Abstract
Congenital myasthenic syndromes (CMS) are caused by mutations in molecules expressed at the neuromuscular junction. We report clinical, structural, ultrastructural, and electrophysiologic features of 4 CMS patients with 6 heteroallelic variants in AGRN, encoding agrin. One was a 7.9-kb deletion involving the N-terminal laminin–binding domain. Another, c.4744G>A — at the last nucleotide of exon 26 — caused skipping of exon 26. Four missense mutations (p.S1180L, p.R1509W, p.G1675S, and p.Y1877D) expressed in conditioned media decreased AChR clusters in C2C12 myotubes. The agrin-enhanced phosphorylation of MuSK was markedly attenuated by p.Y1877D in the LG3 domain and moderately attenuated by p.R1509W in the LG1 domain but not by the other 2 mutations. The p.S1180L mutation in the SEA domain facilitated degradation of secreted agrin. The p.G1675S mutation in the LG2 domain attenuated anchoring of agrin to the sarcolemma by compromising its binding to heparin. Anchoring of agrin with p.R1509W in the LG1 domain was similarly attenuated. Mutations of agrin affect AChR clustering by enhancing agrin degradation or by suppressing MuSK phosphorylation and/or by compromising anchoring of agrin to the sarcolemma of the neuromuscular junction.
Keywords: Genetics
Keywords: Neuromuscular disease
Missense variants in the SEA, LG1, LG2, and LG3 domains in congenital myasthenic syndrome affect clustering of the acetylcholine receptors in a domain-specific manner.
Introduction
Congenital myasthenic syndromes (CMS) are heterogeneous disorders in which neuromuscular transmission is impaired by defects in molecules expressed at the neuromuscular junction (NMJ). The clinical features include fatigable muscle weakness, decreased muscle mass, joint contractures, and minor facial anomalies (1). Defining the genetic basis of a CMS may lead to rational therapy. Defects in more than 30 genes are known to cause CMS. These genes encode subunits of the acetylcholine receptor (AChR), structural proteins for scaffolding AChR clusters, signaling molecules driving AChR clustering, presynaptic molecules involved in release and resynthesis of ACh, synaptic and postsynaptic molecules that facilitate the signal transduction at the NMJ, and glycosylation enzymes (2, 3).
Agrin encoded by AGRN is a heparan sulfate proteoglycan essential for differentiation and maintenance of the NMJ in skeletal muscle. AGRN has 3 alternative exons. Exon A/y is composed of a 12-nucleotide (nt) exon 32, which confers binding to heparin, whereas exons B/z are composed of a 24-nt exon 36 and a 33-nt exon 37, which confers AChR clustering activity (4). Among exons B/z, inclusion of exon 36 confers more potent AChR clustering activity than exon 37. Spinal motor neurons make neural agrin that includes exon 32, as well as exons 36 and/or 37, and transport it to motor nerve terminals. Neural agrin is then released into the synaptic basal lamina, where it binds to the N-terminal domain of the low-density lipoprotein receptor-related protein 4 (LRP4) anchored in the postsynaptic membrane (5, 6). The agrin-LRP4 complex binds to the postsynaptic receptor tyrosine kinase MuSK and activates its kinase domain. This enhances MuSK phosphorylation in concert with a cytoplasmic adaptor protein Dok-7 and promotes clustering of LRP4 and MuSK on the postsynaptic membrane (7). Activated MuSK, in concert with other postsynaptic proteins, acts on rapsyn to concentrate AChR in the postsynaptic membrane, increases synapse-specific gene expression, and promotes postsynaptic differentiation (8, 9). The same signaling pathway is also essential for maintaining the structure of the adult NMJ (10). Neural agrin is critical for synaptic differentiation because agrin-deficient mice die at birth from respiratory failure due to aberrant innervation and lack of postsynaptic differentiation (11, 12). Agrin binds to laminins via its amino (N)-terminal domain (13) and to α-dystroglycan (14) and LRP4 (15) via its carboxy-terminal domains. The carboxy-terminal end of agrin contains 3 laminin G-like (LG) domains; 2 are required for binding to α-dystroglycan, and the last LG domain is required for binding to LRP4 (16). Ten variants of AGRN have been reported in severe forms of CMS with partial characterization of effects of the different variants (17–23).
We here report 4 CMS patients harboring 6 different variants in AGRN. A 7.9-kbp deletion eliminating the laminin-binding domain abrogates binding to laminin. A G-to-A variant at the 3′ end of exon 26 (c.4744G>A) results in skipping of exon 26. Other missense variants p.S1180L, p.R1509W, p.G1675S, and p.Y1877D attenuate agrin-induced AChR clustering in C2C12 myotubes. We also show that defective AChR clustering is caused not only by defects in MuSK phosphorylation, but also by enhanced agrin degradation and/or compromised anchoring of agrin to the sarcolemma and at the NMJ.
Results
Characteristics of the CMS patients.
Patient (Pt.) 1, a 44-year-old woman, was born after a normal pregnancy. She appeared normal at birth and had normal motor development during infancy. However, she developed a waddling gait by age 4 and scoliosis of the spine by age 6. She never ran well and could not keep up with her peers in physical activities. Elacromyography (EMG) studies and an edrophonium test at the age of 9 indicated a defect of neuromuscular transmission. Thymectomy at age 10, several courses of plasmapheresis, varying doses of pyridostigmine, and prednisone therapy between ages 15 and 17 years were ineffective. At age 16, repetitive stimulation of the femoral nerve at 2 Hz revealed a 30% decremental response of the fourth compared with the first evoked muscle action potential in the rectus femoris muscle. At age 17, she had proximal greater than distal limb muscle weakness. The arm elevation time was limited to 45 seconds, and she could not rise from sitting without support. Subsequently, therapy with ephedrine and 3,4-diaminopyridine improved her strength, but her weakness increased with her menses.
Pt. 2, currently 45 years old, was born after 37 weeks of gestation. He appeared normal in the neonatal period but had a waddling gait at age 10 months and could not run. After the age of 2 years, he had increasing difficulty climbing stairs. At age 10, he could walk 1 mile. By age 13, he had mild scoliosis. At age 17, he could walk only one-half block before having to rest. At this age, repetitive stimulation of the femoral nerve at 2 Hz revealed a 35% decremental response in the rectus femoris muscle. Between ages 18 and 37, he had mild bilateral eyelid ptosis and facial weakness, with moderately severe limb muscle weakness, worse proximally than distally. Since age 37, therapy with 6 mg albuterol sulfate per day greatly improved his symptoms.
Pt. 3 is currently 49 years old. During infancy, she required gastrostomy for feeding and a tracheostomy for respiratory support. She began walking at 1 year of age but could not keep up with her peers. By age 10, she had scoliosis that required spine surgery and became wheelchair dependent in her teens. A thigh muscle biopsy for a suspected mitochondrial myopathy was negative. Treatment with pyridostigmine worsened the patient’s weakness, but albuterol improved her strength and endurance. Pregnancy at age 38 increased her symptoms, and in the third trimester, she required ventilatory assistance. Her weakness improved postpartum, but later, it worsened again; she had to use a ventilator for 16 hours per day. Therapy with albuterol stabilized her condition, and 3,4-diaminopyridine after age 41years was of additional benefit. At age 44, repetitive stimulation of the median nerve at 2 Hz revealed a 22% decremental response in the abductor pollicis brevis muscle. She has no similarly affected relatives.
Pt. 4, currently 8 years of age, was born full-term after an uncomplicated pregnancy to third-degree consanguineous parents. He had a weak cry at birth, was in the intensive care unit for 2 months, and was in the hospital for 7 months. During infancy, he needed a tracheostomy and gastrostomy. He walked with support at age 4 years. He has mild intellectual disability, eyelid ptosis, restricted eye movements, mild scoliosis, and generalized hypotonia with proximal and distal limb muscle weakness, as well as episodes of respiratory insufficiency. Repetitive nerve stimulation revealed a 15%–50% decremental response in different muscles. A similarly affected younger sister died at 1 year of age of respiratory failure. The genetic results of this patient were previously reported (24).
Histochemical observations for patient tissues.
Conventional histochemical studies, available in Pts. 2 and 3, revealed no abnormality in the muscle fibers or connective tissue elements. However, the length of the synaptic contacts in transverse sections of frozen muscle was reduced to 6.7 μm and 7.3 μm, respectively (n = 6). The corresponding value for 69 endplates (EPs) of 3 control subjects was 21.3 μm (P < 0.001, 2-tailed Student t test).
Electron microscopy (EM) of patient tissues.
These were done on muscle biopsies of Pts. 1, 2, and 3 and documented in Table 1 and Figure 1. Simple inspection revealed no abnormality in the nerve terminals. In Pts. 1 and 2, the postsynaptic regions were well preserved, but in Pt. 3, they were highly degenerate, displaying numerous disintegrating junctional folds, with globular residues accumulating in a widened synaptic space (Figure 1, A and B). One of 21 nerve terminals observed in Pt. 3 was capped by a degenerating Schwann cell (Figure 1A).
Table 1. Morphometric analysis of single EP regions and α-bungarotoxin binding sites per EP.
Figure 1. Ultrastructural observations in patient (Pt.) 2 and 3.
(A and B) Degeneration and destruction of the junctional folds in Pt. 3. Globular residues of the degenerate folds (asterisk) accumulate in the widened synaptic space. Schwann cell (SC) in A is replaced by debris. (C and D) Localization of AChR on the junctional folds with peroxidase-labeled α-bungarotoxin (black) in a control subject (C), and in Pt. 2 (D). Note attenuated expression of AChR on the incomplete junctional folds at patient endplates. Scale bars: 1 μm.
Morphometry of single EPs.
Performed in Pts. 1, 2, 3, this revealed significantly decreased postsynaptic areas in Pts. 1 and 2 and of the postsynaptic membrane density in Pt. 3. The number of α-bungarotoxin (α-bgt) binding sites per EP was significantly reduced in all 3 patients (Table 1). The expression of AChR on the junctional folds, evaluated in Pts. 1 and 2, was attenuated (Figure 1D) compared with control EPs (Figure 1C).
Microelectrode studies of neuromuscular transmission.
These were done in Pts. 1, 2, and 3. Compared with normal controls, the amplitude of the miniature EP potential (MEPP) was normal in Pt. 1 but was reduced to 54% in Pt. 2, and to 20% in Pt. 3. The amplitude of the miniature EP current (MEPC), measured in Pts. 1 and 2, was mildly reduced in Pt. 2. The quantal content of the EP potential (EPP) in Pt. 3 was decreased to 62% of the corresponding control value (Table 2).
Table 2. Microelectrode studies of neuromuscular transmission.
Variants identified in 4 patients.
Figure 2B shows domains of AGRN expressed at the NMJ, the 6 identified variants in Pts. 1–4, and 10 previously reported mutations. Table 3 indicates mutations in each allele of each patient. Pt. 1 has a 7.9-kbp deletion spanning exons 1 and 2 (Figure 2A). Because the deletion eliminates the AGRN promoter and the translational start site, the mutant agrin is unlikely to be expressed. The rest of the identified variants involved single nucleotides predicting p.S1180L, p.R1509W, p.G1582R, p.G1675S, and p.Y1877D.
Figure 2. Schematic of agrin domains and mutations in CMS.
(A) Genomic structure of the 5′ end of the human AGRN gene with g.950,567_958,457del identified in Pt. 1 are drawn to scale. Transcriptional and translational start sites are indicated. Genomic coordinates are according to GRCh37/hg19. Note that the deletion spans the AGRN promoter region, the transcriptional/translational start sites, and exons 1 and 2, where the secretion signal and the N-terminal agrin domain (NtA) are encoded. (B) Domains according to O00468 (UniProtKB) are drawn to scale. Mutations identified in the current study (red dots) and previous reports (blue dots) according to NM_198576.4 (RefSeq) are indicated to scale. SS, secretion signal peptide; FS, follistatin-like domain; LE, laminin EGF–like domain; S/T, serine/threonine-rich domain; SEA, a sperm protein, enterokinase, and agrin domain; EG, EGF-like domain; and LG, laminin G–like domain. A/y and B/z stand for alternative exons that are included in neuronal isoform (NM_001305275.2). Horizontal bars indicate positions where the indicated binding partners bind. (C) Long and short forms of agrin fragments used in the current studies are shown at the bottom. p.S1180L was introduced into the long form, whereas p.R1509W, p.G1675S, and p.Y1877D were introduced into the short form.
Table 3. Identified mutations in agrin.
c.4744G>A predicting p.G1582R causes aberrant skipping of exon 26.
c.4744G>A in Pt. 2 is at the last nucleotide of exon 26. We previously predicted that 97% of mutations at the last nucleotide of an exon cause aberrant splicing (25). We examined the effect of this mutation on splicing by introducing a minigene spanning exons 24–27 of the human AGRN into HEK293 cells. We found that c.4744G>A caused skipping of 230-nt exon 26 (Figure 3). MaxEntScan::5′ss (26), SD Score (25), and spliceAI (27) indeed predicted that c.4744G>A markedly weakens recognition of the 5′ splice site. Skipping of exon 26 caused a frameshift and introduced a premature termination codon in exon 27 (p.A1506SfsX6), and the transcript is likely degraded by nonsense-mediated mRNA decay (NMD).
Figure 3. c.4744G>A in Pt.
2 at the last nucleotide ofAGRNexon 26 causes aberrant skipping of exon 26. (A) Schematic of pcDNA3.1-AGRN minigene carrying WT sequence or c.4744G>A mutation. The MaxEntScan::5′ss scores (26) and the SD scores (25) of WT and mutant 5′ splice sites are indicated. A probability of donor site loss predicted by SpliceAI (27) is also indicated. RT-PCR of WT and mutant minigene constructs shows that c.4744G>A causes skipping of exon 26 in HeLa cells. Skipping of exon 26 generates a premature termination codon in exon 27 and makes the AGRN transcript susceptible to nonsense-mediated mRNA decay.
Four missense mutations attenuated AChR clustering in C2C12 myotubes.
We next examined the effects of the 4 missense mutations, p.S1180L, p.R1509W, p.G1675S, and p.Y1877D, on AChR clustering in C2C12 myotubes by adding WT and mutant agrin fragments to the culture medium. HEK293 cells transfected with WT and mutant agrin cDNAs secreted agrin fragments in culture medium to similar levels (Figure 4A). We then added the conditioned medium to C2C12 myotubes and found that all mutations reduced the areas of AChR clusters in C2C12 myotubes (Figure 4B and Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.132023DS1). Thus, all 4 missense variants affected AChR-clustering activities, which can account for the reduced numbers of AChRs at the NMJ in Pts. 1, 2, and 3 (Table 1).
Figure 4. Preparation of conditioned medium containing agrin-mycAP and their effects on AChR clustering on C2C12 myotubes.
(A) Secretion of agrin fragments into the conditioned medium of HEK293 cells transfected with AGRN cDNAs. Proteins in the conditioned medium were precipitated with TCA and were immunoblotted with an anti-myc antibody to detect agrin fragments in the conditioned medium (left). Control proteins were isolated from untransfected HEK293 cells. Coomassie brilliant blue staining showing all proteins in the conditioned medium (right panel). Note that no mutation affected agrin expression in the medium. (B) C2C12 myotubes were cultured with control conditioned medium (Control) or conditioned medium containing WT or mutant agrin-mycAP for 18 hours. AChR was visualized with Alexa Fluor 594–conjugated AChR. Scale bar: 20 μm. Blinded morphometric analysis of the AChR signal areas with Metamorph is shown in the right panel. The AChR signal area was normalized by the myotube area and also by the ratio of WT. Mean ± SD are indicated (n = 3 images each in 3 wells). Note that all AGRN mutants reduced AChR clustering in C2C12 myotubes. P < 0.05 by 1-way ANOVA. P < 0.05 by post hoc Tukey test is indicated by a, b, and c.
p.R1509W and p.Y1877D affect agrin-induced MuSK phosphorylation.
We next asked whether these 4 mutations affect induction of MuSK activation. HEK293 cells were transfected with cDNAs with WT LRP4, WT MuSK, and an ATF2-luciferase (ATF2-luc) construct to evaluate MuSK phosphorylation (28). Application of conditioned medium harboring WT or mutant agrin to the transfected HEK293 cells revealed that p.Y1877D, but neither p.S1180L, p.R1509W, nor p.G1675S, reduced ATF2 promoter activity (Figure 5A). We next examined the effects of agrin mutants on agrin-induced MuSK phosphorylation in C2C12 myotubes. Two hours after adding conditioned medium, including WT or mutant agrin, 2 mutations, p.R1509W and p.Y1877D, failed to induce MuSK phosphorylation, whereas the other 2 mutants, p.S1180L and p.G1675S, did induce it (Figure 5B). The discordant effects of p.R1509W in HEK293 and C2C12 cells are likely because p.R1509 is located in a binding domain for α-dystroglycan — and because α-dystroglycan is physiologically clustered with LPR4/MuSK receptors in C2C12 myotubes (29) but not in HEK293 cells.
Figure 5. Effects of agrin mutants on MuSK activation and MuSK phosphorylation.
(A) ATF2-luciferase reporter assay of HEK293 cells. HEK293 cells were transfected with ATF2-luc reporter and phRL-TK Renilla luciferase reporter plasmids with cDNAs for MuSK and LRP4. HEK293 cells were incubated with control conditioned medium (Control) or conditioned medium containing WT or mutant agrin-mycAP for 24 hours. Relative luciferase activity (RLA) was normalized for RLA without MuSK, LRP4, or agrin. Mean ± SD are indicated (n = 4 wells). Note that p.Y1877D reduced ATF-luc activity. One-way ANOVA followed by post hoc Tukey test. P < 0.05 is indicated by a single letter representing each group. (B) MuSK phosphorylation assay of C2C12 myotubes. C2C12 cells were added with control conditioned medium (Control), or conditioned medium containing WT or mutant agrin-mycAP for 2 hours. MuSK phosphorylation was detected by immunoprecipitation with an anti-MuSK antibody followed by immunoblotting with an anti-phosphotyrosine (pTyr) antibody. Quantification of phosphorylated MuSK is shown in the right panel. The ratio of phosphorylated to total MuSK was normalized to that of WT agrin. Mean ± SD (n = 3 independent experiments) are indicated. Note that p.R1509W and p.Y1877D reduced MuSK phosphorylation. One-way ANOVA followed by post hoc Tukey test. P < 0.05 is indicated by a, b, and c.
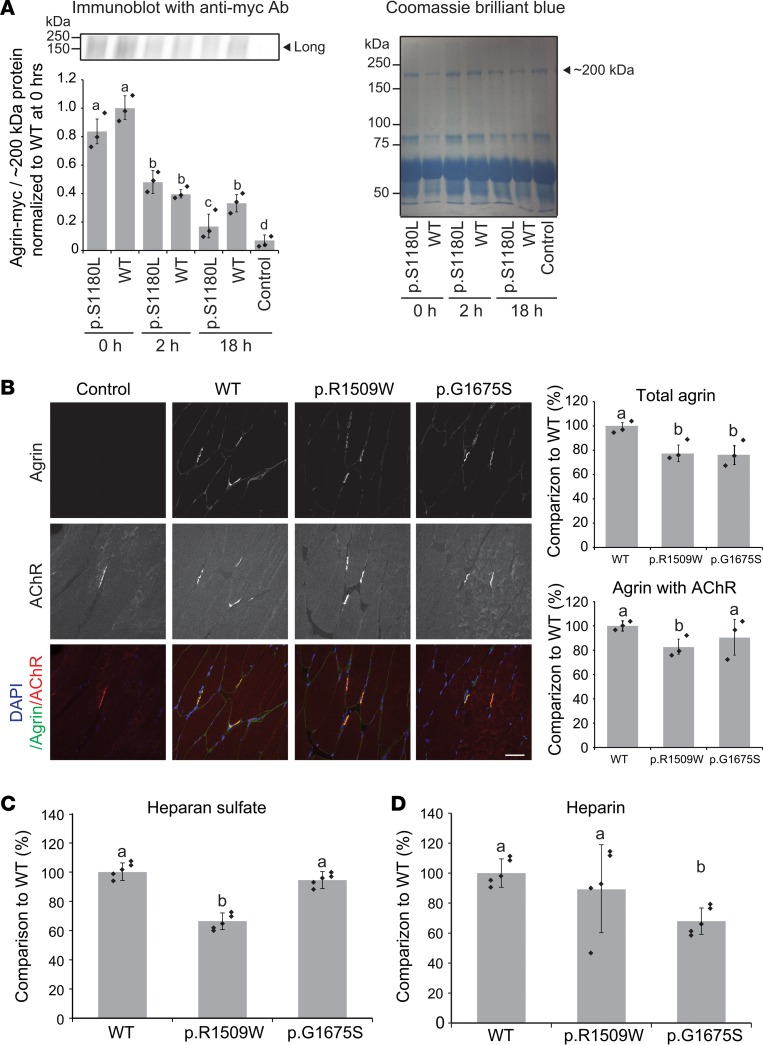
p.S1180L enhances degradation of agrin when added to the culture medium of C2C12 myotubes.
Xi and colleagues previously reported that p.L1176P induced instability of agrin protein and impaired formation of AChR clusters (22). We examined the effect of p.S1180L, which is 4 codons C-terminal to p.L1176P, on the stability of agrin. WT and p.S1180L agrin–myc-tag/alkaline phosphatase (agrin-mycAP) fragments were expressed in HEK293 cells. Purified agrin-mycAP fragments were added to the culture medium of C2C12 myotubes. The amount of agrin-mycAP fragments at 0, 2, and 18 hours were quantified by immunoblotting against myc. This showed that, at 18 hours after adding agrin, the amount of p.S1180L agrin-mycAP was lower than that of WT agrin-mycAP (Figure 6A). Protein degradations of agrin-mycAP fragments at 0, 2, 6, and 18 hours were analyzed by measuring AP enzymatic activity in the medium. The quantitative analysis also showed that the amount of p.S1180L agrin-mycAP was lower than that of WT agrin-mycAP at 6 hours and 18 hours after adding agrin (Supplemental Figure 1B). Thus, S1180L destabilized secreted agrin.
Figure 6. Degradation assay of p.S1180L and binding assays of p.G1509W and p.G1675S to a muscle section and to heparan sulfate/heparin.
(A) Degradation assay of WT and p.S1180L-mutant agrin-mycAP. Purified WT or p.S1180L agrin-mycAP was added to the culture medium of C2C12 myotubes, and the amount of agrin-mycAP was quantified at 0, 2, and 18 hours. The amount of agrin-mycAP was normalized for that of a Coomassie-stained ~200-kDa band and also for the ratio of WT at 0 hours. Mean ± SD (n = 3 independent experiments) are indicated. One-way ANOVA followed by post hoc Tukey test. P < 0.05 is indicated by a, b, c, and d. (B) Overlay of purified WT, p.G1509W, and p.G1675S agrin-mycAP on a section of mouse tibialis anterior muscle. WT agrin was bound to the sarcolemma and especially to the neuromuscular junction (NMJ). p.G1509W and p.G1675S reduced binding to the sarcolemma. p.G1509W also reduced binding to the NMJ. Mean ± SD (n = 3 independent experiments) are indicated in right panels. One-way ANOVA followed by post hoc Tukey test. P < 0.05 is indicated by a and b. (C and D) Plate-binding assay of purified WT, p.G1509W, and p.G1675S agrin-mycAP to heparan sulfate (C) and heparin (D). A total of 80 μL of 50 ng/μL WT and mutant agrin-mycAP in PBS were incubated for 3 hours at room temperature. p.G1509W reduced binding to heparan sulfate and p.G1675S reduced binding to heparin. Mean ± SD (n = 3 wells) are indicated. One-way ANOVA followed by post hoc Tukey test; P < 0.05.
p.G1509W and p.G1675S reduce anchoring of agrin to sarcolemma and the NMJ by compromising binding to glycosaminoglycans.
Agrin binds to extracellular matrix proteins, including proteoglycans, at the NMJ. p.G1509W and p.G1675S are located in the LG1 and LG2 domains, respectively, and both are essential for binding to α-dystroglycan (Figure 2). LG2, where p.G1675S is located, also binds to heparin. We thus examined anchoring of mutant agrin to a section of mouse tibialis anterior (TA) muscle, as well as to purified heparan sulfate and heparin. WT agrin was mostly anchored to the AChR at the NMJ of a muscle section (Figure 6B). Quantification of total agrin signals showed that both p.G1509W and p.G1675S impaired anchoring of agrin to the TA muscle, especially to sarcolemma. Similarly, anchoring to the NMJ was impaired in both mutants, but statistical significance was observed only with p.R1509W. Next, WT and mutant agrin fragments were overlaid on a plastic plate coated either with heparan-sulfate or heparin. The plate-binding assays showed that p.R1509W and p.G1675S reduced binding of agrin to heparan sulfate (Figure 6C and Supplemental Figure 1C) and heparin (Figure 6D and Supplemental Figure 1D), respectively. Both heparan sulfate and heparin are glycosaminoglycans (GAGs). Compared with heparan sulfate, heparin has a higher content of glucosamine (GlcN) N-sulfates and iduronic acid (IdoA). Taken together, p.G1509W and p.G1675S affect binding of agrin to sulfated glycosaminoglycans, which are essential for accumulating agrin at the NMJ.
Discussion
We identified 6 AGRN variants in 4 CMS patients and functionally characterized 1 variant at the RNA level and 4 variants at the protein level. We did not functionally characterize a 7.9-kb deletion spanning AGRN exons 1 and 2 (g.950,567_958,457del) (Figure 2A). This deletion eliminates the AGRN promoter region and the Kozak consensus sequence (5′-GCGCCATGG-3′ at positions 955,548–955,556 according to GRCh37/hg19), which makes it unlikely that the mutant allele generates agrin. However, even if it did, the mutant agrin would lack the signal peptide and the laminin-binding domain (Figure 2A). The p.G76S and p.N105I variants identified in the laminin-binding domain in CMS will hinder formation of AChR clusters in C2C12 myotubes (19), and the laminin-binding domain-lacking agrin is unlikely to be functional.
c.4744G>A at the last nucleotide of AGRN exon 26 predicted p.G1582R. However, it caused skipping of exon 26 and generated a premature termination codon in exon 27 (p.A1506SfsX6) (Figure 3). Exon skipping by c.4744G>A is in line with our previous prediction that 96.8% of single nucleotide variants at the last nucleotide of an exon cause aberrant splicing (25, 30).
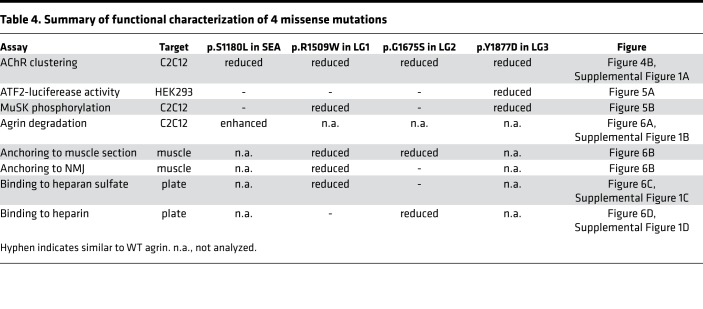
Functional characterization of the 4 missense variants (p.S1180L, p.R1509W, p.G1675S, and p.Y1877D) are summarized in Table 4. When added to the culture medium, each variant failed to induce AChR clustering in C2C12 myotubes (Figure 4B and Supplemental Figure 1A). However, each mutation affected MuSK activation (Figure 5A), MuSK phosphorylation (Figure 5B), and the degradation of agrin (Figure 6A and Supplemental Figure 1B), as well as its anchoring to sarcolemma and the NMJ (Figure 6B) and its binding to heparan sulfate (Figure 6C and Supplemental Figure 1C) and heparin (Figure 6D and Supplemental Figure 1D). Below, we discuss each variant in each domain.
Table 4. Summary of functional characterization of 4 missense mutations.
The SEA domain was named after the 3 proteins (sperm protein, enterokinase, and agrin) in which the domain was identified. An agrin fragment lacking the SEA domain and its upstream domains induces AChR clusters as efficiently as full-length agrin (31) and is able to rescue the perinatal lethal phenotype of Agrn-deficient mice (31). The physiological role of the SEA domain was initially unknown. We also used the SEA-lacking agrin fragment as a short form of agrin in our current study (Figure 2C). Chemical mutagenesis generated a mouse line with a phenylalanine-to-serine substitution at codon 1061 in the SEA domain of mouse agrin, which was equivalent to p.P1175S of human agrin (32). The mutant mouse showed decreased AChR clustering at the NMJ. In addition, the mutant agrin was less glycosylated and less efficiently secreted. Recently, p.L1176P was reported in a CMS patient (22). This variant is located 1 codon C-terminal to the mutation in mouse. This variant impairs AChR clustering in C2C12 myotubes and accelerates degradation of the mutant agrin by increasing its sensitivity to neurotrypsin and other proteases. The p.S1180L residue is positioned 4 codons C-terminal to p.L1176P. We therefore examined the effect of p.S1180L on agrin degradation and found that p.S1180L indeed accelerated agrin degradation (Figure 6A and Supplemental Figure 1B). p.S1180L affected AChR clustering (Figure 4B and Supplemental Figure 1A) but had no effect on MuSK phosphorylation (Figure 5B) in C2C12 myotubes. The AChR clustering assay was performed at 18 hours after adding agrin, whereas the MuSK phosphorylation assay was performed at 2 hours after adding agrin. We also showed that accelerated degradation of p.S1180L was significant at 18 hours but not at 2 hours (Figure 6A and Supplemental Figure 1B). The difference in the incubation time likely accounts for the difference in the AChR clustering and MuSK phosphorylation assays. To summarize, the SEA domain is important for glycosylation and secretion of agrin, and it also protects agrin from degradation by proteases like neurotrypsin.
p.R1509W was the first mutation detected in the first LG domain required for binding to α-dystroglycan (Figure 2B). We found this variant reduced AChR clustering (Figure 4B and Supplemental Figure 1A) and MuSK phosphorylation (Figure 5B) in C2C12 myotubes. However, p.R1509W had no effect on MuSK activation evaluated by the ATF-luc activity in HEK293 cells (Figure 5A). p.R1509W may affect binding of agrin to α-dystroglycan or some other molecules on the cell surface of C2C12 myotubes that might not be sufficiently expressed on HEK293 cells. We also showed that p.R1509W impaired agrin binding to the NMJ and the sarcolemma (Figure 6B), as well as to heparan sulfate (Figure 6C and Supplemental Figure 1C). Thus, R1509 in the first LG domain is essential for anchoring agrin to the NMJ using α-dystroglycan or other muscle-specific cell surface molecule(s).
The second LG domain also binds to α-dystroglycan, along with the first LG domain (33). To date, 5 missense mutations (p.G1675S, ref. 21; p.R1698C, ref. 22; p.G1709R, ref. 17; p.V1727F, ref. 18; and p.A1768P, ref. 20) have been reported in the second LG domain in CMS (Figure 2B). These mutations are dispersed on the predicted LG2 structure generated from the perlecan LG3 domain (33). Three of the 5 mutations have been functionally characterized. The p.R1698C variant renders the secreted mutant agrin unstable and reduces AChR clustering in C2C12 myotubes (22). p.G1709A destabilizes endogenous NMJ when the mutant agrin was injected into rat soleus muscle (17). p.V1727F markedly attenuates MuSK phosphorylation and AChR clustering in C2C12 myotubes (18). p.G1675S was previously reported in CMS without functional characterization (21). We showed that p.G1675S mutation reduces AChR clustering in C2C12 cells (Figure 4B and Supplemental Figure 1A) but had no effect on MuSK activation in HEK293 cells (Figure 5A) or MuSK phosphorylation in C2C12 myotubes (Figure 5B). We also showed that p.G1675S reduced binding of agrin to sarcolemma (Figure 6B) and to heparin (Figure 6D and Supplemental Figure 1D). p.G1675S is likely to impair agrin binding to α-dystroglycan, but low cell surface expression of α-dystroglycan on HEK293 cells masks this impairment, as we discussed with p.R1509W. However, spared MuSK phosphorylation in C2C12 cells of p.G1675S (Figure 5B) cannot be accounted for by this hypothesis. Overexpression of Dok-7 in Agrn-deficient mice compensates for lack of agrin during embryogenesis, but the NMJs rapidly disappear after birth, although MuSK phosphorylation is still driven by Dok-7 (10). A likely explanation is that agrin activates a yet-unidentified pathway or drives an unidentified mechanism to induce the NMJ formation without phosphorylating MuSK. p.G1675S might have compromised the unidentified pathway/mechanism.
AGRN mRNA derived from the spinal motor neurons includes B/z exons composed of alternative exons 36 and 37. Inclusion of exon 36, exon 37, or both encodes 8, 11, or 19 codons, respectively, between S1884 and E1885 according to NM_198576.4 (4). The alternative codons form a loop in the third LG domain (15). p.G1871R was previously reported in a CMS patient but was not functionally characterized (19). p.Y1877D is located at 7 codons N-terminal to the inserted B/z codons (Figure 2B), which confers binding to LRP4 (15). p.Y1877D indeed compromised AChR clustering (Figure 4B and Supplemental Figure 1A) and MuSK phosphorylation (Figure 5B) in C2C12 myotubes, as well as MuSK activation in HEK293 cells transfected with LRP4 and MuSK (Figure 5A). Analysis of p.G1871R corroborates the notion that the third LG domain with B/z exons is essential for inducing AChR clusters by binding to LRP4 and activating MuSK.
Methods
In vitro electrophysiology studies.
External intercostal muscle specimens were obtained from origin to insertion from Pts. 1, 2, and 3. The MEPP and EPP amplitudes and the number of quanta released by nerve impulse (m) were measured as previously reported (34–36). The number of readily releasable quanta (n), estimates of the probability of quantal release (p) (37), and the EP AChR content were determined as previously described (35).
Structural studies.
EPs were localized for EM by an established method (38). Peroxidase-labeled α-bgt was used for the ultrastructural localization of AChR in Pts. 1 and 2 (39). Cholinesterase reactivity at the EPs was visualized in cryosections with nonspecific esterase. The number of AChRs per EP was measured with [125I]α‑bgt, as described (35).
Genetic analysis.
Genomic DNA was isolated from blood from the patients and their parents by standard methods. In Pt. 1, a missense mutation was identified by Medical Neurogenetics LLC and a deletion mutation by Prevention Genetics. In Pt. 2, whole genome sequencing was performed by Macrogen. In Pts. 3 and 4, Sanger sequencing was done in our laboratory. In Pt. 2, RNA was isolated from EP-enriched skeletal muscle specimens by standard methods and converted to cDNA. Sanger sequencing was performed with PCR primers to sequence exons and flanking noncoding regions for genomic DNA and primers with exon-to-exon inclusion for the cDNA. Agrin nucleotides are numbered according to NM_198576.4.
Plasmids.
To analyze splicing of exon 26, we constructed a human AGRN minigene spanning exons 24–27 (positions 984,247–985,417 of chromosome 1, according to GRCh37/hg19) into the pcDNA3.1(+) mammalian expression vector (Addgene) at HindIII and EcoRI sites by amplifying a ~1.2-kb fragment from human genomic DNA extracted from HeLa cell, using a proofreading DNA polymerase (PrimeSTAR polymerase, Takara) (Figure 3). c.4744G>A in Pt. 2 was introduced into the minigene using the QuikChange Site-Directed Mutagenesis Kit (Agilent). Lack of PCR artifacts was verified by sequencing the entire insert.
To generate human long and short forms of agrin-mycAP that retained potency to facilitate AChR clustering, we cloned codons 1130–2068 and codons 1247–2068 of human AGRN cDNA (NP_001292204.1) into pAPtag-5 (GenHunter) at the SnaBI and BglII sites, respectively. The pAPtag-5 contained coding sequences for Igk-originated signal peptide upstream to the insert and the myc-tag/alkaline phosphatase downstream. p.S1180L was introduced into the long form of agrin-mycAP by the QuikChange Site-Directed Mutagenesis kit (Stratagene). Similarly, p. R1509W, p.G1675S, and p.Y1877D were introduced into the short form of agrin-mycAP (Figure 2C). Lack of PCR artifacts was verified by sequencing the entire inserts.
Full-length human LRP4 cDNA (Open Biosystems) was cloned into the EcoRI site of the pcDNA3.1(+) mammalian expression vector. The mouse Musk cDNA in pExpress-1 was purchased from Open Biosystems. ATF2-luc (40) and phRL-TK Renilla luciferase vector (Promega) were used for the luciferase reporter assay.
Reverse transcription PCR (RT-PCR) for splicing analysis of exon 26.
Total RNA was extracted at 48 hours after transfection using Trizol (Thermo Fisher Scientific) following DNase I (Qiagen) treatment. cDNA was synthesized with an oligo-dT(18) primer (Thermo Fisher Scientific) and ReverTra Ace reverse transcriptase (Toyobo). PCR was performed using GoTaq Master Mix (Promega) using primers 5′-TGGATGGTGAGACCCCTGTT-3′ (forward) and 5′-GGCTCTTCTCATCGGCACAG-3′ (reverse). PCR products were resolved on a 2% agarose gel.
Preparation of conditioned media containing secreted agrin-mycAP proteins.
HEK293 cells were cultured in the DMEM supplemented with 10% FCS.
To corroborate that agrin-mycAP is sufficiently secreted in conditioned medium (Figure 4A), WT and mutant agrin-mycAP cDNAs were transfected into HEK293 cells in DMEM with 2% horse serum using Lipofectamine 2000 transfection reagent (Thermo Fischer Scientific). Proteins were precipitated with 20% (w/v) trichloroacetic acid (TCA), followed by 6-acetone washes, and were subjected to immunoblotting with anti-Myc tag antibody (9E10) (Abcam).
To treat C2C12 myotubes with agrin-mycAP for quantifying AChR clustering (Figure 4B and Supplemental Figure 1A), MuSK activation (Figure 5A), and MuSK phosphorylation (Figure 5B), WT and mutant agrin-mycAP cDNAs were similarly transfected into HEK293 cells in DMEM with 2% horse serum. Agrin-mycAP in the conditioned medium was concentrated ~100-fold using an Amicon Ultra-15 filter (Ultracel-100K, MilliporeSigma, UFC910024). The concentrated conditioned medium was resolved by SDS-PAGE and transferred to a polyvinylidene fluoride membrane (Immobilon-P, MilliporeSigma). WT and mutant agrin-mycAP were immunostained with the mouse monoclonal anti–Myc tag antibody (9E10) (1:500, sc-40, Santa Cruz Biotechnology Inc.). We quantified signal intensities of WT and mutant agrin-mycAP to match the concentrations of these agrin-mycAP proteins. We performed immunoblotting again to confirm that the concentrations of WT and mutant agrin-mycAP in the concentrated conditioned medium became similar. The concentrated conditioned medium containing WT and mutant agrin-mycAP were sequentially diluted and were added to C2C12 myotubes to determine a dilution ratio at which AChR clustering activities of WT and mutant agrin-mycAP were within an assayable range. We mostly diluted the conditioned medium to 1:200 for Figure 4B, Figure 5A, and Figure 5B. For Supplemental Figure 1A, we measured AP activity agrin-mycAP in the conditioned medium using LabAssay ALP (Wako) to estimate the concentration of agrin-mycAP in the medium. As a standard, we measured AP activities of a dilutional series of purified agrin-mycAP, the concentration of which was measured by the Pierce 660 nm Protein Assay Kit (Pierce Biotechnology). To quantify degradation of agrin (Figure 6A and Supplemental Figure 1B), anchoring of agrin to a muscle section (Figure 6B), and binding of agrin to heparan sulfate and heparin (Figure 6, C and D and Supplemental Figure 1, C and D), WT and mutant agrin-mycAP cDNAs were transfected into HEK293 cells in serum-free medium. Agrin-mycAP in the conditioned medium of transfected HEK293 cells was concentrated ~100-fold using an Amicon Ultra-15 filter (Ultracel-100K). Agrin-mycAP was then purified using the c-myc–tagged Protein Mild Purification Kit ver. 2 (MBL) and was quantified using the Pierce 660 nm Protein Assay Kit.
Luciferase assays.
HEK293 cells were cultured in the serum-free DMEM and transfected with ATF2-luc and phRL-TK, along with the mouse MuSK and human LRP4 cDNAs using Lipofectamine 2000 transfection reagent (Thermo Fischer Scientific). Cells were cultured for 24 hours in conditioned medium containing WT or mutant agrin-mycAP in a 96-well plate. Cells were lysed with the passive lysis buffer (Promega) and assayed for the luciferase activity using the Dual Luciferase system (Promega). Each experiment was performed in triplicated wells.
MuSK phosphorylation assay and AChR clustering assay with C2C12 myotubes.
C2C12 myoblasts were seeded on a plate coated with collagen I (BD Biosciences) and differentiated in DMEM supplemented with 2% horse serum for 5 days. The differentiated myotubes were treated with conditioned medium containing WT or mutant agrin-mycAP for 2 hours to induce MuSK phosphorylation and for 12 hours to quantify AChR clustering. For the AChR clustering assay, cells were stained with 10 mg/mL Alexa Fluor 594–conjugated α-bgt (1:100, Invitrogen) to label AChR and were fixed with 2% paraformaldehyde. Fluorescence images were observed under an Olympus XL71 fluorescence microscope. The signal areas and intensities of AChR clusters were analyzed with MetaMorph software (Molecular Devices). AChR clusters with the axis length less than 4 μm were excluded from the analysis.
Western blotting.
Agrin-treated C2C12 myotubes were lysed with a buffer containing 50 mM HEPES (pH 7.0) (Dojindo), 150 mM NaCl (191-01665, Wako), 10% glycerol (073-05215, Wako), 1% TritonX-100 (T8787, MilliporeSigma), 1.5 mM MgCl2 (135-01665, Wako), 1 mM EGTA (E4378, MilliporeSigma), 100 mM NaF (S7920, MilliporeSigma), 10 mM sodium pyrophosphate (A17546, Alfa Aesar), 1 μg/μL aprotinin (A1153, MilliporeSigma), 1 μg/μL leupeptin (L2884, MilliporeSigma), 1 μg/μL pepstatin A (P4265, MilliporeSigma), 1 mM PMSF (P7626, MilliporeSigma), and 1 mM sodium orthovanadate (S6508, MilliporeSigma). Cell lysates were subjected to coimmunoprecipitation using 1 μg of anti-MuSK (ABS549, MilliporeSigma) attached to protein G Sepharose beads (GE Healthcare). Total or precipitated proteins were dissolved in 1× Laemmli buffer, separated on a 10% or 7.5% SDS-polyacrylamide gel, and transferred to a polyvinylidene fluoride membrane (Immobilon-P, MilliporeSigma). Membranes were washed in Tris-buffered saline containing 0.05% Tween 20 (TBS-T) and blocked for 1 hour at room temperature in TBS-T with 3% BSA. The membranes were incubated overnight at 4°C either with mouse monoclonal anti–Myc tag (9E10) (1:500, sc-40, Santa Cruz Biotechnology Inc.), anti-phosphotyrosine (1:1000, 4G10, Upstate), anti-MuSK (1:500, ABS549, MilliporeSigma), or anti-β-actin (1:200, sc-47778, Santa Cruz Biotechnology Inc.) antibody. The membranes were washed 3 times for 10 minutes with TBS-T and incubated with secondary goat anti–mouse (1:6000, 7076S, Cell signaling Technology) or donkey anti–rabbit (NA9340V, GE Healthcare) IgG antibody conjugated to horseradish peroxidase (HRP) for 1 hour at room temperature. The blots were detected with Amersham ECL Western blotting detection reagents (GE Healthcare) and quantified with the ImageJ (NIH) program.
Degradation assay of agrin-mycAP in culture medium of C2C12 myotubes.
C2C12 myoblasts were seeded on a plate coated with collagen I (BD Biosciences) and differentiated in DMEM supplemented with 2% horse serum for 5 days. The differentiated myotubes were added with DMEM supplemented with 2% horse serum containing purified 50 ng/mL WT or mutant agrin-mycAP protein for 0, 2, or 18 hours, and the whole medium was harvested by centrifugation at 430 g for 5 minutes at 4°C to remove cell debris. For an assay in Figure 6A, proteins in the conditioned media were precipitated with 20% (w/v) TCA followed by 6-X acetone washes and dissolved in 1× Laemmli buffer. Samples were separated on a 10% or 7.5% SDS-polyacrylamide gel and transferred to a polyvinylidene fluoride membrane (Immobilon-P, MilliporeSigma). Agrin-mycAP was detected by Western blotting; then, the membrane was stained with Coomassie brilliant blue (CBB G-250, 038-17932, Wako). The intensity of immunostained agrin-mycAP at 0, 2, and 18 hours was normalized for the intensity of a Coomassie-stained ~200 kDa band and also for the intensity of WT agrin-mycAP at 0 hours, normalized for the intensity of a Coomassie-stained ~200 kDa band. For an assay in Supplemental Figure 1, differentiated C2C12 myotubes were added with DMEM, supplemented with 2% horse serum containing purified 100 ng/mL WT or mutant agrin-mycAP protein for 0, 2, 6, or 18 hours, and the whole medium was harvested by centrifugation at 430 g for 5 minutes at 4°C to remove cell debris. AP activity in the conditioned medium was measured using LabAssay ALP and was normalized to that at 0 hours.
Overlay of agrin-mycAP on a mouse muscle section.
Frozen sections of the mouse TA muscle were fixed with acetone for 10 minutes at –20°C, washed with PBS several times, and then covered with PBS containing 2% goat serum for 60 minutes. The muscle sections were incubated with purified WT or mutant agrin-mycAP proteins (1:50 of 50 ng/μL agrin protein) in PBS overnight at 4°C in a humidified chamber. After repeated washes with PBS, the sections were incubated with mouse monoclonal anti–Myc tag (9E10) (1:500, sc-40, Santa Cruz Biotechnology Inc.) in PBS overnight at 4°C in a humidified chamber. After this, the sections were incubated with the goat anti–mouse Alexa Fluor 488 secondary antibody (1:100, Thermo Fisher Scientific, A11001) and Alexa Fluor 594–conjugated α-bgt (1:100, Invitrogen, B13423). Residual antibodies were removed with repeated washes in PBS. Finally, the sections were coverslipped with VectaShield mounting medium containing 1.5 μg/mL DAPI (Vector Laboratories) and were visualized using confocal lase microscope system A1Rsi confocal microscope (Nikon). Intensity of agrin signals overlapped with α-bgt signals were also quantified by 2 blinded observers using the MetaMorph software (Molecular Devices).
Binding of agrin-mycAP to heparan sulfate and heparin.
The Immuno plate (Nunc) was coated with 0.15 μg of purified heparan sulfate (Sigma-Aldrich, H7640) or heparin (Sigma-Aldrich, H3149) at 4°C overnight and incubated with a blocking buffer including 1% BSA (Sigma-Aldrich, A9647) in PBS at room temperature for 1 hour. Then, 80 μL of WT and mutant agrin-mycAP in PBS were incubated for 3 hours at room temperature and were washed twice with PBS. Bound AP activity was measured using LabAssay ALP (Wako).
Statistics.
Statistical analyses, including the unpaired Student’s t test, 1-way or 2-way ANOVA, and post hoc Tukey test were performed using the SPSS ver. 23 (IBM Corp). P values less than 0.05 were considered to be statistically significant. The correction was made for multiple comparisons as indicated.
Study approval.
All human studies were approved by the IRB of the Mayo Clinic (no. 334-90) and the Nagoya University Graduate School of Medicine (no. 2007-0598-3).
Author contributions
BO, KO, and AGE conceived the study. BO performed in vitro and cellular studies. MN and SF performed splicing analysis. KO and AGE supervised the project. XS, DS, VB, MAT, LB, AAA, and UY contributed to data collection. DS and XMS contributed to data analysis. BO, KO, and AGE wrote the paper, and all authors confirmed the integrity of the contents.
Supplementary Material
Acknowledgments
We thank Keiko Itano and Harumi Kodama for expert technical assistance. We thank Curt Storlie for advice on correcting statistical analyses for multiple comparisons in Tables 1 and 2. This study was supported by NIH research grant NS109491 from the NINDS to AGE; and by grants-in-aid from JSPS, MHLW, AMED, and NCNP of Japan to BO and KO.
Version 1. 04/09/2020
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(7):e132023.https://doi.org/10.1172/jci.insight.132023.
Contributor Information
Bisei Ohkawara, Email: biseiohkawara@med.nagoya-u.ac.jp.
Duygu Selcen, Email: Selcen.Duygu@mayo.edu.
Mohammad Nazim, Email: m.nazim23@gmail.com.
Vera Bril, Email: vera.bril@uhn.on.ca.
Mark A. Tarnopolsky, Email: tarnopol@mcmaster.ca.
Lauren Brady, Email: bradyla@HHSC.CA.
Sae Fukami, Email: fukami.sae@gmail.com.
Anthony A. Amato, Email: aamato@bwh.harvard.edu.
Uluc Yis, Email: ulyis@yahoo.com.
Kinji Ohno, Email: ohnok@med.nagoya-u.ac.jp.
Andrew G. Engel, Email: age@mayo.edu.
References
- 1.Engel AG, Shen XM, Selcen D, Sine SM. Congenital myasthenic syndromes: pathogenesis, diagnosis, and treatment. Lancet Neurol. 2015;14(4):420–434. doi: 10.1016/S1474-4422(14)70201-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Engel AG. Congenital myasthenic syndromes in 2012. Curr Neurol Neurosci Rep. 2012;12(1):92–101. doi: 10.1007/s11910-011-0234-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Engel AG, Ohno K, Sine SM. Congenital myasthenic syndromes: recent advances. Arch Neurol. 1999;56(2):163–167. doi: 10.1001/archneur.56.2.163. [DOI] [PubMed] [Google Scholar]
- 4.Ohno K, et al. Splicing regulation and dysregulation of cholinergic genes expressed at the neuromuscular junction. J Neurochem. 2017;142 Suppl 2:64–72. doi: 10.1111/jnc.13954. [DOI] [PubMed] [Google Scholar]
- 5.Kim N, et al. Lrp4 is a receptor for Agrin and forms a complex with MuSK. Cell. 2008;135(2):334–342. doi: 10.1016/j.cell.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang B, Luo S, Wang Q, Suzuki T, Xiong WC, Mei L. LRP4 serves as a coreceptor of agrin. Neuron. 2008;60(2):285–297. doi: 10.1016/j.neuron.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohno K, Ohkawara B, Ito M. Agrin-LRP4-MuSK signaling as a therapeutic target for myasthenia gravis and other neuromuscular disorders. Expert Opin Ther Targets. 2017;21(10):949–958. doi: 10.1080/14728222.2017.1369960. [DOI] [PubMed] [Google Scholar]
- 8.McMahan UJ, et al. Agrin isoforms and their role in synaptogenesis. Curr Opin Cell Biol. 1992;4(5):869–874. doi: 10.1016/0955-0674(92)90113-Q. [DOI] [PubMed] [Google Scholar]
- 9.Burden SJ, Yumoto N, Zhang W. The role of MuSK in synapse formation and neuromuscular disease. Cold Spring Harb Perspect Biol. 2013;5(5):a009167. doi: 10.1101/cshperspect.a009167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tezuka T, et al. The MuSK activator agrin has a separate role essential for postnatal maintenance of neuromuscular synapses. Proc Natl Acad Sci USA. 2014;111(46):16556–16561. doi: 10.1073/pnas.1408409111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin W, Burgess RW, Dominguez B, Pfaff SL, Sanes JR, Lee KF. Distinct roles of nerve and muscle in postsynaptic differentiation of the neuromuscular synapse. Nature. 2001;410(6832):1057–1064. doi: 10.1038/35074025. [DOI] [PubMed] [Google Scholar]
- 12.Gautam M, et al. Defective neuromuscular synaptogenesis in agrin-deficient mutant mice. Cell. 1996;85(4):525–535. doi: 10.1016/S0092-8674(00)81253-2. [DOI] [PubMed] [Google Scholar]
- 13.Denzer AJ, Brandenberger R, Gesemann M, Chiquet M, Ruegg MA. Agrin binds to the nerve-muscle basal lamina via laminin. J Cell Biol. 1997;137(3):671–683. doi: 10.1083/jcb.137.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gee SH, Montanaro F, Lindenbaum MH, Carbonetto S. Dystroglycan-alpha, a dystrophin-associated glycoprotein, is a functional agrin receptor. Cell. 1994;77(5):675–686. doi: 10.1016/0092-8674(94)90052-3. [DOI] [PubMed] [Google Scholar]
- 15.Zong Y, et al. Structural basis of agrin-LRP4-MuSK signaling. Genes Dev. 2012;26(3):247–258. doi: 10.1101/gad.180885.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scotton P, et al. Activation of muscle-specific receptor tyrosine kinase and binding to dystroglycan are regulated by alternative mRNA splicing of agrin. J Biol Chem. 2006;281(48):36835–36845. doi: 10.1074/jbc.M607887200. [DOI] [PubMed] [Google Scholar]
- 17.Huzé C, et al. Identification of an agrin mutation that causes congenital myasthenia and affects synapse function. Am J Hum Genet. 2009;85(2):155–167. doi: 10.1016/j.ajhg.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maselli RA, et al. LG2 agrin mutation causing severe congenital myasthenic syndrome mimics functional characteristics of non-neural (z-) agrin. Hum Genet. 2012;131(7):1123–1135. doi: 10.1007/s00439-011-1132-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicole S, et al. Agrin mutations lead to a congenital myasthenic syndrome with distal muscle weakness and atrophy. Brain. 2014;137(Pt 9):2429–2443. doi: 10.1093/brain/awu160. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, et al. A Novel AGRN Mutation Leads to Congenital Myasthenic Syndrome Only Affecting Limb-girdle Muscle. Chin Med J. 2017;130(19):2279–2282. doi: 10.4103/0366-6999.215332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karakaya M, Ceyhan-Birsoy O, Beggs AH, Topaloglu H. A Novel Missense Variant in the AGRN Gene; Congenital Myasthenic Syndrome Presenting With Head Drop. J Clin Neuromuscul Dis. 2017;18(3):147–151. doi: 10.1097/CND.0000000000000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xi J, et al. Novel SEA and LG2 Agrin mutations causing congenital Myasthenic syndrome. Orphanet J Rare Dis. 2017;12(1):182. doi: 10.1186/s13023-017-0732-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rudell JB, Maselli RA, Yarov-Yarovoy V, Ferns MJ. Pathogenic effects of agrin V1727F mutation are isoform specific and decrease its expression and affinity for HSPGs and LRP4. Hum Mol Genet. 2019;28(16):2648–2658. doi: 10.1093/hmg/ddz081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yiş U, et al. Genetic Landscape of Congenital Myasthenic Syndromes From Turkey: Novel Mutations and Clinical Insights. J Child Neurol. 2017;32(8):759–765. doi: 10.1177/0883073817705252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sahashi K, et al. In vitro and in silico analysis reveals an efficient algorithm to predict the splicing consequences of mutations at the 5’ splice sites. Nucleic Acids Res. 2007;35(18):5995–6003. doi: 10.1093/nar/gkm647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11(2-3):377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 27.Jaganathan K, et al. Predicting Splicing from Primary Sequence with Deep Learning. Cell. 2019;176(3):535–548.e24. doi: 10.1016/j.cell.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 28.Ohkawara B, et al. LRP4 third β-propeller domain mutations cause novel congenital myasthenia by compromising agrin-mediated MuSK signaling in a position-specific manner. Hum Mol Genet. 2014;23(7):1856–1868. doi: 10.1093/hmg/ddt578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singhal N, Martin PT. Role of extracellular matrix proteins and their receptors in the development of the vertebrate neuromuscular junction. Dev Neurobiol. 2011;71(11):982–1005. doi: 10.1002/dneu.20953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohno K, Takeda JI, Masuda A. Rules and tools to predict the splicing effects of exonic and intronic mutations. Wiley Interdiscip Rev RNA. 2018;9(1):e1451. doi: 10.1002/wrna.1451. [DOI] [PubMed] [Google Scholar]
- 31.Gesemann M, Denzer AJ, Ruegg MA. Acetylcholine receptor-aggregating activity of agrin isoforms and mapping of the active site. J Cell Biol. 1995;128(4):625–636. doi: 10.1083/jcb.128.4.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bogdanik LP, Burgess RW. A valid mouse model of AGRIN-associated congenital myasthenic syndrome. Hum Mol Genet. 2011;20(23):4617–4633. doi: 10.1093/hmg/ddr396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dempsey CE, Bigotti MG, Adams JC, Brancaccio A. Analysis of α-Dystroglycan/LG Domain Binding Modes: Investigating Protein Motifs That Regulate the Affinity of Isolated LG Domains. Front Mol Biosci. 2019;6:18. doi: 10.3389/fmolb.2019.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elmqvist D, Quastel DM. A quantitative study of end-plate potentials in isolated human muscle. J Physiol (Lond) 1965;178(3):505–529. doi: 10.1113/jphysiol.1965.sp007639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Engel AG, Nagel A, Walls TJ, Harper CM, Waisburg HA. Congenital myasthenic syndromes: I. Deficiency and short open-time of the acetylcholine receptor. Muscle Nerve. 1993;16(12):1284–1292. doi: 10.1002/mus.880161204. [DOI] [PubMed] [Google Scholar]
- 36.Uchitel O, Engel AG, Walls TJ, Nagel A, Atassi MZ, Bril V. Congenital myasthenic syndromes: II. Syndrome attributed to abnormal interaction of acetylcholine with its receptor. Muscle Nerve. 1993;16(12):1293–1301. doi: 10.1002/mus.880161205. [DOI] [PubMed] [Google Scholar]
- 37.Kamenskaya MA, Elmqvist D, Thesleff S. Guanidine and neuromuscular transmission. II. Effect on transmitter release in response to repetitive nerve stimulation. Arch Neurol. 1975;32(8):510–518. doi: 10.1001/archneur.1975.00490500030002. [DOI] [PubMed] [Google Scholar]
- 38. Engel AG, Ohno K, Sine SM. Congenital Myasthenic Syndromes. In: Engel AG, Franzini-Armstrong C, eds. Myology 3rd ed. New York,NY: McGraw Hill; 2004:1801–1844. [Google Scholar]
- 39.Engel AG, Lindstrom JM, Lambert EH, Lennon VA. Ultrastructural localization of the acetylcholine receptor in myasthenia gravis and in its experimental autoimmune model. Neurology. 1977;27(4):307–315. doi: 10.1212/WNL.27.4.307. [DOI] [PubMed] [Google Scholar]
- 40.Ohkawara B, Niehrs C. An ATF2-based luciferase reporter to monitor non-canonical Wnt signaling in Xenopus embryos. Dev Dyn. 2011;240(1):188–194. doi: 10.1002/dvdy.22500. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.