Abstract
Acute graft-versus-host disease (aGVHD) is a T cell–mediated immunological disorder and the leading cause of nonrelapse mortality in patients who receive allogeneic hematopoietic cell transplants. Based on recent observations that protein arginine methyltransferase 5 (PRMT5) and arginine methylation are upregulated in activated memory T cells, we hypothesized that PRMT5 is involved in the pathogenesis of aGVHD. Here, we show that PRMT5 expression and enzymatic activity were upregulated in activated T cells in vitro and in T cells from mice developing aGVHD after allogeneic transplant. PRMT5 expression was also upregulated in T cells of patients who developed aGVHD after allogeneic hematopoietic cell transplant compared with those who did not develop aGVHD. PRMT5 inhibition using a selective small-molecule inhibitor (C220) substantially reduced mouse and human allogeneic T cell proliferation and inflammatory IFN-γ and IL-17 cytokine production. Administration of PRMT5 small-molecule inhibitors substantially improves survival, reducing disease incidence and clinical severity in mouse models of aGVHD without adversely affecting engraftment. Importantly, we show that PRMT5 inhibition retained the beneficial graft-versus-leukemia effect by maintaining cytotoxic CD8+ T cell responses. Mechanistically, we show that PRMT5 inhibition potently reduced STAT1 phosphorylation as well as transcription of proinflammatory genes, including interferon-stimulated genes and IL-17. Additionally, PRMT5 inhibition deregulates the cell cycle in activated T cells and disrupts signaling by affecting ERK1/2 phosphorylation. Thus, we have identified PRMT5 as a regulator of T cell responses and as a therapeutic target in aGVHD.
Keywords: Immunology, Transplantation
Keywords: Bone marrow transplantation, T cells, Tolerance
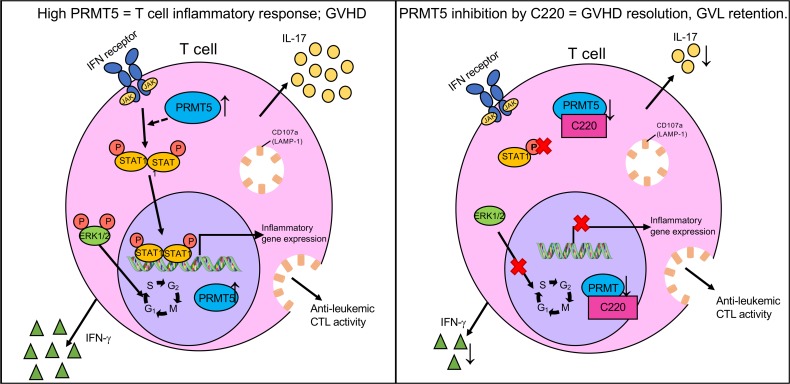
PRMT5 modulates T cell function in graft-versus-host disease.
Introduction
T cell activation via the T cell receptor complex (TCR) involves a complex interaction of signaling networks and subsequent gene transcription pathways that dictate the phenotype of T cell response to antigenic insult. Acute graft-versus-host disease (aGVHD), a T cell–mediated immunological disorder, is a frequent posttransplant complication associated with increased morbidity and mortality in patients who receive allogeneic hematopoietic stem cell transplants (1–3). The pathogenesis of aGVHD involves the recognition of host minor and major histocompatibility complex antigens by immune-competent, donor-derived T cells that mount an inflammatory reaction, initiating T cell alloantigen recognition followed by expansion, migration, and finally end organ damage due to a combination of proinflammatory cytokine secretion (IFN-γ, TNF-α, IL-17) and direct cytotoxic effects (1–3). Th1 cells producing inflammatory cytokines, such as IL-2, IFN-γ, and TNF-α, are considered the crucial subset of T cells that induce aGVHD (4–6). Interestingly, studies have shown that absence of IFN-γ exacerbates aGVHD in part because of increased Th2/Th17 differentiation and Th2/Th17-mediated pathologies while absence of Th17 leads to aggravated Th1-mediated aGVHD (6–10). These studies demonstrate that the contribution of different T cell subsets (Th1, Th2, and Th17) to aGVHD pathogenesis is complex and likely interdependent, involving multiple factors, including preferential expression of chemokine receptors that regulate homing and thereby tissue-specific damage by donor T cells (6–10).
Posttranslational modifications via arginine methylation have been shown to play an important role in several biological processes, including gene transcription, cell cycle progression, and signal transduction (11–13). Arginine (R) methylation is catalyzed by a group of enzymes called protein arginine methyltransferases (PRMTs) that are classified into 3 main groups depending on the type of methyl groups added to the R residues. Type I PRMTs (PRMT1, PRMT3, PRMT4, and PRMT6) catalyze asymmetric dimethylation, type II PRMTs (PRMT5 and PRMT9) catalyze symmetric dimethylation, and type III enzymes (PRMT7) drive monomethylation of R residues (11, 12). All PRMT enzymes utilize S-adenosylmethionine (SAMe) as their methyl donor, transferring a methyl group to terminal guanidine nitrogen atoms of R side chains on histones and other protein substrates (14, 15). PRMT5 is the predominant type II enzyme responsible for the majority of symmetric dimethylation of R residues (11, 13, 16) and is unique among PRMT enzymes in requiring a cofactor, methylosome protein 50, that increases PRMT5’s methyltransferase capabilities by increasing protein substrate affinity (11, 14). Until recently, protein R methylation was detected mostly on nuclear histones and therefore thought to regulate processes such as chromatin remodeling, transcription, cell cycle, and spliceosome assembly (15). However, new research shows that T cells are sensitive to protein R methylation. T cell activation results in global upregulation of R methyltransferase activity (17, 18) that controls signal transduction and T cell function. Disruption of PRMT5 activity by RNA interference results in inhibition of both IL-2–driven and nuclear factor of activated T cells–driven promoter activities and IL-2 secretion (19). Using a conditional knockout model, researchers have shown that PRMT5 plays a critical role in hematopoiesis by impairing proliferation of progenitor cells and cytokine signaling (20). Interestingly, loss of PRMT5 affected thymic cellularity, indicating a role for PRMT5 in early thymocyte development; however, no difference was seen in spleen weight or cellularity (20). T cell–specific deletion of PRMT5 results in defects in peripheral T cell maintenance as well as T cell signal transduction via splicing defects in γc and JAK3 mRNA (21). Our group has shown previously that PRMT5 blockade with a PRMT5 inhibitor suppresses memory T cell responses and reduces inflammation in experimental autoimmune encephalomyelitis mouse models (22). Given that aGVHD is predominantly a T cell–driven inflammatory disease, we sought to investigate the role of PRMT5 in promoting inflammation and wanted to determine the effect of PRMT5 inhibition on aGVHD disease pathogenesis.
In parallel to its role in immune cells, PRMT5 is overexpressed in many leukemias and lymphomas (23–26). Epigenetic changes driven by PRMT5 lead to repression of tumor suppressors, such as ST7, PTPROt, and microRNA-29b, consequently leading to aberrant proliferation and survival of cancer cells (24–28). This finding has led to a surge of interest in developing specific and selective inhibitors of PRMT5 as a therapeutic strategy for its antitumor properties. Targeting PRMT5 has therapeutic benefit in preclinical models of lymphoma and acute myeloid leukemia (AML) (13, 23–27), and currently, PRMT5 inhibition is being evaluated in phase I clinical trials for both solid tumors and advanced hematological malignancies (ClinicalTrials.gov NCT03854227, NCT03573310, NCT02783300, NCT03886831). Acute leukemias and myelodisplastic syndromes (combined with myeloproliferative disorders) are the most common indications for allogeneic transplants, accounting for 72% of allogeneic hematopoietic cell transplantations (HCTs) (29). Therefore, inhibition of PRMT5 in a posttransplant setting could be a novel approach to inhibit inflammatory T cell responses, thereby preventing aGVHD while targeting residual leukemic cells.
C220, a potent, selective PRMT5 inhibitor, is a derivative of a published PRMT5 inhibitor (30) that directly occupies the SAMe binding pocket (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.131099DS1). C220 demonstrates IC50 of 2.4 nM in biochemical assays using histone H4–based peptide as a substrate (Supplemental Figure 1B) and IC50 more than 1 μM against 38 methyltransferases, including other members of the PRMT family (Supplemental Figure 1C). Here, we show that PRMT5 expression and function are upregulated in activated T cells and in T cells of mice and humans with aGVHD. Inhibition of PRMT5 activity using a selective small-molecule inhibitor, C220, results in reduction of alloreactive T cell proliferation and affects T cell signaling, including STAT1 and ERK1/2 phosphorylation and inflammatory IFN-γ and IL-17 cytokine production. PRMT5 inhibition in vivo delays aGVHD and improves survival in mouse models of aGVHD without compromising beneficial graft-versus-leukemia effects. Together, our results demonstrate for the first time to our knowledge a role for PRMT5 in aGVHD pathogenesis and support future studies to investigate PRMT5 inhibitors as a therapeutic strategy for aGVHD.
Results
PRMT5 expression and enzymatic activity are upregulated in activated T cells.
We first set out to investigate the expression and enzymatic function of PRMT5 in activated murine and human T cells. Prmt5 mRNA expression was significantly upregulated in murine T cells stimulated via TCR with CD3/CD28 beads (Figure 1A) as well as T cells stimulated by allogeneic BALB/c bone marrow–derived dendritic cells (BMDCs) mimicking a mode of activation resembling GVHD (Figure 1B). Consistent with mRNA expression, PRMT5 protein expression was also upregulated in murine TCR-stimulated and allostimulated T cells (Figure 1C). The enzymatic activity of PRMT5 was also upregulated in activated T cells, as evidenced by the concordant increased symmetric dimethylation of Arg8 (R8) on histone H3 (H3R8me2s) (Figure 1C). Similar to mouse cells, we observed increased PRMT5 mRNA expression (Figure 1D) and PRMT5 protein and enzymatic activity (Figure 1E) in activated human T cells. Increased enzymatic function was further confirmed by in vitro PRMT5 methyltransferase activity assay, which demonstrated increased specific activity on an H4-Arg3 peptide in nuclear extracts derived from CD3/CD28-activated human T cells compared with control (Figure 1F). To investigate the status of PRMT5 expression/activity in T cells in an inflammatory disease setting, we used the B6 into B6D2F1 (F1) aGVHD mouse model. T cells were isolated from spleens of irradiated B6D2F1 recipient mice that received T cell–depleted bone marrow cells (TCD-BMs, no disease) or mice that received TCD-BM + B6 allogeneic splenocytes (TCD-BM + allo spl, aGVHD mice) around day 25 after transplant. PRMT5 mRNA (Figure 1G) as well as protein expression and enzymatic activity were clearly upregulated in T cells of mice that developed aGVHD compared with mice that did not develop disease (Figure 1H). Together, these data show that activation of T cells results in substantial upregulation of PRMT5 gene and protein expression as well as function, determined by increased enzymatic activity. Last, to evaluate status of PRMT5 in patients with aGVHD, we compared expression of PRMT5 in patients who received an allogeneic stem cell transplant but did not develop aGVHD versus those who developed aGVHD (at time of diagnosis). We found that PRMT5 expression was significantly upregulated in T cells of patients who developed aGVHD as compared with non-aGVHD patients or healthy controls (Figure 1I).
Figure 1. PRMT5 expression and activity is upregulated in activated mouse and human T cells in vitro and in vivo.
Murine B6 T cells were stimulated with either CD3/CD28 Dynabeads or allogeneic BALB/c DCs for 4 days. (A and B) Prmt5 mRNA expression in T cells quantified by real-time PCR (n = 5). Expression relative to unstimulated (US) T cells, with β-actin used as a normalizer. Results are represented as mean ± SD of 2–3 independent experiments. Each symbol represents an individual donor. (C) PRMT5 protein and histone H3R8 symmetric dimethyl protein (H3R8me2s) analyzed by Western blot. One representative Western blot of 3 independent experiments is shown. (D) T cells isolated from healthy human donor (HD) PBMCs were stimulated with CD3/CD28 Dynabeads, with Prmt5 mRNA expression (n = 3 donors) quantified by real-time PCR, and (E) immunoblotted for PRMT5 protein expression and function (n = 2 donors). (F) In vitro PRMT5 methyltransferase activity in nuclear extracts derived from US versus CD3/CD28-stimulated human T cells. Results show specific activity measured in nanograms of dimethylated H4-Arg3 produced per 20 μg nuclear extract. Each symbol represents an individual replicate measurement, with mean ± SD. One representative assay of 3 independent experiments is shown. (G) Splenic T cells isolated from lethally irradiated B6D2F1 mice that received TCD-BM or TCD-BM + B6 allogeneic splenocytes around 25 days after transplant. mRNA expression (n = 9) and (H) protein expression. Two or 3 spleens were pooled from recipient mice to make 1 pooled sample, and 3 pooled samples were analyzed for PRMT5 protein expression by Western blot. (I) Healthy human PBMCs were collected from HDs (n = 7) and from patients after allogeneic bone marrow transplantation with or without GVHD at the time of collection (Supplemental Table 2). Patient samples were matched for day of transplant at which GVHD occurred. PBMCs were stained and analyzed for PRMT5 expression on T cells using mass cytometry; the gating strategy is shown in Supplemental Figure 8. Each symbol represents an individual donor. Data represent mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
PRMT5 inhibition substantially reduces mouse and human allogeneic T cell proliferation and inflammatory cytokine secretion.
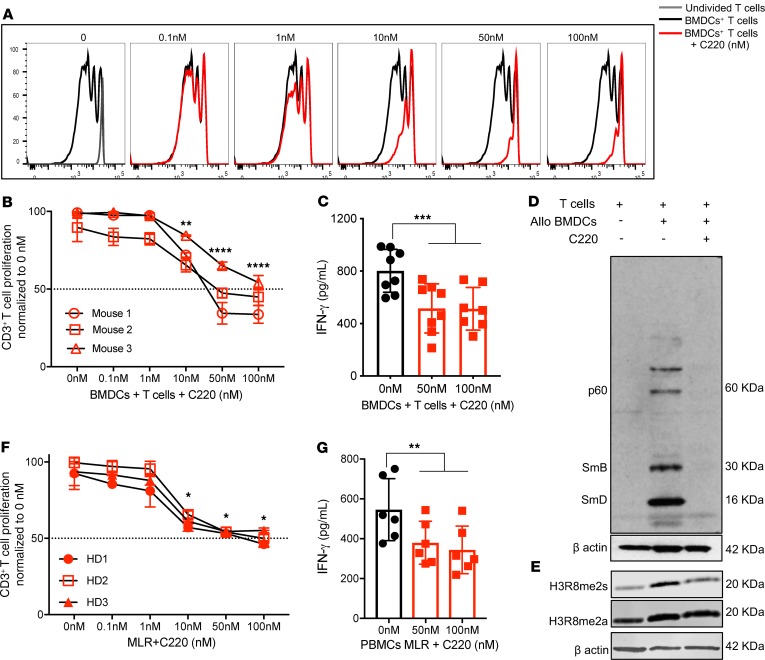
Because PRMT5 expression was upregulated in activated T cells, we sought to determine whether inhibition of PRMT5 would affect T cell proliferation and effector function (as measured by cytokine secretion). B6 T cells were stimulated with allogeneic BMDCs in the presence of increasing concentrations of PRMT5 small-molecule inhibitor (C220). Treatment with C220 significantly impaired both murine T cell proliferation and IFN-γ secretion (Figure 2, A–C, and Supplemental Figure 2). We observed selective reduction in symmetric dimethylation but not asymmetric R methylation (Figure 2, D and E), showing specificity and selectivity of our inhibitor to only PRMT5 and no other members of the PRMT family that mediate asymmetric R methylation. Human PBMCs were stimulated in mixed lymphocyte reaction (MLR) experiments, and similar to the mouse T cells, we observed a dose-dependent decrease in human T cell proliferation, IFN-γ secretion (Figure 2, F and G), and symmetric R dimethylation (Supplemental Figure 3) in response to PRMT5 inhibition.
Figure 2. Inhibition of PRMT5 blunts mouse and human T cell alloreactive response.
Cell trace violet–labeled (CTV-labeled) B6 T cells were stimulated with allogeneic BALB/c BMDCs for 4 days in the presence of increasing concentrations of PRMT5 inhibitor C220. (A) Histogram plots of 1 representative donor. (B) Percentage of alloreactive T cell proliferation normalized to no drug. (C) IFN-γ cytokine in supernatant analyzed by ELISA. B6 T cells were stimulated with BALB/c BMDCs for 4 days, with T cells reisolated and immunoblotted for (D) symmetrically dimethylated arginine residues and (E) H3R8me2s and H3R8me2a. β-actin was used as loading control. One representative Western blot of 3 independent experiments is shown. (F) CTV-labeled CD3+ human T cells were stimulated with allogeneic TCD-PBMCs for 4 days in the presence of increasing concentrations of C220. Percentage of alloreactive T cell proliferation normalized to no drug. (G) IFN-γ cytokine in supernatant analyzed by ELISA. Mean ± SD of biological replicates of 3 mouse/human donors shown. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 compared with allostimulated T cells in the absence of C220.
PRMT5 inhibition improves survival in mouse models of aGVHD.
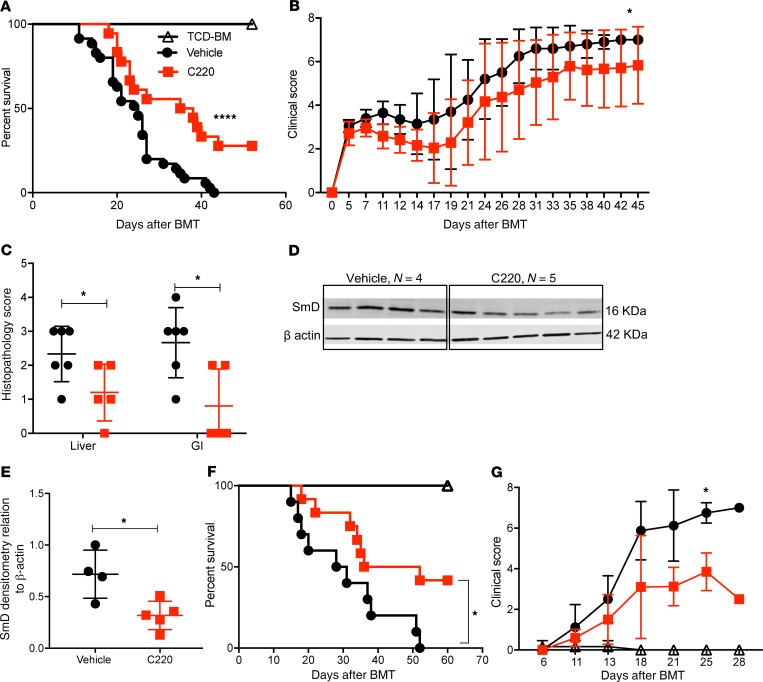
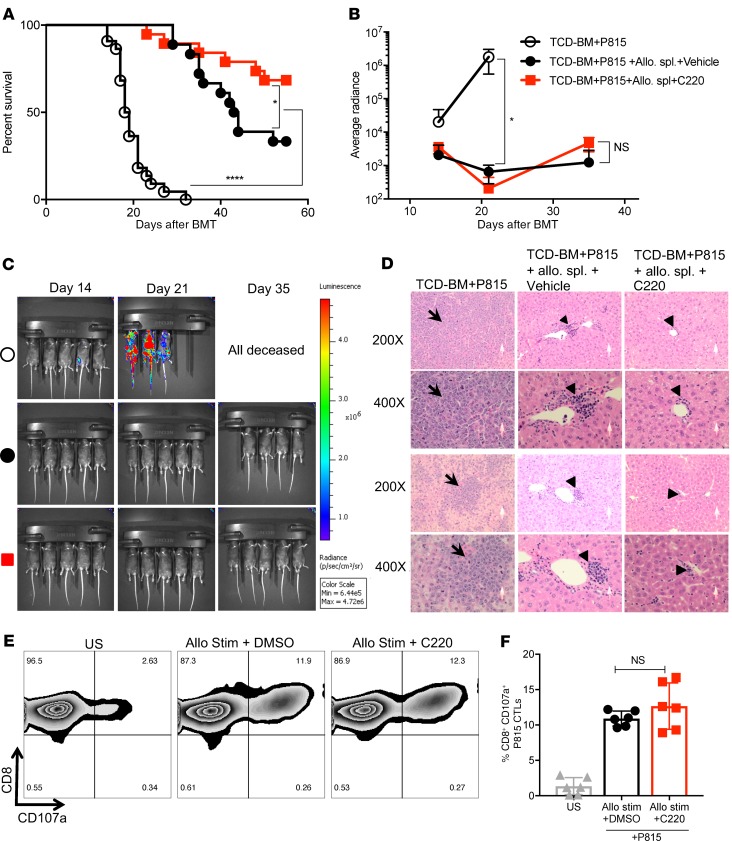
Because PRMT5 regulates T cell proliferation and cytokine secretion, we asked whether administration of PRMT5 inhibitor could improve overall survival and clinical disease severity in mice with aGVHD. Using the B6 into B6D2F1 model, lethally irradiated F1 mice were treated with C220 (2 mg/kg) or vehicle (oral gavage, once weekly) starting at day +7 after infusion of allogeneic B6 splenocytes. PRMT5 inhibition with C220 significantly prolonged survival of allotransplanted recipient mice (Figure 3A) and resulted in reduced aGVHD clinical scores (Figure 3B) and histopathology scores (Figure 3C). The improved pathology score in the C220-treated mice was due to reduced periportal and perivascular inflammation in the liver as well as reduced inflammation and apoptotic bodies in the colon (Supplemental Figure 4). We observed significant downregulation of symmetric dimethylation of SmD protein, a pharmacodynamic marker for PRMT5 inhibition, in splenocytes of mice treated with C220 compared with vehicle (Figure 3, D and E). We further tested the therapeutic effect of PRMT5 inhibition using a mouse model of xenogeneic aGVHD. Briefly, NOD/SCID–γ (NSG) mice were conditioned with 50 cGy and received T cell–depleted PBMCs (10 × 106 cells, TCD-PBMCs) or 15 × 106 to 20 × 106 human PBMCs. Mice that received T cell–replete grafts were given C220 (2 mg/kg, Monday/Wednesday/Friday, starting day 7 after transplant) or vehicle and monitored for survival. Administration of C220 significantly prolonged survival of NSG mice (Figure 3, F and G) compared with vehicle control.
Figure 3. Inhibition of PRMT5 improves survival in mouse models of aGVHD.
Lethally irradiated B6D2F1 recipients received CD45.1+ B6 TCD-BMs (10 × 106 cells) or TCD-BMs + CD45.1+ B6 splenocytes (15 × 106). Recipients of allogeneic splenocytes were treated with C220 (2 mg/kg) or vehicle by oral gavage once weekly starting day 7 after transplant. (A) Kaplan-Meier survival curve. Log-rank test was used to compare survival. Data pooled from 3 independent transplant experiments, with n = 6–8 in TCD-BM + splenocytes group in every transplant experiment; TCD-BM group was included for 2 transplants, with n = 4 each transplant. Total n = 8 for TCD-BM group; n = 18–24 for TCD-BM + splenocytes groups. (B) Clinical GVHD scores. Mann-Whitney U test was used for comparison between groups. B6 into F1 transplant was performed as described above, and a separate cohort of mice were euthanized at day 22 after transplant (after receiving 3 doses of vehicle/C220 on days 7, 14, and 21) and used for C–E. (C) Histopathological assessment of target tissues (liver and gastrointestinal [GI] tract); n = 5 per group. (D) Splenocytes harvested for immunoblotting for pharmacodynamics marker SmD3. (E) Densitometry values normalized to β-actin. Data represent mean ± SD. Each symbol represents an individual donor; n = 4–5. (F) NSG mice were irradiated with 50 cGy on day –1. On day 0, irradiated NSG mice received either TCD-PBMCs (10 × 106, n = 3) or human PBMCs (15 × 106). Mice that received T cell–replete grafts were treated with either C220 (2 mg/kg, n = 12) 3 times weekly or vehicle (n = 10). Survival curve. Log-rank test was used to compare survival. (G) Clinical GVHD scores of xenogeneic GVHD model. Data shown are combined from 2 independent xeno-aGVHD experiments. *P < 0.05; ****P < 0.0001.
PRMT5 inhibition reduces donor Th1 and Th17 responses without adversely affecting engraftment.
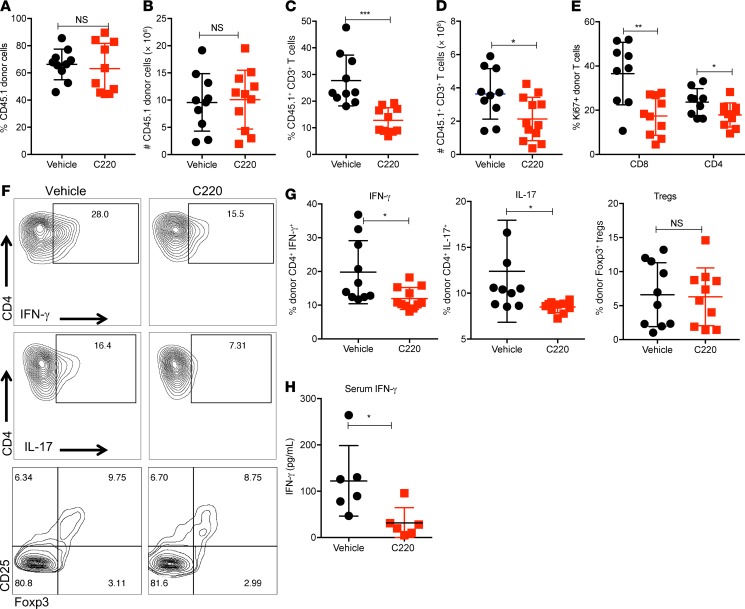
We used the B6 into F1 model to further characterize the impact of PRMT5 inhibition on donor cell engraftment and T cell functions in vivo. Using knockout mouse models, it has previously been shown that while Prmt5 heterozygosity has no effect on hematopoiesis, homozygous loss of Prmt5 leads to bone marrow aplasia and lethal pancytopenia (20). Donor stem cell engraftment is critical to the success of an allogeneic HCT (allo-HCT); therefore, we wanted to evaluate the effect of PRMT5 inhibitor C220 on donor cell engraftment. There was no significant difference in total donor cell chimerism, as evidenced by percentage of CD45.1+ BoyJ donor cells and absolute numbers between vehicle- and C220-treated cohorts, showing PRMT5 inhibition with C220 did not affect donor engraftment (Figure 4, A and B). Concordantly, we did not observe any adverse effects on hematopoiesis in mice that received PRMT5 inhibitor C220 compared with vehicle (Supplemental Figure 5A), confirming that it is possible to pharmacologically inhibit PRMT5 without adversely affecting hematopoiesis. Mice treated with PRMT5 inhibitor had similar absolute numbers of neutrophils (Supplemental Figure 5B), total WBCs, and platelets in circulation (Supplemental Figure 5, C and D) compared with the vehicle-treated counterparts. Interestingly, there were both lower percentage and absolute numbers of CD45.1+CD3+ T cells in mice receiving C220 (Figure 4, C and D). Supporting our in vitro data, we observed fewer Ki-67+ proliferating donor T cells in the spleens of mice receiving C220 compared with vehicle (Figure 4E). We also observed significant reduction in CD4+IFN-γ+ (Th1) as well as a reduction in CD4+IL-17+ donor Th17 cells in mice that received PRMT5 inhibitor compared with vehicle. Importantly, there was no reduction in the forkhead box P3–positive (Foxp3+) Treg compartment in C220-treated mice compared with vehicle, confirming the Treg-sparing effect (22) of PRMT5 inhibition (Figure 4, F and G). Levels of serum IFN-γ were also lower in animals treated with C220 as compared with vehicle (Figure 4H). TNF-α and IL-17 serum levels were undetectable by ELISA. Therefore comparisons could not be made. Altogether, our data suggest that pharmacological inhibition of PRMT5 reduces donor Th1 and Th17 but not Tregs in allotransplanted recipient mice, improving survival while not adversely affecting engraftment.
Figure 4. PRMT5 inhibition reduces donor Th1 and Th17 responses.
B6 into F1 transplant was performed as described in Methods. Mice were sacrificed around day 25 after transplant (n = 5–7 per group) and spleens harvested. (A) Percentage and (B) absolute numbers of CD45.1+ donor cells. (C) Percentage and (D) absolute numbers of CD45.1+CD3+ donor T cells. (E) Percentage of CD45.1+Ki-67+ donor CD4+ and CD8+ T cells. (F) Representative histograms showing intracellular staining for IFN-γ, IL-17, and CD25/Foxp3 on donor CD45.1+CD4+ T cells. (G) Percentage of IFN-γ+, IL-17+, and CD25/Foxp3+ donor CD45.1+CD4+ T cells. (H) Serum IFN-γ analyzed by ELISA. Data shown are combined from 2 independent transplant experiments. Each symbol represents an individual mouse. *P < 0.05; **P < 0.01; ***P < 0.001.
PRMT5 inhibition reduces donor T cell infiltration into GVHD target organs.
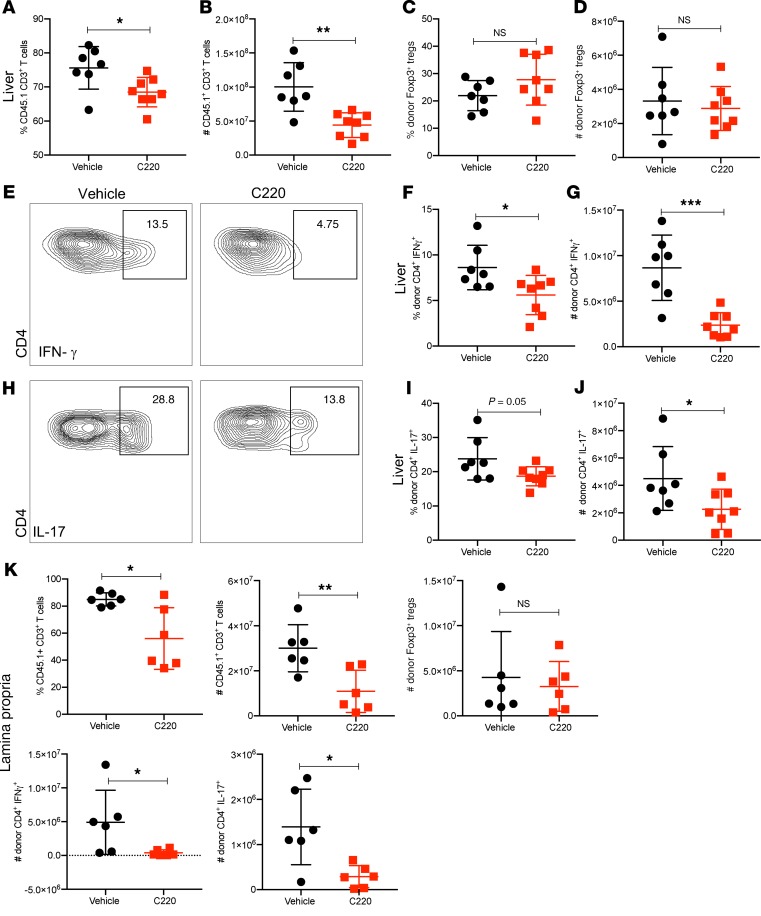
The liver and GI tract represent the major target organs involved in aGVHD pathogenesis, where donor T cell trafficking and cytotoxicity result in organ failure that is ultimately fatal to the recipients of allogeneic hematopoietic stem cell transplants. Therefore, we wanted to evaluate the effects of PRMT5 inhibition on donor T cell infiltration and effector function in the liver and GI tract. Using the B6 into B6D2F1 model, we observed significantly lower percentages and absolute numbers of donor T cells (Figure 5, A and B) in the liver of C220-treated mice compared with vehicle. There was no significant difference in the percentage and numbers of donor Foxp3+ Tregs between mice treated with vehicle and C220 (Figure 5, C and D); however, the percentage and numbers of donor CD4+ T cells secreting IFN-γ (Figure 5, E–G) and IL-17 (Figure 5, H–J) in the liver were significantly lower in mice receiving PRMT5 inhibitor C220 compared with vehicle. Similarly, we observed reduced percentages and absolute numbers of donor T cells as well as reduced numbers of donor CD45.1+CD4+ T cells secreting IFN-γ and IL-17 in the small intestine lamina propria of mice treated with C220 compared with vehicle (Figure 5K).
Figure 5. PRMT5 inhibition reduces donor T cell infiltration into liver and GI tract.
B6 into F1 transplant was performed as described in Methods. Mice were sacrificed around day 25 after transplant (n = 5–7 per group) and liver (A–J) and small intestine of GI tract (K) harvested. Single-cell suspensions were made as described in Methods. (A) Percentage and (B) absolute numbers of CD45.1+CD3+ donor T cells in the liver. (C) Percentage and (D) absolute numbers of Foxp3+ donor CD45.1+CD4+ T cells. (E) Representative histograms showing intracellular staining, (F) percentage, and (G) absolute numbers of donor IFN-γ+CD45.1+CD4+ T cells in the liver. (H) Representative histograms showing intracellular staining, (I) percentage, and (J) absolute numbers of donor IL-17+CD45.1+CD4+ cells in the liver. (K) Percentage and absolute numbers of CD45.1+CD3+ donor T cells and absolute numbers of donor Foxp3+ Tregs, donor IFN-γ+ T cells, and IL-17+CD45.1+CD4+ donor T cells in the lamina propria of the small intestine. One representative experiment of 2 independent transplant experiments is shown. Each symbol represents an individual mouse. *P < 0.05; **P < 0.01; ***P < 0.001.
PRMT5 inhibition maintains graft-versus-leukemia response.
The main goal of an allogeneic transplant is to induce a donor antitumor response to eliminate residual leukemia/lymphoma in the recipient. To investigate whether PRMT5 inhibition retained graft-versus-leukemia (GVL) response, we used a luciferase-transduced murine mastocytoma P815 cell line in the B6 into F1 model. Briefly, lethally irradiated B6D2F1 mice were injected with 10,000–15,000 P815 cells along with B6 TCD-BMs alone or along with allogeneic splenocytes. Recipients of allogeneic splenocytes were treated with vehicle or C220 (2 mg/kg) starting at day 7 after transplant till the end of the study. PRMT5 inhibition retained the beneficial GVL effects, as seen by an improvement in survival (Figure 6A) and decreased luminescence (Figure 6, B and C) compared with vehicle treatment. Cause of death in the recipients of allogeneic splenocytes was confirmed to be aGVHD by histological examination (Figure 6D) and clinical aGVHD scores (Supplemental Figure 6A). Presence/absence of tumor in was confirmed by flow cytometric evaluation of splenic GFP+ P815 cells. In mice that did not receive infusion of allogeneic splenocytes along with the tumor cells, there was a high tumor burden in the spleen (>40% GFP+ P815 cells). In contrast, mice that received allogeneic splenocytes eradicated the tumor with no difference in the percentage of GFP+ P815 cells between mice receiving vehicle or C220, showing retention of beneficial GVL effect (Supplemental Figure 6, B and C). Murine CD8+ T cells were stimulated in vitro with allogeneic BMDCs in the presence of DMSO/C220, and degranulation in response to P815 tumor challenge was analyzed to evaluate the cytotoxic T cell (CTL) capacity of CD8+ T cells. C220-treated CD8+ T cells showed comparable degranulation as measured by CD107a mobilization compared with control (Figure 6, E and F). These results suggest that PRMT5 inhibition does not abrogate CD8+ CTL capacity, an important factor that might contribute to the retention of the GVL effect observed in vivo.
Figure 6. PRMT5 inhibition preserves GVL effect.
Firefly luciferase–transduced P815 cells (10,000 cells) were injected i.v. into lethally irradiated F1 recipients on day 0 along with TCD-BMs and B6 donor splenocytes. Treatment groups included PRMT5 inhibitor C220 or vehicle control. TCD-BMs and P815 cells (leukemia alone) served as the control group. (A) Kaplan-Meier survival curve. Log-rank test was used to compare survival. Data pooled from 3 independent transplant experiments. n = 22 for TCD-BM + P815 alone group; n = 17–19 for TCD-BM + P815 + B6 splenocytes groups. (B and C) Whole-body bioluminescent signal intensity of recipient mice (n = 5 per cohort). Mice were imaged on indicated days. Average radiance expressed as mean ± SD. One representative experiment of 2 is shown. (D) H&E-stained liver sections at original magnification ×200 and ×400 of 2 representative recipients showing leukemic infiltration in the liver in the TCD-BM + P815 only group. There were no leukemic cells in either of the groups that received B6 allogeneic splenocytes. There was lymphocytic infiltration in mice receiving B6 allogeneic splenocytes (triangles). (E) Murine CD45.1 BoyJ CD8+ T cells were stimulated with allogeneic BALB/c BMDCs ± C220 (100 nM) for 5 days. CTL capacity against P815 tumor cells was assessed by flow cytometric evaluation of intracellular CD107a expression. Contour plots of 1 representative donor. (F) Data expressed as mean ± SD of biological duplicates of 3 independent experiments. *P < 0.05; ****P < 0.0001.
PRMT5 inhibition deregulates cell cycle and ERK signaling in activated T cells.
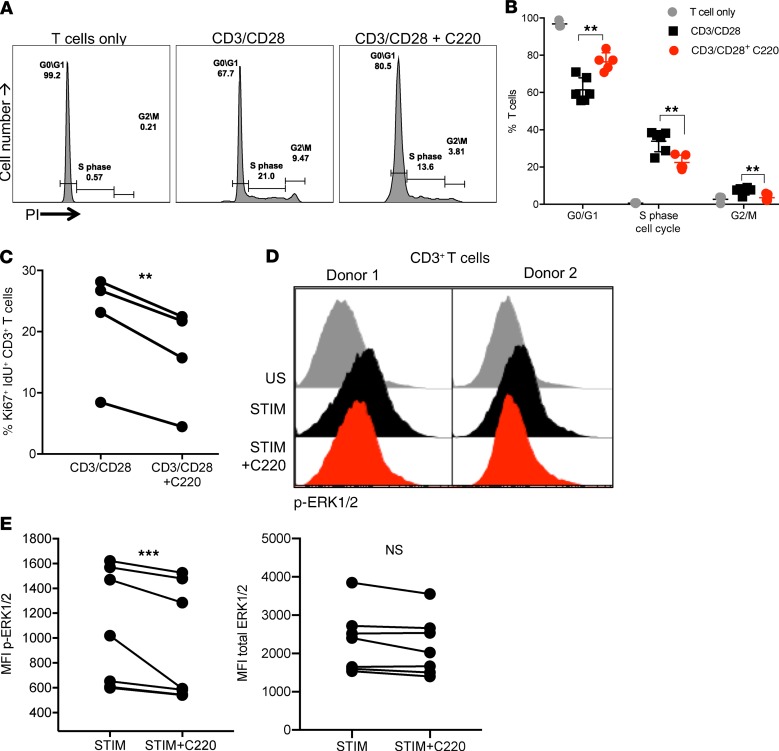
PRMT5 has been shown to promote cell proliferation and play a critical role in G1-to-S cell cycle transition in neoplastic hepatocellular (31) and breast cancer cells (32). Additionally, PRMT5 inhibition in lymphoma leads to transcriptional repression of cyclin D1 (25). Therefore, we hypothesized that deregulation of cell cycle might be a mechanism by which PRMT5 modulates T cell proliferation. To investigate this, murine T cells were stimulated with CD3/CD28 beads in the presence or absence of C220. Inhibition of PRMT5 by C220 resulted in T cell cycle deregulation with a decrease in S phase T cells and a concomitant increase at G0/G1 phase (Figure 7, A and B), demonstrating that PRMT5 regulates G1-to-S transition in activated T cells. CD3/CD28-stimulated human T cells treated with PRMT5 inhibitor C220 also showed a similar decrease in Ki-67 and idoxuridine (IdU) double-positive S phase T cells (Figure 7C). ERK1/2 phosphorylation is affected by PRMT5 in BM cells (20), and ERK1/2 is important for G1-to-S transitions (33, 34). Therefore, we wanted to investigate whether ERK1/2 phosphorylation was affected by PRMT5 in T cells. Here, we show that PRMT5 inhibition resulted in significant downregulation of ERK1/2 phosphorylation (Figure 7, D and E) in T cells that could contribute to cell cycle deregulation.
Figure 7. Inhibition PRMT5 deregulates cell cycle and ERK signaling in mouse and human T cells.
(A) Murine CD3+ T cells stimulated with CD3/CD28 Dynabeads ± C220 for 48 hours. Cell cycle analysis based on PI incorporation. Representative propidium iodide (PI) histogram of 1 experiment. (B) Percentage of T cells in G0/G1, S, and G2/M phases. Mean ± SD of biological triplicates of 2 independent experiments. Significance was determined using 2-tailed t test with Holm-Šídák correction for multiple comparisons. (C) Healthy donor T cells were stimulated with CD3/CD28 beads for 48 hours ± C220 (100 nM). Cells were labeled with IdU for 15 minutes at the end before collection for analysis by mass cytometry. Percentage of Ki-67+Idu+ T cells of 4 individual donors. (D) PBMCs were pretreated with DMSO or C220 (100 nM) for 2 hours and stimulated with PMA/ionomycin for 5 minutes. Phosphorylation of ERK1/2 on CD3+ T cells analyzed by flow cytometry. Histogram of 2 representative donors. (E) MFI of phospho-ERK1/2 and total ERK1/2 in 7 donors. **P < 0.01; ***P < 0.001.
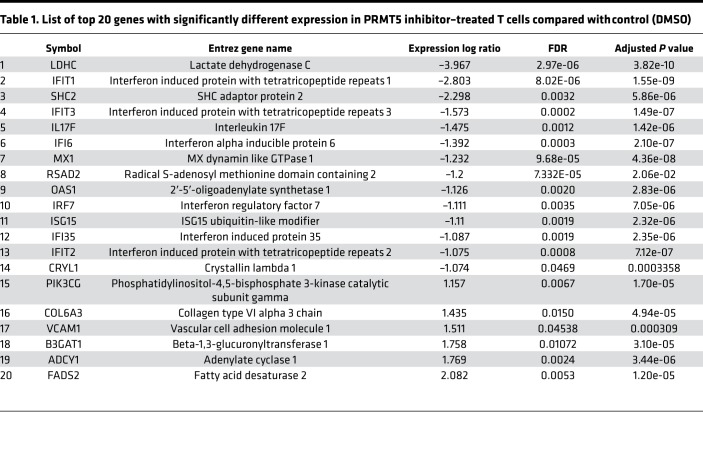
PRMT5 regulates interferon response in T cells.
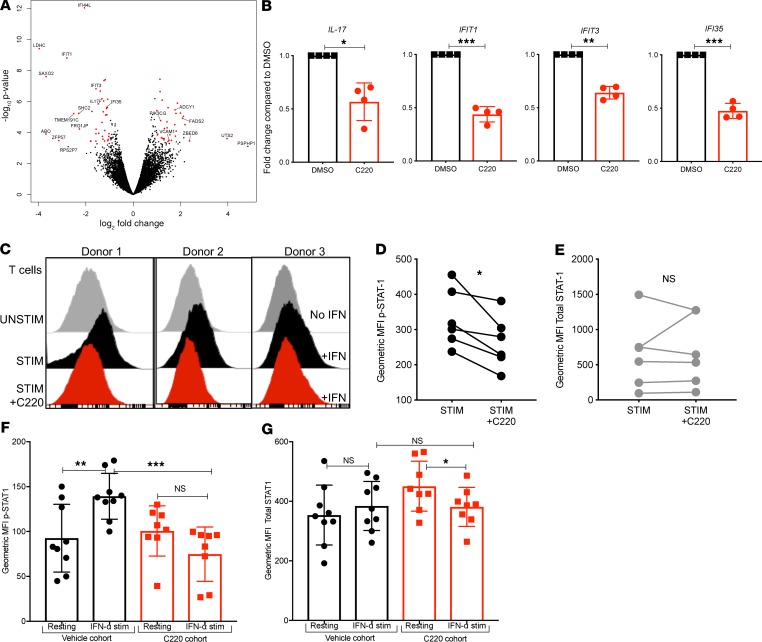
To further elucidate the molecular mechanisms affected in T cells by PRMT5 inhibition, we compared gene expression profiles of CD3/CD28-activated human T cells in the presence or absence of the PRMT5 inhibitor C220. Interestingly, we observed a very strong reduction in expression of interferon-stimulated genes (ISGs) in T cells subjected to PRMT5 inhibition compared with control (DMSO). Additionally, we also observed a strong reduction in IL-17 transcription in PRMT5-inhibited T cells compared with control (Figure 8A). The top genes affected (using Ingenuity Pathway Analysis [IPA] canonical pathway analysis) by PRMT5 inhibition are listed in Table 1. We also validated downregulation of ISGs IFIT1, IFIT3, and IFI35, as well as inflammatory cytokine IL-17, in 4 additional donors by real-time quantitative PCR (qPCR) (Figure 8B). Our results show that PRMT5 inhibition resulted in both decreased IFN-γ secretion (in vitro and in vivo) as well as reduced ISG transcription. It is well known that STAT1 mediates the biological activity of both type I (IFN-α and IFN-β) and type II interferons (IFN-γ) that ultimately results in the transcription of ISGs (35, 36). PRMT5 was originally identified as a Jak2 binding protein (37), and Jak2 is one of the kinases responsible for the phosphorylation of STAT1 in response to interferon cytokine stimulation (38). Therefore, we hypothesized that PRMT5 may regulate STAT1 phosphorylation in response to IFN stimulation. Supporting our hypothesis, we found that PRMT5 inhibition significantly suppressed STAT1 phosphorylation (Figure 8, C and D) without affecting total STAT1 (Figure 8E). We also observed a similar reduction in STAT1 phosphorylation in vivo in mice that received C220 compared with vehicle-treated mice (Figure 8, F and G). Therefore, decreased STAT1 signaling and modulation of interferon response due to PRMT5 inhibition could be a mechanism contributing to reduced pathogenicity of T cells.
Figure 8. PRMT5 regulates interferon response in T cells.
Human T cells were isolated from healthy PBMCs (n = 4) by negative selection. T cells were stimulated CD3/CD28 Dynabeads for 48 hours and treated with vehicle (DMSO) or C220, RNA was isolated, and RNA-Seq was performed. (A) Volcano plot showing the top dysregulated genes. (B) Real-time qPCR validation in 4 additional human donor T cells performed for indicated genes. Fold change compared with DMSO, with gene expression normalized to β-actin. (C) PBMCs were stimulated with CD3/CD28 ± C220 for 48 hours and then rested for 4 hours followed by 15-minute pulse with IFN-α (100 ng/mL). STAT1 phosphorylation on CD3+ T cells was analyzed by flow cytometry. Histogram of 3 representative donors. (D) MFI of p-STAT and (E) MFI total STAT1; n = 6 donors. (F) B6 into F1 transplant was performed, and recipients were treated with C220 or vehicle as described in Methods. Splenocytes were isolated around day 25 after transplant and pulsed with IFN-α (100 ng/mL) for 40 minutes. MFI of p-STAT1 and (G) total STAT1 on CD45.1+CD3+ donor T cells was evaluated by intracellular flow cytometry in both resting and IFN-α–pulsed cells. *P < 0.05; **P < 0.01; ***P < 0.001.
Table 1. List of top 20 genes with significantly different expression in PRMT5 inhibitor–treated T cells compared with control (DMSO).
Discussion
Our results show that PRMT5 mRNA and protein expression as well as enzymatic activity are upregulated in activated T cells. Disruption of PRMT5 results in marked downregulation of T cell proliferation and function in response to allogeneic stimuli both in vitro and in vivo. We demonstrate that PRMT5 inhibition modulated multiple components of T cell signaling machinery, including ERK and STAT1 phosphorylation, culminating in significant downregulation of inflammatory ISGs and IL-17 (Figure 8). PRMT5 inhibition by oral administration of a PRMT5 small-molecule inhibitor significantly improved survival and reduced clinical disease severity in mouse models of aGVHD (Figure 3). Importantly, we show that pharmacological targeting of PRMT5 with a selective small-molecule inhibitor does not adversely affect donor stem cell engraftment and neutrophil recovery after transplant. Neutrophil recovery after allo-HCT is an important parameter that dictates posttransplant outcomes because prolonged periods of neutropenia leave the patient vulnerable to life-threatening infections (39). Additionally, we show that PRMT5 inhibition preserves Foxp3+ Tregs after transplant. Tregs play a central role in the establishment and maintenance of tolerance after allo-HCT (40–42). Therefore, any therapies that can selectively maintain Treg presence and activity while eliminating Th1/Th17 inflammatory alloresponses will provide a superior outcome compared with existing therapies.
The main goal of an allogeneic transplant is to induce a donor antileukemia response to eliminate residual leukemia/lymphoma in the recipient (43). However, because aGVHD and GVL are 2 tightly linked processes, any therapeutic strategy targeting aGVHD must not abrogate the GVL response of the donor allograft. To that end, maintenance of CTL function is critical for the beneficial antineoplastic immune response of the donor allograft (40, 44). Using the P815 GVL model, we show that PRMT5 inhibition retains CD8+ CTL capacity, which might explain why PRMT5 inhibition still preserves the beneficial GVL effect. Further studies are required to demonstrate whether CD4+ and CD8+ T cells have different responses to PRMT5 inhibition. Our studies show that PRMT5 inhibition is a viable posttransplant strategy that can alleviate aGVHD by reducing Th1/Th17 responses and maintaining Treg-mediated tolerance while retaining GVL effect because of maintenance of donor CD8+ CTL capacity.
One of the main steps in aGVHD induction and propagation is the proliferation of donor T cells triggered by DC antigen presentation, the appropriate costimulatory signals, and cytokine environment. This massive expansion begins with T cells entering into and rapidly progressing through the cell cycle (2, 3), and antiproliferative regimens, such as methotrexate, cyclophosphamide, or mycophenolate, are routine prophylactic regimens for aGVHD in the clinic (45). Here, we show that inhibition of PRMT5 significantly reduces T cell proliferation with a concomitant decrease in S phase T cells (Figure 7). The relation between PRMT5 and ERK signaling is complex. PRMT5 has been shown to negatively regulate MAPK signaling in cancer cell lines (33); however, using the inducible Prmt5-KO mouse, researchers have shown that loss of PRMT5 decreases ERK1/2 phosphorylation (20). We extend these findings to T cells and show that inhibition of PRMT5 with a selective small-molecule PRMT5 inhibitor results in decreased ERK1/2 phosphorylation. Targeting the RAS/MEK/ERK pathway (46) using an MEK inhibitor, such as trametinib, has been shown to inhibit alloreactivity and suppress aGVHD by decreasing ERK1/2 phosphorylation and expansion of donor T cells and sparing Tregs (47). Therefore, downregulation of the ERK pathway could be a major mechanism by which inhibition of PRMT5 exerts its protective effects in aGVHD.
The role of cytokines, such as the interferons (type I and II) as well as IL-17, in aGVHD pathogenesis is intricate and depends on a number of factors, including the cellular source (donor or recipient), conditioning regimen (myeloablative vs. reduced intensity), as well as timing of cytokine production (pretransplant, early or late posttransplant) (48–51). Although IFN-γ has been shown to promote aGVHD pathogenesis, it is also important for GVL (48, 52–54). Complete loss of IFN-γ, paradoxically, caused more severe GVHD, potentially because of the loss of antiinflammatory effects of donor-derived IFN-γ and IFN-γ receptor signaling (6, 8, 55, 56). Type I IFNs have been shown to reduce (57, 58) or promote (59) GVHD while playing an important role in contributing to donor-derived GVL effects. Similarly, IL-17–producing Th17 cells have been shown to promote aGVHD; however, absence of IL-17 has been shown to promote aGVHD by augmenting Th1 responses (6, 7, 10, 51, 60, 61). Despite the heterogeneous nature of the contribution of interferons to aGVHD disease progression, there is consensus that inhibition of STAT1, the mediator of biological activity of interferons, can ameliorate aGVHD (59, 62–64). Our results show that inhibition of PRMT5 potently reduces IFN-γ and IL-17 production by activated alloreactive T cells, accompanied by reduced STAT1 phosphorylation, culminating in a reduction in transcription of ISGs. The precise mechanisms by which PRMT5 modulates STAT1 phosphorylation, i.e., whether by modulation of JAK or tyrosine-protein phosphatase activity, is a subject of ongoing investigation in our lab.
We demonstrate that PRMT5 inhibition causes substantial downregulation of inflammatory gene transcription, including ISGs and IL-17. Previous studies have shown that PRMT5-mediated histone tail modifications, particularly symmetric dimethylation of histone arginine residues (H2A, H4R3, H3R2, H3R8), are associated with transcriptional repression (15, 65, 66). PRMT5 is also known to regulate transcription by conferring posttranslational modifications on transcription factors NF-κB (67), p53 (68, 69), and E2F-1 (70). In addition, PRMT5 symmetrically dimethylates multiple members of the spliceosome machinery, such as Sm proteins D1, D3, and B/B′ (21, 71) that regulate posttranscriptional control of mRNA expression. Therefore, determining the exact molecular mechanism underlying the reduction in gene expression — i.e., whether it is due to global gene repression via histone modification or more direct due to modulation of IFN/STAT signaling or due to defects in splicing — requires further investigation. These investigations have the potential to reveal crosstalk networks between epigenetic modulators, such as PRMT5, and signaling pathways influenced by PRMT5 inhibition (such as ERK/STAT1), thereby opening the door to synergistic interactions of future drug combinations.
Epigenetic modifiers are appealing targets for aGVHD not only because of the role epigenetics has in immune regulation but also because many epigenetic modifying agents are used as treatments against hematopoietic neoplasms (72, 73). A variety of epigenetic modifiers are currently being evaluated as potential aGVHD therapeutics in phase I and phase II clinical trials, primarily as combinatorial therapies with other well-established aGVHD treatments (74, 75). DNA methyltransferase inhibitors, such as 5-azacitidine and 5-aza-2′-deoxycytidine (decitabine), have shown great promise in ameliorating GVHD in both preclinical and clinical studies by increasing Treg production and impairing effector T cell proliferation (76–81). Histone deacetylase inhibitors suberoylanilide hydroxamic acid 22 (vorinostat), ITF2357 (givinostat), and LBH589 (panobinostat) have also been very successful in modulating aGVHD because of their multimodal roles in reducing inflammatory cytokine secretion, boosting regulatory T cell production, and suppressing alloreactive antigen-presenting cell activation (64, 82–85).
We and others have shown that targeting PRMT5 has therapeutic benefit in preclinical models of lymphoma and AML (13, 23–27). Here, we propose inhibition of PRMT5 in a posttransplant setting as a “2-for-1 approach” that can inhibit inflammatory T cell responses and maintain tolerance, thereby preventing GVHD and retaining GVL effects while simultaneously targeting residual leukemic cells. Our studies have provided mechanistic insight into the role of PRMT5 in T cell function as well as in regulating aGVHD pathogenesis and reveal PRMT5 as a therapeutic target for aGVHD.
Methods
Mice.
C57BL/6 (B6, H2b), B6.SJL-Ptprca Pepc/BoyJ (CD45.1 B6), B6D2F1 (F1, H2b/d), and BALB/c (H2d) mice were purchased from Jackson ImmunoResearch Laboratories. Female NSG mice were acquired from the NSG mouse colony maintained by the Target Validation Shared Resource (TVSR) at The Ohio State University; breeders (strain 005557) for the colony were received from The Jackson Laboratory. All mice were bred and maintained in an Ohio State University animal care facility. NSG mice were housed in a specific pathogen–free facility in microisolator cages. For transplant experiments, recipient mice were between 12 and 16 weeks of age; for all other experiments, mice were between 8 and 10 weeks of age.
aGVHD murine models.
Only age- and sex-matched mice were used for transplant experiments. Briefly, B6D2F1 mice were irradiated with 1200 cGy administered in 2 fractions (to minimize toxicity) 1 day before transplant. TCD-BMs (10 × 106) plus 15 × 106 total splenocytes from CD45.1 B6 donors were administered via tail vein injection on the day of transplant. T cell depletion from bone marrow cells was carried out by CD90 magnetic bead separation (Miltenyi Biotec). Recipients of allogeneic splenocytes were treated with vehicle or PRMT5 inhibitor C220 2 mg/kg, administered by oral gavage once a week starting day +7 after transplant until the end of the study.
Mouse model of xenogeneic GVHD.
A xenogeneic GVHD model was used based on previous studies (42). One day before transplant, mice were irradiated with 50 cGy. On the day of transplant, mice were injected with 10 × 106 TCD-PBMCs or 15 × 106 to 20 × 106 human PBMCs. Mice receiving T cell–replete grafts were treated with C220 (2 mg/kg) or vehicle administered by oral gavage, 3 times weekly starting day +7 after transplant until the end of the study.
Clinical and histological assessment of aGVHD.
Recipient mice were weighed 2–4 times a week and monitored daily for clinical signs of aGVHD and survival. GVHD scores were performed using a system modified from Cooke et al. (86). Briefly, this scoring system incorporates 5 clinical parameters: weight loss, posture (hunching), activity, fur texture, and skin integrity. Individual mice were ear tagged and graded (on a scale from 0 to 8) twice a week. Mice who reached an aGVHD score of more than or equal to 7 were very sick and were euthanized and their tissues harvested (86, 87). GVHD was also assessed by detailed histopathology analysis of H&E-stained liver and gut tissues using a previously reported scoring system with a range of 0 (absence of signs of GVHD) to 4 (maximal GVHD damage) (88). A separate cohort of mice were euthanized around day 25 (±3 days) after transplant and used for histopathological assessment of target tissues.
PRMT5 inhibitor C220.
PRMT5 inhibitor C220 was synthesized by WuXi Apptech. It was administered in a vehicle consisting of 0.5% methylcellulose, 0.1% Tween-80, and 99.4% sterile water. Mice were given C220 (2 mg/kg) by oral gavage once a week starting day +7 after transplant till the end of the study. For the NSG experiment alone, mice were dosed 3 times weekly. Mice in the control group were treated with the same volume of vehicle as the C220 group.
Cells and cell culture.
Mouse B6 or CD45.1 B6 T cells were isolated from splenocytes using Pan-T Cell Isolation Kit or naive CD8+ T cell isolation kit (Miltenyi Biotec) per the manufacturer’s protocol. All cells were cultured in RPMI 1640, 20% FBS, and 1% pen-strep unless otherwise specified. Healthy donor-derived PBMCs were isolated by Ficoll-Paque PLUS density gradient centrifugation (GE Healthcare). T cells were isolated from PBMCs using Pan-T Cell Isolation Kit (Miltenyi Biotec).
Western blots.
T cells were lysed in RIPA buffer, and Western blot was performed according to standard protocols. For time course experiments, T cells were isolated from mouse spleen or human T cells were isolated from healthy donor leukopaks and stimulated using CD3/CD28 Dynabeads according to the manufacturer’s protocol (Invitrogen, Thermo Fisher Scientific). Cells were lysed in RIPA buffer and immunoblotted using primary Abs against PRMT5 (Abcam), H3R8me2s, and H3R8me2a (EpiGentek polyclonal antibody, catalog A-3716-100). (See Supplemental Table 1 for a complete list of antibodies.)
In vitro T cell proliferation.
CD45.1 B6 T cells and human T cells were labeled with CTV (Invitrogen, Thermo Fisher Scientific) and incubated with allogeneic BALB/c BMDCs or TCD-PBMCs (for human MLR). Cell division was measured by CTV dilution after 4–5 days using LSRII and FACSDiva software (Becton Dickinson).
Cytokine ELISA.
Mouse and human T cells were stimulated as described above. Supernatant cytokines were analyzed by ELISA according to the manufacturer’s protocol (BioLegend). Results are shown as mean ± SD of biological duplicates of 3 donors. Serum was collected by cardiac puncture of mice that were euthanized on day 21 from transplant experiments for cytokine and flow cytometry experiments; cytokines were measured by ELISA (BioLegend).
In vitro PRMT5 methyltransferase assay.
Methylation activity in nuclear extracts derived from indicated human T cells was measured using Epigenase PRMT5 Methyltransferase (Type II-Specific) Activity/Inhibition Assay Kit (EpiGentek) according to the manufacturer’s protocol. Standard curves were generated using Methylated H4-Arg3 standard provided by the manufacturer, with a linear detection range between 0.1 and 2 ng of methylated product. Reactions contained 20 μg of the indicated nuclear extracts (prepared with Nuclear Extraction Kit, OP-0002 from EpiGentek) and were incubated for 120 minutes at 37°C. Absorbance (450 nm with a reference wavelength of 655 nm) was measured with a Tecan Infinite M1000Pro microplate reader (Thermo Fisher Scientific). Each experiment included at least 3 independent replicate measurements per sample, and P values for each sample comparison were calculated using a 2-tailed unpaired t test.
RNA-Seq.
Human T cells were isolated from healthy donor PBMCs (n = 4 donors) by negative selection. T cells were stimulated with CD3/CD28 Dynabeads for 48 hours and treated with either vehicle (DMSO) or C220 (100 nM). RNA was isolated using TRIzol reagent (Invitrogen, Thermo Fisher Scientific) and treated with DNase (QIAGEN Inc.). RNA quality was verified using the Agilent 2100 Bioanalyzer, and the RNA integrity number values were greater than 7 for all samples. Sequencing libraries were generated with poly(A)+ RNA using the TruSeq RNA sample prep kit (Illumina). Libraries underwent paired-end 50-bp sequencing using the Illumina HiSeq2500 sequencer to a depth of 17 million to 20 million passed filter clusters per sample. Demultiplexed and quality-filtered reads were mapped to the human genome GRCh38 using Hierarchical Indexing for Spliced Alignment of Transcripts (89). Raw read counts for each gene were quantified using featureCounts software, with GENCODE v.27 transcript reference (GENCODE annotation) (90). RNAs with less than 20 read counts on average across all samples were excluded from further analysis. Differential gene expression analysis between PRMT5- and DMSO-treated cells was performed using R package edgeR (91). The read counts were normalized using TMM method (92). Differentially expressed genes were selected based on adjusted P value and log2 fold change. Biological pathways and interactome analysis were performed using IPA software (QIAGEN Inc., ref. 93). The data supporting the results of this article are available in the National Center for Biotechnology Information’s Gene Expression Omnibus repository (accession ID GSE145527).
Real-time PCR.
RNA was isolated using TRIzol reagent (Invitrogen, Thermo Fisher Scientific). cDNA was prepared using the SuperScript III cDNA synthesis kit (Thermo Fisher Scientific) and detected by commercially available TaqMan probes for specific genes (Applied Biosystems, Thermo Fisher Scientific) on the 7900HT Fast Real-Time PCR System (Applied Biosystems, Thermo Fisher Scientific). Gene expression was normalized to housekeeping gene β-actin.
Flow cytometry analysis.
Around day 25, cohorts of mice were euthanized, and splenocytes, whole liver, and small intestine were harvested for flow cytometric analysis. Hepatic tissue and lamina propria were digested into a single-cell suspension using a commercial mouse Liver Dissociation Kit (Miltenyi Biotec) and mouse Lamina Propria Tissue Dissociation Kit (Miltenyi Biotec). To select only the donor T cells, a specific gating strategy was used (Supplemental Figure 7). A complete list of Abs used is in Supplemental Table 1. For cytokine evaluation, splenocytes were incubated for 5 hours with eBioscience Cell Stimulation Cocktail (plus protein transport inhibitors, Thermo Fisher Scientific) for T cell stimulation and protein transport inhibition. Cells were then stained with surface Abs, permeabilized, fixed, stained with intracellular Abs, and analyzed within 24 hours. Analysis was performed with a FACS LSRII cytometer with FACSDiva software (Becton Dickinson); data analysis was performed using FlowJo (Tree Star).
IdU incorporation and mass cytometry analysis.
Following healthy donor T cell stimulation as described above, cells were incubated with IdU (MilliporeSigma) at a final concentration of 10 μM for 15 minutes at 37°C. Following IdU incubation cells were fixed using SmartTube Proteomic Stabilizer (SmartTube Inc.) at a 1:1.4 ratio for 10 minutes at room temperature before being stored in a –80°C freezer before staining. Mass cytometry staining and measurement were performed as previously described (94). Briefly surface Ab staining was performed in 100 μL of CSM (1× PBS with 0.5% bovine serum albumin and 0.02% sodium azide) for 50 minutes at room temperature with continuous shaking. Cells were washed twice with CSM, and surface Abs were fixed using 1.5% paraformaldehyde solution (Electron Microscopy Services). Cells were pelleted following surface Ab fixation and permeabilized while vortexing with ice-cold methanol. Cells were washed with PBS and CSM twice before intracellular staining. Intracellular staining was performed in 100 μL of CSM for 50 minutes at room temperature with continuous shaking. Cells were washed twice with CSM and then incubated overnight at 4°C in PBS with 500 nM iridium intercalator pentamethylcyclopentadienyl-Ir(III)-dipyridophenazine (Fluidigm) and 1.6% paraformaldehyde. Excess intercalator was removed with 2 CSM washes and a single wash in PBS. Cells were resuspended in deionized water at approximately 1 million cells/mL. Cell events were acquired on the Helios Mass Cytometer (Fluidigm), and FCS file analysis was performed using the Cytobank data analysis platform. A singlet gate was drawn in Cytobank using cell length by DNA (Ir intercalator) to remove doublets and debris from downstream analysis (Supplemental Figure 8). Cell cycle phases were gated according to previous methods (95, 96). For cell cycle analysis, S phase cells were gated on an IdU and Ki-67 double-positive population.
GVL experiments.
Firefly luciferase–transduced P815 mastocytoma (97, 98) cells (10,000 total) were injected intravenously into F1 recipients on day 0 along with TCD-BM (10 × 106 cells) and B6 donor splenocytes (20 × 106 cells). Treatment groups included vehicle and PRMT5 inhibitor C220 2 mg/kg, administered by oral gavage once a week starting day +7 after transplant. TCD-BMs and P815 cells (leukemia alone) served as the control group. P815-induced leukemic death was defined by the occurrence of either macroscopic tumor nodules in liver and/or spleen or hind limb paralysis. GVHD death was defined by the absence of leukemia and the presence of clinical and histopathological signs of GVHD.
In vivo imaging.
Xenogen IVIS imaging system (Caliper Life Sciences) was used for live animal imaging. Mice were anesthetized using 1.5% isoflurane. XenoLight RediJect D-Luciferin Ultra Bioluminescent Substrate (150 mg/kg body weight; 30 mg/mL in PBS; PerkinElmer) was injected intraperitoneally, and IVIS imaging was performed 10 minutes after substrate injection. Whole-body bioluminescent signal intensity was determined weekly using IVIS Living Image software v4.3.1 (Caliper Life Sciences), and pseudocolor images overlaid on conventional photographs are shown. Data were analyzed and presented as photon counts per area.
Degranulation assay.
CD8 degranulation assay was measured by CD107a mobilization, and experiments were set up based on previously published protocols (40, 99). P815 cells were incubated at 37°C overnight. On the next day, the culture supernatant was removed from the wells, and effector cells were added to the wells. Naive CD8+ T cells isolated from BoyJ CD45.1 splenocytes by negative selection (STEMCELL Technologies) were used as effector T cells in the following setup: (a) CD8+ T cells without any treatment (CD8-resting), (b) CD8+ T cells stimulated with allogeneic BMDCs in the presence of DMSO, or (c) C220 (100 nM) for 5 days. CD8+ T cells were then mixed with P815 cells at effector/target ratio 5:1, centrifuged at 300 g for 3 minutes, and cocultured in the presence of anti-CD107a at 37°C overnight. On the next day, the cells were stained with surface markers CD45.1 and CD8 and analyzed by flow cytometry.
Statistics.
Survival data were analyzed using Kaplan-Meier and log-rank test methods. Differences between continuous variables at a single time point were analyzed using 2-tailed t tests. Data represent mean ± SD. One-way ANOVA with Holm-Šídák post hoc test was used for comparisons of more than 2 groups unless indicated otherwise. Differences between 2 groups over time were analyzed with Mann-Whitney U. All analyses were performed using GraphPad Prism 7.0. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. A P value less than 0.05 was considered significant.
Study approval.
All animal studies were conducted in accordance with the rules and regulations of the Institutional Animal Care and Use Committee at The Ohio State University. Peripheral blood samples from healthy donors and allogeneic transplant patients were obtained following receipt of written informed consent in accordance with the Declaration of Helsinki. Healthy PBMCs were obtained from Versiti, and transplant patient samples were selected from a biorepository study, both approved by the IRB at The Ohio State University.
Author contributions
KJS, NCZ, YG, NES, LNC, and AI performed in vivo murine aGVHD experiments, ex vivo FACS analyses, serum/supernatant ELISAs, Western blots, real-time PCR analyses, in vitro experiments with mouse cells and human PBMCs, and signaling experiments; analyzed the data; and interpreted the results. CK, YG, NES, and RDD performed mass cytometry experiments. GKB was involved in supervision and discussion of results of the mass cytometry experiments. MGA and JEJ performed the in vitro methylation experiments. MP provided the bioinformatics data. MW and YWZ performed the C220 biochemical assay. HL synthesized C220. HKC discussed experimental design, provided data, and edited the manuscript. RAB provided data discussion and edited the manuscript. RG and KV edited the manuscript. PR designed the study, supervised research, interpreted the data, and wrote the manuscript along with NCZ.
Supplementary Material
Acknowledgments
This work was supported by a New Investigator Award from the Leukemia Research Foundation, Pelotonia New Investigator Award, K12 Paul Calabresi Award, and Division of Hematology start-up funds (to PR). Research reported in this publication was supported by the Ohio State University Comprehensive Cancer Center and the National Institutes of Health under grant numbers P30 CA016058 and K12 CA133250. We thank the TVSR at the Ohio State University Comprehensive Cancer Center for providing the NSG mice used in the preclinical studies described herein. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Version 1. 03/19/2020
In-Press Preview
Version 2. 04/23/2020
Electronic publication
Version 3. 04/25/2020
Added brief adstract.
Funding Statement
Paul Calabresi Scholar
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(8):e131099.https://doi.org/10.1172/jci.insight.131099.
Contributor Information
Nina C. Zitzer, Email: zitzer@wisc.edu.
Yandi Gao, Email: yandi.gao@osumc.edu.
Hannah K. Choe, Email: hannah.choe@gmail.com.
Natalie E. Sell, Email: sell.73@osu.edu.
Lotus Neidemire-Colley, Email: neidemire-colley.1@osu.edu.
Anora Ignaci, Email: ignaci.2@osu.edu.
Charuta Kale, Email: kale.28@osu.edu.
Raymond D. Devine, Email: devine.82@osu.edu.
Maria G. Abad, Email: abad-de-partida.1@osu.edu.
Maciej Pietrzak, Email: maciej.pietrzak@osumc.edu.
Min Wang, Email: mwang@preludetx.com.
Hong Lin, Email: hlin@preludetx.com.
Yang W. Zhang, Email: yzhang@preludetx.com.
Gregory K. Behbehani, Email: gregory.behbehani@osumc.edu.
Jane E. Jackman, Email: jackman.14@osu.edu.
Ramiro Garzon, Email: ramiro.garzon@osumc.edu.
Kris Vaddi, Email: kv@preludetx.com.
Robert A. Baiocchi, Email: robert.baiocchi@osumc.edu.
Parvathi Ranganathan, Email: parvathi.ranganathan@osumc.edu.
References
- 1.Ferrara JL, Levine JE, Reddy P, Holler E. Graft-versus-host disease. Lancet. 2009;373(9674):1550–1561. doi: 10.1016/S0140-6736(09)60237-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perkey E, Maillard I. New insights into graft-versus-host disease and graft rejection. Annu Rev Pathol. 2018;13:219–245. doi: 10.1146/annurev-pathol-020117-043720. [DOI] [PubMed] [Google Scholar]
- 3.Zeiser R, Blazar BR. Acute graft-versus-host disease - biologic process, prevention, and therapy. N Engl J Med. 2017;377(22):2167–2179. doi: 10.1056/NEJMra1609337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu J, et al. T-bet is critical for the development of acute graft-versus-host disease through controlling T cell differentiation and function. J Immunol. 2015;194(1):388–397. doi: 10.4049/jimmunol.1401618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fu J, et al. T-bet promotes acute graft-versus-host disease by regulating recipient hematopoietic cells in mice. J Immunol. 2016;196(7):3168–3179. doi: 10.4049/jimmunol.1501020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yi T, et al. Reciprocal differentiation and tissue-specific pathogenesis of Th1, Th2, and Th17 cells in graft-versus-host disease. Blood. 2009;114(14):3101–3112. doi: 10.1182/blood-2009-05-219402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlson MJ, West ML, Coghill JM, Panoskaltsis-Mortari A, Blazar BR, Serody JS. In vitro-differentiated TH17 cells mediate lethal acute graft-versus-host disease with severe cutaneous and pulmonary pathologic manifestations. Blood. 2009;113(6):1365–1374. doi: 10.1182/blood-2008-06-162420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi J, et al. IFNγR signaling mediates alloreactive T-cell trafficking and GVHD. Blood. 2012;120(19):4093–4103. doi: 10.1182/blood-2012-01-403196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delens L, et al. In vitro Th17-polarized human CD4(+) T cells exacerbate xenogeneic graft-versus-host disease. Biol Blood Marrow Transplant. 2019;25(2):204–215. doi: 10.1016/j.bbmt.2018.10.007. [DOI] [PubMed] [Google Scholar]
- 10.Yi T, et al. Absence of donor Th17 leads to augmented Th1 differentiation and exacerbated acute graft-versus-host disease. Blood. 2008;112(5):2101–2110. doi: 10.1182/blood-2007-12-126987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009;33(1):1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poulard C, Corbo L, Le Romancer M. Protein arginine methylation/demethylation and cancer. Oncotarget. 2016;7(41):67532–67550. doi: 10.18632/oncotarget.11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Y, Bedford MT. Protein arginine methyltransferases and cancer. Nat Rev Cancer. 2013;13(1):37–50. doi: 10.1038/nrc3409. [DOI] [PubMed] [Google Scholar]
- 14.Blanc RS, Richard S. Arginine methylation: the coming of age. Mol Cell. 2017;65(1):8–24. doi: 10.1016/j.molcel.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 15.Stopa N, Krebs JE, Shechter D. The PRMT5 arginine methyltransferase: many roles in development, cancer and beyond. Cell Mol Life Sci. 2015;72(11):2041–2059. doi: 10.1007/s00018-015-1847-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parry RV, Ward SG. Protein arginine methylation: a new handle on T lymphocytes? Trends Immunol. 2010;31(4):164–169. doi: 10.1016/j.it.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 17.Blanchet F, Cardona A, Letimier FA, Hershfield MS, Acuto O. CD28 costimulatory signal induces protein arginine methylation in T cells. J Exp Med. 2005;202(3):371–377. doi: 10.1084/jem.20050176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geoghegan V, Guo A, Trudgian D, Thomas B, Acuto O. Comprehensive identification of arginine methylation in primary T cells reveals regulatory roles in cell signalling. Nat Commun. 2015;6:6758. doi: 10.1038/ncomms7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richard S, Morel M, Cléroux P. Arginine methylation regulates IL-2 gene expression: a role for protein arginine methyltransferase 5 (PRMT5) Biochem J. 2005;388(pt 1):379–386. doi: 10.1042/BJ20040373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu F, et al. Arginine methyltransferase PRMT5 is essential for sustaining normal adult hematopoiesis. J Clin Invest. 2015;125(9):3532–3544. doi: 10.1172/JCI81749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inoue M, et al. Arginine methylation controls the strength of γc-family cytokine signaling in T cell maintenance. Nat Immunol. 2018;19(11):1265–1276. doi: 10.1038/s41590-018-0222-z. [DOI] [PubMed] [Google Scholar]
- 22.Webb LM, et al. PRMT5-selective inhibitors suppress inflammatory T cell responses and experimental autoimmune encephalomyelitis. J Immunol. 2017;198(4):1439–1451. doi: 10.4049/jimmunol.1601702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gullà A, et al. Protein arginine methyltransferase 5 has prognostic relevance and is a druggable target in multiple myeloma. Leukemia. 2018;32(4):996–1002. doi: 10.1038/leu.2017.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alinari L, et al. Selective inhibition of protein arginine methyltransferase 5 blocks initiation and maintenance of B-cell transformation. Blood. 2015;125(16):2530–2543. doi: 10.1182/blood-2014-12-619783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chung J, et al. Protein arginine methyltransferase 5 (PRMT5) inhibition induces lymphoma cell death through reactivation of the retinoblastoma tumor suppressor pathway and polycomb repressor complex 2 (PRC2) silencing. J Biol Chem. 2013;288(49):35534–35547. doi: 10.1074/jbc.M113.510669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tarighat SS, et al. The dual epigenetic role of PRMT5 in acute myeloid leukemia: gene activation and repression via histone arginine methylation. Leukemia. 2016;30(4):789–799. doi: 10.1038/leu.2015.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nicholas C, et al. PRMT5 is upregulated in malignant and metastatic melanoma and regulates expression of MITF and p27(Kip1.) PLoS One. 2013;8(9):e74710. doi: 10.1371/journal.pone.0074710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol. 2004;24(21):9630–9645. doi: 10.1128/MCB.24.21.9630-9645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. D’Souza A, Fretham C. Current Uses and Outcomes of Hematopoietic Cell Transplantation (HCT): CIBMTR Summary Slides, 2017. https://www.cibmtr.org/ReferenceCenter/SlidesReports/SummarySlides/pages/index.aspx 2017. Accessed April 3, 2020.
- 30.Bonday ZQ, et al. LLY-283, a potent and selective inhibitor of arginine methyltransferase 5, PRMT5, with antitumor activity. ACS Med Chem Lett. 2018;9(7):612–617. doi: 10.1021/acsmedchemlett.8b00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang H, et al. PRMT5 promotes cell proliferation by inhibiting BTG2 expression via the ERK signaling pathway in hepatocellular carcinoma. Cancer Med. 2018;7(3):869–882. doi: 10.1002/cam4.1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scoumanne A, Zhang J, Chen X. PRMT5 is required for cell-cycle progression and p53 tumor suppressor function. Nucleic Acids Res. 2009;37(15):4965–4976. doi: 10.1093/nar/gkp516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andreu-Pérez P, et al. Protein arginine methyltransferase 5 regulates ERK1/2 signal transduction amplitude and cell fate through CRAF. Sci Signal. 2011;4(190):ra58. doi: 10.1126/scisignal.2001936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meloche S, Pouysségur J. The ERK1/2 mitogen-activated protein kinase pathway as a master regulator of the G1- to S-phase transition. Oncogene. 2007;26(22):3227–3239. doi: 10.1038/sj.onc.1210414. [DOI] [PubMed] [Google Scholar]
- 35.Hu X, Ivashkiv LB. Cross-regulation of signaling pathways by interferon-gamma: implications for immune responses and autoimmune diseases. Immunity. 2009;31(4):539–550. doi: 10.1016/j.immuni.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5(5):375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 37.Pollack BP, Kotenko SV, He W, Izotova LS, Barnoski BL, Pestka S. The human homologue of the yeast proteins Skb1 and Hsl7p interacts with Jak kinases and contains protein methyltransferase activity. J Biol Chem. 1999;274(44):31531–31542. doi: 10.1074/jbc.274.44.31531. [DOI] [PubMed] [Google Scholar]
- 38.Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mackall C, et al. Background to hematopoietic cell transplantation, including post transplant immune recovery. Bone Marrow Transplant. 2009;44(8):457–462. doi: 10.1038/bmt.2009.255. [DOI] [PubMed] [Google Scholar]
- 40.Betts BC, Veerapathran A, Pidala J, Yu XZ, Anasetti C. STAT5 polarization promotes iTregs and suppresses human T-cell alloresponses while preserving CTL capacity. J Leukoc Biol. 2014;95(2):205–213. doi: 10.1189/jlb.0313154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heinrichs J, Bastian D, Veerapathran A, Anasetti C, Betts B, Yu XZ. Regulatory T-cell therapy for graft-versus-host disease. J Immunol Res Ther. 2016;1(1):1–14. [PMC free article] [PubMed] [Google Scholar]
- 42.Hippen KL, et al. Umbilical cord blood regulatory T-cell expansion and functional effects of tumor necrosis factor receptor family members OX40 and 4-1BB expressed on artificial antigen-presenting cells. Blood. 2008;112(7):2847–2857. doi: 10.1182/blood-2008-01-132951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stern M, et al. Sensitivity of hematological malignancies to graft-versus-host effects: an EBMT megafile analysis. Leukemia. 2014;28(11):2235–2240. doi: 10.1038/leu.2014.145. [DOI] [PubMed] [Google Scholar]
- 44.Hartmann N, et al. In vitro-established alloantigen-specific CD8+ CTLs mediate graft-versus-tumor activity in the absence of graft-versus-host disease. Leukemia. 2011;25(5):848–855. doi: 10.1038/leu.2011.16. [DOI] [PubMed] [Google Scholar]
- 45.Ruutu T, et al. Prophylaxis and treatment of GVHD: EBMT-ELN working group recommendations for a standardized practice. Bone Marrow Transplant. 2014;49(2):168–173. doi: 10.1038/bmt.2013.107. [DOI] [PubMed] [Google Scholar]
- 46.Shindo T, Kim TK, Benjamin CL, Wieder ED, Levy RB, Komanduri KV. MEK inhibitors selectively suppress alloreactivity and graft-versus-host disease in a memory stage-dependent manner. Blood. 2013;121(23):4617–4626. doi: 10.1182/blood-2012-12-476218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Itamura H, et al. The MEK inhibitor trametinib separates murine graft-versus-host disease from graft-versus-tumor effects. JCI Insight. 2016;1(10):e86331. doi: 10.1172/jci.insight.86331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu Y, Waller EK. Dichotomous role of interferon-gamma in allogeneic bone marrow transplant. Biol Blood Marrow Transplant. 2009;15(11):1347–1353. doi: 10.1016/j.bbmt.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robb RJ, Hill GR. The interferon-dependent orchestration of innate and adaptive immunity after transplantation. Blood. 2012;119(23):5351–5358. doi: 10.1182/blood-2012-02-368076. [DOI] [PubMed] [Google Scholar]
- 50.Liu Y, et al. The expression of Th17-associated cytokines in human acute graft-versus-host disease. Biol Blood Marrow Transplant. 2013;19(10):1421–1429. doi: 10.1016/j.bbmt.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 51.Zhao XY, Xu LL, Lu SY, Huang XJ. IL-17-producing T cells contribute to acute graft-versus-host disease in patients undergoing unmanipulated blood and marrow transplantation. Eur J Immunol. 2011;41(2):514–526. doi: 10.1002/eji.201040793. [DOI] [PubMed] [Google Scholar]
- 52.Welniak LA, Blazar BR, Anver MR, Wiltrout RH, Murphy WJ. Opposing roles of interferon-gamma on CD4+ T cell-mediated graft-versus-host disease: effects of conditioning. Biol Blood Marrow Transplant. 2000;6(6):604–612. doi: 10.1016/S1083-8791(00)70025-5. [DOI] [PubMed] [Google Scholar]
- 53.Yang YG, Qi J, Wang MG, Sykes M. Donor-derived interferon gamma separates graft-versus-leukemia effects and graft-versus-host disease induced by donor CD8 T cells. Blood. 2002;99(11):4207–4215. doi: 10.1182/blood.V99.11.4207. [DOI] [PubMed] [Google Scholar]
- 54.Yang YG, Wang H, Asavaroengchai W, Dey BR. Role of Interferon-gamma in GVHD and GVL. Cell Mol Immunol. 2005;2(5):323–329. [PubMed] [Google Scholar]
- 55.Burman AC, et al. IFNgamma differentially controls the development of idiopathic pneumonia syndrome and GVHD of the gastrointestinal tract. Blood. 2007;110(3):1064–1072. doi: 10.1182/blood-2006-12-063982. [DOI] [PubMed] [Google Scholar]
- 56.Lu Y, et al. IFN-γ and indoleamine 2,3-dioxygenase signaling between donor dendritic cells and T cells regulates graft versus host and graft versus leukemia activity. Blood. 2012;119(4):1075–1085. doi: 10.1182/blood-2010-12-322891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fischer JC, et al. RIG-I/MAVS and STING signaling promote gut integrity during irradiation- and immune-mediated tissue injury. Sci Transl Med. 2017;9(386):eaag2513. doi: 10.1126/scitranslmed.aag2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robb RJ, et al. Type I-IFNs control GVHD and GVL responses after transplantation. Blood. 2011;118(12):3399–3409. doi: 10.1182/blood-2010-12-325746. [DOI] [PubMed] [Google Scholar]
- 59.Lamarthée B, et al. Donor interleukin-22 and host type I interferon signaling pathway participate in intestinal graft-versus-host disease via STAT1 activation and CXCL10. Mucosal Immunol. 2016;9(2):309–321. doi: 10.1038/mi.2015.61. [DOI] [PubMed] [Google Scholar]
- 60.Cai Y, et al. Adoptively transferred donor IL-17-producing CD4+ T cells augment, but IL-17 alleviates, acute graft-versus-host disease. Cell Mol Immunol. 2018;15(3):233–245. doi: 10.1038/cmi.2016.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kappel LW, et al. IL-17 contributes to CD4-mediated graft-versus-host disease. Blood. 2009;113(4):945–952. doi: 10.1182/blood-2008-08-172155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Capitini CM, et al. Absence of STAT1 in donor-derived plasmacytoid dendritic cells results in increased STAT3 and attenuates murine GVHD. Blood. 2014;124(12):1976–1986. doi: 10.1182/blood-2013-05-500876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ma H, et al. Absence of Stat1 in donor CD4+ T cells promotes the expansion of Tregs and reduces graft-versus-host disease in mice. J Clin Invest. 2011;121(7):2554–2569. doi: 10.1172/JCI43706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Leng C, et al. Reduction of graft-versus-host disease by histone deacetylase inhibitor suberonylanilide hydroxamic acid is associated with modulation of inflammatory cytokine milieu and involves inhibition of STAT1. Exp Hematol. 2006;34(6):776–787. doi: 10.1016/j.exphem.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 65.Dacwag CS, Ohkawa Y, Pal S, Sif S, Imbalzano AN. The protein arginine methyltransferase Prmt5 is required for myogenesis because it facilitates ATP-dependent chromatin remodeling. Mol Cell Biol. 2007;27(1):384–394. doi: 10.1128/MCB.01528-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao Q, et al. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat Struct Mol Biol. 2009;16(3):304–311. doi: 10.1038/nsmb.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wei H, et al. PRMT5 dimethylates R30 of the p65 subunit to activate NF-κB. Proc Natl Acad Sci U S A. 2013;110(33):13516–13521. doi: 10.1073/pnas.1311784110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jansson M, et al. Arginine methylation regulates the p53 response. Nat Cell Biol. 2008;10(12):1431–1439. doi: 10.1038/ncb1802. [DOI] [PubMed] [Google Scholar]
- 69.Durant ST, Cho EC, La Thangue NB. p53 methylation--the Arg-ument is clear. Cell Cycle. 2009;8(6):801–802. doi: 10.4161/cc.8.6.7850. [DOI] [PubMed] [Google Scholar]
- 70.Cho EC, et al. Arginine methylation controls growth regulation by E2F-1. EMBO J. 2012;31(7):1785–1797. doi: 10.1038/emboj.2012.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Friesen WJ, et al. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol Cell Biol. 2001;21(24):8289–8300. doi: 10.1128/MCB.21.24.8289-8300.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pastore F, Levine RL. Epigenetic regulators and their impact on therapy in acute myeloid leukemia. Haematologica. 2016;101(3):269–278. doi: 10.3324/haematol.2015.140822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wouters BJ, Delwel R. Epigenetics and approaches to targeted epigenetic therapy in acute myeloid leukemia. Blood. 2016;127(1):42–52. doi: 10.1182/blood-2015-07-604512. [DOI] [PubMed] [Google Scholar]
- 74.Magenau J, Reddy P. Next generation treatment of acute graft-versus-host disease. Leukemia. 2014;28(12):2283–2291. doi: 10.1038/leu.2014.195. [DOI] [PubMed] [Google Scholar]
- 75.Servais S, et al. Novel approaches for preventing acute graft-versus-host disease after allogeneic hematopoietic stem cell transplantation. Expert Opin Investig Drugs. 2016;25(8):957–972. doi: 10.1080/13543784.2016.1182498. [DOI] [PubMed] [Google Scholar]
- 76.Bashir Q, William BM, Garcia-Manero G, de Lima M. Epigenetic therapy in allogeneic hematopoietic stem cell transplantation. Rev Bras Hematol Hemoter. 2013;35(2):126–133. doi: 10.5581/1516-8484.20130034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cooper ML, et al. Azacitidine mitigates graft-versus-host disease via differential effects on the proliferation of T effectors and natural regulatory T cells in vivo. J Immunol. 2017;198(9):3746–3754. doi: 10.4049/jimmunol.1502399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cruijsen M, et al. Addition of 10-day decitabine to fludarabine/total body irradiation conditioning is feasible and induces tumor-associated antigen-specific T cell responses. Biol Blood Marrow Transplant. 2016;22(6):1000–1008. doi: 10.1016/j.bbmt.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 79.Goodyear OC, et al. Azacitidine augments expansion of regulatory T cells after allogeneic stem cell transplantation in patients with acute myeloid leukemia (AML) Blood. 2012;119(14):3361–3369. doi: 10.1182/blood-2011-09-377044. [DOI] [PubMed] [Google Scholar]
- 80.Pusic I, et al. Maintenance therapy with decitabine after allogeneic stem cell transplantation for acute myelogenous leukemia and myelodysplastic syndrome. Biol Blood Marrow Transplant. 2015;21(10):1761–1769. doi: 10.1016/j.bbmt.2015.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Choi J, et al. In vivo administration of hypomethylating agents mitigate graft-versus-host disease without sacrificing graft-versus-leukemia. Blood. 2010;116(1):129–139. doi: 10.1182/blood-2009-12-257253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Choi S, Reddy P. HDAC inhibition and graft versus host disease. Mol Med. 2011;17(5-6):404–416. doi: 10.2119/molmed.2011.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Choi SW, et al. Histone deacetylase inhibition regulates inflammation and enhances Tregs after allogeneic hematopoietic cell transplantation in humans. Blood. 2015;125(5):815–819. doi: 10.1182/blood-2014-10-605238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Reddy P, et al. Histone deacetylase inhibitor suberoylanilide hydroxamic acid reduces acute graft-versus-host disease and preserves graft-versus-leukemia effect. Proc Natl Acad Sci U S A. 2004;101(11):3921–3926. doi: 10.1073/pnas.0400380101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sun Y, et al. Genome-wide STAT3 binding analysis after histone deacetylase inhibition reveals novel target genes in dendritic cells. J Innate Immun. 2017;9(2):126–144. doi: 10.1159/000450681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cooke KR, et al. An experimental model of idiopathic pneumonia syndrome after bone marrow transplantation: I. The roles of minor H antigens and endotoxin. Blood. 1996;88(8):3230–3239. doi: 10.1182/blood.V88.8.3230.bloodjournal8883230. [DOI] [PubMed] [Google Scholar]
- 87.Zitzer NC, et al. MicroRNA-155 modulates acute graft-versus-host disease by impacting T cell expansion, migration, and effector function. J Immunol. 2018;200(12):4170–4179. doi: 10.4049/jimmunol.1701465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hill GR, Crawford JM, Cooke KR, Brinson YS, Pan L, Ferrara JL. Total body irradiation and acute graft-versus-host disease: the role of gastrointestinal damage and inflammatory cytokines. Blood. 1997;90(8):3204–3213. doi: 10.1182/blood.V90.8.3204. [DOI] [PubMed] [Google Scholar]
- 89.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 91.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40(10):4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Krämer A, Green G, Pollard J, Jr, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30(4):523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bendall SC, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332(6030):687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Behbehani GK, Samusik N, Bjornson ZB, Fantl WJ, Medeiros BC, Nolan GP. Mass cytometric functional profiling of acute myeloid leukemia defines cell-cycle and immunophenotypic properties that correlate with known responses to therapy. Cancer Discov. 2015;5(9):988–1003. doi: 10.1158/2159-8290.CD-15-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Devine RD, Sekhri P, Behbehani GK. Effect of storage time and temperature on cell cycle analysis by mass cytometry. Cytometry A. 2018;93(11):1141–1149. doi: 10.1002/cyto.a.23630. [DOI] [PubMed] [Google Scholar]
- 97.Teshima T, et al. IL-11 separates graft-versus-leukemia effects from graft-versus-host disease after bone marrow transplantation. J Clin Invest. 1999;104(3):317–325. doi: 10.1172/JCI7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tsukada N, Kobata T, Aizawa Y, Yagita H, Okumura K. Graft-versus-leukemia effect and graft-versus-host disease can be differentiated by cytotoxic mechanisms in a murine model of allogeneic bone marrow transplantation. Blood. 1999;93(8):2738–2747. [PubMed] [Google Scholar]
- 99.Yang ZZ, Novak AJ, Ziesmer SC, Witzig TE, Ansell SM. Attenuation of CD8(+) T-cell function by CD4(+)CD25(+) regulatory T cells in B-cell non-Hodgkin’s lymphoma. Cancer Res. 2006;66(20):10145–10152. doi: 10.1158/0008-5472.CAN-06-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.