The lipid environment of the thylakoid membrane chaperones the folding of an integral signal peptidase whose stability is affected by protonmotive force-mimicking pH conditions.
Abstract
Protein folding is a complex cellular process often assisted by chaperones, but it can also be facilitated by interactions with lipids. Disulfide bond formation is a common mechanism to stabilize a protein. This can help maintain functionality amid changes in the biochemical milieu, including those relating to energy-transducing membranes. Plastidic Type I Signal Peptidase 1 (Plsp1) is an integral thylakoid membrane signal peptidase that requires an intramolecular disulfide bond for in vitro activity. We have investigated the interplay between disulfide bond formation, lipids, and pH in the folding and activity of Plsp1. By combining biochemical approaches with a genetic complementation assay using Arabidopsis thaliana plants, we provide evidence that interactions with lipids in the thylakoid membrane have reconstitutive chaperoning activity toward Plsp1. Further, the disulfide bridge appears to prevent an inhibitory conformational change resulting from proton motive force-mimicking pH conditions. Broader implications related to the folding of proteins in energy-transducing membranes are discussed.
INTRODUCTION
Biological membranes form the boundary between cells and their external environment and are responsible for compartmentalization in eukaryotic cells. Known to be much more than a passive barrier, membranes carry out a wide range of essential processes for living cells, including generation and maintenance of electrochemical gradients, perception and transduction of internal and external signals, transport of biomolecules, and synthesis of lipids. Each of these diverse biological processes requires integral and peripherally associated membrane proteins. Due to their tight association with membranes, the structures and functions of membrane proteins can be influenced by properties of the bilayer such lipid composition, fluidity, charge, and thickness. Indeed, there is a vast amount of evidence supporting a crucial role for the lipid bilayer in the function and/or regulation of membrane proteins. First, phospholipids are present in the structures of numerous membrane proteins and complexes (Lee, 2011) including PSII from the cyanobacterium Thermosynechococcus vulcanus (Umena et al., 2011) and that from spinach (Spinacia oleracea; Wei et al., 2016). In many cases, lipids are well-resolved in protein crystal structures and bind in specific grooves, analogous to prosthetic groups (Lee, 2004). Second, specific lipid species are required for the activity (Soom et al., 2001; Lee, 2004), proper folding (Bogdanov et al., 1996), or membrane insertion (van Klompenburg et al., 1998) of a variety of membrane proteins. Finally, membrane lipids can influence transmembrane helix packing (Lee, 2011), induce conformational changes of extra-membranous domains (Hansen et al., 2011), or stimulate oligomerization of membrane proteins (Stangl and Schneider, 2015). These examples highlight the remarkable versatility of membrane lipids.
In addition to lipids, a fundamental aspect of the molecular environment of proteins within energy-transducing membranes is the proton motive force (pmf), which consists of both a proton gradient (ΔpH) and an electrical gradient (Δ). Changes in the magnitude of the pmf and/or the partitioning between Δ and ΔpH have the potential to alter protein structure and function in several ways, for instance, alteration of the protonation state and in turn the polarity of ionizable side chains (Hamsanathan and Musser, 2018). Consistent with this idea is the observation of pmf-driven conformational changes in voltage-gated ion channels (Catterall et al., 2017), as well as pmf-dependent oligomerization of TatA, a membrane protein involved in the transport of folded proteins in bacteria and chloroplasts via the twin-Arg translocon (Hamsanathan and Musser, 2018). Thus, the structure and function of membrane proteins can be influenced not only by lipids in a bilayer but also by a pmf.
Thylakoid membranes are the energy-transducing membranes in chloroplasts and cyanobacteria and are replete with proteins whose structure and function are influenced by lipids and/or a pmf. For example, the structural stability of light-harvesting LHCII trimers in proteoliposomes is significantly enhanced by the galactolipid monogalactosyl diacyl glycerol (MGDG; Seiwert et al., 2017), which comprises up to 50% of the total lipids found in thylakoid membranes (Dörmann and Benning, 2002). Another example is the membrane protein PsbS, which is influenced by both lipids and the pmf. Specifically, insertion of denatured PsbS into liposomes made of thylakoid lipids promotes it to fold into the native conformation (Liu et al., 2016). Furthermore, protonation of two conserved glutamate residues in PsbS in response to acidification of the thylakoid lumen appears to trigger conformational changes leading to its activation (Niyogi et al., 2005). A third example is the thylakoid lumen protein violaxanthin deepoxidase (VDE), in which the catalytic activity (Jahns et al., 2009) and reversible membrane association (Hager and Holocher, 1993) is ΔpH dependent. VDE activity is also stimulated in the presence of liposomes containing MGDG (Latowski et al., 2002) and by exogenous MGDG (Latowski et al., 2000). Although not an integral membrane protein, VDE still responds to the pmf across thylakoids and depends on specific lipids within the membrane. These and other examples make clear the fact that the pmf can serve a broader role than simply providing the energy for ATP synthesis.
The vast majority of chloroplast proteins are encoded in the nuclear genome, synthesized on cytosolic ribosomes, and posttranslationally targeted to the chloroplast (Shi and Theg, 2013). All known soluble thylakoid lumen proteins (Schubert et al., 2002) and several thylakoid membrane proteins (Mant et al., 1994; Michl et al., 1994; Rodrigues et al., 2011) are synthesized as precursors containing bipartite N-terminal transit peptides consisting of a stromal targeting domain and a thylakoid transfer signal (TTS) in tandem. Homologous to bacterial signal peptides (Paetzel et al., 2002), the TTS targets the protein to the thylakoid and is cleaved off on the lumenal side by the integral membrane thylakoidal processing peptidase (TPP), yielding a mature protein (Albiniak et al., 2012). The major isoform of TPP, called Plastidic type I signal peptidase 1 (Plsp1), and its bacterial homolog leader peptidase (LepB)1 are essential for photoautotrophic growth and processing of lumen-targeted proteins in plants and cyanobacteria, respectively (Zhbanko et al., 2005; Shipman-Roston et al., 2010; Hsu et al., 2011). Additionally, Plsp1 can cleave the TTS from numerous TPP substrates in Triton X-100 micelles (Midorikawa et al., 2014). Lipids and/or the pmf likely influence the structure and activity of Plsp1 given that (1) Plsp1 is an integral membrane protein, (2) the majority of the protein resides in the thylakoid lumen (Midorikawa et al., 2014) in which the pH fluctuates during photosynthesis (Shikanai and Yamamoto, 2017), (3) the lumenal domain exhibits a tendency to bind to thylakoid membranes in vitro (Endow et al., 2015), (4) it cleaves signal peptides at or near the surface of the membrane (Dalbey et al., 2012), and (5) exogenous lipids stimulate the activity of the LepB (Tschantz et al., 1995).
Previous biochemical analyses revealed a key role of disulfide bond formation in the structure and function of Plsp1 (Midorikawa et al., 2014). An angiosperm-specific Cys pair (C166 and C286) residing in the lumenal domain of Plsp1 form a noncatalytic disulfide bond required for Plsp1 in vitro activity (Midorikawa et al., 2014). With use of a homology-based structural model, disulfide bond reduction was suggested to alter the conformation of Plsp1 leading to the observed loss of activity (Midorikawa et al., 2014), similar to well-documented examples of conformational differences between oxidized and reduced forms of proteins (Tanaka and Wada, 1988; Choi et al., 2001; Gopalan et al., 2006; Nishii et al., 2015). Thus, it was concluded that disulfide bond formation, often referred to as oxidative folding (Kieselbach, 2013), plays a key role in maintaining the active conformation of Plsp1 (Midorikawa et al., 2014). This was interesting given that Escherichia coli LepB lacks the corresponding Cys pair (Midorikawa et al., 2014) and contains another nonconserved Cys pair that are dispensable for in vitro activity (Sung and Dalbey, 1992; Paetzel et al., 1997). Given that LepB is essential for E. coli viability (Date, 1983; Inada et al., 1989; see below), the nonconserved Cys pair is also likely to be dispensable in vivo.
In this study, we originally sought to determine whether oxidative folding is required for Plsp1 to function in vivo and, if so, elucidate its biological role. Using a genetic complementation assay and a variety of biochemical approaches, we instead discovered that the disulfide bond in Plsp1 is not essential in vivo and that membrane lipids alone facilitate the proper folding and, in turn, activity of Plsp1. We also provide evidence that, while the disulfide bridge is not required for Plsp1 activity in vivo, it assists in the maintenance of catalytic activity in a relatively acidic environment such as that found in the thylakoid lumen during illumination. Our results establish Plsp1 as a membrane protein for which lipids act as reconstitutive lipochaperones. In addition, Plsp1 demonstrates that a protein can use a disulfide bridge to maintain activity in both the stabilizing and destabilizing environments established by the cycle of diurnal energization of the thylakoid membrane. Our work adds Plsp1 to the growing list of integral thylakoid membrane proteins whose structure and function are significantly affected by membrane lipids and also raises the possibility that formation of disulfide bonds is a common mechanism to maintain the proper conformation of proteins in energized membranes, particularly proteins that exist in the thylakoid lumen.
RESULTS
Disulfide Bond Reduction Causes a Conformational Change in Plsp1
Before this study, a structural change in Plsp1 resulting from in vitro disulfide bond reduction had not been directly tested. Two approaches were taken to investigate this issue.
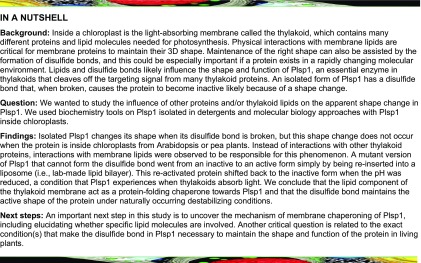
First, we compared the susceptibility of the oxidized and reduced forms of Plsp1 to the protease thermolysin. DTT could not be used as the reductant in this experiment since it inhibited the protease activity of thermolysin (Supplemental Figure 1) due to its ability to chelate the critical Zn2+ cofactor (Cornell and Crivaro, 1972; Pretzer et al., 1992). Instead, TCEP (tris-2-carboxyethylphosphine), which also diminishes the activity of Plsp1 in vitro (Supplemental Figure 1), was used as the reductant. Plsp1 extracted from pea thylakoid membranes with Triton X-100 was treated with or without TCEP, followed by incubation with or without thermolysin. Sample aliquots taken over time were examined by SDS-PAGE and immunoblotting using an antibody raised against the pea ortholog of Plsp1 (Midorikawa et al., 2014). The extracted form of Plsp1 remained relatively stable after 30 min at room temperature regardless of TCEP treatment (Figure 1A). In the presence of thermolysin, Plsp1 was degraded in a time-dependent manner, and this degradation was accelerated if Plsp1 was pretreated with TCEP (Figure 1A). TCEP-treated Plsp1 migrated more slowly than untreated Plsp1 on nonreducing SDS-PAGE (Figure 1B), confirming complete reduction of the disulfide bond (Midorikawa et al., 2014).
Figure 1.
Disulfide Bond Reduction Alters the Structure of Plsp1.
(A) Proteins extracted from pea thylakoids with 0.25% v/v Triton X-100 were pretreated with (+) or without (-) 10 mM TCEP followed by incubation with or without thermolysin. Aliquots taken at 10 (1), 20 (2), and 30 (3) min were analyzed by SDS-PAGE and immunoblotting with an antibody against Pea Plsp1 (top panel). A second gel was loaded with half of the amount of each sample and stained with Coomassie Brilliant Blue after electrophoresis (bottom panel). Input = untreated thylakoid extracts.
(B) An aliquot of the +/− TCEP pretreated thylakoid extracts was analyzed by nonreducing SDS-PAGE and immunoblotting with the antibody against Pea Plsp1. 2SH = reduced Plsp1; S-S = oxidized Plsp1.
(C) Emission spectrum of purified T7-Plsp171-291 (∼0.1 μM) in 1% w/v octyl glucoside after pretreatment with (Blue) or without (Red) 50 mM DTT plotted as fluorescence in counts per second (cps) versus wavelength. Shown are the spectra after subtracting those of buffer blanks. The excitation wavelength was 285 nm.
(D) Processing activity of purified T7-Plsp1 against prPsbO, prPsbP, or prPsbQ. T7-Plsp1 used in (C) was mixed with 35S-Met-labeled substrates after being boiled for 10 min (Δ) or pretreated with 50 mM DTT on ice for 20 min. Reaction mixtures were incubated at ∼25°C for 30 min and analyzed by SDS-PAGE and autoradiography. Arrows denote the processed forms of each substrate tested. TP = translation products.
Our second approach to relate Plsp1’s disulfide to its conformation was based on the intrinsic fluorescence of the three native Trp residues in a purified form of Plsp1 containing an N-terminal T7 tag. When excited with 285 nm light, Plsp1 exhibited strong fluorescence in the range of 300-400 nm with a λmax of 350 ± 2 nm (Figure 1C). After treatment with DTT, the fluorescence spectrum of Plsp1 exhibited a similar shape and λmax (349 ± 2 nm) but with a significant decrease in emission intensities (Figure 1C), which is consistent with an increase in the solvent exposure of Trp side chains. The decrease in fluorescence intensity cannot be attributed to denaturation and aggregation of Plsp1 since the DTT-treated enzyme remains completely soluble (Supplemental Figure 1). Since the quantum yield decreases when a Trp side chain shifts from a buried to a more solvent-exposed local environment (Teale, 1960), reducing the disulfide bond in Plsp1 likely causes a conformational change that increases the exposure of one or more Trp residues to the bulk solvent. As expected, the activity of the enzyme used for the fluorescence experiments is abolished by DTT (Figure 1D). Taken together, the results presented in Figure 1 provide direct evidence that the loss of Plsp1 activity upon disulfide bond reduction can be attributed to a conformational change to an inactive structure.
Plsp1 Cys Residues Form a Disulfide Bond in Intact Chloroplasts
Previous work showed that Cys166 and Cys286 can form an intramolecular disulfide bond in a recombinant form of Plsp1 as well as in Plsp1 in isolated chloroplasts from pea and Arabidopsis (Midorikawa et al., 2014). To confirm that this disulfide bond forms in vivo, we analyzed the in vivo redox state of the Cys in Plsp1 using a thiol labeling assay (Shapiguzov et al., 2016). The endogenous Plsp1 protein in Arabidopsis plants could not be monitored due to a low level of expression. Instead, we transiently expressed T7-tagged Plsp1 in Nicotiana benthamiana leaves to achieve high expression and facilitate detection. A genetic complementation experiment confirmed that T7-Plsp1 is indeed functional (Supplemental Figure 2).
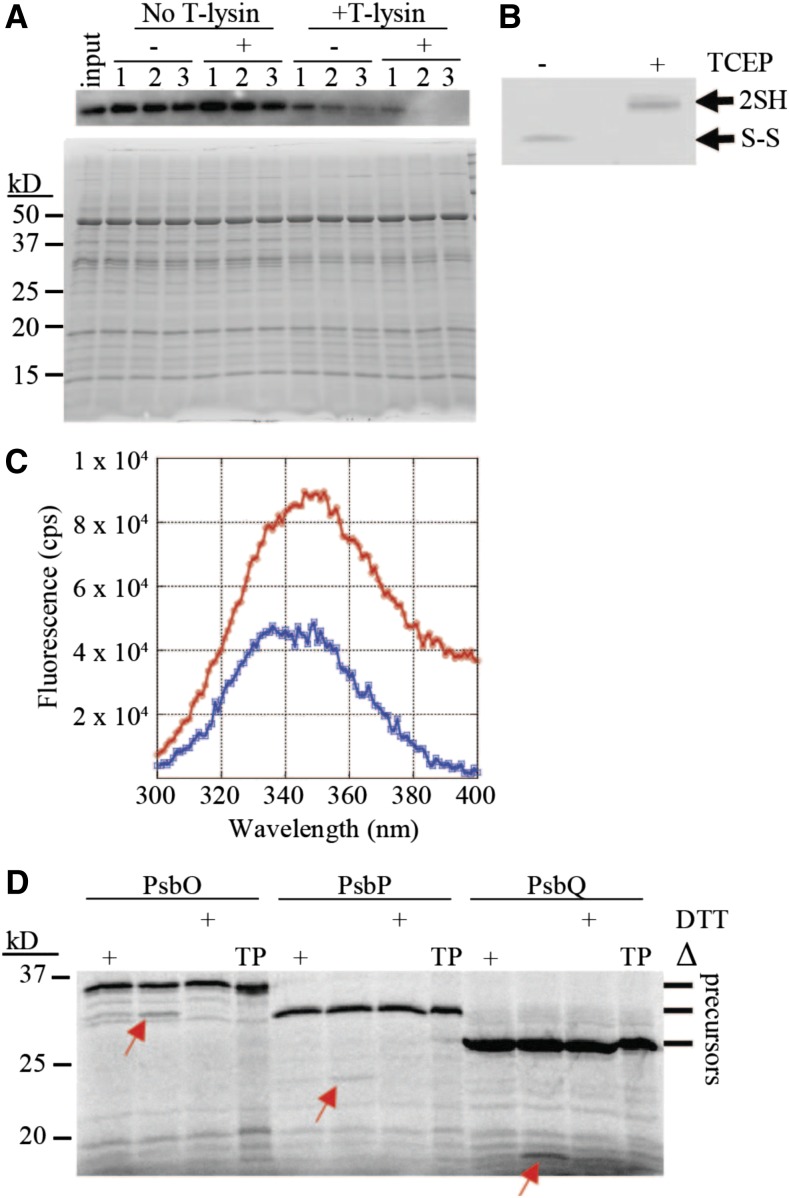
Intact chloroplasts were isolated from leaves after infiltration with Agrobacterium containing a plasmid encoding T7-Plsp1 or after a mock infiltration and were used to probe the redox state of the two Cys residues in Plsp1. After free thiol groups with N-ethyl maleimide (NEM) were blocked, proteins were treated with TCEP followed by addition of methoxypolyethylene glycol maleimide (mPEG-MAL), which confers a large size shift upon labeling the newly exposed thiols. A portion of T7-Plsp1 displays a mobility shift from ∼27 kD to ∼50 kD when TCEP treatment precedes labeling with mPEG-MAL (Figure 2A). This size shift is comparable with that observed when both Cys residues of in vitro translated Plsp1 are labeled with mPEG-MAL (Supplemental Figure 3). We therefore interpret the 50 kD band as T7-Plsp1 with mPEG-MAL on both Cys residues. This size shift was detected regardless of NEM treatment (Figure 2A), which is expected if Plsp1 is initially disulfide bonded. As a control, the single Cys residue in OE23 was labeled by mPEG-MAL, but only when NEM was omitted from the initial chloroplast lysis and membrane solubilization (Figure 2A) confirming that the NEM successfully blocked free thiols in the initial steps of the experiment.
Figure 2.
Plsp1 Cys Form a Disulfide Bond in Chloroplasts.
(A) PEG-MAL labeling of Cys in isolated chloroplast membranes. Thiol labeling was performed as described in Methods. Samples were analyzed by SDS-PAGE and immunoblotting using the α-T7 antibody for detection of T7-Plsp1 (top panel) or the α-OE23 antibody as a control (bottom panel). Unlabeled protein (0). 1 or 2 Cys residues labeled with mPEG-MAL, (1) and (2), respectively. Asterisk indicates unknown immunoreactive protein in N. benthamiana chloroplast membranes.
(B) Coomassie-stained gel of the same samples analyzed in (A).
A minor proportion of T7-Plsp1 was shifted to ∼37 kD (Figure 2A) after TCEP + mPEG-MAL treatment. In addition, a significant proportion of T7-Plsp1 was unlabeled even without NEM treatment. We attribute these results to incomplete mPEG-MAL labeling of thiols and/or disulfide bond reduction by TCEP, and we interpret the ∼37 kD band as T7-Plsp1 with mPEG-MAL on one Cys because the mobility is similar to that of the labeled forms of in vitro translated single Cys Plsp1 mutants (Supplemental Figure 3). Regardless of the technical limitations, it is clear that a significant portion of the T7-Plsp1 population in intact chloroplasts contains an intramolecular disulfide bond.
Processing Activity Is Required for the In Vivo Function of Plsp1
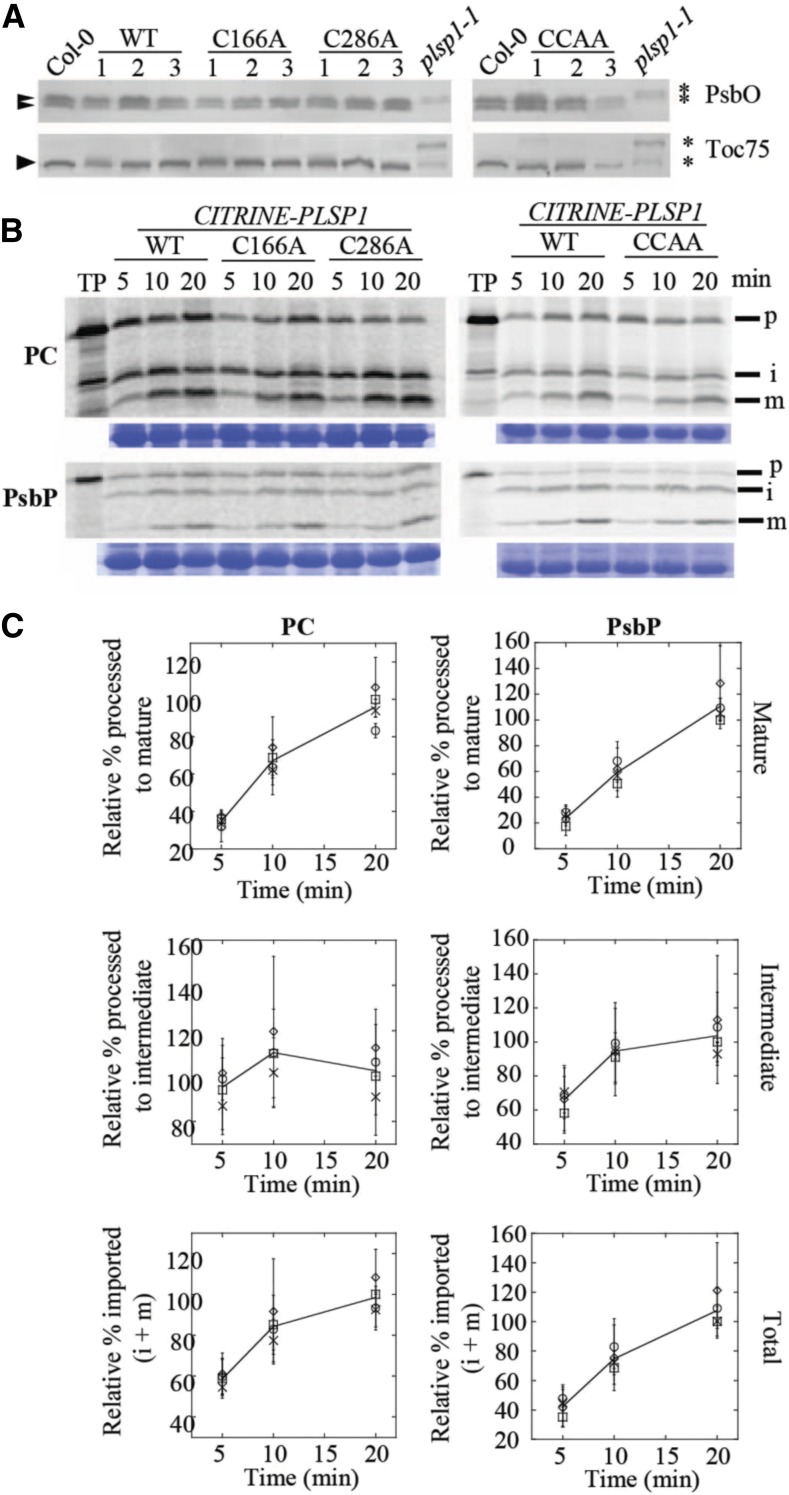
Lack of Plsp1 leads to defective thylakoid development as well as the accumulation of unprocessed forms of several thylakoid lumen proteins (Shipman-Roston et al., 2010; Midorikawa and Inoue, 2013). In addition, overexpression of either of the other two Plsp isoforms (i.e., Plsp2A and Plsp2B) does not rescue the plsp1-1 T-DNA knockout mutant in Arabidopsis (Hsu et al., 2011). These findings led to the conclusions that (1) Plsp1 is the major isoform of the thylakoidal processing peptidase and (2) Plsp1 is responsible for cleaving most, if not all, TTSs in chloroplasts. To test whether Plsp1 directly cleaves thylakoid-transfer signals in vivo, we substituted the catalytic nucleophile Ser142 (Midorikawa et al., 2014) with Ala (i.e., S142A) would disrupt the function of Plsp1 using a genetic complementation approach, as described previously (Endow and Inoue, 2013). For two independent S142A lines, we observed albino hygromycin-resistant seedlings exhibiting a stunted growth phenotype similar to the plsp1-1 mutant (Supplemental Figure 4). Genomic PCR confirmed that the albino hygromycin-resistant seedlings carried the CITRINE-PLSP1 transgene in the plsp1-1 mutant background (Supplemental Figure 4). SDS-PAGE and immunoblotting analysis of total seedling extracts revealed that the Citrine-Plsp1 protein was synthesized in each of the transgenic lines (Supplemental Figure 4). Furthermore, PsbO and Toc75 both accumulated as unprocessed forms in the albino S142A seedlings as in the plsp1-1 mutant (Supplemental Figure 4). Taken together, these data show that expression of a noncatalytic Plsp1 variant cannot rescue the plsp1-1 mutant and also suggest that the other two Plsp isoforms (Plsp2A and 2B) do not compensate for a noncatalytic form of Plsp1, providing a necessary control for the experiments described in Figure 3.
Figure 3.
Expression of Redox-Inactive Citrine-Plsp1 Variants Rescues the plsp1-1 Knockout Mutant.
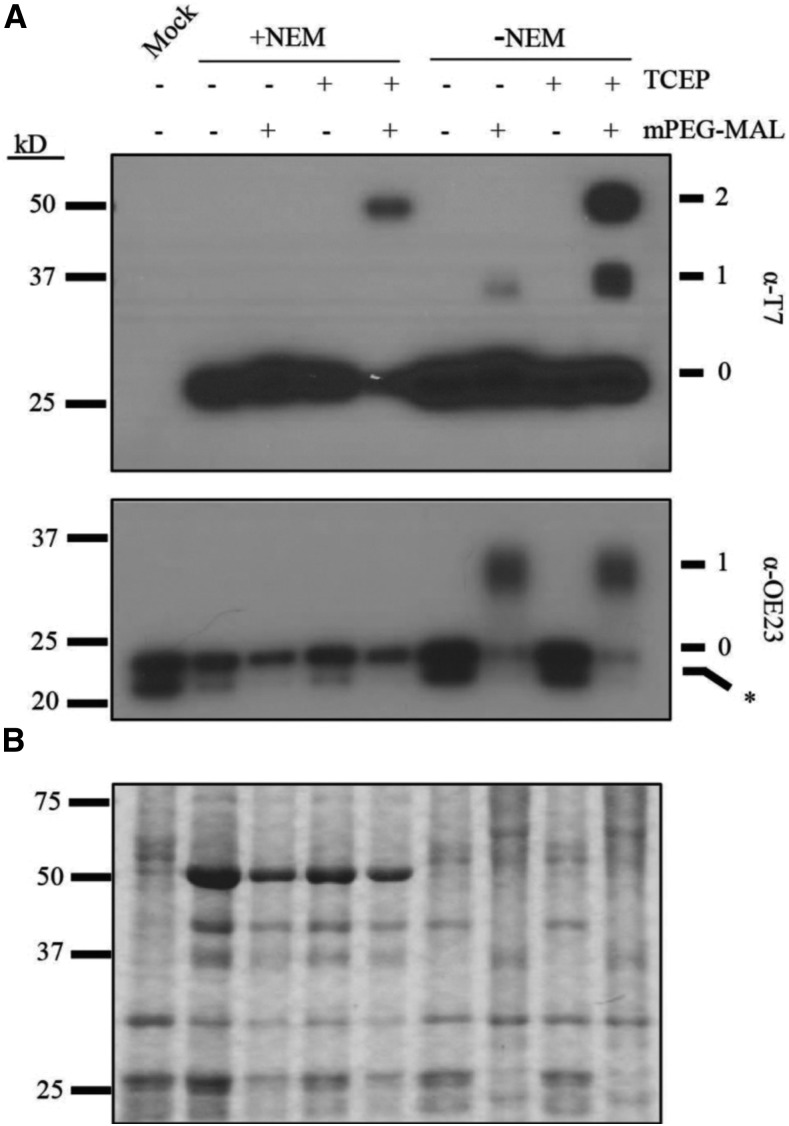
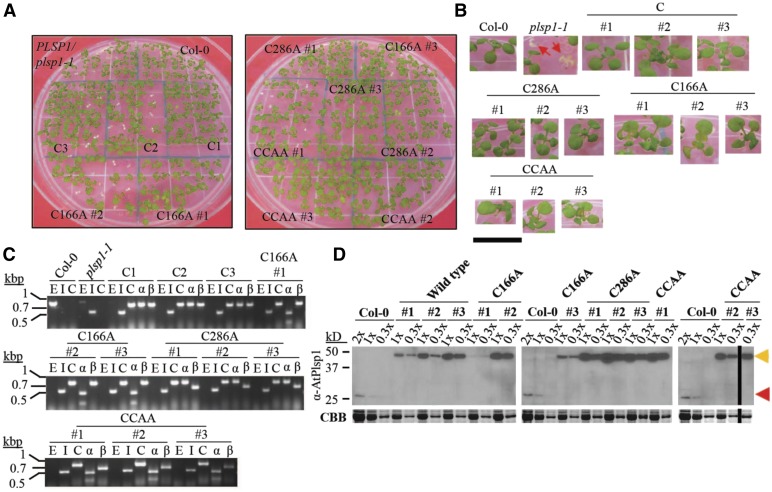
(A) Images of 12-d-old seedlings growing on MS medium supplemented with 1% w/v Suc. C = CITRINE-PLSP1, C166A = CITRINE-PLSP1-C166A; C286A = CITRINE-PLSP1-C286A; CCAA = CITRINE-PLSP1-C166A/C286A.
(B) Close-up images of seedlings from (A). Red arrows indicate plsp1-1 null mutants. Scale bar = 1 cm.
(C) Results of genomic PCR experiment to confirm seedling genotypes. E = PLSP1; I = plsp1-1; C = CITRINE-PLSP1; α = PLSP1-C166A (PstI digestion of C); β = PLSP1-C286A (PvuII digestion of C).
(D) Expression of Citrine-Plsp1 proteins in isolated chloroplasts. Proteins from isolated chloroplasts were analyzed by SDS-PAGE and immunoblotting using the α-AtPlsp1antibody. RbcL bands on duplicate gels stained with Coomassie Brilliant Blue (CBB) are shown below as a loading control. The vertical black bar separates nonadjacent lanes from the same membrane. 1X =1 μg chlorophyll equivalent. Red and orange arrows indicate Plsp1 and Citrine-Plsp1, respectively.
Expression of Redox-Inactive Plsp1 Variants Rescues the plsp1-1 Mutant
To assess whether the in vivo activity of Plsp1 requires a disulfide bond, we tested the functionality of Citrine-tagged Plsp1 variants in which one or both Cys are replaced by Ala. Surprisingly, plants carrying a CITRINE-PLSP1-CA (C166A, C286A, or C166A/C286A) transgene in the plsp1-1 mutant background exhibited a wild type-like phenotype (Figures 3A and 3B). We isolated three independent complemented lines for each construct and checked for the presence of Cys/Ala mutations in PLSP1 by a restriction digest of genomic PCR products (Figure 3C). Chloroplasts from each of the complemented lines expressed Citrine-Plsp1 and lacked the endogenous form of Plsp1 (Figure 3D). Notably, the expression of the Citrine-Plsp1 protein was highly variable across the transgenic lines ranging from close to endogenous Plsp1 levels (e.g., C166A #1) to a level well above the dynamic range of detection (e.g., C286A #2). In addition, the wild-type Citrine-Plsp1 and endogenous Plsp1 displayed redox-dependent mobility on SDS-PAGE, whereas the mobility of the Citrine-Plsp1-CA proteins was not redox dependent (Supplemental Figure 5). This indicates that changing one or both Cys in Plsp1 to Ala prevents disulfide bond formation.
All twelve complemented lines we isolated were homozygous for the plsp1-1 null allele, but five of them were hemizygous for the CITRINE-PLSP1 transgene (Supplemental Figure 5). For each of these lines, the ratio of hygromycin-resistant to-susceptible seedlings was ∼3:1 (Supplemental Figure 5), consistent with segregation for a single transgene insertion. Albino hygromycin-resistant seedlings were never observed. Genomic PCR analysis allowed us to determine that the green wild type-like seedlings were complemented, and the albino seedlings were nontransgenic plsp1-1 mutants. Results of our genetic complementation assays clearly showed that while a disulfide bond is required for in vitro activity, it is not essential for Plsp1 activity in vivo.
Redox-Inactive Plsp1 Variants Partially Complement the E. coli LepB Suppressor Strain FTL85
Because of the unexpected results described in Figure 3, we were also curious whether the disulfide bond in Plsp1 would be dispensable in a nonnative biological context. We decided to use a functional assay with the E. coli LepB suppressor strain called FTL85 in which chromosomal LepB expression is driven by an Ara-inducible promoter (Lüke et al., 2009). Replacing Ara in the medium with Glc causes cell growth to halt due to a lack of a functional type I signal peptidase (Lüke et al., 2009). We exploited this system to test whether Plsp1 could functionally complement LepB suppression in FTL85 cells grown in medium containing Glc (Supplemental Figure 6). FTL85 cells transformed with a plasmid encoding the mature form of Plsp1 (residues 68-291), which contains the transmembrane domain and the C-terminal catalytic domain, could not grow in medium containing Glc (data not shown). The transmembrane domain of Plsp1 may not efficiently target the entire protein to the plasma membrane in E. coli due to differences in the mechanisms of membrane insertion and targeting between Plsp1 and LepB (; Houben et al., 1999; Endow et al., 2015).
To overcome this issue, chimeric constructs were generated in which the catalytic domain of Arabidopsis Plsp1 (i.e., residues 130–291) was genetically fused to the transmembrane domains of E. coli LepB (i.e., residues 1–77). Full-length LepB (residues 1–323) and the empty vector were used as positive and negative controls, respectively. Each of the Plsp1 Cys/Ala mutants as well as the noncatalytic S142A mutant were also prepared, and FTL85 cells carrying the chimeric constructs (termed LepBn-Plsp1c) were grown overnight in medium containing Ara. After dilution of overnight cultures into new medium containing Ara, all strains continued to grow at comparable rates (Supplemental Figure 6A). By contrast, there were significant differences in cell growth rates after dilution into new medium containing Glc (Supplemental Figure 6B). Specifically, wild-type LepBn-Plsp1c supported growth rates comparable with that of the LepB control, but the LepBn-Plsp1c-S142A mutant grew as poorly as the empty vector control (Supplemental Figure 6B). This suggests that the wild-type catalytic domain of Plsp1 can function as a type I signal peptidase in E. coli cells. Although the redox state of Plsp1 was not tested, it could potentially be oxidized by the endogenous periplasmic Dsb system (Herrmann and Riemer, 2014). Interestingly, the LepBn-Plsp1c-Cys/Ala mutants supported intermediate growth rates as compared with the LepB and empty vector controls (Supplemental Figure 6B). This suggests that Plsp1 has only partial functionality in E. coli when it lacks a disulfide bond.
Wild-type and Redox-Inactive Citrine-Plsp1 Variants Have Comparable Activity in Chloroplasts
To qualitatively examine the in vivo activity of Citrine-Plsp1-CA proteins, we checked the sizes of two Plsp1 substrates in isolated chloroplasts. Chloroplasts from wild-type plants and those from all plants complemented with Citrine-Plsp1 accumulated PsbO and Toc75 in their mature/processed forms (Figure 4A). Unprocessed forms of these proteins were detected only in total protein extracts from plsp1-1 mutant seedlings (Figure 4A).
Figure 4.
Citrine-Plsp1-CA Is Active in Chloroplasts.
(A) Size of Plsp1 substrates in chloroplasts isolated from each of the complemented lines shown in Figure 3. Total protein extracts from plsp1-1 null mutants were also used as a control. Proteins were separated on 12% (α-PsbO) or 7.5% (α-Toc75) SDS-PAGE and detected by immunoblotting using antibodies stated at the right. Asterisks indicate unprocessed forms, and arrowheads indicate processed/mature form (S) of each protein.
(B) Time course of in vitro import into isolated chloroplasts. 35S-Met-labeled forms of each precursor protein were incubated with isolated chloroplasts for 5, 10, or 20 min. Intact chloroplasts were reisolated through a 35% Percoll cushion and washed once in import buffer. A portion of the recovered chloroplasts was used for chlorophyll quantification to normalize gel loading, and the remainder was analyzed by SDS-PAGE and autoradiography. The lines used for the experiments are as follows: C #1, C166A #1, C286A #2, CCAA #2. Each import experiment was repeated three times using chloroplasts isolated on different days. RbcL bands on the Coomassie Brilliant Blue–stained gel are shown below as a loading control. P = precursor; i = intermediate; m = mature; PC, plastocyanin.
(C) Quantification of the import products shown in (B). Bands were quantified relative the corresponding wild type (WT) band at the 20-min time point (i.e., wild-type maximum). Show are the means ± sd of three biological replicates. The line in each graph is drawn through the average among all four lines at each time point. Squares, wild type; circles, C166A; diamonds, C286A; X, C166A/C286A.
Because most of our complemented lines expressed Citrine-Plsp1 at high levels (Figure 3D), and high concentrations of Plsp1 exhibit detectable activity despite DTT treatment (Supplemental Figure 7), we sought to rule out the hypothesis that the in vivo processing activity in chloroplasts containing Citrine-Plsp1-CA mutant proteins is low but sufficient for normal thylakoid development. To that end, we quantitatively examined the activities of each Citrine-Plsp1-CA protein in intact chloroplasts using an in vitro protein import assay with radiolabeled precursor proteins. We used the precursors of Arabidopsis PsbP1 and Silene pratensis plastocyanin (Last and Gray, 1989) as model substrates for the two thylakoid lumen protein transport pathways cpTAT and cpSEC1, respectively (Albiniak et al., 2012). For these time-course experiments, we chose one Citrine-Plsp1 wild-type line and one transgenic line representing each Citrine-Plsp1 Cys/Ala mutation. SDS-PAGE and autoradiography of import products showed that all lines imported prPsbP and prPC and processed each to their mature forms at similar rates (Figure 4B). Quantification of signals in the autoradiograms revealed that the rate at which mature PsbP or PC accumulate is comparable between each of the CA mutant lines and the wild-type control (Figure 4C). The total amount of imported PsbP (intermediate + mature) was also comparable over time (Figure 4C), indicating that chloroplasts from all of the tested lines have similar import efficiencies. Considering that the Citrine-Plsp1 expression level varied between the lines used in this experiment, it appears that the in vivo processing rate is not tightly coupled to the level of Plsp1 in the thylakoid. Based on these data, we conclude that even without a disulfide bond, Plsp1 has a normal level of activity in vivo.
Thylakoid Membrane-Bound Plsp1 Activity Is Insensitive to DTT
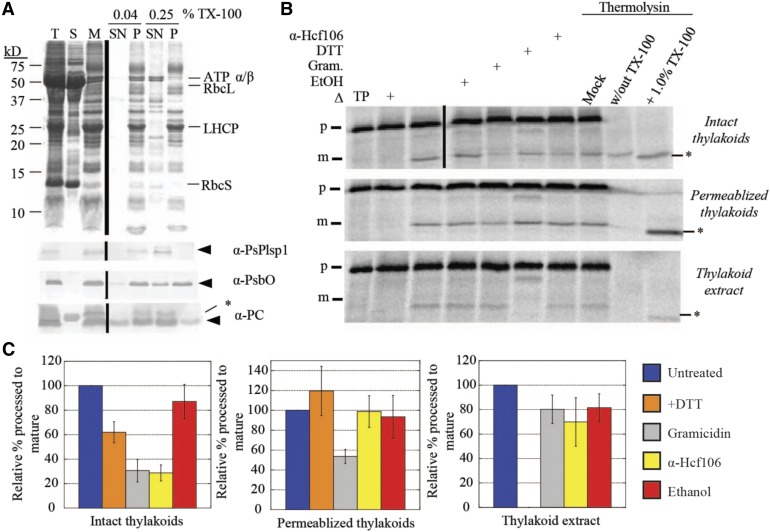
Plsp1 localizes to the thylakoid membrane and presents its catalytic site in the lumen (Kirwin et al., 1988; Shipman and Inoue, 2009). To explain its disulfide-free activity in vivo, we reasoned that either one or more protein–protein interactions in the thylakoid membrane or interactions with the lipid bilayer compensate for a lack of disulfide bond formation. We developed an assay to directly measure Plsp1 activity in native thylakoids using a membrane permeabilization technique (Ettinger and Theg, 1991). Specifically, treatment of isolated thylakoid vesicles with a low concentration of Triton X-100 leads to the release of soluble proteins from the lumen (Ettinger and Theg, 1991). Similarly, we expected that the same low detergent concentration would allow exogenously added substrates to diffuse inside and be processed by Plsp1. Figure 5A (top panel) shows the typical fractionation pattern of proteins from pea chloroplasts after hypotonic lysis followed by treatment of membranes with either a low (0.04%) or high (0.25% v/v) concentration of Triton X-100. The low Triton X-100 treatment released a small amount of the lumenal proteins PsbO and plastocyanin, but Plsp1 remained tightly bound to the membrane (Figure 5A). By contrast, the high Triton X-100 treatment completely solubilized Plsp1 (Figure 5A).
Figure 5.
Thylakoid Membrane-Bound Processing Activity Is Insensitive to DTT.
(A) SDS-PAGE analysis of Pea chloroplast fractions. Proteins were detected by Coomassie Brilliant Blue staining (top panel) or immunoblotting (bottom three panels) with the antibodies indicated at the right of each panel. Arrowheads indicate proteins of interest, and asterisks indicate nonspecific immunoreactive bands. A volume equivalent to 4 μg chlorophyll was loaded in each lane. T = total chloroplasts; S = soluble fraction; M = membranes; SN = supernatant; P = pellet.
(B) Processing activity in Pea chloroplast membrane fractions depicted in (A). The 4 μg chlorophyll equivalents of each of the three fractions were pretreated on ice for 30 min with reaction buffer (control), 1% ethanol (EtOH), 30 μM gramicidin (Gram) in 1% ethanol, 50 mM DTT, or the α-Hcf106 antibody. As a control, one sample of each chloroplast fraction was heat-inactivated (Δ) at 82°C for 10 min before adding the substrate. After 35S-Met-prPsbP was added, each reaction mixture was incubated in the dark at 28°C for 30 min. After the incubation at 28°C, three additional control reactions were treated with 0.5 mM CaCl2, thermolysin (0.1 mg/mL with 0.5 mM CaCl2), or thermolysin (0.1 mg/mL, with CaCl2) in the presence of Triton X-100 (1% v/v) for 40 min on ice. Reaction mixtures were quenched by adding an equal volume of 2× sample buffer containing 30 mM EDTA and boiling for 5 min. Samples were analyzed by SDS-PAGE and autoradiography. Asterisks indicate a degradation product that was observed in some experiments.
(C) ImageJ quantification of mature PsbP bands shown in (B). Values were normalized to the band intensity in the untreated sample and are expressed as the mean ± sd of at least three independent experiments.
Having established reproducible conditions for preparing membrane-bound and detergent-solubilized forms of Plsp1, we tested the effect of DTT on processing activity in each case using prPsbP as the substrate. Figure 5B shows the results of in vitro processing experiments using intact or permeabilized (0.04% Triton X-100) thylakoids and thylakoid extract (0.25% Triton X-100 supernatant). The effects of the different treatments on processing activity in intact thylakoids are consistent with proteins that traverse the thylakoid membrane via the cpTAT pathway, of which PsbP is a well-studied substrate (Braun and Theg, 2008). Specifically, both gramicidin and the antibody against Hcf106 (Figures 5B and 5C) inhibited activity by ∼70% due to their effects on protein transport. Gramicidin forms pores in membranes (Kelkar and Chattopadhyay, 2007), which dissipates the pmf on which cpTAT transport depends, and the α-Hcf106 antibody inhibits cpTAT transport by binding to the Hcf106 subunits of the cpTAT translocon (Rodrigues et al., 2011). Interestingly, DTT inhibited processing activity in intact thylakoids by ∼40%. It is possible that DTT slightly inhibits the processing activity of Plsp1 in intact thylakoids, but we hypothesize that other nonspecific effects are more likely. Resistance to thermolysin confirmed that the processed form of PsbP had been successfully transported into the lumenal compartment of the thylakoid vesicles (Figure 5B).
Processing activity in permeabilized thylakoids relied on simple diffusion of the substrate into the thylakoid vesicles as evidenced by thermolysin susceptibility of processed PsbP and a negligible effect of the α-Hcf106 antibody (Figure 5B). Importantly, DTT did not affect activity in permeabilized thylakoids (Figure 5B), providing further support that the thylakoid membrane-bound form of Plsp1 is active without a disulfide bond. Gramicidin inhibited processing activity by ∼45% (Figures 5B and 5C), which may be due to unspecified direct or indirect effects on the structure of Plsp1. As expected, the activity of solubilized Plsp1 was completely inhibited by DTT (Figures 5B and 5C).
We also used this assay system with Citrine-Plsp1-complemented plants (wild type and C166A/C286A). Permeabilized thylakoids from both genotypes processed prPsbP to the mature form were unaffected by DTT and were completely inhibited by the type I signal peptidase inhibitor Arylomycin A2 (Paetzel, 2014; Supplemental Figure 8). As expected, processing activity in the wild-type thylakoid extracts was completely inhibited by DTT and Arylomycin A2 (Supplemental Figure 8). In comparison, thylakoid extracts containing Citrine-Plsp1-C166A/C286A lacked detectable activity even in the absence of DTT (Supplemental Figure 8). The above results (Figure 5; Supplemental Figure 8), as well as our genetic complementation data (Figure 3; Supplemental Figure 4), provide compelling evidence that Plsp1 requires a disulfide bond for activity only when it is taken out of its native context.
Compensation for Lack of a Disulfide Bond in Plsp1 Is Not due to a Stable Association with PGRL1
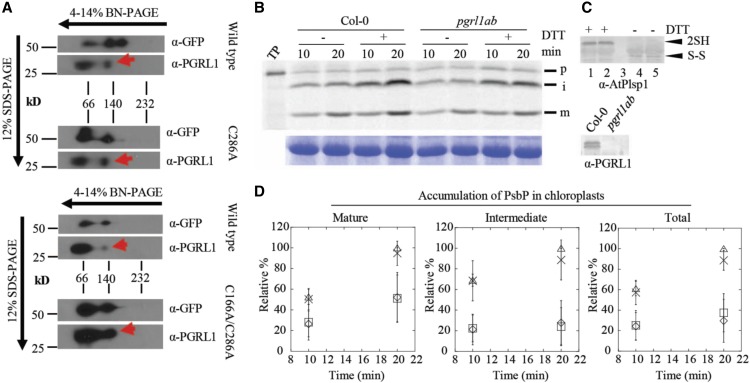
The experiments described in Figure 5 cannot distinguish between effects of protein–protein interactions or lipid interactions. More than 50% of the total Plsp1 pool in thylakoids forms a stable complex with PGRL1 (Endow and Inoue, 2013), a key component of antimycin A-sensitive cyclic electron flow around photosystem I (Strand et al., 2016). All Plsp1 substrates examined accumulate as their mature forms in chloroplasts lacking PGRL1 (Endow and Inoue, 2013), but Plsp1 is presumably oxidized in the mutant chloroplasts. Two-dimensional blue-native/SDS-PAGE followed by immunoblotting showed that a portion of the Citrine-Plsp1 and PGRL1 pools comigrate despite changing the Cys in Plsp1 to Ala (Figure 6A), suggesting that the stable association does not require a disulfide bond in Plsp1. We therefore hypothesized that the interaction with PGRL1 might keep the reduced form of Plsp1 catalytically active.
Figure 6.
Association with PGRL1 Does Not Compensate for Lack of Cys in Plsp1.
(A) 2D-Blue Native (BN)/SDS-PAGE analysis of Arabidopsis chloroplasts complemented with Citrine-Plsp1, Citrine-Plsp1-C286A, or Citrine-Plsp1-C166A/C286A. Total chloroplasts equivalent to 5 μg (left) or 7.5 μg chlorophyll were solubilized with 1.3% w/v n-dodecyl-β-D-maltoside and run on 4-14% Blue Native-PAGE gels. Lanes were then excised, heated at ∼80°C for 30 min in denaturing buffer (65 mM Tris-HCl pH 6.8; 3.3% SDS; 570 mM β-ME), and overlayed on 12% SDS-PAGE gels. After electrophoresis, proteins were blotted to polyvinylidene difluoride membranes. Membranes were cut at ∼37-kD and probed with the GFP antibody (top) or the PGRL1 antibody (bottom). Red arrows indicate the population of PGRL1 that comigrates with Citrine-Plsp1.
(B) Analysis of processing activity during in vitro import. Chloroplasts pretreated on ice for 30 min with or without 50 mM DTT were mixed with 35S-Met-prPsbP and MgATP (3 mM final) and were incubated in the light at 20°C for 10 or 20 min. Intact chloroplasts were reisolated as described in Figure 3B and analyzed by autoradiography (top panel). RbcL bands from the Coomassie Brilliant Blue–stained gels is shown as a loading control (bottom panel).
(C) Immunoblotting analysis of chloroplasts used for in vitro import experiments. For top panel, an aliquot of chloroplasts treated with or without 50 mM DTT before in vitro import were mixed with nonreducing (no β-ME) sample loading buffer and analyzed by SDS-PAGE and immunoblotting using the antibody against Arabidopsis Plsp1. ”2SH” and ”S-S” indicate the reduced and oxidized forms of Plsp1, respectively. 1 and 4 = Col-0; 2 and 5 = pgrl1ab; 3 = blank. For bottom panel, chloroplasts used for import experiments were analyzed by SDS-PAGE and immunoblotting using the PGRL1 antibody.
(D) Quantification of import products shown in (C). Bands were quantified relative to the corresponding band (S) in the Col-0 + DTT sample at 20 min, which was set as 100%. Data are expressed as the mean ± sd of three independent experiments. Squares, wild type - DTT; triangles, wild type + DTT; diamonds, pgrl1ab - DTT; X, pgrl1ab + DTT.
To test this hypothesis, prPsbP was imported into Col-0 wild-type chloroplasts or those from the pgrl1ab knock out mutant (Endow and Inoue, 2013) after pretreatment of chloroplasts with or without DTT. If PGRL1 maintains Plsp1 activity, we expected that PsbP would either (1) accumulate exclusively as the intermediate form or (2) be processed to the mature form more slowly after treatment with DTT in pgrl1ab chloroplasts. However, both the intermediate and mature forms of PsbP accumulated at similar rates in the two genotypes regardless of DTT treatment (Figures 6B and 6C). The total amount of imported PsbP was also similar between both genotypes (Figure 6C). Immunoblotting confirmed that the pgrl1ab mutant chloroplasts lacked detectable PGRL1 protein and that the DTT pretreatment of chloroplasts completely reduced Plsp1 (Figure 6D). Notably, DTT pretreatment increased the total amount of PsbP that was imported (Figure 6C), an effect first observed for import of the ferredoxin precursor into pea chloroplasts (Pilon et al., 1992). This is most likely due to reductive enhancement of components of the import apparatus (Bölter et al., 2015). Based on these data, we concluded that the association with PGRL1 does not compensate for the lack of a disulfide bond in Plsp1.
A Lipid Bilayer Mitigates the Effect of DTT on Plsp1 Activity
Having shown that membrane-embedded Plsp1 is not stabilized against reductants by interaction with PGRL1, we sought to test whether this structural stability was imparted by the membrane environment itself. As pointed out by Hamsanathan and Musser (2018), it is well known that membrane proteins can lose their activities upon removal from membranes. We asked if this would translate into solubilized Plsp1 maintaining its structure in the presence of reductants upon reconstitution into membranes. As a first attempt to answer this question, we reconstituted purified Plsp1 into lipid vesicles prepared from E. coli total lipid extract. As seen in Supplemental Figure 9, this led to a Plsp1 sample that retained ∼40% of its activity upon reduction with DTT. Encouraged by these results and recognizing the different chemical natures of the lipids in E. coli and thylakoids, we repeated these experiments using liposomes composed of MGDG, digalactosyl diacyl glycerol (DGDG), sulfoquinovosyl diacyl glycerol (SQDG), and phosphatidyl glycerol (PG), the four major diacylglycerolipids found in thylakoid membranes (Webb and Green, 1991). Notably, the lipid ratios we used (19.8% MGDG, 54.9% DGDG, 14.8% SQDG, 10.5% PG) are similar to those used in a previous study of reconstituted photosystem II sub-complexes (Zhou et al., 2009) but differ significantly from those found in intact thylakoids (51.4% MGDG, 21.6% DGDG, 5.3% SQDG, 21.6% PG; Essigmann et al., 1998). The primary reason for this is that MGDG, which is the predominant lipid in thylakoids, does not form bilayers on its own and can promote liposome fusion and aggregation if used above ∼30 mol% (Latowski et al., 2000).
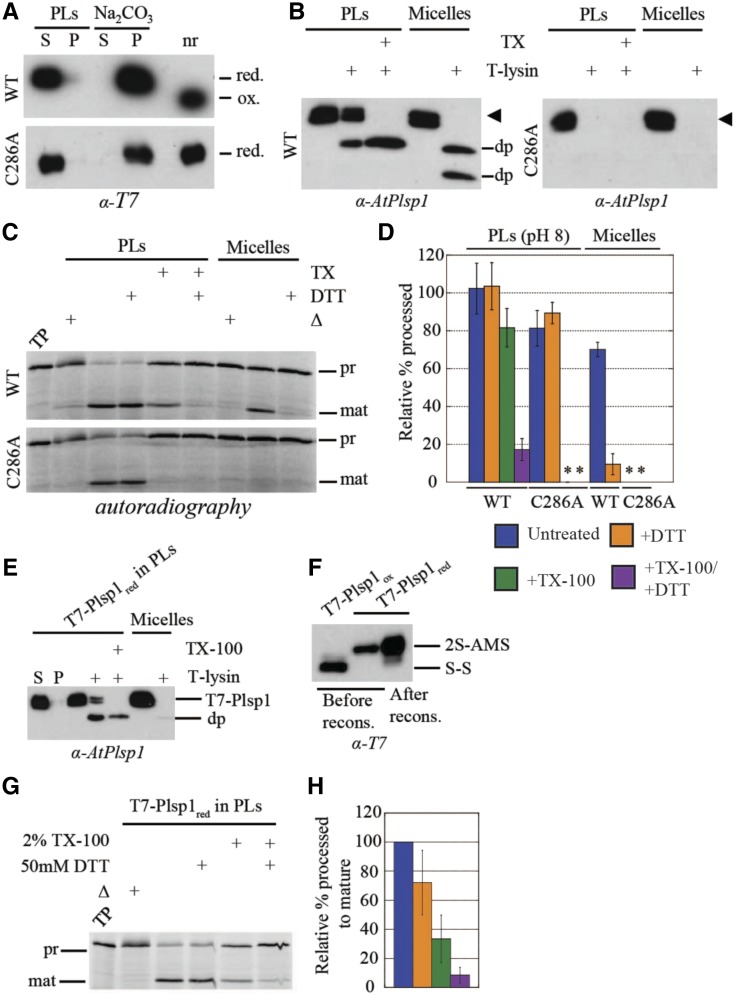
After detergent removal, Plsp1 was recovered in the pelleted liposomes and remained so after treatment of the proteoliposomes with sodium carbonate (Figure 7A). Under nonreducing conditions, Plsp1 exhibited a faster migration on SDS-PAGE, indicating the presence of the disulfide bond (Figure 7A). These results provide evidence that Plsp1 was successfully reconstituted into liposomes and that it was in the expected redox state.
Figure 7.
Reconstitution into Thylakoid Lipid Vesicles Recovers the Activity of Prereduced and Redox-Inactive Plsp1.
(A) T7-Plsp1 (wild type or C286A) was purified in 0.25% Triton X-100 (micelles, final pH∼7.5) and reconstituted into liposomes to yield proteoliposomes (PLs) and were then divided into soluble (S) and pellet (P) fractions by centrifugation as described in Methods. The soluble fraction was treated with 0.1 M Na2CO3 for 5 min on ice and then subjected to ultracentrifugation to yield soluble and pellet fractions; nr = aliquot run in non-reducing sample buffer. Samples were analyzed by SDS-PAGE and immunoblotting.
(B) PLs at pH 8.0 were subjected to treatment with thermolysin (10 μg/mL with 0.5 mM CaCl2) with or without 2% Triton X-100 (TX) at 25°C for 40 min. Samples were analyzed by SDS-PAGE and immunoblotting. The arrowhead indicates the T7-Plsp1 doublet, and “dp” indicates degradation products observed after thermolysin treatment.
(C) PLs at pH 8.0 were pretreated with or without 50 mM DTT and/or 2% Triton X-100 (TX) for 30 min on ice followed by incubation with 35S-prPsbP for 30 min at 25°C. As a control, 60 nM purified T7-Plsp1 pretreated with or without 50 mM DTT was included. Samples were analyzed by SDS-PAGE and autoradiography. Δ = Boiled for 10 min before adding substrate; TP = translation product.
(D) Quantification of mature PsbP bands in C relative to the TP on the same gel. Shown are the means ± sd from three independent experiments. WT =wild type. Asterisks indicate undetectable activity.
(E) T7-Plsp1 in 1% octyl glucoside was treated with 50 mM DTT followed by reconstitution into liposomes made from thylakoid lipids as described in Methods except that 50 mM DTT was present throughout the reconstitution procedure. The recovered PLs were subjected to thermolysin treatment as in (B). S = soluble fraction after resuspending pelleted liposomes; P = pellet fraction after resuspending pelleted liposomes.
(F) T7-Plsp1 in 1% octyl glucoside was treated with or without DTT followed by TCA precipitation and incubation in the presence of 10 mM AMS. An aliquot of the DTT-treated reconstituted (recons.) T7-Plsp1 was also precipitated and treated with 10 mM AMS. 2S-AMS = reduced and AMS-labeled form of Plsp1; S-S = oxidized form of Plsp1.
(G) PLs described in E were incubated with 35S-prPsbP as in (C), and samples were analyzed by SDS-PAGE and autoradiography.
(H) Quantification of mature PsbP bands in G relative to the untreated control. Shown are the means ± sd from two independent reconstitution experiments.
T7-Plsp1 exhibited differential sensitivity to thermolysin in the presence and absence of Triton X-100 (Figure 7B). In the absence of Triton X-100, a portion of wild-type Plsp1 in proteoliposomes was completely resistant to thermolysin, and a portion was truncated to a ∼22 kD degradation product (Figure 7B, left panel). The degradation product is similar in size to that observed when endogenous Plsp1 is subject to thermolysin treatment (Shipman-Roston et al., 2010; Endow et al., 2015) and likely represents an N-terminal truncation up to the transmembrane region with the catalytic domain of Plsp1 facing inside the vesicle lumen. The portion that is completely resistant to thermolysin likely represents Plsp1 with the tightly folded oxidized catalytic domain facing the bulk solvent. This is comparable with the observation that Plsp1 can associate with the cis face of isolated thylakoids in vitro such that the catalytic domain is resistant to thermolysin (Endow et al., 2015). In the presence of Triton X-100, full-length Plsp1 was completely removed by thermolysin, and a similar ∼22 kD degradation product representing the tightly-folded catalytic domain was observed (Figure 7B, left panel). Purified Plsp1 in 0.25% Triton X-100 was degraded by thermolysin with degradation products of ∼22 kD and ∼20 kD appearing in most experiments (Figure 7B, left panel). The observation that reconstituted Plsp1 is not completely degraded after bilayer solubilization may be due to differences in Plsp1 folding or thermolysin activity in 2% versus 0.25% Triton X-100, or the presence of lipids in the solubilized proteoliposomes. Indeed, membrane protein conformation can be significantly stabilized by 2% Triton X-100 as was the case for the E. coli outer membrane protein PhoE (de Cock et al., 1996).
We performed processing assays using reconstituted Plsp1 and purified Plsp1 in Triton X-100 to test whether the lipid bilayer mitigates the effect of DTT on Plsp1 activity.
Interestingly, reconstituted Plsp1 exhibited processing activity that was insensitive to DTT (Figures 7C and 7D). Upon solubilization of proteoliposomes with Triton X-100, the processing activity of wild-type Plsp1 was inhibited by DTT to a similar extent as the nonreconstituted form (Figures 7C and 7D). These results are consistent with those obtained from in vitro processing assays using permeabilized thylakoids and thylakoid extracts from the plsp1-1-complemented Arabidopsis plants (Supplemental Figure 8). These data indicate that integration into liposomes composed of native thylakoid diacyl lipids renders oxidized Plsp1 activity insensitive to reducing agents.
Chaperoning Activity of the Lipid Environment Mediates Recovery of Activity in Nondisulfide Bonded Plsp1
We were curious whether the lipid environment only allowed for the maintenance of Plsp1 activity in the presence of reductant, or whether it would promote the reconstitutive folding of the protein from an inactive conformation. To this end, we reconstituted Plsp1 into proteoliposomes after its inactivation by reduction of the detergent-solubilized form. Remarkably, Plsp1 regained significant activity after reconstitution, and this was minimally affected by DTT (Figures 7G and 7H). In addition, this form of Plsp1 was sensitive to thermolysin similar to the oxidized form (Figure 7E) and remained in a reduced state after the reconstitution procedure (Figure 7F). Although solubilization of proteoliposomes with Triton X-100 slightly decreased the activity of the oxidized form of Plsp1 (Figure 7D), the decrease in activity was significantly greater for the reduced form of Plsp1 (Figure 7H). Interestingly, DTT inhibited activity when the proteoliposomes had been solubilized with Triton X-100 (Figures 7G and 7H) despite the fact the Plsp1 was fully reduced at the end of the reconstitution procedure (Figure 7F). Our interpretation is that a portion of the reconstituted Plsp1 had become reoxidized in between resuspension of proteoliposomes in buffer and the addition of substrate (i.e., ∼3.5 h). These results indicate that reduced Plsp1, which had adopted an inactive conformation in its reduced state, had been refolded upon incorporation into proteoliposomes, suggesting that the lipid environment provides reconstitutive chaperoning activity directed toward this protein.
We also tested the effect of reconstitution into proteoliposomes of the C286A mutant of Plsp1 described above. With one of the two cysteines missing, this protein cannot form a disulfide bond, and consequently, displayed no processing activity in its solubilized form (Midorikawa et al., 2014; see also Supplemental Figure 10). As with wild-type Plsp1, the C286A mutant protein was recovered in the proteoliposome pellet after reconstitution and was not extracted by carbonate washing (Figure 7A). As expected, its mobility on SDS-PAGE was not altered by treatment with a reducing agent, indicating a lack of a disulfide bond (Figure 7A). Remarkably, the reconstituted C286A mutant exhibited processing activity in the proteoliposomes, indicating that it was folded by its association with the membrane bilayer. This activity was, of course, insensitive to DTT, but was completely abolished by solubilization of the liposomes with Triton X-100 (Figures 7C and 7D). In aggregate, our experiments with proteoliposomes indicate that integration into liposomes composed of native thylakoid diacyl lipids renders Plsp1 activity insensitive to reducing agents and can also facilitate reconstitutive folding from an inactive to an active conformation in the absence of a disulfide bond.
The Disulfide Bridge in Plsp1 May Impart Conformational Stability during the Diurnal Light/Dark Cycle
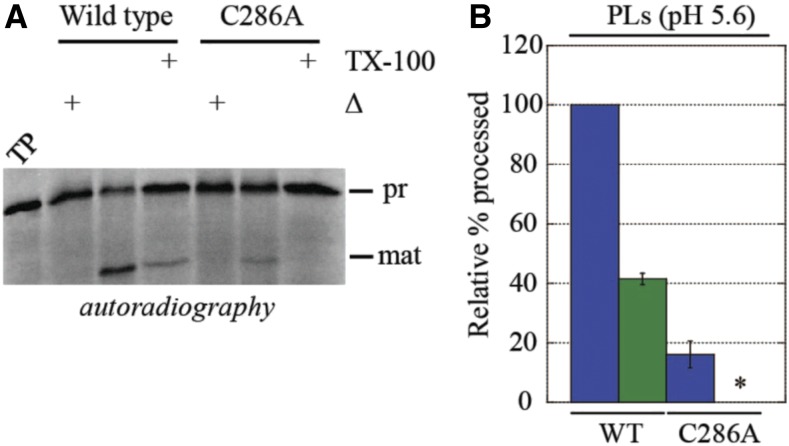
Our experiments suggest that the disulfide bridge that stabilizes the active conformation of Plsp1 may be superfluous while it is in a membrane environment. Yet, the placement of the bridging cysteines is invariant in all the angiosperms (Midorikawa et al., 2014), suggesting some evolutionary pressure to form the disulfide in these plants. We recognized that Plsp1’s environment is not static and that it likely changes during the day/night cycle of membrane energization. Specifically, the thylakoid membrane goes from a nonenergized to an energized state with the imposition of the pmf in the light, and the lumen pH changes from ∼8 in the dark to ∼6 or less under illumination (Kieselbach, 2013). In testing the importance of this, we found that the peptidase activity of T7-Plsp1-C286A in proteoliposomes at pH 8.0 was similar to that of wild-type Plsp1 (Figures 7C and 7D). When assayed at pH 5.6, however, the activity of the C286A variant in proteoliposomes was significantly lower than that of the wild-type and was abolished upon solubilization of the liposomes with Triton X-100 (Figures 8A and 8B). This suggests that the disulfide bond in Plsp1 may not be required at night (in the dark) but provides structural stability to offset the destabilizing effects of, at the least, the pH component of the pmf developed in the light during the day.
Figure 8.
Disulfide Bond Formation Enhances the Activity of Reconstituted Plsp1 at Low pH.
(A) Proteoliposomes (PLs) containing T7-Plsp1 (wild type, or WT, or C286A) were resuspended in MES liposome buffer at pH 5.6 and were assayed for processing activity.
(B) Quantification of mature PsbP bands in (A) relative to wild type -TX-100. Shown are the means ± sd from three independent experiments. Blue = untreated PLs; Green = PLs + 2% TX-100. An asterisk denotes no detectable activity.
We note that the disulfide bond is not essential for Plsp1 in vivo activity in our overexpression lines grown under a diurnal cycle (Figure 3A). It is possible that the growth conditions used for the plants described in Figure 3 (i.e., constant low light intensity during the light period) may not be sufficient to necessitate a disulfide bond for structural stabilization of Plsp1. Growth under dynamic conditions, which are closer to what plants experience in nature, produces a more variable pmf in terms of the magnitude and composition (Armbruster et al., 2017), and such variability in the pmf may be sufficient to distinguish between plants expressing wild-type and redox-inactive forms of Plsp1. To test this hypothesis, we grew all of our Citrine-Plsp1 overexpression lines on Murashige and Skoog (MS) medium under a fluctuating light regime similar to that described previously (Suorsa et al., 2012). Unfortunately, there was no clear phenotypic difference between any of our overexpression lines grown under the fluctuating light conditions (Supplemental Figure 11). As expected, a small proportion of albino seedlings were observed in some cases and correspond to the hemizygous lines described earlier (Figure 3A; Supplemental Figure 5). Regardless of the genotype, all of the wild type-like plants grew noticeably larger under the fluctuating light regime (Supplemental Figure 11) similar to previously published results (Suorsa et al., 2012). These results indicate that the fluctuating light regime we tested does not obviously compromise the function of redox-inactive forms of Plsp1.
DISCUSSION
Plsp1 Forms a Nonregulatory Disulfide Bond in the Thylakoid Lumen
Approximately 40% of the confirmed soluble lumen proteome, as well as the lumenal domains of several thylakoid membrane proteins, possess redox-active Cys residues, which are either known or predicted to form disulfide bonds (Hall et al., 2010; Brooks et al., 2013; Karamoko et al., 2013; Shapiguzov et al., 2016). The results of our in vivo thiol-labeling assay indicate that the two conserved Cys residues in Plsp1 form a disulfide bond in the thylakoid lumen (Figure 2A), adding it to this growing list. The classic view of disulfide bonds is that they enhance the structural stability of proteins (Thornton, 1981; Betz, 1993), but they can also play a role in allosteric regulation, as is the case for most enzymes involved in carbon fixation (Schmidt et al., 2006; Balsera et al., 2014). Midorikawa et al. (2014) hypothesized that the disulfide bond in Plsp1 is reversible in vivo and acts to regulate activity similar to the proposed paradigm of lumenal redox regulation (Gopalan et al., 2006; Karamoko et al., 2013; Simionato et al., 2015). However, our experimental results now show that this interpretation is incorrect since (1) Plsp1 is functional in vivo without a disulfide bond (Figure 3) and (2) the activity of Plsp1 in proteoliposomes is unaffected by DTT (Figure 7D).
Oxidation of Plsp1 is expected to occur in the thylakoid lumen due to the nature of its thylakoid targeting mechanism. Thylakoid transport of Plsp1 is mediated by the cpSEC1 pathway (Endow et al., 2015), which can accommodate only unfolded polypeptides (Albiniak et al., 2012). The presence of a disulfide bond between C166 and C286 would introduce a large loop in Plsp1, which would likely make the protein incompatible with the cpSecY translocon. Although some oxidized lumen proteins are transported in a folded conformation by the cpTAT pathway (Schubert et al., 2002; Hall et al., 2010) and may be oxidized in the stroma or the lumen, we predict that membrane translocation must precede oxidation for all cpSEC1 substrates that possess a disulfide bridge.
Lipids Have a Major Effect on Plsp1 Structure and Function
One of the first pieces of evidence supporting the hypothesis that lipids restore activity of redox-inactive forms of Plsp1 was the observation that the activity of Plsp1 in permeabilized thylakoid vesicles is insensitive to reducing agents (Figures 5C and 5D; Supplemental Figure 8). Additional experiments showed that although the stable association between Plsp1 and PGRL1 is not affected by changing one or both Cys in Plsp1 to Ala (Figure 6A), this interaction does not have a role in maintaining the active conformation of Plsp1 (Figures 6B and 6D). By contrast, reconstitution into liposomes rendered Plsp1 activity insensitive to DTT. Further, it restored the activity of previously reduced wild type protein (Figures 7G and 7H), as well as that of the single Cys Plsp1 mutant (Figures 7C and 7D). The effect of lipids appears to be completely reversible since solubilization of proteoliposomes with Triton X-100 abolished the activity of a reconstituted single Cys Plsp1 (Figures 7C and 7D) as well as the activity of the Cys-less Plsp1 mutant from isolated thylakoids (Supplemental Figure 8). Our interpretation of these data is that interactions between Plsp1 and membrane lipids were necessary to prevent the inhibitory conformational change and were also sufficient to shift the enzyme from an inactive into a catalytically active form (Figure 8). Activity-inducing conformational changes stimulated by association with membrane lipids are also seen in a class of proteins called bacteriocins (van der Goot et al., 1991; Mel and Stroud, 1993) and in the K+ channel Kir2.2 (Hansen et al., 2011). More extreme cases are seen in the thylakoid membrane protein PsbS and in certain bacterial outer membrane β-barrel proteins that can be folded from completely denatured forms into their native conformations in the presence of liposomes (Kleinschmidt, 2015; Liu et al., 2016). Taken together, the above-mentioned results lead us to conclude that thylakoid lipids possess reconstitutive folding/chaperone activity toward the lumenal catalytic domain of Plsp1 and that the native folding occurs without, and perhaps before, disulfide bond formation (Figure 9).
Figure 9.
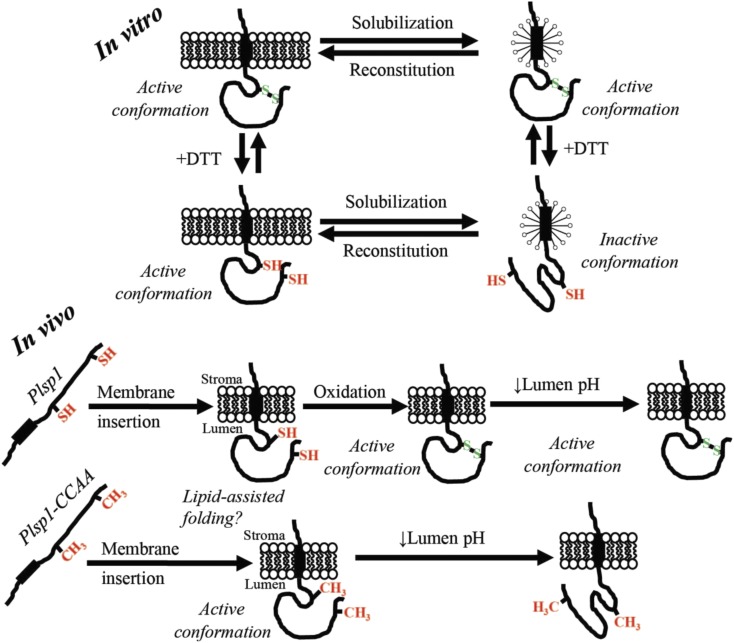
In Vitro and In Vivo Models.
For in vitro, Plsp1 is solubilized in from membranes in an active form due to an intramolecular disulfide bond. Reduction of this disulfide bond causes an inactivating conformational change in detergent micelles that is prevented by association with a lipid bilayer. Reintegration of Plsp1 into a bilayer causes reconstitutive folding back into the active state. For in vivo, Plsp1 is targeted to thylakoids in a reduced form. Once the catalytic domain traverses the membrane, reconstitutive folding into the catalytically active form is assisted by bilayer lipids. In the case of the wild-type protein, oxidation to form the disulfide bond then takes place and stabilizes the structure. Structural stabilization by the disulfide bond allows Plsp1 to remain optimally active amid fluctuations in the lumen pH.
How do lipids make Plsp1 insensitive to the effects of reducing the disulfide bond? The soluble catalytic domain of E. coli LepB spontaneously inserts into membranes via its proposed hydrophobic membrane association surface (van Klompenburg et al., 1998; Bhanu and Kendall, 2014). A similar hydrophobic surface is revealed in the predicted structure of Plsp1 and may be responsible for the propensity of Plsp1 to associate with isolated thylakoid membranes (Endow et al., 2015). Insertion of the catalytic domain of Plsp1 into a membrane may rigidify the tertiary structure and prevent inactivating conformational changes resulting from disulfide bond reduction. However, there are still likely to be some conformational differences between oxidized and reduced Plsp1 despite being integrated into a membrane since the protease susceptibility of the wild type and single Cys variant in proteoliposomes are different (Figure 7B). Another possible explanation is related to signal peptide binding. Type I signal peptidases possess a specific groove into which a signal peptide binds (Paetzel, 2014). Midorikawa et al. (2014) hypothesized that the C-terminal region can sterically interfere with binding of a thylakoid transfer signal (TTS) when Plsp1 is in a reduced state. If this is true, perhaps the catalytic domain of Plsp1 inserts into the membrane such that the TTS binding groove becomes inaccessible to the C-terminal segment. An analogous example of intramolecular inhibition can be seen in Rv1827 from Mycobacterium tuberculosis in which phosphorylation of an N-terminal Thr residue causes this region of the protein to occlude a surface involved in interactions with binding partners (Nott et al., 2009).
Our results clearly indicate that lipids play an important role in the folding and activity of Plsp1, but they do not reveal the mechanism by which a membrane facilitates a reversible transition from an inactive to a catalytically active conformation. In E. coli, the multi-pass lactose permease (LacY) requires interactions with phosphatidylethanolamine (PE) to attain proper membrane topology and folding of a specific periplasmic domain (Bogdanov et al., 1996). Once LacY is properly folded, interactions with PE are no longer required (Bogdanov et al., 2002). The mechanism of reconstitutive folding of Plsp1 is likely different since Plsp1 only has a single transmembrane alpha helix, and the stimulatory effects of lipids on the single Cys Plsp1 variant are reversed by solubilization with Triton X-100 (Figures 7C and 7D). Thus, the interaction between lipids and Plsp1 in membranes is likely stable as opposed to the transient nature of the interaction between PE and LacY (Bogdanov and Dowhan, 1999). Perhaps the mechanism with Plsp1 is similar to that of the membrane insertion of the pore-forming domain of colicin A (Lakey et al., 1991). Specifically, interactions with negatively charged lipid head groups are proposed to induce rearrangements of several amphipathic helices leading to exposure of two hydrophobic helices and subsequent membrane insertion (Lakey et al., 1991). Another possibility, albeit completely speculative, is that the C-terminal region of Plsp1 occupies the TTS binding groove when the disulfide bond is lacking, as suggested above, but is displaced by lipids during membrane insertion.
There are many examples of proteins that require specific lipid molecules for activity (Latowski et al., 2000; Hansen et al., 2011), structural stability (Seiwert et al., 2017), proper folding (Dowhan et al., 2004), or membrane insertion (Mel and Stroud, 1993). In some cases, this is due to properties of the lipid head group such as charge (Mel and Stroud, 1993; Hansen et al., 2011), and in others it is related to biophysical properties of the lipid (Latowski et al., 2004; Seiwert et al., 2017). Although our current data do not reveal an effect of a specific lipid molecule, we noted already that liposomes made from thylakoid lipids are significantly more effective in maintaining Plsp1 activity than are those made from E. coli lipids. Similarly, Plsp1 variants lacking the internal disulfide bridge were only able to partially complement the E. coli LepB suppression strain (i.e., FTL85), suggesting that the membrane lipid composition has a significant influence on Plsp1 structure and function in vivo as well as in vitro. The most abundant lipid found in thylakoid membranes (MGDG) is a nonbilayer forming lipid (Dörmann and Benning, 2002) and is important for the structure and function of various thylakoid proteins (Latowski et al., 2000; Seiwert et al., 2017). MGDG is also the only nonbilayer forming lipid used in our liposomes. PE is another nonbilayer forming lipid that is abundant in E. coli plasma membranes, mediates membrane insertion of LepB (van Klompenburg et al., 1998), and is required for maximal activity of LepB reconstituted into proteoliposomes (Wang et al., 2004). Interestingly, membrane binding of LepB can also be facilitated by MGDG albeit to a lesser extent than PE (van den Brink-van der Laan et al., 2001). It would be interesting to test whether nonbilayer forming lipids are also important for the activity and redox-dependency of Plsp1 in proteoliposomes by replacing MGDG with PE or phosphatidylcholine (a bilayer-forming lipid; van Klompenburg et al., 1998; Latowski et al., 2004).
Does Oxidative Folding Stabilize the Structure of Plsp1 during Diurnal Changes in the Energetic State of the Thylakoid Membrane?
In liposome reconstitution experiments, we found that a single Cys Plsp1 mutant has comparable activity to the wild-type form at pH 8.0 but lower comparative activity at pH 5.6 (Figures 8A and 8B). This suggests that the oxidized form of Plsp1 is more stable than the reduced form under moderately acidic conditions, despite the fact that the membrane alone is sufficient to facilitate proper reconstitutive folding (Figure 7). The lumen pH drops when thylakoids are energized and can fluctuate in response to changes in light intensity (Shikanai and Yamamoto, 2017). Therefore, we hypothesize that the disulfide bond helps maintain Plsp1 structure and activity in response to changes in lumen acidity (Figure 9). Changes in pH can alter protein and/or lipid head group protonation states, which can disrupt intramolecular interactions or interactions between Plsp1 and lipid head groups that may be important for reconstitutive folding.
Despite the compelling results from our in vitro experiments, we were still unable to detect a phenotypic difference between plants overexpressing wild-type and redox-inactive forms of Plsp1 when grown under either low (Figure 3A) or fluctuating (Supplemental Figure 11) light conditions. One possibility is that the redox-inactive forms of Plsp1 have low but sufficient activity when expressed above a certain threshold, which could be the case in our constitutive overexpression lines. Another possibility is that the single fluctuating light regime we tested was still insufficient to discriminate between our Citrine-Plsp1 wild-type and mutant overexpression lines. Variations to the experiment could include shortening the intervals between high and low light periods, increasing the light intensity during the high light period, and/or manipulating the photoperiod. One additional alternative would be to reduce the in vivo expression of Citrine-Plsp1 variants with a native expression system and repeat our complementation experiments. These and other experiments should be the subject of future work.
It is interesting to consider whether our data are specific to Plsp1 or whether they apply to other proteins in general. One thought is that an effect of pH fluctuations on activity and/or folding may apply more often in proteins found in energy-transducing membranes. Such membranes give rise to the possibility that the protein will be found in rather different environments depending on the state of energization, which itself depends on external factors, such as availability of substrates for oxidative electron transport and light for photosynthetic electron transport. Hamsanathan and Musser (2018) recently considered explicitly that the native environment of a protein embedded in an energetic membrane would necessarily include the pmf. Indeed, an influence of an electric field on protein structural stability has been observed experimentally (Bekard and Dunstan, 2014) and by simulations (Jiang et al., 2019). Extramembranous effects of the pmf necessarily include the pH values in the flanking compartments, i.e., the thylakoid lumen in the case of Plsp1. The importance of the membrane for the reconstitutive folding of Plsp1 is noteworthy given that the active site is most likely near the membrane/lumen interface (Dalbey et al., 2012).
Many enzymes have evolved to function in extreme environments such as high temperatures, low pH, and high salt concentrations (D'Amico et al., 2003; van den Burg, 2003). Under such circumstances, those proteins exhibit properties adapted to the particular conditions encountered, and in fact, those conditions must prevail for maximal activity. For example, the maximum in vitro activities of alpha amylases from a psychrophile and a thermophile were observed near the temperatures at which the host organisms are adapted to growing (D'Amico et al., 2003). This reflects the fact that the conditions under which the proteins are evolved to handle are relatively stable. This contrasts to the proteins residing in the energy-transducing membranes in chloroplasts and photosynthetic bacteria, which can exist in the presence of a variable pmf (i.e., magnitude and composition) resulting from rapidly changing light conditions during the day (Armbruster et al., 2017; Shikanai and Yamamoto, 2017) and darkness at night. Such proteins must be able to adapt to a more variable environment than those that exist under stable but harsh environments. The oxidative folding of Plsp1 may be an example of such an adaptation to a changing energy environment. Other examples may include STN7, another thylakoid membrane protein that is likely thermodynamically stabilized by a lumenal disulfide bond (Shapiguzov et al., 2016). While the disulfide bond in STN7 was proposed to maintain the conformation necessary for its interaction with the cytochrome b6f complex rather than simply enhancing conformational stability (Shapiguzov et al., 2016), this was not investigated under energizing and nonenergizing conditions.
A puzzling feature of oxidative folding of Plsp1 is the fact that it is not conserved throughout all photosynthetic organisms but is apparently restricted to the angiosperms (Midorikawa et al., 2014). Homologs of Plsp1 are found in the bacterial plasma membrane and the mitochondrial inner membrane and cleave signal sequences from proteins targeted to the periplasm and intermembrane space, respectively (Paetzel et al., 2002). In contrast with these energy-transducing membranes, thylakoids store a major proportion of their total pmf as a ΔpH (Cruz et al., 2001). While it is tempting to speculate that oxidative folding evolved in Plsp1 as a general adaptation to functioning in a compartment that undergoes more drastic pH changes, we recognize that lumen acidification is likely a feature of all organisms that perform oxygenic photosynthesis, not just the angiosperms. One possibility is that all nonangiosperm Plsp1 orthologs possess a mechanism other than oxidative folding to stabilize the structure amid pH changes. Another possibility is that conservation of Cys in angiosperm Plsp1 orthologs is related to the differences in pmf regulation between angiosperms and other members of the green lineage (Shikanai and Yamamoto, 2017). While the pmf composition can vary depending on environmental conditions (Shikanai and Yamamoto, 2017), perhaps the contribution of ΔpH is more variable in angiosperms compared with other photosynthetic organisms.
Conclusion
The role of lipids in protein structure and function has gained increasing attention in recent decades. An important role of lipids in reconstitutive folding and structural stability may be a feature of many integral and peripheral proteins in all membranes, but structural stabilization by disulfide bonds may be most common in thylakoid membranes. This may be due to the fact that the thylakoid is the only major energy-transducing membrane, which supports a substantial and highly dynamic pH gradient. Additionally, the apparent lack of major chaperones in the thylakoid lumen (Kieselbach and Schröder, 2003; Peltier et al., 2002; Schubert et al., 2002) may necessitate other means to stabilize proteins under periods of stress. While not discussed here, the electrical component of the pmf can also influence protein functions (Bekard and Dunstan, 2014; Jiang et al., 2019). Overall, we suggest our work has the potential to prompt and inform future discussions about the role of lipids and electrochemical gradients in protein structure and function.
METHODS
Antibodies
The pea Plsp1 antibody was from crude antiserum described previously (Midorikawa et al., 2014). The antibody against the extreme C terminus of Arabidopsis Plsp1 was as described (Shipman and Inoue, 2009). The antibodies against OE23 and Toc75 were as described previously (Shipman-Roston et al., 2010), and that against Hcf106 was as described (Rodrigues et al., 2011). The OE33 antibody was a gift from Dr. Hsou-Min Li (Institute of Molecular Biology, Academia Sinica), that against PGRL1 was from AgriSera (AS194311, 1:3000 dilution), that against GFP was from Santa Cruz Biotech (sc-8334, 1:2000 dilution), and the polyclonal T7 antibody was obtained from Millipore (AB3790, 1:1000 dilution). Immunoblots were developed either by enhanced chemiluminescence using LuminataTM Crescendo Western HRP Substrate or colorimetrically using the reagents 5-bromo-4-chloro-3-indolyl phosphate and nitroblue tetrazolium.
Plant Materials
The pgrl1ab double mutant was generated by a cross between SAIL_443E10 and SALK_059233 (DalCorso et al., 2008). Arabidopsis (Arabidopsis thaliana) seeds, ecotype Columbia-0 and the plsp1-1 T-DNA mutant (SALK_106199), were obtained from the Arabidopsis Biological Resource Center (https://abrc.osu.edu/).
DNA Constructs
The pUNI51 plasmid containing the coding sequence of Arabidopsis PsbP1 (At1g06680) was obtained from the Arabidopsis Biological Resource Center. The plasmid containing the sequence encoding the Silene pratensis plastocyanin precursor protein was as described (Last and Gray, 1989).
The cDNAs encoding Plsp1 variants for expression in planta were prepared by long flanking homology PCR as described previously (Endow and Inoue, 2013) using the primers listed in Supplemental Table 1 with PLSP11-70-CITRINE-PLSP171-291 in pMDC32 as the template. PCR products were recombined into pDONR207 (Addgene) by a BP reaction using Gateway™ BP Clonase™ II Enzyme mix (Invitrogen). Gateway™ LR Clonase™ II Enzyme mix (Invitrogen) was then used for recombination into pMDC32 (Curtis and Grossniklaus, 2003). Each plasmid was confirmed by DNA sequencing and transformed into Agrobacterium GV3101 cells for plant transformation.
The cDNAs encoding the transmembrane helices of LepB with a His6 tag (LepB1-34-His6-41-77) and different variants of Plsp1130-291 (i.e., the catalytic domain) were amplified from plasmid DNA by PCR using primers described in Supplemental Table 1. The PCR products were fused by long-flanking homology PCR to produce LepB1-34(His6)41-77-Plsp1130-291 (referred heretofore as LepBn-Plsp1c) and were further amplified by an additional PCR step before ligation. The cDNA encoding full-length Escherichia coli LepB with a His6 tag between transmembrane helices 1 and 2 (LepB1-34-His6-41-323) was also amplified by PCR using primers described in Supplemental Table 1. PCR products were ligated into the BsaI sites of pASK-IBA33plus and were propagated in E. coli DH5α cells. Each construct was confirmed by DNA sequencing.
Transient Expression in Nicotiana benthamiana
Agrobacterium cells carrying the P19 suppressor (Voinnet et al., 2003) in pBIN69 or PLSP11-70-T7-PLSP171-291 in pMDC32 were grown overnight at ∼28°C in Luria-Bertani (LB) broth with Kanamycin (25 μg/mL), Rifampicin (35 μg/mL), and Gentamycin (25 μg/mL). Cell cultures were then diluted in LB with Kanamycin (25 μg/mL) and grown to an OD600 of ∼0.2. Cells were pelleted at 2500 × g and resuspended in 10 mM MES-KOH pH 5.6, 1 mM MgCl2, 0.2% w/v Glc, and 150 μM acetosyringone (induction medium) to an OD600 of ∼0.4. T7-Plsp1 and P19 cell cultures were mixed at a ratio of 4:1 and incubated with moderate shaking at ∼28°C for 2 h. Cells were pelleted again at 2500 × g and resuspended in 5% w/v Suc and 300 μM acetosyringone (infiltration medium). Nicotiana benthamiana leaves were coinfiltrated using a syringe. After leaf infiltration, the plants were held in dark overnight and returned to a growth chamber (12-h light per day, 20°C, 400-Watt metal halide bulbs) for 1 to 3 d before isolating chloroplasts or thylakoids.
Chloroplast Isolation
Nicotiana benthamiana chloroplasts were isolated from leaves 2 to 4 d after infiltration with Agrobacterium cells. Leaves were homogenized in At grinding buffer (50 mM HEPES-KOH, pH 8; 0.33 M sorbitol; 2 mM EDTA; and 0.5% [w/v] BSA) and filtered through two layers of Miracloth (Millipore). Pelleted chloroplasts (3000 × g, 4°C, 3 min) were resuspended in a small volume of At grinding buffer and were carefully layered on top of a 40%/80% (v/v) Percoll (GE Healthcare) step gradient (24 mL 40% [v/v] Percoll on top of 6 mL 80% [v/v] Percoll, both in At grinding buffer). After centrifugation at 4000 × g for 10 min, the green band at the interface (intact chloroplasts) was collected and diluted with ∼25 mL of import buffer (50 mM HEPES-KOH or 50 mM Tricine-KOH, 0.33 M sorbitol, pH 8). After centrifugation at 2600 × g for 5 min, the intact chloroplasts were resuspended in import buffer, and a small volume was used for chlorophyll quantification as described (Arnon, 1949). The remainder was centrifuged again at 2600 × g for 5 min, and the pellet was resuspended in import buffer to 1 mg Chl/mL.
Arabidopsis chloroplasts were isolated from plants grown on phytoagar plates containing MS with Gamborg’s vitamins (Caisson Laboratories), supplemented with 1% (w/v) Suc, in a manner similar to that for N. benthamiana chloroplast isolation. After filtration through miracloth and centrifugation, chloroplasts were resuspended in At grinding buffer and layered on top of a 50% (v/v) continuous Percoll gradient. The green band near the bottom of the gradient after centrifugation at 8000 × g for 10 min was collected into ∼25 mL of import buffer, and chloroplasts were pelleted and washed in import buffer as above. Chlorophyll quantification and resuspension to 1 mg Chl/mL were performed as described above.
Chloroplasts from Pisum sativum plants (Little Marvel) were isolated from 11- to 14-d-old plants exactly as was done for Arabidopsis chloroplasts with the following exception: pea grinding buffer (50 mM HEPES-KOH pH 8; 0.33 M sorbitol; 2 mM EDTA; 1 mM MgCl2; 1 mM MnCl2; and 0.1% [w/v] BSA) was used in place of At grinding buffer.
Protease Treatments of Thylakoid Extracts
Pea chloroplasts isolated as described above were lysed hypotonically at ∼1 mg Chl/mL in 1 mM Tricine-NaOH, pH 7, and 5 mM MgCl2 (Buffer A) for 10 min on ice in the dark. Membranes were pelleted at 5000 × g for 5 min and were washed three times with 10 mM Tricine-NaOH, pH 7, and 5 mM MgCl2 0.3 M Suc (Buffer B). Washed thylakoids were resuspended in buffer C (50 mM Tricine-NaOH, pH 7; 5 mM MgCl2; and 15 mM NaCl) to ∼2 mg Chl/mL. An equal volume of buffer containing 0.5% v/v Triton X-100 was added, and the sample was incubated for 10 min on ice in the dark. After centrifugation at 100,000 × g, 4°C, 8 min (TLS55 rotor), the light-green supernatant (thylakoid extract) was transferred to a new tube and used immediately for experiments. Thylakoid extracts were pretreated with or without 10 mM TCEP (Thermoscientific) for 20 min on ice in the dark. Aliquots of each were then mixed with buffer containing CaCl2 (0.5 mM final) with or without thermolysin (0.1 μg/mL final). Each reaction mixture was incubated at 25°C. Aliquots taken at 10, 20, and 30 min were mixed with an equal volume of 2× reducing (0.2 M β-ME) sample buffer containing 20 mM EDTA and were boiled for 5 min. Samples were then analyzed by SDS-PAGE and immunoblotting or Coomassie Brilliant Blue staining.
Affinity Purification of T7-Plsp1
T7-Plsp171-291 (T7-Plsp1) was extracted from N. benthamiana thylakoids after transient expression of PLSP11-70-T7-PLSP171-291 in leaves. Leaves were homogenized in pea grinding buffer with 244 μM PMSF, filtered through four layers of Miracloth, and the filtrate was centrifuged at 2500 × g for 4 min at 4°C. The resulting pellet was washed once with 50 mM Tricine-NaOH (pH 7) and 5 mM sorbitol and three times with 50 mM Tricine-NaOH (pH 7), 5 mM MgCl2, and 0.1 M sorbitol. For extraction with Triton X-100, crude thylakoids were resuspended in buffer C (see above) at ∼2 mg Chl/mL and mixed with an equal volume of the same buffer containing 0.5% v/v Triton X-100. For extraction with octyl glucoside, crude thylakoids were resuspended in buffer C at ∼1 mg Chl/mL, and solid octyl glucoside was added to 1% w/v. After mixing at 4°C in the dark for 10-30 min, the supernatant containing T7-Plsp1 was obtained by centrifugation at 30,000 × g for 30 min at 4°C. T7-Plsp1 was then bound to T7•Tag Antibody Agarose (Millipore) for 1 to 2 h at 4°C on a BioRad EconoPac 25-mL column. The column was washed once with buffer C containing the appropriate detergent (0.25% v/v Triton X-100 or 1% w/v octyl glucoside) and was eluted with 0.1 M Gly-HCl pH 2.2 containing the same detergent. Elution fractions were collected into 4 M Tris-HCl (pH 9.5; 20 μL/mL elution) to give a final pH of ∼7.5. Each fraction was analyzed by SDS-PAGE and Coomassie Brilliant Blue staining or immunoblotting using the indicated antibodies. Purified T7-Plsp1 was divided into numerous aliquots, which were frozen with liquid N2 and stored at −80°C.
Synthesis of Radiolabeled Proteins
Proteins were synthesized from plasmid DNA templates in the presence of 35S (EasyTag™ EXPRESS35S Protein Labeling Mix, Perkin Elmer) using rabbit reticulocyte lysate (TnT® Quick Coupled Transcription/Translation System, Promega) according to the manufacturer’s guidelines. Reaction mixtures were quenched with an equal volume of 50 mM L-Met, 15 mM L-Cys in 0.1 M HEPES-KOH (pH 8), and 0.66 M sorbitol and were used for experiments on the same day.
Fluorescence Analysis of Plsp1
Aliquots of purified T7-Plsp1 in 1% w/v octyl glucoside were thawed on ice. Emission spectra of 150 μL samples of T7-Plsp1, T7-Plsp1 with 50 mM DTT, and the corresponding buffer blanks all prepared in elution buffer neutralized with Tris-HCl were collected using a Fluorolog spectrofluorometer (Horiba Scientific). The final concentration of T7-Plsp1 in each sample was ∼0.1 μM as determined by comparison with a BSA standard curve on a Coomasie-stained SDS-PAGE gel. Samples were excited at 285 nm, and slit widths were set to 5 nm for both excitation and emission. T7-Plsp1 with 50 mM DTT was incubated on ice for 20 min before collecting the emission spectrum. Spectra of buffer blanks were subtracted from those of each T7-Plsp1 sample to give corrected emission spectra.
Thiol Labeling Assay
Intact chloroplasts were isolated from Nicotiana benthamiana leaves after Agrobacterium-mediated transient expression of Plsp11-70-T7-Plsp171-291 as described above or after infiltration with infiltration medium (mock). Chloroplasts from mock-infiltrated leaves were subject to mock treatments at each step. Chloroplasts containing T7-Plsp1 were hypotonically lysed in the presence or absence of 0.1 M NEM and washed two times. Washed chloroplast membranes were then solubilized for 30 min at 28°C with 0.2 M Bis-Tris-HCl (pH 6.5) and 2% (w/v) SDS (labeling buffer) with or without 0.1 M NEM. Proteins were precipitated with an equal volume of 20% (w/v) TCA/80% (v/v) acetone, and pellets were washed 3 to 4 times with 1 mL 80% (v/v) acetone. Protein pellets were then resuspended in labeling buffer with or without 2 mM TCEP and incubated for 20 min at 28°C. Labeling buffer with or without methoxypolyethylene glycol maleimide (mPEG-MAL, Sigma) was then added to 10 mM final, and samples were then incubated for another 2 h at 28°C. Proteins were again precipitated as above and were resuspended in 2X sample buffer (0.1 M Tris-HCl, pH 6.8; 4% [w/v] SDS; 20% [v/v] glycerol; 0.2% [w/v] bromophenol blue; 0.2 M β-ME). Sample were analyzed by SDS-PAGE and immunoblotting.
Generation and Identification of Transgenic Arabidopsis Plants
Stable transgenic Arabidopsis plants were obtained by floral dip transformation of heterozygotes for the plsp1-1 T-DNA insertion using Agrobacterium tumefaciens cell cultures carrying the indicated transgene in pMDC32. T1 seeds were surface sterilized with 35% bleach and 0.02% v/v Triton X-100 and then screened on MS medium containing 25 or 50 μg/mL hygromycin (Calbiochem). Hygromycin-resistant seedlings (i.e., visible true leaves, roots penetrating the agar medium, wild type-like appearance) were transferred to soil after 2 to 3 weeks and were grown at 16 h light per day, 20°C. Soil-grown plants were screened by PCR using whole leaf DNA extracts. Selected plants were propagated to the T3 or T4 generation and used for experiments.
Arabidopsis in Vitro Import Assay
Intact chloroplasts were isolated as described above from 21- to 24-d-old Arabidopsis plants grown on MS agar medium at ∼20°C under ∼60 μE light with a 12-h light per day regime. The 50 μL import reactions containing 3 mM MgATP, 10 μL radiolabeled precursor proteins, and chloroplasts at 0.22 mg Chl/mL were prepared on ice in the dark. Upon addition of the radiolabeled proteins, each sample was immediately moved to ambient light (10–30 μE) at 20° to 22°C and incubated for 5, 10, or 20 min. Start times were staggered such that all import reactions would end at the same time. Samples were then immediately moved to ice in the dark and layered on top of 35% Percoll cushions in import buffer to reisolate intact chloroplasts. After centrifugation at 900 × g, 5 min, 4°C, all but ∼50 μL of each supernatant was removed. Import buffer (100 μL) was added, and the green pellets were resuspended by pipetting. Each 100 μL sample was then transferred to a new tube and centrifuged again at ∼2000 × g, 5 min, 4°C. After discarding the supernatants, each green pellet was resuspended in 2× sample buffer and heated at ∼82°C for 5 min. The leftover ∼50 μL solutions were used for chlorophyll quantification. Samples were analyzed by SDS-PAGE and autoradiography with the loading normalized based on total chlorophyll. Radioactive bands were quantified using ImageJ 1.5i (National Institutes of Health). Signals were normalized to that of the Citrine-Plsp1 wild-type sample at 20 min.
Total Protein Extraction
Whole seedlings were frozen and homogenized under liquid N2 followed by suspension and boiling for ∼15 min in extraction buffer (0.1 M Tris-HCl, pH 6.8; 4% [w/v] SDS; 15% [v/v] glycerol; 10 mM EDTA; 2% [v/v] β-ME). After centrifugation to remove tissue debris, four volumes of 100% [v/v] acetone were added to the soluble extract for precipitation at −20°C. Precipitated proteins were collected by centrifugation, washed with 80% [v/v] acetone, and resuspended in 8 M urea buffered with 0.1 M sodium phosphate and 10 mM Tris-HCl, pH 8.6. Total proteins were quantified by the Bradford assay (Bradford, 1976) using BSA as the standard.
In Vitro Processing Assays
A typical 10 μL processing assay consisted of 1 μL radiolabeled protein, enzyme sample (purified Plsp1 or crude thylakoid extract), and enzyme buffer with or without various additions such as DTT. Preincubations without substrate were typically performed on ice for ∼30 min. Reaction mixtures were incubated at 25° to 30°C for 30 min and quenched by addition of 10 μL of 2× sample buffer. After boiling for 5 min, samples were analyzed by SDS-PAGE and autoradiography.
Permeabilization of thylakoid membranes was performed as described previously (Ettinger and Theg, 1991). Washed thylakoids were prepared as done for protease treatment experiments described above. Samples at ∼1 mg Chl/mL were mixed with an equal volume of buffer C (see above) containing 0.08% v/v Triton X-100 and incubated on ice in the dark for 10 min. Thylakoid extracts were prepared as described above. For processing assays using permeabilized thylakoids (A) or thylakoid extracts (B), 8 μL of enzyme sample was mixed with 1 μL of buffer containing 0.08% v/v Triton X-100 (A) or 0.5% v/v Triton X-100 (B) and 1 μL of radiolabeled substrate protein. For pretreatments, the buffer contained the indicated chemical at a 10× concentration. Samples were incubated at ∼25°C for 30 min in the dark.
Reconstitution of T7-Plsp1 into Liposomes
Purified T7-Plsp1 in 0.25% (v/v) Triton X-100 was reconstituted into liposomes composed of E. coli total lipid extract or a mixture of thylakoid lipids (19.8 mol% MGDG, 54.9 mol% DGDG, 14.8 mol% SQDG, 10.5 mol% PG). All lipids were from Avanti Polar Lipids. Plant MGDG (840523P), plant DGDG (840524P), SQDG (840525P), and L-α-PG were from chicken egg (841138C). Small unilamellar vesicles (SUVs) were prepared by resuspending dried lipids at 4 mg/mL in liposome buffer (50 mM Tris-HCl, pH 8; 50 mM KCl; 10% [v/v] glycerol) and sonicating with a probe sonicator until optical clarity was achieved. Metal fragments from the sonicator tip were removed by centrifugation at 17,000 × g for 2 min. SUVs were mixed with Triton X-100 at a mass ratio of 2:1 to saturate the vesicles with detergent (Rigaud and Lévy, 2003). This mixture was incubated on ice for 15 min before adding purified T7-Plsp1 in 0.25% (v/v) Triton X-100 at a lipid to protein mass ratio of ∼80:1. After mixing at 4°C for 30 min, the mixture was added to an amount of BioBeads SM-2 (BioRad) equal to approximately ten times the mass of Triton X-100 present. The supernatant was added to a second batch of BioBeads after 1 h of mixing at 4°C and to a third batch after 2 h of mixing at 4°C. The third BioBeads incubation was done at 4°C for 18–20 h. BioBeads were removed using a Promega spin column, and the flow through was centrifuged at 250,000 × g for 30 min at 4°C. The transparent pellet was resuspended in 100 μL of liposome buffer and then centrifuged at 16,000 × g for 2 min to remove any aggregates. The supernatant was used for in vitro processing assays, thermolysin treatments, or a carbonate wash (0.1 M Na2CO3).
For reconstitution of prereduced Plsp1, T7-Plsp1 in 1% (w/v) octyl glucoside was reduced with 50 mM DTT, and 50 mM DTT was present throughout the reconstitution procedure to maintain Plsp1 in a reduced state. Proteoliposomes were pelleted as described above and resuspended in 70 μL of liposome buffer followed by centrifugation at 16,000 × g for 2 min to remove any aggregates. The supernatant was used for in vitro processing assays and thermolysin treatments. To confirm the redox state of Plsp1 before and after reconstitution, an aliquot was precipitated with 10% (w/v) trichloroacetic acid followed by incubation in SDS-PAGE sample buffer (without β-ME) containing the thiol-specific reagent 4-Acetamido-4'-Maleimidylstilbene-2,2'-Disulfonic Acid (AMS) at a concentration of 10 mM.
Growth of Arabidopsis Plants under Fluctuating Light
Arabidopsis seeds were sown on MS agar medium (1% [w/v] Sucrose, 3 mM MES-KOH, 1× strength MS, 0.7% [w/v] phytoagar, pH 5.7) and exposed to light after 2 d at 4°C in the dark. For growth under fluctuating light, plants were grown under alternating low (∼40 μE) and high (∼300 μE) light intensity with 10-min intervals. As a control, a duplicate set of plants was grown under constant low light (∼40 μE) during the day period. All plants were exposed to a 12-h photoperiod at ∼22°C.
Assay for Functionality of Plsp1 Catalytic Domain Variants in E. coli Cells
Cells of the LepB suppressor strain FTL85, which contains LepB driven by an Ara-inducible promoter (Lüke et al., 2009), were a kind gift of Yuanyuan Chen and Ross Dalbey (Ohio State University). The full length LepB construct and the different LepBn-Plsp1c constructs (wild type, C166A, C286A, C166A/C286A, S142) were transformed into FTL85 cells for a functional growth assay similar to that describe previously (Lüke et al., 2009). As a control, the empty pASK-IBA33plus vector was also transformed into FTL85 cells. Then 10 mL of LB containing ampicillin (100 μg/mL), kanamycin (50 μg/mL), and 0.05% (w/v) Ara in a 50-mL tube was inoculated with FTL85 carrying each construct in pASK-IBA33plus or the empty vector. Cultures were grown overnight (14–18 h) at 37°C, 225 rpm. For each culture, two 50-mL tube containing 10 mL LB with antibiotic and either 0.05% (w/v) Ara or 0.05% (w/v) Glc was prepared. Then 100 μL of overnight culture was added to each tube, and 0.5 mL of the new culture was taken for the initial OD600 measurement. Tubes were placed on a shaker at 37°C, 225 rpm, and 0.5 mL of culture was sampled every hour to measure OD600. After 5 h at 37°C, 100 μL of each culture was subcultured into a new tube containing preprepared fresh media identical to the original tube. Tubes were returned to the shaker at 37°C, 225 rpm, for an additional 5 h with samples taken each hour until 10 h of total culture time.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AT3G24590 (Arabidopsis Plsp1), AT1G06680 (Arabidopsis PsbP1), AT5G66570 (Arabidopsis PsbO1), AT3G46740 (Arabidopsis Toc75-III), AT1G06430 (Arabidopsis FtsH8), NM_001324194.1 (Zea mays PsbQ), X02965.1 (Silene pratensis plastocyanin), MK301208.1 (Citrine), AT4G22890 (PGRL1A), AT4G11960 (PGRL1B).
Supplemental Data
Supplemental Figure 1. Justification for using TCEP as a reductant, and Plsp1 solubility +/− DTT.
Supplemental Figure 2. T7-Plsp1 is functional in vivo.
Supplemental Figure 3. PEG-MAL labeling of in vitro translated 10His-Plsp168-291.
Supplemental Figure 4. Plsp1 requires the catalytic nucleophile Ser142 for in vivo functionality.
Supplemental Figure 5. Analysis of albino plants from hemizygous complemented lines and Citrine-Plsp1 mobility on SDS-PAGE
Supplemental Figure 6. Substitution of conserved Cys with Ala partially disrupts Plsp1 functionality in E. coli cells.
Supplemental Figure 7. The amount of inhibition of Plsp1 activity by DTT is dependent on the enzyme concentration.
Supplemental Figure 8. Processing activity of Citrine-Plsp1-CA is lost upon extraction from thylakoids.
Supplemental Figure 9. Reconstitution into E. coli lipid vesicles partially maintains Plsp1 activity under reducing conditions.
Supplemental Figure 10. Substituting both Cys in Plsp1 for Ala abolishes in vitro activity.
Supplemental Figure 11. Growth of Arabidopsis plants under fluctuating light.
Supplemental Table. List of primers used in this study.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
This work was supported by the Office of Basic Energy Sciences of the U.S. Department of Energy (DOE) (grant DE-SC0017035 to S.M.T. and grant DE-FG02-08ER15963 to K.I.), University of California Davis Department of Plant Sciences Graduate Student Research Assistantship (to L.J.M.), and a Henry A. Jastro Graduate Research Award (to L.J.M.).
Author Contributions
L.J.M. performed all other experiments; L.J.M., K.I., and S.M.T. designed the experiments; L.J.M. and S.M.T. wrote the article; J.F. performed the tryptophan fluorescence analysis described in Figure 1; J.K.E. cloned the LepB-Plsp1 chimeric constructs and performed the functional assay in E. coli cells;
References
- Albiniak A.M., Baglieri J., Robinson C.(2012). Targeting of lumenal proteins across the thylakoid membrane. J. Exp. Bot. 63: 1689–1698. [DOI] [PubMed] [Google Scholar]
- Armbruster U., Correa Galvis V., Kunz H.H., Strand D.D.(2017). The regulation of the chloroplast proton motive force plays a key role for photosynthesis in fluctuating light. Curr. Opin. Plant Biol. 37: 56–62. [DOI] [PubMed] [Google Scholar]
- Arnon D.I.(1949). Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta Vulgaris. Plant Physiol. 24: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsera M., Uberegui E., Schürmann P., Buchanan B.B.(2014). Evolutionary development of redox regulation in chloroplasts. Antioxid. Redox Signal. 21: 1327–1355. [DOI] [PubMed] [Google Scholar]
- Bekard I., Dunstan D.E.(2014). Electric field induced changes in protein conformation. Soft Matter 10: 431–437. [DOI] [PubMed] [Google Scholar]
- Betz S.F.(1993). Disulfide bonds and the stability of globular proteins. Protein Sci 2: 1551–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhanu M.K., Kendall D.A.(2014). Fluorescence spectroscopy of soluble E. coli SPase I Δ2-75 reveals conformational changes in response to ligand binding. Proteins 82: 596–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanov M., Dowhan W.(1999). Lipid-assisted protein folding. J. Biol. Chem. 274: 36827–36830. [DOI] [PubMed] [Google Scholar]
- Bogdanov M., Heacock P.N., Dowhan W.(2002). A polytopic membrane protein displays a reversible topology dependent on membrane lipid composition. EMBO J. 21: 2107–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanov M., Sun J., Kaback H.R., Dowhan W.(1996). A phospholipid acts as a chaperone in assembly of a membrane transport protein. J. Biol. Chem. 271: 11615–11618. [DOI] [PubMed] [Google Scholar]
- Bölter B., Soll J., Schwenkert S.(2015). Redox meets protein trafficking. Biochim. Biophys. Acta 1847: 949–956. [DOI] [PubMed] [Google Scholar]
- Bradford M.M.(1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254. [DOI] [PubMed] [Google Scholar]
- Braun N.A., Theg S.M.(2008). The chloroplast Tat pathway transports substrates in the dark. J. Biol. Chem. 283: 8822–8828. [DOI] [PubMed] [Google Scholar]
- Brooks M.D., Sylak-Glassman E.J., Fleming G.R., Niyogi K.K.(2013). A thioredoxin-like/β-propeller protein maintains the efficiency of light harvesting in Arabidopsis. Proc. Natl. Acad. Sci. USA 110: E2733–E2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterall W.A., Wisedchaisri G., Zheng N.(2017). The chemical basis for electrical signaling. Nat. Chem. Biol. 13: 455–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H., Kim S., Mukhopadhyay P., Cho S., Woo J., Storz G., Ryu S.E.(2001). Structural basis of the redox switch in the OxyR transcription factor. Cell 105: 103–113. [DOI] [PubMed] [Google Scholar]
- Cornell N.W., Crivaro K.E.(1972). Stability constant for the zinc-dithiothreitol complex. Anal. Biochem. 47: 203–208. [DOI] [PubMed] [Google Scholar]
- Cruz J.A., Sacksteder C.A., Kanazawa A., Kramer D.M.(2001). Contribution of electric field (Delta psi) to steady-state transthylakoid proton motive force (pmf) in vitro and in vivo. control of pmf parsing into Delta psi and Delta pH by ionic strength. Biochemistry 40: 1226–1237. [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U.(2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amico S., Marx J.C., Gerday C., Feller G.(2003). Activity-stability relationships in extremophilic enzymes. J. Biol. Chem. 278: 7891–7896. [DOI] [PubMed] [Google Scholar]
- Dalbey R.E., Wang P., van Dijl J.M.(2012). Membrane proteases in the bacterial protein secretion and quality control pathway. Microbiol. Mol. Biol. Rev. 76: 311–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DalCorso G., Pesaresi P., Masiero S., Aseeva E., Schünemann D., Finazzi G., Joliot P., Barbato R., Leister D.(2008). A complex containing PGRL1 and PGR5 is involved in the switch between linear and cyclic electron flow in Arabidopsis. Cell 132: 273–285. [DOI] [PubMed] [Google Scholar]
- Date T.(1983). Demonstration by a novel genetic technique that leader peptidase is an essential enzyme of Escherichia coli. J. Bacteriol. 154: 76–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Cock H., van Blokland S., Tommassen J.(1996). In vitro insertion and assembly of outer membrane protein PhoE of Escherichia coli K-12 into the outer membrane. Role of Triton X-100. J. Biol. Chem. 271: 12885–12890. [DOI] [PubMed] [Google Scholar]
- Dörmann P., Benning C.(2002). Galactolipids rule in seed plants. Trends Plant Sci. 7: 112–118. [DOI] [PubMed] [Google Scholar]
- Dowhan W., Mileykovskaya E., Bogdanov M.(2004). Diversity and versatility of lipid-protein interactions revealed by molecular genetic approaches. Biochim. Biophys. Acta 1666: 19–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endow J.K., Inoue K.(2013). Stable complex formation of thylakoidal processing peptidase and PGRL1. FEBS Lett. 587: 2226–2231. [DOI] [PubMed] [Google Scholar]
- Endow J.K., Singhal R., Fernandez D.E., Inoue K.(2015). Chaperone-assisted post-translational transport of plastidic type I signal peptidase 1. J. Biol. Chem. 290: 28778–28791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essigmann B., Güler S., Narang R.A., Linke D., Benning C.(1998). Phosphate availability affects the thylakoid lipid composition and the expression of SQD1, a gene required for sulfolipid biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 95: 1950–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettinger W.F., Theg S.M.(1991). Physiologically active chloroplasts contain pools of unassembled extrinsic proteins of the photosynthetic oxygen-evolving enzyme complex in the thylakoid lumen. J. Cell Biol. 115: 321–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalan G., He Z., Battaile K.P., Luan S., Swaminathan K.(2006). Structural comparison of oxidized and reduced FKBP13 from Arabidopsis thaliana. Proteins 65: 789–795. [DOI] [PubMed] [Google Scholar]
- Hager A., Holocher K.(1993). Localization of the xanthophyll-cycle enzyme violaxanthin de-epoxidase within the thylakoid lumen and abolition of its mobility by a (light-dependent) pH decrease. Planta 192: 581–589. [Google Scholar]
- Hall M., Mata-Cabana A., Akerlund H.E., Florencio F.J., Schröder W.P., Lindahl M., Kieselbach T.(2010). Thioredoxin targets of the plant chloroplast lumen and their implications for plastid function. Proteomics 10: 987–1001. [DOI] [PubMed] [Google Scholar]
- Hamsanathan S., Musser S.M. (2018). The Tat protein transport system: Intriguing questions and conundrums. FEMS Microbiol. Lett. 365: fny123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen S.B., Tao X., MacKinnon R.(2011). Structural basis of PIP2 activation of the classical inward rectifier K+ channel Kir2.2. Nature 477: 495–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann J.M., Riemer J.(2014). Three approaches to one problem: protein folding in the periplasm, the endoplasmic reticulum, and the intermembrane space. Antioxid Redox Signal 21: 438–456. [DOI] [PubMed] [Google Scholar]
- Houben E., de Gier J.W., van Wijk K.J.(1999). Insertion of leader peptidase into the thylakoid membrane during synthesis in a chloroplast translation system. Plant Cell 11: 1553–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu S.C., Endow J.K., Ruppel N.J., Roston R.L., Baldwin A.J., Inoue K.(2011). Functional diversification of thylakoidal processing peptidases in Arabidopsis thaliana. PLoS One 6: e27258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada T., Court D.L., Ito K., Nakamura Y.(1989). Conditionally lethal amber mutations in the leader peptidase gene of Escherichia coli. J. Bacteriol. 171: 585–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahns P., Latowski D., Strzalka K.(2009). Mechanism and regulation of the violaxanthin cycle: the role of antenna proteins and membrane lipids. Biochim. Biophys. Acta 1787: 3–14. [DOI] [PubMed] [Google Scholar]
- Jiang Z., You L., Dou W., Sun T., Xu P.(2019). Effects of an electric field on the conformational transition of the protein: A molecular dynamics simulation study. Polymers (Basel) 11: 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamoko M., Gabilly S.T., Hamel P.P.(2013). Operation of trans-thylakoid thiol-metabolizing pathways in photosynthesis. Front Plant Sci 4: 476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelkar D.A., Chattopadhyay A.(2007). The gramicidin ion channel: A model membrane protein. Biochim. Biophys. Acta 1768: 2011–2025. [DOI] [PubMed] [Google Scholar]
- Kieselbach T.(2013). Oxidative folding in chloroplasts. Antioxid Redox Signal 19: 72–82. [DOI] [PubMed] [Google Scholar]
- Kieselbach T., Schröder W.P.(2003). The proteome of the chloroplast lumen of higher plants. Photosynth. Res. 78: 249–264. [DOI] [PubMed] [Google Scholar]
- Kirwin P.M., Elderfield P.D., Williams R.S., Robinson C.(1988). Transport of proteins into chloroplasts. Organization, orientation, and lateral distribution of the plastocyanin processing peptidase in the thylakoid network. J. Biol. Chem. 263: 18128–18132. [PubMed] [Google Scholar]
- Kleinschmidt J.H.(2015). Folding of β-barrel membrane proteins in lipid bilayers - Unassisted and assisted folding and insertion. Biochim. Biophys. Acta 1848: 1927–1943. [DOI] [PubMed] [Google Scholar]
- Lakey J.H., Massotte D., Heitz F., Dasseux J.L., Faucon J.F., Parker M.W., Pattus F.(1991). Membrane insertion of the pore-forming domain of colicin A. A spectroscopic study. Eur J Biochem 196: 599–607. [DOI] [PubMed] [Google Scholar]
- Last D.I., Gray J.C.(1989). Plastocyanin is encoded by a single-copy gene in the pea haploid genome. Plant Mol. Biol. 12: 655–666. [DOI] [PubMed] [Google Scholar]
- Latowski D., Akerlund H.E., Strzałka K.(2004). Violaxanthin de-epoxidase, the xanthophyll cycle enzyme, requires lipid inverted hexagonal structures for its activity. Biochemistry 43: 4417–4420. [DOI] [PubMed] [Google Scholar]
- Latowski D., Kostecka A., Strzałka K.(2000). Effect of monogalactosyldiacylglycerol and other thylakoid lipids on violaxanthin de-epoxidation in liposomes. Biochem. Soc. Trans. 28: 810–812. [PubMed] [Google Scholar]
- Latowski D., Kruk J., Burda K., Skrzynecka-Jaskier M., Kostecka-Gugala A., Strzalka K.(2002). Kinetics of violaxanthin de-epoxidation by violaxanthin de-epoxidase, a xanthophyll cycle enzyme, is regulated by membrane fluidity in model lipid bilayers. Eur J Biochem 269: 4656–4665. [DOI] [PubMed] [Google Scholar]
- Lee A.G.(2004). How lipids affect the activities of integral membrane proteins. Biochim. Biophys. Acta 1666: 62–87. [DOI] [PubMed] [Google Scholar]
- Lee A.G.(2011). Biological membranes: The importance of molecular detail. Trends Biochem. Sci. 36: 493–500. [DOI] [PubMed] [Google Scholar]
- Liu C., Gao Z., Liu K., Sun R., Cui C., Holzwarth A.R., Yang C.(2016). Simultaneous refolding of denatured PsbS and reconstitution with LHCII into liposomes of thylakoid lipids. Photosynth. Res. 127: 109–116. [DOI] [PubMed] [Google Scholar]
- Lüke I., Handford J.I., Palmer T., Sargent F.(2009). Proteolytic processing of Escherichia coli twin-arginine signal peptides by LepB. Arch. Microbiol. 191: 919–925. [DOI] [PubMed] [Google Scholar]
- Mant A., Nielsen V.S., Knott T.G., Møller B.L., Robinson C.(1994). Multiple mechanisms for the targeting of photosystem I subunits F, H, K, L, and N into and across the thylakoid membrane. J. Biol. Chem. 269: 27303–27309. [PubMed] [Google Scholar]
- Mel S.F., Stroud R.M.(1993). Colicin Ia inserts into negatively charged membranes at low pH with a tertiary but little secondary structural change. Biochemistry 32: 2082–2089. [DOI] [PubMed] [Google Scholar]
- Michl D., Robinson C., Shackleton J.B., Herrmann R.G., Klösgen R.B.(1994). Targeting of proteins to the thylakoids by bipartite presequences: CFoII is imported by a novel, third pathway. EMBO J. 13: 1310–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midorikawa T., Endow J.K., Dufour J., Zhu J., Inoue K.(2014). Plastidic type I signal peptidase 1 is a redox-dependent thylakoidal processing peptidase. Plant J 80: 592–603. [DOI] [PubMed] [Google Scholar]
- Midorikawa T., Inoue K.(2013). Multiple fates of non-mature lumenal proteins in thylakoids. Plant J 76: 73–86. [DOI] [PubMed] [Google Scholar]
- Nishii W., Kukimoto-Niino M., Terada T., Shirouzu M., Muramatsu T., Kojima M., Kihara H., Yokoyama S.(2015). A redox switch shapes the Lon protease exit pore to facultatively regulate proteolysis. Nat. Chem. Biol. 11: 46–51. [DOI] [PubMed] [Google Scholar]
- Niyogi K.K., Li X.P., Rosenberg V., Jung H.S.(2005). Is PsbS the site of non-photochemical quenching in photosynthesis? J. Exp. Bot. 56: 375–382. [DOI] [PubMed] [Google Scholar]
- Nott T.J., Kelly G., Stach L., Li J., Westcott S., Patel D., Hunt D.M., Howell S., Buxton R.S., O’Hare H.M., Smerdon S.J.(2009). An intramolecular switch regulates phosphoindependent FHA domain interactions in Mycobacterium tuberculosis. Sci. Signal. 2: ra12. [DOI] [PubMed] [Google Scholar]
- Paetzel M.(2014). Structure and mechanism of Escherichia coli type I signal peptidase. Biochim. Biophys. Acta 1843: 1497–1508. [DOI] [PubMed] [Google Scholar]
- Paetzel M., Karla A., Strynadka N.C., Dalbey R.E.(2002). Signal peptidases. Chem. Rev. 102: 4549–4580. [DOI] [PubMed] [Google Scholar]
- Paetzel M., Strynadka N.C., Tschantz W.R., Casareno R., Bullinger P.R., Dalbey R.E.(1997). Use of site-directed chemical modification to study an essential lysine in Escherichia coli leader peptidase. J. Biol. Chem. 272: 9994–10003. [DOI] [PubMed] [Google Scholar]
- Peltier J.B., Emanuelsson O., Kalume D.E., Ytterberg J., Friso G., Rudella A., Liberles D.A., Söderberg L., Roepstorff P., von Heijne G., van Wijk K.J.(2002). Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction. Plant Cell 14: 211–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon M., de Kruijff B., Weisbeek P.J.(1992). New insights into the import mechanism of the ferredoxin precursor into chloroplasts. J. Biol. Chem. 267: 2548–2556. [PubMed] [Google Scholar]
- Pretzer D., Schulteis B., Vander Velde D.G., Smith C.D., Mitchell J.W., Manning M.C.(1992). Effect of zinc binding on the structure and stability of fibrolase, a fibrinolytic protein from snake venom. Pharm. Res. 9: 870–877. [DOI] [PubMed] [Google Scholar]
- Rigaud J.L., Lévy D.(2003). Reconstitution of membrane proteins into liposomes. Methods Enzymol. 372: 65–86. [DOI] [PubMed] [Google Scholar]
- Rodrigues R.A., Silva-Filho M.C., Cline K.(2011). FtsH2 and FtsH5: Two homologous subunits use different integration mechanisms leading to the same thylakoid multimeric complex. Plant J 65: 600–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt B., Ho L., Hogg P.J.(2006). Allosteric disulfide bonds. Biochemistry 45: 7429–7433. [DOI] [PubMed] [Google Scholar]
- Schubert M., Petersson U.A., Haas B.J., Funk C., Schröder W.P., Kieselbach T.(2002). Proteome map of the chloroplast lumen of Arabidopsis thaliana. J. Biol. Chem. 277: 8354–8365. [DOI] [PubMed] [Google Scholar]
- Seiwert D., Witt H., Janshoff A., Paulsen H.(2017). The non-bilayer lipid MGDG stabilizes the major light-harvesting complex (LHCII) against unfolding. Sci. Rep. 7: 5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiguzov A., Chai X., Fucile G., Longoni P., Zhang L., Rochaix J.D.(2016). Activation of the Stt7/STN7 kinase through dynamic interactions with the cytochrome b6f complex. Plant Physiol. 171: 82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L.X., Theg S.M.(2013). The chloroplast protein import system: From algae to trees. Biochim. Biophys. Acta 1833: 314–331. [DOI] [PubMed] [Google Scholar]
- Shikanai T., Yamamoto H.(2017). Contribution of cyclic and pseudo-cyclic electron transport to the formation of proton motive force in chloroplasts. Mol. Plant 10: 20–29. [DOI] [PubMed] [Google Scholar]
- Shipman R.L., Inoue K.(2009). Suborganellar localization of plastidic type I signal peptidase 1 depends on chloroplast development. FEBS Lett. 583: 938–942. [DOI] [PubMed] [Google Scholar]
- Shipman-Roston R.L., Ruppel N.J., Damoc C., Phinney B.S., Inoue K.(2010). The significance of protein maturation by plastidic type I signal peptidase 1 for thylakoid development in Arabidopsis chloroplasts. Plant Physiol. 152: 1297–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simionato D., Basso S., Zaffagnini M., Lana T., Marzotto F., Trost P., Morosinotto T.(2015). Protein redox regulation in the thylakoid lumen: The importance of disulfide bonds for violaxanthin de-epoxidase. FEBS Lett. 589: 919–923. [DOI] [PubMed] [Google Scholar]
- Soom M., Schönherr R., Kubo Y., Kirsch C., Klinger R., Heinemann S.H.(2001). Multiple PIP2 binding sites in Kir2.1 inwardly rectifying potassium channels. FEBS Lett. 490: 49–53. [DOI] [PubMed] [Google Scholar]
- Stangl M., Schneider D.(2015). Functional competition within a membrane: Lipid recognition vs. transmembrane helix oligomerization. Biochim. Biophys. Acta 1848: 1886–1896. [DOI] [PubMed] [Google Scholar]
- Strand D.D., Fisher N., Davis G.A., Kramer D.M.(2016). Redox regulation of the antimycin A sensitive pathway of cyclic electron flow around photosystem I in higher plant thylakoids. Biochim. Biophys. Acta 1857: 1–6. [DOI] [PubMed] [Google Scholar]
- Sung M., Dalbey R.E.(1992). Identification of potential active-site residues in the Escherichia coli leader peptidase. J. Biol. Chem. 267: 13154–13159. [PubMed] [Google Scholar]
- Suorsa M., Järvi S., Grieco M., Nurmi M., Pietrzykowska M., Rantala M., Kangasjärvi S., Paakkarinen V., Tikkanen M., Jansson S., Aro E.M.(2012). PROTON GRADIENT REGULATION5 is essential for proper acclimation of Arabidopsis photosystem I to naturally and artificially fluctuating light conditions. Plant Cell 24: 2934–2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Wada K.(1988). The status of cysteine residues in the extrinsic 33 kDa protein of spinach photosystem II complexes. Photosynth. Res. 17: 255–266. [DOI] [PubMed] [Google Scholar]
- Teale F.W.J.(1960). The ultraviolet fluorescence of proteins in neutral solution. Biochem. J. 76: 381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton J.M.(1981). Disulphide bridges in globular proteins. J. Mol. Biol. 151: 261–287. [DOI] [PubMed] [Google Scholar]
- Tschantz W.R., Paetzel M., Cao G., Suciu D., Inouye M., Dalbey R.E.(1995). Characterization of a soluble, catalytically active form of Escherichia coli leader peptidase: Requirement of detergent or phospholipid for optimal activity. Biochemistry 34: 3935–3941. [DOI] [PubMed] [Google Scholar]
- Umena Y., Kawakami K., Shen J.R., Kamiya N.(2011). Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473: 55–60. [DOI] [PubMed] [Google Scholar]
- van den Brink-van der Laan E., Dalbey R.E., Demel R.A., Killian J.A., de Kruijff B.(2001). Effect of nonbilayer lipids on membrane binding and insertion of the catalytic domain of leader peptidase. Biochemistry 40: 9677–9684. [DOI] [PubMed] [Google Scholar]
- van den Burg B.(2003). Extremophiles as a source for novel enzymes. Curr. Opin. Microbiol. 6: 213–218. [DOI] [PubMed] [Google Scholar]
- van der Goot F.G., González-Mañas J.M., Lakey J.H., Pattus F.(1991). A ‘molten-globule’ membrane-insertion intermediate of the pore-forming domain of colicin A. Nature 354: 408–410. [DOI] [PubMed] [Google Scholar]
- van Klompenburg W., Paetzel M., de Jong J.M., Dalbey R.E., Demel R.A., von Heijne G., de Kruijff B.(1998). Phosphatidylethanolamine mediates insertion of the catalytic domain of leader peptidase in membranes. FEBS Lett. 431: 75–79. [DOI] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D.(2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33: 949–956. [DOI] [PubMed] [Google Scholar]
- Wang Y., Bruckner R., Stein R.L.(2004). Regulation of signal peptidase by phospholipids in membrane: characterization of phospholipid bilayer incorporated Escherichia coli signal peptidase. Biochemistry 43: 265–270. [DOI] [PubMed] [Google Scholar]
- Webb M.S., Green B.R.(1991). Biochemical and biophysical properties of thylakoid acyl lipids. Biochim. Biophys. Acta 1060: 133–158. [Google Scholar]
- Wei X., Su X., Cao P., Liu X., Chang W., Li M., Zhang X., Liu Z.(2016). Structure of spinach photosystem II-LHCII supercomplex at 3.2 Å resolution. Nature 534: 69–74. [DOI] [PubMed] [Google Scholar]
- Zhbanko M., Zinchenko V., Gutensohn M., Schierhorn A., Klösgen R.B.(2005). Inactivation of a predicted leader peptidase prevents photoautotrophic growth of Synechocystis sp. strain PCC 6803. J. Bacteriol. 187: 3071–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Liu S., Hu Z., Kuang T., Paulsen H., Yang C.(2009). Effect of monogalactosyldiacylglycerol on the interaction between photosystem II core complex and its antenna complexes in liposomes of thylakoid lipids. Photosynth. Res. 99: 185–193. [DOI] [PubMed] [Google Scholar]