Backtracking/arrest of RNA polymerase II commonly occurs during transcript elongation and reactivation of the enzyme is a prerequisite for plant growth.
Abstract
Transcript elongation factors associate with elongating RNA polymerase II (RNAPII) to control the efficiency of mRNA synthesis and consequently modulate plant growth and development. Encountering obstacles during transcription such as nucleosomes or particular DNA sequences may cause backtracking and transcriptional arrest of RNAPII. The elongation factor TFIIS stimulates the intrinsic transcript cleavage activity of the polymerase, which is required for efficient rescue of backtracked/arrested RNAPII. A TFIIS mutant variant (TFIISmut) lacks the stimulatory activity to promote RNA cleavage, but instead efficiently inhibits unstimulated transcript cleavage by RNAPII. We could not recover viable Arabidopsis (Arabidopsis thaliana) tfIIs plants constitutively expressing TFIISmut. Induced, transient expression of TFIISmut in tfIIs plants provoked severe growth defects, transcriptomic changes and massive, transcription-related redistribution of elongating RNAPII within transcribed regions toward the transcriptional start site. The predominant site of RNAPII accumulation overlapped with the +1 nucleosome, suggesting that upon inhibition of RNA cleavage activity, RNAPII arrest prevalently occurs at this position. In the presence of TFIISmut, the amount of RNAPII was reduced, which could be reverted by inhibiting the proteasome, indicating proteasomal degradation of arrested RNAPII. Our findings suggest that polymerase backtracking/arrest frequently occurs in plant cells, and RNAPII-reactivation is essential for correct transcriptional output and proper growth/development.
INTRODUCTION
In eukaryotes, the abundance of functional mRNAs is precisely controlled in a spatially and temporally defined manner. Besides post-transcriptional mechanisms (e.g., control of splicing or mRNA stability), synthesis of pre-mRNAs by RNA polymerase II (RNAPII) is accurately regulated. For a considerable time, it was assumed that the transcription of protein-coding genes is controlled exclusively during transcriptional initiation. However, the elongation stage of RNAPII transcription also turned out to be highly dynamic and tightly regulated. As a distinguishing feature, the carboxy-terminal domain of the large subunit of RNAPII (RNAPII-CTD) is modified during subsequent steps of transcript synthesis. Thus, residues within conserved heptapeptide repeats (e.g., S2, S5) of the RNAPII-CTD are differentially phosphorylated in the course of transcriptional elongation (Hajheidari et al., 2013; Harlen and Churchman, 2017).
The importance of regulating the progression of RNAPII is also reflected by existence of a variety of transcript elongation factors (TEFs). Although transcript elongation is generally processive, it represents a rather discontinuous process, involving pausing, backtracking, and even transcriptional arrest, requiring the action of certain TEFs to stimulate RNAPII progression. TEFs also enable efficient transcript synthesis in the chromatin context, because nucleosomes represent major obstacles to transcriptional elongation. Consequently, TEFs act as histone chaperones, modify histones within transcribed regions, or regulate catalytic properties of RNAPII to ensure that elongation occurs efficiently (Sims et al., 2004; Selth et al., 2010; Chen et al., 2018).
Among the modulators of RNAPII activity is TFIIS, which facilitates RNAPII transcription through blocks to elongation (Wind and Reines, 2000; Fish and Kane, 2002). TFIIS deeply inserts a highly conserved, β-hairpin of its C-terminal domain into the RNAPII complex approaching the polymerase active site to reactivate arrested RNAPII (Kettenberger et al., 2003). In addition to its RNA polymerization activity, RNAPII has a relatively weak intrinsic RNA nuclease activity. Structural studies demonstrated that reactivation of arrested RNAPII is accomplished by TFIIS-induced extensive conformational changes in the elongation complex (Kettenberger et al., 2003). Consequently, the backtracked/arrested RNA is mobilized, and two invariant acidic side chains positioned at the tip of the TFIIS hairpin complement the enzyme active site. Thereby, TFIIS stimulates the weak intrinsic nuclease activity of RNAPII, resulting in cleavage of the backtracked/arrested RNA, and generating a new RNA 3′ end at the active site that allows transcription to resume (Wang et al., 2009; Cheung and Cramer, 2011). Consistent with these structural studies, earlier in vitro transcription analyses demonstrated that the TFIIS-promoted RNA cleavage stimulated transcript elongation by RNAPII (Izban and Luse, 1992; Jeon et al., 1994).
In view of the importance of TFIIS for transcript elongation in vitro, it was surprising that yeast (Saccharomyces cerevisiae) TFIIS (Dst1) is dispensable for cells grown under normal conditions, suggesting that the slower, unstimulated RNA cleavage activity of RNAPII is sufficient for transcription when yeast cells are grown at standard conditions (Wind and Reines, 2000; Fish and Kane, 2002; Svejstrup, 2007). Alternatively, the apparently nonessential function of TFIIS in yeast could imply that transcription in vivo is a smoother process than in vitro, so that transcript cleavage and escape from arrest are not important.
Expression of a mutant version of TFIIS in wild-type yeast cells caused severe dominant-negative growth defects and expression in ∆dst1 cells even proved to be lethal (Sigurdsson et al., 2010). In this mutant version (termed TFIISmut), the two conserved acidic residues at the tip of the hairpin were changed to Ala residues, which resulted in a loss of the transcript cleavage stimulatory activity. Beyond that, this mutation efficiently inhibited the intrinsic RNAPII transcript cleavage reaction (Jeon et al., 1994; Kettenberger et al., 2004; Sigurdsson et al., 2010; Imashimizu et al., 2013). The experiments in yeast further suggested that transcriptional elongation problems frequently occur in vivo and require reactivation of backtracked/arrested RNAPII (Sigurdsson et al., 2010).
Several TEFs including TFIIS were found to associate with elongating RNAPII in plant cells, forming the transcript elongation complex (TEC; Antosz et al., 2017). Moreover, various studies particularly in the Arabidopsis (Arabidopsis thaliana) model have demonstrated that different TEFs are critical for proper plant growth and development (van Lijsebettens and Grasser, 2014). Hence, RNAPII-associated TEFs (e.g., FACT, PAF1C, SPT4/SPT5, Elongator) were found to modulate diverse processes such as hormone signaling, circadian clock, and developmental transitions (He et al., 2004; Oh et al., 2004; Lolas et al., 2010; Nelissen et al., 2010; Dürr et al., 2014; Ma et al., 2018). However, similar to the situation in yeast, TFIIS seems to be nonessential in Arabidopsis under normal growth conditions, as mutant plants deficient in TFIIS or plants overexpressing TFIIS show basically wild-type appearance (Grasser et al., 2009; Mortensen and Grasser, 2014).
Interestingly, despite their apparent wild-type, phenotype tfIIs mutants exhibit defects in seed dormancy (Grasser et al., 2009; Liu et al., 2011), which are caused by reduced transcript levels of DELAY OF GERMINATION1 (Liu et al., 2011; Mortensen and Grasser, 2014). This gene is expressed seed-specifically and is a known quantitative trait locus for seed dormancy in Arabidopsis (Bentsink et al., 2006). Expression of the TFIISmut variant in Arabidopsis wild type plants resulted in a range of distinct developmental defects including leaf serration and reduced stem elongation. Viable transformants could not be recovered when it was attempted to express TFIISmut in tfIIs mutant background (Dolata et al., 2015). This finding indicates that in Arabidopsis, the endonucleolytic RNA cleavage activity of RNAPII is required for viability, as it was observed in yeast (Sigurdsson et al., 2010).
To further study the role of the intrinsic nuclease activity of RNAPII in Arabidopsis in the absence of endogenous TFIIS, we established an inducible expression system of TFIISmut in the tfIIs mutant background. Induction of TFIISmut expression initially caused marked growth defects and ultimately lethality. Using this inducible system, we investigated the effects of transient expression of TFIISmut and transcript cleavage inhibition on plant growth and cell proliferation as well as on gene expression pattern and genome-wide RNAPII distribution.
RESULTS
Induced Expression of TFIISmut Severely Impairs Growth and Cell Proliferation of tfIIs Plants
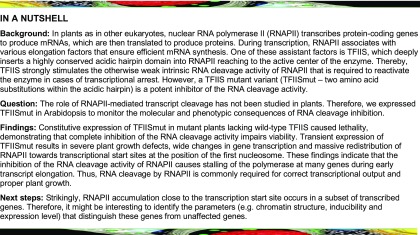
To study the effect of TFIISmut expression in plants, the two invariant acidic residues (D359 and E360 in Arabidopsis TFIIS; Figures 1A and 1B) positioned at the tip of the highly conserved hairpin of the C-terminal domain were changed to Ala. To that end the genomic TFIIS sequence was mutagenized and expressed under control of the native promoter in tfIIs plants. Consistent with a previous study (Dolata et al., 2015), we were unable to obtain viable, homozygous tfIIs plants harboring the TFIISmut construct. This confirmed that the intrinsic nuclease activity of RNAPII is essential in Arabidopsis.
Figure 1.
Induced Expression of TFIISmut in tfIIs Mutant Background Severely Affects Plant Growth.
(A) Amino acid sequence alignment of the C-terminal part of TFIIS from Arabidopsis (At), rice (Os), human (Hs), and the yeast S. cerevisiae (Sc). Asterisks indicate conserved residues and the invariant acidic residues (D359, E360 indicated in red) that were mutated in this work (the corresponding residues were mutated in other studies [Jeon et al., 1994; Kettenberger et al., 2004; Sigurdsson et al., 2010; Imashimizu et al., 2013]).
(B) Cartoon representation of the computational structure model of domains II and III of Arabidopsis TFIIS. The model (C-score = 0.82) was generated using the I-TASSER server (Yang and Zhang, 2015). Domain II is indicated in green, the interdomain linker in orange, domain III in yellow, and the position of the two invariant acidic hairpin residues (changed to Ala residues in TFIISmut) is highlighted in red and marked by an arrow.
(C) Growth of Arabidopsis plants expressing TFIISmut is severely impaired. Seven-DAS plants of three independent plant lines each expressing GFP-TFIISmut or GFP-TFIIS were grown in comparison to the Col-0 wild type on MS medium under inductive conditions (+β-estradiol) or mock-treated (ethanol).
To further analyze the endonucleolytic RNA cleavage by RNAPII, we employed transient expression of TFIISmut (and for comparison, TFIIS) based on a β-estradiol–inducible system (Brand et al., 2006) schematically presented in Supplemental Figure 1A. The sequences encoding TFIISmut/TFIIS were N-terminally fused to GFP to visualize expression of the proteins. The tfIIs plants were transformed with these constructs and three independent transgenic lines each were further characterized by PCR-based genotyping (Supplemental Figure 1B). Analysis of leaves and roots of these plants at different times after addition of β-estradiol (or mock-treated with ethanol) by confocal fluorescence microscopy revealed comparable induction kinetics for GFP-TFIISmut and GFP-TFIIS (Supplemental Figure 2). Nuclear GFP fluorescence was detectable after 3 h of induction. After longer induction periods (6 h, 24 h), the fluorescence signal was observed in a larger number of cells with increasing intensity. These analyses showed that both GFP-TFIISmut and GFP-TFIIS are efficiently expressed upon β-estradiol induction.
To examine the effect of induced TFIISmut expression on plant growth, seeds of three independent lines—each inducibly expressing GFP-TFIISmut or GFP-TFIIS—were sown on Murashige and Skoog (MS) medium supplemented with β-estradiol or mock-treated. Images taken 7 d after stratification (7 DAS) demonstrated that GFP-TFIISmut plants were severely affected in growth. Many seeds were unable to establish seedling growth at all, whereas the growth of controls is indistinguishable from that of Columbia-0 (Col-0) wild-type plants (Figure 1C). These observations confirm the deleterious effects of the expression of GFP-TFIISmut in the tfIIs background on plant growth, whereas plants expressing TFIIS (Col-0; tfIIs expressing GFP-TFIIS) or plants lacking TFIIS (tfIIs) grow comparably.
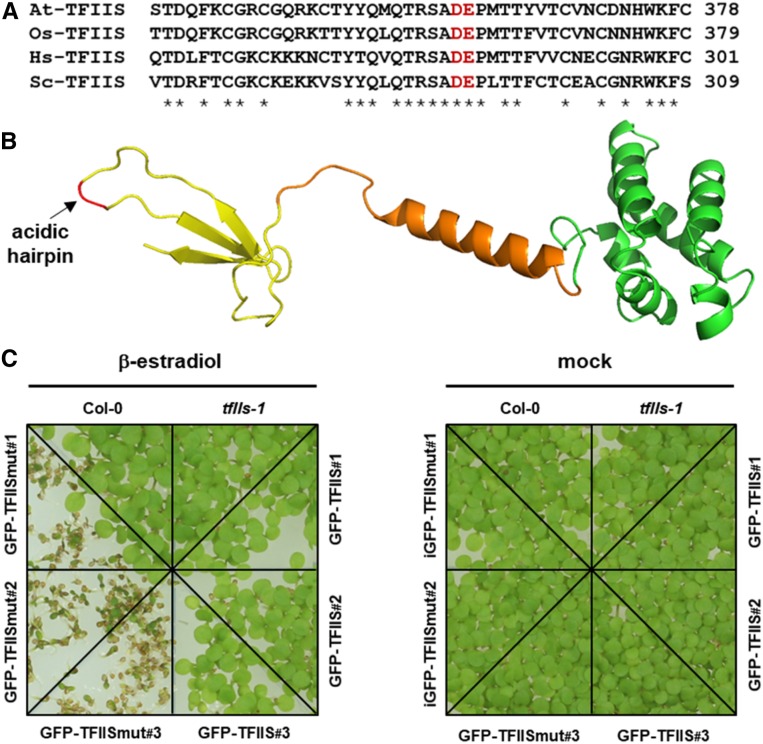
To further evaluate the effects of temporary TFIISmut expression on plants, we measured the root growth of seedlings. Seedlings at 8 DAS were transferred onto medium supplemented with β-estradiol or ethanol, and growth of the primary root was scored at different times after induction. In comparison to non-induced plants, a clear inhibition of root growth was observed at different times after induction with GFP-TFIISmut plants, but not with GFP-TFIIS (Figure 2A). Quantification of absolute and relative root elongation (Figures 2B and 2C) demonstrated that root growth of GFP-TFIISmut plants was distinctly decreased.
Figure 2.
Plants Expressing TFIISmut Exhibit Reduced Growth of the Primary Root and Decreased Cell Proliferation.
(A) Inhibition of root growth upon expression of TFIISmut. Eight-DAS plants were transferred onto MS plates containing β-estradiol to induce expression of GFP-TFIISmut and GFP-TFIIS, or mock-treated. Images were taken at the indicated times. White arrow (and black line) indicates the position of the root tip at the time of transfer (0 h). Root elongation was measured to determine absolute (B) and relative (C) elongation.
(B) Dots represent mean values ± sd and dotted lines represent linear regression (n = 6).
(C) Bars represent mean values ± sd and asterisks indicate the outcome of the Student’s t test: *P < 0.05, **P < 0.01, ***P < 0.001.
(D) CLSM analysis of GFP fluorescence of root tips of 5-DAS plants induced (or mock-treated) for 24 h to express GS-TFIIS or GS-TFIISmut and harboring the pCYCB1;1-GFP reporter.
(E) Bars represent mean values ± sd of the number of mitotic, GFP-expressing cells (n = 5 plants per line/condition). Different characters (a, b) indicate the outcome of a multicomparison Tukey’s test (P < 0.05).
Employing the cell cycle marker pCYCB1;1-GFP that allows detection of cells at the G2-M phase of the cell cycle (Ubeda-Tomás et al., 2009), we examined whether reduced cell proliferation might be a cause of decreased root growth. To allow visualization of the cell cycle marker, the pCYCB1;1-GFP construct was introduced into plants that harbor inducible-expression cassettes of TFIISmut/TFIIS fused to a GS-tag (Bürckstümmer et al., 2006); see below for details). When compared with GS-TFIIS, β-estradiol–induced expression of GS-TFIISmut resulted in a significantly reduced number of mitotic cells (Figures 2D and 2E), indicating that decreased cell proliferation contributed to reduced root growth.
Like TFIIS, TFIISmut Interacts with the TEC, But Exhibits Reduced Nuclear Mobility
Using protein affinity purification in combination with mass spectrometry (AP-MS), we examined the association of TFIISmut and TFIIS with the RNAPII-TEC and compared the composition of the complex. Both proteins fused to an N-terminal GS-tag were expressed under control of the β-estradiol–inducible system in Arabidopsis PSB-D suspension cultured cells and affinity-purified using IgG-chromatography. This method was successfully employed before to isolate various nuclear protein complexes (Nelissen et al., 2010; Dürr et al., 2014; Antosz et al., 2017). Both fusion proteins were efficiently isolated from protein extracts that showed a comparable band pattern as analyzed by SDS-PAGE (Supplemental Figure 3A). Co-purified proteins were examined initially using immunoblotting with different RNAPII-specific antibodies (Supplemental Figure 3B). Both GS-TFIISmut and GS-TFIIS co-purified with RNAPII, as observed before for Arabidopsis TFIIS (Antosz et al., 2017). The S2P modified version co-purifies ∼3-fold more prominently with GS-TFIISmut than with GS-TFIIS. Further analysis of the co-purified proteins by AP-MS revealed that in addition to TFIIS and RNAPII subunits, various RNAPII-associated TEFs were detected (Table 1). Among the identified TEFs were characteristic TEC-components such as PAF1C, FACT, and SPT4/SPT5. These results indicated that both TFIIS and TFIISmut interact with the RNAPII-TEC that exhibits a comparable composition.
Table 1. Known TEC Components Co-purifying with GS-TFIIS and GS-TFIISmut.
| TFIISa | TFIISmuta | Interactorb | Arabidopsis Genome Initiative | Complex |
|---|---|---|---|---|
| 3748 / 3 | 2882 / 3 | TFIIS | AT2G38560 | TFIIS |
| 1487 / 3 | 1763 / 3 | NRPB1 | AT4G35800 | Polymerase II |
| 1378 / 3 | 1319 / 3 | NRPB2 | AT4G21710 | Polymerase II |
| 466 / 3 | 551 / 3 | NRP(B/D/E)3a | AT2G15430 | Polymerase II |
| 462 / 3 | 474 / 3 | NRP(A/B/C/D)5 | AT3G22320 | Polymerase II |
| 216 / 3 | 149 / 2 | NRPB7 | AT5G59180 | Polymerase II |
| —c | 105 / 2 | NRP(B/D/E)9a | AT3G16980 | Polymerase II |
| 904 / 3 | 859 / 3 | CDC73 | AT3G22590 | PAF1C |
| 504 / 3 | 416 / 3 | VIP4, LEO1 | AT5G61150 | PAF1C |
| 553 / 3 | 408 / 3 | ELF7, PAF1 | AT1G79730 | PAF1C |
| 438 / 3 | 408 / 2 | VIP3, SKI8 | AT4G29830 | PAF1C |
| 327 / 3 | 245 / 3 | VIP5, RTF1 | AT1G61040 | PAF1C |
| 342 / 2 | 546 / 3 | SPT6L | AT1G65440 | SPT6 |
| 364 / 3 | 649 / 3 | SPT5-2 | AT4G08350 | SPT4/SPT5 |
| 435 / 2 | 544 / 3 | SPT16 | AT4G10710 | FACT |
| 211 / 2 | 261 / 3 | SSRP1 | AT3G28730 | FACT |
| 218 / 2 | 171 / 2 | ELP1, ELO2 | AT5G13680 | Elongator |
The numbers indicate the respective average MASCOT (http://www.matrixscience.com/) score and how many times the proteins were detected in three independent affinity purifications. The proteins that were detected in fewer than two out of three affinity purifications are not listed.
TEC components previously identified as interactors of elongating Arabidopsis RNAPII (Antosz et al., 2017) are listed here.
Blank cell indicates not identified.
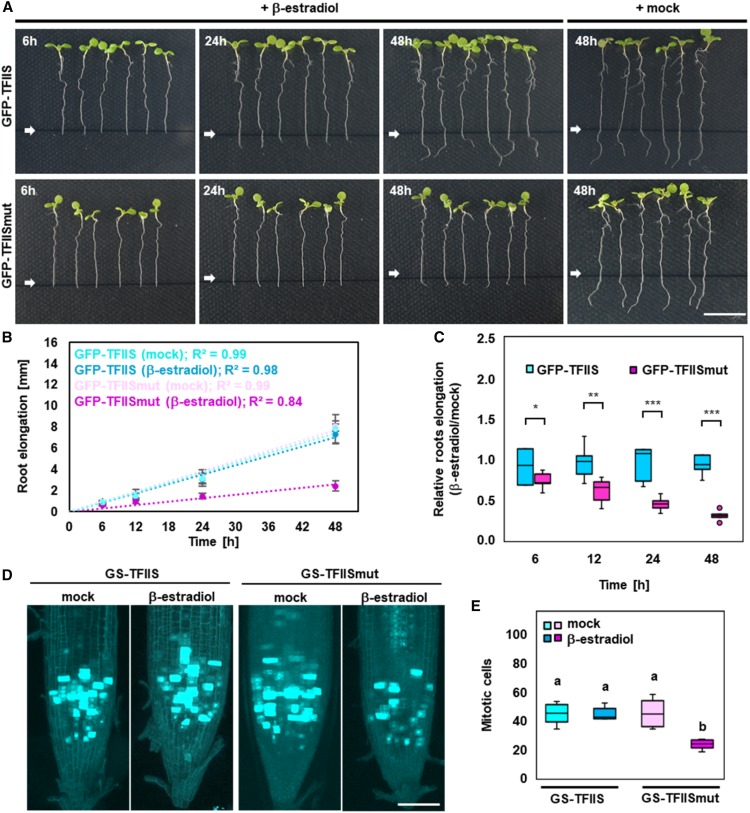
In view of the TFIISmut-induced inhibition of RNAPII-mediated transcript cleavage that results in transcriptional arrest, elongation complexes containing TFIISmut may display reduced nuclear dynamics. To examine whether TFIIS and TFIISmut associate with complexes of different mobility, we studied their dynamics in Arabidopsis cells. GFP-TFIISmut and GFP-TFIIS were expressed in PSB-D cells and analyzed by fluorescence recovery after photobleaching (FRAP). FRAP measurements demonstrated that GFP-TFIISmut displayed a clearly lower mobility in nuclei (Figure 3A). Further quantification of the data revealed that GFP-TFIISmut redistributed more slowly after photobleaching than GFP-TFIIS, resulting in an increased half-time of the protein (t1/2 = 0.74 s versus t1/2 = 0.26 s), while the mobile fraction (Mf = 69.7% versus Mf = 97.7%) of GFP-TFIISmut is reduced relative to that of GFP-TFIIS (Figures 3B and 3C). Taken together, our results suggest that TFIISmut associates properly with the RNAPII-TEC, but it exhibits a reduced nuclear mobility, which may be due to RNAPII stalling upon association with TFIISmut.
Figure 3.
Nuclear Mobility of TFIISmut Is Markedly Lower Than That of TFIIS.
(A) to (C) FRAP analysis of Arabidopsis PSB-D cells induced for 24 h to express TFIISmut or TFIIS. Mean fluorescence recovery curves ± sd (full-scale normalization) representing 40 prebleach and 50 postbleach time points (A). Calculated t1/2 (B) and Mf (C) represented as box plots with *P < 0.05 according to Student’s t test.
Marked Changes in the Transcriptome Induced by the Expression of TFIISmut
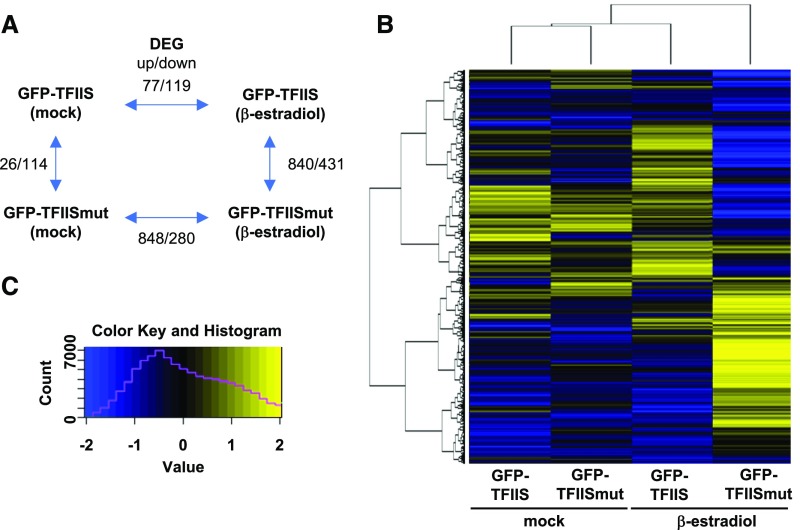
To explore whether the presence of TFIISmut (relative to that of TFIIS) results in an altered transcriptome, changes in mRNA abundance upon induced expression of TFIISmut versus TFIIS were analyzed by RNA sequencing (RNA-seq). cDNAs prepared from polyadenylated mRNAs of total RNA samples of 9-DAS GFP-TFIISmut and GFP-TFIIS seedlings either after 24 h of β-estradiol induction or mock-treated were analyzed by next-generation sequencing. Differentially expressed genes (DEGs; log2fold change (FC) > 1, P < 0.001) were identified between the different plant lines and depending on induction conditions. Modest transcriptomic changes (<200 DEGs) were observed between the mock controls and upon induction of GFP-TFIIS, whereas induction of GFP-TFIISmut resulted in more severe (> 1,100 DEGs) transcriptomic alterations (Figure 4A). Among the DEGs dependent on GFP-TFIISmut expression, upregulation of genes was more prominent than downregulation. Visualization of the transcriptomic changes using a heat map representing all transcribed genes illustrates the relative similarity between the mock-treated samples and GFP-TFIIS induction, whereas the induction of GFP-TFIISmut yields a very different transcriptomic pattern (Figures 4B and 4C). Gene ontology analysis of DEGs indicated that stress-responsive genes are prominently upregulated, while genes involved in cell metabolism are preferentially downregulated (Supplemental Table 1)—which may reflect the fact that TFIISmut expression provokes plant stress responses.
Figure 4.
Expression of TFIISmut Leads to Major Transcriptomic Changes.
(A) Schematic representation of the number of DEGs (up/down) between the analyzed plant lines/conditions.
(B) Heat map (n = 15,836) of the transcriptomic changes between the analyzed plant lines/conditions. Only genes with FPKM ≥ 5 in at least one line/condition were considered and the hierarchical clustering is indicated on the left.
(C) Heat map legend with yellow-blue gradient indicating the log2FPKM [−2; 2]. The magenta line indicates the number of values in a given color range.
Expression of TFIISmut Results in Massive Redistribution of RNAPII over Transcribed Regions
Next, we examined the recruitment of GFP-TFIISmut and GFP-TFIIS to transcribed regions. Using chromatin immunoprecipitation (ChIP) with GFP-specific antibodies, their association with two relatively long genes (At3g02260, At1g49090) was analyzed whose expression (according to the RNA-seq data) was unaffected by the applied induction conditions. The association with different regions of the genes was quantified by qPCR. GFP-TFIISmut and GFP-TFIIS associated with sites around the transcription start site (TSS) and within the transcribed region, but not with non-transcribed regions down-stream of the polyadenylation site (pA; Supplemental Figure 4). Moreover, within the transcribed regions, an increased association of GFP-TFIISmut relative to that of GFP-TFIIS was observed, which may imply perturbed transcriptional elongation in the presence of GFP-TFIISmut. This might be evoked by the accumulation of backtracked/arrested elongation complexes (see below), and is consistent with the observed reduced mobility of TFIISmut-containing complexes.
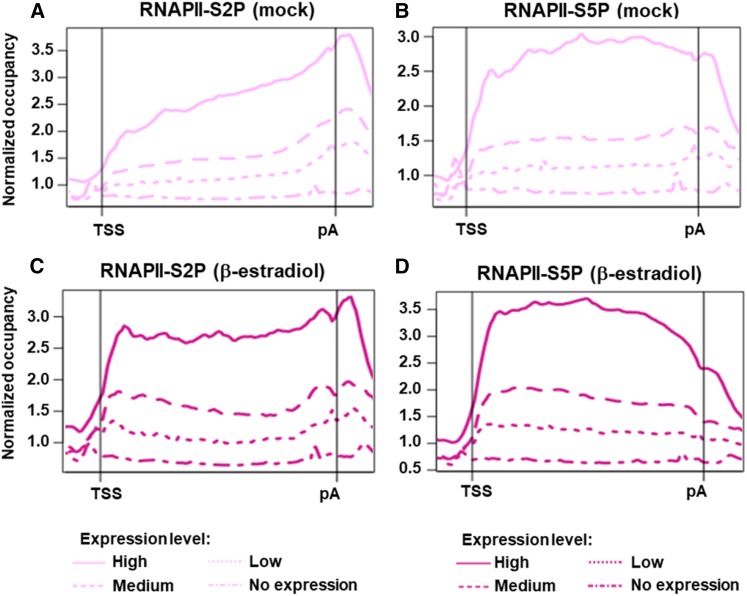
The genomic distribution of elongating RNAPII has not been thoroughly studied in plants. Therefore, the association of RNAPII was analyzed genome-wide by ChIP sequencing (ChIP-seq) using antibodies specific for the phosphorylated forms of the RNAPII-CTD, modified at positions 2 and 5 (S2P and S5P). The role of TFIISmut was examined by analyzing tfIIs plants induced to express GFP-TFIISmut after a 24-h β-estradiol treatment or mock-treated for the same time. Analysis of the ChIP-seq data revealed that, in the mock-treated samples, RNAPII occupancy (S2P and S5P) correlated with gene expression levels, as determined by RNA-seq (Figures 5A and 5B) illustrating the basic accordance of both data sets. Generally, we observed RNAPII accumulation over the transcribed regions of active genes. In the mock control, the distribution of RNAPII-S2P showed an enrichment from the 5′ to the 3′ end with maximal accumulation around the pA (Figure 5A). Compared with that, RNAPII-S5P was more uniformly distributed over the transcribed region (Figure 5B). Genes that were found to be associated with decreased amounts of RNAPII upon GFP-TFIISmut expression tended to show reduced transcript levels, while increased RNAPII association generally was reflected by elevated transcript levels (Supplemental Figures 5 and 6), illustrating the expected connection between RNAPII occupancy and transcriptional outcome. Additionally, the induced expression of GFP-TFIISmut was accompanied by a clear redistribution of both RNAPII-S2P and –S5P toward the TSS (Figures 5C and 5D). We also noticed that RNAPII redistribution differed between long and short genes. For RNAPII-S2P, the redistribution toward the TSS is clearly more pronounced for long genes (TSS-pA >2 kb) than for short genes (TSS-pA < 1 kb), while the differences are less marked for RNAPII-S5P (Supplemental Figure 7).
Figure 5.
The Genomic Occupancy of Elongating RNAPII Is Related to the Corresponding Transcript Levels and the Distribution within Transcribed Regions Is Altered upon Expression of TFIISmut.
(A) to (D) As indicated, gene-averaged profiles represent the distribution of RNAPII-S2P ([A] and [C]) and RNAPII-S5P ([B] and [D]) over transcribed regions (TSS – pA) according to the ChIP-seq data of 10-DAS plants induced for 24 h to express GFP-TFIISmut ([C] and [D]; dark magenta) or mock-treated ([A] and [C]; light magenta). Profiles represent genes with no (18,754), low (3,488), medium (6,975), or high (3,488) transcript levels based on the FPKM values determined by RNA-seq.
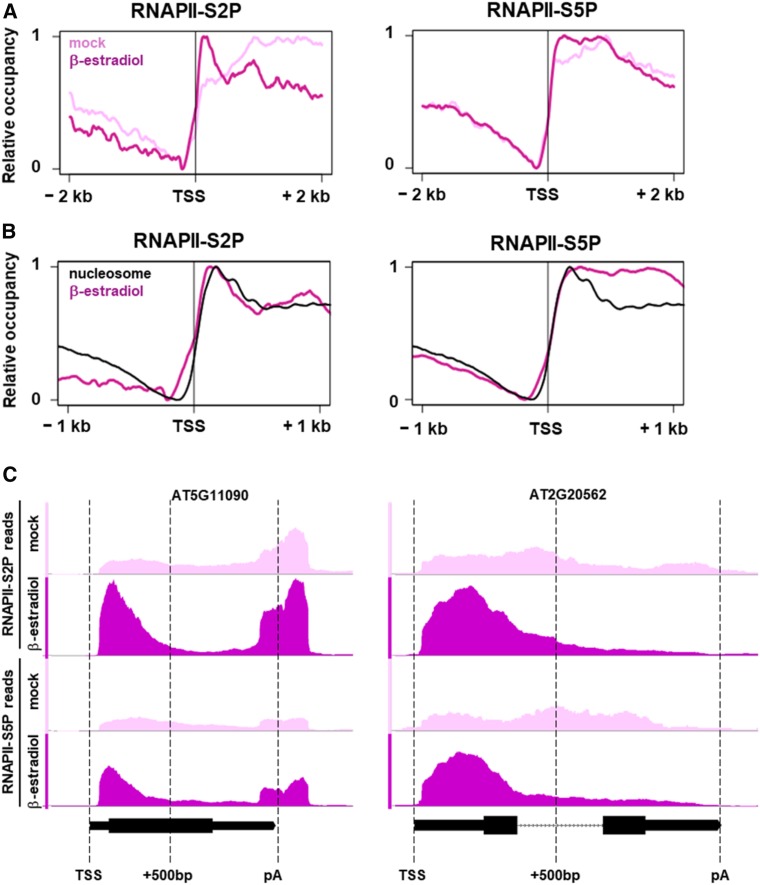
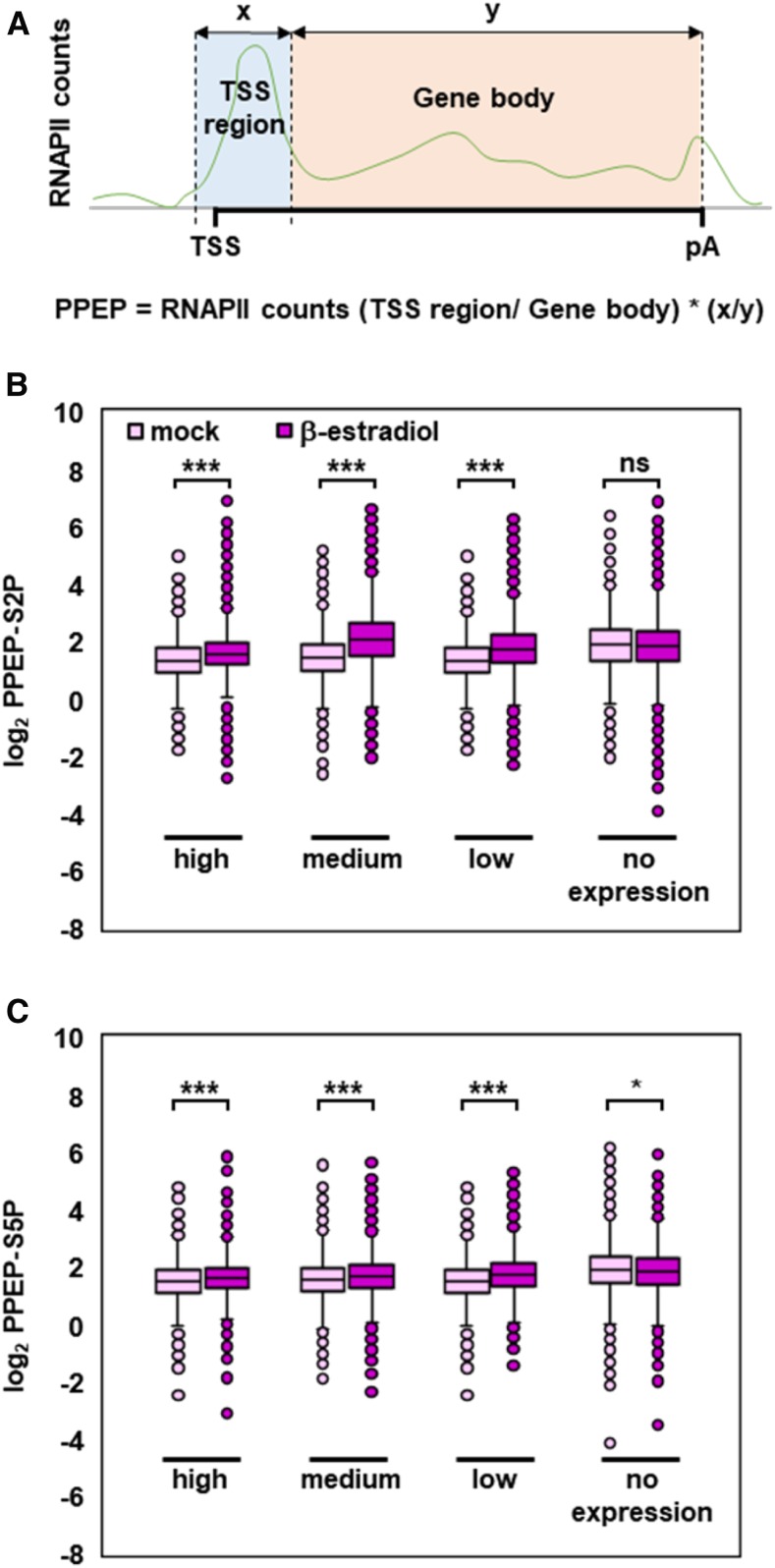
Further analysis of the global RNAPII distribution around the TSS upon GFP-TFIISmut expression demonstrated that the peak, particularly of RNAPII-S2P, overlaps with that of the nucleosome (+1) downstream of the TSS (Figures 6A and 6B). For this comparison, we reanalyzed data determined using the micrococcal nuclease digestion followed by deep sequencing (MNase-seq) approach (Li et al., 2014). The likely stalling of RNAPII at this position downstream of the TSS is also observed for S5P-modified RNAPII, albeit to a lesser extent (Figures 6A and 6B). In view of the changes in the RNAPII profile upon expression of TFIISmut, we quantified the accumulation of RNAPII at the TSS. We calculated the promoter proximal enrichment of RNAPII (PPEP) for RNAPII-S2P (PPEP-S2P) and RNAPII-S5P (PPEP-S5P) as a measure for the association of RNAPII just downstream of the TSS (0 bp to 500 bp) relative to the rest of the transcribed region. Of ∼17,000 genes, 731 and 659 genes upon TFIISmut expression showed significantly increased PPEP-S2P and PPEP-S5P, respectively (Z-score < −2). Genes with significantly decreased PPEP-S2P were not detected, whereas only two genes showed decreased PPEP-S5P (Z-score > 2). Examples of individual genes illustrate the accordance of massive RNAPII redistribution and calculated PPEP values (Figure 6C). To examine whether increased PPEP values correlate with gene expression levels, analyzed genes were subdivided into four classes of expression according to above-described RNA-seq data. Increased PPEP-S2P and PPEP-S5P were observed with genes expressed at high, medium and low levels, but not with non-expressed genes (Figure 7). The magnitude of PPEP changes essentially conforms with the expression level of the gene classes. These analyses demonstrate that upon induced expression of GFP-TFIISmut, RNAPII accumulates close to the TSS, as though in the absence of its transcript cleavage activity RNAPII would undergo frequently transcriptional arrest during early elongation.
Figure 6.
In the Presence of TFIIS, Elongating RNAPII Accumulates Downstream of the TSS at a Position Overlapping with the +1 Nucleosome.
(A) Gene-averaged profiles of the RNAPII-S2P and RNAPII-S5P distribution around the TSS according to ChIP-seq data of 10-DAS plants induced for 24 h to express GFP-TFIISmut (dark magenta) or mock-treated (light magenta). Profiles represent 33,574 and 33,560 loci for RNAPII-S2P and RNAPII-S5P, respectively.
(B) The black line represents the nucleosome occupancy of 14-DAS plants calculated from MNase-seq data (Li et al., 2014) that was combined with the profiles of RNAPII-S2P and RNAPII-S5P after induced GFP-TFIISmut expression.
(C) ChIP-seq profiles of RNAPII-S2P and RNAPII-S5P at the At5g11090 and At2g20562 loci with increased PPEP values upon β-estradiol versus mock treatment. Plots were generated with the Integrative Genomics Browser using representative biological replicates. In gene models, thin bars represent UTRs; thick bars represent exons; dotted lines represent introns.
Figure 7.
TFIISmut-Induced Changes in PPEP Are Related To Transcript Levels.
(A) Scheme depicting the promoter proximal region (0 to +500 bp, blue) and the rest of the transcribed region to pA (orange), as well as calculation of PPEP.
(B) and (C) Changes in PPEP-S2P (B) and PPEP-S5 (C) upon TFIISmut expression versus mock treatment (dark magenta versus bright magenta, respectively) were calculated for genes with no (18,754), low (3,488), medium (6,975), or high (3,488) transcript levels based on FPKM values determined by RNA-seq. PPEP-S2P and PPEP-S5 values are visualized by box plots; asterisks indicate the outcome of the Student’s t test: *P < 0.05, ***P < 0.001.
Proteasomal Degradation of Arrested RNAPII
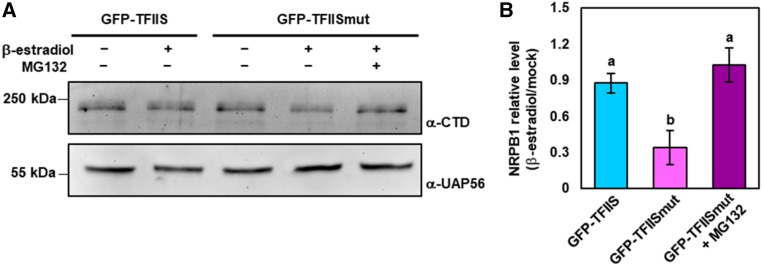
As a last resort, arrested yeast RNAPII is specifically targeted for proteasomal degradation (Somesh et al., 2005). We therefore examined whether a similar mechanism may exist in plants. Protein extracts of plants induced to express GFP-TFIIS and GFP-TFIISmut and corresponding controls were analyzed by immunoblotting using an antibody directed against NRPB1, the large subunit of RNAPII. To suppress protein synthesis, the plants were additionally treated with cycloheximide. Immunoblot analyses revealed that the amount of NRPB1 upon expression of GFP-TFIISmut was reduced by ∼50% relative to control samples, while this effect was not observed upon expression of GFP-TFIIS (Figure 8A). To examine whether the decrease of NRPB1 levels was mediated by the proteasome, GFP-TFIISmut plants were additionally treated with the proteasome inhibitor MG132. In the presence of MG132, the amount of NRPB1 was not reduced (Figures 8A and 8B) and remained at the level of the controls. These findings indicated that similar to the situation in yeast, expression of TFIISmut resulted in increased transcriptional arrest of RNAPII in Arabidopsis and the arrested polymerases were removed by proteasome-mediated degradation.
Figure 8.
Upon TFIISmut Expression, the Amount of the Large Subunit NRPB1 of RNAPII Is Decreased by Proteasomal Degradation.
(A) Ten-DAS plants inducibly expressing GFP-TFIIS or GFP-TFIISmut were treated for 24 h with the inducer β-estradiol (or mock-treated) and the proteasome inhibitor MG132 as indicated. All plants were additionally co-treated with cycloheximide. Total protein extracts of these plants were analyzed by immunoblotting using an antibody directed against the NRPB1-CTD, and as loading control, an antibody directed against the RNA helicase UAP56.
(B) Band intensities were quantified using the software ImageJ (https://imagej.nih.gov/ij/). Colored bars represent mean values ± sd of three independent experiments. Data were statistically analyzed using one-way ANOVA and different characters (a, b) indicate the outcome of a multicomparison Tukey’s test (P < 0.05).
DISCUSSION
Transcript elongation by RNAPII is a discontinuous process and transcription problems leading to backtracking/arrest of the enzyme occur relatively frequently. There are various causes disturbing transcriptional elongation, including certain DNA sequences that are more difficult to transcribe than others, packaging of the DNA into nucleosomes/chromatin, other DNA binding proteins, or topological constraints (Svejstrup, 2007; Gómez-Herreros et al., 2012; Palangat and Larson, 2012). While RNAPI and RNAPIII have a strong intrinsic RNA cleavage activity to reverse transcriptional arrest, the cleavage activity of RNAPII requires assistance by TFIIS to enable rapid backtrack recovery (Ruan et al., 2011; Lisica et al., 2016). In light of this, it seems surprising that TFIIS is not essential for the growth of yeast cells (Archambault et al., 1992; Nakanishi et al., 1992) and that the growth of Arabidopsis tfIIs mutant plants under standard conditions is largely indistinguishable from that of wild-type plants (Grasser et al., 2009).
The TFIIS acidic hairpin is highly conserved (Figure 1A) and the inhibitory effect of mutation of the two invariant acidic hairpin residues on the RNA cleavage activity of RNAPII has been demonstrated comprehensively (Jeon et al., 1994; Kettenberger et al., 2004; Sigurdsson et al., 2010; Imashimizu et al., 2013). Thus, it may be expected that plant acidic hairpin mutant TFIISmut comparably inhibits transcript cleavage by RNAPII. Consistently, expression of TFIISmut causes various growth and developmental defects in wild-type Arabidopsis plants (Dolata et al., 2015). The observation that viable tfIIs plants constitutively expressing TFIISmut cannot be recovered suggests that the RNA cleavage activity of RNAPII is critical for viability in plants, as it is in yeast (Sigurdsson et al., 2010). In wild-type plants, endogenous TFIIS competes with TFIISmut for RNAPII binding, resulting in partial suppression of transcript cleavage activity, whereas in tfIIs plants in the absence of endogenous TFIIS, TFIISmut efficiently inhibits cleavage activity. Inducible expression of TFIISmut in tfIIs plants confirmed the detrimental effects on Arabidopsis growth and viability.
Comparative characterization of protein interactions of TFIISmut and TFIIS revealed that both proteins interact preferentially with the elongating RNAPII as components of the TEC. Notably, FRAP measurements demonstrated that in Arabidopsis nuclei TFIISmut displayed a lower mobility (fluorescence recovery, Mf) than TFIIS. This is consistent with the more prominent association of TFIISmut with arrested RNAPII. Transcriptome analyses demonstrated that induced expression of TFIISmut resulted in substantial changes in mRNA abundance. Expression of TFIIS caused only minor transcriptomic changes, as expected from the mild phenotype of tfIIs plants and their moderate differences in mRNA abundance relative to wild-type plants (Grasser et al., 2009). In the presence of TFIISmut, we observed a preferential downregulation of genes related to cell metabolism as well as an upregulation of stress-responsive genes. Both effects may be a consequence of the fact that transient TFIISmut expression results in frequent transcriptional arrest, provoking severe plant stress symptoms along with corresponding transcriptional reprogramming (Baena-González and Sheen, 2008; Less et al., 2011; Cohen and Leach, 2019), eventually compromising cell viability.
In plants, genome-wide distribution of transcribing RNAPII is not well investigated, but global run-on sequencing (GRO-seq) and native elongating transcript sequencing analyses indicated certain differences relative to yeast and mammals (Hetzel et al., 2016; Zhu et al., 2018). Thus, RNAPII was found to display characteristic accumulation in proximity to pAs. Our ChIP-seq experiments using antibodies directed against elongating RNAPII (CTD S2P, S5P) generally showed an enrichment over the transcribed region of active genes consistent with earlier analyses of individual loci by ChIP-qPCR (Ding et al., 2011; Dürr et al., 2014), and association of RNAPII is in good agreement with transcript levels of the corresponding genes. For RNAPII-S2P, an accumulation over the transcribed region of genes toward the pA was observed that is consistent with the above-mentioned enrichment of RNAPII at pAs (Hetzel et al., 2016; Zhu et al., 2018). In general, the distribution pattern of RNAPII-S2P along transcribed regions is similar to that observed in yeast and mammals (Hajheidari et al., 2013; Jeronimo et al., 2016; Harlen and Churchman, 2017). On the other hand, Arabidopsis RNAPII-S5P was relatively evenly distributed over transcribed regions, whereas, in yeast and mammals, RNAPII-S5P peaks at the TSS and levels decline toward the pA (Hajheidari et al., 2013; Jeronimo et al., 2016; Harlen and Churchman, 2017).
Induced expression of TFIISmut resulted in a clear redistribution of elongating RNAPII. Regarding the distribution of both RNAPII-S2P and –S5P, we observed a substantial shift within transcribed regions toward the TSS (PPEP). In yeast, the density of RNAPII became progressively lower within the transcribed region upon TFIISmut expression, suggesting that TFIISmut inhibits transcript elongation (Sigurdsson et al., 2010). Recent studies demonstrated that co-expression of mouse TFIISmut, along with the endogenous TFIIS in human cells, impaired the ability of 5′ paused RNAPII to resume elongation due to inhibition of the RNA cleavage activity (Sheridan et al., 2019). In addition, it was found that the expression of TFIISmut in human cells resulted in slower elongation rates of RNAPII and in relative depletion of polymerases toward the end of genes (Zatreanu et al., 2019). The importance of TFIIS for release of stalled RNAPII to proceed elongation was also demonstrated in Drosophila (Adelman et al., 2005). Neither the distribution of elongating RNAPII mapped in the absence of TFIIS in this study, nor GRO-seq analyses (Hetzel et al., 2016), indicated RNAPII pausing during early elongation in plants in the manner of that exemplified by mammals. Promoter-proximal pausing of RNAPII was recently reported at a subset of Arabidopsis genes based on GRO-seq and native elongating transcript sequencing experiments (Zhu et al., 2018). However, in this study a rather broad peak of RNAPII accumulation was detected ∼+300 down-stream of the TSS, whereas the well-characterized promoter-proximal pausing in mammals occurs in a narrow region between +25 and +50 (Core and Adelman, 2019). Therefore, it remains to be clarified whether the accumulation of plant RNAPII down-stream of the TSS indeed represents RNAPII pausing comparable to that described in mammals or whether it represents a slowing-down of transcriptional elongation possibly due to obstacles to transcription (see also below). Consistent with the lack of compelling evidence for RNAPII pausing in Saccharomyces cerevisiae, Caenorhabditis elegans, and plants, orthologs of the subunits of the important pausing regulatory complex NELF are not encoded in genomes of organisms including yeast, worms and plants. This implies a functional connection between NELF and promoter-proximal RNAPII pausing and its regulated escape into productive elongation (van Lijsebettens and Grasser, 2014; Core and Adelman, 2019).
The observed TFIISmut-induced PPEP in Arabidopsis distinctly overlaps with the position of the +1 nucleosome at the 5′ end of the transcribed region. The +1 nucleosome is most tightly positioned and often contains histone variants such as H2A.Z, features that are apparently also relevant in plants (Jiang and Pugh, 2009; Struhl and Segal, 2013; Jarillo and Piñeiro, 2015). Nucleosomes in general represent obstacles to transcript elongation by RNAPII (Kulaeva et al., 2013; Teves et al., 2014) and passage of the +1 nucleosome appears to be particularly critical for the efficiency of transcription (Nock et al., 2012; Weber et al., 2014). This could be related to the elongation rate of RNAPII, which is slowest near promoters (Danko et al., 2013). Various studies have demonstrated that TFIIS can partially relieve the nucleosomal barrier to transcript elongation (Kireeva et al., 2005; Ujvári et al., 2008; Nock et al., 2012; Ishibashi et al., 2014). The stimulatory effect of TFIIS on chromatin transcription likely is related to the fact that RNAPII frequently backtracks upon encountering the nucleosome barrier, which can lead to nucleosome-induced RNAPII arrest (Kireeva et al., 2005; Weber et al., 2014). Therefore, the accumulation of RNAPII upon expression of Arabidopsis TFIISmut at the position of the +1 nucleosome may reflect that the enzyme in the absence of its RNA cleavage activity cannot efficiently pass the nucleosomal barrier, resulting in a significant fraction of arrested RNAPII. Because permanently stalled RNAPII complexes obstruct transcription of affected genes, in yeast and mammals arrested RNAPII becomes ubiquitinated and degraded by the proteasome (Wilson et al., 2013). The induced expression of TFIISmut resulted in reduced levels of RNAPII that can be reverted by the proteasome inhibitor MG132, suggesting that the proteasomal degradation pathway of arrested RNAPII is essentially conserved in plants. However, expression of TFIISmut efficiently inhibits the RNA cleavage activity of RNAPII, resulting in prevailing transcriptional arrest. Consequently, the degradation machinery is not capable anymore to sufficiently remove arrested RNAPII complexes, causing profound transcription problems that may impair plant growth and eventually compromise viability. In conclusion, our analyses indicate that backtracking of RNAPII frequently occurs in plant cells during transcriptional elongation on chromatin templates, and that they underscore the importance of reactivating backtracked/arrested RNAPII for efficient transcript elongation as a prerequisite for correct transcriptional output and proper plant growth/development.
METHODS
Plasmid Constructions
The required gene or cDNA sequences were amplified by PCR with KAPA DNA polymerase (PeqLab) using Arabidopsis (Arabidopsis thaliana) genomic DNA or cDNA as template, and the primers (providing also the required restriction enzyme cleavage sites) listed in Supplemental Table 2. The PCR fragments were inserted into suitable plasmids using standard methods or Gateway cloning technology. All plasmid constructions were checked by DNA sequencing, and details of the plasmids generated in this work are summarized in Supplemental Table 3.
Plant Material, Transformation, and Documentation
Arabidopsis (Col-0) was grown at 21°C under long-day conditions (16-h photoperiod per day, 100 μmol m−2 s−1 using white fluorescent tubes (Master Eco; Philips) on MS medium (Dürr et al., 2014; Antosz et al., 2017). The T-DNA insertion line tfIIs-1 was reported in Grasser et al. (2009). After sowing, seeds were stratified in darkness for 48 h at 4°C before incubation in a plant incubator (Percival Scientific). Agrobacterium-mediated transformation of Col-0 or tfIIs plants and characterization of obtained transgenic lines was performed as previously described by Dürr et al. (2014) and Antosch et al. (2015). Plant phenotypes including root growth in the presence of β-estradiol were documented as described by Lolas et al. (2010) and Dürr et al. (2014). For other experiments, plants were initially grown vertically on MS medium with 1% (w/v) phyto agar and then transferred onto MS medium supplemented with 2 μM of β-estradiol (Sigma-Aldrich). Additionally, transferred plants were covered with sterile metal grids, submerged in liquid MS medium supplemented with 2 μM of β-estradiol, and vacuum-infiltrated for 10 min. Excess liquid MS medium was subsequently removed, and plants were incubated for the desired induction time.
PCR-Based Genotyping and qPCR
To distinguish among plants being wild-type, heterozygous, or homozygous for the T-DNA insertions, genomic DNA was isolated from leaves. Genomic DNA was used for PCR analysis with Taq DNA polymerase (PeqLab) and primers specific for DNA insertions and target genes (Supplemental Table 2). DNA precipitated during ChIP assay was analyzed by qPCR with locus-specific primers (Supplemental Table 2). Five microliters of precipitated DNA were used for amplification by PCR using KAPA SYBR FAST Universal reagents (PeqLab) and a Mastercycler ep realplex2 (Eppendorf). Data was analyzed after normalization to the input as described in Dürr et al. (2014).
Antibodies
For ChIP and/or immunoblotting, the following primary antibodies were used: α-RNAPII-CTD-S2P (cat. no. ab5095; Abcam), α-RNAPII-CTD-S5P (cat. no. ab1791; Abcam), α-RNAPII-CTD (cat. no. ab817; Abcam), GFP-Trap (ChromoTeq), α-GS (Sigma-Aldrich), and α-UAP56 (Kammel et al., 2013). Antibodies were used at dilutions of 1:2,000 (immunoblots) and 1:100 (ChIP).
AP-MS Analysis of Proteins
Arabidopsis-suspension–cultured PSB-D cells were maintained and transformed as previously described by van Leene et al. (2015). Isolation of GS-tagged proteins, purification, and MS analyses were essentially performed as previously described by Antosz et al. (2017). In brief, after sonification of 15 g of cells in extraction buffer, the unspecific endonuclease Benzonase (50 U/mL extract) was added to degrade DNA and RNA. GS-tagged proteins were affinity-purified using IgG-coupled magnetic beads and eluted proteins were analyzed by SDS-PAGE and digested with trypsin. Peptides were separated by reversed-phase chromatography, and the liquid chromatography system was coupled on-line to a maXis plus UHR-QTOF System (Bruker Daltonics) via a nanoflow electrospray source (Bruker Daltonics). Data-dependent acquisition of MS/MS spectra and processing of the data was performed as previously described by Antosz et al. (2017). The experimental background of contaminating proteins isolated with the unfused GS-tag, or which co-purify nonspecifically independent of the used bait protein, was subtracted.
Plant Protein Extraction and Immunoblotting
Whole protein extracts from PSB-D cultures were prepared using the procedure described above, whereas the whole protein extracts from Arabidopsis seedlings were prepared as described by Tsugama et al. (2011). After separation by SDS-PAGE, proteins were electro-transferred onto a PVDF membrane and detected by chemiluminescence (Antosch et al., 2015; Antosz et al., 2017).
Light Microscopy and FRAP
For cell imaging, roots (5 DAS to 10 DAS) and fragments of Arabidopsis leaves (5 DAS) were mounted in water on objective slides with coverslips. Confocal laser scanning microscopy (CLSM) was performed using a SP8 microscope (Leica) equipped with 40×/1.3 oil or 63×/1.3 glycerol objectives as previously described by Dürr et al. (2014) and Pfab et al. (2018). GFP was excited using an Argon laser at 488 nm and mCherry/propidium iodide was excited using a laser (DPSS) at 561 nm. GFP emission was detected with a hybrid detector at 500 nm to 550 nm. For FRAP, PSB-D cells were used and a rectangle region-of-interest area of 9 μm2 was defined for bleaching performed at 100% laser power with eight bleaching pulse iterations. Fifty prebleach and 100 postbleach images were taken with 1% laser power. The raw measurements were processed using the software easyFRAP with double full-scale normalization (Rapsomaniki et al., 2012). Obtained normalized values were used to determine t1/2 and Mf. All pictures were generated using the software X-Las (Leica).
Transcript Profiling by RNA-Seq
Total RNA was isolated from 10-DAS seedlings grown vertically on sterile plates with MS medium with or without 24-h β-estradiol induction using the RNeasy R Mini Plant kit (Qiagen). cDNA libraries were prepared using a TruSeq Stranded mRNA Sample Preparation Kit (Illumina) and quality was assessed using a Bioanalyzer 2100 (Agilent Technologies). Sequencing was performed using a HiSeq 1000 (Illumina) instrument. Raw reads were trimmed using the tool Trimmomatic (Bolger et al., 2014) with a minimum average quality score >30 and a minimum length of 70 bp. Processed reads were mapped to the Arabidopsis reference transcriptome assembly (The Arabidopsis Information Resource 10) using the softwares TopHat 2.1.1 (Kim et al., 2013) and BowTie v2.3.4 (Langmead and Salzberg, 2012), allowing up to two base mismatches per read. Reads mapped to multiple locations were discarded. For the analysis of DEGs, data was transformed logarithmically and normalized using a quantile normalization method. DEGs were determined by the software DESeq2 (Love et al., 2014) with a threshold |log2 Fragments Per Kilobase of transcript per Million (FPKM)| > 2 and an adjusted P-value < 0.001 using Benjamini and Hochberg’s method (Benjamini and Hochberg, 1995) for multiple testing. DEG analysis and heat maps were generated using R environment (v3.2.2; https://www.r-project.org/). Genes with FPKM ≥ 5 were considered transcriptionally active.
ChIP-qPCR and ChIP-Seq
Seedlings (10 DAS) grown on MS medium were treated with 2 µM of β-estradiol or ethanol (mock) for 24 h before harvesting. After cross linking with 1% (v/v) formaldehyde, plant material was further processed and used for immunoprecipitation as described in Dürr et al. (2014) and Antosz et al. (2017). For ChIP-qPCR, 5 μL of precipitated DNA (diluted 1:200 for input and 1:10 for GFP-trap and the control without antibody) was analyzed by qPCR with locus-specific primers (Supplemental Table 2).
For ChIP-seq, libraries were generated using the DNA SMART ChIP-Seq kit (Clontech) and quality was assessed using a model no. 2100 Bioanalyzer (Agilent Technologies). Sequencing was performed with the HiSeq 1000 (Illumina). Raw reads were trimmed with the tool Trimmomatic (Bolger et al., 2014) and mapped to the Arabidopsis reference genome (The Arabidopsis Information Resource) using the software BowTie v2.3.4. Nucleosome occupancy was determined from MNase-seq data (Li et al., 2014).
Data processing was performed using “bamCoverage” from the DeepTools suite (Ramírez et al., 2014) to calculate read density per nucleotide and three biological replicates were merged before the calculation. Meta-analysis of relative RNAPII distribution over transcribed loci (TSS-pA) and additional regions was performed using “computeMatrix” from the DeepTools suite (with maxThreshold 500). RNAPII density was additionally determined around the TSS (−2000; 2000) and pA (−2,000; 2,000). The software suite HTSeq (Anders et al., 2015) was used to determine RNAPII read counts within gene bodies. Obtained raw values were normalized using the program DEseq2 (Love et al., 2014) and averaged for three biological replicates. FC between studied conditions was calculated and transformed logarithmically. Significantly different occupancy was determined with a threshold |log2FC| > 1 and P-value < 0.1 adjusted using the Benjamini–Hochberg method for multiple testing. RNAPII-S2P and RNAPII-S5P profiles over single genes were visualized using the Integrative Genomics Browser (http://software.broadinstitute.org/software/igv/) with standard settings.
To determine PPEP, RNAPII read counts from three biological replicates were summed up and the number of tags was determined for each gene in the gene body (+500 bp to pA) and in the promoter-proximal region (TSS to +500 bp). Genes with ≥10 or more tags in the promoter-proximal region were further analyzed and the number of tags were calculated in the promoter proximal region relative to the gene body region. Obtained values were compared relatively between studied conditions and transformed logarithmically resulting in log2 PPEP values for which Z-scores were calculated (Juntawong et al., 2014). Changes in PPEP were considered significantly different for the |Z-score| > 2 between studied conditions.
Accession Numbers
Sequence data of TFIIS (At2g38560) and RNAPII-NRPB1 (At4g35800) can be found in the GenBank/EMBL libraries.
RNA-seq and ChIP-seq data were deposited in the NCBI Gene Expression Omnibus under the accession numbers GSE129072 and GSE128316, respectively.
Supplemental Data
Supplemental Figure 1. β-estradiol–induced expression of GFP-TFIIS and GFP-TFIISmut in tfIIs plants.
Supplemental Figure 2. Analysis of the induction of GFP-TFIIS and GFP-TFIISmut by CLSM.
Supplemental Figure 3. Association TFIIS and TFIISmut with RNAPII.
Supplemental Figure 4. TFIISmut accumulates within transcribed regions.
Supplemental Figure 5. RNAPII-S2P and RNAPII-S5P occupancy correlates with transcript levels.
Supplemental Figure 6. Genome-wide correlation of RNAPII-S2P/-S5P occupancy and transcript levels.
Supplemental Figure 7. Redistribution of RNAPII-S2P/-S5P upon TFIISmut expression in relation to gene length.
Supplemental Table 1. Overrepresented GO terms among the DEGs upon TFIISmut expression.
Supplemental Table 2. Oligonucleotide primers used in this study and in construction of plasmids.
Supplemental Table 3. Plasmids used in this study.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Simon Obermeyer for TFIIS structure modelling, Simon Mortensen for contributions during early stages of the project, Eduard Hochmuth for recording mass spectra, and Peter Doerner and Arp Schnittger for the pCYCB1;1-GFP marker line. This work was supported by the German Research Foundation (grant GR1159/14-2 to K.D.G. and SFB960 to T.D.), the Swiss National Foundation (Sinergia grant CRSII3_154471 to Y.P.), and the EC Marie Curie Research Training Network “Chromatin in Plants – European Training and Mobility” (CHIP-ET FP7-PEOPLE-2013-ITN607880 to K.D.G.).
AUTHOR CONTRIBUTIONS
W.A. and K.D.G. designed the research and wrote the article; W.A., J.D., and K.B. performed the research; W.A., J.D., K.B., A.B., Y.P., T.D., and K.D.G. analyzed the data; A.B., Y.P., T.D., and K.D.G. provided the reagents and the tools for the analysis; all authors commented on the article and contributed to the writing.
Footnotes
Articles can be viewed without a subscription.
References
- Adelman K., Marr M.T., Werner J., Saunders A., Ni Z., Andrulis E.D., Lis J.T.(2005). Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol. Cell 17: 103–112. [DOI] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., Huber W.(2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antosch M., Schubert V., Holzinger P., Houben A., Grasser K.D.(2015). Mitotic lifecycle of chromosomal 3xHMG-box proteins and the role of their N-terminal domain in the association with rDNA loci and proteolysis. New Phytol. 208: 1067–1077. [DOI] [PubMed] [Google Scholar]
- Antosz W., et al. (2017). The composition of the Arabidopsis RNA polymerase II transcript elongation complex reveals the interplay between elongation and mRNA processing factors. Plant Cell 29: 854–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archambault J., Lacroute F., Ruet A., Friesen J.D.(1992). Genetic interaction between transcription elongation factor TFIIS and RNA polymerase II. Mol. Cell. Biol. 12: 4142–4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González E., Sheen J.(2008). Convergent energy and stress signaling. Trends Plant Sci. 13: 474–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y.(1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B 57: 289–300. [Google Scholar]
- Bentsink L., Jowett J., Hanhart C.J., Koornneef M.(2006). Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 17042–17047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B.(2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B.(2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand L., Hörler M., Nüesch E., Vassalli S., Barrell P., Yang W., Jefferson R.A., Grossniklaus U., Curtis M.D.(2006). A versatile and reliable two-component system for tissue-specific gene induction in Arabidopsis. Plant Physiol. 141: 1194–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürckstümmer T., Bennett K.L., Preradovic A., Schütze G., Hantschel O., Superti-Furga G., Bauch A.(2006). An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat. Methods 3: 1013–1019. [DOI] [PubMed] [Google Scholar]
- Chen F.X., Smith E.R., Shilatifard A.(2018). Born to run: Control of transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 19: 464–478. [DOI] [PubMed] [Google Scholar]
- Cheung A.C.M., Cramer P.(2011). Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature 471: 249–253. [DOI] [PubMed] [Google Scholar]
- Cohen S.P., Leach J.E.(2019). Abiotic and biotic stresses induce a core transcriptome response in rice. Sci. Rep. 9: 6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core L., Adelman K.(2019). Promoter-proximal pausing of RNA polymerase II: A nexus of gene regulation. Genes Dev. 33: 960–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danko C.G., Hah N., Luo X., Martins A.L., Core L., Lis J.T., Siepel A., Kraus W.L.(2013). Signaling pathways differentially affect RNA polymerase II initiation, pausing, and elongation rate in cells. Mol. Cell 50: 212–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Avramova Z., Fromm M.(2011). Two distinct roles of ARABIDOPSIS HOMOLOG OF TRITHORAX1 (ATX1) at promoters and within transcribed regions of ATX1-regulated genes. Plant Cell 23: 350–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolata J., Guo Y., Kołowerzo A., Smoliński D., Brzyżek G., Jarmołowski A., Świeżewski S.(2015). NTR1 is required for transcription elongation checkpoints at alternative exons in Arabidopsis. EMBO J. 34: 544–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürr J., Lolas I.B., Sørensen B.B., Schubert V., Houben A., Melzer M., Deutzmann R., Grasser M., Grasser K.D.(2014). The transcript elongation factor SPT4/SPT5 is involved in auxin-related gene expression in Arabidopsis. Nucleic Acids Res. 42: 4332–4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish R.N., Kane C.M.(2002). Promoting elongation with transcript cleavage stimulatory factors. Biochim. Biophys. Acta 1577: 287–307. [DOI] [PubMed] [Google Scholar]
- Gómez-Herreros F., de Miguel-Jiménez L., Millán-Zambrano G., Peñate X., Delgado-Ramos L., Muñoz-Centeno M.C., Chávez S.(2012). One step back before moving forward: Regulation of transcription elongation by arrest and backtracking. FEBS Lett. 586: 2820–2825. [DOI] [PubMed] [Google Scholar]
- Grasser M., Kane C.M., Merkle T., Melzer M., Emmersen J., Grasser K.D.(2009). Transcript elongation factor TFIIS is involved in Arabidopsis seed dormancy. J. Mol. Biol. 386: 598–611. [DOI] [PubMed] [Google Scholar]
- Hajheidari M., Koncz C., Eick D.(2013). Emerging roles for RNA polymerase II CTD in Arabidopsis. Trends Plant Sci. 18: 633–643. [DOI] [PubMed] [Google Scholar]
- Harlen K.M., Churchman L.S.(2017). The code and beyond: Transcription regulation by the RNA polymerase II carboxy-terminal domain. Nat. Rev. Mol. Cell Biol. 18: 263–273. [DOI] [PubMed] [Google Scholar]
- He Y., Doyle M.R., Amasino R.M.(2004). PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes Dev. 18: 2774–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzel J., Duttke S.H., Benner C., Chory J.(2016). Nascent RNA sequencing reveals distinct features in plant transcription. Proc. Natl. Acad. Sci. USA 113: 12316–12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imashimizu M., Kireeva M.L., Lubkowska L., Gotte D., Parks A.R., Strathern J.N., Kashlev M.(2013). Intrinsic translocation barrier as an initial step in pausing by RNA polymerase II. J. Mol. Biol. 425: 697–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi T., Dangkulwanich M., Coello Y., Lionberger T.A., Lubkowska L., Ponticelli A.S., Kashlev M., Bustamante C.(2014). Transcription factors IIS and IIF enhance transcription efficiency by differentially modifying RNA polymerase pausing dynamics. Proc. Natl. Acad. Sci. USA 111: 3419–3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izban M.G., Luse D.S.(1992). The RNA polymerase II ternary complex cleaves the nascent transcript in a 3′→5′ direction in the presence of elongation factor SII. Genes Dev. 6: 1342–1356. [DOI] [PubMed] [Google Scholar]
- Jarillo J.A., Piñeiro M.(2015). H2A.Z mediates different aspects of chromatin function and modulates flowering responses in Arabidopsis. Plant J. 83: 96–109. [DOI] [PubMed] [Google Scholar]
- Jeon C., Yoon H., Agarwal K.(1994). The transcription factor TFIIS zinc ribbon dipeptide Asp-Glu is critical for stimulation of elongation and RNA cleavage by RNA polymerase II. Proc. Natl. Acad. Sci. USA 91: 9106–9110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeronimo C., Collin P., Robert F.(2016). The RNA polymerase II CTD: The increasing complexity of a low-complexity protein domain. J. Mol. Biol. 428: 2607–2622. [DOI] [PubMed] [Google Scholar]
- Jiang C., Pugh B.F.(2009). Nucleosome positioning and gene regulation: Advances through genomics. Nat. Rev. Genet. 10: 161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juntawong P., Girke T., Bazin J., Bailey-Serres J.(2014). Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: E203–E212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammel C., Thomaier M., Sørensen B.B., Schubert T., Längst G., Grasser M., Grasser K.D.(2013). Arabidopsis DEAD-box RNA helicase UAP56 interacts with both RNA and DNA as well as with mRNA export factors. PLoS One 8: e60644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettenberger H., Armache K.-J., Cramer P.(2003). Architecture of the RNA polymerase II-TFIIS complex and implications for mRNA cleavage. Cell 114: 347–357. [DOI] [PubMed] [Google Scholar]
- Kettenberger H., Armache K.-J., Cramer P.(2004). Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol. Cell 16: 955–965. [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L.(2013). TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kireeva M.L., Hancock B., Cremona G.H., Walter W., Studitsky V.M., Kashlev M.(2005). Nature of the nucleosomal barrier to RNA polymerase II. Mol. Cell 18: 97–108. [DOI] [PubMed] [Google Scholar]
- Kulaeva O.I., Hsieh F.-K., Chang H.-W., Luse D.S., Studitsky V.M.(2013). Mechanism of transcription through a nucleosome by RNA polymerase II. Biochim. Biophys. Acta 1829: 76–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L.(2012). Fast gapped-read alignment with BowTie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Less H., Angelovici R., Tzin V., Galili G.(2011). Coordinated gene networks regulating Arabidopsis plant metabolism in response to various stresses and nutritional cues. Plant Cell 23: 1264–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Liu S., Wang J., He J., Huang H., Zhang Y., Xu L.(2014). ISWI proteins participate in the genome-wide nucleosome distribution in Arabidopsis. Plant J. 78: 706–714. [DOI] [PubMed] [Google Scholar]
- Lisica A., Engel C., Jahnel M., Roldán É., Galburt E.A., Cramer P., Grill S.W.(2016). Mechanisms of backtrack recovery by RNA polymerases I and II. Proc. Natl. Acad. Sci. USA 113: 2946–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Geyer R., van Zanten M., Carles A., Li Y., Hörold A., van Nocker S., Soppe W.J.(2011). Identification of the Arabidopsis REDUCED DORMANCY 2 gene uncovers a role for the polymerase associated factor 1 complex in seed dormancy. PLoS One 6: e22241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lolas I.B., Himanen K., Grønlund J.T., Lynggaard C., Houben A., Melzer M., van Lijsebettens M., Grasser K.D.(2010). The transcript elongation factor FACT affects Arabidopsis vegetative and reproductive development and genetically interacts with HUB1/2. Plant J. 61: 686–697. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S.(2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S.(2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Gil S., Grasser K.D., Mas P.(2018). Targeted recruitment of the basal transcriptional machinery by LNK clock components controls the circadian rhythms of nascent RNAs in Arabidopsis. Plant Cell 30: 907–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen S.A., Grasser K.D.(2014). The seed dormancy defect of Arabidopsis mutants lacking the transcript elongation factor TFIIS is caused by reduced expression of the DOG1 gene. FEBS Lett. 588: 47–51. [DOI] [PubMed] [Google Scholar]
- Nakanishi T., Nakano A., Nomura K., Sekimizu K., Natori S.(1992). Purification, gene cloning, and gene disruption of the transcription elongation factor S-II in Saccharomyces cerevisiae. J. Biol. Chem. 267: 13200–13204. [PubMed] [Google Scholar]
- Nelissen H., et al. (2010). Plant elongator regulates auxin-related genes during RNA polymerase II transcription elongation. Proc. Natl. Acad. Sci. USA 107: 1678–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nock A., Ascano J.M., Barrero M.J., Malik S.(2012). Mediator-regulated transcription through the +1 nucleosome. Mol. Cell 48: 837–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S., Zhang H., Ludwig P., van Nocker S.(2004). A mechanism related to the yeast transcriptional regulator Paf1c is required for expression of the Arabidopsis FLC/MAF MADS box gene family. Plant Cell 16: 2940–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palangat M., Larson D.R.(2012). Complexity of RNA polymerase II elongation dynamics. Biochim. Biophys. Acta 1819: 667–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfab A., Bruckmann A., Nazet J., Merkl R., Grasser K.D.(2018). The adaptor protein ENY2 is a component of the deubiquitination module of the Arabidopsis SAGA transcriptional co-activator complex but not of the TREX-2 complex. J. Mol. Biol. 430: 1479–1494. [DOI] [PubMed] [Google Scholar]
- Ramírez F., Dündar F., Diehl S., Grüning B.A., Manke T.(2014). deepTools: A flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42: W187-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapsomaniki M.A., Kotsantis P., Symeonidou I.E., Giakoumakis N.N., Taraviras S., Lygerou Z.(2012). easyFRAP: An interactive, easy-to-use tool for qualitative and quantitative analysis of FRAP data. Bioinformatics 28: 1800–1801. [DOI] [PubMed] [Google Scholar]
- Ruan W., Lehmann E., Thomm M., Kostrewa D., Cramer P.(2011). Evolution of two modes of intrinsic RNA polymerase transcript cleavage. J. Biol. Chem. 286: 18701–18707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selth L.A., Sigurdsson S., Svejstrup J.Q.(2010). Transcript elongation by RNA polymerase II. Annu. Rev. Biochem. 79: 271–293. [DOI] [PubMed] [Google Scholar]
- Sheridan R.M., Fong N., D’Alessandro A., Bentley D.L.(2019). Widespread backtracking by RNA Pol II Is a major effector of gene activation, 5′ pause release, termination, and transcription elongation rate. Mol. Cell 73: 107–118.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdsson S., Dirac-Svejstrup A.B., Svejstrup J.Q.(2010). Evidence that transcript cleavage is essential for RNA polymerase II transcription and cell viability. Mol. Cell 38: 202–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims R.J. III, Belotserkovskaya R., Reinberg D.(2004). Elongation by RNA polymerase II: The short and long of it. Genes Dev. 18: 2437–2468. [DOI] [PubMed] [Google Scholar]
- Somesh B.P., Reid J., Liu W.F., Søgaard T.M., Erdjument-Bromage H., Tempst P., Svejstrup J.Q.(2005). Multiple mechanisms confining RNA polymerase II ubiquitylation to polymerases undergoing transcriptional arrest. Cell 121: 913–923. [DOI] [PubMed] [Google Scholar]
- Struhl K., Segal E.(2013). Determinants of nucleosome positioning. Nat. Struct. Mol. Biol. 20: 267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svejstrup J.Q.(2007). Contending with transcriptional arrest during RNAPII transcript elongation. Trends Biochem. Sci. 32: 165–171. [DOI] [PubMed] [Google Scholar]
- Teves S.S., Weber C.M., Henikoff S.(2014). Transcribing through the nucleosome. Trends Biochem. Sci. 39: 577–586. [DOI] [PubMed] [Google Scholar]
- Tsugama D., Liu S., Takano T.(2011). A rapid chemical method for lysing Arabidopsis cells for protein analysis. Plant Methods 7: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda-Tomás S., Federici F., Casimiro I., Beemster G.T., Bhalerao R., Swarup R., Doerner P., Haseloff J., Bennett M.J.(2009). Gibberellin signaling in the endodermis controls Arabidopsis root meristem size. Curr. Biol. 19: 1194–1199. [DOI] [PubMed] [Google Scholar]
- Ujvári A., Hsieh F.-K., Luse S.W., Studitsky V.M., Luse D.S.(2008). Histone N-terminal tails interfere with nucleosome traversal by RNA polymerase II. J. Biol. Chem. 283: 32236–32243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leene J., et al. (2015). An improved toolbox to unravel the plant cellular machinery by tandem affinity purification of Arabidopsis protein complexes. Nat. Protoc. 10: 169–187. [DOI] [PubMed] [Google Scholar]
- van Lijsebettens M., Grasser K.D.(2014). Transcript elongation factors: Shaping transcriptomes after transcript initiation. Trends Plant Sci. 19: 717–726. [DOI] [PubMed] [Google Scholar]
- Wang D., Bushnell D.A., Huang X., Westover K.D., Levitt M., Kornberg R.D.(2009). Structural basis of transcription: Backtracked RNA polymerase II at 3.4 Ångström resolution. Science 324: 1203–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber C.M., Ramachandran S., Henikoff S.(2014). Nucleosomes are context-specific, H2A.Z-modulated barriers to RNA polymerase. Mol. Cell 53: 819–830. [DOI] [PubMed] [Google Scholar]
- Wilson M.D., Harreman M., Svejstrup J.Q.(2013). Ubiquitylation and degradation of elongating RNA polymerase II: The last resort. Biochim. Biophys. Acta 1829: 151–157. [DOI] [PubMed] [Google Scholar]
- Wind M., Reines D.(2000). Transcription elongation factor SII. BioEssays 22: 327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Zhang Y.(2015). I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 43 (W1): W174–W181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatreanu D., Han Z., Mitter R., Tumini E., Williams H., Gregersen L., Dirac-Svejstrup A.B., Roma S., Stewart A., Aguilera A., Svejstrup J.Q.(2019). Elongation factor TFIIS prevents transcription stress and R-loop accumulation to maintain genome stability. Mol. Cell 76: 57–69.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Liu M., Liu X., Dong Z.(2018). RNA polymerase II activity revealed by GRO-seq and pNET-seq in Arabidopsis. Nat. Plants 4: 1112–1123. [DOI] [PubMed] [Google Scholar]