Abstract
Background
The T1 Mapping and Extracellular volume (ECV) Standardization (T1MES) program explored T1 mapping quality assurance using a purpose-developed phantom with Food and Drug Administration (FDA) and Conformité Européenne (CE) regulatory clearance. We report T1 measurement repeatability across centers describing sequence, magnet, and vendor performance.
Methods
Phantoms batch-manufactured in August 2015 underwent 2 years of structural imaging, B0 and B1, and “reference” slow T1 testing. Temperature dependency was evaluated by the United States National Institute of Standards and Technology and by the German Physikalisch-Technische Bundesanstalt. Center-specific T1 mapping repeatability (maximum one scan per week to minimum one per quarter year) was assessed over mean 358 (maximum 1161) days on 34 1.5 T and 22 3 T magnets using multiple T1 mapping sequences. Image and temperature data were analyzed semi-automatically. Repeatability of serial T1 was evaluated in terms of coefficient of variation (CoV), and linear mixed models were constructed to study the interplay of some of the known sources of T1 variation.
Results
Over 2 years, phantom gel integrity remained intact (no rips/tears), B0 and B1 homogenous, and “reference” T1 stable compared to baseline (% change at 1.5 T, 1.95 ± 1.39%; 3 T, 2.22 ± 1.44%). Per degrees Celsius, 1.5 T, T1 (MOLLI 5s(3s)3s) increased by 11.4 ms in long native blood tubes and decreased by 1.2 ms in short post-contrast myocardium tubes. Agreement of estimated T1 times with “reference” T1 was similar across Siemens and Philips CMR systems at both field strengths (adjusted R2 ranges for both field strengths, 0.99–1.00). Over 1 year, many 1.5 T and 3 T sequences/magnets were repeatable with mean CoVs < 1 and 2% respectively. Repeatability was narrower for 1.5 T over 3 T. Within T1MES repeatability for native T1 was narrow for several sequences, for example, at 1.5 T, Siemens MOLLI 5s(3s)3s prototype number 448B (mean CoV = 0.27%) and Philips modified Look-Locker inversion recovery (MOLLI) 3s(3s)5s (CoV 0.54%), and at 3 T, Philips MOLLI 3b(3s)5b (CoV 0.33%) and Siemens shortened MOLLI (ShMOLLI) prototype 780C (CoV 0.69%). After adjusting for temperature and field strength, it was found that the T1 mapping sequence and scanner software version (both P < 0.001 at 1.5 T and 3 T), and to a lesser extent the scanner model (P = 0.011, 1.5 T only), had the greatest influence on T1 across multiple centers.
Conclusion
The T1MES CE/FDA approved phantom is a robust quality assurance device. In a multi-center setting, T1 mapping had performance differences between field strengths, sequences, scanner software versions, and manufacturers. However, several specific combinations of field strength, sequence, and scanner are highly repeatable, and thus, have potential to provide standardized assessment of T1 times for clinical use, although temperature correction is required for native T1 tubes at least.
Keywords: T1 mapping, Standardization, Calibration, Phantom, Repeatability, Extracellular volume
Introduction
T1 mapping aids clinicians in the assessment and diagnosis of myocardial disease. However, measurement needs to be stable over time with transferable values. Knowledge of normal reference ranges would benefit from not requiring local healthy subject scanning, and the pooling of multi-scanner datasets would have advantages such as increasing available sample sizes for the detection of subtle effects or subgroup analysis and increasing result robustness and generalizability, lowering the chance of unforeseen bias when compared to single-center data [1]. Combining results however introduces sequence, magnet, and field strength bias [2]. The field of T1 mapping would therefore benefit from a “T1 standard” to enable cross-center T1 mapping data pooling and delivery [3]—like the international normalized ratio (INR) which makes it possible to adjust the dosing of vitamin K antagonists regardless of which laboratory has performed the test [4].
The T1 Mapping and Extracellular volume (ECV) Standardization (T1MES) phantom program was established to explore T1 mapping quality assurance at 1.5 T and 3 T and understand the feasibility of delivering a “T1 standard” [5]. The first step toward the goal was development and mass-production of a phantom [5] and its European Union Conformité Européenne (CE) and United States Food and Drug Administration (FDA) regulatory clearance. In September 2015, cardiovascular magnetic resonance (CMR) centers worldwide joined the T1MES consortium and committed to submit a minimum of 12 months of center-specific T1 mapping data. Data submitted have now been analyzed to explore phantom performance.
We report phantom data at 1 and 2 years using various T1 mapping sequences, temperature sensitivity, and include platform performance, although we emphasize that comparison of different T1 methods and systems was not the main aim, rather investigating long-term stability towards the “T1 standard”. This involved modeling some of the potential sources of the T1 variation longitudinally and between T1MES centers to identify the most influential factors.
Methods
The development and description of the T1MES phantom (Fig. 1) has been previously reported [5]. Briefly, the T1MES phantom was designed to be field-strength specific (i.e., separate 1.5 T and 3 T models). Each phantom contains four tubes representing human native blood/myocardial T1 and T2 values (i.e., pre-gadolinium-based contrast agent [GBCA] values) and five tubes representing human post-GBCA blood/myocardial values. While the main aim of the present study was the collection and analysis of the multi-center data (see the “Methods part 2—Multi-center phantom testing” section), some other tests were applied to a small number of the phantoms during the 2 years to explore the utility of T1MES as a quality assurance device, and these tests are described here first (“Methods part 1—Evaluation of the phantom”). Imaging biomarker terms used follow the recommendations of the Quantitative Imaging Biomarkers Alliance (QIBA) of the Radiological Society of North America (RSNA) [6].
Fig. 1.
Left panel: Exemplar high-resolution (0.42 mm isotropic) imaging conducted at 3 T on 5 bottles (15E031; 15E033; 15E034 shown here; 30E017; 30E018 shown here) sampled out of the original batch in August 2017 (this is 2 years post manufacture) confirmed their structural integrity. The whole length of the phantom was imaged (three exemplar slices only shown here, represented by the green dashed lines). The internal tubes are labeled. Surrounding the tubes is a speckled pattern due to high density polyethylene (HDPE) macrobeads in an agarose/NiCl2 mixture . Confluent bright patches between tubes represent patches of agarose/NiCl2 mixture due to displacement of macrobeads. There were no signs of structural deterioration of the phantoms 2 years after manufacture. Middle panel: The nine tubes are supported on a translucent resin base composed of unsaturated polyester/styrene. The matrix fill is packed with compact HDPE pellets and agarose/NiCl2 mixture. Right panel: The outer physical appearance (front and back surfaces) of a phantom (30E018) at 2 years post manufacture (the plastic packaging wrap around the bottle cap dates back to the time of manufacture). HDPE, high-density polyethylene; NiCl2, nickel chloride; PE, polyethylene; PVC, polyvinyl chloride
Methods part 1—Evaluation of the phantom
Structural integrity
Gel integrity and aging were checked at each submission time point for participating sites through the manual inspection of localizers that formed part of the minimum dataset requirement for participation. In addition, a high-resolution, isotropic, three-dimensional (3D) gradient echo sequence (0.42 mm3) was run on four phantoms (three 1.5 T phantoms; one 3 T phantom) at baseline (October 2015) and at 2 years post manufacturing in each case using a 3 T MAGNETOM Skyra (Siemens Healthineers, Erlangen, Germany; software syngo MR D13C). The sequence acquired two overlapping slabs (due to scanner software constraints), each with two directions of phase encoding, a slow repetition time (repetition time, TR = 17 ms), and narrow sampling bandwidth (250 Hz/pixel) for better signal-to-noise ratio (SNR). This sequence had weak T1 and T2 image contrast and was only for structural examination.
“Reference” rT1 and rT2 data
Baseline (October 2015) “reference” T1 and T2 values (rT1, rT2) were acquired at the Royal Brompton Hospital CMR Unit using basic single-slice TR = 10 s inversion recovery spin echo (IRSE, 8 inversion times [TI] from 25 to 3200 ms) and single-slice repetition time (TR) = 10 s SE (8 echo times [TE] from 10 to 640 ms) [5] respectively. These sequences were identically repeated at 2 years on the same three 1.5 T phantoms and on the same three 3 T phantoms sampled from the production batch. The identifying serial numbers of the three 1.5 T phantoms were 15E031, 15E033, and 15E034, and these phantoms were scanned on a 1.5 T MAGNETOM Avanto [Siemens Healthineers; software syngo MR B17A]. The three 3 T phantoms were 30E001, 30E017, and 30E018, and they were scanned on a 3 T MAGNETOM Skyra [Siemens Healthineers; software syngo MR D13C].
Separate rT1 and rT2 data were acquired on the same 3 T phantom (30E021) at the German National Metrology Institute, Physikalisch-Technische Bundesanstalt (PTB) over a period of 1041 days (64 scans) commencing September 2015 (3 T MAGNETOM Verio (Siemens Healthineers; software syngo MR B17A). Sequences used for rT1 and rT2 were respectively basic single-slice TR = 8000 ms IRSE (IRSE, 7 TI from 25 to 4800 ms) and single-slice TR = 3000 ms SE (5 TE from 24 to 400 ms).
Temperature sensitivity
The following three methods were used:
First, controlled-temperature experiments over the range 10–30 °C were conducted at the United States National Institute of Standards and Technology (NIST) on six loose T1MES tubes at 1 year (Fig. 2i). T1 and T2 were measured at 10, 17, 20, 23, and 30 °C on an VnmrJ4 small-bore scanner operating at 1.5 T (Varian Medical Systems, Palo Alto, California, USA) in a temperature-controlled environment using a fiber optic temperature probe. T1 was measured by IRSE (TR = 10 s, TI = 50–3000 ms) and T2 by SE (TR = 10 s, TE = 15–960 ms).
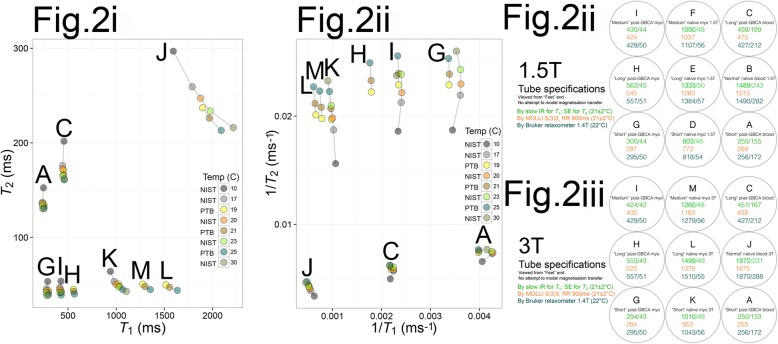
Fig. 2.
Temperature experiments (i) performed at two national metrology institutes: the US National Institute of Standards and Technology (NIST) laboratory after 1 year on six loose tubes from T1MES and at Physikalisch-Technische Bundesanstalt (PTB) on phantom 30E012. ii and iii indicate to which field-strength device the tested tubes belong. At NIST, T1 and T2 were measured on a Varian/Agilent small bore scanner operating at 1.5 T in a temperature-controlled environment. Temperatures were measured using a fiber optic probe. At PTB, T1 and T2 were measured on a 3 T MAGNETOM Verio scanner (Siemens Healthineers; software syngo MR B17A). The phantom was always stored, moved, and scanned while resting in a Styrofoam box to ensure that the temperatures picked at bottle hull reflects the tube temperature. At scan time, the box was placed in the head coil (12 ch) of the PTB 3 T scanner. Temperatures were measured using a Pt100 resistance thermometer. Similar to temperature dependency results immediately after phantom manufacture, [5] short-T1 tubes (modeling post-GBCA myocardium and blood) are more stable with temperature than very long-T1 tubes (native blood) where T1 increases more significantly with temperature
Second, controlled-temperature experiments at 19, 21, and 25 °C were conducted at the PTB laboratory on T1MES phantom 30E012 at 1 year (also Fig. 2i). T1 and T2 were measured on a 3 T MAGNETOM Verio scanner (Siemens Healthineers; software syngo MR B17A) using a Pt100 resistance thermometer. T1 was measured by IRSE (TR = 8000 ms, TI = 25–4800 ms) and T2 by SE (TR = 3000 ms, TE = 24–400 ms).
Third, for each T1MES phantom scan at all centers, temperature was measured using liquid crystal thermometers adhered to every phantom. These measurements were pooled and analyzed to derive temperature-correction algorithms (see Statistical Analysis).
B0 and B1 uniformity
These uniformities and the fundamental distortion of B1 by water dielectric permittivity especially at 3 T had been tested at baseline (October 2015, previously reported [5]). These uniformities were mapped later to check against “cracking” of the gel and subsequent impact of air gaps on B0 in particular, while potential “clumping” of the plastic beads over time might in theory affect the B1 [5]. We therefore considered it prudent to check whether anything unexpected occurred over the long term.
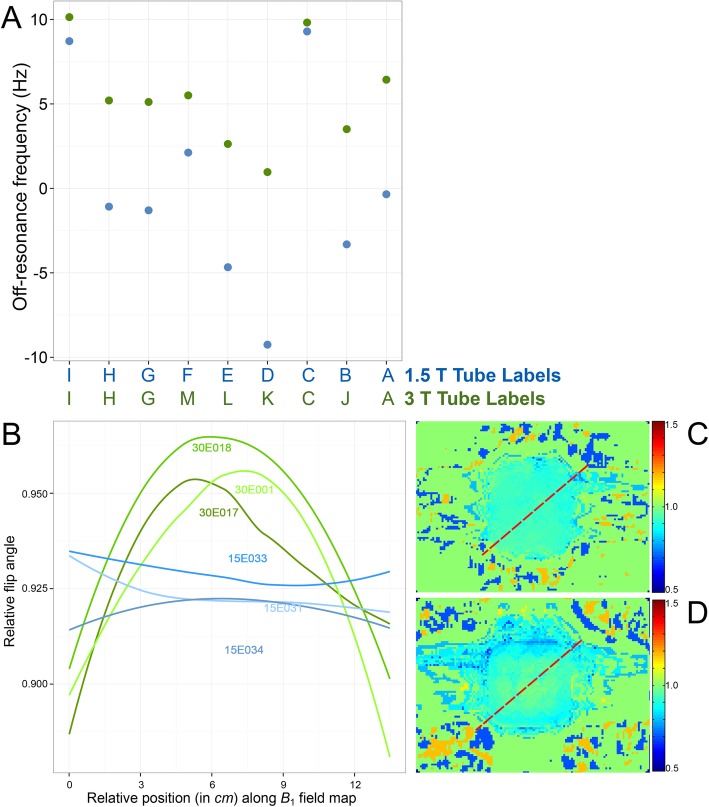
B0 uniformity was therefore mapped at 2 years in six phantoms, in the transverse slice, midway along the length of the tubes, using a multi-echo gradient echo sequence, based on the phase difference between known TEs [7]. A frequency range of ± 50 Hz across the phantom was considered acceptable, based on published T1 mapping off-resonance sensitivity [8]. B1 homogeneity was similarly evaluated using flip angle (FA) maps (double angle method using FA 60° and 120° [θ1, 2 × θ1] with long TR [8 s], and 4 ms sinc [− 3π to + 3π] slice excitation profiles to minimize error due to FA variation through the slice).
Methods part 2—Multi-center phantom testing
Serial, multi-center T1 mapping data
The T1MES user manual (10.6084/m9.figshare.c.3610175_D1.v1) defined strict scanning instructions (scanning and shim volume strictly at isocenter, use of same supporting materials, etc.). Each contributed T1MES dataset (localizers, sets of inversion recovery images, and inline scanner-generated T1 maps, Fig. 3) underwent initial quality assurance, checking orientation, and isocenter (through visual inspection of localizers and maps and semi-automatically by inspecting metadata contained in Digital Imaging and COmmunications in Medicine [DICOM] headers “ImagePositionPatient” and “ImageOrientationPatient”) and to exclude image artifacts. All Siemens sequences except MyoMaps product variant and all Philips (Philips Healthcare, Best, the Netherlands) sequences except CardiacQuant product variant were prototypes. Any tubes with artifacts detected by operator inspection of the submitted T1 maps were excluded from the analysis. Software version changes were captured automatically from DICOM headers (“StationName” and “SoftwareVersion”).
Fig. 3.
Representative maps exactly as they were submitted by collaborating sites, showing the 3 commonly used T1 mapping sequences appraised in T1MES: MOLLI 5s(3s)3s [448B], ShMOLLI 5b(1b)1b(1b)1b [1041B], and SASHA VE 11A
The T1 measurements from T1MES datasets (directly using only the parametric maps submitted, not by any T1 fitting applied centrally to the submitted sets of T1 recovery images) were carried out using a bespoke MATLAB pipeline (The MathWorks Inc., Natick, Massachusetts, USA, R2012b) assembled in collaboration with the US National Institutes of Health. From the data, T1 for each of the nine tubes was measured in identically sized regions of interest (ROI) occupying the central 50% by area of each tube (accommodating ~ 40 independent pixels) and collated in a dedicated research electronic data capture instrument (REDCap [9, 10]).
Methods part 3—Statistical analysis
Analysis was performed using R (version 3.0.1, R Foundation for Statistical Computing, Vienna, Austria). Descriptive data are expressed as mean ± standard deviation (SD) and standard error of the mean (SEM) as appropriate. Distribution of data were assessed on histograms and using the Shapiro-Wilk test.
Temperature sensitivity
Linear regression equations were used to relate temperature (predictor variable in degrees Celsius) and the response variable, phantom T1, by the formula: T1 = Intercept + (β ∗ [Temperature − 21 ° C]), with β representing the temperature correction, and 21 °C our arbitrarily chosen temperature for cross-center comparison.
Correlation with rT1 times
Correlations between estimated and rT1 times were derived using linear regression. Tests for significant inter-sequence and cross-vendor correlation differences (setting null value to 0.001) were conducted with alpha 0.01 and confidence level 0.95 [11].
T1 repeatability
After considering the normal values for native myocardial T1 reported in the published literature (e.g., in [12–16] as mean ± 1SD, though a 95% reference range is approximately ± 2SD), where 1 SD of the mean native myocardial T1 is generally ~ 20–30 ms at 1.5 T and ~ 50 ms at 3 T, we arbitrarily pre-defined as repeatable (and suitable for clinical/research use), T1 mapping approaches where the estimated variance of serial T1 data did not exceed ½ of the above in vivo 1SD. For T1 mapping at 1.5 T, this was ≤ 10 ms, i.e., CoV ≤ 1%; for T1 mapping at 3 T ≤ 25 ms, i.e., CoV ≤ 2%.
The CoV between serial repeat T1MES scans was calculated as the ratio of the SD to the mean. We appraised CoV as a compound measure of all causes of change in the estimated T1 of all nine tubes before and after temperature correction. We also appraised CoV after temperature correction separately for the four native and five post-GBCA tubes. Sequence-specific differences between the nine temperature-adjusted CoVs were calculated using paired t test with P value adjustment for multiple comparisons by the Bonferroni method (taking two-tailed P < 0.01 as significant).
Sources of T1 variation
Using temperature-adjusted T1 values of the “Medium” native myocardium tubes (tubes “F” and “M” respectively), we constructed linear mixed models to study the interplay of some known sources of T1 variation in multi-center phantom data. We did this separately for 1.5 T and 3 T phantom data. Considering temperature-adjusted T1 time as the response variable of interest, we examined the influence of phantom ID with and without the added effect of phantom age, as the combined fixed effect. With this, we then tested the following random effects:
-
i)
Main effects and interactions of scanner vendor/scanner model (Siemens, Philips or General Electric [GE; General Electric Healthcare, Waukesha, Wisconsin, USA]; e.g., for Siemens: MAGNETOM Aera vs. Avanto vs. Espree, etc.);
-
ii)
Main effects and interaction of sequence/scanner software version (considering all submitted variants of native modified Look-Locker inversion recovery [MOLLI] [17] sequences, shortened MOLLI [18] [ShMOLLI], native saturation-recovery single-shot acquisition [19] [SASHA], and saturation method using adaptive recovery times for cardiac T1 mapping [20] [SMART]; e.g., for Philips: R4.1.3SP2 vs. R5.1.7SP2 vs. R5.2.0SP2, etc.).
The response variable T1 fitted a normal probability distribution, so we estimated model parameters using maximum likelihood. ANOVA function using a type II Wald chi-square test evaluated the significance of fixed effects in the model. To compare models, Akaike and Bayesian information criteria (AIC, BIC) with the “smaller-is-better” criterion as well as chi-square values from inter-model ANOVA tests were used. The formulas used for model fitting and more definitions of the applied statistical tests are provided in Table 3.
Table 3.
Linear mixed models for 1.5 T and 3 T multi-center temperature-adjusted T1 mapping data (normalized to 21 °C, considering the “Medium” native myocardium tubes “F” and “M” respectively). The best model is “A5” at 1.5 T and “A3” at 3 T
| Model fitting formulas | AIC | BIC | Log likelihood | χ2 | P value | Best model | ||
|---|---|---|---|---|---|---|---|---|
| 1.5 T model | A1: T1 ~ sequence + (1|ID) | 5850.3 | 5920.8 | − 2909.1 | “ref” | “ref” | “ref” | |
| A2: T1 ~ sequence + (1|ID/age) [worse fit with age] | 5852.3 | 5927.2 | − 2909.1 | 0.0 | 1.000 | – | ||
| A3: T1 ~ sequence × software + (1|ID) | 5725.4 | 5883.9 | − 2826.7 | 164.9 | < 0.0001 | – | ||
| A4: T1 ~ sequence × software + vendor + (1|ID) [vendor non-contributory to fit] | 5725.4 | 5883.9 | − 2826.7 | 0.0 | 1.000 | – | ||
| A5: T1 ~ sequence × software + model + (1|ID) | 5723.4 | 5890.8 | − 2823.7 | 5.9 | 0.051 | A5 | ||
| A6: T1 ~ sequence × software + vendor × model + (1|ID) [vendor non-contributory to fit] | 5723.4 | 5890.8 | − 2823.7 | 0.0 | 1.000 | – | ||
| Final model (A5) parameters | Variance | SD | χ2 | Pvalue | ||||
| Random effects | ID | 2547.2 | 50.47 | “ref” | “ref” | |||
| Fixed effects | Sequence (β range − 32.3 to 455.6a) | / | / | 1101.0 | < 0.0001 | |||
| Software (β range 385.1 to 522.3a) | / | / | 63.8 | < 0.0001 | ||||
| Model (β range − 161.8 to 16.0a) | / | / | 11.2 | 0.011 | ||||
| Sequence: Software (β range − 161.8 to − 6.9a) | / | / | 149.6 | < 0.0001 | ||||
| 3 T model | A1: T1 ~ sequence + (1|ID) | 4390.4 | 4447.0 | − 2181.2 | “ref” | “ref” | “ref” | |
| A2: T1 ~ sequence + (1|ID/age) [worse fit with age] | 4392.4 | 4453.1 | − 2181.2 | 0.0 | 1.000 | – | ||
| A3: T1 ~ sequence × software + (1|ID) | 4238.3 | 4339.4 | − 2094.2 | 174.1 | < 0.0001 | A3 | ||
| A4: T1 ~ sequence × software + vendor + (1|ID) [vendor non-contributory to fit] | 4238.3 | 4339.4 | − 2094.2 | 0.0 | 1.000 | – | ||
| A5: T1 ~ sequence × software + model + (1|ID) | 4239.2 | 4348.4 | − 2092.6 | 3.1 | 0.212 | – | ||
| A6: T1 ~ sequence × software + vendor × model + (1|ID) [vendor non-contributory to fit] | 4239.2 | 4348.4 | − 2092.6 | 0.0 | 1.000 | – | ||
| Final model (A3) parameters | Variance | SD | χ2 | P-value | ||||
| Random effects | ID | 372.4 | 19.3 | “ref” | “ref” | |||
| Fixed effects | Sequence (β range − 114.5 to 38.5a) | / | / | 732.8 | < 0.0001 | |||
| Software (β range − 203.0 to 32.7a) | / | / | 46.0 | < 0.0001 | ||||
| Sequence: Software (β range − 48.8 to 267.3a) | / | / | 189.0 | < 0.0001 | ||||
The symbol (1|ID) in the model formulas refers to the random effect of individual phantoms (by identity number). At 1.5 T models, A5 and A6 have equal AIC/BIC but given the lack of statistically significant χ2 improvement from A5 to A6 (P = 1.0); A5 is considered the most parsimonious model
Age refers to phantom age at scanning since date of manufacture, AIC Akaike information criterion, BIC Bayesian information criterion, χ2 chi-square, ref reference, SD standard deviation
aComplete list of β coefficients for model A5 at 1.5 T and A3 at 3 T are provided in Supplementary Tables 7 and 8 respectively
Software upgrades
To explore whether software upgrades resulted in an abrupt “step” change in the temperature-adjusted T1 reads, we performed piece-wise linear regression to check for any segmented relationship between the covariates “scan day” and “tube T1” (considering tube “F” at 1.5 T and “M” at 3 T) [21]. For any broken-line relationship discovered, we defined slope parameters and break points where the linear relation/s changed and temporally correlated these with DICOM software metadata.
Results part 1—Evaluation of the phantom
Structural integrity
Individual inspection of all the localizers submitted by sites with each phantom dataset revealed no visible gel rips or tears down any of the tubes. At baseline and at 2 years following batch manufacturing (Supplementary Movie 1), phantoms were free of air bubbles and susceptibility artifacts at both field strengths. High-resolution imaging showed no evidence of gel rips or tears down any of the tubes, and the gels were intact in the mid slice—the piloted location for serial mapping (Fig. 1, left panel). T1 maps collected through the midline of the phantom, using the specified T1MES scan setup, were free from off-resonance artifacts.
“Reference” rT1 and rT2 data
Phantom measurements (averaged across all tubes) collected at the Royal Brompton Hospital showed that temperature-corrected rT1 and rT2 values at 2 years were stable compared to baseline. At 1.5 T, the 2-year temperature-adjusted %T1 change was 1.95 ± 1.39% SD, 0.37% SEM. At 3 T, the 2-year temperature-adjusted %T1 change was 2.22 ± 1.44% SD, 0.25% SEM. Three tesla measurements at PTB showed a 2-year temperature-adjusted %T1 change of 0.80 ± 0.49%, SEM 0.16% (Supplementary Figure S1A). Although not the aim of this work, all reference T2 parameters remained stable with relative < 1% change at PTB (temperature-adjusted %T2 change over 2 years at 3 T = 1.65 ± 0.26% | 0.09%, Supplementary Figure S1B) and < 2% change over 2 years at Royal Brompton Hospital.
Sequences from Siemens and Philips at 1.5 T collected at Royal Brompton Hospital showed strong correlation with rT1 times with small offsets, as shown in Fig. 4 (left panel). There were no statistically significant differences between the rT1 correlations for a given sequence when comparing Siemens to Philips platforms (Supplementary Table 1, panel C) or when individual sequences were compared on Philips (Supplementary Table S1, panel B). However, on Siemens, rT1 correlations for SASHA were significantly stronger than for both MOLLI and ShMOLLI (Supplementary Table S1, panel A).
Fig. 4.
T1 times in the nine tubes for the three main sequence types (modified Look-Locker with inversion recovery (MOLLI), shortened MOLLI (ShMOLLI), saturation recovery single-shot acquisition (SASHA)) at 1.5 T (left) and 3 T (right) split according to vendor (Siemens, Philips). At 3 T, the contributed Philips MOLLI 5b(1b)1b(1b)1b was missing the iterative/data dropping steps in map creation as per Siemens ShMOLLI, so these data are not shown. Measured T1 times by the 3 sequences (mean of multiple centers, no temperature correction) are represented by symbols. The line of identity, i.e., “reference” rT1 times by slow inversion recovery is represented as a discontinuous gray line. For each sequence, correlations between absolute measurements of T1 and rT1 times are shown. Correlations for the much sparser GE data (MOLLI, ShMOLLI, SMART each n = 1) are not reported. Vertical error bars within each shape represent standard error of the mean. aR, adjusted R2; GE, General Electric Healthcare
Sequences from Siemens and Philips at 3 T showed strong correlation with rT1 times but with visibly imperfect absolute measurement of T1 across tubes of longer T1 times as shown in Fig. 4 (right panel). The correlations between absolute measurements of T1 and rT1 were similar for MOLLI and SASHA on Philips (Supplementary Table S2, panel B). On Siemens, the correlation between absolute measurements of T1 and rT1 was significantly stronger for SASHA when compared to MOLLI and ShMOLLI (Supplementary Table S2, panel A). The correlation between absolute measurements of T1 and rT1 for SASHA was significantly stronger on Siemens compared to Philips (Supplementary Table S2, panel C). Comparison of errors at 3 T relative to 1.5 T showed no significant difference (average SEM 5.49 vs. 4.04 respectively, P = 0.428).
Temperature sensitivity
Temperature experiments at both NIST and PTB using spin echo, long TR, and sequences (Fig. 2i) consistently showed that short-T1 tubes (modeling post-GBCA myocardium and blood) were more stable with temperature than very long-T1 tubes (native blood), where T1 increased more significantly with temperature. T1 increased by ~ 11 ms/°C for a typical MOLLI 5s(3s)3s 1.5 T dataset in tube “B” (normal native blood). Conversely, T1 decreased by ~ 1.2 ms/°C in tube “G” (short post-GBCA myocardium). Temperature corrections for each of the nine tubes at both field strengths from the pooled multi-center analysis are presented separately (Supplementary Table S3) from temperature sensitivity experiments of NIST and PTB due to the latter’s use of bespoke standard operating procedures.
B0 and B1 uniformity
B0 uniformity at 2 years was delivered to within ± 9 Hz at 1.5 T and ± 10 Hz at 3 T (Fig. 5a–b). At 2 years after manufacture, the B1 field distortion caused by the phantom, continued to be adequately flattened to within 10% of the B1 at the center of the phantom, at both 1.5 T and 3 T (Fig. 5c–d).
Fig. 5.
aB0 field homogeneity at 2 years post manufacture across the nine phantom compartments as a measure of off-resonance in hertz at 1.5 T (blue, averaged for three phantoms) and 3 T (green, averaged for another three phantoms). These are extremely small shifts in frequency (e.g., 10 Hz = 0.08 ppm at 3 T) and should not be regarded as significantly different between the tube compartments. b Diagonal profiles of the B1 field at 2 years post manufacture as per red discontinuous lines (right panels) in six phantoms: three at 1.5 T scanned on 1.5 T MAGNETOM Avanto (Siemens Healthineers; software syngo MR B17A); example field map (c) and three at 3 T scanned on 3 T MAGNETOM Skyra (Siemens Healthineers; software syngo MR D13C); example field map (d)
Results part 2—Multi-center phantom testing
Thirty-four of the magnet systems appraised were 1.5 T and 22 were 3 T (see Supplementary Data File and Supplementary Table S4) with manufacturers being 35 Siemens, 18 Philips, and 3 GE. Scan frequencies are reported in the Supplementary Data File. The planned program was 1 year, but 24 centers voluntarily extended, resulting in mean longitudinal data of 358 days, and the longest 1161 days.
Quality assurance of the contributed DICOM image files identified significant protocol deviations or sequence reconstruction failures in nine submissions, the majority of which took place at the start of the study (examples in Supplementary Fig. S2). These were excluded with subsequent advice sent to centers permitting later inclusion in most cases, except where a tube artifact affecting > 25% of a single center’s series led to exclusion of that tube from pooled analysis (see Supplementary Data File).
Fifty-two magnets non-exclusively contributed MOLLI datasets, 16 ShMOLLI, and 12 SASHA, and 2 magnets contributed SMART T1 maps. Center-, session-, and sequence-specific T1 mapping contributions are detailed in Supplementary Data File. During the project, there were three software upgrades at centers: two Siemens and one GE (details in Supplementary Fig. S4).
T1 repeatability
For serial multi-center data, temperature-unadjusted CoV per tube, per phantom, per sequence, and per magnet are detailed in the Supplementary Data File. Temperature-adjusted CoV for the various native sequences together with inter-sequence and cross-platform differences, at 1.5 T and 3 T, are summarized in Tables 1 and 2 respectively and data for post-GBCA in Supplementary Tables S5 and S6. Results of native and post-GBCA sequence/scanner software version repeatability are provided in Tables 1 and 2 and in Supplementary Tables S5 and S6 respectively.
Table 1.
Temperature-adjusted (normalized to 21 °C) native T1 and CoV (%) at 1.5 T summarized by vendor and sequence
| Summary table ofx̅CoV ofT1(%) across 4 tubes |
Order of tube IDs follows their orientation in the scanned bottle
|
|||||
| Siemens | MOLLI 5s(3s)3s [448B] | 0.27 | ||||
| Philips | MOLLI 3s(3s)5s | 0.54 | ||||
| Siemens | SASHA | 0.56 | ||||
| Siemens | ShMOLLI 5b(1b)1b(1b)1b [1041B] | 0.64 | ||||
| Philips | SASHA | 0.92 | ||||
| Philips | ShMOLLI 5b(1b)1b(1b)1bc | 1.04 | ||||
| General Electric | MOLLI 5b(1b)1b(1b)1bd | 1.28 | ||||
| General Electric | SMART | 3.00 | ||||
| Platform | Sequence [prototype, #] | x̅’ CoV ofT1(%), globalx̅± SD ofT1according to native tube ID [tube rT1 by slow IR] | ||||
| F [1090 ms] | E [1333 ms] | D [803 ms] | B [1489 ms] | |||
| Siemens | MOLLI MyoMaps product 5s(3s)3s [5] | 1.07, 1041 ± 23 | 1.38, 1273 ± 35 | 0.85, 771 ± 6 | 1.45, 1538 ± 46 | |
| MOLLI 3b(3s)3b(3s)5b [448, 3] | 0.87, 1019 ± 11 | 1.08, 1225 ± 17 | 0.65, 765 ± 6 | 1.19, 1462 ± 21 | ||
| MOLLI 5b(3s)3b [448, 3] | 0.18, 1005 ± 13 | 0.53, 1217 ± 24 | 0.26, 762 ± 6 | 0.74, 1464 ± 23 | ||
| MOLLI 5b(3s)3b [448B, 2] | 0.65, 1052 ± 7 | 0.82, 1289 ± 10 | 0.61, 784 ± 4 | 1.00, 1555 ± 16 | ||
| MOLLI 5b(3s)3b [780, 2] | 0.70, 1048 ± 14 | 1.18, 1276 ± 20 | 0.61, 776 ± 6 sic. | 1.15, 1536 ± 24 | ||
| MOLLI 5b(3s)3b [780B, 4] | 0.76, 1043 ± 9 | 0.99, 1279 ± 17 | 0.67, 776 ± 6 | 1.45, 1550 ± 37 | ||
| MOLLI 5s(3s)3s [448, 2] | 0.89, 1035 ± 10 | 1.51, 1261 ± 25 | 1.04, 770 ± 9 | 1.44, 1538 ± 24 | ||
| MOLLI 5s(3s)3s [448B, 1]a | 0.14, 1051 ± 1 | 0.38, 1282 ± 5 | 0.17, 783 ± 1 | 0.38, 1553 ± 6 | ||
| MOLLI 5s(3s)3s [780B, 2] | 0.81, 1043 ± 15 | 0.69, 1277 ± 15 | 0.42, 771 ± 6 | 0.90, 1546 ± 25 | ||
| MOLLI 5s(3s)3s [1041, 1] | 1.11, 1045 ± 12 | 0.69, 1285 ± 9 | 0.81, 772 ± 6 | 0.96, 1560 ± 15 | ||
| MOLLI 5s(3s)3s [1041B, 1] | 1.27, 1033 ± 13 | 1.80, 1232 ± 22 | 0.66, 771 ± 5 | 1.47, 1494 ± 22 | ||
| ShMOLLI 5b(1b)1b(1b)1b [448, 1] | 1.01, 989 ± 10 | 1.39, 1223 ± 17 | 0.60, 729 ± 4 | 1.50, 1510 ± 23 | ||
| ShMOLLI 5b(1b)1b(1b)1b [448C, 1] | 0.61, 983 ± 6 | 0.64, 1218 ± 8 | 0.63, 727 ± 5 | 1.57, 1507 ± 22 | ||
| ShMOLLI 5b(1b)1b(1b)1b [780B, 3] | 2.16, 945 ± 34 | 3.29, 1147 ± 62 | 0.98, 711 ± 11 | 3.70, 1392 ± 85 | ||
| ShMOLLI 5b(1b)1b(1b)1b [1048, 1] | 0.70, 972 ± 7 | 0.71, 1208 ± 9 | 0.68, 715 ± 5 | 1.01, 1489 ± 15 | ||
| ShMOLLI 5b(1b)1b(1b)1b [1041B, 1]a | 0.59, 978 ± 6 | 0.67, 1201 ± 8 | 0.43, 721 ± 3 | 0.86, 1501 ± 13 | ||
| SASHA [4] | 0.39, 1104 ± 17 | 0.65, 1362 ± 27 | 0.47, 814 ± 8 | 0.73, 1522 ± 30 | ||
| Philips | MOLLI CardiacQuant product 5s(3s)3s [2] | 0.53, 1025 ± 13 | 0.86, 1265 ± 19 | 0.50, 760 ± 7 | 1.04, 1516 ± 28 | |
| MOLLI 3b(3s)3b(3s)5b [2] | 0.47, 1006 ± 17 | 0.94, 1247 ± 39 | 0.46, 751 ± 6 | 1.25, 1503 ± 46 | ||
| MOLLI 3s(3s)3s(3s)5s [1] | 0.94, 1026 ± 10 | 1.66, 1251 ± 21 | 0.69, 765 ± 5 | 2.15, 1450 ± 31 | ||
| MOLLI 3s(3s)5s [1]a | 0.20, 936 ± 2 | 1.11, 1100 ± 12 | 0.27, 722 ± 2 | 0.59, 1323 ± 8 | ||
| MOLLI 5b(3s)3b(3s)2b [1] | 6.78, 1168 ± 79 | 6.32, 1374 ± 87 | 3.04, 816 ± 25 | 6.53, 1665 ± 109 | ||
| MOLLI 5b(3s)3b [1] | 1.18, 1027 ± 12 | 1.48, 1265 ± 19 | 0.67, 758 ± 5 | 1.16, 1522 ± 18 | ||
| MOLLI 5 s(3s)3s [3] | 0.89, 1012 ± 17 | 1.35, 1233 ± 31 | 1.20, 756 ± 9 | 1.76, 1456 ± 58 | ||
| ShMOLLI 5b(1b)1b(1b)1bc [1] | 0.89, 1027 ± 9 | 1.37, 1270 ± 17 | 0.50, 753 ± 4 | 1.41, 1522 ± 21 | ||
| SASHA [2] | 1.02, 1057 ± 44 | 0.89, 1336 ± 84 | 0.87, 798 ± 30 | 0.89, 1505 ± 33 | ||
| General Electric | MOLLI 5b(3s)5b [1] | 11.18, 653 ± 73 | 7.69, 853 ± 66 | 8.45, 591 ± 50 | 6.68, 1319 ± 88 | |
| MOLLI 5b(1b)1b(1b)1bd [1]a | 1.40, 575 ± 8 | 1.76, 642 ± 11 | 0.78, 543 ± 4 | 1.19, 1002 ± 12 | ||
| SMART [1] | 2.05, 1063 ± 22 | 4.76, 1273 ± 61 | 0.95, 798 ± 8 | 4.22, 1430 ± 60 | ||
The term sequence refers to either MOLLI, ShMOLLI, SASHA, or SMART
MOLLI/ShMOLLI protocol nosology has the number of inversions per experiment as the total count of numbers outside brackets, image cycles are outside brackets, pause cycles are within brackets, and cycle lengths defined in terms of either heart beats (b) or seconds (s)
CoV coefficient of variation, ID identity code, GBCA gadolinium-based contrast agent, GE General Electric, MOLLI modified Look-Locker inversion recovery, rT1 “reference” slow inversion recovery T1, SASHA saturation-recovery single-shot acquisition, SD standard deviation, ShMOLLI shortened MOLLI, SMART saturation method using adaptive recovery times for cardiac T1 mapping, T Tesla
aDenotes the T1 mapping sequence|software combination with lowest overall CoV% for a given vendor where multiples exist. P values for differences in CoV between sequences/vendors are reported for this highly repeatable sequence where multiples exist. Less favorable CoVs (> 1%, see the “T1 repeatability” section) are in italics. Post-GBCA tubes are not shown here as their data are reported separately in relation to post-GBCA sequences in Supplementary Table 5
bDenotes the number of different magnets submitting that particular sequence from which the average CoVs were derived
cUsing iterative/data dropping steps in map creation as per ShMOLLI
dIn the absence of iterative/data dropping steps in map creation as per ShMOLLI
x̅ = average CoV across the 4 native tubes for a given sequence
x̅’ = where more than one sequence type was submitted, individual CoVs were then averaged to derive x̅’ CoV; while for single sequence submissions x̅’ CoV is from the global mean T1 ± SD for that one sequence
Table 2.
Temperature-adjusted (normalized to 21 °C) native T1 and CoV (%) at 3 T summarized by vendor and sequence
| Summary table ofx̅CoV ofT1(%) across 4 tubes |
Order of tube IDs follows their orientation in the scanned bottle
|
|||||
| Philips | MOLLI 3b(3s)5b | 0.33 | ||||
| Siemens | ShMOLLI 5b(1b)1b(1b)1b [780C] | 0.69 | ||||
| Siemens | MOLLI 5b(3s)3b [448B] | 0.95 | ||||
| Siemens | SASHA | 1.29 | ||||
| Philips | SASHA | 3.76 | ||||
| GE | SMART | 3.76 sic. | ||||
| Platform | Sequence [prototype, #] | x̅’ CoV ofT1(%), globalx̅± SD ofT1according to native tube ID [tube rT1 by slow IR] | ||||
| M [1260 ms] | L [1499 ms] | K [1010 ms] | J [1872 ms] | |||
| Siemens | MOLLI MyoMaps product 5s(3s)3s [4] | 1.28, 1182 ± 20 | 1.61, 1367 ± 29 | 1.02, 955 ± 13 | 2.42, 1831 ± 62 | |
| MOLLI 3b(3s)3b(3s)5b [448, 1] | 1.16, 1092 ± 13 | 1.92, 1252 ± 24 | 0.56, 889 ± 5 | 1.58, 1724 ± 27 | ||
| MOLLI 5b(3s)3b [448B, 1]a | 0.36, 1233 ± 4 | 0.78, 1449 ± 11 | 0.76, 986 ± 7 | 0.86, 2012 ± 17 | ||
| MOLLI 5b(3s)3b [780B, 2] | 0.99, 1204 ± 14 | 1.13, 1410 ± 18 | 0.74, 970 ± 9 | 1.50, 1912 ± 28 | ||
| MOLLI 5s(3s)3s [780B, 1] | 1.46, 1222 ± 18 | 2.36, 1435 ± 34 | 1.76, 980 ± 17 | 3.04, 1916 ± 58 | ||
| ShMOLLI 5b(1b)1b(1b)1b [780B, 2] | 1.17, 1155 ± 27 | 1.62, 1361 ± 35 | 0.80, 922 ± 17 | 2.45, 1882 ± 41 | ||
| ShMOLLI 5b(1b)1b(1b)1b [780C, 1]a | 0.78, 1109 ± 9 | 0.89, 1322 ± 12 | 0.64, 888 ± 6 | 1.51, 1892 ± 29 | ||
| SASHA [2] | 2.04, 1284 ± 30 | 1.15, 1534 ± 27 | 0.70, 1025 ± 13 | 1.27, 1949 ± 35 | ||
| Philips | MOLLI 3b(3s)3b(3s)5b [4] | 1.64, 1147 ± 52 | 2.42, 1343 ± 80 | 1.05, 923 ± 29 | 3.90, 1785 ± 115 | |
| MOLLI 3b(3s)5b [1]a | 0.01, 1055 ± 1 | 0.49, 1217 ± 6 | 0.05, 862 ± 1 | 0.78, 1710 ± 13 | ||
| MOLLI 3s(3s)5s [1] | 5.41, 1175 ± 64 | 3.72, 1392 ± 52 | 7.13, 937 ± 67 | 3.56, 1833 ± 65 | ||
| MOLLI 5b(3s)3b [3] | 4.18, 1098 ± 48 | 5.44, 1286 ± 73 | 4.37, 868 ± 42 | 6.33, 1784 ± 106 | ||
| MOLLI 5s(3s)3s [6] | 2.40, 1131 ± 70 | 2.41, 1320 ± 108 | 2.60, 906 ± 34 | 2.71, 1727 ± 131 | ||
| MOLLI 5b(1b)1b(1b)1bc [2] | 2.98, 948 ± 97 | 4.53, 1082 ± 137 | 0.96, 794 ± 60 | 6.29, 1286 ± 282 | ||
| SASHA [2] | 3.11, 1290 ± 46 | 3.40, 1826 ± 171 | 2.11, 1054 ± 38 | 6.41, 2157 ± 267 | ||
| General Electric | SMART [1] | 2.85, 1150 ± 33 | 4.13, 1227 ± 51 | 2.04, 973 ± 20 | 6.03, 1419 ± 86 | |
Less favorable CoVs (> 2%, see the “T1 repeatability” section) are in italics. Post-GBCA tubes are not shown here as their data are reported separately in relation to post-GBCA sequences in Supplementary Table 6
sic = text is quoted exactly as it stands in the original, i.e., this is not a typo. Abbreviations as in Table 1
aDenotes the T1 mapping sequence|software combination with lowest overall CoV% for a given vendor where multiples exist
bDenotes the number of different magnets submitting that particular sequence from which the average CoVs were derived
cIn the absence of iterative/data dropping steps in map creation as per ShMOLLI
x̅ = average CoV across the 4 native tubes for a given sequence
x̅’ = where more than one sequence type was submitted, individual CoVs were then averaged to derive x̅’ CoV; while for single sequence submissions x̅’ CoV is from the global mean T1 ± SD for that one sequence
Based on these temperature-adjusted data, mean (x̅) CoV were generally higher (implying poorer repeatability) at 3 T than at 1.5 T and for the much sparser GE data over Siemens/Philips. Over 1 year, many 1.5 T and 3 T sequences/magnets were repeatable with x̅ CoV < 1% and < 2% respectively. For sequences optimized for native T1 mapping applied to T1MES native tubes, we observed excellent repeatability for several sequences, for example, Siemens MOLLI 5s(3s)3s 448B (x̅ CoV = 0.27%) and Philips MOLLI 3s(3s)5s (x̅ CoV 0.54%) at 1.5 T (Table 1) and Philips MOLLI 3b(3s)5b (x̅ CoV 0.33%) and Siemens ShMOLLI (x̅ CoV 0.69%) at 3 T (Table 2).
Among sequences optimized for post-GBCA T1 mapping applied to T1MES post-GBCA tubes, excellent repeatability was observed for several sequences, for example, Siemens ShMOLLI 1041B (x̅ CoV 0.21%) and Siemens MOLLI 4s(1s)3s(1s)2s 448B (x̅ CoV 0.26%) at 1.5 T (Supplementary Table S5) and Siemens MOLLI 4b(1s)3b(1s)2b 448B (x̅ CoV 0.10%) and Siemens ShMOLLI (x̅ CoV 0.28%) at 3 T (Supplementary Table S5).
Sources of T1 variation
Linear mixed models which excluded centers that had experienced a software change during the time of data collection (Table 3 and Supplementary Tables S7 and S8) indicated that temperature-adjusted T1 differs significantly between sequences and software versions at both field strengths (P < 0.001 all) and between magnet models at 1.5 T (P = 0.011). Notably, phantom age had no significant effect on T1 (model A2).
Software upgrades
The two software upgrades on Siemens both occurred towards the end of longitudinal data submissions (Supplementary Fig. S4A, 15E031; 4B, 30E017), so the paucity of data points post-upgrade events precluded statistical testing for significant T1 shifts. For the software upgrade on GE, however, differences between pre- and post-linear regression slopes (Supplementary Fig. S4C, 30E012) indicate a marginally significant T1 shift event (P = 0.024).
Discussion
To our knowledge, this is the first multi-center study using a wide range of T1 mapping methods to study the interplay of some of the known sources of measured T1 variation. The basis of this work is the T1MES phantom, and a major aim of this work was to evaluate how this performed in use at multiple centers. The phantom appears sufficiently robust for T1 mapping quality assurance purposes. Based on our data, estimated phantom T1 values may show substantial variation between different T1 mapping sequences, scanner software versions, and potentially also scanner models, and temperature correction, at least for native T1 tubes, is necessary to achieve the desired repeatability. Inspection of the Supplementary Table also suggests that there are occasional performance variations producing outliers even within a given T1 mapping sequence prototype/software combination across different magnets. In spite of this variation, however, several specific combinations of field strength, sequence, and scanner generally exhibit excellent repeatability. Given the choices, the CMR community may prefer to standardize and use combinations with high repeatability for future clinical and research use, e.g., CoV < 1% at 1.5 T and < 2% at 3 T, while seeking to optimize less repeatable combinations. Less data were available for GE in T1MES, so the observed variability requires verification in a larger more representative sample.
Agreement with “reference” slow scanning, rT1 times was slightly greater for SASHA compared to MOLLI or ShMOLLI, which both slightly underestimated T1, mainly from the known T2-related underestimation which is larger when measuring the myocardium [13–15], (Fig. 4, Supplementary Tables S1 and S2) and not due to magnetization transfer, which is negligible in these agar phantoms [22].
The phantom data presented here suggest that the “T1 standard” framework remains possible, but the wide variety of different sequence options, vendors, and field strengths distributed the data over too many categories for reliable modeling, unlike the “locked-down” approach mentioned below which does have that strong advantage. Further work will explore the transferability of clinical measurement based on in vitro phantom calibration.
T1MES is but one of a number of phantom objects that have been used or proposed to support T1 mapping quality assurance (elaborated in Table 4). Some alternatives that target native and post-GBCA myocardial and blood relaxation times in support of T1 mapping work are (1) Brompton phantom by Vassiliou et al. [23], (2) Hypertrophic Cardiomyopathy Registry [24] phantom (HCMR) by Piechnik et al., and (3) International Society for Magnetic Resonance in Medicine (ISMRM)/NIST MR imaging phantom [25].
Table 4.
Comparison of recently reported phantoms for cardiac T1 mapping quality assurance
| Dedicated T1 device | Combined T1/T2 device | |||
|---|---|---|---|---|
| Phantom | HCMR (cardiac specific) | Brompton (cardiac specific) | T1MES (cardiac specific) | ISMRM/NIST system phantom (not cardiac specific) |
| Field strength specificity | ☒ | ☒ | ☑ | ☒ |
| Tube ingredients | NiCl2-doped agar + carrageenan | NiCl2-doped agar | NiCl2-doped agar | NiCl2-doped water, MnCl2-doped agar |
| Structure | 9 T1/T2 tubes in an amber PVC sealed jar that is ~ 25% smaller in total volume compared to T1MES. | Air-filled box containing 4 duplicate T1/T2 glued glass tubes (total 8) requires cylindrical MRI test bottles on either side for B1 field/reference frequency calibration. | 9 plastic T1/T2 tubes in an amber PVC sealed jar with inter-tube gaps packed with a carefully specified agarose/ HDPE plastic macrobead/NiCl2-doped fill that flattened the B1 field and provided sufficient B0 homogeneity to obviate the need for side phantoms. | A layer of 14 T1 spheres and a separate layer of 14 T2 spheres so no unified T1/T2 compartment representing the relaxation parameters of the human heart. |
| Cardiac T1/T2coverage | Health and disease, 9 biologies but limited T2 coverage (57 and 75 ms). | Health only, 4 biologies: (1) native myocardium, (2) native blood, (3) post-GBCA myocardium, and (4) post-GBCA blood. | Health and disease, 9 biologies and broad T2 coverage (1.5 T, 44, 48, 50, 155, 189, and 243 ms). Also see Supplementary Table 3. | Health and disease, but of the 14 T1 spheres (ranging from 21 to 2038 ms), half are not useful to cardiac T1 mapping as T1 times are too short for either native or post-GBCA myocardium/blood. Half the T2 spheres (ranging from 11 to 581 ms) are also anti-physiological for cardiac mapping. Also, the T1/T2 ratio is not representative of cardiac tissue. |
| MT coverage | – | – | – | – |
| Regulatory clearance | – | – | FDA, CE-mark | – |
| Developers | Piechnik et al. | Vassiliou et al. | Captur et al. | NIST/ISMRM |
CE Conformitée-Europeen, FDA Food and Drug Administration, HDPE high-density polyethylene, MT magnetization transfer, PVC poly vinyl chloride
At 1 year, CoV for T1 tubes of the Vassiliou [26] phantom on a single Siemens scanner ranged from 1.0 to 3.6% using only the native MOLLI 5s(3s)3s sequence, compared to 0.27 to 3.0% in T1MES (for 1.5 T MOLLI 5s(3s)3s [448B] on Siemens to SMART on GE respectively). This within sequence precision heterogeneity, as already alluded to by Kellman et al. [27], linked to protocol modifications within MOLLI, may partly explain some of the higher CoV for specific MOLLI sequences compared to ShMOLLI where centers obligatorily scanned using a fixed 5b(1b)1b(1b)1b sequence with conditional fitting. As mentioned above for the dispersion of T1MES results, this “real-world” heterogeneity of T1 mapping sequences highlights that while T1 mapping research and innovation calls for multiple flexible prototypes, the resultant diversity poses a nontrivial challenge to multi-center standardization. Conversely, the more “locked-down” approach like that adopted for ShMOLLI, albeit less editable by external researchers, potentially facilitates standardization.
The design of phantoms for this purpose (although often seen as trivial) is challenging: to have a large sufficient ROI in each sample tube and a good number of sample tubes in the main jar of the phantom, the overall size of the phantom increases to the point that B1 non-uniformity at 3 T due to the dielectric permittivity of any water-based phantom becomes problematic. While of course B1 is non-uniform in vivo, one purpose of phantom tests is to eliminate uncertainty or at least enable controlled testing of factors such as B1 rather than introduce their own errors. Conversely, if the phantom is made very small, then the truly acquired pixel size of in vivo T1 mapping methods (which obviously must not be modified or adapted [28] for the phantom scans) becomes important compared to the sample tube inner diameters leading to questions over SNR [29] and the impact of Gibbs artifact, for example.
Another obstacle for relaxation time phantoms (on top of basic water T1 and T2 temperature and pressure variations, gel instabilities [30, 31], small leakages, dehydration, or ion migration effects over time) is the differing temperature dependence of each species of paramagnetic ion’s relaxivity (r1, r2) and furthermore frequency dispersion in all of these physical properties between 60 MHz and 120 MHz [32–36]. Efforts to use alternative chemistry have so far not provided the range of T1 and T2 needed for this application.
The results confirmed, as is widely known in the field, that the T1 measured by even nominally identical mapping sequences can differ significantly between software revisions. Scanner software upgrades are not uncommon in centers, and we encountered three such events among contributing partners. There was a measurable shift on at least one system, with potentially important implications for the field as alluded to in the 2017 consensus statement on multi-parametric mapping by the Society for Cardiovascular Magnetic Resonance (SCMR) [37]. Future work using phantoms at scale, across vendors and over time, would ideally continue monitoring center-specific data before and after potential shift events to more fully understand their impact on T1 allowing the community to set up mitigation approaches.
Clearly, sources of T1 variance and drift over time and between centers are numerous. Some T1 variance is related to differences between sequences and magnet platforms: to the scanner itself (equipment drift or equipment shifts from updates of software, replacement of parts, routine service recalibrations, etc.), to the environment (temperature, pressure, humidity), and to the operator (phantom positioning inside scanner bore). In a phantom study, some T1 variance may additionally have been due to intra-phantom differences (liquid/agar status over time collectively contributing to phantom aging) and inter-phantom manufacturing variations. In T1MES, we mitigated potential inter-phantom manufacturing variations by exercising extreme caution in design and prototyping and by the strict batch manufacturing processes to medical device grade standards. Drifts in the system setup (long-term stability) would tend to be “adjusted” out by the shim and center frequency (CF) and transmitter B1 reference (TxREF) adjustments that could otherwise bias estimated T1. Any receiver coil or gain changes would most likely cancel out of T1 maps as constant across the series of input images to T1 derivation (appearing in A and B maps instead). It is considered unlikely that gradient performance would drift significantly, and any significant sudden changes in that are also unlikely even when the eddy-current compensation is adjusted on routine service visits. However, we have shown how unforeseen software changes could potentially cause “step-wise” changes in the estimated T1 records. To directly model the interplay between all these potential sources of T1 variation in longitudinal phantom studies would be a daunting task, and we considered only a subset of these sources in our analyses.
Though B0 and B1 homogeneity tests in T1MES (Fig. 5) were stable, we did measure slightly greater field inhomogeneity at the phantom edge and corners compared to the core (phantom corners and edges are the zones which our T1MES data flagged up as areas of least B0/B1 homogeneity). The tubes with the highest T1 times are the tubes most susceptible to such field inhomogeneities, and this suggests that a future iteration of T1MES could be further optimized by rearrangement to position such tubes in the most uniform regions.
The results of this study are relevant to clinicians or researchers choosing sequences for T1 mapping, and we highlight 3 take home messages as follows:
-
i.
Sequence/software combinations in Tables 1 and 2, with a CoV ≤ 1% and 2% respectively, are sufficiently repeatable for clinical/research use (see rationale in the “Methods part 3—Statistical analysis” and “T1 repeatability” sections). For the native myocardium T1MES tube at 1.5 T (reference T1 ~ 1037 ms by MOLLI), a ≤ 1% CoV is well within ± 1SD of normal values for T1, meaning that small-magnitude biological changes (e.g., diffuse myocardial fibrosis), will be resolvable with high precision [37]. By contrast, CoVs of 3 or 6% at 1.5 T (variance of 31 and 62 ms respectively) will undermine a center’s ability to resolve small-magnitude changes, and they may also impact the resolution of large-magnitude biological changes [37] (like amyloid, iron overload, Fabry disease, acute myocardial injury). Our data indicate that many of the investigated sequences for Siemens and Philips at 1.5 T exhibited CoVs ≤ 1%. At 3 T, MOLLI (Siemens and Philips), ShMOLLI (Siemens), and SASHA (Siemens) exhibited CoVs ≤ 2%. The absence of GE from this list is due to limited participation.
-
ii.
For developers, the highlighting of sequence performance may encourage work to refine sequences in some cases.
-
iii.
The prospect of using phantom calibration to remove the need for local reference ranges [37] is potentially becoming more tangible than before, yet more work is needed to deliver such calibration.
These data may facilitate the use of T1 mapping as a useful clinical biomarker. T1 standardization will also be important to facilitate clinical research. At the present time, there are 25 registered clinical research studies using T1 mapping (clinicaltrials.gov accessed January 2019).
Limitations
Presented results are not intended to compare the SNR obtained in the phantom setup in different machines but are aimed at studying long-term stability. In spite of our outreach to advertise enrollment at study start, GE centers are under-represented. The scarcity of multi-center data for many of the specific T1 mapping sequence, software, and field-strength combinations has limited our ability to deliver a “T1 standard”. Eight centers enrolled, received a phantom, but then could not deliver the serial phantom scans, and four centers did not meet the 1 year minimum data submission requirement. Each center provided legitimate justifications for the missing data, and furthermore, we highlight that contributions to T1MES were entirely voluntary, with no center receiving remuneration for any staff or scanner time invested in the program.
In total, 3 magnets deviated from the prescribed nominal intervals in some of their initially submitted datasets. Phantom repeatability studies represent an in vitro experiment. Some sequences and scanners that performed well in vitro may exhibit greater variability in the face of biological tissue (magnetization transfer, flow, motion), patient characteristics (fast/slow heart rates, heart rate variability, breath-hold length, implanted metal devices), and scanning process (non-isocentering, scanning in multiple planes). The T1MES phantom does not capture magnetization transfer. The biomarker performance that was measured here accounts for T1 mapping in vitro. While this is obviously related to diagnostic performance in vivo, cardiac motion, etc., introduce yet another degree of freedom, and different hardware/software combinations might deal with this differently. In addition, CoV as a single metric may not be the best way to pick a pulse sequence for repeatability when the data is only derived from a static phantom. A very precise (reproducible) T1 sequence that requires long breatholds or is heart rate-variability-sensitive might perform very well with the T1MES phantom setup but perform poorly in patients with varying biology, e.g., that cannot hold their breath well or who are in atrial fibrillation. The phantom has no intracellular/extracellular component, so any researcher seeking to model “ECV” using pre- and post-GBCA T1 mapping values derived from T1MES (see example data in Supplementary Table S9) must bear this in mind.
The phantom design, ROI size, and coil setup instructions all aimed to make the fundamental image noise SNR an irrelevant factor in the CoV results. The artificially high and “clean” SNR of a phantom setup (if without phantom-related distortions, as we have shown in the “B0 and B1 uniformity” section) is clearly not directly an indicator of satisfactory in vivo performance. It contains many sequence-related factors that may have different consequences in vivo [29], and such SNR comparisons are not the aim of this work. Ideally, participating sites would have proved this by sending multiple T1 maps per session. However, contributions were already voluntary, and further demands on sites could not be supported. For the same reason, it was not feasible to demand long reference T1/T2 data for each participating site, and this was never part of the original enrollment commitment. As some indication of the typical short-term thermal noise random jitter, the raw (temperature-unadjusted) T1 values from some sites that did anyway submit several repeated T1 maps each session are plotted in Supplementary Fig. S3.
Conclusion
The T1MES phantom developed to CE/FDA manufacturing standards for T1 mapping is a robust quality assurance device. T1 mapping can now be quality assured on a multi-center scale, fulfilling a key requirement for the use of T1 mapping in clinical decision-making or as a surrogate endpoint in drug trials. A good number of T1 measurement sequence/scanner model/vendor/field strength combinations are remarkably repeatable, but some combinations less so, with coefficients of variation exceeding 1–2% over 1 to 2 years. Given the alternatives, we recommend that combinations with poor repeatability are deprioritized for clinical and research use. In spite of the use of a large number of different sequence prototypes and products in this study, a number of agreements and encouraging results are reported. Phantom calibration of T1 mapping which obviates the need for local reference ranges, enabling the establishment of a “T1 standard” to facilitate multi-center T1 studies, comes closer, with further work required to address this.
Supplementary information
Additional file 1: Supplementary Movie 1. High-resolution imaging of phantom 30E017 at baseline (October 2015, Left panel) and at two years post manufacturing (Right panel). (MP4 69,750 kb)
Additional file 2: Supplementary file containinglist of Supplementary Tables and Figures referenced in the main text.
Additional file 3. Supplementary data file reporting all the center-, session- and sequence-specific T1 mapping contributions to the T1MES program.
Acknowledgements
We thank all other members and contributors of the T1MES consortium listed in the Supplementary Material, and we also thank staff at Siemens Healthineers, Philips Healthcare, and General Electric Healthcare for their expert review of the final manuscript. Dr. Matthias Friedrich served as a JCMR Guest Editor for this manuscript.
Abbreviations
- 3D
Three-dimensional
- AIC
Akaike information criteria
- BIC
Bayesian information criteria
- CE
Conformité Européenne
- CF
Center frequency
- CMR
Cardiovascular magnetic resonance
- CoV
Coefficient of variation
- DICOM
Digital Imaging and COmmunications in Medicine
- ECV
Extracellular volume
- FA
Flip angle
- FDA
Food and Drug Administration
- GBCA
Gadolinium-based contrast agent
- GE
General Electric Healthcare
- HCMR
Hypertrophic Cardiomyopathy Registry
- INR
International normalized ratio
- IRSE
Inversion recovery spin echo
- ISMRM
International Society for Magnetic Resonance in Medicine
- MOLLI
Modified Look-Locker inversion recovery
- NIST
National Institute of Standards and Technology
- PTB
Physikalisch-Technische Bundesanstalt
- QIBA
Quantitative Imaging Biomarkers Alliance
- REDCap
Research electronic data capture
- ROI
Region of interest
- RSNA
Radiological Society of North America
- rT1
“Reference” T1
- rT2
“Reference” T2
- SASHA
Saturation recovery single-shot acquisition
- SCMR
Society for Cardiovascular Magnetic Resonance
- SD
Standard deviation
- ShMOLLI
Shortened MOLLI
- SMART
Saturation method using adaptive recovery times for cardiac T1 mapping
- SNR
Signal-to-noise ratio
- T
Tesla
- T1MES
T1 mapping and extracellular volume standardization
- TE
Echo time
- TR
Repetition time
- TxREF
Transmitter B1 reference
Authors’ contributions
JCM together with PG and GC conceived the project. PG provided continuous expert physics support for the project, oversaw the experimental testing of phantoms from prototype out to 2 years, scanned the phantom, expertly reviewed the manuscript and analysis, and assisted with all aspects of the project. GC and PG designed the prototypes and final phantom. GC curated the phantom data, managed the consortium, developed the analytical pipeline (with help from PK and RB), organized and analyzed the data, scanned the phantom including experimental tests, performed the statistical analysis, wrote the manuscript, and built the figures. JCM expertly reviewed the manuscript and assisted with all aspects of the project. AB, YY, RJE, GB, CT, LR, and NF analyzed the data. RB, BI, and KK performed the PTB and NIST experiments respectively. RB and PK provided analytical support and scanned the phantom. DMH, AG, DAB, RZ, TD, and JJ provided expert appraisal of the manuscript and analytical approaches. JPG, LEMP, CBL, CBD, HJL, RS, SWL, CB, SV, AF, CdL, FDC, MV, PC, DD, AJT, FPM, SP, DM, MAM, SMG, ML, JJ, and RN scanned the phantom. MS and NC provided expert advice. All authors reviewed the manuscript. The authors read and approved the final manuscript.
Funding
This program was funded by the following grants to GC: European Association of Cardiovascular Imaging (EACVI) of the European Society of Cardiology (ESC), the UK National Institute of Health Research (NIHR) Biomedical Research Center (BRC) at University College London (UCL, #BRC/199/JM/101320), and the Barts Charity (#1107/2356/MRC0140). GC is supported by the National Institute for Health Research Rare Diseases Translational Research Collaboration (NIHR RD-TRC) and by the NIHR UCL Hospitals Biomedical Research Center. JCM is directly and indirectly supported by the UCL Hospitals NIHR BRC and Biomedical Research Unit at Barts Hospital respectively. This work was supported by the NIHR infrastructure at Leeds.
CBD is supported directly and indirectly by the NIHR Biomedical Research Centre at University Hospitals Bristol NHS Foundation Trust and the University of Bristol. The views expressed in this publication are those of the author(s) and not necessarily those of the NHS, the National Institute for Health Research, or the Department of Health and Social Care.
NIST disclaimer: Contribution of the National Institute of Standards and Technology not subject to copyright in the United States. Certain commercial instruments and software are identified to specify the experimental study adequately. This does not imply endorsement by NIST or that the instruments and software are the best available for the purpose.
PTB disclaimer: Certain commercial instruments and software are identified to specify the experimental study adequately. This does not imply endorsement by PTB or that the instruments and software are the best available for the purpose.
Availability of data and materials
Datasets collected for the current study are available from the corresponding author upon reasonable request.
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
An industrial group (Resonance Health) had been funded to mass-produce the described T1MES phantoms to CE mark/FDA standards to permit international distribution in the T1MES academic program. WP is an employee of Resonance Health. AG is an employee of Siemens Healthineers. PH is an employee of Philips Healthcare. No financial or non-financial competing interests exist for any of the other authors.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Gabriella Captur, Email: gabriella.captur@ucl.ac.uk.
Abhiyan Bhandari, Email: abhiyan.bhandari.16@ucl.ac.uk.
Rüdiger Brühl, Email: ruediger.bruehl@ptb.de.
Bernd Ittermann, Email: bernd.ittermann@ptb.de.
Kathryn E. Keenan, Email: kathryn.keenan@nist.gov
Ye Yang, Email: yeyang1222@zju.edu.cn.
Richard J. Eames, Email: eames.richard@gmail.com
Giulia Benedetti, Email: giulietta.benedetti@gmail.com.
Camilla Torlasco, Email: camilla.torlasco@gmail.com.
Lewis Ricketts, Email: lewis.ricketts.13@ucl.ac.uk.
Redha Boubertakh, Email: r.boubertakh@qmul.ac.uk.
Nasri Fatih, Email: n.fatih@ucl.ac.uk.
John P. Greenwood, Email: j.greenwood@leeds.ac.uk
Leonie E. M. Paulis, Email: leonie.paulis@mumc.nl
Chris B. Lawton, Email: chris.lawton@uhbristol.nhs.uk
Chiara Bucciarelli-Ducci, Email: c.bucciarelli-ducci@bristol.ac.uk.
Hildo J. Lamb, Email: h.j.lamb@lumc.nl
Richard Steeds, Email: rick.steeds@uhb.nhs.uk.
Steve W. Leung, Email: steve.leung@uky.edu
Colin Berry, Email: colin.berry@glasgow.ac.uk.
Sinitsyn Valentin, Email: vsini@mail.ru.
Andrew Flett, Email: drflett@gmail.com.
Charlotte de Lange, Email: charlotte.de.lange@rr-research.no.
Francesco DeCobelli, Email: decobelli.francesco@hsr.it.
Magalie Viallon, Email: magalie.viallon@creatis.insa-lyon.fr.
Pierre Croisille, Email: croisille@creatis.insa-lyon.fr.
David M. Higgins, Email: david.higgins@philips.com
Andreas Greiser, Email: andreas.greiser@siemens-healthineers.com.
Wenjie Pang, Email: wenjiep@resonancehealth.com.
Christian Hamilton-Craig, Email: christian.hamilton-craig@health.qld.gov.au.
Wendy E. Strugnell, Email: wendy.strugnell@health.qld.gov.au
Tom Dresselaers, Email: tom.dresselaers@uzleuven.be.
Andrea Barison, Email: dr.andrea.barison@gmail.com.
Dana Dawson, Email: dana.dawson@abdn.ac.uk.
Andrew J. Taylor, Email: andrew.taylor@bakeridi.edu.au
François-Pierre Mongeon, Email: francois-pierre.mongeon@icm-mhi.org.
Sven Plein, Email: s.plein@leeds.ac.uk.
Daniel Messroghli, Email: daniel.messroghli@charite.de.
Mouaz Al-Mallah, Email: mouaz74@gmail.com.
Stuart M. Grieve, Email: stuart.michael.grieve@gmail.com
Massimo Lombardi, Email: massimo.lombardi@grupposandonato.it.
Jihye Jang, Email: jjang1@bidmc.harvard.edu.
Michael Salerno, Email: ms5pc@hscmail.mcc.virginia.edu.
Nish Chaturvedi, Email: n.chaturvedi@ucl.ac.uk.
Peter Kellman, Email: kellmanp@nhlbi.nih.gov.
David A. Bluemke, Email: dbluemke@rsna.org
Reza Nezafat, Email: rnezafat@bidmc.harvard.edu.
Peter Gatehouse, Email: p.gatehouse@rbht.nhs.uk.
James C. Moon, Email: j.moon@ucl.ac.uk
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12968-020-00613-3.
References
- 1.Stonnington CM, Tan G, Klöppel S, Chu C, Draganski B, Jack CR, Chen K, Ashburner J, Frackowiak RSJ. Interpreting scan data acquired from multiple scanners: a study with Alzheimer’s disease. Neuroimage. 2008;39:1180–1185. doi: 10.1016/j.neuroimage.2007.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nordin S, Kozor R, Baig S, Abdel-Gadir A, Medina-Menacho K, Rosmini S, Captur G, Tchan M, Geberhiwot T, Murphy E, Lachmann R, Ramaswami U, Edwards NC, Hughes D, Steeds RP, Moon JC. Cardiac phenotype of prehypertrophic Fabry disease. Circ Cardiovasc Imaging. 2018;11:e007168. doi: 10.1161/CIRCIMAGING.117.007168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keenan KE, Biller JR, Delfino JG, Boss MA, Does MD, Evelhoch JL, Griswold MA, Gunter JL, Hinks RS, Hoffman SW, Kim G, Lattanzi R, Li X, Marinelli L, Metzger GJ, Mukherjee P, Nordstrom RJ, Peskin AP, Perez E, Russek SE, Sahiner B, Serkova N, Shukla-Dave A, Steckner M, Stupic KF, Wilmes LJ, Wu HH, Zhang H, Jackson EF, Sullivan DC. Recommendations towards standards for quantitative MRI (qMRI) and outstanding needs. J Magn Reson Imaging. 2019;0. 10.1002/jmri.26598 Epub ahead of print. [DOI] [PMC free article] [PubMed]
- 4.Lecompte T, Samama MM. Monitoring of treatment with vitamin K antagonists: a plea for “INR” (international normalized ratio) Rev Prat. 1990;40:563–565. [PubMed] [Google Scholar]
- 5.Captur G, Gatehouse P, Keenan KE, Heslinga FG, Bruehl R, Prothmann M, Graves MJ, Eames RJ, Torlasco C, Benedetti G, Donovan J, Ittermann B, Boubertakh R, Bathgate A, Royet C, Pang W, Nezafat R, Salerno M, Kellman P, Moon JC. A medical device-grade T1 and ECV phantom for global T1 mapping quality assurance-the T1 Mapping and ECV Standardization in cardiovascular magnetic resonance (T1MES) program. J Cardiovasc Magn Reson. 2016;18:58–70. doi: 10.1186/s12968-016-0280-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang EP, Raunig DL, Kessler LG, Kondratovich M, Obuchowski NA, Sullivan DC, Reeves AP, McShane LM, Wahl RL, Barboriak DP, Gatsonis C, Guimaraes AR. Metrology standards for quantitative imaging biomarkers. Radiology. 2015;277:813–825. doi: 10.1148/radiol.2015142202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hernando D, Kellman P, Haldar JP, Liang Z. Robust water/fat separation in the presence of large field inhomogeneities using a graph cut algorithm. Magn Reson Med. 2010;63:79–90. doi: 10.1002/mrm.22177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kellman P, Herzka DA, Arai AE, Hansen MS. Influence of off-resonance in myocardial T1-mapping using SSFP based MOLLI method. J Cardiovasc Magn Reson. 2013;15:63–73. doi: 10.1186/1532-429X-15-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)-a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42:377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Captur G, Stables RH, Kehoe D, Deanfield J, Moon JC. Why democratize bioinformatics? BMJ Innov. 2016;2:166–171. doi: 10.1136/bmjinnov-2016-000129. [DOI] [Google Scholar]
- 11.Diedenhofen B, Musch J. Cocor: a comprehensive solution for the statistical comparison of correlations. PLoS One. 2015;10:e0121945. doi: 10.1371/journal.pone.0121945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu JM, Liu A, Leal J, McMillan F, Francis J, Greiser A, Rider OJ, Myerson S, Neubauer S, Ferreira VM, Piechnik SK. Measurement of myocardial native T1 in cardiovascular diseases and norm in 1291 subjects. J Cardiovasc Magn Reson. 2017;19:74–84. doi: 10.1186/s12968-017-0386-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosmini S, Bulluck H, Captur G, Treibel TA, Abdel-Gadir A, Bhuva AN, Culotta V, Merghani A, Fontana M, Maestrini V, Herrey AS, Chow K, Thompson RB, Piechnik SK, Kellman P, Manisty C, Moon JC. Myocardial native T1 and extracellular volume with healthy ageing and gender. Eur Heart J Cardiovasc Imaging. 2018;19:615–621. doi: 10.1093/ehjci/jey034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roy C, Slimani A, de Meester C, Amzulescu M, Pasquet A, Vancraeynest D, Vanoverschelde J-L, Pouleur A-C, Gerber BL. Age and sex corrected normal reference values of T1, T2 T2* and ECV in healthy subjects at 3T CMR. J Cardiovasc Magn Reson. 2017;19:72–82. doi: 10.1186/s12968-017-0371-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Granitz M, Motloch LJ, Granitz C, Meissnitzer M, Hitzl W, Hergan K, Schlattau A. Comparison of native myocardial T1 and T2 mapping at 1.5T and 3T in healthy volunteers : reference values and clinical implications. Wien Klin Wochenschr. 2018. 10.1007/s00508-018-1411-3 [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 16.Higgins D, Keeble C, Juli C, Dawson D, Waterton J. Reference range determination for imaging biomarkers: myocardial T1. J Magn Reson Imaging. 2019;50:771–8. [DOI] [PubMed]
- 17.Messroghli DR, Radjenovic A, Kozerke S, Higgins DM, Sivananthan MU, Ridgway JP. Modified Look-Locker inversion recovery (MOLLI) for high-resolution T1 mapping of the heart. Magn Reson Med. 2004;52:141–146. doi: 10.1002/mrm.20110. [DOI] [PubMed] [Google Scholar]
- 18.Piechnik SK, Ferreira VM, Dall’Armellina E, Cochlin LE, Greiser A, Neubauer S, Robson MD. Shortened Modified Look-Locker inversion recovery (ShMOLLI) for clinical myocardial T1-mapping at 1.5 and 3T within a 9 heartbeat breathhold. J Cardiovasc Magn Reson. 2010;12:69–79. doi: 10.1186/1532-429X-12-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chow K, Kellman P, Spottiswoode BS, Nielles-Vallespin S, Arai AE, Salerno M, Thompson RB. Saturation pulse design for quantitative myocardial T1 mapping. J Cardiovasc Magn Reson. 2015;17:84–94. doi: 10.1186/s12968-015-0187-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Slavin GS, Stainsby JA. True T1 mapping with SMART1 Map (saturation method using adaptive recovery times for cardiac T1 mapping): a comparison with MOLLI. J Cardiovasc Magn Reson. 2013;15(Suppl 1):P3. doi: 10.1186/1532-429X-15-S1-P3. [DOI] [Google Scholar]
- 21.Muggeo VM. Segmented: an R package to fit regression models with broken-line relationships. R News. 2008;20–25:1–73. [Google Scholar]
- 22.Robson MD, Piechnik SK, Tunnicliffe EM, Neubauer S. T1 measurements in the human myocardium: the effects of magnetization transfer on the SASHA and MOLLI sequences. Magn Reson Med. 2013;70:664–670. doi: 10.1002/mrm.24867. [DOI] [PubMed] [Google Scholar]
- 23.Vassiliou VS, Heng EL, Gatehouse PD, Donovan J, Raphael CE, Giri S, Babu-narayan SV, Gatzoulis MA, Pennell DJ, Prasad SK, Firmin DN. Magnetic resonance imaging phantoms for quality-control of myocardial T1 and ECV mapping: specific formulation, long-term stability and variation with heart rate and temperature. J Cardiovasc Magn Reson. 2016;18:1–12. [DOI] [PMC free article] [PubMed]
- 24.Kramer CM, Appelbaum E, Desai MY, Desvigne-Nickens P, DiMarco JP, Friedrich MG, Geller N, Heckler S, Ho CY, Jerosch-Herold M, Ivey EA, Keleti J, Kim D-Y, Kolm P, Kwong RY, Maron MS, Schulz-Menger J, Piechnik S, Watkins H, Weintraub WS, Wu P, Neubauer S. Hypertrophic Cardiomyopathy Registry: the rationale and design of an international, observational study of hypertrophic cardiomyopathy. Am Heart J. 2015;170:223–230. doi: 10.1016/j.ahj.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keenan KE, Stupic K, Boss M, Russek S, Chenevert T, Prasad P, Reddick W, Cecil K, Zheng J, Hu P, Jackson E. Proceedings of the 24th annual meeting of ISMRM, Singapore. 2016. Multi-site, multi-vendor comparison of T1 measurement using ISMRM/NIST system phantom. [Google Scholar]
- 26.Vassiliou V, Heng E, Donovan J, Greiser A, Babu-Narayan SV, Gatzoulis MA, Firmin D, Pennell DJ, Gatehouse P, Prasad SK. Longitudinal stability of gel T1 MRI phantoms for quality assurance of T1 mapping. J Cardiovasc Magn Reson. 2015;17(Suppl 1):W28. doi: 10.1186/1532-429X-17-S1-W28. [DOI] [Google Scholar]
- 27.Kellman P, Hansen MS. T1-mapping in the heart: accuracy and precision. J Cardiovasc Magn Reson. 2014;16:2–12. doi: 10.1186/1532-429X-16-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kellner E, Dhital B, Kiselev VG, Reisert M. Gibbs-ringing artifact removal based on local subvoxel-shifts. Magn Reson Med. 2016;76:1574–1581. doi: 10.1002/mrm.26054. [DOI] [PubMed] [Google Scholar]
- 29.Kouwenhoven M. On the impact of slice thickness definition and slice profile in 2D bSSFP on SNR and T1 mapping in cardiac MRI for multi-center trials. J Cardiovasc Magn Reson. 2019;2019:22–23. [Google Scholar]
- 30.Mitchell MD, Kundel HL, Axel L, Joseph PM. Agarose as a tissue equivalent phantom material for NMR imaging. Magn Reson Imaging. 1986;4:263–266. doi: 10.1016/0730-725X(86)91068-4. [DOI] [PubMed] [Google Scholar]
- 31.Yoshida A, Kato H, Kuroda M, Hanamoto K. Development of a phantom compatible for MRI and hyperthermia using carrageenan gel–relationship between T 1 and T 2 values and NaCl concentration. Int J Hyperth. 2004;8:803–814. doi: 10.1080/0265673042000199268. [DOI] [PubMed] [Google Scholar]
- 32.Koenig SH, Brown RD. Relaxation of solvent protons by paramagnetic ions and its dependence on magnetic field and chemical environment: implications for NMR imaging. Magn Reson Med. 1984;1:478–495. doi: 10.1002/mrm.1910010407. [DOI] [PubMed] [Google Scholar]
- 33.Jiang Y, Ma D, Keenan KE, Stupic KF, Gulani V, Griswold MA. Repeatability of magnetic resonance fingerprinting T1 and T2 estimates assessed using the ISMRM/NIST MRI system phantom. Magn Reson Med. 2017;78:1452–1457. doi: 10.1002/mrm.26509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laukien G, Schlüter JZ. Impulstechnishe messungen der spin-gitter und spin-spin-relaxationszeiten von protonen in waessrigen loesungen paramagnetischer Ionen. Physik. 1956;146:113–114. doi: 10.1007/BF01326008. [DOI] [Google Scholar]
- 35.Hausser R, Laukien G. Messung und deutung der temperaturabhängigkeit der kernmagnetischen protonenrelaxation in wäßrigen lösungen von Ionen der I. Gruppe der Übergangselemente. Z Phys. 1959;153:394–411. doi: 10.1007/BF01337607. [DOI] [Google Scholar]
- 36.Morgan LO, Nolle AW. Proton spin relaxation in aqueous solutions of paramagnetic ions. II. Cr+++, Mn++, Ni++, cu++, and Gd+++ J Chem Phys. 1959;31:365–368. doi: 10.1063/1.1730360. [DOI] [Google Scholar]
- 37.Messroghli DR, Moon JC, Ferreira VM, Grosse-Wortmann L, He T, Kellman P, Mascherbauer J, Nezafat R, Salerno M, Schelbert EB, Taylor AJ, Thompson R, Ugander M, van Heeswijk RB, Friedrich MG. Clinical recommendations for cardiovascular magnetic resonance mapping of T1, T2, T2* and extracellular volume: a consensus statement by the Society for Cardiovascular Magnetic Resonance (SCMR) endorsed by the European Association for Cardiovascular Imaging. J Cardiovasc Magn Reson. 2017;19:75–85. doi: 10.1186/s12968-017-0389-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Movie 1. High-resolution imaging of phantom 30E017 at baseline (October 2015, Left panel) and at two years post manufacturing (Right panel). (MP4 69,750 kb)
Additional file 2: Supplementary file containinglist of Supplementary Tables and Figures referenced in the main text.
Additional file 3. Supplementary data file reporting all the center-, session- and sequence-specific T1 mapping contributions to the T1MES program.
Data Availability Statement
Datasets collected for the current study are available from the corresponding author upon reasonable request.