Highlights
-
•
The TP1 and TP2 peptides, derived from the TMUV E protein, inhibited TMUV infection.
-
•
TP2 exhibited cross-inhibitory activity against JEV.
-
•
The TP1 and TP2 peptides blocked TMUV antibody-dependent enhancement (ADE) in duck peripheral blood lymphocytes.
-
•
Both peptides interacted with the surface of TMUV, leading to release of viral RNA and interfering with virus:cell binding.
Keywords: Tembusu virus, Envelope protein, Inhibitory peptides, Antiviral
Abstract
The outbreak and spread of Tembusu virus (TMUV) has caused very large losses in the waterfowl-breeding industry since 2010. The viral envelope (E) protein, the principal surface protein of viral particles, plays a vital role in viral entry and fusion. In this study, two peptides derived from domain II (DII) and the stem of the TMUV envelope protein, TP1 and TP2, respectively, were tested for their antiviral activity. TP1 and TP2 inhibited TMUV infection in BHK-21 cells, and their 50% inhibitory concentrations (IC50) were 14.19 mg/L and 7.64 mg/L, respectively. Viral inhibition assays in different cell lines of avian origin showed that the inhibitory effects of TP1 and TP2 are not cell type dependent. Moreover, TP2 also exhibited inhibitory activity against Japanese encephalitis virus (JEV) infection. The two peptides inhibited antibody-mediated TMUV infection of duck peripheral blood lymphocytes. Co-immunoprecipitation assays and indirect enzyme-linked immunosorbent assays (ELISAs) indicated that both peptides interact with the surface of the TMUV virion. RNase digestion assays confirmed the release of viral RNA following incubation with TP1, while incubation with TP1 or TP2 interfered with the binding between TMUV and cells. Taken together, these results show that TP1 and TP2 may be developed into antiviral treatments against TMUV infection.
1. Introduction
Tembusu virus (TMUV), a member of the Flaviviridae, is a newly emerging pathogen of avian origin that causes paralysis and ovarian haemorrhage in mainly ducks and geese. TMUV-infected ducks and geese exhibit a rapid drop in egg production and daily food intake. The sudden outbreak and fast spread of TMUV have caused heavy economic losses in the major waterfowl-breeding provinces of China since 2010. Concerningly, the range of TMUV hosts is expanding. In addition to ducks and geese, chickens, pigeons and house sparrows can be infected by TMUV. Therefore, TMUV has a severe impact on development of the domestic poultry industry (Chen et al., 2016, Chen et al., 2017b; Zhang et al., 2017).
The whole TMUV genome encodes three structural proteins and seven nonstructural proteins. Among these viral proteins, the envelope (E) protein is located on the surface of mature virions and forms head-to-tail homodimers (Kuhn et al., 2002). The E protein consists of an ectodomain (containing three domains: DI, DII and DIII) and a stem-transmembrane domain that anchors the protein to the membrane. The first step in the process by which TMUV enters host cells is binding of the E protein to cellular receptors on the cell surface (Rodenhuis-Zybert et al., 2010). To date, several cellular receptors have been identified; these include glucose-regulated protein 78 (GRP78) on BHK-21 cells, heat shock protein A9 on DF-1 cells and heparin sulfate on both BHK-21 and duck embryo fibroblasts (DEFs) (Zhao et al., 2018b; Liu et al., 2017; Wu et al., 2019). These receptors play an important role in the first step of TMUV entry and viral attachment. Subsequently, attached TMUV enters cells by the clathrin-mediated endocytosis pathway, after which endocytic vesicles carrying the virus are delivered to endosomes (Smit et al., 2011). Under the low-pH conditions of the endosome, the E homodimer dissociates into monomeric proteins, exposing the fusion loop in DII. Subsequently, the fusion loop inserts into the target membrane, and three copies of the E protein interact to form the E protein trimer. DIII then folds back against the trimer, directing the E stem anchor towards the fusion loop. The energy released by these conformational changes induces fusion between the viral membrane and host cell membrane. Finally, a fusion pore is formed, and delivery of the nucleocapsid into the cytosol initiates replication (Pierson and Kielian, 2013).
Previous studies have shown that artificially synthesized small peptides based on a protein domain can bind intact proteins and inhibit their biological activity through interfering with proper protein folding. Envelope protein-derived peptides were reported to bind the envelope protein and inhibit viral infection (Hall and Frieden, 1989). Jiang et al. (1993) reported that a synthetic peptide corresponding to the envelope protein of human immunodeficiency virus type I (HIV-I) could block virion infectivity. Subsequently, other synthetic inhibitory peptides of the fusion proteins of paramyxoviruses, herpesviruses, orthomyxoviruses, retroviruses and coronaviruses have also been identified (Porotto et al., 2007; Okazaki and Kida, 2004; Markosyan et al., 2004; Yao and Compans, 1996; Bosch et al., 2003).
To date, several peptides corresponding to different domains of the flavivirus E protein have been shown to inhibit flavivirus infection. DN59, an inhibitory peptide corresponding to the stem region of dengue virus (DENV) E protein, almost completely inhibited DENV infection in LLC-MK2 cells and exhibited cross-inhibitory activity against West Nile virus (WNV) infection. The WN83 inhibitor, a direct mimic of DII of the WNV E protein, reproducibly inhibited WNV infectivity at a low-μM concentration (Hrobowski et al., 2005). The inhibitory peptide Z2, which was synthesized based on the stem region of the Zika virus (ZIKV) E protein, could penetrate the placental barrier of pregnant ICR mice and inhibit vertical transmission of ZIKV in pregnant C57BL/6 mice (Yu et al., 2017). The peptide P5, derived from the stem region of the Japanese encephalitis virus (JEV) E protein, could block JEV and ZIKV infection in vitro. P5 also protected mice from JEV-induced lethality and reduced ZIKV-induced histopathological damage by decreasing viral load (Chen et al., 2017a). Moreover, DN59 and 1OAN1 could inhibit antibody-dependent enhancement (ADE)-mediated DENV infection in vitro, suggesting peptide inhibitors as a strategy to inhibit flavivirus ADE (Nicholson et al., 2011).
Although multiple peptide inhibitors corresponding to the E proteins of other flaviviruses have been studied, inhibitory peptides against TMUV have not been reported. In this study, the peptides TP1 and TP2 derived from DII and the stem region of the TMUV E protein, respectively, were synthesized, and their ability to prevent TMUV infection was investigated. The data demonstrated that TP1 inhibited TMUV infection through destroying the integrity of the viral particles, while both TP1 and TP2 exerted inhibitory effects by interfering with the binding of TMUV to cells. Additional results suggested that TP2 cross-inhibited JEV infection. These findings provide new insight into the development of drugs against TMUV.
2. Materials and methods
2.1. Cells and viruses
TMUV strain JS804 (GenBank accession No. JF895923) was isolated by our laboratory (Huang et al., 2013). JEV strain SA14-14-2 (GenBank accession No. AF315119) was obtained from Dr. Xiuli Feng at Nanjing Agricultural University. TMUV strain JS804 and JEV strain SA14-14-2 were propagated in baby hamster kidney cells (BHK-21 cells) grown in RPMI-1640 medium (HyClone, South Logan, USA) containing 10% foetal calf serum (FCS, HyClone, South Logan, USA). Viral titres in BHK-21 cells were determined by plaque assay as previously described (Zhao et al., 2015). Specific-pathogen-free (SPF) ducks were purchased from the Harbin Veterinary Research Institute in China. This study was approved by the Animal Care and Use Committee of Jiangsu Province. Monoclonal antibody against the E protein of TMUV strain JS804 was produced by our laboratory (Niu et al., 2013). Anti-β-actin monoclonal antibody was obtained from Cowin Biotechnology Company (Cowin, Beijing, China). HRP-labelled goat anti-mouse IgG antibody was purchased from Zsbio Biotechnology Company (Beijing, China).
2.2. Peptide synthesis
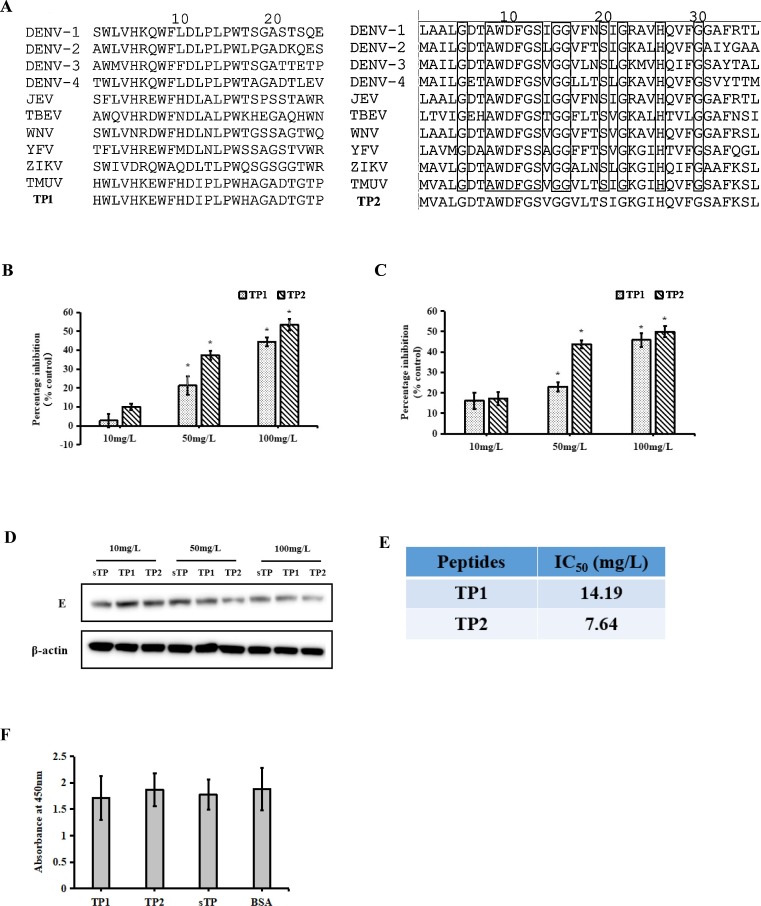
The peptides TP1 (SWLVNRDWFHDLNLPWTGSSAGTWQ) and TP2 (MVALGDTAWDFGSVGGVLTSIGKGIHQVFGSAFKSL), the negative peptide sTP (MTPLTRWHSSGEEHHLVTKRMEVATWLVGAAPGQGEWIRSI, Panya et al., 2015), TP1-biotin, TP2-biotin and sTP-biotin at a 95% purity (Fig. 1 A) were synthesized by the Nanjing Genscript Biotechnology Company.
Fig. 1.
Inhibitory effects of TP1 and TP2 against TMUV infection in BHK-21 cells. A, Sequence alignment of TP1 and TP2 with the corresponding regions from other flaviviruses: four DENV serotypes, JEV, TBEV, WNV, YFV and ZIKV. The black boxes indicates conserved amino acid residues. B–D, The inhibitory effects of the peptides were detected by qRT-PCR (B), a plaque assay (C), and western blotting (D). E, IC50 values of the two peptides. F, Toxicity of TP1 and TP2 in BHK-21 cells. The data are expressed as the means ± SDs of results from three independent experiments. The asterisk indicates a statistically significant difference (P < 0.05).
2.3. Inhibition of TMUV infection
The peptides were solubilized in dimethyl sulfoxide (DMSO) and diluted in serum-free RPMI-1640 medium. TMUV at a concentration of approximately 200 TCID50 (0.2 mL) was added to equal volumes of peptide solutions at different concentrations. Then, 0.4 mL of each mixture was incubated at 37 °C for 1 h and added to BHK-21 cells grown in 12-well plates. After incubation at 37 °C for 1 h, the supernatants were removed, and the cells were washed with chilled PBS twice. Then, fresh RPMI-1640 medium containing 5% FCS was added. At 24 h post-infection, the culture supernatants were collected, and viral titres were determined by qRT-PCR and plaque assay. The TMUV titres of the culture supernatants of infected cells treated with peptides was compared with those of infected control cells treated with negative peptide, which was set as 100% infection. The percentage inhibition was calculated as follows:
2.4. RNA extraction and qRT-PCR
Viral RNA was extracted with an AxyPrep Body Fluid Viral DNA/RNA Miniprep Kit (Axygen, Union City, USA) according to the manufacturer's protocol. First-strand cDNA was synthesized using HiScript II Q Select RT SuperMix for qPCR (+gDNA wiper, Vazyme, Nanjing, China). The primers used for qRT-PCR were NS2AF primer (forward, 5′-GCAGCCCAGGAGATTTTGAG-3′) and NS2AR primer (reverse, 5′-CTAACGCAACGCCAAGCA-3′).
2.5. Plaque assay
The viral titres in the supernatants of cultured cells were determined by a plaque assay as described previously (Zhao et al., 2015). Briefly, ten-fold serial dilutions of the supernatants were used to inoculate a monolayer of BHK-21 cells. After 1 h of incubation at 37 °C, the cells were washed with PBS and covered with cell culture medium containing 1% agarose. After 72 h of incubation at 37 °C, the cells were stained with 0.1% neutral red to visualize the plaques.
2.6. Western blotting
The amounts of TMUV E protein in the cell lysates were detected by western blotting as described previously (Zhao et al., 2019). Briefly, TMUV-infected cells were lysed using cell lysis buffer containing 1 mM phenylmethylsulfonyl fluoride (Beyotime, Shanghai, China). Samples containing equal amounts of protein were separated by SDS-PAGE, and the proteins were then electrophoretically transferred to a PVDF membrane which was blocked at 37 °C for 2 h with 2% ECL Prime blocking agent (GE, Buckinghamshire, UK) in PBS containing 0.05% Tween-20. The membrane was then incubated with anti-TMUV or anti-β-actin monoclonal antibody at 4 °C overnight. After washing three times with PBST, the membrane was probed with HRP-labelled goat anti-mouse IgG antibody at 37 °C for 1 h. The membrane washing procedure was repeated three times, and the proteins were detected using the BeyoECL Plus Kit (Beyotime, Shanghai, China).
2.7. Toxicity assay
Cells were treated with 150 mg/L TP1 or TP2 for 24 h at 37 °C. To determine whether the two peptides are toxic to cells, cellular viability was detected with a CCK-8 kit (Dojindo, Kumamoto, Japan) according to the manufacturer’s instructions.
2.8. Assay to determine the 50% inhibitory concentration (IC50)
The IC50 values of the two peptides were measured as previously described (Panya et al., 2015). Briefly, BHK-21 monolayer cells were grown in a 96-well plate. Two-fold serial dilutions of the peptides were mixed with TMUV at a concentration of 200 TCID50 and then incubated at 37 °C for 1 h before their infection of BHK-21 cells. After 1 h of incubation at 37 °C, supernatants were removed, and the cells were covered with a mixture of 1% agarose and RPMI-1640 medium containing 5% FCS. The cells were further incubated for 48 h and then stained with 0.1% neutral red to visualize plaques. The TMUV plaques were counted and analysed by comparison with control cells infected with TMUV treated with negative peptide (set as 0% inhibition). The nonlinear regression function of GraphPad Prism software (GraphPad Software, La Jolla, USA) was used to calculate IC50 values of the peptides against TMUV.
2.9. ADE inhibition assays
Duck anti-TMUV serum was prepared and stored in our laboratory. The neutralizing antibody titre of the antiserum was assessed by the plaque reduction neutralization test as previously described (Zhao et al., 2018a). Duck peripheral blood lymphocytes were isolated using a duck peripheral blood lymphocyte isolation kit (Haoyang, Tianjin, China) according to the manufacturer’s protocol. Briefly, 5 mL of sterile peripheral blood with anticoagulant was obtained from ducks and then diluted with 5 mL of RPMI-1640 medium. Ten millilitres of separation medium was added to the centrifuge tube. The diluted whole blood was carefully layered onto the separation medium. The tube was centrifuged with a horizontal rotor at 400 ×g for 30 min at 4 °C. Cells at the interface were carefully transferred into a new centrifuge tube containing 10 mL of washing buffer and mixed gently. The tube was centrifuged with a horizontal rotor at 250 ×g for 10 min at 4 °C. The cell pellet was then washed twice at 4 °C with washing buffer. Finally, the cells were resuspended in RPMI-1640 medium containing 10% FCS and inoculated into a 12-well plate. ADE inhibition assays were carried out as previously described (Nicholson et al., 2011). Briefly, inactivated duck anti-TMUV serum with a neutralizing antibody titre of 1:64 was diluted 1:100 with RPMI-1640 medium. Then, 0.2 mL of diluted inactivated duck anti-TMUV serum was incubated with an equal volume of virus at a concentration of 200 TCID50 for 1 h at 37 °C. Then, the serum-virus mixtures were incubated with peptides at different concentrations for an additional 1 h. The mixtures were added to duck peripheral blood lymphocytes and incubated for 48 h at 37 °C. The cultures were harvested, and viral titres were determined by plaque assay and qRT-PCR.
2.10. Co-immunoprecipitation assay
A co-immunoprecipitation assay was conducted as previously described (Zhao et al., 2018b). Briefly, TMUV was incubated with biotin-peptide on a rocker at 4 °C overnight, followed by incubation with anti-biotin antibody (Sigma, St. Louis, USA) for 5 h. Subsequently, protein A/G agarose beads (Beyotime, Shanghai, China) were added to the mixture and incubated for 3 h. The beads were washed with PBS five times and collected for qRT-PCR detection.
2.11. Indirect enzyme-linked immunosorbent assay (ELISA)
Ninety-six-well ELISA plates were pre-coated with purified TMUV at 4 °C overnight. After washing with PBST three times, the plates were blocked with PBST containing 1% BSA at 37 °C for 2 h. After washing three times, 0.1 mL of diluted biotin-peptides was added to each well and incubated at 37 °C for 1 h. The plates were washed with PBST three times, followed by the addition of 0.1 mL of HRP-labelled streptavidin diluted 1:5000. After incubation at 37 °C for 1 h and three further washes, tetramethyl benzidine (TMB) substrate was added and incubated for 15 min at room temperature. The reaction was stopped with the addition of 2 M H2SO4, and the plates were read by a BioTek microplate reader.
2.12. RNase digestion assay
An RNase digestion assay was carried out as previously described (Lok et al., 2017; Yu et al., 2017). Briefly, TMUV at a concentration of approximately 4 × 105 TCID50 was incubated with TP1 or TP2 at 37 °C for 1 h, after which the released genomic RNA was digested with micrococcal nuclease (NEB, Ipswich, USA) for 1 h at 37 °C. Genomic RNA was extracted and detected by qRT-PCR as described above.
2.13. Viral binding inhibition assay
A viral binding inhibition assay was conducted as described previously (Costin et al., 2010). Confluent BHK-21 cell monolayers were washed once with chilled PBS. TMUV and peptides were co-incubated at 37 °C for 1 h before their addition to the monolayers and incubation for 2 h at 37 °C. The monolayers were washed with chilled PBS three times and then harvested for RNA extraction and detection.
2.14. Statistical analysis
Statistical analyses were performed using Statistical Package for Social Science (SPSS) statistical software. Differences with a P value <0.05 were considered statistically significant.
3. Results
3.1. Peptides TP1 and TP2 exerted inhibitory effects against TMUV infection
TMUV was incubated with TP1 or TP2 before its infection of BHK-21 cells. At 24 h post-inoculation, TP1- or TP2-mediated inhibition of TMUV infection was assessed by qRT-PCR (Fig. 1B), plaque assay (Fig. 1C), and western blotting (Fig. 1D). The qRT-PCR results showed that both peptides inhibited TMUV infection in a dose-dependent manner. Incubation with TP1 or TP2 at a concentration of 10 mg/L led to slightly decreased virus production, but these differences were not statistically significant. However, incubation with 50 mg/L and 100 mg/L TP1 or TP2 caused a significant decrease in virus production. Compared with negative peptide treatment, the virus production following 50 mg/L and 100 mg/L TP1 treatment was decreased by 21.3% and 44.6%, respectively. Similarly, 50 mg/L and 100 mg/L TP2 treatment reduced virus production by 37.4% and 53.4%, respectively. The results of the plaque assay and western blot analysis were similar to those obtained by qRT-PCR. In addition, the IC50 values suggested that TP2 derived from the stem region exhibited a stronger inhibitory effect than TP1 derived from DII (Fig. 1E). As expected, the negative peptide did not show an inhibitory effect. A toxicity assay was conducted to exclude the possibility that the antiviral effects of the two peptides were due to underlying cytotoxicity. The results showed no significant differences between BHK-21 cells with or without the peptides, suggesting that the two peptides are non-toxic at the indicated concentrations (Fig. 1F). These results suggest that TP1 and TP2 can effectively inhibit TMUV infection.
3.2. Peptides TP1 and TP2 inhibited TMUV infection in different cell types
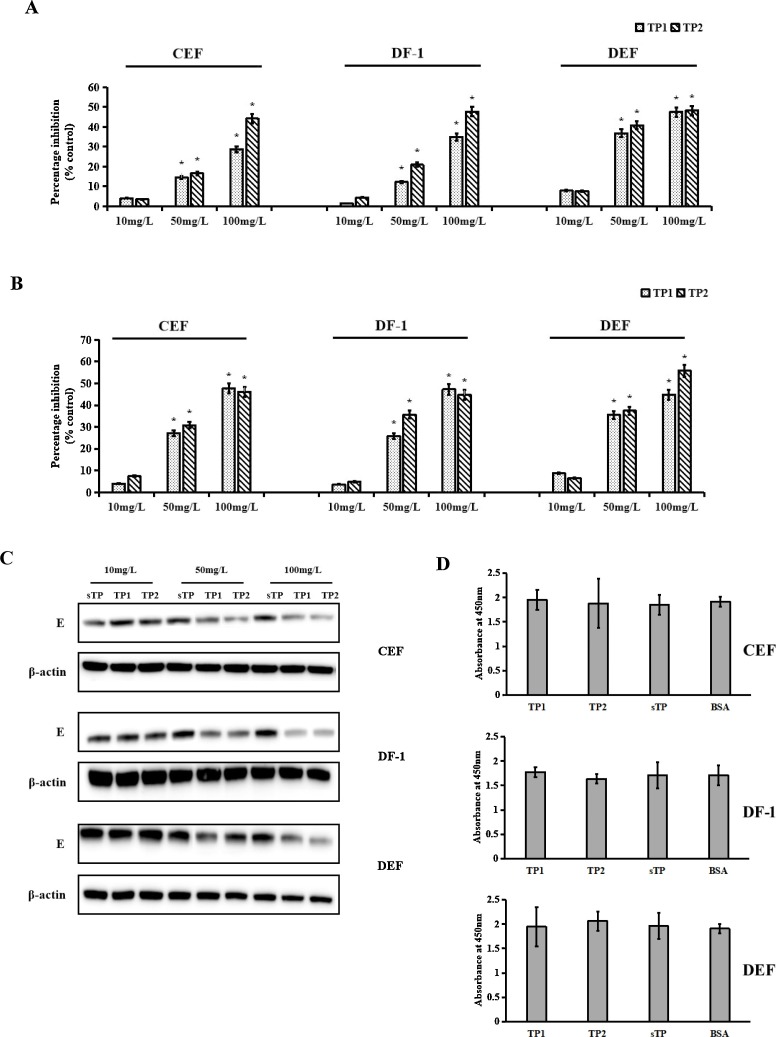
To determine whether the inhibitory effects of TP1 and TP2 are cell type dependent, different cell types of avian origin (chicken embryo fibroblasts (CEFs), DF-1 cells (immortalised CEFs) and duck embryo fibroblasts (DEFs)) were used to test the effects of TP1 and TP2 on TMUV infection. At 24 h post-infection, qRT-PCR, plaque assays, and western blotting were conducted to detect virus production with peptide treatment. As shown in Fig. 2 , in the presence of 50 mg/L and 100 mg/L TP1 or TP2, virus production in the three cell types was significantly decreased (Fig. 2A). The results of the plaque assay were similar to those obtained by qRT-PCR. Compared with negative peptide treatment, treatment with 50 mg/L and 100 mg/L TP1 reduced virus production by 27.17% and 47.68% in CEFs, 25.76% and 35.63% in DF-1 cells and 35.54% and 44.83% in DEFs, respectively. However, 50 mg/L and 100 mg/L TP2 decreased virus production by 30.74% and 46.13% in CEFs, 35.63% and 44.68% in DF-1 cells and 37.42% and 55.86% in DEFs, respectively (Fig. 2B). The antiviral effects of TP1 and TP2 were also confirmed by western blotting (Fig. 2C). Moreover, TP1 and TP2 showed no cytotoxic effect on CEFs, DF-1 cells or DEFs (Fig. 2D). These results indicated that the peptides exhibited similar inhibitory effects in different cell types.
Fig. 2.
Inhibitory effects of TP1 and TP2 against TMUV infection in different cell types. Virus production was determined by qRT-PCR (A), a plaque assay (B), and western blotting (C). D, The toxicity of TP1 and TP2 in different cells. Data are the means ± SDs of triplicate experiments. The asterisk indicates a statistically significant difference (P < 0.05).
3.3. Peptide TP2 cross-inhibited JEV infection
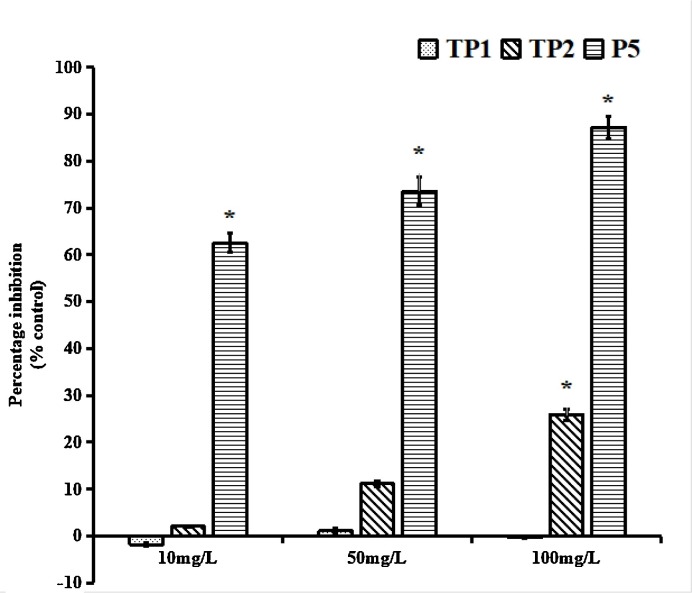
Because the E proteins among flaviviruses share sequence homology, whether the peptides would inhibit JEV infection was investigated. The inhibitory effects of the peptides were determined by the plaque assay at 24 h post-infection. TP2 efficiently protected against JEV infection, demonstrating that TP2 could cross-inhibit JEV infectivity. However, TP1 did not exhibit inhibitory activity against JEV (Fig. 3 ).
Fig. 3.
Effect of TP1 and TP2 against JEV. The inhibitory activities of TP1 and TP2 at different concentrations against JEV were tested. At 24 h post-infection, the culture supernatants were harvested, and the JEV titres were determined by plaque assay. The peptide P5 (AWDFGSIGGVFNSIGKAVHQV, Chen et al., 2017a) was used as the positive control. The data presented are from three separate experiments. * p < 0.05 vs. negative peptide treatment.
3.4. Peptides TP1 and TP2 blocked TMUV ADE
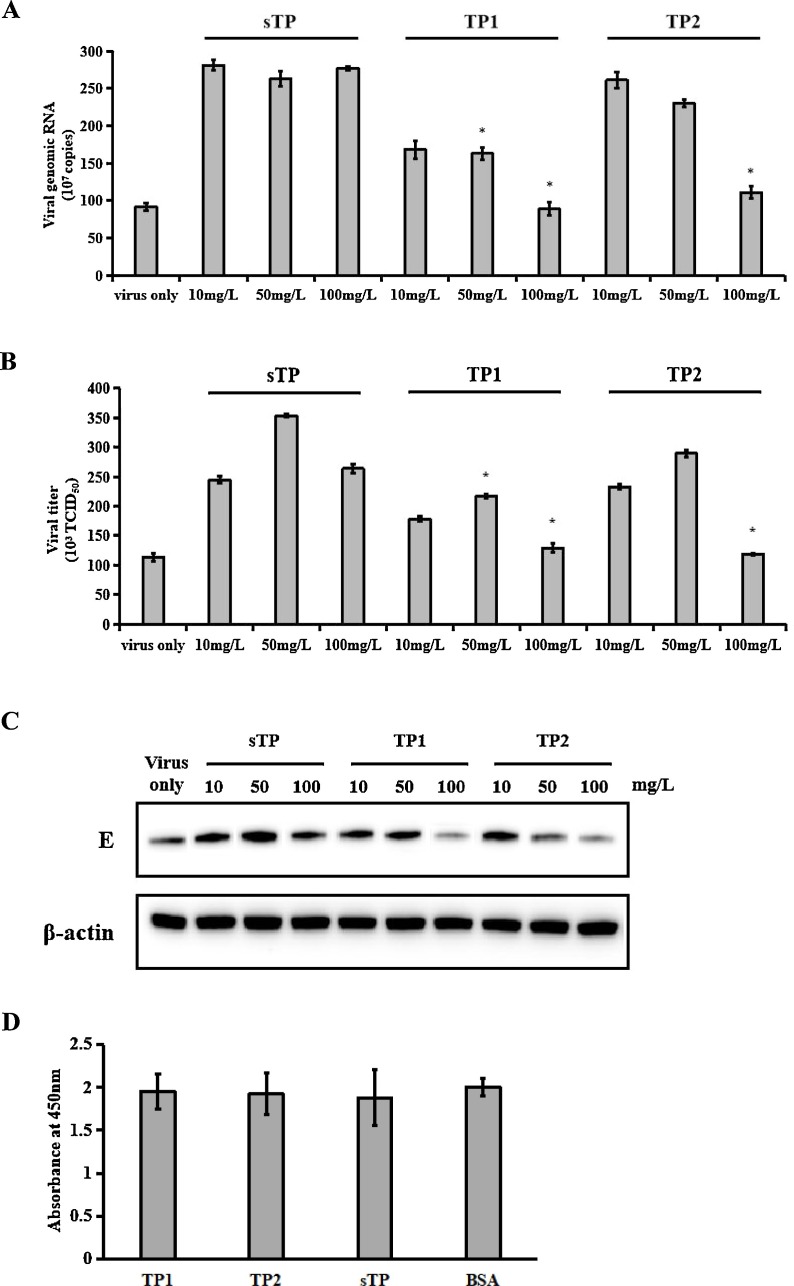
Previous studies have shown that antibodies can mediate flavivirus infection to facilitate viral entry, leading to enhanced infection and severe disease outcomes, in a process known as ADE (Halstead, 2003;Hawkes, 1964). Liu et al. (2013) reported TMUV ADE when anti-TMUV antibody levels were low. Earlier studies in our laboratory indicated that duck anti-TMUV serum could enhance infection in duck peripheral blood lymphocytes, with this effect peaking with anti-TMUV serum (with a neutralizing antibody titre of 1:64) diluted 1:100 (data not shown). As shown in Fig. 4 , anti-TMUV serum diluted 1:100 facilitated infection in duck peripheral blood lymphocytes. However, incubation of anti-TMUV serum mixed with TP1 or TP2 inhibited ADE in a concentration-dependent manner. Although the inhibitory peptides did not block ADE completely, treatment with TP1 or TP2 led to a significant reduction in virus production compared with that following treatment with the negative peptide. Although TP1 and TP2 inhibited TMUV ADE, the two peptides showed no cytotoxic effect on duck peripheral blood lymphocytes (Fig. 4C).
Fig. 4.
The peptides TP1 and TP2 inhibited ADE in duck peripheral blood lymphocytes. TP1 and TP2 blocked the infection of duck peripheral blood lymphocytes in the presence of a maximally enhancing dilution (1:100) of anti-TMUV serum. Virus production was determined by qRT-PCR (A), a plaque assay (B), and western blotting (C). D, The toxicity of TP1 and TP2 in duck peripheral blood lymphocytes. The data are expressed as the means ± SDs of results from three independent experiments. * p < 0.05 vs. negative peptide treatment.
3.5. Peptides TP1 and TP2 bound TMUV
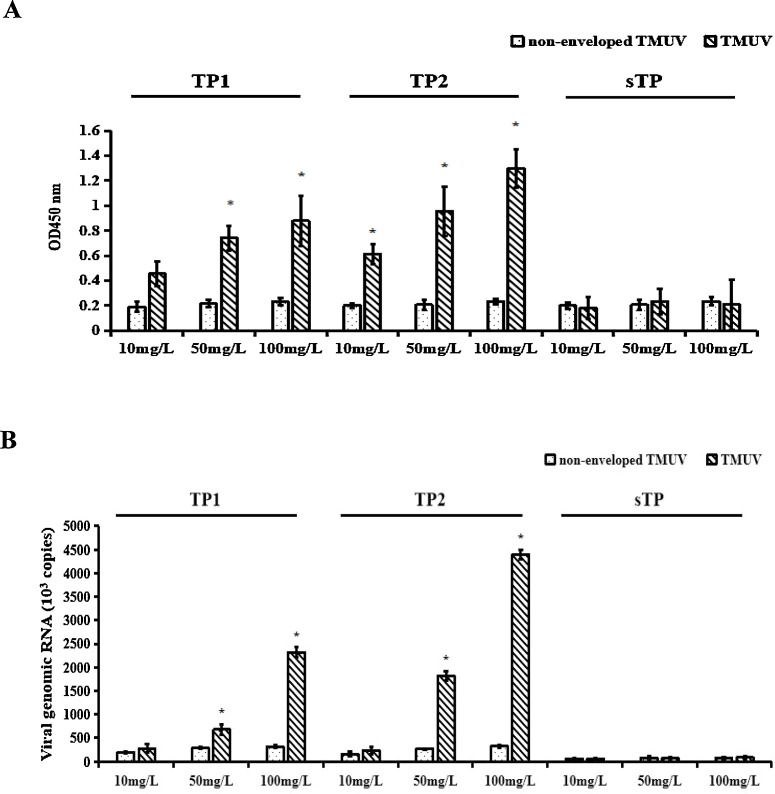
Several studies have shown that the interaction between inhibitory peptides and the virion causes a reduction in infection (Zhang et al., 2012). In this study, the binding of peptides to TMUV was examined by the addition of biotin-peptides at different concentrations to an ELISA plate pre-coated with purified TMUV that were then detected using HRP-labelled streptavidin. Biotin-TP1 or biotin-TP2 at increasing concentrations increased the signals indicating binding to the TMUV that had been coated on the plates in a concentration-dependent manner, but signals from negative peptide and non-enveloped TMUV were very low (Fig. 5 A). The binding between TMUV and the peptides was further confirmed by co-immunoprecipitation assays (Fig. 5B). These results verified the direct interaction between biotin-peptides and TMUV.
Fig. 5.
The peptides TP1 and TP2 interacted with TMUV. The peptide inhibitors were allowed to bind TMUV, and their TMUV-binding activity was tested by indirect ELISA (A) and a co-immunoprecipitation assay (B). Non-enveloped TMUV (TMUV treated with chloroform) incubated with peptide inhibitors in the same manner served as a negative control. The data presented are from three separate experiments. The asterisk indicates a statistically significant difference (P < 0.05).
3.6. TP1 released viral RNA from virions
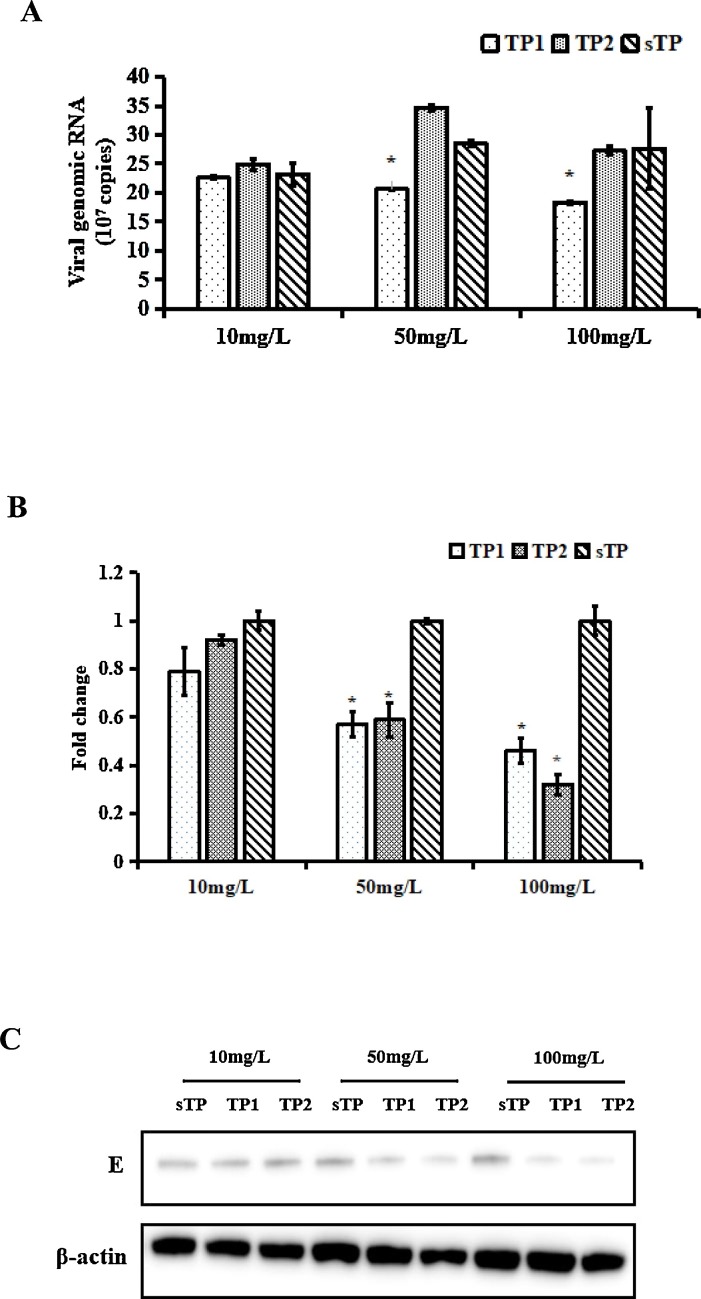
Lok et al. (2012) reported that the inhibitory peptide DN59 could inhibit DENV by interacting with virions and inducing the release of viral genomic RNA. Here, the release of viral genomic RNA induced by the interaction between TMUV and the peptides was investigated by exposing peptide-treated virions to RNase digestion. The RNase digestion assay showed that genomic RNA released from TMUV virions co-incubated with TP1 at increasing doses was sensitive to micrococcal nuclease. Approximately 33.7% of the genomic RNA released by TMUV virions treated with 100 mg/L TP1 was degraded by RNase. However, the genomic RNA released by virions treated with negative peptide or TP2 was protected from RNase digestion (Fig. 6 A).
Fig. 6.
Study of the mechanism by which TP1 and TP2 blocked TMUV infection. A, Degradation of genomic RNA released from TMUV due to TP1 or TP2 in an RNase digestion assay. B and C, Pre-incubation of TMUV with either TP1 or TP2 reduced the binding of TMUV to BHK-21 cells compared to that of TMUV pre-incubated with negative peptide. B, Viral genomic RNA was detected by qRT-PCR. The data were analysed using the comparative Ct (ΔΔCT) method. GAPDH was chosen as a reference gene for internal control. Negative peptide treatment was used as a reference for each comparison. Data are the means ± SDs of triplicate experiments. The asterisk indicates a statistically significant difference (P < 0.05). C, Cells lysates were subjected to western blotting to study the levels of TMUV E proteins. β-Actin was used as a loading control.
3.7. Peptides TP1 and TP2 interfered with the binding between TMUV and cells
To explore the mechanism of the inhibitory effects of TP2, a virus binding inhibition assay was performed to determine whether the peptides interfered with the binding between TMUV and cells. TMUV was incubated with TP1 or TP2 before its addition to BHK-21 cells at 37 °C for 1 h to allow viral attachment. Unbound virus was repeatedly washed with chilled PBS, and the amount of TMUV remaining on the cells was then detected by qRT-PCR and western blotting. Compared to the RNA of virus treated with negative peptide, relative TMUV RNA was significantly decreased after TP1 or TP2 treatment. The results of western blot analysis were consistent with those of qRT-PCR and suggested that the binding of TMUV to the cells was reduced after TP1 or TP2 bound TMUV (Fig. 6B and C).
4. Discussion
At present, several commercial inactivated and attenuated live vaccines have been approved by the Chinese Ministry of Agriculture for TMUV prevention. However, ducks and geese in some farms can still be infected by TMUV because of the lack of immunization or failure to induce adequate antibodies after immunization. However, there are no effective and specific treatments for TMUV infection. The need to develop treatments to control the spread of TMUV is urgent. In this study, the peptide TP1, derived from DII of the TMUV E protein, and peptide TP2, corresponding to a sequence in the stem region of the TMUV E protein, were highly effective in inhibiting TMUV infection, indicating that TP1 and TP2 are potential candidates for TMUV treatment.
The stem region of the E protein is highly conserved among flaviviruses. Previous studies reported that peptide inhibitors derived from the stem region exhibited cross-antiviral activity. DN59, derived from the stem region of the DENV E protein, had inhibitory effects against DENV as well as WNV (Hrobowski et al., 2015). P5, corresponding to the stem region of the JEV E protein, effectively blocked ZIKV infection with an IC50 at the micromolar level (Chen et al., 2017a). In this study, the TP2 peptide exhibited antiviral activity against TMUV as well as JEV. Since the corresponding stem regions of TMUV, JEV and other flaviviruses are highly conserved (Fig. 1), other flaviviruses may be inhibited by TP2. Therefore, TP2, TP2 derivatives or analogous peptides probably act as broad-spectrum flavivirus inhibitors.
In this study, co-immunoprecipitation assays and indirect ELISA indicated that TP1 or TP2 could bind TMUV in a concentration-dependent manner. The direct binding of TP1 or TP2 to TMUV, but not to target cells, might explain why both peptides exhibited similar inhibitory effects in the different cell types BHK-21 cells, CEFs, DF-1 cells and DEFs.
The mechanism by which flavivirus infection is inhibited by peptide inhibitors is not clearly understood. Previous studies reported that once DN59 bound the virion, it likely inserted itself between the E protein ectodomain and the membrane during the dynamic “breathing” process of the viral particle (Johnson, 2003; Lok et al., 2008). This insertion resulted in the formation of holes in the viral membrane or disrupted the membrane bilayer structure. Subsequently, negatively charged genomic RNA escaped from the viral particles. Similarly, the binding of Z2 to the E protein inactivated ZIKV by disrupting the integrity of the ZIKV membrane (Yu et al., 2017). An RNase digestion assay showed that TMUV treated with TP1, but not TP2, was sensitive to RNase digestion, indicating disruption of the viral membrane and the escape of genomic RNA from viral particles. However, the underlying mechanism by which TP1 disrupts the viral membrane is still unclear. As described in previous studies, possible explanations are hydrophobic and electrostatic effects or viral dynamic “breathing”, but determining this mechanism lies beyond the scope of this study.
The RNase digestion assay showed that TP2 treatment did not destroy the integrity of TMUV. Therefore, a virus binding inhibition assay was conducted to investigate the mechanism by which TP2 inhibited TMUV infection. The results demonstrated that treatment with either TP1 or TP2 prevented TMUV from binding cells. Costin et al. (2010) reported that the surface of DENV particles changed from smooth to rough and that DENV particles lost their icosahedral symmetry after incubation with the peptides DN57opt and 1OAN1. This structural deformation probably interfered with cell binding. Alhoot et al. (2013) demonstrated that the inhibitory peptides DET2 and DET4 caused structural abnormalities and altered the arrangement of the viral E protein, which interfered with viral binding and entry. TP1 and TP2 are likely to interfere with viral attachment by a mechanism similar to that described above. Nevertheless, the idea that the decrease in TMUV attachment after incubation with TP1 was due to the release of the genome cannot be excluded. Thus, further research to elucidate the precise mechanism of the inhibitory effects of TP1 and TP2 is needed.
The antibody induced by an inactivated vaccine may enhance TMUV infection through ADE. This phenomenon increases the challenge in developing an effective and safe vaccine to provide protection against TMUV. TP1 and TP2, which show inhibitory activity against ADE, are probably favourable for the inoculation of infected ducks and geese without the concern of ADE. In addition, unlike most small-molecule antiviral drugs, the nontoxicity of TP1 and TP2 also ensures the safety of these peptide inhibitors for antiviral therapy. However, why TP1 and TP2 inhibit TMUV ADE is still unclear. Yu et al. (2017) reported that the inhibitory peptide Z2 interacted with the E protein of ZIKV and caused viral genomic RNA release, eventually resulting in the inactivation of ZIKV virions. It is presumed that the binding of TP1 to TMUV inactivates TMUV virions and hence relieves ADE-mediated TMUV infection. In addition, TP1 and TP2 were shown to interfere with the cellular binding of TMUV. It is possible that these two inhibitory peptides block ADE by reducing viral binding to cells. These conjectures still require further experimental confirmation.
5. Conclusions
This is the first report on peptide inhibitors of TMUV infection derived from the E protein. TP1 and TP2 exhibited effective anti-TMUV activity in a concentration-dependent manner. Additionally, TP2 cross-inhibited JEV infection in BHK-21 cells. Furthermore, both peptides could block ADE in vitro. TP1 functions through a mechanism that involves release of the viral genome and interference with cellular TMUV binding, while TP2 might be an interference factor between TMUV and the cell. These findings suggest that TP1 and TP2 could be further developed as novel treatments against TMUV.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgements
The study was supported by National Natural Science Foundation of China (No. 31802222), National Key Research and Development Program of China (2017YFD0500804). The authors also acknowledge the financial support from China Scholarship Council program (File No. 201908320137).
Contributor Information
Yin Li, Email: muziyin08@163.com.
Peng Zhao, Email: zhaopeng@sdau.edu.cn.
References
- Alhoot M.A., Rathinam A.K., Wang S.M., Manikam R., Sekaran S.D. Inhibition of dengue virus entry into target cells using synthetic antiviral peptides. Int. J. Med. Sci. 2013;10:719–729. doi: 10.7150/ijms.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch B.J., van der Zee R., de Haan C.A., Rottier P.J. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Liu Y., Wang S., Sun J., Wang P., Xin Q., Zhang L., Xiao G., Wang W. Antiviral activity of peptide inhibitors derived from the protein E stem against Japanese encephalitis and Zika viruses. Antiviral Res. 2017;141:140–149. doi: 10.1016/j.antiviral.2017.02.009. [DOI] [PubMed] [Google Scholar]
- Chen S., Luo G., Yang Z., Lin S., Chen S., Wang S., Goraya M.U., Chi X., Zeng X., Chen J.L. Avian Tembusu virus infection effectively triggers host innate immune response through MDA5 and TLR3-dependent signaling pathways. Vet. Res. 2016;47:74. doi: 10.1186/s13567-016-0358-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Wang L., Chen J., Zhang L., Wang S., Goraya M.U., Chi X., Na Y., Shao W., Yang Z., Zeng X., Chen S., Chen J.L. Avian interferon-inducible transmembrane protein family effectively restricts avian tembusu virus infection. Front. Microbiol. 2017;8:672. doi: 10.3389/fmicb.2017.00672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costin J.M., Jenwitheesuk E., Lok S.M., Hunsperger E., Conrads K.A., Fontaine K.A., Rees C.R., Rossmann M.G., Isern S., Samudrala R., Michael S.F. Structural optimization and de novo design of dengue virus entry inhibitory peptides. PLoS Negl. Trop. Dis. 2010;4:e721. doi: 10.1371/journal.pntd.0000721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J.G., Frieden C. Protein fragments as probes in the study of protein folding mechanisms: differential effects of dihydrofolate reductase fragments on the refolding of the intact protein. Proc. Natl. Acad. Sci. U S A. 1989;86:3060–3064. doi: 10.1073/pnas.86.9.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstead S.B. Neutralization and antibody-dependent enhancement of dengue viruses. Adv. Virus Res. 2003;60:421–467. doi: 10.1016/s0065-3527(03)60011-4. [DOI] [PubMed] [Google Scholar]
- Hawkes R.A. Enhancement of the infectivity of arboviruses by specific antisera produced in domestic fowls. Aust. J. Exp. Biol. Med. Sci. 1964;42:465–482. doi: 10.1038/icb.1964.44. [DOI] [PubMed] [Google Scholar]
- Hrobowski Y.M., Garry R.F., Michael S.F. Peptide inhibitors of dengue virus and West Nile virus infectivity. Virol. J. 2005;2:49. doi: 10.1186/1743-422X-2-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Han K., Zhao D., Liu Y., Zhang J., Niu H., Zhang K., Zhu J., Wu D., Gao L., Li Y. Identification and molecular characterization of a novel flavivirus isolated from geese in China. Res. Vet. Sci. 2013;94:774–780. doi: 10.1016/j.rvsc.2012.11.014. [DOI] [PubMed] [Google Scholar]
- Jiang S., Lin K., Strick N., Neurath A.R. Inhibition of HIV-1 infection by a fusion domain binding peptide from the HIV-1 envelope glycoprotein GP41. Biochem. Biophys. Res. Commun. 1993;195:533–538. doi: 10.1006/bbrc.1993.2078. [DOI] [PubMed] [Google Scholar]
- Johnson J.E. Virus particle dynamics. Adv. Protein Chem. 2003;64:197–218. doi: 10.1016/s0065-3233(03)01005-2. [DOI] [PubMed] [Google Scholar]
- Kuhn R.J., Zhang W., Rossmann M.G., Pletnev S.V., Corver J., Lenches E., Jones C.T., Mukhopadhyay S., Chipman P.R., Strauss E.G., Baker T.S., Strauss J.H. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002;108:717–725. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Huang X., Zhao D., Han K., Liu Y., Yang J., Bi K., Li Y. Identification of heat shock protein A9 as a Tembusu virus binding protein on DF-1 cells. Virus Res. 2017;227:110–114. doi: 10.1016/j.virusres.2016.09.020. [DOI] [PubMed] [Google Scholar]
- Liu Z., Ji Y., Huang X., Fu Y., Wei J., Cai X., Zhu Q. An adapted duck Tembusu virus induces systemic infection and mediates antibody-dependent disease severity in mice. Virus Res. 2013;176:216–222. doi: 10.1016/j.virusres.2013.06.010. [DOI] [PubMed] [Google Scholar]
- Lok S.M., Costin J.M., Hrobowski Y.M., Hoffmann A.R., Rowe D.K., Kukkaro P., Holdaway H., Chipman P., Fontaine K.A., Holbrook M.R., Garry R.F., Kostyuchenko V., Wimley W.C., Isern S., Rossmann M.G., Michael S.F. Release of dengue virus genome induced by a peptide inhibitor. PLoS One. 2012;7 doi: 10.1371/journal.pone.0050995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lok S.M., Kostyuchenko V., Nybakken G.E., Holdaway H.A., Battisti A.J., Sukupolvi-Petty S., Sedlak D., Fremont D.H., Chipman P.R., Roehrig J.T., Diamond M.S., Kuhn R.J., Rossmann M.G. Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins. Nat. Struct. Mol. Biol. 2008;15:312–317. doi: 10.1038/nsmb.1382. [DOI] [PubMed] [Google Scholar]
- Markosyan R.M., Bates P., Cohen F.S., Melikyan G.B. A study of low pH-induced refolding of Env of avian sarcoma and leukosis virus into a six-helix bundle. Biophys. J. 2004;87:3291–3298. doi: 10.1529/biophysj.104.047696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson C.O., Costin J.M., Rowe D.K., Lin L., Jenwitheesuk E., Samudrala R., Isern S., Michael S.F. Viral entry inhibitors block dengue antibody-dependent enhancement in vitro. Antiviral Res. 2011;89:71–74. doi: 10.1016/j.antiviral.2010.11.008. [DOI] [PubMed] [Google Scholar]
- Niu H., Huang X., Han K., Liu Y., Zhao D., Zhang J., Liu F., Li T., Zhou X., Li X., Li Y. Development of double antibody sandwich ELISA for detection of duck or goose flavivirus. J. Int. Agri. 2013;12:1638–1643. [Google Scholar]
- Okazaki K., Kida H. A synthetic peptide from a heptad repeat region of herpesvirus glycoprotein B inhibits virus replication. J. Gen. Virol. 2004;85:2131–2137. doi: 10.1099/vir.0.80051-0. [DOI] [PubMed] [Google Scholar]
- Panya A., Sawasdee N., Junking M., Srisawat C., Choowongkomon K., Yenchitsomanus P.T. A peptide inhibitor derived from the conserved ectodomain region of DENV membrane (M) protein with activity against dengue virus infection. Chem. Biol. Drug Des. 2015;86:1093–1104. doi: 10.1111/cbdd.12576. [DOI] [PubMed] [Google Scholar]
- Pierson T.C., Kielian M. Flaviviruses: braking the entering. Curr. Opin. Virol. 2013;3:3–12. doi: 10.1016/j.coviro.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porotto M., Carta P., Deng Y., Kellogg G.E., Whitt M., Lu M., Mungall B.A., Moscona A. Molecular determinants of antiviral potency of paramyxovirus entry inhibitors. J. Virol. 2007;81:10567–10574. doi: 10.1128/JVI.01181-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodenhuis-Zybert I.A., Wilschut J., Smit J.M. Dengue virus life cycle: viral and host factors modulating infectivity. Cell. Mol. Life Sci. 2010;67:2773–2786. doi: 10.1007/s00018-010-0357-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit J.M., Moesker B., Rodenhuis-Zybert I., Wilschut J. Flavivirus cell entry and membrane fusion. Viruses. 2011;3:160–171. doi: 10.3390/v3020160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Wu Z., Wu Y., Wang T., Wang M., Jia R., Zhu D., Liu M., Zhao X., Yang Q., Wu Y., Zhang S., Liu Y., Zhang L., Yu Y., Pan L., Chen S., Cheng A. Heparin sulfate is the attachment factor of duck Tembus virus on both BHK21 and DEF cells. Virol. J. 2019;16:134. doi: 10.1186/s12985-019-1246-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Q., Compans R.W. Peptides corresponding to the heptad repeat sequence of human parainfluenza virus fusion protein are potent inhibitors of virus infection. Virology. 1996;223:103–112. doi: 10.1006/viro.1996.0459. [DOI] [PubMed] [Google Scholar]
- Yu Y., Deng Y.Q., Zou P., Wang Q., Dai Y., Yu F., Du L., Zhang N.N., Tian M., Hao J.N., Meng Y., Li Y., Zhou X., Fuk-Woo Chan J., Yuen K.Y., Qin C.F., Jiang S., Lu L. A peptide-based viral inactivator inhibits Zika virus infection in pregnant mice and fetuses. Nat. Commun. 2017;8:15672. doi: 10.1038/ncomms15672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Hunke C., Yau Y.H., Seow V., Lee S., Tanner L.B., Guan X.L., Wenk M.R., Fibriansah G., Chew P.L., Kukkaro P., Biukovic G., Shi P.Y., Shochat S.G., Grüber G., Lok S.M. The stem region of premembrane protein plays an important role in the virus surface protein rearrangement during dengue maturation. J. Biol. Chem. 2012;287:40525–40534. doi: 10.1074/jbc.M112.384446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Chen S., Mahalingam S., Wang M., Cheng A. An updated review of avian-origin Tembusu virus: a newly emerging avian Flavivirus. J. Gen. Virol. 2017;98:2413–2420. doi: 10.1099/jgv.0.000908. [DOI] [PubMed] [Google Scholar]
- Zhao D., Han K., Huang X., Zhang L., Wang H., Liu N., Tian Y., Liu Q., Yang J., Liu Y., Li Y. Screening and identification of B-cell epitopes within envelope protein of tembusu virus. Virol. J. 2018;15:142. doi: 10.1186/s12985-018-1052-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D., Huang X., Liu Y., Han K., Zhang J., Yang J., Xie X., Li Y. Domain I and II from newly emerging goose tembusu virus envelope protein functions as a dominant-negative inhibitor of virus infectivity. Res. Vet. Sci. 2015;98:121–126. doi: 10.1016/j.rvsc.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D., Liu Q., Han K., Wang H., Yang J., Bi K., Liu Y., Liu N., Tian Y., Li Y. Identification of glucose-regulated protein 78 (GRP78) as a receptor in BHK-21 cells for duck tembusu virus infection. Front. Microbiol. 2018;9:694. doi: 10.3389/fmicb.2018.00694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D., Yang J., Han K., Liu Q., Wang H., Liu Y., Huang X., Zhang L., Li Y. The unfolded protein response induced by tembusu virus infection. BMC Vet. Res. 2019;15:34. doi: 10.1186/s12917-019-1781-4. [DOI] [PMC free article] [PubMed] [Google Scholar]