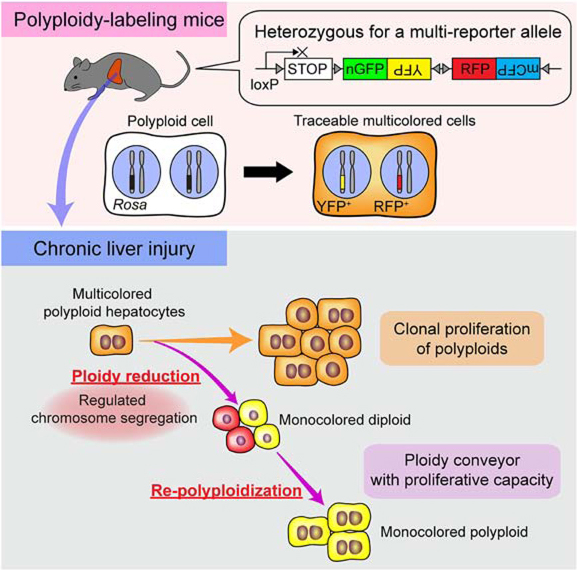
Summary
The identity of cellular populations that drive liver regeneration after injury is the subject of intense study, and the contributions of polyploid hepatocytes to organ regeneration and homeostasis have not been systematically assessed. Here we developed a multicolor reporter allele system to genetically label and trace polyploid cells in situ. Multicolored polyploid hepatocytes undergo ploidy reduction and subsequent re-polyploidization after transplantation, providing direct evidence for the hepatocyte ploidy conveyor model. Marker segregation revealed that ploidy reduction rarely involves chromosome missegregation in vivo. We also traced polyploid hepatocytes in several different liver injury models and found robust proliferation in all settings. Importantly, ploidy reduction was seen in all injury models studied. We therefore conclude that polyploid hepatocytes have extensive regenerative capacity in situ and routinely undergo reductive mitoses during regenerative responses.
Keywords: hepatocyte, polyploidy, lineage tracing, ploidy reduction, ploidy conveyor
Graphical Abstract
eTOC blurb
Polyploid cells are common in several mammalian tissues. Using a genetic system to label and trace polyploid cells in vivo, Matsumoto et al. showed that polyploid hepatocytes are major contributors to regeneration of chronically injured livers. Polyploid hepatocytes continuously proliferate, and dynamically decrease and increase their ploidy while remaining proliferative.
Introduction
Mammalian somatic cells typically have diploid genomes, but polyploid cells with more than two sets of chromosomes are abundantly found in tissues such as the liver, skeletal and cardiac muscle, and bone marrow (Pandit et al., 2013). Physiological polyploidization occurs in a developmentally programmed manner in hepatocytes, cardiomyocytes, and megakaryocytes (Ovrebo and Edgar, 2018). Polyploidization is also enhanced in some disease conditions such as liver cirrhosis and myocardial infarction (Gjelsvik et al., 2019; Ovrebo and Edgar, 2018). Given that polyploidy is known to have advantages for evolution and adaptation in plants, fungi and insects (Schoenfelder and Fox, 2015; Van de Peer et al., 2017), polyploidy in mammalian somatic cells may also be beneficial. However, it is still unclear how exactly polyploid cells are involved in mammalian physiology.
The hepatocyte, the main liver parenchymal cell, is one of the cell types that most frequently exhibits polyploidy. More than 50% and 90% of hepatocytes in human and rodent adult liver are polyploid, respectively (Duncan et al., 2012b; Wilkinson et al., 2019). Polyploidization of hepatocytes occurs mainly through cytokinesis failure and starts around weaning in mice (Duncan, 2013; Gentric and Desdouets, 2014). The number of polyploid hepatocytes increases with age in humans and rodents (Duncan, 2013; Gentric and Desdouets, 2014), as well as pathological conditions such as chronic hepatitis (Gentric et al., 2015; Toyoda et al., 2005).
It has been hypothesized that polyploidy in hepatocytes may enhance metabolism and protein synthesis/secretion (Orr-Weaver, 2015; Ovrebo and Edgar, 2018; Pandit et al., 2013). Polyploid hepatocytes may also buffer hepatic dysfunction by generating aneuploid progeny genetically adapted to stress (Duncan et al., 2012a). Moreover, redundant genome of polyploids are thought to be less impacted by genotoxic damage (Pandit et al., 2013). Some advantages of polyploids such as adaptation and genomic damage insensitivity are well established in non-mammalian organisms (Schoenfelder and Fox, 2015).
In addition, polyploid cells could potentially contribute to tissue regeneration by proliferation. In some cell types polyploidization induces cell cycle arrest or cellular senescence (Ganem and Pellman, 2007), and can be involved in terminal cell differentiation (Pandit et al., 2013). Conversely, other polyploid cell types including hepatocytes and fibroblasts can proliferate (Guidotti et al., 2003; Miyaoka et al., 2012; Ohshima and Seyama, 2013; Tanaka et al., 2015; Wilkinson et al., 2019). We have demonstrated that polyploid hepatocytes can undergo many rounds of cell division after transplantation into Fah−/− mice (Overturf et al., 1999; Overturf et al., 1997). We have also previously shown that polyploid hepatocytes can reduce their ploidy via multipolar mitosis (Duncan et al., 2010). This multipolar reductive mitosis might enhance effective organ regeneration by generating more than two cells in one cell division. On the other hand, some studies have shown that polyploid hepatocytes exhibit senescence-associated changes following one or two rounds of cell divisions (Gentric et al., 2012; Sigal et al., 1999). Additionally, it has been suggested that diploid hepatocytes adjacent to the central vein of the liver lobule act as stem cells during both homeostasis and injury responses (Wang et al., 2015). Therefore, it is still controversial whether polyploids continuously proliferate to regenerate the chronically damaged liver. It also remains unknown whether the multipolar reductive mitoses seen in tissue culture and during transplantation play a part in physiologic regeneration.
In the present study, to evaluate the contribution of polyploids and their ploidy reduction to organ regeneration, we established a mouse model to genetically label polyploid cells using multicolored reporter mice. Tracing polyploid hepatocytes in various chronically injured livers showed that polyploid hepatocytes did not only proliferate via multiple cycles of cell division while remaining polyploid, but also generated ploidy-reduced cells with extensive proliferative potential. Those ploidy-reduced cells underwent re-polyploidization in the process of regeneration, indicating dynamic ploidy alterations during injury responses. We also found that reporter chromosomes did not segregate randomly, but distributed in a manner that genetically resembles completion of cytokinesis followed by mitosis. Our findings shed light on the role of polyploids and ploidy dynamics in organ regeneration. No evidence for a diploid population of stem/progenitor cells was found.
Results
Polyploid cells are genetically labeled and traceable in mice heterozygous for a multicolor reporter allele
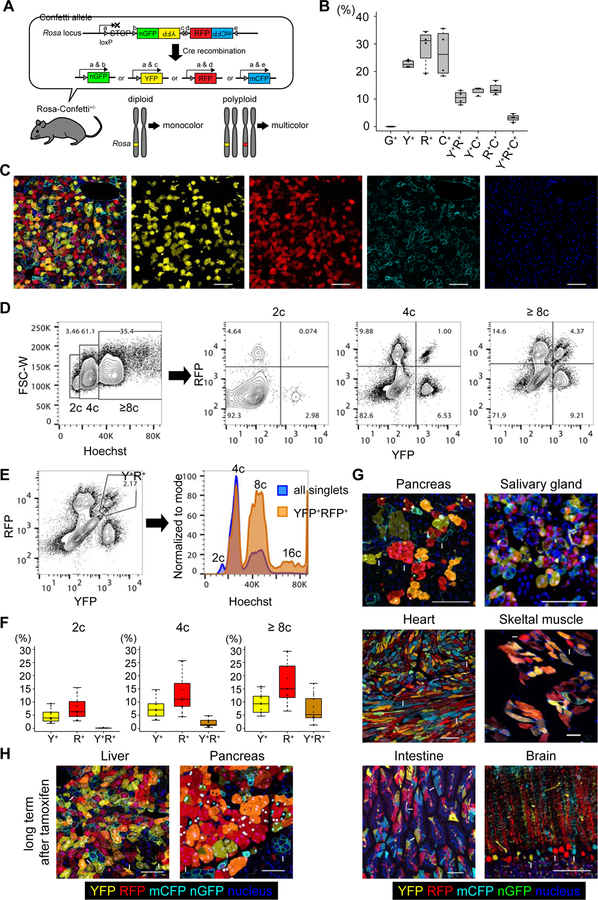
To trace the fate of polyploid hepatocytes in vivo, Rosa-Confetti multicolor reporter mice (Snippert et al., 2010) were used. The Rosa-Confetti allele consists of a floxed stop cassette followed by four different reporter genes, which are the nuclear green (nGFP), cytoplasmic yellow (YFP), cytoplasmic red (RFP), and membrane-bound cyan (mCFP) fluorescent proteins (Figure 1A). After Cre recombination only one fluorescent protein is stochastically expressed from each Rosa-Confetti allele. In germline heterozygous Rosa-Confetti mice, one of the two sets of chromosomes (2c) harbors a Rosa-Confetti allele, and thus, diploid cells can express only one reporter, whereas polyploids can be labeled by co-expressing multiple colors (Figure 1A).
Figure 1. Genetic labeling of polyploid hepatocytes using heterozygous Rosa-Confetti mice.
(A) Scheme of genetic labeling in Rosa-Confetti+/− mice. Arrowheads indicate loxP sites. (B) Labeling frequencies of hepatocytes by Confetti reporter genes. Livers of four Ubc-CreERT2/Rosa-Confetti+/− mice were analyzed by microscopy after a 3-week tamoxifen washout period. Frequencies of the different colors among total labeled hepatocytes were calculated. (C) Representative microscopic images of the liver of Ubc-CreERT2/Rosa-Confetti+/− mice. Arrowheads indicate YFP+RFP+mCFP+ tricolored cells. (D, E) Representative FACS plots of hepatocytes isolated from Ubc-CreERT2/Rosa-Confetti+/− mice. The percentages of cells with each ploidy and their fractions based on reporter expressions are shown. mCFP was not analyzed as it is too dim to detect on flow cytometry. Note that frequencies of YFP+ and/or RFP+ cells shown in FACS plots are lower than Figure 1B because unlabeled cells are included in the denominator in FACS plots whereas only labeled cells are analyzed on microscopy in Figure 1B. (F) Labeling frequencies of diploid and polyploid hepatocytes estimated by flow cytometry. Box plots show the median and interquartile range (IQR) between 25th and 75th percentile, and whiskers show the lowest data within 25th percentile – 1.5 × IQR and the highest data within 75th percentile + 1.5 × IQR. (G) Representative microscopic images of various organs of Ubc-CreERT2/Rosa-Confetti+/− mice. Mice were analyzed after a 3-week tamoxifen washout period. (H) Representative microscopic images of the liver and pancreas of old Ubc-CreERT2/Rosa-Confetti+/− mice. Tissues were harvested more than 18 months after tamoxifen administration. Some representative multicolored (arrowheads) and monocolored (arrows) cells are indicated in (G) and (H). Single-color images of (G) and (H) are shown in Figure S1. G+, nGFP+; Y+, YFP+; R+, RFP+; C+, mCFP+. Scale bars: 100 μm. See also Figures S1 and S2.
To induce temporally controlled Cre recombination, the Rosa-Confetti mice were crossed with Ubc-CreERT2 transgenics that express tamoxifen-inducible Cre recombinase. Tamoxifen was administered to heterozygous Ubc-CreERT2/Rosa-Confetti+/− mice after the physiological polyploidization period of hepatocytes at 4–8 weeks of age (Gentric and Desdouets, 2014). No labeled cells were detectable without tamoxifen administration (not shown), confirming strictly regulated Cre activity by tamoxifen. After a 3-week tamoxifen washout period, hepatocytes were stochastically labeled with YFP, RFP or mCFP, while almost no hepatocytes were labeled with nGFP (Figure 1B). This scarcity of nGFP-labeled cells is consistent with previous reports (Snippert et al., 2010), and might result from GFP-specific cytotoxicity (Taghizadeh and Sherley, 2008). Of note, many hepatocytes were co-labeled with multiple colors (Figures 1B and 1C). Flow cytometric analysis of isolated hepatocytes showed that all multicolored hepatocytes were polyploid, whereas diploids were monocolored (Figures 1D, 1E, and 1F). Multicolored hepatocytes were preferentially located in pericentral zone 3 compared to periportal zone 1, which is consistent with a biased distribution of diploid hepatocytes to the periportal area (Figures S1A and S1B) (Tanami et al., 2017). Importantly, multi-color labeling was seen not only in hepatocytes, but also found in many other cell types including pancreatic acinar cells, cardiomyocytes, skeletal muscle cells, and salivary gland epithelia (Figures 1G and S2A). In contrast, intestinal epithelia and brain cortex neurons which are mostly diploid were exclusively monocolored (Figures 1G and S2A). Interestingly, multicolored polyploid hepatocytes and pancreatic acinar cells were observed more than 18 months after tamoxifen administration (Figures 1H and S2B). This proves that polyploid epithelial cells generated in young animals can persist for their entire lifespan. Together these findings show that mice heterozygous for a multicolor reporter allele can be used to trace polyploids in vivo, and that polyploidy status can be tracked based on the genetic labeling of multiple reporter genes.
Clonal tracing of transplanted polyploids demonstrates extensive proliferation and ploidy reduction
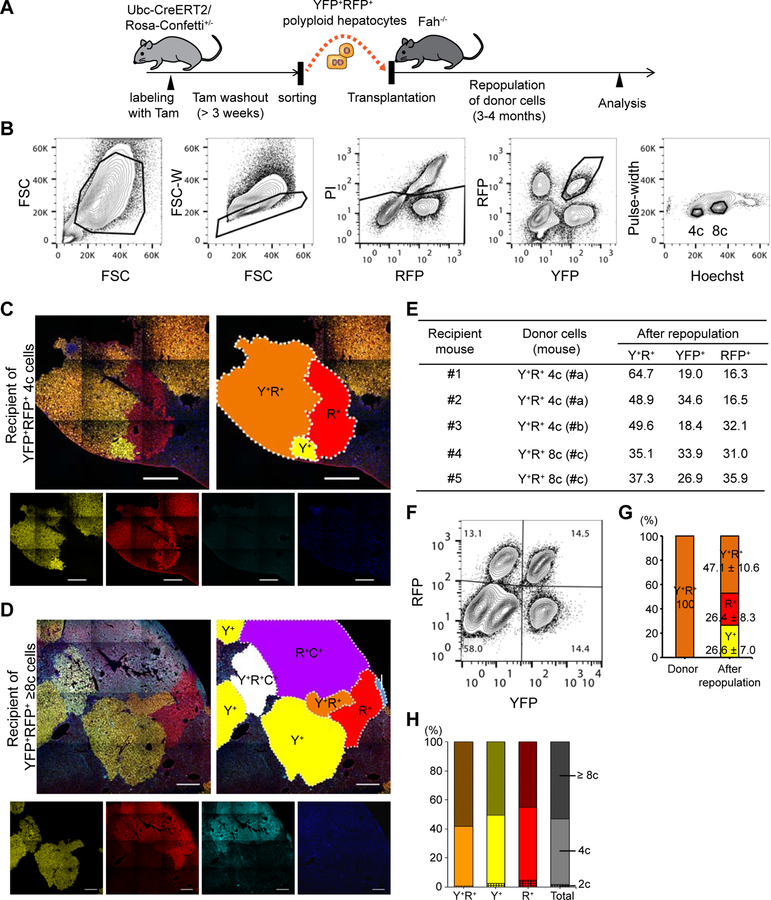
We previously followed the fate of bulk populations of polyploid hepatocytes during transplantation (Duncan et al., 2010), but clonal analysis was lacking. To study the behavior of individual polyploid hepatocytes, YFP+RFP+ multicolored polyploid hepatocytes from Ubc-CreERT2/Rosa-Confetti+/− mice were isolated by fluorescence-activated cell sorting (FACS), and transplanted into Fah−/− recipient mice in low numbers (< 10 thousand cells per mouse, Figures 2A and 2B). As mCFP was too dim to be detected by flow cytometry, it was not used as a marker for FACS. Many large single cell derived clones (200+ cells) consisting of YFP+RFP+ multicolored cells were detected in the repopulated livers, indicating that polyploid hepatocytes underwent multiple cycles of cell division while remaining polyploid. Importantly, YFP+ and RFP+ monocolored clones were frequently found directly adjacent to YFP+RFP+ bicolored nodules. These monocolored satellites derived from pure bicolored populations are suggestive of ploidy reduction (Figure 2C). Of note, in livers transplanted with YFP+RFP+ 8c hepatocytes, some repopulated nodules stochastically co-expressed mCFP, and clonal proliferation of tricolored 8c cells was seen (Figures 2D and S3). Immediately adjacent to the tricolored clones, bicolored and monocolored nodules were found (Figures 2D and S3). This indicates that even octaploid cells can clonally proliferate, and that ploidy-reduced hepatocytes contribute to liver regeneration.
Figure 2. Repopulation of Fah-deficient liver by ploidy-reduced cells derived from transplanted polyploid cells.
(A) Experimental scheme of transplantation of multicolored polyploid hepatocytes into Fah−/− mice. (B) Sorting strategy for multicolored polyploid hepatocytes. Hepatocytes were sorted based on both their DNA content (Hoechst) and fluorescent reporter expression. Note that doublets were strictly excluded by narrow gating with the pulse-width parameter. (C, D) Representative microscopic images of the recipient livers repopulated with YFP+RFP+ 4c and 8c donors (C and D, respectively). Stitched images are shown. Each clonally repopulated area is indicated in the schematic diagrams. Y+, YFP+; R+, RFP+; C+, mCFP+. Scale bars: 500 μm. (E) Details of transplantation. Frequencies of reporter expression of donor-derived cells are shown as percentages of total reporter-positive (YFP+ or RFP+) cells. (F) Representative FACS plots of hepatocytes isolated from the recipient liver. The percentages of each fraction are shown. (G) Frequencies of YFP+ and/or RFP+ cells before and after repopulation. (H) Ploidy distribution of reporter-positive donor derived cells after repopulation. Values are shown as means ± standard deviations (SD, n = 5, G and H). See also Figures S3 and S4.
Flow cytometric analysis also detected many YFP+ or RFP+ monocolored cells emerging in the repopulated livers (Figures 2E, 2F and 2G). The ploidy of these monocolored cells was distributed from 2c to 8c and more (Figure 2H). The loss of reporter expression suggests that transplanted polyploid cells underwent ploidy reduction during proliferation. The close similarity between the frequencies of YFP+ and RFP+ monocolored cells is consistent with the feature of reductive mitosis generating ploidy-reduced daughters in pairs (Duncan et al., 2010).
Contamination of the original bicolored donor cell population by small amounts of monocolored cells could be an alternative explanation for the emergence of monocolored cells if these contaminants proliferated more than the bicolored cells. In fact, although we strictly excluded doublet cells using pulse-width gating in FACS, the average purity of sorted cells validated by reporter gene expression on flow cytometry and microscopy was only 98.8 ± 1.8% and 98.5 ± 1.5%, respectively. To evaluate the proliferation efficiency of bicolored and monocolored cells, the size of each regenerative clone was examined. The ratio of the average size of YFP+ or RFP+ monocolored clones to that of bicolored clones in the same mouse was only 0.75 ± 0.33 (6 mice), dismissing the possibility that monocolored cells overgrew during repopulation. To further assess the potential influence of contamination, we intentionally contaminated multicolored polyploids with sex-mismatched monocolored cells before transplantation (Figure S4A). In mice repopulated with mixtures of YFP+RFP+ male polyploids (85%) and YFP+ and RFP+ monocolored female cells (5% and 10%, respectively), the proportion of YFP+ and RFP+ monocolored cells increased up to 24.2 ± 4.5% and 29.8 ± 4.2%, respectively (Figure S4B). Quantitative PCR for Y chromosome DNA extracted from sorted cells revealed that 76.1 ± 14.4% and 63.4 ± 9.4% of those YFP+ and RFP+ monocolored cells were male (Figure S4C). This proves that majority of the monocolored cells originated from the male bicolored polyploids and not from female contaminating monocolored cells. The same result was found when the polyploid cells were female and the monocolored cells were male (Figures S4D and S4E). Taken together, these findings confirmed that polyploid hepatocytes generated ploidy-reduced cells with proliferative capacity during liver regeneration.
Ploidy-reduced cells can undergo re-polyploidization and re-polyploidized cells proliferate for liver regeneration
In the Fah−/− recipient livers transplanted with YFP+RFP+ 4c cells, most (96.6 ± 0.9%) of their monocolored derivatives exhibited polyploidy after repopulation. This suggests that ploidy-reduced cells underwent re-polyploidization during subsequent mitoses.
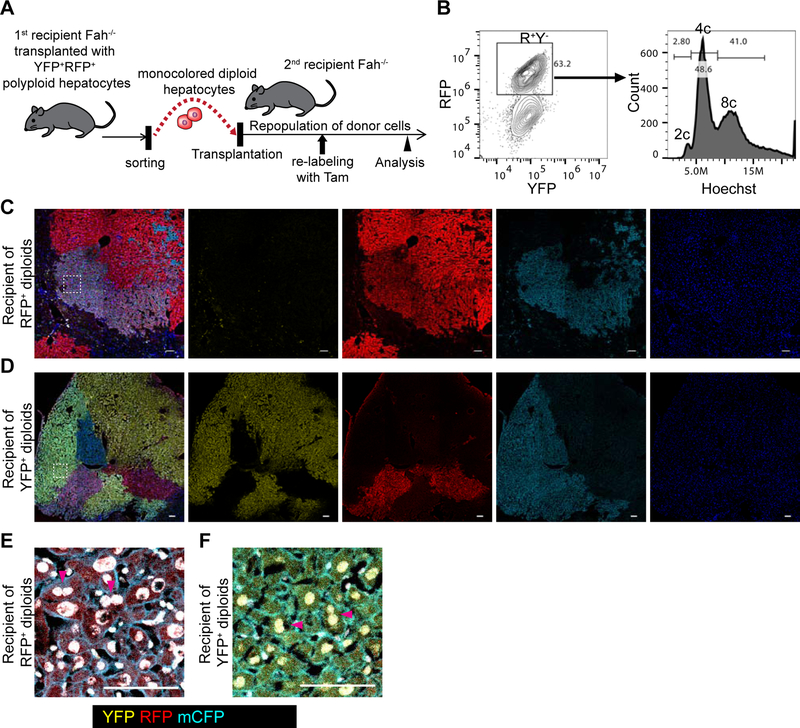
To directly examine whether ploidy-reduced cells undergo re-polyploidization, we sorted YFP+ or RFP+ monocolored diploid cells from mice repopulated by YFP+RFP+ bicolored polyploids, and serially transplanted them into secondary Fah−/− recipients (Figure 3A). Flow cytometric analysis showed that most of the cells derived from donor diploids re-polyploidized during repopulation (Figure 3B). To examine the proliferative capacity of these re-polyploidized cells, we administered tamoxifen to secondary recipient mice in the middle of the repopulation period (Figure 3A). As Cre recombination of Rosa-Confetti allele is reversible, re-polyploidization will generate a second chromosome that can be recombined de novo. A second pulse of tamoxifen can generate a second color in previously monocolored cells. Microscopic analysis indeed showed clonal repopulation by multicolored cells (Figures 3C and 3D), demonstrating the abundant proliferation potential of re-polyploidized hepatocytes even in the presence of competing diploid donors. The re-polyploidized cells frequently had binucleated as well as large polyploid nuclei, suggesting that re-polyploidization resulted from incomplete cytokinesis as in physiological hepatocyte polyploidization (Figures 3E and 3F). Taken together, these findings revealed that ploidy-reduced cells undergo re-polyploidization during proliferation, and that re-polyploidized cells also contribute to liver regeneration along with diploids.
Figure 3. Re-polyploidization of ploidy-reduced cells during liver regeneration.
(A) Experimental scheme for tracing of ploidy-reduced cells in serial recipient mice. (B) Representative FACS plot of a recipient mouse transplanted with ploidy-reduced cells. A mouse serially transplanted with RFP+ monocolored diploid hepatocytes which were originally derived from YFP+RFP+ polyploids is shown. (C, D) Representative microscopic images of the recipient livers repopulated with YFP+ or RFP+ ploidy-reduced 2c cells. Stitched images are shown. (E, F) High-magnification images of (C) and (D). Areas are indicated by dotted line in (C) and (D). Nuclei are shown in white pseudocolor. Scale bars: 100 μm.
Polyploid hepatocytes undergo multiple rounds of cell division in several liver injury models
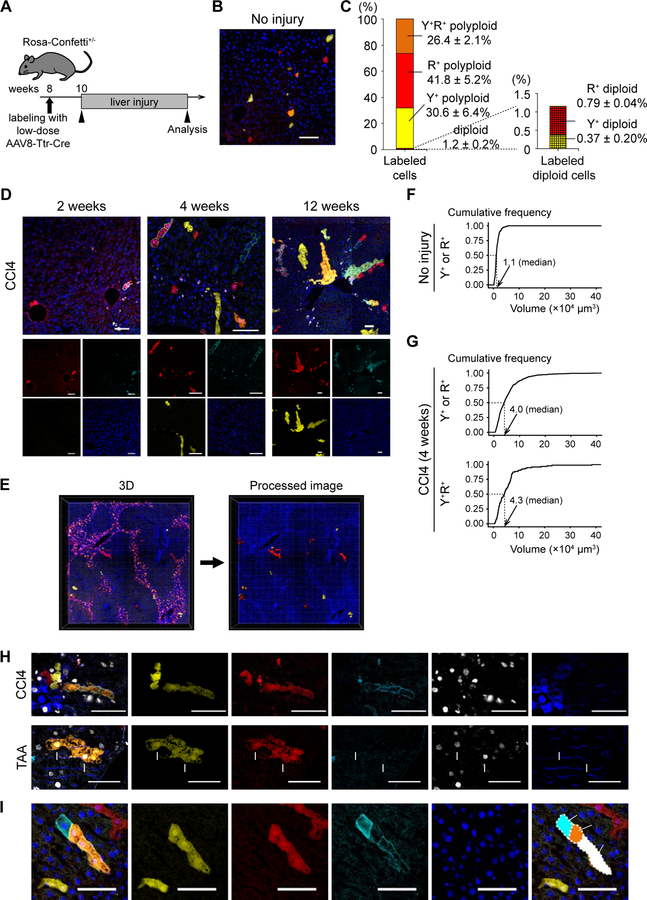
Our previous studies of hepatocyte ploidy kinetics (Duncan et al., 2009; Duncan et al., 2010) and the experiments described above relied on liver repopulation of Fah−/− mice. To determine whether similar phenomena (proliferation of polyploids and ploidy reduction) also play a role in hepatic injury responses in situ, we performed lineage tracing using multicolor reporter mice in the context of different liver injury models. Heterozygous Rosa-Confetti mice were injected with low-dose AAV8-Ttr-Cre (≤ 5×108 vg) to induce sparse single-cell labeling specifically of hepatocytes (Figures 4A and 4B). Flow cytometric analysis showed that 1.7 ± 1.1% of hepatocytes were labeled with YFP and/or RFP, and most (≈ 98.8%) of the labeled cells exhibited polyploidy under this condition (Figure 4C). Mice were then subjected to liver injury starting 2 weeks after hepatocyte labeling. In livers chronically damaged by carbon tetrachloride (CCl4), labeled cells including multicolored cells proliferated for up to 12 weeks, and were detectable as clonal areas (Figure 4D). Clonal expansion of each labeled hepatocyte was further evaluated by 3D image analysis of thick sections (> 150 μm) to eliminate sampling bias caused by thin sections (Figure 4E). Whereas the average volume of a single labeled hepatocyte before injury was 1.1×104 μm3 (SD 0.9×104), the median volume of each clone derived from a single labeled cell was 4.0 ×104 μm3 (IQR 2.0–6.9×104) after 4-week CCl4 injury (Figures 4F and 4G). Although the extent of expansion of each clone varied from clone to clone, the majority (57.3%, 382 of 666) of labeled clones had a volume more than 3 times as large as that of a single hepatocyte. YFP+RFP+ bicolored cells alone also exhibited a similar expansion profile of clonally-proliferating cells (Figure 4G). These findings suggested that many polyploid hepatocytes underwent multiple cycles of cell division and continuously proliferated to regenerate the chronically-injured liver.
Figure 4. Tracing of single cell-derived clonal expansions in Rosa-Confetti+/− mice.
(A) Scheme to trace sparsely-labeled hepatocytes. (B) Representative image of the sparsely-labeled liver. (C) Distribution of ploidy and reporter gene expression of sparsely-labeled hepatocytes. mCFP was not analyzed due to its dim signal. Values are shown as means ± SD (n = 3). (D) Microscopic images of representative sparsely-labeled livers after CCl4 injury. The livers were injured with CCl4 for the indicated time. (E) 3D reconstructed images of the sparsely-labeled liver injured with CCl4. YFP, RFP, and Hoechst (nuclei) signals were imaged. mCFP, which was quenched by the tissue clearing process, was not detected in 3D imaging. YFP+ and/or RFP+ cells were extracted for analysis using Imaris software, and the processed image is shown in the right panel. (F, G) Cumulative frequency graphs of the volumes of clonal areas. The data of untreated control livers (F) and CCl4-injured livers (G) are shown. More than 350 clones in 3 or more mice were analyzed. The median values are indicated. (H) Immunofluorescence for Ki67 and GS. Representative results of the livers injured with CCl4 for 4 weeks or TAA for 3 months are shown. Ki67+ multicolored hepatocytes are indicated by arrowheads or arrows. (I) Representative images of a clonal expansion suggesting ploidy reduction. Images were stitched to generate large composite images. Y+, YFP+; R+, RFP+; C+, mCFP+; C, central veins; P, portal veins. Scale bars: 100 μm. See also Figure S5.
To further examine the involvement of polyploids in liver regeneration, sparsely-labeled hepatocytes were also traced in three additional chronic damage models including Fah deficiency (Grompe et al., 1993), thioacetamide (TAA)-induced injury (Schnur et al., 1999), and 3,5-diethoxycarbonyl-1,4-dihydrocollidine (DDC)-induced injury (Preisegger et al., 1999). Labeled cells including multicolored cells clonally proliferated in all of these damage models (Figure S5A). 3D image analysis confirmed that proliferation was not restricted to a rare subset, but occurred in the majority of labeled cells (Figures S5B and S5C).
While proliferation of polyploid hepatocytes was seen in all four models of liver injury examined here, their mechanisms of hepatotoxicity are quite different (Liu et al., 2013). Not unexpectedly, clonal analysis of polyploid hepatocytes also revealed injury specific differences. Ki67+ proliferating hepatocytes including multicolored cells were predominantly located in pericentral areas in CCl4-damaged livers, whereas they were distributed across all lobular zones in TAA-injury (Figures 4H, S5D, and S5E). Morphological analysis of the 3D images showed that regenerating clones expanded in a prolate (sausage-like) ellipsoidal shape in CCl4- and TAA-induced liver injuries, whereas they had an oblate (disc-like) ellipsoidal shape in DDC-induced livers (Figure S5F). Consistently, regenerating hepatocytes tended to proliferate along hepatic cords in CCl4- and TAA-damaged livers (Figure S5G). These findings suggest that polyploid hepatocytes are an important source of liver regeneration, and that their clonal characteristics are influenced by injury specific local signals.
Polyploid hepatocytes generate ploidy-reduced cells during regeneration of injured liver
Given continuous cell division of polyploid hepatocytes in various liver injury models, we next sought to examine whether ploidy reduction occurs not only during liver repopulation after transplantation but also during physiologic damage responses. Interestingly, although 26.4 ± 2.1% of labeled cells were bicolored (YFP+RFP+) in the sparsely-labeled liver without injury (Figure 4C), YFP+RFP+ clones were noticeably less frequent (11.1 ± 1.2%) after 4-week CCl4 injury. This decrease in bicolored cells is consistent with allele segregation during ploidy reduction. In addition, 14.8 ± 3.3% of proliferating multicolored clones in the livers injured with CCl4 for 4 weeks contained monocolored cells, as expected from reductive mitosis (Figure 4I).
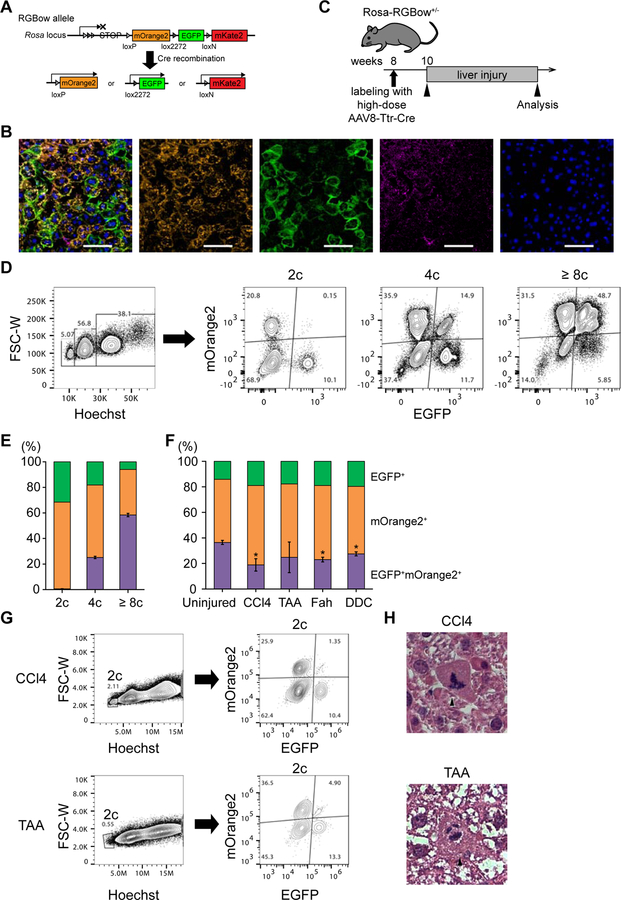
To further examine whether the decrease in the percentage of multicolored cells was caused by ploidy reduction, we carefully quantified the percentage of multicolored hepatocytes in livers with high labeling efficiency. For this experiment, Rosa-RGBow mice were used (He et al., 2016). Similar to the Rosa-Confetti reporter, each RGBow allele contains three kinds of reporter genes (mOrange2, EGFP, and mKate2), and expresses only one of them after Cre recombination (Figure 5A). Each Rosa-RGBow allele can undergo Cre recombination only once, and therefore the labeling color is irreversible even with continuous Cre expression (Figure 5A). As expected Rosa-RGBow+/− mice injected with AAV8-Ttr-Cre as neonates had only monocolored hepatocytes at 4–8 weeks of age even after the physiological polyploidization period of age (Figure S6). This confirmed that recombined RGBow alleles never undergo recombination again.
Figure 5. Ploidy reduction in various kinds of chronically injured livers.
(A) Schema of the Rosa-RGBow allele. Arrowheads indicate loxP and lox variant sites. Note that all three reporter genes are expressed in a membrane bound manner. (B) Representative microscopic images of Rosa-RGBow+/− livers. Mice were labeled with AAV8-Ttr-Cre at 8 weeks of age, and analyzed 2 weeks later. A rare mKate2+ cell is indicated by arrowheads. Scale bars: 100 μm. (C) Scheme of tracing experiments using Rosa-RGBow mice. (D) Representative FACS plots of hepatocytes isolated from uninjured labeled Rosa-RGBow+/− mice. The percentages of cells with each ploidy and their fractions based on reporter expressions are shown. All live hepatocytes regardless of mKate2 expression were analyzed because mKate2+ cells are quite rare (< 1% of labeled cells). (E, F) Frequencies of EGFP+ and/or mOrange+ cells among labeled cells in uninjured (E) and injured (F) Rosa-RGBow+/− mice. Error bars show SD of the frequency of EGFP+mOrange+ bicolored cells (n ≥ 3 in each group). Asterisks indicate statistical significance between uninjured and injured mice (p < 0.01). (G) Representative FACS plots of hepatocytes isolated from Rosa-RGBow+/− liver injured with CCl4 and TAA. (H) Representative hematoxylin and eosin staining images of CCl4- and TAA-injured livers. Hepatocytes undergoing multipolar mitosis are indicated by arrowheads. The livers were harvested after 4 weeks of injury in CCl4 model, and after 3 months of injury in TAA, Fah deficiency, and DDC models. Scale bars: 20 μm. See also Figure S6.
Administration of AAV8-Ttr-Cre (6×1010 vg) into 8-week-old Rosa-RGBow+/− mice after polyploidization efficiently induced hepatocyte-specific Cre recombination. About 75% of hepatocytes were labeled by one or more reporter gene (Figures 5B and 5C). As mKate2 was rarely expressed (< 1% of labeled cells) probably due to the long distance between loxN sites (Zheng et al., 2000), we limited our FACS analysis to the expression of mOrange2 and EGFP. Flow cytometric analysis at two or more weeks after AAV administration confirmed that mOrange2+EGFP+ bicolored hepatocytes were exclusively (> 99.9%) polyploid and no diploids were seen (Figures 5D and 5E). Similar frequencies of reporter expressions were consistently observed under non-injury conditions (Figure 5F).
Highly labeled Rosa-RGBow+/− mice were then subjected to liver injury. As seen in the sparsely labeled Rosa-Confetti mice, the percentage of mOrange2+EGFP+ bicolored hepatocytes significantly decreased after 4-week CCl4 injury (Figure 5F). A decrease in bicolored hepatocytes was also observed in the other injury models, including Fah deficiency and TAA, DDC toxicity (Figure 5F). Potential mechanisms for this consistently observed depletion of bicolored cells include ploidy reduction of polyploid cells and/or overgrowth of monocolored diploid cells. Importantly, small numbers of bicolored diploid cells consistently emerged in the livers injured with CCl4 or TAA (Figure 5G). mOrange2+EGFP+ bicolored cells accounted for 2.3 ± 1.2% (n = 4) and 6.8 ± 3.6% (n = 5) of labeled diploids after CCl4 or TAA injury, respectively (Figure 5G), whereas bicolored diploids were never (<< 1%) observed without injury. In addition, histological analysis readily identified multipolar mitotic structures in the CCl4- and TAA-injured livers (Figure 5H). These findings strongly suggest that multipolar mitoses and ploidy reduction occurred in these injured livers.
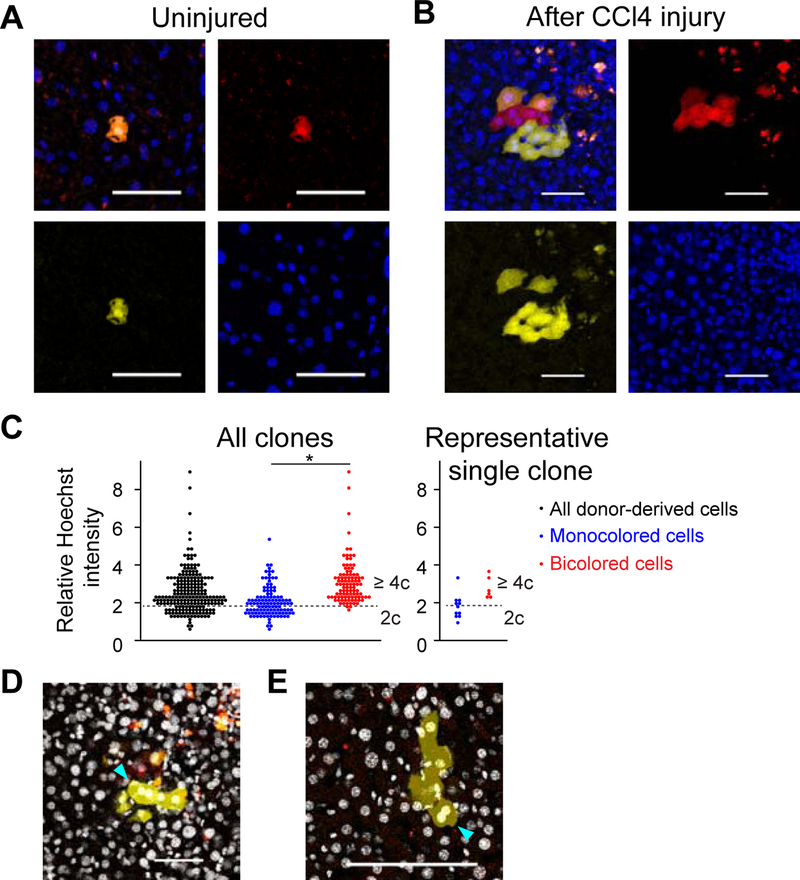
To further validate ploidy reduction in chronic liver injury, YFP+RFP+ bicolored polyploid hepatocytes were transplanted in low numbers into uninjured, wild-type mice, and subsequently traced after CCl4 injury. Partial hepatectomy and microscopic analysis prior to injury demonstrated that all labelled donor hepatocytes (> 100 cells analyzed) expressed both YFP and RFP at baseline (Figure 6A). Strikingly, however, YFP+ or RFP+ monocolored cells were observed at very high frequency in the regenerating clones after CCl4 injury (Figure 6B). Microscopic cytometry of nuclear Hoechst intensity in regenerating clones containing both bicolored and monocolored cells demonstrated a characteristic distribution of nuclear DNA content. The nuclei of monocolored cells had significantly lower DNA content than those of bicolored cells (relative Hoechst intensities; monocolored: 2.1 ± 0.8; bicolored: 3.1 ± 1.2; p < 0.01, Figures 6C and S7A). Although some monocolored cells exhibited higher ploidy presumably due to re-polyploidization, the reduced ploidy of most monocolored cells is consistent with ploidy reduction. In addition, coexistence of bicolored polyploids and ploidy-reduced monocolored cells was detected in 80% (8 out of 10) of regenerating clones analyzed, indicating that most of monocolored cells resulted from ploidy reduction (Figure 6C). Mixed clones consisting of bicolored polyploids and ploidy-reduced monocolored daughter cells sometimes also contained binucleated monocolored cells whose two nuclei each exhibited reduced ploidy, indicating re-polyploidization by failed cytokinesis after ploidy reduction (Figures 6D, 6E, S7B, and S7C). Further microscopic cytometry of the sparsely-labeled livers injured with CCl4 also suggested ploidy reduction, which started by 2 weeks after the initiation of CCl4 administration, and subsequent re-polyploidization by 4 weeks during CCl4 injury (Figures S7D and S7E). Taken together, these findings indicated that polyploid hepatocytes undergo reductive mitosis during regeneration in a variety of liver injuries.
Figure 6. Ploidy reduction and re-polyploidization during CCl4 injury.
(A, B) Representative microscopic images of wild-type mouse livers transplanted with YFP+RFP+ bicolored polyploid hepatocytes. Images of uninjured (A) and CCl4-injured livers (B) are shown. Scale bars: 50 μm. (C) Image cytometry of regenerating clones. Ten clones which contain both bicolored and monocolored cells were analyzed (> 200 cells, 3 mice). Sum of nuclear Hoechst intensity of each donor-derived cell was normalized by the mean Hoechst intensity of the background, and shown as a relative Hoechst intensity. A dotted line that divides polyploid from diploid is set in the same position as in Figure S7A. Result of a representative clone containing ploidy-reduced monocolored cells is shown in the right panel. *p < 0.01 (D, E) Binucleated monocolored cells with ploidy-reduced nuclei. Binucleated cells observed in two clones harboring both bicolored and monocolored cells are indicated by arrowheads. Ploidy of each nucleus in both clones was analyzed in (C). (D) is the same clone as that shown in (B). Nuclei stained with Hoechst are visualized by white pseudocolor. Serial z-stack images are shown in Figures S7B and S7C. The livers were harvested after 6 weeks of CCl4-induced injury. See also Figure S7.
Chromosomes segregate non-randomly during ploidy reductive mitoses
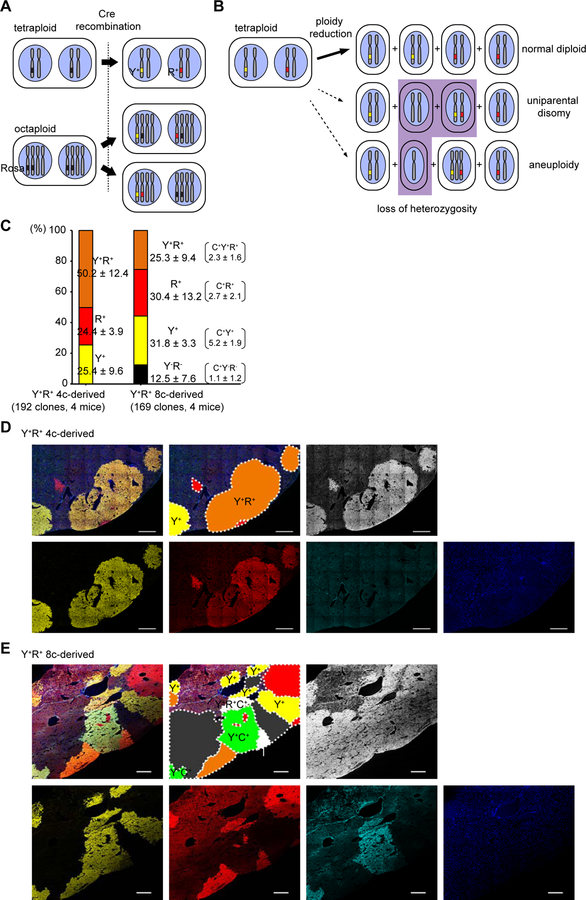
We previously demonstrated frequent aneuploidy in ploidy-reduced hepatocytes in vitro (Duncan et al., 2010). In cancer cell lines multipolar reductive mitosis is reported to frequently result in daughter cell death or cell cycle arrest, probably due to extensive chromosome missegregation (Ganem et al., 2009). Our finding that ploidy-reduced cells can continuously proliferate to regenerate the liver in vivo raised the question whether ploidy reduction produces cells that are resistant massive aneuploidy, or healthy, proliferative daughters via regulated chromosomal segregation.
To evaluate the frequency of chromosomal missegregation after ploidy reduction, Fah−/− recipient livers transplanted with YFP+RFP+ multicolored polyploids as shown in Figure 2 were stained for Fah. As donor cells are homozygous for normal Fah alleles, all donor-derived regenerative clones selected in Fah−/− livers must be positive for Fah. On the other hand, each YFP+RFP+ bicolored tetraploid harbors one YFP+, one RFP+ and two wild-type copies of chromosome 6 (Figure 7A), and thus random chromosome segregation into ploidy-reduced daughters (Figure 7B; dashed arrows) would generate a high frequency (~1/4) of non-colored Fah+ daughters without Rosa-Confetti alleles. Importantly, all of Fah+ regenerative clones derived from bicolored tetraploids (n = 192, 4 mice) were labeled with YFP and/or RFP, and no clones lacked Rosa-Confetti alleles (Figures 7C and 7D). In marked contrast, 12.5% of Fah+ regenerative clones derived from YFP+RFP+ octaploids, which can harbor unlabelled Rosa-Confetti alleles (Figure 7A), lacked both YFP and RFP expression (Figures 7C and 7E). These findings support a model of chromosome segregation, in which the chromosomes of the two nuclei in a binucleated polyploid cell remain separated in the ploidy reduced daughters, hence keeping a pair of paternal and maternal genomes each (Figure 7B; bold arrow). Such a process will yield mostly euploid daughters, consistent with the high regenerative capacity demonstrated here.
Figure 7. Fidelity of chromosome segregation among proliferating progenies after ploidy reduction.
(A) Scheme of possible Cre recombinations that create YFP+RFP+ bicolored cells. Representative patterns of labeled tetraploid and octaploid cells are shown. (B) Scheme of chromosome segregation during ploidy reduction from YFP+RFP+ tetraploid cells. Representative patterns of ploidy reduction generating normal diploid (top, bold arrow) or abnormal daughters with LOH (bottom, dashed arrows) are depicted. (C) Frequencies of YFP and RFP positivity among Fah+ regenerative clones in the livers transplanted with multicolored polyploids. Some 8c-derived clones co-expressed mCFP, and the frequencies of mCFP+ clones are shown in parentheses. Values are shown as means ± SD (n = 4). (D, E) Representative microscopic images of the recipient livers repopulated with YFP+RFP+ 4c and 8c donor-derived cells (D and E, respectively). Stitched images are shown. Each clonally repopulated area is indicated in the schematic diagrams. Y+, YFP+; R+, RFP+; C+, mCFP+, N, no color. Scale bars: 500 μm.
Discussion
In the present study, we developed a novel genetic system to trace polyploid cells in vivo utilizing multicolored reporter mice, and visualized proliferation of polyploids with dynamic ploidy alterations. We showed that polyploid hepatocytes significantly contribute to liver regeneration in multiple settings by generating abundant numbers of daughter cells. Polyploid hepatocytes underwent multiple cycles of mitosis while remaining polyploid, and expanded in all areas of the hepatic lobule in an injury-specific manner. In addition, proliferating polyploids consistently underwent ploidy reductive mitoses at a readily measurable frequency. Transplantation experiments had previously demonstrated the remarkable plasticity of hepatocyte ploidy including ploidy reduction and subsequent re-polyploidization, a phenomenon referred to as the “ploidy conveyor” (Duncan et al., 2010). Ploidy reductive mitosis has also been shown in other eukaryote systems, notably the fly gut (Lucchetta and Ohlstein, 2017). Importantly, we showed that both ploidy-reduced and re-polyploidized cells expanded extensively despite the abundant presence of competing diploid hepatocytes. Although some studies have reported the existence of liver stem cells in special subsets of diploid hepatocytes (Font-Burgada et al., 2015; Wang et al., 2015), our findings here do not support a special stem-cell like role for any diploid liver cell in either homeostasis or injury. Moreover, although it is unclear whether a specific subset of polyploid cells such as cells with high telomerase expression have higher proliferative capacity than the others (Lin et al., 2018), clonal proliferation of the majority of abundant polyploid hepatocytes in various kinds of injured livers (Figures 4G and S5C) suggests that the bulk of polyploid hepatocytes participate in liver regeneration.
While polyploidization of mammalian cells is frequently observed under physiological or pathological conditions, information on ploidy reduction is limited. Multipolar mitoses of polyploids which subsequently result in ploidy reduction have been mostly reported in malignant cells (Chan, 2011; Kalatova et al., 2015; Storchova and Pellman, 2004). Some reports have shown that ~2–10% of mitoses in tetraploid cancer cells are multipolar (Ganem et al., 2009; Telentschak et al., 2015; Vitale et al., 2010). However, it is still controversial whether multipolar mitosis of polyploid cancer cells generates viable ploidy-reduced cells (Ganem et al., 2009; Telentschak et al., 2015). Multipolar mitosis and ploidy reduction in non-cancerous cells have been shown to a limited extent using mammalian fibroblasts in vitro (Martin and Sprague, 1969; Pera and Rainer, 1973) and hepatocytes in vitro and in vivo (Duncan et al., 2010). Importantly, the previous work on mouse hepatocytes involved extensive proliferation after transplantation, arguably a non-physiologic phenomenon. Our findings here demonstrate ploidy reduction of polyploid hepatocytes without transplantation in various chronic liver injuries. We also show that ploidy reduction is not a terminal event and that ploidy-reduced hepatocytes contribute significantly to liver regeneration. Importantly, we also demonstrate for the first time that ploidy-reduced hepatocytes efficiently re-polyploidize. Re-polyploidization after ploidy reduction (the ploidy conveyor) had previously only been inferred (Duncan et al., 2010).
The robustness of the ploidy reduction phenomenon and continuous proliferation of its progeny raises the question of its chromosomal stability. Multipolar mitosis, the mechanism observed by live-cell imaging during ploidy reduction of hepatocytes in vitro (Duncan et al., 2010) is associated with a high frequency of aneuploidy. In many cell types this triggers the mitosis checkpoint and cell death. In fact, multipolar mitosis results in total incapability of proliferation in some cancer cell lines (Ganem et al., 2009). Here we analyzed the segregation of chromosome 6 during ploidy reduction, and did not detect any non-colored clones indicative of loss of heterozygosity (LOH) of chromosome 6 among regenerative clones derived from bicolored tetraploids (Figure 7). Hence, the segregation of chromosomes into ploidy-reduced daughter cells was not random. One possible mechanism for this pattern is that the chromosomes of each nucleus in a binucleated polyploid remain associated with each other (Figure 7B). In this sense, the observed chromosome segregation resembled the outcome that would be obtained from completion of cytokinesis followed by mitosis. Our findings are also consistent with recent report showing that proper chromosome segregation is maintained in vivo environment (Knouse et al., 2018). Future studies are needed to determine the exact mechanistic details of non-random chromosome segregation during ploidy reduction.
Although LOH was not observed in large repopulation clones derived from polyploid cells, a small number of bicolored diploid cells in the livers injured with CCl4 or TAA was consistently found (Figure 5G). This suggests that multipolar mitosis is not always faithful and may induce aneuploidy in some daughters. As aneuploidy interferes with cellular proliferation (Santaguida and Amon, 2015), these cells may have reduced fitness and not proliferate extensively during injury. Why does the liver tolerate even a low level multipolar reductive mitosis at risk of chromosome missegregation? Polyploidy-mediated genetic diversity is one possible advantage during organ regeneration. In fact, polyploidy has been shown to drive adaptation to stress via aneuploidy in yeast and the mammalian livers (Duncan et al., 2012a; Selmecki et al., 2015). However, the genomic diversity elicited in proliferating polyploids may not only be beneficial but could also have harmful side effects like carcinogenesis (Duncan et al., 2010; Sansregret et al., 2018; Tanaka et al., 2018; Telentschak et al., 2015). Thus, especially in the early phase of liver injuries chromosomal plasticity in proliferating polyploids would benefit regeneration by producing injury-resistant, viable progeny, whereas prolonged chromosomal instability might cumulatively enhance the risk of carcinogenesis.
While we focused on polyploid hepatocytes, our genetic system can be used to label and trace polyploid cells in any tissue. Polyploid cells can be seen in multiple endodermal tissues such as the pancreas and salivary glands as well as muscles. Given our results with polyploid hepatocytes, polyploid cells in other regenerative tissues may also be important contributors to regeneration and be capable of ploidy reduction.
In conclusion, our data provide solid evidence that hepatocyte ploidy dynamically changes during liver regeneration, and that polyploid hepatocytes and their ploidy-reduced daughters contribute to regeneration of chronically injured liver. Although it is still unclear how ploidy reduction and ploidy fidelity of polyploids are regulated and to what extent polyploids are involved in the regeneration of other organs, targeting ploidy dynamics may represent a novel promising strategy for enhancing regeneration and preventing carcinogenesis in polyploid organs.
STAR Methods
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Markus Grompe (grompem@ohsu.edu). This study did not generate new unique reagents.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
Wild-type C57BL/6, Ubc-CreERT2 (Ruzankina et al., 2007), Rosa-Confetti (Snippert et al., 2010), and Rosa-RGBow (He et al., 2016) mice were obtained from The Jackson Laboratory. Ubc-CreERT2 and Rosa-Confetti were backcrossed with C57BL/6 more than 6 times and were maintained on the C57BL/6 background until transplantation into Fah−/− mice. Fah−/− mice on the C57BL/6 background (Grompe et al., 1993) were maintained on 8 mg/L of 2-(2-nitro-4-trifluoromethylbenzoyl)-1,3-cyclo-hexanedione (NTBC; Yecuris Corp., Portland, OR) in drinking water until transplantation. All mice were immune competent, and bred and housed under conventional conditions at the animal care facility at Oregon Health & Science University. All animal experiments were approved by the Oregon Health & Science University Institutional Animal Care and Use Committee (Portland, OR), and performed in accordance with the approved protocols.
METHOD DETAILS
Cre recombination
Tamoxifen (Sigma-Aldrich, St Louis, MO) was dissolved in in corn oil (Sigma-Aldrich, 20 mg/ml) and intraperitoneally injected into 8-week-old or older adult Ubc-CreERT2/Rosa-Confetti+/− mice at a dose of 150 mg/kg body weight 3 to 5 times. Mice were processed for experiments after a tamoxifen washout period of at least 3 weeks. For sparse labeling of Rosa-Confetti mice, AAV8-Ttr-Cre (Malato et al., 2011) was retro-orbitally injected into 8-week-old mice at a dose of less than 2 × 109 vector genomes (vg) per mouse. To induce cre recombination in Rosa-RGBow mice, 6 × 1010 vg of AAV8-Ttr-Cre were injected into 8-week-old mice. A 2-week interval after AAV injection was allowed for cre recombination to occur before analysis or injury initiation.
Hepatocyte isolation and transplantation
Primary hepatocytes were isolated by digestion of the liver with a two-step collagenase perfusion method (Overturf et al., 1996). Briefly, the liver was perfused via portal vein with Ca/Mg-free Hanks’ solution (HBSS) containing 0.5 mM EGTA, followed by perfusion with Ca/Mg-containing HBSS with 0.1 mg/ml collagenase type II (Worthington). Primary hepatocytes were collected by a series of centrifugation steps at 50 g × 2 minutes. Collected cells were kept in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum and 10 mM HEPES until FACS sorting. Sorted cells were resuspended in phosphate-buffered saline and transplanted into Fah−/− mice or wild-type C57BL/6 mice by intra-portal or intra-splenic injection. NTBC was withdrawn from Fah−/− recipient mice following transplantation and cycled until the liver was repopulated (Overturf et al., 1996).
Liver injury inductions
CCl4 (Sigma-Aldrich) was diluted in corn oil (1:4, v/v) and intraperitoneally injected twice a week at a dose of 5 ml/kg body weight (Beer et al., 2008). Thioacetamide (Sigma-Aldrich) was diluted in drinking water (0.03%, w/v) and administered to mice continuously (Schnur et al., 1999). For 3,5-diethoxycarbonyl-1,4-dihydrocollidine (DDC) liver injury, chow containing 0.1% (w/w) DDC (Sigma-Aldrich) was continuously fed to mice (Preisegger et al., 1999). To induce liver injury caused by Fah deficiency, Fah knockout mice were put on cycling withdrawal of NTBC (14 days off and 4 days on) (Grompe et al., 1993).
Flow cytometry and FACS
For detection of hepatocyte ploidy, dissociated cells (2 × 106/ml) were incubated with 15 mg/ml Hoechst 33342 (ThermoFisher Scientific, Waltham, MA) and 5 mM reserpine (Sigma-Aldrich) for 30 min at 37 °C. Dead cells were excluded by staining with propidium iodide (Sigma-Aldrich) or SYTOX Red Dead Cell Stain (ThermoFisher Scientific). DNA content was quantified with Hoechst 33342. Cells were sorted with a Cytopeia inFluxV-GS (Becton Dickenson, Franklin Lakes, NJ) using a 140 μm nozzle at a flow rate of approximately 1,000 cells/sec, or analysed with a BD LSRFortessa (Becton Dickenson) or CytoFLEX S (Beckman Coulter Life Sciences, Indianapolis, IN). Data were analysed using FlowJo software (Becton Dickenson).
Histology, immunofluorescence, and imaging
Liver tissue was fixed with 4% paraformaldehyde (Sigma-Aldrich), and embedded in paraffin or optimum cutting temperature compound (Leica Instruments, Nussloch, Germany). Paraffin-embedded tissues were sectioned and stained with hematoxylin and eosin. For immunofluorescence, frozen sections were treated with TrueBlack lipofusin autofluorescence quencher (Biotium, Fremont, CA), and stained with primary and secondary antibodies. Nuclei were counterstained with Hoechst 33342 or RedDot2 (Biotium). Primary and secondary antibodies included rat anti-Ki67 (ThermoFisher Scientific), rabbit anti-GS (abcam), rat anti-CD31 (Becton Dickenson), rabbit anti-Fah (Overturf et al., 1996), Alexa Fluor 647-conjugated anti-rat (Jackson ImmunoResearch), Alexa Fluor 647-conjugated anti-rabbit (Jackson ImmunoResearch), and DyLight 405-conjugated anti-rabbit (Jackson ImmunoResearch). For 3D imaging analysis, fixed tissues were sectioned into 250-μm thick slices using a vibratome (VT1000S, Leica). Tissue clearing was performed using Ce3D (Li et al., 2017) with some modifications. Briefly, sectioned tissues were incubated at room temperature on a rotor for 16–20 hours in the modified Ce3D medium consisted with N-methylacetamide (ThermoFisher Scientific), Omnipaque 350 (GE Healthcare), Triton X-100 (0.1% v/v, ThermoFisher Scientific) and 1-thioglycerol (0.5% v/v, Sigma-Aldrich). Fluorescent images were obtained with a Zeiss LSM 780 confocal microscope (Carl Zeiss AG, Jena, Germany), and analyzed using Imaris (Bitplane Scientific Software) and Zeiss Zen software (Carl Zeiss AG). Tiled images were automatically taken and stitched by Zeiss Zen software to generate large composite images. For analysis of sparsely-labeled regenerative clones in 3D images, each labeled clone was identified using surface function in Imaris. Background signals were subtracted and objects were smoothed by Gaussian filtering with parameters automatically set by the software. Values of volume and ellipsoid axis lengths of each clone were obtained for quantitative analysis. For image cytometric analysis, sum of nuclear Hoechst intensity of each nucleus was obtained using Zeiss Zen software, and normalized by the mean Hoechst intensity of the background.
Quantification of Y chromosome copy number
DNA was isolated from sorted hepatocytes using the MasterPure Complete DNA/RNA Purification kit (Lucigen, Middleton, WI). Quantitative real-time PCR was performed on LightCycler 96 System (Roche, Basel, Switzerland) using FastStart Essential DNA Green Master (Roche). The Y chromosome copy number was determined by amplification of the Sry gene using forward primer 5’-CTCATCGGAGGGCTAAAGTG-3’ and reverse primer 5’-AAGCTTTGCTGGTTTTTGGA-3’ (D’Hulst et al., 2013). As an internal control, exon 5 of the Fah gene was amplified using forward primer 5’-GGACTTCTACTCTTCTCGGCA-3’ and reverse primer 5’-CAATTTGGCAACAGCGCATTC-3’. As Fah−/− mice lack the target region of this primer set, only Fah-positive cells derived from donor hepatocytes were detected by this PCR assay. The Y chromosome copy number was normalized to the Fah gene, and the ratio of male cells was calculated based on standard curve prepared with defined percentages of male and female DNA.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis was performed by Student’s t test using R software (version 3.4.3, R Foundation for Statistical Computing). A value of p < 0.05 was considered to be statistically significant.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat anti-Ki67 | ThermoFisher Scientific | Cat# 14-5698-80; RRID:AB_10853185 |
| Rabbit anti-GS | Abcam | Cat# ab49873; RRID: AB_880241 |
| Rat anti-CD31 | BD Biosciences | Cat# 550274; RRID: AB_393571 |
| Rabbit anti-Fah | Overturf et al., 1996 | N/A |
| Alexa Fluor 647-conjugated anti-rat | Jackson ImmunoResearch Labs | Cat# 212-606-168; RRID: AB_2339249 |
| DyLight 405-conjugated anti-rabbit | Jackson ImmunoResearch Labs | Cat# 711-475-152; RRID:AB_2340616 |
| Alexa Fluor 647-conjugated anti-rabbit | Jackson ImmunoResearch Labs | Cat# 711-606-152; RRID:AB_2340625 |
| Bacterial and Virus Strains | ||
| AAV8-Ttr-Cre | Malato et al., 2011 | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Tamoxifen | Sigma-Aldrich | Cat# T5648 |
| Thioacetamide (TAA) | Sigma-Aldrich | Cat# 163678 |
| 3,5-diethoxycarbonyl-1,4-dihydro-collidin (DDC) | Sigma-Aldrich | Cat# 137030 |
| carbon tetrachloride (CCl4) | Sigma-Aldrich | Cat# 270652 |
| corn oil | Sigma-Aldrich | Cat# C8267 |
| 2-(2-nitro-4-trifluoromethylbenzoyl)-1,3-cyclo-hexanedione (NTBC) | ArkPharm | Cat# AK-55736 |
| Collagenase, type 2 | Worthington Biochemical | Cat# LS004176 |
| Hoechst 33342 | ThermoFisher Scientific | Cat# H3570 |
| reserpine | Sigma-Aldrich | Cat# 83580 |
| propidium iodide | Sigma-Aldrich | Cat# 287075 |
| SYTOX Red Dead Cell Stain | ThermoFisher Scientific | Cat# S34859 |
| TrueBlack Lipofuscin Autofluorescence Quencher | Biotium | Cat# 23007 |
| RedDot2 Far-Red Nuclear Stain | Biotium | Cat# 40061 |
| N-methylacetamide | ThermoFisher Scientific | Cat# 11409056 |
| Omnipaque 350 | GE Healthcare | Cat# 542Y |
| Triton X-100 | ThermoFisher Scientific | Cat# BP151-100 |
| 1-thioglycerol | Sigma-Aldrich | Cat# M1753 |
| MasterPure Complete DNA/RNA Purification kit | Lucigen | Cat# MC85200 |
| FastStart Essential DNA Green Master | Roche | Cat# 06402712001 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Wild-type C57BL/6 | The Jackson Laboratory | Stock# 000664; RRID: IMSR_JAX:000664 |
| Mouse: B6.Cg-Ndor1Tg(UBC-cre/ERT2)1Ejb/2J (Ubc-CreERT2) | The Jackson Laboratory, Ruzankina et al., 2007 | Stock# 008085; RRID: IMSR_JAX:008085 |
| Mouse: B6.129P2-Gt(ROSA)26Sortm1(CAG-Brainbow2.1)Cle/J (Rosa-Confetti) | The Jackson Laboratory, Snippert et al., 2010 | Stock# 017492; RRID: IMSR_JAX:017492 |
| Mouse: B6;129S4-Gt(ROSA)26Sortm4(CAG-mOrange2,-EGFP,-mKate2)Zjh/J (Rosa-RGBow) | The Jackson Laboratory, He et al., 2016 | Stock# 028583; RRID: IMSR_JAX:028583 |
| Fah−/− | Grompe et al., 1993 | N/A |
| Oligonucleotides | ||
| Primers for Sry: Forward: CTCATCGGAGGGCTAAAGTG |
D’Hulst et al., 2013 | N/A |
| Primers for Sry: Forward: AAGCTTTGCTGGTTTTTGGA |
D’Hulst et al., 2013 | N/A |
| Primers for Fah: Forward: GGACTTCTACTCTTCTCGGCA |
This paper | N/A |
| Primers for Fah: Forward: CAATTTGGCAACAGCGCATTC |
This paper | N/A |
| Software and Algorithms | ||
| R | https://www.r-project.org/ | Version 3.4.3 |
| beeswarm | R | Version 0.2.3 |
| ggplot2 | R | Version 2.2.1 |
| ImageJ | https://imagej.nih.gov/ij/ | Version 1.52 |
| ZEN 2.3 (blue edition) | Zeiss | Version 2.3.69.1000 |
| Imaris | Bitplane | Version 9.2.1 |
| FlowJo | Becton-Dickenson | Version 10.6.0 |
| Other | ||
| Zeiss LSM 780 | Zeiss | N/A |
| Cytopeia inFluxV-GS | Becton-Dickenson | N/A |
| BD LSRFortessa | Becton-Dickenson | N/A |
| CytoFLEX S | Beckman Coulter | N/A |
TABLE WITH EXAMPLES FOR AUTHOR REFERENCE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-Snail | Cell Signaling Technology | Cat#3879S; RRID: AB_2255011 |
| Mouse monoclonal anti-Tubulin (clone DM1A) | Sigma-Aldrich | Cat#T9026; RRID: AB_477593 |
| Rabbit polyclonal anti-BMAL1 | This paper | N/A |
| Bacterial and Virus Strains | ||
| pAAV-hSyn-DIO-hM3D(Gq)-mCherry | Krashes et al., 2011 | Addgene AAV5; 44361-AAV5 |
| AAV5-EF1a-DIO-hChR2(H134R)-EYFP | Hope Center Viral Vectors Core | N/A |
| Cowpox virus Brighton Red | BEI Resources | NR-88 |
| Zika-SMGC-1, GENBANK: KX266255 | Isolated from patient (Wang et al., 2016) | N/A |
| Staphylococcus aureus | ATCC | ATCC 29213 |
| Streptococcus pyogenes: M1 serotype strain: strain SF370; M1 GAS | ATCC | ATCC 700294 |
| Biological Samples | ||
| Healthy adult BA9 brain tissue | University of Maryland Brain & Tissue Bank; http://medschool.umaryland.edu/btbank/ | Cat#UMB1455 |
| Human hippocampal brain blocks | New York Brain Bank | http://nybb.hs.columbia.edu/ |
| Patient-derived xenografts (PDX) | Children’s Oncology Group Cell Culture and Xenograft Repository | http://cogcell.org/ |
| Chemicals, Peptides, and Recombinant Proteins | ||
| MK-2206 AKT inhibitor | Selleck Chemicals | S1078; CAS: 1032350-13-2 |
| SB-505124 | Sigma-Aldrich | S4696; CAS: 694433-59-5 (free base) |
| Picrotoxin | Sigma-Aldrich | P1675; CAS: 124-87-8 |
| Human TGF-β | R&D | 240-B; GenPept: P01137 |
| Activated S6K1 | Millipore | Cat#14-486 |
| GST-BMAL1 | Novus | Cat#H00000406-P01 |
| Critical Commercial Assays | ||
| EasyTag EXPRESS 35S Protein Labeling Kit | Perkin-Elmer | NEG772014MC |
| CaspaseGlo 3/7 | Promega | G8090 |
| TruSeq ChIP Sample Prep Kit | Illumina | IP-202-1012 |
| Deposited Data | ||
| Raw and analyzed data | This paper | GEO: GSE63473 |
| B-RAF RBD (apo) structure | This paper | PDB: 5J17 |
| Human reference genome NCBI build 37, GRCh37 | Genome Reference Consortium | http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human/ |
| Nanog STILT inference | This paper; Mendeley Data | http://dx.doi.org/10.17632/wx6s4mj7s8.2 |
| Affinity-based mass spectrometry performed with 57 genes | This paper; and Mendeley Data | Table S8; http://dx.doi.org/10.17632/5hvpvspw82.1 |
| Experimental Models: Cell Lines | ||
| Hamster: CHO cells | ATCC | CRL-11268 |
| D. melanogaster: Cell line S2: S2-DRSC | Laboratory of Norbert Perrimon | FlyBase: FBtc0000181 |
| Human: Passage 40 H9 ES cells | MSKCC stem cell core facility | N/A |
| Human: HUES 8 hESC line (NIH approval number NIHhESC-09-0021) | HSCI iPS Core | hES Cell Line: HUES-8 |
| Experimental Models: Organisms/Strains | ||
| C. elegans: Strain BC4011: srl-1(s2500) II; dpy-18(e364) III; unc-46(e177)rol-3(s1040) V. | Caenorhabditis Genetics Center | WB Strain: BC4011; WormBase: WBVar00241916 |
| D. melanogaster: RNAi of Sxl: y[1] sc[*] v[1]; P{TRiP.HMS00609}attP2 | Bloomington Drosophila Stock Center | BDSC:34393; FlyBase: FBtp0064874 |
| S. cerevisiae: Strain background: W303 | ATCC | ATTC: 208353 |
| Mouse: R6/2: B6CBA-Tg(HDexon1)62Gpb/3J | The Jackson Laboratory | JAX: 006494 |
| Mouse: OXTRfl/fl: B6.129(SJL)-Oxtrtm1.1Wsy/J | The Jackson Laboratory | RRID: IMSR_JAX:008471 |
| Zebrafish: Tg(Shha:GFP)t10: t10Tg | Neumann and Nuesslein-Volhard, 2000 | ZFIN: ZDB-GENO-060207-1 |
| Arabidopsis: 35S::PIF4-YFP, BZR1-CFP | Wang et al., 2012 | N/A |
| Arabidopsis: JYB1021.2: pS24(AT5G58010)::cS24:GFP(-G):NOS #1 | NASC | NASC ID: N70450 |
| Oligonucleotides | ||
| siRNA targeting sequence: PIP5K I alpha #1: ACACAGUACUCAGUUGAUA | This paper | N/A |
| Primers for XX, see Table SX | This paper | N/A |
| Primer: GFP/YFP/CFP Forward: GCACGACTTCTTCAAGTCCGCCATGCC | This paper | N/A |
| Morpholino: MO-pax2a GGTCTGCTTTGCAGTGAATATCCAT | Gene Tools | ZFIN: ZDB-MRPHLNO-061106-5 |
| ACTB (hs01060665_g1) | Life Technologies | Cat#4331182 |
| RNA sequence: hnRNPA1_ligand: UAGGGACUUAGGGUUCUCUCUAGGGACUUAGGGUUCUCUCUAGGGA | This paper | N/A |
| Recombinant DNA | ||
| pLVX-Tight-Puro (TetOn) | Clonetech | Cat#632162 |
| Plasmid: GFP-Nito | This paper | N/A |
| cDNA GH111110 | Drosophila Genomics Resource Center | DGRC:5666; FlyBase:FBcl0130415 |
| AAV2/1-hsyn-GCaMP6- WPRE | Chen et al., 2013 | N/A |
| Mouse raptor: pLKO mouse shRNA 1 raptor | Thoreen et al., 2009 | Addgene Plasmid #21339 |
| Software and Algorithms | ||
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| Bowtie2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Samtools | Li et al., 2009 | http://samtools.sourceforge.net/ |
| Weighted Maximal Information Component Analysis v0.9 | Rau et al., 2013 | https://github.com/ChristophRau/wMICA |
| ICS algorithm | This paper; Mendeley Data | http://dx.doi.org/10.17632/5hvpvspw82.1 |
| Other | ||
| Sequence data, analyses, and resources related to the ultra-deep sequencing of the AML31 tumor, relapse, and matched normal. | This paper | http://aml31.genome.wustl.edu |
| Resource website for the AML31 publication | This paper | https://github.com/chrisamiller/aml31SuppSite |
Highlights.
A genetic system to trace polyploid cells in any organ in vivo was developed.
Polyploid hepatocytes regenerate injured livers and frequently reduce their ploidy.
Ploidy-reduced progeny proliferates and re-polyploidizes in subsequent mitoses.
Chromosome segregation during ploidy reduction is not random but faithful.
Acknowledgements
The AAV-Ttr-Cre construct was kindly provided by Dr. Holger Willenbring of UCSF. We thank Pamela Canaday (Flow Cytometry Shared Resource at OHSU) for sorting and Stefanie Kaech Petrie (Advanced Light Microscopy Core at OHSU) for microscopy assistance. This work was supported by Japan Society for the Promotion of Science Overseas Research Fellowships, KANAE Foundation for the Promotion of Medical Science the 46th Kanae Foreign Study Grants, and National Institutes of Health grant CA190144.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
Oregon Health and Science University and Dr. Grompe have a significant financial interest in Yecuris, Inc a company that may have a commercial interest in the results of this research and technology. This potential conflict of interest has been reviewed and managed by Oregon Health and Science University.
DATA AND CODE AVAILABILITY
This study did not generate any unique datasets or code.
References
- Beer S, Komatsubara K, Bellovin DI, Kurobe M, Sylvester K, and Felsher DW (2008). Hepatotoxin-induced changes in the adult murine liver promote MYC-induced tumorigenesis. PLoS One 3, e2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JY (2011). A clinical overview of centrosome amplification in human cancers. Int J Biol Sci 7, 1122–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Hulst C, Parvanova I, Tomoiaga D, Sapar ML, and Feinstein P (2013). Fast quantitative real-time PCR-based screening for common chromosomal aneuploidies in mouse embryonic stem cells. Stem Cell Reports 1, 350–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan AW (2013). Aneuploidy, polyploidy and ploidy reversal in the liver. Semin Cell Dev Biol 24, 347–356. [DOI] [PubMed] [Google Scholar]
- Duncan AW, Hanlon Newell AE, Bi W, Finegold MJ, Olson SB, Beaudet AL, and Grompe M (2012a). Aneuploidy as a mechanism for stress-induced liver adaptation. J Clin Invest 122, 3307–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan AW, Hanlon Newell AE, Smith L, Wilson EM, Olson SB, Thayer MJ, Strom SC, and Grompe M (2012b). Frequent aneuploidy among normal human hepatocytes. Gastroenterology 142, 25–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan AW, Hickey RD, Paulk NK, Culberson AJ, Olson SB, Finegold MJ, and Grompe M (2009). Ploidy reductions in murine fusion-derived hepatocytes. PLoS Genet 5, e1000385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan AW, Taylor MH, Hickey RD, Hanlon Newell AE, Lenzi ML, Olson SB, Finegold MJ, and Grompe M (2010). The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature 467, 707–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Font-Burgada J, Shalapour S, Ramaswamy S, Hsueh B, Rossell D, Umemura A, Taniguchi K, Nakagawa H, Valasek MA, Ye L, et al. (2015). Hybrid Periportal Hepatocytes Regenerate the Injured Liver without Giving Rise to Cancer. Cell 162, 766–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Godinho SA, and Pellman D (2009). A mechanism linking extra centrosomes to chromosomal instability. Nature 460, 278–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, and Pellman D (2007). Limiting the proliferation of polyploid cells. Cell 131, 437–440. [DOI] [PubMed] [Google Scholar]
- Gentric G, Celton-Morizur S, and Desdouets C (2012). Polyploidy and liver proliferation. Clin Res Hepatol Gastroenterol 36, 29–34. [DOI] [PubMed] [Google Scholar]
- Gentric G, and Desdouets C (2014). Polyploidization in liver tissue. Am J Pathol 184, 322–331. [DOI] [PubMed] [Google Scholar]
- Gentric G, Maillet V, Paradis V, Couton D, L’Hermitte A, Panasyuk G, Fromenty B, Celton-Morizur S, and Desdouets C (2015). Oxidative stress promotes pathologic polyploidization in nonalcoholic fatty liver disease. J Clin Invest 125, 981–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjelsvik KJ, Besen-McNally R, and Losick VP (2019). Solving the Polyploid Mystery in Health and Disease. Trends Genet 35, 6–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grompe M, al-Dhalimy M, Finegold M, Ou CN, Burlingame T, Kennaway NG, and Soriano P (1993). Loss of fumarylacetoacetate hydrolase is responsible for the neonatal hepatic dysfunction phenotype of lethal albino mice. Genes Dev 7, 2298–2307. [DOI] [PubMed] [Google Scholar]
- Guidotti JE, Brégerie O, Robert A, Debey P, Brechot C, and Desdouets C (2003). Liver cell polyploidization: a pivotal role for binuclear hepatocytes. J Biol Chem 278, 19095–19101. [DOI] [PubMed] [Google Scholar]
- He M, Tucciarone J, Lee S, Nigro MJ, Kim Y, Levine JM, Kelly SM, Krugikov I, Wu P, Chen Y, et al. (2016). Strategies and Tools for Combinatorial Targeting of GABAergic Neurons in Mouse Cerebral Cortex. Neuron 91, 1228–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalatova B, Jesenska R, Hlinka D, and Dudas M (2015). Tripolar mitosis in human cells and embryos: occurrence, pathophysiology and medical implications. Acta Histochem 117, 111–125. [DOI] [PubMed] [Google Scholar]
- Knouse KA, Lopez KE, Bachofner M, and Amon A (2018). Chromosome Segregation Fidelity in Epithelia Requires Tissue Architecture. Cell 175, 200–211 e213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Germain RN, and Gerner MY (2017). Multiplex, quantitative cellular analysis in large tissue volumes with clearing-enhanced 3D microscopy (Ce3D). Proc Natl Acad Sci U S A 114, E7321–E7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Nascimento EM, Gajera CR, Chen L, Neuhofer P, Garbuzov A, Wang S, and Artandi SE (2018). Distributed hepatocytes expressing telomerase repopulate the liver in homeostasis and injury. Nature 556, 244–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Meyer C, Xu C, Weng H, Hellerbrand C, ten Dijke P, and Dooley S (2013). Animal models of chronic liver diseases. Am J Physiol Gastrointest Liver Physiol 304, G449–468. [DOI] [PubMed] [Google Scholar]
- Lucchetta EM, and Ohlstein B (2017). Amitosis of Polyploid Cells Regenerates Functional Stem Cells in the Drosophila Intestine. Cell Stem Cell 20, 609–620 e606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malato Y, Naqvi S, Schürmann N, Ng R, Wang B, Zape J, Kay MA, Grimm D, and Willenbring H (2011). Fate tracing of mature hepatocytes in mouse liver homeostasis and regeneration. J Clin Invest 121, 4850–4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GM, and Sprague CA (1969). Parasexual cycle in cultivated human somatic cells. Science 166, 761–763. [DOI] [PubMed] [Google Scholar]
- Miyaoka Y, Ebato K, Kato H, Arakawa S, Shimizu S, and Miyajima A (2012). Hypertrophy and unconventional cell division of hepatocytes underlie liver regeneration. Curr Biol 22, 1166–1175. [DOI] [PubMed] [Google Scholar]
- Ohshima S, and Seyama A (2013). Establishment of proliferative tetraploid cells from normal human fibroblasts. Front Oncol 3, 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver TL (2015). When bigger is better: the role of polyploidy in organogenesis. Trends Genet 31, 307–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overturf K, Al-Dhalimy M, Finegold M, and Grompe M (1999). The repopulation potential of hepatocyte populations differing in size and prior mitotic expansion. Am J Pathol 155, 2135–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overturf K, al-Dhalimy M, Ou CN, Finegold M, and Grompe M (1997). Serial transplantation reveals the stem-cell-like regenerative potential of adult mouse hepatocytes. Am J Pathol 151, 1273–1280. [PMC free article] [PubMed] [Google Scholar]
- Overturf K, Al-Dhalimy M, Tanguay R, Brantly M, Ou CN, Finegold M, and Grompe M (1996). Hepatocytes corrected by gene therapy are selected in vivo in a murine model of hereditary tyrosinaemia type I. Nat Genet 12, 266–273. [DOI] [PubMed] [Google Scholar]
- Ovrebo JI, and Edgar BA (2018). Polyploidy in tissue homeostasis and regeneration. Development 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit SK, Westendorp B, and de Bruin A (2013). Physiological significance of polyploidization in mammalian cells. Trends Cell Biol 23, 556–566. [DOI] [PubMed] [Google Scholar]
- Pera F, and Rainer B (1973). Studies of multipolar mitoses in euploid tissue cultures. I. Somatic reduction to exactly haploid and triploid chromosome sets. Chromosoma 42, 71–86. [DOI] [PubMed] [Google Scholar]
- Preisegger KH, Factor VM, Fuchsbichler A, Stumptner C, Denk H, and Thorgeirsson SS (1999). Atypical ductular proliferation and its inhibition by transforming growth factor beta1 in the 3,5-diethoxycarbonyl-1,4-dihydrocollidine mouse model for chronic alcoholic liver disease. Lab Invest 79, 103–109. [PubMed] [Google Scholar]
- Ruzankina Y, Pinzon-Guzman C, Asare A, Ong T, Pontano L, Cotsarelis G, Zediak VP, Velez M, Bhandoola A, and Brown EJ (2007). Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell 1, 113–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansregret L, Vanhaesebroeck B, and Swanton C (2018). Determinants and clinical implications of chromosomal instability in cancer. Nat Rev Clin Oncol 15, 139–150. [DOI] [PubMed] [Google Scholar]
- Santaguida S, and Amon A (2015). Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat Rev Mol Cell Biol 16, 473–485. [DOI] [PubMed] [Google Scholar]
- Schnur J, Nagy P, Sebestyen A, Schaff Z, and Thorgeirsson SS (1999). Chemical hepatocarcinogenesis in transgenic mice overexpressing mature TGF beta-1 in liver. Eur J Cancer 35, 1842–1845. [DOI] [PubMed] [Google Scholar]
- Schoenfelder KP, and Fox DT (2015). The expanding implications of polyploidy. J Cell Biol 209, 485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmecki AM, Maruvka YE, Richmond PA, Guillet M, Shoresh N, Sorenson AL, De S, Kishony R, Michor F, Dowell R, et al. (2015). Polyploidy can drive rapid adaptation in yeast. Nature 519, 349–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigal SH, Rajvanshi P, Gorla GR, Sokhi RP, Saxena R, Gebhard DR, Reid LM, and Gupta S (1999). Partial hepatectomy-induced polyploidy attenuates hepatocyte replication and activates cell aging events. Am J Physiol 276, G1260–1272. [DOI] [PubMed] [Google Scholar]
- Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD, et al. (2010). Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143, 134–144. [DOI] [PubMed] [Google Scholar]
- Storchova Z, and Pellman D (2004). From polyploidy to aneuploidy, genome instability and cancer. Nat Rev Mol Cell Biol 5, 45–54. [DOI] [PubMed] [Google Scholar]
- Taghizadeh RR, and Sherley JL (2008). CFP and YFP, but not GFP, provide stable fluorescent marking of rat hepatic adult stem cells. J Biomed Biotechnol 2008, 453590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Goto H, Inoko A, Makihara H, Enomoto A, Horimoto K, Matsuyama M, Kurita K, Izawa I, and Inagaki M (2015). Cytokinetic Failure-induced Tetraploidy Develops into Aneuploidy, Triggering Skin Aging in Phosphovimentin-deficient Mice. J Biol Chem 290, 12984–12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Goto H, Nishimura Y, Kasahara K, Mizoguchi A, and Inagaki M (2018). Tetraploidy in cancer and its possible link to aging. Cancer Sci 109, 2632–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanami S, Ben-Moshe S, Elkayam A, Mayo A, Bahar Halpern K, and Itzkovitz S (2017). Dynamic zonation of liver polyploidy. Cell Tissue Res 368, 405–410. [DOI] [PubMed] [Google Scholar]
- Telentschak S, Soliwoda M, Nohroudi K, Addicks K, and Klinz FJ (2015). Cytokinesis failure and successful multipolar mitoses drive aneuploidy in glioblastoma cells. Oncol Rep 33, 2001–2008. [DOI] [PubMed] [Google Scholar]
- Toyoda H, Bregerie O, Vallet A, Nalpas B, Pivert G, Brechot C, and Desdouets C (2005). Changes to hepatocyte ploidy and binuclearity profiles during human chronic viral hepatitis. Gut 54, 297–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y, Mizrachi E, and Marchal K (2017). The evolutionary significance of polyploidy. Nat Rev Genet 18, 411–424. [DOI] [PubMed] [Google Scholar]
- Vitale I, Senovilla L, Jemaà M, Michaud M, Galluzzi L, Kepp O, Nanty L, Criollo A, Rello-Varona S, Manic G, et al. (2010). Multipolar mitosis of tetraploid cells: inhibition by p53 and dependency on Mos. EMBO J 29, 1272–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Zhao L, Fish M, Logan CY, and Nusse R (2015). Self-renewing diploid Axin2(+) cells fuel homeostatic renewal of the liver. Nature 524, 180–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson PD, Delgado ER, Alencastro F, Leek MP, Roy N, Weirich MP, Stahl EC, Otero PA, Chen MI, Brown WK, et al. (2019). The polyploid state restricts hepatocyte proliferation and liver regeneration. Hepatology 69, 1242–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Sage M, Sheppeard EA, Jurecic V, and Bradley A (2000). Engineering mouse chromosomes with Cre-loxP: range, efficiency, and somatic applications. Mol Cell Biol 20, 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.