A detailed screen of the Solanum lycopersicum cv M82 × Solanum pennellii ac. LA716 introgression line (IL) population revealed ILs with altered leaf trichome densities, abnormal proportions of different trichome types, and aberrant trichome morphologies leading to the identification of previously unexplored genomic regions with roles in trichome formation in tomato.
Abstract
The aerial epidermis of plants plays a major role in environmental interactions, yet the development of the cellular components of the aerial epidermis—trichomes, stomata, and pavement cells—is still not fully understood. We have performed a detailed screen of the leaf epidermis in two generations of the well-established Solanum lycopersicum cv M82 × Solanum pennellii ac. LA716 introgression line (IL) population using a combination of scanning electron microscopy (SEM) techniques. Quantification of trichome and stomatal densities in the ILs revealed four genomic regions with a consistently low trichome density. This study also found ILs with abnormal proportions of different trichome types and aberrant trichome morphologies. This work has led to the identification of new, unexplored genomic regions with roles in trichome formation in tomato. This study investigated one interval in IL2-6 in more detail and identified a new function for the transcription factor SlMixta-like in determining trichome patterning in leaves. This illustrates how these SEM images, publicly available to the research community, provide an important dataset for further studies on epidermal development in tomato and other species of the Solanaceae family.
INTRODUCTION
The epidermis is the external cell layer of plant organs and, in aerial tissues, consists of three types of specialized cells: trichomes, stomata, and pavement cells. Trichomes, commonly referred to as “hairs,” are outgrowths that can have different sizes and shapes, and their morphology has been used commonly for taxonomic purposes (Payne, 1978). In tomato (Solanum lycopersicum) and related species, trichomes are multicellular and have been classified into seven different types according to size, morphology, and their metabolic profiles when they bear glandular heads (Luckwill, 1943; Simmons and Gurr, 2005). Stomata are pores surrounded by two specialized guard cells in which turgor is regulated to control gas exchange between the plant and the atmosphere (Hetherington and Woodward, 2003). Stomata, in contrast with trichomes, have conserved morphology and function in plants (Chater et al., 2017). Pavement cells are relatively unspecialized epidermal cells that ensure an adequate patterning of trichomes and stomata across the leaf surface. In tomato leaves, pavement cells have a characteristic undulated shape, like pieces of a jigsaw puzzle, a shape that is not uncommon in other dicotyledonous plants (Vőfély et al., 2019). These different cell types develop from a single cell layer, the protodermis, and therefore are developmentally linked, as suggested by studies in tobacco (Nicotiana tabacum) and tomato (Glover et al., 1998; Glover, 2000). The importance of correct spacing of epidermal cell types and the potentially limited size of the pool of protodermal cells implies cross talk in the regulation of different cell fates in the epidermis.
From a developmental point of view, most studies of trichome formation have focused on the model plant Arabidopsis (Arabidopsis thaliana), which produces only one type of unicellular, nonglandular trichome, contributing to the establishment of a detailed model for initiation and development at the cellular and molecular levels (Szymanski et al., 1998; Pattanaik et al., 2014). The transcriptional regulation of trichome initiation in Arabidopsis involves the formation of an MYB–basic helix-loop-helix–tryptophan-aspartic acid repeat protein (WD40); the MBW complex, that induces trichome formation. The main MYB-domain transcription factor forming part of this complex is GLABROUS1 (Larkin et al., 1994), although MYB23 and MYB82 can perform the same function redundantly (Kirik et al., 2005; Liang et al., 2014). Three basic helix-loop-helix factors can form part of this MBW complex: GLABRA3 (Payne et al., 2000), ENHANCER OF GLABRA3 (Zhang et al., 2003), and TRANSPARENT-TESTA8, which plays this role in leaf margins (Maes et al., 2008). Additionally, a factor with WD40 repeats has been identified as part of the complex: TRANSPARENT TESTA GLABRA1 (TTG1; Walker et al., 1999). This complex activates the expression of downstream genes necessary for trichome development. One such target is the gene encoding a WRKY transcription factor, TTG2. TTG2 has been suggested to be recruited to the MBW complex itself through interaction with the WD40 protein TTG1 (Pesch et al., 2014; Lloyd et al., 2017). Alternatively, TTG1 and TTG2 may interact downstream of the MBW complex to narrow the target genes responding to its transcriptional regulation (Lloyd et al., 2017). Among other targets of the MBW complex, GL2 encodes a Homeodomain Leucine zipper (HD-Zip) IV transcription factor essential for the correct morphogenesis of mature trichomes (Szymanski et al., 1998). Furthermore, the MBW complex can be activated by a hierarchical cascade of C2H2 zinc finger transcription factors, including GLABROUS INFLORESCENCE STEM proteins (GIS, GIS2, and GIS3) and ZINC FINGER PROTEIN5, ZINC FINGER PROTEIN6, and ZINC FINGER PROTEIN8 (Gan et al., 2006, 2007; Zhou et al., 2011, 2013; Sun et al., 2015). By contrast, negative regulation of trichome initiation involves the expression of small R3 MYB transcription factors in the trichome initial cell, including TRYPTICON (TRY), CAPRICE, ENHANCER OF TRYPTICON AND CAPRICE1, ENHANCER OF TRYPTICON AND CAPRICE2, and ENHANCER OF TRYPTICON AND CAPRICE3 (ETC1, ETC2, and ETC3), and TRICHOMLESS1/2 (TCL1 and TCL2). These R3 MYB proteins can move to neighboring cells and compete with GL1 in the formation of the MBW complex (Wada et al., 1997; Schnittger et al., 1999; Kirik et al., 2004a, 2004b; Wester et al., 2009). However, this model does not apply to tobacco, tomato, or related species (Serna and Martin, 2006), where multicellular trichome formation is controlled by different regulatory proteins (Lloyd et al., 2017).
In tomato, the focus of research has been on glandular trichomes and the metabolites they secrete (Schilmiller et al., 2008; McDowell et al., 2011). A number of different transcription factors involved in trichome formation have been identified. SlMx1 (for S. lycopersicum Mixta, a Mixta-type transcription factor; Brockington et al., 2013) was shown to control trichome initiation in tomato, while also regulating cuticle deposition and carotenoid content in fruit (Ewas et al., 2016). Mixta itself was first identified in snapdragon (Antirrhinum majus), where it is expressed in petals and is required for formation of conical cells that provide sheen and brightness to petals (Noda et al., 1994). Mixta was also shown to induce multicellular trichome formation when ectopically expressed in tobacco and in snapdragon (Glover et al., 1998; Payne et al., 1999; Martin et al., 2002). Mixta-type transcription factors are known regulators of trichome initiation in several other species such as sweet wormwood (Artemisia annua; Yan et al., 2018), cotton (Gossypium hirsutum; Wu et al., 2018), and poplar (Populus trichocarpa; Plett et al., 2010). Two Leucine HD-ZIP transcription factors have been identified as regulators of trichome development in tomato: Woolly (Yang et al., 2011) and Cutin Deficient2 (Nadakuduti et al., 2012). Woolly controls trichome initiation and specifically the morphogenesis of long, glandular type-I trichomes (Yang et al., 2011) and does so by forming a complex with a small cyclin, SlCycB2, which positively regulates mitotic divisions in multicellular trichomes (Gao et al., 2017). Cutin Deficient2 regulates cuticle deposition and the formation of glandular type-VI trichomes (Nadakuduti et al., 2012), and the development of this trichome type is also regulated by the bHLH transcription factor SlMyc1 (Xu et al., 2018). A C2H2 zinc finger transcription factor, Hair, was identified as a regulator of the formation of both type-I and type-VI trichomes and may control trichome initiation (Chang et al., 2018). Finally, the tomato homolog of TRYPTICHON (SlTry) is functionally equivalent to TRY when ectopically expressed in Arabidopsis, but its native function in tomato remains uncharacterized (Tominaga-Wada et al., 2013). In general, our current understanding of the regulation of trichome initiation and development in tomato is much more limited than for Arabidopsis.
The use of wild tomato species as a source of genetic variation has resulted in the identification of important quantitative trait loci for many traits (Rick and Chetelat, 1995), and has been undertaken traditionally by screening near-isogenic introgression lines (ILs), generated by successive backcrosses of the offspring of a cross between a wild relative species and a cultivated crop back to the cultivated parent (Eshed and Zamir, 1995). The most widely used IL population for tomato is the S. lycopersicum cv M82 × Solanum pennellii ac. LA716 IL population, which has been extensively curated and genotyped (Chitwood et al., 2013). Comprehensive analysis of this IL population has led to the identification of loci of interest that include tolerance to pathogens (Smart et al., 2007; Sharlach et al., 2013), abiotic stress (Frary et al., 2011; Rigano et al., 2014), primary metabolism (de Oliveira Silva et al., 2018), and morphogenesis (Chitwood et al., 2013; Ron et al., 2013). With respect to trichomes, extensive work on characterizing trichome secretion in the ILs revealed quantitative trait loci involved in the synthesis of acyl sugars and terpenoids (Schilmiller et al., 2010), and a visual assessment of trichome phenotypes in the IL population aided in the identification of Hair as a regulator of trichome formation (Chang et al., 2018). However, the aerial epidermis of the ILs has not been characterized fully and a detailed understanding of the degree of variability present in the population is lacking. Differences in trichome and stomatal densities and trichome types reported for S. lycopersicum and S. pennellii (Heichel and Anagnostakis, 1978; Simmons and Gurr, 2005; McDowell et al., 2011) support the use of the S. lycopersicum cv M82 × S. pennellii ac. LA716 ILs as a platform for investigating trichome development in tomato.
We have performed a comprehensive analysis of the leaf epidermis of two generations of the S. lycopersicum cv M82 × S. pennellii ac. LA716 ILs by a combination of scanning electron microscopy (SEM) techniques. The outputs of this study provide a large data resource for further research into cellular development and led to the identification of unexplored genomic regions associated with the determination of stomatal and trichome density in leaves, as well as trichome morphogenesis. The synteny between the tomato genome and important closely related crops like potato (Solanum tuberosum) and eggplant (Solanum melongena) as well as with more distant relatives in the Solanaceae family like pepper (Capsicum sp.) and tobacco means that this mapped resource of natural variation in trichome development could be indicative of genomic regions conferring effects on trichome formation in these other species, which will be of importance in identifying traits impacting drought tolerance (Galdon-Armero et al., 2018) and insect resistance (Schilmiller et al., 2010), among others.
RESULTS
Identification of Genomic Regions Involved in the Determination of Leaf Trichome Density
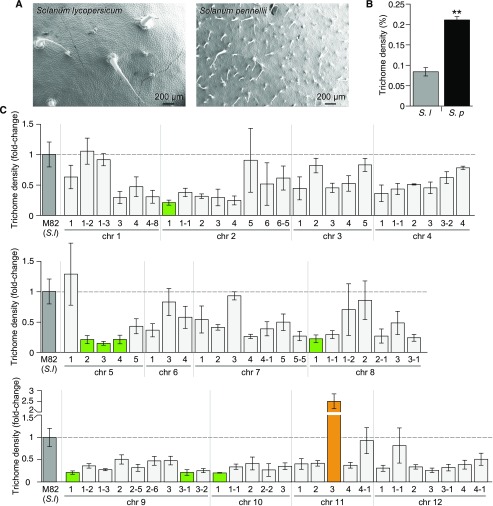
We evaluated the adaxial leaf epidermis of fully expanded first true leaves of the IL population, and we identified ILs consistently showing differences for specific parameters over two generations. We measured trichome density in the parental lines S. lycopersicum cv M82 and S. pennellii ac. LA716 (Figures 1A and 1B) and in the ILs over two generations (Figure 1C and Supplemental Figure 1).
Figure 1.
Trichome Densities of the S. lycopersicum cv M82 × S. pennellii ac. LA716 ILs.
(A) SEMs of the adaxial epidermis of a fully expanded leaf of S. lycopersicum (cv M82; left) and. S. pennellii (ac. LA716; right).
(B) Trichome density of the first fully expanded true leaf of S. lycopersicum cv M82 (gray bar) and S. pennellii ac. LA716 (black bar). Values are expressed as mean ± se (n = 3). Stars indicate significant differences (P-value < 0.01) according to t tests.
(C) Trichome density of the first generation of ILs, grouped according to the chromosomal location of the introgressed S. pennellii genomic region. Values are mean ± se (n = 3 to 4) of the relative value of trichome density compared with M82 values (medium gray bars). Significant differences were determined using the Dunnett’s test. Green bars indicate ILs with significantly lower trichome densities than M82 (P-value < 0.05) and orange bars indicate ILs with significantly higher trichome densities than M82 (P-value < 0.05). Light gray bars indicate other ILs.
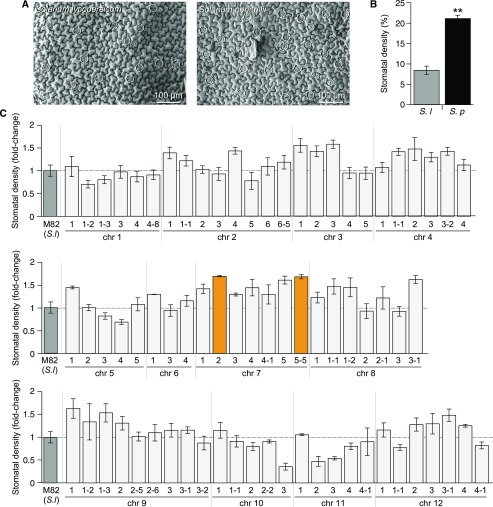
We found that trichome density on the adaxial surface of the first true leaves was threefold higher in S. pennellii ac. LA716 than in S. lycopersicum cv M82 (Figures 1A and 1B). This is in agreement with previous reports of trichomes in these Solanum species (Simmons and Gurr, 2005). We also measured stomatal density in S. lycopersicum and S. pennellii and found a twofold higher density in S. pennellii (Figures 2A and 2B), which contrasts with previous reports of stomatal density in S. pennellii, where they were lower than in S. lycopersicum (Heichel and Anagnostakis, 1978; Chitwood et al., 2013). This discrepancy may reflect differences between the developmental phase of the leaves assessed, measurement of density on the basis of stomata per unit area rather than per total cell number (stomatal index), or differences in environmental conditions during cultivation of the plants. The relatively large differences between the parental species, particularly in trichome density, supported the use of S. pennellii as a donor species to study epidermal development in tomato.
Figure 2.
Stomatal Density of the S. lycopersicum cv M82 × S. pennellii ac. LA716 ILs.
(A) SEMs of the adaxial epidermis of a fully expanded leaf of S. lycopersicum (cv M82; left) and S. pennellii (ac. LA716; right). White circles indicate stomatal locations.
(B) Stomatal density of the first fully expanded true leaf of S. lycopersicum cv M82 (gray bar) and S. pennellii ac. LA716 (black bar). Values are expressed as mean ± se (n = 3). Stars indicate significant differences (P-value < 0.01) according to a t test.
(C) Stomatal density of the first generation of ILs, grouped according to the chromosomal location of the introgressed S. pennellii genomic region. Values show the mean ± se (n = 3 to 4) of the relative value of stomatal density compared with M82 values (medium gray bars). Significant differences were determined using the Dunnett’s test. Orange bars indicate ILs with a significantly higher stomatal density than M82 (P-value < 0.05). Light gray bars indicate any other ILs.
We observed four ILs with low trichome density over the two generations (Figure 1C and Supplemental Figure 1): IL5-3 and IL5-4 on chromosome 5; IL7-2 on chromosome 7; IL9-3-1 on chromosome 9; and IL10-1 and IL10-3 on chromosome 10. IL11-3 had a significantly higher trichome density only in the first generation. Although not significantly different from M82 in the second generation, IL11-3 did average the highest value for trichome density out of all IL lines.
Identification of Genomic Regions Involved in the Determination of Leaf Stomatal Density
We measured the stomatal density of the S. lycopersicum cv M82 × S. pennellii ac. LA716 ILs (Figures 2A and 2B) over two generations (Figure 2C and Supplemental Figure 2). No IL showed consistently lower or higher stomatal density than M82, over either generation, suggesting that genetic factors are relatively unimportant in determining differences in stomatal density between these two tomato species.
Identification of Genomic Regions Involved in the Determination of Trichome Types
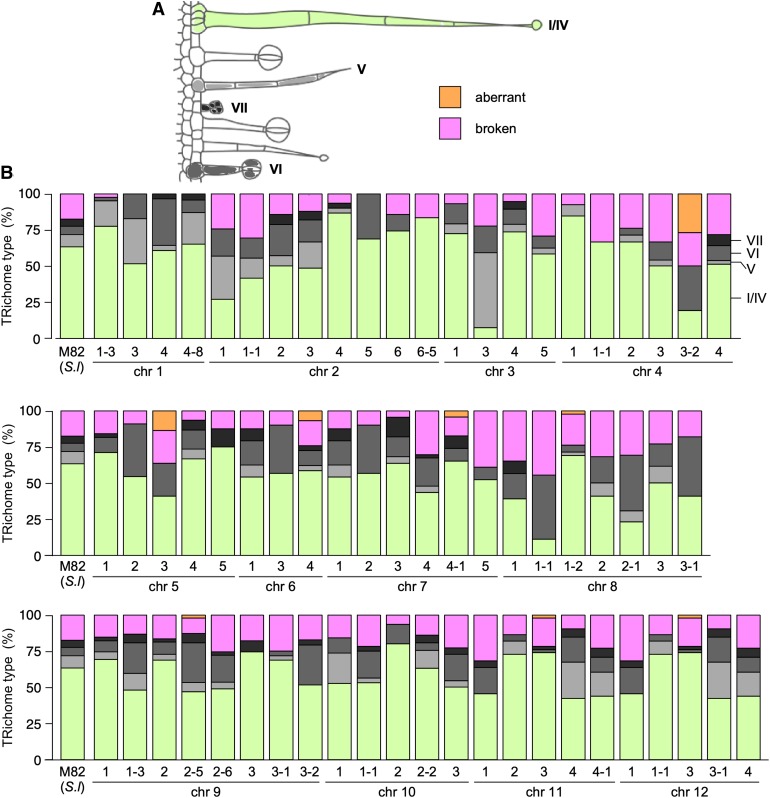
Trichomes were classified according to Luckwill (1943), although we grouped type I and type IV together based on their similar morphology and metabolic profiles as proposed by McDowell et al. (2011), and all nonglandular trichomes as type V (Figure 3 and Supplemental Figure 3). Although we examined leaves of the IL population over two generations, trichomes were extensively damaged by the use of chemical fixation and critical point drying of the samples in the first generation (Supplemental Figure 3). We therefore focused our analysis of trichome types on the data from the second generation (Figure 3). In M82, the main trichome type was type I/IV, accounting for 63% of the total. The other types of trichomes found in S. lycopersicum were observed in smaller percentages (5% to 10%). We used the distribution in M82 as a standard for comparison with the rest of the ILs. For most lines, type-I/IV trichomes were the most abundant, although IL2-1, IL3-3, IL4-3-2, IL8-1-1, and IL8-2-1 showed substantial reductions in this type of trichome. This reduction in type-I/IV trichomes was compensated by an increase in nonglandular type-V trichomes in IL2-1 and IL3-3. For 21 of the ILs, we did not observe any type-V trichomes on the adaxial surface of the first true leaf. Simmonds et al. (2005) reported that there were no type-V trichomes in S. pennellii and some or all of these 21 intervals may contain the genetic information required to specify the formation of these nonglandular trichomes in tomato, which were reported by Vendemiatti et al. (2017) to be typical of adult vegetative phase leaves. Type-VI trichomes were generally observed at low frequency, while type-VI trichomes were the most common type in IL8-1-1, IL8-2-1, and IL8-3-1. The high number of type-VI trichomes in IL8-1-1 could be due to the presence in that interval of the S. pennellii allele of MYC1 (Solyc08g005050.2.1), shown to be required for formation of type-VI trichomes in tomato (Xu et al., 2018). A total of six ILs (IL2-4, IL2-6-5, IL4-1, IL4-1-1, IL5-5, and IL9-3) showed no type-VI trichomes in the assessed tissue. Type-VII trichomes were the rarest and were absent on the first true leaves of 31 ILs (out of the 67 sampled in the second generation). It should be noted, however, that the absence of any one type of trichome from the adaxial epidermis of the first true leaf did not imply a complete absence of this trichome type in other tissues, as they were found in the epidermis over vascular tissues.
Figure 3.
Trichome Type Distribution of the S. lycopersicum cv M82 × S. pennellii ac. LA716 ILs.
(A) Classification scheme of trichome types in ILs, according to Luckwill (1943).
(B) Lines were classified according to the chromosomal region introgressed from S. pennellii and each bar represents an IL, and the height of each color section represents the proportion as a percentage of each type of trichome. Light green represents type-I/IV trichomes; light gray represents type-V trichomes; medium gray represents type-VI trichomes; dark gray represents type-VII trichomes; orange represents damaged trichomes; and magenta represents aberrant trichomes.
Identification of Genomic Regions Involved in Trichome Morphogenesis and Spatial Patterning
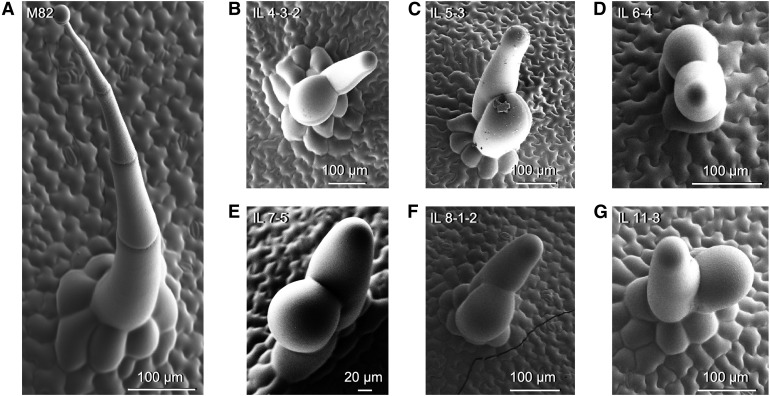
For each line under study, trichomes showing aberrant morphology were recorded (Figure 4 and Supplemental Figure 4). The most common type of aberrant trichome found in the population consisted of two swollen cells emerging from a type-I–like multicellular base. This type of trichome was observed in six ILs (IL4-3-2, IL5-3, IL6-4, IL7-5, IL8-1-2, and IL11-3; Figures 4B to 4G). Trichomes with aberrant division patterns were also found in three ILs (IL1-1-2, IL5-1, and IL7-4-1; Supplemental Figure 4A to 4C). In IL10-2, we observed another type of aberrant trichome consisting of branched, multicellular, nonglandular trichomes (Supplemental Figure 4D). In every IL with aberrant trichomes, the aberrant forms always appeared alongside wild-type trichomes of the same type, in both generations (Figure 3 and Supplemental Figure 3).
Figure 4.
Aberrant Trichome Morphologies in the IL Population.
(A) SEM micrograph of a representative type-I trichome on the leaf surface of M82. Micrographs of representative aberrant trichomes are shown for ILs.
(B) IL4-3-2.
(C) IL5-3.
(D) IL6-4.
(E) IL7-5.
(F) IL8-1-2.
(G) IL11-3.
(B) to (G) All sharing a similar swollen and forked appearance. Scale bars are shown in each micrograph.
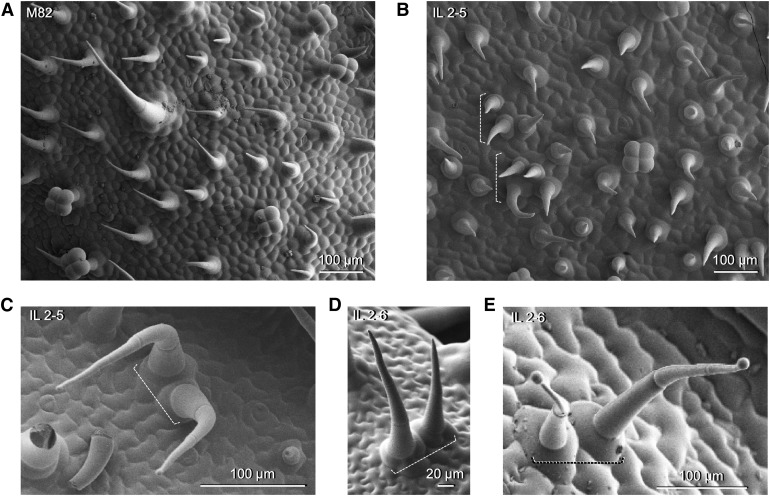
Finally, we observed unusual clusters of trichomes on the adaxial surface of leaves in IL2-5 and IL2-6 (Figure 5). In young leaves, trichomes in M82 are equally distributed on the leaf surface and they are oriented in the same direction (Figure 5A). However, trichomes in IL2-5 were clustered in groups of up to four trichomes and they were randomly oriented (Figure 5B). In mature, fully expanded leaves, we could still observe clusters of two trichomes in IL2-5 and IL2-6, and these observations were made for both glandular and nonglandular trichomes (Figure 5). Trichomes in these clusters were found oriented in more random directions with respect to each other than in M82.
Figure 5.
Abnormal Trichome Clusters in the IL Population.
(A) SEM micrograph of the adaxial surface of a young leaf of M82.
(B) SEM micrograph of the adaxial surface of a young leaf of IL2-5.
(C) SEM micrograph of a cluster of two type-IV trichomes in IL2-5.
(D) SEM micrograph of a cluster of two type-V trichomes in IL2-6.
(E) SEM micrograph of a cluster of two type-IV trichomes in IL2-6.
(A) to (E) Scale bars are shown for each micrograph. White dashed brackets indicate clusters of trichomes.
Characterization of Candidate Genes Underlying the Trichome Cluster Phenotype in IL2-5 and IL2-6
As a proof-of-principle, we characterized the genetic basis for trichome clustering on leaves observed in IL2-5 and IL2-6. We screened candidate genes contained in the genomic region of chromosome 2 covered by IL2-5 and IL2-6 (Supplemental Figure 5A). The most likely bin to contain a gene responsible for the phenotype was bin 2-L, which corresponds to the overlapping region between IL2-5 and IL2-6 displaying the clustered phenotype, but excludes IL2-6-5, which did not show trichome clustering (Chitwood et al., 2013). Bin 2-K was also searched, as the borders of the IL regions are sometimes not very well defined. The genes were filtered based on their annotation (Fernandez-Pozo et al., 2015), with a preference for those genes with annotation terms related to trichome development. SlMixta-like, found in bin 2-K, was selected as a gene encoding a protein linked to trichome development. SlMixta-like is expressed in trichomes (Balcke et al., 2017) and its expression is substantially lower in leaves of IL2-6 compared with other ILs (Supplemental Figures 5B and 5C). SlMixta-like is an ortholog of the snapdragon Mixta-like2 gene (AmMYBML2) and of the petunia (Petunia hybrida) gene (PhMyb1) involved in conical cell formation in flowers and fruit (Perez-Rodriguez et al., 2005; Baumann et al., 2007; Lashbrooke et al., 2015).
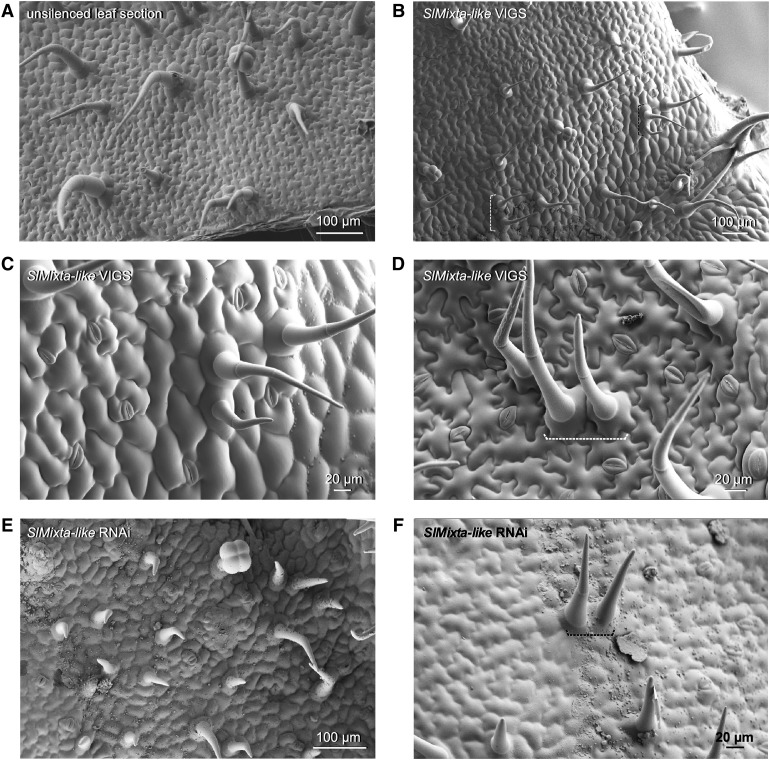
An effective reduction of SlMixta-like expression in tissues silenced by Virus Induced Gene Silencing (VIGS) in tomato seedlings (Supplemental Figure 6) resulted in the development of trichome clusters on the adaxial (Figures 6B and 6C) and abaxial (Figure 6D) leaf surface of silenced regions, similar to the clusters observed in IL2-5 and IL2-6.
Figure 6.
Virus-Induced Gene Silencing of SlMixta-like in Tomato Leaves.
(A) SEM micrograph of the adaxial surface of an unsilenced region of a leaf.
(B) SEM micrograph of the adaxial surface of leaf sections where SlMixta-like was silenced.
(C) Close-up view of a cluster of trichomes on the adaxial surface of leaf tissue where SlMixta-like was silenced.
(D) Close-up view of a cluster of trichomes on the abaxial surface of leaf tissue where SlMixta-like was silenced.
(E) SEM micrograph of the adaxial surface of a leaf from a SlMixta-like RNAi line.
(F) Close-up view of a cluster of trichomes on the adaxial surface of a leaf from a SlMixta-like RNAi line.
(A) to (F) Scale bars are shown in each micrograph. White or black dashed brackets indicate clusters of trichomes.
SlMixta-like (Solyc02g088190) function has been described in tomato lines in which its expression had been lowered via RNA interference (RNAi; Lashbrooke et al., 2015). Silencing of SlMixta-like had two main effects: (1) flattening of conical cells in the epidermis of the fruit; and (2) a reduction of cuticle thickness in fruit. Although silencing also had some pleiotropic effects on vegetative growth, including slower growth and curling and wrinkling of leaves, no effect was reported on conical cells in petals nor on flower trichomes, which suggested that SlMixta-like might have a highly specialized function in fruit in tomato, compared with the role of its ortholog PhMyb1 in petunia (Baumann et al., 2007). To confirm the phenotype of low SlMixta-like expression in tomato, we examined the leaves of an RNAi line, kindly supplied to us by Asaph Aharoni (Lashbrooke et al., 2015). We found numerous examples of clustered trichomes (Figures 6E and 6F), confirming that low expression of SlMixta-like is associated with aberrant spacing of trichomes resulting in trichome clustering.
Analysis of Knock-out Alleles of SlMixta-like in Tomato
Four independent Slmixta-like mutant lines were generated by CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats and CRISPR-Associated nuclease) genome editing, and are designated ko no. 1 to no. 4. We targeted two sequences in the SlMixta-like gene, one in the 5′ untranslated region (5′UTR) and the other in the second exon, using two different single guide RNAs (sgRNAs; Supplemental Figure 7A). Both sgRNAs were effective and deletions were observed in both regions (Supplemental Figures 7B and 7C). The mixta-like line ko no. 1 was selected for further analysis as the 2-bp deletion in the coding sequence caused a translational frameshift that introduced an early stop codon. If translated, this genome-edited allele would lead to the production of a truncated 34-amino acid protein, compared with the 417 amino acids of the wild-type allele (Supplemental Figure 7C). Homozygous Slmixta-like ko no. 1 T1 plants were used for detailed analysis. We also analyzed the epidermis of two additional independent ko lines, no.3 and no. 4, for functional confirmation. Both ko no. 3 and ko no. 4 had the same 1-bp insertion, causing a frameshift and a predicted truncated protein (Supplemental Figure 7B).
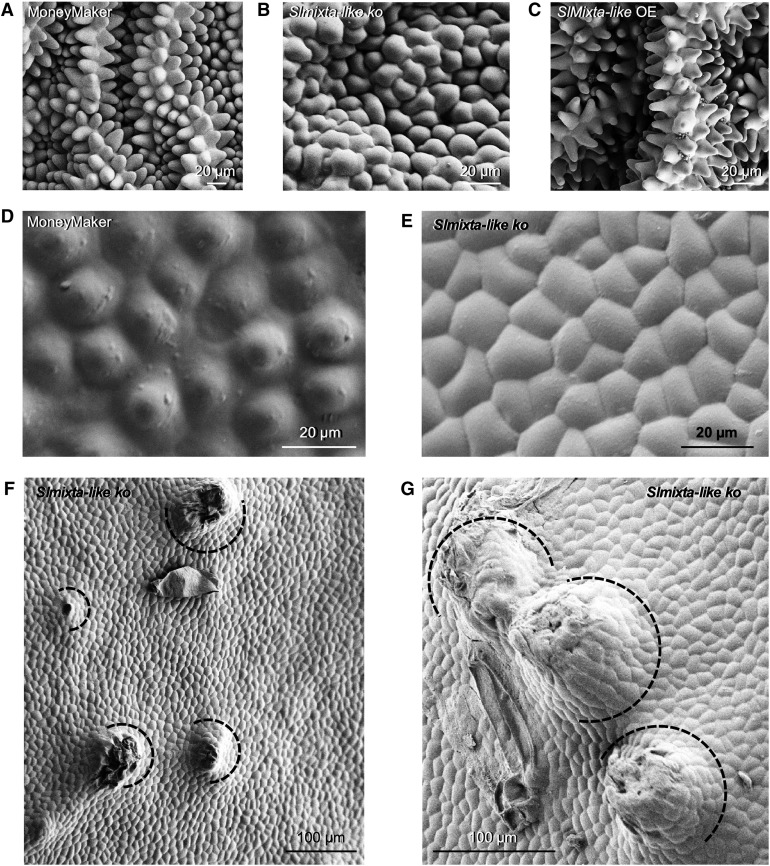
Conical cells in petals were flattened in Slmixta-like ko no. 1, compared with control petals (Figures 7A and 7B), although upon close inspection they were not completely flat, and very similar to the petal epidermal cells of the petunia phmyb1 mutant (Baumann et al., 2007). A similar situation was observed on the fruit surface. Conical cells were flatter in Slmixta-like ko no. 1 (Figure 7E), as shown previously by Lashbrooke et al. (2015) for their RNAi lines. Interestingly, fruits of Slmixta-like ko no. 1 also had protuberances all over their surface, some of them with trichomes on (Figures 7F and 7G). This gave a very rough feeling to the tomato skin when touched with bare hands, indicative of altered cuticular wax production.
Figure 7.
Effect of SlMixta-like on the Formation of Conical Cells in Tomato.
(A) SEM micrograph of conical cells in petals of a MoneyMaker plant.
(B) SEM micrograph of conical cells in petals of the Slmixta-like ko line no. 1, showing a flatter shape.
(C) SEM micrograph of conical cells in petals of a SlMixta-like OE plant, showing more elongated and pointed shape.
(D) SEM micrograph of the conical cells in the epidermis of a mature green fruit of a MoneyMaker plant.
(E) SEM micrograph of the conical cells in the epidermis of a mature green fruit of the SlMixta-like KO line no. 1, showing much flatter cells. Scale bars are shown for each micrograph.
(F) SEM micrograph of the epidermis of a mature green fruit of the Slmixta-like ko line no. 1.
(G) Close-up view of protuberances on the surface of mature green fruits of the Slmixta-like ko line no. 1.
(F) and (G) Black semicircles indicate multicellular protuberances.
(A) to (G) Scale bars are shown in each micrograph.
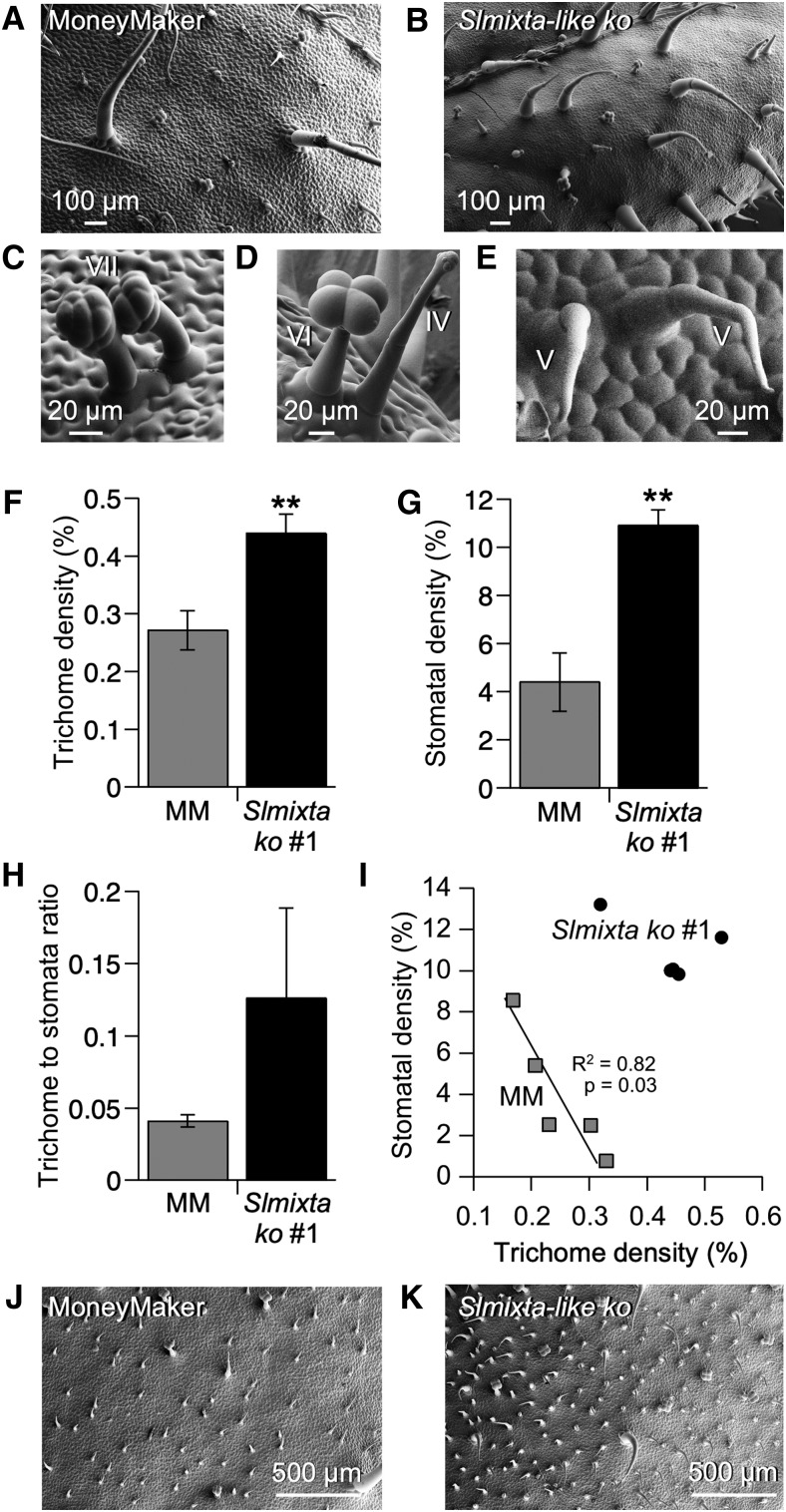
A higher abundance of trichomes was observed on the adaxial surface of Slmixta-like ko no. 1 leaves (Figures 8A and 8B). Clusters of two trichomes were also observed for type I/IV (Figure 8B), type-VII (Figure 8C), type-VI (Figure 8D), and type-V (Figure 8E) trichomes.
Figure 8.
Analysis of the Leaf Epidermis of Slmixta-like ko Line No. 1.
(A) SEM micrograph of the adaxial surface of the first fully expanded leaf of S. lycopersicum cv MoneyMaker.
(B) Representative SEM micrograph of the adaxial surface of the first fully expanded leaf of a Slmixta-like ko line no. 1.
(C) Close-up view of a cluster of two type-VII trichomes in Slmixta-like ko line no. 1.
(D) Close-up view of a cluster of a type-VI and a type-IV trichome in Slmixta-like ko line no. 1.
(E) Close-up view of a cluster of two type-V trichomes in Slmixta-like ko line no. 1.
(A) to (E) Scale bars are shown in each micrograph.
(F) Trichome density in control leaves (gray bar) and leaves from Slmixta-like ko line no. 1 (black bar).
(G) Stomatal density in control leaves (gray bar) and leaves from ko line no. 1 (black bar).
(H) Trichome-to-stomata ratio of control tomato leaves (gray bar) and ko line no. 1 (black bar).
(I) Correlation between trichome and stomatal density in the assessed leaves. Gray squares correspond to values from control leaves; black circles correspond to Slmixta-like ko line no. 1. A significant correlation was only observed in control leaves. Correlation index and P-values are shown in the graph.
For (F), (G), and (H), values are expressed as mean ± se (n = 5). Two stars indicate a P-value < 0.01 according to a t test between values corresponding to each line.
(J) SEM micrograph of the adaxial surface of a mature leaf (metamer > 5) of S. lycopersicum cv MoneyMaker.
(K) SEM micrograph of the adaxial surface of a mature leaf (metamer > 5) of Slmixta-like ko line no. 1.
(J) and (K) Scale bars are shown in each micrograph.
When trichome and stomatal densities were quantified, Slmixta-like ko no. 1 showed a significantly higher trichome density compared with leaves from the parental cultivar MoneyMaker (Figure 8F). Stomatal density was also significantly higher in Slmixta-like ko no. 1 (Figure 8G), but consequently, the trichome-to-stomata ratio was not statistically significantly different between wild type and Slmixta-like ko no. 1 (Figure 8H). Interestingly, the previously reported correlation between trichome and stomatal density was maintained when the data-points for control MoneyMaker leaves were used, while this correlation was not observed when the data-points for Slmixta-like ko no. 1 were included or when all data-points for both lines were taken together (Figure 8I). We confirmed that the increase in trichome density was also observed in adult-phase leaves (metamer > 6) of Slmixta-like ko no. 1 compared with MoneyMaker (Figures 8J and 8K).
We analyzed two other ko lines (no. 3 and no. 4) to confirm our characterization of SlMixta-like function in leaves (Supplemental Figure 8). Both lines showed higher trichome density than the control MoneyMaker leaves (Supplemental Figure 8A), confirming our observations for Slmixta-like ko no. 1. Stomatal density was not different from the control in Slmixta-like ko no. 3 but was higher in Slmixta-like ko no. 4, in line with the observations for Slmixta-like ko no. 1 (Supplemental Figure 8B).
The trichomes on the leaves of Slmixta-like ko no. 1 plants were classified into types according to Luckwill (1943) and Simmons and Gurr (2005). Type-I/IV trichome density was significantly higher in Slmixta-like ko no. 1 compared with control MoneyMaker leaves (Supplemental Figure 9B), but the densities of other trichome types were not affected significantly. When the percentages of each trichome type were compared, the percentage of type-I and type-IV trichomes was significantly different between Slmixta-like ko no. 1 and control lines (Supplemental Figures 9B and 9C). No aberrant trichomes were observed in any of the Slmixta-like ko lines.
No significant differences in abundance of specific types of trichomes were observed in Slmixta-like ko no. 3 or no. 4 except for a greater abundance of type-VI trichomes in Slmixta-like ko no. 4 (Supplemental Figures 9D and 9E). When percentages were considered, no significant differences were found in trichome types between any of the Slmixta-like ko lines and wild-type controls.
Analysis of the Leaf Epidermis in SlMixta-like Overexpression Lines
Seven independent SlMixta-like overexpression (OE) lines were generated, driven by the Cauliflower Mosaic Virus 35S promoter. The abundance of the SlMixta-like transcript in the leaves of four of these OE lines was analyzed by RT-qPCR, and three lines (OEs no. 1 to no. 3) with significantly higher expression levels of SlMixta-like (Supplemental Figure 10) were analyzed phenotypically in the T1 generation. We also analyzed an SpMixta-like OE line (Supplemental Figure 11) to compare the functionality of the SlMixta-like proteins in S. lycopersicum and S. pennellii.
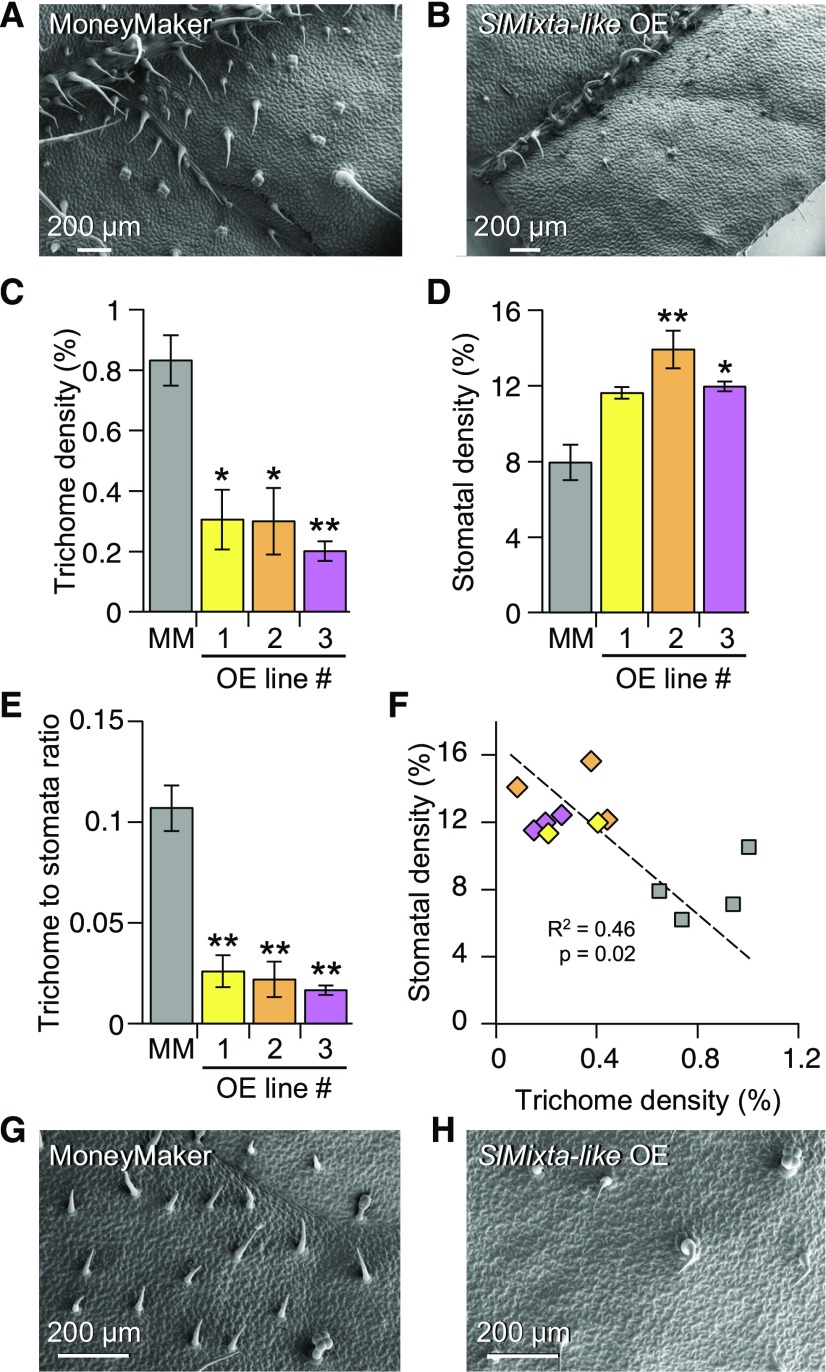
We observed a low trichome number on the adaxial surface of SlMixta-like OE lines (Figures 9A to 9C). When quantified, the three OE lines had significantly lower trichome densities compared with the MoneyMaker parent (Figure 9C). Alongside the change in trichome density, we observed an increase in stomatal density in all three OE lines (Figure 9D), although this was statistically significant for only OE lines no. 2 and no. 3 (Figure 9D). The OE lines had a lower trichome-to-stomata ratio when compared with MoneyMaker (Figure 9E), and a negative association between trichome and stomatal density was observed when all data-points were considered (Figure 9F). In the SpMixta-like OE line, trichome density was lower than in the MoneyMaker control (Supplemental Figure 11B), implying that SlMixta-like and SpMixta-like have very similar functionalities and that it is their different levels of expression in M82 and IL2-6, respectively, that determine the trichome clustering phenotype of IL2-6. Confirming this hypothesis, comparisons of the transcripts of SlMixta-like and SpMixta-like suggested that the lower expression of SpMixta-like might result from the use of a different and less efficient TATA box and transcriptional start site than SlMixta-like (Supplemental Figure 11D).
Figure 9.
Analysis of the Leaf Epidermis of SlMixta-like OE Lines.
(A) SEM micrograph of the adaxial surface of the first fully expanded leaf of S. lycopersicum cv MoneyMaker.
(B) Representative SEM micrograph of the adaxial surface of the first fully expanded leaf of a SlMixta-like OE line.
(A) and (B) Scale bars are shown in each micrograph.
(C) Trichome density in control leaves (gray bar) and leaves from three independent OE lines (yellow, orange, and magenta bars).
For (A), (B), and (C), n = 4 for control leaves, n = 2 for OE no. 1 and n = 3 for OE no. 2 and no. 3.*P-value < 0.05; **P-value < 0.01 according to a t test between the corresponding OE line and the control value.
(D) Stomatal density in control leaves (gray bar) and leaves from three independent OE lines (yellow, orange, and magenta bars).
(E) Trichome-to-stomata ratio in control leaves (gray bar) and leaves from three independent OE lines (yellow, orange, and magenta bars).
(F) Correlation between trichome and stomatal density in the assessed leaves. Gray squares correspond to values from control leaves, while diamonds correspond to OE lines (yellow, OE no. 1 values; orange, OE no. 2 values; magenta, OEno. 3 values). Correlation index and P-value are shown in the graph.
(G) SEM micrograph of the adaxial surface of a mature leaf (metamer > 5) of S. lycopersicum cv MoneyMaker.
(H) SEM micrograph of the adaxial surface of a mature leaf (metamer > 5) of a SlMixta-like OE line.
(A), (B), (G), and (H) Scale bars are shown in each micrograph.
We scored trichome types according to the categories of Luckwill (1943) and Simmons and Gurr (2005) in our OE lines to determine whether the observed changes in trichome density affected specific trichome types. No differences in the density of type-I/IV or -VII trichomes were observed between parental and OE lines. However, the density of type-V trichomes was lower in OE no. 2 and OE no. 3 lines compared with the MoneyMaker parent. Similarly, the density of type-VI trichomes was lower in OE no. 1 and OE no. 3 compared with the parental line (Supplemental Figure 12). The percentage of each type of trichome relative to the total trichome number showed no differences between MoneyMaker and OE lines (Supplemental Figure 12). In OE lines no. 2 and no. 3, which had the highest expression of SlMixta-like, swollen aberrant trichomes were also observed (Supplemental Figure 12).
It has been reported that the predominance of type-I/IV trichomes on the first true leaf of tomato represents a juvenile trait and that leaves from higher metamers (>5) represent the adult phase and carry predominantly type-V trichomes (Vendemiatti et al., 2017). To investigate whether SlMixta-like affected trichome density in the adult phase, we compared a leaflet from an older leaf metamer (>6) from Slmixta-like ko no. 1, and an SlMixta-like OE line to wild-type (MoneyMaker). Slmixta-like ko no. 1 showed a higher density of trichomes, predominantly of type V, compared with MoneyMaker (Figures 8J and 8K) and the OE line showed lower trichome density compared with MoneyMaker (Figures 9G and 9H), confirming that SlMixta-like exerts its negative effect on trichome density/patterning in leaves during both the juvenile and adult vegetative phases.
DISCUSSION
Natural Variation in Epidermal Development in the S. lycopersicum cv M82 x S. pennellii ac. LA716 ILs
Through comparison to S. pennellii ac. LA716 and S. lycopersicum cv M82 (Figures 1A and 2A) we identified four ILs that showed consistent trichome phenotypes over two generations, suggesting stable genetic components regulating trichome density in these ILs. Interestingly, most of the ILs analyzed here showed lower trichome density than M82, and these differences were consistent in both generations of the IL population (Figure 1B and Supplemental Figure 1B). This was unexpected, given the values observed for the parents (Figures 1A to 1C). However, the complexity of trichome development, which involves cell wall expansion, cell division, and differentiation (Glover, 2000), and the negative and positive control exerted by different regulatory factors (Tian et al., 2017), could well be impaired by small changes in the activity or expression of alleles introgressed in the ILs. To the best of our knowledge, no genes currently known to be involved in trichome development map to the genomic regions corresponding to the four ILs identified. Further analysis of these ILs will be necessary to determine the factors involved in the determination of trichome density.
We assessed trichomes on the adaxial surface of the first true leaf of two generations of the ILs (Supplemental Figure 13), using chemical fixation coupled with room temperature SEM for the first generation (Supplemental Figure 3) and cryo-SEM for the second generation (Figure 3). The most abundant trichome type in M82 found on the adaxial surface of the first true leaf and on most ILs was type I/IV (Figure 3), in contrast to previous reports, which ranked type-VI trichomes as the most common glandular trichomes (Bergau et al., 2015). However, the density of type-I trichomes has been reported to be a juvenility trait, being very high in the first leaf—the leaf we assessed (Figure 3 and Supplemental Figure 3; Vendemiatti et al., 2017). We observed five lines with a low proportion of type-I/IV trichomes (Figure 3). In the case of IL2-1 and IL3-3, this was compensated by an increase in the percentage of type-V trichomes, in contrast to the observations of Vendemiatti et al. (2017), making these good candidate lines for further research into the control of the juvenile phase in vegetative development of tomato. In the case of IL8-1-1 and IL8-2-1, the observed reduction in type-I/IV trichomes was accompanied by an increased proportion of type-VI trichomes. This could indicate that different types of trichomes are determined by different regulatory mechanisms but that these mechanisms might be interlinked to compensate for alterations in the distribution between trichome types, in agreement with previous reports in tomato (Li et al., 2004; Yang et al., 2011) and tobacco (Payne et al., 1999).
Type-VI trichomes were relatively rare in the sampled leaves, but absent only in six ILs of the population. IL4-1 had no type-VI trichomes (Figure 3). However, although we could not find type-VI trichomes on the adaxial surface of the first true leaves, they were present on stems and major veins, suggesting that IL4-1 influences tissue-specific regulation of epidermal development, which has been reported in other species such as Arabidopsis (Schnittger et al., 1998) and snapdragon (Glover et al., 1998). Type-VII trichomes were rare in the interveinal leaf tissue of all lines, and indeed were completely absent in almost half of the lines (Figure 3). This type of trichome was, however, commonly found on major and minor veins, confirming that their number is higher in epidermis overlying vascular tissues.
We found several ILs with aberrant trichome morphologies (Figure 4 and Supplemental Figure 4) and unusual epidermal patterning (Figure 5). The most common aberrant morphology consisted of a multicellular base similar to that found in type-I trichomes, with two swollen cells emerging from this base (Figure 4). These structures were reminiscent of the aberrant type-I trichomes observed in the odorless-2 (Kang et al., 2010) and dialytic mutants (Chang et al., 2016). The swollen appearance of these trichomes was similar to that observed in the hairless mutant (Kang et al., 2016), which carries a partial deletion of the highly conserved Specifically-Rac1 Associated (SRA1) gene, encoding a subunit of the WAVE regulatory complex, and is thus compromised in the organization of the actin cytoskeleton in trichomes, or the inquieta mutant, which harbors an altered copy of the Actin-Related Protein 2/3 Complex Subunit 2a (ARPC2a) (Jeong et al., 2017). It is possible that the observed phenotypes are due to alterations in the processes of cytoskeletal reorganization or cell elongation in trichomes. In fact, the ARPC2a gene maps to the IL 11-3 region of tomato (Figure 4G). The Actin-Related Protein 3 (ARP3) and Actin-Related Protein 2/3 Complex Subunit 3 (ARPC3) genes encode proteins that are part of the complex required for actin filament formation (Goley and Welch, 2006), and map to the IL4-3-2 and IL7-5 regions, respectively (Figures 4B and 4E). All of these aberrant trichomes appear amid wild-type–looking trichomes (Figure 3 and Supplemental Figure 3), suggesting phenotypes caused by small changes in expression or functionality of S. pennellii alleles in the S. lycopersicum genetic background rather than gain- or loss-of-function differences between the two species. We also observed branched, multicellular, nonglandular trichomes in IL10-2 that lacked a multicellular base (Supplemental Figure 4D). This structure resembles the type-V–like trichomes observed in SlCycB2 or Woolly RNAi lines (Yang et al., 2011). Satisfyingly, this IL delimits a genomic region that covers CycB2, and carries the SpCycB2 allele, indicating that the S. pennellii allele might have reduced activity compared with its S. lycopersicum counterpart. Finally, we observed an unusual clustering of trichomes in IL2-5 and IL2-6 (Figure 5). Trichomes are generally evenly distributed over the leaf surface in tomato (Figure 5A), although the mechanisms by which this patterning is determined are unclear. In Arabidopsis, trichome patterning is mediated by small MYB transcription factors that inhibit trichome initiation in cells adjacent to newly formed trichomes (Hauser, 2014), but the mechanism in tomato and related species is not understood (Tominaga-Wada et al., 2013).
We also measured the stomatal density for each IL line (Figure 2 and Supplemental Figure 2). We identified no ILs with consistently higher or lower stomatal density over either generation. Interestingly, most of the ILs showed a higher stomatal density than the parental line M82 (Figure 2 and Supplemental Figure 2). These observations suggested that natural variation may be relatively unimportant in determining stomatal density in S. lycopersicum and S. pennellii, and that environmental conditions may be more relevant. However, Slmixta-like genome-edited lines did impact stomatal density, although we did not detect this as a phenotype in IL2-6. Analysis across the ILs suggested a possible developmental link between the two epidermal cell types, perhaps involving an early commitment of cell fate to trichome formation that prevents stomatal formation thereafter as inferred for tobacco (Glover et al., 1998) and tomato under drought conditions (Galdon-Armero et al., 2018). The absence of consistent stomatal phenotypes in the ILs highlights the importance of environmental and developmental factors in the control of stomatal density.
To confirm that the S. lycopersicum cv M82 × S. pennellii ac. LA716 IL population could be used to identify new biological functions in trichome initiation and distribution on leaves, we investigated the genetic basis of trichome clustering in IL2-5 and IL2-6. Molecular analysis suggested that this was likely a function of low expression of the SlMixta-like gene, as confirmed by RNaseq and RT-qPCR analyzes (Supplemental Figures 5B and 5C), reflecting a newly defined role for this transcription factor in ensuring patterning and density of trichomes in tomato leaves. The predicted amino acid sequences of SlMixta-like and SpMixta-like are highly similar, especially over their DNA binding domains (Supplemental Figure 11A), suggesting similar functionalities, which were confirmed by overexpression of both alleles in S. lycopersicum, where they both suppressed trichome density on leaves (Supplemental Figures 11B and 11C). A comparison of the sequences of the 5′UTRs and proximal promoter regions of SlMixta-like and SpMixta-like genes revealed 6 bp missing at the start of transcription in SpMixta-like compared with SlMixta-like (Supplemental Figure 11D). A single SpMixta-like transcript was identified in all the RNA-seq databases, and appeared to have an alternative start of transcription to SlMixta-like. The absence of these 6 bp in the SpMixta-like 5′UTR may result in the use of this alternative transcriptional start site compared with SlMixta-like and account for the relatively low level of expression of SpMixta-like in leaves.
SlMixta-like has been defined as a positive regulator of conical cells and cutin biosynthesis in tomato fruit by Lashbrooke et al., (2015), which we confirmed using Slmixta-like ko plants generated by CRISPR/Cas9 genome editing (Figure 7). The tomato skin of Slmixta-like ko no. 1 was very rough to the touch, even though the cells on the surface were flatter. This rough feeling of the fruit was observed in all the different ko mutants generated (Supplemental Figure 7B), and at all different stages of fruit development. This effect could be due to defects in cuticle formation given the role of SlMixta-like in controlling cutin synthesis and deposition (Lashbrooke et al., 2015), although after examining the tomato surface of Slmixta-like ko lines under SEM, protrusions on the epidermis were observed (Figures 7F and 7G) that were not present on the surface of MoneyMaker fruit. Some of these protrusions had trichomes on top that might also impact the texture of the fruit. The cause of the formation of these multicellular outgrowths was unclear; whether they were caused directly by the absence of SlMixta-like activity or whether they are a mechanical consequences of the altered, flatter fruit surface needs further investigation.
We also observed that petals of Slmixta-like ko lines had much flatter conical cells compared with MoneyMaker petals, while SlMixta-like OE petals had more elongated conical cells (Figures 7B and 7C), in agreement with reports on the function of SlMixta-like orthologs, including PhMYB1 in petunia, AtMYB16 in Arabidopsis and AmMixta-like2 in snapdragon (Baumann et al., 2007). The appearance of the petal epidermal cells in Slmixta-like ko plants was not completely flat (Figure 7B), suggesting that other factors are necessary for initiation of conical cells in tomato petals, in a similar way to the distinct functions of Mixta and Mixta-like2 in snapdragon (Baumann et al., 2007). This function of SlMixta-like was not reported by Lashbrooke et al., (2015) using RNAi silencing of SlMixta-like in tomato, suggesting that even low levels of expression may be sufficient to fulfill this function of SlMixta-like in petals.
SlMixta-like is a Negative Regulator of Trichome Initiation
In Slmixta-like ko lines, trichome density was increased compared with control MoneyMaker plants (Figures 8A and 8B). Consistent with these observations, trichome density was reduced ∼3-fold when SlMixta-like was overexpressed in the three independent lines analyzed (Figures 9A and 9B). Therefore, SlMixta-like must function as a negative regulator of trichome initiation in tomato leaves. This function contrasts with the functions of other Mixta and Mixta-like proteins. In tomato, SlMx1, which belongs to a separate phylogenetic clade of MYB subgroup-9 proteins (Brockington et al., 2013), is a positive regulator of trichome density (Ewas et al., 2016) and SlMixta-like itself positively regulates conical cell development in fruit (Lashbrooke et al., 2015) and petals (Figure 7). In other species, most Mixta-like genes have been characterized as positive regulators of conical cell or trichome formation (Brockington et al., 2013). Examples are PhMyb1 in petunia, AmMixta-like2 in snapdragon, and AtMYB16 in Arabidopsis, which all encode close homologs of SlMixta-like and positively regulate conical cell development in their respective species (Baumann et al., 2007). Interestingly, AtMYB16 regulates branching of trichomes and wax deposition in vegetative tissues in Arabidopsis (Oshima et al., 2013) pointing to a dual function in vegetative and reproductive organs. Multiple functionalities of Mixta/Mixta-like transcription factors in different cell types have also been reported in snapdragon (Perez-Rodriguez et al., 2005)
In terms of trichome regulation, most Mixta-like proteins are positive regulators. This is the case of poplar PtMYB186 (Plett et al., 2010), AaMixta1 in A. annua (Shi et al., 2018), GhMYB25 in cotton (Machado et al., 2009), and MtMYBML3 in Medicago truncatula (Gilding and Marks, 2010). However, some genes in this family are also negative regulators of trichome development. In Monkey flower (Mimulus guttatus), the expression of MgMYBML8 correlated negatively with trichome density (Scoville et al., 2011). In Arabidopsis, MYB106 acts as both a positive and negative regulator of trichome development, affecting branching of trichomes (Gilding and Marks, 2010). Recently, a Mixta-like protein in cucumber (Cucumis sativus), CsMYB6, was described as a negative regulator of trichome development on the fruit surface (Yang et al., 2018), acting in a very similar fashion to SlMixta-like in tomato. This underscores the pleiotropic effects of Mixta/Mixta-like proteins in plants and supports a role for SlMixta-like as a negative regulator of trichome initiation in leaves. It is important to note that SlMixta-like OE lines were not completely glabrous (Figures 9B and 9C) and Slmixta-like ko lines were not extremely pubescent (Figures 8B and 8F), both showing a change in trichome density of ∼2-fold compared with leaves of their parental lines (Figures 8 and 9). This suggests that the function of this gene is not sufficient to inhibit trichome formation completely, and it is probably part of a network that also involves positive regulators of trichome development in tomato, such as SlMx1 (Ewas et al., 2016) and Woolly (Yang et al., 2011).
SlMixta-like appears to play a role in epidermal patterning and spatial distribution of trichomes in leaves. This new role of SlMixta-like was identified in the screen of the IL population by the presence of clusters of two or three misoriented trichomes in IL2-5 and IL2-6. The lower expression of SlMixta-like in leaves of the ILs (Chitwood et al., 2013), especially in IL2-6 (Supplemental Figures 5B and 5C), suggested the involvement of this gene in the clustered trichome phenotype, and that this difference might contribute to the higher trichome density in S. pennellii compared with S. lycopersicum (M82; Figure 1C). Its role in preventing trichome clustering was confirmed by transient silencing in tomato, which gave rise to clustered trichomes (Figure 6). The consequences of low or no expression of SlMixta-like were reminiscent of loss-of-function mutants of a group of negative trichome regulators in Arabidopsis (TRY, CPC, ETC1, ETC2, ETC3, TCL1, and TCL2). Analysis of SlMixta-like OE and ko lines showed that OE lines did not have any clusters, but that ko lines had trichome clusters that affected all trichome types (Figure 8). However, any mechanism for determination of epidermal patterning involving a Mixta-related protein is probably distinct from the regulation of trichome patterning in Arabidopsis, where the small R3 MYBs prevent trichome initiation in cells adjacent to a newly initiated trichome (Wada et al., 1997; Schnittger et al., 1999; Ishida et al., 2008) by cell-to-cell movement and competitive inhibition of the MBW complex. Indeed, it is unlikely that SlMixta-like can move between cells because of the size of the protein. The existence of discrete sectors of revertant, fully conical cells, where the dTph1 transposon has excised from the orthologous PhMYB1 gene in petunia, also argues strongly for a cell-autonomous function of SlMixta-like (Baumann et al., 2007). It is possible that this inhibitory function of SlMixta-like is indirect, involving competition for DNA binding sites with trichome-initiating Mixta transcription factors such as SlMx1. Alternatively, other proteins may interact with SlMixta-like to prevent trichome clustering, similar to trichome clustering mutants in Arabidopsis: for example, try cpc double mutants have a much higher clustering frequency than the single mutants (Schellmann et al., 2002).
We tried to determine whether the effects of SlMixta-like gain- or loss-of-function were specific for particular trichome types (Supplemental Figures 8 and 9). In SlMixta-like OE lines, type-I/IV trichome density was unaffected, while type-V and type-VI trichome densities were specifically reduced (Supplemental Figure 9). However, when the percentage of each trichome type was compared between OE and control lines, no significant differences were apparent (Figure 9). The observed densities of type-V and type-VI trichomes were probably the result of the lower numbers of trichomes in OE lines and the high frequency of broken trichomes may also have masked effects of SlMixta-like on type-V and type-VI trichomes. In Slmixta-like ko mutants, cluster formation was trichome type-independent (Figure 8), suggesting that patterning and spatial distribution of trichomes are determined before type specification takes place, as suggested in studies of type-VI trichome development in tomato species (Bergau et al., 2015). Analyzing the effect of the mutation on trichome density, the increase in Slmixta-like ko lines affected type I/IV in ko line no. 1 and type VI in ko line no. 4 in lines grown at different times of the year, suggesting that initiation and trichome type determination are independent processes. If trichome initiation takes place before trichome identity has been determined (Bergau et al., 2015) and SlMixta-like negatively influences trichome initiation, this difference could be the result of independent mechanisms for trichome type determination.
SlMixta-like is Essential for Maintenance of the Negative Association between Trichome and Stomatal Development
In SlMixta-like OE lines, the decrease in trichome density (Figure 9C) is accompanied by an increase in stomatal density (Figure 9D). In fact, this change caused a significant decrease in the trichome-to-stomata ratio (Figure 9E), which has been suggested to be an important physiological parameter in tomato. A significant negative correlation between trichome density and stomatal density was observed when all data-points were considered together (Figure 9F). This kind of relationship has been observed before in tobacco and tomato (Glover et al., 1998; Glover, 2000) although it may be limited to certain species or dependent on environmental conditions, such as water availability (Galdon-Armero et al., 2018). In Slmixta-like ko mutants, the increase in trichome density was not accompanied by a decrease in stomatal density (Figure 8G and Supplemental Figure 8B) and consequently, the trichome-to-stomata ratio was not affected significantly (Figure 8H). More importantly, the inverse association between trichome and stomatal densities was not observed in Slmixta-like ko no. 1 (Figure 8I), although it was for the MoneyMaker parent, suggesting that SlMixta-like may be key to negatively linking trichome and stomatal development in leaves of tomato. The observations made of SlMixta-like OE and ko lines suggest that SlMixta-like plays a role in the mechanism controlling trichome and stomatal densities. SlMixta-like may therefore influence the dynamics of cell specialization in the leaf epidermis as well as functioning in petal and fruit epidermal development.
METHODS
Plant Material
We grew two generations of the Solanum lycopersicum cv M82 × Solanum pennellii ac. LA716 ILsEshed and Zamir, 1995) to assess epidermal cells on the adaxial leaf epidermis. The first generation consisted of 74 ILs grown under the same conditions in the greenhouse at the John Innes Centre in cereal mix (loam, peat, and grit), with an average temperature of 20°C to 22°C and supplemental lighting by sodium lamps to maintain 16 h of light each day, and all seeds were sown simultaneously in October 2016. The second generation consisted of 67 ILs, which were grown under greenhouse conditions at the John Innes Centre, with an average temperature of 20°C to 22°C. These lines were grown successively from October 2015 to February 2016, with approximately six ILs phenotyped per week. For both generations, plants were grown for 4 weeks, until the first true leaf was fully expanded. Samples of the terminal leaflet of the first fully expanded true leaf were used for analysis (Supplemental Figure 13). For each line, 3 to 4 plants in each generation were phenotyped.
SlMixta-like RNAi lines were kindly supplied by Asaph Aharoni (Lashbrooke et al., 2015).
SEM
Trichome phenotypes were assessed using a model no. Supra 55 VP SEM (Zeiss). Two different ways of sample fixation were used to preserve the structure of trichomes and other epidermal structures. Chemical fixation was used to screen the first generation of ILs. Chemical fixation of the samples was achieved by vacuum-infiltrating them with 2.5% (w/v) glutaraldehyde in 0.1-M cacodylate (pH 7.4) buffer and dehydrated through an ethanol series. Samples were dried in a model no. CPD300 critical‐point dryer (Leica Microsystems), where water was replaced by liquid CO2 and then evaporated at the critical point for CO2, removing all the liquid without damaging the structures of the sample. Dried samples were glued to a SEM stub and gold-coated before imaging in the main chamber of the microscope, at high-vacuum and room-temperature conditions.
For cryo-fixation, plant samples were glued to a cryo-stage and then submerged in liquid nitrogen in a vacuum-generating environment to reduce frosting of water vapor. The samples were then introduced in the microscope preparation chamber, where any frost was removed by sublimation at –95°C for 3.5 min. The samples were then sputter-coated with platinum for 150 s and transferred to the main microscope chamber, at –125°C. Imaging took place in the main chamber, where the electron beam was active, and the secondary and back-scattered electrons could be perceived by the detectors. This fixation protocol is destructive, and samples cannot be stored for further imaging, but ensures integrity of most trichomes. Cryo-fixation was used to screen the second generation of ILs.
Quantification of Epidermal Structures
For each leaf sample, 8 to 15 micrographs of 0.3 mm2 were generated. In every case, the same leaf (first fully expanded leaf) and the same part of each leaflet (intervein space close to the central vein; Supplemental Figure 13) were used for assessment of stomatal, trichome, and pavement cell numbers. These three cell types were manually quantified in micrographs at a relatively low magnification (×600) using the software ImageJ v1.49 (National Institutes of Health). Trichome and stomatal densities were calculated as a percentage of total epidermal cells and expressed as a fold change of each line with respect to the M82 values obtained in the corresponding generation. We classified trichomes in different groups according to Luckwill (1943), except that type-I and type-IV trichomes were grouped together under type I/IV (McDowell et al., 2011) and all nonglandular trichomes were classified as type V. For trichome density values, all trichomes were considered together. Trichome-to-stomata ratios were calculated by dividing trichome density by stomatal density. Aberrant trichome morphologies were recorded and SEM pictures were taken at the most appropriate magnification. All micrographs used in this study are available in the BioStudies database (http://www.ebi.ac.uk/biostudies; Sarkans et al., 2018) under accession number S-BSST262.
Selection of Candidate Genes
The genomic region introgressed from S. pennellii in IL2-5 and IL2-6 was clearly defined using the high-resolution bin map created by Chitwood et al., (2013). The genes corresponding to bins 2-K and 2-L (Supplemental Figure 5A) and their annotations were retrieved from the SolGenomics Network database (Fernandez-Pozo et al., 2015), and candidate genes were selected based on their annotation (hormone response, cytoskeleton formation, expansins, and transcriptional regulators). Their expression level was assessed using RNA-seq data from the ILs (Chitwood et al., 2013). SlMixta-like (Solyc02g088190) was selected as a candidate gene from this region.
Generation of Genetic Constructs
For the VIGS assays, 300-bp fragments of the SlMixta-like coding sequence were selected using the VIGS tool on the Sol Genomics Network website (Fernandez-Pozo et al., 2015) for amplification and insertion into a pTRV2-Del/Ros1-GW plasmid (Orzaez et al., 2009). This pTRV2 version is a modification of the original one described by Liu et al. (2002), and includes a recombinant fragment targeting Delila and Rosea1, followed by a Gateway cassette. This allowed silencing of the candidate gene together with silencing of the Del/Ros1 genes overexpressed in high-anthocyanin tomato plants, which led to formation of green tissue (compared with purple, unsilenced tissue) that could be used as a visual marker for silenced sectors. The fragment was amplified from S. lycopersicum cv MicroTom cDNA (F: GGGGACAAGTTTGTACAAAAAAGCAGGCTTATTAGCTTATATCGAAGAA and R: GGGGACCACTTTGTACAAGAAAGCTGGGTAGGTCACTGGGTCGATCCC) and cloned using the Gateway system (Katzen, 2007). For the generation of SlMixta-like and SpMixta-like OE lines, the complete coding sequence of SlMixta-like and SpMixta-like were amplified from S. lycopersicum cv MoneyMaker or S. pennellii cDNA (F:GGGGACAAGTTTGTACAAAAAAGCAGGCTTAATGGGTCGATCTCCGTGT/GGGGACAAGTTTGTACAAAAAAGCAGGCTTAATGGGTCGATCTCCGTGT and R:GGGGACCACTTTGTACAAGAAAGCTGGGTATTAGAACATAGATGAATC/GGGGACCACTTTGTACAAGAAAGCTGGGTATTAGAACATAGATGAATC) and introduced in the vector pJAM1502 (Luo et al., 2007), which contains a double Cauliflower Mosaic Virus 35S promoter preceding the SlMixta-like coding sequences, using the Gateway recombination system (Katzen, 2007).
For the generation of SlMixta-like CRISPR/Cas9 constructs, two specific sgRNAs were selected using the CRISPRdirect website (Naito et al., 2015) with NGG as the PAM sequence. The 20-nt–long guide sequences were incorporated in the pICSL002218A plasmid (TSLSynBio), including the sgRNA backbones, the Cas9 expression cassette, and the Neomycin Phosphotransferase II expression cassette, using the Golden Gate successive digestion-ligations with BsaI for sgRNA1 (F: ATTGAAGAAGAGCAAGATGAGAT and R: AAACATCTCATCTTGCTCTTCTT) and BbsI for sgRNA2 (F: GTGATTGTAATTTGCAGTTTCTGTCCCGTT and R: CTAAAACGGGACAGAAACTGCAAATTACAA).
VIGS Assays
Seeds from Del/Ros1-overexpressing plants were grown under greenhouse conditions for 7 to 10 d before agroinfiltration. Agrobacterium (Agrobacterium tumefaciens) strain AGL1 was transformed by electroporation with the pTRV1 plasmid and the pTRV2-Del/Ros1-SlMixta-like plasmid and grown in selective medium containing kanamycin (50 μg/mL), ampicillin (100 μg/mL), and rifampicin (50 μg/mL). Liquid cultures were grown to an OD600 = 0.5 to 0.8, and the cells were pelleted and resuspended in a buffer of 10 mM of MgCl2 and 10 mM of MES at pH 5.6 with 200 μM of acetosyringone to a final OD600 = 0.1. The pTRV1 and pTRV2 solutions were mixed 2:1, and Del/Ros1-overexpressing plantlets were vacuum-infiltrated using a chemical duty pump (Merck) and a vacuum desiccator for 10 min. pTRV1 and pTRV2-Del/Ros1-GW empty vectors were also infiltrated as a positive control for silencing. After infiltration, plants were grown for 3 weeks; and fully expanded leaves, where the silencing was obvious due to presence of purple and green sectors, were sampled for trichome phenotyping. Green (silenced) and purple (unsilenced) sectors of the same leaf were imaged using a cryo-SEM and compared.
Stable Tomato Transformation
S. lycopersicum cv MoneyMaker plants were transformed with the OE and CRISPR/Cas9 constructs using Agrobacterium strain AGL1 following a standard transformation protocol with cotyledons as initial explants (Fillatti et al., 1987). Transformants carrying the 35Spro:SlMixta-like construct were validated by PCR (F: CTATCCTTCGCAAGACCCTTC and R: CACTGGGTCGATCCCCATTT) and addition of SlMixta-like in CRISPR/Cas9 transformants was verified by amplification and sequencing of a SlMixta-like fragment including both expected cut sites (F: TTGAAGCACCATCAATTTTCTTT and R: TGAATCGCATACCAGCTTTG).
cDNA Synthesis
cDNA was synthesized using RNA extracted from leaf samples. Contaminant genomic DNA was eliminated by treating the samples with Amplification Grade DNase I (Merck). The reaction was performed in a volume of 10 μL with 1× DNase I buffer and 1 unit of DNase I, both supplied in the commercial kit. The digestion of genomic DNA was performed at room temperature (∼25°C) for 15 min. DNase I was inactivated by addition of 50 mM of EDTA to a final concentration of ∼5 mM of EDTA and heating at 70°C for 10 min. cDNA was then synthesized using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems). Reactions were performed in a volume of 20 μL using up to 2 μg of RNA as a template for retro transcription to single-stranded DNA. The reaction mix consisted of 1× retro transcription buffer, 4 mM of dNTPs, 1× Random Primers, 1 unit of MultiScribe Reverse Transcriptase, and 1 unit of RNase inhibitor (all from Applied Biosystems). The reaction consisted of three steps, a 10-min step at 25°C, a 120-min step at 37°C, and a 5-min step at 85°C to inactivate the reverse transcriptase. cDNA was then stored at −20°C until required.
qPCR
qPCR was used to determine the abundance of specific transcripts in the cDNA made from the RNA samples. The qPCR reactions were performed using the SYBR Green JumpStart Ready Kit (Sigma-Aldrich) in volumes of 25 μL including 1× SYBR buffer (1.25 unit of Taq polymerase, 10 mM of Tris-HCl, 50 mM of KCl, 3.5 mM of MgCl2, and 0.2 mM of dNTPs), 1 mM of each primer, and 5 μL of template cDNA, diluted 1:10 from the final volume after cDNA synthesis. At least three technical replicates were assayed for each reaction. The qPCR reaction program consisted of an initial denaturation step at 94°C for 2 min followed by 40 cycles with an initial denaturation step at 94°C for 15 s followed by an annealing and extension step at 61.2°C for 1 min. After each cycle, the fluorescence of the SYBR dye bound to the double-stranded DNA (dsDNA) was measured and plotted. After the 40 cycles, the samples were subjected to a multistep temperature gradient coupled with fluorescence readings to build a melting curve and ensure specificity of the primers. The thermocycler used was a CFX96 Touch (Bio-Rad), and the background value and Ct values were automatically calculated by its software (Bio-Rad). The abundance of SlMixta-like transcripts was determined using the following primers: F: GTCGGCCATAGCCACTCATT and R: CACTGGGTCGATCCCCATTT, and the cDNA made with RNA samples from VIGS assays, OE lines, and IL2-6; SlGAPDH (Solyc05g014470); F:GGCTGCAATCAAGGAGGAA and R:ATGCTTGACCTGCTGTCACC or SlACT (Solyc03g078400). F:GGGGGCTATGAATGCACGGT and R:GGCAATGCATCAGGCACCTC were used as reference genes to estimate expression levels, retrieved from Expósito-Rodríguez et al. (2008) and the mean of their Ct value was used as the reference Ct of the sample. Expression level was estimated as 2−ΔCt, where ΔCt was calculated as the difference between the Ct value of the target gene in the sample and the reference Ct value for the sample. At least three biological replicates were used for each comparison.
Statistical Analysis
To assess differences in trichome and stomatal density in the IL population, we performed an ANOVA test followed by a post-hoc Dunnett’s test, suitable for comparing multiple lines to a control line. For other pair-wise comparisons in the article, we used a two-tailed t test. For the trichome type distributions, we made qualitative observations without determining significance. The analyses were performed using the software R (v3.2.2; R Core Team). Details of all statistical analyses are provided in the Supplemental Data Set.
Accession Numbers
Sequence data from this article can be found under the following accession numbers in the Sol Genomics Network Database: SlMixta-like (Solyc02g088190), SpMixta-like (Sopen02g032880).
Supplemental Data
Supplemental Figure 1. Trichome density of the second generation of the S. lycopersicum cv M82 × S. pennellii (ac. LA716) ILs.
Supplemental Figure 2. Stomatal density of the second generation of the S. lycopersicum cv M82 × S. pennellii (ac. LA716) ILs.
Supplemental Figure 3. Trichome type distribution of the first generation of S. lycopersicum cv M82 × S. pennellii (ac. LA716) ILs.
Supplemental Figure 4. Aberrant trichomes found in the IL population.
Supplemental Figure 5. Identification of SlMixta-like as a candidate gene for the trichome patterning phenotype.
Supplemental Figure 6. Expression level of SlMixta-like in unsilenced and silenced tissue.
Supplemental Figure 7. Description of the SlMixta-like gene-edited lines.
Supplemental Figure 8. Analysis of the leaf epidermis of additional Slmixta-like ko lines no. 3 and no. 4.
Supplemental Figure 9. Differences in trichome types in control tomato leaves and Slmixta-like ko leaves.
Supplemental Figure 10. Expression level of SlMixta-like in leaves from control untransformed tomato plants and three independent SlMixta-like OE (35Spro:SlMixta-like) lines.
Supplemental Figure 11. Functional analysis of SpMixta-like.
Supplemental Figure 12. Differences in trichome types in control, untransformed tomato leaves, and three independent SlMixta-like OE (35Spro:SlMixta-like) lines.
Supplemental Figure 13. Explanation of sampling method used for epidermal analysis of tomato seedlings.
Supplemental Data Set. Statistical analyses used for the data shown in each Supplemental figure.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
This work was supported by the John Innes Foundation (Rotation Studentship to J.G.-A.) and the United Kingdom Biotechnology and Biological Sciences Research Council (grants BB/J004596/1 and BB/P012523/1 to the John Innes Centre).
AUTHOR CONTRIBUTIONS
J.G.-A., J.L., and C.M. designed the experiments and analysis; J.G.-A., L.A.-R., and M.D. performed the experiments; J.G.-A., L.A.-R., M.D., and C.M. carried out data analyses; J.G.-A. and C.M. wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Balcke G.U., Bennewitz S., Bergau N., Athmer B., Henning A., Majovsky P., Jiménez-Gómez J.M., Hoehenwarter W., Tissier A.(2017). Multi-omics of tomato glandular trichomes reveals distinct features of central carbon metabolism supporting high productivity of specialized metabolites. Plant Cell 29: 960–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann K., Perez-Rodriguez M., Bradley D., Venail J., Bailey P., Jin H., Koes R., Roberts K., Martin C.(2007). Control of cell and petal morphogenesis by R2R3 MYB transcription factors. Development 134: 1691–1701. [DOI] [PubMed] [Google Scholar]
- Bergau N., Bennewitz S., Syrowatka F., Hause G., Tissier A.(2015). The development of type VI glandular trichomes in the cultivated tomato Solanum lycopersicum and a related wild species S. habrochaites. BMC Plant Biol. 15: 289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockington S.F., Alvarez-Fernandez R., Landis J.B., Alcorn K., Walker R.H., Thomas M.M., Hileman L.C., Glover B.J.(2013). Evolutionary analysis of the MIXTA gene family highlights potential targets for the study of cellular differentiation. Mol. Biol. Evol. 30: 526–540. [DOI] [PubMed] [Google Scholar]
- Chang J., Yu T., Gao S., Xiong C., Xie Q., Li H., Ye Z., Yang C.(2016). Fine mapping of the dialytic gene that controls multicellular trichome formation and stamen development in tomato. Theor. Appl. Genet. 129: 1531–1539. [DOI] [PubMed] [Google Scholar]
- Chang J., et al. (2018). Hair, encoding a single C2H2 zinc-finger protein, regulates multicellular trichome formation in tomato. Plant J. 96: 90–102. [DOI] [PubMed] [Google Scholar]
- Chater C.C.C., Caine R.S., Fleming A.J., Gray J.E.(2017). Origins and evolution of stomatal development. Plant Physiol. 174: 624–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood D.H., Kumar R., Headland L.R., Ranjan A., Covington M.F., Ichihashi Y., Fulop D., Jiménez-Gómez J.M., Peng J., Maloof J.N., Sinha N.R.(2013). A quantitative genetic basis for leaf morphology in a set of precisely defined tomato introgression lines. Plant Cell 25: 2465–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira Silva F.M., et al. (2018). The genetic architecture of photosynthesis and plant growth-related traits in tomato. Plant Cell Environ. 41: 327–341. [DOI] [PubMed] [Google Scholar]
- Eshed Y., Zamir D.(1995). An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141: 1147–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewas M., Gao Y., Wang S., Liu X., Zhang H., Nishawy E.M.E., Ali F., Shahzad R., Ziaf K., Subthain H., Martin C., Luo J.(2016). Manipulation of SlMXl for enhanced carotenoids accumulation and drought resistance in tomato. Sci. Bull. (Beijing) 61: 1413–1418. [Google Scholar]
- Expósito-Rodríguez M., Borges A.A., Borges-Pérez A., Pérez J.A.(2008). Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol. 8: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Pozo N., Menda N., Edwards J.D., Saha S., Tecle I.Y., Strickler S.R., Bombarely A., Fisher-York T., Pujar A., Foerster H., Yan A., Mueller L.A.(2015). The Sol Genomics Network (SGN)—from genotype to phenotype to breeding. Nucleic Acids Res. 43: D1036–D1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillatti J., Kiser J., Ronald R., Comai L.(1987). Efficient transfer of glyphosate tolerance gene into tomato using a binary Agrobacterium tumefaciens vector. Biotechnology 5: 726–730. [Google Scholar]
- Frary A., Keles D., Pinar H., Gol D., Doganlar S.(2011). NaCl tolerance in Lycopersicon pennellii introgression lines: QTL related to physiological responses. J. Biol. Plant 55: 461–468. [Google Scholar]
- Galdon-Armero J., Fullana-Pericas M., Mulet P.A., Conesa M.A., Martin C., Galmes J.(2018). The ratio of trichomes to stomata is associated with water use efficiency in Solanum lycopersicum (tomato). Plant J. 96: 607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Y., Kumimoto R., Liu C., Ratcliffe O., Yu H., Broun P.(2006). GLABROUS INFLORESCENCE STEMS modulates the regulation by gibberellins of epidermal differentiation and shoot maturation in Arabidopsis. Plant Cell 18: 1383–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Y., Liu C., Yu H., Broun P.(2007). Integration of cytokinin and gibberellin signalling by Arabidopsis transcription factors GIS, ZFP8 and GIS2 in the regulation of epidermal cell fate. Development 134: 2073–2081. [DOI] [PubMed] [Google Scholar]
- Gao S., Gao Y., Xiong C., Yu G., Chang J., Yang Q., Yang C., Ye Z.(2017). The tomato B-type cyclin gene, SlCycB2, plays key roles in reproductive organ development, trichome initiation, terpenoids biosynthesis and Prodenia litura defense. Plant Sci. 262: 103–114. [DOI] [PubMed] [Google Scholar]
- Gilding E.K., Marks M.D.(2010). Analysis of purified glabra3-shapeshifter trichomes reveals a role for NOECK in regulating early trichome morphogenic events. Plant J. 64: 304–317. [DOI] [PubMed] [Google Scholar]
- Glover B.J.(2000). Differentiation in plant epidermal cells. J. Exp. Bot. 51: 497–505. [DOI] [PubMed] [Google Scholar]
- Glover B.J., Perez-Rodriguez M., Martin C.(1998). Development of several epidermal cell types can be specified by the same MYB-related plant transcription factor. Development 125: 3497–3508. [DOI] [PubMed] [Google Scholar]
- Goley E.D., Welch M.D.(2006). The ARP2/3 complex: An actin nucleator comes of age. Nat. Rev. Mol. Cell Biol. 7: 713–726. [DOI] [PubMed] [Google Scholar]
- Hauser M.T.(2014). Molecular basis of natural variation and environmental control of trichome patterning. Front Plant Sci 5: 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heichel G.H., Anagnostakis S.L.(1978). Stomatal response to light of Solanum pennellii, Lycopersicon esculentum, and a graft-induced chimera. Plant Physiol. 62: 387–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetherington A.M., Woodward F.I.(2003). The role of stomata in sensing and driving environmental change. Nature 424: 901–908. [DOI] [PubMed] [Google Scholar]
- Ishida T., Kurata T., Okada K., Wada T.(2008). A genetic regulatory network in the development of trichomes and root hairs. Annu. Rev. Plant Biol. 59: 365–386. [DOI] [PubMed] [Google Scholar]
- Jeong N.-R., Kim H., Hwang I.-T., Howe G.A., Kang J.-H.(2017). Genetic analysis of the tomato inquieta mutant links the ARP2/3 complex to trichome development. J. Plant Biol. 60: 582–592. [Google Scholar]
- Kang J.-H., Campos M.L., Zemelis-Durfee S., Al-Haddad J.M., Jones A.D., Telewski F.W., Brandizzi F., Howe G.A.(2016). Molecular cloning of the tomato Hairless gene implicates actin dynamics in trichome-mediated defense and mechanical properties of stem tissue. J. Exp. Bot. 67: 5313–5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J.-H., Liu G., Shi F., Jones A.D., Beaudry R.M., Howe G.A.(2010). The tomato odorless-2 mutant is defective in trichome-based production of diverse specialized metabolites and broad-spectrum resistance to insect herbivores. Plant Physiol. 154: 262–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzen F.(2007). Gateway recombinational cloning: A biological operating system. Expert Opin. Drug Discov. 2: 571–589. [DOI] [PubMed] [Google Scholar]
- Kirik V., Lee M.M., Wester K., Herrmann U., Zheng Z., Oppenheimer D., Schiefelbein J., Hulskamp M.(2005). Functional diversification of MYB23 and GL1 genes in trichome morphogenesis and initiation. Development 132: 1477–1485. [DOI] [PubMed] [Google Scholar]
- Kirik V., Simon M., Huelskamp M., Schiefelbein J.(2004a). The ENHANCER OF TRY AND CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Dev. Biol. 268: 506–513. [DOI] [PubMed] [Google Scholar]
- Kirik V., Simon M., Wester K., Schiefelbein J., Hulskamp M.(2004b). ENHANCER of TRY and CPC 2 (ETC2) reveals redundancy in the region-specific control of trichome development of Arabidopsis. Plant Mol. Biol. 55: 389–398. [DOI] [PubMed] [Google Scholar]
- Larkin J.C., Oppenheimer D.G., Lloyd A.M., Paparozzi E.T., Marks M.D.(1994). Roles of the GLABROUS1 and TRANSPARENT TESTA GLABRA genes in Arabidopsis trichome development. Plant Cell 6: 1065–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashbrooke J., et al. (2015). The tomato MIXTA-Like transcription factor coordinates fruit epidermis conical cell development and cuticular lipid biosynthesis and assembly. Plant Physiol. 169: 2553–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Zhao Y., McCaig B.C., Wingerd B.A., Wang J., Whalon M.E., Pichersky E., Howe G.A.(2004). The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 16: 126–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G., He H., Li Y., Ai Q., Yu D.(2014). MYB82 functions in regulation of trichome development in Arabidopsis. J. Exp. Bot. 65: 3215–3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Dinesh-Kumar S.P.(2002). Virus-induced gene silencing in tomato. Plant J. 31: 777–786. [DOI] [PubMed] [Google Scholar]
- Lloyd A., Brockman A., Aguirre L., Campbell A., Bean A., Cantero A., Gonzalez A.(2017). Advances in the MYB-bHLH-WD Repeat (MBW) pigment regulatory model: Addition of a WRKY factor and co-option of an anthocyanin MYB for betalain regulation. Plant Cell Physiol. 58: 1431–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckwill L.(1943). The Genus Lycopersicon; an Historical, Biological, and Taxonomic Survey of the Wild and Cultivated Tomatoes. (Aberdeen: The University Press; ). [Google Scholar]
- Luo J., et al. (2007). Convergent evolution in the BAHD family of acyl transferases: Identification and characterization of anthocyanin acyl transferases from Arabidopsis thaliana. Plant J. 50: 678–695. [DOI] [PubMed] [Google Scholar]
- Machado A., Wu Y., Yang Y., Llewellyn D.J., Dennis E.S.(2009). The MYB transcription factor GhMYB25 regulates early fibre and trichome development. Plant J. 59: 52–62. [DOI] [PubMed] [Google Scholar]
- Maes L., Inzé D., Goossens A.(2008). Functional specialization of the TRANSPARENT TESTA GLABRA1 network allows differential hormonal control of laminal and marginal trichome initiation in Arabidopsis rosette leaves. Plant Physiol. 148: 1453–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C., Bhatt K., Baumann K., Hailing J., Zachgo S., Roberts K., Schwarz-Sommer Z., Glover B., Perez-Rodrigues M.(2002). The mechanics of cell fate determination in petals. Phil. Trans. R. Soc. Lond. B. 357: 809–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell E.T., et al. (2011). Comparative functional genomic analysis of Solanum glandular trichome types. Plant Physiol. 155: 524–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadakuduti S.S., Pollard M., Kosma D.K., Allen C. Jr., Ohlrogge J.B., Barry C.S.(2012). Pleiotropic phenotypes of the sticky peel mutant provide new insight into the role of CUTIN DEFICIENT2 in epidermal cell function in tomato. Plant Physiol. 159: 945–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito Y., Hino K., Bono H., Ui-Tei K.(2015). CRISPRdirect: Software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 31: 1120–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda K., Glover B.J., Linstead P., Martin C.(1994). Flower colour intensity depends on specialized cell shape controlled by a Myb-related transcription factor. Nature 369: 661–664. [DOI] [PubMed] [Google Scholar]
- Orzaez D., Medina A., Torre S., Fernández-Moreno J.P., Rambla J.L., Fernández-Del-Carmen A., Butelli E., Martin C., Granell A.(2009). A visual reporter system for virus-induced gene silencing in tomato fruit based on anthocyanin accumulation. Plant Physiol. 150: 1122–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima Y., Shikata M., Koyama T., Ohtsubo N., Mitsuda N., Ohme-Takagi M.(2013). MIXTA-like transcription factors and WAX INDUCER1/SHINE1 coordinately regulate cuticle development in Arabidopsis and Torenia fournieri. Plant Cell 25: 1609–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanaik S., Patra B., Singh S.K., Yuan L.(2014). An overview of the gene regulatory network controlling trichome development in the model plant, Arabidopsis. Front Plant Sci 5: 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne C.T., Zhang F., Lloyd A.M.(2000). GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156: 1349–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne T., Clement J., Arnold D., Lloyd A.(1999). Heterologous myb genes distinct from GL1 enhance trichome production when overexpressed in Nicotiana tabacum. Development 126: 671–682. [DOI] [PubMed] [Google Scholar]
- Payne W.W.(1978). A glossary of plant hair terminology. Brittonia 30: 239–255. [Google Scholar]
- Perez-Rodriguez M., Jaffe F.W., Butelli E., Glover B.J., Martin C.(2005). Development of three different cell types is associated with the activity of a specific MYB transcription factor in the ventral petal of Antirrhinum majus flowers. Development 132: 359–370. [DOI] [PubMed] [Google Scholar]
- Pesch M., Dartan B., Birkenbihl R., Somssich I.E., Hülskamp M.(2014). Arabidopsis TTG2 regulates TRY expression through enhancement of activator complex-triggered activation. Plant Cell 26: 4067–4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plett J.M., Wilkins O., Campbell M.M., Ralph S.G., Regan S.(2010). Endogenous overexpression of Populus MYB186 increases trichome density, improves insect pest resistance, and impacts plant growth. Plant J. 64: 419–432. [DOI] [PubMed] [Google Scholar]
- Rick C.M., Chetelat R.T.(1995). Utilization of Related Wild Species for Tomato Improvement.. (Leuven: International Society for Horticultural Science; ), pp. 21–38. [Google Scholar]
- Rigano M.M., Arena C., Di Matteo A., Sereno S., Frusciante L., Barone A.(2014). Eco-physiological response to water stress of drought-tolerant and drought-sensitive tomato genotypes. Plant Biosys. 150: 682–691. [Google Scholar]
- Ron M., Dorrity M.W., de Lucas M., Toal T., Hernandez R.I., Little S.A., Maloof J.N., Kliebenstein D.J., Brady S.M.(2013). Identification of novel loci regulating interspecific variation in root morphology and cellular development in tomato. Plant Physiol. 162: 755–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkans U., Gostev M., Athar A., Behrangi E., Melnichuk O., Ali A., Minguet J., Rada J.C., Snow C., Tikhonov A., Brazma A., McEntyre J.(2018). The BioStudies database-one stop shop for all data supporting a life sciences study. Nucleic Acids Res. 46: D1266–D1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellmann S., Schnittger A., Kirik V., Wada T., Okada K., Beermann A., Thumfahrt J., Jürgens G., Hülskamp M.(2002). TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J. 21: 5036–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller A.L., Last R.L., Pichersky E.(2008). Harnessing plant trichome biochemistry for the production of useful compounds. Plant J. 54: 702–711. [DOI] [PubMed] [Google Scholar]
- Schilmiller A.L., Miner D.P., Larson M., McDowell E., Gang D.R., Wilkerson C., Last R.L.(2010). Studies of a biochemical factory: Tomato trichome deep expressed sequence tag sequencing and proteomics. Plant Physiol. 153: 1212–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittger A., Folkers U., Schwab B., Jürgens G., Hülskamp M.(1999). Generation of a spacing pattern: The role of triptychon in trichome patterning in Arabidopsis. Plant Cell 11: 1105–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittger A., Jürgens G., Hülskamp M.(1998). Tissue layer and organ specificity of trichome formation are regulated by GLABRA1 and TRIPTYCHON in Arabidopsis. Development 125: 2283–2289. [DOI] [PubMed] [Google Scholar]
- Scoville A.G., Barnett L.L., Bodbyl-Roels S., Kelly J.K., Hileman L.C.(2011). Differential regulation of a MYB transcription factor is correlated with transgenerational epigenetic inheritance of trichome density in Mimulus guttatus. New Phytol. 191: 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serna L., Martin C.(2006). Trichomes: Different regulatory networks lead to convergent structures. Trends Plant Sci. 11: 274–280. [DOI] [PubMed] [Google Scholar]
- Sharlach M., et al. (2013). Fine genetic mapping of RXopJ4, a bacterial spot disease resistance locus from Solanum pennellii LA716. Theor. Appl. Genet. 126: 601–609. [DOI] [PubMed] [Google Scholar]
- Shi P., et al. (2018). The roles of AaMIXTA1 in regulating the initiation of glandular trichomes and cuticle biosynthesis in Artemisia annua. New Phytol. 217: 261–276. [DOI] [PubMed] [Google Scholar]
- Simmons A.T., Gurr G.M.(2005). Trichomes of Lycopersicon species and their hybrids: Effects on pests and natural enemies. Agric. For. Entomol. 7: 265–276. [Google Scholar]
- Smart C.D., Tanksley S.D., Mayton H., Fry W.E.(2007). Resistance to Phytophthora infestans in Lycopersicon pennellii. Plant Dis. 91: 1045–1049. [DOI] [PubMed] [Google Scholar]
- Sun L., Zhang A., Zhou Z., Zhao Y., Yan A., Bao S., Yu H., Gan Y.(2015). GLABROUS INFLORESCENCE STEMS3 (GIS3) regulates trichome initiation and development in Arabidopsis. New Phytol. 206: 220–230. [DOI] [PubMed] [Google Scholar]
- Szymanski D.B., Jilk R.A., Pollock S.M., Marks M.D.(1998). Control of GL2 expression in Arabidopsis leaves and trichomes. Development 125: 1161–1171. [DOI] [PubMed] [Google Scholar]
- Tian N., Liu F., Wang P., Zhang X., Li X., Wu G.(2017). The molecular basis of glandular trichome development and secondary metabolism in plants. Plant Gene 12: 1–12. [Google Scholar]
- Tominaga-Wada R., Nukumizu Y., Sato S., Wada T.(2013). Control of plant trichome and root-hair development by a tomato (Solanum lycopersicum) R3 MYB transcription factor. PLoS One 8: e54019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendemiatti E., Zsögön A., Silva G.F.F.E., de Jesus F.A., Cutri L., Figueiredo C.R.F., Tanaka F.A.O., Nogueira F.T.S., Peres L.E.P.(2017). Loss of type-IV glandular trichomes is a heterochronic trait in tomato and can be reverted by promoting juvenility. Plant Sci. 259: 35–47. [DOI] [PubMed] [Google Scholar]
- Vőfély R.V., Gallagher J., Pisano G.D., Bartlett M., Braybrook S.A.(2019). Of puzzles and pavements: A quantitative exploration of leaf epidermal cell shape. New Phytol. 221: 540–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T., Tachibana T., Shimura Y., Okada K.(1997). Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 277: 1113–1116. [DOI] [PubMed] [Google Scholar]
- Walker A.R., Davison P.A., Bolognesi-Winfield A.C., James C.M., Srinivasan N., Blundell T.L., Esch J.J., Marks M.D., Gray J.C.(1999). The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11: 1337–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wester K., Digiuni S., Geier F., Timmer J., Fleck C., Hülskamp M.(2009). Functional diversity of R3 single-repeat genes in trichome development. Development 136: 1487–1496. [DOI] [PubMed] [Google Scholar]
- Wu H., Tian Y., Wan Q., Fang L., Guan X., Chen J., Hu Y., Ye W., Zhang H., Guo W., Chen X., Zhang T.(2018). Genetics and evolution of MIXTA genes regulating cotton lint fiber development. New Phytol. 217: 883–895. [DOI] [PubMed] [Google Scholar]
- Xu J., van Herwijnen Z.O., Dräger D.B., Sui C., Haring M.A., Schuurink R.C.(2018). SlMYC1 regulates type VI glandular trichome formation and terpene biosynthesis in tomato glandular cells. Plant Cell 30: 2988–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan T., et al. (2018). A novel HD-ZIP IV/MIXTA complex promotes glandular trichome initiation and cuticle development in Artemisia annua. New Phytol. 218: 567–578. [DOI] [PubMed] [Google Scholar]
- Yang C., Li H., Zhang J., Luo Z., Gong P., Zhang C., Li J., Wang T., Zhang Y., Lu Y., Ye Z.(2011). A regulatory gene induces trichome formation and embryo lethality in tomato. Proc. Natl. Acad. Sci. USA 108: 11836–11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Cai Y., Liu X., Dong M., Zhang Y., Chen S., Zhang W., Li Y., Tang M., Zhai X., Weng Y., Ren H.(2018). A CsMYB6-CsTRY module regulates fruit trichome initiation in cucumber. J. Exp. Bot. 69: 1887–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Gonzalez A., Zhao M., Payne C.T., Lloyd A.(2003). A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130: 4859–4869. [DOI] [PubMed] [Google Scholar]
- Zhou Z., An L., Sun L., Zhu S., Xi W., Broun P., Yu H., Gan Y.(2011). Zinc finger protein5 is required for the control of trichome initiation by acting upstream of zinc finger protein8 in Arabidopsis. Plant Physiol. 157: 673–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Sun L., Zhao Y., An L., Yan A., Meng X., Gan Y.(2013). Zinc Finger Protein 6 (ZFP6) regulates trichome initiation by integrating gibberellin and cytokinin signaling in Arabidopsis thaliana. New Phytol. 198: 699–708. [DOI] [PubMed] [Google Scholar]