,946,101 | G | A | 49 | 0.83 | 40 | CDS | At5g51560 | non-synonymous | G | S | — |
AF, allele frequency; AM, amino acid inqem2; AR, amino acid in the reference genome (svp flc ft tsf soc1); CDS, coding sequence; Chr, chromosome; Gen ID, gene identifier; M, nucleotide in qem2; N, number of reads supporting the mutation; Pos, position of the mutated nucleotide; R, nucleotide in the reference genome (svp flc ft tsf soc1); Region, region of the locus where the mutation was identified; Score (maximum 40); Sh, SHORE Score (maximum 40); Type, type of mutation (non-synonymous or synonymous); Dashes indicate no gene name.
Phenotypic Characterization of chr4 and Its Effects on Gene Expression during Floral Induction
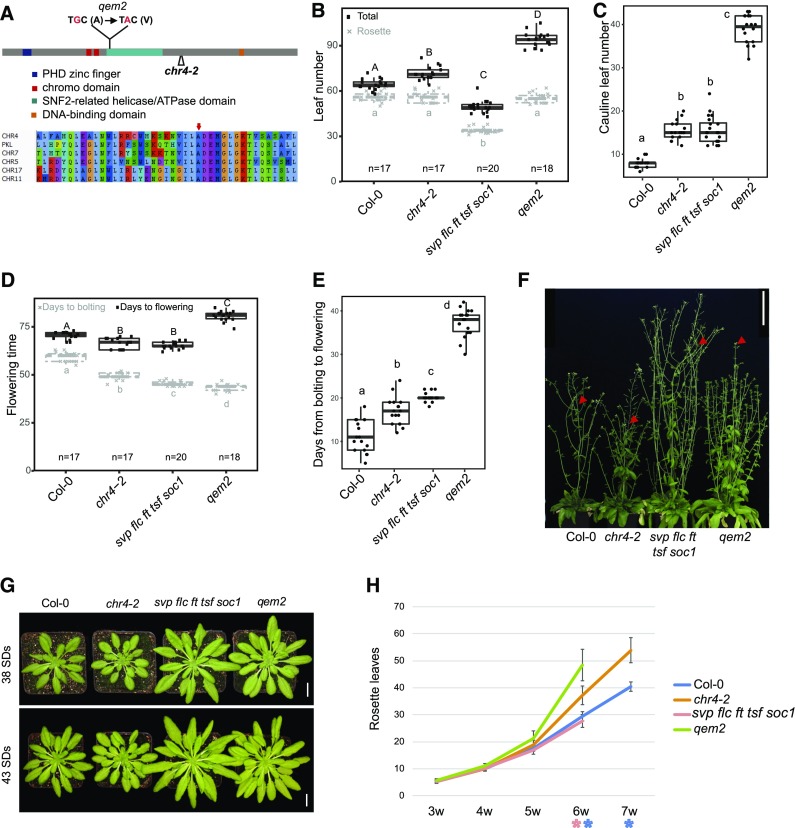
The qem2 mutant contains a mutation in the SNF2-related helicase/ATPase domain of CHR4, resulting in the substitution of a conserved alanine residue by valine (ala713val; Figure 3A). To analyze the chr4 mutant phenotype in the Col-0 background, we characterized the T-DNA insertion allele chr4-2 (SAIL_783_C05), containing a T-DNA insertion within the coding sequence between the helicase/ATPase domain and the DNA binding domain (Figure 3A). The T-DNA insertion also causes a reduction in CHR4 mRNA levels (Supplemental Figure 3).
Figure 3.
Characterization of CHR4.
(A) Schematic representation of the CHR4 locus showing the position of the mutation in qem2 and the T-DNA insertion site (chr4-2). The CHR4 protein domains are illustrated: A plant homeodomain (PHD) zinc finger (blue), a chromo domain (red), a SNF2-related helicase/ATPase domain (green), and a DNA binding domain (yellow). The EMS-induced protein sequence change is located within the SNF2-related helicase/ATPase domain.
(B to E) Leaf number (B), cauline leaf number (C), days to bolting and flowering (D), and number of days from bolting to flowering (E) of Col-0, chr4-2, svp flc ft tsf soc1, and qem2 plants grown under SDs. At least 17 plants were analyzed for each genotype. The data were compared with one-way ANOVA using Tukey’s honest significant difference as a post-hoc test. Different letters indicate significant differences (P ≤ 0.05). Whiskers represent a distance of 1.5 times the interquartile range.
(F) Twelve-week–old plants growing in SDs. Red arrows indicate first open flower. Scale bar = 10 cm.
(G) Rosettes of Col-0, chr4-2, svp flc ft tsf soc1, and qem2 plants after 38 d and 43 d of growth in SDs. Scale bar = 1 cm.
(H) Rosette leaf number of Col-0, chr4-2, svp flc ft tsf soc1, and qem2 plants grown under SDs from 3 weeks to 7 weeks. Eighteen plants were analyzed for each genotype. Error bars represent sd of the mean. The asterisks indicate significant differences (P-value < 0.05) between Col-0 and chr4-2 (blue) or svp flc ft tsf soc1 and qem2 (red).
We compared the leaf number, bolting time, and flowering time of qem2 and chr4-2 with those of their respective progenitors under LDs (Supplemental Figures 4A and 4B) and SDs (Figures 3B to 3E). The qem2 mutant formed ∼20 more rosette leaves and 30 more cauline leaves than the quintuple mutant under both LDs and SDs (Figures 3B and 3C; Supplemental Figures 4A and 4B). Despite having more rosette leaves, the bolting time of qem2 was similar to that of its progenitor (Figure 3D), whereas time to first open flower was markedly delayed in qem2 (Figures 3D and 3F), which is consistent with the increased number of cauline leaves. The phenotypic difference between Col-0 and chr4-2 was less severe than that between qem2 and the quintuple mutant. Under LDs, chr4-2 and Col-0 initiated a similar number of leaves (Supplemental Figure 4B). Under SDs, chr4-2 and Col-0 had a similar rosette leaf number, but chr4-2 bolted earlier and produced more cauline leaves (Figures 3B to 3E). CHR4 function appeared to be more important for flowering control in the quintuple mutant background, suggesting it might preferentially regulate flowering via the GA and aging pathways.
The chr4-2 and qem2 mutants bolted slightly earlier than their progenitors but initiated a similar number or more rosette leaves (Figures 3B and 3D), suggesting that they might have a shorter plastochron and initiate rosette leaves more rapidly. To determine the plastochron, we counted rosette leaves weekly until the plants bolted under SDs. Early in rosette development, chr4-2 and qem2 produced leaves at a similar rate as their progenitors, but later in rosette development, the mutants produced leaves more rapidly than the progenitors, leading to a steep increase in leaf number (Figures 3G and 3H). More rapid leaf initiation can be related to an enlarged SAM (Barton, 2010); therefore, we compared the SAMs of chr4-2 and qem2 to those of Col-0 and the quintuple mutant, respectively, after 4 and 5 weeks of growth under SDs (Supplemental Figure 5). The SAMs of plants carrying either chr4 mutant allele were larger than those of their progenitors, but this was most pronounced for qem2 compared with the quintuple mutant (Supplemental Figure 5).
The transition to flowering in Arabidopsis can be conceptualized as two sequential steps in which the inflorescence meristem acquires different identities. After the transition from a vegetative meristem, the inflorescence meristem (I1) initially forms cauline leaves and axillary branches, and after transition from I1 to I2, it initiates floral primordia (Ratcliffe et al., 1999). Rosette leaf number and days to bolting can be used as a proxy for the I1 transition, whereas the number of cauline leaves produced on the flowering stem and days to the first open flower indicate when the I1 to I2 transition occurs. Cauline leaves can be distinguished from rosette leaves due to their smaller size and more pointed shape, so that the increased number of leaves on the inflorescence stem can be explained by a delayed I2 transition rather than by enhanced internode elongation between rosette leaves. Compared with Col-0, chr4-2 is not delayed in the transition from vegetative meristem to I1 but is delayed in the transition from I1 to I2. By contrast, compared with the quintuple mutant, qem2 mutants were strongly delayed in both the transition to I1 and to I2 (Figures 3B to 3E).
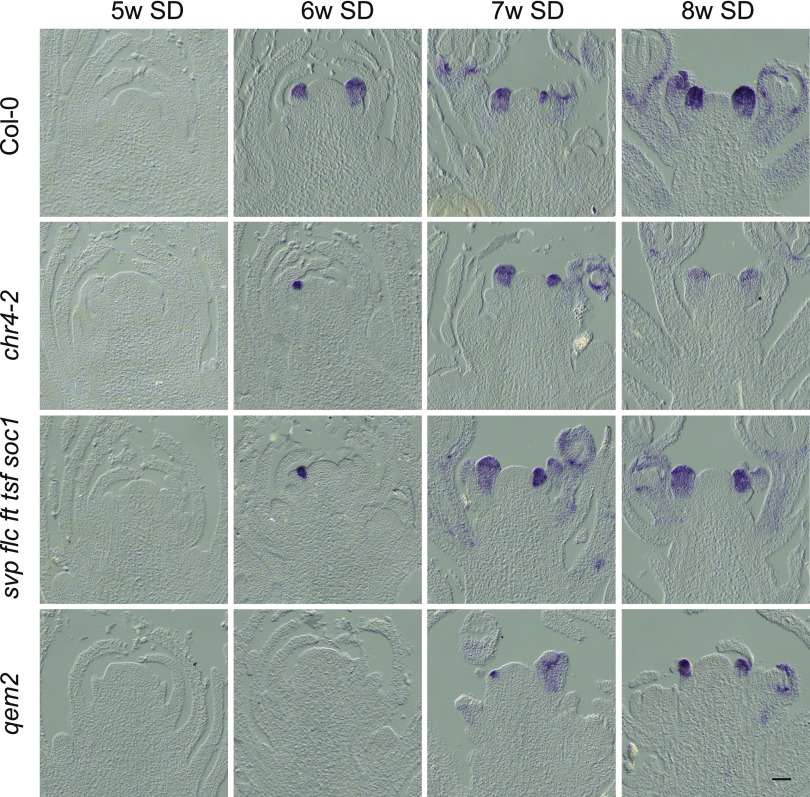
In Arabidopsis, AP1 confers floral meristem identity and is a marker for the I1 to I2 transition; therefore, we performed in situ hybridizations to monitor the appearance of AP1 mRNA through a developmental time course (Figure 4). At 5 weeks after germination, no AP1 expression was detected in any of the genotypes, indicating that the plant meristems were still vegetative. AP1 mRNA was detected at 6 weeks in Col-0 and chr4-2. In qem2 mutants, AP1 mRNA appeared more than 1 week later than in the quintuple mutant, which is consistent with observation that more cauline leaves formed in qem2 (Figure 4).
Figure 4.
Temporal and Spatial Patterns of Expression of the Floral Meristem Identity Gene AP1 in Col-0, chr4-2, svp flc ft tsf soc1, and qem2.
In situ hybridization analysis of AP1 mRNA accumulation in the SAMs of plants under SDs. The genotypes analyzed are shown together with the number of weeks (w) after germination when material was harvested. For each time point and genotype, three independent apices were examined with similar results. Scale bar = 50 μm.
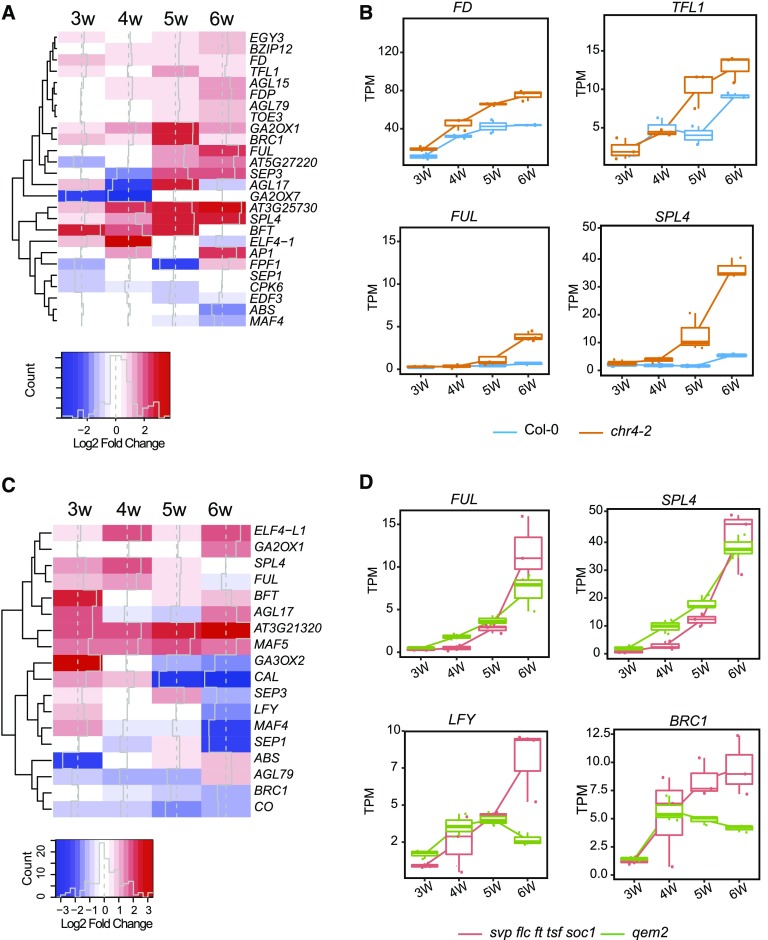
We then performed RNA-seq along a developmental time course to identify the genome-wide effects of CHR4 on gene expression during the floral transition. We examined the transcriptomes of shoot apices of Col-0, the quintuple mutant, chr4-2, and qem2 plants grown for 3, 4, 5, or 6 weeks under SDs and compared the chr4-2 and qem2 transcriptomes to those of Col-0 and the quintuple mutant, respectively (Supplemental Data Set 2). The analysis focused on 237 genes previously reported to regulate the floral transition in Arabidopsis (Bouché et al., 2016). In total, 26 of these genes were significantly differentially expressed (adjusted P-value < 0.05 and log2FC |1|) between chr4-2 and Col-0 (Figure 5A), and 18 were DEGs between qem2 and the quintuple mutant (Figure 5C). Nine genes were common to the two lists (AGL79, BRANCHED1, FUL, SEP3, AGL17, SPL4, BROTHER OF FT AND TFL1 [BFT], EARLY FLOWERING4 [ELF4], and MAF4). The expression of SPL4, which encodes a component of the age-dependent flowering pathway, increased at several time points in the chr4 and qem2 mutants compared with their respective progenitors (Figures 5A to 5D). In particular, SPL4 was most highly expressed in 4-week–old qem2 and in 5-week–old chr4-2 plants (Figures 5B to 5D). FUL was also more highly expressed in both mutants at later time points (Figures 5B and 5D) and is a direct target of SPL9, SPL15, and SPL3 during the floral transition (Wang et al., 2009; Yamaguchi et al., 2009; Hyun et al., 2016). Indeed, a corresponding small increase in mRNA levels of SPL9 and SPL15 was also observed in the CHR4 mutants (Supplemental Data Set 2). The earlier increase in expression of SPL4, SPL9, and SPL15 is consistent with the earlier bolting observed in the mutants, as qem2 bolted ∼2-d and chr4-2 ∼10-d earlier than their respective progenitors (Figure 3D).
Figure 5.
Transcriptional Changes in chr4 Mutants.
(A) Transcriptional profile comparisons represented as a heatmap to highlight genes implicated in flowering time control that are significantly upregulated (red) or downregulated (blue) in chr4-2 compared with wild type. Gene expression changes are represented as log2-fold change.
(B) Box plots from RNA-seq data showing FD, TFL1, FUL, and SPL4 transcript levels in apices of chr4-2 and Col-0 under SDs. The Y-axis shows TPM. The X-axis shows time of sampling as weeks after sowing. Whiskers represent distance from the lowest to the largest data point.
(C) Transcriptional profile comparisons represented as a heatmap to highlight genes implicated in flowering time control that are significantly upregulated (red) or downregulated (blue) in qem2 compared with svp flc ft tsf soc1.
(D) Box plots from RNA-seq data showing FUL, SPL4, LFY, and BRANCHED1 (BRC1) transcript levels shown as TPM in apices of qem2 and svp flc ft tsf soc1 under SDs. The Y-axis shows TPM. The X-axis shows time of sampling as weeks after sowing. Whiskers represent distance from the lowest to the largest data point.
We detected elevated expression of TERMINAL FLOWER1 (TFL1) in chr4-2 (Figures 5A and 5B) and BFT in qem2 (Figure 5C). Overexpression of TFL1 and BFT, which both encode proteins related to phosphatidylethanolamine binding proteins, reduces AP1 and LFY expression and delays floral organ initiation (Ratcliffe et al., 1998; Yoo et al., 2010). Consistent with this finding, LFY mRNA was also less abundant in qem2 (Figure 5D). During the inflorescence meristem transition from I1 to I2, increased LFY activity induces floral meristem identity by directly activating AP1 transcription and reducing GA levels, such that SPL9 recruits DELLA proteins to the regulatory region of AP1 (Weigel et al., 1992; Wagner et al., 1999; Yamaguchi et al., 2014). Therefore, in the absence of CHR4 function, attenuated LFY transcription likely contributes to a delay in the transition to the I2 phase, as reflected by the increased number of cauline leaves in qem2.
CHR4 Protein Localization In Planta and Identification of In Vivo Protein Interactors of CHR4
Chromatin remodelers are often recruited to target genes by specific transcription factors. Therefore, to further understand its mode of action during the floral transition, we identified proteins that interact with CHR4. We used the transgenic plants described above that express a fusion of VENUS fluorescent protein and CHR4 expressed from its native promoter (pCHR4:CHR4-VENUS). We analyzed the expression pattern of this CHR4-VENUS protein by confocal microscopy and compared it to the results of in situ hybridization analysis of CHR4 mRNA. CHR4-VENUS was localized to the nucleus and its spatial pattern was similar to the mRNA pattern detected by in situ hybridization in the SAM, floral organs, and young leaves (Supplemental Figure 6).
To identify protein interactors, we immunoprecipitated CHR4-VENUS protein from inflorescence tissue and 5-week–old SD apical-enriched tissue using anti-GFP antibodies and used p35S-YFP transgenic plants as a negative control. Proteins that specifically co-immunoprecipitated with CHR4-VENUS were identified by protein mass spectrometry (MS; see Methods). In total, 136 and 342 proteins were significantly (false discovery rate, FDR = 0.01) enriched in inflorescences and 5-week–old SD apex enriched tissue, respectively. The CHR4-interacting proteins in inflorescences included the floral homeotic MADS-domain transcription factors AP1, SEP3, PI, and AP3 (Table 2; Supplemental Data Set 3). The reciprocal experiment of immunoprecipitating AP1 was performed with gAP1:GFP plants and CHR4 was detected among the coimmunoprecipitated proteins (Supplemental Data Set 3 and Supplemental Figure 7). Taken together, these results confirm the previous finding that CHR4 could be co-immunoprecipitated with AGAMOUS, AP3, PI, SEP3, and AP1 (Smaczniak et al., 2012). Moreover, SEP1 and SEP2 were also found here to be interaction partners of CHR4 in inflorescence tissues (Table 2; Supplemental Data Set 3; Supplemental Figure 7). In addition to floral homeotic proteins, other MADS-domain proteins were found to interact with CHR4 in inflorescences, including AGL6 and the fruit- and ovule-specific protein SHATTERPROOF2 (SHP2; Favaro et al., 2003; Table 2; Supplemental Data Set 3; Supplemental Figure 7).
Table 2. List of CHR4 Interacting Proteins.
| Gene ID | Name | SAMs with Younger Leaves at 5w-SD-Stage | |||
|---|---|---|---|---|---|
| No. of Unique Peptides (IP1-IP2-IP3) | Sequence Coverage (%) (IP1-IP2-IP3) | Log2 Ratio | P-Value | ||
| AT5G44800 | CHR4 | 142 (128-130-114) | 59.6 (55.2-55.9-53.7) | 10.41 | 1.43e-05 |
| Transcription factors | |||||
| AT1G69120 | AP1 | — | — | — | — |
| AT5G20240 | PI | — | — | — | — |
| AT3G54340 | AP3 | — | — | — | — |
| AT5G15800 | SEP1 | — | — | — | — |
| AT3G02310 | SEP2 | — | — | — | — |
| AT2G45650 | AGL6 | — | — | — | — |
| AT2G42830 | SHP2 | — | — | — | — |
| AT3G13960 | GRF5 | 7 (6-5-4) | 18.1 (15.9-13.9-9.6) | 6.05 | 2.16e-04 |
| AT4G37740 | GRF2 | 6 (6-5-3) | 17.4 (17.4-15.3-9.5) | 5.19 | 1.90e-05 |
| AT5G43270 | SPL2 | — | — | — | — |
| AT1G02065 | SPL8 | — | — | — | — |
| AT1G27360 | SPL11 | — | — | — | — |
| AT5G50670 | SPL13 | 5 (5-4-2) | 19.2 (19.2-15-6.7) | 4.84 | 2.20e-03 |
| AT2G28550 | TOE1 | 5 (5-5-3) | 15.4 (15.4-15.4-8.7) | 4.01 | 1.89e-03 |
| AT3G02150 | TCP13 | 5 (4-4-3) | 18 (18-18-10.4) | 4.21 | 2.20e-02 |
| Chromatin remodeler | |||||
| AT2G46020 | BRM | 37 (30-31-16) | 24.2 (19.3-19.7-10.8) | 2.68 | 8.95e-04 |
| AT1G08600 | ATRX | 23 (19-22-6) | 13.9 (11.7-13.3-5.5) | 5.23 | 9.14e-05 |
| AT5G04240 | ELF6 | 10 (8-10-1) | 11.7 (7.8-11.7-0.8) | 3.09 | 2.20e-03 |
| AT2G28290 | SYD | 27 (21-21-15) | 11.1 (7.9-7.9-5.9) | 3.09 | 1.19e-03 |
| AT2G25170 | PKL | 19 (17-17-14) | 19.8 (16.7-17.4-14.8) | 2.71 | 5.48e-04 |
| AT3G12810 | PIE1 | 18 (14-15-9) | 11.5 (9.9-9.8-6.8) | 3.23 | 6.26e-03 |
| AT5G18620 | CHR17 | 17 (15-15-8) | 44.1 (42.4-42.4-25.3) | 2.85 | 5.34e-04 |
| AT3G06400 | CHR11 | 12 (11-10-10) | 45.1 (41.9-41.7-30.3) | 2.90 | 6.86e-04 |
| AT3G48430 | REF6 | 27 (22-25-18) | 23.4 (18.8-21-17) | 2.92 | 2.49e-03 |
| AT5G11530 | EMF1 | 10 (7-8-3) | 10.4 (6.9-8.3-3.5) | 5.12 | 1.12e-04 |
| AT2G06210 | ELF8 | 14 (12-13-9) | 15.9 (11.8-13.6-11.2) | 2.72 | 2.50e-03 |
| AT5G53430 | SDG29 | 5 (4-5-2) | 8 (7-8-4.5) | 4.40 | 8.19e-03 |
| AT4G02020 | SWN | 3 (2-1-2) | 4.8 (3-1.3-3) | 2.40 | 5.04e-03 |
| General transcriptional coregulators | |||||
| AT3G07780 | OBE1 | 14 (13-13-8) | 31.3 (29.7-31.3-20.8) | 6.47 | 7.02e-05 |
| AT5G48160 | OBE2 | 23 (21-17-9) | 41.5 (40.8-30.1-19.3) | 5.16 | 3.03e-03 |
| AT1G15750 | TPL | 12 (11-10-9) | 31.7 (27-26.9-24.4) | 3.93 | 3.17e-04 |
| AT1G80490 | TPR1 | 9 (8-7-6) | 25.4 (23.6-22.5-17.5) | 4.59 | 2.03e-03 |
| AT3G16830 | TPR2 | 8 (7-6-4) | 13.5 (12.6-10.2-5.1) | 3.57 | 1.99e-02 |
| AT2G32950 | COP1 | 7 (6-7-2) | 12.7 (11.7-12.7-4.3) | 3.99 | 4.02e-03 |
| AT2G46340 | SPA1 | 10 (7-10-3) | 13.2 (9.2-13.2-3.6) | 2.72 | 3.80e-02 |
| AT1G43850 | SEU | 12 (11-10-6) | 18.1 (16.9-12.9-9.9) | 3.69 | 2.49e-03 |
| Inflorescence under LDs | |||||
| AT5G44800 | CHR4 | 117 (114-99-114) | 51.4 (51.4-49.7-51.2) | 8.76 | 1.09e-04 |
| Transcription factors | |||||
| AT1G69120 | AP1 | 12 (8-3-7) | 34 (21.5-10.2-24.2) | 3.55 | 1.24e-02 |
| AT5G20240 | PI | 8 (6-3-8) | 31.7 (25.5-11.5-31.7) | 6.31 | 3.36e-03 |
| AT3G54340 | AP3 | 7 (7-5-7) | 31.5 (31.5-19-31.5) | 4.56 | 4.63e-02 |
| AT5G15800 | SEP1 | 2 (2-1-2) | 23.5 (23.5-17.1-21.9) | 4.06 | 2.63e-02 |
| AT3G02310 | SEP2 | 3 (2-1-2) | 32.8 (23.6-17.2-22) | 4.05 | 4.47e-03 |
| AT2G45650 | AGL6 | 3 (3-2-3) | 10.3 (10.3-10.3-10.3) | 3.78 | 9.16e-03 |
| AT2G42830 | SHP2 | 4 (4-3-4) | 29.3 (29.3-24-29.3) | 4.88 | 6.01e-03 |
| AT3G13960 | GRF5 | 4 (3-3-4) | 13.6 (9.1-9.1-13.6) | 3.62 | 3.28e-02 |
| AT4G37740 | GRF2 | 1 (1-1-1) | 3.2 (3.2-3.2-3.2) | 1.39 | 2.17e-01 |
| AT5G43270 | SPL2 | 4 (4-4-4) | 17.2 (17.2-17.2-17.2) | 5.30 | 3.32e-03 |
| AT1G02065 | SPL8 | 4 (4-2-4) | 18.3 (18.3-12-18.3) | 3.93 | 1.40e-02 |
| AT1G27360 | SPL11 | 8 (5-2-7) | 27 (17.8-10.2-21.9) | 5.57 | 1.53e-03 |
| AT5G50670 | SPL13 | 4 (3-1-4) | 13.9 (11.1-3.1-13.9) | 3.68 | 4.96e-03 |
| AT2G28550 | TOE1 | — | — | — | — |
| AT3G02150 | TCP13 | 6 (5-3-5) | 18.3 (15.5-10.1-15.5) | 3.96 | 5.85e-03 |
| Chromatin remodeler | |||||
| AT2G46020 | BRM | 24 (13-12-18) | 13 (8.6-8.4-10) | 2.57 | 2.36e-02 |
| AT1G08600 | ATRX | 28 (20-18-24) | 18.3 (13.8-13.3-16.3) | 4.64 | 2.59e-03 |
| AT5G04240 | ELF6 | 4 (4-1-4) | 5.8 (5.8-0.7-5.8) | 2.63 | 5.22e-02 |
| AT2G28290 | SYD | 21 (19-12-20) | 8 (7.6-4.6-8) | 3.70 | 4.76e-02 |
| AT2G25170 | PKL | 26 (23-18-24) | 23.6 (23-17.4-20.9) | 3.49 | 1.38e-02 |
| AT3G12810 | PIE1 | 7 (4-3-6) | 4.5 (3.2-2.6-4.1) | 1.89 | 1.04e-01 |
| AT5G18620 | CHR17 | 14 (11-13-12) | 37.4 (35.6-32.6-37.2) | 2.97 | 1.49e-02 |
| AT3G06400 | CHR11 | 17 (13-9-14) | 41.3 (39.2-32-39.1) | 2.34 | 9.72e-03 |
| AT3G48430 | REF6 | 33 (27-17-29) | 28.8 (24.8-12.9-24.5) | 2.21 | 1.39e-02 |
| AT5G11530 | EMF1 | 7 (6-3-7) | 8.5 (7.8-2.9-8.5) | 3.31 | 1.96e-02 |
| AT2G06210 | ELF8 | — | — | — | — |
| AT5G53430 | SDG29 | 7 (4-1-5) | 10.6 (7-2.5-7.2) | 1.99 | 3.08e-02 |
| AT4G02020 | SWN | 3 (2-2-2) | 4.2 (2.5-2.5-2.5) | 1.52 | 3.13e-02 |
| General transcriptional coregulators | |||||
| AT3G07780 | OBE1 | 12 (7-6-8) | 26 (17.8-14.7-20.7) | 4.12 | 3.47e-03 |
| AT5G48160 | OBE2 | 11 (9-5-11) | 26.3 (21.3-12.9-26.3) | 3.73 | 6.56e-03 |
| AT1G15750 | TPL | — | — | — | — |
| AT1G80490 | TPR1 | — | — | — | — |
| AT3G16830 | TPR2 | — | — | — | — |
| AT2G32950 | COP1 | 4 (3-3-3) | 6.7 (5.6-5.6-5.6) | 1.17 | 1.63e-01 |
| AT2G46340 | SPA1 | 3 (1-1-2) | 4.4 (1.4-1.4-2.6) | 1.75 | 9.65e-03 |
| AT1G43850 | SEU | 7 (6-2-6) | 9.8 (9.7-3.6-8.6) | 3.55 | 2.64e-03 |
Dashes indicate no data.
Other classes of transcription factors involved in the floral transition were identified in CHR4 complexes. Notably, SPL2, SPL8, and SPL11 were found to be interaction partners in inflorescences, whereas SPL13 was identified as a partner in inflorescences and enriched apices (Table 2; Supplemental Data Set 3; Supplemental Figure 7). Furthermore, TARGET OF EARLY ACTIVATION TAGGED1 (TOE1), an AP2-domain transcription factor that represses the floral transition (Aukerman and Sakai, 2003), also interacted with CHR4 in enriched apices. A further list of transcription factors and chromatin remodelers identified as CHR4 interactors is provided in Table 2 and Supplemental Data Set 3.
These experiments demonstrated that CHR4 associates in vivo with several transcription factors of the MADS, SPL, and AP2 classes that contribute to the floral transition and floral meristem identity.
Genome-wide Effects of CHR4 on Histone Modifications and Gene Expression
Proteins from the CHD3 group that includes CHR4 can participate in different chromatin remodeling pathways and either repress or activate gene expression, depending on the factors with which they associate. For example, PKL associates with genes enriched in trimethylation of histone H3 lysine 27 (H3K27me3), which is related to gene repression (Zhang et al., 2008, 2012), and maintains this epigenetic state (Carter et al., 2018). In addition, PKL reduces H3K27me3 at specific target genes in particular tissues and environments (Jing et al., 2013). Changes in H3K27me3 and H3K4me3 were also reported in the rice (Oryza sativa) mutant of a CHR4 homologue (Hu et al., 2012). To test whether CHR4 regulates gene expression by influencing histone modifications, we compared global H3K27me3 and H3K4me3 levels in Col-0 and chr4-2 plants (Supplemental Figure 8). No clear difference in the global frequency of these histone marks was observed between the two genotypes, suggesting that CHR4 does not affect the total accumulation of these histone modifications.
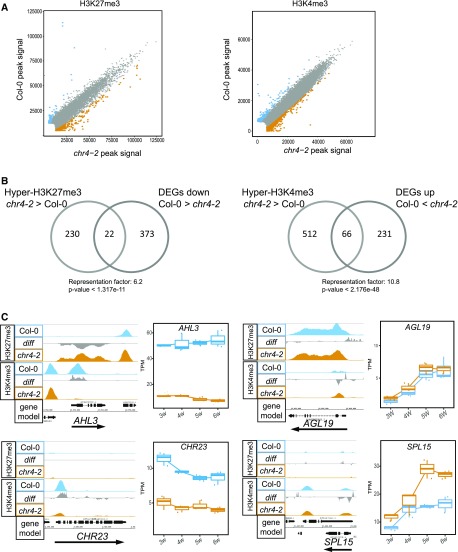
To test whether CHR4 affects the deposition of these histone marks at specific loci, we performed chromatin immunoprecipitation sequencing (ChIP-seq) to compare genome-wide H3K27me3 and H3K4me3 levels in Col-0 and chr4-2. H3K27me3 and H3K4me3 ChIP-seq experiments were performed on three biological replicates for each genotype (see Methods). In total, 10,194 H3K27me3-marked regions and 15,992 H3K4me3-marked regions were identified in the two genotypes (Supplemental Data Set 4). Quantitative comparison with DANPOS2 (Chen et al., 2013) revealed a subset of regions with significant differences (FDR < 0.05) in H3K27me3 or H3K4me3 levels between Col-0 and chr4-2. In total, 857 regions were differentially marked with H3K27me3 and 1,032 regions were differentially marked with H3K4me3 (Supplemental Data Set 4). Notably, hypermethylated as well as hypomethylated regions were identified in chr4-2 (Figure 6A). The genes differentially marked with H3K27me3 included regulators of key hormonal pathways involved in the floral transition, such as GIBBERELLIN3-OXIDASE1 and GIBBERELLIN3-OXIDASE4, which encode GA biosynthesis enzymes. Genes encoding components of auxin signaling (ETTIN and AUXIN RESISTANT1) and an enzyme that catabolizes cytokinin (CYTOKININ OXIDASE5) were also differentially marked with H3K27me3 in chr4-2 (Supplemental Data Set 4). Genes differentially marked with H3K4me3 included the regulators of the floral transition SPL15, FLORAL TRANSITION AT THE MERISTEM1 (Torti et al., 2012), and JUMONJI DOMAIN-CONTAINING PROTEIN30 (Jones et al., 2010; Yan et al., 2014; Supplemental Data Set 4). In addition, 39 genes differentially marked by both H3K27me3 and H3K4me3 were detected, including the flowering-time regulators miR156D and AGL19 (Figure 6C; Supplemental Data Set 4).
Figure 6.
Histone Modification Variation in chr4-2.
(A) Scatterplots showing H3K27me3 and H3K4me3 enrichment between Col-0 and chr4-2 in apices of 5-week–old plants grown under SDs. Blue and orange dots represent significantly more highly methylated regions at FDR = 0.05 in Col-0 and chr4-2, respectively.
(B) Venn diagram showing the overlap between DEGs and genes differentially marked by H3K27me3 and H3K4me3.
(C) H3K27me3 and H3K4me3 profiles and expression of AHL3, AGL19, CHR23, and SPL15.
We also examined the extent to which the differentially marked genes were also differentially expressed. H3K27me3 is associated with gene repression, and therefore, genes with higher H3K27me3 levels in chr4-2 compared with Col-0 were expected to be expressed at lower levels in chr4-2 than in Col-0. Indeed, a significant overrepresentation (Representation factor: 6.2, P-value < 1.317e-11) of downregulated genes was observed among those marked with increased levels of H3K27me3 in chr4-2 (Figure 6B). Among the downregulated and hypermethylated genes in chr4-2 was AHL3, encoding an AT-hook protein that regulates vascular tissue boundaries in roots (Figure 6C; Zhou et al., 2013). By contrast, H3K4me3 is associated with gene activation and therefore, genes marked with higher H3K4me3 levels in chr4-2 compared with Col-0 were expected to be expressed at higher levels. Indeed, a significant overrepresentation (Representation factor: 10.8, P-value < 2.176e-48) of upregulated genes between those marked with higher levels of H3K4me3 was observed (Figure 6B). Among the upregulated and hypermethylated genes in chr4-2 are CHR23, which is involved in stem-cell maintenance at the SAM (Sang et al., 2012) and SPL15, a promoter of the floral transition at the shoot meristem (Figure 6C; Hyun et al., 2016). Moreover, spl15 produced fewer cauline leaves than the wild type (Schwarz et al., 2008), indicating a premature transition to the I2 phase of flower initiation. On the other hand, plants expressing a miR156-resistant transcript of SPL15, which leads to an increase in SPL15 protein accumulation, produced more cauline leaves than the wild type (Hyun et al., 2016), indicating a delay in the transition to the I2 phase of flower initiation, as observed in qem2 mutants.
In conclusion, CHR4 affects H3K27me3 and H3K4me3 levels at a subset of loci in the genome, and changes in both histone modifications in chr4-2 are significantly correlated with changes in gene expression. Notably, a significant increase in H3K4me3 was detected at the SPL15 locus, and a higher level of SPL15 mRNA was found in chr4-2; these findings are consistent with the premature bolting and delay in the transition to the I2 phase of flower initiation observed in chr4-2.
DISCUSSION
We performed an enhanced genetic screen to identify regulators of the floral transition, and in particular, to focus on endogenous flowering pathways at the shoot meristem. To this end, we generated a quintuple mutant background strongly impaired in floral responses to environmental stimuli. Mutagenesis of these plants identified a chromatin remodeler, CHR4, which plays important roles in the floral transition, especially in response to endogenous flowering pathways and during the transition from forming cauline leaves with axillary branches (I1) to forming floral primordia (I2).
The Quintuple Mutant Is Strongly Impaired in Environmental Flowering Responses and Flowers via Endogenous Pathways
The quintuple mutant showed strongly reduced flowering responses to long photoperiods and high ambient temperature. This insensitivity is consistent with the loss of function of FT and TSF, which confer photoperiodic responses, and the loss of function of FT, TSF, and SVP, which are involved in responses to high ambient temperature (Yamaguchi et al., 2005; Kumar et al., 2012; Fernández et al., 2016). Therefore, the floral transition in the quintuple mutant is likely promoted by endogenous flowering pathways. In support of this conclusion, RNA-seq analysis detected higher mRNA levels of several SPL genes in the mutant versus Col-0. Some of these genes, such as SPL15 and SPL4, are negatively regulated by miR156, which decreases in abundance as plants proceed from the juvenile to the adult phase (Wu and Poethig, 2006; Gandikota et al., 2007; Hyun et al., 2016). Therefore, these SPLs were previously considered to be components of an age-related flowering pathway (Wang et al., 2009; Hyun et al., 2017). However, the mRNA of SPL8, which is not regulated by miR156 but has overlapping functions with the miR156-targeted SPL genes (Xing et al., 2010), also increased in abundance in the quintuple mutant, suggesting a broader deregulation of this class of transcription factors in this genetic background.
Transcriptome profiling of the quintuple mutant also detected differential expression of genes encoding enzymes involved in GA biosynthesis, such as GA20ox2. Higher GA20ox2 mRNA expression was detected in the quintuple mutant compared with Col-0 under SDs. The accumulation of GA4 under SDs in Col-0 plants coincides with the floral transition and increased abundance of the mRNAs of floral meristem identity genes such as LFY (Eriksson et al., 2006). Although the GA biosynthesis pathway is complex and includes many enzymatic steps (Yamaguchi, 2008), GA20ox2 appears to be important for controlling the floral transition, especially under SDs (Rieu et al., 2008; Plackett et al., 2012; Andrés et al., 2014). SVP reduces GA20ox2 transcript levels and GA levels at the shoot apex as part of the mechanism by which it represses flowering (Andrés et al., 2014). We therefore propose that increased GA20ox2 transcription in the quintuple mutant contributes to its higher GA levels and earlier floral transition under SDs. In support of this notion, the qem1 mutation was found to be an allele of GA20ox2 and to delay flowering of the quintuple mutant.
The proposed role for SPLs and GA in causing early flowering of the quintuple mutant is consistent with the previous finding that SPL proteins mediate some of the effects of GA during reproductive development (Porri et al., 2012; Yu et al., 2012; Yamaguchi et al., 2014; Hyun et al., 2016) and that SPL8 regulates several GA-mediated developmental processes (Zhang et al., 2007). Furthermore, SPL9 and SPL15 interact with DELLA proteins, which are negative regulators of GA responses that are degraded in the presence of GA (Davière and Achard, 2013). SPL15 promotes the transcription of target genes that induce flowering, such as FUL and miR172b, and activation of these genes by SPL15 is repressed by interaction with DELLAs (Hyun et al., 2016). In Col-0, the role of SPL15 in flowering is particularly important under SDs, when floral induction occurs independently of environmental cues and is dependent on endogenous processes such as the GA pathway (Hyun et al., 2019). By contrast, the DELLA-SPL9 interaction can negatively or positively affect transcription, depending on the target genes and the developmental context (Yamaguchi et al., 2009; Yu et al., 2012). Taken together, these results demonstrate that the floral transition in the sensitized quintuple mutant background involves the interdependent functions of GA and SPL proteins.
A Chromatin Remodeler Was Identified as a Regulator of the Floral Transition in the Sensitized Screen
The genetic framework for flowering-time control in Arabidopsis is based on analysis of late-flowering mutants identified after mutagenesis of early-flowering accessions (Koornneef et al., 1998). However, important regulators were not identified in these screens, but were readily found as early-flowering mutants from mutagenizing late-flowering lines (Michaels and Amasino, 1999) or as late-flowering suppressor mutants after mutagenesis of transgenic plants or mutants requiring vernalization (Chandler et al., 1996; Onouchi et al., 2000). Here, we extended this approach by mutagenizing a quintuple mutant background that flowered almost independently of environmental cues. Until recently, the molecular characterization of mutations isolated in such complex backgrounds using classical genetic approaches would have been extremely time-consuming and laborious, but this process has been simplified by the implementation of bulk-segregant analysis after backcrossing the mutant to the progenitor followed by whole-genome resequencing (Abe et al., 2012; Hartwig et al., 2012; Schneeberger, 2014).
The second characterized mutation identified in the quintuple mutant background, qem2, is an allele of CHR4. This gene encodes a chromatin remodeler that was previously identified as a member of protein complexes that include AP1 and other MADS-box transcription factors (Smaczniak et al., 2012), but its role in flowering had not been demonstrated genetically. Nevertheless, several chromatin modifiers and remodelers contribute to the regulation of the floral transition (Farrona et al., 2008), such as BRAHMA (BRM), a member of the SWI/SNF complex involved in nucleosome sliding and/or eviction, and the H3K27me3-specific histone demethylase RELATIVE OF EARLY FLOWERING6 (REF6), which acts cooperatively with BRM to regulate gene expression during floral development (Farrona et al., 2004; Lu et al., 2011; Wu et al., 2012; Li et al., 2016; Richter et al., 2019). Also, the SWI2/SNF2-RELATED1 complex protein PHOTOPERIOD-INSENSITIVE EARLY FLOWERING1 (PIE1) is involved in H2A.Z deposition and delays the floral transition (Noh and Amasino, 2003; March-Díaz et al., 2008; Coleman-Derr and Zilberman, 2012). Interestingly, PKL and PIE1 were previously proposed to act in the same pathway to define and maintain genomic domains with elevated H3K27me3 levels, suggesting that CHR4 may contribute at different levels within this process (Carter et al., 2018). Taken together, MS identified several proteins in association with CHR4 that are involved in regulating histone modifications as well as multiple transcription factors with specific roles in floral meristem identity or the floral transition, suggesting that CHR4 functions in different multimeric complexes that regulate flowering.
CHR4 Affects the Expression of Flowering Genes by Modulating H3K4me3 and H3K27me3 Levels and Affects Different Stages of the Floral Transition
The most closely related protein to CHR4 is another CHD3-like family member, PKL, which orchestrates deposition of H3K27me3 and facilitates nucleosome retention (Zhang et al., 2008, 2012; Jing et al., 2013; Carter et al., 2018). In rice, loss of function of the CHR4 homologue CHR729 results in changes in the abundance of H3K27me3 and H3K4me3 at ∼56% and 23%, respectively, of loci marked by these modifications (Hu et al., 2012). Similarly, we observed variation in H3K27me3 or H3K4me3 levels at a subset of loci marked by these modifications in chr4-2, indicating a conserved function between rice and Arabidopsis. Notably, we observed higher levels of H3K4me3 at the SPL15 locus in chr4-2 versus the wild type.
The floral transition is considered to be a dual-step process: In the first step, the inflorescence meristem produces cauline leaves and axillary branches (I1); and in the second step, it forms floral primordia (I2; Ratcliffe et al., 1999). Detailed phenotypic analysis of chr4 mutants showed that CHR4 affects both these phases but with opposite effects. The chr4 mutation accelerates the transition from the vegetative meristem to I1 but delays the I1 to I2 transition. The premature transition to I1 was reflected by earlier bolting, and this correlated with increased abundance of SPL15, SPL4, and FUL mRNA expression. These genes are associated with early bolting and flowering, and SPL15 in particular caused premature bolting when its expression was increased by mutations that rendered its mRNA insensitive to miR156 (Hyun et al., 2016). SPL15 also promotes the meristematic transition from vegetative to inflorescence meristem (Hyun et al., 2016). Moreover, spl15 mutants produced fewer cauline leaves than the wild type (Schwarz et al., 2008), whereas rSPL15 transgenic plants produced more cauline leaves (Hyun et al., 2016), indicating that SPL15 extends the I1 phase. We propose that the higher expression of SPL15 in chr4 promotes earlier bolting and extends the I1 phase. This increased activity of SPL15 could also be enhanced in chr4 by increased activity of the GA biosynthetic pathway, as the resulting reduction in DELLA activity would be predicted to allow SPL15 to more effectively activate transcription of its target genes, leading to premature bolting and more cauline leaves.
Mutant chr4 plants also produced more cauline leaves and required more time to open the first flowers than their progenitors, indicating a delay in the I2 transition. These mutants also exhibited higher levels of TFL1 and BFT mRNAs; the overexpression of these genes delays the I2 transition by repressing AP1 and LFY expression (Ratcliffe et al., 1998; Yoo et al., 2010). Consistent with this conclusion, the onset of AP1 transcription occurred later in qem2 than in the quintuple mutant progenitor, and LFY mRNA was less abundant in qem2 than in the quintuple mutant in the RNA-seq time-course at week 6 in SDs. The chr4 mutant phenotype is strongly enhanced in the quintuple mutant background, probably explaining why chr4 was recovered in the sensitized mutant screen but was not previously identified by mutagenesis of Col-0 plants, where it exhibited a strong effect only under SDs. We propose that CHR4 contributes to the floral transition in response to GA signaling and that the increased dependency of the quintuple mutant on the GA pathway to promote flowering increases the impact of CHR4 loss of function on the floral transition. Similarly, the stronger phenotype of chr4-2 in Col-0 under SDs than LDs is consistent with a specific role in the floral transition mediated by GA.
In conclusion, the combination of forward genetics and functional gene characterization identified CHR4 as a regulator of different stages of the floral transition. Immunoprecipitation of CHR4 suggested that it acts in distinct protein complexes that contain different transcription factors as well as other CHR proteins. The contribution of CHR4 within distinct complexes presumably explains its pleiotropic effects, even during flowering, where it affects both bolting and floral identity during the transition from I1 to I2. Our genome-wide analyses represent the first step in understanding the mechanism through which CHR4 affects these phenotypes by identifying genes whose expression is altered by H3K27me3 or H3K4me3 in chr4 mutants. Further studies are now required to link the specific protein complexes in which CHR4 contributes to histone changes on defined targets. Attempts to perform ChIP-seq on pCHR4:CHR4-VENUS lines did not succeed, but pursuing this approach in the future would define the genome-wide sites with which CHR4 associates and help define its effects on the histone marks at direct target genes. Such approaches would help determine the mechanisms by which CHR4 regulates gene expression and allow this mechanism to be compared with that of PKL, which cooperates with PIE1 and CLF at target genes to maintain elevated H3K27me3 levels (Carter et al., 2018).
METHODS
Plant Materials, Growth Conditions, and Phenotypic Analysis
For all studies, Arabidopsis (Arabidopsis thaliana) Columbia (Col-0) ecotype was used as the wild type. To construct the svp-41 flc-3 ft-10 tsf-1 soc1-2 quintuple mutant, svp-41 flc-3 FRI plants (Mateos et al., 2015) were first crossed to svp-41 ft-10 tsf-1 soc1-2 ful-2 plants (Andrés et al., 2014). The F1 plants were self-fertilized and the F2 progeny were genotyped for each mutation except ful-2, which was scored phenotypically. Approximately 1,000 F2 plants were grown in soil under LD conditions and DNA was extracted from those that flowered later than Col-0. Genotyping was performed to identify plants that carried all mutations, lacked the FRI introgression, and were homozygous for FUL in the F3 generation. chr4-2 corresponds to SAIL_783_C05. Homozygous mutant plants were selected by PCR using specific primers (Supplemental Data Set 5).
Seeds were immersed in 0.1% melt universal agarose (Bio-Budget Technologies) for 3 d at 4°C in darkness for stratification. Plants were grown in soil under controlled conditions of LDs (16-h light/8-h dark) and SDs (8-h light/16-h dark) at 21°C or 27°C. The light intensity was 150 μmol⋅m−2⋅s−1 under all conditions. The growth-chamber is equipped with fluorescent tube bulbs (F17T8/TL841 ALTO-T8; Philips) to supply wavelengths from 430 to 650 nm, and supplemented with LEDs to provide light in the far-red spectrum. As a proxy for flowering time, the number of rosette and cauline leaves on the main shoot was counted as well as the number of days to bolting and first flower opening.
Ethylmethanesulfonate Treatment of Seeds
For ethylmethanesulfonate (EMS) treatment, 200 mg (∼10,000) seeds of the quintuple mutant were wrapped in Miracloth (EMD Millipore) and immersed in 0.1% (v/v) KCl solution on a shaker at 4°C for 14 h. The seeds were washed with double distilled water and treated with 100 mL of 30-mM EMS diluted in double distilled water on a magnetic stirrer in a fume hood overnight (8 h to 9 h). The seeds were washed twice with 100 mL of 100-mM sodium thiosulfate for 15 min and three times with 500 mL of double distilled water for 30 min. After washing, the seeds were immersed in 2 L of 1% (w/v) agarose. Approximately 50 seeds in 10 mL of agarose were sown as the M1 generation in 9 × 9 cm pots using plastic pipettes. The M1 plants were grown and self-fertilized, and seeds were harvested in bulks of 50 M1 plants. One-hundred-and-forty-six M2 bulked families were screened for plants showing altered flowering time.
GA Treatment
The GA4 stock (cat. no. G7276-5MG; Sigma-Aldrich) was prepared in 100% ethanol with a final concentration of 1 mM. GA treatments were performed by spraying 2-week–old plants under SDs with either a GA solution (10 μM of GA4 and 0.02% [v/v] Silwet 77; Loveland Industries) or a mock solution (1% [v/v] ethanol and 0.02% [v/v] Silwet 77). Spraying was performed twice weekly until the plants bolted.
Selection of Mutants and Sequencing
Approximately 10 M2 generation seeds from each M1 plant were sown. Screening for potential mutants was initially performed under LD greenhouse conditions, and all plants were grown together with the quintuple mutant and Col-0 plants as a reference. Individuals that flowered later or earlier than the quintuple mutant in the M2 population were selected. These M2 putative mutants were self-fertilized and rescreened in the M3 generation. Approximately 24 M3 progeny of each potential mutant were grown under the same conditions to test the heritability of the phenotype. M3 plants were backcrossed to the quintuple mutants to generate BC1F1 seeds. The BC1F2 offspring of such a cross formed the isogenic mapping population. Approximately 70 plants showing the mutant phenotype were selected from a population of ∼300 BC1F2 plants. One leaf sample of each selected plant was harvested and pooled. Leaf material from the quintuple plants was also harvested as a control. Genomic DNA was extracted from both pools and sent for Illumina sequencing with a depth of ∼80-fold coverage. Reads were aligned to The Arabidopsis Information Resource (TAIR10) reference genome (ftp://ftp.arabidopsis.org/home/tair) using the software tool SHORE (Schneeberger et al., 2009). The program SHOREmap (Schneeberger et al., 2009; Sun and Schneeberger, 2015) was used to identify polymorphisms, and those present in ∼100% of reads in the identified mutant but absent from the progenitor were identified as candidates for the causal mutation.
In Situ Hybridization
In situ hybridization was performed as described in Bradley et al. (1993), with minor modifications. Instead of Pronase, proteinase K (1 mg/mL in 100 mM of Tris at pH 8, and 50 mM of EDTA) was used for protease treatment by incubating at 37°C for 30 min. Post-hybridization washes were performed in 0.1× saline sodium citrate instead of the original 2× saline sodium citrate with 50% (w/v) formamide. The sequences of primers used to generate the probes are listed in Supplemental Data Set 5. For each genotype and time point, three independent apices were analyzed.
RNA Extraction and RNA-Seq Analysis
Total RNA was extracted from 15 shoot apices after removing all visible leaves under a binocular for each of the three independent biological replicates using an RNeasy Plant Mini Kit (Qiagen) and treated with DNase (Ambion) to remove residual genomic DNA. Library for sequencing was prepared using a TruSeq Library Preparation Kit (Illumina) according to the manufacturer’s protocol. Sequencing was performed using the HiSeq3000 platform (Illumina) in 150-bp single reads. For each sample, ∼15,000,000 reads were generated. The software tool FastQC was used to assess quality control parameters (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). To estimate expression levels, the RNA-seq reads were mapped to the Arabidopsis TAIR10 (Lamesch et al., 2012) reference genome using the software TopHat2 under default settings (Kim et al., 2013), except that only a single alignment was permitted per read and the coverage-based junction search was disabled (settings: −g 1 –no-coverage-search). The program Samtools was used to sort and index BAM alignment files and to calculate BAM file statistics (Li et al., 2009). The software HTSeq was used to tabulate the number of reads mapping to each genomic feature, with counts tabulated only for genes that completely overlapped a given feature (Anders et al., 2015). We used the Wald test implemented in the program DESeq2 (https://bioconductor.org/packages/release/bioc/html/DESeq2.html) to detect DEGs for pair-wised comparison. To visualize the expression levels of candidate genes, the expression level for each gene was calculated as transcripts per million (TPM).
ChIP-Seq Experiment and Data Analysis
Three independent biological replicates for each genotype were generated. For each sample, 1 g of plant material was used per biological replicate. Material was collected from plants grown in SD at 21°C for 5 weeks (5 h to 6 h after lights on). Using jeweler’s forceps, leaves with elongated petioles were removed to obtain SAM-enriched tissues. ChIP experiments were performed following a protocol from Kaufmann et al. (2010) with minor modifications. Samples were sonicated in a water bath Bioruptor (Diagenode) four times for 5 min each of 15 s on and 15 s off, with a 1-min incubation between each sonication treatment. After the preclearing step, the sample was split into three aliquots: The first aliquot was incubated with anti-H3K27me3 antibody (cat. no. 39155, lot no. 25,812,014; Active Motif), the second one was incubated with anti-H3K4me3 antibody (cat. no. 17–614, lot no. 1973237; Millipore), and the third one with anti-H3 antibody (cat. no. ab1791; Abcam). Samples were prepared for Illumina sequencing using the Ovation Ultralow V2 DNA-Seq Library Preparation Kit (Tecan Genomics) according to the manufacturer’s protocol. H3K27me3 and H3K4me3 enrichment was tested by ChIP-quantitative PCR before and after library preparation.
Libraries were analyzed on the Bioanalyzer (Agilent Technologies) and quantified with the Qubit Fluorometric Quantification (Invitrogen ) before sequencing on the HiSeq3000 (Illumina). Samples were sequenced in a 150-bp single reads’ run.
FASTQ files were mapped to the Arabidopsis genome TAIR10 using the software BowTie (Langmead et al., 2009) with default parameters. Clonal reads were removed using a customized Python (https://www.python.org/) script. Reproducibility between biological replicates was assessed using the Spearman correlation for the genome-wide read distribution at each pair of replicates using the software deepTools (Ramírez et al., 2014). The “multiBamSummary” function was used with default parameters except for “bin size,” which was set to 1 kb and the “plotCorrelation” function of deepTools2 in Galaxy (http://deeptools.ie-freiburg.mpg.de/; Supplemental Figure 9). H3K27me3- and H3K4me3-modified regions were identified with the tool DANPOS2 (Chen et al., 2013). The “Dpeak” function in DANPOS2 was used with default parameters, except for the parameter −l (read extension length), which was set to 300 bp, the mean size of the DNA in the samples after sonification. Genomic regions were associated with genes if located within the start and the end of the gene using a customized Python script.
Plasmid Construction
Cloning of the CHR4 locus was performed based on polymerase incomplete primer extension (Klock and Lesley, 2009) with modifications for large fragments and multiple inserts. All PCR amplifications were performed with Phusion Enzyme (New England BioLabs) following the manufacturer’s recommendations. The constructs pCHR4:CHR4-pDONR207 (18.4 kb) and pCHR4:CHR4:9AV-pDONR207 (19 kb) were generated as follows: Primers Q810 and Q811 were used to amplify the CHR4 promoter (3.6 kb) and the PCR products were cloned into pDONR207 by BP reaction to generate the pCHR4-pDONR207 construct. The primer pairs Q058 and Q814, and Q815 and Q816 were used to amplify a fragment containing 9xala-VENUS (9AV; 0.7 kb) and the 3′UTR of CHR4 (3.8 kb), respectively. Overlap PCR with primers Q058 and Q816 was performed to fuse the amplicons. The primers Q817 and Q818 were used to linearize the construct pCHR4-pDONR207. The amplicons were mixed with linearized pCHR4-pDONR207 to construct the plasmid pCHR4:9AV:3′URTCHR4-pDONR207. The obtained plasmid was linearized with primers Q835 and Q836 and mixed with the coding sequence of CHR4 (8.5 kb) amplified with primers Q819 and Q820 to construct the plasmid pCHR4:CHR4:9AV-pDONR207 (called pCHR4:CHR4-VENUS in the text). All primers used for molecular cloning are listed in Supplemental Data Set 5. Subsequently, the plasmids were cloned into the binary vector pEarleyGate301 (Earley et al., 2006) by LR reaction and transformed into Escherichia coli DH5-α-cells before being transformed into Agrobacterium tumefaciens GV3101 cells (Van Larebeke et al., 1974).
Plant Transformation and Selection
Plants (Col-0 and svp flc ft tsf soc1) were transformed by the floral-dip method (Clough and Bent, 1998). Transformants were selected by spraying twice with Basta (Bayer Crop Science). The progenies were grown on plates with 1× Murashige and Skoog medium (Murashige and Skoog, 1962) containing Suc and 10 μg mL−1 of phosphinotricin (PPT) to test for segregation and to select for single locus insertion lines and homozygosity in the following generations. Alternatively, the nondestructive PPT leaf assay was used to assess resistance to PPT. One young leaf per plant was harvested and placed on a plate with 1× Murashige and Skoog without Suc with 10 μg mL−1 of PPT. The plates were incubated for 4 d.
Confocal Microscopic Analyses
To visualize VENUS expression in shoot meristems, the method of Kurihara et al. (2015) was used with minor modifications. Shoot apices were collected and placed in ice-cold 4% paraformaldehyde (Sigma-Aldrich) prepared in phosphate-buffered saline at pH 7.0. The samples were vacuum-infiltrated twice for 10 min each time, transferred to fresh 4% (v/v) paraformaldehyde, and stored at 4°C overnight. The next day, the samples were washed in phosphate-buffered saline twice for 10 min each and cleared with ClearSee (10% [w/v] xylitol, 15% [w/v] sodium deoxycholate, and 25% [w/v] urea; Kurihara et al., 2015) at room temperature for ∼1 week. The samples were then transferred to fresh ClearSee solution with 0.1% (v/v) SCRI Renaissance 2200 (Renaissance Chemicals) and incubated in the dark overnight. The shoot meristems were imaged by confocal laser scanning microscopy (LSM780; Zeiss) using settings optimized to visualize VENUS fluorescent proteins (laser wavelength, 514 nm; detection wavelength, 517 nm to 569 nm) and Renaissance 2200 (laser wavelength, 405 nm; detection wavelength, 410 nm to 510 nm).
Sample Preparation and Liquid Chromatography with Tandem MS Data Acquisition
Three independent biological replicates for each genotype (gCHR4-VENUS and p35S-YFP), each consisting of 1 g of plant material, were generated. For inflorescence tissues, plants were grown in LD at 21°C, whereas SAM-enriched tissue samples were collected from plants growing in SD at 21°C for 5 weeks (5 h to 6 h after lights on). Using jeweler’s forceps, leaves with elongated petioles were removed to obtain SAM-enriched tissues. Nuclei were isolated according to a protocol by Kaufmann et al. (2010). Samples were sonicated in a Bioruptor (Diagenode) water bath four times, 5 min each of 15 s on and 15 s off, with a 1-min incubation between each sonication treatment. Sonicated samples were centrifuged twice at 4°C for 10 min. The supernatants were transferred to a clean tube. After adding 40 μL of GFP-trap Agarose beads (gta-20; Chromotek) and 10 μL of Benzonase, the samples were incubated at 4°C for 2 h. After incubation, the GFP-trap beads were washed four times with 1 mL of wash buffer (750 μL 5 M NaCl and 1.25 mL of Tris-HCl at pH 7.4, in 25 mL of water). Immunoprecipitated samples enriched with GFP-trap beads were submitted to on-bead digestion. In brief, dry beads were redissolved in 25 μL of digestion buffer 1 (50 mM of Tris at pH 7.5, 2 M of urea, 1 mM of DTT, and 5 μg μL−1 trypsin) and incubated for 30 min at 30°C in a Thermomixer (Eppendorf) with 400 rpm. Next, the beads were pelleted, and the supernatant was transferred to a fresh tube. Digestion buffer 2 (50 mM of Tris at pH 7.5, 2 M of urea, and 5 mM of chloroacetamide) was added to the beads. After mixing and centrifugation, the supernatant was collected and combined with the previous one. The combined supernatants were incubated overnight in the dark at 32°C in a Thermomixer (Eppendorf) at 400 rpm. The digestion was stopped by adding 1 μL of trifluoroacetic acid and the samples were desalted with C18 Empore disk membranes (3M) according to the StageTip protocol (Rappsilber et al., 2003).
Dried peptides were redissolved in 2% (v/v) acetonitrile (ACN) and 0.1% (v/v) trifluoroacetic acid (10 μL) for analysis, and measured without dilution. The samples were analyzed using an EASY-nLC 1200 (Thermo Fisher Scientific) coupled to a Q Exactive Plus mass spectrometer (Thermo Fisher Scientific). Peptides were separated on 16-cm frit-less silica emitters (0.75 μm of inner diameter; New Objective), packed in-house with reversed-phase ReproSil-Pur C18 AQ 1.9-μm resin (Dr. Maisch). Peptides (0.5 μg) were loaded onto the column and eluted for 115 min using a segmented linear gradient of 5% to 95% solvent B (0 min: 5% B; 0 min to 5 min → 5% B; 5 min to 65 min → 20% B; 65 min to 90 min → 35% B; 90 min to 100 min → 55%; 100 min to 105 min → 95%; 105 min to 115 min → 95%; solvent A 0% ACN, 0.1% FA; solvent B 80% ACN, 0.1% FA) at a flow rate of 300 nL min−1. MS were acquired in data-dependent acquisition mode using the TOP15 method, according to which per full scan the 15 most abundant precursors are selected for MS/MS fragmentation. MS spectra were acquired in the Orbitrap analyzer (Thermo Fisher Scientific) with a mass range of 300 m/z to 1,750 m/z at a resolution of 70,000 full width at half maximum (FWHM) and a target value of 3 × 106 ions. Precursors were selected with an isolation window of 1.3 m/z. Higher-energy collision dissociation fragmentation was performed at a normalized collision energy of 25. Tandem MS (MS/MS) spectra were acquired with a target value of 105 ions at a resolution of 17,500 FWHM, a maximum injection time of 55 ms, and a fixed first mass of m/z 100. Peptides with a charge of +1, >6, or with an unassigned charge state were excluded from fragmentation for MS/MS. Dynamic exclusion for 30 s prevented repeated selection of precursors.
Data Analysis
Raw data were processed using MaxQuant software (v1.5.7.4, http://www.maxquant.org/; Cox and Mann, 2008) with label-free quantification (LFQ) and Intensity Based Absolute Quantification enabled (Tyanova et al., 2016). MS/MS spectra were searched by the Andromeda search engine against a combined database containing Arabidopsis sequences (TAIR10_pep_20101214; ftp://ftp.arabidopsis.org/home/tair/Proteins/_TAIR10_protein_lists/) and sequences of 248 common contaminant proteins and decoy sequences. Trypsin specificity was required and a maximum of two missed cleavages allowed. Minimal peptide length was set to seven amino acids. Carbamidomethylation of Cys residues was set as “fixed” and oxidation of Met and protein N-terminal acetylation as “variable” modifications. Peptide-spectrum matches and proteins were retained if they were below an FDR of 1%. Statistical analysis of the MaxLFQ values was performed using the program Perseus (v1.5.8.5, http://www.maxquant.org/). Quantified proteins were filtered for reverse hits and hits “identified by site,” and MaxLFQ values were log2-transformed. After grouping the samples by condition, only proteins that had two valid values in one of the conditions were retained for subsequent analysis. Two-sample t tests were performed with a permutation-based FDR of 5%. Alternatively, quantified proteins were grouped by condition and only hits that had three valid values in one of the conditions were retained. Missing values were imputed from a normal distribution (0.3 width, 2.0 downshift, separately for each column). Volcano plots were generated in Perseus using an FDR of 1% and an S0 = 1. The Perseus output was exported and further processed using Microsoft Excel. ANOVA tables are shown in Supplemental Data Set 6.
Accession Numbers
The sequence of the genes and loci described here can be obtained from TAIR (ftp://ftp.arabidopsis.org/home/tair) using the following gene identifiers: CHR4 (AT5G44800), SVP (AT2G22540), FLC (AT5G10140), SOC1 (AT2G45660), FT (AT1G65480), TSF (AT4G20370), GA20ox2 (AT5G51810), and SPL15 (AT3G57920).
The Illumina sequencing data have been deposited to the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) with the dataset identifier GSE140728. The MS proteomics data have been deposited to the ProteomeXchange Consortium via the Proteomics Identification Database (Vizcaíno et al., 2016) partner repository (http://www.proteomexchange.org/) with the dataset identifier PXD016457.
Supplemental Data
Supplemental Figure 1. svp flc ft tsf soc1 probably flowers as a result of endogenous pathways.
Supplemental Figure 2. Molecular genetic analysis of qem1.
Supplemental Figure 3. CHR4 expression in Col-0 and chr4-2.
Supplemental Figure 4. CHR4 loss-of-function phenotype in LDs.
Supplemental Figure 5. SAM size.
Supplemental Figure 6. CHR4 expression profile and protein localization.
Supplemental Figure 7. Volcano plot of protein–protein interactions.
Supplemental Figure 8. Global accumulation H3K27me3 and H3K4me3 marks in Col-0 and chr4-2.
Supplemental Figure 9. Spearman correlation for ChIP-seq samples.
Supplemental Table. Candidate SNPs annotated in genes by SHOREmap for qem1.
Supplemental Data Set 1. Whole-genome expression profiling experiments comparing the profiles of the genotypes Col-0 and svp flc ft tsf soc1 grown for 3, 4, 5, or 6 weeks under SD conditions.
Supplemental Data Set 2. Whole-genome expression profiling experiments comparing the profiles of the genotypes Col-0 versus chr4-2 and svp flc ft tsf soc1 versus qem2 grown for 3, 4, 5, or 6 weeks under SD conditions.
Supplemental Data Set 3. Immunoprecipitation-MS results for CHR4-VENUS and AP1-GFP pull-down: list of CHR4-interacting proteins.
Supplemental Data Set 4. Comparative analysis of H3K27me3 and H3K4me3 ChIP-seq results in Col-0 and chr4-2 obtained with DANPOS2.
Supplemental Data Set 5. List of primers used in the study.
Supplemental Data Set 6. ANOVA tables.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Anne Harzen for support in the MS experiments. We thank René Richter, Franziska Turck, and John Chandler for critical comments to the article. This work was supported by the European Molecular Biology Organization (AFL-2017-74 to A.P.), the China Scholarship Council (201804910196 to X.Y.), the Deutsche Forschungsgemeinschaft under Germany´s Excellence Strategy (DFG EXC 2048/1 Project ID: 390686111 to G.C.), and is supported by a core grant from the Max Planck Society.
AUTHOR CONTRIBUTIONS
S.Q., A.P., and G.C. conceived and designed the experiments; S.Q., A.P., X.Y., and F.A. performed the experiments; S.Q., A.P., K.S., H.S., B.S., S.C.S., and H.N. analyzed the data; A.P., Q.S., and G.C. wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Abe A., et al. (2012). Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 30: 174–178. [DOI] [PubMed] [Google Scholar]
- Abe M., Kobayashi Y., Yamamoto S., Daimon Y., Yamaguchi A., Ikeda Y., Ichinoki H., Notaguchi M., Goto K., Araki T.(2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056. [DOI] [PubMed] [Google Scholar]
- Abe M., Kosaka S., Shibuta M., Nagata K., Uemura T., Nakano A., Kaya H.(2019). Transient activity of the florigen complex during the floral transition in Arabidopsis thaliana. Development 146: 146. [DOI] [PubMed] [Google Scholar]
- Aichinger E., Villar C.B., Farrona S., Reyes J.C., Hennig L., Köhler C.(2009). CHD3 proteins and polycomb group proteins antagonistically determine cell identity in Arabidopsis. PLoS Genet. 5: e1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Airoldi C.A., McKay M., Davies B.(2015). MAF2 is regulated by temperature-dependent splicing and represses flowering at low temperatures in parallel with FLM. PLoS One 10: e0126516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An H., Roussot C., Suárez-López P., Corbesier L., Vincent C., Piñeiro M., Hepworth S., Mouradov A., Justin S., Turnbull C., Coupland G.(2004). CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131: 3615–3626. [DOI] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., Huber W.(2015). HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés F., Coupland G.(2012). The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13: 627–639. [DOI] [PubMed] [Google Scholar]
- Andrés F., Porri A., Torti S., Mateos J., Romera-Branchat M., García-Martínez J.L., Fornara F., Gregis V., Kater M.M., Coupland G.(2014). SHORT VEGETATIVE PHASE reduces gibberellin biosynthesis at the Arabidopsis shoot apex to regulate the floral transition. Proc. Natl. Acad. Sci. USA 111: E2760–E2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukerman M.J., Sakai H.(2003). Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S., Sureshkumar S., Lempe J., Weigel D.(2006). Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genet. 2: e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton M.K.(2010). Twenty years on: The inner workings of the shoot apical meristem, a developmental dynamo. Dev. Biol. 341: 95–113. [DOI] [PubMed] [Google Scholar]
- Bouché F., Lobet G., Tocquin P., Périlleux C.(2016). FLOR-ID: An interactive database of flowering-time gene networks in Arabidopsis thaliana. Nucleic Acids Res. 44: D1167–D1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman J.L., Eshed Y.(2000). Formation and maintenance of the shoot apical meristem. Trends Plant Sci. 5: 110–115. [DOI] [PubMed] [Google Scholar]
- Bradley D., Carpenter R., Sommer H., Hartley N., Coen E.(1993). Complementary floral homeotic phenotypes result from opposite orientations of a transposon at the plena locus of Antirrhinum. Cell 72: 85–95. [DOI] [PubMed] [Google Scholar]
- Carter B., Bishop B., Ho K.K., Huang R., Jia W., Zhang H., Pascuzzi P.E., Deal R.B., Ogas J.(2018). The chromatin remodelers PKL and PIE1 act in an epigenetic pathway that determines H3K27me3 homeostasis in Arabidopsis. Plant Cell 30: 1337–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler J., Wilson A., Dean C.(1996). Arabidopsis mutants showing an altered response to vernalization. Plant J. 10: 637–644. [DOI] [PubMed] [Google Scholar]
- Chen K., Xi Y., Pan X., Li Z., Kaestner K., Tyler J., Dent S., He X., Li W.(2013). DANPOS: Dynamic analysis of nucleosome position and occupancy by sequencing. Genome Res. 23: 341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F.(1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Coleman-Derr D., Zilberman D.(2012). Deposition of histone variant H2A.Z within gene bodies regulates responsive genes. PLoS Genet. 8: e1002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collani S., Neumann M., Yant L., Schmid M.(2019). FT modulates genome-wide DNA-binding of the bZIP transcription factor FD. Plant Physiol. 180: 367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier L., Vincent C., Jang S., Fornara F., Fan Q., Searle I., Giakountis A., Farrona S., Gissot L., Turnbull C., Coupland G.(2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033. [DOI] [PubMed] [Google Scholar]
- Cox J., Mann M.(2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26: 1367–1372. [DOI] [PubMed] [Google Scholar]
- Davière J.M., Achard P.(2013). Gibberellin signaling in plants. Development 140: 1147–1151. [DOI] [PubMed] [Google Scholar]
- Deng W., Ying H., Helliwell C.A., Taylor J.M., Peacock W.J., Dennis E.S.(2011). FLOWERING LOCUS C (FLC) regulates development pathways throughout the life cycle of Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 6680–6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S.(2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629. [DOI] [PubMed] [Google Scholar]
- Eriksson S., Böhlenius H., Moritz T., Nilsson O.(2006). GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. Plant Cell 18: 2172–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrona S., Coupland G., Turck F.(2008). The impact of chromatin regulation on the floral transition. Semin. Cell Dev. Biol. 19: 560–573. [DOI] [PubMed] [Google Scholar]
- Farrona S., Hurtado L., Bowman J.L., Reyes J.C.(2004). The Arabidopsis thaliana SNF2 homolog AtBRM controls shoot development and flowering. Development 131: 4965–4975. [DOI] [PubMed] [Google Scholar]
- Favaro R., Pinyopich A., Battaglia R., Kooiker M., Borghi L., Ditta G., Yanofsky M.F., Kater M.M., Colombo L.(2003). MADS-box protein complexes control carpel and ovule development in Arabidopsis. Plant Cell 15: 2603–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández V., Takahashi Y., Le Gourrierec J., Coupland G.(2016). Photoperiodic and thermosensory pathways interact through CONSTANS to promote flowering at high temperature under short days. Plant J. 86: 426–440. [DOI] [PubMed] [Google Scholar]
- Ferrándiz C., Gu Q., Martienssen R., Yanofsky M.F.(2000). Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 127: 725–734. [DOI] [PubMed] [Google Scholar]
- Fu X., Li C., Liang Q., Zhou Y., He H., Fan L.M.(2016). CHD3 chromatin-remodeling factor PICKLE regulates floral transition partially via modulating LEAFY expression at the chromatin level in Arabidopsis. Sci. China Life Sci. 59: 516–528. [DOI] [PubMed] [Google Scholar]
- Galvão V.C., Collani S., Horrer D., Schmid M.(2015). Gibberellic acid signaling is required for ambient temperature-mediated induction of flowering in Arabidopsis thaliana. Plant J. 84: 949–962. [DOI] [PubMed] [Google Scholar]
- Galvão V.C., Horrer D., Küttner F., Schmid M.(2012). Spatial control of flowering by DELLA proteins in Arabidopsis thaliana. Development 139: 4072–4082. [DOI] [PubMed] [Google Scholar]
- Gandikota M., Birkenbihl R.P., Höhmann S., Cardon G.H., Saedler H., Huijser P.(2007). The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. 49: 683–693. [DOI] [PubMed] [Google Scholar]
- Hartwig B., James G.V., Konrad K., Schneeberger K., Turck F.(2012). Fast isogenic mapping-by-sequencing of ethyl methanesulfonate-induced mutant bulks. Plant Physiol. 160: 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Liu D., Zhong X., Zhang C., Zhang Q., Zhou D.X.(2012). CHD3 protein recognizes and regulates methylated histone H3 lysines 4 and 27 over a subset of targets in the rice genome. Proc. Natl. Acad. Sci. USA 109: 5773–5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun Y., Richter R., Coupland G.(2017). Competence to flower: Age-controlled sensitivity to environmental cues. Plant Physiol. 173: 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun Y., Richter R., Vincent C., Martinez-Gallegos R., Porri A., Coupland G.(2016). Multi-layered regulation of SPL15 and cooperation with SOC1 integrate endogenous flowering pathways at the Arabidopsis shoot meristem. Dev. Cell 37: 254–266. [DOI] [PubMed] [Google Scholar]
- Hyun Y., Vincent C., Tilmes V., Bergonzi S., Kiefer C., Richter R., Martinez-Gallegos R., Severing E., Coupland G.(2019). A regulatory circuit conferring varied flowering response to cold in annual and perennial plants. Science 363: 409–412. [DOI] [PubMed] [Google Scholar]
- Jaeger K.E., Wigge P.A.(2007). FT protein acts as a long-range signal in Arabidopsis. Curr. Biol. 17: 1050–1054. [DOI] [PubMed] [Google Scholar]
- Jang S., Torti S., Coupland G.(2009). Genetic and spatial interactions between FT, TSF and SVP during the early stages of floral induction in Arabidopsis. Plant J. 60: 614–625. [DOI] [PubMed] [Google Scholar]
- Jing Y., Guo Q., Lin R.(2019a). The chromatin-remodeling factor PICKLE antagonizes polycomb repression of FT to promote flowering. Plant Physiol. 181: 656–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing Y., Guo Q., Zha P., Lin R.(2019b). The chromatin-remodelling factor PICKLE interacts with CONSTANS to promote flowering in Arabidopsis. Plant Cell Environ. 42: 2495–2507. [DOI] [PubMed] [Google Scholar]
- Jing Y., Zhang D., Wang X., Tang W., Wang W., Huai J., Xu G., Chen D., Li Y., Lin R.(2013). Arabidopsis chromatin remodeling factor PICKLE interacts with transcription factor HY5 to regulate hypocotyl cell elongation. Plant Cell 25: 242–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M.A., Covington M.F., DiTacchio L., Vollmers C., Panda S., Harmer S.L.(2010). Jumonji domain protein JMJD5 functions in both the plant and human circadian systems. Proc. Natl. Acad. Sci. USA 107: 21623–21628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky I., Shukla V.K., Ahn J.H., Dagenais N., Christensen S.K., Nguyen J.T., Chory J., Harrison M.J., Weigel D.(1999). Activation tagging of the floral inducer FT. Science 286: 1962–1965. [DOI] [PubMed] [Google Scholar]
- Kaufmann K., Muiño J.M., Østerås M., Farinelli L., Krajewski P., Angenent G.C.(2010). Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP). Nat. Protoc. 5: 457–472. [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L.(2013). TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klock H.E., Lesley S.A.(2009). The Polymerase Incomplete Primer Extension (PIPE) method applied to high-throughput cloning and site-directed mutagenesis. Methods Mol. Biol. 498: 91–103. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y., Kaya H., Goto K., Iwabuchi M., Araki T.(1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962. [DOI] [PubMed] [Google Scholar]
- Koornneef M., Alonso-Blanco C., Blankestijn-de Vries H., Hanhart C.J., Peeters A.J.(1998). Genetic interactions among late-flowering mutants of Arabidopsis. Genetics 148: 885–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M., Hanhart C.J., van der Veen J.H.(1991). A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 229: 57–66. [DOI] [PubMed] [Google Scholar]
- Kumar S.V., Lucyshyn D., Jaeger K.E., Alós E., Alvey E., Harberd N.P., Wigge P.A.(2012). Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484: 242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara D., Mizuta Y., Sato Y., Higashiyama T.(2015). ClearSee: a rapid optical clearing reagent for whole-plant fluorescence imaging. Development 142: 4168–4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara D., Mizuta Y., Sato Y., Higashiyama T.(2015). ClearSee: A rapid optical clearing reagent for whole-plant fluorescence imaging. Development 142: 4168–4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P., et al. (2012). The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 40: D1202–D1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L.(2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Ryu H.S., Chung K.S., Posé D., Kim S., Schmid M., Ahn J.H.(2013). Regulation of temperature-responsive flowering by MADS-box transcription factor repressors. Science 342: 628–632. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Yoo S.J., Park S.H., Hwang I., Lee J.S., Ahn J.H.(2007). Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev. 21: 397–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., et al. (2016). Concerted genomic targeting of H3K27 demethylase REF6 and chromatin-remodeling ATPase BRM in Arabidopsis. Nat. Genet. 48: 687–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Liu C., Shen L., Wu Y., Chen H., Robertson M., Helliwell C.A., Ito T., Meyerowitz E., Yu H.(2008). A repressor complex governs the integration of flowering signals in Arabidopsis. Dev. Cell 15: 110–120. [DOI] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Proc G.P.D.; 1000 Genome Project Data Processing Subgroup. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F., Cui X., Zhang S., Jenuwein T., Cao X.(2011). Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat. Genet. 43: 715–719. [DOI] [PubMed] [Google Scholar]
- March-Díaz R., García-Domínguez M., Lozano-Juste J., León J., Florencio F.J., Reyes J.C.(2008). Histone H2A.Z and homologues of components of the SWR1 complex are required to control immunity in Arabidopsis. Plant J. 53: 475–487. [DOI] [PubMed] [Google Scholar]
- Mateos J.L., Madrigal P., Tsuda K., Rawat V., Richter R., Romera-Branchat M., Fornara F., Schneeberger K., Krajewski P., Coupland G.(2015). Combinatorial activities of SHORT VEGETATIVE PHASE and FLOWERING LOCUS C define distinct modes of flowering regulation in Arabidopsis. Genome Biol. 16: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J., Warthmann N., Küttner F., Schmid M.(2007). Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr. Biol. 17: 1055–1060. [DOI] [PubMed] [Google Scholar]
- Melzer S., Lens F., Gennen J., Vanneste S., Rohde A., Beeckman T.(2008). Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana. Nat. Genet. 40: 1489–1492. [DOI] [PubMed] [Google Scholar]
- Michaels S.D., Amasino R.M.(1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S.D., Amasino R.M.(2001). Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13: 935–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T., Skoog F.(1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15: 473–497. [Google Scholar]
- Noh Y.S., Amasino R.M.(2003). PIE1, an ISWI family gene, is required for FLC activation and floral repression in Arabidopsis. Plant Cell 15: 1671–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogas J., Kaufmann S., Henderson J., Somerville C.(1999). PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc. Natl. Acad. Sci. USA 96: 13839–13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onouchi H., Igeño M.I., Périlleux C., Graves K., Coupland G.(2000). Mutagenesis of plants overexpressing CONSTANS demonstrates novel interactions among Arabidopsis flowering-time genes. Plant Cell 12: 885–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Oh D.H., Dassanayake M., Nguyen K.T., Ogas J., Choi G., Sun T.P.(2017). Gibberellin signaling requires chromatin remodeler pickle to promote vegetative growth and phase transitions. Plant Physiol. 173: 1463–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plackett A.R., et al. (2012). Analysis of the developmental roles of the Arabidopsis gibberellin 20-oxidases demonstrates that GA20ox1, -2, and -3 are the dominant paralogs. Plant Cell 24: 941–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porri A., Torti S., Romera-Branchat M., Coupland G.(2012). Spatially distinct regulatory roles for gibberellins in the promotion of flowering of Arabidopsis under long photoperiods. Development 139: 2198–2209. [DOI] [PubMed] [Google Scholar]
- Posé D., Verhage L., Ott F., Yant L., Mathieu J., Angenent G.C., Immink R.G., Schmid M.(2013). Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature 503: 414–417. [DOI] [PubMed] [Google Scholar]
- Ramírez F., Dündar F., Diehl S., Grüning B.A., Manke T.(2014). deepTools: A flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42: W187–W191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J., Ishihama Y., Mann M.(2003). Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75: 663–670. [DOI] [PubMed] [Google Scholar]
- Ratcliffe O.J., Amaya I., Vincent C.A., Rothstein S., Carpenter R., Coen E.S., Bradley D.J.(1998). A common mechanism controls the life cycle and architecture of plants. Development 125: 1609–1615. [DOI] [PubMed] [Google Scholar]
- Ratcliffe O.J., Bradley D.J., Coen E.S.(1999). Separation of shoot and floral identity in Arabidopsis. Development 126: 1109–1120. [DOI] [PubMed] [Google Scholar]
- Richter R., Kinoshita A., Vincent C., Martinez-Gallegos R., Gao H., van Driel A.D., Hyun Y., Mateos J.L., Coupland G.(2019). Floral regulators FLC and SOC1 directly regulate expression of the B3-type transcription factor TARGET OF FLC AND SVP 1 at the Arabidopsis shoot apex via antagonistic chromatin modifications. PLoS Genet. 15: e1008065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieu I., et al. (2008). The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J. 53: 488–504. [DOI] [PubMed] [Google Scholar]
- Sang Y., Silva-Ortega C.O., Wu S., Yamaguchi N., Wu M.F., Pfluger J., Gillmor C.S., Gallagher K.L., Wagner D.(2012). Mutations in two non-canonical Arabidopsis SWI2/SNF2 chromatin remodeling ATPases cause embryogenesis and stem cell maintenance defects. Plant J. 72: 1000–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M., Uhlenhaut N.H., Godard F., Demar M., Bressan R., Weigel D., Lohmann J.U.(2003). Dissection of floral induction pathways using global expression analysis. Development 130: 6001–6012. [DOI] [PubMed] [Google Scholar]
- Schneeberger K.(2014). Using next-generation sequencing to isolate mutant genes from forward genetic screens. Nat. Rev. Genet. 15: 662–676. [DOI] [PubMed] [Google Scholar]
- Schneeberger K., Ossowski S., Lanz C., Juul T., Petersen A.H., Nielsen K.L., Jørgensen J.E., Weigel D., Andersen S.U.(2009). SHOREmap: Simultaneous mapping and mutation identification by deep sequencing. Nat. Methods 6: 550–551. [DOI] [PubMed] [Google Scholar]
- Schwarz S., Grande A.V., Bujdoso N., Saedler H., Huijser P.(2008). The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol. Biol. 67: 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle I., He Y., Turck F., Vincent C., Fornara F., Kröber S., Amasino R.A., Coupland G.(2006). The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 20: 898–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon C.C., Burn J.E., Perez P.P., Metzger J., Edwards J.A., Peacock W.J., Dennis E.S.(1999). The FLF MADS box gene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11: 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaczniak C., et al. (2012). Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc. Natl. Acad. Sci. USA 109: 1560–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth A., Schmid M.(2011). Regulation of flowering time: All roads lead to Rome. Cell. Mol. Life Sci. 68: 2013–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-López P., Wheatley K., Robson F., Onouchi H., Valverde F., Coupland G.(2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120. [DOI] [PubMed] [Google Scholar]
- Sun H., Schneeberger K.(2015). SHOREmap v3.0: Fast and accurate identification of causal mutations from forward genetic screens. Methods Mol. Biol. 1284: 381–395. [DOI] [PubMed] [Google Scholar]
- Torti S., Fornara F., Vincent C., Andrés F., Nordström K., Göbel U., Knoll D., Schoof H., Coupland G.(2012). Analysis of the Arabidopsis shoot meristem transcriptome during floral transition identifies distinct regulatory patterns and a leucine-rich repeat protein that promotes flowering. Plant Cell 24: 444–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S., Temu T., Cox J.(2016). The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11: 2301–2319. [DOI] [PubMed] [Google Scholar]
- Valverde F., Mouradov A., Soppe W., Ravenscroft D., Samach A., Coupland G.(2004). Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303: 1003–1006. [DOI] [PubMed] [Google Scholar]
- Van Larebeke N., Engler G., Holsters M., Van den Elsacker S., Zaenen I., Schilperoort R.A., Schell J.(1974). Large plasmid in Agrobacterium tumefaciens essential for crown gall-inducing ability. Nature 252: 169–170. [DOI] [PubMed] [Google Scholar]
- Vizcaíno J.A., et al. (2016). 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44 (D1): D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D., Sablowski R.W., Meyerowitz E.M.(1999). Transcriptional activation of APETALA1 by LEAFY. Science 285: 582–584. [DOI] [PubMed] [Google Scholar]
- Wang J.W., Czech B., Weigel D.(2009). miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138: 738–749. [DOI] [PubMed] [Google Scholar]
- Weigel D., Alvarez J., Smyth D.R., Yanofsky M.F., Meyerowitz E.M.(1992). LEAFY controls floral meristem identity in Arabidopsis. Cell 69: 843–859. [DOI] [PubMed] [Google Scholar]
- Whittaker C., Dean C.(2017). The FLC locus: A platform for discoveries in epigenetics and adaptation. Annu. Rev. Cell Dev. Biol. 33: 555–575. [DOI] [PubMed] [Google Scholar]
- Wigge P.A., Kim M.C., Jaeger K.E., Busch W., Schmid M., Lohmann J.U., Weigel D.(2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059. [DOI] [PubMed] [Google Scholar]
- Wilson R.N., Heckman J.W., Somerville C.R.(1992). Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol. 100: 403–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Poethig R.S.(2006). Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133: 3539–3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M.F., Sang Y., Bezhani S., Yamaguchi N., Han S.K., Li Z., Su Y., Slewinski T.L., Wagner D.(2012). SWI2/SNF2 chromatin remodeling ATPases overcome polycomb repression and control floral organ identity with the LEAFY and SEPALLATA3 transcription factors. Proc. Natl. Acad. Sci. USA 109: 3576–3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S., Salinas M., Höhmann S., Berndtgen R., Huijser P.(2010). miR156-targeted and nontargeted SBP-box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell 22: 3935–3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Hu T., Zhao J., Park M.Y., Earley K.W., Wu G., Yang L., Poethig R.S.(2016). Developmental functions of miR156-regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes in Arabidopsis thaliana. PLoS Genet. 12: e1006263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A., Kobayashi Y., Goto K., Abe M., Araki T.(2005). TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol. 46: 1175–1189. [DOI] [PubMed] [Google Scholar]
- Yamaguchi A., Wu M.F., Yang L., Wu G., Poethig R.S., Wagner D.(2009). The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev. Cell 17: 268–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N., Winter C.M., Wu M.F., Kanno Y., Yamaguchi A., Seo M., Wagner D.(2014). Gibberellin acts positively then negatively to control onset of flower formation in Arabidopsis. Science 344: 638–641. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S.(2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59: 225–251. [DOI] [PubMed] [Google Scholar]
- Yan Y., Shen L., Chen Y., Bao S., Thong Z., Yu H.(2014). A MYB-domain protein EFM mediates flowering responses to environmental cues in Arabidopsis. Dev. Cell 30: 437–448. [DOI] [PubMed] [Google Scholar]
- Yoo S.J., Chung K.S., Jung S.H., Yoo S.Y., Lee J.S., Ahn J.H.(2010). BROTHER OF FT AND TFL1 (BFT) has TFL1-like activity and functions redundantly with TFL1 in inflorescence meristem development in Arabidopsis. Plant J. 63: 241–253. [DOI] [PubMed] [Google Scholar]
- Yu S., Galvão V.C., Zhang Y.C., Horrer D., Zhang T.Q., Hao Y.H., Feng Y.Q., Wang S., Schmid M., Wang J.W.(2012). Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA promoter binding-like transcription factors. Plant Cell 24: 3320–3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Bishop B., Ringenberg W., Muir W.M., Ogas J.(2012). The CHD3 remodeler PICKLE associates with genes enriched for trimethylation of histone H3 lysine 27. Plant Physiol. 159: 418–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Rider S.D. Jr., Henderson J.T., Fountain M., Chuang K., Kandachar V., Simons A., Edenberg H.J., Romero-Severson J., Muir W.M., Ogas J.(2008). The CHD3 remodeler PICKLE promotes trimethylation of histone H3 lysine 27. J. Biol. Chem. 283: 22637–22648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Schwarz S., Saedler H., Huijser P.(2007). SPL8, a local regulator in a subset of gibberellin-mediated developmental processes in Arabidopsis. Plant Mol. Biol. 63: 429–439. [DOI] [PubMed] [Google Scholar]
- Zhou J., Wang X., Lee J.Y., Lee J.Y.(2013). Cell-to-cell movement of two interacting AT-hook factors in Arabidopsis root vascular tissue patterning. Plant Cell 25: 187–201. [DOI] [PMC free article] [PubMed] [Google Scholar]