Abstract
Rapid and reliable screening of SARS-CoV-2 is fundamental to assess viral spread and limit the pandemic we are facing. In this study, we compared direct rRT-PCR method (without RNA extraction) using SeeGene AllplexTM 2019-nCoV rRT-PCR with the RealStar® SARS-CoV-2 rRT-PCR kit (Altona Diagnostics). Furthermore, we assessed the impact of swab storage media composition on PCR efficiency. We show that SeeGene and Altona’s assays provide similar efficiency. Importantly, we provide evidence that RNA extraction can be successfully bypassed when samples are stored in UTM medium or in molecular water but not when samples are stored in saline solution and in Hanks medium.
Keywords: COVID19, SARS-CoV-2, Coronavirus, Virus detection, Direct rRTPCR, RNA extraction
1. Background
Viral detection is key to isolate positive patients and stop viral transmission [[1], [2], [3], [4], [5], [6]] in our battle against the 2020 SARS-CoV-2 pandemic. However, there are many challenges associated with ramping up testing capacity, including shortage in the chain of supplies for extraction reagents [7]. This situation called for alternative protocols to ensure the continuity of testing in laboratories.
The standard procedure for testing SARS-CoV-2 from oro-nasopharyngeal swabs in the clinical laboratory of the Centre Intégré Universitaire de Santé et Services Sociaux de la Mauricie et Centre du Québec (CIUSSS-MCQ) is based on RNA extraction followed by viral genes detection with a multiplex assay using the Altona RealStar® SARS-CoV-2 rRT-PCR Kit RUO (thereafter Altona). Due to an increased demand in SARS-CoV-2 screenings, alternative protocols with similar sensitivity were needed [[8], [9], [10], [11]]. Thus, we compared the sensitivity of detection from Health Canada approved SeeGene Allplex™ 2019-nCoV rRT-QPCR Assay (thereafter SeeGene) and Altona, with and without RNA extraction. Samples originated from remaining swabs media and RNA extracts from symptomatic individuals that had been previously screened using the CIUSS-MCQ routine method. Hence, samples were declared negative or positive before these experiments.
2. Methods
2.1. Specimens
Swabs from the oro- and naso-pharyngeal area from symptomatic individuals were placed in 2 ml of UTM (Remel RE12569), or Hanks medium (0,14 M NaCl; 5 mM KCl, 0,4 mM MgSO4, 0,3 mM Na2HPO4, 0,4 mM KH2PO4, 1 mM CaCl2 and 6 mM glucose from Fisher; and Phenol Red 1% from Sigma), molecular -grade water (Fisher bioreagents), or saline (BioMérieux). Specimens were inactivated at 95 °C for 5 min and stored at 4 °C.
2.2. Standard operating procedure using RNA extraction and Altona
RNA was extracted using Abbott mSample Preparation Systems DNA kit on m2000sp instrument (Abbott). rRT-PCR protocol was automated: prepared on the Abbott m2000sp and detected on the m2000rt using the Altona kit (Altona diagnostics). Ct values from FAM (E gene), Cy5 (S gene) and Joe/HEX (internal control) were acquired.
2.3. Alternative rRT-PCR using SeeGene
SeeGene was performed as per manufacturer’s instruction. Ct from FAM (E gene), Cal Red 610 (RDRP gene), Quasar 670 (N gene) and HEX (internal control) were acquired.
Samples were considered positive when a signal was detected at Ct<40 for any gene. A sample was considered negative if the internal control was amplified but not the viral genes. A specimen was considered invalid when there was no amplification of the internal control.
3. Results and discussion
3.1. Detection of SARS-CoV-2 with SeeGene compared to standard procedure
RNA stored at −80 °C extracted from 65 SARS-CoV-2-positive and 23 -negative specimens were amplified using SeeGene’s kit. 23 negative samples were confirmed, 64 out of 65 positive samples were confirmed positive (96,92 %), one sample detected at Ct = 35.6 (gene S only) with Altona’s kit was missed using SeeGene’s kit (Fig. 1 ).
Fig. 1.
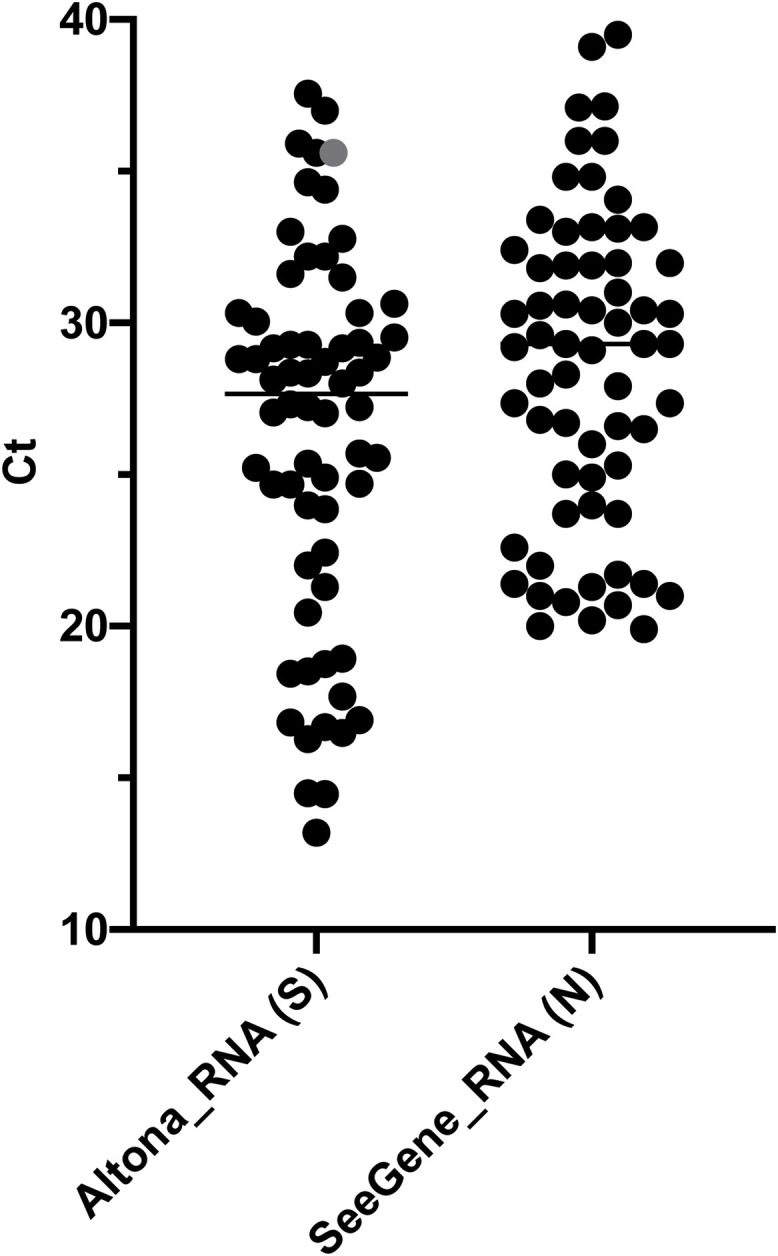
Detection of SARS-CoV-2 with SeeGene compared to standard procedure.
Data are presented as scattered dot plots with means. Each dot represents 1 Ct value of 65 reactions, each corresponding to a specimen. Purified RNA from 65 SARS-CoV2+ specimens as validated by standard procedure (Altona, S) were amplified using SeeGene (SeeGene_RNA). The most sensitive targets, i.e. positive in the highest number of specimens, are shown (S for Altona and N for SeeGene). The grey dot represents one sample detected at Ct = 35.6 during standard procedure and missed using SeeGene.
3.2. rRT-PCR directly from swabs: without RNA extraction
The swab storage media for respiratory virus detection can have important impact on the rRT-PCR efficiency, when this media is used directly for PCR reaction. To assess the feasibility of direct rRT-PCR using different media we performed the following experiments.
3.2.1. Detection of SARS-CoV-2 from swabs stored in Hanks medium
95 positive specimens stored in Hanks medium were randomly chosen for direct rRT-PCR in two separate experiments. 21 (22,3%) reactions did not show amplification neither of viral genes nor of the internal control suggesting rRT-PCR inhibition and 69,15% (n = 65) were positive following analysis (Fig. 2 ). In comparison, 30 samples in which the RNA was extracted of 30 tested were positive. Hence, swabs in Hanks are not appropriate samples for direct rRT-PCR.
Fig. 2.
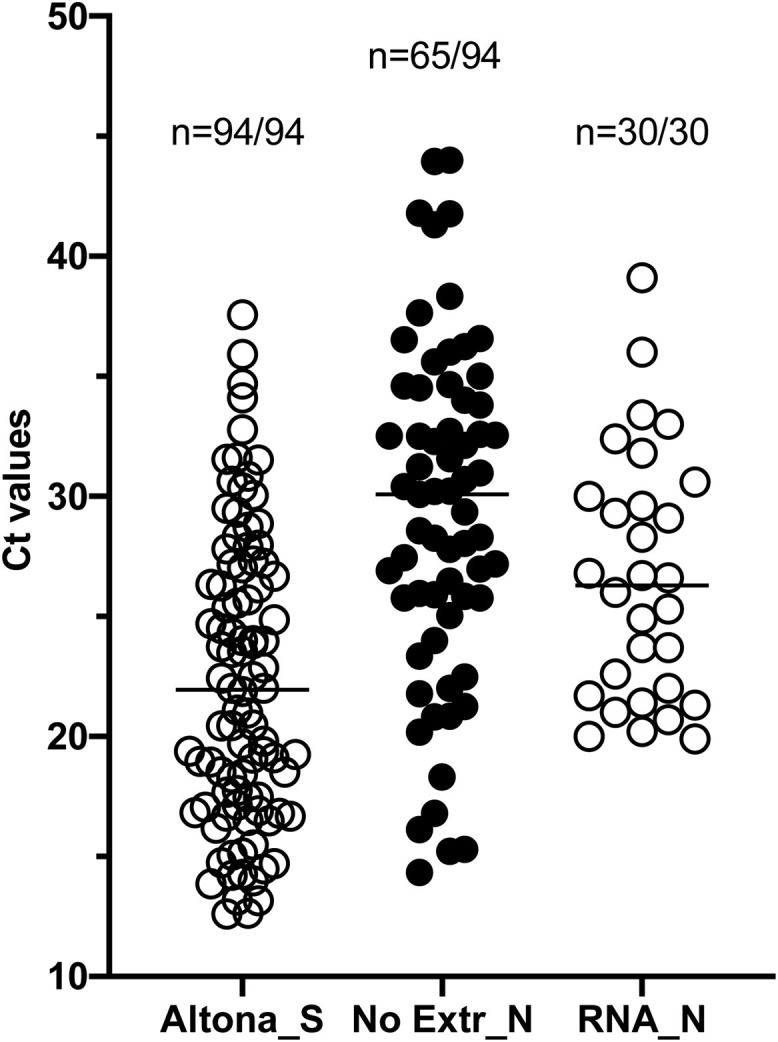
Detection of SARS-CoV-2 from swabs stored in Hanks medium. Data are depicted as scattered dot plots with means. Each dot represents 1 Ct value corresponding to 1 specimen. 94 SARS-CoV2+ specimens as validated by standard procedure (Altona, S, n = 94) stored in Hanks medium at −80 °C were used for direct rRT-PCR using SeeGene (No Extr_N, n = 65/94). RNA isolated from these specimens and stored at −80 °C were separately used as template for SeeGene rRT-PCR (RNA_N, n = 30/30). The most sensitive target is shown. 29 positive specimens were not detected by direct rRT-PCR (30,85%).
3.2.2. Detection of SARS-CoV-2 using direct rRT-PCR from swabs stored in UTM medium
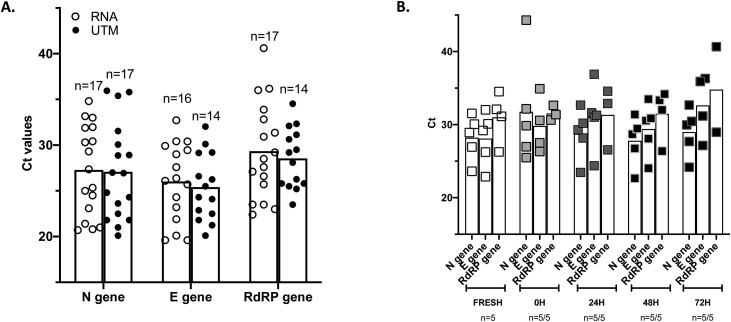
During this study, some specimens were collected in UTM medium during routine screening. Seventeen of these swabs tested positive following standard procedure stored at −80 °C, were used for direct rRT-PCR in two independent experiments. 100 % were detected using SeeGene and Ct means were equivalent whether or not RNA was extracted before the rRT-PCR (Fig. 3 A). Next, we measured the stability of the detection in 5 additional samples by amplifying from fresh (stored at 4 °C), frozen at −80 °C overnight, thawed and stored at 4 °C for 3 days (T0, T24, T48 and T72) (Fig. 3B). Ct values remained constant across the experiment and all samples could be detected at all timepoints. Our results suggest that UTM medium can be used for direct rRT-PCR and that storage at 4 °C for a few days before testing does not lead to a significant loss of detection.
Fig. 3.
Detection of SARS-CoV-2 using direct rRT-PCR from swabs stored in UTM medium (without RNA extraction). Results are plotted as bar with mean graphs and scattered dot plots. Each dot represents one reaction corresponding to one specimen. A. Seventeen SARS-CoV-2+ swabs collected and stored in UTM at −80 °C were randomly chosen and directly amplified (UTM) or amplified from RNA eluates (RNA). ‘n’ stands for the number of SARS-CoV-2+ specimens. Results for all three targets amplified by SeeGene multiplex assay are shown. The N gene is successfully detected in all the samples. B. Five positive specimens stored in UTM at −80 °C were randomly chosen, thawed and kept at 4 °C for 3 days. Levels of SARS-CoV-2 RNA was assessed at each time point. All 5 samples were detected by amplification of the N gene at all timepoint.
3.2.3. Detection of SARS-CoV-2 from saline or molecular water
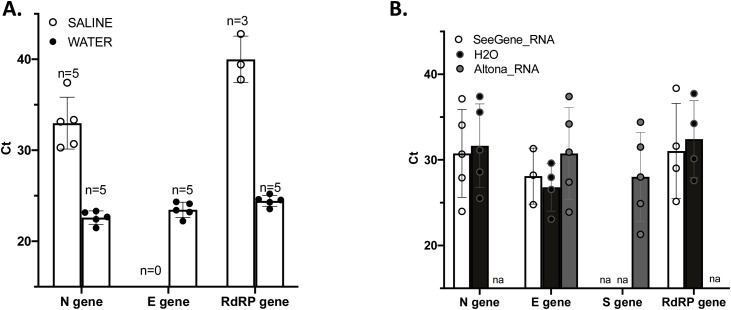
Then, we assessed easily available and cheap storage media: saline and molecular grade water. First, we used extracted and purified RNA from five positive specimens (Ct between 12,61 and 14,22) that we diluted using saline or water and used for rRT-PCR. All five samples were detected by the N target when diluted in saline or in water. However, when samples were stored in saline, Ct for the N gene were above 30 compared to a mean of 22 when RNA was diluted in water following SeeGene. The E gene could not be amplified from saline dilutions (Fig. 4 a). These results suggest that saline is not compatible with direct rRT-PCR. We assessed the sensitivity in water and diluted a positive swab by performing five serial dilutions (each 1:10) from this swab. These dilutions were also used for RNA extraction and amplification using the standard procedure. The undiluted sample and 4 of 5 dilutions showed detectable levels of using N gene and S gene, with both Altona and SeeGene. Ct values of the E gene and of the N gene were similar whether RNA was extracted or not, and whether SeeGene or Altona was used (Fig. 4b).
Fig. 4.
Detection of SARS-CoV-2 in saline or molecular water samples. Results are plotted as bar graphs with standard deviation. A. Each dot represents one reaction corresponding to one specimen. One hundredth dilution of RNA purified from 5 positive specimens with high Ct values were performed in molecular water and in saline water. All 5 diluted samples were detected, however, samples diluted in saline water required 10 more Ct before being detected. B. Detection of SARS-CoV-2 by direct rRT-PCR from swabs stored in molecular water. Each dot represents one reaction corresponding to one dilution of the same specimen that contained RNAse inhibitor. The specimen collected in water was diluted 5 times following a serial dilution of 1:10 and used for RNA extraction and amplification using the routine procedure in comparison to SeeGene. Ct values from amplification of extracted RNA from a swab stored in water by standard procedure (Altona_RNA, S and E genes) and using SeeGene’s kit (SeeGene_RNA; N, RdRP and E genes), or amplified directly from swab and dilutions in water medium (H20; N and E gene) using SeeGene’s kit are plotted. ‘na’ means ‘not applicable’.
3.2.4. Detection of SARS-CoV-2 using specimens collected in molecular water and stored at −80C
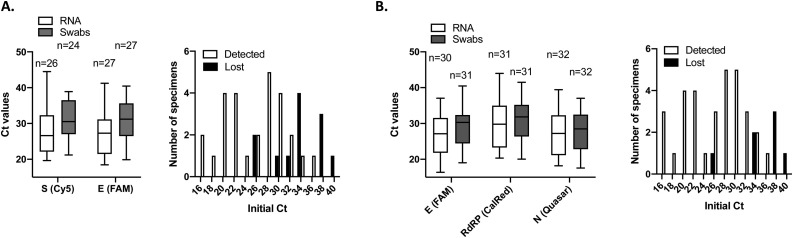
Thirty-nine swabs collected in water and stored at −80 °C for one week, with characterized Ct values (ranging between 16,3 and 39,7) were used as a template for direct rRT-PCR and for RNA extraction and amplification, using Altona (Fig. 5 A) and SeeGene (Fig. 5B). Twenty-seven of them could be amplified post-thawing when RNA or swabs were used with Altona’s multiplex assay. Thirty-two could be detected from RNA eluates or directly from swabs with SeeGene’s multiplex assay (Fig. 5b). The initial Ct mean (before freezing) of samples that were lost following thawing was significantly higher than the Ct mean of samples that could be amplified (25,7 vs 33,7; p < 0.0001; unpaired t-test) using Altona and SeeGene (25,97 vs 35,68; p < 0.0001; unpaired t-test). This suggests that storing specimens in water at −80°C can lead to a significant loss in sensitivity for low viral load samples. Direct rRT-PCR was as efficient as RNA extraction followed by rRT-PCR to detect positive samples using both methods. In addition, Ct values of the internal control was significantly higher using direct rRT-PCR compared to rRT-PCR from purified RNA with Altona (p = 0.0002, paired t-test), suggesting PCR inhibition when RNA is not purified which was not observed with SeeGene. From this experiment, we conclude that RNA extraction can be successfully skipped when swabs are collected in molecular water. However, storing samples at −80 °C collected in water before screening could impact the sensitivity and a decreased detection of patients with lower viral loads.
Fig. 5.
Detection of SARS-CoV-2 testing using specimens collected in molecular water stored at −80C. Results are plotted as box and violin graph with range and histograms of distribution. A. 39 samples with a declared positive status collected and stored in water at −80 °C were used for RNA extraction and amplification, or for direct amplification using Altona’s kit. Using RNA or directly the wab as a template, 27 were detected post-thawing. Internal control was amplified efficiently from both types of template, but Ct values were significantly higher when swabs were used as direct templates. 12 samples could not be detected post-thawing. Their Ct mean before freezing was significantly higher than the Ct mean of samples that could be amplified (25,69 vs 33,7; p < 0.0001; unpaired t-test). B. The same 39 positive swabs were used for RNA extraction and amplification or direct amplification using SeeGene’s kit. Using RNA or swabs as templates, 32 were detected post-thawing. Internal controls were successfully amplified using both RNA and swabs as templates. 7 samples were lost upon thawing and the Ct mean of these samples before freezing was significantly higher compared to the initial Ct mean of samples that were successfully (25,97 vs 35,68; p < 0.0001; unpaired t-test).
4. Conclusion
Our results suggest that: i) SeeGene and Altona kits provide similar efficiency, ii) direct rRT-PCR without RNA extraction is possible if samples are collected in UTM or molecular water; ii), specimens stored in water should be screened rapidly, iii) RNA extraction is necessary if samples are in saline water or Hanks medium.
Declaration of Competing Interest
None.
Acknowledgments
The authors would like to thank Dr Marco Bergevin for kindly providing a calibrated CFX96 Thermal cycler machine waiting for ours to be calibrated and the SeeGene committee from the Province of Quebec for insightful discussions.
Contributor Information
Natacha Merindol, Email: Natacha.Merindol@uqtr.ca.
Geneviève Pépin, Email: Genevieve.Pepin3@uqtr.ca.
Caroline Marchand, Email: caroline_marchand_labo@ssss.gouv.qc.ca.
Marylène Rheault, Email: marylene_rheault@ssss.gouv.qc.ca.
Christine Peterson, Email: christine_peterson@ssss.gouv.qc.ca.
André Poirier, Email: andre_poirier_chrtr@ssss.gouv.qc.ca.
Claudia Houle, Email: claudia_houle_chrtr@ssss.gouv.qc.ca.
Hugo Germain, Email: Hugo.Germain@uqtr.ca.
Alexis Danylo, Email: Alexis_Danylo@ssss.gouv.qc.ca.
References
- 1.Coronaviridae Study Group of the International Committee on Taxonomy of V The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5(4):536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tang Y.W., Schmitz J.E., Persing D.H., Stratton C.W. The laboratory diagnosis of COVID-19 infection: current issues and challenges. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.00512-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323(11) doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin Y.H., Cai L., Cheng Z.S., Cheng H., Deng T., Fan Y.P. A rapid advice guideline for the diagnosis and treatment of 2019 novel coronavirus (2019-nCoV) infected pneumonia (standard version) Mil. Med. Res. 2020;7(1):4. doi: 10.1186/s40779-020-0233-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan J.F., Yip C.C., To K.K., Tang T.H., Wong S.C., Leung K.H. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-polymerase chain reaction assay validated in vitro and with clinical specimens. J. Clin. Microbiol. 2020;58(5) doi: 10.1128/JCM.00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Babiker A., Myers C.W., Hill C.E., Guarner J. SARS-CoV-2 testing. Am. J. Clin. Pathol. 2020;153(6) doi: 10.1093/ajcp/aqaa052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rhoads D.D., Cherian S.S., Roman K., Stempak L.M., Schmotzer C.L., Sadri N. Comparison of Abbott ID Now, Diasorin Simplexa, and CDC FDA EUA methods for the detection of SARS-CoV-2 from nasopharyngeal and nasal swabs from individuals diagnosed with COVID-19. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.00760-20. pii: JCM.00760-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X., Yao H., Xu X., Zhang P., Zhang M., Shao J. Limits of detection of six approved RT-PCR kits for the novel SARS-coronavirus-2 (SARS-CoV-2) Clin. Chem. 2020 doi: 10.1093/clinchem/hvaa099. pii: hvaa099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moran A., Beavis K.G., Matushek S.M., Ciaglia C., Francois N., Tesic V. The detection of SARS-CoV-2 using the Cepheid Xpert Xpress SARS-CoV-2 and Roche cobas SARS-CoV-2 assays. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.00821-20. pii: JCM.00772-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waggoner J.J., Stittleburg V., Pond R., Saklawi Y., Sahoo M.K., Babiker A. Triplex real-time RT-PCR for severe acute respiratory syndrome coronavirus 2. Emerg. Infect. Dis. 2020;26(7) doi: 10.3201/eid2607.201285. [DOI] [PMC free article] [PubMed] [Google Scholar]