SUMMARY
One approach to magnetogenetics uses radiofrequency (RF) waves to activate transient receptor potential channels (TRPV1 and TRPV4) that are coupled to cellular ferritins. The mechanisms underlying this effect are unclear and controversial. Theoretical calculations suggest that the heat produced by RF fields is likely orders of magnitude weaker than needed for channel activation. Using the FeRIC (Ferritin iron Redistribution to Ion Channels) system, we have un-covered a mechanism of activation of ferritin-tagged channels via a biochemical pathway initiated by RF disturbance of ferritin and mediated by ferritin-associated iron. We show that, in cells expressing TRPVFeRIC channels, RF increases the levels of the labile iron pool in a ferritin-dependent manner. Free iron participates in chemical reactions, producing reactive oxygen species and oxidized lipids that ultimately activate the TRPVFeRIC channels. This biochemical pathway predicts a similar RF-induced activation of other lipid-sensitive TRP channels and may guide future magnetogenetic designs.
In Brief
Radio-frequency (RF) fields activate TRPV channels coupled to endogenous ferritins. Herná ndez-Morales et al. show that ferritins transduce RF into biochemical signals responsible for channel activation. The interaction between RF and ferritin causes the increase of free iron, reactive oxygen species, and oxidized lipids, all of them TRPV actuators.
Graphical Abstract

INTRODUCTION
Magnetic control of ion channels promises remote and cell-specific stimulation or inhibition of neurons or other cells without limitation on tissue depth or requirements of invasive surgeries. Unlike optical fibers or implants, used in optogenetics and electrophysiological techniques, magnetic fields below gigahertz frequencies penetrate tissues with little attenuation (Young et al., 1980). There have been several independent reports of experimental evidence of magnetic control of transient receptor potential channels, TRPV1 and TRPV4, that are tagged with magnetic nanoparticles using either static (Stanley et al., 2015, 2016; Wheeler et al., 2016) or radiofrequency (RF) waves (Chen et al., 2015; Huang et al., 2010; Hutson et al., 2017; Munshi et al., 2017; Stanley et al., 2012, 2015, 2016). While the mechanisms of both static and RF-induced channel activation remain unclear and controversial (Barbic, 2019; Duret et al., 2019; Kole et al., 2019; Meister, 2016; Wang et al., 2019; Wheeler et al., 2016, 2019; Xu et al., 2019), the present study uses only RF waves.
TRPV1 and TRPV4 belong to a family of transient receptor potential channels that are non-selective cation channels. TRPV1 can be activated by vanilloids and heat (above 43°C) (Caterina et al., 1997); TRPV4 can be activated by heat (above 34°C) and mechanical force (Liedtke et al., 2000; Strotmann et al., 2000). The channels’ magnetic sensitivity is reportedly induced by attaching them to either exogenous magnetic nanoparticles (Chen et al., 2015; Huang et al., 2010; Munshi etal., 2017; Stanley et al., 2012) or ferritins (Hutson et al., 2017; Stanley et al., 2015, 2016; Wheeler et al., 2016). In one ferritin-based approach, chimeric anti-GFP-TRPV1 and GFP-tagged ferritin were co-expressed to generate a linker between TRPV1 and ferritin via GFP (Stanley et al., 2015, 2016). In another approach, TRPV1 and TRPV4 were fused with the ferritin-binding domain 5 (D5) of kininogen-1, which resulted in an endogenous ferritin iron redistribution to ion channels (FeRIC) (Hutson et al., 2017). Both Stanley et al. and Hutson et al. reported the use of RF to modulate cytosolic Ca2+ concentration in vitro and to generate physiological effects in vivo. For example, mice expressing anti-GFP-TRPV1 and GFP-tagged ferritin in glucose-sensing hypothalamic neurons have shown increased production of insulin upon stimulation with RF waves (Stanley et al., 2016). Further-more, combining a similar approach with a Ca2+-activated RhoA protein in HEK293 cells promoted cell migration upon stimulation with pulsed static magnetic fields (Mosabbir and Truong, 2018). In another example, RF stimulation of chicken embryos expressing TRPV1FeRIC or TRPV4FeRIC in cardiac neural crest cells has phenocopied congenital heart defects mimicking those of human patients (Hutson et al., 2017). Despite these experimental data, the mechanism underlying these phenomena remains unknown. Heating has been suggested as the stimulus produced by the interaction between RF and ferritin that activates the channels. One theoretical consideration, assuming that ferritin stores up to ~2,400 iron atoms and that those atoms exhibit independent magnetic domains (paramagnetism), indicates that this mechanism is not plausible (Meister, 2016). Another theoretical calculation, assuming that ferritins are at the maximum iron storage level (4,500 iron atoms) and that the iron atoms exhibit a single magnetic domain (superparamagnetism) or are clustered with each cluster having a different magnetic domain, indicates that the interaction between DC magnetic fields (0.1–2 T) and ferritin could generate heat that is sufficient to activate temperature-sensitive ion channels (Barbic, 2019). Currently, there is no theoretical indication supporting that an interaction between weak RF waves (<1 mT, ~100 MHz) and ferritin could generate the necessary heat for channel activation.
Besides their sensitivity to heat, both TRPV1 and TRPV4 are also sensitive to reactive oxygen species (ROS), lipids, and oxidized lipids. Here, we show that ROS production and lipid oxidation induced by RF and mediated by ferritin-associated iron can activate ferritin-tagged ion channels.
RESULTS
RF Stimulation of TRPV1FeRIC and TRPV4FeRIC Channels Increases Cytosolic Ca2+ Levels
Using the FeRIC system, we tested the effect of RF stimulation on the cytosolic Ca2+ levels in cells expressing GCaMP6 or expressing GCaMP6 plus TRPV. The cells expressing TRPV were identified as mCherry positive (mCherry+) cells (Figure 1A; Figure S1A). RF waves (180 MHz, 12 μT) were generated with a solenoid coil covering a cell culture dish. Data were quantified as the change in GCaMP6 fluorescence divided by baseline fluorescence (ΔF/F0) and the area under the curve (AUC). GCaMP6 data are presented as the averages (± SEM) from all TRPV-expressing cells. A cell was considered responsive to RF when the GCaMP6 ΔF/F0 increased 10 times over the standard deviation of its baseline fluorescence. RF stimulation did not change significantly the GCaMP6 AUC in Neuro2a (N2a) cells expressing GCaMP6 plus wild-type TRPV4 (TRPV4WT) compared with cells expressing GCaMP6 (Figures 1B–1D). Conversely, RF stimulation of TRPV4FeRIC-expressing cells increased the GCaMP6 AUC and the cell responsiveness compared with non-stimulated cells or cells expressing GCaMP6 (AUC = GCaMP6 RF: −0.54 ± 1.6; TRPV4FeRIC no RF: −0.56 ± 1.01; TRPV4FeRIC RF: 57.7 ± 9, p < 0.0001) (Figures 1B–1D). This effect was inhibited with the TRPV4 antagonist GSK 2193874 (GSK219, 100 nM) and in the absence of extracellular Ca2+ (Figures 1B–1D; Table S1). Next, we generated a TRPV4FeRIC mutant (M680D/DK675) whose Ca2+ permeability is abolished (Voets et al., 2002). In N2a cells expressing TRPV4M680D/DK675FeRIC, RF did not increase the GCaMP6 DF/F0 relative to GCaMP6-expressing cells (Figures 1B–1D; Table S1). Similarly, as previously reported (Hutson et al., 2017), in HEK293T cells expressing TRPV1FeRIC (Figure S1A), RF stimulation increased significantly the GCaMP6 AUC (Figures S1B and S1C; no RF: 4.3 ± 0.86; RF: 26 ± 5.1, p < 0.0001) and the cell responsiveness (Figure S1D; 1.95-fold change, p < 0.001) relative to those of non-stimulated cells. This effect was abolished by the TRPV1 antagonist SB 366791 (SB366, 10 mM) and in the absence of extracellular Ca2+ (Figures S1B–S1D). Furthermore, in HEK293T cells expressing the non-conductive mutant TRPV1E637KFeRIC (García-Martınez et al., 2000), RF did not increase the GCaMP6 DF/F0 or the cell responsiveness (Figures S1B–S1DE; Table S1). In all experiments, the functional expression of TRPV4 and TRPV1 was corroborated with the specific agonists GSK 1016790A (GSK101, 1 μM) and capsaicin (1 μM), respectively (Figures 1B and S1B). These results indicate that RF waves activate TRPV4FeRIC and TRPV1FeRIC and that this effect depends on Ca2+ influx through the channel pore.
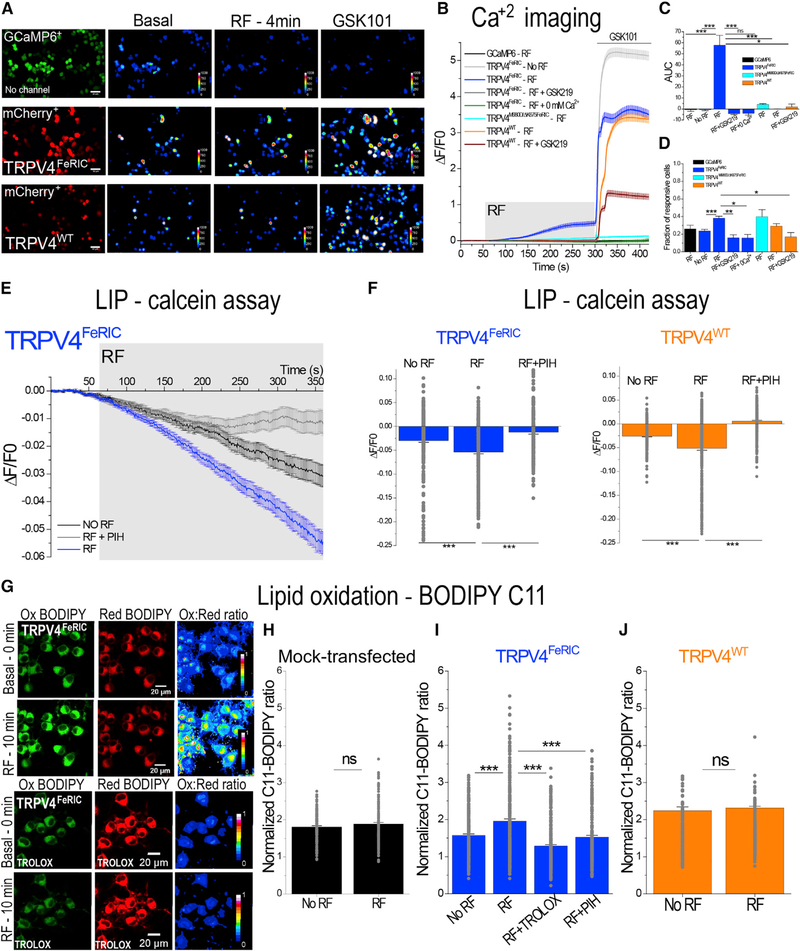
Figure 1. RF Increases the Cytosolic Ca2+ Concentration, the Labile Iron Pool (LIP), and the Levels of Lipid Oxidation in TRPV4FeRIC-Expressing N2a Cells.
(A) Leftmost column: representative images of N2a cells expressing GCaMP6 or GCaMP6 plus TRPV4FeRIC or TRPV4WT (mCherry+). Columns on the right: pseudo-color images of GCaMP6 fluorescence before (Basal) and after RF stimulation (12 μT), following 1 μM GSK101. Scale bars, 50 μm.
(B) Average changes (± SEM) in GCaMP6 ΔF/F0 in N2a cells expressing GCaMP6 or GCaMP6 plus TRPV4FeRIC, TRPV4M680D/ΔK675FeRIC, or TRPV4WT following exposure to RF (4 min, gray rectangle) and next GSK101 (bar). In separate series, cells were imaged without RF stimulation (no RF), in the absence of extracellular Ca2+ (0 mM Ca2+), or in the presence of GSK219.
(C) Averages of the GCaMP6 AUC (± SEM) for the period of RF stimulation.
(D) Cell responsiveness (± SEM) for data in (C).
(E) Average changes (± SEM) in calcein ΔF/F0 in N2a cells expressing TRPV4FeRIC imaged in the absence of RF (NO RF) or in the presence of RF (RF). In separate series, cells were first incubated with PIH and then imaged upon RF stimulation (RF + PIH).
(F) Average changes (± SEM) of calcein ΔF/F0 in cells expressing TRPV4FeRIC or TRPV4WT in the absence of RF, in the presence of RF, or in the presence on RF plus PIH.
(G) Representative confocal images of N2a cells expressing TRPV4FeRIC and labeled with BODIPY 581/591 C11 (BODIPY C11). The emissions of Ox and Red BODIPY C11 forms were collected at 490–520 nm and 570–620 nm, respectively.Pseudo-color images indicate the ratio of the fluorescence of the Ox to Red forms of BODIPY C11. Cells were imaged at the beginning of the experiment (Basal - 0 min) and after 10 min in the presence of RF or in the presence of RF plus TROLOX. Scale bars, 20 μm.
(H–J) Normalized BODIPY C11 Ox/Red ratio (± SEM) of (H) mock-transfected cells or cells expressing (I) TRPV4FeRIC or (J) TRPV4WT imaged after 10 min in the absence or in the presence of RF. In separate experiments, cells were pretreated with TROLOX or PIH following stimulation with RF. Data were normalized to the basal BODIPY C11 ratio obtained at the beginning of every single experiment. GCaMP6 data correspond to 3–13 independent experiments with 59–488 cells analyzed. For calcein data, 189–408 cells from 4–6 independent experiments were analyzed. For BODIPY C11 data, 228–314 cells from 4–8 independent experiments were analyzed. For this and the following figures, data correspond to the averages from all TRPV-expressing cells, and significance was determined using Student”s t test (n = 2) or one-way ANOVA (n > 3) followed by Tukey”s multiple comparisons test. For nonparametric data, significance was determined using Kruskal-Wallis ANOVA followed by Dunn”s multiple comparisons test. Where applicable, *p < 0.05, **p < 0.001, or ***p < 0.0001 was considered a statistically significant difference.
RF Stimulation Increases the Labile Iron Pool (LIP)
Ferritin is the main cellular iron storage protein (Arosio et al., 2017), and it has been reported that RF waves disturb its capacity to handle iron (Céspedes and Ueno, 2009; Céspedes et al., 2010). Next, we tested the effects of RF exposure on the LIP in cells loaded with calcein (Figure 1E). Calcein fluorescence is quenched following iron chelation, allowing the estimation of the LIP. Data were quantified as the change in calcein fluorescence (ΔF/F0) (Figures 1E and 1F; Table S2). The change of calcein ΔF/F0 over 5 min in TRPV4FeRIC- and TRPV4WT-expressing cells was significantly larger in the presence of RF relative to that in the absence of RF (TRPV4FeRIC: 2.2-fold change; TRPV4WT: 2-fold change; p < 0.0001) (Figures 1E and 1F). Moreover, treating cells with the iron chelator pyridoxal isonicotinoyl hydrazone (PIH; 100 mM) abolished the effect of RF on calcein ΔF/F0 in both TRPV4FeRIC- and TRPV4WT-expressing cells (Figures 1E and 1F). Likewise, RF stimulation of TRPV1FeRIC- and TRPV1WT-expressing HEK293T cells significantly changed the calcein ΔF/F0 compared with non-stimulated cells (TRPV1FeRIC: 2.8-fold change; TRPV1WT: 1.2-fold change; p < 0.001) (Figures S1E and S1F). To test the effect of RF on ferritin iron directly, we examined the levels of iron in a ferritin solution with the ferrozine-based assay. Ferrozine chelates the Fe2+ ion, forming a compound that was detected by spectroscopy. Application of RF to the ferritin solution significantly increased the concentration of the soluble Fe2+ ion (1.67-fold change, p < 0.05) compared with that in non-stimulated ferritins (Figure S2A). However, the concentration of the total iron (Fe2+ plus Fe3+) did not change following RF stimulation (Figure S2B). These results indicate that RF increases the levels of the LIP in both TRPV4FeRIC- and TRPV4WT-expressing cells.
RF Stimulation Increases Lipid Oxidation in Cells Expressing FeRIC Channels
The increase of the LIP may increase the levels of ROS via, e.g., redox reactions, or react with membrane lipids (Peterson et al., 1978; Schafer et al., 2000). Thus, we examined the levels of ROS using the 2′,7′-dichlorofluorescin diacetate (DCFDA), which is a non-fluorescent compound that is converted to the fluorescent DCF following its oxidation by ROS. RF stimulation of mock-transfected cells (Figures S2C, S2F, and S2J) or cells expressing TRPV4WT (Figure S2E) or TRPV1WT (Figure S2I) channels did not significantly change the DCF fluorescence relative to non-stimulated cells. Conversely, RF stimulation of cells expressing TRPV4FeRIC (Figure S2D), TRPV1FeRIC (Figure S2G), or TRPV1E637KFeRIC (Figure S2H) significantly increased DFC fluorescence (TRPV4FeRIC:1.14-fold change; TRPV1FeRIC: 1.22-fold change; TRPV1E637KFeRIC: 1.37-fold change; p < 0.05) relative to non-stimulated cells. We postulate that, in TRPVFeRIC-expressing cells, the increase of LIP and ROS may trigger the oxidation of lipids, because those channels reallocate ferritins in close proximity to the cell membrane (Hutson et al., 2017). Lipid oxidation was evaluated with BODIPY 581/591 C11 (BOD-IPY C11) (Drummen et al., 2002), which is a fatty acid that shifts its fluorescence emission from ~590 to ~510 nm upon oxidation.Cells were labeled with BODIPY C11, and oxidized (Ox) and reduced (Red) fluorescence were acquired simultaneously (Figure 1G). Data were quantified as the Ox/Red ratio of BODIPY C11. In TRPV4FeRIC-expressing cells, RF stimulation significantly increased the Ox/Red ratio of BODIPY C11 compared with non-stimulated cells (1.25-fold change; p < 0.0001) (Figure 1I). This RF effect was inhibited when TRPV4FeRIC cells were treated with the antioxidant TROLOX (100 μM) (−1.22-fold change; p < 0.0001) and with PIH ( 1.03-fold change; p < 0.0001) (Figure 1I). Conversely, in mock-transfected (Figure 1H) or TRPV4WT-expressing (Figure 1J) cells, RF did not change the Ox/Red ratio of BODIPY C11 compared with that in non-stimulated cells (mock-transfected: 1.04-fold change, p = 0.22; TRPV4WT:1.03-fold change, p = 0.44). Similarly, RF stimulation of TRPV1FeRIC-(Figure S1G) or TRPV1E637KFeRIC-expressing (Figure S1H) HEK293T cells significantly increased the Ox/Red ratio of BODIPY C11 (TRPV1FeRIC: 1.12-fold change, p < 0.001; TRPV1E637KFeRIC: 1.15-fold change, p < 0.0001) compared with that in non-stimulated cells but did not change that in TRPV1WT-expressing cells (−1.04-fold change, p = 0.42) (Figure S1I). These results indicate that, in cells expressing FeRIC channels, RF produces iron-dependent oxidation of lipids. Since TRPV1E637KFeRIC is a non-conductive channel, these results suggest that RF-induced ROS increase and lipid oxidation do not depend on cation flux through the channel. Interestingly, although RF increases the LIP levels in cells expressing FeRIC and WT channels (Figures 1E, 1F, S1E, and S1F), the RF-induced BODIPY C11 oxidation was observed only in cells expressing FeRIC channels, suggesting that the localization of ferritin near the membrane is needed to produce measurable lipid oxidation.
LIP, Lipid Oxidation, and ROS Are Involved in the RF-Induced Activation of TRPVFeRIC
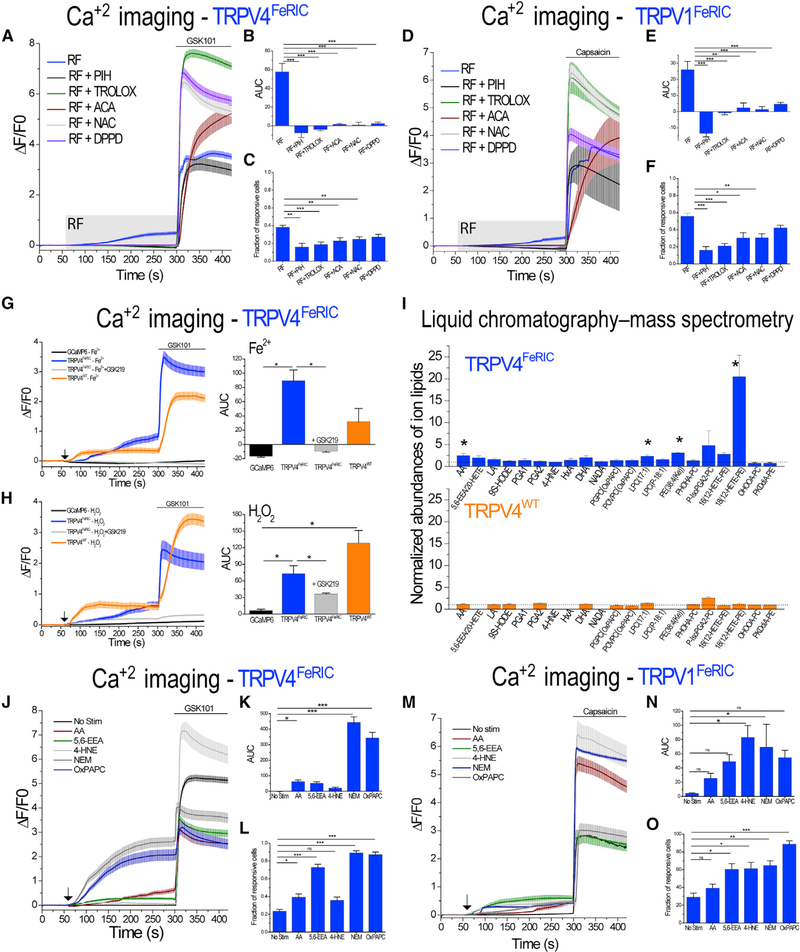
Next, we examined whether the increase of the LIP, ROS and oxidized lipids might be responsible for the activation of FeRIC channels. TRPV4FeRIC-expressing N2a cells were stimulated with RF in the absence or in the presence of TROLOX, PIH, N-acetyl-L-cysteine (NAC; a ROS inhibitor, 500 μM), N-(p-amylcinnamoyl) anthranilic acid (ACA; an inhibitor of the phospholipase A2, 50 μM), or N,N′-diphenyl-p-phenylenedi-amine (DPPD; an inhibitor of lipid peroxyl radicals, 1 μM) (Figures 2A–2C). Treatment of TRPV4FeRIC-expressing cells with all those substances inhibited the RF-induced effect on GCaMP6 AUC relative to non-treated cells (non-treated: 57.7 ± 9; PIH: −7.5 ± 5.06; TROLOX: −4 ± 1.3; ACA: 1.4 ± 0.9; NAC: 0.9 ± 2.82; DPPD: 2.4 ± 1.6; p < 0.0001) (Figures 2A–2C). Nevertheless, in the presence of all aforementioned substances, TRPV4FeRIC cells were activated with GSK101 (Figure 2A), ruling out the direct inhibition of the channels. Likewise, in TRPV1FeRIC-expressing HEK293T cells, the treatment with TROLOX, PIH, ACA, NAC, and DPPD inhibited the RF-induced changes of GCaMP6 AUC (non-treated: 26 ± 5.1; PIH: −13.5 ± 2.2; TROLOX: −0.8 ± 1.4; ACA: 2.3 ± 2.9; NAC: 1.3 ± 1.8; DPPD: 4.6 ± 1.1; p < 0.0001) relative to non-treated cells (Figures 2D–2F). Next, we tested whether stimulation with iron and a ROS generator activates TRPV4. In N2a cells expressing TRPV4FeRIC and TRPV4WT, stimulation with ferrous ammonium sulfate (Fe2+) (Figure 2G) and H2O2 (Figure 2H) increased the GCaMP6 AUC (TRPV4FeRIC-Fe2+: 89.4 ± 15.3; TRPV4WT-Fe2+: 31.8 ± 18.6; TRPV4FeRIC-H2O2: 72.8 ± 14.5; TRPV4WT-H2O2: 128.6 ± 22.4). Both Fe2+- and H2O2-induced GCaMP6 ΔF/F0 increases were suppressed by GSK219 (Figures 2G and 2H; Table S1).
Figure 2. Iron, ROS, Lipids, and Oxidized Lipids Are Involved in RF-Induced Activation of TRPVFeRIC.
(A and D) Average changes (± SEM) in GCaMP6 ΔF/F0 in (A) N2a cells expressing TRPV4FeRIC or (D) HEK293T cells expressing TRPV1FeRIC following exposure to RF (12 μT, gray rectangle) and next chemical agonists (bar). In separate series, cells were incubated for 15–60 min before RF and agonist stimulation with PIH, TROLOX, ACA, NAC, or DPPD.
(B and E) Averages of the GCaMP6 AUC (± SEM) for the period of RF stimulation. (C and F) Cell responsiveness (± SEM) for data in (B) and (E), respectively.
(G and H) Average changes (± SEM) in GCaMP6 ΔF/F0 in N2a cells expressing GCaMP6 or expressing GCaMP6 plus TRPV4FeRIC or TRPV4WT following exposure to (G) Fe2+ (100 μM) or (H) H2O2 (100 μM) (black arrows) and next GSK101 (bar). Insets: GCaMP6 AUC (± SEM) for the period from 61 to 290 s. In separate series, cells were incubated for 15 min before stimulation with GSK219.
(I) Normalized abundances of lipids detected in N2a cells expressing TRPV4FeRIC or TRPV4WT that were stimulated with RF, as measured by LC-MS. Ion abundances were normalized to the average ion abundance for samples that were not stimulated with RF. Data indicate means (± SEM) from 3–4 separate experiments each for RF and no RF conditions.
(J and M) Average changes (± SEM) in GCaMP6 ΔF/F0 in cells expressing (J) TRPV4FeRIC or (M) TRPV1FeRIC following exposure to AA, 5,6-EEA, 4-HNE, NEM, and OxPAPC (black arrows) and next agonists (bar).
(K and N) Averages of the GCaMP6 AUC (± SEM) for the period from 61 to 290 s.
(L and O) Cell responsiveness (± SEM) for data in (K) and (N), respectively.
GCaMP6 data correspond to 3–7 independent experiments with 62–276 cells analyzed. For LC-MS data, the statistical test applied was Student”s t test. Where applicable, *p < 0.05, **p < 0.001, or ***p < 0.0001 was considered a statistically significant difference.
See Table S3 for lipid name abbreviations.
To identify lipids and oxidized lipids involved in the RF-induced activation of FeRIC channels, we analyzed lipids extracted from TRPV4FeRIC- and TRPV4WT-expressing cells using liquid chromatography coupled to high-resolution mass spectrometry (LC-MS) (Figure 2I). Several lipids and oxidized lipids were found in TRPV4-expressing cells (Figure 2I; Table S3). Interestingly, whereas in cells expressing TRPV4WT, RF stimulation did not change the levels of the identified lipids relative to non-stimulated cells, in TRPV4FeRIC-expressing cells, RF increased the levels of oxidized lipids (Figure 2I; Table S3). The lipids that showed a significant increase in RF-stimulated TRPV4FeRIC-expressing cells were arachidonic acid (AA), lysophospholipids LPC(17:1) and PE(38:4(Ke)), and oxidized phospholipid 18(12-HETE-PE) (Figure 2I; Table S3), which are potential TRPV agonists (Ciardo and Ferrer-Montiel, 2017; Taberner et al., 2015). Moreover, some lipids associated with lipid oxidation processes such as 5,6-epoxy-eicosatrienoic acid (5,6-EEA), prostaglandin A1 (PGA1), 4-hydroxynonenal (4-HNE), and hepoxilin (HxA), among others, were found only in RF-stimulated TRPV4FeRIC-expressing cells (Figure 2I; Table S3). Corroborating the increased lipid oxidation, some common lipids, e.g., glycero-phospholipids, were significantly decreased in FeRIC-expressing samples stimulated with RF relative to non-stimulated cells (Table S3). To corroborate that FeRIC channels are activated by some of those lipids, cells expressing FeRIC channels were stimulated with AA (15 μM), added directly or through its production by activating the phospholipase A2 with N-ethylmaleimide (NEM; 100 μM), OxPAPC (a mixture of oxidation products of phosphati-dylcholine, 30 μg/mL), 5,6-EEA (300 nM), and 4-HNE (100 μM). In cells expressing TRPV4FeRIC and TRPV1FeRIC, all those lipids significantly increased the GCaMP6 ΔF/F0 relative to non-stimulated cells (Figures 2J–2O; Table S1). These results support that oxidized lipids are key players in the RF-induced activation of TRPV1FeRIC and TRPV4FeRIC.
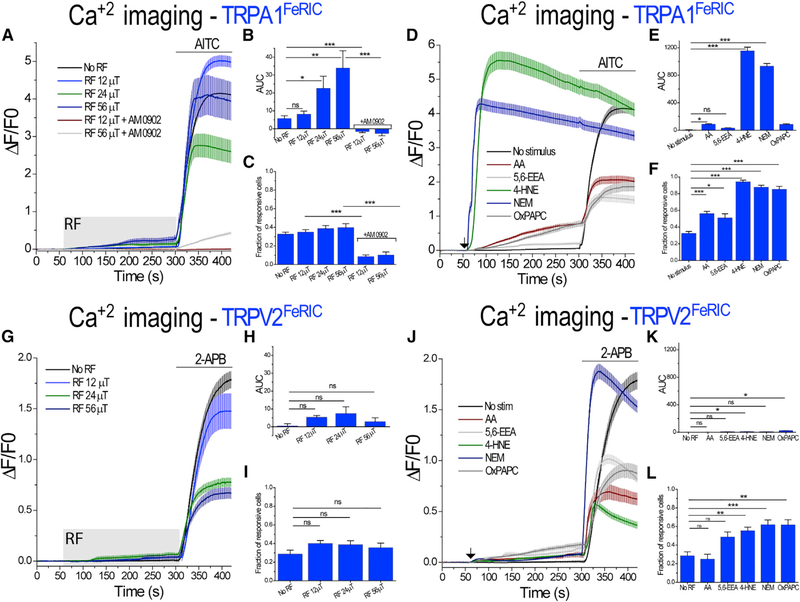
RF Activates TRPA1FeRIC Channels
We reason that, if the RF-induced activation of TRPV1FeRIC and TRPV4FeRIC is mediated by LIP and oxidized lipids, FeRIC should enable RF to modulate other ion channels with similar sensitivity to these signaling molecules. Rodent TRPA1, which is activated by noxious cold (~18°C) (Story et al., 2003), was selected because of its high sensitivity to ROS (Andersson et al., 2008) and oxidized lipids (Ciardo and Ferrer-Montiel, 2017; Taberner et al., 2015; Zygmunt and Högestätt, 2014). We fused rat TRPA1 with the D5 at the carboxyl terminus (Figures S3A and S3B). In TRPA1FeRIC-expressing N2a cells, stimulation with RF at 12 μT did not change the GCaMP6 AUC (no RF: 5.7 ± 1.4; RF: 8.15 ± 1.7, p = 0.29). However, RF at 24 and 56 μT significantly increased the GCaMP6 AUC in TRPA1FeRIC-expressing N2a cells relative to non-stimulated cells (RF at 24 μT: 22.7 ± 6.7, p < 0.05; RF at 56 μT: 34.03 ± 9.6, p < 0.001) (Figures 3A–3C). Moreover, the RF-induced activation of TRPA1FeRIC was abolished with the TRPA1 antagonist AM 0902 (1μM) (Figures 3A–3C). In all experiments, functional expression of TRPA1FeRIC was corroborated with the TRPA1 agonist AITC (100 μM) (Figure 3A). Next, we tested the sensitivity of TRPA1FeRIC to lipids, oxidized lipids, and ROS generators. All these compounds increased the GCaMP6 ΔF/F0 and responsiveness in TRPA1FeRIC-expressing cells (Figures 3D–3F and S4A; Table S1). On the other hand, rat TRPV2 is activated by noxious heat (above 50°C) (Caterina et al., 1999) and mechanical stimulus (Katanosaka et al., 2018; Muraki et al., 2003), but is much less sensitive to lipids and oxidized lipids (Ciardo and Fer-rer-Montiel, 2017; Taberner et al., 2015). We fused rat TRPV2 with the D5 at the carboxyl terminus (Figures S3C and S3D). RF stimulation (at 12, 24, and 56 μT) of TRPV2FeRIC-expressing N2a cells did not increase the GCaMP6 AUC relative to non-stimulated cells (no RF: 0.5 ± 1.2; RF at 12 μT: 5.5 ± 1.0; RF at 24 μT: 7.5 ± 3.7; RF at 56 μT: 3.0 ± 2.1, p = 0.15) (Figures 3G–3I; Table S1). However, RF stimulation for 1 h of cells expressing TRPV2FeRIC significantly increased the DFC fluorescence relative to non-stimulated cells (1.12-fold change, p < 0.0001) (Figures S2J and S2K), ruling out that the lack of RF-induced activation of TRPV2FeRIC is related to the lack of the RF-induced increase of ROS levels. The absence of RF activation of TRPV2FeRIC is consistent with the weak or no response to the aforementioned lipids, oxidized lipids (Figures 3J–3L) and ROS generators (Figure S4B).
Figure 3. RF-Induced Increments of Cytosolic Ca2+ in TRPA1FeRIC-Expressing N2a Cells.
(A) Average changes (± SEM) in GCaMP6 ΔF/F0 in N2a cells expressing GCaMP6 plus TRPA1FeRIC imaged in the absence of RF or in the presence of RF (gray rectangle) at different strengths (12, 24, and 56 μT) and next AITC (bar). For experiments including AM 0902, cells were incubated before RF stimulation.
(B) Average changes of the GCaMP6 AUC (± SEM) for the period of RF stimulation.
(C) Cell responsiveness (± SEM) for data in (B).
(D) Average changes (± SEM) in GCaMP6 ΔF/F0 in cells expressing TRPA1FeRIC following exposure to AA, 5,6-EEA, 4-HNE, NEM, OxPAPC (black arrow), and next AITC (bar).
(E) Average changes of the GCaMP6 AUC (± SEM) for the period from 61 to 290 s.
(F) Cell responsiveness (± SEM) for data in (E).
(G) Average changes (± SEM) in GCaMP6 ΔF/F0 in N2a cells expressing GCaMP6 or expressing GCaMP6 plus TRPV2FeRIC imaged in the absence of RF or upon exposure to RF (gray rectangle) at different strengths (12, 24, and 56 μT) and next 2-APB.
(H) Average changes of the GCaMP6 AUC (± SEM) for the period of RF stimulation.
(I) Cell responsiveness (± SEM) for data in (H).
(J) Average changes (± SEM) in GCaMP6 ΔF/F0 in cells expressing TRPV2FeRIC following exposure to AA, 5,6-EEA, 4-HNE, NEM, OxPAPC (black arrow) and next 2-APB (bar).
(K) Average changes of the GCaMP6 AUC (± SEM) for the period from 61 to 290 s.
(L) Cell responsiveness for data in (K).
GCaMP6 data correspond to 3–9 independent experiments with 88–180 cells analyzed. Where applicable, *p < 0.05, **p < 0.001, or ***p < 0.0001 was considered a statistically significant difference.
See Table S3 for lipid name abbreviations.
Ferritin, but Not TRPV4 Temperature Sensitivity, Is Involved in RF-Induced FeRIC-Channel Activation
Previously, we confirmed that activation of TRPV1FeRIC requires endogenous ferritin (Hutson et al., 2017). Here, we corroborate that ferritin is necessary for RF-mediated lipid oxidation and activation of TRPV4FeRIC. The ferritin heavy polypeptide 1 gene (Fth1) was knocked out in N2a cells (N2aFth1KO) with the clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein 9 (Cas9) (Figure 4A). We first examined the cytosolic Ca2+ levels in N2aFth1KO cells. RF stimulation of N2aFth1KO cells expressing TRPV4FeRIC induced GCaMP6 ΔF/F0 increments that were significantly smaller compared with those in wild-type N2a cells expressing TRPV4FeRIC (−9.2-fold change, p < 0.05), which was partially rescued by co-expressing FTH1 (ferritin heavy chain 1, −1.6-fold change, p = 0.3) (Figures 4B–4E; Table S1). Similar results were observed in N2aFth1KO cells expressing TRPA1FeRIC (Figure S4C; Table S1). Next, we corroborated that ferritin is necessary for the RF-induced LIP increase. The change of calcein ΔF/F0 in N2aFth1KO cells expressing either TRPV4FeRIC (Figure 4F) or TRPV4WT (Figure 4H) was not significantly different in RF-stimulated cells relative to non-stimulated cells (TRPV4FeRIC: 1.1-fold change, p = 0.65; TRPV4WT: 1.1-fold change, p = 0.48). The RF-induced LIP increase was rescued in N2aFth1KO cells by the co-expression of FTH1. RF stimulation of Na2Fth1KO cells expressing FTH1 plus TRPV4FeRIC (Figure 4G) or TRPV4WT (Figure 4I) significantly changed the calcein ΔF/F0 (TRPV4FeRIC: 2.4-fold change; TRPV4WT:1.5-fold change, p < 0.001) relative to non-stimulated cells. Finally, we corroborated that ferritin is necessary for RF-induced lipid oxidation. In TRPV4FeRIC-expressing N2aFth1KO cells, RF did not change the Ox/Red ratio of BODIPY C11 compared with that for non-stimulated cells (no RF: 1.84 ± 0.03; RF: 1.74 ± 0.04, p = 0.35) (Figure 4J). However, expression of TRPV4FeRIC plus FTH1 in N2aFth1KO cells significantly increased the Ox/Red ratio of BODIPY C11 upon RF exposure (no RF: 1.74 ± 0.04; RF: ± 0.03, p < 0.001) (Figure 4K). To examine whether the temperature change is involved in RF-induced activation of TRPV4FeRIC, we generated the mutant TRPV4ΔTFeRIC (Y555A/S556A), whose temperature sensitivity was abolished (Duret et al., 2019; Voets et al., 2002) (Figures S4D and S4E). RF stimulation of TRPV4ΔTFeRIC-expressing cells increased the GCaMP6 ΔF/F0 that was not significantly different compared with that of RF-stimulated TRPV4FeRIC-expressing cells (−1.1-fold change, p = 0.76) (Figures 4B–4E). Taken together, these results indicate that RF activation of FeRIC channels through the increase of the LIP, ROS, and oxidized lipids requires endogenous ferritins. Although further studies are needed, these results also suggest that temperature is not a major player in the RF activation of FeRIC channels.
Figure 4. Ferritin Is Necessary for RF-Induced Activation of FeRIC Channels, Increments of LIP, and Increase of Lipid Oxidation.
(A) Western blot analysis of lysates from N2aWT and two colonies of Na2Fth1KO cells. The knockout of Fth1 was corroborated by stimulating the cells with 100 μM FeCl3.
(B) Average changes (± SEM) in GCaMP6 ΔF/F0 in N2aWT or N2aFth1KO cells expressing TRPV4FeRIC or expressing TRPV4FeRIC plus FTH1 or TRPV4DTFeRIC following exposure to RF (12 μT, gray rectangle) and next GSK219 (bar).
(C) Zoom-in of the average changes (± SEM) in GCaMP6 ΔF/F0 corresponding to the period of RF stimulation.
(D) Average changes of the GCaMP6 AUC (± SEM) for the period of RF stimulation.
(E) Cell responsiveness (± SEM) for data in (D).
(F–I) Time course of the average changes (± SEM) in calcein ΔF/F0 in N2aFth1KO cells expressing (F) TRPV4FeRIC or (G) TRPV4FeRIC plus FTH1 and (H) TRPV4WT or (I) TRPV4WT plus FTH1 imaged in the absence of RF or in the presence of RF.
(J and K) Average changes (± SEM) of BODIPY C11 fluorescence ratio (Ox/Red) in N2aFth1KO cells expressing (J) TRPV4FeRIC or (K) TRPV4FeRIC plus FTH1 imaged in the absence or in the presence of RF.
(L) The cartoon illustrates the biochemical mechanism of RF-induced activation of FeRIC channels. (1) Stimulation with RF (2) increases the levels of LIP and ROS in a ferritin-dependent manner. (3) ROS and LIP react with membrane lipids producing oxidized lipids. (4) ROS, lipids, and oxidized lipids activate FeRIC channels. GCaMP6 data correspond to 5–13 independent experiments with 80–464 cells analyzed. For calcein, data correspond to 155–467 cells analyzed from 4–8 independent experiments. BODIPY C11 data correspond to 161–284 cells analyzed from 4–6 independent experiments. Where applicable, *p < 0.05, **p < 0.001, or ***p < 0.0001 was considered a statistically significant difference.
DISCUSSION
Bioactive iron participates in several redox reactions, producing ROS, hydroxyl free radicals, lipid radicals, and oxidized lipids (Ayala et al., 2014; Catalá , 2009; Graf et al., 1984). Notably, ROS, several lipids, and oxidized lipids are detected by TRPV channels (Andersson et al., 2008; Ciardo and Ferrer-Montiel, 2017; Taberner et al., 2015; Zygmunt and Högestätt, 2014). Our results indicate that RF stimulation elevates the LIP, ROS, and oxidized lipids, which ultimately activates FeRIC channels (Figure 4L).
The key for this signaling pathway to occur is the spatial proximity between ferritin, membrane lipids, and TRPV channels, which promotes the production of ROS, lipids, and oxidized lipids near the channels. Although RF also elevates the LIP in cells that express the wild-type TRPV channels, it likely does not reach the cell membrane lipids and produce sufficient signaling lipids to activate the channels. The gradual increase of intracellular Ca2+ levels over a period of a few minutes and the limited responsiveness (30%–40%) upon RF stimulation of FeRIC-expressing cells are also consistent with the characteristics of channel activation by signaling lipids. Further, the oxidation mechanism predicts RF-induced activation in other ferritin-tagged ion channels that are sensitive to ROS, lipids, and oxidized lipids, but it will not affect ferritin-tagged channels insensitive to those chemical actuators. We verified this prediction with TRPA1 and TRPV2, respectively (Andersson et al., 2008; Ciardo and Ferrer-Montiel, 2017; Taberner et al., 2015; Zygmunt and Högestätt, 2014). Although the evidence presented here indicates that the mechanism for the ferritin-dependent oxidation of membrane lipids is involved in the RF-induced activation of FeRIC channels, further studies are necessary to determine whether it is valid for other ferritin-tagged ion channels—e.g., anti-GFP-TRPV1/GFP-ferritin (Stanley et al., 2015, 2016)—that have reported sensitivity to RF waves. Although these approaches all couple TRP channels with ferritins, they are different and complex in terms of their components. For instance, whereas our FeRIC channels use endogenous ferritins, some other channels are chimeras of TRP channels with ferritin subunits. It is unknown whether the chimeric ferritins handle Fe2+ atoms in a manner similar to that of endogenous ferritins or whether their interaction with RF increases the LIP, ROS, and oxidized lipids. In addition, heating-dependent mechanisms, via the magneto-caloric or the Einstein-de Hass effects (Barbic, 2019), have been proposed as possibly responsible for staticfield activation of temperature-sensitive channels. Our results indicate that the ablation of temperature sensitivity of FeRIC channels does not prevent their RF-induced activation. While the oxidation mechanism does not require direct temperature activation of the channels, the mechanism does not exclude the possibility of localized heating or synergy of lipid oxidation and heating mechanisms.
Magnetogenetics approaches have been developed as promising tools to control neuronal excitability (Stanley et al., 2016; Wheeler et al., 2016). There has been a long line of research searching for naturally occurring biological molecules that are sensitive to magnetic fields (Hore and Mouritsen, 2016; Johnsen and Lohmann, 2005, 2008; Krishnan et al., 2018). Other recent studies utilized genetically encoded TRP channels tagged with ferritins, actuated by magnetic fields. It is necessary to make a distinction between approaches using static (Stanley et al., 2016; Wheeler et al., 2016) and RF (Hutson et al., 2017; Stanley et al., 2016) fields as the underlying mechanisms, which are likely different, and especially when the efficacy of these technologies is still being investigated (Kole et al., 2019; Wang et al., 2019; Xu et al., 2019). The focus of the present study was to gain insight into the molecular basis of the RF-induced activation of FeRIC channels. However, we have not examined the control of neuronal membrane potential with FeRIC technology. This is the next step to validate FeRIC technology as a tool for remotely controlling neuronal excitability.
In conclusion, our study identified a mechanism for RF-induced activation of ion channels via iron-induced redox reactions; mainly, lipid oxidation. The mechanism predicted a verifiable phenomenon and pointed out potential directions for designing and optimizing magnetic control of ion channels.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will fulfilled by the Lead Contact, Chunlei Liu (chunlei.liu@berkeley.edu). Plasmids will be made available upon request and will require a material transfer agreement from C.L.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
Where indicated, the HEK293T cell line (ATCC Cat# CRL-3216, RRID:CVCL_0063) or Neuro2a cell line (N2a, ATCC Cat# CCL-131, RRID:CVCL_0470) were used. Cell lines were obtained from the UCB Cell Culture Facility (University of California Berkeley). Cell identity and negative Mycoplasma contamination were verified by the UCB Cell Culture Facility. Cells were maintained in Dulbeccos’s Modified Eagle Medium (DMEM, GIBCO) supplemented with 10% fetal bovine serum (FBS, hyclone) and 100 units/mL penicillin, and 100 mg/mL streptomycin at 37°C and 5% CO2. For described experiments, cell lines were employed between the passages 5 to 30.
METHOD DETAILS
Fth1 deletion in Neuro2a cells
Ferritin heavy chain (Fth1) was deleted in N2a cells using commercially available double nickase (CRISPR/Cas9) plasmids. Neuro2a cells were transfected with the Fth1 CRISPR/Cas9 KO plasmid plus the HDR plasmid (Santa Cruz Biotechnology Inc.). The Fth1 KO plasmid uses 20-nucleotides guide RNA sequences targeting the mouse Fth1 gene. The HDR plasmid incorporated the puromycin resistance gene and the red fluorescent protein (RFP) for selection of cells where the Fth1 gene have knocked out. Cells were transfected with both Fth1 KO and HDR plasmids. 24 h later, cells were selected in DMEM supplemented medium containing puromycin (5mg/mL) for 7–14 days. Surviving cells were harvested, washed, and sorted with flow cytometry to select the RFP positive (RFP+). Following, RFP+ cells were serially diluted into a 96-well plate. Wells containing single cells were expanded and screened by RT-PCR for full-length Fth1 mRNA. Deletion was confirmed by RT-PCR and western blot for two cell colonies (#1 and #4). Following, the RFP+ cell colony #4, corroborated as knocked out for Fth1, was selected for propagation (Figure 4A). For imaging experiments, the RFP gene, flanked by two LoxP sites, was removed by transfecting the Na2Fth1KO cells with the Cre plasmid. Next, cells were sorted by flow cytometry and those cells negative to RFP expression were propagated.
Plasmids
The constructs TRPV1FeRIC, TRPV1WT, TRPV4FeRIC, TRPV4WT, and human FTH1 were obtained as described previously (Hutson et al., 2017). The mCherry red fluorescent protein included into the PLVX-IRES-mCherry constructs was used as a reporter of the TRPV-channels expression. To generate the TRPA1FeRIC construct, we used the full-length rat TRPA1 cDNA, which was a gift from David Julius (UCSF). To generate the TRPA1FeRIC construct, PCR primers were designed to introduce novel 5′Spe I sites and 3′ Not I sites into their respective locations outside the open reading frame using PCR. The PCR product TRPA1 with Spe I and Not I sites was subcloned within the multiple cloning site of the PLVX-IRES-mCherry containing the human domain 5 (D5) of Kininogen-1 (FeRIC) obtained previously (Hutson et al., 2017) (Figures S3A and S3B). To generate the TRPV2FeRIC construct, we used the full-length rat TRPV2 cDNA, which was a gift from David Julius (UCSF). To generate the TRPV2FeRIC construct, PCR primers were designed to introduce novel 5′Spe I sites and 3′ Not I sites into their respective locations outside the open reading frame using PCR. The PCR product TRPV2 with novel 5′Spe I sites and 3′ Not I sites were introduced into their respective locations outside the open reading frame using PCR. This product was subcloned into Spe I and Not I sites within the multiple cloning site of the PLVX-IRES-mCherry containing the human domain5 of Kininogen-1 (FeRIC) obtained previously (Hutson et al., 2017) (Figures S3C and S3D). To obtain the non-conductive TRPV1FeRIC channel, a single point mutation at E637K was inserted (García-Martınez et al., 2000). To obtain the non-conductive TRPV4FeRIC channel, a single point mutation (M680D) and a deletion (DK675) were inserted (Duret et al., 2019; Voets et al., 2002). TRPA1FeRIC, TRPV2FeRIC, and TRPV4M680D/DK675FeRIC constructs were generated at QB3 Macrolab (UC Berkeley). TRPV1E637KFeRIC construct was generated at GenScript®. All completed constructs were sequence-verified by the Molecular Cell Biology Sequencing Facility (UC Berkeley) and analyzed using MacVector 13.0 and Serial Cloner 2.6.
RF coils
Two RF emitting coils with diameters of 5 cm or 25 cm were used. The smaller 5 cm coil was made of two loops of wire, and the larger 25 cm coil was made of a single loop of copper tape. The coils were connected in series with tuning capacitors forming an LC circuit, and were tuned to a resonance frequency of about 180 MHz. The larger coil distributed the series capacitance around the loop. Matching capacitors were connected in parallel to the resonant circuits to match the coils’ impedances to 50 ohms. RF signal was generated by a broadband (35 MHz to 4.4 GHz) signal generator (RF Pro Touch, Red Oak Canyon LLC) and amplified using a 5W amplifier (Amplifier Research, model 5U100, 500 kHz to 1000 MHz). The magnetic field produced by the coils was calculated by measuring the voltage induced in a separate loop of wire connected to a high input impedance PC-based USB oscilloscope (Analog Arts model SA985) and then applying Faraday’s law. Magnetic field strength was about 12 mT at the position of cell culture dishes. For RF stimulation of TRPA1FeRIC and TRPV2FeRIC channels, the magnetic field strength was increased up to 24 and 56 μT. For an RF strength of 12 μT, the corresponding electric field at the center of cell culture dish was about 31 V/m.
Ca2+ imaging
Epifluorescence imaging experiments were conducted as previously described (Hutson et al., 2017). HEK293T were plated on glass-bottom 35-mm dishes coated with 0.01% Poly-L-lysine (SIGMA-Aldrich #P4832). For coating the plates, 200 μL of Poly-L-lysine was placed over the glass-bottom dishes and were incubated at 37°C for 1 h. Next, dishes were washed three times with sterile HBSS (ThermoFisher # 14025092). N2a cells (wild-type and Fth1KO) were plated on non-coated glass-bottom 35-mm dishes. Cells were cultured in DMEM supplemented medium. After 18–24 h, cells were transfected using the Lipofectamine LTX Plus reagent (ThermoFisher #15338030) with GCaMP6 (GCaMP6 medium, Addgene cat.40754) and ion channels (TRPV1FeRIC, TRPV1ΔTFeRIC, TRPV1E637KFeRIC TRPV1WT, TRPV4FeRIC, TRPV4ΔTFeRIC, TRPV4M680D/ΔK675FeRIC, TRPV4WT, TRPA1FeRIC, or TRPV2FeRIC). For transfection, OptiMEM free serum medium (ThermoFisher #31985088) was used to prepare the DNA/Lipofectamine LTX mix. Transfection mix had the following composition per each 35-mm dish: 300 μL OptiMEM, 4 μL Lipofectamine LTX, 3 μL PLUS reagent, 0.7 μg TRP DNA, and 0.7 μg of GCaMP6 DNA. Because the expression of TRP channels in N2aFh1KO cells or TRPV1 in HEK293T showed effects in the health in a fraction of cell population, EGTA (pH 8, ThermoFisher Scientific, #NC0997810) was added to dishes after transfection to achieve 1 mM final concentration. Using the free online calculator, it was estimated that 1 mM of EGTA decreases the Ca2+ concentration in the DMEM culture medium from 1.8 to 0.8 mM (https://somapp.ucdmc.ucdavis.edu/pharmacology/bers/maxchelator/CaEGTA-TS.htm). Cytosolic levels of Ca2+ were monitored by fluorescence imaging of cells positive for GCaMP6. Cells expressing either FeRIC, mutated FeRIC, or wild-type channels were identified as those cells with mCherry+ expression. Experiments were conducted using an upright AxioExaminer Z-1 (Zeiss) equipped with a camera (Axiocam 702 mono) controlled by Zen 2.6 software. Excitation light was delivered from a LED unit (33 W/cm2; Colibri 7 Type RGB-UV, Zeiss). mCherry was excited at 590/27 nm and emission was collected at 620/60 nm. GCaMP6 was excited at 469/38 nm and emission was collected at 525/50 nm. Illumination parameters were adjusted to prevent overexposure and minimize GCaMP6 photobleaching. All the experiments corresponding to a series were done under the same illumination settings. Images were captured with a W “Plan-Apochromat” 20x/1.0 DIC D = 0.17 M27 75mm lens at 1 frame/s. Unless stated otherwise, experiments were carried out at room temperature (20 – 22°C) using the Live Cell Imaging Solution (Invitrogen, A14291DJ) that contains (in mM) 140 NaCl, 2.5 KCl, 1 MgCl2, 1.8 CaCl2, 20 HEPES, pH 7.3. Live Cell Imaging Solution was supplemented with 10 mM D-glucose. At the beginning of imaging experiments, a 35-mm glass-bottom dish containing the cells was washed three times with 1 mL of the Live Cell Imaging Solution. Next, the dish was placed onto the microscope stage and the cells were localized with transmitted illumination (bright field). Next, with reflected illumination, the fluorescence signals from mCherry and GCaMP6 were corroborated and the field of interest was selected. Preferred fields were those with isolated and healthy cells. A thermocouple coupled to a thermistor readout (TC-344C, Warner Instruments) was placed inside the plate in direct contact with the Live Cell Imaging Solution. The temperature of the Live Cell Imaging Solution was monitored during the experiment (Temperature initial: 22.09°C; Temperature final: 22.03°C; ΔT: Ȓ0.6°C; n = 305). Cells were rested for about 10 min before imaging. RF was delivered using a custom-built RF-emitting coil that fits the 35-mm tissue culture dish, as described above. Unless stated otherwise, cells were stimulated with RF waves at 12 μT. For each experiment, cells were imaged for the first 60 s with no stimulus (Basal) following RF exposure for 4 min. After RF stimulation, cells were exposed to TRPV agonists by pipetting 1 mL of the agonist diluted in Live Cell Imaging Solution at twice the final concentration. In the second series of experiments, cells were exposed to diverse chemical compounds (ion channels antagonists, iron chelators, antioxidants, lipid peroxyl inhibitors, and ROS inhibitors) for 10–60 min and then stimulated with RF and the agonists. Images were analyzed using ImageJ FIJI (National Institutes of Health) and a customized code programmed with MATLAB.
Calcein assay – cytosolic free iron level (LIP)
Epifluorescence imaging experiments were conducted as described above for Ca2+ imaging. N2a or HEK293T cells were plated on glass-bottom 35-mm dishes and cultured in DMEM supplemented medium. After 24 h, cells were transfected using the Lipofectamine LTX Plus reagent with TRPV channels. Cytosolic levels of free iron were monitored by fluorescence imaging of cells loaded with calcein-AM (C3100MP, Thermo Fisher Scientific) which is a cell permeable iron chelator. Once in the cytosol, calcein-AM is hydrolyzed by the endogenous esterases producing the fluorescent and impermeable calcein form. N2a cells expressing TRPV channels were loaded with calcein (500 nM) for 20 min at 37°C. Afterward, cells were washed three times with Live Cell Imaging Solution and incubated at 37°C for 30 min. Calcein has a peak emission of 515 nm and following iron chelation, its fluorescence is quenched. Cells expressing the TRPV channels were identified as those mCherry+ cells (excitation at 590/27 nm/emission at 620/60 nm). Calcein was excited at 469/38 nm and emission was collected at 525/50 nm. Images were captured with a 20x lens at 1 frame/s. Experiments were carried out at room temperature (20 – 22°C) using the Live Cell Imaging Solution (Invitrogen, A14291DJ) supplemented with 10 mM D-glucose.
BODIPY® 581/591 C11 assay – Lipid oxidation
Confocal microscopy experiments were conducted as follows. HEK293T were plated on glass-bottom 35-mm dishes coated with 0.01% Poly-L-lysine (SIGMA-Aldrich #P4832). N2a cells (wild-type and Fth1KO) were plated on non-coated glass-bottom 35-mm dishes. Cells were cultured in DMEM supplemented medium. After 24 h, cells were transfected using the Lipofectamine LTX Plus reagent with the Flag versions of TRPV1FeRIC, TRPV1WT, TRPV4FeRIC, or TRPV4WT. For TRPV1E637KFeRIC, cells were transfected with the original construct containing the mCherry reporter. In particular, when HEK293T cells were transfected with TRPV1FeRIC or TRPV1WT, culture medium was supplemented with EGTA to reach a final concentration of about 500 μM. Twenty-four hours after transfection, cells were labeled with the lipid oxidation sensor BODIPY® 581/591 C11 (Image-iT® Lipid Peroxidation Kit, Thermo Fisher Scientific, D3861) at 1 μM in Live Cell Imaging Solution for 30 min at 37°C. Next, cells were washed three times with Live Cell Imaging Solution. BODIPY® 581/591 C11 imaging was carried out in a phenol red-free culture Medium (OptiMEM, Thermo Fisher Scientific, #11058021). Cells were scanned in a laser scanning microscope (Zeiss 710). The 20x objective lens was used for examine the fluorescence of BODIPY® 581/591 C11. In all experiments, cells were scanned under the same microscope settings, i.e., laser power and pinhole (1 airy unit). The green and red fluorescence of BODIPY™ C11 was acquired simultaneously using double wavelength excitation (laser lines 488 and 568 nm) and emission (490–530 and 570–610 nm). The red and green fluorescence signals correspond to the reduced (Red) and oxidized (Ox) forms of BODIPY® 581/591 C11, respectively. Cells labeled with BODIPY 581/591 C11 were imaged at the beginning of the experiment (basal, t = 0 min) and after 10 min in the absence or in the presence of RF. In a separate series of experiments, cells were simultaneously labeled with BODIPY 581/591 C11 and treated with the iron chelator PIH or TROLOX. Experiments were conducted at 37°C in an incubation chamber with temperature-control.
Lipid extraction
N2a cells were plated on 100-mm dishes and cultured in DMEM supplemented medium. After 24 h, cells were transfected using the Lipofectamine LTX Plus reagent with TRPV4FeRIC or TRPV4WT. 24 h after transfection, DMEM culture medium was replaced with opti-MEM medium (Invitrogen, 11058021) which is buffered with HEPES and sodium bicarbonate. The RF and No RF samples were processed simultaneously. For RF groups, RF was applied at 12 μT with the 25-cm RF coil for 30 min. For No RF groups, samples were maintained in the same room, but placed where RF was negligible. Experiments were conducted at room temperature (20–22°C). Next, cells were slightly trypsinized (trypsin 0.05% for 1 min at 37°C) and quantified. A total of 10 × 106 cells was taken from each sample and transferred to glass tubes. Next, lipids were extracted following the Folch method (Folch et al., 1957). Samples were washed with cold 0.3% ammonium acetate solution following centrifugation at 311 × g for 5 min at 4°C. Cell pellet was suspended with 1.5 mL of a cold solution of methanol and chloroform (ratio 2:1 v/v). Suspension was vigorously vortexed and incubated on ice for 40 min. Next, 0.38 mL of water was added to separate the aqueous and organic layers. The suspension was vortexed and incubated on ice for 10 min. Samples were centrifuged at 311 × g for 5 min at 4°C. The lower layer (organic) was transferred to a new tube. Samples were dried under nitrogen. Samples were reconstituted in 50 μL of isopropanol and transferred to a liquid chromatography autosampler vial with glass insert.
High-resolution liquid chromatography-mass spectrometry (LC-MS)
LC-MS measurements were performed using a liquid chromatography system (1200 series, Agilent) that was connected in line with a mass spectrometer (LTQ-Orbitrap-XL, Thermo Fisher Scientific) equipped with an electrospray ionization source. The LC was equipped with a C18 column (Restek). High-resolution mass spectra were acquired using the negative ion mode. Data acquisition was performed using Xcalibur software (version 2.0.7, Thermo). Measured ion mass-to-charge ratio (m/z) values were searched against LipidMAPS and PubChem databases to identify lipids.
Ferrozine assay – Ferrous (Fe2+) and total iron levels (Fe2+ + Fe3+)
Fe2+ levels of a solution of horse spleen ferritin (SIGMA-Aldrich, F4503) were monitored by absorption spectrophotometry of the Fe2+-ferrozine complex. All solutions were freshly prepared with the assay buffer (5M ammonium acetate, pH 7). Ferritin was prepared at 3 mg/mL, ferrozine (SIGMA-Aldrich, 160601) was prepared at 4 mM, and L-Ascorbic acid was prepared at 1M. Ferritin samples (100 μL) were transferred to a 96 well plate. To measure ferrous iron (Fe2+), 5 μL of assay buffer was added to each sample. To measure total iron (Fe2+ plus Fe3+), 5 μL of 1M of L-Ascorbic acid was added to each sample. L-Ascorbic acids acts as the reducing agent, allowing the measurement of total iron. Samples were mixed and incubated for 30 min at room temperature. Next, 100 μL of 4 mM ferrozine was added to each sample. Samples were mixed and incubated for 1-h protected from light. Absorbance was measured at 550 nm with a plate reader (EnVision Plate reader 21049). The RF and No RF ferritin samples were processed simultaneously. For RF groups, RF was applied at 12 μT with the 25-cm RF coil during the entire period of the experiment (1.5-h). For No RF groups, samples were maintained in the same room, but placed where RF was negligible. The calibration curve was obtained with Fe2+ standards from 0 to 50 μM, freshly prepared every time the assay was conducted.
DCFDA/H2DCFDA – Cellular ROS assay
The levels of cellular ROS were monitored by measuring the fluorescence of 2′, 7’ –dichlorofluorescein (DCF) by spectrophotometry. Experiments were conducted following the manufacturer directions (DCFDA / H2DCFDA - Cellular ROS Assay Kit, ab113851). N2a cells were plated at 1.5 × 104 cells per well on a 96 well plate and cultured in DMEM supplemented medium. After 24 h, cells were transfected using the Lipofectamine LTX Plus reagent with the Flag-TRPV4FeRIC or Flag-TRPV4WT. For TRPV1E637KFeRIC and TRPV2FeRIC, cells were transfected with the original constructs containing the mCherry reporter. 24 h after transfection, cells were stained with 25 μM 2°,7’ –dichlorofluorescin diacetate (DCFDA) for 1-h in the absence or in the presence of RF. For RF groups, RF was applied at 12 mT with the 25-cm RF coil during the entire period of the experiment (1-h). For No RF groups, samples were maintained in the same room, but placed where RF was negligible. DCFDA is a non-fluorescent and cell permeable compound. Once in the cytosol, DCFDA is deacetylated by endogenous esterases to a non-fluorescent compound that can be converted to DCF following its oxidation by ROS. DCF is a highly fluorescent compound which emission (excitation at 495 nm/emission at 529 nm) was detected with a plate reader (EnVision Plate reader 21049). Each experimental point is the average (±SEM) of triplicate readings. All samples were processed simultaneously at room temperature (20 – 22°C). Positive controls were obtained with cells treated with 10 μM H2O2 during the 1-h staining period with DCFDA.
QUANTIFICATION AND STATISTICAL ANALYSIS
Ca2+ imaging analysis
Cytosolic Ca2+ levels were monitored in cells expressing GCaMP6 medium and either FeRIC or wild-type TRP channels. The fluorescence intensity of GCaMP6 was acquired at 1 Hz. For analysis, images were background subtracted and filtered with the “Median” command (Radius = 2 pixels) using the software ImageJ FIJI. Next, files were saved using the NIfTI-1 data format. GCaMP6 fluorescence was computed in a cell-based analysis with a customized MATLAB (Release 2018b, MathWorks Inc., Natick, Massachusetts) code. The maximum intensity projection was performed along the time axis to get the maximum intensity signal of cells expressing GCaMP6 in the field of view. In experiments where cells were stimulated with agonists, the maximum intensity projection corresponds to the cells which expressed functional ion channels. The watershed algorithm (MATLAB implemented function: Watershed transform) (Meyer, 1994) was used to identify and label the cells to generate a cell-based mask for each experiment. The algorithm does not contain the motion correction component because the spatial movement of cells during the time-lapse acquisition was negligible. The GCaMP6 fluorescence intensity was measured for each masked-cell of the time-lapse acquisition (421 s). GCaMP6 fluorescence signal is presented as ΔF/F0, where F0 is the basal fluorescence averaged over 1–61 s before the start of stimulation and DF is the change in fluorescence over basal values. For analysis, the GCaMP6 fluorescence measurements corresponding to the first 5 frames were discarded because of the appearance of an inconsistent artifact. For each masked-cell, the data from 6–61 s were fit with a mono-exponential curve [f(t; a, b) = a*exp(b*t)]. The fitted curve was used to correct the GCaMP6 photobleaching effect over the entire acquisition period. The masked-cells that showed abnormal behavior, observed as the value for the growth factor (b) above 0.002, were excluded from the analysis. Masked-cells were considered responsive when the averaged ΔF/F0 over time during the period from 62 to 290 s (RF-stimulation) increased 10 times over the standard deviation of the GCaMP6 ΔF/F0 of the basal session. For each experimental group, the change of GCaMP6 ΔF/F0 and the corresponding AUC (t = 61 – 300 s) of all analyzed masked-cells, that correspond to all TRPV-expressing cells, were averaged. The plots of the GCaMP6 ΔF/F0 changes correspond to the data obtained from all TRPV-expressing cells, including both responsive and non-responsive cells. The responsiveness was estimated for each independent experiment as the ratio of the responsive masked-cells relative to the total analyzed masked-cells. The GCaMP6 AUC (t = 62 – 300 s) was estimated for all TRPV-expressing cells. Data are presented as the average changes of GCaMP6 ΔF/F0, the average GCaMP6 AUC (±SEM), and the fraction of cells responsive to either RF or lipids/oxidized lipids. For each experimental condition, more than 3 independent experiments were conducted. The MATLAB code that we used for the Ca2+ imaging analysis is available at the Data and code availability section.
Calcein imaging analysis
The calcein fluorescence was monitored in cells expressing either FeRIC or wild-type TRP channels. Calcein fluorescence intensity was acquired at 1 Hz. Images were analyzed using ImageJ FIJI. Regions of interest (ROIs) were placed over those expressing mCherry+. Fluorescence intensity of calcein was measured for each image of the time-lapse acquisition. For analysis, the calcein fluorescence measurements corresponding to the first 30 frames were discarded because of the appearance of an inconsistent artifact. Calcein fluorescence signal is presented as ΔF/F0, where F0 is the basal fluorescence averaged over 30 s before the start of stimulation and DF is the change in fluorescence over basal values. For each ROI, the first 30 s were fit with a mono-exponential curve [f(t; a, b) = a*exp(b*t)]. The fitted curve was used to correct the calcein photobleaching effect over the entire acquisition period. Data correspond to the average of calcein ΔF/F0 (±SEM) obtained from all TRPV-expressing cells. For each experimental condition, more than 3 independent experiments were conducted.
BODIPY® 581/591 C11 imaging analysis
For BODIPY® 581/591 C11 assay cells were transfected with the Flag versions of TRPV1FeRIC, TRPV1WT, TRPV4FeRIC, or TRPV4WT. For TRPV1E637KFeRIC, cells were transfected with the original construct containing the mCherry reporter. The fluorescence intensity of images was analyzed using the software ImageJ FIJI. Images were background subtracted using the ImageJ FIJI plugin “Rolling Ball Background Subtraction’” (50 pixels). Next, images were filtered with the “Median”” command (Radius = 2 pixels). Regions of interest (ROIs) were placed over isolated cells. For each ROI, we measured the intensity fluorescence for the oxidized (490–530 nm) and the reduced (570–610 nM) dye forms. For each ROI, the oxidized:reduced (Ox/Red) ratio was calculated using Microcal OriginPro 2020 software (OriginLab). Data were normalized with respect to the average ratio of Ox/Red of basal acquisition scanned at the beginning of every single experiment. Data correspond to the averages obtained from all imaged cells.
High-resolution liquid chromatography-mass spectrometry (LC-MS) analysis
Data acquisition and analysis were performed using Xcalibur software (version 2.0.7, Thermo). Measured ion mass-to-charge ratio (m/z) values were searched against LipidMAPS and PubChem databases to identify lipids. Three independent experiments were performed for RF and No RF conditions. Lipid ion abundance normalization was relative to the average ion abundance for the control samples.
Ferrozine assay analysis
Fe2+ levels of a solution of ferritin were monitored by absorption spectrophotometry of the Fe2+-ferrozine complex. Absorbance was measured at 550 nm with a plate reader (EnVision Plate reader 21049). Each point is the average (±SEM) of triplicate readings. The averaged absorbance was calculated using Microcal OriginPro 2020 software (OriginLab). Data were normalized with respect to the average absorbance of No RF experimental groups.
DCFDA-DCF assay analysis
Intracellular ROS levels were monitored by the detection of the DCF fluorescence (excitation at 495 nm/emission at 529 nm). DCF fluorescence was detected with a plate reader (EnVision Plate reader 21049). Each experimental point is the average (±SEM) of triplicate readings. The averaged DCF emission was calculated using Microcal OriginPro 2020 software (OriginLab). Data were normalized with respect to the average DCF emission of No RF experimental groups. Data correspond to the averages obtained from all cells.
Statistical analysis
For all experiments, data correspond to the averages (±SEM) obtained from all TRPV-expressing cells. All experiments were repeated a minimum of three times. Differences in continuous datasets were analyzed using Microcal OriginPro 2020 software (OriginLab). The hypothesis testing was performed using a one-way ANOVA, followed by Tukey”s post hoc test. When only two experimental groups were compared, the statistical probe applied was Student”s t test. For statistical comparison of the fraction of cells responsive to RF or lipids, each cell was binarily labeled. Responsive cells were assigned with the value 1 and non-responsive cells were assigned with the value 0. Next, for each experimental group, the binary data from all the independent experiments were grouped and the hypothesis testing was performed using the nonparametric Kruskal-Wallis ANOVA followed by Dunn”s multiple comparisons test. Data are means ± SEM, where applicable *p < 0.05, **p < 0.001, or ***p < 0.0001 was considered a statistically significant difference.
DATA AND CODE AVAILABILITY
The code generated during this study is available at GitHub (https://github.com/LiuCLab/FeRIC/blob/master/FeRIC_Ca_imaging.m). The cDNA sequences of FeRIC channels are available at GenBank (https://www.ncbi.nlm.nih.gov/nuccore/) with the following identifiers: TRPV1FeRIC: MT025941, TRPV4FeRIC: MT025942, TRPV2FeRIC: MT025943, and TRPA1FeRIC: MT025944. The statistical analysis of the fraction of cells responsive to RF and the Ca2+ imaging analysis files are available at Mendeley Data (10.17632/swdz7cjh49.2).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| MAX Efficiency DH5α Competent Cells | Thermo Fisher Scientific | 18258012 |
| One Shot TOP10 Chemically Competent E. coli | Thermo Fisher Scientific | C404006 |
| Chemicals | Peptides | and Recombinant Proteins |
| Opti-MEM I Reduced Serum Medium, no phenol red | Thermo Fisher Scientific | 11058021 |
| Calcein, AM, cell-permeant dye | Thermo Fisher Scientific | C3100MP |
| Live Cell Imaging Solution | Thermo Fisher Scientific | A14291DJ |
| Lipofectamine LTX Reagent with PLUS Reagent | Thermo Fisher Scientific | A12621 |
| Poly-L-lysine solution | Sigma-Aldrich | P4832 |
| Puromycin Dihydrochloride | Thermo Fisher Scientific | A1113802 |
| Penicillin-Streptomycin (10,000 U/mL) | Thermo Fisher Scientific | 15140122 |
| Fetal Bovine Serum, qualified, heat inactivated, United States | Thermo Fisher Scientific | 16140071 |
| DMEM, high glucose, GlutaMAX Supplement | Thermo Fisher Scientific | 10566016 |
| Trypsin-EDTA (0.25%), phenol red | Thermo Fisher Scientific | 25200056 |
| GSK1016790A | Sigma-Aldrich | G0798 |
| Ammonium iron(II) sulfate hexahydrate | Sigma-Aldrich | 203505 |
| Trolox | TOCRIS | 6002 |
| Allyl isothiocyanate | Sigma-Aldrich | 377430 |
| 2-APB | Sigma-Aldrich | 100065 |
| 4-Hydroxynonenal | Sigma-Aldrich | 393204 |
| oxPAPC Oxidized 1-palmitoyl-2-arachidonoyl-snglycero-3-phosphocholine | Avanti polar lipids | 870604 |
| Ferritin from equine spleen | Sigma-Aldrich | F4503 |
| Ferrozine, 3-(2-Pyridyl)-5,6-diphenyl-1,2,4-triazine-p,p′-disulfonic acid monosodium salt hydrate | Sigma-Aldrich | 160601 |
| N,N′-Diphenyl-p-phenylenediamine | Sigma-Aldrich | 292265 |
| N-Acetyl-L-cysteine | Sigma-Aldrich | A7250 |
| Arachidonic acid | TOCRIS | 2756 |
| GSK 2193874 | TOCRIS | 5106 |
| Capsaicin | Sigma-Aldrich | M2028 |
| SB 366791 | TOCRIS | 1615 |
| AM 0902 | TOCRIS | 5914 |
| N-(p-Amylcinnamoyl) anthranilic Acid (ACA) | Santa Cruz Biotechnology | sc-200734 |
| Pyridoxal isonicotinoyl hydrazone (PIH) | abcam | ab145871 |
| N-Ethylmaleimide | Santa Cruz Biotechnology | sc-202719A |
| 5′,6′-Epoxyeicosatrienoic acid | Santa Cruz Biotechnology | sc-221066A |
| EGTA Buffer 0.5M, pH 8.0 | Thermo Fisher Scientific | NC0997810 |
| Critical Commercial Assays | ||
| Ferritin heavy chain CRISPR/Cas9 KO Plasmid (m) | Santa Cruz Biotechnology | Inc. sc-420420 |
| Ferritin heavy chain HDR Plasmid (m) | Santa Cruz Biotechnology | Inc. sc-420420-HDR |
| Image-iT® Lipid Peroxidation Kit | Thermo Fisher Scientific | D3861 |
| DCFDA / H2DCFDA | abcam | ab113851 |
| Deposited Data | ||
| TRPV1FeRIC cDNA sequence | Hutson et al., 2017 | GenBank: MT025941 |
| Statistical analysis of the fraction of cells responsive to RF | This paper | Mendeley, Data: 10.17632/swdz7cjh49.2 |
| Ca2+ imaging analysis data | This paper | Mendeley, Data: 10.17632/swdz7cjh49.2 |
| TRPV2FeRIC cDNA sequence | This paper | GenBank: MT025943 |
| TRPV4FeRIC cDNA sequence | Hutson et al., 2017 | GenBank: MT025942 |
| TRPA1FeRIC cDNA sequence | This paper | GenBank: MT025944 |
| Experimental Models: Cell Lines | ||
| Neuro-2a | ATCC | CCL-131 |
| HEK293T | Clontech | 632180 |
| Oligonucleotides | ||
| Reverse primer mouse Fth1 | This paper | 5′-agtcagcttagctctcatc-3′ |
| Forward primer mouse Fth1 | This paper | 5′-gccaccatgaccaccgcgtc-3′ |
| Forward rat TRPA1-1 | This paper | 5′-atgaagcgcagcttgagga-3′ |
| Forward rat TRPA1-3031 | This paper | 5′-aatagacccaggcacggc-3′ |
| Forward rat TRPV2-1 | This paper | 5′-atgacttcagcctccagc-3′ |
| Forward rat TRPV2-2042 | This paper | 5′-agaaacatcgtgaaggga-3′ |
| Forward human FTH1 | Hutson et al., 2017 | 5′-gccgccatgacgaccgcgt-3′ |
| Reverse human FTH1 | Hutson et al., 2017 | 5′-ccgaggcttagctttcatt-3′ |
| Forward eEF1a promoter | Hutson et al., 2017 | 5′-ccatttcaggtgtcgtga-3′ |
| Recombinant DNA | ||
| Rat TRPV4 (TRPV4WT) | Hutson et al., 2017 | n/a |
| pLVX-Ef1a-TRPV4WT-IRES-mCherry | ||
| Rat TRPV4FeRIC | Hutson et al., 2017 | n/a |
| pLVX-Ef1a-TRPV4FeRIC-IRES-mCherry | ||
| Rat TRPV4ΔTFeRIC | This paper | n/a |
| Y555A/S556A | ||
| pLVX-Ef1a-TRPV4DTFeRIC-IRES-mCherry | ||
| Human FTH1 | Hutson et al., 2017 | n/a |
| PCDNA 3.1-FTH1 | ||
| Human FTH1mCherry | Hutson et al., 2017 | n/a |
| pLVX-FTH1mCherry-N1 | ||
| Rat TRPV2FeRIC | This paper | n/a |
| pLVX-Ef1a-TRPV2FeRIC-IRES-mCherry | ||
| Rat TRPA1FeRIC | This paper | n/a |
| pLVX-Ef1a-TRPA1FeRIC-IRES-mCherry | ||
| Mouse TRPV1 (TRPV1WT) | Hutson et al., 2017 | n/a |
| pLVX-Ef1a-TRPV1WT-IRES-mCherry | ||
| Mouse TRPV1FeRIC | Hutson et al., 2017 | n/a |
| pLVX-Ef1a-TRPV1FeRIC-IRES-mCherry | ||
| Mouse TRPV1DTFeRIC | Hutson et al., 2017 | n/a |
| N628K/N652T/Y653T | ||
| pLVX-Ef1a-TRPV1DTFeRIC-IRES-mCherry | ||
| Cre Vector | Santa Cruz Biotechnology Inc. | sc-418923 |
| GCaMP6m | Addgene | #40754 |
| Rat TRPV4M680AFeRIC | This paper | n/a |
| pLVX-Ef1a-TRPV4M680AFeRIC-IRES-mCherry | ||
| Rat TRPV1E637KFeRIC | This paper | n/a |
| pLVX-Ef1a-TRPV1E637KFeRIC-IRES-mCherry | ||
| Software and Algorithms | ||
| ZEN 2.6 | Zeiss | https://www.zeiss.com/microscopy/us/downloads.html |
| Xcalibur | Thermo Fisher Scientific | https://www.thermofisher.com/order/catalog/product/OPTON-30965 |
| ImageJ | NIH | https://imagej.nih.gov/ij/index.html |
| SERIAL CLONER 2.6 | SerialBasics | http://serialbasics.free.fr/Home/Home.html |
| MATLAB R2018b | MathWorks, USA | https://www.mathworks.com/products/matlab.html |
| OriginPro 2020 | OriginLab, MA | https://www.originlab.com/ |
| BioRender | BioRender | https://biorender.com/ |
| Adobe Illustrator | Adobe | https://www.adobe.com/creativecloud.html |
| Ca2+ data analysis code | This paper | https://github.com/LiuCLab/FeRIC/blob/master/FeRIC_Ca_imaging.m |
Highlights.
Radio-frequency (RF) waves activate ferritin-tagged channels via a biochemical pathway
RF interacts with ferritins, increasing the levels of free iron (labile iron pool)
Free iron produces reactive oxygen species and oxidizes membrane lipids
RF activates ferritin-tagged channels via iron-induced lipid oxidation
ACKNOWLEDGMENTS
We thank Arman Hashemizadeh (University of California, Berkeley) for constructing the RF coils. We thank Kelly Pegram and Eric Benner (Duke University) for providing N2aFth1KO cells. We thank David Julius (University of California [UC] San Francisco) for providing TRPA1 and TRPV2 plasmids. We thank Holly Aaron (CRL Molecular Imaging Center, UC Berkeley) for her assistance in confocal microscopy. We thank Anthony Iavarone (California Institute for Quantitative Biosciences, UC Berkeley) for conducting the LC-MS experiments and for his assistance with the lipid profile analysis. We thank Richard Kramer and Evan Miller (UC Berkeley) for critical review of the manuscript. Research reported in this publication was, in part, supported by the National Institute of Neurological Disorders and Stroke of the National Institutes of Health (NIH) under award number R01NS110554. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
DECLARATION OF INTERESTS
C.L. shares ownership of a patent application (WO2016004281 A1 PCT/ US2015/038948) relating to the use of FeRIC for cell modulation and treatments. All other authors declare that they have no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.02.070.
REFERENCES
- Andersson DA, Gentry C, Moss S, and Bevan S. (2008). Transient receptor potential A1 is a sensory receptor for multiple products of oxidative stress. J. Neurosci 28, 2485–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arosio P, Elia L, and Poli M. (2017). Ferritin, cellular iron storage and regulation. IUBMB Life 69, 414–422. [DOI] [PubMed] [Google Scholar]
- Ayala A, Muñ oz MF, and Argu€elles S. (2014). Lipid peroxidation: production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid. Med. Cell. Longev 2014, 360438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbic M. (2019). Possible magneto-mechanical and magneto-thermal mechanisms of ion channel activation in magnetogenetics. eLife 8, e45807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalá A. (2009). Lipid peroxidation of membrane phospholipids generates hydroxy-alkenals and oxidized phospholipids active in physiological and/or pathological conditions. Chem. Phys. Lipids 157, 1–11. [DOI] [PubMed] [Google Scholar]
- Caterina MJ, Schumacher MA, Tominaga M, Rosen TA, Levine JD, and Julius D. (1997). The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature 389, 816–824. [DOI] [PubMed] [Google Scholar]
- Caterina MJ, Rosen TA, Tominaga M, Brake AJ, and Julius D. (1999). A capsaicin-receptor homologue with a high threshold for noxious heat. Nature 398, 436–441. [DOI] [PubMed] [Google Scholar]
- Céspedes O, and Ueno S. (2009). Effects of radio frequency magnetic fields on iron release from cage proteins. Bioelectromagnetics 30, 336–342. [DOI] [PubMed] [Google Scholar]
- Céspedes O, Inomoto O, Kai S, Nibu Y, Yamaguchi T, Sakamoto N, Akune T, Inoue M, Kiss T, and Ueno S. (2010). Radio frequency magnetic field effects on molecular dynamics and iron uptake in cage proteins. Bio-electromagnetics 31, 311–317. [DOI] [PubMed] [Google Scholar]
- Chen R, Romero G, Christiansen MG, Mohr A, and Anikeeva P. (2015). Wireless magnetothermal deep brain stimulation. Science 347, 1477–1480. [DOI] [PubMed] [Google Scholar]
- Ciardo MG, and Ferrer-Montiel A. (2017). Lipids as central modulators of sensory TRP channels. Biochim Biophys Acta Biomembr 1859 (9 Pt B), 1615–1628. [DOI] [PubMed] [Google Scholar]
- Drummen GPC, van Liebergen LCM, Op den Kamp JAF, and Post JA (2002). C11-BODIPY(581/591), an oxidation-sensitive fluorescent lipid peroxidation probe: (micro)spectroscopic characterization and validation of methodology. Free Radic. Biol. Med 33, 473–490. [DOI] [PubMed] [Google Scholar]
- Duret G, Polali S, Anderson ED, Bell AM, Tzouanas CN, Avants BW, and Robinson JT (2019). Magnetic entropy as a proposed gating mechanism for magnetogenetic ion channels. Biophys. J 116, 454–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch J, Lees M, and Sloane Stanley GH (1957). A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem 226, 497–509. [PubMed] [Google Scholar]
- García-Martínez C, Morenilla-Palao C, Planells-Cases R, Merino JM, and Ferrer-Montiel A. (2000). Identification of an aspartic residue in the P-loop of the vanilloid receptor that modulates pore properties. J. Biol. Chem 275, 32552–32558. [DOI] [PubMed] [Google Scholar]
- Graf E, Mahoney JR, Bryant RG, and Eaton JW (1984). Iron-catalyzed hydroxyl radical formation. Stringent requirement for free iron coordination site. J. Biol. Chem 259, 3620–3624. [PubMed] [Google Scholar]
- Hore PJ, and Mouritsen H. (2016). The radical-pair mechanism of magneto-reception. Annu. Rev. Biophys 45, 299–344. [DOI] [PubMed] [Google Scholar]
- Huang H, Delikanli S, Zeng H, Ferkey DM, and Pralle A. (2010). Remote control of ion channels and neurons through magnetic-field heating of nano-particles. Nat. Nanotechnol 5, 602–606. [DOI] [PubMed] [Google Scholar]
- Hutson MR, Keyte AL, Hernández-Morales M, Gibbs E, Kupchinsky ZA, Argyridis I, Erwin KN, Pegram K, Kneifel M, Rosenberg PB, et al. (2017). Temperature-activated ion channels in neural crest cells confer maternal fever–associated birth defects. Sci. Signal 10, eaal4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen S, and Lohmann KJ (2005). The physics and neurobiology of magnetoreception. Nat. Rev. Neurosci 6, 703–712. [DOI] [PubMed] [Google Scholar]
- Johnsen S, and Lohmann KJ (2008). Magnetoreception in animals. Phys. Today 61, 29–35. [Google Scholar]
- Katanosaka K, Takatsu S, Mizumura K, Naruse K, and Katanosaka Y. (2018). TRPV2 is required for mechanical nociception and the stretch-evoked response of primary sensory neurons. Sci. Rep 8, 16782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kole K, Zhang Y, Jansen EJR, Brouns T, Bijlsma A, Calcini N, Yan X, Lantyer ADS, and Celikel T. (2019). Assessing the utility of Magneto to control neuronal excitability in the somatosensory cortex. Nat. Neurosci. Published online September 30, 2019 10.1038/s41593-019-0474-4. [DOI] [PubMed] [Google Scholar]
- Krishnan V, Park SA, Shin SS, Alon L, Tressler CM, Stokes W, Banerjee J, Sorrell ME, Tian Y, Fridman GY, et al. (2018). Wireless control of cellular function by activation of a novel protein responsive to electromagnetic fields. Sci. Rep 8, 8764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liedtke W, Choe Y, Martí-Renom MA, Bell AM, Denis CS, Sali A, Hudspeth AJ, Friedman JM, and Heller S. (2000). Vanilloid receptor-related osmotically activated channel (VR-OAC), a candidate vertebrate osmoreceptor. Cell 103, 525–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister M. (2016). Physical limits to magnetogenetics. eLife 5, e17210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F. (1994). Topographic distance and watershed lines. Signal Processing 38, 113–125. [Google Scholar]
- Mosabbir AA, and Truong K. (2018). Genetically encoded circuit for remote regulation of cell migration by magnetic fields. ACS Synth. Biol 7, 718–726. [DOI] [PubMed] [Google Scholar]
- Munshi R, Qadri SM, Zhang Q, Castellanos Rubio I, Del Pino P, and Pralle A. (2017). Magnetothermal genetic deep brain stimulation of motor behaviors in awake, freely moving mice. eLife 6, e27069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraki K, Iwata Y, Katanosaka Y, Ito T, Ohya S, Shigekawa M, and Imaizumi Y. (2003). TRPV2 is a component of osmotically sensitive cation channels in murine aortic myocytes. Circ. Res 93, 829–838. [DOI] [PubMed] [Google Scholar]
- Peterson DA, Gerrard JM, Rao GH, Krick TP, and White JG (1978). Ferrous iron mediated oxidation of arachidonic acid: studies employing nitroblue tetrazolium (NBT). Prostaglandins Med. 1, 304–317. [DOI] [PubMed] [Google Scholar]
- Schafer FQ, Qian SY, and Buettner GR (2000). Iron and free radical oxidations in cell membranes. Cell. Mol. Biol 46, 657–662. [PMC free article] [PubMed] [Google Scholar]
- Stanley SA, Gagner JE, Damanpour S, Yoshida M, Dordick JS, and Friedman JM (2012). Radio-wave heating of iron oxide nanoparticles can regulate plasma glucose in mice. Science 336, 604–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley SA, Sauer J, Kane RS, Dordick JS, and Friedman JM (2015). Remote regulation of glucose homeostasis in mice using genetically encoded nanoparticles. Nat. Med 21, 92–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley SA, Kelly L, Latcha KN, Schmidt SF, Yu X, Nectow AR, Sauer J, Dyke JP, Dordick JS, and Friedman JM (2016). Bidirectional electromagnetic control of the hypothalamus regulates feeding and metabolism. Nature 531, 647–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Story GM, Peier AM, Reeve AJ, Eid SR, Mosbacher J, Hricik TR,Earley TJ, Hergarden AC, Andersson DA, Hwang SW, et al. (2003). ANKTM1, a TRP-like channel expressed in nociceptive neurons, is activated by cold temperatures. Cell 112, 819–829. [DOI] [PubMed] [Google Scholar]
- Strotmann R, Harteneck C, Nunnenmacher K, Schultz G, and Plant TD (2000). OTRPC4, a nonselective cation channel that confers sensitivity to extracellular osmolarity. Nat. Cell Biol 2, 695–702. [DOI] [PubMed] [Google Scholar]
- Taberner FJ, Fernández-Ballester G, Fernández-Carvajal A, and Ferrer-Montiel A. (2015). TRP channels interaction with lipids and its implications in disease. Biochim. Biophys. Acta 1848, 1818–1827. [DOI] [PubMed] [Google Scholar]
- Voets T, Prenen J, Vriens J, Watanabe H, Janssens A, Wissenbach U, Bödding M, Droogmans G, and Nilius B. (2002). Molecular determinants of permeation through the cation channel TRPV4. J. Biol. Chem 277, 33704–33710. [DOI] [PubMed] [Google Scholar]
- Wang G, Zhang P, Mendu SK, Wang Y, Zhang Y, Kang X, Desai BN, and Zhu JJ (2019). Revaluation of magnetic properties of Magneto. Nat. Neurosci Published online September 30, 2019 10.1038/s41593-019-0473-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler MA, Smith CJ, Ottolini M, Barker BS, Purohit AM, Grippo RM, Gaykema RP, Spano AJ, Beenhakker MP, Kucenas S, et al. (2016). Genetically targeted magnetic control of the nervous system. Nat. Neurosci 19, 756–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler MA, Deppmann CD, Patel MK, and Güler AD (2019). Reply to: Magneto is ineffective in controlling electrical properties of cerebellar Purkinje cells, assessing the utility of Magneto to control neuronal excitability in the somatosensory cortex and revaluation of magnetic properties of Magneto. Nat. Neurosci. Published online September 30, 2019 10.1038/s41593-019-0472-6. [DOI] [PubMed] [Google Scholar]
- Xu F-X, Zhou L, Wang X-T, Jia F, Ma K-Y, Wang N, Lin L, Xu F-Q, and Shen Y. (2019). Magneto is ineffective in controlling electrical properties of cerebellar Purkinje cells. Nat. Neurosci. Published online September 30, 2019 10.1038/s41593-019-0475-3. [DOI] [PubMed] [Google Scholar]
- Young JH, Wang M-T, and Brezovich IA (1980). Frequency/depth-penetration considerations in hyperthermia by magnetically induced currents. Electron. Lett 16, 358–359. [Google Scholar]
- Zygmunt PM, and Högestätt ED (2014). TRPA1. Handb. Exp. Pharmacol 222, 583–630. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The code generated during this study is available at GitHub (https://github.com/LiuCLab/FeRIC/blob/master/FeRIC_Ca_imaging.m). The cDNA sequences of FeRIC channels are available at GenBank (https://www.ncbi.nlm.nih.gov/nuccore/) with the following identifiers: TRPV1FeRIC: MT025941, TRPV4FeRIC: MT025942, TRPV2FeRIC: MT025943, and TRPA1FeRIC: MT025944. The statistical analysis of the fraction of cells responsive to RF and the Ca2+ imaging analysis files are available at Mendeley Data (10.17632/swdz7cjh49.2).