Antibiotic resistant bacteria are a significant threat to human health, with one estimate suggesting they will cause 10 million worldwide deaths per year by 2050, surpassing deaths due to cancer1. Since new antibiotic development can take a decade or longer, it is imperative to effectively use currently available drugs. Antibiotic combination therapy offers promise for treating highly resistant bacterial infections, but the factors governing the sporadic efficacy of such regimens have remained unclear. Dogma suggests that antibiotics ineffective as monotherapy can be effective in combination2. Here, using carbapenem-resistant Enterobacteriaceae (CRE) clinical isolates, we revealed the underlying basis for the majority of effective combinations to be heteroresistance. Heteroresistance is a poorly understood mechanism of resistance reported for different classes of antibiotics3–6 in which only a subset of cells are phenotypically resistant7. Within an isolate, the subpopulations resistant to different antibiotics were distinct, and over 88% of CRE isolates exhibited heteroresistance to multiple antibiotics (“multiple heteroresistance”). Combinations targeting multiple heteroresistance were efficacious, whereas those targeting homogenous resistance were ineffective. Two pan-resistant Klebsiella isolates were eradicated by combinations targeting multiple heteroresistance, highlighting a rational strategy to identify effective combinations that employs existing antibiotics and could be clinically implemented immediately.
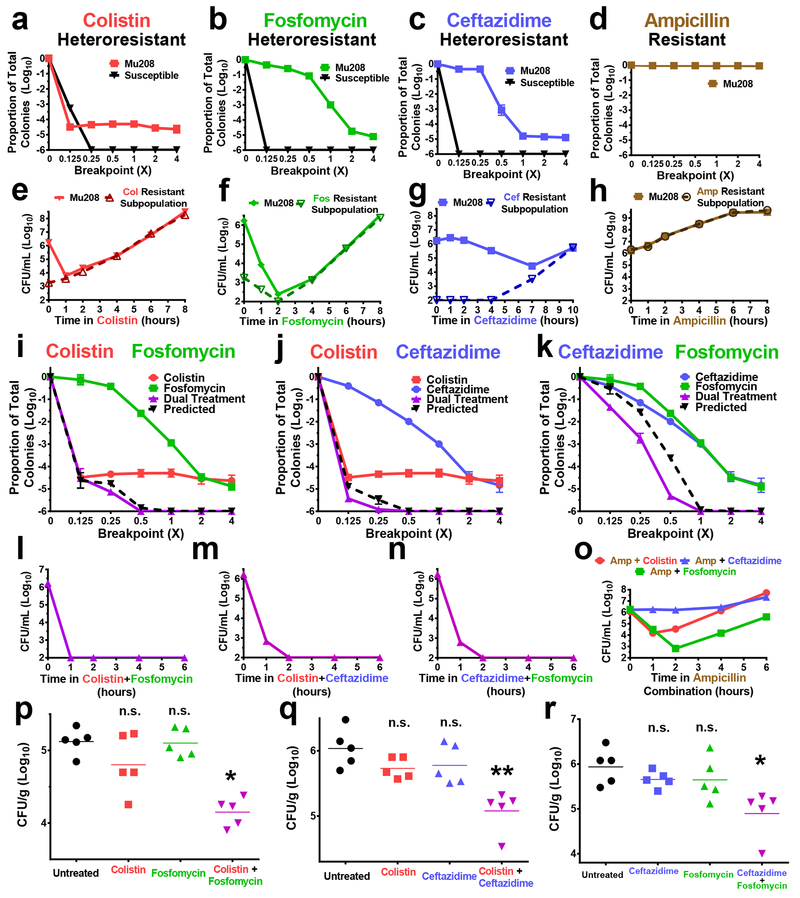
Among antibiotic-resistant bacteria, carbapenem-resistant Enterobacteriaceae (CRE; including Enterobacter spp., Klebsiella spp., Escherichia spp.) have emerged over the last two decades as an “urgent” public health threat8, with a mortality rate up to 30% for invasive infections9. Some CRE isolates are resistant to all available antibiotics and there is a lack of therapeutic options to treat such infections10. We identified an Enterobacter cloacae (Mu208) clinical isolate exhibiting heteroresistance to the last-line antibiotic, colistin, from the Georgia Emerging Infections Program’s (GA EIP) Multi-site Gram-negative Surveillance Initiative (MuGSI) for CRE11. Roughly 4 logs of Mu208 cells were killed by a concentration of colistin below the clinical breakpoint, the concentration of an antibiotic at which bacterial growth correlates with clinical resistance, and at which growth restriction correlates with clinical susceptibility and treatment success (Fig. 1a). However, a resistant subpopulation survived (Fig. 1a). Population analysis profile (PAP), in which dilutions of bacteria are plated on increasing concentrations of a respective antibiotic (Supplementary Figure 1a), revealed that this colistin resistant subpopulation survived at 4-fold the colistin breakpoint, and had a minimum inhibitory concentration (MIC) at least 32-fold greater than the susceptible cells in the population (Fig. 1a). In contrast, all the cells of a representative susceptible isolate were killed at a concentration of colistin below the breakpoint (Fig. 1a).
Figure 1. Enterobacter clinical isolate Mu208 is heteroresistant to multiple antibiotics but killed by their combinations.
a-d, Population analysis profiles (PAPs) of Mu208 and representative susceptible isolates plated on the indicated antibiotics at concentrations relative to their breakpoint. Resistance status of Mu208 to each antibiotic is indicated. Proportion of total colonies was calculated compared to growth on drug-free plates, e-h, Mu208 was treated with (e) colistin (16 μg/ml), (f) fosfomycin (256 μg/ml), (g) ceftazidime (128 μg/ml), or (h) ampicillin (128 μg/ml) at concentrations at or above their breakpoints to ensure killing of the antibiotic susceptible populations. Bacteria were plated at the indicated timepoints for enumeration of total (solid line) and resistant (dashed line) cells. i-k, PAPs of Mu208 plated on concentrations of the indicated single antibiotics or two-drug combinations (purple) relative to their breakpoints. The proportion of surviving colonies on single drug PAPs were multiplied to determine predicted additive killing (black dashed line), l-o, Mu208 was treated with (l) colistin+fosfomycin, (m) colistin+ceftazidime, (n) fosfomycin+ceftazidime, or (o) colistin, fosfomycin, or ceftazidime combinations with ampicillin (same concentrations of each drug as in e-h), and plated at the indicated timepoints to enumerate bacterial levels, p-r, Mice were infected with Mu208 intraperitoneally and treated with indicated drug combinations starting at 4 hours post infection. Peritoneal lavage was harvested at 24 hours post infection and CFU were quantified. Data shown as mean ± s.d. with n=3 (a-o) or as geometric mean with n=5 (p-r). n.s., not significant (p) p = 0.389, 0.802; (q) p = 0.087, 0.246; (r) p = 0.278, 0.286), * p < 0.05, ** p < 0.01, two-sided Mann-Whitney test.
Interestingly, PAPs using other antibiotics indicated that Mu208 also exhibited heteroresistance to antibiotics from distinct classes: fosfomycin (Fig. 1b) and ceftazidime (beta-lactam; Fig. 1c). In this study, we define heteroresistance (HR) using PAP as the survival of a subpopulation at an antibiotic concentration at least 2-fold above the breakpoint (Supplementary Fig. 1b). For both fosfomycin (Fig. 1b) and ceftazidime (Fig. 1c), a resistant subpopulation survived at 4-fold the breakpoint (Fig. 1a). In contrast, Mu208 displayed homogenous resistance to ampicillin, indicated by the lack of killing at any of the concentrations tested (Fig. 1d). These data demonstrate that Mu208 exhibits heteroresistance to multiple antibiotics, or “multiple heteroresistance”.
We monitored Mu208 growth kinetics in the presence of colistin, fosfomycin, or ceftazidime and observed for each after an initial period of killing of the susceptible cells, the resistant subpopulation rapidly replicated in the respective antibiotic (Fig. 1e–g, Supplementary Fig. 2a–c). This was consistent with heteroresistance and indicated that the resistant cells are not persisters, which do not robustly replicate in antibiotic12–14. Subsequently, upon subculture in the absence of antibiotic, the frequency of each resistant subpopulation returned to the baseline levels observed prior to antibiotic treatment (Supplementary Fig. 3a–c), consistent with unstable heteroresistance6,15. Exposure to ampicillin did not lead to the initial reduction in bacterial levels observed with the other antibiotics, consistent with Mu208 exhibiting homogenous resistance to this drug (Fig. 1h, Supplementary Fig. 2d, Supplementary Figure 3d). Together, these data are consistent with heteroresistance and show that the colistin, fosfomycin, and ceftazidime resistant subpopulations are not persisters and are not due to a stable mutation.
The colistin, fosfomycin, and ceftazidime resistant subpopulations displayed differing PAP curves (Fig. 1a–c) and kinetics of killing and growth (Fig. 1e–g) in their respective antibiotics, suggesting they were independent. We therefore investigated if the colistin, fosfomycin, and ceftazidime resistant subpopulations were specifically enriched by growth in the presence of their respective antibiotic. Colistin led to an increase in the frequency of the colistin resistant subpopulation, but did not alter the frequencies of the fosfomycin or ceftazidime resistant subpopulations (Supplementary Fig. 4a). Similarly, fosfomycin and ceftazidime led to increased frequencies of their resistant subpopulations with no effect on the other subpopulations (Supplementary Fig. 4b,c).
To further test the independence of these cells, we generated deletion mutants lacking a gene in each resistance pathway. phoQ is required for colistin heteroresistance in E. cloacae6,16. While the colistin resistant subpopulation was undetected in the phoQ mutant (Supplementary Fig. 4d), the frequencies of the fosfomycin and ceftazidime resistant subpopulations were unaffected. FosA enzymes are transferases that catalyze the inactivation of fosfomycin17 and AmpR confers beta-lactam resistance (i.e. ceftazidime) by regulating the expression of beta-lactamases18. Deletion offosA led to a reduction in the fosfomycin resistant subpopulation (Supplementary Fig. 4d), while deletion of ampR led to the reduction of the ceftazidime resistant subpopulation to undetectable levels (Supplementary Fig. 4d). Neither of these targeted mutations had effects on the other two resistant subpopulations as assayed by PAP or broth microdilution (BMD) (Supplementary Fig. 4d, 4e). In addition, none of these genes affected persister formation, as the phoQ, fosA, and ampR mutants harbored similar numbers (Supplementary Fig. 5). These data indicate that Mu208 harbors three distinct subpopulations, resistant to three different antibiotics, highlighting a diverse repertoire of phenotypic resistance mechanisms in an isolate exhibiting multiple heteroresistance.
Colistin, fosfomycin, and ceftazidime each kill a large proportion of Mu208 cells but are unable to prevent subsequent outgrowth (Fig. 1e–g). Since the subpopulations resistant to each antibiotic are at least in some cases independent (Supplementary Fig. 4), we hypothesized that combinations of these drugs would lead to killing equal to the product of the magnitudes of killing for each individual drug. Indeed, PAPs with two drugs (“dual PAPs”; using increasing concentrations of two antibiotics, each at the same multiple of their respective breakpoints) demonstrated that the killing by each dual antibiotic combination closely matched the magnitude predicted from the individual treatments (Fig. 1i–k). This indicated that killing was additive. In a time-kill experiment, the colistin/fosfomycin, colistin/ceftazidime, and fosfomycin/ceftazidime combinations each killed Mu208 within two hours (Fig. 1l–n), preventing the outgrowth observed with the individual drugs (Fig. 1e–g). These results were further supported by agar diffusion experiments. Whereas resistant colonies were visible within the zone of inhibition for both fosfomycin and ceftazidime (by Etest or disk diffusion; Supplementary Fig. 6a–f), the resistant colonies were absent at the interface of two antibiotic disks (colistin+fosfomycin, colistin+ceftazidime, or fosfomycin+ceftazidime)(Supplementary Fig. 6g–i). In both dual PAP (Fig. 1i–k) and time-kill experiments (Fig. 1l–n), the activity of colistin, fosfomycin, or ceftazidime was not enhanced by combination with ampicillin. This demonstrated that efficacy correlated with antibiotic combinations involving two drugs to which the isolate exhibited heteroresistance, but that a similar effect was not observed when the isolate was heteroresistant to one drug and homogenously resistant to the other (Supplementary Fig. 7, Fig. 1o). Together, these data demonstrate that combinations of antibiotics to which an isolate exhibits multiple heteroresistance are effective.
We next tested the efficacy of antibiotic combinations targeting multiple heteroresistance using an in vivo mouse model of peritonitis. After infection with Mu208 and subsequent monotherapy with colistin, fosfomycin, or ceftazidime, we observed that CFU recovered from these mice at 24 hours were similar to untreated mice (Fig. 1 p–r). In contrast, all three combinations (colistin+fosfomycin, Fig. 1p; colistin+ceftazidime, Fig. 1q; ceftazidime+fosfomycin, Fig. 1r) resulted in significant reduction of CFU. We tested another Enterobacter clinical isolate, Mu866, which was heteroresistant to colistin and ceftazidime but susceptible to fosfomycin (Supplementary Fig. 8a). Monotherapy with either colistin or ceftazidime was ineffective, whereas the combination of the two antibiotics led to a significant decrease in bacterial load (Supplementary Fig. 8b). The reduction was similar to that achieved by fosfomycin monotherapy, indicating that combination therapy targeting multiple heteroresistance was as effective as monotherapy targeting susceptibility (Supplementary Fig. 8b). We next tested a highly drug-resistant clinical isolate of Klebsiella pneumoniae, Mul343, which was not susceptible to any antibiotics tested including ceftazidime, but exhibited heteroresistance to colistin and fosfomycin (Supplementary Fig. 8c). Colistin and fosfomycin monotherapy failed to reduce bacterial levels in vivo while the combination of both drugs led to a significant decrease (Supplementary Fig. 8d). Finally, as a control, we used another K. pneumoniae isolate (Mul251) that was homogenously resistant to colistin but heteroresistant to fosfomycin and ceftazidime (Supplementary Fig. 8e). While the combination of fosfomycin and ceftazidime led to in vivo efficacy, neither of the two combinations including colistin reduced bacterial levels (Supplementary Fig. 8f). Taken together, these data showed that antibiotic combinations to which an isolate exhibits multiple heteroresistance are effective while those including two drugs to which an isolate exhibits heteroresistance and homogenous resistance, are ineffective.
It was unclear whether multiple heteroresistance was an infrequent or common phenomenon. Since combination antibiotic regimens are often employed when treating strains that are resistant to most or all available drugs, we interrogated a collection of 104 clinical isolates (Supplementary Table 1) of multidrug resistant CRE from the GA MuGSI surveillance program. We tested for heteroresistance using PAP, investigating a total of 11,648 conditions for the proportion of surviving bacteria (104 isolates x 16 antibiotics x 7 drug concentrations).
Heteroresistance was observed for each antibiotic tested and the proportion of isolates exhibiting heteroresistance varied widely for different drugs, ranging from 72.1% for fosfomycin to 1.0% for ampicillin (Fig. 2a, Supplementary Table 2). Surprisingly, 97.1% of the isolates were heteroresistant to at least one of the 16 antibiotics, and 86.5% exhibited heteroresistance to at least two drugs (Fig. 2b). These data indicate that heteroresistance is widespread, in line with a recent study by Nicoloff el al.19, and suggest that combination antibiotic regimens targeting multiple heteroresistance may be applicable to a large proportion of CRE.
Figure 2. Multiple heteroresistance is common in carbapenem-resistant Enterobacteriaceae (CRE).
a, One hundred and four CRE clinical isolates from a surveillance network in Georgia, USA were screened for heteroresistance to 16 antibiotics using the population analysis profile (PAP) method. Percentages of isolates heteroresistant to each antibiotic are listed, highest in red and lowest in green. Pip/Tazo; piperacillin/tazobactam, Trimeth/Sulfa; trimethoprim/sulfamethoxazole, b, Isolates classified by the number of antibiotics to which they are heteroresistant out of the 16 tested (none were heteroresistant to more than 7) with the percentage heteroresistant to more than one antibiotic indicated by central grey ring, c, Percentage of clinical susceptibility testing results (for 104 isolates and 16 antibiotics) classified as resistant (black) or susceptible (light grey). Those designated heteroresistant by PAP are indicated by central blue ring, d, Representation of all the PAPs of 104 isolates on 16 antibiotics, with lines indicating average bacterial survival at each concentration for all drug-isolate interactions, when segregated into 4 groupings: those classified as resistant by clinical testing and PAP (black circles, “Resistant (R)”), resistant by clinical testing and heteroresistant by PAP (black squares, “Heteroresistant (R)”), susceptible by clinical testing and heteroresistant by PAP (dark grey triangles, “Heteroresistant (S)”), and susceptible by clinical testing and PAP (light grey inverted triangles, “Susceptible (S)”). Data represented as mean ± SD. **** p < 1e-17 using two-tailed Welch’s t-test of average logs killing at 1x breakpoint concentration for Heteroresistant (R) vs. Heteroresistant (S) (t = 9.01).
Clinical antimicrobial susceptibility testing largely relies on liquid media-based diagnostics that assay for growth of the entire population of bacteria in the presence of different antibiotics. This approach only classifies isolate/antibiotic pairs as resistant or susceptible, but cannot differentiate heteroresistance. Sixty-four percent of the isolate/antibiotic interactions were classified as resistant by clinical testing and 35.7% as susceptible (Fig. 2c). However, PAP revealed that the bacteria exhibited heteroresistance in 21.4% of the interactions (Fig. 2c). Strikingly, 23.3% of the interactions classified as resistant by clinical testing were actually heteroresistance, as well as 17.5% of those classified as susceptible in the clinic (Fig. 2c). When we plotted the average of the PAP curves for isolate/antibiotic interactions classified as resistant by clinical testing, those also designated resistant by PAP were not killed at the breakpoint concentration (Fig. 2d). However, those demonstrated to be heteroresistant by PAP exhibited an average of 2 logs of killing at the breakpoint concentration (Fig. 2d). The average PAP curve for isolates classified as susceptible by clinical testing but heteroresistant by PAP had 4 logs of killing at the breakpoint, while those classified as susceptible by both clinical testing and PAP demonstrated 6 logs of killing at the breakpoint (Fig. 2d). Previous studies investigating the number of resistant cells influencing susceptibility testing may be explained by our findings20. These data reveal that: 1) a significant portion of the isolate/antibiotic interactions currently designated resistant by clinical testing are due to heteroresistance, 2) some heteroresistance is undetected and classified as susceptibility, and 3) detected heteroresistance is on average due to resistant subpopulations with higher frequencies (~1 in 100 cells), whereas undetected heteroresistance is associated with less frequent resistant subpopulations (~1 in 10,000; Fig. 2d).
We next investigated the efficacy of targeting multiple heteroresistance in a combination screen using a subset of 8 isolates (3 Enterobacter, 4 Klebsiella, 1 Escherichia) chosen due to the variety of antibiotics to which they exhibit heteroresistance. We tested all 120 distinct combinations of 16 antibiotics by plating the 8 isolates on breakpoint concentrations of each antibiotic pair for a total of 960 tests (Fig. 3a). We first focused on combinations of two antibiotics to which individual isolates were classified as resistant by clinical testing; 90 distinct antibiotic pairs across the 8 isolates (Supplementary File 1), for a total of 313 interactions. Among such isolate/antibiotic interactions, the majority led to less than a log of killing, but many combinations killed over 3 logs of bacteria (Fig. 3b). When the combinations (classified as resistant by clinical testing) were segregated into those involving two antibiotics to which a given isolate was designated resistant by PAP (RxR), or those to which an isolate was classified heteroresistant by PAP (HRxHR), only 4.4% of RxR interactions led to 1 log or more of killing, while 97.2% of HRxHR combinations killed one log or more bacteria and 33.3% killed at least 5 logs (Fig. 3c, d). For RxHR combinations, the reduction in bacterial levels closely mirrored the reduction observed using only the antibiotic to which a given isolate was heteroresistant (“HR alone”)(Supplementary Fig. 9a). This indicated that the antibiotic to which an isolate exhibited homogenous resistance did not significantly contribute to the reduction in bacterial levels (Fig. 1o, Supplementary Fig. 7). Additionally, monotherapy with a single drug to which a strain was heteroresistant resulted in significantly less killing than combination therapy with two drugs targeting heteroresistance (Supplementary Fig. 9a). As expected, antibiotic combinations involving drugs to which an isolate was classified as susceptible by clinical testing were highly effective in reducing bacterial levels (Supplementary Fig. 9b). Targeting heteroresistance classified as susceptible by clinical testing (HR(S)) resulted in greater killing than targeting heteroresistance classified as resistant (HR(R))(Supplementary Fig. 9c). Together, these data highlight that antibiotic combinations targeting multiple heteroresistance lead to increased bacterial killing compared to combinations targeting homogenous resistance.
Figure 3. Efficacy of antibiotic combinations is largely dependent on multiple heteroresistance.
a, Schematic of the antibiotic combination screen. Eight representative clinical isolates of CRE (4 K. pneumoniae, 3 E. cloacae, 1 E. coli) were treated with 16 antibiotics at the clinical breakpoint concentration alone, or in all 120 possible combinations. Amk, amikacin; Gen, gentamicin; Tob, tobramycin; Amp, ampicillin; Azt, aztreonam; Cfz, cefazolin; Cpm, cefepime; Cft, ceftazidime; Mer, meropenem; PTz, piperacillin/tazobactam; Cip, ciprofloxacin; Col, colistin; Fos, fosfomycin; Tet, tetracycline; Tig, tigecycline; SXT, trimethoprim/sulfamethoxazole, b, Graph of the number of antibiotic combination/isolate interactions resulting in the indicated number of logs of killing for the subset of cases in which an isolate was classified by clinical testing as resistant to both drugs (RxR)(n=313). c,e, Isolates classified as resistant by clinical testing, and designated by PAP as either resistant (RxR) or heteroresistant (HRxHR) to both drugs, categorized by the number of logs killing observed compared to antibiotic-free control (percentages are shown). In (e), only combinations including both an aminoglycoside and beta-lactam are shown. d,f, Antibiotic combinations designated as resistant by clinical testing, and either resistant (RxR; n=l 17) or heteroresistant (HRxHR; n=36) to both drugs by PAP, categorized by the number of logs of killing when compared to an antibiotic free control, expressed in number of total treatments. In (f), only combinations including both an aminoglycoside and beta-lactam are shown (RxR, n=22; HRxHR, n=10). **** p < 0.0001, two-sided Mann-Whitney U test of logs killing, binned in 1 log increments.
Aminoglycoside/beta-lactam combinations have been reported to exhibit synergy and have historically been among the most frequently used clinically21–26. We analyzed the dataset for combinations involving one of 3 aminoglycosides (amikacin, gentamicin, tobramycin) and one of 7 beta-lactams (ampicillin, aztreonam, cefazolin, cefepime, ceftazidime, meropenem or the beta-lactam/beta-lactamase inhibitor, piperacillin/tazobactam). Similar to the data including all antibiotics, 90.0% of these HRxHR combinations reduced bacterial levels by 4 logs or more (Fig. 3e, f) while only 9.1% of RxR aminoglycoside/beta-lactam combinations reduced bacterial levels by one log (Fig. 3e, f). This indicates that even the efficacy of aminoglycoside/beta-lactam combinations seems to depend on multiple heteroresistance, and that these combinations are not inherently more effective than others.
The standard method for identifying effective antibiotic combinations is the checkerboard assay which tests for the activity of a combination of two antibiotics that is greater than what would be predicted using the MIC for each drug alone27. Such combinations are described as being synergistic, and have fractional inhibitory concentration (FIC) values below 0.527,28. We performed checkerboard assays on the isolates used in the dual PAP screen (Fig. 3a), testing all of the combinations that included two drugs to which a respective isolate was classified as resistant by clinical diagnostic testing (Supplementary Fig. 10a). Only HRxHR combinations led to FIC scores below 0.5 (53.3% of HRxHR interactions were below FIC=0.5) and were classified as synergistic (Supplementary Fig. 10b). In addition, the three effective combinations that target Mu208 multiple heteroresistance were classified as synergistic by checkerboard (Supplementary Fig. 10c–e), whereas combinations including ampicillin were not (Supplementary Fig. 10f–h). These results indicate that for the isolates and antibiotics tested here, RxR combinations were not classified as synergistic, and that multiple heteroresistance may explain a significant proportion of combinations previously identified as synergistic.
To further evaluate the potential utility of combination regimens targeting multiple heteroresistance, we tested this approach against two pan-resistant K. pneumoniae isolates (Nevada-2016 which caused a lethal infection10, and AR0040). Despite being classified as pan-resistant, Nevada-2016 was determined to be heteroresistant to fosfomycin and sulfamethoxazole/trimethoprim (SXT)(Fig. 4a, b). Dual PAP revealed that fosfomycin and SXT in combination led to additive killing of Nevada-2016 (Fig. 4b). We next performed time-kill experiments which indicated that while fosfomycin or SXT alone led to significant initial killing of Nevada-2016, bacterial growth was observed by 24 hours (Fig. 4c). Only the combination of both drugs killed the bacteria and prevented their outgrowth through 48 hours (Fig. 4c, d). In contrast, the combination of amikacin and piperacillin/tazobactam, to which Nevada-2016 exhibits homogenous resistance and heteroresistance, respectively, did not lead to killing beyond that of piperacillin/tazobactam alone (Supplementary Fig. 11a). AR0040 was heteroresistant to amikacin and piperacillin/tazobactam (Fig. 4e, f) and we observed that either antibiotic alone led to killing of some bacteria, but only their combination led to eradication of the culture and prevented subsequent growth (Fig. 4f–h). In contrast, the combination of SXT and fosfomycin, to which AR0040 is resistant and heteroresistant, respectively, did not lead to eradication of the bacteria (Supplementary Fig. 11b). Furthermore, we determined that AR0040 displayed unstable heteroresistance to both amikacin and piperacillin/tazobactam, since after selection in each antibiotic and subsequent passage in antibiotic-free media, the frequency of the resistant subpopulation returned to baseline (Supplementary Fig. 12a,b). To investigate whether the efficacy of the combination of these two drugs extended to in vivo infections, we infected mice with a lethal dose of AR0040 and subsequently treated with each monotherapy or the combination. Only dual treatment with both amikacin and piperacillin/tazobactam rescued mice from lethal infection, whereas the monotherapies were ineffective (Fig. 4i). These data highlight that antibiotic combinations targeting multiple heteroresistance can effectively combat isolates deemed pan-resistant by clinical testing and which had been thought to be untreatable.
Figure 4. Eradication of pan-resistant Klebsiella by antibiotic combinations targeting multiple heteroresistance.
a, Antibiogram of Nevada-2016 as determined by clinical testing (left; using VITEK or E-test), as well as an updated representation of the antibiogram that includes heteroresistance as detected by PAP (right). aFosfomycin breakpoints are not established for Klebsiella isolates by CLSI, however we used the uropathogenic E. coli (UPEC) breakpoint (256μg/mL) for determining heteroresistance by PAP. b, PAPs of Nevada-2016 using fosfomycin (Fos; breakpoint 256 μg/mL), trimethoprim/sulfamethoxazole (SXT; breakpoint 4/76μg/mL), or their combination (“dual treatment”; purple). Predicted survival for an additive interaction (dashed black line) was determined by multiplying the survival after each single drug treatment, c, Nevada-2016 was treated with the indicated antibiotics at their breakpoint concentration and plated for enumeration of surviving bacteria at the indicated timepoints over a 48 hour period, d, Images and blanked optical densities (OD) of Nevada-2016 after 48 hour culture in the indicated single or combination antibiotic regimens (fosfomycin, Fos; trimethoprim/sulfamethoxazole, SXT). Media without bacteria (− Ctrl) and bacteria in media without antibiotics (+ Ctrl) are included as controls. Experiment was conducted twice with similar results, e-h, AR0040 was used in experiments mirroring those for Nevada-2016 (a-d). e, Antibiograms from clinical testing (left) and a modified version to indicate heteroresistance (right), f, PAPs of AR0040 using amikacin (Amk; breakpoint 64μg/mL), piperacillin/tazobactam (PTz; breakpoint 256/4 μg/mL), or their combination, g, h, AR0040 was treated as was Nevada-2016 in c and d, but with Amk, PTz, or Amk and PTz. i, Mice were infected intraperitoneally with AR0040 and then treated with PBS, amikacin (12.5 mg/kg), piperacillin/tazobactam (80/10 mg/kg), or their combination (dual treatment), every 12 hours beginning 30 minutes post infection. Survival was monitored for 150 hours. Data shown as mean ± s.d. with n=3 (b,c,f,g) or as survival with n=5 (i). ** p < 0.01, two-sided log-rank test.
There has been no clear basis for effective antibiotic combination therapy and some studies have concluded that these regimens are not superior to monotherapy29,30. While our data indicate that antibiotic combinations to which an isolate exhibits homogenous resistance can be synergistic, this occurs in a minority of cases (Fig. 3c). Our data predict frequent failure of such combination therapies, but greatly enhanced efficacy of combinations to which isolates display multiple heteroresistance. Aminoglycoside-beta lactam combinations have historically been among the most frequently used21–26. It is interesting to speculate that the relatively high prevalence of heteroresistance to some aminoglycosides and beta-lactams, and therefore the increased chance that combination therapy with these antibiotics would target multiple heteroresistance, may explain why this combination of classes became favored clinically.
Improved diagnostics would prevent the misclassification of heteroresistance as susceptibility, and caution against monotherapy in those cases. Rather than designating many instances of heteroresistance as homogenous resistance, and essentially excluding the respective drugs as treatment options, they would also highlight antibiotics to which isolates are heteroresistant as potential components of combination regimens. In the latter case, quantifying the frequency of the resistant subpopulation in heteroresistance would be critical. This would allow the hierarchical selection of combination regimens that include antibiotics to which the resistant subpopulation is least frequent, thereby facilitating the greatest magnitude of bacterial killing. Our data suggest that targeting multiple heteroresistance represents a rational strategy to use clinically approved antibiotics when monotherapy would fail.
Methods
Bacterial isolates.
Enterobacter cloacae isolates Mu208 and Mu866, as well as Klebsiella pneumoniae isolates Mul251 and Mul343, were isolated from patients at Atlanta area hospitals. The following isolates were used as controls: Mul 176 (resistant to colistin, fosfomycin, ceftazidime, ampicillin), Mu819 (susceptible to colistin and fosfomycin), Mu712 (susceptible to ceftazidime), andMu661 (susceptible to ampicillin). Carbapenem-resistant Enterobacteriaceae (CRE) were collected between 2013 and 2015 by the Georgia EIP MuGSI as described previously9. MuGSI collects isolates in Georgia from 27 labs serving 184 medical facilities, representing a surveillance population of 4 million people11. We are grateful to the CDC for providing pan-resistant Klebsiella pneumoniae isolates AR0040 and Nevada-2016.
Clinical susceptibility testing.
All isolates were tested for their clinical MIC and resistance designation in the clinical microbiology lab at Emory University Hospital. Twelve of the 16 drugs were tested by automated Vitek 2 (Biomerieux, Marc’-l’Étoile, France) using the GN74 susceptibility card. Antibiotics that were not on the GN74 panel (colistin, fosfomycin, ampicillin, ciprofloxacin) were tested using the respective Etest gradient strips as is standard protocol in this clinical laboratory. Etests were used as recommended (Biomerieux, Marc’-l’Étoile, France)31. Isolates were classified as resistant to colistin, ampicillin, or ciprofloxacin if colonies appeared in the zone of clearing. Susceptibility results were interpreted by a licensed clinical microbiologist.
Bacterial culture.
Bacteria were struck onto Mueller Hinton (MH) agar plates from frozen glycerol stocks. Single colonies were inoculated into MH broth and incubated at 250rpm, 37°C overnight. Colony forming units (CFU) were determined by serial dilutions of bacteria in phosphate buffered saline (PBS) plated onto MH agar plates at 37°C. CFU were determined at the lowest distinguishable dilution.
Population analysis profile.
Population analysis profiles (PAPs) were conducted as described previously6, with some modifications. Briefly, solid agar plates were made for each antibiotic at 7 concentrations containing 0, 0.125, 0.25, 0.5, 1, 2 and 4 times the breakpoint, using Mueller Hinton agar. Breakpoint concentrations for Enterobacteriaceae from CLSI (amikacin 64ug/mL, gentamicin 16ug/mL, tobramycin 16ug/mL, ampicillin 32 ug/mL, aztreonam 16ug/mL, cefazolin 32ug/mL, cefepime 16ug/mL, ceftazidime 16ug/mL, meropenem 4ug/mL, piperacillin/tazobactam 128/4 ug/mL, ciprofloxacin lug/mL, fosfomycin 256ug/mL (breakpoint for UPEC; 25mg/L glucose-6-phosphate added to the media), tetracycline 16ug/mL, trimethoprim/sulfamethoxazole 4/76 ug/mL), FDA (tigecycline 8ug/mL), or EUCAST (colistin 4ug/mL) were used. Dual PAPs were made similarly with 6 concentrations of antibiotics tested, with both drugs in each plate at the same multiple of their respective breakpoint (0, 0.125, 0.25, 0.5, 1, 2, 4 times the breakpoint). Isolates to be tested were grown up overnight in Mueller Hinton broth from a single colony isolated from a frozen stock. Serial microdilutions were plated at each concentration of antibiotic. Colonies were enumerated after overnight growth at 37°C. An isolate was classified as resistant if the number of colonies that grew at the breakpoint concentration were at least 50% of those that grew on antibiotic free plates. If an isolate was not resistant, it was classified as heteroresistant if the number of colonies that grew at 2 or 4 times the breakpoint were at least 0.0001% (1 in 106) of those that grew on antibiotic free plates. If isolates were neither classified as resistant or heteroresistant, they were classified as susceptible.
Time kills.
Time kills were conducted as previously described6. Briefly, 106 CFU/mL bacteria from an overnight culture were inoculated into 2mL MH media in culture tubes (Globe Scientific) with and without antibiotic(s). The following concentrations were used: colistin 16 μg/mL, ceftazidime 128 μg/mL, fosfomycin 256 μg/mL (+25mg/L Glucose-6-Phosphate (G6P)), ampicillin 128 μg/mL (Fig. 1e–h, l–o and Supplementary Fig. 2–4), fosfomycin 256 μg/mL (+25mg/L G6P), trimethoprim 4 μg/mL sulfamethoxazole 76 μg/mL, amikacin 64 μg/mL, and piperacillin 256 μg/mL tazobactam 4 μg/mL (Fig. 4c, d, g, h). Cultures were incubated at 37°C shaking at 250RPM for 1-48 hours. CFU were determined by serial diluting bacteria in PBS prior to plating on MH agar plates at various timepoints in the assay (0, 1, 2, 4, 6, 8, 10, 24, and 48 hour timepoints were used for timekills). In Figure 1e–h, serially diluted bacteria were also plated on MH agar plates with antibiotic added (colistin 16 μg/mL, ceftazidime 128 μg/mL, fosfomycin 256 μg/mL(+25mg/L G6P), and ampicillin 128 μg/mL). In Fig. 4d, h culture tubes were imaged with a Canon EOS Rebel T3i.
Cloning and mutagenesis.
Mu208 ΔphoQ, ΔfosA, and ΔampR were generated as previously described by lambda red recombination32. Briefly, a kanamycin cassette flanked by FRT sites with homology to the regions directly upstream and downstream, respectively, of the phoQ,fosA or ampR open reading frame (ORF) was inserted into the respective ORF in Mu208 with pKD46-tet, prior to it being removed with a Flp recombinase encoded on the temperature sensitive plasmid, PCP20.
Persister assay
Mu208, ΔphoQ, ΔfosA, and ΔampR from overnight cultures were resuspended to 109 CFU/mL in MH with 900μg/mL kanamycin and incubated at 37°C at 250RPM. At 0, 1, 2, 4, and 6 hours, aliquots were washed 3x in sterile PBS, serially diluted, and plated on MH plates. Plates were incubated at 37°C overnight and colonies were enumerated to determine the level of persisters.
Disk Diffusion and Etest
An overnight culture of Mu208 was adjusted to 0.5 McFarland standard (1.5×108 CFU/mL) and plated onto MH agar plates. For disk diffusion, 6mm blank paper discs (BBL) were sterilely added to the plates. Discs were treated with 0.1mg colistin, 1mg fosfomycin, or O.lmg ceftazidime. Plates used for fosfomycin treatment contained 25mg/L G6P. For dual disk diffusion, the same protocol was used, but discs were placed 1cm apart. For Etests (BioMerieux), test strips were sterilely added to the plates. Disk diffusion and Etest plates were incubated at 37°C for 20 hours.
Antibiotic resistance stability assay
For antibiotic treatments, bacteria were grown in the presence of a given drug at 37°C at 250RPM for 20 hours. The following concentrations were used: colistin 16 μg/mL, ceftazidime 128 μg/mL, fosfomycin 256 μg/mL (+25mg/L G6P), ampicillin 128 μg/mL, amikacin 64 μg/mL, and piperacillin/tazobactam 128/4 μg/mL. Antibiotic-free subcultures were performed by diluting overnight cultures 1:1,000 and incubating for 20 hours at 37°C at 250RPM, allowing for 10 generations of growth without drug. Bacteria were serially diluted in PBS and plated on MH with and without the respective antibiotic to determine the proportion of resistant bacteria.
Checkerboard assay.
5×105 CFU bacteria were used to inoculate 100μL MH in a 96 well plate (Falcon flat bottom tissue culture plate) with and without drug, which was subsequently incubated at 37°C for 20-24 hours. Breathe-easy sealing membranes (RPI) were amended prior to incubation. Drug was diluted by 1:1 serial diluting antibiotic with fresh media prior to bacterial inoculation. Minimum inhibitory concentration (MIC) was determined by finding the lowest concentration of drug that did not have turbidity in the well. All fosfomycin antibiotic assays contained 25mg/L glucose-6-phosphate (G6P). The drugs were diluted 1:1 by serial diluting antibiotic with fresh media. One drug was serial diluted right to left in a 96 well plate, and the second drug was serially diluted top to bottom in a 96 well plate. Half of the final volume of the checkerboard plate was added from each of the two serial diluted drug plates to make the checkerboard plate. 5×105 CFU was then inoculated, a breathe-easy strip was amended, and the plate was incubated at 37°C for 20-24 hours. The checkerboard screen involved checkerboards of Mu208, Mu772, Mu308, Mu638, Mul309, Mul 197, Mu827, and Mul343 with the drugs ampicillin, tetracycline, meropenem, trimethoprim sulfamethoxazole, ciprofloxacin, cefepime, tobramycin, and fosfomycin. Using the clinical susceptibility testing designation via antibiograms for the 8 isolates, all RxR interactions were setup for these drugs to these 8 isolates. In total 86 synergy plates, 28 unique drug combinations, 1792 unique drug doses, and 5504 unique drug dose+bacterial combinations were performed using this method. MICs for each isolate and drug were determined the day of the checkerboard experiment to ensure similar drug concentrations for FIC determination. FIC was calculated as previously described (FIC=(MIC DrugA with DrugB / MIC DrugA alone)+ (MIC DrugB with DrugA / MIC DrugB alone)), with the synergistic FIC≤0.527,28.
Antibiotic combination screen.
The combination screen (Fig. 3a) included 8 representative isolates (3 Enterobacter, 4 Klebsiella, 1 Escherichia) from the 104 MuGSI CRE isolates tested for heteroresistance (Fig. 2a, b), chosen due to the variety of antibiotics to which they were classified as heteroresistant. Mueller Hinton agar plates were made using each of the 16 antibiotics at the breakpoint concentrations mentioned above. In addition, all 120 distinct two-drug combinations were tested (Fig. 3a), using the breakpoint concentrations for both drugs. Log killing for each combination was calculated by dividing the enumerated CFU on the two-drug combination plate by the CFU on antibiotic free plates. Each antibiotic combination/isolate interaction was tested in duplicate, and the enumerated CFU on each plate were averaged.
Mouse infections
Female C57BL/6 mice were obtained from Jackson Laboratory (Bar Harbor, ME). Six week old mice were infected intraperitoneally with ~2×108 CFU of Mu208, ~2×108 CFU Mu866, ~1×108 CFU Mu1251, ~1×108 CFU Mul343, or ~2×108 CFU AR0040, in 100uL sterile PBS. Antibiotic treatments were initiated at 4 hours post infection and given at 6 hour increments, except in the lethal infections, where antibiotic therapy was initiated 30 minutes post infection and given at 12 hour increments. Antibiotic treatments were given in 100uL PBS at lOmg/kg colistin, 200mg/kg fosfomycin, 20mg/kg ceftazidime, 6.25 mg/kg amikacin, or 80/10 mg/kg piperacillin/tazobactam as indicated. For infections where CFU were quantified, mice were sacrificed at 24 hours post infection and peritoneal lavages were collected with 3mL of sterile PBS which was subsequently plated on solid media for colony enumeration. Experiments which assayed for survival were closely monitored, with mouse weights being taken every 6 hours and mice euthanized if moribund or below 80% starting weight. All experiments were conducted in compliance with approved protocols and guidelines of the Emory University Institutional Animal Care and Use Committee (IACUC). Sample size, as reported in the figure legends, was determined by allowing for significance by the Mann-Whitney test (n > 4) while minimizing the number of animals used, thus five mice were used per group for all experiments. No randomization or blinding was done in the animal studies.
Statistical analyses.
All data presented is from measurements taken from distinct samples. All experiments were repeated at least twice to ensure reproducibility. This excludes large screens, such as those in Figure 2, 3, and Supplementary Fig. 10. Statistical significance between two groups was performed by appropriate two-tailed t-test, after assessment of variance using the f-test. Data for histograms (Figs. 3b, d, f, 4g, h) was binned by whole logs (Fig. 3; <1, 1, 2, 3, 4, 5, >6) or by increments of FIC (Supplementary Fig. 10; >2, 1, 0.75, 0.5, 0.25, <0.1). Error bars represent standard error of the mean from biological replicates. These data were analyzed for significance by Mann-Whitney U test. Statistical analysis was performed using Graphpad Prism software.
Supplementary Material
Acknowledgements
We would like to thank Arash Grakoui, Hannah Ratner and William Shafer for critical reading of the manuscript. We are grateful to Chris Bower and the GA EIP/MuGSI staff for providing CRE isolates. D. A H. is supported by a postdoctoral research fellowship from the Cystic Fibrosis Foundation. E.X.S. is supported by T32 training grant AIl06699 from the National Institutes of Health (NIH). D.S.W. is supported by a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Disease award and NIH grant AI141883. The GA EIP is funded by the Centers for Disease Control and Prevention. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH, CDC, or the Department of Veterans Affairs.
Footnotes
Data Availability
All data needed to evaluate the conclusions in the paper are present in the paper or the supplementary materials. Any additional data can be requested from the corresponding author.
Competing Interests
V.I.B., D.A.H., and D.S.W. are listed authors on a provisional patent that has been filed related to the work described here.
References
- 1.Review on Antimicrobial Resistance. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations, <https://amr-review.org/Publications.html> (2014).
- 2.Micek ST et al. Empiric combination antibiotic therapy is associated with improved outcome against sepsis due to Gram-negative bacteria: a retrospective analysis. Antimicrob Agents Chemother 54, 1742–1748, doi: 10.1128/AAC.01365-09 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexander ΗE & Leidy G Mode of action of streptomycin on type B Hemophilis influenzae: Nature of resistant variants. JExp Med 85, 607–621 (1947). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ott JL, Turner JR & Mahoney DF Lack of correlation between beta-lactamase production and susceptibility to cefamandole or cefoxitin among spontaneous mutants of Enterobacteriaceae. Antimicrob Agents Chemother 15, 14–19 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sogaard P & Gahrn-Hansen B Population analysis of susceptibility to ciprofloxacin and nalidixic acid in Staphylococcus, Pseudomonas aeruginosa, and Enterobacteriaceae. Acta Pathol Microbiol Immunol ScandB 94, 351–356 (1986). [DOI] [PubMed] [Google Scholar]
- 6.Band VI et al. Antibiotic failure mediated by a resistant subpopulation in Enterobacter cloacae. Nat Microbiol 1, 16053, doi: 10.1038/nmicrobiol.2016.53 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.El-Halfawy ΟM & Valvano MA Antimicrobial heteroresistance: an emerging field in need of clarity. Clin Microbiol Rev 28, 191–207, doi: 10.1128/CMR.00058-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.United States Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States. (2013).
- 9.Guh AY et al. Epidemiology of Carbapenem-Resistant Enterobacteriaceae in 7 US Communities, 2012-2013. JAMA 314, 1479–1487, doi: 10.1001/jama.2015.12480 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen L, Todd R, Kiehlbauch I, Walters M & Kallen A Vol. 66 33 (Morbidity and Mortality Weekly Report, 2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.United States Centers for Disease Control and Prevention. Multi-site Gram-negative Surveillance Initiative, <https://www.cdc.gov/hai/eip/mugsi.html> (2016).
- 12.Brauner A, Fridman O, Gefen O & Balaban NQ Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat Rev Microbiol 14, 320–330, doi: 10.1038/nrmicro.2016.34 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Keren L, Minami S, Rubin E & Lewis K Characterization and transcriptome analysis of Mycobacterium tuberculosis persisters. MBio 2, e00100–00111, doi: 10.1128/mBio.00100-11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wakamoto Y et al. Dynamic persistence of antibiotic-stressed mycobacteria. Science 339, 91–95, doi: 10.1126/science. 1229858 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Napier BA, Band V, Burd EM & Weiss DS Colistin heteroresistance in Enterobacter cloacae is associated with cross-resistance to the host antimicrobial lysozyme. Antimicrob Agents Chemother 58, 5594–5597, doi: 10.1128/AAC.02432-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang KN et al. Colistin heteroresistance in Enterobcater cloacae is regulated by PhoPQ-dependent 4-amino-4-deoxy-l-arabinose addition to lipid A. Mol Microbiol, doi: 10.1111/mmi.l4240 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang TY, Lu PL & Tseng SP Update on fosfomycin-modified genes in Enterobacteriaceae. J. Microbiol. Immunol. Infect 52, 9–21, doi: 10.1016/j.jmii.2017.10.006 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Normark S Beta-Lactamase induction in gram-negative bacteria is intimately linked to peptidoglycan recycling. Microb Drug Resist 1, 111–114, doi: 10.1089/mdr.1995.1.111 (1995). [DOI] [PubMed] [Google Scholar]
- 19.Nicoloff H, Hjort K, Levin BR & Andersson DI The high prevalence of antibiotic heteroresistance in pathogenic bacteria is mainly caused by gene amplification. Nat Microbiol 4, 504–514, doi: 10.1038/s41564-018-0342-0 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Ballestero-Tellez M, et al. Role of inoculum and mutant frequency on fosfomycin MIC discrepancies by agar dilution and broth microdilution methods in Enterobacteriaceae. Clin Microbiol Infect 23, 325–331, doi: 10.1016/j.cmi.2016.12.022. (2017). [DOI] [PubMed] [Google Scholar]
- 21.Paul M, Benuri-Silbiger L, Soares-Weiser K & Leibovici L Beta lactam monotherapy versus beta lactam-aminoglycoside combination therapy for sepsis in immunocompetent patients: systematic review and meta-analysis of randomised trials. BMJ 328, 668, doi: 10.1136/bmj.38028.520995.63 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leibovici L et al. Monotherapy versus beta-lactam-aminoglycoside combination treatment for gram-negative bacteremia: a prospective, observational study. Antimicrob Agents Chemother 41, 1127–1133 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siegman-Igra Y, Ravona R, Primerman H & Giladi M Pseudomonas aeruginosa bacteremia: an analysis of 123 episodes, with particular emphasis on the effect of antibiotic therapy. Int JInfect Dis 2, 211–215 (1998). [DOI] [PubMed] [Google Scholar]
- 24.Crabtree TD, Pelletier SJ, Gleason TG, Pruett TL & Sawyer RG Analysis of aminoglycosides in the treatment of gram-negative infections in surgical patients. Arch Surg 134, 1293–1298; discussion 1298-1299 (1999). [DOI] [PubMed] [Google Scholar]
- 25.Montravers P et al. Diagnostic and therapeutic management of nosocomial pneumonia in surgical patients: results of the Eole study. Crit Care Med 30, 368–375 (2002). [DOI] [PubMed] [Google Scholar]
- 26.Grasela TH et al. A nationwide survey of antibiotic prescribing patterns and clinical outcomes in patients with bacterial pneumonia. DICP 24, 1220–1225 (1990). [DOI] [PubMed] [Google Scholar]
- 27.Odds FC Synergy, antagonism, and what the chequerboard puts between them. J Antimicrob Chemother 52, 1, doi: 10.1093/jac/dkg301 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Doern CD When does 2 plus 2 equal 5? A review of antimicrobial synergy testing. J Clin Microbiol 52, 4124–4128, doi: 10.1128/JCM.01121-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamma PD, Cosgrove SE & Maragakis LL Combination therapy for treatment of infections with gram-negative bacteria. Clin Microbiol Rev 25, 450–470, doi: 10.1128/CMR.05041-11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paul M et al. Combination therapy for carbapenem-resistant Gram-negative bacteria. J Antimicrob Chemother 69, 2305–2309, doi: 10.1093/jac/dku168 (2014). [DOI] [PubMed] [Google Scholar]
- 31.bioMerieux. Etest application guide. 2012.
- 32.Datsenko KA & Wanner BL One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97, 6640–6645, doi: 10.1073/pnas.120163297 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.