Abstract
The purpose of testing for any communicable disease is to support clinicians in the diagnosis and management of individual patients and to describe transmission dynamics. The novel coronavirus is formally named SARS-CoV-2 and the clinical disease state resulting from an infection is known as COVID-19. Control of the COVID-19 pandemic requires clinicians, epidemiologists, and public health officials to utilise the most comprehensive, accurate and timely information available to manage the rapidly evolving COVID-19 environment. High performance sport is a unique context that may look towards comprehensive testing as a means of risk mitigation. Characteristics of the common testing options are discussed including the circumstances where additional testing may be of benefit and considerations for the associated risks. Finally, a review of the available technology that could be considered for use by medical staff at the point of care (PoC) in a high-performance sporting context is included.
Keywords: Sports medicine, Novel coronavirus, Surveillance, Point of care testing, SARS-CoV-2, Infectious disease
1. Introduction
COVID-19, the clinical disease state resulting from an infection with SARS-CoV-2 has impacted all sectors of society, including sport. Sport is an important part of society and has a key role to play in restoring normality after the initial wave of COVID-19. In the current environment, sport organisations and athletes are faced with complex decisions regarding the resumption of training and sporting activities, particularly in high performance/professional sport where athletes are effectively returning to work. Unique and intrinsic characteristics of high performance/professional sport include sharing facilities and equipment and close and prolonged physical contact between teammates and opponents. These factors coupled with maximal levels of physical exertion, which likely increase aerosol generation, are conducive to the spread of infectious diseases. The overriding priority for high performance/professional sport, must ensure that any return to activity does not endanger athletes, staff and the public. A proactive risk management strategy including protocols for a possible case of COVID-19 must be in place.
In situations of potential elevated local prevalence, early detection of COVID-19 is a crucial prerequisite for effective isolation, treatment and tracing of contacts. Testing of asymptomatic persons has been proposed in other higher risk situations as an effective means to mitigate workforce shortages such as in healthcare.1 Large scale testing and isolation of asymptomatic individuals has the potential to substantially reduce the prevalence of COVID-19. Mass polymerase chain reaction (PCR) screening of 300 people was undertaken in an Italian town and all positive cases were isolated. As a result, people with COVID-19 symptoms reduced by 90% in 10 days.2 Structuring testing for early identification of asymptomatic carriage as well as probable and confirmed cases is the best approach to mitigate increased risks associated with the high-performance sporting environment in areas of widespread local transmission.
In the absence of a national or international eradication strategy it must be accepted that the occurrence of sporadic cases appearing within the sporting environment is inevitable. The capacity to quickly identify and effectively manage sporadic cases to contain transmission should be the rate limiting step in determining if, when and how, high performance/professional sporting activity should proceed in the absence of COVID-19 eradication. Integration of devices that can be made available at the point of care (PoC) to facilitate the rapid diagnosis of COVID-19 can be a consideration.3
Point of care testing alone is not a sufficient risk mitigation strategy and careful planning of an outbreak monitoring program in collaboration with Local Public Health Authorities is required. Testing facilities and protocols need to be aligned to the resources available and skill sets of the attending physicians. A comprehensive approach to the management of a potential infectious disease outbreak in the high-performance sporting environment requires monitoring of the recent exposure and illness status of all individuals within the team. An assessment of the capacity to effectively track, diagnose and contain infection should be performed in conjunction with input from an expert infectious diseases physician.
2. Overview of test options
SARS-CoV-2 can be identified from clinical samples utilising molecular tests (e.g. PCR, next generation sequencing (NGS), Crisper Cas 12 and 13), serology testing, rapid antigen testing, and viral culture with electron microscopy. PCR is the recommended modality for diagnosing viral respiratory infections.4 Some public health laboratories use NGS on positive PCR samples to obtain the genetic sequence of SARS-CoV-2 to monitor patterns of transmission and mutations.5 PCR is considered ideal for identifying the presence of viral respiratory tract infections in most circumstances and is combined with clinical findings to provide a final diagnosis of COVID-19.6
PCR identifies the presence of SARS-CoV-2 genetic material in a clinical sample obtained from nasopharyngeal or throat swabs.4 It should be noted that any clinical sample can be PCR tested with appropriate extraction methods. PCR tests for respiratory viral infections have a high sensitivity and specificity.7 However, currently available data for SARS-CoV-2 detection from clinical samples by PCR testing suggests higher false negative rates than is usually observed for validated PCR tests of well characterised pathogens.8
Methods used for PCR testing for SARS-CoV-2 vary between laboratories in different countries or regions. Important differences to note include the sequence of targets chosen, the number of targets used for a given assay and Ribonucleic acid (RNA) purification and extraction methods. Commercial assays adopted by public health laboratories help to standardise the testing process and will be refined over time with emerging evidence about each testing kits performance.9 Sample collection, transport, storage and patient selection are other important factors to consider for test performance.
A positive PCR test for SAR-CoV-2 confirms the presence of the virus. In an asymptomatic individual, this can indicate either asymptomatic carriage or the individual in the incubation period, who may progress to develop COVID-19. Epidemiological reports from the Diamond Princess cruise ship demonstrated 50% of PCR positive cases were asymptomatic (n = 320 asymptomatic cases; symptomatic cases, n = 314; total, n = 634),10 with increasing reports of asymptomatic transmission globally.11, 12, 13, 14, 15 All patients testing positive for SARS CoV-2 are likely important sources of viral transmission with 44–55% of transmissions occurring in the asymptomatic stage16 and is likely to contribute to the higher reproduction number observed.
3. Brief description of the test and characteristics
In an outpatient setting, samples obtained for the detection of SARS-CoV-2 can either be collected from the nasopharynx and/or throat swabs, saliva17 or sputum. Although optimal sample type for diagnosis was initially debated, nasopharyngeal swabs are more sensitive than throat swabs to detect SARS-CoV-2.18 Comparison of various specimen types, with bronchoalveolar lavage fluid specimens showing the highest true positive rate (93%), followed by sputum (72%), nasal swabs (63%), fibro-bronchoscope brush biopsy (46%), pharyngeal swabs (32%), faeces (29%), and blood (1%) with urine specimens not testing positive by PCR.19 Nasopharyngeal swabs likely represent the best practical means of testing for COVID-19 in the outpatient setting.3
Concerns of sexual transmission from men given the combination of high levels of angiotensin converting enzyme 2 (ACE-2) expression in the testicles and persistent viral excretion observed from immune privileged tissues have been investigated. SARS-CoV-2 has not been observed in the semen of infected individuals or from testicular biopsy from patients at various stages of illness.20 There is no clinical indication for PCR testing of semen currently.
The probability of correctly obtaining a positive test result in an infected individual is estimated to be highest at the time of symptom onset and gradually declines over the subsequent 3–4 weeks.16, 18 However, in the pre-symptomatic period, it is estimated that PCR testing will be positive in the 2–3 days prior to the onset of symptoms.16 This window from exposure to first symptoms (the incubation period) presents as an opportunity for primary prevention for team members who would have been in close contact. Relying on symptomology delays intervention and allows the virus to be communicated to contacts of the infected individuals at the start of the infectious period. The effectiveness of this strategy will depend on the pre-test probability of the patient having the infection which is a function of the current community prevalence.
False negative rates have the potential to undermine any testing and screening program and a cautious approach should be utilised given reports of false negative results as high as 34%.8 Repeating the test 24 h later can reduce the risk of false negative18 and should be a consideration for assessing symptomatic patients when there is a high index of suspicion or if the test results are being relied on to remove an individual from isolation. Parallel screening methods can improve sensitivity of clinical testing. Therefore, a clinical review in conjunction with PCR testing of symptomatic individuals represents best practice. Known symptoms and vectors of transmissions should also be investigated at the time of PCR testing to reduce the reliance on a single test.
The probability of obtaining a false positive PCR test for SARS-COV-2 has not been described in the literature to date. Cross reactivity with other Deoxyribonucleic acid (DNA)/RNA present in the sample is unlikely given the design used for primers utilises a comprehensive genetic analysis to account for cross reactivity.21 While cross reactivity is not expected with commercial and public health laboratory assays, this possibility is yet to be formally established in the post marketing data. False positives in PCR testing for respiratory viruses in general are thought to be rare when testing is conducted in an ideal setting. The causes of the small number of false positives are likely a result of sample and test contamination as opposed to the intrinsic testing process.22 Given the extensive measures currently in place by the Federal, State and Territory Governments in Australia, the potential public health consequences and the overall low probability of a false positive, these risks are currently considered manageable.
Co-infection of SARS-CoV-2 is common and should be considered, particularly for pathogens with an approved therapy (e.g. influenza). An analysis of 116 SARS-CoV-2 positive samples in Wuhan, People's Republic of China, found 21% of samples were positive for at least one additional respiratory pathogen.23 The identification of co-infections can alter management decisions and identify individuals who may have a more complicated recovery phase.
4. Ancillary testing options
Next generation sequencing (NGS) platforms such as the MinIon are currently transitioning from the research only context to a potential PoC tool. NGS platforms (e.g. MinIon) can be used to generate whole genome sequences for detailed epidemiological analysis or perform nanopore targeted sequencing, and have been successfully implemented as part of a field laboratory set up to investigate Ebola and Zika virus outbreaks.24, 25, 26 Combining NGS with PCR has demonstrated improved diagnostic performance and reduced false negative rates compared to PCR alone.27
Serological tests aim to detect the presence of antibodies from blood samples. Laboratory based serological studies have demonstrated an increase in total antibodies followed by Immunoglobulin M (IgM) and Immunoglobulin G (IgG) respectively. The median seroconversion time following illness onset, as measured by a laboratory based enzyme-linked immunosorbent assay (ELISA) assay, results in a positive at 10 days for IgM and 12 days for IgG.28 30% of individuals who have recovered from a confirmed SARS CoV-2 infection (i.e. COVID-19) do not produce detectable levels of antibodies.29 The PoC lateral flow device intends to identify the same antibodies (i.e. IgM and IgG). Currently, there is no data available for accuracy of these kits. It is not expected that they will be capable of similar accuracy as the laboratory-based ELISA assays. Therefore, PoC lateral flow devices should not be relied upon to confirm a negative result in the acute phase of illness. The primary role for laboratory based serological testing is to understand patterns of immunity, and previous exposure.
Antigen tests have been previously used for influenza as a PoC test, however their implementation has been limited due to poor sensitivity and negative predictive value.30 It is unlikely that a rapid antigen test will provide any risk mitigation benefit in the high performance/professional sports context.
5. Lung imaging
Lung imaging has been used as an adjunct to molecular testing to identify COVID-19 cases. In the People's Republic of China and Italy, chest CT has been shown to have high sensitivity and moderate specificity in hospitalised patients.8, 31 Currently, it is unclear when radiographic changes occur during COVID-19 or the severity of the patient condition this applies to. In the hospital setting it has been observed that people infected with SARS-CoV-2 can present with radiological and clinical features suggestive of COVID-19 but an initial negative PCR test. The time to positive PCR test result in these patients was between 2 and 8 days.32 There has been reports of chest CT abnormalities that were identified preceding the onset of symptoms in adults33 and in paediatric patients.34
In order to minimise radiation exposure of athletes who are well enough to be managed as an outpatient, lung ultrasound should be considered as an alternative to chest CT. Lung ultrasound may have a role in diagnosis and clinical monitoring of unwell patients.35 In the high performance/professional sport environment, team physicians are often skilled in PoC ultrasound and there is no risk of radiation exposure to the athlete. It would be uncommon for high performance/professional athletes to have pre-existing lung pathology that would result in an abnormal lung ultrasound at baseline. This raises the pre-test probability that an irregular pleural line or the presence of B lines should trigger further investigation or suspicion of a false negative PCR test. Lung ultrasound has been used to help diagnose and monitor COVID-19 in an outpatient setting.36, 37, 38
6. Strategic approach to testing
PCR tests that can be used on portable and automated analysers are being developed and imported under emergency authorisation schemes. In a high performance/professional sport environment, these devices can enable broader testing with shorter turnaround times to identify asymptomatic carriage when there is potential for increased but occult outbreak risk. The current options available are summarised in Table 1 .
Table 1.
Potential near patient or point of care (PoC) molecular testing options for high performance sporting context.
 |
Two potential units that could be deployed practically in a high-performance sport context include the bioMérieux BIOFIRE® FILMARRAY® system and the Cepheid GeneXpert system. Both systems fit easily on a benchtop and integrate sample preparation, reverse transcription and quantitative polymerase chain reaction (qPCR) interpretation into a sealed cartridge-based system which takes 45 min to test result. The BioFire® FilmArray® assay currently tests for 21 targets from a single sample and will add SARS-CoV-2 later this year. The GeneXpert tests 1-2 targets per test. Although both systems are scalable, that allows testing of multiple samples at a time, the GeneXpert is a smaller integrated system that allows screening multiple samples for a single target. The accuracy of these tests is widely accepted with units being utilised in public hospitals, particularly in emergency departments.40
In the United States of America, systems need to be Clinical Laboratory Improvement Amendments (CLIA) waived or have a corresponding complexity of testing appropriate for the intended use. In Australia, registered physicians can use diagnostic tests that are listed on the Australian Register of Therapeutic Goods (ARTG) and hold approval as an in vitro diagnostic device (IVD). Approved tests can be carried out providing they are conducted in line with the manufacturer's description of the intended use and there is a documented policy conforming to the Guidelines for Point of Care Testing (PoCT) First Edition 2015.41 However, additional standards apply if the physician intends on billing Medicare (in Australia) for diagnostic services.
Infectious disease testing requires additional consideration for biosafety at each stage of testing. These will vary depending on the equipment and context chosen for testing. The manufacturer's documentation regarding biosafety must always be followed. If testing is conducted outside of a laboratory setting, it is recommended that any manual handling of samples should be conducted inside of an appropriately sized, negative pressure flexible film isolator compliant with clause 10.7.2 of Australian/New Zealand Standard TM, Safety in laboratories “Part 3: Microbiological safety and containment (AS/NZS 2243.3:2010)42” in conjunction with appropriate personal protective equipment.
7. Case definitions for high performance sport
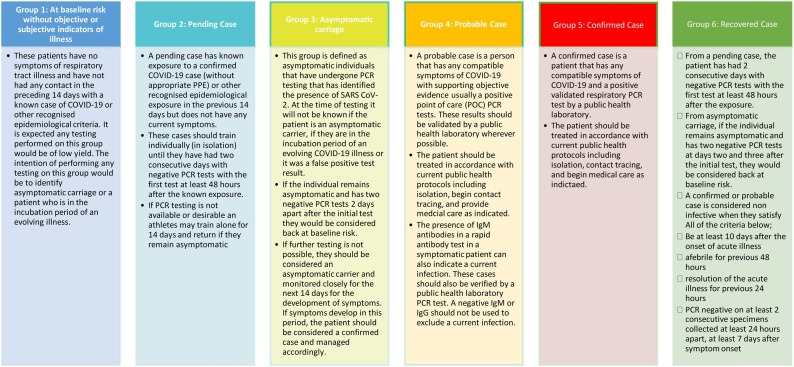
All persons within the high-performance sport environment will fit into one of six case definitions proposed for PCR screening protocol in high performance sport (Fig. 1 ).
Fig. 1.
Proposed case definitions for a PCR screening protocol in high performance sport.
Case definitions are mouldable to the context and resources available. Given limited resources available during travel with sporting teams (internationally and domestic), it may not be possible to perform sufficient testing in a timely manner to fully understand the transmission dynamics occurring, even in symptomatic individuals. In the absence of sufficient testing capacity, the team physician should consider modifying the case definitions (listed in Fig. 1) to assume the presence of the illness persisting until it has been reasonably excluded by time in isolation or a suitable sequence of tests. It is important to keep a documented record of modified case definitions in each context.
The aim of utilising COVID-19 tests combined with symptoms and epidemiological exposure is to stratify all members of the team into categories according to their risk assessment. Management strategies should prioritise the health status of the individual, wider team, and the community. Risk assessment of all team members is a dynamic process and will change as a situation unfolds. All athletes and personnel need to be re-evaluated upon identifying a positive COVID-19 case.
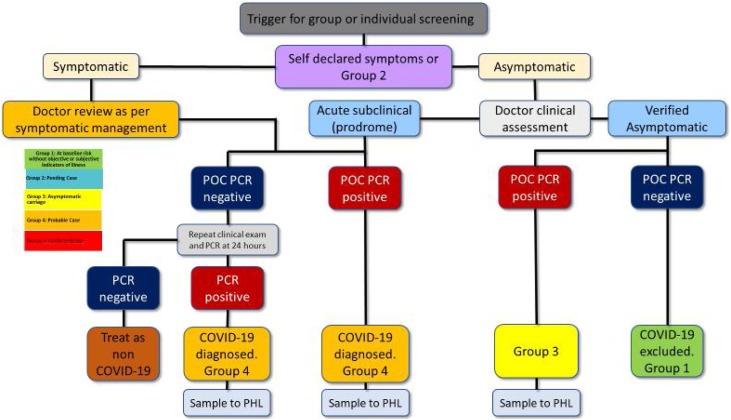
Case definitions listed in Fig. 1 can be used as a template for approaching management and testing decisions in a high-performance sport environment with sufficient testing capacity. Fig. 2 illustrates a proposed decision-making tree of the team physician responsible for testing. Risk identification and mitigation is illustrated in Table 2 . Depending on resources available and the current community prevalence, PCR could be utilised as an entry tool into the high performance/professional sporting environment, as a surveillance tool to randomly sample individuals or be triggered by high risk exposures.
Fig. 2.
Management algorithm for the approach to asymptomatic or minimally symptomatic PCR positive patients. POC PCR, point of care polymerase chain reaction; PHL, public health laboratory.
Table 2.
Risk identification and mitigation.
| Outcome | Risk | Risk if there is no PoC test | Risk if PoC test is implemented | Risk Difference | Relevant Risk Mitigation Strategy |
|---|---|---|---|---|---|
| False negative COVID-19 | False negative is not able to be confirmed by the public health lab | 100% of COVID cases will continue to interact with the community actively spreading the infection as a test would not have occurred. | Up to 30% of cases tested could be a false negative | Risk is lower by completing PoC testing | Patient should be isolated and retested next day if there are clinical concerns for a false negative |
| False negative that can be corrected by the public health laboratory | Could not occur as the public laboratory does not need to confirm its own result | The chances of this occurring are unknown but expected to be small with units listed in Table 1. | This should be small given the similar automated tests are used in the public hospital setting | Consider sending a proportion of negative samples to a public health laboratory for confirmation. | |
| False positive COVID-19 | If a positive is identified and the public health lab is unable to confirm this | Could not occur as no confirmation test is done | Unnecessary diagnosis, further investigation and isolation Unnecessary intensity of clinical monitoring Stigma attached to having COVID-19 Initiating therapeutics assuming positive result |
False positive rates are expected to be small Treatment is currently only symptomatic treatment and isolation, no pharmacological risk currently |
Informed consent within the team prior to doing test Careful maintenance and cleaning of working space to limit contamination May require additional support from team doctor/mental health professional |
| False positive that can be corrected by the public health laboratory | Could not occur | Consider probability that a low positive sample degrades prior to reference laboratory testing Confusion and distrust in PoC test |
Discordant PoC to public health laboratory results could occur. The chances of this occurring are unknown | Reference laboratory testing is a risk mitigation strategy Review for sources of contamination in testing workflow Complete additional negative quality controls Option of re-testing same individuals 24 hours later |
8. Conclusion
Doctors involved in high performance sport may be required to diagnose and manage possible outbreaks as a result of the COVID-19 pandemic. All symptomatic individuals should be tested for COVID-19. Testing of asymptomatic individuals may be considered in circumstances where community prevalence is increased, where there has been recent high-risk exposure or as a part of a surveillance project. Currently PCR is the only way of identifying asymptomatic or pre-symptomatic carriers who are responsible for approximately 44%–55% of transmissions of COVID-19. Some of this risk can be mitigated by implementing PCR based screening protocols in conjunction with well-structured case definitions detailing how to approach the management of asymptomatic PCR positive patients. Setting up a PCR testing process for high performance sport needs to consider many factors beyond the simple purchasing of equipment required for testing. Factors including biosafety, minimising sources of error in the chosen workflow, quality control and regulatory issues have been reviewed.
Practical implications
-
•
PCR testing is the only means of identifying individuals who are asymptomatic carriers of SARS-CoV-2, however the false negative rates in this period is unclear.
-
•
Asymptomatic patients likely account for 44–55% of SARS-CoV-2 transmissions and has significant implications for community transmission.
-
•
High performance/professional sport may look to allocate resources to dedicated PCR testing as a part of medical service provision to its athletes for improved time to test result or expanded indications for testing.
-
•
Any expanded testing framework should prioritise circumstances of elevated pre-test probability. Regular surveillance of well people may be useful in larger groups.
-
•
Implementing PoC PCR should only occur after due consideration of the local regulatory frameworks; capacity to manage identified cases and reporting obligations, capacity to implement appropriate quality standards, and biosafety at all stages of testing.
-
•
Portable PCR testing in sport is potentially useful for teams when time to test result is crucial or for teams travelling internationally where access to testing is limited.
-
•
Portable PCR testing in sport is likely only suitable for a subset of high performance/professional sport considering the initial and ongoing cost, requirement for a doctor with an appropriate skill set and access to sufficient personal protective equipment (PPE).
Financial support
Dr Mathew Mooney was supported by University of Canberra and Australian Institute of Sport as an industry funded PhD scholar.
Author contribution
All authors contributed to all items in the ICMJE contributorship guidelines.
Acknowledgements
The authors wish to acknowledge the contributions of Dr Nicholas Coatsworth for his technical advice and contributions to manuscript development.
Footnotes
Rapid response papers and have not undergone the full peer review process.
References
- 1.Black JRM, Bailey C, Przewrocka J, Dijkstra KK, Swanton C. COVID-19: the case for health-care worker screening to prevent hospital transmission. The Lancet. [DOI] [PMC free article] [PubMed]
- 2.Day M. Covid-19: identifying and isolating asymptomatic people helped eliminate virus in Italian village. BMJ. 2020;368:m1165. doi: 10.1136/bmj.m1165. [DOI] [PubMed] [Google Scholar]
- 3.Tang Y.-W., Schmitz J.E., Persing D.H., Stratton C.W. The Laboratory Diagnosis of COVID-19 infection: current issues and challenges. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.00512-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hadaya J., Schumm M., Livingston E.H. Testing individuals for coronavirus disease 2019 (COVID-19) JAMA. 2020 doi: 10.1001/jama.2020.5388. [DOI] [PubMed] [Google Scholar]
- 5.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. The Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, Kang H, Liu X, Tong Z. Combination of RT-qPCR testing and clinical features for diagnosis of COVID-19 facilitates management of SARS-CoV-2 outbreak. J Med Virol. [DOI] [PMC free article] [PubMed]
- 7.Anderson T.P., Werno A.M., Barratt K., Mahagamasekera P., Murdoch D.R., Jennings L.C. Comparison of four multiplex PCR assays for the detection of viral pathogens in respiratory specimens. J Virol Methods. 2013;191(2):118–121. doi: 10.1016/j.jviromet.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ai T, Yang Z, Hou H, Zhan C, Chen C, Lv W, et al. Correlation of Chest CT and RT-PCR Testing in Coronavirus Disease 2019 (COVID-19) in China: A Report of 1014 Cases. Radiology.0(0):200642. [DOI] [PMC free article] [PubMed]
- 9.Chan J.F.-W., Yip C.C.-Y., To K.K.-W., Tang T.H.-C., Wong S.C.-Y., Leung K.-H. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-polymerase chain reaction assay validated in vitro and with clinical specimens. J Clin Microbiol. 2020 doi: 10.1128/JCM.00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mizumoto K., Kagaya K., Zarebski A., Chowell G. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Eurosurveillance. 2020;25(10):2000180. doi: 10.2807/1560-7917.ES.2020.25.10.2000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bai Y., Yao L., Wei T., Tian F., Jin D.-Y., Chen L. Presumed Asymptomatic Carrier Transmission of COVID-19. JAMA. 2020 doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rothe C., Schunk M., Sothmann P., Bretzel G., Froeschl G., Wallrauch C. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N Engl J Med. 2020;382(10):970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tong Z.-D., Tang A., Li K., Li P., Wang H., Yi J. Potential presymptomatic transmission of SARS-CoV-2, Zhejiang Province China, 2020. Emerging Infect Dis. 2020;26(5) doi: 10.3201/eid2605.200198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye F., Xu S., Rong Z., Xu R., Liu X., Deng P. Delivery of infection from asymptomatic carriers of COVID-19 in a familial cluster. Int J Infect Dis. 2020 doi: 10.1016/j.ijid.2020.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimball A. Asymptomatic and presymptomatic SARS-CoV-2 infections in residents of a long-term care skilled nursing facility—King County, Washington, March 2020. MMWR Morbidity and mortality weekly report. 2020:69. doi: 10.15585/mmwr.mm6913e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He X., Lau E.H.Y., Wu P., Deng X., Wang J., Hao X. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nature Med. 2020 doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- 17.Azzi L., Carcano G., Gianfagna F., Grossi P., Gasperina D.D., Genoni A. SALIVA IS A RELIABLE TOOL TO DETECT SARS-CoV-2. J Infect. 2020 doi: 10.1016/j.jinf.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wikramaratna P., Paton R.S., Ghafari M., Lourenco J. Estimating false-negative detection rate of SARS-CoV-2 by RT-PCR. medRxiv. 2020 doi: 10.2807/1560-7917.ES.2020.25.50.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang W., Xu Y., Gao R., Lu R., Han K., Wu G. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020 doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song C., Wang Y., Li W., Hu B., Chen G., Xia P. Detection of 2019 novel coronavirus in semen and testicular biopsy specimen of COVID-19 patients. medRxiv. 2020 2020.03.31.20042333. [Google Scholar]
- 21.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25(3):2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Curran T., McCaughey C., Ellis J., Mitchell S.J., Feeney S.A., Watt A.P. False-positive PCR results linked to administration of seasonal influenza vaccine. J Med Microbiol. 2012;61(3):332–338. doi: 10.1099/jmm.0.036178-0. [DOI] [PubMed] [Google Scholar]
- 23.Kim D., Quinn J., Pinsky B., Shah N.H., Brown I. Rates of co-infection between SARS-CoV-2 and other respiratory pathogens. JAMA. 2020 doi: 10.1001/jama.2020.6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quick J., Grubaugh N.D., Pullan S.T., Claro I.M., Smith A.D., Gangavarapu K. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nature Protocols. 2017;12(6):1261. doi: 10.1038/nprot.2017.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoenen T., Groseth A., Rosenke K., Fischer R.J., Hoenen A., Judson S.D. Nanopore sequencing as a rapidly deployable Ebola outbreak tool. Emerging Infect Dis. 2016;22(2):331. doi: 10.3201/eid2202.151796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quick J., Loman N.J., Duraffour S., Simpson J.T., Severi E., Cowley L. Real-time, portable genome sequencing for Ebola surveillance. Nature. 2016;530(7589):228–232. doi: 10.1038/nature16996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang M., Fu A., Hu B., Tong Y., Liu R., Gu J. Nanopore target sequencing for accurate and comprehensive detection of SARS-CoV-2 and other respiratory viruses. medRxiv. 2020 doi: 10.1002/smll.202002169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lou B., Li T., Zheng S., Su Y., Li Z., Liu W. Serology characteristics of SARS-CoV-2 infection since the exposure and post symptoms onset. medRxiv. 2020 doi: 10.1183/13993003.00763-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu F., Wang A., Liu M., Wang Q., Chen J., Xia S. 2020. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. [Google Scholar]
- 30.Green D.A., StGeorge K. Rapid antigen tests for influenza: rationale and significance of the FDA reclassification. J Clin Microbiol. 2018;56(10):e00711–e00718. doi: 10.1128/JCM.00711-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caruso D., Zerunian M., Polici M., Pucciarelli F., Polidori T., Rucci C. Chest CT features of COVID-19 in Rome, Italy. Radiology. 2012;2020:37. doi: 10.1148/radiol.2020201237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie X, Zhong Z, Zhao W, Zheng C, Wang F, Liu J. Chest CT for Typical 2019-nCoV Pneumonia: Relationship to Negative RT-PCR Testing. Radiology.0(0):200343. [DOI] [PMC free article] [PubMed]
- 33.Lin C., Ding Y., Xie B., Sun Z., Li X., Chen Z. Asymptomatic novel coronavirus pneumonia patient outside Wuhan: The value of CT images in the course of the disease. Clin Imaging. 2020;63:7–9. doi: 10.1016/j.clinimag.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu X., Zhang L., Du H., Zhang J., Li Y.Y., Qu J. SARS-CoV-2 infection in children. N Engl J Med. 2020;382(17):1663–1665. doi: 10.1056/NEJMc2005073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buonsenso D, Pata D, Chiaretti A. COVID-19 outbreak: less stethoscope, more ultrasound. The Lancet Respiratory Medicine. [DOI] [PMC free article] [PubMed]
- 36.Buonsenso D., Piano A., Raffaelli F., Bonadia N., Donati K.D.G., Franceschi F. novel coronavirus disease-19 pnemoniae: a case report and potential applications during COVID-19 outbreak. Eur Rev Med Pharmacol Sci. 2020;24:2776–2780. doi: 10.26355/eurrev_202003_20549. [DOI] [PubMed] [Google Scholar]
- 37.Buonsenso D., Pata D., Chiaretti A. COVID-19 outbreak: less stethoscope, more ultrasound. Lancet Respir Med. 2020 doi: 10.1016/S2213-2600(20)30120-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang Y., Wang S., Liu Y., Zhang Y., Zheng C., Zheng Y. 2020. A preliminary study on the ultrasonic manifestations of peripulmonary lesions of non-critical novel coronavirus pneumonia (COVID-19) Available at SSRN 3544750. [Google Scholar]
- 40.Wahrenbrock M.G., Matushek S., Boonlayangoor S., Tesic V., Beavis K.G., Charnot-Katsikas A. Comparison of cepheid Xpert Flu/RSV XC and BioFire film array for detection of Influenza A, Influenza B, and Respiratory Syncytial Virus. J Clin Microbiol. 2016;54(7):1902–1903. doi: 10.1128/JCM.00084-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Council NPAA . First ed. 2015. Guidelines for Point of Care Testing. In: Health Do, editor. [Google Scholar]
- 42.Safety in laboratories . Standards Australia; Standards New Zealand; Sydney, N.S.W: 2010. Part 3: Microbiological safety and containment. [Google Scholar]