Abstract
Due to their crucial biochemical roles, membrane proteins are important drug targets. Although it is clear that lipids can influence membrane protein function, the chemistry of lipid binding remains difficult to study because protein-lipid interactions are polydisperse, competitive, and transient. Furthermore, detergents, which are often used to solubilize membrane proteins in micelles, may disrupt lipid interactions that occur in bilayers. Here, we present two new approaches to quantify protein-lipid interactions in bilayers and understand how membrane proteins remodel their surrounding lipid environment. First, we used mass spectrometry (MS) to measure the exchange of lipids between lipoprotein nanodiscs with and without an embedded membrane protein. Shifts in the lipid distribution towards the membrane protein nanodiscs revealed lipid binding, and titrations allowed measurement of the optimal lipid composition for the membrane protein. Second, we used native or nondenaturing MS to ionize membrane protein nanodiscs with heterogeneous lipids. Ejecting the membrane protein complex with bound lipids in the mass spectrometer revealed enrichment of specific lipids around the membrane protein. Both new approaches showed that the E. coli ammonium transporter AmtB prefers phosphatidylglycerol lipids overall but has a minor affinity for phosphatidylcholine lipids.
Graphical Abstract
Membrane proteins are important drug targets for a range of diseases and play critical biochemical roles as transporters, receptors, and enzymes. Although membrane proteins are often studied outside of their natural membrane environment in detergent micelles, there is growing evidence that lipids can be essential for their structure, function, and stability.1,2 However, the molecular mechanisms of protein-lipid interactions are often unclear. Great strides have been made in computational modeling of lipid bilayer systems,3–5 but protein-lipid interactions remain difficult to study experimentally.
One central question is how membrane proteins remodel their surrounding lipid environment. In other words, how do the lipids surrounding the membrane protein differ from the bulk lipid bilayer? Because protein-lipid interactions are often transient and polydisperse, they are difficult to capture by traditional structural biology techniques. NMR and EPR have been successful for probing protein-lipid interactions but generally require labelled lipids.6,7 Liquid chromatography-mass spectrometry (LC-MS) can identify lipids that co-purify with membrane proteins in detergents8 or styrene maleic acid lipid particles (SMALPs)9 and uncover naturally-bound lipids. However, fast exchange of lipids in micelles and SMALPs may disrupt some natural interactions.10,11 Native MS has also been used to study lipid binding but largely probes small numbers of tightly-bound lipids in micelles.12–14
Here, we present two new experimental approaches to study how membrane proteins remodel the surrounding lipid bilayer in lipoprotein nanodiscs, which are nanoscale lipid bilayers encircled by two membrane scaffold protein (MSP) belts.15 First, we measure how membrane proteins shift the equilibrium lipid distribution between nanodiscs with and without embedded membrane proteins as lipids exchange between the two populations. Second, we perform native MS on membrane proteins in mixed lipid nanodiscs and eject membrane protein-lipid complexes with collisional activation in the mass spectrometer to quantify the bound lipid composition. We demonstrate both methods with AmtB, a trimeric E. coli ammonium transporter. Prior research has shown that AmtB binds phosphatidylglycerol (PG) lipids and that PG is important for its activity.16,17 Our results from both lipid exchange-mass spectrometry (LX-MS) and native MS reveal that the lipid annulus around AmtB is preferentially enriched in PG lipids but also has an affinity for phosphatidylcholine (PC) lipids.
Prior research has shown that lipids slowly exchange between nanodiscs, ultimately leading two distinct populations of “empty” nanodiscs with different lipids but without embedded membrane proteins to equilibrate to the same lipid composition.18 Lipid exchange likely follows a monomer diffusion model, where the rate-limiting step is diffusion of a monomeric lipid out of the nanodisc into free solution rather than a collision between two nanodiscs.
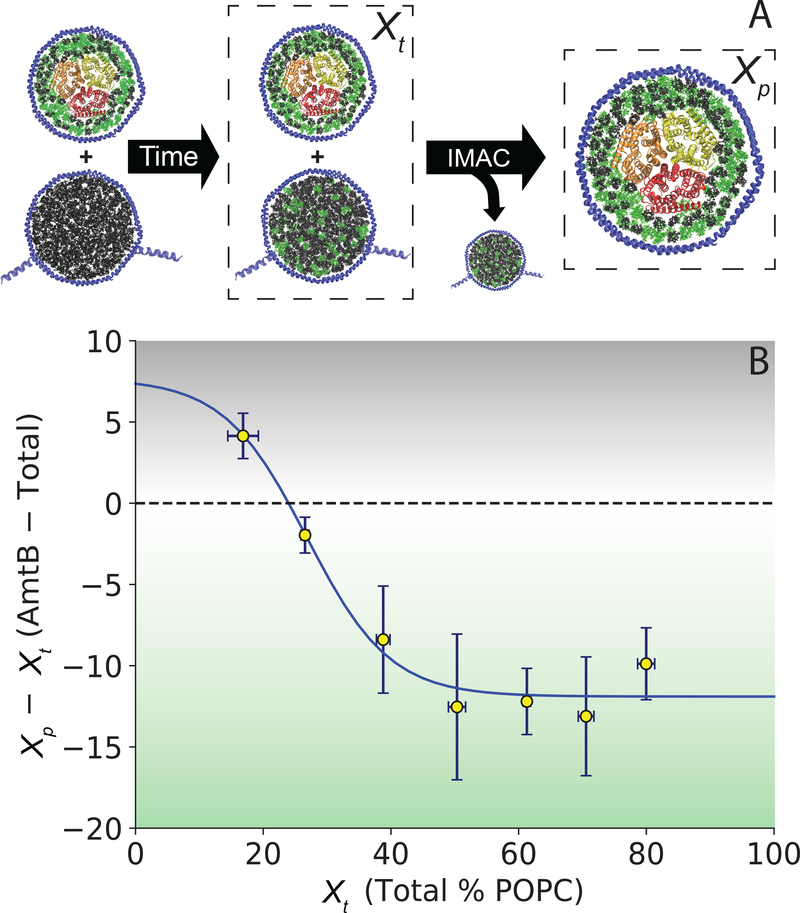
We hypothesized that embedding an integral membrane protein in one population of nanodiscs would shift the equilibrium distribution of lipids between the two populations. In other words, binding of one lipid type to the membrane protein would enrich the membrane protein nanodiscs in the bound lipid type according to Le Chatelier’s principle. Assuming the two lipid types are sufficiently interchangeable, this experiment is analogous to equilibrium dialysis except that no dialysis membrane is used. Instead, the two populations of nanodiscs are mixed for exchange and then separated prior to analysis using a polyhistidine tag on the empty nanodisc population. A similar concept has been used to study lipid enrichment by adding peripheral membrane proteins directly to preformed peptide nanodiscs, which exchange lipids much faster than MSP nanodiscs.19,20
Here, we incorporated AmtB into untagged nanodiscs with 50/50 palmitoyl-oleoyl-phosphatidylcholine (POPC)/palmitoyl-oleoyl-phosphatidylglycerol (POPG). Polyhistidine-tagged empty nanodiscs without AmtB were also prepared at various POPC/POPG ratios. Tagged empty nanodiscs were mixed with untagged AmtB nanodiscs, and the mixture was allowed to exchange at room temperature (21–23 °C) over 2.8 days. After exchange, an equimolar mixture of PC and PG lipids with different tails was added as internal standards, and the total lipid mole percentage of POPC (tagged and untagged), Xt, was measured by direct infusion MS. The amount of POPC and POPG relative to internal standards were measured in positive and negative mode respectively and then compared. Then, the tagged empty nanodiscs were removed by passing the mixture over immobilized metal affinity chromatography (IMAC) beads, and the untagged AmtB nanodiscs were measured alone to determine the lipid mole percentage of POPC in the membrane protein nanodiscs, Xp. Preliminary experiments confirmed that the exchange was at equilibrium within experimental error by 2.8 days, but a full examination of kinetics was not performed due to limited amounts of sample. Control experiments without AmtB showed that empty nanodiscs equilibrated to a nearly uniform lipid composition between tagged and untagged nanodiscs (Figure S1). Full experimental details and method validation can be found in the Supporting Information.
Comparing the total lipid composition (tagged and untagged) with the AmtB nanodiscs (untagged) revealed clear inhomogeneities in lipid distributions. Figure 1 shows the difference in POPC mole percentage between the AmtB nanodiscs and the total (Xp − Xt) as a function of Xt. At high total POPC, AmtB shifted the equilibrium to acquire more POPG in the AmtB nanodiscs, giving a negative Xp − Xt. Conversely, at low POPC levels, AmtB shifted the equilibrium to enrich the AmtB nanodiscs in POPC, giving a positive Xp − Xt. In other words, AmtB remodeled the local lipid environment towards a more optimal lipid distribution, which resulted in an unequal partitioning of lipids between empty nanodiscs and AmtB nanodiscs.
Figure 1.
A) Schematic of LX-MS between AmtB nanodiscs and empty POPC nanodiscs. B) Fit (blue) of the mole percentage of POPC difference between AmtB nanodiscs and total lipids (Xp − Xt) vs. the total mole percentage of POPC (Xt) shows the x-intercept or isolipid point at 24 ± 2% POPC. The black-shaded region highlights preference of AmtB for bound POPC, whereas the green-shaded region highlights preference of AmtB for bound POPG. White shading corresponds to slight or no enrichment.
After establishing that AmtB remodels the local membrane environment, we next sought to determine the optimal POPC/POPG ratio for the protein. We fit Xt versus Xp − Xt to a sigmoidal curve and determined the x-intercept where Xp − Xt = 0. At this mole percentage, which we have termed the isolipid point, the lipid composition of the membrane protein nanodiscs is the same as the empty nanodiscs. More importantly, the composition of the annular lipids should match the bulk lipid bilayer at the isolipid point. For AmtB, we found the isolipid point to be 24/76 ± 2% POPC/POPG.
Thus, LX-MS demonstrates that AmtB has a clear preference for POPG and will remodel its surrounding lipid membrane to enrich in POPG. However, it does have some affinity for POPC and will enrich the surrounding membrane in POPC if not enough is present. Overall, the optimal ratio for AmtB is 24/76 POPC/POPG.
To further investigate the lipid selectivity of AmtB, we developed a new approach to eject membrane proteins from mixed lipid nanodiscs with native MS and measure the composition of lipids that remain bound to the protein. Klassen and coworkers have previously used a catch-and-release assay to quantify binding and lipid selectivity for soluble proteins specifically bound to glycolipids in nanodiscs.21 Furthermore, Morgner and coworkers previously ejected integral membrane proteins from mixed lipid nanodiscs with LILBID-MS, but they were not able to resolve bound lipids.22 Here, we studied an integral membrane protein with less specific annular lipid interactions using native electrospray MS. We used the same AmtB nanodiscs described above and focused on nanodiscs with 50/50 POPC/POPG lipids.
Integral membrane proteins can be challenging to eject efficiently from nanodiscs by collision induced dissociation (CID). Prior research has shown that AmtB tends to retain many lipids during ejection from POPC nanodiscs under normal conditions.23,24 We observed that AmtB is also ejected with many bound lipids in 50/50 POPC/POPG nanodiscs (Figure S2). Supercharging reagents have been shown to modulate the stability of membrane protein-nanodisc complexes for native MS.25 Specifically, propylene carbonate and glycerol carbonate stabilize and destabilize AmtB POPC nanodiscs, respectively, in positive ionization mode. Thus, to improve ejection and investigate more tightly bound lipids, we added the supercharging reagent glycerol carbonate to the solution prior to electrospray ionization. Collisional activation was applied from 60 to 200V in 20V increments (Figure 2A), and spectra were deconvolved and summed using MetaUniDec.26,27 Additional details are provided in the Supporting Methods. A representative deconvolved zero-charge mass spectrum is shown in Figure 2B as a function of the number of lipids bound to AmtB. A bimodal distribution of bound lipids was observed with a distribution around 20–25 bound lipids at lower collision voltages and another distribution around 3 lipids at higher collision voltages (Figure S2).
Figure 2.
A) Schematic for ejection of AmtB from mixed lipid nanodiscs using glycerol carbonate during native MS. B) Summed deconvolved mass spectrum (60–200V) of AmtB trimer ejected from 50/50 POPC/POPG nanodiscs. The black-shaded region highlights bound lipids enriched in POPC, whereas the green-shaded region highlights bound lipids enriched in POPG. White shading corresponds to slight or no enrichment. C) The average mass of bound lipids (60–200V) shows lipid enrichment where it deviates from the expected average mass of 50% POPC/POPG, indicated by a dashed line.
We measured the average mass of lipids bound to AmtB by subtracting the mass of the AmtB trimer from each peak and dividing the difference by the number of bound lipids. Without lipid enrichment, the average lipid mass should be equal to the average mass of POPC and POPG, which is 754.5 Da. Instead, we discovered that each of two distributions has a distinct lipid enrichment (Figure 2C). The first six bound lipids showed average masses that were closer to the mass of POPC (760 Da). The shift towards higher mass indicates that the six lipids that bind tightest during native MS are enriched in POPC. Conversely, the next thirty bound lipids showed lower average masses that are closer to the mass of POPG (749 Da). The lower average masses indicate that lipids in the second distribution are enriched in POPG.
To examine larger numbers of bound lipids, activated glycerol carbonate spectra were combined with partially-activated spectra collected without additives, which show ejected AmtB that retains dozens of bound lipids but has lost both MSP belts.23,24 We also collected non-activated spectra with added propylene carbonate, which largely preserves the intact nanodisc complex and retains both MSP belts (Figure S2).25 By combining data collected under these three conditions, we were able to detect the AmtB trimer bound to 0 through 175 lipids (Figure S3). At higher numbers of bound lipids, the average lipid masses shift toward the bulk 50/50 mixture and show progressively less enrichment. Controls with 100% POPC AmtB nanodiscs showed no enrichment (Figure S4 and S5).
Thus, native MS of AmtB ejected from POPC/POPG nanodiscs shows that AmtB remodels the surrounding lipid environment to become enriched in POPG. However, there are a few binding sites around the protein where POPC is preferred and remains tightly bound to the protein, likely through ionic interactions.
In conclusion, LX-MS and native MS provide complementary pictures of how AmtB remodels its local membrane environment in nanodiscs. LX-MS reveals that AmtB prefers POPG over POPC with an optimal lipid composition (the isolipid point) of 24/76 POPC/POPG. Although AmtB prefers more POPG bound, there is still an affinity for POPC, as shown by the increase in POPC content in AmtB nanodiscs when the overall amount of POPC is low. Native MS of AmtB ejected from 50/50 POPC/POPG nanodiscs also shows that AmtB is enriched in POPG but that there are a small number of tightly bound lipids enriched in POPC. Together, these data indicate that there are likely two types of binding sites around AmtB, a small number that prefer POPC and a larger number that prefer POPG. Future work will explore the binding strengths and stoichiometries of these binding sites in more detail.
Overall, these two novel methods are broadly useful for probing lipid selectivity of membrane proteins in a controlled lipid bilayer environment. Neither experiment requires labelled lipids, which avoids potential distortions to lipid binding and opens a wide range of lipids for study. Ultimately, we expect both LX-MS and native MS with mixed lipid nanodiscs will provide unique insights into the chemistry and biophysics of membrane protein-lipid interactions occurring in lipid membranes.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Maria Reinhardt-Szyba, Kyle Fort, and Alexander Makarov at Thermo Fisher Scientific for support on the Q-Exactive HF instrument. The pMSP1D1 and pMSP1E3D1 plasmids were gifts from Stephen Sligar (Addgene plasmid nos. 20061 and 20066). The authors thank Stephen Sligar and Mark McLean for helpful discussions and their early work on lipid exchange. This work was funded by the National Institute of General Medical Sciences and National Institutes of Health (R35 GM128624). Equipment funding was provided by the National Science Foundation (CHE-1845230). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
ASSOCIATED CONTENT
Supporting Information. The Supporting Information is available free of charge via the Internet at http://pubs.acs.org. Supplemental figures and methods for protein expression and purification, nanodisc assembly, native MS of membrane protein nanodiscs, lipid exchange, MS for lipid exchange quantification, analysis of MS data, and method validation.
REFERENCES
- 1.Overduin M; Esmaili M Memtein: The fundamental unit of membrane-protein structure and function. Chem. Phys. Lipids 2019, 218, 73–84. [DOI] [PubMed] [Google Scholar]
- 2.Brown MF Soft Matter in Lipid-Protein Interactions. Annu. Rev. Biophys. 2017, 46, 379–410. [DOI] [PubMed] [Google Scholar]
- 3.Corradi V; Sejdiu BI; Mesa-Galloso H; Abdizadeh H; Noskov SY; Marrink SJ; Tieleman DP Emerging Diversity in Lipid-Protein Interactions. Chem. Rev. 2019, 119, 5775–5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Enkavi G; Javanainen M; Kulig W; Rog T; Vattulainen I Multiscale Simulations of Biological Membranes: The Challenge To Understand Biological Phenomena in a Living Substance. Chem. Rev. 2019, 119, 5607–5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manna M; Nieminen T; Vattulainen I Understanding the Role of Lipids in Signaling Through Atomistic and Multiscale Simulations of Cell Membranes. Annu. Rev. Biophys. 2019, 48, 421–439. [DOI] [PubMed] [Google Scholar]
- 6.Marsh D Protein modulation of lipids, and vice-versa, in membranes. Biochim. Biophys. Acta 2008, 1778, 1545–1575. [DOI] [PubMed] [Google Scholar]
- 7.Yeagle PL Non-covalent binding of membrane lipids to membrane proteins. Biochim. Biophys. Acta 2014, 1838, 1548–1559. [DOI] [PubMed] [Google Scholar]
- 8.Bechara C; Nöll A; Morgner N; Degiacomi MT; Tampé R; Robinson CV A subset of annular lipids is linked to the flippase activity of an ABC transporter. Nat. Chem. 2015, 7, 255–262. [DOI] [PubMed] [Google Scholar]
- 9.Dorr JM; Koorengevel MC; Schafer M; Prokofyev AV; Scheidelaar S; van der Cruijsen EA; Dafforn TR; Baldus M; Killian JA Detergent-free isolation, characterization, and functional reconstitution of a tetrameric K+ channel: the power of native nanodiscs. Proc. Natl. Acad. Sci. U.S.A. 2014, 111, 18607–18612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hazell G; Arnold T; Barker RD; Clifton LA; Steinke N-J; Tognoloni C; Edler KJ Evidence of Lipid Exchange in Styrene Maleic Acid Lipid Particle (SMALP) Nanodisc Systems. Langmuir 2016. [DOI] [PubMed] [Google Scholar]
- 11.Cuevas Arenas R; Danielczak B; Martel A; Porcar L; Breyton C; Ebel C; Keller S Fast Collisional Lipid Transfer Among Polymer-Bounded Nanodiscs. Sci. Rep. 2017, 7, 45875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frick M; Schmidt C Mass spectrometry-A versatile tool for characterising the lipid environment of membrane protein assemblies. Chem. Phys. Lipids 2019, 221, 145–157. [DOI] [PubMed] [Google Scholar]
- 13.Robinson CV Mass spectrometry: From plasma proteins to mitochondrial membranes. Proc. Natl. Acad. Sci. U.S.A. 2019, 116, 2814–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calabrese AN; Radford SE Mass spectrometry-enabled structural biology of membrane proteins. Methods 2018, 147, 187–205. [DOI] [PubMed] [Google Scholar]
- 15.Bayburt TH; Grinkova YV; Sligar SG Self-assembly of discoidal phospholipid bilayer nanoparticles with membrane scaffold proteins. Nano Lett. 2002, 2, 853–856. [Google Scholar]
- 16.Laganowsky A; Reading E; Allison TM; Ulmschneider MB; Degiacomi MT; Baldwin AJ; Robinson CV Membrane proteins bind lipids selectively to modulate their structure and function. Nature 2014, 510, 172–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mirandela GD; Tamburrino G; Hoskisson PA; Zachariae U; Javelle A The lipid environment determines the activity of the Escherichia coli ammonium transporter AmtB. FASEB J. 2019, 33, 1989–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakano M; Fukuda M; Kudo T; Miyazaki M; Wada Y; Matsuzaki N; Endo H; Handa T Static and dynamic properties of phospholipid bilayer nanodiscs. J. Am. Chem. Soc. 2009, 131, 8308–8312. [DOI] [PubMed] [Google Scholar]
- 19.Barnaba C; Sahoo BR; Ravula T; Medina-Meza IG; Im S-C; Anantharamaiah GM; Waskell L; Ramamoorthy A Cytochrome-P450-Induced Ordering of Microsomal Membranes Modulates Affinity for Drugs. Angew. Chem. Int. Ed. 2018, 57, 3391–3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barnaba C; Ravula T; Medina-Meza IG; Im S-C; Anantharamaiah GM; Waskell L; Ramamoorthy A Lipid-exchange in nanodiscs discloses membrane boundaries of cytochrome-P450 reductase. Chem. Commun. 2018, 54, 6336–6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y; Liu L; Daneshfar R; Kitova EN; Li C; Jia F; Cairo CW; Klassen JS Protein-glycosphingolipid interactions revealed using catch-and-release mass spectrometry. Anal. Chem. 2012, 84, 7618–7621. [DOI] [PubMed] [Google Scholar]
- 22.Henrich E; Peetz O; Hein C; Laguerre A; Hoffmann B; Hoffmann J; Dötsch V; Bernhard F; Morgner N Analyzing native membrane protein assembly in nanodiscs by combined non-covalent mass spectrometry and synthetic biology. eLife 2017, 6, e20954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marty MT; Hoi KK; Gault J; Robinson CV Probing the Lipid Annular Belt by Gas-Phase Dissociation of Membrane Proteins in Nanodiscs. Angew. Chem. Int. Ed. Engl. 2016, 55, 550–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reid DJ; Keener JE; Wheeler AP; Zambrano DE; Diesing JM; Reinhardt-Szyba M; Makarov A; Marty MT Engineering Nanodisc Scaffold Proteins for Native Mass Spectrometry. Anal. Chem. 2017, 89, 11189–11192. [DOI] [PubMed] [Google Scholar]
- 25.Keener JE; Zambrano DE; Zhang G; Zak CK; Reid DJ; Deodhar BS; Pemberton JE; Prell JS; Marty MT Chemical additives enable native mass spectrometry measurement of membrane protein oligomeric state within intact nanodiscs. J. Am. Chem. Soc. 2019, 141, 1054–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marty MT; Baldwin AJ; Marklund EG; Hochberg GK; Benesch JL; Robinson CV Bayesian deconvolution of mass and ion mobility spectra: from binary interactions to polydisperse ensembles. Anal. Chem. 2015, 87, 4370–4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reid DJ; Diesing JM; Miller MA; Perry SM; Wales JA; Montfort WR; Marty MT MetaUniDec: High-Throughput Deconvolution of Native Mass Spectra. J. Am. Soc. Mass Spectrom. 2019, 30, 118–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.