Abstract
BackgroundHuman bocavirus (HBoV) is a newly identified human parvovirus that was originally identified in the respiratory secretions of children with respiratory tract disease. To further investigate the epidemiological profile and clinical characteristics of HBoV infection, we screened infants and children <2 years of age (hereafter referred to as “children”) for HBoV
MethodsChildren for whom respiratory specimens submitted to a diagnostic laboratory tested negative for respiratory syncytial virus, parainfluenza viruses (types 1–3), influenza A and B viruses, and adenovirus, as well as asymptomatic children, underwent screening for HBoV by use of polymerase chain reaction (PCR). Respiratory specimens were obtained from the children from 1 January 2004 through 31 December 2004
ResultsTwenty-two (5.2%) of the 425 children who had a respiratory specimen submitted to the diagnostic laboratory and 0 of the 96 asymptomatic children were found to be positive for HBoV by PCR (P=.02). Fever, rhinorrhea, cough, and wheezing were observed in ⩾50% of the HBoV-positive children. Of the 17 children who had chest radiography performed, 12 (70.6%) had abnormal findings. HBoV appeared to have a seasonal distribution. Nucleotide polymorphisms were detected in the viral capsid protein (VP) 1/VP2 genes. Two distinct HBoV genotypes circulated during the study period
ConclusionsHBoV is circulating in the United States and is associated with both upper and lower respiratory tract disease in infants and young children
Among children, respiratory tract infections are a leading cause of morbidity and mortality worldwide [1]. Respiratory syncytial virus (RSV), influenza viruses, parainfluenza viruses, adenoviruses, rhinoviruses, coronaviruses, and the recently discovered human metapneumovirus likely account for a majority of respiratory tract illnesses. However, for a substantial proportion of respiratory tract illnesses in children, a pathogen cannot be identified, even when sensitive detection methods, such as polymerase chain reaction (PCR), are used [2–4]. This finding suggests that unidentified pathogens may be circulating and causing disease in children
In 2005, Allander et al. [5] discovered a previously unidentified human parvovirus in respiratory secretions obtained from individuals with clinical features of respiratory tract illness. This novel virus was discovered with a technique that included viral purification (with filtration and ultracentrifugation), PCR amplification of purified viral DNA with primers containing random hexamer-binding sites, and subsequent cloning and sequencing of the amplified DNA products. Sequence and phylogenetic analyses demonstrated that this new virus was a parvovirus and was closely related to, although distinct from, bovine parvovirus and minute virus of canines, which are members of the genus Bocavirus of the family Parvoviridae, subfamily Parvovirinae. This new virus was designated “human bocavirus” (HBoV)
Parvoviruses are nonenveloped, single-stranded DNA viruses that are among the smallest viruses. The family Parvoviridae contains 2 subfamilies: the Densovirinae subfamily, which infects insects, and the Parvovirinae subfamily, which infects vertebrates [6]. Within the subfamily Parvovirinae are the genus Erythrovirus (which contains parvovirus B19, which was previously thought to be the only human pathogenic parvovirus), the genus Dependovirus and the genus Bocavirus [7]. Within the genus Bocavirus HBoV is clearly distinct from both bovine parvovirus and minute virus of canines, having only ∼42% amino acid identity within the major open reading frames with the 2 viruses [5]
The complete genomic sequences of 2 strains of HBoV have been elucidated. The 5217-base viral genome contains 3 open reading frames encoding (in the 5′–3′ orientation) NS1 (a gene encoding a nonstructural protein), NP-1 (the function of which is unknown), and the viral capsid protein (VP) 1/VP2 overlapping genes, which likely encode the structural proteins of the HBoV virion. Overall, there is relatively little heterogeneity in the HBoV genome of the 2 isolates studied to date, although most of the nucleotide polymorphisms appear to be in the VP1/VP2 genes [5]
Overall, Allander et al. [5] found that 17 (3.1%) of 540 respiratory specimens tested positive for HBoV. These specimens were collected from November 2003 through October 2004 and were obtained from children hospitalized on a pediatric infectious diseases ward at Karolinska University Hospital in Stockholm, Sweden. No other respiratory virus was detected in specimens obtained from 14 of these children. A majority of these children had fever, and, of the 7 children who had chest radiography performed, 6 had abnormal findings [5]. HBoV has been detected in Australia and Japan. Of 324 Australian children with acute respiratory tract infection who had respiratory specimens submitted to a diagnostic laboratory, 18 (5.6%) tested positive for HBoV. These specimens were collected from May through August (i.e., autumn and winter in the Southern Hemisphere) in 2004 [8]. Eighteen (5.7%) of 318 Japanese children with lower respiratory tract infection tested positive for HBoV [9]. The findings of the aforementioned studies suggested that HBoV may be a common human virus; however, because none of these initial studies included a control group of asymptomatic children, it is difficult to draw any conclusions about causation. To determine whether HBoV circulated in New Haven, Connecticut, and to further define the epidemiological profile and clinical characteristics associated with HBoV in young children, we screened respiratory specimens obtained from both symptomatic and asymptomatic infants and children (hereafter referred to as “children”) for HBoV
Materials and Methods
Clinical specimensAll respiratory specimens that were screened for HBoV were obtained from children <2 years of age. Two groups of children were screened for HBoV. The first group of children had respiratory specimens submitted to the Clinical Virology Laboratory, Yale–New Haven Hospital (New Haven, Connecticut). These respiratory specimens (nasal swabs, respiratory secretions, and bronchoalveolar lavage fluid samples) were submitted to the Clinical Virology Laboratory from 1 January 2004 through 31 December 2004 and were stored at −20°C after the addition of an equal volume of “viral freezing media” (2× Dulbecco’s modified Eagle medium, 200 mmol/L MgS04, and 100 mmol/L HEPES [pH 7.5]). Specimens originated from children seen in the emergency department, inpatient wards, intensive care units, and the hospital-affiliated outpatient urgent care clinic and were submitted at the discretion of the medical teams. These specimens tested negative for RSV, parainfluenza viruses (types 1–3), influenza A and B viruses, and adenovirus by means of direct immunofluorescence assay (DFA). The second group of children was comprised of asymptomatic control subjects who were seen at the Yale–New Haven Hospital Pediatric Primary Care Center for routine well-child visits. Nasal wash specimens, obtained after the parent or guardian of the child provided written, informed consent, were collected according to standard protocols [10] and were stored at −20°C after the addition of an equal volume of viral freezing media
Nucleic acid purification and PCR screeningNucleic acids from each respiratory specimen were extracted by use of QIAamp nucleic acid purification kits (Qiagen), according to the manufacturer’s protocol. DNA was subsequently screened for the presence of HBoV by means of PCR performed with HotStar Taq polymerase (Qiagen), according to the manufacturer’s specification. The primers used in the screening of the respiratory specimens were identical to those described elsewhere by Allander et al. [5]. The forward primer 5′-GACCTCTGTAAGTACTATTAC-3′ and the reverse primer 5′-CTCTGTGTTGACTGAATACAG-3′ target a portion of the HBoV NP-1 gene and produce a 354-bp amplicon that corresponds to nt 2351–2704 of the HBoV genome (GenBank accession number DQ000495). PCR amplification cycles were performed at 95°C for 15 min, followed by 35 cycles at 94°C for 1 min, 54°C for 1 min, and 72°C for 2 min, and then by a final extension cycle at 72°C for 10 min [5]. Each set of PCRs contained appropriate negative controls. The authenticity of each amplicon was confirmed by DNA sequencing. For phylogenetic analysis, regions of the VP1/VP2 genes were amplified and sequenced. The forward primer 5′-GGACCACAGTCATCAGAC-3′ and the reverse primer 5′-CCACTACCATCGGGCTG-3′ target nt 4370–4387 and nt 5172–5189, respectively, of the HBoV genome, and PCR amplification yields a predicted amplicon of 819 bases. The PCR amplification cycle that was used is the same as the one outlined above. Each HBoV-positive specimen was screened for human metapneumovirus by reverse-transcriptase PCR [11]
DNA sequencing and phylogenetic analysisSequencing was performed using an Applied Biosystems 3730 XL DNA Analyzer at the W. M. Keck Biotechnology Resource Laboratory, Yale University School of Medicine. Each nucleotide polymorphism that was present in only 1 strain of HBoV was confirmed by sequencing of an amplicon product from an independent PCR. Phylogenetic analysis was performed using Lasergene MegAlign software (version 5.05; DNAstar), with use of the Clustal W alignment method
Clinical dataThe medical records of all HBoV-positive children were reviewed. The demographic and clinical characteristics of each HBoV-positive child were recorded on a standard collection form. Each HBoV-positive specimen was assigned a numerical code, and none of the data presented in the current study includes protected health information. Specimen collection and collection of clinical data were approved by the Yale University Human Investigation Committee
Statistical analysisFisher’s exact test was used to determine whether the difference in the percentage of HBoV-positive specimens between the 2 groups was statistically significant
Results
Overall, 1271 DFA-negative respiratory specimens collected from children <2 years of age were obtained from the Clinical Virology Laboratory. We screened every third sample for HBoV. Twenty-two (5.2%) of 425 samples tested positive for HBoV. None of the 96 respiratory specimens obtained from asymptomatic children tested positive for HBoV; this difference between the 2 groups was statistically significant (P=.02). Each of the HBoV-positive specimens tested positive for both the NP-1 gene and the VP1/VP2 genes by PCR. None of these HBoV-positive specimens tested positive for human metapneumovirus
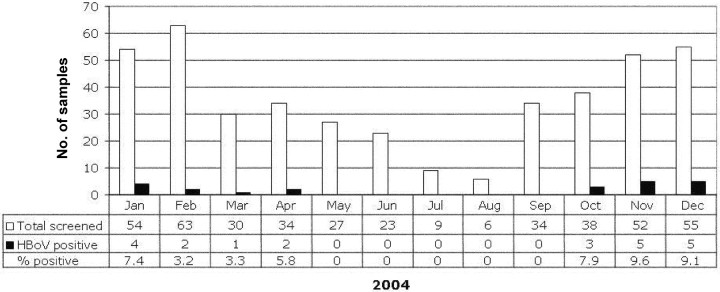
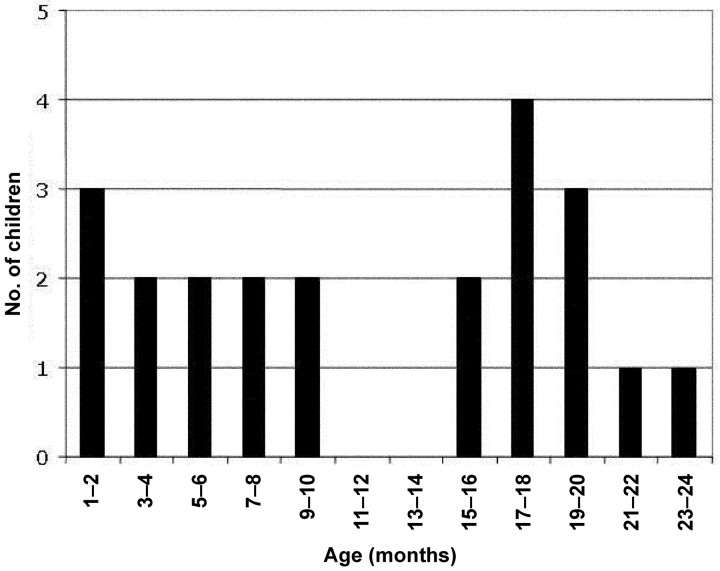
The monthly distribution of HBoV-positive specimens is displayed in figure 1. HBoV was detected in January–April and in October–December. No HBoV-positive specimens were obtained in May–August. During the months in which HBoV was detected, the range of the percentage of HBoV-positive specimens was 3.3% (in March) to 9.6% (in November). Overall, 79 (82.3%) of the 96 specimens collected from asymptomatic children were obtained during months in which HBoV was detected in specimens obtained from symptomatic patients. The age distribution of HBoV-positive children is shown in figure 2. In general, the presence of HBoV was not limited to a specific subpopulation (e.g., young infants) among the children <2 years of age who had screening performed. The youngest HBoV-positive individual was 40 days of age at the time that the respiratory specimen was obtained
Figure 1.
Distribution of human bocavirus (HBoV)–positive specimens in 2004, by month. The no. of respiratory specimens submitted to the Clinical Virology Laboratory at Yale–New Haven Hospital (New Haven, Connecticut) (white bars) the no. of HBoV-positive specimens (black bars), and the percentage of HBoV-positive specimens collected in each month are shown
Figure 2.
Distribution of human bocavirus–positive children, by age
The clinical characteristics of 20 of the 22 HBoV-positive children are shown in table 1. Medical records were available for 21 of the 22 HBoV-positive children. One child whose respiratory specimen was subsequently found to be culture positive for adenovirus was not included in the analysis of clinical characteristics of HBoV infection. Nearly two-thirds of the HBoV-positive children were males. Rhinorrhea (in 18 [90%] of 20 children), fever (in 14 [70%] of 20 children), cough (in 14 [70%] of 20 children), and wheezing (in 10 [50%] of 20 children) were present in ⩾50% of the children. Six (30%) of 20 children had hypoxia (SaO2, <90%). Of the 17 children who had chest radiography performed, 12 (70.6%) had abnormal findings. Eight of these children had radiographic findings consistent with lung hyperinflation, peribronchial cuffing, or atelectasis; 4 children had evidence of lobar infiltrates, and 1 child had evidence of pleural effusion. Fifteen (75%) of the 20 children had comorbidities, and 12 (80%) of these 15 patients had a history of prematurity or underlying lung disease or both. Complete blood counts were performed for 15 of the 20 children. The mean WBC count was 11,700 cells/μL (range, 7700–15,800 cells/μL), and the mean platelet count was 449,000 cells/μL (range, 245,000–680,000 cells/μL). Bacterial cultures of blood or of blood and urine were obtained for 2 patients (patients 3141 and 4176, respectively). All culture results were negative. Rashes were not reported for any patient
Table 1.
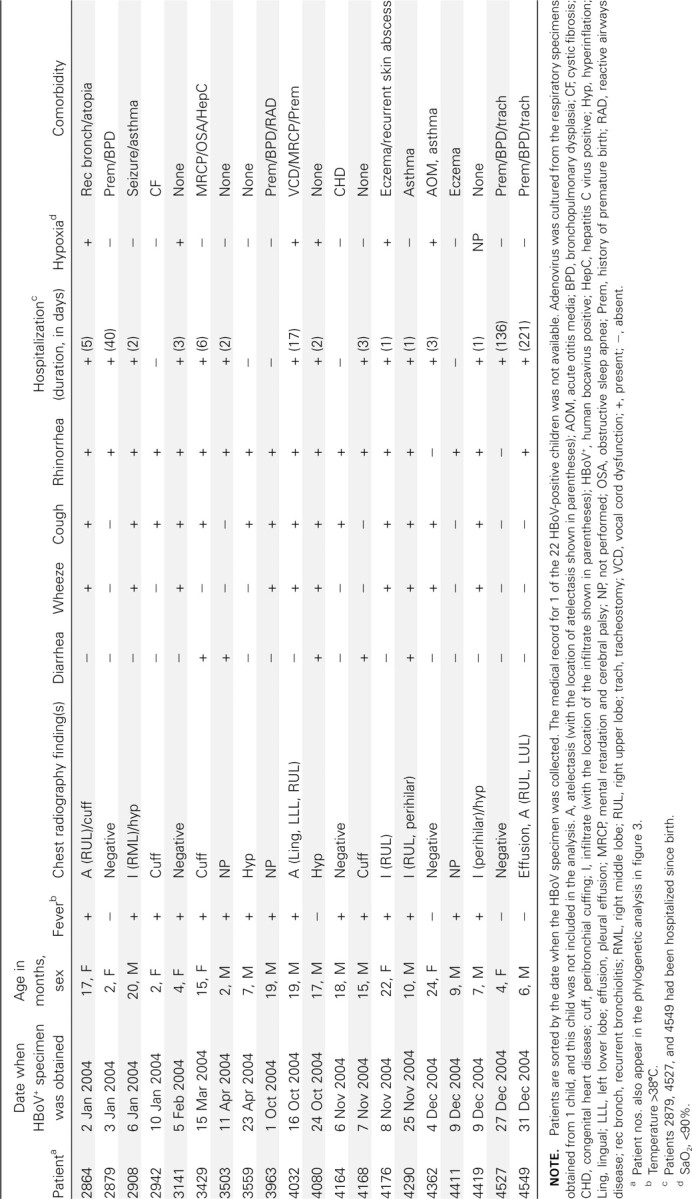
Clinical characteristics associated with human bocavirus (HBoV) infection in children <2 years of age
Five children had diarrhea, and 3 of these children (patients 3429, 3503, and 3559) ultimately had “viral gastroenteritis” diagnosed. For 2 of the 3 children who had viral gastroenteritis diagnosed (patients 3429 and 3503), a stool specimen tested negative for rotavirus antigen. A stool specimen obtained from patient 4168 tested positive for Clostridium difficile toxin
Nosocomial infection occurred in at least 3 children. These children (patients 2879, 4527, and 4549), who were 1, 4, and 6 months of age at the time that their HBoV-positive specimen was obtained, had been hospitalized since birth. The HBoV-positive specimens obtained from the child who was 4 months of age (patient 4527) and the child who was 6 months of age (patient 4549) were collected 4 days apart, and both specimens were collected in late December of 2004. These children received care in separate rooms in the same respiratory care unit, although they received care from the same medical and nursing personnel
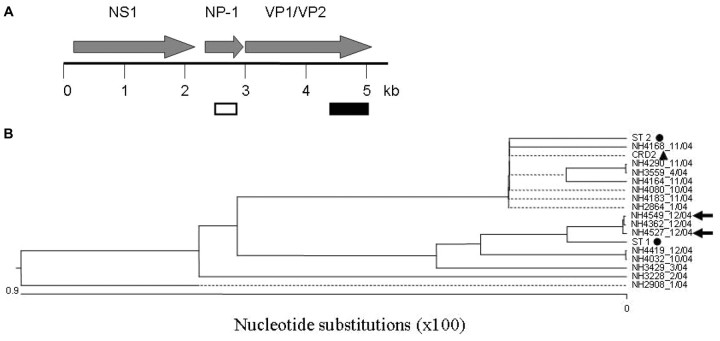
Rare nucleotide polymorphisms were observed in the amplified 354-base portion of the NP-1 gene of the New Haven HBoV isolates (a map of the HBoV genome is shown in figure 3A) (GenBank accession numbers DQ652146–DQ652167). The maximum number of nucleotide changes observed for any given sequence, compared with the sequences of the reference HBoV genome (reference strain ST1 [GenBank accession number DQ000495]), was 1. Three strains contained a G→A change at nt 2471, and 4 strains contained a G→A change at nt 2516 (data not shown). Single-nucleotide polymorphisms, which occurred in single-isolate sequences, were observed at nt 2585 and nt 2638 of the HBoV genome. None of these nucleotide changes resulted in changes in the predicted amino acid sequence
Figure 3.
A Map of the human bocavirus (HBoV) genome. The putative open reading frames of the HBoV genome are shown above the map. A portion of the NP-1 gene (white box) was targeted during screening for HBoV in respiratory specimens. Phylogenetic analysis was based on sequences of the 3′ third of the viral capsid protein (VP) 1/VP2 genes (nt 4370–5189 of the HBoV genome) (black box). B Phylogenetic analysis of HBoV isolates. The sequences of the New Haven (NH) isolates, the patient nos., and the month of acquisition of each HBoV-positive specimen (month/year) are shown. HBoV isolates recovered from 2 children with nosocomial infection whose respiratory specimens were collected 4 days apart are denoted by arrows. &cirf;, Initial strains of HBoV (ST1 and ST2) identified by Allander et al. [5]; ▴, HBoV isolate from St Louis, Missouri (CRD2). For figure clarity, not all New Haven isolates were included in the phylogenetic analysis. The HBoV sequences that were omitted were identical to at least one of the strains shown in the figure
Sequence analysis of the VP1/VP2 genes of nearly one-half of the HBoV-positive specimens revealed that the greatest frequency of nucleotide polymorphism occurred in the 3′ third of the gene. Therefore, the sequences of the 3′ third of the VP1/VP2 gene of all HBoV-positive specimens were obtained (nt 4370–5189 of the HBoV genome) (GenBank accession numbers DQ652168–DQ652182). Phylogenetic analysis of these sequences, which is shown in figure 3B revealed that at least 2 distinct genotypes circulated during the study period. The nucleotide sequences of viruses of one genotype were either identical or nearly identical to those of ST1, which is 1 of the 2 HBoV isolates identified by Allander et al. [5]. Likewise, nucleotide sequences of viruses of the second genotype were either identical to or nearly identical to those of ST2 (GenBank accession number DQ000496), the second HBoV isolate identified by Allander et al. [5]. The majority of the nucleotide polymorphisms did not translate into changes in the predicted amino acid sequences. HBoV isolates ST1 and ST2 differ at nt 4823, resulting in either a serine or threonine in the predicted amino acid sequence. The polymorphism at nt 4823 was noted in the New Haven isolates. For the 2 children with nosocomial infection whose samples were obtained 4 days apart, the nucleotide sequences were identical in both NP1 and VP1/VP2 (figure 3B, arrows). The sequences of these 2 isolates were also identical to those of other isolates that circulated during the same time of year and to those of isolates that circulated earlier in the year
Discussion
HBoV was detected in 5.2% of children who were <2 years of age and for whom a respiratory specimen submitted to a diagnostic virology laboratory was found to have DFA results that were negative for RSV, parainfluenza viruses (types 1–3), influenza A and B viruses, and adenovirus. The percentage of HBoV-positive specimens was similar to that noted in the limited numbers of clinical studies of HBoV infection [5, 8, 9]. It is unlikely that our findings were the result of PCR contamination. Each HBoV-positive specimen tested positive at least twice by use of independent PCR primers and reactions. Furthermore, each set of PCRs included appropriate negative controls
Several lines of evidence indicate that HBoV was the agent responsible for the disease observed in HBoV-positive children. All but one of the 22 HBoV-positive children tested negative for other respiratory viruses. Although the DFA and PCR screening tests that were used do not screen for all known respiratory viruses (such common viruses as rhinoviruses and coronaviruses were not included in the screening), and although the sensitivity of DFA for adenovirus is only 70% (one child whose respiratory specimen ultimately tested positive for adenovirus in cell culture was found to be negative for adenovirus by DFA), it is unlikely that another respiratory virus was present in 21 HBoV-positive specimens. Furthermore, none of the 96 asymptomatic children who had screening performed tested positive for HBoV. One potential explanation for the absence of HBoV in the asymptomatic children was that those children were selected and underwent screening during months (May to September) of the year-long study period in which HBoV apparently was not circulating. However, this was not the case. The respiratory specimens of 82.3% of asymptomatic children were collected in January–April and in October–December, the months during which HBoV was detected in specimens collected from symptomatic children
The screening of respiratory specimens submitted to a diagnostic laboratory has potential shortcomings, not the least of which is that there are no specific criteria used for sample collection. However, there are potential benefits to this approach. Screening is not limited to hospitalized children, as has been the case in other studies of HBoV infection [5]. Because respiratory specimens are often collected and screened for reasons other than the detection of respiratory tract disease (e.g., for the identification of the source of a fever), other manifestations of HBoV-associated disease not limited to the respiratory tract may be detected. Indeed, 5 of the HBoV-positive children had diarrhea, and 3 of these 5 children (2 children were hospitalized) had viral gastroenteritis diagnosed. Our study was not designed to evaluate the potential role of HBoV in gastrointestinal disease. However, both bovine parvovirus and minute virus of canines, the other 2 members of the genus Bocavirus (HBoV is the third member of this genus), cause enteric disease in their natural host [12, 13]. The potential role of HBoV in enteric disease will require further investigation
Our data suggest that HBoV may have a seasonal distribution. All of the HBoV-positive specimens were collected in fall, winter, and early spring (although only a relatively few specimens obtained in July and August were screened, and, therefore, it is not possible to draw any conclusions about seasonality from our data ). Nonetheless, this finding is in agreement with the findings of other studies of HBoV. In the 1-year study by Allander et al. [5], 16 of the 17 HBoV-positive specimens were collected from December 2003 through April 2004. Of the 18 HBoV-positive specimens identified in a Japanese study, 17 were collected from January through May [9]. Likewise, HBoV was detected in Australian children during the fall and winter months [8]. Erythema infectiosum (i.e., fifth disease), caused by parvovirus B19 (which, before the discovery of HBoV, was the only known human parvovirus to be associated with a specific disease), is also seasonal, although occurrence of the disease peaks in the spring and summer [14]
Nosocomial infection was observed in at least 3 children who had been hospitalized since birth. Two of these children, whose HBoV-positive specimens were obtained 4 days apart, were apparently infected with the same strain of virus. This strain was detected in other children in the same and in different periods of the year. These children were hospitalized on the same hospital ward at the same time. Because the mode of transmission of HBoV is unknown, it is impossible to determine, at this point, how these children became infected with HBoV
Our initial sequencing efforts focused on a portion of the NP-1 gene of the New Haven isolates of HBoV. Rare nucleotide changes were observed in NP-1, and none of these polymorphisms resulted in amino acid changes. On the basis of the sequencing of 2 isolates, Allander et al. [5] found that nucleotide polymorphisms were most common in the VP1/VP2 gene of HBoV. Therefore, we hypothesized that sequencing of the VP1/VP2 gene of the New Haven isolates would allow for the most robust phylogenetic analysis. We sequenced the VP1/VP2 gene of ∼50% of the isolates and found that the majority of nucleotide changes occurred in the 3′ third of the VP1/VP2 cassette. Therefore, we sequenced the 3′ third of the VP1/VP2 gene of all New Haven isolates. Although nucleotide polymorphisms were infrequent, there appeared to be at least 2 strains of HBoV circulating. A polymorphism in VP1/VP2 was observed at nt 4823 in the HBoV genome. This polymorphism resulted in either a serine or a threonine in the predicted amino acid sequence. This polymorphism was also present in ST1 and ST2, the original isolates identified by Allander et al. [5]. Our findings indicate that the 2 genotypes of HBoV identified in New Haven are remarkably similar to the genotypes of the initial strains of HBoV identified in Sweden, and this suggests that the nucleotide sequences (of the NP-1 and VP1/VP2 genes, at least) are highly conserved among viruses circulating in different locations
In conclusion, our data demonstrate that HBoV is circulating in the United States. Furthermore, our data suggest that HBoV is the etiological agent responsible for both upper and lower respiratory tract disease in infants and young children. Additional studies are required to completely define the epidemiological profile of this newly recognized pathogen
Acknowledgments
We thank Dr. George Miller, John F. Enders Professor of Pediatric Infectious Diseases, for his continued support, intellectual and scientific input, and exchange and critical review of the data. We thank the staff of the Clinical Virology Laboratory, Yale–New Haven Hospital, for their assistance in this study. We thank nurse Nancy Holabird for her assistance in the enrollment and sampling of asymptomatic children
Footnotes
(See the editorial commentary by McIntosh, on pages 1197–9, and the major article by Manning et al, on pages 1283–90.)
Potential conflicts of interests: none reported
Financial support: The Patrick and Catherine Weldon Donaghue Medical Research Foundation (grant to J.S.K.); Public Health Service, National Institutes of Health (NIH; training grant T32HD 07094 to D.K.); NIH (grants AI68280 [to M.V.] and AI001703 [to E.D.S.]). The study was funded in part by the Division of Clinical Research within the National Center for Research Resources, NIH (General Clinical Research Center grant M01 RR00125 to Yale University)
References
- 1.Murray C, Lopez A, Mathers C, Stein C. Global programme on evidence for health policy. Geneva: World Health Organization; 2001. The Global Burden of Disease 2000 Project: aims, methods and data sources. [Google Scholar]
- 2.Louie JK, Hacker JK, Gonzales R, et al. Characterization of viral agents causing acute respiratory infection in a San Francisco University Medical Center clinic during the influenza season. Clin Infect Dis. 2005;41:822–8. doi: 10.1086/432800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies HD, Matlow A, Petric M, Glazier R, Wang EE. Prospective comparative study of viral, bacterial and atypical organisms identified in pneumonia and bronchiolitis in hospitalized Canadian infants. Pediatr Infect Dis J. 1996;15:371–5. doi: 10.1097/00006454-199604000-00017. [DOI] [PubMed] [Google Scholar]
- 4.Wright AL, Taussig LM, Ray CG, Harrison HR, Holberg CJ. The Tucson Children’s Respiratory Study. II. Lower respiratory tract illness in the first year of life. Am J Epidemiol. 1989;129:1232–46. doi: 10.1093/oxfordjournals.aje.a115243. [DOI] [PubMed] [Google Scholar]
- 5.Allander T, Tammi MT, Eriksson M, Bjerkner A, Tiveljung-Lindell A. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci USA. 2005;102:12891–6. doi: 10.1073/pnas.0504666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berns KI. Parvoviridae: the viruses and their replication. In: Fields BN, Knipe DM, Howley PM, editors. Fields virology. Vol. 2. Philadelphia: Lippincott-Raven; 1996. pp. 2173–98. [Google Scholar]
- 7.International Committee on Taxonomy of Viruses Virus index database. Available at: http://www.danforthcenter.org/iltab/ictvnet/asp/iVirusIndex.asp. Accessed 20 September 2006.
- 8.Sloots TP, McErlean P, Speicher DJ, Arden KE, Nissen MD. Evidence of human coronavirus HKU1 and human bocavirus in Australian children. J Clin Virol. 2006;35:99–102. doi: 10.1016/j.jcv.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma X, Endo R, Ishiguro N, et al. Detection of human bocavirus in Japanese children with lower respiratory tract infections. J Clin Microbiol. 2006;44:1132–4. doi: 10.1128/JCM.44.3.1132-1134.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeVincenzo JP, Hall CB, Kimberlin DW, et al. Surveillance of clinical isolates of respiratory syncytial virus for palivizumab (Synagis)–resistant mutants. J Infect Dis. 2004;190:975–8. doi: 10.1086/423213. [DOI] [PubMed] [Google Scholar]
- 11.Esper F, Boucher D, Weibel C, Martinello RA, Kahn JS. Human metapneumovirus infection in the United States: clinical manifestations associated with a newly emerging respiratory infection in children. Pediatrics. 2003;111:1407–10. doi: 10.1542/peds.111.6.1407. [DOI] [PubMed] [Google Scholar]
- 12.Mochizuki M, Hashimoto M, Hajima T, et al. Virologic and serologic identification of minute virus of canines (canine parvovirus type 1) from dogs in Japan. J Clin Microbiol. 2002;40:3993–8. doi: 10.1128/JCM.40.11.3993-3998.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Storz J, Leary JJ, Carlson JH, Bates RC. Parvoviruses associated with diarrhea in calves. J Am Vet Med Assoc. 1978;173:624–7. [PubMed] [Google Scholar]
- 14.Young NS, Brown KE. Parvovirus B19. N Engl J Med. 2004;350:586–97. doi: 10.1056/NEJMra030840. [DOI] [PubMed] [Google Scholar]