Members of the photosensory-signaling pathway orchestrate the expression of the circadian clock gene CCA1 to regulate plant responses to daily changes in the light environment.
Abstract
The circadian clock provides a time-keeping mechanism that synchronizes various biological activities with the surrounding environment. Arabidopsis (Arabidopsis thaliana) CIRCADIAN CLOCK ASSOCIATED1 (CCA1), encoding a MYB-related transcription factor, is a key component of the core oscillator of the circadian clock, with peak expression in the morning. The molecular mechanisms regulating the light induction and rhythmic expression of CCA1 remain elusive. In this study, we show that two phytochrome signaling proteins, FAR-RED ELONGATED HYPOCOTYL3 (FHY3) and its paralog FAR-RED IMPAIRED RESPONSE1 (FAR1), are essential for the light-induced expression of CCA1. FHY3 and FAR1 directly bind to the CCA1 promoter and activate its expression, whereas PHYTOCHROME INTERACTING FACTOR5 (PIF5) directly binds to its promoter and represses its expression. Furthermore, PIF5 and TIMING OF CAB EXPRESSION1 physically interact with FHY3 and FAR1 to repress their transcriptional activation activity on CCA1 expression. These findings demonstrate that the photosensory-signaling pathway integrates with circadian oscillators to orchestrate clock gene expression. This mechanism might form the molecular basis of the regulation of the clock system by light in response to daily changes in the light environment, thus increasing plant fitness.
INTRODUCTION
The circadian clock generates and maintains ∼24-h rhythms that help organisms anticipate and synchronize various developmental and physiological activities with the diurnal light/dark changes in the environment, thus enhancing plant fitness (Michael et al., 2003; Dodd et al., 2005). In the model plant species Arabidopsis (Arabidopsis thaliana), the central oscillator of the clock is believed to be composed of a series of transcriptional feedback loops, in which two morning-expressed single MYB-related transcription factors, CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), directly repress the expression of evening clock genes, such as TIMING OF CAB EXPRESSION1 (TOC1; also known as PSEUDO-RESPONSE REGULATOR1 [PRR1]), EARLY FLOWERING4 (ELF4), LUX ARRHYTHMO (LUX; also known as PHYTOCLOCK1), PRR7, and PRR5, by directly binding to the evening element motifs in their promoters (Alabadí et al., 2001; Hazen et al., 2005; Perales and Más, 2007; Li et al., 2011; Kamioka et al., 2016). In turn, the expression of CCA1 and LHY is repressed in a sequential manner by PRR9 (morning-expressed), PRR7 (midday-expressed), PRR5 (afternoon-expressed), and then TOC1 (evening-expressed) from noon until about midnight (Nakamichi et al., 2010; Huang et al., 2012). In addition, other components of the clock, such as CCA1 HIKING EXPEDITION (CHE), LUX, BROTHER OF LUX ARRHYTHMO (also known as NOX), PROTEIN ARGININE METHYL TRANSFERASE5 (PRMT5), ELF3, and ELF4, also participate in the regulation of CCA1 (Kikis et al., 2005; Pruneda-Paz et al., 2009; Deng et al., 2010; Dai et al., 2011; Helfer et al., 2011). However, the detailed molecular mechanisms remain largely unknown.
A key feature of the clock is that it has an intrinsic ability to reset its activity to synchronize with the surrounding environment. Light is a major signal for resetting the clock through the informational input pathway. Cryptochromes and phytochromes, which are photoreceptors for blue/UV-A and red/far-red light, respectively, are required for transducing the light signal to the central clock (Somers et al., 1998; Yanovsky et al., 2000). CCA1 and LHY expression is induced by light, allowing them to initiate and set the phase of various rhythmic activities (Wang et al., 1997; Kikis et al., 2005). Two TCP transcription factors (TCP20 and TCP22) that are directly involved in light-induced activation of CCA1 expression at dawn have been identified recently (Wu et al., 2016). In addition, the phytochrome-interacting factor (PIF) family of transcription factors was reported to mediate the connection between photosynthate signaling and the clock by direct binding to the promoters of CCA1 and LHY in a sucrose-dependent manner (Shor et al., 2017). However, there are conflicting reports on the roles of PIFs in regulating the clock (Martínez-García et al., 2000; Viczián et al., 2005; Leivar et al., 2009; Nusinow et al., 2011). Moreover, whether these transcription factors are directly involved in connecting phytochrome-mediated light signaling to the clock has not been resolved. Thus, the molecular mechanisms by which light activates CCA1 expression and resets the clock remain poorly understood.
The phytochrome signaling intermediate FAR-RED ELONGATED HYPOCOTYL3 (FHY3) plays an important role in gating red light signaling to the clock during the daytime (Allen et al., 2006). FHY3 and its paralog FAR-RED IMPAIRED RESPONSE1 (FAR1) are transposase-derived transcription factors that directly activate the expression of the evening gene ELF4 (Lin et al., 2007; Li et al., 2011). In this study, we show that FHY3 and FAR1 are also required for the light induction and normal rhythmic expression of CCA1 by directly binding to its promoter and activating its expression. In addition, we show that their activity is antagonized by PIF5 and TOC1 through physical interactions. Our results expand our understanding of the biological roles of FHY3 and FAR1 and provide important insights into the molecular mechanisms regulating CCA1 activation and resetting of the clock by light signals.
RESULTS
FHY3 and FAR1 Are Required for Light-Induced CCA1 Expression
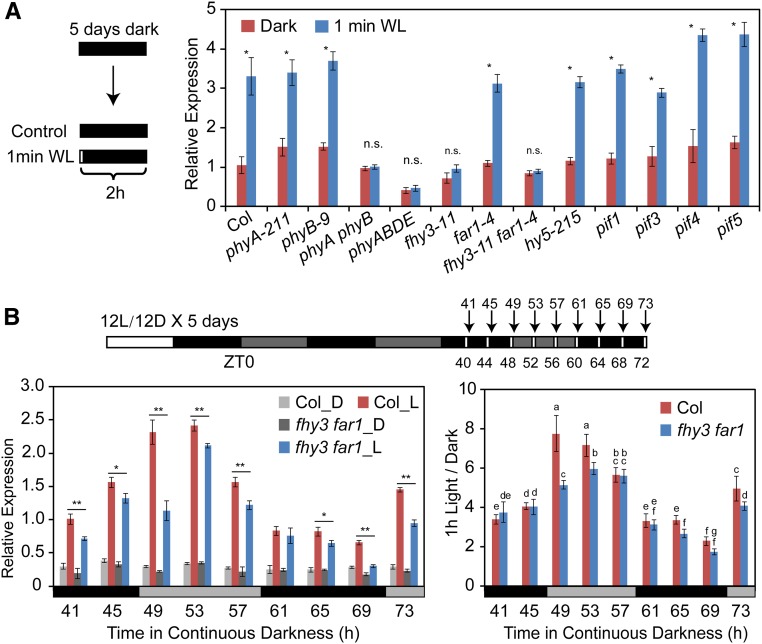
CCA1 expression is quickly induced and initiates its oscillation when dark-grown seedlings are exposed to light (Kikis et al., 2005). To identify the signaling components involved in light-induced CCA1 expression, we examined the effects of light treatment on 5-d-old etiolated seedlings including the wild type (Col), various light signaling mutants (phyA-211, phyB-9, phyA phyB, phyABDE, fhy3-11, far1-4, fhy3-11 far1-4, hy5-215, pif1, pif3, pif4, pif5, pif4 pif5, and pifq), and transgenic line 35S:PIF5-HA. Etiolated seedlings were given a brief light exposure (1 min of white light) and returned to darkness for 2 h prior to harvesting for RNA extraction. qRT-PCR showed that 1 min of white light exposure was sufficient to induce CCA1 expression in wild-type seedlings (ecotype Columbia-0 [Col-0]) as well as in phyA-211, phyB-9, hy5, and pif-related seedlings. However, light-induced CCA1 expression was severely compromised in the fhy3-11 single mutant, fhy3 far1 double mutant, phyA phyB double mutant, phyABDE quadruple mutant, and 35S:PIF5-HA transgenic plants (Figure 1A; Supplemental Figure 1). These observations indicate that phytochromes (primarily phyA and phyB), FHY3, and FAR1 play important roles in the rapid induction of CCA1 expression by light, whereas PIF5 likely plays a repressive role in light-induced CCA1 expression.
Figure 1.
FHY3 and FAR1 Are Required for Light-Induced CCA1 Expression.
(A) qRT-PCR analysis showing the light-induced regulation of CCA1 expression in various light signaling-related mutants. Five-day-old dark-grown Arabidopsis seedlings were treated with a 1-min pulse of white light (WL) and incubated in the dark for 2 h before harvesting (*, P < 0.05, Student’s t test; n.s. no significance). Values are means ± sd (n = 3 technical replicates). Two independent biological replicates (see Methods) showed similar results.
(B) qRT-PCR analysis showing the effects of light treatment at different ZTs on the expression of CCA1. Wild-type and fhy3 far1 seedlings were grown in 12L:12D conditions for 5 d before being transferred to continuous darkness. Beginning at ZT44, seedlings were exposed to light for 1 h at different time points (ZT44 to ZT72) and immediately harvested for RNA extraction. Seedlings grown in the dark at the corresponding time points were used as controls. The CCA1 expression level was normalized to PP2A (*, P < 0.05 and **, P < 0.01, Student’s t test; left panel). The ratio of CCA1 expression in seedlings subjected to 1 h of light treatment versus dark-grown seedlings was used to evaluate the effects of light treatment at different time points (right panel). Different letters indicate significant differences by one-way ANOVA with SAS software (P < 0.05). Two independent biological replicates showed similar results.
Next, we investigated whether light-induced activation of CCA1 is regulated by the clock. Arabidopsis seedlings were clock entrained (grown under a 12-h-light/12-h-dark [12L:12D] cycle for 5 d) and then released into continuous darkness to maintain CCA1 at a steady low level. The seedlings were then exposed to white light for 1 h at various time points (Zeitgeber time 40 [ZT40], ZT44, ZT48, ZT52, ZT56, ZT60, ZT64, ZT68, and ZT72) and harvested immediately after the light treatment (at ZT41, ZT45, ZT49, ZT53, ZT57, ZT61, ZT65, ZT69, and ZT73). qRT-PCR showed that in wild-type seedlings, CCA1 expression increased more significantly when the light treatment was given during the subjective early day (ZT49 and ZT53) versus the subjective night (ZT61, ZT65, and ZT69; all P < 0.001) and that the induction was obviously compromised in the fhy3 far1 mutant at some time points (Figure 1B). These findings suggest that light-induced CCA1 expression is also subjected to a gating effect of the clock, which is consistent with the finding that FHY3 plays an important role in gating red light input to the circadian clock during the subjective day (Allen et al., 2006).
FHY3 and FAR1 Directly Bind to the CCA1 Promoter and Activate Its Expression
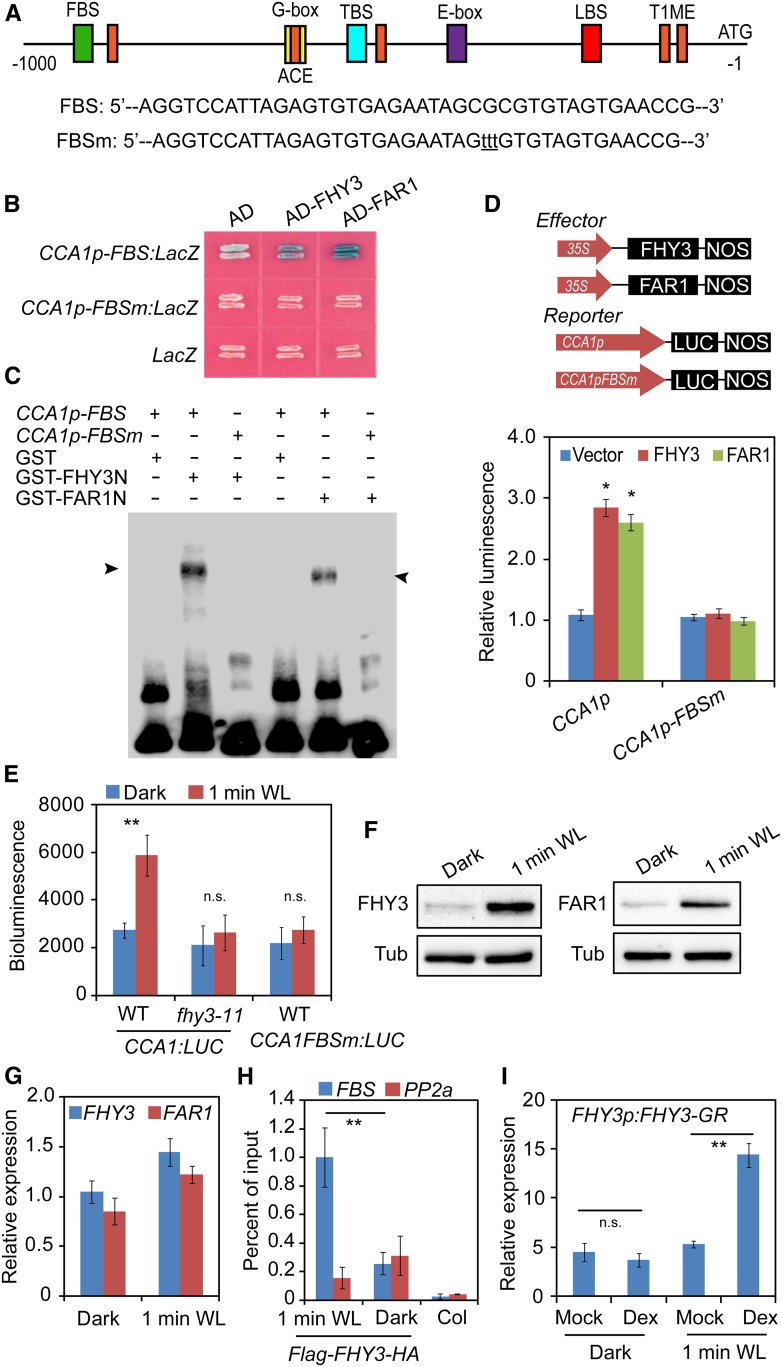
We previously showed that FHY3 and FAR1 are associated with the CCA1 promoter in vivo (Li et al., 2011), suggesting that CCA1 might be a direct downstream target of FHY3 and FAR1. Bioinformatic analysis of the cis-elements in the CCA1 promoter revealed that, besides the known TOC1 binding sites, G-box element, ACE element, CHE binding site, and LUX binding site, there is a putative FHY3/FAR1 binding site (FBS; with the sequence CACGCGC, nucleotides −694 to −700; Figure 2A). Thus, we performed a yeast one-hybrid assay and an electrophoretic mobility shift assay (EMSA) to determine whether FHY3 and FAR1 directly bind to the CCA1 promoter. Both assays showed that indeed FHY3 and FAR1 directly bound to the FBS element, whereas mutations in the FBS element abolished the DNA binding activity of FHY3 and FAR1 (Figures 2B and 2C), indicating that the binding is specific. Next, we performed a transient expression assay to examine the regulatory effect of FHY3 and FAR1 on CCA1 expression in Nicotiana benthamiana leaf cells. Both FHY3 and FAR1 activated the expression of the CCA1p:LUC reporter (Figure 2D). Furthermore, a mutation in the FBS motif in the CCA1 promoter abolished this activation by FHY3 and FAR1. These observations suggest that FHY3 and FAR1 positively regulate CCA1 expression by directly binding to the FBS motif in its promoter.
Figure 2.
FHY3 and FAR1 Directly Bind to the CCA1 Promoter and Activate Its Expression.
(A) Schematic representation of the positions and nucleotide sequences of various cis-elements in the CCA1 promoter. The mutations within the FBS element are shown below the diagram in lowercase letters. LBS, LUX binding site; TBS, TCP binding site (CHE binding site); T1ME, TOC1 binding site.
(B) Yeast one-hybrid assay showing that FHY3 and FAR1 directly bind to the CCA1 promoter. The LacZ reporter gene was driven by the CCA1 promoter with a wild-type or mutated FBS element. Mutation of the FBS site in the CCA1 promoter abolished the binding.
(C) EMSA showing that GST-FHY3N (the first 200 amino acids of FHY3) and GST-FAR1N (the first 200 amino acids of FAR1) specifically bind to the biotin-labeled CCA1p-FBS probe. The arrowheads indicate GST-FHY3N and GST-FAR1N.
(D) Transient expression assay showing that FHY3 and FAR1 activate CCA1 expression in N. benthamiana leaf cells (*, P < 0.05, Student’s t test). Values are means ± sd (n = 3 technical replicates). Three independent biological replicates showed similar results.
(E) Bioluminescence assays showing the activities of CCA1 promoters with a wild-type or mutated FBS motif in wild-type or fhy3-11 seedlings (**, P < 0.01, Student’s t test; n.s., no significance). Values are means ± sd (n = 3 technical replicates). Three independent biological replicates showed similar results.
(F) Immunoblot assay showing increased accumulation of FHY3 and FAR1 protein in seedlings treated with 1 min of white light (WL). Five-day-old dark-grown or white light-treated 35S:Flag-FHY3-HA and 35S:Flag-FAR1-HA transgenic seedlings were collected for immunoblot analysis. Anti-Flag antibodies were used to detect the FHY3 or FAR1 protein. Tubulin (Tub) was used as an internal control.
(G) qRT-PCR analysis showing the expression levels of FHY3 and FAR1 in dark-grown or 1-min white light-treated seedlings. Values are means ± sd (n = 3 technical replicates). Two independent biological replicates showed similar results.
(H) ChIP-qPCR assay showing a significant enrichment of FHY3 on the CCA1 promoter by 1 min of white light exposure. The PP2A amplicon was used as a negative control (**, P < 0.01, Student’s t test). An independent biological replicate showed similar results.
(I) qRT-PCR analysis of CCA1 expression in FHY3p:FHY3-GR transgenic seedlings. Five-day-old dark-grown seedlings were treated with 20 μM Dex or DMSO (Mock) for 2 h before being exposed to 1 min of white light (**, P < 0.01, Student’s t test; n.s., no significance). Values are means ± sd (n = 3 technical replicates). Three independent biological replicates showed similar results.
Next, we investigated whether this direct binding of FHY3 to the CCA1 promoter is required for the rapid induction of CCA1 expression by light. We generated transgenic plants expressing a luciferase (LUC) reporter gene driven by wild-type (CCA1p) and FBS mutated forms of the CCA1 promoter (CCA1p-FBSm). As expected, the CCA1p-LUC reporter gene was rapidly induced by 1 min of white light treatment in the wild-type background but not in the fhy3 background (Figure 2E). However, the LUC reporter gene driven by the CCA1 promoter with mutated FBS (CCA1p-FBSm) lost the response to light (Figure 2E). These observations indicate that the direct interaction between FHY3 and the CCA1 promoter is indispensable for the induction of CCA1 by light. Immunoblot analysis showed that the accumulation of FHY3 and FAR1 was significantly enhanced by 1 min of white light treatment (Figure 2F), although the FHY3 transcript level was only mildly upregulated (Figure 2G). Consistent with this finding, a chromatin immunoprecipitation (ChIP) assay showed that the enrichment of FHY3 on the CCA1 promoter substantially increased in response to light treatment (Figure 2H).
To further investigate the effect of FHY3 on CCA1 induction, we treated FHY3p:FHY3-GR fhy3-4 transgenic plants (Lin et al., 2007; FHY3 protein fused with a dexamethasone-inducible [Dex] glucocorticoid receptor [GR]) with DMSO or Dex for 2 h, exposed them to white light for 1 min, and incubated them in the dark for 2 h before tissue harvest. qRT-PCR showed that a brief (1-min) exposure to white light after Dex treatment (but not DMSO treatment) induced CCA1 expression; however, Dex treatment alone did not induce CCA1 expression (Figure 2I). Immunoblot assays showed that FHY3 protein levels were similar in DMSO- and Dex-treated samples (Supplemental Figure 2), indicating that both the nuclear localization of FHY3 (triggered by Dex treatment) and light treatment are required for light-induced CCA1 expression.
PIF3 and PIF5 Directly Bind to the CCA1 Promoter
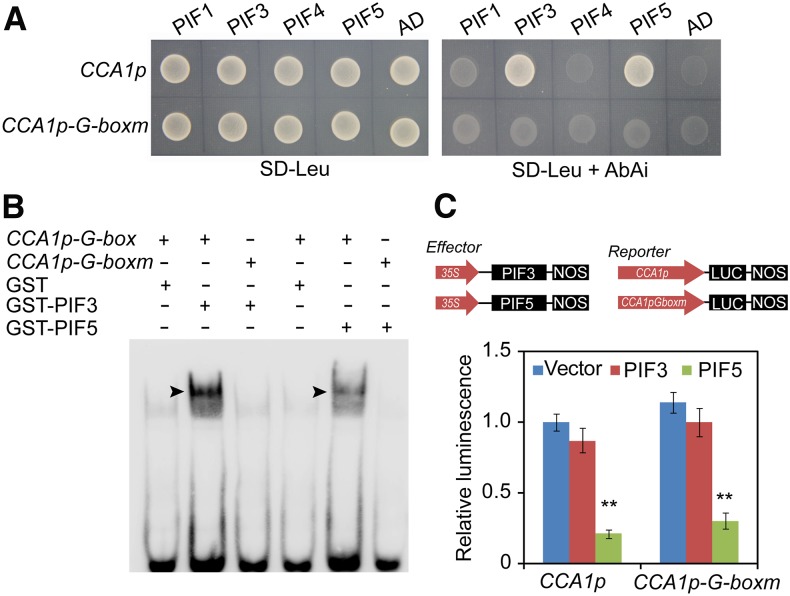
Given the presence of a G-box element in the CCA1 promoter and the in vivo binding of PIF proteins to the CCA1 promoter (Shor et al., 2017), we also investigated whether the PIFs directly bind to the CCA1 promoter. A yeast one-hybrid assay showed that only PIF3 and PIF5, but not PIF1 and PIF4, specifically bind to the G-box element in the CCA1 promoter (Figure 3A). We further confirmed the binding of PIF3 and PIF5 to the CCA1 promoter by EMSA (Figure 3B). Transient expression assays showed that PIF5, but not PIF3, significantly repressed the expression of the CCA1p:LUC reporter in N. benthamiana leaf cells (Figure 3C). Interestingly, the repressive effect of PIF5 was still present even when the G-box motif in the CCA1 promoter was mutated (Figure 3C). This observation suggests that the repressive effect of PIF5 on CCA1 expression is independent of its DNA binding activity.
Figure 3.
PIF5 Directly Binds to the CCA1 Promoter and Represses Its Expression.
(A) Yeast one-hybrid assay showing that PIF3 and PIF5 directly bind to the CCA1 promoter. PIF3 and PIF5, but not PIF1 and PIF4, activated the AbAr reporter gene driven by the wild-type CCA1 promoter but not the AbAr reporter gene driven by the CCA1 promoter with a mutated G-box element. Empty vector expressing the activation domain (AD) alone was used as the negative control.
(B) EMSA showing that GST-PIF3 and GST-PIF5 bHLH (DNA binding domain) specifically bind to the biotin-labeled CCA1p-G-box (right) probe. The arrowheads indicate GST-PIF3 and GST-PIF5 bHLH proteins.
(C) Transient expression assay showing that PIF5 represses CCA1 expression in N. benthamiana leaf cells (**, P < 0.05, Student’s t test). Values are means ± sd (n = 3 technical replicates). Three independent biological replicates showed similar results.
FHY3 and PIF5 Are Required for the Normal Rhythmic Expression of CCA1
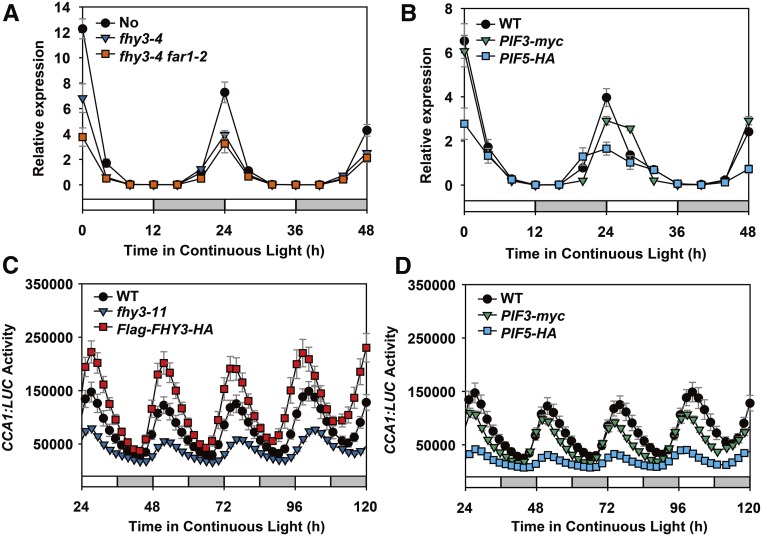
We then investigated the roles of FHY3, FAR1, and PIFs in regulating the rhythmic expression of CCA1 in planta by comparing the diurnal CCA1 expression patterns in the wild type (Nossen [No-0] and Col-0 ecotypes), fhy3-4 single mutant, fhy3-4 far1-2 double mutant, and several pif-related lines (pif3, pif5, pif4 pif5, pifq, 35S:PIF3-myc, and 35S:PIF5-HA). The seedlings were grown in 12L:12D conditions for 7 d before being transferred to continuous light conditions. qRT-PCR revealed that under free-running conditions, the amplitude of CCA1 expression was significantly reduced in the fhy3-4, fhy3-4 far1-2, and 35S:PIF5-HA overexpression transgenic plants but not in the pif mutants or 35S:PIF3-myc transgenic plants (Figures 4A and 4B; Supplemental Figure 3). Similarly, LHY expression level was also reduced in fhy3-11 and 35S:PIF5-HA transgenic plants under these conditions (Supplemental Figure 4). To confirm these observations, we introduced the CCA1:LUC reporter (Salomé and McClung, 2005) into the fhy3-11, 35S:Flag-FHY3-HA, 35S:PIF3-myc, and 35S:PIF5-HA backgrounds and found that the activity of CCA1:LUC was severely reduced in the fhy3-11 background but increased in the 35S:Flag-FHY3-HA background under continuous light conditions, compared with the wild type (Figure 4C). In addition, CCA1:LUC expression was notably reduced in the 35S:PIF5-HA transgenic background but appeared to be only slightly reduced in the 35S:PIF3-myc transgenic background (Figure 4D). These observations further support our conclusion that FHY3 and FAR1 positively regulate CCA1 expression, while PIF5 negatively regulates CCA1 expression, under diurnal cycle conditions.
Figure 4.
FHY3 and FAR1 Activate and PIF5 Represses the Rhythmic Expression of CCA1.
(A) and (B) qRT-PCR analysis showing changes in the cyclic expression of CCA1 in fhy3-4 and fhy3-4 far1-2 (A) and 35S:PIF3-myc and 35S:PIF5-HA (B) plants. Seedlings were entrained at 22°C in 12L:12D conditions for 7 d before being released to continuous light conditions. Values are means ± sd (n = 3 technical replicates). Two independent experiments were performed, with similar results.
(C) and (D) Bioluminescence assays showing expression of the CCA1:LUC reporter in wild-type, fhy3-11, 35S:Flag-FHY3-HA, 35S:PIF3-myc, and 35S:PIF5-HA plants. Seedlings carrying the CCA1:LUC reporter were grown under 12L:12D conditions for 7 d before being transferred to continuous white light. Values are means ± sd (n = 3 technical replicates). Two independent biological replicates showed similar results.
TOC1 and PIF5 Interact with FHY3
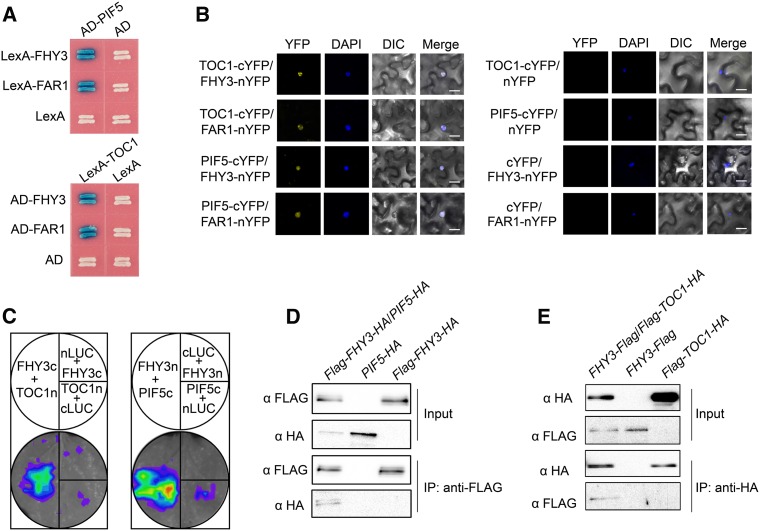
Considering the direct binding of FHY3, FAR1, PIF5, and TOC1 (this study and Li et al., 2011) to the CCA1 promoter, we speculated that FHY3 (and probably FAR1), PIF5 (and probably PIF3), and TOC1 coordinately regulate CCA1 expression through the formation of higher-order protein complex(es). To test this possibility, we conducted pair-wise protein-protein interaction studies using a yeast two-hybrid assay. Both FHY3 and FAR1 interacted with PIF1, PIF3, PIF5, and TOC1 but not with other PRR family members (PRR9, PRR7, and PRR5; Figure 5A; Supplemental Figures 5 and 6). Domain deletion analysis revealed that the C-terminal domain of PIF5 (including the basic helix-loop-helix [bHLH] motif) and the central linker domain of TOC1 are responsible for the interaction with FHY3 (Supplemental Figures 7A and 7B). Conversely, the central transposase domain and C-terminal SWIM domain of FHY3 are required for the interactions with PIF5 and TOC1 (Supplemental Figure 7C). The in vivo interaction of FHY3 with PIF5 and TOC1 was further confirmed using a bimolecular fluorescence complementation (BiFC) assay and a luciferase complementation imaging (LCI) assay (Figures 5B and 5C).
Figure 5.
FHY3 Interacts with TOC1 and PIF5.
(A) Yeast two-hybrid assay showing that FHY3 and FAR1 interact with PIF5 and TOC1.
(B) BiFC assay showing interactions between FHY3 and TOC1, FAR1 and TOC1, FHY3 and PIF5, and FAR1 and PIF5 in N. benthamiana leaf epidermal cells. FHY3 and FAR1 were fused to the N-terminal fragment of YFP (nYFP), and TOC1 and PIF5 were fused to the C-terminal fragment of YFP (cYFP). Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). DIC, differential interference contrast. Bars = 20 µm.
(C) LCI assay showing that FHY3 interacts with TOC1 and PIF5 in planta. The C-terminal half of firefly LUC (cLUC) was fused to FHY3 or PIF5, and the N-terminal half of firefly LUC (nLUC) was fused to FHY3 or TOC1.
(D) and (E) Coimmunoprecipitation assays showing that FHY3 associates with PIF5 and TOC1 in planta.
(D) Protein extracts from seedlings expressing 35S:Flag-FHY3-HA/35S:PIF5-HA, 35S:Flag-FHY3-HA, or 35S:PIF5-HA were immunoprecipitated with anti-Flag antibodies and detected by immunoblot analysis using anti-Flag or anti-HA antibodies.
(E) Protein extracts from seedlings expressing 35S:FHY3-Flag/35S:Flag-TOC1-HA, 35S:FHY3-Flag, or 35S:Flag-TOC1-HA were immunoprecipitated with anti-HA antibodies and detected by immunoblot analysis using anti-HA or anti-Flag antibodies.
To further confirm PIF5-FHY3 interaction in planta, we performed coimmunoprecipitation (Co-IP) using 35S:Flag-FHY3-HA/35S:PIF5-HA double transgenic plants (generated by crossing 35S:Flag-FHY3-HA and 35S:PIF5-HA transgenic plants). Anti-Flag antibodies precipitated PIF5-HA along with Flag-FHY3-HA (Figure 5D). To confirm the TOC1-FHY3 interaction in vivo, we generated 35S:FHY3-Flag and 35S:Flag-TOC1-HA transgenic plants. The 35S:FHY3-Flag transgene successfully rescued the long-hypocotyl phenotype of the fhy3-11 mutant under continuous far-red light conditions, suggesting that the FHY3-Flag fusion protein is biologically functional (Supplemental Figure 8A). Similarly, 35S:Flag-TOC1-HA transgenic seedlings displayed shorter hypocotyls than the wild-type plants (Supplemental Figure 8B), like the previously reported TOC1 overexpression lines (Más et al., 2003), suggesting that the Flag-TOC1-HA fusion protein is biologically functional. We crossed 35S:FHY3-Flag and 35S:Flag-TOC1-HA transgenic plants to produce 35S:FHY3-Flag/35S:Flag-TOC1-HA double transgenic plants. In a Co-IP assay using anti-HA antibodies, FHY3-Flag protein was pulled down together with Flag-TOC1-HA protein (Figure 5E). Together, these results support the physical interaction of FHY3 with PIF5 and TOC1 in planta.
TOC1 and PIF5 Repress the Transcriptional Activation Activity of FHY3
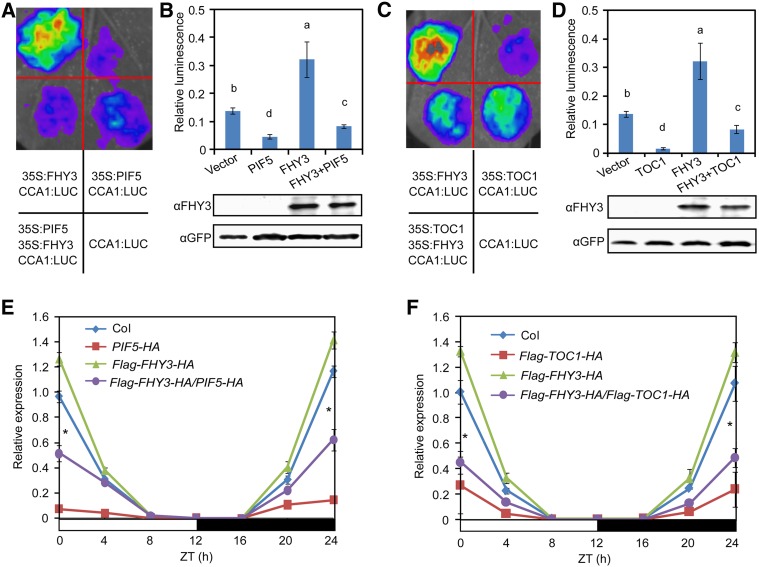
We performed a transient expression assay in N. benthamiana leaves to test the effects of FHY3-PIF3, FHY3-PIF5, and FHY3-TOC1 interactions on CCA1 transcription. FHY3 effectively induced CCA1p:LUC reporter gene expression, whereas coexpression of PIF5 or TOC1, but not PIF3, with FHY3 led to significantly less induction of the CCA1p:LUC reporter gene (Figures 6A to 6D; Supplemental Figure 9), indicating that both PIF5 and TOC1, but not PIF3, suppress the transcriptional activation activity of FHY3. Notably, the repressive activity of PIF5 on the activation of CCA1 expression by FHY3 was still observed when the G-box motif in the CCA1 promoter was mutated (Supplemental Figure 10), suggesting that PIF5 might repress FHY3 activity via a direct protein-protein interaction.
Figure 6.
TOC1 and PIF5 Repress the Transcriptional Activation Activity of FHY3 on CCA1 Expression.
(A) and (B) PIF5 suppresses the activation activity of FHY3 on CCA1 expression in N. benthamiana leaves. Relative LUC activity was normalized to REN activity (LUC/REN). Different letters indicate significant differences by one-way ANOVA with SAS software (P < 0.05). Values are means ± sd (n = 3 technical replicates). Three independent biological replicates showed similar results.
(C) and (D) TOC1 suppresses the activation activity of FHY3 on CCA1 expression in N. benthamiana leaves. Different letters indicate significant differences by one-way ANOVA with SAS software (P < 0.05). Values are means ± sd (n = 3 technical replicates). Three independent biological replicates showed similar results.
(E) CCA1 expression is reduced in 35S:Flag-FHY3-HA/35S:PIF5-HA seedlings compared with its parental line 35S:Flag-FHY3-HA.
(F) CCA1 expression is reduced in 35S:Flag-FHY3-HA/35S:Flag-TOC1-HA seedlings compared with its parental line 35S:Flag-FHY3-HA.
For (E) and (F), seedlings were grown at 22°C in 12L:12D conditions for 7 d before being harvested for RNA extraction (*, P < 0.05, Student’s t test). Values are means ± sd (n = 3 technical replicates). Two independent biological replicates showed similar results.
To further investigate the effects of FHY3-PIF5 and FHY3-TOC1 interactions on the rhythmic expression of CCA1, we examined the expression of CCA1 in the double transgenic plants 35S:Flag-FHY3-HA/35S:Flag-TOC1-HA and 35S:Flag-FHY3-HA/35S:PIF5-HA. qRT-PCR analysis showed that the amplitude of CCA1 expression was significantly reduced in these double transgenic plants, although the transcript levels of FHY3, TOC1, and PIF5 in these plants were comparable to those in their respective parental plants (Figures 6E and 6F; Supplemental Figures 11A to 11C). These observations support that notion that TOC1 and PIF5 play a suppressive role in FHY3-induced CCA1 expression.
FHY3, PIF5, and TOC1 Coordinately Regulate CCA1 Expression during the Diurnal Light/Dark Cycle
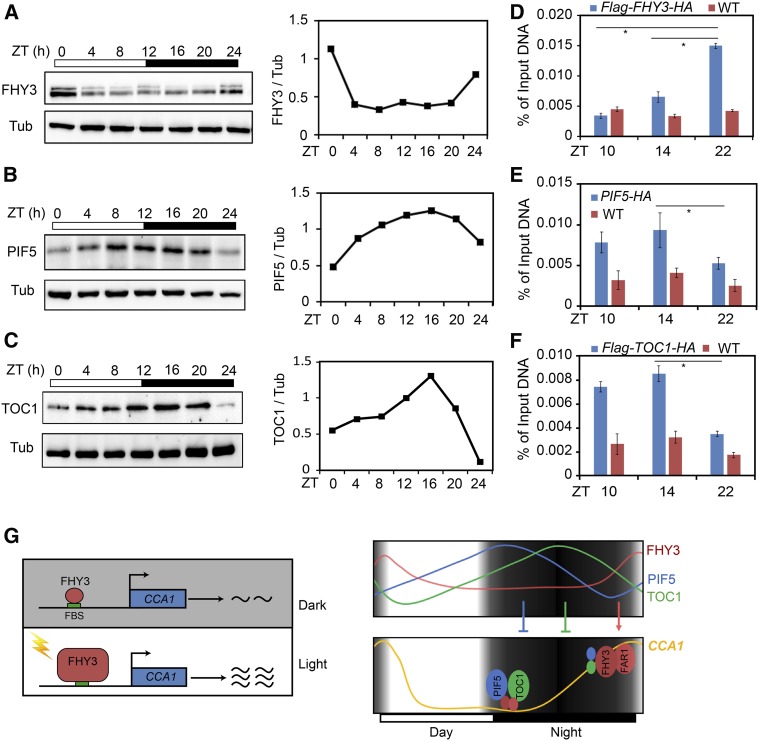
We previously showed that FHY3 protein levels oscillate and peak at dawn under diurnal light/dark cycle conditions (Li et al., 2011). To verify this finding, we performed an immunoblot assay using 35S:Flag-FHY3-HA transgenic plants and found that the protein level of FHY3 was indeed diurnally regulated, with peak accumulation detected at dawn (Figure 7A). Consistent with the finding that the accumulation of PIF5 mRNA is regulated by the clock (Nusinow et al., 2011), our immunoblot analysis showed that in the 35S:PIF5-HA transgenic lines, PIF5 protein accumulated from daytime to dusk, peaked at ZT8 to ZT16, and gradually declined before dawn (Figure 7B). Similarly, TOC1 protein accumulation increased in the early evening and declined before dawn in the 35S:Flag-TOC1-HA transgenic lines (Figure 7C). To confirm the diurnal accumulation patterns of these proteins, we generated PIF5p:PIF5-myc transgenic plants (in which the PIF5-myc transgene was driven by its endogenous promoter). Immunoblot analysis of FHY3, PIF5, and TOC1 proteins in the FHY3p:FHY3-YFP (Lin et al., 2008), PIF5p:PIF5-myc, and TOC1p:TOC1-YFP (Más et al., 2003) transgenic plants revealed similar accumulation patterns for these proteins to those observed in their respective overexpression lines (Supplemental Figure 12). To further determine whether the association of FHY3, PIF5, and TOC1 with the CCA1 promoter is consistent with their accumulation patterns, we performed a time-course ChIP assay. The maximum binding of FHY3 to the CCA1 promoter occurred at predawn (ZT22), whereas the maximum binding of PIF5 and TOC1 to the CCA1 promoter occurred postdusk (ZT14; Figures 7D to 7F). These observations indicate that the dynamic accumulation patterns of FHY3, PIF5, and TOC1 are consistent with their DNA binding activities to the CCA1 promoter.
Figure 7.
FHY3-TOC1 and FHY3-PIF5 Interactions Coordinately Regulate CCA1 Expression.
(A) to (C) Left panels show immunoblot assays showing the oscillation of FHY3 (A), PIF5 (B), and TOC1 (C) protein levels under diurnal cycle conditions. Tubulin (Tub) was used as an internal control. Five-day-old, 12L:12D-entrained 35S:Flag-FHY3-HA, 35S:PIF5-HA, and 35S:Flag-TOC1-HA seedlings were harvested at the indicated time points. Anti-Flag antibodies (1:4000; MBL) were used to detect FHY3 and TOC1. Anti-HA antibodies (1:5000; MBL) were used to detect PIF5. Right panels show estimates of FHY3, PIF5, and TOC1 protein levels using ImageJ software based on the immunoblot results.
(D) to (F) ChIP-qPCR assay showing that FHY3, PIF5, and TOC1 associate with the CCA1 promoter at ZT10, ZT14, and ZT22. Chromatin was extracted from 35S:Flag-FHY3-HA (D), 35S:PIF5-HA (E), and 35S:Flag-FHY3-HA (F) seedlings and precipitated using anti-HA antibodies. No-antibody precipitates and PP2A served as negative controls. Asterisks indicate significant differences between the indicated means with P < 0.05 by Student’s t test. Values are means ± sd (n = 3 technical replicates). Two independent biological replicates showed similar results.
(G) Schematic diagram illustrating how FHY3 mediates the light-induced expression of CCA1 and that the role of FHY3/FAR1 in activating CCA1 expression is antagonistically regulated by TOC1 and PIF5. After light exposure, the accumulation of FHY3 increases, and it binds to the CCA1 promoter to activate its transcription. Under diurnal cycles, TOC1 and PIF5 accumulate from day to midnight, and they repress the activity of FHY3 and FAR1 before midnight. At predawn, the protein levels of PIF5 and TOC1 decrease, thus lifting their repression of FHY3, leading to increased CCA1 expression at dawn.
Feedback Regulation of CCA1 Expression
As the expression of several PIF genes (PIF1, PIF4, and PIF5) is regulated by the clock (Yamashino et al., 2003; Nozue et al., 2007; Nusinow et al., 2011), we examined the effects of the cca1-1 mutation and CCA1 overexpression (CCA1-OX) on FHY3 and PIF5 expression. Although the transcript level of FHY3 did not show an obvious rhythmic pattern, it was obviously reduced in the cca1-1 mutant and increased in the CCA1-OX line compared with the wild-type plants (Supplemental Figure 13A), suggesting that CCA1 positively regulates FHY3 expression. In addition, as CCA1 can physically interact with FHY3 (Li et al., 2011), we also examined the effect of CCA1 on the transcriptional activity of FHY3. Indeed, CCA1 repressed the transcriptional activation activity of FHY3 on CCA1 (Supplemental Figure 13B). This observation is consistent with the finding that the constitutive expression of CCA1 disrupts its rhythmic expression pattern (Wang and Tobin, 1998). Notably, the expression level of PIF5 was also obviously reduced in the cca1-1 mutant during the subjective day but markedly increased in the CCA1-OX background from day to night (Supplemental Figure 13C). These observations suggest that CCA1 expression is also subjected to feedback regulation by FHY3, PIF5, and itself.
DISCUSSION
We previously demonstrated that Arabidopsis FHY3 and FAR1, two signaling intermediates of the phytochrome pathway, are essential for activating the expression of the evening gene ELF4 and that their activity is negatively regulated by CCA1 and LHY through physical interactions (Li et al., 2011). In this study, we obtained multiple lines of evidence showing that FHY3 and FAR1 also play important roles in the light-induced activation of CCA1 expression. First, we showed that CCA1 expression in dark-grown seedlings is activated by a brief exposure of light (1 min) and that this induction is significantly compromised in the fhy3 single and fhy3 far1 double mutant backgrounds (Figures 1A and 1B). Second, we showed that FHY3 and FAR1 can directly bind to the CCA1 promoter through the FBS site (Figures 2B and 2C). Third, we showed that FHY3 and FAR1 can activate CCA1 expression in a transient expression assay (Figure 2D). Fourth, we showed that FHY3 protein accumulation increased in the light (Figure 2F). Consistent with this finding, a ChIP-PCR assay revealed that the in vivo binding of FHY3 to the CCA1 promoter is stronger in the light than in the dark (Figure 2H). These results convincingly demonstrate that FHY3 and FAR1 play positive roles in light-induced CCA1 expression.
Moreover, we demonstrated that FHY3 and FAR1 physically interact with other light signaling intermediates (such as PIF5) and key components of the central oscillator (such as TOC1) to coordinately regulate the normal rhythmic patterns of CCA1 and LHY expression. Both qRT-PCR and CCA1:LUC reporter assays showed that under free-running conditions, the amplitude of CCA1 and LHY expression was significantly reduced in fhy3-4, fhy3-4 far1-2, and 35S:PIF5-HA overexpression plants but increased in the 35S:Flag-FHY3-HA background under continuous light conditions (Figure 4; Supplemental Figure 4). These findings suggest that FHY3 and FAR1 positively regulate CCA1 and LHY expression, whereas PIF5 negatively regulates their expression.
We also showed that PIF5 and TOC1 physically interact with FHY3 and FAR1 and repress their transcriptional activation activity (Figures 5 and 6). In addition, FHY3, PIF5, and TOC1 proteins displayed distinct oscillation patterns under diurnal day/night cycle conditions. Peak accumulation of FHY3 was detected at dawn, which resembles the expression pattern of CCA1 (Figure 7A; Supplemental Figure 11A). PIF5 protein accumulation peaked at ZT8 to ZT16 and then gradually declined before dawn (Figure 7B; Supplemental Figure 11B). Similarly, TOC1 protein accumulated in the early evening and declined at predawn (Figure 7C; Supplemental Figure 11C). These observations collectively suggest that decreased accumulation of PIF5 and TOC1 and the concomitant increase in FHY3 (and probably FAR1) accumulation at dawn are required to lift the repressive activity of TOC1 and PIF5 on FHY3, thus allowing FHY3/FAR1 to activate CCA1 expression at dawn (Figure 7G). This model is consistent with the observation that PIF5 still repressed the transcriptional activation activity of FHY3 on CCA1 expression even when its binding site (the G-box) was mutated (Supplemental Figure 10).
It is worth noting that the current clock model in Arabidopsis is mainly based on negative feedback loops formed by transcriptional repressors (Harmer, 2009). Two sets of activator and coactivator systems were subsequently identified for the core clock genes. Two midday-expressed MYB-like transcription factors, REVEILLE4 (RVE4) and RVE8, form a complex with LNK1 (NIGHT LIGHT-INDUCIBLE AND CLOCK-REGULATED1) and LNK2 and activate the expression of TOC1, PRR5, and the evening complex genes (Farinas and Mas, 2011; Rawat et al., 2011; Hsu et al., 2013; Xie et al., 2014). Another study identified a complex composed of TCP20, TCP22, and their coactivators LWD1 and LWD2 that targets the promoters of PRR9 and CCA1 to activate their expression (Wang et al., 2011; Wu et al., 2016). The difficulties in identifying positive regulators of clock genes using forward genetics approaches may lie in genetic redundancy, and thus, more diversified approaches are needed to tackle this challenge. In this study, we showed that FHY3 and FAR1 are required for light-induced CCA1 expression (Figure 1A). Moreover, we showed that the activation activity of FHY3 and FAR1 to confer the normal rhythmic expression pattern of CCA1 is gated by the circadian clock (Figure 1B) and regulated by their protein-protein interactions with PIF5 and TOC1 (Figure 7G). These findings provide valuable insight into the molecular mechanisms regulating the circadian clock in Arabidopsis and likely other plants as well.
Notably, previous reports indicated that multiple PIFs proteins (PIF1, PIF3, PIF4, and PIF5) associate with the CCA1 and LHY promoters in vivo based on ChIP-PCR (Shor et al., 2017). However, evidence supporting the direct binding of PIF proteins to these promoters is currently lacking. Our yeast one-hybrid assay showed that only PIF3 and PIF5, but not PIF1 and PIF4, directly bind to the CCA1 promoter through the G-box motif (Figures 3A and 3B). The underlying reasons for this discrepancy remain unknown at this stage. Our finding that only PIF5, but not PIF3, represses CCA1 expression is consistent with the earlier reports that these PIF proteins have both shared and distinct DNA binding targets, thus conferring both shared and distinct biological roles for these PIFs (Jeong and Choi, 2013; Pfeiffer et al., 2014). Furthermore, the expression of PIF1, PIF4, and PIF5, but not PIF3, is regulated by the circadian clock (Yamashino et al., 2003; Nozue et al., 2007; Nusinow et al., 2011). Thus, exploring the different roles of PIF1, PIF3, and PIF4 in regulating CCA1 expression and the circadian clock represents an interesting avenue for future research.
Two scenarios have been proposed for the role of TOC1 in repressing CCA1 expression: TOC1 directly associates with the CCA1 promoter to repress its transcription (Gendron et al., 2012) or acts indirectly through interactions with other DNA binding factors (such as CHE; Pruneda-Paz et al., 2009). However, these two scenarios are not mutually exclusive. Indeed, TOC1 interacts with PIF3 and PIF4 and represses the transcriptional activation activities of PIF3 and PIF4 on cobound downstream growth-related genes to mediate the circadian gating of diurnal and thermoresponsive growth (Soy et al., 2016; Zhu et al., 2016). Our results show that TOC1 represses CCA1 expression by physically interacting with and suppressing the transcriptional activation activity of FHY3. Interestingly, our initial yeast two-hybrid assay showed that PIF5 also physically interacted with TCP20, a previously identified activator of CCA1 expression (Supplemental Figure 14), suggesting that PIF5 might also regulate (probably repress) the activity of TCP20. Future efforts to elucidate the functional relationships between FHY3 (and FAR1), PIF5, and TOC1 with the TCP-LWD1 complex should provide additional insights into the multilayered regulation of CCA1 expression.
We previously reported that FHY3 and FAR1 are positive regulators of ELF4, a key evening gene (Li et al., 2011). The finding that FHY3 and FAR1 also act as positive regulators of morning genes (CCA1 and LHY) is intriguing. CCA1 and LHY directly bind to the promoters of evening genes (such as TOC1 and ELF4) to repress their expression (Alabadí et al., 2001; Li et al., 2011). Thus, the regulatory relationship between FHY3/FAR1, CCA1/LHY, and ELF4 is consistent with the previously described type 1 incoherent feedforward loop model (Alon, 2007). According to this model, the two arms of the feedforward loop act in an opposite manner to regulate gene Z: X (in this case, FHY3 and FAR1) activates Z (in this case, ELF4) but also activates Y (in this case, CCA1 and LHY) to repress Z (ELF4; Supplemental Figure 15). At dawn, light promotes the accumulation of FHY3 and FAR1, which activate the expression of both morning genes and evening genes, but the expression of evening genes (ELF4) at dawn is repressed by the products of morning genes (CCA1 and LHY) and other regulators (such as PIF5), resulting in the repression of evening genes at dawn. In addition, we showed that the activation activity of FHY3 and FAR1 on ELF4 and CCA1 expression is regulated by their protein-protein interaction with HY5, CCA1, and LHY (Li et al., 2011) and PIF5 and TOC1 (this study), respectively. Finally, we showed that the expression of FHY3 and PIF5 is also regulated by CCA1 (Supplemental Figure 13) and that CCA1 also represses the transcriptional activation activity of FHY3 on CCA1 itself (Supplemental Figure 13B). Therefore, it is apparent that key components of the light input pathway and the central oscillator form multiple interlocking feedforward loop circuits to generate the proper temporal expression patterns for the clock genes. Although it is a daunting task, it would be rewarding to uncover the transcriptional networks and different types of feedforward loops that constitute the molecular bases of the biological clock using a combination of mathematical modeling and experimental approaches.
METHODS
Plant Materials and Growth Conditions
The wild-type Arabidopsis (Arabidopsis thaliana) plants used in this study were of the Col-0 ecotype unless otherwise indicated. The fhy3-4, far1-2, fhy3-4 far1-2, FHY3p:FHY3-YFP, FHY3p:FHY3-GR/fhy3-4, 35S:Flag-FHY3-HA, and 35S:Flag-FAR1-HA plants were in the No-0 ecotype background and were previously described by Lin et al. (2007, 2008) and Li et al. (2011). The pif1-1, pif3-3, pif4-2, pif5-3, pif4 pif5, and pifq mutants (Leivar et al., 2008), 35S:PIF3-myc (Feng et al., 2008), 35S:PIF5-HA (de Lucas et al., 2008), and TOC1p:TOC1-YFP (Más et al., 2003) were in the Col-0 ecotype background. cca1-1 and CCA1-OX were in Wassilewskija-2 ecotype background (Green and Tobin, 1999; Wang and Tobin, 1998). fhy3-11 (SALK_002711) and far1-4 (SALK_031652) was obtained from the ABRC. The CCA1:LUC reporter line was previously described by Salomé and McClung (2005). The 35S:Flag-TOC1-HA, 35S:FHY3-Flag, and PIF5p:PIF5-myc transgenic plants (all in the Col-0 ecotype background) were generated in this study (see below). The 35S:Flag-FHY3-HA/35S:Flag-TOC1-HA and 35S:Flag-FHY3-HA/35S:PIF5-HA double transgenic lines were obtained by crossing 35S:Flag-FHY3-HA with the 35S:Flag-TOC1-HA and 35S:PIF5-HA lines, respectively. Plants were grown on Murashige and Skoog medium containing 2% (w/v) sucrose and 0.75% (w/v) agar under continuous light or 12L:12D conditions (75 μmol m−2 s−1) in a Percival growth chamber (Percival Scientific; cool-white fluorescent bulb at 22°C).
Plasmid Construction
All plasmids were constructed using an In-Fusion HD cloning kit (Clontech). To generate the CCA1p-FBS:LacZ and CCA1p-FBSm:LacZ reporter constructs, oligonucleotides were synthesized as two complementary oligo primers with an EcoRI site overhang at the 5′ end and an XhoI site overhang at the 3′ end (Supplemental Data Set 1). The oligo primers were annealed, and the double-stranded oligonucleotides were ligated into the EcoRI-XhoI sites of the pLacZi2μ vector (Lin et al., 2007). The CCA1 promoter fragment (1.1 kb from the ATG site) was cloned into the pAbAi vector (Clontech) digested with HindIII and XhoI, creating CCA1p-AbAi. For mutagenesis of the FBS and G-box sites in the CCA1 promoter, primers harboring mutation sites and overlapping with the cis-elements were used to amplify the CCA1 promoter fragments containing the mutated cis-elements. The two PCR products were used as the templates for another round of overlapping PCR to obtain the mutated full-length CCA1 promoter. AD-FHY3, AD-FAR1, AD-PIF3, AD-PIF5, LexA-FHY3, LexA-FAR1, and various deletion constructs of LexA-FHY3 were previously described by Liu et al. (2017) and Xie et al. (2017). AD-TOC1 and AD-TOC1 were generated by subcloning the full-length TOC1 coding sequence (CDS) into the pEG202 and pB42AD vectors, respectively. Various deletions of TOC1 and PIF5 were PCR-amplified and inserted into pEG202 or pB42AD to generate various domain deletion forms of LexA-TOC1 and AD-PIF5. To obtain the wild type, FBS mutated, and G-box muted CCA1 promoter-driven luciferase construct, the amplified CCA1p-WT, CCA1pFBSm, and CCA1p-G-boxm were individually subcloned into the pPZP221-ELF4:LUC vector (Li et al., 2011) through PstI/BamHI sites.
To generate 35S:FHY3-Flag transgenic plants, the FHY3 CDS was amplified and subcloned into pCAMBIA1300-221-Flag (Ren et al., 2014) through the XbaI site to generate the 35S:FHY3-Flag construct. To generate 35S:Flag-TOC1-HA transgenic plants, the full-length CDS of TOC1 was digested with EcoRI and SalI. Fragments of 3×Flag, TOC1, and 3×HA were ligated together and inserted into the pSAT6-MCS vector (Tzfira et al., 2005) digested with BglII and KpnI to produce the pSAT6-Flag-TOC1-HA construct. The expression cassette of 35S:Flag-TOC1-HA was released by PI-PspI digestion and inserted into the pRCS2-OCS-Bar vector (Tzfira et al., 2005) to produce the pRCS2-Flag-TOC1-HA construct. To generate PIF5p:PIF5-myc transgenic plants, the genomic region of PIF5 was amplified and inserted into the pSPYNE-35S vector digested with HindIII/SalI to generate the PIF5p:PIF5-myc construct. The 35S:FHY3-Flag, 35S:Flag-TOC1-HA, and PIF5p:PIF5-myc constructs were transformed into Arabidopsis via Agrobacterium tumefaciens-mediated transformation (Clough and Bent, 1998). Positive transgenic lines (at least 10 independent lines) were selected on Murashige and Skoog medium based on kanamycin (50 mg/L) or hygromycin (50 mg/L) resistance and subjected to immunoblot analysis.
Yeast Assays
Yeast one-hybrid and yeast two-hybrid assays were performed as described previously (Liu et al., 2017).
Gene Expression Analysis
The seedling samples were harvested, frozen immediately in liquid nitrogen, and stored at −80°C until use. Two biological replicates were set up for each time point. For each replicate, 30 to 40 seedlings were harvested. The samples were ground in a grinder with a 3-mm steel ball in liquid nitrogen. Total RNA was extracted from the seedlings using Trizol (Invitrogen) following the manufacturer’s protocols. The first-strand cDNA was synthesized from 1 μg of RNA using reverse transcriptase (Tiangen, FastQuant RT Kit) following digestion with gDNase from the kit to remove genomic DNA contamination. The cDNA was diluted 1:10 and subjected to qPCR using SuperReal PreMix Plus (Tiangen) and a 7500 Real Time PCR System (Applied Bio-systems) cycler according to the manufacturer’s instructions. The following thermal cycling profile was used: 95°C for 15 min, ∼40 cycles of 95°C for 10 s and 60°C for 32 s, followed by a melt curve ranging from 65 to 95°C with increments of 0.5°C for 5 s. The comparative CT method was used to determine relative gene expression levels, with the expression of PP2A used as the internal control. Efficiency calculations assume amplicon doubling during every cycle when measuring differences in expression. Mean values of were calculated from three technical repeats. Primers are listed in Supplemental Data Set 1. All experiments were replicated two or three times with similar results.
ChIP
35S:Flag-FHY3-HA and 35S:PIF5-HA transgenic seedlings were used in the ChIP assays as previously described by Liu et al. (2017). Briefly, ∼2 g of seedling tissue was cross-linked for 10 min in 1% (v/v) formaldehyde solution under a vacuum. The cross-linked chromatin complex was isolated using nuclear lysis buffer (50 mM Tris-HCl at pH 8.0, 10 mM EDTA, 1% [w/v] SDS, 1 mM PMSF, and 1× protease inhibitor cocktail), diluted fivefold in ChIP dilution buffer (16.7 mM Tris-HCl at pH 8.0, 167 mM NaCl, 1.1% [v/v] Triton X-100, 1.2 mM EDTA, 1 mM PMSF, and 1× protease inhibitor cocktail), and sheared by sonication. The sonicated chromatin complex was immunoprecipitated using anti-HA antibodies (2 μL; Cali-Bio). The beads were washed with low-salt buffer (50 mM Tris-HCl at pH 8.0, 2 mM EDTA, 150 mM NaCl, and 1% [v/v] Triton X-100), high-salt buffer (50 mM Tris-HCl at pH 8.0, 2 mM EDTA, 500 mM NaCl, and 1% [v/v] Triton X-100), LiCl buffer (10 mM Tris-HCl at pH 8.0, 1 mM EDTA, 0.25 M LiCl, 0.5% [v/v] Nonidet P-40, and 0.5% [w/v] deoxycholate), and TE buffer (10 mM Tris-HCl at pH 8.0 and 1 mM EDTA) and eluted with elution buffer (1% [w/v] SDS and 0.1 M NaHCO3). After reverse cross-linking, the DNA was precipitated with phenol/chloroform/isoamyl alcohol and analyzed by ChIP-qPCR. Primers used for ChIP-qPCR are listed in Supplemental Data Set 1.
EMSA
EMSA was performed using a LightShift Chemiluminescent EMSA kit (Pierce) according to the manufacturer’s instructions. GST-FHY3N, GST-FAR1N, and GST-PIF5 bHLH fusion proteins were described previously (Liu et al., 2017; Xie et al., 2017). The oligonucleotide sequences of biotin-labeled probes are listed in Supplemental Data Set 1. Briefly, biotin-labeled probes were incubated for 20 min with the expressed proteins in binding buffer at room temperature. The DNA-protein complexes were separated on 6% (w/v) native polyacrylamide gels, and the signal was detected using the Biostep Celvin S420 system.
BiFC Assay
The CDSs of FHY3 and FAR1 were amplified and cloned into the pSPYNE-35S vector digested with BamHI/SalI to generate FHY3-nYFP and FAR1-nYFP. The CDSs of TOC1 and PIF5 were subcloned into pSPYCE-35S to generate TOC1-cYFP and PIF5-cYFP. The nYFP- and cYFP-related constructs were transformed into A. tumefaciens strain EHA105. Agrobacterium cultures containing the combination of nYFP and cYFP constructs were incubated for 2 h and infiltrated into 3-week-old Nicotiana benthamiana leaves. Reconstitution of YFP fluorescence was observed with a confocal microscope (Zeiss, LSM 700) with the following YFP filter setup: excitation at 515 nm and emission at 525 to 560 nm.
Co-IP Assay
For Co-IP assays using Arabidopsis seedlings, total proteins were homogenized in extraction buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 10 mM MgCl2, 0.1% [v/v] Tween 20, 1 mM PMSF, and 1× protease inhibitor cocktail) and centrifuged twice at 12,000g. The cleared extract was mixed with anti-Flag or anti-HA magnetic agarose beads (MBL) and incubated overnight at 4°C. After washing five times with Co-IP washing buffer (100 mM NaCl and 20 mM Tris-HCl at pH 7.6), the magnetic agarose beads were resuspended in extraction buffer. For immunoblot analysis, samples were separated by SDS-PAGE, and the target protein was detected using anti-Flag (1:4000) or anti-HA (1:5000) antibody (MBL; M185-7 or M180-7, respectively).
LCI Assay
The firefly LCI assays were performed using N. benthamiana leaves. The CDSs of FHY3 and TOC1 were ligated into the KpnI/SalI sites of the p1300-35S-cLUC vector (Chen et al., 2008). For the nLUC-FHY3 and nLUC-PIF5 constructs, the CDSs of FHY3 and PIF5 were ligated into the KpnI/SalI sites of the p1300-35S-cLUC vector (Chen et al., 2008). Both the nLUC- and cLUC-fused proteins were coinfiltrated into N. benthamiana leaves via Agrobacterium-mediated coinfiltration. The infiltrated plants were incubated for 3 d and examined using the NightSHADE LB985 Plant Imaging System (Berthold).
Transient Expression in N. benthamiana Leaf Cells
The CCA1 promoter was amplified and cloned into the pGreenII 0800-LUC vector (Hellens et al., 2005) to generate CCA1p:LUC. The CDSs of FHY3, PIF3, PIF5, and TOC1 were amplified and subcloned into the SPYNE vector (Walter et al., 2004) through the BamHI/SalI sites. For transient expression, the effector and reporter constructs were coinfiltrated into N. benthamiana leaves via Agrobacterium-mediated coinfiltration. Firefly luciferase and Renilla luciferase activities were quantified 3 d after transformation. To measure firefly luciferase activity, 40 μL of Lar II was added to the protein extract, and the luminescence was measured for 5 s. To measure Renilla luciferase activity, 40 μL of Stop and Glow solution was added, and the luminescence was again measured for 5 s using a Berthold LB942 luminometer.
Bioluminescence Assay
The CCA1:LUC reporter line (kindly provided by Rob McClung) was crossed into different mutant and transgenic backgrounds (fhy3-11, 35S:Flag-FHY3-HA, 35S:PIF3-myc, and 35S:PIF5-HA). Homozygous seedlings carrying the CCA1:LUC reporter were selected and used for the bioluminescence assay. Seedlings were entrained for 10 d in 12L:12D cycles (22°C) before being released into continuous light (22°C) conditions for LUC measurements. After spraying with 1 mM luciferin (Goldbio), the bioluminescence generated from the CCA1:LUC reporter was recorded with a Top-Count luminometer.
Statistical Analysis
All statistics were calculated using SPSS software. To determine statistical significance, we employed independent t tests between two groups and one-way ANOVA among various genotypes. A value of P < 0.05 was considered to be statistically significant. All sample sizes and significance thresholds are indicated in the figure legends. The results of statistical analyses are shown in Supplemental Data Set 2.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: FHY3 (AT3G22170), FAR1 (AT4G15090), CCA1 (AT2G46830), PIF1 (AT2G20180), PIF3 (AT1G09530), PIF4 (AT2G43010), PIF5 (AT3G59060), and TOC1 (AT5G61380).
Supplemental Data
Supplemental Figure 1. Light-induced CCA1 expression in PIF-related mutants.
Supplemental Figure 2. Immunoblot analysis of FHY3 protein levels in FHY3p:FHY3-GR transgenic plants with and without Dex treatment in both the dark and light.
Supplemental Figure 3. Diurnal expression pattern of CCA1 in pif3, pif5, pif4 pif5 and pifq mutants.
Supplemental Figure 4. qRT-PCR analysis showing the changes of LHY expression in wild-type (Col), fhy3-11, and 35S:PIF5-HA seedlings.
Supplemental Figure 5. Yeast two-hybrid assay showing that PIF1, PIF3, and PIF5 physically interact with FHY3 and TOC1.
Supplemental Figure 6. Yeast two-hybrid assay showing that FHY3 and FAR1 interact with TOC1, but not with PRR9, PRR7, or PRR5.
Supplemental Figure 7. Mapping of the interacting domains of PIF5, TOC1, and FHY3 using yeast two-hybrid assays.
Supplemental Figure 8. Phenotypic and molecular characterization of 35S:FHY3-Flag and 35S:Flag-TOC1-HA transgenic plants.
Supplemental Figure 9. Transient expression assay in N. benthamiana leaves showing the effects of co-expressing PIF3 and FHY3 on the expression of LUC Reporter driven by wild type CCA1 promoter.
Supplemental Figure 10. Transient expression assay in N. benthamiana leaves showing the effects of co-expressing PIF5 and FHY3 on the expression of LUC reporter driven by CCA1 promoter with a mutated G-box.
Supplemental Figure 11. qRT-PCR analysis of the expression levels of FHY3, TOC1, and PIF5.
Supplemental Figure 12. Immunoblots showing the oscillation patterns of FHY3, PIF5, and TOC1 under diurnal cycle conditions.
Supplemental Figure 13. Feedback regulation between FHY3, PIF5, and CCA1.
Supplemental Figure 14. Yeast two-hybrid assay showing that PIF5 interacts with TCP20.
Supplemental Figure 15. Putative structure of a type 1 incoherent feedforward loop (I1-FFL) composed of FHY3/FAR1, CCA1/LHY, and ELF4.
Supplemental Data Set 1. Primers used in this study.
Supplemental Data Set 2. Statistical report of t test and ANOVA results for the data presented in each figure.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
pifQ Gramene: pif1 pif3 pif4 pif5
pifQ Araport: pif1 pif3 pif4 pif5
Acknowledgments
We thank Rob McClung and Steve Kay for kindly providing the CCA1-LUC reporter line and the TOC1p:TOC1-YFP transgenic line, respectively. We also thank Jigang Li (China Agricultural University) for providing purified anti-FHY3 antibodies and Qiguang Xie (Henan University) and Wei Huang (South China Agricultural University) for comments and valuable suggestions on this work. This work was supported by the National Natural Science Foundation of China (grants 31430008 to H.W., 31500239 to Y.L., and 31570285 to X.X.) and the China Postdoctoral Science Foundation (grant 2014M560142).
AUTHOR CONTRIBUTIONS
H.Y.W. and Y.L. designed the research and wrote the article; H.Y.W., X.X., and P.F.D. supervised the work; Y.L., M.M., G.L., L.Y., Y.X., H.W., X.M., Q.L., and H.X. performed the experiments and analyzed the data.
References
- Alabadí D., Oyama T., Yanovsky M.J., Harmon F.G., Más P., Kay S.A.(2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883. [DOI] [PubMed] [Google Scholar]
- Allen T., Koustenis A., Theodorou G., Somers D.E., Kay S.A., Whitelam G.C., Devlin P.F.(2006). Arabidopsis FHY3 specifically gates phytochrome signaling to the circadian clock. Plant Cell 18: 2506–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon U.(2007). Network motifs: Theory and experimental approaches. Nat. Rev. Genet. 8: 450–461. [DOI] [PubMed] [Google Scholar]
- Chen H., Zou Y., Shang Y., Lin H., Wang Y., Cai R., Tang X., Zhou J.M.(2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146: 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F.(1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Dai S., Wei X., Pei L., Thompson R.L., Liu Y., Heard J.E., Ruff T.G., Beachy R.N.(2011). BROTHER OF LUX ARRHYTHMO is a component of the Arabidopsis circadian clock. Plant Cell 23: 961–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M., Davière J.M., Rodríguez-Falcón M., Pontin M., Iglesias-Pedraz J.M., Lorrain S., Fankhauser C., Blázquez M.A., Titarenko E., Prat S.(2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484. [DOI] [PubMed] [Google Scholar]
- Deng X., Gu L., Liu C., Lu T., Lu F., Lu Z., Cui P., Pei Y., Wang B., Hu S., Cao X.(2010). Arginine methylation mediated by the Arabidopsis homolog of PRMT5 is essential for proper pre-mRNA splicing. Proc. Natl. Acad. Sci. USA 107: 19114–19119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd A.N., Salathia N., Hall A., Kévei E., Tóth R., Nagy F., Hibberd J.M., Millar A.J., Webb A.A.R.(2005). Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309: 630–633. [DOI] [PubMed] [Google Scholar]
- Farinas B., Mas P.(2011). Functional implication of the MYB transcription factor RVE8/LCL5 in the circadian control of histone acetylation. Plant J. 66: 318–329. [DOI] [PubMed] [Google Scholar]
- Feng S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron J.M., Pruneda-Paz J.L., Doherty C.J., Gross A.M., Kang S.E., Kay S.A.(2012). Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. USA 109: 3167–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R.M., Tobin E.M.(1999). Loss of the circadian clock-associated protein 1 in Arabidopsis results in altered clock-regulated gene expression. Proc. Natl. Acad. Sci. USA 96: 4176–4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer S.L.(2009). The circadian system in higher plants. Annu. Rev. Plant Biol. 60: 357–377. [DOI] [PubMed] [Google Scholar]
- Hazen S.P., Schultz T.F., Pruneda-Paz J.L., Borevitz J.O., Ecker J.R., Kay S.A.(2005). LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc. Natl. Acad. Sci. USA 102: 10387–10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfer A., Nusinow D.A., Chow B.Y., Gehrke A.R., Bulyk M.L., Kay S.A.(2011). LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr. Biol. 21: 126–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens R.P., Allan A.C., Friel E.N., Bolitho K., Grafton K., Templeton M.D., Karunairetnam S., Gleave A.P., Laing W.A.(2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P.Y., Devisetty U.K., Harmer S.L.(2013). Accurate timekeeping is controlled by a cycling activator in Arabidopsis. eLife 2: e00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Pérez-García P., Pokhilko A., Millar A.J., Antoshechkin I., Riechmann J.L., Mas P.(2012). Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336: 75–79. [DOI] [PubMed] [Google Scholar]
- Jeong J., Choi G.(2013). Phytochrome-interacting factors have both shared and distinct biological roles. Mol. Cells 35: 371–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamioka M., Takao S., Suzuki T., Taki K., Higashiyama T., Kinoshita T., Nakamichi N.(2016). Direct repression of evening genes by CIRCADIAN CLOCK-ASSOCIATED1 in the Arabidopsis circadian clock. Plant Cell 28: 696–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikis E.A., Khanna R., Quail P.H.(2005). ELF4 is a phytochrome-regulated component of a negative-feedback loop involving the central oscillator components CCA1 and LHY. Plant J. 44: 300–313. [DOI] [PubMed] [Google Scholar]
- Leivar P., Monte E., Oka Y., Liu T., Carle C., Castillon A., Huq E., Quail P.H.(2008). Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 18: 1815–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Tepperman J.M., Monte E., Calderon R.H., Liu T.L., Quail P.H.(2009). Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell 21: 3535–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., et al. (2011). Coordinated transcriptional regulation underlying the circadian clock in Arabidopsis. Nat. Cell Biol. 13: 616–622. [DOI] [PubMed] [Google Scholar]
- Lin R., Ding L., Casola C., Ripoll D.R., Feschotte C., Wang H.(2007). Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318: 1302–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Teng Y., Park H.J., Ding L., Black C., Fang P., Wang H.(2008). Discrete and essential roles of the multiple domains of Arabidopsis FHY3 in mediating phytochrome A signal transduction. Plant Physiol. 148: 981–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Xie Y., Wang H., Ma X., Yao W., Wang H.(2017). Light and ethylene coordinately regulate the phosphate starvation response through transcriptional regulation of PHOSPHATE STARVATION RESPONSE1. Plant Cell 29: 2269–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-García J.F., Huq E., Quail P.H.(2000). Direct targeting of light signals to a promoter element-bound transcription factor. Science 288: 859–863. [DOI] [PubMed] [Google Scholar]
- Más P., Alabadí D., Yanovsky M.J., Oyama T., Kay S.A.(2003). Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell 15: 223–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael T.P., Salomé P.A., Yu H.J., Spencer T.R., Sharp E.L., McPeek M.A., Alonso J.M., Ecker J.R., McClung C.R.(2003). Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science 302: 1049–1053. [DOI] [PubMed] [Google Scholar]
- Nakamichi N., Kiba T., Henriques R., Mizuno T., Chua N.H., Sakakibara H.(2010). PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22: 594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozue K., Covington M.F., Duek P.D., Lorrain S., Fankhauser C., Harmer S.L., Maloof J.N.(2007). Rhythmic growth explained by coincidence between internal and external cues. Nature 448: 358–361. [DOI] [PubMed] [Google Scholar]
- Nusinow D.A., Helfer A., Hamilton E.E., King J.J., Imaizumi T., Schultz T.F., Farré E.M., Kay S.A.(2011). The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475: 398–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales M., Más P.(2007). A functional link between rhythmic changes in chromatin structure and the Arabidopsis biological clock. Plant Cell 19: 2111–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer A., Shi H., Tepperman J.M., Zhang Y., Quail P.H.(2014). Combinatorial complexity in a transcriptionally centered signaling hub in Arabidopsis. Mol. Plant 7: 1598–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda-Paz J.L., Breton G., Para A., Kay S.A.(2009). A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323: 1481–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat R., Takahashi N., Hsu P.Y., Jones M.A., Schwartz J., Salemi M.R., Phinney B.S., Harmer S.L.(2011). REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock. PLoS Genet. 7: e1001350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y., et al. (2014). GLUTELIN PRECURSOR ACCUMULATION3 encodes a regulator of post-Golgi vesicular traffic essential for vacuolar protein sorting in rice endosperm. Plant Cell 26: 410–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé P.A., McClung C.R.(2005). PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell 17: 791–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shor E., Paik I., Kangisser S., Green R., Huq E.(2017). PHYTOCHROME INTERACTING FACTORS mediate metabolic control of the circadian system in Arabidopsis. New Phytol. 215: 217–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers D.E., Devlin P.F., Kay S.A.(1998). Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282: 1488–1490. [DOI] [PubMed] [Google Scholar]
- Soy J., Leivar P., González-Schain N., Martín G., Diaz C., Sentandreu M., Al-Sady B., Quail P.H., Monte E.(2016). Molecular convergence of clock and photosensory pathways through PIF3-TOC1 interaction and co-occupancy of target promoters. Proc. Natl. Acad. Sci. USA 113: 4870–4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T., Tian G.W., Lacroix B., Vyas S., Li J., Leitner-Dagan Y., Krichevsky A., Taylor T., Vainstein A., Citovsky V.(2005). pSAT vectors: A modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol. Biol. 57: 503–516. [DOI] [PubMed] [Google Scholar]
- Viczián A., Kircher S., Fejes E., Millar A.J., Schäfer E., Kozma-Bognár L., Nagy F.(2005). Functional characterization of phytochrome interacting factor 3 for the Arabidopsis thaliana circadian clockwork. Plant Cell Physiol. 46: 1591–1602. [DOI] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J.(2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438. [DOI] [PubMed] [Google Scholar]
- Wang Y., Wu J.F., Nakamichi N., Sakakibara H., Nam H.G., Wu S.H.(2011). LIGHT-REGULATED WD1 and PSEUDO-RESPONSE REGULATOR9 form a positive feedback regulatory loop in the Arabidopsis circadian clock. Plant Cell 23: 486–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.Y., Kenigsbuch D., Sun L., Harel E., Ong M.S., Tobin E.M.(1997). A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell 9: 491–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.Y., Tobin E.M.(1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93: 1207–1217. [DOI] [PubMed] [Google Scholar]
- Wu J.F., Tsai H.L., Joanito I., Wu Y.C., Chang C.W., Li Y.H., Wang Y., Hong J.C., Chu J.W., Hsu C.P., Wu S.H.(2016). LWD-TCP complex activates the morning gene CCA1 in Arabidopsis. Nat. Commun. 7: 13181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q., et al. (2014). LNK1 and LNK2 are transcriptional coactivators in the Arabidopsis circadian oscillator. Plant Cell 26: 2843–2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Liu Y., Wang H., Ma X., Wang B., Wu G., Wang H.(2017). Phytochrome-interacting factors directly suppress MIR156 expression to enhance shade-avoidance syndrome in Arabidopsis. Nat. Commun. 8: 348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashino T., Matsushika A., Fujimori T., Sato S., Kato T., Tabata S., Mizuno T.(2003). A link between circadian-controlled bHLH factors and the APRR1/TOC1 quintet in Arabidopsis thaliana. Plant Cell Physiol. 44: 619–629. [DOI] [PubMed] [Google Scholar]
- Yanovsky M.J., Mazzella M.A., Casal J.J.(2000). A quadruple photoreceptor mutant still keeps track of time. Curr. Biol. 10: 1013–1015. [DOI] [PubMed] [Google Scholar]
- Zhu J.Y., Oh E., Wang T., Wang Z.Y.(2016). TOC1-PIF4 interaction mediates the circadian gating of thermoresponsive growth in Arabidopsis. Nat. Commun. 7: 13692. [DOI] [PMC free article] [PubMed] [Google Scholar]