Abstract
Background
T. spiralis aspartic protease has been identified in excretion/secretion (ES) proteins, but its roles in larval invasion are unclear. The aim of this study was to characterize T. spiralis aspartic protease-2 (TsASP2) and assess its roles in T. spiralis invasion into intestinal epithelial cells (IECs) using RNAi.
Methodology/Principal findings
Recombinant TsASP2 (rTsASP2) was expressed and purified. The native TsASP2 of 43 kDa was recognized by anti-rTsASP2 serum in all worm stages except newborn larvae (NBL), and qPCR indicated that TsASP2 transcription was highest at the stage of intestinal infective larvae (IIL). IFA results confirmed that TsASP2 was located in the hindgut, midgut and muscle cells of muscle larvae (ML) and IIL and intrauterine embryos of the female adult worm (AW), but not in NBL. rTsASP2 cleaved several host proteins (human hemoglobin (Hb), mouse Hb, collagen and IgM). The proteolytic activity of rTsASP2 was host-specific, as it hydrolyzed mouse Hb more efficiently than human Hb. The enzymatic activity of rTsASP2 was significantly inhibited by pepstatin A. The expression levels of TsASP2 mRNA and protein were significantly suppressed by RNAi with 5 μM TsASP2-specific siRNA. Native aspartic protease activity in ML crude proteins was reduced to 54.82% after transfection with siRNA. Larval invasion of IECs was promoted by rTsASP2 and inhibited by anti-rTsASP2 serum and siRNA. Furthermore, cell monolayer damage due to larval invasion was obviously alleviated when siRNA-treated larvae were used. The adult worm burden, length of adult worms and female fecundity were clearly reduced in mice challenged using siRNA-treated ML relative to the PBS group,
Conclusions
rTsASP2 possesses the enzymatic activity of native aspartic protease and facilitates T. spiralis invasion of host IECs.
Author summary
Trichinellosis has been regarded as a re-emerging or emerging disease, and it is distributed worldwide. Studies investigating T. spiralis ES protein are beneficial to explore potential molecular targets for anti-T. spiralis vaccines. The functions of aspartic protease have been studied in other parasites and demonstrated to be crucial for their parasitism in the host. However, the functions of T. spiralis aspartic protease have not been reported. Here, we expressed and purified a T. spiralis aspartic protease-2 (TsASP2). Our results showed that TsASP2 was expressed in all T. spiralis developmental stages other than NBL and located in the hindgut, midgut and muscle cells of ML and IIL, as well as in areas surrounding embryos within the female uterus. The rTsASP2 possessed aspartic protease activity and functioned to cleave hemoglobin, collagen and IgM. Silencing of the TsASP2 gene could significantly decrease the aspartic protease activity in muscle larva crude proteins, larval invasion of IECs and worm development in the host. We conclude that TsASP2 plays an important role in T. spiralis penetration into host intestinal epithelial cells and could be a candidate vaccine target molecule against T. spiralis infection.
Introduction
As a pathogen of worldwide food-borne zoonosis, Trichinella parasite has been found to infect more than 100 mammalian species [1]. Humans acquire trichinellosis through the ingestion of raw or poorly cooked meat containing the infective larvae of Trichinella [2]. Outbreaks of trichinellosis have been reported in many counties worldwide, especially in developing countries [3,4]. In China, 15 outbreaks were recorded from 2004 to 2009 due to raw or undercooked pork food [5]. Trichinellosis has not only become a public health concern but also threatened porcine animal production and food safety [6]. Thus, trichinellosis has been regarded as a re-emerging or emerging disease worldwide and has gained increasing attention [7]. These concerns have promoted the exploration of anti-Trichinella vaccines, especially to identify molecules that play key roles in T. spiralis invasion of intestinal epithelium [8].
Muscle larvae (ML) of T. spiralis dwell in skeletal muscles of hosts. When the contaminated meat is ingested, the ML are liberated from the muscles by digestive enzymes in the stomach [9]. After activation by bile and enteral contents, the larvae develop into intestine infective larvae (IIL). The IIL invade the small intestinal epithelium where they undergo four molts to mature into adult worms (AW). The newborn larvae (NBL) are shed by female AW after mating and then enter the venules and lymphatic vessels, eventually penetrating into the skeletal muscle via the bloodstream [10]. The mechanism of T. spiralis penetration into intestinal epithelium is critical for T. spiralis to complete its lifecycle in the host and seems to be orchestrated by several T. spiralis protein molecules. Thus, studies on the characterization and functions of these T. spiralis proteins will be very valuable for the development of an anti-Trichinella vaccine.
Proteinases released by T. spiralis play an essential role in parasite invasion [11,12]. Several serine proteinases have been identified and demonstrated to be involved in various adaptive functions, such as tissue invasion and immune evasion, [13–15]. In another study, proteinases produced by T. spiralis adult worms could cleave fibrinogen and plasminogen, and this hydrolytic activity might be related to the activity of a serine or aspartyl proteinase [16]. These studies revealed that the characterization of proteinases derived from T. spiralis would provide critical information for explorations of anti-T. spiralis vaccines.
Aspartic protease, classified as clan AA in the MEROPS database, was the first protease type to be described [17]. The aspartic protease family, including pepsins, renins, cathepsins D and E, and chymosins [18], is characterized by a typical Asp-Thr (Ser)-Gly sequence, and the protein hydrolytic activity is closely associated with an Asp residue in the clefts of the active sites [19]. The proteolytic function of aspartic proteases is optimal under acidic conditions (pH 3.0–4.0) [12]. Many aspartic proteases have been demonstrated to play key roles in the degradation of host hemoglobin and other proteins, especially in hematophagous parasites. An aspartic protease named Na-APR-1/2 originating from hookworms can efficiently cleave hemoglobin, collagen and serum albumin from human and dog [20,21]. A cathepsin D-like aspartic protease from Opisthorchis viverrini has been shown to digest bovine serum albumin (BSA) and hemoglobin [22]. The proteolytic activity and probable functions of other parasites such as Schistosoma mansoni, Necator americanus and Onchocerca volvulus, which can also secrete aspartic protease, have also been investigated [23–25]. An aspartic protease has been identified in T. spiralis ES proteins [11], but its function in T. spiralis invasion into intestinal epithelium is not clear.
Since it was first conducted in Caenorhabditis elegans [26], RNA interference (RNAi) has been widely applied to identify gene function in parasites. The application of RNAi to downregulate target molecules to reduce protein expression can affect specific gene functions during some developmental stages of parasites. Recently, RNAi was used to identify some important protein functions of parasites, such as Clonorchis sinensis enolase [27], ATPase RNA helicase and trehalose-6-phosphate phosphatase in Brugia malayi [28,29], nematode Setaria digitata-specific protein [30], and calcium-regulated heat-stable protein of 24 kDa and type V collagen in Schistosoma japonicum [31,32]. Nevertheless, the functions of only a few T. spiralis genes have been ascertained by RNAi, including the Nudix hydrolase, serine protease inhibitor (TsSPI) and paramyosin genes [7,33,34].
Four aspartic proteases were identified in the draft genome of T. spiralis, all of which are bilobal enzymes. The similarities among four aspartic proteases ranged from 15.6% to 81.8%. However, only T. spiralis aspartic protease 2 (TsASP2; GenBank: 339237490) from a T. spiralis muscle larva cDNA library has been demonstrated to be present in T. spiralis excretion/secretion (ES) proteins [11]; however, its function has remained unclear. Since the proteases in ES proteins are first exposed to host intestinal epithelium cells (IECs), they are likely to play a major role or participate in larval invasion of IECs. Therefore, TsASP2 was selected and expressed in the present study. We further investigated the functions of TsASP2 in T. spiralis larval invasion of host IECs, and a specific TsASP2 siRNA sequence was designed and electroporated into muscle larvae to elucidate the gene functions.
Methods and materials
Ethics statement
This study was carried out according to the National Guidelines for Experimental Animal Welfare (Minister of Science and Technology, People’s Republic of China, 2006). The animal experiment procedure was approved by the institutional Life Science Ethics Committee, Zhengzhou University (No. SCXK 2017–0001).
Parasites, experimental cells and animals
The T. spiralis isolate (ISS534) used in this study was originally obtained from domestic pigs in Nanyang (Henan province, China) and maintained in our laboratory by serial passages in BALB/c mice. Specific pathogen-free (SPF) BALB/c mice aged 5 weeks old were purchased from the Experimental Animal Center of Henan Province. Normal IECs were obtained from mouse small intestines and used for the invasion assay at passage 8 [10]. The IECs were cultured as previously described [35].
Worm collection and protein preparation
T. spiralis ML were collected from infected mice at 42 days post-infection (dpi) by digestion of carcasses with 0.75% pepsin and 1% HCl as previously described [36]. The IIL were obtained from small intestines of infected mice at 6 hours post-infection (hpi). AW were isolated from duodenum and jejunum of infected mice at 3 and 6 dpi. The newborn larvae (NBL) were obtained from female adult worms cultured as previously described [37]. The ML ES antigens and crude soluble antigens of AW, NBL, ML and IIL were prepared as previously reported [10]. Briefly, the worms were first homogenized using a high-speed tissue grinder (KZ-II Servicebio) for 1 min, and the worm fragments were further homogenized by ultrasonication (99 3-s cycles, 100 W, 0°C). The supernatant containing crude proteins was collected after centrifugation at 15,000 g for 1 h at 4°C. To collect the ES proteins, the larvae were washed with sterile saline and then cultured in RPMI-1640 medium at a density of 5000 worms/ml for 18 h at 37°C in 5% CO2. After the media containing ES proteins were filtered with a 0.22-μm membrane, the ultrafiltration tubes were used to concentrate the ES proteins. The concentration of these proteins was measured using the Bradford method.
Expression of recombinant TsASP2 protein in Escherichia coli
The entire CDS sequence of TsASP2 encoding aspartic protease (spanning Gly-17 to Ser-406) without the signal peptide was amplified by PCR and cloned into the expression vector pQE80L (His tag) and pMAL-c2x (MBP tag) using the Bam HI and Hind III site. The recombinant plasmids were transferred into BL21 (DE3). The rTsASP2 was induced with 0.1 mM IPTG at 16°C for 20 h. The Ni-NTA-Sefinose resin (for His-tagged protein) and Amylose Pre-packed Column (for MBP-tagged protein) (NEB, China) were used to purify the rTsASP2. The rTsASP2 with a His tag was purified under denaturing conditions and then refolded and used in subsequent immunization experiments. The rTsASP2 with an MBP tag was used for its functional characterization. SDS-PAGE was applied to analyze the purified rTsASP2, and the Bradford method was used to determine the rTsASP2 concentration.
Preparation of anti-rTsASP2 serum
Fifteen BALB/c mice were used to produce anti-rTsASP2 serum. First, the mice were immunized subcutaneously with 20 μg rTsASP2 emulsified with complete Freund’s adjuvant. Three boost immunizations were further carried out every 2 weeks by injecting the same amount rTsASP2 emulsified with incomplete Freund’s adjuvant. Blood samples were collected from immunized mice on day 7 after last immunization, and sera were isolated.
Western blot analysis
Western blot analysis was carried out according to previous studies [38]. First, ES protein and crude protein samples from ML, IIL, AW and NBL were separated by SDS-PAGE on a 12% acrylamide separation gel and subsequently transferred onto polyvinylidene difluoride (PVDF) membrane. Second, the membranes were blocked with 5% skim milk in Tris-buffered saline containing 0.05% Tween-20 (TBST). The membranes were washed three times with TBST to remove the residual skim milk and then incubated with 1:100 dilutions of anti-rTsASP2 serum at 37°C for 1 h. Following another wash with TBST, the membranes were incubated with 1:5 000 dilutions of HRP-conjugated goat anti-mouse IgG (Southern Biotechnology, USA) at 37°C for 1 h. Finally, the membranes were stained with 3, 3'-diaminobenzidine tetrahydrochloride (DAB; Sigma) as a substrate, which was terminated by washing the membranes with deionized water [35,39].
qPCR
Total RNAs of different T. spiralis phases (ML, IIL, 3-day AW, 6-day AW and NBL) were extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). All RNA samples were pre-treated with DNase I (Thermo Fisher Scientific, San Francisco, CA, USA) before use. Transcription of the TsASP2 gene at different worm stages was measured by qPCR as previously described [40]. The qPCR experiment was performed on a 7500 Fast Real-time PCR System (Applied Biosystems). The specific primers (across introns) for TsASP2 gene amplification included forward 5'-AATTCAACCCGTCCGTCTCC-3' and reverse 5'-TTCCAACTTGCGGCCATAGT-3'. The TsASP2 transcription level was normalized by subtracting the transcription level of GAPDH (GenBank: AF452239). The data were calculated according to the comparative Ct (2-ΔΔCt) method [40]. Each experiment was performed with three replicates of each sample.
Immunofluorescent assay (IFA)
IFA was carried out to confirm the expression of TsASP2 at diverse T. spiralis stages. Various T. spiralis worm stages (ML, IIL, AW and NBL) were fixed in paraformaldehyde and embedded in paraffin. The 2-μm-thick sections were prepared using a microtome. After blocking with 5% normal goat serum, the sections were incubated with anti-rTsASP2 serum (1:50 dilutions) at 37°C for 1 h. After washing with PBS, they were incubated with FITC-labeled goat anti-mouse IgG (1:100 dilution, Santa Cruz, USA) and observed under a fluorescence microscope (Olympus, Japan) [41].
Cleavage of Hb and other proteins by rTsASP2
Hemoglobin (Hb) from mice and human was collected by lysis of fresh erythrocytes as previously described [20]. Approximately 2 μg Hb was incubated with 0.8 μg rTsASP2 in pH 2.5–5.5 buffer solution, and the hydrolysates were detected by SDS-PAGE and staining with Coomassie brilliant blue. To compare the hydrolysis efficiency of rTsASP2 for different Hb, hydrolysis experiments were further conducted with different incubation times (5 min, 30 min, 90 min and 4 h). Other proteins (collagen IV, human IgM and IgG) were also used as substrates to evaluate the cleavage function and specificity of rTsASP2. Anti-rTsASP2 serum (heated at 56°C for 35 min or not heated) at a 1:25 dilution or 0.8 μg pepstatin A was pre-incubated with rTsASP2 for 1 h, followed by incubation with mouse Hb for 2 h to detect the enzyme activity.
Enzymatic activity of rTsASP2
The fluorescent substrate MCA-Gly-Lys-Pro-Ile-Leu-Phe-Phe-Arg-Leu-Lys (DNP) -D-Arg-amide (synthesized by Sangon, Shanghai) was used to assess the enzymatic activity of rTsASP2 [42]. The total reaction volume was 100 μl, including 20 μg/ml rTsASP2 and 5 μM fluorescent substrate. After the enzyme and substrate were mixed for 30 min, the reaction termination fluid (35% methyl alcohol, 30% ethyl alcohol, 35% ddH2O) was added, and the fluorescence intensity was continuously detected by spectrophotofluorometry (Synergy H1, BioTek, USA) using an excitation wavelength of 320 nm and emission wavelength of 390 nm, respectively. To determine the optimal pH, the reaction was carried out using assay buffers with different pH values: 0.2 mol/L Gly-HCl buffer (pH 2.0–3.0), 0.2 mol/L HAc-NaAc (pH 3.5–5.5), 0.2 mol/L Na2HPO4-NaH2PO4 (pH 6.0). The relative enzymatic activity was calculated by setting the highest enzyme activity as 100% relative activity. Different concentrations (1 mM, 10 mM, 100 mM, and 200 mM) of Fe2+, Zn2+, Cu2+, Mn2+ and Mg2+ were added to the assay buffer to evaluate the influence of metal ions on rTsASP2 enzyme activity. Inhibitors (pepstatin A, PMSF, 1, 10-phenanthrolin, AEBSF, EDTA and E-64) were pre-incubated with rTsASP2 for 30 min, and then the effects on rTsASP2 enzyme activity were evaluated. Reaction buffers without addition of metal ions and inhibitors were used as respective controls.
Electroporation of T. spiralis ML with siRNA
The TsASP2-specific siRNA was designed using siDirect version 2.0 according to the complete cDNA encoding TsASP2. Three siRNA sequences were used in the present study, including a TsASP2-specific siRNA, a control siRNA carrying the scrambled sequence and another T. spiralis aspartic protease-1 (TsASP1) siRNA sequence to control for specificity. All siRNAs were synthesized by Sangon Biotech (Shanghai, China), and the sequence information is listed in Table 1.
Table 1. The sequences of the siRNAs.
| siRNA’ name | sense(5'-3') | antisense(5'-3') |
|---|---|---|
| TsASP1- siRNA | GUCAACAUUCAAAGAAUAUTT | AUAUUCUUUGAAUGUUGACTT |
| TsASP2- siRNA | CAUGAUUGAGCAAAAUCUUTT | AAGAUUUUGCUCAAUCAUGTT |
| Control siRNA | AUCGGCUACCAAGUCAUACTT | GUAUGACUUGGUAGCCGAUTT |
The ML was obtained from infected mice at 35 dpi and washed three times with PBS. Approximately 2500 ML worms were treated with 5 μM siRNA in electroporation buffer. The siRNA was delivered into ML by electroporation (125 V, 20 ms) with a Gene Pulse Xcell System (Bio-Rad, USA), after which the worms were cultured in DMEM for 1–7 days.
Analysis of TsASP2 mRNA and protein expression after siRNA transfection
qPCR was performed to analyze TsASP2 mRNA transcription in siRNA-treated ML as described above. The TsASP2 protein expression in treated worms was also evaluated by western blot analysis [34]. In brief, the crude proteins extracted from siRNA-treated ML were separated by SDS-PAGE and then transferred onto a PVDF membrane. Anti-rTsASP2 serum (1:100) was first used to recognize the membrane and then visualized using an enhanced chemiluminescent kit (Beyotime Biotech, China). The membrane was washed with stripping buffer (Beyotime Biotech, China) and then incubated with mouse anti-GAPDH IgG for quantitative protein control.
RNAi effect on the enzymatic activity of aspartic protease
Crude protein extracts were obtained from approximately 2000 ML treated with siRNA or PBS. The enzymatic activity assay was carried out in a 100-μl reaction mixture with 100 μg crude protein and a final concentration of 5 μM fluorogenic substrate in sodium format, pH 3.5 and incubated at 37°C for 30 min. After the reaction was terminated, the fluorescence from substrate hydrolysis was measured as described above.
RNAi effect on in vitro larval penetration
The effect of RNAi on in vitro IEC penetration by T. spiralis was also assessed. C2C12 was insensitive to T. spiralis penetration and used as negative control. The rTsASP2 protein was found to facilitate in vitro T. spiralis invasion of IEC. Briefly, the ML were activated into IIL with 5% swine bile, and the IEC cell monolayers (grown to confluence in 6-well culture plates) were overlaid with 100 IIL mixed with 2 ml of DMEM semisolid medium [10]. Different concentrations of rTsASP2 proteins were added to the medium to investigate the effects of the rTsASP2 protein on larval invasion. The different dilutions (1:50–1:800) of anti-rTsASP2 serum, infection serum and normal serum were then added to the medium. The IILs that had penetrated into the IECs were counted by microscopy after being cultivated for 2 h at 37°C. The penetrated and unpenetrated worms were examined and counted as previously reported [10,34]. Subsequently, the IEC cell monolayer capped by 100 RNAi treated or untreated larvae was also observed to evaluate the RNAi effect on larval invasion. The larval invasion rate was compared between experiments, and the larval invasive ability was assessed.
To confirm the RNAi effect on larval invasion, dead or damaged cells were also counted [43]. Briefly, after incubation, monolayers were stained with 10 μg/ml propidium iodide (PI) for 10 min and washed three times with PBS. The numbers of dead or damaged cells (stained red) were determined by fluorescence microscopy (Olympus, IX53, Japan) and NIH Image software. Dead or damaged cells was counted for 3 monolayers for each group. A total of 10 microscope fields of each monolayer were captured, and the mean number of dead (damage) cells was determined.
To detect the remaining TsASP2 in monolayers, the IECs were grown to confluence on coverslips. Following incubation, the slide was first stained with PI and then fixed with 4% paraformaldehyde for 15 min. The monolayers were probed using anti-rTsASP2 serum (1:20 dilutions) for 1 h at 37°C. Positive and negative controls were also probed using infection serum and normal serum instead of anti-rTsASP2 serum. After washing three times with PBS, the coverslips were incubated with FITC-conjugated goat anti-mouse IgG (1:100) for 1 h at 37°C. Next, the coverslips were rinsed three times with PBS and mounted for further fluorescence microscopy observation.
RNAi effect on larval development and survival
To evaluate the infectivity of siRNA-treated larvae, the larval challenge infection experiment was performed. Thirty mice were equally divided into 3 groups, and each mouse in the different groups was orally infected with 300 larvae treated with TsASP2 siRNA, control siRNA or PBS. Adult worms at 6 dpi were collected from each group, and the parasite burden was ascertained. The fecundity of female AW was assessed by counting the newborn larva production by each female worm for 72 h. The morphology of AW and NBL was observed and imaged under a microscope (OLYMPUS IX53), and the length of the worms was measured using the measuring tool provided with the image software (CellSens Standard).
Statistical analysis
Data analysis was performed with the aid of SPSS 19.0 software. The data are presented as the mean ± standard deviation (SD). The Chi square test was used to compare the percentage of larval invasion in the different groups. One-way ANOVA was used to compare the data from different groups in the following experiment: the relative TsASP2 transcription or expression levels, enzymatic activity of aspartic protease from siRNA-treated larvae, length of worms and damaged cell numbers of IEC destroyed by T. spiralis. P < 0.05 was regarded as a statistically significant difference.
Results
Expression of rTsASP2
The 1170-bp CDS sequence without a signal peptide of TsASP2 was amplified, which encodes 406 amino acids. After BL21 (DE3) containing the two different recombinant plasmids (pQE-80L/TsASP2 and pMAL-c2x/TsASP2) was induced with IPTG, the fusion protein was expressed as 43.4 kDa (His tag) (S1 Fig) for the former recombinant protein and 86.4 kDa (containing the 43 kDa MBP tag) for the other one (Fig 1).
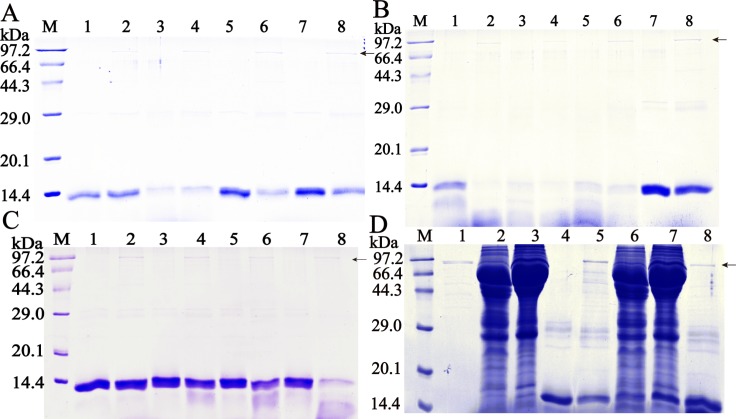
Fig 1. SDS-PAGE analysis of rTsASP2.
M: protein marker; lane 1: lysates of recombinant bacteria incorporating pMAL-C2X/TsASP2 without induction; lane 2: lysates of recombinant bacteria incorporating pMAL-C2X/TsASP2 after induction; lane 3: lysate supernatant of recombinant bacteria incorporating pMAL-C2X/TsASP2 after induction; lane 4: sediment of recombinant bacteria incorporating pMAL-C2X/TsASP2 after induction; lane 5: purified rTsASP2. The arrow represents the band of rTsASP2 (86.4 kDa).
Western blot and qRT-PCR analysis of TsASP2 expression in various stages
Western blot analysis showed that the native TsASP2 protein of 43 kDa was recognized by anti-rTsASP2 serum (Fig 2A) in all worm stages except NBL, and the other bands that were also recognized may have been protein isoforms of TsASP2. TsASP2 gene transcription in these stages was further detected by qPCR. The results showed that the transcription level of TsASP2 gene was highest at the IIL stage and lowest at the NBL stage (Fig 2B). The TsASP2 transcription level was significantly higher in the IIL stage than the other stages (F = 3.719, P < 0.01), while the transcription level was significant lower in NBL stage than the other stages (F = 4.007, P < 0.0001). The relatively low expression of the TsASP2 gene in the NBL stage could explain why TsASP2 protein in NBL proteins was not recognized by anti-TsASP2 serum by western blot analysis.
Fig 2.
Western blot (A) and qPCR (B) analysis of TsASP2 protein and mRNA expression in different T. spiralis stages. A: Anti-rTsASP2 serum recognized native TsASP2 in different T. spiralis stage crude proteins, including ML (lane 1), 6-h IIL (lane 2), 3-d AW (lane 3), 6-d AW (lane 4), but not NBL (lane 5), and ML ES (lane 6) and IIL ES (lane 7). B: The TsASP2 mRNA expression levels in different T. spiralis stages were assessed by qPCR. Asterisks indicate a statistically significant difference compared with the ML stage (*P < 0.05).
Expression and location of TsASP2 in different stages
The IFA results confirmed the expression of TsASP2 in different life cycle stages of T. spiralis. Immunofluorescent staining of hindgut, midgut and muscle cells of ML and IIL was observed, as well as around the embryos of AW, but not NBL (Fig 3).
Fig 3. Immunofluorescent analysis of TsASP2 location in various worm phases.
Intense green staining was observed in the midgut, hindgut and muscle cells of ML and 6-h IIL, as well as around intrauterine embryos of female adults; no immuno-staining was observed in NBL; ML recognition by infection serum as a positive control and normal serum as a negative control. The cell nuclei were stained red with propidium iodide (PI). Scale bars: 50 μm.
Cleavage of different protein by rTsASP2
As Hb protein has frequently been used as a substrate to investigate the proteolytic roles of aspartic protease, the enzymatic activity of rTsASP2 was first confirmed by the cleavage of Hb protein from human and mouse. The results showed that human and mouse Hb were hydrolyzed by rTsASP2 at pH 2.5–5.5. Hb could be degraded by self-hydrolysis at an acidic pH, especially for mouse Hb at pH 2.5–4.5. After comparison to the patterns of self-hydrolysis at the same pH, the optimal pH was determined for the degradation of different Hbs. rTsASP2 could degrade mouse Hb most efficiently at pH 2.5, while it degraded human Hb at an optimal pH of 4.5. Furthermore, the degradation efficiency of Hbs hydrolyzed by rTsASP2 was observed (Fig 4). Degradation of mouse Hb was detected at 30 min (lane 4) after incubation with rTsASP2, and more cleavage fragments were observed after 4 h of incubation (lane 8). However, cleavage of human Hb was not observed at 4 h after incubation with rTsASP2 at the optimal pH 4.5 (S2 Fig). Both the heated anti-rTsASP2 serum and pepstatin A could inhibit the hydrolytic activity of rTsASP2 on mouse Hb (Fig 4D).
Fig 4. Hemoglobin degradation.
A-B Hydrolysis of Hb by rTsASP2 at different pH values. A: human Hb; B: mouse Hb; M: protein marker; lanes 1, 3, 5 and 7: Hb alone; lanes 2, 4, 6 and 8: Hb+ rTsASP2; lanes 1 and 2: pH 2.5; lanes 3 and 4: pH 3.5; lanes 5 and 6: pH 4.5; lanes 7 and 8: pH 5.5. C: Hydrolysis efficiency effect of rTsASP2 on mouse Hb (pH 2.5). M: protein marker; lanes 1, 3, 5 and 7: Hb alone; lanes 2, 4, 6 and 8: Hb+ rTsASP2; lanes 1 and 2: 5 min; lanes 3 and 4: 30 min; lanes 5 and 6: 90 min; lanes 7 and 8: 4 h. D: Inhibition effect of anti-rTsASP2 serum and pepstatin A on rTsASP2 hydrolysis of mouse Hb (pH 2.5). M: protein marker; lane 1: rTsASP2; lane 2: anti-rTsASP2 serum; lane 3: heated anti-rTsASP2 serum; lane 4: Hb; lane 5: Hb +rTsASP2; lane 6: anti-rTsASP2 serum pre-incubated with rTsASP2 +Hb; lane 7: heated anti-rTsASP2 serum pre-incubated with rTsASP2+ Hb; lane 8: pepstatin A pre-incubated with rTsASP2 + Hb. The arrow represents the band of rTsASP2 (86.4 kDa).
To investigate the putative proteolytic activity of rTsASP2, several other proteins (collagen, IgM, IgG and albumin) were used as the substrate for the enzymatic catalysis assay. Fig 5 shows that collagen (A) and IgM (B) were also hydrolyzed by rTsASP2 at pH 2.5–3.5, while no degradation of IgG and albumin was observed (S3 Fig and S4 Fig).
Fig 5. Digestion of collagen IV and IgM.
The substrate including collagen IV (A) and IgM (B) was incubated with rTsASP2. The band of approximately 98-kDa collagen IV was degraded (arrow), and degraded IgM fragments were observed compared with untreated IgM. M: protein marker; lanes 1 and 3: substrates in buffer alone; lanes 2 and 4: substrate + rTsASP2; lane 5: purified rTsASP2; lanes 1 and 3: pH 2.5; lanes 2 and 4: pH 3.5.
The enzymatic activity of rTsASP2 was further assessed by using synthetic fluorogenic peptide as a substrate (Fig 6). The maximum activity of rTsASP2 was detected at pH 3.0, although the enzyme showed a relatively broad pH range (pH 2.0–5.5) for hydrolysis of the substrate. Different metal irons have different effects on rTsASP2 catalytic activity. The enzymatic activity was clearly inhibited by the Fe2+ at 1 mM; it was also inhibited by Cu2+ in a dose-dependent manner. However, no obvious changes in hydrolytic activity were observed following the addition of Zn2+ or Mn2+ to the assay environment. Conversely, Mg2+ could enhance the proteolytic activity of rTsASP2, also in a dose-dependent manner. Under optimal assay conditions, rTsASP2 enzymatic activity was significantly inhibited by pepstatin A.
Fig 6. Enzymatic activity assay via cleavage of fluorescence substrate.
A: The optimal pH of rTsASP2 enzymatic activity. B: The enzymatic activities were assayed at 15–55°C. C: The effects of metal ions on enzymatic activities. The assays were carried out under different metal ion concentrations. D: The effects of various inhibitors on enzymatic activities, where the concentration of inhibitors was 1 mM of PMSF, 1 mM of 1,10-phenanthrolin, 1 mM of AEBSF, 10 μM of E64, 1 mM of EDTA, and 10 μM of pepstatin A. All the enzymatic activities were expressed as the relative activity of the highest reaction in each experiment.
TsASP2 mRNA and protein expression level after silencing the TsASP2 gene
After 5 μM TsASP2 siRNA was delivered into worms for 5 days, the relative expression of TsASP2 mRNA and protein was reduced by 36.42% and 35.21% compared with the PBS group, respectively (Fig 7) (P < 0.05). Another siRNA of T. spiralis aspartic protease (TsASP1 siRNA) did not reduce the TsASP2 expression. Likewise, no obvious changes in TsASP1 expression were detected in worms treated with TsASP2 siRNA (S5 Fig).
Fig 7.
Effects of TsASP2 RNAi on TsASP2 mRNA expression (A) and TsASP2 protein expression (B). qPCR (A) and western blot (B) analysis of relative TsASP2 expression levels after T. spiralis muscle larvae were transfected with 5 μM TsASP2 siRNA for 1–7 d. Asterisks indicate a statistically significant difference compared with the control siRNA and PBS groups (*P < 0.05).
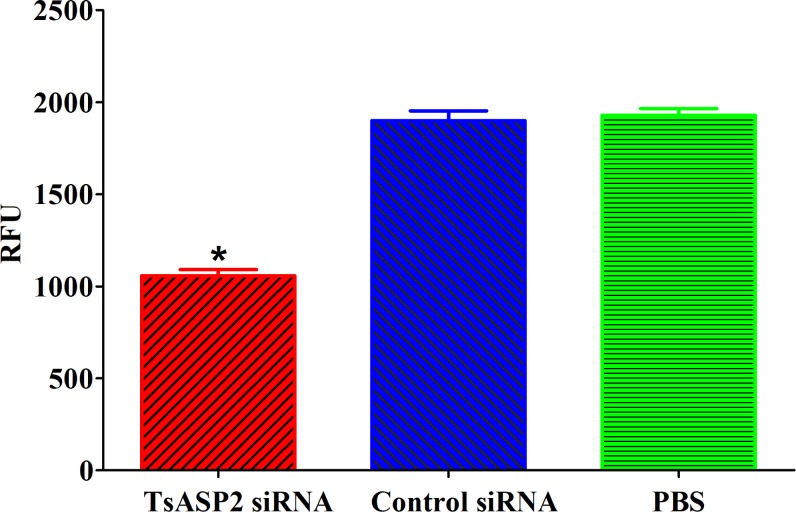
RNAi-mediated reduction of aspartic protease activity
After silencing the TsASP2 gene in T. spiralis ML, we investigated the activity of aspartic protease in crude protein from siRNA treated-ML using the synthetic fluorogenic peptide as the substrate. The results showed that the enzymatic activity was reduced in the TsASP2 siRNA-treated group by 54.82% compared with the PBS group. However, the enzymatic activity in the control siRNA-treated group was similar to the PBS group, suggesting that TsASP2 expression was closely related to aspartic protease in crude protein from ML (Fig 8).
Fig 8. Effects of TsASP2 RNAi on aspartic protease enzymatic activities of ML.
Aspartic protease activity in crude proteins of T. spiralis ML treated with siRNA was detected with a fluorogenic substrate. Asterisks indicate a statistically significant difference compared with the control siRNA and PBS groups (*P < 0.05).
RNAi effect on larval penetration of IEC
After incubation with IEC cell monolayers in semisolid medium for 2 hours, IIL invasion and migration in the monolayers were assessed (Fig 9A). The percentage of larval penetration was dose-dependently related to rTsASP2, exhibiting an increasing trend along with the increasing concentration of rTsASP2 protein (F = 353.945, P< 0.0001) (Fig 9B). When the medium was replenished with 1:100 dilutions of anti-rTsASP2 serum, infection serum or normal serum and incubated for 2 hours, the invasion rate was 35.66, 34.33 and 50.17%, respectively (χ2 = 29.085, P < 0.0001). The anti-rTsASP2 serum (1:50 to 1:100 dilutions) inhibited larval penetration into the monolayer to a greater extent than normal serum (P < 0.0001) (Fig 9C).
Fig 9. The invasion process of IECs by T. spiralis worms and RNAi inhibition on larval invasion of IEC.
A1: The larva that invaded the IEC monolayer was mobile, and its migrating trail was observed. A2: The non-invaded larva was coiled. A3: Non-invaded larva in the C2C12 monolayer. B: Promotion of larval penetration of IECs by different concentrations of rTsASP2 protein, where significant differences (P < 0.05) are marked with asterisks (*) relative to the blank control group without rTsASP2. C: Inhibition of larval invasion of IECs by different dilutions of anti-rTsASP2 serum and infection serum, where significant differences (P < 0.05) are marked with asterisks (*) relative to the normal serum group. D: Larval penetration of IECs by worms treated with siRNA. Scale bar: 1 mm.
Additionally, silencing of TsASP2 with TsASP2 siRNA significantly suppressed larval invasion of IEC, exhibiting a 62.54% decrease when the worms were treated with 5 μM TsASP2-siRNA (χ2 = 13.926, P < 0.0001) (Fig 9D). No apparent reduction of larval penetration was observed when the worms were treated with control siRNA.
TsASP2 protein is present in damaged cells invaded by larvae
After the invasion assay, TsASP2 protein was detected in remnants of damaged cells (stained with PI) using anti-rTsASP2 serum. In addition, secreted proteins from T. spiralis IIL larvae were also recognized in damaged cells using infection serum but not normal serum (Fig 10).
Fig 10. Microscopy of damaged cells following larval penetration.
The IEC monolayer was stained with PI (red), and the antigen present on the damaged cells was detected with anti-rTsASP2 serum and infection serum and visualized as green fluorescence. No immunostaining was observed with normal serum. Scale bar: 50 μm.
RNAi effect of cell damage on the IEC monolayer
Larvae treated with TsASP2 siRNA, control siRNA or PBS were used in the invasion assay. Compared with the PBS or control siRNA group, the damaged cells were significant reduced in each monolayer of the TsASP2 siRNA group, implying an important role of TsASP2 in invading IECs (P < 0.0001) (Fig 11).
Fig 11. The number of damaged cells.
The number of damaged cells caused by larvae treated with TsASP2 siRNA, control siRNA or PBS is expressed as the mean ± SD in an area of 3.02 mm2 (10×objective) for three monolayers. Asterisks indicate a statistically significant difference compared with the control siRNA and PBS groups (*P <0.05).
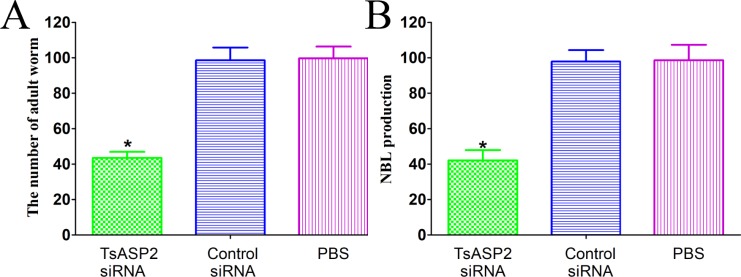
RNAi effect on larval infectivity and female worm fecundity
Compared with the PBS group, mice that were orally infected with larvae treated with TsASP2 siRNA displayed a 56.36% reduction in adult worm burden (F = 260.322, P < 0.0001) (Fig 12A). The female worms collected from the TsASP2 siRNA-treated group produced fewer NBL within 72 h than those from the other two groups (F = 195.828, P < 0.0001) (Fig 12B). In addition, the adult worms and NBL from the three groups were morphologically observed under a microscope and their lengths measured. The length of adult worms was significantly shorter compared with the PBS group (Ffemale = 58.706, Fmale = 41.308, P < 0.0001), and the length of NBL showed no significant differences among the three groups (Fig 13).
Fig 12.
Adult worm burdens (A) and newborn larva produced by females (B) recovered from mice infected with worms treated with TsASP2 siRNA. Asterisks indicate a statistically significant difference compared with the control siRNA and PBS groups (*P <0.05).
Fig 13. The lengths of different T. spiralis stage worms in mice infected with muscle larvae treated with TsASP2 siRNA.
A: Male adults; B: female adults; C: NBL. Asterisks indicate a statistically significant difference compared with the control siRNA and PBS groups (*P <0.05).
Discussion
Aspartic proteases have been found in many nematodes [24,44,45] and shown to play a key role in worm invasion and survival. Park et al. [11] characterized a T. spiralis aspartic protease of 45 kDa, but its function remained unclear.
In the present study, rTsASP2 was expressed using two different plasmids (pQE-80L and PMAL-C2X) in a prokaryotic expression system. The rTsASP2 carrying a His-tagged protein was expressed in pQE-80L as an inclusion body (S1 Fig) and did not show any protease activity, so it was used only to immunize mice to obtain anti-rTsASP2 serum. In contrast, the rTsASP2 co-expressed with the MBP tag was mainly used to investigate its enzymatic activity and biological function.
The expression of TsASP2 in various developmental stages of T. spiralis was obviously different. Native TsASP2 protein in crude proteins of all the worm stages except NBL was detected by anti-rTsASP2 serum. Furthermore, no immuno-staining was observed in NBL by IFA, which confirmed the low level of TsASP2 expression in this stage, suggesting that TsASP2 protein expression in the NBL stage was too low to be detected by Western blotting and IFA. The IFA results revealed that TsASP2 was located in the hindgut, midgut and muscle cells of ML and IIL, suggesting that TsASP2 might participate in nutrient intake. The aspartic proteases of other parasites are also located in the intestine and have been suggested to have essential functions in parasite nutrition [21,23]. The strong staining encircling the embryos suggested that TsASP2 might also be involved in female reproduction.
The primary function of aspartic protease is to digest hemoglobin [46]. In our study, rTsASP2 hydrolyzed human and murine Hb. Similar to other aspartic proteases, rTsASP2 also cleaves Hbs at an acidic pH (pH 2.5–4.5), with diverse pH values for different Hbs. The different optimal pH of aspartic protease activity could be related to different substrates [47,48]. The host-specific cleavage of Hbs by aspartic protease has been verified in previous studies [20,21]. Similarly, rTsASP2 cleaved mouse Hbs more efficiently than human Hbs. The high degradation efficiency of murine Hbs was likely due to the passaging of the T. spiralis in mice in our laboratory for more than 30 years. In addition, rTsASP2 could also hydrolyze IgM and collagens at an acidic pH, which may be associated with immune evasion, degradation of host proteins and larval migration through host tissues [49,50]. Given the highest TsASP2 expression level in the IIL stage, TsASP2 might play a key role in T. spiralis invasion of intestinal epithelium.
The enzymatic activity of rTsASP2 was further characterized by hydrolyzing the fluorescent substrate at various condition (as shown in Fig 6). The optimal pH for rTsASP2 was 3.0, which suggested that rTsASP2 had hydrolytic activity under acidic conditions similar to its optimal pH for cleaving Hb. The enzymatic activity of rTsASP2 was inhibited by Fe2+ and Cu2+ but enhanced by Mg2+, and it was not sensitive to Zn2+ and Mn2+. Previous studies have demonstrated that the activity of aspartic protease (ASP) from parasites is sensitive to copper, such as ASP from Trichomonas vaginalis (TV-CatD) and Plasmodium falciparum (plasmepsin II AP) [51,52]. In another study on Metschnikowia pulcherrima ASP [53], the cations Fe2+ and Mg2+ were found to be insensitive to ASP, while Mn2+ and Zn2+ had a slight inhibitory effect on enzymatic activity. The distinct properties of TsASP2 from other ASPs were likely due to their origination from different organism species. Nevertheless, elucidation of the effects of metal ions on enzymatic activity of ASP requires further investigation.
The enzymatic activity of rTsASP2 was clearly inhibited by pepstatin A (a common ASP inhibitor), suggesting that TsASP2 is a kind of typical aspartic protease [52]. Additionally, PMSF could also suppress 68.5% of the enzymatic activity, which differed from ASP purified form Plasmodium vivax. No obvious inhibition was detected using other enzyme inhibitors, similar to other aspartic proteases [54,55]. The proteolytic activities of TsASP2 were further confirmed by the RNAi assay. At five days after the ML were treated with 5 μM TsASP2 siRNA, the TsASP2 mRNA and protein expression levels were significantly reduced, suggesting that the TsASP2 gene was successfully silenced. Furthermore, aspartic protease activity was significantly reduced in siRNA-treated worm crude proteins, demonstrating that TsASP2 protein expression was associated with the proteolytic activities of T. spiralis aspartic protease.
In previous studies, protease enzymatic activities have been suggested to play key roles in T. spiralis infection, especially larval invasion of host IECs. The results of the larval in vitro invasion assay demonstrated that TsASP2 promoted larval penetration of IECs, and the percent of invaded larvae was dose-dependent on rTsASP2. It has been reported that anti-serum against Na-APR-2 (a hookworm aspartic protease) can inhibit the migration of the parasite through skin [20]. Similarly, larval invasion of IECs was significant inhibited when anti-rTsASP2 serum was added to the medium, supporting a facilitating function of TsASP2 in T. spiralis invasion into IECs. Our previous studies have shown that recombinant serine proteases also have a promoting role in larval invasion of IECs, whereas antibodies against serine protease have an inhibitory effect, suggesting that various proteases participate in T. spiralis larval invasion and development [15, 56–59].
RNAi is commonly applied to downregulate target molecules and investigate the biological functions of target proteins [27,34]. After downregulating TsASP2 in the ML stage, and then activating the knockdown ML into IIL, the transfected larvae showed a 62.54% reduction of larval invasion. After larval invasion, the damaged cells could be clearly detected by PI staining and then counted by microscopy. The damaged cells were significantly reduced when IECs were co-incubated with larvae treated with TsASP2 siRNA. Previous studies have suggested that the evaluation of cell damage after invasion is an objective indicator for assessing the invasion efficiency [43,60]. Our results revealed that silencing TsASP2 expression could impede T. spiralis invasion into IECs in vitro, validating that TsASP2 participated in larval penetration of host intestinal epithelium.
The TsASP2 function in T. spiralis penetration of IECs was further confirmed by silencing the TsASP2 gene with RNAi. Previous results have demonstrated that silencing of some T. spiralis genes with RNAi can impair T. spiralis worm viability [7] or inhibit its development and reproductive capacity [33, 34,61]. Silencing of the expression of these genes in T. spiralis results in reduced parasite viability and infectivity, such as impaired T. spiralis molting or invasion. Our results revealed low enteral adult worm burdens, and NBL production of female worms was reduced when TsASP2 was silenced, indicating that enteral larval growth, development and female fecundity were suppressed. Silencing of the TsASP2 gene also impeded other T. spiralis developmental stages, as adult worms recovered from mice infected with siRNA-treated larvae were shorter than those recovered from the control and PBS groups. Specific gene silencing by RNAi has been widely applied for gene function identification in other parasites [62,63], and the results of the present study indicated that TsASP2 played a crucial role in T. spiralis invasion of IEC, and silencing of the TsASP2 gene significantly reduced larval infectivity and development in mice.
In conclusion, TsASP2 was highly expressed in the IIL stage of T. spiralis, mainly located in hindgut, midgut and muscle cells of ML and IIL and around intrauterine embryos of female adults. The TsASP2 has the native aspartic protease activities to cleave Hbs, IgM and collagen under acidic condition, and the proteolytic activity was host-specific. Silencing of TsASP2 gene by RNAi could significantly reduce the TsASP2 protein expression, which inhibited the native aspartic protease activities and larval invasion of host’ enterocytes. The results indicated that TsASP2 plays an important role in the T. spiralis invasion and it could be a candidate vaccine target molecular against T. spiralis infection.
Supporting information
(DOCX)
(DOCX)
rTsASP2 has no degradation on IgG from human (A) and mice (B).
(DOCX)
rTsASP2 has no degradation on albumins from human (A) and bovine (B).
(DOCX)
qPCR (A) and Western blot (B) analysis of the expression levels of TsASP1 and TsASP2 in ML transfected using TsASP1 siRNA or TsASP2 siRNA.
(DOCX)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This study was supported by grants of the National Natural Science Foundation of China (No. U1704284 and 81871673). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bruschi F (2012) Trichinellosis in developing countries: is it neglected? J Infect Dev Ctries 6: 216–222. 10.3855/jidc.2478 [DOI] [PubMed] [Google Scholar]
- 2.Jiang P, Zhang X, Wang LA, Han LH, Yang M, et al. (2016) Survey of Trichinella infection from domestic pigs in the historical endemic areas of Henan province, central China. Parasitol Res 115:4707–4709. [DOI] [PubMed] [Google Scholar]
- 3.Pozio E, Gomez Morales MA, Dupouy-Camet J (2003) Clinical aspects, diagnosis and treatment of trichinellosis. Expert Rev Anti Infect Ther 1: 471–482. 10.1586/14787210.1.3.471 [DOI] [PubMed] [Google Scholar]
- 4.Gottstein B, Pozio E, Nockler K (2009) Epidemiology, diagnosis, treatment, and control of trichinellosis. Clin Microbiol Rev 22: 127–145. 10.1128/CMR.00026-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cui J, Wang ZQ, Xu BL (2011) The epidemiology of human trichinellosis in China during 2004–2009. Acta Trop 118: 1–5. 10.1016/j.actatropica.2011.02.005 [DOI] [PubMed] [Google Scholar]
- 6.Pozio E (2007) World distribution of Trichinella spp. infections in animals and humans. Vet Parasitol 149: 3–21. 10.1016/j.vetpar.2007.07.002 [DOI] [PubMed] [Google Scholar]
- 7.Chen X, Yang Y, Yang J, Zhang Z, Zhu X (2012) RNAi-mediated silencing of paramyosin expression in Trichinella spiralis results in impaired viability of the parasite. PLoS One 7: e49913 10.1371/journal.pone.0049913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bai X, Hu X, Liu X, Tang B, Liu M (2017) Current Research of Trichinellosis in China. Front Microbiol 8: 1472 10.3389/fmicb.2017.01472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Capo V, Despommier DD (1996) Clinical aspects of infection with Trichinella spp. Clin Microbiol Rev 9: 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu J, Yang F, Yang DQ, Jiang P, Liu RD, et al. (2018) Molecular characterization of Trichinella spiralis galectin and its participation in larval invasion of host's intestinal epithelial cells. Vet Res 49: 79 10.1186/s13567-018-0573-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park JN, Park SK, Cho MK, Park MK, Kang SA, et al. (2012) Molecular characterization of 45 kDa aspartic protease of Trichinella spiralis. Vet Parasitol 190: 510–518. 10.1016/j.vetpar.2012.06.029 [DOI] [PubMed] [Google Scholar]
- 12.Balasubramanian N, Nascimento G, Ferreira R, Martinez M, Simoes N (2012) Pepsin-like aspartic protease (Sc-ASP155) cloning, molecular characterization and gene expression analysis in developmental stages of nematode Steinernema carpocapsae. Gene 500: 164–171. 10.1016/j.gene.2012.03.062 [DOI] [PubMed] [Google Scholar]
- 13.Liu MY, Wang XL, Fu BQ, Li CY, Wu XP, et al. (2007) Identification of stage-specifically expressed genes of Trichinella spiralis by suppression subtractive hybridization. Parasitology 134: 1443–1455. 10.1017/S0031182007002855 [DOI] [PubMed] [Google Scholar]
- 14.Cwiklinski K, Meskill D, Robinson MW, Pozio E, Appleton JA, et al. (2009) Cloning and analysis of a Trichinella pseudospiralis muscle larva secreted serine protease gene. Vet Parasitol 159: 268–271. 10.1016/j.vetpar.2008.10.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun GG, Ren HN, Liu RD, Song YY, Qi X, et al. (2018) Molecular characterization of a putative serine protease from Trichinella spiralis and its elicited immune protection. Vet Res 49: 59 10.1186/s13567-018-0555-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Todorova VK, Stoyanov DI (2000) Partial characterization of serine proteinases secreted by adult Trichinella spiralis. Parasitol Res 86: 684–687. 10.1007/pl00008552 [DOI] [PubMed] [Google Scholar]
- 17.Szecsi PB (1992) The aspartic proteases. Scand J Clin Lab Invest Suppl 210: 5–22. [PubMed] [Google Scholar]
- 18.Tcherepanova I, Bhattacharyya L, Rubin CS, Freedman JH (2000) Aspartic proteases from the nematode Caenorhabditis elegans. Structural organization and developmental and cell-specific expression of asp-1. J Biol Chem 275: 26359–26369. 10.1074/jbc.M000956200 [DOI] [PubMed] [Google Scholar]
- 19.Tang J, Wong RN (1987) Evolution in the structure and function of aspartic proteases. J Cell Biochem 33: 53–63. 10.1002/jcb.240330106 [DOI] [PubMed] [Google Scholar]
- 20.Williamson AL, Brindley PJ, Abbenante G, Datu BJ, Prociv P, et al. (2003) Hookworm aspartic protease, Na-APR-2, cleaves human hemoglobin and serum proteins in a host-specific fashion. J Infect Dis 187: 484–494. 10.1086/367708 [DOI] [PubMed] [Google Scholar]
- 21.Williamson AL, Brindley PJ, Loukas A (2003) Hookworm cathepsin D aspartic proteases: contributing roles in the host-specific degradation of serum proteins and skin macromolecules. Parasitology 126: 179–185. 10.1017/s0031182002002706 [DOI] [PubMed] [Google Scholar]
- 22.Suttiprapa S, Mulvenna J, Huong NT, Pearson MS, Brindley PJ, et al. (2009) Ov-APR-1, an aspartic protease from the carcinogenic liver fluke, Opisthorchis viverrini: functional expression, immunolocalization and subsite specificity. 41: 1148–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morales ME, Rinaldi G, Gobert GN, Kines KJ, Tort JF, et al. (2008) RNA interference of Schistosoma mansoni cathepsin D, the apical enzyme of the hemoglobin proteolysis cascade. Mol Biochem Parasitol 157: 160–168. 10.1016/j.molbiopara.2007.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jolodar A, Fischer P, Buttner DW, Miller DJ, Schmetz C, et al. (2004) Onchocerca volvulus: expression and immunolocalization of a nematode cathepsin D-like lysosomal aspartic protease. Exp Parasitol 107: 145–156. 10.1016/j.exppara.2004.06.006 [DOI] [PubMed] [Google Scholar]
- 25.Pearson MS, Bethony JM, Pickering DA, de Oliveira LM, Jariwala A, et al. (2009) An enzymatically inactivated hemoglobinase from Necator americanus induces neutralizing antibodies against multiple hookworm species and protects dogs against heterologous hookworm infection. FASEB J 23: 3007–3019. 10.1096/fj.09-131433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, et al. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811. 10.1038/35888 [DOI] [PubMed] [Google Scholar]
- 27.Wang X, Chen W, Tian Y, Huang Y, Li X, et al. (2014) RNAi-mediated silencing of enolase confirms its biological importance in Clonorchis sinensis. Parasitol Res 113: 1451–1458. 10.1007/s00436-014-3785-0 [DOI] [PubMed] [Google Scholar]
- 28.Singh M, Singh PK, Misra-Bhattacharya S (2012) RNAi mediated silencing of ATPase RNA helicase gene in adult filarial parasite Brugia malayi impairs in vitro microfilaria release and adult parasite viability. J Biotechnol 157: 351–358. 10.1016/j.jbiotec.2011.12.003 [DOI] [PubMed] [Google Scholar]
- 29.Kushwaha S, Singh PK, Shahab M, Pathak M, Bhattacharya SM (2012) In vitro silencing of Brugia malayi trehalose-6-phosphate phosphatase impairs embryogenesis and in vivo development of infective larvae in jirds. PLoS Negl Trop Dis 6: e1770 10.1371/journal.pntd.0001770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Somarathne M, Gunawardene Y, Chandrasekharan NV, Dassanayake RS (2018) Development of siRNA mediated RNA interference and functional analysis of novel parasitic nematode-specific protein of Setaria digitata. Exp Parasitol 186: 42–49. 10.1016/j.exppara.2018.02.001 [DOI] [PubMed] [Google Scholar]
- 31.Zou X, Jin YM, Liu PP, Wu QJ, Liu JM, et al. (2011) RNAi silencing of calcium-regulated heat-stable protein of 24 kDa in Schistosoma japonicum affects parasite growth. Parasitol Res 108: 567–572. 10.1007/s00436-010-2099-0 [DOI] [PubMed] [Google Scholar]
- 32.Yang Y, Jin Y, Liu P, Shi Y, Cao Y, et al. (2012) RNAi silencing of type V collagen in Schistosoma japonicum affects parasite morphology, spawning, and hatching. Parasitol Res 111: 1251–1257. 10.1007/s00436-012-2959-x [DOI] [PubMed] [Google Scholar]
- 33.Wang ZQ, Zhang SB, Jiang P, Liu RD, Long SR, et al. (2015) The siRNA-mediated silencing of Trichinella spiralis nudix hydrolase results in reduction of larval infectivity. Parasitol Res 114: 3551–3557. 10.1007/s00436-015-4650-5 [DOI] [PubMed] [Google Scholar]
- 34.Yang F, Yang DQ, Song YY, Guo KX, Li YL, et al. (2019) In vitro silencing of a serine protease inhibitor suppresses Trichinella spiralis invasion, development, and fecundity. Parasitol Res 118: 2247–2255. 10.1007/s00436-019-06344-4 [DOI] [PubMed] [Google Scholar]
- 35.Yang W, Li LG, Liu RD, Sun GG, Liu CY, et al. (2015) Molecular identification and characterization of Trichinella spiralis proteasome subunit beta type-7. Parasit Vectors 8: 18 10.1186/s13071-014-0626-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang P, Wang ZQ, Cui J, Zhang X (2012) Comparison of artificial digestion and Baermann's methods for detection of Trichinella spiralis pre-encapsulated larvae in muscles with low-level infections. Foodborne Pathog Dis 9: 27–31. 10.1089/fpd.2011.0985 [DOI] [PubMed] [Google Scholar]
- 37.Yang Y, Lacour SA, Laine-Prade V, Versille N, Grasset-Chevillot A, et al. (2015) Trichinella spiralis newborn larvae: characterization of a stage specific serine proteinase expression, NBL1, using monoclonal antibodies. Parasitology 142: 783–790. 10.1017/S0031182014001851 [DOI] [PubMed] [Google Scholar]
- 38.Liu RD, Qi X, Sun GG, Jiang P, Zhang X, et al. (2016) Proteomic analysis of Trichinella spiralis adult worm excretory-secretory proteins recognized by early infection sera. Vet Parasitol 231: 43–46. 10.1016/j.vetpar.2016.10.008 [DOI] [PubMed] [Google Scholar]
- 39.Zhang NZ, Liu JY, Li WH, Li L, Qu ZG, et al. (2016) Cloning and characterization of thioredoxin peroxidases from Trichinella spiralis. Vet Parasitol 231:53–58. [DOI] [PubMed] [Google Scholar]
- 40.Liu RD, Wang ZQ, Wang L, Long SR, Ren HJ, et al. (2013) Analysis of differentially expressed genes of Trichinella spiralis larvae activated by bile and cultured with intestinal epithelial cells using real-time PCR. Parasitol Res 112: 4113–4120. 10.1007/s00436-013-3602-1 [DOI] [PubMed] [Google Scholar]
- 41.Sun GG, Song YY, Jiang P, Ren HN, Yan SW, et al. (2018) Characterization of a Trichinella spiralis putative serine protease. Study of its potential as sero-diagnostic tool. PLoS Negl Trop Dis 12: e0006485 10.1371/journal.pntd.0006485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu LN, Wang ZQ, Zhang X, Jiang P, Qi X, et al. (2015) Characterization of Spirometra erinaceieuropaei Plerocercoid Cysteine Protease and Potential Application for Serodiagnosis of Sparganosis. PLoS Negl Trop Dis 9: e0003807 10.1371/journal.pntd.0003807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.ManWarren T, Gagliardo L, Geyer J, McVay C, Pearce-Kelling S, et al. (1997) Invasion of intestinal epithelia in vitro by the parasitic nematode Trichinella spiralis. Infect Immun 65: 4806–4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schulmeister A, Heyers O, Morales ME, Brindley PJ, Lucius R, et al. (2005) Organization and functional analysis of the Schistosoma mansoni cathepsin D-like aspartic protease gene promoter. Biochim Biophys Acta 1727: 27–34. 10.1016/j.bbaexp.2004.11.004 [DOI] [PubMed] [Google Scholar]
- 45.Geier G, Banaj HJ, Heid H, Bini L, Pallini V, et al. (1999) Aspartyl proteases in Caenorhabditis elegans. Isolation, identification and characterization by a combined use of affinity chromatography, two-dimensional gel electrophoresis, microsequencing and databank analysis. Eur J Biochem 264: 872–879. 10.1046/j.1432-1327.1999.00679.x [DOI] [PubMed] [Google Scholar]
- 46.Brindley PJ, Kalinna BH, Wong JY, Bogitsh BJ, King LT, et al. (2001) Proteolysis of human hemoglobin by schistosome cathepsin D. Mol Biochem Parasitol 112: 103–112. 10.1016/s0166-6851(00)00351-0 [DOI] [PubMed] [Google Scholar]
- 47.Stoknes I, Rustad T (1995) Purification and characterization of a multicatalytic proteinase from Atlantic salmon (Salmo salar) muscle. Comp Biochem Physiol B Biochem Mol Biol 111: 587–596. 10.1016/0305-0491(95)00030-c [DOI] [PubMed] [Google Scholar]
- 48.Cao Y, Shi Y, Qiao H, Yang Y, Liu J, et al. (2014) Distribution of lethal giant larvae (Lgl) protein in the tegument and negative impact of siRNA-based gene silencing on worm surface structure and egg hatching in Schistosoma japonicum. Parasitol Res 113: 1–9. 10.1007/s00436-013-3620-z [DOI] [PubMed] [Google Scholar]
- 49.Kong Y, Chung YB, Cho SY, Kang SY (1994) Cleavage of immunoglobulin G by excretory-secretory cathepsin S-like protease of Spirometra mansoni plerocercoid. Parasitology 109 (Pt 5): 611–621. [DOI] [PubMed] [Google Scholar]
- 50.Song CY, Chappell CL (1993) Purification and partial characterization of cysteine proteinase from Spirometra mansoni plerocercoids. J Parasitol 79: 517–524. [PubMed] [Google Scholar]
- 51.He S, Zhu L, Liu F, Liu Q, Shao Y, et al. (2018) Functions of the Vasa gene in Schistosoma japonicum as assessed by RNA interference. Gene 638: 13–19. 10.1016/j.gene.2017.09.054 [DOI] [PubMed] [Google Scholar]
- 52.Mancilla-Olea MI, Ortega-Lopez J, Figueroa-Angulo EE, Avila-Gonzalez L, Cardenas-Guerra RE, et al. (2018) Trichomonas vaginalis cathepsin D-like aspartic proteinase (Tv-CatD) is positively regulated by glucose and degrades human hemoglobin. Int J Biochem Cell Biol 97: 1–15. 10.1016/j.biocel.2018.01.015 [DOI] [PubMed] [Google Scholar]
- 53.Theron LW, Bely M, Divol B (2017) Characterisation of the enzymatic properties of MpAPr1, an aspartic protease secreted by the wine yeast Metschnikowia pulcherrima. J Sci Food Agric 97: 3584–3593. 10.1002/jsfa.8217 [DOI] [PubMed] [Google Scholar]
- 54.Valdivieso E, Dagger F, Rascon A (2007) Leishmania mexicana: identification and characterization of an aspartyl proteinase activity. Exp Parasitol 116: 77–82. 10.1016/j.exppara.2006.10.006 [DOI] [PubMed] [Google Scholar]
- 55.Sharma A, Eapen A, Subbarao SK (2005) Purification and characterization of a hemoglobin degrading aspartic protease from the malarial parasite Plasmodium vivax. J Biochem 138: 71–78. 10.1093/jb/mvi105 [DOI] [PubMed] [Google Scholar]
- 56.Ren HN, Guo KX, Zhang Y, Sun GG, Liu RD, et al. (2018) Molecular characterization of a 31 kDa protein from Trichinella spiralis and its induced immune protection in BALB/c mice. Parasit Vectors 11: 625 10.1186/s13071-018-3198-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang B, Wang ZQ, Jin J, Ren HJ, Liu LN, et al. (2013) Cloning, expression and characterization of a Trichinella spiralis serine protease gene encoding a 35.5 kDa protein. Exp Parasitol 134:148–154. 10.1016/j.exppara.2013.03.004 [DOI] [PubMed] [Google Scholar]
- 58.Cui J, Wang L, Sun GG, Liu LN, Zhang SB, et al. (2015) Characterization of a Trichinella spiralis 31 kDa protein and its potential application for the serodiagnosis of trichinellosis. Acta Trop 142:57–63. 10.1016/j.actatropica.2014.10.017 [DOI] [PubMed] [Google Scholar]
- 59.Xu J, Liu RD, Long SR, Song YY, Jiang P, et al. (2020) Characterization of a chymotrypsin-like enzyme from Trichinella spiralis and its facilitation of larva penetration into the host's enteral epithelial cells. Res Vet Sci 128:1–8. 10.1016/j.rvsc.2019.10.018 [DOI] [PubMed] [Google Scholar]
- 60.Butcher BA, Gagliardo LF, ManWarren T, Appleton JA (2000) Larvae-induced plasma membrane wounds and glycoprotein deposition are insufficient for Trichinella spiralis invasion of epithelial cells. Mol Biochem Parasitol 107: 207–218. 10.1016/s0166-6851(00)00189-4 [DOI] [PubMed] [Google Scholar]
- 61.Zhang SB, Jiang P, Wang ZQ, Long SR, Liu RD, et al. (2016) DsRNA-mediated silencing of Nudix hydrolase in Trichinella spiralis inhibits the larval invasion and survival in mice. Exp Parasitol 162: 35–42. 10.1016/j.exppara.2016.01.005 [DOI] [PubMed] [Google Scholar]
- 62.Misra S, Gupta J, Misra-Bhattacharya S (2017) RNA interference mediated knockdown of Brugia malayi UDP-Galactopyranose mutase severely affects parasite viability, embryogenesis and in vivo development of infective larvae. Parasit Vectors 10: 34 10.1186/s13071-017-1967-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.You H, Liu C, Du X, Nawaratna S, Rivera V, et al. (2018) Suppression of Schistosoma japonicum Acetylcholinesterase Affects Parasite Growth and Development. Int J Mol Sci 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
rTsASP2 has no degradation on IgG from human (A) and mice (B).
(DOCX)
rTsASP2 has no degradation on albumins from human (A) and bovine (B).
(DOCX)
qPCR (A) and Western blot (B) analysis of the expression levels of TsASP1 and TsASP2 in ML transfected using TsASP1 siRNA or TsASP2 siRNA.
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.