Abstract
Changes in maternal immunity during pregnancy can result in an altered immune state, and as a natural perturbation, this provides an opportunity to understand functional interactions of the immune system in vivo. We report characterization of maternal peripheral immune phenotypes for 33 longitudinally sampled normal pregnancies, using clinical measurements of complete blood counts and major immune cell populations, as well as high parameter flow cytometry for 30 leukocyte antigens characterizing 79 cell populations, and monitoring of 1305 serum proteins using the SomaLogic platform. Cellular analyses characterized transient changes in T cell polarization and more persistent alterations in T and B cell subset frequencies and activation. Serum proteomic analysis identified a potentially novel set of 7 proteins that are predictive of gestational age: DDR1, PLAU, MRC1, ACP5, ROBO2, IGF2R, and GNS. We further show that gestational age can be predicted from the parameters obtained by complete blood count tests and clinical flow cytometry characterizing 5 major immune cell populations. Inferring gestational age from this routine clinical phenotyping data could be useful in resource-limited settings that lack obstetric ultrasound. Overall, both the cellular and proteomic analyses validate previously reported phenotypic immunological changes of pregnancy and uncover potentially new alterations and predictive markers.
Keywords: Immunology, Reproductive Biology
Keywords: Cellular immune response, Obstetrics/gynecology, Proteomics
Longitudinal phenotyping of maternal peripheral blood using high parameter flow cytometry, serum proteomics, and routine clinical tests predicts gestational age.
Introduction
Epidemiological observations show that maternal immunity is perturbed during normal human pregnancy. Increased risk of severe disease is reported in pregnancy for defined infections, including listeriosis, malaria, and varicella (1–3). Worse outcomes of influenza infection, and diminished antibody responses to influenza vaccination, are observed in pregnant women (4, 5). On the other hand, specific maternal autoimmune disorders, such as rheumatoid arthritis and multiple sclerosis, may regress during pregnancy, whereas symptoms of systemic lupus erythematosus and myasthenia gravis may worsen (6–8). The immunological changes during pregnancy that underlie these effects are complex and remain poorly understood. Immunological accommodation of the semiallogeneic fetus is mediated primarily by the dedicated tissues of maternal decidua and fetal placenta, which exhibit multiple features of opposing selective forces in viviparity, including imprinting (9). Maternal peripheral immunity is also clearly altered, with changes in systemic IL-4 levels and frequencies of Th2 cells described in normal pregnancy over 25 years ago (10, 11). Subsequent work has continued to define the changes in peripheral immune phenotypes to add to our understanding of the physiologically significant modifications of maternal immunity that occur during normal pregnancy (12, 13). This understanding could address the clinical susceptibilities or resistance to some pathologies during pregnancy or aid the design of maternal immunization strategies.
Given the broad nature of peripheral maternal immunological changes during pregnancy, high dimensional phenotyping is particularly appropriate. Systems biology analyses of intracellular signaling responses measured by mass cytometry, and combined with other modalities, have been used to define molecular clocks of gestational age (GA) (14, 15). In addition to biological understanding of the changes during gestation, these clocks could be used to detect pathologies of pregnancy, such as preterm birth. To complement these data, we have conducted a multimodal high dimensional analysis of longitudinally sampled individuals in the context of normal pregnancies. Here, we identified changes in maternal peripheral immunity, focusing on immune cell populations and the serum proteome. This analysis allowed us to validate previous findings, identify potentially novel changes throughout pregnancy, and determine models predictive of GA that use minimal sets of serum proteins or routinely acquired clinical phenotyping data.
Results
Study design.
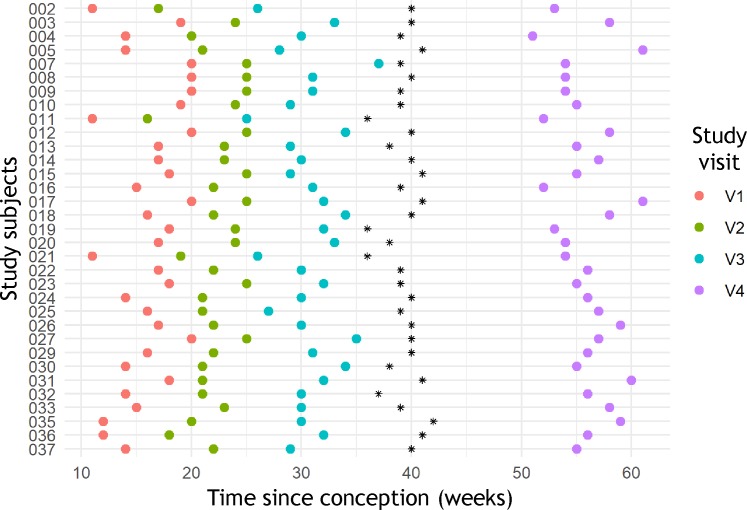
For 33 pregnancies with vaginal delivery of healthy singletons, 4 peripheral blood samples were obtained from 3 time points throughout gestation as well as once after delivery. The distribution of sampling time points and cohort characteristics are summarized in Figure 1 and Table 1. For all 132 samples immune cell phenotypes were characterized by high parameter flow cytometry, and the serum proteome was analyzed using the SomaLogic platform. Measurements were also obtained from complete blood count (CBC) tests and low parameter clinical flow cytometry quantifying 5 major immune cell populations.
Figure 1. Distribution of time points sampled in the study population.
For all 33 individuals the time points sampled are shown for 3 visits during gestation, and 1 visit after parturition, with parturition marked as a black asterisk.
Table 1. Epidemiological and physiological characteristics of the 33 individuals studied.
Immune cell population phenotypes.
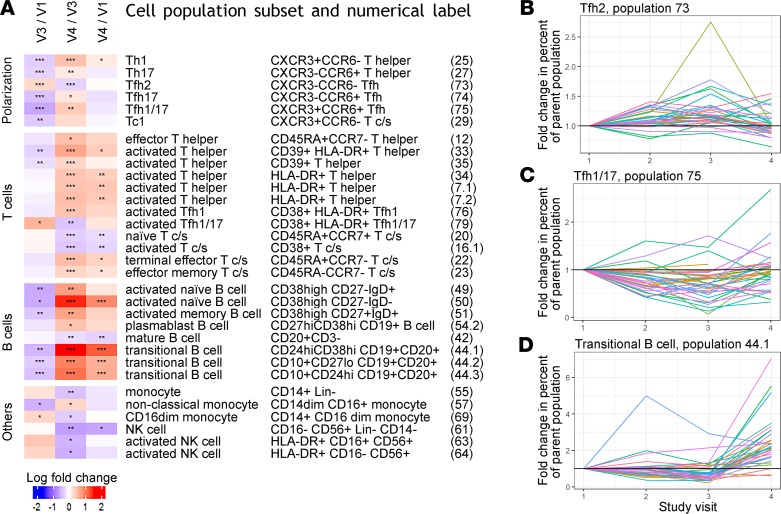
Peripheral blood immune cell phenotypes were studied by high parameter flow cytometry to quantify 79 cellular subsets (Supplemental Tables 1 and 2; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.134838DS1). Populations that significantly changed in frequency were identified by longitudinal paired tests comparing between visit 1 (early gestation), visit 3 (late gestation), or visit 4 (postpartum). We detected 32 population subsets that differed significantly in at least 1 of these comparisons (fold change > 1.2 or < 0.8, q < 0.05) (Figure 2A). In some cases, the same population was identified by more than one of the parallel staining panels used, and technical validation was provided by detection of similar changes in these populations. An example is the increase in HLA-DR+CD4+ cells after parturition, which was detected by 3 panels as populations 7.1, 7.2, and 34. Among the 32 populations, some changed during gestation and rebounded after parturition, so no difference was observed between early gestation and postpartum, representing transient gestation-associated perturbations. Other populations did differ between the early gestation and postpartum time points, indicating persistent perturbations because of either gestation or parturition (Figure 2A).
Figure 2. Changes in peripheral blood immune populations throughout pregnancy.
(A) Using all 33 subjects in our study, longitudinal comparisons between visits 1 and 3 (early and late gestation), visits 3 and 4 (late gestation and postpartum), or visits 1 and 4 (early gestation and postpartum) identified 32 subsets of immune cell populations with fold change greater than 1.2 or less than 0.8 and significant differences for at least 1 comparison using Wilcoxon’s signed-rank paired tests (*FDR < 0.05, **FDR < 0.01, and ***FDR < 0.001). T c/s, CD8+ T cells (cytotoxic/suppressors). (B) For representative populations that demonstrated different patterns of change, fold changes compared with visit 1 are shown for all 4 visits studied. Type 2 CD4+ Tfh cells increased during gestation and declined after parturition. (C) Type 1/17 Tfh cells decreased during gestation and rebounded after parturition. (D) Transitional B cells declined significantly during gestation but increased much more strongly after parturition. See A for FDR-adjusted P values.
Transient gestation-associated changes in T cell polarization.
A transient bias in T cell polarization was observed during gestation that resolved rapidly after parturition. Notably, Th1 and Th17 cell frequencies decreased during gestation, between visits 1 and 3 (Figure 2A). This occurred not only for CD4+ T helper cells generally but also for CD4+CXCR5+ T follicular helper (Tfh) populations. The Tfh compartment showed significantly decreased frequencies of cells with type 1/17 phenotype, defined by CXCR3 and CCR6 expression, and significantly increased frequencies of type 2 cells lacking both of these markers (Figure 2, B and C). The shift in polarization was also observed for CD8+ T cells, with significantly decreased frequencies of CXCR3+CCR6– type 1 cytotoxic T cells during gestation (Figure 2A). CXCR3 expression by CD8+ T cells can enable recruitment to sites of inflammation, for example by virus-specific cells in acute infection (16). Almost all the T cell polarization phenotypes that changed significantly during gestation, between visits 1 and 3, changed significantly in the opposite direction between visits 3 and 4, which spanned parturition, so significant differences did not remain between the early gestation and postpartum time points (Figure 2A).
Persisting perturbation postparturition.
In contrast to the rapidly resolving polarization changes, other differences in T and B cell populations were observed between the extreme time points of our study, the early gestation and postpartum visits 1 and 4 (Figure 2A). Strikingly, the longitudinal profile of these persistent perturbations differed between T and B cell populations. T cell subsets showed few changes between early and late gestation, other than in polarization, but after parturition numerous populations differed compared with either time point during gestation. During this period, CD8+ T cells skewed from naive CCR7+CD45RA+ cells to terminal effector CCR7–CD45RA+ or effector memory CCR7–CD45RA– cells (Figure 2A). Changes in activation differed between CD4+ and CD8+ T cells, with the frequency of HLA-DR+ activated CD4+ cells increasing and CD38+ activated CD8+ cells decreasing (Figure 2A). In contrast to these T cell changes, B cell populations that differed between visits 1 and 4 did show significant change before the end of gestation. Frequencies of transitional and activated naive B cells decreased between visits 1 and 3, before rebounding more strongly during parturition, resulting in frequencies significantly higher postpartum compared with early gestation (Figure 2, A and D). Together these observations delineate impacts of pregnancy on the immune system that persist from early gestation to beyond 10 weeks after parturition.
Serum protein characterization.
Peripheral blood serum proteins were measured using the SomaLogic platform to quantify 1305 proteins, in samples matching those used for flow cytometry. Longitudinal paired tests comparing between early and late gestation (visits 1 and 3) identified 434 proteins that differed significantly (q < 0.05). We observed no correlations between the cell populations and serum proteins that were identified to change during this period (Supplemental Figure 1). This indicates that the circulating proteins and cell phenotypes behave differently and represent independent data sets. We also detected no evidence that either the cell populations or serum proteins identified to change during gestation differed significantly between individuals in correlation with parity, maternal age, prior miscarriage, duration of gestation, body mass index, or blood pressure (Supplemental Table 3). This is likely due to comparison of a small number of pregnancies with little variation in parameters such as duration of gestation.
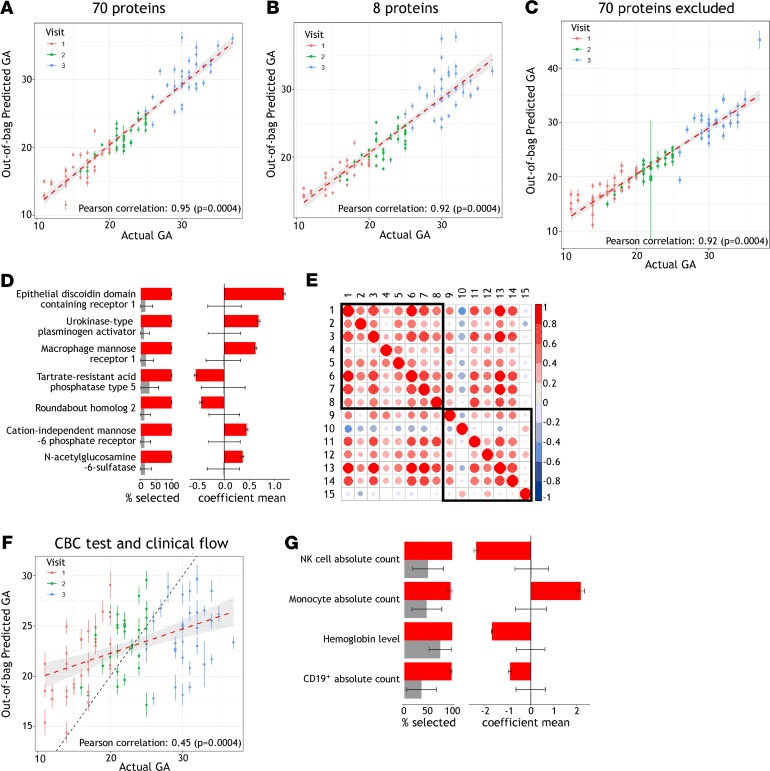
Two other studies have recently been reported that used a similar approach for serum proteomic measurement throughout normal pregnancy (17, 18). Romero et al. found that 10% of the proteins analyzed changed in abundance as a function of GA, and Aghaeepour et al. used elastic net (EN) modeling to identify 74 proteins that could predict GA, as well as a reduced subset of 8, which was similarly effective (17, 18). We first set out to validate these findings, by testing whether the reported protein sets could be used to predict GA, using the measurements for these proteins in our cohort. Excluding 4 proteins that failed quality control parameters in our assay, we used the 70- and 8-protein sets Aghaeepour et al. defined to generate EN models predicting GA. The models were trained on 80% of randomly selected samples and tested on the remaining 20%. For both protein sets, highly significant correlations were seen between observed and predicted GA (r > 0.9, P = 0.0004) (Figure 3, A and B; and Supplemental Figure 2). Thus our results validate the serum proteins reported to predict GA in an independent data set.
Figure 3. Serum proteins and cell populations predictive of GA.
(A and B) EN models generated from both a set of 70 proteins, and a subset of 8 proteins previously identified to predict GA, demonstrated significant correlation between observed and predicted GA when using serum proteomic data from 3 time points during pregnancy for the 33 women in our study. Predicted GA is shown with mean and standard deviation of 100 model iterations randomly sampling training and test sets, with red dashed line indicating a linear regression with 95% confidence interval. (C) Excluding these 70 proteins and selecting from the remaining 1124 serum proteins measured, EN models still showed significant correlation between observed and predicted GA. (D) For the 7 proteins newly identified to predict GA, frequency of selection and average weights differed significantly from null models and are plotted in red showing mean and standard deviation from the 100 model iterations randomly sampling training and test sets. In gray are shown null models with mean and standard deviation from random permutation of sample labels 25 times and 100 random samplings of the training and test sets. (E) The relationship between new and previously identified proteins predicting GA was assessed by pairwise Pearson’s correlations of protein relative intensities during gestation in our data set. Eight proteins previously identified to predict GA by Aghaeepour et al., and the 7 proteins identified in this study, are numbered 1–8 and 9–15, respectively, and defined as follows: 1, chorionic somatomammotropin hormone; 2, prolactin; 3, macrophage colony-stimulating factor 1 receptor; 4, polymeric immunoglobulin receptor; 5, proto-oncogene tyrosine-protein kinase receptor Ret; 6, glypican-3; 7, granulins; 8, α-fetoprotein; 9, macrophage mannose receptor 1; 10, tartrate-resistant acid phosphatase type 5; 11, N-acetylglucosamine-6-sulfatase; 12, cation-independent mannose-6-phosphate receptor; 13, epithelial discoidin domain-containing receptor 1; 14, urokinase-type plasminogen activator; and 15, roundabout homolog 2. (F) Standard clinical phenotyping measurements from CBC tests and clinical flow cytometry can also be used to predict GA. For the 33 subjects sampled at 3 time points during pregnancy, 19 routinely acquired clinical parameters were used to generate EN models for which predicted and observed GA correlated significantly, with black dashed line marking equal GA. (G) Frequency of selection and average weights are shown for the significant model features, plotted as in D.
Identification of more clinically tractable predictors of GA.
Given the predictive analyses possible using serum protein measurements, and the changes in cell population frequencies observed throughout gestation, we sought to predict GA using more accessible features, such as a minimal protein set or routinely available clinical data. Excluding the 70 proteins Aghaeepour et al. defined, we repeated generation of an EN model and found that prediction of GA could still be observed (r > 0.9, P = 0.0004) (Figure 3C). Seven proteins selected with highest average weight were all significantly different from null models: DDR1, PLAU, MRC1, ACP5, ROBO2, IGF2R, and GNS (Figure 3D). In pairwise comparisons with the set of 8 proteins Aghaeepour et al. identified, the 7 proteins newly identified showed some correlations but also substantial differences, indicating that these protein sets behave somewhat differently and do not directly mark each other (Figure 3E). From this comprehensive serum analysis, we therefore identify a novel set of 7 proteins that can be quantified to predict GA.
Given the widespread changes throughout gestation that we observed in cell population frequencies by high parameter flow cytometry, we investigated whether GA could be predicted from immune population frequencies determined at the much lower resolution of standard clinical phenotyping. We applied the same EN modeling approach used for predictive analysis with proteomic data, using all 33 subjects and the 3 time points during gestation. Models were generated using routine clinical phenotyping measurements from CBC tests and clinical flow cytometry that additionally quantified 5 major populations of T, B, and NK cells (Supplemental Table 4). Using 19 parameters from both CBC tests and clinical flow cytometry, models predicting GA were identified with significant correlation between predicted and observed GA (r = 0.45, P = 0.0004) (Figure 3F). Features that associated with GA were increase in absolute counts of monocytes, decrease in B and NK cell absolute counts, as well as decrease in hemoglobin level (Figure 3G). We did detect significant changes in total monocyte and NK populations by high parameter flow cytometry, where specifically CD14+CD16dim monocytes and activated NK cells were altered most as a fraction of their parent populations (Figure 2A). Changes in peripheral NK cell frequencies throughout gestation are not well established in previous studies, although CD56bright NK cells show extraordinary enrichment to dominate the decidual leukocyte population at implantation; these cells are transcriptionally distinct from circulating NK cells (19–21). An increase with GA has previously been reported for the total monocyte population (20). This population can be quantified by a CBC test without clinical flow cytometry, and indeed, when using only 9 parameters measured from the CBC test alone, models predicting GA could still be identified where monocyte counts and hemoglobin levels were the dominant features (r = 0.35, P = 0.0012) (Supplemental Figure 3). These results support the idea that simple, cost-effective, routine clinical phenotyping measurements could be used to predict GA.
Discussion
We report high dimensional analyses characterizing immune cell populations and the serum proteome, in maternal peripheral blood sampled longitudinally during normal pregnancy. In both the immune populations and serum proteome, we validate previously reported phenotypic changes, and add new details and markers to the changes in maternal peripheral immunity that have been identified. Expanding the changes defined adds to our understanding of the physiologically significant modifications of maternal immunity that occur during normal pregnancy and may aid the design of maternal immunization strategies.
During gestation, transient T cell polarization from Th1/17 to Th2 was observed, not only for CD4+ T helper cells generally but also for CXCR5+ Tfh cells specifically and also CD8+ T cell subsets. T cell polarization has long been described to alter during pregnancy, with changes documented in Th1/2/17 subsets of the broad T helper peripheral CD4+ population, although sometimes with contradictory results (10, 22, 23). Our data support a peripheral polarization bias from Th1/17 to Th2 but that the effects are transient, with the postpartum time point no different from early gestation. Our novel observations that Tfh and even CD8+ cells show this polarization could potentially modify how these polarization shifts are thought to alter responses to viral infections and autoantigens, which have often focused on Th cells (24–26). Tfh cells are important for vaccine responses, and the changes in this population may be particularly relevant for optimization of maternal vaccination (27).
Other cellular phenotypic changes showed differences in comparisons between early gestation and postpartum time points. The dynamics of T cell changes, observed particularly in frequencies of CD8+ cell subsets, and more widely in terms of activation status, was consistent with previous reports that broad CD4+ or CD8+ T cell populations show persistent alteration relative to time points from early gestation, in measurements of cell counts and in vitro cytokine production (20). Although immune activation has been implicated in the onset of parturition, the observation that more than 10 weeks after parturition CD4+ T cell activation is still increased compared with our early gestation time point could potentially affect responses in diverse scenarios, such as infection, autoimmunity, and pathologies associated with chronic inflammation (28–30). It is possible the changes in cell populations could affect transfer of passive immunity to breastfed neonates because CD8+ T cells particularly have been observed to survive in intestinal Peyer’s patches of nursed infants (31). B cell populations showed a different profile of changes, with population frequencies and activation states decreasing during gestation before rebounding more strongly after parturition, to result in levels that were higher postpartum compared with early gestation. Our observations are consistent with previous reports that transitional B cells increase within days after parturition (32). Changes throughout the B cell compartment could derive from endocrine factors because hormones such as human chorionic gonadotropin have been implicated in modifying B cell differentiation and function (33). These changes in B cells could contribute to mechanistic understanding of some of the specific alterations in immunity that are observed during pregnancy, such as reduced response to influenza vaccination (4). In addition to changes in population frequencies, it will be important to understand alterations in cellular functional responsiveness during pregnancy because intracellular signaling markers at endogenous levels and in response to stimulation have been found to dominate modeling of GA (14).
We applied an EN modeling approach to validate sets of 70 and 8 serum protein sets previously reported to predict GA and to identify a novel set of 7 serum proteins that can be used to predict GA. The serum protein marker of GA with highest average weight was epithelial discoidin domain-containing receptor 1 (DDR1), which is known to be involved in mammary gland development (34). DDR1 has previously been associated with birthweight in the context of disparate genome-wide approaches. Analyses of selection in primate evolution, and differential methylation between pre- and full-term birth in cord blood, both implicated the region encoding DDR1 (35, 36). Others among our identified markers of GA have also been previously associated with pregnancy. Urokinase-type plasminogen activator (PLAU) is involved in extravillous trophoblast migration during placentation, detected to be upregulated in normal pregnancy, and associated with pregnancy disorders (37–39). Cation-independent mannose-6-phosphate receptor, also known as insulin like growth factor 2 receptor (IGF2R), was one of the first genes identified to be imprinted in mice, although in humans only a minority of individuals show biased expression from the maternal allele (40, 41). This modeling of GA is restricted to the period from 11 to 37 weeks for which samples were collected; it would be of significant interest to characterize earlier time points of gestation with prospectively collected cohorts.
In addition to the set of 7 serum proteins, we also show that GA can be predicted from the routinely acquired clinical data of CBC tests and clinical flow cytometry for 5 major immune populations. The major features selected in these models were monocyte counts and hemoglobin levels. These models may be investigated in pregnancies with pathologies, to potentially shed light on the mechanisms conferring risk. Preterm labor, for example, has been shown to be predicted by cell-free RNA, white blood cell counts, and corticotrophin-releasing hormone, but premature rupture of membranes remains the most accurate indicator of delivery within 48 hours (42, 43). Although much larger studies will have to be done before these observations can be adapted clinically, the ability to infer GA from standard CBC tests and clinical flow cytometry measurements could be most valuable in resource-poor settings. In these circumstances obstetric ultrasound is often not possible to monitor fetal growth for detection of abnormalities that indicate increased risk of neonatal mortality (44).
Methods
Subjects and clinical phenotyping.
Subjects were included in this study with a pregnancy of GA between 10 and 20 weeks, estimated by ultrasound or last menstrual period, and maternal age between 18 and 45 years. Exclusion criteria were a hemoglobin reading of less than 8 g/dL or any medical condition adversely affecting the immune system or requiring immunomodulating medications. In total 33 individuals were studied, of which 76% described their ethnicity as white and 21% reported prior miscarriage. Subjects were analyzed at 4 time points, with clinical phenotyping, high parameter flow cytometry, and serum proteomic analysis performed for all 132 samples. Clinical phenotyping was performed in the NIH clinical center and comprised CBC tests and clinical flow cytometry, together measuring counts and frequencies for platelets, CD4+ T cells, CD8+ T cells, CD19+ cells, NK cells, neutrophils, and monocytes, as well as hemoglobin levels (Supplemental Table 4).
High parameter flow cytometry.
PBMCs were isolated by separation with Ficoll-Paque PLUS and cryopreserved according to Center for Human Immunology (CHI) protocols (45). High parameter flow cytometry was performed using the Human Immunology Project Consortium (HIPC) panels as previously described (46, 47). Briefly, 5 parallel 10-color panels with a total of 30 unique markers enable detection of 79 subsets of PBMCs represented as a fraction of their parent population (Supplemental Tables 1 and 2). Staining was performed using the HIPC lyophilized antibody plates, each with up to 10 samples in addition to controls, after an additional incubation with LIVE/DEAD Fixable Blue Dead Cell Stain (Thermo Fisher Scientific). Acquisition was performed with a Becton Dickinson LSRFortessa, using DIVA 8 software, acquiring 250,000 cells for each sample. Subsequent analysis to determine population frequencies used FlowJo version 9.6.2. Compensation performed with unstained cells and Becton Dickinson compensation beads was used to aid acquisition monitoring. Subsequently a final compensation matrix was calculated using FlowJo during postacquisition analysis. Gates used to define all 79 populations are shown for a representative sample (Supplemental Figures 4–8). For 1 sample, subject 008 at visit 4, high parameter flow cytometry data were not obtained.
Serum proteomic analysis.
Peripheral blood serum was isolated using SST tubes and cryopreserved according to CHI protocols (45). Serum proteomic analysis used the SOMAscan 1.3k Assay (SomaLogic). This is an aptamer-based assay able to detect 1305 protein analytes, optimized for analysis of human serum (48, 49). Briefly, aptamers are short single-stranded DNA sequences modified to confer specific binding to target proteins and can be highly multiplexed for discovery of biomarker signatures. The proteins quantified include cytokines, hormones, growth factors, receptors, kinases, proteases, protease inhibitors, and structural proteins. A complete list of analytes measured can be found at http://somalogic.com/wp-content/uploads/2017/06/SSM-045-Rev-2-SOMAscan-Assay-1.3k-Content.pdf The assay was performed according to manufacturer specifications, with data then inspected using a web tool and subjected to quality control procedures as previously described, after which results from 1194 somamers were used for analyses (50, 51).
Data and code availability.
The data used in this study, comprising CBC tests, clinical flow cytometry, high parameter flow cytometry, and serum proteomic analysis by SomaLogic, as well as the R code and documentation to reproduce the results of our analyses, are available at https://github.com/kotliary/pregnancy
Statistics.
Statistical analysis was performed using R/Bioconductor. Longitudinal changes in PBMC populations or serum proteins were evaluated by Wilcoxon’s signed-rank paired test, and P values were corrected for multiple comparisons with Benjamini-Hochberg method. Modeling of GA using serum proteins or cell populations was performed by EN method using the eNetXplorer package varying parameter α between 0 (ridge regression) and 1 (lasso regression) with step 0.1 (52). For each α the regularization parameter λ was selected by a crossvalidation procedure optimizing correlation between observed and predicted GA. For this crossvalidation the modeling was run 100 times, each time using a random 80% of samples as the training set and the remaining 20% as the test set. The best parameter α was chosen for models with highest average correlation between observed and predicted GA. To estimate significance of model coefficients, eNetXplorer computes null models, randomly permuting sample labels 25 times, each time with 100 random samples of training and test sets as for the model.
Study approval.
For this study volunteers were enrolled in 10-I-0205, an NIH protocol approved and monitored by the National Institute of Allergy and Infectious Diseases/NIH Institutional Review Board (Bethesda, Maryland, USA). All subjects provided informed consent prior to their participation in the study.
Author contributions
LB, SMO, HZ, and RS recruited subjects and collected samples. JC, A Biancotto, and A Babyak performed experimental assays. RA, YK, FC, and KLH analyzed the data. RA, YK, YB, and JST wrote the manuscript. SMH, YB, JST, and CSZ conceived and oversaw the study.
Supplementary Material
Acknowledgments
We thank Andrew Martins for constructive criticism of the manuscript. This research was supported by the Intramural Research Program of the NIH, National Institute of Allergy and Infectious Diseases, and the Trans-NIH Center for Human Immunology, Autoimmunity, and Inflammation (AI001226-01).
Version 1. 03/12/2020
In-Press Preview
Version 2. 04/09/2020
Electronic publication
Version 3. 04/09/2020
Electronic publication
Footnotes
YK’s present address is: Biometric Research Program, Division of Cancer Treatment and Diagnosis, National Cancer Institute, NIH, Rockville, Maryland, USA.
A Biancotto’s present address is: Precision Immunology, Immunology & Inflammation Research Therapeutic Area, Sanofi, Cambridge, Massachusetts, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(7):e134838.https://doi.org/10.1172/jci.insight.134838.
Contributor Information
Richard Apps, Email: richard.apps@nih.gov.
Yuri Kotliarov, Email: yuri.kotliarov@nih.gov.
Foo Cheung, Email: foo.cheung@nih.gov.
Kyu Lee Han, Email: kyulee.han@nih.gov.
Jinguo Chen, Email: Jinguo.Chen@nih.gov.
Huizhi Zhou, Email: huizhi.zhou@nih.gov.
Rongye Shi, Email: rongye.shi@nih.gov.
Sharon M. Osgood, Email: so140o@nih.gov.
Yasmine Belkaid, Email: ybelkaid@niaid.nih.gov.
Steven M. Holland, Email: SHOLLAND@niaid.nih.gov.
John S. Tsang, Email: john.tsang@nih.gov.
References
- 1.Avelino MM, Campos D, Parada JB, Castro AM. Risk factors for Toxoplasma gondii infection in women of childbearing age. Braz J Infect Dis. 2004;8(2):164–174. doi: 10.1590/s1413-86702004000200007. [DOI] [PubMed] [Google Scholar]
- 2.Shulman CE, Dorman EK. Importance and prevention of malaria in pregnancy. Trans R Soc Trop Med Hyg. 2003;97(1):30–35. doi: 10.1016/S0035-9203(03)90012-5. [DOI] [PubMed] [Google Scholar]
- 3.Harger JH, et al. Risk factors and outcome of varicella-zoster virus pneumonia in pregnant women. J Infect Dis. 2002;185(4):422–427. doi: 10.1086/338832. [DOI] [PubMed] [Google Scholar]
- 4.Schlaudecker EP, Ambroggio L, McNeal MM, Finkelman FD, Way SS. Declining responsiveness to influenza vaccination with progression of human pregnancy. Vaccine. 2018;36(31):4734–4741. doi: 10.1016/j.vaccine.2018.05.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klein SL, Passaretti C, Anker M, Olukoya P, Pekosz A. The impact of sex, gender and pregnancy on 2009 H1N1 disease. Biol Sex Differ. 2010;1(1):5. doi: 10.1186/2042-6410-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elenkov IJ, et al. IL-12, TNF-alpha, and hormonal changes during late pregnancy and early postpartum: implications for autoimmune disease activity during these times. J Clin Endocrinol Metab. 2001;86(10):4933–4938. doi: 10.1210/jcem.86.10.7905. [DOI] [PubMed] [Google Scholar]
- 7.Runmarker B, Andersen O. Pregnancy is associated with a lower risk of onset and a better prognosis in multiple sclerosis. Brain. 1995;118(pt 1):253–261. doi: 10.1093/brain/118.1.253. [DOI] [PubMed] [Google Scholar]
- 8.Imrie HJ, McGonigle TP, Liu DT, Jones DR. Reduction in erythrocyte complement receptor 1 (CR1, CD35) and decay accelerating factor (DAF, CD55) during normal pregnancy. J Reprod Immunol. 1996;31(3):221–227. doi: 10.1016/0165-0378(96)00977-1. [DOI] [PubMed] [Google Scholar]
- 9.Moffett A, Loke C. Immunology of placentation in eutherian mammals. Nat Rev Immunol. 2006;6(8):584–594. doi: 10.1038/nri1897. [DOI] [PubMed] [Google Scholar]
- 10.Saito S, Sakai M, Sasaki Y, Tanebe K, Tsuda H, Michimata T. Quantitative analysis of peripheral blood Th0, Th1, Th2 and the Th1:Th2 cell ratio during normal human pregnancy and preeclampsia. Clin Exp Immunol. 1999;117(3):550–555. doi: 10.1046/j.1365-2249.1999.00997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin H, Mosmann TR, Guilbert L, Tuntipopipat S, Wegmann TG. Synthesis of T helper 2-type cytokines at the maternal-fetal interface. J Immunol. 1993;151(9):4562–4573. [PubMed] [Google Scholar]
- 12.Pazos M, Sperling RS, Moran TM, Kraus TA. The influence of pregnancy on systemic immunity. Immunol Res. 2012;54(1-3):254–261. doi: 10.1007/s12026-012-8303-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zmora N, Bashiardes S, Levy M, Elinav E. The role of the immune system in metabolic health and disease. Cell Metab. 2017;25(3):506–521. doi: 10.1016/j.cmet.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 14.Aghaeepour N, et al. An immune clock of human pregnancy. Sci Immunol. 2017;2(15):eaan2946. doi: 10.1126/sciimmunol.aan2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghaemi MS, et al. Multiomics modeling of the immunome, transcriptome, microbiome, proteome and metabolome adaptations during human pregnancy. Bioinformatics. 2019;35(1):95–103. doi: 10.1093/bioinformatics/bty537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slütter B, Pewe LL, Kaech SM, Harty JT. Lung airway-surveilling CXCR3(hi) memory CD8(+) T cells are critical for protection against influenza A virus. Immunity. 2013;39(5):939–948. doi: 10.1016/j.immuni.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aghaeepour N, et al. A proteomic clock of human pregnancy. Am J Obstet Gynecol. 2018;218(3):347.e1–347.e14. doi: 10.1016/j.ajog.2017.12.208. [DOI] [PubMed] [Google Scholar]
- 18.Romero R, et al. The maternal plasma proteome changes as a function of gestational age in normal pregnancy: a longitudinal study. Am J Obstet Gynecol. 2017;217(1):67.e1–67.e21. doi: 10.1016/j.ajog.2017.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moffett-King A. Natural killer cells and pregnancy. Nat Rev Immunol. 2002;2(9):656–663. doi: 10.1038/nri886. [DOI] [PubMed] [Google Scholar]
- 20.Kraus TA, et al. Characterizing the pregnancy immune phenotype: results of the Viral Immunity and Pregnancy (VIP) study. J Clin Immunol. 2012;32(2):300–311. doi: 10.1007/s10875-011-9627-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koopman LA, et al. Human decidual natural killer cells are a unique NK cell subset with immunomodulatory potential. J Exp Med. 2003;198(8):1201–1212. doi: 10.1084/jem.20030305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santner-Nanan B, et al. Systemic increase in the ratio between Foxp3+ and IL-17-producing CD4+ T cells in healthy pregnancy but not in preeclampsia. J Immunol. 2009;183(11):7023–7030. doi: 10.4049/jimmunol.0901154. [DOI] [PubMed] [Google Scholar]
- 23.Saito S, Nakashima A, Shima T, Ito M. Th1/Th2/Th17 and regulatory T-cell paradigm in pregnancy. Am J Reprod Immunol. 2010;63(6):601–610. doi: 10.1111/j.1600-0897.2010.00852.x. [DOI] [PubMed] [Google Scholar]
- 24.Jamieson DJ, Theiler RN, Rasmussen SA. Emerging infections and pregnancy. Emerging Infect Dis. 2006;12(11):1638–1643. doi: 10.3201/eid1211.060152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ostensen M, Villiger PM. Immunology of pregnancy-pregnancy as a remission inducing agent in rheumatoid arthritis. Transpl Immunol. 2002;9(2–4):155–160. doi: 10.1016/s0966-3274(02)00017-5. [DOI] [PubMed] [Google Scholar]
- 26.Figueiredo AS, Schumacher A. The T helper type 17/regulatory T cell paradigm in pregnancy. Immunology. 2016;148(1):13–21. doi: 10.1111/imm.12595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hill DL, et al. The adjuvant GLA-SE promotes human Tfh cell expansion and emergence of public TCRβ clonotypes. J Exp Med. 2019;216(8):1857–1873. doi: 10.1084/jem.20190301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Romero R, Dey SK, Fisher SJ. Preterm labor: one syndrome, many causes. Science. 2014;345(6198):760–765. doi: 10.1126/science.1251816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kotas ME, Medzhitov R. Homeostasis, inflammation, and disease susceptibility. Cell. 2015;160(5):816–827. doi: 10.1016/j.cell.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simons KH, de Jong A, Jukema JW, de Vries MR, Arens R, Quax PHA. T cell co-stimulation and co-inhibition in cardiovascular disease: a double-edged sword. Nat Rev Cardiol. 2019;16(6):325–343. doi: 10.1038/s41569-019-0164-7. [DOI] [PubMed] [Google Scholar]
- 31.Cabinian A, et al. Transfer of maternal immune cells by breastfeeding: maternal cytotoxic T lymphocytes present in breast milk localize in the Peyer’s patches of the nursed infant. PLoS One. 2016;11(6):e0156762. doi: 10.1371/journal.pone.0156762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lima J, et al. Characterization of B cells in healthy pregnant women from late pregnancy to post-partum: a prospective observational study. BMC Pregnancy Childbirth. 2016;16(1):139. doi: 10.1186/s12884-016-0927-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fettke F, et al. Maternal and fetal mechanisms of B cell regulation during pregnancy: human chorionic gonadotropin stimulates B cells to produce IL-10 while alpha-fetoprotein drives them into apoptosis. Front Immunol. 2016;7:495. doi: 10.3389/fimmu.2016.00495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vogel WF, Aszódi A, Alves F, Pawson T. Discoidin domain receptor 1 tyrosine kinase has an essential role in mammary gland development. Mol Cell Biol. 2001;21(8):2906–2917. doi: 10.1128/MCB.21.8.2906-2917.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plunkett J, et al. An evolutionary genomic approach to identify genes involved in human birth timing. PLoS Genet. 2011;7(4):e1001365. doi: 10.1371/journal.pgen.1001365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fernando F, et al. The idiopathic preterm delivery methylation profile in umbilical cord blood DNA. BMC Genomics. 2015;16:736. doi: 10.1186/s12864-015-1915-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye Y, et al. Role of plasminogen activator inhibitor Type 1 in pathologies of female reproductive diseases. Int J Mol Sci. 2017;18(8):E1651. doi: 10.3390/ijms18081651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brenner B. Haemostatic changes in pregnancy. Thromb Res. 2004;114(5-6):409–414. doi: 10.1016/j.thromres.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 39.Chakraborty C, Gleeson LM, McKinnon T, Lala PK. Regulation of human trophoblast migration and invasiveness. Can J Physiol Pharmacol. 2002;80(2):116–124. doi: 10.1139/y02-016. [DOI] [PubMed] [Google Scholar]
- 40.Xu Y, Goodyer CG, Deal C, Polychronakos C. Functional polymorphism in the parental imprinting of the human IGF2R gene. Biochem Biophys Res Commun. 1993;197(2):747–754. doi: 10.1006/bbrc.1993.2542. [DOI] [PubMed] [Google Scholar]
- 41.Barlow DP, Stöger R, Herrmann BG, Saito K, Schweifer N. The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature. 1991;349(6304):84–87. doi: 10.1038/349084a0. [DOI] [PubMed] [Google Scholar]
- 42.Hill JL, et al. Prediction of preterm birth in symptomatic women using decision tree modeling for biomarkers. Am J Obstet Gynecol. 2008;198(4):468.e1–7; discussion 468.e7. doi: 10.1016/j.ajog.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 43.Ngo TTM, et al. Noninvasive blood tests for fetal development predict gestational age and preterm delivery. Science. 2018;360(6393):1133–1136. doi: 10.1126/science.aar3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wanyonyi SZ, Mutiso SK. Monitoring fetal growth in settings with limited ultrasound access. Best Pract Res Clin Obstet Gynaecol. 2018;49:29–36. doi: 10.1016/j.bpobgyn.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 45. Center for Human Immunology, NIH. Standard Operating Procedures. Center for Human Immunology website. https://chi.niaid.nih.gov/web/new/our-research/sop.html Accessed March 25, 2020.
- 46.Maecker HT, McCoy JP, Nussenblatt R. Standardizing immunophenotyping for the Human Immunology Project. Nat Rev Immunol. 2012;12(3):191–200. doi: 10.1038/nri3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Finak G, et al. Standardizing flow cytometry immunophenotyping analysis from the Human ImmunoPhenotyping Consortium. Sci Rep. 2016;6:20686. doi: 10.1038/srep20686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gold L, et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One. 2010;5(12):e15004. doi: 10.1371/journal.pone.0015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rohloff JC, et al. Nucleic acid ligands with protein-like side chains: modified aptamers and their use as diagnostic and therapeutic agents. Mol Ther Nucleic Acids. 2014;3:e201. doi: 10.1038/mtna.2014.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Candia J, et al. Assessment of variability in the SOMAscan assay. Sci Rep. 2017;7(1):14248. doi: 10.1038/s41598-017-14755-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheung F, et al. Web tool for navigating and plotting SomaLogic ADAT files. J Open Res Softw. 2017;5:20. doi: 10.5334/jors.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Candia J, Tsang JS. eNetXplorer: an R package for the quantitative exploration of elastic net families for generalized linear models. BMC Bioinformatics. 2019;20(1):189. doi: 10.1186/s12859-019-2778-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used in this study, comprising CBC tests, clinical flow cytometry, high parameter flow cytometry, and serum proteomic analysis by SomaLogic, as well as the R code and documentation to reproduce the results of our analyses, are available at https://github.com/kotliary/pregnancy