Putatively dosage-sensitive genes exhibit coordinated transcriptional responses to ploidy level change in Arabidopsis (Arabidopsis thaliana) consistent with gene duplicate retention being driven by selection on gene balance.
Abstract
The gene balance hypothesis postulates that there is selection on gene copy number (gene dosage) to preserve the stoichiometric balance among interacting proteins. This presupposes that gene product abundance is governed by gene dosage and that gene dosage responses are consistent for interacting genes in a dosage-balance-sensitive network or complex. Gene dosage responses, however, have rarely been quantified, and the available data suggest that they are highly variable. We sequenced the transcriptomes of two synthetic autopolyploid accessions of Arabidopsis (Arabidopsis thaliana) and their diploid progenitors, as well as one natural tetraploid and its synthetic diploid produced via haploid induction, to estimate transcriptome size and dosage responses immediately following ploidy change. Similar to what has been observed in previous studies, overall transcriptome size does not exhibit a simple doubling in response to genome doubling, and individual gene dosage responses are highly variable in all three accessions, indicating that expression is not strictly coupled with gene dosage. Nonetheless, putatively dosage balance-sensitive gene groups (Gene Ontology terms, metabolic networks, gene families, and predicted interacting proteins) exhibit smaller and more coordinated dosage responses than do putatively dosage-insensitive gene groups, suggesting that constraints on dosage balance operate immediately following whole-genome duplication and that duplicate gene retention patterns are shaped by selection to preserve dosage balance.
INTRODUCTION
Gene duplication is prevalent in eukaryotic genomes, occurring with a frequency similar to that of single-nucleotide substitutions (Lynch and Conery, 2000, 2003; Tasdighian et al., 2017) and is a major contributor to genetic diversity and the evolution of novel traits (Lynch and Conery, 2000). Most gene duplicates, however, are eventually pseudogenized and/or deleted from the genome, with an estimated half-life for duplicated genes in plants of 17 million years (Lynch and Conery, 2003). Following whole-genome duplication (WGD, polyploidy) the majority of duplicated gene pairs (homoeologs) return to single copy in the process of fractionation (Langham et al., 2004; Schnable et al., 2011, Wendel et al., 2018). A minority of duplicates from both small-scale duplication (SSD) and WGD, however, escape this decay process and are preserved over much longer periods of time. In Arabidopsis (Arabidopsis thaliana), for example, ∼25 percent of genes are retained in duplicate from the α-WGD approximately 32 to 43 million years ago (Blanc and Wolfe, 2004; Barker et al., 2009; Edger et al., 2018).
The retention or loss of redundant genes is not random. Certain classes of genes are preferentially retained in duplicate following WGD (Blanc and Wolfe, 2004), and many of these same genes exhibit minimal duplication via SSD (e.g., tandem duplication, transposition; Freeling, 2009). This pattern, in which some classes of genes preferentially retain duplicates originating from WGD but retain few duplicates derived from SSD is referred to as “reciprocal retention” (Tasdighian et al., 2017). Among the various models that have been proposed to explain the long-term retention of duplicated genes (e.g., neofunctionalization, subfunctionalization, selection on absolute dosage, selection on relative gene dosage; Panchy et al., 2016), only the gene balance hypothesis (GBH) provides an explanation for reciprocal retention (Papp et al., 2003; Freeling, 2009; Birchler and Veitia, 2012). The GBH predicts that there is a fitness cost in disrupting the stoichiometric balance between at least some proteins involved in coordinated interaction networks (e.g., protein complexes and signaling cascades). By duplicating every gene in the network, WGD is thought to preserve this balance, and any subsequent gene losses would disrupt it. As a consequence, genes in these networks are retained together through the diploidization process via purifying selection to preserve balance. Conversely, duplicates arising from SSD disrupt balance in dosage balance-sensitive networks, and selection acts to purge them. The range of functional stoichiometries (and, therefore, dosage sensitivities) is likely to be more constrained for some proteins than others (e.g., bridge proteins versus peripheral proteins in heteromeric complexes) and for some interaction networks than others (e.g., signal transduction complexes versus metabolic pathways; Birchler et al., 2016), explaining why some genes exhibit reciprocal retention and others do not.
Three main lines of evidence support the GBH (Edger and Pires, 2009; Freeling, 2009; Tasdighian et al., 2017; Hou et al., 2018): (1) signaling cascades, regulatory networks, and protein complexes that are known to be disrupted by unbalanced changes in protein abundance tend to exhibit reciprocal retention patterns; (2) reciprocally retained genes exhibit greater selective constraint on sequence evolution (lower ratio of the number of nonsynonymous substitutions per nonsynonymous site to the number of synonymous substitutions per synonymous site) and less divergence in expression patterns than nonreciprocally retained genes; and (3) reciprocally retained genes often exhibit deleterious phenotypes when over- or underexpressed—this last piece of evidence is often cited as the ultimate proof needed to demonstrate dosage sensitivity and confirm the GBH. However, demonstrating that a deleterious phenotype is induced by over- or underexpressing a gene provides evidence for dosage sensitivity at the protein level, but it does not necessarily follow that there exists dosage sensitivity at the level of gene copy number. Gene dosage differences alone do not produce the deleterious phenomena associated with imbalance; the genes must be transcribed and translated. If gene copy number is decoupled from the final protein concentration at the point of interaction (e.g., multisubunit complex assembly), selection on preservation of gene copy number loses its power as an explanation for gene retention. Decoupling can occur through such diverse mechanisms as differential expression of genes encoding members of a dosage-balance-sensitive complex, differential stability of mRNAs encoding members of the complex, differential translation of those mRNAs, or differential stability of proteins.
Such decoupling is evident in response to polyploidy because not all genes show identical expression responses following duplication—whether measured at the level of transcript abundance (e.g., Guo et al., 1996; Riddle et al., 2006; Stupar et al., 2007; Yu et al., 2010; Hou et al., 2018; Pirrello et al., 2018; Robinson et al., 2018; additional references in Doyle and Coate, 2019) or protein abundance (Birchler and Newton, 1981; Yao et al., 2011; Zhu et al., 2012; Soltis et al., 2016; Deng et al., 2017; Fan et al., 2017; Wang et al., 2017; Yan et al., 2017). Consequently, WGD does not necessarily preserve protein dosage balance for all genes, and the extent to which dosage responses following WGD are coordinated among genes encoding interacting proteins is unknown. Conversely, if all genes in a complex are dosage compensated (no change in expression with change in copy number), protein stoichiometry would be unaltered by SSD or the loss of duplicates from WGD, and there would be no selection to drive reciprocal retention. To affect balance at the protein level, gene copy number minimally should be “felt” at the level of the transcriptome. For the GBH to have explanatory power as a force maintaining gene copy number, maintenance of transcriptomic balance is necessary, though not sufficient.
Therefore, the GBH predicts that (1) genes in reciprocally retained gene networks (protein complexes, metabolic pathways, etc.) exhibit changes in expression in response to WGD (they are not dosage compensated) and (2) that these changes are similar for all genes in the network (what we refer to as “coordinated responses”). Our previous study examined the relationship between duplication history and gene dosage responses at the level of transcription in Glycine neoallopolyploids (Coate et al., 2016). We showed that genes in reciprocally retained GO terms and metabolic pathways showed more coordinated dosage responses than genes in nonreciprocally retained networks, consistent with gene dosage sensitivity. The Coate et al. (2016) study, however, was complicated by the fact that the observed expression patterns were the net result of WGD and hybridization, as well as by approximately 0.5 million years of post-WGD evolution. Additionally, Coate et al. (2016) only measured relative expression levels (transcript concentrations) rather than absolute dosage responses. In fact, there remains very little data about the immediate dosage responses to “pure” doubling (autopolyploidy; Spoelhof et al., 2017; Visger et al., 2019) and whether or not these dosage responses are consistent with the GBH.
Long-term patterns of gene retention and loss as predicted by the GBH rely on simple assumptions that can be tested in synthetic polyploids. First, there should be low variation in transcript abundance among individuals for genes that encode proteins in dosage-balance-sensitive complexes (Lemos et al., 2004; Birchler et al., 2005; Coate et al., 2016). This is because the stoichiometry of the complex would be disrupted when low-expressing alleles for some subunits are combined with high-expressing alleles for others. Second, gene duplication should immediately alter gene expression and do so in a coordinated fashion for genes encoding dosage-balance-sensitive proteins (Birchler and Newton, 1981). Synthetic polyploids allow us to see the instantaneous effects of gene duplication on gene expression, thereby testing these assumptions. This study, therefore, builds upon past work by using diploid/synthetic autotetraploid pairs of Arabidopsis (accessions C24 and Wassilewskija [Ws]) and a tetraploid/synthetic diploid pair (Warschau [Wa]) to quantify transcriptome size, expression variance, and gene dosage responses in the first generations post-WGD in the absence of hybridization. We test whether there is an intrinsic, heritable difference between genes that are reciprocally retained and those that are not and find that reciprocally retained gene groups immediately exhibit smaller and more coordinated dosage responses to changes in genome dosage (both WGD and genome halving) than their nonreciprocally retained counterparts.
RESULTS
Classes of Genes Grouped by Gene Ontology and by Metabolic Pathway Exhibit Patterns of Reciprocal Retention
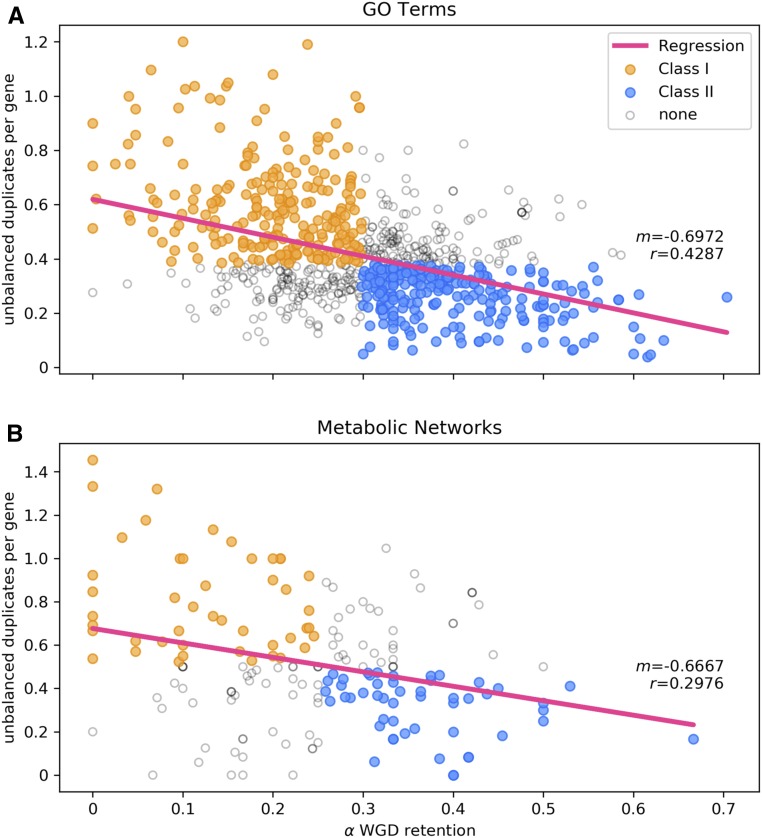
Arabidopsis genes were categorized as singletons, WGD duplicates, or SSD duplicates (including tandem, proximal, or transposed duplicates) according to Wang et al. (2013). We then tested whether functionally related gene groups—gene ontologies (GO) or metabolic pathways (Schläpfer et al., 2017)—exhibited patterns of reciprocal retention. As previously observed (Freeling, 2009; Coate et al., 2016; Tasdighian et al., 2017), we found that both GO terms and metabolic pathways with high retention following WGD tended to have lower retention of SSD (linear regression for GO terms, slope = −0.6972, R2 = 0.1839, F = 175.05, df = 1 and 777, P < 0.001; linear regression for metabolic networks, slope = 0.6667, R2 = 0.0886, F = 17.31, df = 1 and 178, P < 0.001; Figures 1A and 1B). To test whether the GBH explains these patterns of reciprocal retention, we grouped GO terms or networks into those that are putatively dosage insensitive (class I; lower than median WGD retention and higher than median SSD [unbalanced] duplication; Figure 1, yellow) and those that are putatively dosage sensitive (class II; higher than median WGD retention and lower than median SSD; Figure 1, blue) following the methods of Coate et al. (2016). Note that assignment to class I or class II was based entirely on patterns of duplicate retention. If selection on dosage balance explains these patterns, we would expect GO terms and networks in class II to also exhibit predictable patterns of expression. Namely, we predict that genes in these groups should exhibit coordinated expression response to ploidy change and low expression level variance among individuals within a species. In the following analyses, we assess whether class II gene groups meet these predictions.
Figure 1.
Reciprocal Relationship between Small-Scale and Whole-Genome Duplications.
The number of unbalanced duplicates (duplicates produced by tandem, proximal, or TE-mediated duplication) per gene (y axis) versus fraction of genes retaining duplicates from the 𝛼 WGD (x axis) for GO classes and AraCyc metabolic pathways.
(A) GO.
(B) Metabolic pathways.
Yellow data points indicate gene groups assigned to class I (putatively dosage balance insensitive; higher-than-median numbers of unbalanced duplicates per gene and lower-than-median percentages of 𝛼 WGD retention), and blue data points indicate groups assigned to class II (putatively dosage balance sensitive; lower-than-median numbers of unbalanced duplicates per gene and higher-than-median percentages of 𝛼 WGD retention).
Doubling the Genome Does Not Result in Twice the Total Amount of Transcripts per Cell
The GBH depends on there being a strong correlation between gene dosage and transcript abundance (Coate et al., 2016). If gene dosage and transcript abundance are perfectly correlated for all genes, then WGD would maintain a constant number of transcripts (transcriptome size) per genome, resulting in a doubling of total transcripts per cell. We measured transcriptome size per genome and per cell to assess how closely transcript abundance correlates with gene copy number overall.
Both synthetic tetraploids (C24 and Ws) exhibited small but significant deviations in mRNA transcriptome size per genome relative to their diploid progenitors (P < 0.001; one-sample t test). Interestingly, the direction of change differed for the two accessions, with C24 exhibiting a reduction in transcripts per genome (0.79-fold ± 0.10 SD) and Ws exhibiting an increase in transcripts per genome (1.19-fold ± 0.06 SD). As with Ws, the natural tetraploid (Wa) exhibited slightly more transcripts per genome than its derived diploid (1.15-fold ± 0.10 SD; P < 0.001; one-sample t test). Thus, in none of the three accessions did genome doubling produce a simple doubling of transcripts, indicating that individual gene dosage responses deviate on average from a simple 1:1 dosage response.
Notably, both synthetic tetraploids also exhibited reduced levels of endopolyploidy relative to their diploid progenitors (C24, t = 8.253, df = 5, P < 0.001; Ws, t = 3.80, df = 4, P = 0.019; two-sample t test), such that mRNA transcriptome size per cell was, on average, significantly less than doubled in both accessions (P < 0.001; one-sample t test). The size of the mRNA transcriptome per cell relative to the diploid progenitor was 1.16 ± 0.14 for C24 and 1.77 ± 0.09 for Ws. Thus, variable dosage responses and reduced endoreduplication interact to produce a smaller-than-expected transcriptome per cell on average (though the effect in any single cell or cell type was not measured here). The natural tetraploid, Wa, also exhibited a reduced level of endopolyploidy relative to its derived diploid, but the reduction was not significant (t = 1.177, df = 7, P = 0.278; two-sample t test) and less extreme than in the derived tetraploids (average ploidy in Wa tetraploids was 1.83-fold higher than in diploids, compared to 1.46-fold higher in C24 and 1.49-fold higher in Ws). As a consequence, the derived Wa diploid transcriptome per cell was roughly one-half of the average natural tetraploid transcriptome (tetraploid:diploid, 2.11-fold ± 0.18 SD).
Individual Gene Dosage Responses Are Highly Variable, and Many Genes Are Dosage Compensated
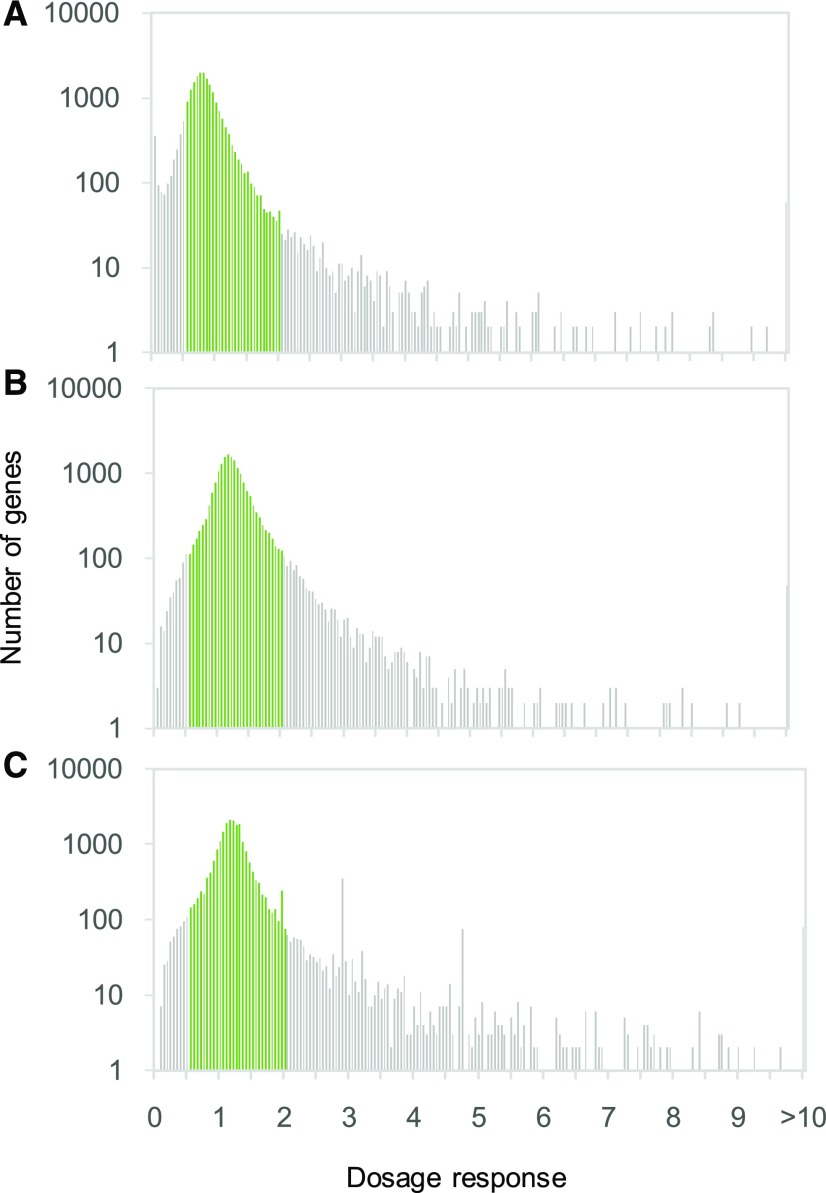
By quantifying transcriptome size, we were able to estimate absolute dosage responses at individual loci (fold change in expression with a doubling of gene copy number). In all three accessions, dosage responses (change in transcripts per gene copy) were unimodally distributed around the estimate of overall transcriptome size but with extreme values in each direction ranging from near silencing of expression with a doubling of gene copy number (a strong negative dosage effect) to a greater than 88-fold increase with a doubling in gene copy number (Figure 2). 9.1%, 9.8% and 13.4% of genes deviated more than twofold from a 1:1 dosage response in Ws, Wa, and C24, respectively.
Figure 2.
Distribution of Gene Dosage Responses (Transcripts per Genome in the Tetraploid Divided by Transcripts per Genome in the Diploid) in Three Arabidopsis thaliana Accessions.
(A) C24 (n = 21,260 genes).
(B) Ws (n = 19,594 genes).
(C) Wa (n = n = 22,325 genes).
Additionally, many genes exhibited responses to WGD or genome halving consistent with dosage compensation (a change in expression that compensates for change in gene copy number, resulting in no change in expression per cell). For example, in Ws, the 95% confidence interval for transcripts per genome overlapped with 0.5 (dosage compensation) for 4114 out of 19,594 genes for which we were able to estimate dosage responses A dosage response of 1 indicates equal expression per gene copy or doubled expression per cell in tetraploids versus diploids (a 1:1 dosage response). Dosage responses that differ by more than twofold from a 1:1 dosage response are shown in gray (C24, n = 2843 genes, 13.4% of total; Ws, n = 1789, 9.1% of total; Wa, n = 2198, 9.8% of total). The x axis is cut off at 10 for display purposes, but 59, 48, and 79 genes exhibit dosage responses >10 in C24, Ws, and Wa, respectively (maximum value = 88.7 in Ws). (21%). 891 out of 21,260 genes (4.2%) and 7061 out of 22,325 genes (31%) were dosage compensated in C24 and Wa, respectively. This is relevant because dosage compensation decouples duplication from protein abundance, making gene dosage invisible to selection to maintain balance. Thus, individual gene dosage responses are variable, and a large fraction of genes do not behave in a strictly dosage-dependent manner. Consequently, although the simplest way in which selection for maintaining balance among interacting proteins could drive reciprocal retention is if all genes exhibit 1:1 dosage responses (a 1:1 correspondence between transcript abundance and gene copy number, regardless of the mechanism of copy number change), this is not the case, regardless of whether the comparison is between synthetic polyploids and their natural diploid progenitors (C24 and Ws) or between a natural polyploid (Wa) and its synthetically derived diploid.
Putatively Dosage-Balance-Sensitive Gene Classes Exhibit Coordinated Dosage Responses
Selection on dosage balance could still explain the reciprocal pattern of retention even given the lack of a uniform relationship between dosage and expression if all genes in a connected network whose products interact in a dosage-balance-sensitive manner have comparable, or coordinated, dosage responses (Coate et al., 2016). We tested if there are coordinated transcriptional responses to genome doubling for reciprocally retained gene groups. Following the methods of Coate et al. (2016), for a given functional class (GO term) or metabolic pathway, we calculated the mean and coefficient of variation (SD divided by the mean) of dosage responses for all included genes.
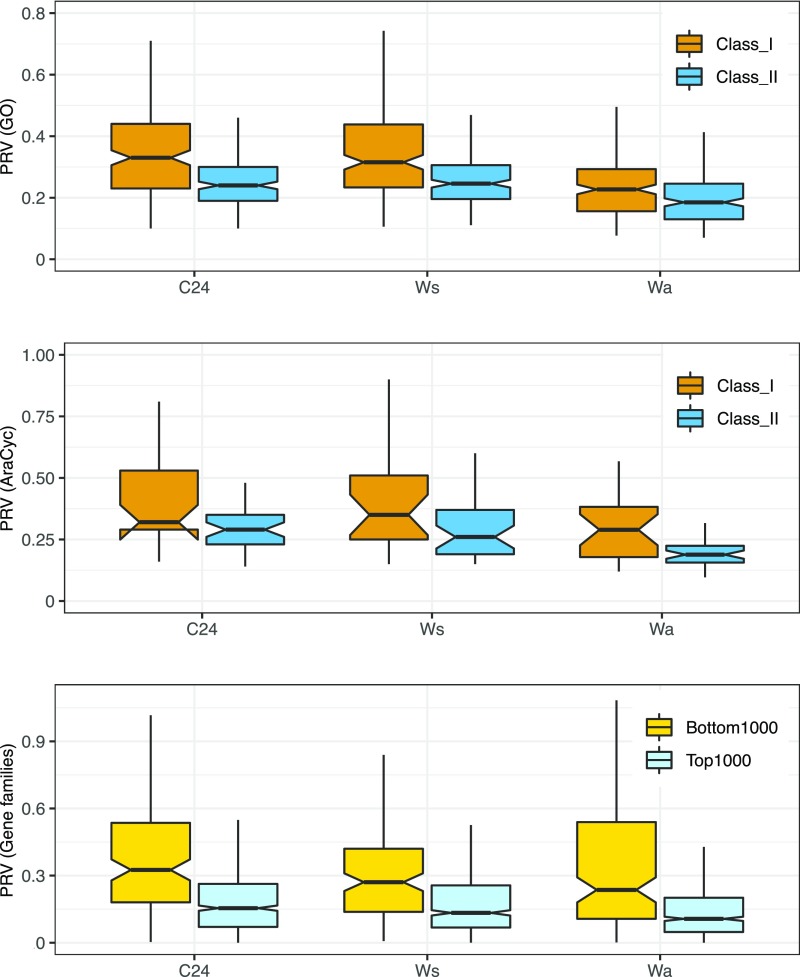
The coefficient of variation, which we refer to as the polyploid response variance (PRV), is a measure of the degree to which the dosage responses of genes within a network are correlated—a low PRV indicates strong coordination of dosage responses, whereas a high PRV indicates uncoordinated or variable dosage responses (Coate et al., 2016). We then looked to see if putatively dosage-sensitive (class II; reciprocally retained) metabolic pathways or GO terms exhibit lower PRV than putatively insensitive (class I; not reciprocally retained) pathways or GO terms. Consistent with the GBH, PRV is lower for class II than for class I across all three polyploid-diploid pairs (though the difference is not significant for metabolic pathways in C24; (Table 1; Figures 3A and 3B; Supplemental Data Set).
Table 1. Summary Statistics and Kruskal-Wallis Tests for Differences in PRV by Class.
| Grouping | Accession | N (Class I) | N (Class II) | Mean (SD) Class I | Mean (SD) Class II | X2 | df | P |
|---|---|---|---|---|---|---|---|---|
| GO |
C24 | 185 | 198 | 0.369 (0.208) | 0.261 (0.114) | 44.47 | 1 | 2.58−11 |
| Ws | 185 | 191 | 0.348 (0.189) | 0.267 (0.133) | 26.341 | 1 | 2.86−07 | |
| Wa | 189 | 198 | 0.239 (0.107) | 0.199 (0.090) | 16.718 | 1 | 4.34−05 | |
| AraCyc |
C24 | 29 | 41 | 0.428 (0.229) | 0.342 (0.223) | 3.3058 | 1 | 0.0690 |
| Ws | 25 | 37 | 0.511(0.567) | 0.262 (0.174) | 6.7835 | 1 | 0.0092 | |
| Wa | 26 | 41 | 0.301 (0.153) | 0.200 (0.076) | 8.740 | 1 | 0.0031 | |
| Gene |
C24 | 141 | 652 | 0.407 (0.327) | 0.209 (0.211) | 62.531 | 1 | 2.62−15 |
| Ws | 127 | 618 | 0.334 (0.283) | 0.192 (0.187) | 39.95 | 1 | 2.60−10 | |
| Wa | 149 | 650 | 0.356 (0.339) | 0.166 (0.188) | 54.2 | 1 | 1.81−13 | |
| S-PPI | C24 | 7692 | 501 | 0.309 (0.318) | 0.223 (0.219) | 29.227 | 1 | 6.44−08 |
| Ws | 7416 | 484 | 0.236 (0.227) | 0.204 (0.193) | 9.0861 | 1 | 0.0026 | |
| Wa | 8377 | 520 | 0.367 (0.466) | 0.242 (0.361) | 34.85 | 1 | 3.56−09 |
Summary statistics and Kruskal-Wallis tests for differences in PRV by class for GO, metabolic pathways (AraCyc), Tasdighian et al. (2017) orthogroups (gene families), or Dong et al. (2019) structure-based protein-protein interactions (S-PPI). N, number of functional groups included in the analysis.
Figure 3.
PRV by Dosage Sensitivity Class in C24, Ws, and Wa.
(A) GO terms.
(B) Metabolic pathways.
(C) Reciprocal retention ranking of gene families (Tasdighian et al., 2017).
Putatively dosage-sensitive gene families (class II/top 1000) show lower average PRV than dosage-insensitive gene families (class I/bottom 1000).
The variance associated with expression estimates in RNA sequencing (RNA-seq) experiments is inversely correlated with expression level (variance is lower for highly expressed genes than for genes expressed at lower levels; Conesa et al., 2016; Mortazavi et al., 2008). Consequently, estimates of dosage response (fold change of expression in tetraploid versus diploid) are expected to be more variable at low expression levels, potentially inflating estimates of PRV. Indeed, when PRV is plotted against mean expression level for GO terms not assigned to class I or class II (to minimize the influence of differences in gene dosage sensitivity), we see a weak negative correlation (GO terms with higher expression levels have lower PRV; Supplemental Figure 2A). Therefore, we checked to see if the lower PRVs for class II are a function of higher mean expression levels. For these and all other non-normally distributed data in the study, we used the nonparametric Kruskal-Wallis test to test for differences in means and found that class II GO terms have lower expression, on average, than class I terms in all three accessions (P < 0.005, Kruskal-Wallis tests; Supplemental Figure 2B). This indicates that the smaller PRVs are not a function of higher expression and that observed differences in PRV may in fact be offset by differences in expression, thereby underestimating the degree to which coordination is higher in class II versus class I interaction networks.
We also examined if genes exhibiting extreme dosage responses (Figure 1) might explain the observed differences in PRV between class I and class II groups. We repeated the analysis after first excluding genes that exhibited >fivefold changes in expression per genome (tetraploid/diploid ≥ 5 or ≤ 0.2). With these genes removed from the data set, PRV was still significantly higher for class I GO terms than for class II GO terms in each accession (P < 0.01; Kruskal-Wallis). Thus, differences in PRV between class I and class II are not driven by differences in expression level or by the subset of genes showing extreme dosage responses.
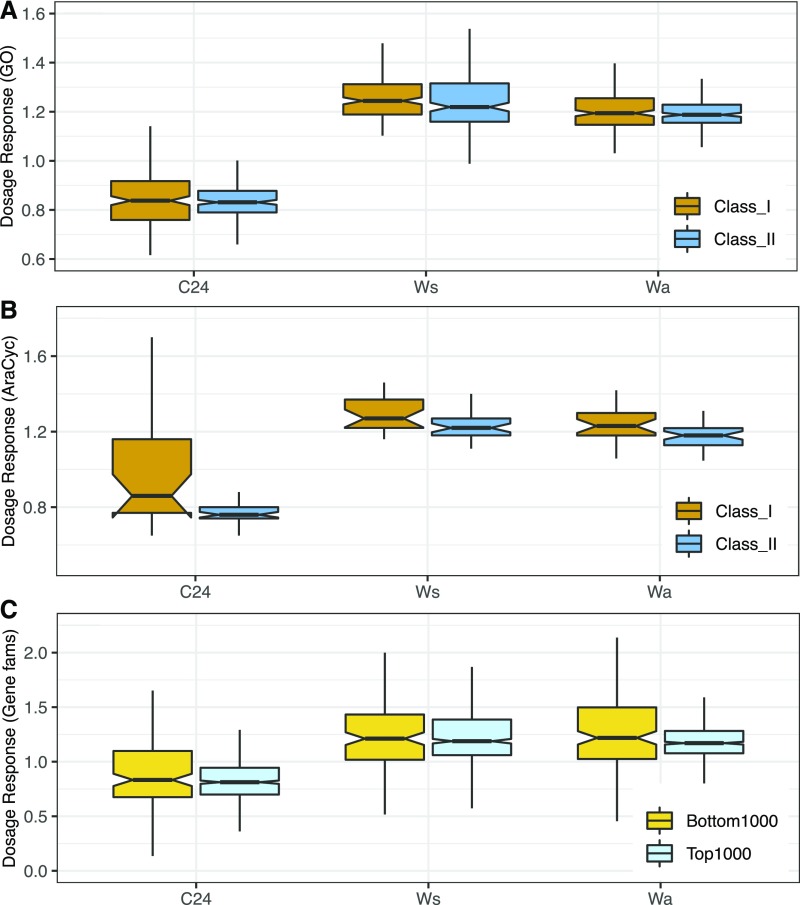
Absolute dosage responses (fold change in expression between tetraploids and diploids) were also smaller on average in putatively dosage sensitive gene groups (class II GO terms and metabolic pathways) than in putatively insensitive groups (class I GO terms and metabolic pathways). However, the difference was significant in only a subset of the comparisons (Table 2; Figures 4A and 4B; Supplemental Data Set).
Table 2. Summary Statistics and Kruskal-Wallis Tests for Differences in Dosage Responses by Class.
| Grouping | Accession | N (Class I) | N (Class II) | Mean (SD) Class I | Mean (SD) Class II | X2 | df | P |
|---|---|---|---|---|---|---|---|---|
| GO | C24 | 185 | 197 | 0.847 (0.116) | 0.827 (0.078) | 0.717 | 1 | 0.397 |
| Ws | 185 | 191 | 1.233 (0.117) | 1.206 (0.135) | 13.867 | 1 | 0.002 | |
| Wa | 189 | 198 | 1.214 (0.107) | 1.197 (0.066) | 0.351 | 1 | 0.554 | |
| AraCyc | C24 | 29 | 41 | 0.936 (0.236) | 0.799 (0.113) | 6.602 | 1 | 0.010 |
| Ws | 25 | 37 | 1.453 (0.582) | 1.230 (0.078) | 6.561 | 1 | 0.010 | |
| Wa | 26 | 41 | 1.250 (0.139) | 1.176 (0.063) | 7.852 | 1 | 0.005 | |
| Gene families | C24 | 141 | 652 | 1.162 (1.274) | 0.848 (0.326) | 2.946 | 1 | 0.086 |
| Ws | 127 | 618 | 1.735 (4.161) | 1.267 (0.504) | 0.015 | 1 | 0.903 | |
| Wa | 149 | 650 | 1.880 (5.343) | 1.240 (0.529) | 2.653 | 1 | 0.103 | |
| S-PPI | C24 | 7692 | 501 | 0.971 (1.264) | 0.822 (0.259) | 0.168 | 1 | 0.682 |
| Ws | 7416 | 484 | 1.346 (1.015) | 1.322 (0.425) | 3.720 | 1 | 0.054 | |
| Wa | 8377 | 520 | 1.274 (1.045) | 1.300 (1.087) | 0.295 | 1 | 0.587 |
Summary statistics and Kruskal-Wallis tests for differences in dosage response by class for GO, metabolic pathways (AraCyc), Tasdighian et al. (2017) orthogroups (gene families), or Dong et al. (2019) structure-based protein-protein interactions (S-PPI). N, number of functional groups included in the analysis.
Figure 4.
Dosage Responses (Fold Change in Expression per Genome between Tetraploids and Diploids) by Dosage Sensitivity Class in C24, Ws, and Wa.
(A) GO terms.
(B) Metabolic networks.
(C) Reciprocal retention ranking of gene families (Tasdighian et al., 2017).
Reciprocally Retained Gene Families Exhibit Coordinated Expression Responses
Although there is a moderately strong pattern of reciprocal retention for GO terms (Figure 2), Tasdighian et al. (2017) have correctly pointed out that GO terms are sufficiently generic and that many likely include both dosage-balance-sensitive and -insensitive genes. They argue that dosage sensitivity is better defined at the level of gene families as opposed to broad functional groupings. We therefore assessed if their 1000 most reciprocally retained gene families also exhibit lower PRV (more coordinated dosage responses) than do their 1000 least reciprocally retained gene families. We found coordinated expression responses consistent with the expectations of the GBH (Table 1; Figure 3C).
Notably, the difference in PRV was more pronounced in this comparison than in the comparison of class I versus class II GO terms or metabolic pathways, consistent with the Tasdighian et al. (2017) assertion that dosage balance sensitivity is better characterized at the level of gene families and not necessarily a shared property of all genes of a broad functional category. In contrast to GO terms and metabolic pathways, however, we did not observe smaller dosage responses in the top 1000 gene families than in the bottom 1000 gene families (Kruskal-Wallis tests—C24, χ2 = 2.95, df = 1, P = 0.086; Ws, χ2 = 0.01, df = 1, P = 0.903; Wa, χ2 = 2.65, df = 1, P = 0.103; Table 2; Figure 4C).
Dosage-Sensitive Gene Classes Exhibit Less Variable Expression Levels across Accessions
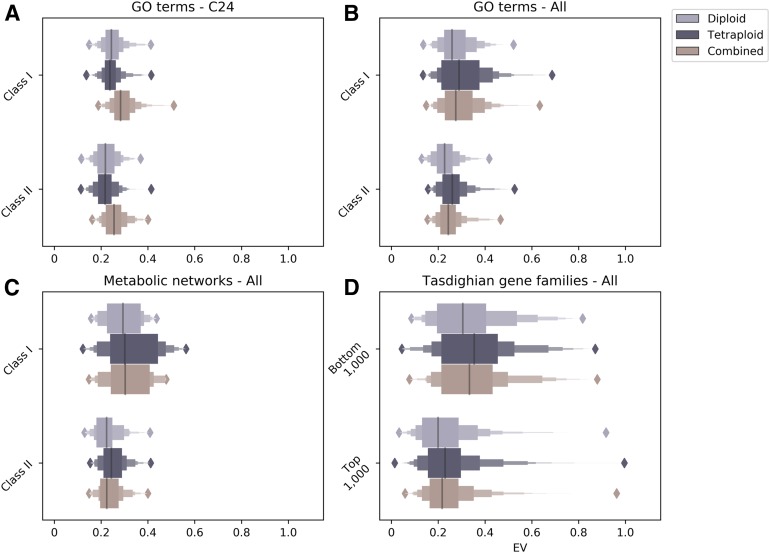
If dosage-sensitive gene classes are under selection for coordinated expression of gene products, then these genes should exhibit similar expression levels across individuals to avoid expression imbalances resulting from recombining alleles (Coate et al., 2016). Consistent with this expectation, expression variance (EV) across individuals within an accession was smaller for class II GO terms than for class I GO terms, and this was true whether we considered diploids, tetraploids, or all individuals combined (P < 0.0001; Kruskal-Wallis tests; Figure 5A).
Figure 5.
EV by Dosage Sensitivity Class in Diploids, Tetraploids, and Diploids and Tetraploids Combined.
(A) EV among individuals (biological replicates) of C24 for GO.
(B) EV among accessions for GO terms.
(C) EV among accessions for metabolic pathways.
(D) EV among accessions for reciprocal retention ranking of gene families (Tasdighian et al., 2017).
The data are presented as boxen plots, with the median shown by the vertical black line. The widest box encompasses the lower to upper fourths of the data (as with a traditional box plot), the next narrowest box encompasses the lower to upper eighths of the data, then the lower and upper sixteenths, etc.
There is likely also some natural gene flow among Arabidopsis accessions (Platt et al., 2010) that would impose the same constraints on interaccession EV. Additionally, constraint on EV within accessions would be expected to slow expression level divergence among accessions. Consequently, we further predicted that interaccession EV would be lower for class II than for class I groupings (GO terms, metabolic networks, and gene families), and this was in fact the case (Table 3; Figures 5B and 5C; Supplemental Data Set). In all groupings, this was true if we looked at EV among diploids, tetraploids, or diploids and tetraploids combined (Table 3).
Table 3. Summary Statistics and Kruskal-Wallis Tests for Differences in EV by Class.
| Grouping | Ploidy | N (Class I) | N (Class II) | Mean (SD) Class I | Mean (SD) Class II | X2 | df | P |
|---|---|---|---|---|---|---|---|---|
| GO | Diploid | 174 | 190 | 0.274 (0.072) | 0.230 (0.055) | 33.396 | 1 | 7.52 × 10−09 |
| Tetraploid | 174 | 190 | 0.304 (0.102) | 0.260 (0.062) | 16.007 | 1 | 6.31 × 10−05 | |
| All | 174 | 190 | 0.291 (0.087) | 0.247 (0.056) | 23.605 | 1 | 1.18 × 10−06 | |
| AraCyc | Diploid | 26 | 37 | 0.292 (0.084) | 0.228 (0.060) | 9.01 | 1 | 0.0027 |
| Tetraploid | 26 | 37 | 0.326 (0.124) | 0.251 (0.058) | 6.11 | 1 | 0.0135 | |
| All | 26 | 37 | 0.312 (0.101) | 0.238 (0.056) | 8.43 | 1 | 0.0037 | |
| Gene families | Diploid | 77 | 501 | 0.327 (0.167) | 0.224 (0.123) | 30.16 | 1 | 3.98 × 10−8 |
| Tetraploid | 77 | 501 | 0.356 (0.175) | 0.247 (0.133) | 31.495 | 1 | 2.00 × 10−8 | |
| All | 77 | 501 | 0.344 (0.162) | 0.238 (0.110) | 34.276 | 1 | 4.78 × 10−9 | |
| Diploid | 5228 | 398 | 0.247 (0.141) | 0.202 (0.104) | 36.44 | 1 | 1.57E-09 | |
| Tetraploid | 5228 | 398 | 0.260 (0.169) | 0.252 (0.112) | 4.2141 | 1 | 0.04009 | |
| All | 5228 | 398 | 0.253 (0.145) | 0.228 (0.090) | 2.1145 | 1 | 0.1459 |
Summary statistics and Kruskal-Wallis tests for differences in expression variance (EV) by class for GO, metabolic pathways (AraCyc), Tasdighian et al. (2017) orthogroups (gene families), or Dong et al. (2019) structure-based protein-protein interactions (S-PPI). N, number of functional groups included in the analysis.
Dosage-Sensitive-Predicted Interacting Protein Pairs Exhibit Coordinated Expression Responses
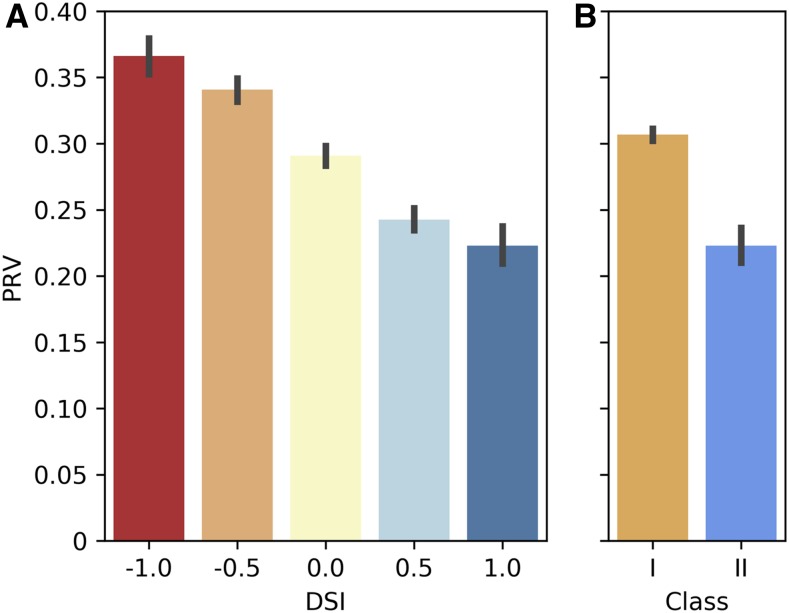
Though Tasdighian et al. (2017) argue that dosage sensitivity is better characterized at the level of gene families rather than broader functional groups (e.g., GO terms), ultimately, dosage sensitivity presumably results from the need for stoichiometric balance between interacting proteins. In many cases, interacting proteins are members of the same gene family, but this is not always the case. We therefore next focused our analysis of expression patterns on protein-protein interactions. Using the top 1% ranked structure-based predicted protein-protein interactions from Dong et al. (2019), we assessed whether the genes encoding interacting protein pairs exhibit a more coordinated expression pattern than random pairs of proteins. Surprisingly, on average, they did not. When separated by duplication history, however, we found that putatively dosage-balance-sensitive protein pairs exhibit significantly lower PRV than do putative dosage-insensitive protein pairs (one or both encoding genes have lost their duplicate from the α-WGD and/or retain duplicates from SSD; class I; Table 1; Figure 6). This reinforces the notion that not all protein-protein interactions are dosage sensitive but that those protein-protein interactions that are dosage sensitive have evolved to maintain coordinated gene dosage responses. Looking at diploids and tetraploids separately, class II protein-protein interactions also exhibit lower EV (Table 3).
Figure 6.
PRV by Dosage Sensitivity for Predicted Interacting Pairs of Proteins.
(A) PRV by dosage sensitivity index (DSI). For each interacting protein pair, the duplication history of the encoding genes was used to calculated DSI, which is equal to WGD retention (1 if both genes have retained their α duplicate, 0.5 if 1 out of 2 has, 0 if neither has) minus small scale duplication (1 if both have been duplicated by small-scale events, 0.5 if 1 out of 2 has, 0 if neither has). A DSI of 1 is evidence that the interaction is dosage sensitive, and decreasing values of DSI suggest decreasing levels of dosage sensitivity.
(B) PRV by class. Class II is defined as DSI = 1, and class I is everything else. PRV is calculated as described for GO terms and metabolic pathways.
DISCUSSION
Although there is growing experimental support for selection on relative gene dosage (dosage balance) as a significant driver of the biased patterns of gene retention and loss following polyploidy, there remains a scarcity of studies testing the connection between gene dosage responses and gene dosage sensitivity necessary for the GBH to explain reciprocal retention (Springer et al., 2010; Coate et al., 2016; Tasdighian et al., 2017). Importantly, because the GBH assumes that selection operates to maintain relatively constant protein amounts among network members, it presupposes that gene dosage affects protein levels. Examining the immediate transcriptional response to genome doubling, therefore, allows us to measure the extent to which expression level is driven by copy number and assess the potential for selection on gene dosage balance to shape the long-term evolutionary fate of genes.
We first estimated overall mRNA transcriptome size and found that it is not exactly doubled or halved with a doubling or halving of the genome and that most genes do not exhibit simple 1:1 gene dosage responses. Hou et al. (2018) also observed slightly less than 1:1 increases in expression in a separate Arabidopsis ploidy series. Similar deviations from a simple 1:1 dosage response have been observed in leaves of maize (Zea mays; Guo et al., 1996), leaves of allotetraploid relatives of soybean (Glycine max; Coate and Doyle, 2010), sepals of autotetraploid Arabidopsis (Robinson et al., 2018), and leaves of allotetraploid Tolmiea (Visger et al., 2019). Nonlinear transcriptional responses to changes in gene dosage have also been observed following SSDs. For example, Konrad et al. (2018) observed greater than twofold increases in expression following segmental duplication in Caenorhabditis elegans. In contrast, dosage compensation (minimal change in expression with gene doubling) has been observed in Drosophila yakuba, Drosophila melanogaster, yeast, and mammals (Qian et al., 2010; Zhou et al., 2011; Rogers et al., 2017). Zhou et al. (2011), for example, observed no differences in expression for 79% of 207 copy number variants in D. melanogaster.
Because alleles share a common genomic address, they likely share more similar cis-regulatory environments than do paralogs. Consequently, one might expect gene expression to be tightly correlated with allelic dosage. Yet Springer et al. (2010) showed that 20% of allelic deletions did not result in a halving of protein abundance in diploid yeast (Saccharomyces cerevisiae), with 3% of genes exhibiting dosage compensation. Thus, many genes deviate from a simple 1:1 relationship between gene dosage and transcript abundance, whether dosage is altered via allelic deletion/duplication, SSD, or WGD. Furthermore, transcriptional responses to WGD vary considerably across lines generated from independent polyploidy events (Pignatta et al., 2010; Yu et al., 2010), perhaps resulting from rapid cis-regulatory evolution and/or transposable element (TE) dynamics as observed, for example, in Capsella (Steige et al., 2015). Therefore, because WGD does not increase the abundance of all transcripts equally, this necessitates an assessment of whether or not stoichiometry is preserved by WGD for putatively dosage-sensitive gene networks in the face of variable dosage responses.
Despite the observed disconnect between gene dosage and gene product amount, selection could act on gene dosage if genes in connected networks exhibit coordinated expression responses. Having estimated transcriptome size responses in both synthetic polyploid-natural diploid pairs and a synthetic diploid-natural polyploid pair, we asked whether genes in reciprocally retained networks exhibit coordinated dosage responses. If dosage sensitivity explains long term retention patterns, then there must be mechanisms to facilitate their coregulation (Papp et al., 2003) and, by extension, their coordinated responses to WGD.
Our data are consistent with this hypothesis. Reciprocally retained and, therefore, putatively dosage sensitive, gene groups (GO terms, metabolic pathways, gene families, and predicted protein-protein interactions) exhibit less variable expression levels within and across accessions as well as more coordinated responses to changes in whole genome dosage. This pattern is consistent with our previous studies in allopolyploid Glycine dolichocarpa (Coate et al., 2016), extending expression-level support for the GBH to synthetic autopolyploids, which are putatively isogenic with their diploid progenitor. Thus, it appears that coordinated regulation within dosage-sensitive networks is both independent of, and robust to, hybridization and the resulting novel regulatory combinations. A limitation of our previous study (Coate et al., 2016) is that it relied on natural tetraploids, whose expression patterns reflect approximately 0.5 million years (Bombarely et al., 2014) of independent evolution that could obscure immediate responses to genome doubling. The GBH, however, explains reciprocal retention as an “instant and neutral byproduct, a spandrel, of purifying selection” (Freeling, 2009). For this to be true, coordinated expression responses need to be an instantaneous response to WGD. The comparison of induced polyploids to their isogenic diploid parents in this study enabled us to assess if this is true and demonstrates that reciprocally retained gene groups do, in fact, exhibit a higher degree of coordination in their dosage responses immediately following WGD than do other genes.
It has been widely speculated that dosage constraints preserve duplicates in the short term but that over longer evolutionary time periods, selection on gene dosage balance is relaxed, enabling the retained duplicates to subsequently subfunctionalize or neofunctionalize (Birchler et al., 2005; Freeling and Thomas, 2006; Birchler and Veitia, 2007; Coate and Doyle, 2011; Schnable et al., 2012; Conant et al., 2014; Gout and Lynch, 2015; Coate et al., 2016). Under this scenario, one might expect to see more coordinated dosage responses among reciprocally retained gene networks in nascent polyploids (where genes are under purifying selection to preserve dosage) than in older polyploids (where genes may be under relaxed selection on gene dosage with some having begun to diverge in function). Intriguingly, however, the degree to which dosage responses are more coordinated among class II networks than among class I networks is not discernibly more pronounced in the synthetic autotetraploids (this study) versus natural allotetraploids (Coate et al., 2016). This could suggest that for most genes, selection on gene dosage does not relax appreciably for more than a half-million years. This is consistent with observations that whole-genome duplicates tend to diverge in expression more slowly than expected (Rodgers-Melnick et al., 2012; Tasdighian et al., 2017) and to diverge in expression and/or function more slowly than do small scale duplicates (Hakes et al., 2007; Wang et al., 2011; Rodgers-Melnick et al., 2012; Qiao et al., 2018; Defoort et al., 2019). If this is the case, performing equivalent analyses on older polyploids would help to resolve the timeline for when relaxation of selection on gene dosage occurs (e.g., cotton [Gossypium hirsutum], formed by allopolylpoidy 1to 2 million years ago).
Alternatively, or in addition, the lack of a stronger pattern in synthetic polyploids could be the result of deleterious (unbalanced) dosage responses arising at some loci in the nascent polyploids that are subsequently “corrected” by selection in polyploid lineages that survive the initial shift in genome dosage. We demonstrate that class II gene groups show more coordinated dosage responses than do class I groups, but there is still considerable variation in dosage responses within class II groups, some of which could represent unbalanced and, therefore, deleterious expression patterns that are rectified by purifying selection over subsequent generations.
Our study expanded the scope of Coate et al. (2016), which looked at GO and metabolic pathways, by also assessing the top and bottom dosage-sensitive gene families from Tasdighian et al. (2017), which those authors argue reveals a clearer pattern because dosage sensitivity is better measured at the level of gene families than in broad functional groups where direct interactions between genes are less certain. Consistent with their assertion, we observed highly significant reductions in both PRV and EV in the top 1000 gene families relative to the bottom 1000 gene families (Figures 3 and 5; Tables 1 and 3), and the differences were generally more pronounced than those observed between class II and class I GO terms or metabolic pathways.
Likewise, with the recent publication of an Arabidopsis predicted protein-protein interaction network (Dong et al., 2019), we were also able to investigate the GBH using pairs of genes whose products are predicted to directly interact as opposed to the indirect estimates provided by GO terms, metabolic networks, or gene families for which the gene products do not necessarily have direct interactions. In all cases, we found a strong, consistent pattern of coordinated gene dosage responses across dosage-sensitive groups, pathways, and interacting protein pairs.
A prediction of the GBH is that genes in dosage-balance-sensitive networks will be coregulated, and Papp et al. (2003) provided evidence that this is in fact the case in yeast. We extend this observation to show that these genes are not only coregulated within and across genomes at a given ploidy level, but that they are coregulated in terms of their response to WGD. One possible explanation for this surprising observation is that connected genes, unlike unconnected genes, have evolved to share cis-regulatory element(s) (e.g. transcription factor binding sites) and thus are regulated by the same complement of transcription factors. This would explain why connected genes tend to be coregulated and show coordinated dosage responses, driven by natural selection to preserve balance across the entire connected group. There is no reason why class I gene groups (GO terms, metabolic pathways, etc.) should share cis elements, since their expression is not coordinated, and thus it is reasonable that they should show less-coordinated expression responses to WGD. Consistent with this hypothesis, Taggart and Li (2018) demonstrated that protein subunits in complexes with fixed subunit stoichiometry are produced in proportion to their dosage and concluded that their expression levels are hardwired by cis-regulatory sequences.
A related explanation could be that dosage-balance-sensitive gene groups reside in common chromatin contexts that coordinate expression. Though Arabidopsis generally lacks canonical topologically associating domains (TADs; Liu et al., 2017), it does have various other chromatin interaction domains, including local chromatin loops (Liu et al., 2017), an intra- and interchromosomal structure termed the KNOT (Grob et al., 2014; Grob and Grossniklaus, 2017), analogs to A and B compartments (Grob et al., 2014; Zhang et al., 2019), “positive strips,” and TAD-like structures (Wang et al., 2015), all of which correlate with specific expression profiles. Nuclear pore complexes are subnuclear compartments that are thought to be involved in organizing chromatin domains and thereby regulating transcription (Sun et al., 2019). Selection could favor the arrangement of genes from dosage-balance-sensitive complexes into common chromatin domains, potentially mediated by nuclear pore complexes, to ensure coregulation. Xie et al. (2019) showed that TADs and A/B compartments are largely conserved across related Brassica species. To the extent that these structures also persist after WGD events, these too could facilitate coordinated gene dosage responses. Notably, Xie et al. (2019) found that duplicates retained from the whole-genome triplication event in Brassica were more likely to be colocalized in 3D chromatin domains. Zhang et al. (2019) showed that most genes remained in compact or loose chromatin domains following autopolyploidy in Arabidopsis but that 12% (perhaps enriched for class I genes) shifted domains. Thus, colocalization in chromatin domains is associated with both coregulation and elevated duplicate retention following WGD. These observations are consistent with the notion that dosage-balance-sensitive genes have evolved to be coregulated via colocalization in shared chromatin domains, which in turn favors retention of balanced gene duplicates.
TEs can also provide an innate mechanism of expression coordination following polyploidization. Zhang et al. (2015) showed that WGD induces methylation in class II TEs, which suppresses expression not only of TEs but of nearby genes. They proposed that this could minimize deleterious gene dosage effects. Perhaps selection has favored the arrangement of genes in dosage-balance-sensitive gene networks in close proximity to TEs because this resulted in less disruption of dosage balance following previous episodes of gene and genome duplication. This TE-based mechanism would be consistent with our observation that putatively dosage-sensitive GO terms and metabolic networks (but not gene families or interacting protein pairs) tend to show smaller average dosage responses (Figure 4). It has been proposed that partial dosage compensation is due to selection to minimize disruption of balance by minimizing transcriptional change in response to change in gene dosage. Katju and Bergthorsson (2019) explain that this could be due to the relatively high metabolic cost of duplicating genes that produce large numbers of transcripts. Likewise, Qian et al. (2010) describe expression reduction as a special class of subfunctionalization that could help explain the retention of duplicates.
These two studies provide a useful framework for why dosage-sensitive genes have evolved to have smaller dosage responses (to minimize disruptions to balance from SSDs) and therefore, as a corollary, smaller dosage responses offer further evidence that these genes are dosage sensitive. Qian et al. (2010) proposed that selection favors regulatory mutations that reduce expression. However, we observe smaller dosage responses for class II genes in the first generations post-WGD, making it unlikely that postduplication mutations are the cause. Epigenetic suppression resulting from the methylation of TEs could, therefore, be a plausible mechanism. It would be interesting to determine, therefore, if class II genes are preferentially located in the vicinity of hypermethylated TEs. In a preliminary analysis, we quantified the distance from each gene to its nearest TE in the Col-0 genome (https://www.arabidopsis.org/download_files/Genes/TAIR10_genome_release/TAIR10_transposable_elements/TAIR10_Transposable_Elements.txt) and found that putatively dosage-balance-sensitive genes (defined either by assignment to class II GO terms or by only retaining duplicates from WGD and not SSD) were actually farther from TEs on average than were putatively dosage-insensitive genes. This was true if we considered all TEs or only class II (DNA) TEs. Thus, we do not find evidence to support the hypothesis that dosage responses are smaller or more coordinated in class II genes as a result of clustering near TEs. Nonetheless, there may be clustering near particular TE families that are preferentially methylated. Alternatively, the orientation of TEs near genes (e.g., 5′ or 3′), rather than their absolute distance, may exert a greater influence on gene expression. It will be useful to examine this and other features of the surrounding chromatin in more detail.
Finally, while our study indicates that reciprocally retained gene groups exhibit transcriptional responses consistent with the GBH, it does not address whether these coordinated transcriptional responses produce coordination at the level of protein abundance. Multiple layers of posttranscriptional gene regulation could potentially result in imbalance at the protein level despite maintenance of balance at the gene dosage and/or transcriptional levels (e.g., Walley et al., 2016). However, dosage-sensitive genes tend to exhibit tighter, more coordinated regulation of transcription and translation than do other genes (Gsponer et al., 2008; Vavouri et al., 2009), meaning that transcriptional dosage responses are likely to be a reasonable proxy for protein dosage responses. Nonetheless, performing similar analyses to those presented here but that incorporate ribosome profiling (Taggart and Li, 2018) and/or quantitative proteomic data would be necessary to assess fully whether protein dosage is sufficiently linked with gene dosage for selection to act on gene copy number to preserve protein balance. However, any influence of gene dosage on protein abundance is presumably mediated by transcription, so the fact that the expected patterns are observed at the level of transcription attests to the efficacy of even these more indirect approaches and provides an important layer of support for the GBH.
METHODS
Plant Material
Gene dosage responses to ploidy change were quantified in two naturally occurring diploid Arabidopsis (Arabidopsis thaliana) accessions (C24 and Ws) and colchicine-induced autotetraploids of the same accessions, as well as in one natural tetraploid accession (Wa) and a synthetic diploid generated by the Tailswap haploid induction system (Ravi and Chan, 2010). C24 and Ws tetraploids were generated in the lab of Dr. Luca Comai as described for Col-0 and Ler in Pignatta et al. (2010). For both accessions, diploid and tetraploid lines were derived from the same colchicine-treated plant (generation C1). Ploidy level was determined in C2 plants produced by selfing C1. Seeds from selfed C2 plants (C3 generation) were obtained from the Comai lab. C3 plants were subsequently selfed to bulk seed, and all plants used in this study were of the C4 generation (third generation post-colchicine-treatment). Wa diploids (dihaploids) were also generated in the lab of Luca Comai. Tetraploid Wa was crossed with the Tailswap haploid inducer to produce diploid (dihaploid) seed (H1 generation) as described in (Ravi and Chan, 2010). Ploidy was determined in H1 plants via chromosome spreads (Ravi and Chan, 2010). Diploid and tetraploid H1s derived from the same Tailswap cross were then selfed. We obtained H2 seed, which we subsequently selfed to bulk H3 seed. All Wa plants used in this study were grown from H3 seed (second generation post-haploid-induction). Seeds were sown on Sunshine #4 potting mix, cold stratified for 4 d, and grown in a growth chamber with 16/8 h light/dark cycles at 21°C/18°C, respectively, with approximately 125 µmol/m2/s light intensity provided by full spectrum fluorescent bulbs.
DNA/RNA Coextraction
All tissue was collected 1 to 2 h into the growth chamber light cycle in order to minimize variance in gene expression due to circadian effects. Tissue was harvested from rosette leaves at the 10- to 12-leaf stage and prior to bolting to minimize differences in developmental stage. Fully expanded leaves were harvested to ensure that they had reached their final levels of endopolyploidy. Tissue was flash frozen in liquid nitrogen followed by immediate nucleic extraction or storage at −80°C until extractions were performed. DNA and RNA were coextracted using Qiagen AllPrep universal kits. Extractions were performed on three to four individuals per accession using ∼80 mg of leaf tissue per extraction. Nucleic acid yields were quantified by Qubit using DNA high-sensitivity and RNA broad-range assays (Life Technologies). The size of the total RNA transcriptome (total RNA per unit of DNA) was estimated by the ratio of RNA to DNA.
Flow Cytometry
Base ploidy level was confirmed and degree of endopolyploidy quantified by flow cytometry. Fifty to seventy-five milligrams of fully expanded leaf tissue collected from plants at the same stage as used for nucleic acid extraction was chopped with a razor blade in 600 µL Aru buffer (Arumuganathan and Earle, 1991). Suspended nuclei were filtered through a 20 µm CellTrics filter (Partec), treated with RNase (0.01 µg/100 mL of sample), and stained with propidium iodide (0.001 µg/100 mL of sample). Samples were analyzed on a FACSCanto II (BD Biosciences) flow cytometer to obtain counts of nuclei per ploidy level. Average ploidy level was determined by multiplying the fraction of events at a given ploidy level by the value of that ploidy level (i.e., 2, 4, 8, 16, 32, or 64) and summing the values for all ploidy levels. Flow cytometry data were consistent with each line being euploid at the expected ploidy level, but euploidy was further verified from the RNA-seq data as described below.
RNA-seq
RNA-seq libraries were generated for each sample from 1 to 2 µg of extracted RNA. Libraries were generated and analyzed from three to five individuals (biological replicates) per cytotype per accession: three replicates of diploid Ws; four replicates each of diploid C24, tetraploid C24, and tetraploid Ws; and five replicates each of diploid and tetraploid Wa. To enable estimation of mRNA transcriptome size per unit of DNA, each sample was spiked with ERCC mix 1 in proportion to the DNA/RNA ratio determined above, as described in Robinson et al. (2018). Libraries were generated using Illumina TruSeq stranded library prep kits. Libraries were multiplexed with 8 to 12 samples per lane and 100-bp single-end sequences were generated on an Illumina HiSeq 250 at the Cornell Biotechnology Resource Center’s genomics facility.
RNA-seq Data Processing and Analysis
Raw FASTQ files were trimmed and filtered to remove low-quality reads and technical sequences using Trimmomatic (Bolger et al., 2014) with the following settings: ILLUMINACLIP, TruSeq3SE.fa:2:30, 10; LEADING, 3; TRAILING, 3; SLIDINGWINDOW, 4:15; MINLEN, 36. Filtered reads were aligned with HISAT2 (Pertea et al., 2016) to the Arabidopsis reference sequence (TAIR10) and to the ERCC reference. HTSeq (Anders et al., 2015) was used to determine read counts per gene.
Autotetraploids periodically produce aneuploid offspring. To confirm that the tetraploid individuals we sequenced were not aneuploid, we calculated fold change in relative expression (transcripts per million; TPM) per gene for every pairwise comparison of biological replicates (individual plants; Supplemental Figures 1A and 1B). If one individual was aneuploid for a given chromosome, we would expect to see a coordinated increase or decrease in TPM for genes on that chromosome, reflected in a shift in fold change of expression relative to the other biological replicates (Supplemental Figure 1C). No chromosome or chromosomal segment exhibits such a shift, consistent with flow-cytometry-based estimates of genome size, indicating that all tetraploid individuals were euploid.
Fold changes in expression between ploidy levels and differentially expressed genes were identified using DESeq2 (Love et al., 2014). Fold-changes (FCs; tetraploid/diploid) were calculated per transcriptome and per genome. Per transcriptome, FC was calculated using the standard DESeq2 procedure, which normalizes for Arabidopsis library size (total count of reads mapped to the Arabidopsis reference). To estimate FC per genome, Arabidopsis read counts were normalized by ERCC library size. ERCC-specific size factors were estimated by DESeq2 using the estimateSizeFactors function on ERCC read counts, and these size factors were then used to normalize DESeq2-based analysis of Arabidopsis read counts. FC per transcriptome is a measure of change in transcript concentration (what fraction of the total transcriptome is composed of transcripts from the gene in question). FC per genome is a measure of relative expression per gene copy or gene dosage response (change in expression per change in gene copy number).
Relative mRNA transcriptome size per genome (tetraploid/diploid) was estimated individually based on the FC estimates for each gene in the RNA-seq data set according to the equation:
Reported values of transcriptome size per genome are the average of these individual estimates. Relative mRNA transcriptome size per cell was estimated by multiplying transcriptome size per genome by relative mean ploidy level (mean ploidytetraploid/mean ploidydiploid).
Glossary of Terms
Gene Groupings
AraCyc is the database of Arabidopsis metabolic pathways from the Plant Metabolic Network database. S-PPI is the set of structure-based protein-protein interactions from Dong et al. (2019). Gene families are as circumscribed by Tasdighian et al. (2017).
Duplication History-Based Dosage Sensitivity Classes
Class I are gene groupings that are putatively dosage balance insensitive based on duplication history. Specifically, these groups have lower-than-median retention of duplicates from the 𝛼-WGD and higher-than-median levels of duplication via small-scale events (tandem, proximal or TE-mediated duplications).
Class II are gene groupings that are putatively dosage balance sensitive based on duplication history. Specifically, these groups have higher-than-median retention of duplicates from the 𝛼-WGD and lower-than-median levels of duplication via small-scale events.
Metrics of Polyploid Expression Responses
Absolute dosage response is the fold change in expression per genome between tetraploids and diploids. A value of 1 indicates a 1:1 dosage response. A value of 0.5 indicates dosage compensation.
PRV is the coefficient of variation (SD/mean) for absolute dosage responses within a gene group (GO term, metabolic pathway, etc.). This variable measures the degree to which the dosage responses of genes within a network are correlated.
EV is the average coefficient of variation of relative expression (TPM) across accessions (C24, Ws, Wa) for all genes in a gene group. EV was calculated for diploids and tetraploids separately and for both ploidy levels together (“combined”).
All scripts for data processing are available on GitHub (https://github.com/barneypotter24/ploidy-seq).
Accession Numbers
Sequence data (FASTQ files from RNA-seq experiments) from this article can be found in the National Center for Biotechnology Information Sequence Read Archive as BioProject PRJNA606953.
Supplemental Data
Supplemental Figure 1. Assessment of polyploid lines for aneuploidy.
Supplemental Figure 2. Expression level versus PRV for GO terms.
Supplemental Data Set. RNA-seq and flow cytometry data.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Luca Comai’s lab for generating and providing seed for the Arabidopsis lines used in this work. This work was supported by the National Science Foundation (grant 1257522 to J.E.C. and J.J.D., Reed College Summer Undergraduate Research Fellowship to B.I.P., and XSEDE allocation TG-BIO170018 to J.E.C., M.J.S., and B.I.P.)
AUTHOR CONTRIBUTIONS
J.E.C., J.J.D., B.I.P., and M.J.S. designed the experiment; J.E.C., B.I.P., and M.J.S. performed the research. J.E.C., B.I.P., and M.J.S. analyzed the data. J.E.C., B.I.P., M.J.S., and J.J.D. wrote the article.
References
- Anders S., Pyl P.T., Huber W.(2015). HTSeq: A Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumuganathan K., Earle E.(1991). Nuclear DNA content of some important plant species. Plant Mol. Biol. Report. 9: 208–218. [Google Scholar]
- Barker M.S., Vogel H., Schranz M.E.(2009). Paleopolyploidy in the Brassicales: Analyses of the Cleome transcriptome elucidate the history of genome duplications in Arabidopsis and other Brassicales. Genome Biol. Evol. 1: 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J.A., Johnson A.F., Veitia R.A.(2016). Kinetics genetics: Incorporating the concept of genomic balance into an understanding of quantitative traits. Plant Sci. 245: 128–134. [DOI] [PubMed] [Google Scholar]
- Birchler J.A., Newton K.J.(1981). Modulation of protein levels in chromosomal dosage series of maize: The biochemical basis of aneuploid syndromes. Genetics 99: 247–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J.A., Riddle N.C., Auger D.L., Veitia R.A.(2005). Dosage balance in gene regulation: Biological implications. Trends Genet. 21: 219–226. [DOI] [PubMed] [Google Scholar]
- Birchler J.A., Veitia R.A.(2007). The gene balance hypothesis: From classical genetics to modern genomics. Plant Cell 19: 395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J.A., Veitia R.A.(2012). Gene balance hypothesis: Connecting issues of dosage sensitivity across biological disciplines. Proc. Natl. Acad. Sci. USA 109: 14746–14753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G., Wolfe K.H.(2004). Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16: 1679–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B.(2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bombarely A., Coate J.E., Doyle J.J.(2014). Mining transcriptomic data to study the origins and evolution of a plant allopolyploid complex. PeerJ 2: e391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coate J.E., Doyle J.J.(2010). Quantifying whole transcriptome size, a prerequisite for understanding transcriptome evolution across species: An example from a plant allopolyploid. Genome Biol. Evol. 2: 534–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coate J.E., Doyle J.J.(2011). Divergent evolutionary fates of major photosynthetic gene networks following gene and whole genome duplications. Plant Signal. Behav. 6: 594–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coate J.E., Song M.J., Bombarely A., Doyle J.J.(2016). Expression-level support for gene dosage sensitivity in three Glycine subgenus Glycine polyploids and their diploid progenitors. New Phytol. 212: 1083–1093. [DOI] [PubMed] [Google Scholar]
- Conant G.C., Birchler J.A., Pires J.C.(2014). Dosage, duplication, and diploidization: Clarifying the interplay of multiple models for duplicate gene evolution over time. Curr. Opin. Plant Biol. 19: 91–98. [DOI] [PubMed] [Google Scholar]
- Conesa A., Madrigal P., Tarazona S., Gomez-Cabrero D., Cervera A., McPherson A., Szcześniak M.W., Gaffney D.J., Elo L.L., Zhang X., Mortazavi A.(2016). A survey of best practices for RNA-seq data analysis. Genome Biol. 17: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defoort J., Van de Peer Y., Carretero-Paulet L.(2019). The evolution of gene duplicates in angiosperms and the impact of protein-protein interactions and the mechanism of duplication. Genome Biol. Evol. 11: 2292–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng M., Dong Y., Zhao Z., Li Y., Fan G.(2017). Dissecting the proteome dynamics of the salt stress induced changes in the leaf of diploid and autotetraploid Paulownia fortunei. PLoS One 12: e0181937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S., et al. (2019). Proteome-wide, structure-based prediction of protein-protein interactions/new molecular interactions viewer. Plant Physiol. 179: 1893–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J.J., Coate J.E.(2019). Polyploidy, the nucleotype, and novelty: The impact of genome doubling on the biology of the cell. Int. J. Plant Sci. 180: 1–52. [Google Scholar]
- Edger P.P., et al. (2018). Brassicales phylogeny inferred from 72 plastid genes: A reanalysis of the phylogenetic localization of two paleopolyploid events and origin of novel chemical defenses. Am. J. Bot. 105: 463–469. [DOI] [PubMed] [Google Scholar]
- Edger P.P., Pires J.C.(2009). Gene and genome duplications: The impact of dosage-sensitivity on the fate of nuclear genes. Chromosome Res. 17: 699–717. [DOI] [PubMed] [Google Scholar]
- Fan G., Wang L., Dong Y., Zhao Z., Deng M., Niu S., Zhang X., Cao X.(2017). Genome of Paulownia (Paulownia fortunei) illuminates the related transcripts, miRNA and proteins for salt resistance. Sci. Rep. 7: 1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M.(2009). Bias in plant gene content following different sorts of duplication: Tandem, whole-genome, segmental, or by transposition. Annu. Rev. Plant Biol. 60: 433–453. [DOI] [PubMed] [Google Scholar]
- Freeling M., Thomas B.C.(2006). Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res. 16: 805–814. [DOI] [PubMed] [Google Scholar]
- Gout J.-F., Lynch M.(2015). Maintenance and loss of duplicated genes by dosage subfunctionalization. Mol. Biol. Evol. 32: 2141–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grob S., Grossniklaus U.(2017). Chromosome conformation capture-based studies reveal novel features of plant nuclear architecture. Curr. Opin. Plant Biol. 36: 149–157. [DOI] [PubMed] [Google Scholar]
- Grob S., Schmid M.W., Grossniklaus U.(2014). Hi-C analysis in Arabidopsis identifies the KNOT, a structure with similarities to the flamenco locus of Drosophila. Mol. Cell 55: 678–693. [DOI] [PubMed] [Google Scholar]
- Gsponer J., Futschik M.E., Teichmann S.A., Babu M.M.(2008). Tight regulation of unstructured proteins: From transcript synthesis to protein degradation. Science 322: 1365–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M., Davis D., Birchler J.A.(1996). Dosage effects on gene expression in a maize ploidy series. Genetics 142: 1349–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakes L., Pinney J.W., Lovell S.C., Oliver S.G., Robertson D.L.(2007). All duplicates are not equal: The difference between small-scale and genome duplication. Genome Biol. 8: R209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J., et al. (2018). Global impacts of chromosomal imbalance on gene expression in Arabidopsis and other taxa. Proc. Natl. Acad. Sci. USA 115: E11321–E11330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katju V., Bergthorsson U.(2019). Old trade, new tricks: Insights into the spontaneous mutation process from the partnering of classical mutation accumulation experiments with high-throughput genomic approaches. Genome Biol. Evol. 11: 136–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konrad A., Flibotte S., Taylor J., Waterston R.H., Moerman D.G., Bergthorsson U., Katju V.(2018). Mutational and transcriptional landscape of spontaneous gene duplications and deletions in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 115: 7386–7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langham R.J., Walsh J., Dunn M., Ko C., Goff S.A., Freeling M.(2004). Genomic duplication, fractionation and the origin of regulatory novelty. Genetics 166: 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos B., Meiklejohn C.D., Hartl D.L.(2004). Regulatory evolution across the protein interaction network. Nat. Genet. 36: 1059–1060. [DOI] [PubMed] [Google Scholar]
- Liu C., Cheng Y.-J., Wang J.-W., Weigel D.(2017). Prominent topologically associated domains differentiate global chromatin packing in rice from Arabidopsis. Nat. Plants 3: 742–748. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S.(2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Conery J.S.(2000). The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155. [DOI] [PubMed] [Google Scholar]
- Lynch M., Conery J.S.(2003). The evolutionary demography of duplicate genes. J. Struct. Funct. Genomics 3: 35–44. [PubMed] [Google Scholar]
- Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B.(2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- Panchy N., Lehti-Shiu M., Shiu S.-H.(2016). Evolution of gene duplication in plants. Plant Physiol. 171: 2294–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B., Pál C., Hurst L.D.(2003). Dosage sensitivity and the evolution of gene families in yeast. Nature 424: 194–197. [DOI] [PubMed] [Google Scholar]
- Pertea M., Kim D., Pertea G.M., Leek J.T., Salzberg S.L.(2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11: 1650–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignatta D., Dilkes B.P., Yoo S.Y., Henry I.M., Madlung A., Doerge R.W., Jeffrey Chen Z., Comai L.(2010). Differential sensitivity of the Arabidopsis thaliana transcriptome and enhancers to the effects of genome doubling. New Phytol. 186: 194–206. [DOI] [PubMed] [Google Scholar]
- Pirrello J., et al. (2018). Transcriptome profiling of sorted endoreduplicated nuclei from tomato fruits: How the global shift in expression ascribed to DNA ploidy influences RNA-Seq data normalization and interpretation. Plant J. 93: 387–398. [DOI] [PubMed] [Google Scholar]
- Platt A., et al. (2010). The scale of population structure in Arabidopsis thaliana. PLoS Genet. 6: e1000843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W., Liao B.-Y., Chang A.Y.-F., Zhang J.(2010). Maintenance of duplicate genes and their functional redundancy by reduced expression. Trends Genet. 26: 425–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao X., Yin H., Li L., Wang R., Wu J., Wu J., Zhang S.(2018). Different modes of gene duplication show divergent evolutionary patterns and contribute differently to the expansion of gene families involved in important fruit traits in pear (Pyrus bretschneideri). Front Plant Sci 9: 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi M., Chan S.W.(2010). Haploid plants produced by centromere-mediated genome elimination. Nature 464: 615–618. [DOI] [PubMed] [Google Scholar]
- Riddle N.C., Kato A., Birchler J.A.(2006). Genetic variation for the response to ploidy change in Zea mays L. Theor. Appl. Genet. 114: 101–111. [DOI] [PubMed] [Google Scholar]
- Robinson D.O., Coate J.E., Singh A., Hong L., Bush M., Doyle J.J., Roeder A.H.K.(2018). Ploidy and size at multiple scales in the Arabidopsis sepal. Plant Cell 30: 2308–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers-Melnick E., Mane S.P., Dharmawardhana P., Slavov G.T., Crasta O.R., Strauss S.H., Brunner A.M., Difazio S.P.(2012). Contrasting patterns of evolution following whole genome versus tandem duplication events in Populus. Genome Res. 22: 95–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers R.L., Shao L., Thornton K.R.(2017). Tandem duplications lead to novel expression patterns through exon shuffling in Drosophila yakuba. PLoS Genet. 13: e1006795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schläpfer P., Zhang P., Wang C., Kim T., Banf M., Chae L., Dreher K., Chavali A.K., Nilo-Poyanco R., Bernard T., Kahn D., Rhee S.Y.(2017). Genome-wide prediction of metabolic enzymes, pathways, and gene clusters in plants. Plant Physiol. 173: 2041–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable J.C., Springer N.M., Freeling M.(2011). Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc. Natl. Acad. Sci. USA 108: 4069–4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable J.C., Wang X., Pires J.C., Freeling M.(2012). Escape from preferential retention following repeated whole genome duplications in plants. Front Plant Sci 3: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis D.E., Misra B.B., Shan S., Chen S., Soltis P.S.(2016). Polyploidy and the proteome. Biochim. Biophys. Acta 1864: 896–907. [DOI] [PubMed] [Google Scholar]
- Spoelhof J.P., Soltis P.S., Soltis D.E.(2017). Pure polyploidy: Closing the gaps in autopolyploid research. J Syst Evol 55: 340–352. [Google Scholar]
- Springer M., Weissman J.S., Kirschner M.W.(2010). A general lack of compensation for gene dosage in yeast. Mol. Syst. Biol. 6: 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steige K.A., Reimegård J., Koenig D., Scofield D.G., Slotte T.(2015). Cis-regulatory changes associated with a recent mating system shift and floral adaptation in Capsella. Mol. Biol. Evol. 32: 2501–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupar R.M., Bhaskar P.B., Yandell B.S., Rensink W.A., Hart A.L., Ouyang S., Veilleux R.E., Busse J.S., Erhardt R.J., Buell C.R., Jiang J.(2007). Phenotypic and transcriptomic changes associated with potato autopolyploidization. Genetics 176: 2055–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Shi Y., Yildirim E.(2019). The nuclear pore complex in cell type-specific chromatin structure and gene regulation. Trends Genet. 35: 579–588. [DOI] [PubMed] [Google Scholar]
- Taggart J.C., Li G.-W.(2018). Production of protein-complex components is stoichiometric and lacks general feedback regulation in eukaryotes. Cell Syst. 7: 580–589.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasdighian S., Van Bel M., Li Z., Van de Peer Y., Carretero-Paulet L., Maere S.(2017). Reciprocally retained genes in the angiosperm lineage show the hallmarks of dosage balance sensitivity. Plant Cell 29: 2766–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavouri T., Semple J.I., Garcia-Verdugo R., Lehner B.(2009). Intrinsic protein disorder and interaction promiscuity are widely associated with dosage sensitivity. Cell 138: 198–208. [DOI] [PubMed] [Google Scholar]
- Visger C.J., Wong G.K.-S., Zhang Y., Soltis P.S., Soltis D.E.(2019). Divergent gene expression levels between diploid and autotetraploid Tolmiea relative to the total transcriptome, the cell, and biomass. Am. J. Bot. 106: 280–291. [DOI] [PubMed] [Google Scholar]
- Walley J.W., Sartor R.C., Shen Z., Schmitz R.J., Wu K.J., Urich M.A., Nery J.R., Smith L.G., Schnable J.C., Ecker J.R., Briggs S.P.(2016). Integration of omic networks in a developmental atlas of maize. Science 353: 814–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Liu C., Roqueiro D., Grimm D., Schwab R., Becker C., Lanz C., Weigel D.(2015). Genome-wide analysis of local chromatin packing in Arabidopsis thaliana. Genome Res. 25: 246–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Tan X., Paterson A.H.(2013). Different patterns of gene structure divergence following gene duplication in Arabidopsis. BMC Genomics 14: 652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wang X., Tang H., Tan X., Ficklin S.P., Feltus F.A., Paterson A.H.(2011). Modes of gene duplication contribute differently to genetic novelty and redundancy, but show parallels across divergent angiosperms. PLoS One 6: e28150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Fan G., Dong Y., Zhai X., Deng M., Zhao Z., Liu W., Cao Y.(2017). Implications of polyploidy events on the phenotype, microstructure, and proteome of Paulownia australis. PLoS One 12: e0172633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel J.F., Lisch D., Hu G., Mason A.S.(2018). The long and short of doubling down: Polyploidy, epigenetics, and the temporal dynamics of genome fractionation. Curr. Opin. Genet. Dev. 49: 1–7. [DOI] [PubMed] [Google Scholar]
- Xie T., Zhang F.-G., Zhang H.-Y., Wang X.-T., Hu J.-H., Wu X.-M.(2019). Biased gene retention during diploidization in Brassica linked to three-dimensional genome organization. Nat. Plants 5: 822–832. [DOI] [PubMed] [Google Scholar]
- Yan L., Fan G., Deng M., Zhao Z., Dong Y., Li Y.(2017). Comparative proteomic analysis of autotetraploid and diploid Paulownia tomentosa reveals proteins associated with superior photosynthetic characteristics and stress adaptability in autotetraploid Paulownia. Physiol. Mol. Biol. Plants 23: 605–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H., Kato A., Mooney B., Birchler J.A.(2011). Phenotypic and gene expression analyses of a ploidy series of maize inbred Oh43. Plant Mol. Biol. 75: 237–251. [DOI] [PubMed] [Google Scholar]
- Yu Z., Haberer G., Matthes M., Rattei T., Mayer K.F., Gierl A., Torres-Ruiz R.A.(2010). Impact of natural genetic variation on the transcriptome of autotetraploid Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 107: 17809–17814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Zheng R., Wang Y., Zhang Y., Hong P., Li G., Fang Y. (2019). The effects of Arabidopsis genome duplication on the chromatin organization and transcriptional regulation. Nucleic Acids Res 47: 7857–7869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Liu Y., Xia E.-H., Yao Q.-Y., Liu X.-D., Gao L.-Z.(2015). Autotetraploid rice methylome analysis reveals methylation variation of transposable elements and their effects on gene expression. Proc. Natl. Acad. Sci. USA 112: E7022–E7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Lemos B., Dopman E.B., Hartl D.L.(2011). Copy-number variation: The balance between gene dosage and expression in Drosophila melanogaster. Genome Biol. Evol. 3: 1014–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Soltis P.S., Soltis D.E., Chen S., Koh J.(2012). Proteomics and mass spectrometry of Tragopogon polyploid evolution. J. Biomol. Tech. 23: S50. [Google Scholar]