Abstract
Although human endogenous retroviruses (HERVs) represent a substantial proportion of the human genome and some HERVs, such as HERV-K(HML-2), are reported to be involved in neurological disorders, little is known about their biological function. We report that RNA from an HERV-K(HML-2) envelope gene region binds to and activates human Toll-like receptor (TLR) 8, as well as murine Tlr7, expressed in neurons and microglia, thereby causing neurodegeneration. HERV-K(HML-2) RNA introduced into the cerebrospinal fluid (CSF) of either C57BL/6 wild-type mice or APPPS1 mice, a mouse model for Alzheimer’s disease (AD), resulted in neurodegeneration and microglia accumulation. Tlr7-deficient mice were protected against neurodegenerative effects but were resensitized toward HERV-K(HML-2) RNA when neurons ectopically expressed murine Tlr7 or human TLR8. Transcriptome data sets of human AD brain samples revealed a distinct correlation of upregulated HERV-K(HML-2) and TLR8 RNA expression. HERV-K(HML-2) RNA was detectable more frequently in CSF from individuals with AD compared with controls. Our data establish HERV-K(HML-2) RNA as an endogenous ligand for species-specific TLRs 7/8 and imply a functional contribution of human endogenous retroviral transcripts to neurodegenerative processes, such as AD.
Keywords: Immunology, Neuroscience
Keywords: Alzheimer’s disease, Innate immunity, Neurodegeneration
Human endogenous retrovirus K(HML-2) RNA induces neurodegeneration through Toll-like receptor signaling and correlates with upregulated Toll-like receptor 8 expression in Alzheimer’s disease.
Introduction
Neurodegenerative diseases are characterized by progressive loss of neurons, but the mechanisms underlying the deleterious spread of neuronal injury remain unclear. Toll-like receptors (TLRs) play a crucial role in regulating immunity against both pathogens and host-derived molecules (1). These membrane-bound receptors are composed of an ectodomain constituted by leucin-rich repeats, which is involved in ligand binding and receptor activation; a transmembrane domain; and a cytoplasmic Toll/IL-1 receptor (TIR) domain. The TIR domain interacts with adapters, such as MyD88, which couple to downstream protein kinases, leading to the activation of transcription factors, such as NF-κB, and ultimately inducing genes involved in inflammation (2). Contribution of TLRs to central nervous system (CNS) damage has been reported in several mouse models of neurological disorders, including Alzheimer’s disease (AD) (3–6). However, the endogenous ligands involved in TLR activation, associated signaling pathways, and cellular mechanisms by means of which TLRs lead to tissue injury in the different pathological contexts remain elusive.
The genomes of vertebrates contain endogenous retroviruses, which are remnants of ancestral germline infections by exogenous retroviruses (7). Although human endogenous retroviruses (HERVs) comprise a substantial proportion of the human genome and some HERVs have been suggested to be involved in neurodegenerative processes, their biological functions and pathophysiological relevance are poorly understood. The multicopy HERV-K(HML-2) group comprises numerous human-specific proviruses that are transcriptionally active in the brain (8–10) and encodes various former retroviral proteins and retrovirus-like particles (11, 12). Although HERV-K(HML-2) RNA is detected in various human tissue and cell types, transcription patterns of specific HERV-K(HML-2) proviruses differ considerably between tissue and cell types and interindividually, the latter due to presence/absence of alleles of HERV-K(HML-2) elements (9, 13, 14). Deregulated HERV-K(HML-2) transcription has been reported for various diseases, including neurological disorders (15, 16). HERV-derived RNA is present in blood, cerebrospinal fluid (CSF), and brain samples of patients with various CNS diseases (16–18). Nonetheless, a direct signaling role of HERV RNA in cellular activation has not been demonstrated to date. Subsets of HERV-K(HML-2) proviruses encode, on the RNA level, the sequence 5′-GUUGUGU-3′ within the envelope (env) gene while an evolutionarily younger HERV-K(HML-2) subset harbors a mutated sequence (5′-GUUGCGU-3′) in that position (see below). Notably, these motifs resemble the core of the GU-rich sequence that is responsible for species-specific recognition via TLR7 and TLR8 (19, 20).
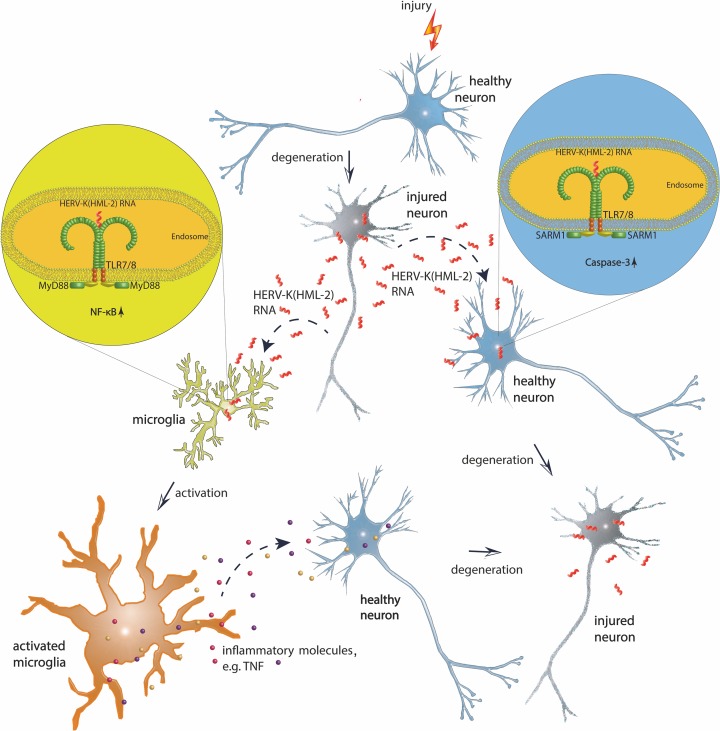
We report that extracellular HERV-K(HML-2) RNA is a potent activator of human TLR8 (hTLR8) and murine Tlr7 (mTlr7). Although HERV-K(HML-2) RNA induces an inflammatory response in microglia, stimulation of hTLR8 and mTlr7 in neurons leads to apoptosis in these cells. Injection of HERV-K(HML-2) transcripts into the CSF of mice induces neurodegeneration dependent on hTLR8- and mTlr7. Correspondingly, we observed a higher frequency of HERV-K(HML-2) RNA presence in CSF and a parallel upregulation of HERV-K(HML-2) and TLR8 RNA expression in brains of patients with AD . Thus, our data implicate a contribution of HERV-K(HML-2) transcripts through TLR signaling to neurodegenerative processes, such as AD.
Results
Extracellular HERV-K(HML-2) RNA activates Tlr7 and TLR8.
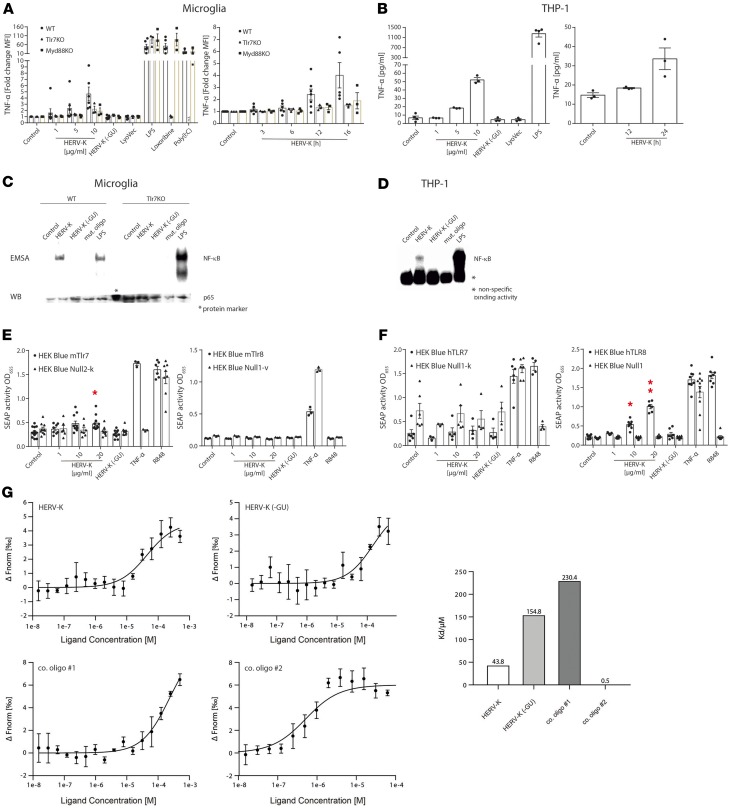
TLR7 and TLR8 respond to GU- or U-rich single-stranded RNA (ssRNA) and short interfering RNAs (19, 21). Among the known RNA ligands for TLR7 and TLR8 is the 20-nucleotide ssRNA40 derived from the exogenous retrovirus HIV (19, 20). We found that a subset of HERV-K(HML-2) proviruses (referred to as HERV-K in the following) contains sequences similar to ssRNA40, specifically a GUUGUGU motif within the env gene (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.131093DS1) that is responsible for TLR7 and TLR8 activation (19, 20). Thus, we postulated that HERV-K RNA acts as an endogenous signaling activator of TLR7 and TLR8. We investigated the response of mTlr7-expressing microglia and macrophages (6) to HERV-K RNA, using a synthetic 22-nucleotide containing the GUUGUGU motif (HERV-K) matching the particular HERV-K env region. Following incubation with HERV-K, both murine microglia (Figure 1A) and bone marrow-derived macrophages (BMDMs, Supplemental Figure 2A) released proinflammatory molecules, such as TNF-α (Figure 1A and Supplemental Figure 2A) and CXCL1 (Supplemental Figure 2B), in a dose- and time-dependent fashion. This response required mTlr7 and MyD88 (Figure 1A and Supplemental Figure 2B). The HERV-K RNA effect was dependent on the GU-rich core because a control oligoribonucleotide matching a sequence located upstream of the GUUGUGU motif within the env region of HERV-K, HERV-K (-GU), did not activate microglia or macrophages (Figure 1A and Supplemental Figure 2, A and B). The TLR ligands lipopolysaccharide (LPS, Tlr4), loxoribine (Tlr7), and poly(I:C) (Tlr3) served as positive controls for TLR-mediated cytokine/chemokine induction. The response of Tlr2/Tlr4-deficient microglia was similar to that of wild-type cells after exposure to HERV-K RNA, excluding the possibility of contamination of the HERV-K oligoribonucleotide with LPS or Tlr2 ligands (Supplemental Figure 2C). Human-derived macrophages also responded to HERV-K RNA by TNF-α release in a sequence-, dose-, and time-dependent manner (Figure 1B). To test whether the canonical TLR/NF-κB pathway is involved in HERV-K RNA–induced signaling, we analyzed microglia and BMDMs treated with HERV-K RNA by electrophoretic mobility shift assay (Figure 1C and Supplemental Figure 2D). HERV-K RNA induced NF-κB activation, comparable to the positive control LPS and dependent on Tlr7 (Figure 1C and Supplemental Figure 2D), suggesting that HERV-K RNA directly activates Tlr7. Likewise, human macrophages responded to HERV-K RNA by NF-κB activation, although to a much lesser extent than to the one LPS induced (Figure 1D). Specificity of HERV-K RNA–induced NF-κB activation was supported by detection of supershifted transcription factor subunits p50 and p65 and IκB kinase phosphorylation by Western blot (Supplemental Figure 2, E and F).
Figure 1. HERV-K(HML-2)–derived oligoribonucleotides activate microglia and macrophages via Tlr7 and TLR8.
(A) Microglia from C57BL/6 (wild-type, WT), Tlr7-KO, or MyD88-KO mice and (B) THP-1 macrophages were incubated for 12 hours with various doses of HERV-K(HML-2) oligoribonucleotide containing a GUUGUGU motif present in the env region (HERV-K, left) or with 5 μg/mL of HERV-K for various durations (right). Untreated cells (control) and control oligoribonucleotide, HERV-K (-GU), 10 μg/mL, served as negative controls. LPS (100 ng/mL), loxoribine (1 mM), poly(I:C) (100 ng/mL), and LyoVec served as further controls. TNF-α amounts in culture supernatants were determined by immuno multiplex assay. Data are pooled from 3 independent experiments. (C) Microglia and (D) THP-1 macrophages were incubated for 2 hours with 5 μg/mL HERV-K, HERV-K (-GU), or mutant oligoribonucleotide or 1 μg/mL LPS. Protein lysates were assayed for NF-κB activation by electrophoretic mobility shift assay (EMSA), while a parallel Western blot probed with p65 antibody confirmed equal loading of probes. One representative experiment of 3 independent experiments is shown. HEK-Blue cells coexpressing murine (E) or human (F) TLR7 or TLR8 and an NF-κB/AP1-inducible secreted embryonic alkaline phosphatase (SEAP) reporter gene were incubated for 48 hours with various HERV-K doses, HERV-K (-GU) (20 μg/mL), R848 (100 ng/mL, TLR7/8 agonist), or TNF-α (100 ng/mL, SEAP induction). HEK-Blue–null cells served as negative controls. Data are pooled from 3–7 independent experiments. Results are presented as mean ± SEM. *P < 0.05, and **P < 0.01 over HERV-K dose compared with control (1-way ANOVA, Bonferroni’s post hoc analysis). (G) Binding affinity measurements of TLR8 protein and oligoribonucleotides using microscale thermophoresis. TLR8-RNA interaction was monitored by titrating RNA from 500 μM to 30 nM [HERV-K, HERV-K (-GU), control oligoribonucleotide 1] and 62.5 μM to 3.8 nM (control oligoribonucleotide 2) against 50 nM RED-Tris-NTA–labeled TLR8 measured with NanoTemper Monolith NT.115 (left). KD values were calculated from dose response curves, which were calculated from titration experiments (right). Results are presented as mean ± SD. n = 3.
To systematically validate the ability of HERV-K RNA to activate TLR signaling in both murine and human cells and to distinguish between responses mediated by TLR7 and/or phylogenetically and structurally highly related TLR8 (19), we used human embryonic kidney 293 (HEK293) cells stably coexpressing murine (Figure 1E) or human (Figure 1F) TLR7 or TLR8 and an NF-κB/activator protein 1–inducible secreted embryonic alkaline phosphatase reporter gene. As expected, HERV-K RNA induced NF-κB in HEK293 cells expressing mTlr7 in a sequence-dependent manner. In contrast, HEK293 cells expressing hTLR7 did not respond to HERV-K RNA. Instead, HERV-K RNA induced NF-κB in HEK293 cells expressing hTLR8 in a sequence-specific fashion, while cells expressing mTlr8 did not respond to HERV-K RNA.
Interaction between HERV-K oligoribonucleotide and hTLR8 protein yielded a KD of 43.8, confirming direct binding of HERV-K RNA harboring the GU-rich motif to hTLR8, as assessed by microscale thermophoresis (Figure 1G). Comparatively, interaction between HERV-K (-GU) RNA and hTLR8 resulted in a KD of 154.8, while sequence-modified control oligoribonucleotides bound to the receptor with a KD of 230.4 and a KD of 0.5.
Taken together, HERV-K RNA serves as a ligand for mTlr7 and hTLR8, thereby inducing the canonical TLR pathway in microglia and macrophages.
HERV-K RNA causes neuronal injury via TLR signaling.
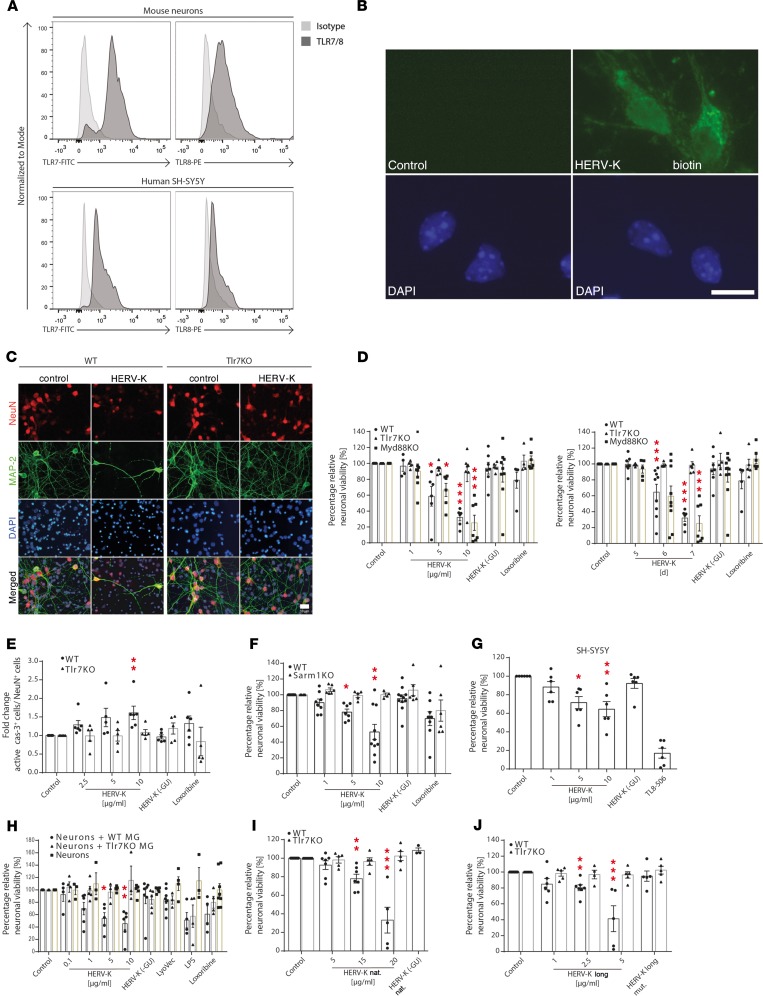
TLR7 and TLR8 are primarily expressed in immune cells (19) but also detected in murine neurons (6, 22). We tested whether these receptors are expressed in human neurons. As is the case for murine neurons, both TLR7 and TLR8 were readily detectable, by FACS, in neurons derived from the human neuroblastoma cell line SH-SY5Y (Figure 2A). Biotinylated HERV-K oligoribonucleotide entered neurons without transfection (Figure 2B). To investigate whether extracellular HERV-K RNA might affect primary neurons, cortical neurons from wild-type and Tlr7-deficient mice were incubated with HERV-K oligoribonucleotides containing or lacking the GUUGUGU motif, HERV-K or HERV-K (-GU). HERV-K RNA induced neuronal injury in a dose- and time-dependent fashion, exceeding the activity of the established Tlr7 ligand loxoribine (Figure 2, C and D). HERV-K RNA–induced neurotoxicity was sequence specific because deletion of the GU motif in the control oligoribonucleotide eliminated injurious effects (Figure 2D). Further, Tlr7-deficient neurons were unaffected by HERV-K RNA treatment (Figure 2, C and D). To determine the molecular process of these neurotoxic effects, neurons were analyzed for the expression of active caspase-3, a downstream effector of apoptosis. HERV-K RNA treatment caused an increased expression of active caspase-3 in wild-type but not in Tlr7-deficient neurons (Figure 2E). This apoptosis induction was observed using HERV-K RNA doses as low as 0.01 μg/mL over a period of up to 12 days (Supplemental Figure 3). Because the canonical Tlr7-mediated signaling pathway involves MyD88, we analyzed the response of MyD88-mutant cells to HERV-K RNA (Figure 2D). Similar to wild-type neurons, MyD88-deficient neurons underwent cell death in response to HERV-K RNA, indicating that MyD88 is not required for HERV-K oligoribonucleotide-induced neuronal injury. Correspondingly, HERV-K RNA did not induce NF-κB activation in neurons (Supplemental Figure 4).
Figure 2. Extracellular HERV-K RNA induces neurotoxicity through Tlr7.
(A) Neurons and SH-SY5Y cells were analyzed for TLR7/TLR8 expression by FACS. (B) Neurons were incubated with 5 μg/mL biotinylated HERV-K or PBS (control) for 12 hours and stained with streptavidin–Alexa Fluor 488 and DAPI. Scale bar: 10 μm. (C) WT and Tlr7-KO neurons were incubated with 5 μg/mL HERV-K for 7 days or left untreated (control). Cells were immunostained with NeuN, MAP-2, and DAPI. Scale bar: 10 μm. (D) WT, Tlr7-KO, and Myd88-KO neurons were incubated with various HERV-K doses for 7 days (left) or with 5 μg/mL HERV-K for various durations (right) (P = 0.0001 over dose and time, WT groups; P < 0.0001, P = 0.0006 over dose and time, respectively, Myd88-KO groups; n.s. over Tlr7-KO, all groups). *P < 0.05, and ***P < 0.001 (n = 4–11). (E) WT and Tlr7-KO neurons were incubated with various HERV-K doses for 5 days and immunostained for active caspase-3, NeuN, and DAPI. Caspase-3–positive cells were quantified (P = 0.0005 over WT groups; n.s. over Tlr7-KO groups). **P < 0.01 (n = 5–6). (F) WT and Sarm1-KO neurons were incubated with HERV-K for 7 days (P = 0.0006 over WT groups; n.s. over Sarm1-KO groups). *P < 0.05, and **P < 0.01 (n = 4–10). (G) SH-SY5Y cells were incubated with HERV-K for 3 days (P = 0.0015 over all groups). *P < 0.05, and **P < 0.01 (n = 6). (H) WT neurons and neuron/microglia (+WT MG or +Tlr7-KO MG) cocultures were incubated with HERV-K for 3 days (P = 0.009 over neurons + WT MG groups; n.s. over neurons + Tlr7-KO MG groups; n.s. over neurons all groups). *P < 0.05, and **P < 0.01 (n = 5–6). (I) WT and Tlr7-KO neurons were incubated with nonphosphorothioated HERV-K oligoribonucleotide for 7 days (P < 0.0001 over WT groups, n.s. over Tlr7-KO groups). **P < 0.01, and ***P < 0.001 (n = 3–12). (J) WT and Tlr7-KO neurons were incubated with 50-nucleotide HERV-K oligoribonucleotide (HERV-K long) or with scrambled 50-nucleotide HERV-K (HERV-K long mut.) for 7 days (P < 0.0001 over WT groups and n.s. over Tlr7-KO). **P < 0.01, and ***P < 0.001 (n = 4–11). (D–J) Unless stated otherwise, relative neuronal viability was assessed. Controls were 10 μg/mL HERV-K (-GU) oligoribonucleotide, 1 mM loxoribine, 1 mg/mL LPS, or 10 ng/mL TL8-506. Results are presented as mean ± SEM. P values for all groups: Kruskal-Wallis test; P values for indicated relevant groups compared with control: Kruskal-Wallis test with Dunn’s post-hoc analysis.
Sterile alpha and TIR domain-containing 1 (SARM1) is a member of the TIR domain–containing adapter protein family and mediates axonal degeneration (23). To test the role of SARM1 in HERV-K RNA–triggered neurotoxicity, wild-type and Sarm1-deficient neurons were incubated with HERV-K oligoribonucleotide. Contrary to wild-type neurons, Sarm1-deficient neurons were resistant to HERV-K RNA (Figure 2F), indicating that SARM1 is a crucial mediator for neuronal injury induced by HERV-K/TLR signaling.
Relevant contamination of analyzed murine neurons with glia was ruled out by immunostaining (6). To confirm the cell-autonomous nature of HERV-K RNA–induced neurotoxicity and to investigate HERV-K RNA–induced effects in human-derived neurons, SH-SY5Y cells were tested. SH-SY5Y cells incubated with HERV-K RNA underwent sequence-specific cell death similar to that observed in primary murine neurons (Figure 2G). Likewise, HERV-K RNA did not induce NF-κB activation in SH-SY5Y cells (Supplemental Figure 4C). Thus, data from highly purified neurons and cell line SH-SY5Y indicate that microglia are not required for HERV-K RNA–induced neurotoxicity, despite the ability of HERV-K RNA to induce an inflammatory response in microglia (Figure 1A and Supplemental Figure 2B). Nevertheless, because microglia can mediate neuronal injury through TLRs (4), we compared HERV-K RNA–mediated effects on purified neurons with cocultures of neurons and microglia from wild-type or Tlr7-deficient mice (Figure 2H) after 3 days of incubation, when HERV-K RNA–induced neuronal loss was not observed in the experiments above (Figure 2D). In the presence of wild-type microglia, HERV-K RNA–induced neurotoxicity occurred 3 days earlier and at lower HERV-K doses compared with toxic effects observed for purified neurons, pointing to an additional impact on neuronal survival mediated by an inflammatory environment. This microglia-enhanced neurotoxicity required microglial Tlr7 (Figure 2H).
Although we used HERV-K oligoribonucleotide stabilized by phosphorothioate linkages throughout the study, we showed that unmodified HERV-K RNA also induced Tlr7-dependent neurotoxicity (Figure 2I). The specificity of HERV-K RNA as the causative agent of neurotoxicity was confirmed using a longer HERV-K oligoribonucleotide (50-nucleotide, HERV-K long, Figure 2J), which induced sequence-specific and Tlr7-dependent neuronal injury. This neurotoxicity was enhanced compared with that observed when using the 22-oligoribonucleotide (Figure 2D).
Taken together, HERV-K RNA induces neuronal apoptosis via TLR and SARM1 signaling. This cell-autonomous neurotoxicity can be enhanced by microglia in response to HERV-K RNA.
HERV-K transcripts are released from injured neurons and induce further neurodegeneration.
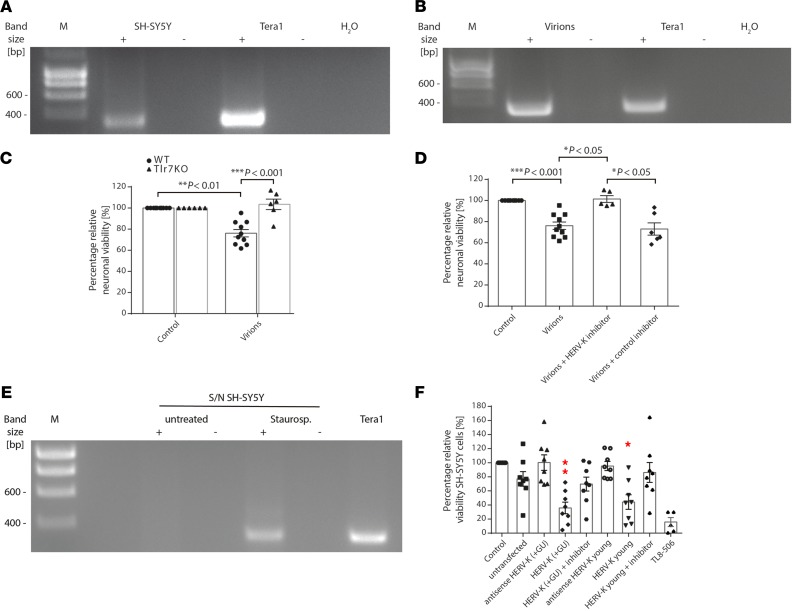
We sought to determine whether endogenous full-length HERV-K transcripts are capable of inducing neuronal injury. Analysis of SH-SY5Y cells by reverse transcription PCR (RT-PCR) showed that human-derived neurons harbor HERV-K transcripts containing the sequence GUUGUGU (Figure 3A). The human teratocarcinoma cell line Tera-1, which transcribes HERV-K at a high level and produces HERV-K–encoded retroviral particles containing full-length HERV-K transcripts, some of which harbor GU-rich sequences (12, 24), served as a positive control. Retroviral particles released from Tera-1 cells were isolated, and the presence of HERV-K transcripts in retroviral particles was shown by endpoint RT-PCR (Figure 3B). Subsequently, murine neurons were incubated with virions containing HERV-K transcripts, which induced neurotoxicity dependent on neuronal Tlr7 (Figure 3C). To confirm that this neurotoxicity was specifically induced by endogenous HERV-K transcripts, we made use of a locked nucleic acid–based antisense oligonucleotide (referred to as HERV-K inhibitor in the following) with a sequence complementary to the HERV-K env region containing the GUUGUGU motif, which sequesters the target ssRNA in highly stable heteroduplexes (25). Indeed, neurons were protected against cell death induced by virion particles in the presence of HERV-K inhibitor (Figure 3D), whereas neurons without inhibitor treatment underwent cell death in response to virions. In contrast, a negative control inhibitor consisting of a scrambled RNA sequence failed to provide protective effects (Figure 3D).
Figure 3. Endogenous HERV-K transcripts trigger neurodegeneration.
(A) SH-SY5Y cells and (B) virion particles isolated from Tera-1 cells were analyzed by RT-PCR using primers amplifying an HERV-K env region harboring the motif GUUGUGU. Tera-1 cells served as a positive control. (C) WT and Tlr7-KO neurons were incubated with 10 μL suspension of Tera-1 virions or left untreated (control) for 6 days. Relative neuronal viability was determined (P < 0.0001 over all groups, Kruskal-Wallis test) (n = 6–10). (D) WT neurons were incubated with 10 μL suspension of Tera-1 virions for 6 days, with or without HERV-K inhibitor. A nonspecific inhibitor served as a negative control. Relative neuronal viability was assessed (P < 0.0001 over all groups, Kruskal-Wallis test) (n = 5–10). (E) Supernatants (S/N) from untreated and apoptotic neurons treated with staurosporine were analyzed by RT-PCR for HERV-K expression. (F) For 4 days,20 μg/mL RNA from untransfected SH-SY5Y cells (control) or SH-SY5Y cells overexpressing HERV-K RNA harboring a GUUGUGU motif [HERV-K (+ GU)], a GUUGCGU motif (HERV-K young), or the respective antisense sequence were used for incubation of SH-SY5Y cells with or without HERV-K inhibitor. Untreated SH-SY5Y cells served as negative control, and TL8-506 served as positive control. Relative neuronal viability was assessed (P = 0.0001 over all groups, Kruskal-Wallis test). Results are presented as mean ± SEM. *P < 0.05, and **P < 0.01 compared with control (n = 5). Indicated P values were determined by Kruskal-Wallis test with Dunn’s post hoc analysis.
Injured tissues release molecules, thereby activating innate immunity (1, 5). We hypothesized that HERV-K RNA is released during neuronal injury and subsequently acts on neighboring neurons. Consequently, these neurons could accelerate neuronal damage by releasing neurotoxic HERV-K RNA. To test whether injured human neurons release HERV-K RNA, apoptosis was induced in SH-SY5Y cells by adding staurosporine. Culture supernatants were separated from cell remnants by centrifugation and analyzed for the presence of HERV-K RNA by RT-PCR (Figure 3E). Whereas HERV-K RNA was not detected in supernatants from untreated neurons, HERV-K RNA was observed in supernatants from apoptotic neurons. To confirm that endogenous HERV-K transcripts from human neurons are capable of inducing neuronal injury, total RNA from native SH-SY5Y cells and SH-SY5Y cells overexpressing a 785-nucleotide covering the HERV-K env region and containing the GUUGUGU motif — HERV-K (+GU) — or the sequence GUUGCGU — HERV-K young — the latter being present in an evolutionarily younger HERV-K subset (Supplemental Figure 1B), was isolated and incubated with freshly plated SH-SY5Y cells (Figure 3F). Similar to synthetic HERV-K oligoribonucleotides, RNA isolated from cells overexpressing HERV-K env transcripts induced SH-SY5Y cell death, comparable to the TLR8 agonist TL8-506. Neurotoxicity was, at least in part, induced by HERV-K transcripts containing either the GUUGUGU or the GUUGCGU motif. These overexpressed transcripts exceeded neurotoxicity induced by RNA from untransfected SH-SY5Y cells, and RNA isolated from SH-SY5Y cells overexpressing the respective RNA in antisense failed to induce comparable cell death (Figure 3F). The presence of HERV-K inhibitor protected neurons against HERV-K transcript–induced cell death (Figure 3F).
Taken together, HERV-K transcripts released from both injured murine and human neurons trigger neurodegeneration.
HERV-K RNA in CSF induces neurodegeneration via mTlr7 and hTLR8 in vivo.
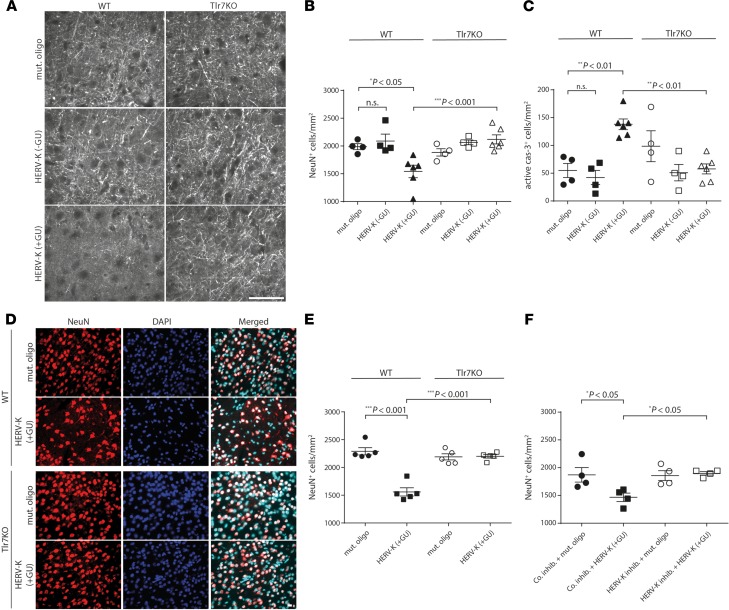
To evaluate the role of HERV-K RNA as a Tlr7 activator in neurons in vivo, we injected mice intrathecally with HERV-K oligoribonucleotide containing the GUUGUGU motif [HERV-K (+GU)], control oligoribonucleotide lacking this motif [HERV-K (-GU)], or mutant oligoribonucleotide with reduced GU content. HERV-K (+GU) RNA induced axonal injury (Figure 4A) and neuronal loss (Figure 4B) in wild-type cerebral cortex 3 days after injection, whereas HERV-K (-GU) and mutant oligoribonucleotide affected neither axon integrity nor neuronal numbers. In contrast, Tlr7-deficient mice were protected from HERV-K (+GU) RNA–induced neurodegeneration (Figure 4, A and B). Immunohistochemical analysis confirmed caspase-3–positive apoptosis induction exclusively in the cerebral cortex of HERV-K (+GU)–treated wild-type animals (Figure 4C). HERV-K (+GU) RNA–induced neurodegeneration increased with time, with immunohistochemistry revealing a loss of 22.4% of neurons in the cerebral cortex after 3 days (Figure 4B) and 31.9% of neurons after 2 weeks (Figure 4, D and E). A single injection of an HERV-K (+GU) dose as low as 0.001 μg triggered neurodegenerative effects after 4 weeks (Supplemental Figure 5). In contrast, Tlr7-deficient mice displayed no neuronal damage/loss during the entire observation period. Similarly, intrathecal injection of an HERV-K oligoribonucleotide containing the evolutionarily younger motif GUUGCGU resulted in loss of cortical neurons dependent on mTlr7 (Supplemental Figure 6). To confirm HERV-K RNA as the causative factor, we injected wild-type mice intrathecally with HERV-K inhibitor or nonspecific control inhibitor before intrathecal injection of HERV-K (+GU) RNA or mutant oligoribonucleotide. Immunohistochemical analysis of the cerebral cortex after 3 days revealed that pretreatment with HERV-K inhibitor abolished HERV-K (+GU) RNA–induced neurodegeneration, whereas pretreatment with control inhibitor did not protect neurons (Figure 4F).
Figure 4. Intrathecal administration of HERV-K RNA causes neurodegeneration through Tlr7.
Ten micrograms of HERV-K, HERV-K (-GU), or mutant oligoribonucleotide was injected intrathecally into WT [HERV-K, n = 5–6; HERV-K (-GU), n = 4; mutant oligoribonucleotide, n = 4–5] or Tlr7-KO mice [HERV-K, n = 5–6; HERV-K (-GU), n = 4; mutant oligoribonucleotide, n = 4–5] mice. (A–C) After 3 days, brain sections were immunostained with neurofilament antibody (A, scale bar: 50 μm), NeuN antibody (B), or active caspase-3 antibody (C) and DAPI. NeuN-positive (B) and caspase-3–positive (C) cortical cells were quantified. (D and E) After 14 days, brain sections were immunostained with NeuN antibody and DAPI (D, scale bar: 10 μm). NeuN-positive cortical cells were quantified (E). (F) HERV-K inhibitor or nonspecific inhibitor (co. inhib.; 125 pmol) was injected intrathecally into WT mice. After 16 hours, mice were injected intrathecally with 10 μg of HERV-K or mutant oligoribonucleotide (HERV-K inhibitor: HERV-K, n = 4; mut. oligo, n = 4; co inhib: HERV-K, n = 4; mut. oligo, n = 4). After further 3 days, brain sections were immunostained with NeuN antibody. NeuN-positive cortical cells were quantified. Results are presented as mean ± SEM. One-way ANOVA test yielded P = 0.0007 (B), P = 0.0003 (C), P < 0.0001 (E), and P = 0.0145 (F) over all groups. Indicated P values were determined by 1-way ANOVA with Bonferroni’s post hoc test. n.s., not significant.
To analyze whether extracellular HERV-K contributes to microglial accumulation and amyloid-β (Aβ) plaque pathology — besides neuronal loss and neurofibrillary tangles, 2 major hallmarks of AD (26) — we injected amyloid-prone APPPS1 mice (27) intrathecally with HERV-K RNA combined with or without HERV-K inhibitor treatment. No significant changes with regard to Aβ plaque after HERV-K treatment were observed (Supplemental Figure 7A). However, we detected a 66.6% increase in microglial numbers at 120 days of age for APPPS1 mice injected with HERV-K RNA (Supplemental Figure 7B). Also, HERV-K oligoribonucleotide induced a loss of 31.9% of neurons in the APPPS1 cerebral cortex (Supplemental Figure 7C). Pretreatment with HERV-K inhibitor before HERV-K injection abolished both increased microglial numbers and neurodegenerative effects (Supplemental Figure 7, B and C). Of note, studies on extracellular HERV-K in APPPS1 mice have to be considered with caution because APPPS1 mice lack HERV-K and HERV-K expression must be regarded in a human-specific context. Nevertheless, our data from APPPS1 mice corroborate our findings on HERV-K RNA–induced neurodegeneration and microglial activation.
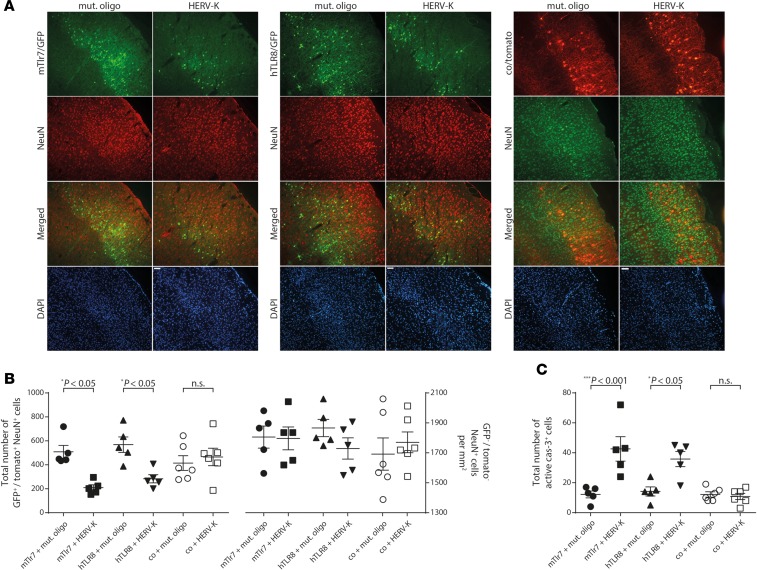
To further link TLR expression with HERV-K RNA–induced neurodegeneration, we reintroduced mTlr7 into the cerebral cortex of Tlr7-deficient mice by in utero electroporation. Because hTLR8 also responds to HERV-K RNA (Figure 1) and because TLR signaling is highly conserved in different species, including humans and mice (2), we also transfected Tlr7-deficient mice with hTLR8. Tlr7-deficient mouse embryos were electroporated with plasmids encoding GFP-mTlr7 or GFP-hTLR8 or with control vector expressing red fluorescent protein (RFP) (tdTomato) only at embryonic day 14.5. At postnatal day 19, HERV-K oligoribonucleotide containing the GUUGUGU motif (HERV-K) or mutant oligoribonucleotide was administered intrathecally, and cortices were analyzed 3 days later (Figure 5). When mTlr7 or hTLR8 was reintroduced into HERV-K RNA–treated Tlr7-deficient cortices, we observed a 58.9% and 50.1%, respectively, loss of GFP-positive neurons compared with control oligoribonucleotide–injected mice (Figure 5, A and B). Likewise, reintroduction of mTlr7 and hTLR8 resulted in increased numbers of active caspase-3–positive cells in the cerebral cortex of HERV-K RNA–treated mice (Figure 5C). Unelectroporated, NeuN-positive neurons were not affected by HERV-K RNA (Figure 5B). Injection of HERV-K RNA had no effect on neurons or caspase-3–positive cells transfected with control vector (Figure 5, A–C). These results confirm that neurodegenerative activity of intrathecal HERV-K RNA requires neuronal mTlr7 or hTLR8 and demonstrate a functional role for a human neuronal TLR, namely hTLR8, in HERV-K RNA–induced neurodegeneration in vivo.
Figure 5. Transfection of murine Tlr7 or human TLR8 restores neurodegenerative effects of HERV-K RNA in vivo.
(A) Tlr7-KO mouse embryos (E14.5) were electroporated in utero with an expression vector for murine Tlr7 (Tlr7/GFP, left), an expression vector for human TLR8 (TLR8/GFP, middle) or control (co/RFP [tdTomato], right). EGFP was coexpressed from an IRES cassette. At postnatal day 19, 10 μg of HERV-K RNA (Tlr7 vector, n = 5; TLR8 vector, n = 5; control vector, n = 6) or mutant oligoribonucleotide (Tlr7 vector, n = 5; TLR8 vector, n = 5; control vector, n = 6) were administered intrathecally. Cerebral cortex was analyzed by immunostaining with NeuN antibody 3 days later. Scale bar: 50 μm. (B) Quantitation of GFP+/Tomato+ NeuN+ cells (left) and GFP−/Tomato– NeuN+ cells (right) in the electroporated cortex. (C) Sections of the electroporated cortex were immunostained with active caspase-3 antibody, and caspase-3+ cells were quantified. Results are presented as mean ± SEM. One-way ANOVA test yielded (B) P = 0.0014 (left) and n.s. (right) and (C) P < 0.0001 over all groups. Indicated P values were determined by 1-way ANOVA with Bonferroni’s post hoc test. n.s., not significant.
Correlation of upregulated HERV-K and TLR8 expression in AD.
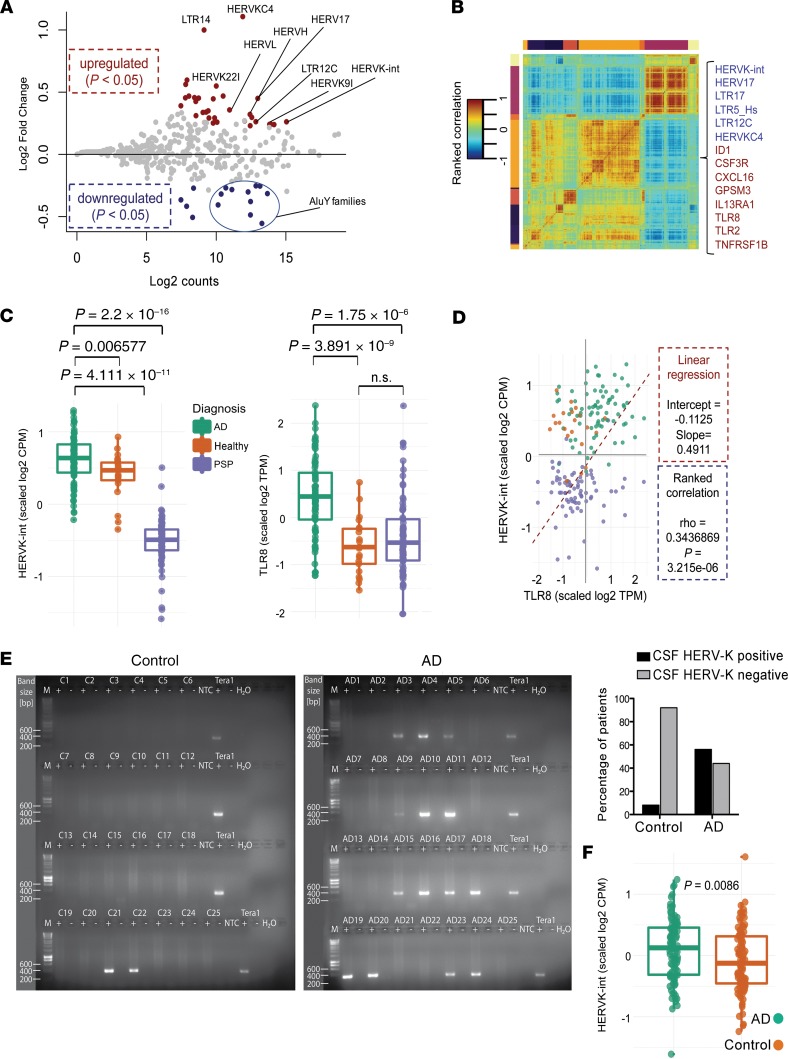
Differential expression of HERV-K in brains of individuals with neurodegenerative diseases, including AD, was previously observed (17, 28). To determine whether HERV-K transcription correlates with TLR7/8 expression in AD patients, we analyzed transcriptome data sets (29) as provided by the Mayo Clinic Brain Bank (https://www.synapse.org), which included transcriptome data of 376 brain samples from the temporal cortex (TCX) and cerebellum of AD patients and control individuals (age-matched healthy individuals and patients with progressive supranuclear palsy [PSP, ref. 30] as an unrelated neurodegenerative disease control). Using an unbiased strategy, we first determined expression values of transposable element (TE) families, restricting the analysis to data sets with high RNA integrity number (>7). Global expression analysis of brain samples revealed that upregulated TE families were mostly (~90%) primate-specific elements (present only in human and primate genomes), while the rest of the detected expression derived from ancient TEs, found also in other mammals/vertebrates (Supplemental Figure 8A). In the human brain, the most abundantly expressed TEs identified were non–long terminal repeat (non-LTR) elements, such as SINE-VNTR-Alus (SVA) and the retrotranspositionally active L1_Hs (31), followed by endogenous retrovirus groups HERV-E_a and HERV-K (Supplemental Figure 8, A and B). While investigating TE expression in AD and controls, we observed differential HERV-K transcription in TCX but not in cerebellum. Thus, for detailed analysis, we established a TCX data frame of AD samples (n = 82) and a combined control of age-matched healthy and PSP samples (n = 94) (Supplemental Figure 8C). In AD, we identified HERV-K as the most expressed and differentially upregulated TE (Figure 6A and Supplemental Figure 8C). Further, expression of AluY elements in AD samples was reduced, while, despite their high overall expression, L1 and SVA were not among the differentially expressed TEs (Figure 6A).
Figure 6. Correlated upregulated HERV-K and TLR8 expression in AD.
(A) Differentially expressed TEs in TCX of AD patients. MA plot (Bland–Altman plot) shows differential TE expression (log2 fold change) in TCX of 176 postmortem samples of AD patients (n = 82) versus controls (aging healthy individuals and patients with PSP, n = 94). Each dot represents 1 TE family. Dots are colored if the P value from DESeq2/limma statistics was less than 0.05. Note that the most differentially up- and downregulated TEs in AD patients versus controls include HERV-K-int — as designated in Repbase, i.e., HERV-K(HML-2) — HERVK9I, and various AluY subfamilies. (B) Correlated expression of HERV-K-int with immune response genes. Clustered pairwise correlation matrix generated by weighted gene coexpression network analysis across 176 brain samples (see A, including ~2400 genes and 17 TE families, Spearman’s rank correlation, Euclidian distance). In a highlighted cluster, TEs (blue) including HERV-K-int/LTR5_Hs, HERV-K(HML-2), are coexpressed with immune response genes (red), including TLR8 and TLR2. (C) Jitter box plots visualize Z score–transformed, unbiased relative expression of (left) HERV-K-int/HERV-K(HML-2) (log2 counts per million; CPM) and (right) TLR8 (log2 transcripts per million; TPM) in TCX samples of AD patients, PSP patients, and controls. Every dot represents 1 sample. P values were determined by Wilcoxon’s test with further adjustment for multiple corrections. n.s., not significant. (D) Correlation analysis of HERV-K-int/HERV-K(HML-2) and TLR8 expression. Scatter plot shows scaled log2-transformed expression of HERV-K-int/HERV-K(HML-2) (CPM) and TLR8 (TPM) (color codes defined as in C). Linear regression analysis (dashed line) indicates correlation between HERV-K-int/HERV-K(HML-2) and TLR8 expression in AD. Rho values were obtained from pairwise-ranked correlation analysis. (E) CSF from AD patients (n = 25) and controls (n = 25) was assayed by RT-PCR using primers specific for the HERV-K env region encompassing the TLR7/8 recognition motif (left). Frequency of HERV-K detection in CSF samples from controls and AD patients (right). (F) Presence of HERV-K(HML-2) in AD CSF and age-controlled samples. Jitter box plots represent the average of Z score–transformed counts of expressed HERV-K(HML2) loci (n = 98) in AD (n = 6) and control (n = 6) CSF. Every dot represents a distinct HERV-K(HML-2) locus. Significance was determined by Wilcoxon’s test. The box plots depict the median (thick line), and the length of each box represents the interquartile range.
To identify genes with expression patterns similar to HERV-K, we produced a pairwise correlation matrix generated by weighted gene coexpression network analysis (WGCNA) across the 176 TCX samples described above, including approximately 2400 genes and 17 TE families. WGCNA assigned HERV-K expression to a correlation cluster with several immune response genes, including TLR8 and TLR2 (Figure 6B). Both HERV-K and TLR8 expression were upregulated in AD versus control (Figure 6C), and TLR8 exhibited the highest correlation value relative to HERV-K (Figure 6D), when compared with statistically significant values for TLR2, TLR5, and TLR7 (Supplemental Figure 8D). Reanalysis of further previously published RNA-Seq data sets derived from brains of AD patients and control subjects, GSE53697 (32) and GSE104705 (33), corroborated distinct upregulation of both HERV-K and TLR8 expression in AD (Supplemental Figure 8E).
To investigate whether endogenous HERV-K RNA is released in AD, we examined CSF of AD patients and control CSF (for patient details, see Supplemental Table 1) for the presence of HERV-K transcripts by RT-PCR. HERV-K RNA was detected in CSF of 14 out of 25 (56%) AD patients, whereas only 2 out of 25 (8%) control individuals were positive for HERV-K RNA (P = 0.0006) (Figure 6E). Sanger-based amplicon-sequencing of amplified RT-PCR products from HERV-K RNA–positive CSF supported RT-PCR products as the targeted HERV-K env gene region and indicated transcripts with the sequence 5′-GUUGCGU-3′ to be the dominant variant based on sequence peak heights (data not shown). Next, we compared CSF-derived small RNA-Seq data sets from AD patients and healthy controls (34) with regard to TE expression, including HERVs. Small RNA from LTR5_Hs/HERV-K was enriched exclusively in patients with AD (Figure 6F), further supporting an association between HERV-K activation and AD.
Discussion
The human endogenous retrovirus group HERV-K(HML-2) is present at various loci of the human genome and is transcriptionally active. Several HERV-K(HML-2) (HERV-K) proviruses in the human genome formed recently, specifically a few million years ago in ancestral primate species, or only in the human lineage, with some of the latter proviruses being allelic in the human population (14, 35). Many of these proviruses are relatively intact, expressing viral RNA and proteins (10). Exogenous infections sustained by different viral species, including HIV and herpesviruses, as well as inflammatory factors such as NF-κB, are suggested to modulate HERV-K transcription (36, 37). However, stimuli that may trigger endogenous retrovirus reactivation and transcription, thus promoting viral RNA expression, which may further modulate the pathological status in CNS diseases, remain poorly understood (28). Further studies of endogenous retroviruses (ERVs) in animal models combined with investigations of human brain tissue may help in deciphering these aspects. Principal mechanisms by which endogenous retroviruses can injure the brain may be based on release of virus-encoded proteins or induction of potential immunopathogenic molecules from activated host immune cells (17, 38). By contrast, our current study indicates a potentially novel function for human endogenous retroviral RNA transcripts in neurodegeneration. On the basis of sequence similarity to known neurotoxicity-triggering TLR7 and TLR8 ligands derived from exogenous retroviruses (39), we asked whether a similar GU-rich sequence present in the HERV-K env gene can also activate TLR signaling. Indeed, we found that HERV-K–derived RNA containing this motif binds directly to human TLR8 and induces signaling through both human TLR8 and murine Tlr7. Following up on this, we investigated human-derived cell systems in terms of neuronal injury and modeled TLR-mediated neurodegeneration by delivering HERV-K RNA extracellularly to neurons from mice that lack HERV-K, instead harboring phylogenetically different ERVs (40). Exposure of neurons to HERV-K RNA harboring GU-rich sequences induced neurodegeneration through intrinsic murine Tlr7 and human TLR8 signaling. The essential feature of that signal is the mislocalization of an endogenous RNA into the extracellular space, potentially released during cellular injury initiated by a disease-causing factor that gains access to TLRs located in neuronal endosomes (6). Accordingly, apoptotic neurons released HERV-K RNA, which in turn induced neurodegeneration. Under physiological conditions, interaction between these HERV-K RNA transcripts and neuronal Tlr7/TLR8 may be prevented by location of these components in separate cellular compartments. However, precise local concentrations of extracellularly functional HERV-K RNA in brain parenchyma at the site of injury/pathology in neurodegenerative processes are not known and difficult to estimate. Thus, the HERV-K RNA concentrations used were based on previous studies on RNA-mediated neurotoxicity (6, 39) and may be considered supraphysiological, especially because neurotoxicity observed in our current studies occurred within days and weeks whereas neurodegeneration in the context of CNS disorders may evolve over multiple decades. Although our data present the first proof of principle demonstration to our knowledge of HERV-K RNA–mediated CNS injury, future HERV-K expression studies on the human brain will be required to determine pathophysiological concentrations of HERV-K transcripts, their cellular sources, and spatial-anatomical distribution.
Although we cannot rule out that the observed neurodegenerative effects are not specific for HERV-K RNA, but can be caused by any RNA species containing the TLR7/TLR8 recognition motif, and that presence of that motif in HERV-K is incidental, several aspects should be considered regarding a specific role for HERV-K RNA in this context. First, among HERVs the sequence motifs GUUGUGU and GUUGCGU were detected in HERV-K as a regular sequence within the env gene. Second, several differently sized nucleotides, 22-nt, 50-nt, and 785-nt, derived from the concerned HERV-K env region, as well as endogenous retroviral particle–derived full-length HERV-K transcripts, were capable of inducing neurodegeneration. Third, HERV-K RNA is present and stable in CSF. Finally, HERV-K inhibitor reduced neurotoxicity of lysates from dying neurons applied to neurons. Given that locked nucleic acid–based (LNA-based) RNA inhibitors, such as the one used, are highly sequence specific, up to the level of single-nt mismatches (25); our results indicate that HERV-K RNA is indeed involved in the induction and/or progression of neurodegeneration.
Substantial reduction of the GU content in the HERV-K sequence motif 5′-GUUGUGU-3′ abolished Tlr7 activation in neurons. However, the ability to stimulate Tlr7 depended on a GU-rich element rather than on the exact GUUGUGU motif because HERV-K RNA transcript subsets that contain slight motif variations (GUUGCGU) effectively engaged Tlr7. Several HERV-K proviruses harboring GUUGCGU are among the evolutionarily youngest proviruses (Supplemental Figure 1); that is, such proviruses were formed in the genome relatively recently. We found that HERV-K RNA inhibitor reduced neurotoxicity of virions, harboring RNA from several HERV-K loci, including loci containing the TLR7/8 recognition motifs (12, 24). Considering the sequence specificity of the LNA-based RNA inhibitor, our data strongly indicate that HERV-K RNA physiologically expressed in humans can induce neurodegeneration.
Although microglia released inflammatory molecules in response to HERV-K RNA and enhanced HERV-K RNA–induced neurodegeneration, they were not required for the observed injurious effects. Because in utero electroporation at embryonic stage E14.5 targets only neurons and not glia (6), only those neurons newly expressing mTlr7 or hTLR8 were susceptible to HERV-K RNA, and neighboring neurons lacking these TLRs were unaffected. These results indicate that HERV-K RNA activates an intrinsic cell death pathway in neurons. Further, neuronal injury induced by HERV-K RNA required SARM1, an established regulator of neurodegeneration (23) that negatively regulates TLR-mediated signaling pathways, including NF-κB activation (41). Accordingly, we found that HERV-K RNA does not induce NF-κB activation in neurons, and MyD88 is not involved in HERV-K RNA–mediated neurodegeneration. However, the exact signaling cascade linking Tlr7 and TLR8 signaling with caspase-3 activation and the relative contribution of an inflammatory response through microglia in promoting HERV-K RNA–induced neurodegeneration in vivo remain unclear and should be investigated in future studies, e.g., using microglia-depleted CNS model systems.
HERV expression has been associated with several neurological diseases, although an etiological demonstration that HERVs cause or contribute to particular diseases is lacking so far. Our analysis of transcriptome data sets derived from a large cohort of AD patients and control individuals identified substantial correlation of upregulated HERV-K and TLR8 expression (TLR7 expression being less correlated) in AD brains. Notably, this correlation was observed in TCX but not in cerebellum. Also, we observed that the most active TEs in human TCX are phylogenetically young, primate- and/or human-specific HERV-derived elements (e.g., HERV-E, LTR5_HS/HERV-K, SVA) and some potentially mutagenic long interspersed nuclear elements from subfamilies L1_Hs, L1_PA2, and L1_PA3. SVA and HERV-K can form active chromatin regions in the brain and thus affect neighbor gene expression. Because they are polymorphic in the human population, they might be considered factors for susceptibility to human-specific neurological diseases, such as AD (42, 43). Confirming previous reports (44), we detected an overall high expression of potentially mutagenic L1_Hs elements in the investigated brain samples. However, unlike HERV-K, L1_Hs was not differentially upregulated in patients with AD. Also, HERV-K RNA encompassing the TLR7/8 recognition motif was detected, by RT-PCR, more often in CSF of AD patients than in control individuals, and these data were corroborated by reanalysis of small RNA-Seq data sets derived from patients with AD. Thus, although the observed TLR8 and HERV-K upregulation in patients with AD can be both a cause and a consequence of the disease, our data may have implications for AD patients with increased HERV-K levels in CSF. Because HERV-K RNA can induce CNS injury, blocking functional effects of HERV-K transcripts may affect the course of AD. In our study, an HERV-K RNA–specific inhibitor protected neurons from neurotoxicity HERV-K transcripts induced. The HERV-K inhibitor also prevented neurodegeneration and microglial activation in response to intrathecal HERV-K in APPPS1 mice. Nevertheless, our observation of neurodegeneration and microglial accumulation, but no impact on the Aβ plaque burden, in this model points to a more complex role for HERV-K in AD.
Our data provide what may be the first conclusive evidence for a role of HERV RNA as a signaling molecule for immune receptor activation, thereby providing novel insight into the biological mode of HERV action, and for an injurious role of HERV-derived transcripts in the brain. Further research including more differentiated patient cohorts will be required to investigate the probably complex role of HERV-K in the CNS, to establish clinical consequences of neurodegeneration triggered by HERV-K through TLRs in CNS disorders, and to identify means to prevent neurodegenerative consequences from HERV-K expression.
Methods
Reagents.
The following phosphothiorate-modified oligoribonucleotides (Purimex DNA/RNA-Oligonucleotide, Integrated DNA Technologies, with “s” depicting phosphorothioate bond modification) were used: 22-nt HERV-K(HML-2) [HERV-K, HERV-K (+GU)], 5′-CsCsUsUsUsAsCsAsAsAsGsUsUsGsUsGsUsAsAsAsGsC-3′; HERV-K upstream of GUUGUGU motif, 5′-AsAsUsCsUsCsUsAsCsCsCsCsAsAsGsAsCsCsAsAsAsA-3′ [HERV-K (-GU)]; 50-nt HERV-K, 5′-GsUsCsUsAsAsCsAsGsUsUsCsCsUsUsUsAsCsAsAsAsGsUsUsGsUsGsUsAsAsAsGsCsCsCsCsCsUsUsAsUsAsUsGsCsUsAsGsUsU-3′ (HERV-K long); scrambled 50-nt HERV-K, 5′-UsUsGsGsGsAsUsUsCsCsUsAsAsUsAsAsUsUsCsAsGsUsAsAsUsCsUsAsCsUsCsCsUsUsUsCsCsAsAsCsUsGsAsUsGsGsGsCsAsU-3′ (HERV-K long mutant); oligoribonucleotides with modified sequence, 5′-GsGsUsCsAsUsAsCsUsGsUsAsGsAsAsUsUsAsAsCsCsU-3′ and 5′-UsGsAsGsGsUsAsGsUsAsGsGsAsAsGsUsGsUsGsGsUsA-3′ (control oligoribonucleotide 1 and 2, respectively); mutant oligoribonucleotide, 5′-UsGsAsGsGsUsAsGsAsAsGsGsAsUsAsUsAsAsGsGsAsU-3′; and HERV-K young, 5′-CsCsUsUsUsAsCsAsAsAsGsUsUsGsCsGsUsAsAsAsGsC-3′. For some experiments, unmodified oligoribonucleotides were used. Loxoribine, R848, TL8-506, and polyinosine-polycytidylic acid were purchased from InvivoGen. LPS was obtained from List Biological Labs.
Mice and cell lines.
Tlr7-KO, Myd88-KO, Tlr2/4-KO, (provided by S. Akira, Osaka University, Osaka, Japan), APPPS1, and C57BL/6 mice were housed and bred under specific pathogen–free conditions at the Forschungseinrichtungen für Experimentelle Medizin Charité animal facility. Sarm1-KO mice (University of Cambridge, Cambridge, United Kingdom) and their wild-type counterparts (C57BL/6) were maintained as previously described (45). All animal experiments complied with the Animal Research: Reporting of In Vivo Experiments guidelines.
Immortalized BMDMs (University of Massachusetts Medical School, Worcester, Massachusetts, USA) (46) and Tera-1 cells (Universität des Saarlandes, Saarbrücken, Germany) (12) were maintained as previously described. SH-SY5Y cells (Markus Höltje, Charité – Universitätsmedizin, Berlin, Germany), HEK-Blue TLR7/TLR8 reporter cells, and HEK-Blue Null1, Null1-v, Null1-k, and Null2-k lines (InvivoGen) were cultured in DMEM with 10% heat-inactivated fetal calf serum and penicillin (100 U/mL)/streptomycin (100 μg/mL) (Thermo Fisher Scientific). THP-1 cells (Elisabeth Kowenz-Leutz, Max Delbrück Center for Molecular Medicine, Berlin, Germany) were cultured in RPMI-1640 with 0.05 mM 2-mercaptoethanol, 10% heat-inactivated fetal calf serum, and penicillin (100 U/mL)/streptomycin (100 μg/mL). Four days before use, THP-1 cells were differentiated into macrophages by incubation with 100 ng/mL PMA for 48 hours. Cells were cultured for another 2 days in PMA-free medium and were grown at 37°C in humidified air with 5% CO2.
Immunocytochemistry and immunohistochemistry.
Immunostaining was performed as described previously (5) using the following antibodies: NeuN (MAB377, clone A60), neurofilament (MAB5262, clone RT97), microtubule-associated protein 2 (MAB3418, clone AP20) (all 1:1000, Merck Millipore), active caspase-3 (9661, Asp175, Cell Signaling Technology, 1:300), and Iba1 (019-19741, Wako, 1:500). IB4 was obtained from Invitrogen, Thermo Fisher Scientific. DAPI was purchased from Roche. Aβ immunostaining with 4G8 antibody (BioLegend) and morphometric analysis were performed as described previously (47). An Olympus BX51 microscope and a Leica TCS SL confocal laser-scanning microscope were used.
Toxicity assays.
Oligoribonucleotides and other reagents were added to neuronal cultures for the indicated periods. Inhibition of HERV-K transcripts was achieved by using an inhibitor oligoribonucleotide with phosphorothioate backbone (5′-TACACAACTTTGTAAA-3′, Exiqon). A nonspecific inhibitor oligoribonucleotide (5′-ACGTCTATACGCCCA-3′) was used as a negative control. Neurons were seeded in 24-well plates (5 × 105 per well). After 24 hours, cells were incubated with HERV-K inhibitor (10 nM) or nonspecific inhibitor in the presence of Tera-1 virion particles for the indicated periods. For each condition, experiments were performed in duplicate. NeuN-positive cells were counted by analyzing 6 high-power fields per coverslip. Viability of control cells was set to 100%. Numbers of NeuN-positive cells observed for each condition were compared with control. Results were expressed as relative neuronal viability.
Intrathecal injection.
Intrathecal injection of 6- to 8-week-old male C57BL/6 and Tlr7-KO mice with 10 μg RNA was performed as described previously (6, 48).
APPPS1 mice, 4 weeks old, were intrathecally injected with 10 μg HERV-K RNA with or without 125 pmol HERV-K inhibitor treatment given intrathecally 16 hours beforehand. Sham-operated mice received PBS instead of RNA. Injections were repeated every 4 weeks until the end of the experiment (120 days of age).
After transcardial perfusion with 4% paraformaldehyde, brains were removed and cryoprotected in 30% sucrose. Neuronal survival and microglial numbers were analyzed by blinded quantification counting cortical NeuN-/active caspase-3–positive cells and Iba1-positive cells, respectively, in 6 fields (original magnification, ×60) of 5 representative sections of each brain.
In utero electroporation.
The plasmid pCAG-mTLR7-IRES-EGFP was constructed by subcloning the mTlr7 coding sequence from pUNO-TLR7-HA (Invitrogen, Thermo Fisher Scientific) into pCAG-IRES-EGFP, as previously described (6). The plasmid pCAG-hTLR8-IRES-EGFP was generated by blunt ligation of pUNO-hTLR8 (InvivoGen) digested by AgeI and NheI into EcoRV-digested pCAG-IRES-EGFP. pCAG-IRES-tdTomato served as the control vector. Plasmids were purified using an endotoxin-free plasmid purification kit (QIAGEN) and eluted into nuclease-free water. Before electroporation, plasmids were diluted to 2 μg/μL with water and mixed with 1 μL fast green per 20 μL DNA. In utero electroporation was carried out as described (49) with minor adjustments. E14.5 mice were deeply anesthetized (isoflurane, DeltaSelect, and oxygen, 1 L/min). The uterus with embryos was exposed by performing a ~2-cm midline incision in the ventral abdomen peritoneum. Plasmid DNA was injected into the lateral ventricle using a glass capillary and a pico spritzer. Electroporation was performed by applying 6 electric square pulses of 35 V each, with 50-ms duration and 950-ms intervals, using an ECM830 electroporator (BTX Harvard Apparatus). The uterus was returned to the abdominal cavity; the body wall was sutured. One-third of the litter received control (tdTomato) plasmid. At P19, animals were intrathecally injected with RNA, as described above.
Overexpression of HERV-K by lentiviral transduction.
We utilized lentiviral overexpression in SH-SY5Y cells for HERV-K RNA transcription. Within a lentiviral shuttle vector (FUGW) (50), a 785-nt HERV-K sequence portion (corresponding to nt 7291–8076 in GenBank accession number AF074086.2) (51) was subcloned downstream of a nucleus-targeting GFP (NLS-GFP) reporter gene that was controlled by a strong synthetic CAG promoter. The HERV-K sequence contained either the TLR7 recognition motif 5′-GTTGTGT-3′ f(CAG)-NLS-GFP-HML2(TLR)-w or an engineered mutated motif 5′-GTTGCGT-3′: f(CAG)-NLS-GFP-HML2(noTLR)-w. HERV-K sequences with wild-type or mutated TLR7 recognition motif were subcloned in reverse-complement orientation into the lentiviral vector: f(CAG)-NLS-GFP-asHML2(TLR)-w, f(CAG)-NLS-GFP-asHML2(noTLR)-w. Viral production was performed by the Charité Viral Core Facility (http://vcf.charite.de) (50) using the following lentivirus constructs: BL-1177 f(CAG)-NLS-GFP-HML2(TLR)-w, BL-1178 f(CAG)-NLS-GFP-HML2(noTLR)-w, BL-1179 f(CAG)-NLS-GFP-asHML2(TLR)-w, and BL-1180 f(CAG)-NLS-GFP-asHML2(noTLR)-w.
Primary neurons and microglia.
Cultures of purified cortical neurons and microglia and cocultures were generated as described previously (4).
Isolation of virions from Tera-1 cells.
Retroviral particles released from Tera-1 cells (12, 24, 52) were isolated using Total Exosome Isolation Kit (Invitrogen, Thermo Fisher Scientific) according to the manufacturer’s instructions. Briefly, supernatant was centrifuged at 2000 g for 30 minutes at 4°C. Then, 0.5 volume of exosome isolation reagent was added and mixed before incubating overnight at 4°C. Samples were centrifuged at 10,000 g for 1 hour at 4°C. Supernatant was removed. The pellet containing virions was resuspended in PBS.
RNA extraction.
Total cellular RNA was isolated using the RNeasy Mini Kit (QIAGEN) following the manufacturer’s protocol, using 5 × 106 cells. After centrifugation of cells at 300 g for 5 minutes, supernatant was removed, and 600 μL disrupting buffer was added to the cell pellet. Lysate was homogenized using QIAshredder spin columns (QIAGEN). One volume of 70% (v/v) ethanol was added to the homogenized lysate and transferred into an RNeasy spin column. After 3 washing steps, RNA was eluted by 2 centrifugation steps using 40 μL RNase-free water.
Total RNA from supernatant of cultured cells was isolated using the mirVana Paris RNA Purification Kit (Thermo Fisher Scientific). Supernatant was centrifuged (10 minutes at 1200 g) before the isolation procedure to remove cell remnants. An equal volume of 2× denaturating solution was added and incubated on ice for 5 minutes. One volume of acid-phenol:chloroform (Thermo Fisher Scientific) was added, vortexed for 60 seconds, and centrifuged for 5 minutes. The aqueous phase was mixed with 1.25 volumes of pure ethanol. The mixture was pipeted onto a filter cartridge and washed 3 times. RNA was eluated into nuclease-free water preheated to 95°C.
Total RNA from CSF was isolated using the QIAamp Viral RNA Mini Kit (QIAGEN) according to the manufacturer’s protocol. Then, 500 μL CSF was mixed with 1 equal volume of lysis buffer and incubated at room temperature for 10 minutes, then loaded onto the QIAamp Mini column. After 2 washing steps, RNA was eluted into 40 μL of AVE Buffer (QIAGEN).
Following RNA isolation, RNA preparations were DNase treated using TURBO DNA-free Kit (Invitrogen, Thermo Fisher Scientific) to remove contaminating DNA according to the manufacturer’s protocol. Ten micrograms of RNA was incubated with 1 μL TURBO DNase in 1× reaction buffer for 30 minutes at 37°C. Next, 0.1 volume of DNase Inactivation Reagent was added to the mixture and incubated for 5 minutes at room temperature followed by centrifugation at 10,000 g for 1.5 minutes. Supernatant was transferred to a fresh tube and used for subsequent experiments.
HERV-K–specific RT-PCR.
For RT-PCR, 11.5 μL of RNA isolated from CSF was used in a 20-μL reaction volume using Moloney Murine Leukemia Virus Reverse Transcriptase (MMLV-RT; Promega). RT mixture was incubated for 60 minutes at 37°C followed by an inactivation step for 10 minutes at 70°C. With each sample, RT+ reaction (including MMLV-RT) and RT– reaction (without MMLV-RT) were set up from the same master mix. The presence of HERV-K RNA was evaluated by endpoint RT-PCR using GoTaq DNA Polymerase (Promega) and 1 μL of the generated cDNA in a 20-μL total volume reaction following the manufacturer’s protocol. Each reaction mixture contained 0.5 μM of HERV-K env–specific primers each (HERV 101F: 5′-TCTACCCTTGGGAATGGGGA-3′; HERV 499R: 5′-AGCAGAATACGGTGTTGCCA-3′). Amplification was performed using a Bio-Rad C1000 Touch Thermal Cycler at 95°C for 5 minutes followed by 40 cycles of 95°C for 50 seconds, 48°C for 50 seconds, 72°C for 1 minute, and a final elongation step for 10 minutes at 72°C. Then, 10 μL of the amplification product was electrophoresed in a 1.5% agarose gel.
Multiplex immunoassay.
Microglia, BMDMs, or THP-1 macrophages were incubated for indicated durations with HERV-K or control oligoribonucleotide complexed to the transfection agent LyoVec (InvivoGen). Supernatants were evaluated using ProcartaPlex Multiplex Immunoassay (Invitrogen, Thermo Fisher Scientific) following the manufacturer’s protocol. Samples were analyzed on a Bio-Plex System (Bio-Rad).
Flow cytometry.
After fixation and permeabilization (Cytofix/Cytoperm Kit, BD Biosciences), SH-SY5Y cells and neurons were incubated for 30 minutes at 4°C with fluorescein isothiocyanate–conjugated anti-TLR7 (IMG-665C, Imgenex), phycoerythrin-conjugated anti-TLR8 (44C143, Abcam), Alexa Fluor 647 anti–β-tubulin (MA3-226000-A647, Thermo Fisher Scientific), or isotype (1:100). Fcγ receptors were blocked before cell surface and intracellular staining. FACSCanto II (BD Biosciences) and FlowJo software (Tree Star) were used for analysis.
EMSAs.
EMSAs were performed as described previously (53). Cells were washed with ice-cold PBS and transferred to a tube. Cells were centrifuged (10,000 g at 4°C for 15 seconds), resuspended in WholeCellExtract buffer (20 mM HEPES, 450 mM NaCl, 20% glycerin, 1 mM MgCl2, 0.5 mM EDTA, 0.1 mM EGTA, 1% NP-40, pH 7–8), and incubated at 4°C and 1000 rpm for 30 minutes on a thermo shaker, followed by another centrifugation at 14,000 rpm at 4°C for 10 minutes. Supernatant was transferred into a new tube. Protein concentration was determined, with lysates frozen in liquid nitrogen and stored at –80°C. For EMSAs, cell extracts (10 μg) were incubated for 30 minutes at 25°C with radioactively labeled oligonucleotides (5′-gatcCAGGGCTGGGGATTCCCCATCTCCACAGG-3′, 5′-gatcCCTGTGGAGATGGGGAATCCCCAGCCCTG-3′) and supplemented with 2× EMSA buffer (40 mM HEPES pH 7.9, 120 mM KCl, 8% Ficoll, 2 mM DTT), 2 μg BSA, 2 μg poly(dI-dC), and water (volume 20 μL). For super shift experiments, cell extracts were preincubated with 1 μg antibody for 30 minutes on ice. Probes were analyzed by electrophoresis (5% polyacrylamide gel in 0.5× Tris-Borat-EDTA buffer).
HEK-Blue reporter assay.
One day after seeding 50,000 cells/well in a 96-well plate, HEK-Blue SEAP reporter 293 cells expressing mTlr7, mTlr8, hTLR7, or hTLR8 and control cells were transfected with HERV-K or control oligoribonucleotide. Cells were stimulated with indicated agents dissolved in 90% HEK-Blue Detection reagent (InvivoGen) and 10% culture medium. SEAP reporter activation was detected at 655 nm using Varioskan Flash (Thermo Fisher Scientific). For each condition, 4 wells were individually measured. The average was used for evaluation.
Microscale thermophoresis.
Purified polyhistidine-tagged TLR8 protein (LSBio) was delivered in Tris/HCL buffer with 50% glycerol and was labeled following the manufacturer’s protocol (RED-Tris-NTA dye, NanoTemper Technologies). TLR8 (2 μM) was diluted 1:10 in microscale thermophoresis (MST) buffer (50 mM Tris/HCl pH 7.4, 150 mM NaCl, 10 mM MgCl2, 0.05% polysorbate [Tween] 20, 0.06% n-Dodecyl-β-Maltoside) to a final concentration of 200 nM. RED-Tris-NTA was dissolved in 1× PBS-Tween buffer to a concentration of 100 nM, mixed 1:1 with TLR8, and incubated on ice for 30 minutes. MST was performed at 10°C. To prevent nucleotide oligomerization, each oligoribonucleotide was incubated for 5 minutes at 80°C and placed on ice until the experiment. RNA was titrated by serial water dilution. For each measurement, 5 μL of RNA was mixed with 5 μL RED-Tris-NTA–labeled TLR8 and incubated on ice for 5 minutes before samples were loaded onto standard glass capillaries (Monolith NT.115 MST power = medium; LED power = 100%). Data were analyzed using NanoTemper MO Affinity Analysis V2.3 software and visualized using SigmaPlot V13.0.
RNA-Seq data analysis.
RNA-Seq data sets were analyzed for gene and TE expression. The Mayo data set (https://www.synapse.org) included 376 individuals. Data sets of cerebellum and TCX samples derived from AD patients, PSP patients, and controls, as well as covariate files containing clinical information, were downloaded from the Synapse portal and were converted from bam to FASTQ format using the bamtofastq function from bedtools. Following quality control, 45 samples were excluded from further analysis because of low quality (RNA integrity number ≤ 7). Additionally, the following RNA-Seq data sets were analyzed: dorsolateral prefrontal cortex samples from 9 AD patients and 8 control individuals (GSE53697) (32), 8 samples from the lateral temporal lobe of young healthy individuals, 10 samples from the lateral temporal lobe of healthy aged individuals, and 12 samples from the lateral temporal lobe from aged AD patients (GSE104705) (33). Data were downloaded as raw sequence reads in sequence read archive format and converted to FASTQ files with SRA toolkit. Due to variable quality scores, 2–5 terminal nt were removed from the sequencing reads depending on quality control scores of FASTQ files. To analyze RefSeq genes and TEs simultaneously, coordinates of TE loci were downloaded from UCSC Genome Browser. For further analysis, only those TE loci were considered that are not in the vicinity of coding sequences (|5 kb|). Gene track format (gtf) of protein-coding genes and TEs were combined into one gtf file. To estimate gene expression levels, we indexed the soft-masked hg19/GRCh37 genome providing the transcriptome model (combined gtf of RefSeq genes and TEs) using STAR default parameters. For read mapping, we used our defined settings (–alignIntronMin 20 –alignIntronMax 1000000 –chimSegmentMin 15 –chimJunctionOverhangMin 15 –outFilterMultimapNmax 20) for STAR splice mapper (54). Reads with low map quality score (<30) were not used. To obtain uniquely mapped read counts of genes and TEs with RefSeq annotations, we used featureCounts. For TE expression, multimapping reads counted only if they mapped exclusively within a TE family. Gene expression levels were calculated as TPM counts over a gene (defined as any transcript located between transcription start and end sites). To analyze differential TE expression, we counted uniquely mapped reads over given coordinates and calculated CPM of mappable reads. Reads per kilobase per million values were calculated as average expression of values for both TE loci and genes only if pairwise comparison was performed. To calculate differential gene expression, we used the generalized fold change algorithm, which calculates normalization constant and variance to extract fold changes from unreplicated RNA-Seq data. To calculate log2 fold changes and empirical Bayes-corrected P values for each expressed TE family or locus, we subjected normalized counts (using DESeq2) to limma/Voom analysis. For comparative analyses, data sets from different layouts were never merged into one data frame. Differential expression of HERV-K was statistically significant between AD and control TCX samples but not cerebellum samples. After this filter step, the TCX data frame consisted of 82 AD and 94 control (PSP and age-matched healthy individual) samples (29). Also, TE expression was analyzed with regard to evolutionary age of TE families by modifying the curated TE classification from Repbase as follows: TEs of New World monkeys (Platyrrhini), Old World monkeys (Catarrhini), and hominoids (Hominoidae) were merged into one group as “primate specific.” Because it was not annotated correctly in Repbase, HERV-H was moved from the “eutherian” to the “primate-specific” group. To compare relative expression of an individual TE family or a gene across the data frame in a pairwise manner, the row-wise Z score of DESeq2-normalized counts was used. Significance was calculated using Wilcoxon’s test.
Small RNA-Seq data from CSF samples of AD patients and controls were provided by Julie A. Saugstad (Oregon Health & Science University, Portland, Oregon, USA) in FASTQ format. Detailed data information is available on the exRNA Research Portal (http://exRNA.org/; ref. 34). We analyzed small RNA-Seq raw data by mapping high-quality reads (map quality score > 30) to the human reference genome using bowtie2 with the following parameters: --local -p 8 -q --phred33 -D 20 -R 3 -N 0 -L 8 -i S,1,0.50. hg19 coordinates annotated by Repeatmasker showing more than 5 uniquely mapped reads were reported.
Pathway analysis of differentially expressed genes.
Canonical pathways and biological functions of differentially expressed genes in data sets were investigated using Kyoto Encyclopedia of Genes and Genomes pathway and Gene Ontology tools. Overrepresentation of a biological pathway was assessed by Fisher’s exact test and corrected for multiple testing using the Benjamini-Hochberg procedure. The ratio was calculated as the number of genes from the data set that map to a particular pathway divided by the number of total genes included in the pathway.
Gene correlational network analysis.
To identify significant correlation gene networks, we applied WGCNA. This algorithm was used to determine gene sets coexpressed in the transcriptome of the 176 TCX samples above and identified approximately 2400 genes and 17 TE families, including HERV-K, as highly expressed. The metamodel identified probable significant networks. In a significant coexpression module, HERV-K was detected and then used as a probe to identify HERV-K–associated coexpression clusters. Genes correlated beyond the provided correlation thresholds were extracted from the HERV-K cluster. Finally, a pairwise-ranked correlation matrix of identified genes and TEs was generated. Row-wise Z scores of DESeq2-normalized counts were displayed as scatter plots, significance of differential expression was calculated by Wilcoxon’s test, and rho value and significance of correlation were calculated using cor.test built-in R function.
Statistics.
Data are expressed as mean ± SEM or ± SD, as indicated. Statistical differences over all groups were determined using nonparametric Kruskal-Wallis test or 1-way ANOVA, as indicated. Statistical differences between selected groups were determined by Kruskal-Wallis test with Dunn’s post hoc analysis or 1-way ANOVA with Bonferroni’s post hoc analysis. Statistical differences between AD patients and controls were determined using 2-tailed Student’s t test, Fisher’s exact test, or Wilcoxon’s test, as indicated. Statistical differences were considered significant at P < 0.05.
Study approval.
Studies on patients and control individuals complied with all relevant ethical regulations and were approved by institutional review boards (Ethikkommission Charité, Dementia competence network). Written informed consent was obtained from all participants prior to inclusion in the study. Mouse experiments conformed to the guidelines approved by the Landesamt für Gesundheit und Soziales, Berlin, Germany.
Author contributions
SL, JM, ZI, and KR conceived the study and wrote the manuscript. MC, OP, DG, GK, PS, VT, FLH, TW, HS, and C Scheidereit planned the experiments. PD, AGN, M Singh, MH, M Szczepek, CK, KD, C Schipke, RA, OD, BCR, and TT planned and carried out the experiments.
Supplementary Material
Acknowledgments
This work was supported by Deutsche Forschungsgemeinschaft (DFG) LE 2420/2-1, SFB-TRR167/B3, NeuroCure Exc 257 (to SL), SFB740/B6, SFB1078/B6 (to PS), DFG Cluster of Excellence “Unifying Concepts in Catalysis” (EXC 314 - Research Field D/E) (to GK and PS), and European Research Council, ERC Advanced (ERC-2011-ADG 294742) (to ZI). JM is supported by DFG.
Published data in raw format were provided by Kendall Van Keuren-Jensen, Translational Genomics Research Institute, Phoenix, Arizona, USA, and Julie A. Saugstad, Oregon Health & Science University, Portland, Oregon, USA. Access to RNA-Seq data sets was supported by Vikas Bansal, Deutsches Zentrum für Neurodegenerative Erkrankungen in der Helmholtz-Gemeinschaft, Tübingen, Germany.
Version 1. 04/09/2020
Electronic publication
Footnotes
RA’s present address is: Department of Anatomy, Histology and Embryology, Faculty of Medicine, University of Szeged, Szeged, Hungary.
M Singh’s present address is: Department of Molecular Biology and Genetics, Cornell University, Ithaca, New York, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(7):e131093.https://doi.org/10.1172/jci.insight.131093.
Contributor Information
Paul Dembny, Email: paul.dembny@charite.de.
Andrew G. Newman, Email: andrew.newman@charite.de.
Manvendra Singh, Email: Manvendra.Singh@mdc-berlin.de.
Michael Hinz, Email: m.Hinz@mdc-berlin.de.
Michal Szczepek, Email: michal.szczepek@charite.de.
Christina Krüger, Email: christina.krueger@charite.de.
Robert Adalbert, Email: radalbert@rvc.ac.uk.
Omar Dzaye, Email: omar.dzaye@charite.de.
Thorsten Trimbuch, Email: thorsten.trimbuch@charite.de.
Thomas Wallach, Email: thomas.wallach@charite.de.
Gunnar Kleinau, Email: gunnar.kleinau@charite.de.
Katja Derkow, Email: katjaderkow@aol.de.
Claus Scheidereit, Email: scheidereit@mdc-berlin.de.
Harald Stachelscheid, Email: Harald.Stachelscheid@charite.de.
Douglas Golenbock, Email: douglas.Golenbock@umassmed.edu.
Oliver Peters, Email: oliver.peters@charite.de.
Frank L. Heppner, Email: frank.heppner@charite.de.
Patrick Scheerer, Email: patrick.scheerer@charite.de.
Victor Tarabykin, Email: victor.tarabykin@charite.de.
Klemens Ruprecht, Email: klemens.ruprecht@charite.de.
Jens Mayer, Email: Jens.Mayer@uks.eu.
Seija Lehnardt, Email: seija.lehnardt@charite.de.
References
- 1.Kono H, Rock KL. How dying cells alert the immune system to danger. Nat Rev Immunol. 2008;8(4):279–289. doi: 10.1038/nri2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11(5):373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 3.Owens T. Toll-like receptors in neurodegeneration. Curr Top Microbiol Immunol. 2009;336:105–120. doi: 10.1007/978-3-642-00549-7_6. [DOI] [PubMed] [Google Scholar]
- 4.Lehnardt S, et al. Activation of innate immunity in the CNS triggers neurodegeneration through a Toll-like receptor 4-dependent pathway. Proc Natl Acad Sci U S A. 2003;100(14):8514–8519. doi: 10.1073/pnas.1432609100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lehnardt S, et al. A vicious cycle involving release of heat shock protein 60 from injured cells and activation of toll-like receptor 4 mediates neurodegeneration in the CNS. J Neurosci. 2008;28(10):2320–2331. doi: 10.1523/JNEUROSCI.4760-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lehmann SM, et al. An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat Neurosci. 2012;15(6):827–835. doi: 10.1038/nn.3113. [DOI] [PubMed] [Google Scholar]
- 7.Dewannieux M, Heidmann T. Endogenous retroviruses: acquisition, amplification and taming of genome invaders. Curr Opin Virol. 2013;3(6):646–656. doi: 10.1016/j.coviro.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 8.Hohn O, Hanke K, Bannert N. HERV-K(HML-2), the best preserved family of HERVs: endogenization, expression, and implications in health and disease. Front Oncol. 2013;3:246. doi: 10.3389/fonc.2013.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flockerzi A, et al. Expression patterns of transcribed human endogenous retrovirus HERV-K(HML-2) loci in human tissues and the need for a HERV Transcriptome Project. BMC Genomics. 2008;9:354. doi: 10.1186/1471-2164-9-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanke K, Hohn O, Bannert N. HERV-K(HML-2), a seemingly silent subtenant - but still waters run deep. APMIS. 2016;124(1-2):67–87. doi: 10.1111/apm.12475. [DOI] [PubMed] [Google Scholar]
- 11.Boller K, et al. Human endogenous retrovirus HERV-K113 is capable of producing intact viral particles. J Gen Virol. 2008;89(pt 2):567–572. doi: 10.1099/vir.0.83534-0. [DOI] [PubMed] [Google Scholar]
- 12.Ruprecht K, et al. Human endogenous retrovirus family HERV-K(HML-2) RNA transcripts are selectively packaged into retroviral particles produced by the human germ cell tumor line Tera-1 and originate mainly from a provirus on chromosome 22q11.21. J Virol. 2008;82(20):10008–10016. doi: 10.1128/JVI.01016-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marchi E, Kanapin A, Magiorkinis G, Belshaw R. Unfixed endogenous retroviral insertions in the human population. J Virol. 2014;88(17):9529–9537. doi: 10.1128/JVI.00919-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wildschutte JH, Williams ZH, Montesion M, Subramanian RP, Kidd JM, Coffin JM. Discovery of unfixed endogenous retrovirus insertions in diverse human populations. Proc Natl Acad Sci U S A. 2016;113(16):E2326–E2334. doi: 10.1073/pnas.1602336113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnston JB, Silva C, Holden J, Warren KG, Clark AW, Power C. Monocyte activation and differentiation augment human endogenous retrovirus expression: implications for inflammatory brain diseases. Ann Neurol. 2001;50(4):434–442. doi: 10.1002/ana.1131. [DOI] [PubMed] [Google Scholar]
- 16.Douville R, Liu J, Rothstein J, Nath A. Identification of active loci of a human endogenous retrovirus in neurons of patients with amyotrophic lateral sclerosis. Ann Neurol. 2011;69(1):141–151. doi: 10.1002/ana.22149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Douville RN, Nath A. Human endogenous retroviruses and the nervous system. Handb Clin Neurol. 2014;123:465–485. doi: 10.1016/B978-0-444-53488-0.00022-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeong BH, Lee YJ, Carp RI, Kim YS. The prevalence of human endogenous retroviruses in cerebrospinal fluids from patients with sporadic Creutzfeldt-Jakob disease. J Clin Virol. 2010;47(2):136–142. doi: 10.1016/j.jcv.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 19.Heil F, et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303(5663):1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 20.Forsbach A, et al. Identification of RNA sequence motifs stimulating sequence-specific TLR8-dependent immune responses. J Immunol. 2008;180(6):3729–3738. doi: 10.4049/jimmunol.180.6.3729. [DOI] [PubMed] [Google Scholar]
- 21.Hornung V, et al. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat Med. 2005;11(3):263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- 22.Ma Y, et al. Toll-like receptor 8 functions as a negative regulator of neurite outgrowth and inducer of neuronal apoptosis. J Cell Biol. 2006;175(2):209–215. doi: 10.1083/jcb.200606016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osterloh JM, et al. dSarm/Sarm1 is required for activation of an injury-induced axon death pathway. Science. 2012;337(6093):481–484. doi: 10.1126/science.1223899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhardwaj N, Montesion M, Roy F, Coffin JM. Differential expression of HERV-K (HML-2) proviruses in cells and virions of the teratocarcinoma cell line Tera-1. Viruses. 2015;7(3):939–968. doi: 10.3390/v7030939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kauppinen S, Vester B, Wengel J. Locked nucleic acid: high-affinity targeting of complementary RNA for RNomics. Handb Exp Pharmacol. 2006;(173):405–422. doi: 10.1007/3-540-27262-3_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Querfurth HW, LaFerla FM. Alzheimer’s disease. N Engl J Med. 2010;362(4):329–344. doi: 10.1056/NEJMra0909142. [DOI] [PubMed] [Google Scholar]
- 27.Radde R, et al. Abeta42-driven cerebral amyloidosis in transgenic mice reveals early and robust pathology. EMBO Rep. 2006;7(9):940–946. doi: 10.1038/sj.embor.7400784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manghera M, Ferguson J, Douville R. Endogenous retrovirus-K and nervous system diseases. Curr Neurol Neurosci Rep. 2014;14(10):488. doi: 10.1007/s11910-014-0488-y. [DOI] [PubMed] [Google Scholar]
- 29.Hodes RJ, Buckholtz N. Accelerating Medicines Partnership: Alzheimer’s Disease (AMP-AD) knowledge portal aids Alzheimer’s drug discovery through open data sharing. Expert Opin Ther Targets. 2016;20(4):389–391. doi: 10.1517/14728222.2016.1135132. [DOI] [PubMed] [Google Scholar]
- 30.Golbe LI. Progressive supranuclear palsy. Semin Neurol. 2014;34(2):151–159. doi: 10.1055/s-0034-1381736. [DOI] [PubMed] [Google Scholar]
- 31.Coufal NG, et al. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460(7259):1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheckel C, et al. Regulatory consequences of neuronal ELAV-like protein binding to coding and non-coding RNAs in human brain. Elife. 2016;5:e10421. doi: 10.7554/eLife.10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nativio R, et al. Dysregulation of the epigenetic landscape of normal aging in Alzheimer’s disease. Nat Neurosci. 2018;21(4):497–505. doi: 10.1038/s41593-018-0101-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saugstad JA, et al. Analysis of extracellular RNA in cerebrospinal fluid. J Extracell Vesicles. 2017;6(1):1317577. doi: 10.1080/20013078.2017.1317577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subramanian RP, Wildschutte JH, Russo C, Coffin JM. Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology. 2011;8:90. doi: 10.1186/1742-4690-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grandi N, Tramontano E. Human endogenous retroviruses are ancient acquired elements still shaping innate immune responses. Front Immunol. 2018;9:2039. doi: 10.3389/fimmu.2018.02039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mortelmans K, Wang-Johanning F, Johanning GL. The role of human endogenous retroviruses in brain development and function. APMIS. 2016;124(1-2):105–115. doi: 10.1111/apm.12495. [DOI] [PubMed] [Google Scholar]
- 38.Li W, et al. Human endogenous retrovirus-K contributes to motor neuron disease. Sci Transl Med. 2015;7(307):307ra153. doi: 10.1126/scitranslmed.aac8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lehmann SM, et al. Extracellularly delivered single-stranded viral RNA causes neurodegeneration dependent on TLR7. J Immunol. 2012;189(3):1448–1458. doi: 10.4049/jimmunol.1201078. [DOI] [PubMed] [Google Scholar]
- 40.Stocking C, Kozak CA. Murine endogenous retroviruses. Cell Mol Life Sci. 2008;65(21):3383–3398. doi: 10.1007/s00018-008-8497-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carty M, Goodbody R, Schröder M, Stack J, Moynagh PN, Bowie AG. The human adaptor SARM negatively regulates adaptor protein TRIF-dependent Toll-like receptor signaling. Nat Immunol. 2006;7(10):1074–1081. doi: 10.1038/ni1382. [DOI] [PubMed] [Google Scholar]
- 42.Fuentes DR, Swigut T, Wysocka J. Systematic perturbation of retroviral LTRs reveals widespread long-range effects on human gene regulation. Elife. 2018;7:e35989. doi: 10.7554/eLife.35989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pontis J, et al. Hominoid-specific transposable elements and KZFPs facilitate human embryonic genome activation and control transcription in naive human ESCs. Cell Stem Cell. 2019;24(5):724–735.e5. doi: 10.1016/j.stem.2019.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun W, Samimi H, Gamez M, Zare H, Frost B. Pathogenic tau-induced piRNA depletion promotes neuronal death through transposable element dysregulation in neurodegenerative tauopathies. Nat Neurosci. 2018;21(8):1038–1048. doi: 10.1038/s41593-018-0194-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gilley J, Orsomando G, Nascimento-Ferreira I, Coleman MP. Absence of SARM1 rescues development and survival of NMNAT2-deficient axons. Cell Rep. 2015;10(12):1974–1981. doi: 10.1016/j.celrep.2015.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roberson SM, Walker WS. Immortalization of cloned mouse splenic macrophages with a retrovirus containing the v-raf/mil and v-myc oncogenes. Cell Immunol. 1988;116(2):341–351. doi: 10.1016/0008-8749(88)90236-5. [DOI] [PubMed] [Google Scholar]
- 47.Vom Berg J, et al. Inhibition of IL-12/IL-23 signaling reduces Alzheimer’s disease-like pathology and cognitive decline. Nat Med. 2012;18(12):1812–1819. doi: 10.1038/nm.2965. [DOI] [PubMed] [Google Scholar]
- 48.Hoffmann O, et al. TLR2 mediates neuroinflammation and neuronal damage. J Immunol. 2007;178(10):6476–6481. doi: 10.4049/jimmunol.178.10.6476. [DOI] [PubMed] [Google Scholar]
- 49.Saito T. In vivo electroporation in the embryonic mouse central nervous system. Nat Protoc. 2006;1(3):1552–1558. doi: 10.1038/nprot.2006.276. [DOI] [PubMed] [Google Scholar]
- 50.Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295(5556):868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 51.Mayer J, Sauter M, Rácz A, Scherer D, Mueller-Lantzsch N, Meese E. An almost-intact human endogenous retrovirus K on human chromosome 7. Nat Genet. 1999;21(3):257–258. doi: 10.1038/6766. [DOI] [PubMed] [Google Scholar]
- 52.Boller K, et al. Evidence that HERV-K is the endogenous retrovirus sequence that codes for the human teratocarcinoma-derived retrovirus HTDV. Virology. 1993;196(1):349–353. doi: 10.1006/viro.1993.1487. [DOI] [PubMed] [Google Scholar]
- 53.Hinz M, et al. Signal responsiveness of IkappaB kinases is determined by Cdc37-assisted transient interaction with Hsp90. J Biol Chem. 2007;282(44):32311–32319. doi: 10.1074/jbc.M705785200. [DOI] [PubMed] [Google Scholar]
- 54.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.