Abstract
Septic cardiomyopathy is a life-threatening organ dysfunction caused by sepsis. Ribonuclease 1 (RNase 1) belongs to a group of host-defense peptides that specifically cleave extracellular RNA (eRNA). The activity of RNase 1 is inhibited by ribonuclease-inhibitor 1 (RNH1). However, the role of RNase 1 in septic cardiomyopathy and associated cardiac apoptosis is completely unknown. Here, we show that sepsis resulted in a significant increase in RNH1 and eRNA serum levels compared with those of healthy subjects. Treatment with RNase 1 resulted in a significant decrease of apoptosis, induced by the intrinsic pathway, and TNF expression in murine cardiomyocytes exposed to either necrotic cardiomyocytes or serum of septic patients for 16 hours. Additionally, treatment of septic mice with RNase 1 resulted in a reduction in cardiac apoptosis, TNF expression, and septic cardiomyopathy. These data demonstrate that eRNA plays a crucial role in the pathophysiology of the organ (cardiac) dysfunction in sepsis and that RNase and RNH1 may be new therapeutic targets and/or strategies to reduce the cardiac injury and dysfunction caused by sepsis.
Keywords: Immunology, Inflammation
Keywords: Anesthesiology, Innate immunity, Pharmacology
Extracellular RNA contributes to sepsis-associated cardiac dysfunction and can be targeted by RNAase or RNH1 to reduce cardiac injury in a murine sepsis model.
Introduction
Sepsis is one of the most common causes of death on intensive care units (ICUs) and is defined as a life-threatening organ dysfunction secondary to an infection (1, 2). In addition to respiratory failure, septic cardiomyopathy is a common organ dysfunction in septic shock and is clearly associated with prolonged hospital stays and increased mortality (3, 4). Septic cardiomyopathy is a functional restriction of the heart that results in a ventricular dilation and reduced contractility (5, 6). To date, however, no specific therapeutic strategies are available.
The tissue injury associated with sepsis results in the release of a variety of damage-associated molecular patterns (DAMPs) (7, 8). Extracellular RNA (eRNA) belongs to this heterogeneous group of DAMPs and includes several types of RNA present in the extracellular environment, including microRNA (miRNA), transfer RNA (tRNA), small interfering RNA (siRNA), and long noncoding RNA (lncRNA). eRNA is released as a result of tissue injury and necrotic cell death from a variety of cells, including cardiomyocytes (9), and may play a role in differentiation, chromatin modification, and inflammation as well as tissue injury and repair (10–14). However, it is unknown whether eRNA plays a role in septic cardiomyopathy and cardiac apoptosis. Playing a decisive role in septic cardiomyopathy (15), apoptosis represents a programmed, caspase-mediated cell death that is triggered by intrinsic or extrinsic signaling pathways.
In 1988, Steven Benner described eRNA as an intercellular “communicator” and hypothesized that the activity of eRNA is modulated by endogenous ribonucleases (RNases) (16). RNases belong to a group of host-defense peptides, which are ubiquitously expressed in several tissues and body fluids. As a part of the innate immune system, RNases modulate local and systemic inflammatory responses induced by exogenous pathogens and endogenous danger molecules (17). Specifically, RNase 1 cleaves eRNA and, therefore, modulates and/or limits the levels of circulating eRNA (17). As an endogenous modulator of RNase 1, the human ribonuclease-inhibitor 1 (RNH1) is ubiquitously expressed in a variety of tissues and binds to and inactivates RNase 1 with a high affinity (18, 19). RNH1 and RNase 1 have complementary structures (20). Primarily induced by oxidative stress (20), RNH1 regulates the activity of RNases and, thus, plays a crucial role in the immunomodulatory and host-defense functions of RNases (21). However, the respective roles of RNase 1 and RNH1 in septic cardiomyopathy and cardiac apoptosis are unknown.
The aim of this reverse-translational study was to investigate (a) the levels and kinetics of RNH1 and the eRNA profile in the serum of patients with sepsis and healthy subjects (control), (b) the potential of RNase 1 to reduce apoptosis in cardiomyocytes, and (c) whether the application of RNase 1 attenuates septic cardiomyopathy and cardiac apoptosis in a murine model of polymicrobial sepsis.
Results
RNH1 is elevated in serum of septic patients.
We recently reported that serum levels of RNase 1 are substantially elevated in the early phase of sepsis (22). Because RNH1 inhibits the enzymatic activity of RNase 1, serum levels of RNH1 in septic patients (n = 21) were investigated on the day of and 3 days after diagnosis. Moreover, serum levels of RNH1 were analyzed in age- and sex-matched healthy subjects (n = 10). The characteristics of the study population according to the groups are shown in Supplemental Table 1 (supplemental material available online with this article; https://doi.org/10.1172/jci.insight.131571DS1). In healthy subjects, the RNH1 concentrations were below the detection limit of the ELISA used (1.563 ng/mL; n = 10; Figure 1A). In contrast, a mean concentration of 4.22 ± 1.00 ng/mL RNH1 was detected in serum of septic patients at time of diagnosis (Figure 1A), which rose further to 5.29 ± 1.36 ng/mL 3 days after diagnosis of sepsis (p < 0.05 when compared with healthy subjects; Figure 1A).
Figure 1. RNH1 and total extracellular RNA serum levels.
(A) RNH1 levels of healthy subjects (n = 10) and ICU patients with sepsis on the day of diagnosis (Sepsis DD) and 3 days after diagnosis (Sepsis D3; both n = 21) are displayed. A 1-way ANOVA followed by Bonferroni test was used for multiple comparisons. Data are presented as dot plot with the mean ± SEM. (B) Total eRNA levels from serum of healthy subjects (n = 10) and septic patients (n = 21) 3 days after diagnosis are demonstrated. An unpaired t test (2-tailed) was used for statistical analysis. Data are presented as dot plot with the mean ± SEM. (C) The eRNA size distribution from serum of healthy subjects (n = 10) and septic patients 3 days after diagnosis (n = 21) are presented in an electropherogram and a gel image. The first peak (between 10 and 100 nt) of the electropherograms represented the ladder. §P < 0.05 versus control/healthy. RNH1, ribonuclease-inhibitor 1; eRNA, extracellular RNA.
Levels of eRNA are elevated in serum of septic patients.
After demonstrating elevated serum levels of RNase 1 and of its inhibitor RNH1 in patients with sepsis, the levels of eRNA in serum of septic patients (n = 21) and healthy volunteers (n = 10) were measured 3 days after diagnosis. The different sizes of eRNA presented in serum of septic patients and healthy volunteers were also investigated. A mean concentration of 59.64 ± 4.92 ng/μL eRNA was measured in healthy subjects. In contrast, in serum of septic patients 3 days after diagnosis a significantly increased mean concentration of 105.6 ± 4.85 ng/μL eRNA was detected (p < 0.05 when compared with healthy subjects; Figure 1B). The electropherograms from serum of septic patients 3 days after diagnosis (n = 21) showed a higher variety of eRNA compared with heathy subjects (n = 10; Figure 1C). In healthy volunteers, a more uniform eRNA size distribution was observed, with the highest sample intensity in serum of healthy volunteers between 100 and 500 nt. In contrast, serum of septic patients showed a much higher variance in eRNA size distribution and a much higher sample intensity, which is caused by the higher concentration of eRNA in serum of septic patients (Figure 1C).
RNase 1 reduces the caspase-3 activation and apoptosis induced by necrotic cardiomyocytes.
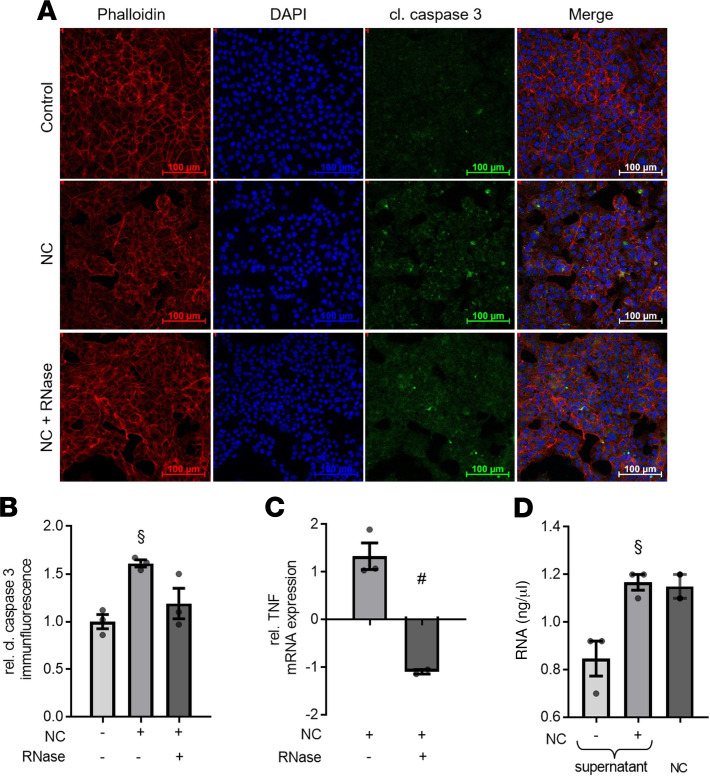
As demonstrated in earlier studies, septic cardiomyopathy is associated with cardiac apoptosis and necrotic cell death (23, 24). We investigated the potential induction of apoptosis by eRNA released from necrotic cardiomyocytes (NC) by exposing murine cardiomyocytes to NC. We also investigated whether RNase 1 reduces the degree of apoptosis caused by the eRNA released from NC. Cardiomyocytes exposed to NC, in the absence of RNase 1 treatment, showed a significant increase of cleaved/activated caspase-3 when compared with unstimulated cells measured by immunofluorescence (p < 0.01; Figure 2, A and B). Treatment with RNase 1, however, resulted in a decrease in caspase-3 activation (p < 0.05; Figure 2, A and B). Because eRNA leads to an increased expression of TNF via binding to TLR-3 and -7 (23–25), TNF mRNA expression was investigated in this setting. We found that cardiomyocytes stimulated with NC in the presence of RNase 1 showed a significant reduction in the expression of TNF mRNA when compared with cardiomyocytes challenged with NC in the absence of RNase 1 (p < 0.05; Figure 2C). Additionally, we investigated the amount of total eRNA released from 105 NC/mL and from the cell supernatants of unstimulated and/or stimulated cardiomyocytes. In cell supernatant of untreated cardiomyocytes, the mean concentration of eRNA was 0.85 ± 0.07 ng/mL. In contrast, we found significantly increased eRNA concentrations in the supernatant of cardiomyocytes exposed to NC for 16 hours (1.17 ± 0.03 ng/mL) as well as in 105 necrotic cells/mL (1.15 ± 0.07 ng/mL; p < 0.05; Figure 2D).
Figure 2. RNase 1 treatment results in decrease of cleaved caspase-3 immunofluorescence and TNF mRNA expression in murine cardiomyocytes exposed to necrotic cardiomyocytes.
(A) Cardiomyocytes exposed to 105 necrotic cardiomyocytes (NC) for 16 hours in presence (RNase + NC) or absence of 2.8 U/mL RNase 1 were stained with Phalloidin, DAPI, and anti-cleaved caspase-3 and compared with unstimulated cells (control). Phalloidin represents the cytosol (red), DAPI stained represents the nuclei (blue), and green immunofluorescence represents the cleaved caspase-3 (cl. caspase 3) expression. Scale bars: 100 μm. (B) Quantification of cleaved caspase 3 immunofluorescence (all n = 3). A 1-way ANOVA followed by Bonferroni test was used for multiple comparisons. Data are presented as dot plot with the mean ± SEM. (C) Relative TNF mRNA expression of cardiomyocytes exposed to NC in presence of RNase 1 compared with cardiomyocytes stimulated in absence of RNase 1 (both n = 3). An unpaired t test (2-tailed) was used for statistical analysis. Data are presented as dot plot with the mean ± SEM. (D) Total extracellular RNA in cell supernatant of cardiomyocytes exposed to NC for 16 hours and untreated cells (both n = 3) as well as eRNA level in 105 NC/mL (n = 2). A 1-way ANOVA followed by Bonferroni test was used for multiple comparisons. Data are presented as dot plot with the mean ± SEM. §P < 0.05 versus control; #P < 0.05 versus NC. NC, necrotic cardiomyocytes; RNase, ribonuclease 1.
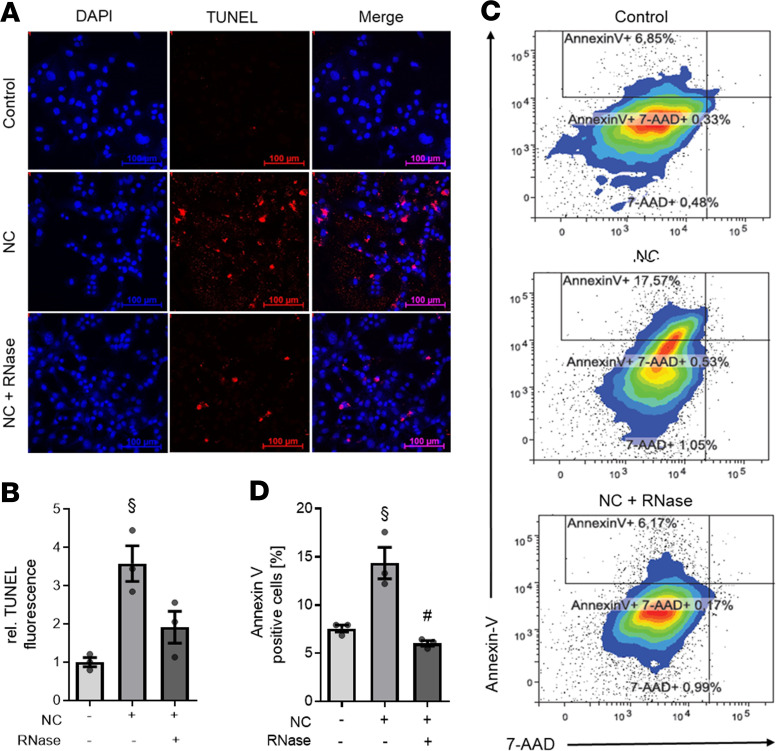
Next, we aimed to confirm that apoptosis was the predominant type of cell death in cardiomyocytes exposed to NC by TUNEL labeling. Unstimulated cardiomyocytes (control) did not show any TUNEL fluorescence (Figure 3A). However, in cardiomyocytes exposed to NC for 16 hours, there was an increased number of TUNEL+ cells (significant increase of a mean 3.58 ± 0.47 relative TUNEL fluorescence compared with unstimulated cells; P < 0.001; Figure 3, A and B). In contrast, the exposure of cardiomyocytes to necrotic cells in the presence of RNase 1 resulted in a smaller number of TUNEL+ cells (mean 1.91 ± 0.42 relative TUNEL fluorescence compared with cardiomyocytes exposed to necrotic cells in absence of RNase; Figure 3, A and B). The amount of apoptosis was further quantified and/or confirmed using flow cytometry. Relatively few unstimulated cells were positive for annexin V (7.57% ± 0.36%), 7-aminoactinomycin D (7-AAD) (0.97% ± 0.30%), and annexin V plus 7-AAD (0.54% ± 0.12%). The main part of the population was represented by living cells (Figure 3C). Cardiomyocytes exposed to 1 × 105 NC contained 13.36% ± 2.17% annexin V, 1.26% ± 0.12% 7-AAD and 0.51% ± 0.01% annexin V+ and 7-AAD+ cells (Figure 3C). However, exposure of NC in the presence of RNase 1 resulted in 7.55% ± 1.09% annexin V, 0.82% ± 0.21% 7-AAD, and 0.96% ± 0.77% annexin V+ and 7-AAD+ cells (Figure 3C). The amount of annexin V+ cells significantly increased in cardiomyocytes exposed to NC compared with unstimulated cells (p < 0.001; Figure 3D), which was significantly diminished by the addition of RNase 1 (p < 0.001; Figure 3D).
Figure 3. RNase 1 treatment of cardiomyocytes exposed to necrotic cardiomyocytes results in decreased apoptosis.
Cardiomyocytes exposed to 105 necrotic cardiomyocytes in presence (RNase + NC) or absence (NC) of 2.8 U/mL RNase 1 for 16 hours compared with unstimulated cells (control) were analyzed (all n = 3). (A) Apoptotic cells were labeled with TUNEL (red) and the nuclei of cardiomyocytes stained with DAPI (blue). Scale bars: 100 μm. (B) Quantification of TUNEL fluorescence. A 1-way ANOVA followed by Bonferroni test was used for multiple comparisons. Data are presented as dot plot with the mean ± SEM. (C) Treated HL-1 cells were stained with annexin V and 7-AAD and analyzed by flow cytometry. Gate I (bottom left) displays the living cells. The annexin V+ cells are shown in Gate II (top left) and represent the apoptotic cardiomyocytes. The double-positive cells are demonstrated in gate III (top right) and in gate IV (bottom right), where the 7-AAD+ or necrotic cells are located. (D) The percentage of annexin V+ (apoptotic) cells is displayed. A 1-way ANOVA followed by Bonferroni test was used for multiple comparisons. Data are presented as dot plot with the mean ± SEM. §P < 0.05 versus control; #P < 0.05 versus NC. NC, necrotic cardiomyocytes; RNase, ribonuclease 1.
RNase 1 reduces the caspase-3 activation induced by serum of septic patients.
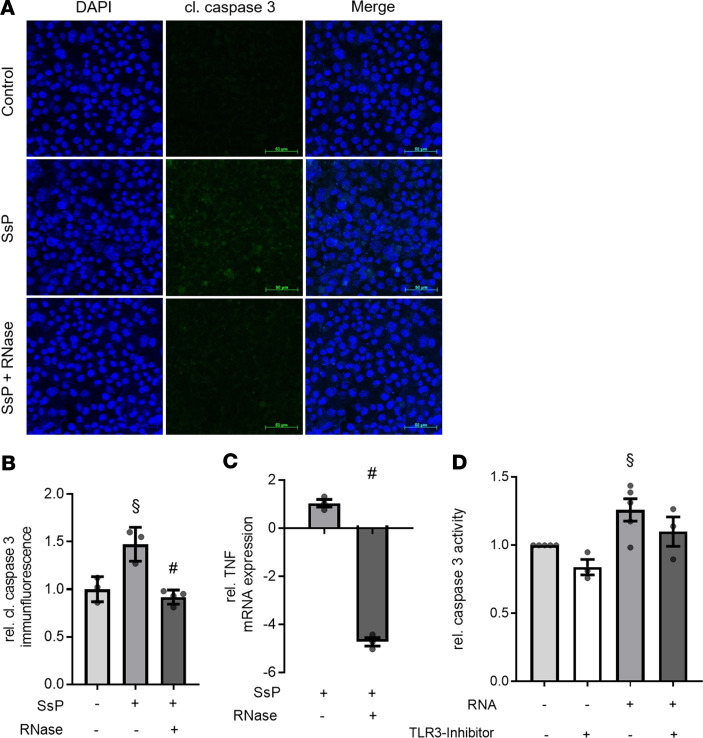
After demonstrating serum levels of RNH1 as well as total eRNA are significantly elevated in serum of septic patients (both P < 0.05), we investigated caspase-3 activation and TNF mRNA expression in murine cardiomyocytes challenged with serum of septic patients, as a reverse translational approach. Specifically, we aimed to confirm the potential protective/antiapoptotic effect of RNase 1 in this setting. When compared with unstimulated cells, cardiomyocytes exposed to serum of septic patients (in the absence of RNase 1) showed a significant increase in the cleavage of caspase-3 as well as an increase in the expression of TNF mRNA (p < 0.05; Figure 4, A–C). In contrast, RNase 1 attenuated the increases in both caspase-3 activation and TNF mRNA expression in cardiomyocytes challenged with serum of septic patients (p < 0.05; Figure 4, A–C).
Figure 4. RNase 1 treatment or TLR3 inhibition results in decrease of caspase-3 activation and TNF expression in murine cardiomyocytes exposed to serum of septic patients or RNA.
(A) Cardiomyocytes exposed to 5 % serum of patients with sepsis (SsP) for 16 hours in presence (RNase + SsP; n = 4) or absence of 2.8 U/mL RNase 1 (n = 3) were stained with DAPI and anti-cleaved caspase-3 and compared with unstimulated cells (control; n = 3). DAPI stained the nuclei (blue) and the green immunofluorescence represents the cleaved caspase-3 (cl. caspase 3) expression. Scale bars: 50 μm. (B) Quantification of cleaved caspase-3 immunofluorescence. A 1-way ANOVA followed by Bonferroni test was used for multiple comparisons. Data are presented as dot plot with the mean ± SEM. (C) Relative TNF mRNA expression of cardiomyocytes exposed to SsP in presence of RNase 1 compared with cardiomyocytes stimulated in absence of RNase 1 (both n =3). An unpaired t test (2-tailed) was used for statistical analysis. Data are presented as dot plot with the mean ± SEM. (D) Relative caspase activity was analyzed in cardiomyocytes exposed to 100 ng/mL eRNA and unstimulated cells for 16 hours in presence (both n = 3) or absence (both n = 5) of 2 μM TLR3-Inhibitor using Caspase Glo assay. A 1-way ANOVA followed by Bonferroni test was used for multiple comparisons. Data are presented as dot plot with the mean ± SEM. §P < 0.05 versus control; #P < 0.05 versus SsP. SsP, serum of septic patients; RNase, ribonuclease 1.
TLR3 inhibition reduces the caspase-3 activation induced by eRNA.
After demonstrating that apoptosis and TNF expression were induced by NC and serum of septic patients, which both contain eRNA, caspase-3 activation in cardiomyocytes challenged with eRNA isolated from cardiomyocytes was investigated. A significantly increased relative caspase-3 activation was detected in cardiomyocytes exposed to eRNA, when compared with unstimulated cardiomyocytes (1.26 ± 0.08; p < 0.05; Figure 4D). Furthermore, we wanted to investigate whether caspase-3 activation in cardiomyocytes exposed to eRNA is caused by binding to TLR3. We found a decreased relative caspase-3 activation in cardiomyocytes stimulated with eRNA in the presence of a TLR3 inhibitor (CU CPT 4a) compared with cardiomyocytes exposed to eRNA in the absence of the TLR3 inhibitor (1.10 ± 0.11; Figure 4D).
RNase 1 improves the cardiovascular function and reduces cardiac apoptosis in murine polymicrobial sepsis.
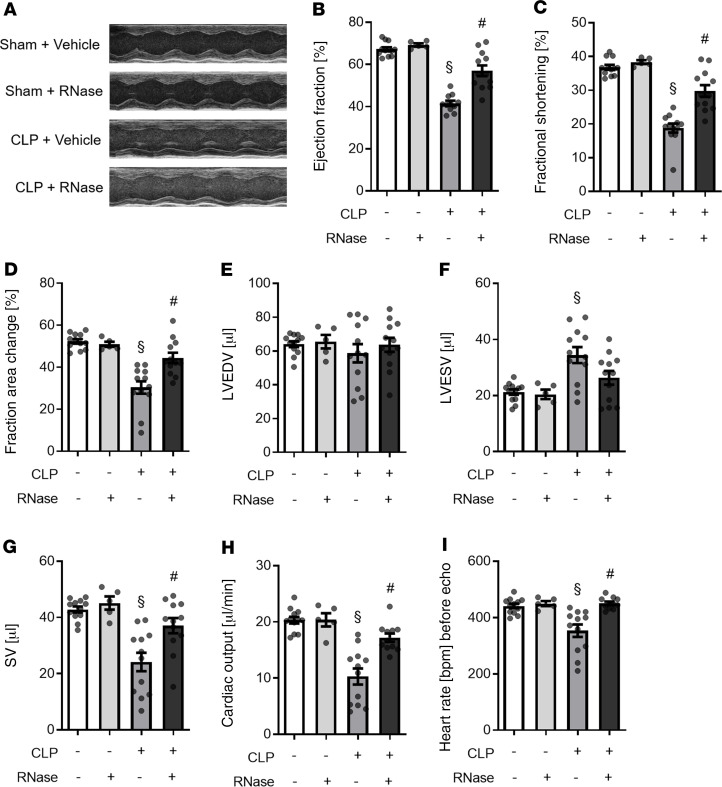
After demonstrating that the exposure of murine cardiomyocytes in the presence of RNase 1 resulted in reduced apoptosis (when challenged with NC or serum of septic patients), we next investigated the effect of RNase 1 on cardiac function and cardiac apoptosis in a murine model of polymicrobial sepsis. When compared with sham-operated mice, cecal ligation and puncture–induced (CLP-induced) sepsis resulted in a substantial cardiac dysfunction (measured as a reduction of ejection fraction [EF], fractional shortening [FS], and fractional area change [FAC] by echocardiography) (all p < 0.001; Figure 5). Treatment with RNase 1, however, significantly attenuated the cardiac dysfunction associated with CLP-sepsis, as indicated by higher values of EF, FS (both P < 0.001; Figure 5), and FAC (p < 0.01; Figure 5) compared with vehicle-treated CLP mice (control). To analyze the cardiac dysfunction in sepsis and the potential beneficial effects of RNase 1, we further investigated the left ventricular end-diastolic volume (LVEDV), left ventricular end-systolic volume (LVESV), and stroke volume (SV) as well as the cardiac output and the heart rate before echocardiography. Of significance, no change in LVEDV was detected between the groups (Figure 5E). In CLP-sepsis, a significant increased LVESV was measured, when compared with sham-operated mice (p < 0.01; Figure 5). RNase 1 treatment of CLP animals, however, resulted in a decrease of LVESV (when compared with vehicle-treated CLP-sepsis). Accordingly, when compared with sham-operated mice, CLP-sepsis resulted in a significant decreased SV, heart rate, and cardiac output, which was significantly diminished by RNase 1 treatment (all p < 0.01; Figure 5). Sham-operated animals treated with RNase 1 showed no changes in any of the preceding parameters, when compared with sham-operated animals treated with vehicle (Figure 5).
Figure 5. RNase 1 treatment of mice with polymicrobial sepsis resulted in an improved heart function.
(A) Representative echocardiography images of each group are shown. (B) The ejection fraction, (C) fractional shortening, (D) fraction area change, (E) left ventricular end-diastolic volume (LVEDV), (F) left ventricular end-systolic volume (LVESV), (G) stroke volume (SV), (H) cardiac output and (I) heart rate before echocardiography of sham-operated mice treated with vehicle (n = 12) or RNase 1 (n = 5) and mice with polymicrobial sepsis induced by CLP treated with vehicle or RNase 1 were analyzed (both n = 12). Individual values are plotted as mean ± SEM. A 1-way ANOVA followed by Bonferroni test was used for multiple comparisons (graph B–I). §P < 0.05 versus sham; #P < 0.05 versus CLP + vehicle. CLP, cecal ligation and puncture; RNase, ribonuclease 1.
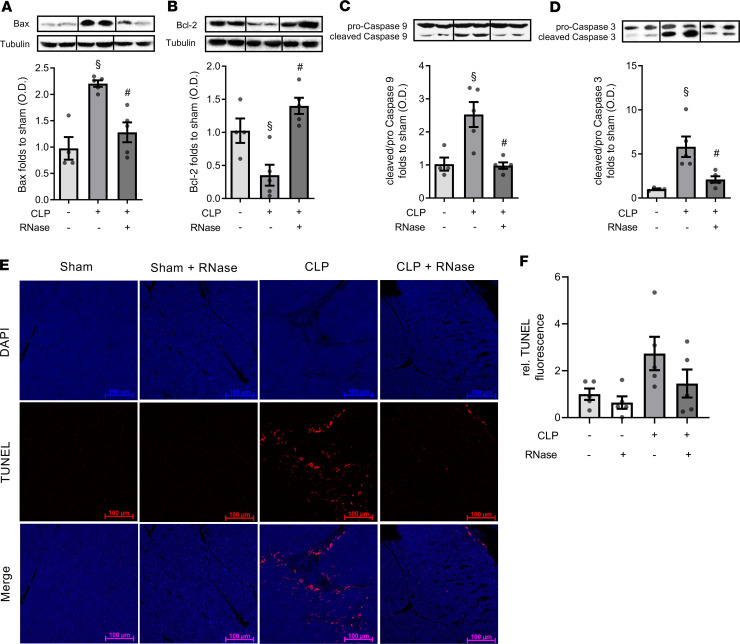
To gain a better understanding of the underlying signaling events involved in the observed beneficial effects of RNase 1 on sepsis-associated cardiomyopathy, we investigated the relevant apoptosis pathways in cardiac tissue at 24 hours after onset of sepsis (e.g., the time point at which cardiac function was evaluated). When compared with sham-operated animals, CLP-sepsis resulted in a significant increase in Bax levels as well as caspase-9 and caspase-3 activation (p < 0.001; p < 0.01; p < 0.01; Figure 6). In contrast to vehicle-treated animals, the treatment of CLP mice with RNase 1, however, significantly attenuated the increases in Bax levels and the associated activation of caspase-9 and caspase-3 associated with sepsis (p < 0.01; p < 0.01; p < 0.05; Figure 6). Furthermore, CLP-sepsis resulted in a decrease in Bcl-2 levels in the heart (p < 0.05; Figure 6), an effect that was also significantly attenuated by RNase 1 treatment (p < 0.01; Figure 6). To confirm that apoptosis is induced by CLP in the heart, we analyzed the number of apoptotic cells in heart tissue by TUNEL labeling. When compared with sham-operated animals, CLP-sepsis resulted in an increase in relative TUNEL fluorescence (mean 2.73 ± 0.71 relative TUNEL fluorescence; Figure 6, E and F). The treatment of CLP-sepsis with RNase 1 resulted in a decrease of relative TUNEL fluorescence compared with vehicle-treated CLP animals (mean 1.45 ± 0.60 relative TUNEL fluorescence; Figure 6, E and F). However, treatment of sham-operated mice with RNase 1 did not result in an increase in TUNEL fluorescence (mean 0.64 ± 0.26 relative TUNEL fluorescence; Figure 6, E and F).
Figure 6. RNase 1 treatment of mice with polymicrobial sepsis resulted in a decreased cardiac apoptosis.
Heart tissue of sham-operated mice (n = 4) or mice with polymicrobial sepsis induced by CLP treated with vehicle or RNase 1 (both n = 5) were analyzed by Western blot. To analyze the intrinsic apoptosis signaling pathway, (A) Bax and (B) Bcl-2 levels as well as (C) caspase-9 and (D) caspase-3 activation were investigated. A 1-way ANOVA followed by Bonferroni test was used for multiple comparisons. Data are presented as dot plot with the mean ± SEM. Representative immunoblots are presented above the bar charts. The quantified OD of bands was corrected for the corresponding tubulin bands and normalized using the related sham band. Heart tissue of sham-operated mice treated with vehicle or RNase or mice with polymicrobial sepsis induced by CLP also treated with vehicle or RNase 1 (all n = 5) were analyzed by TUNEL labeling. (E) Apoptotic cells were labeled with TUNEL (red) and the nuclei of cardiomyocytes stained with DAPI (blue). Scale bars: 100 μm. (F) Quantification of TUNEL fluorescence. A 1-way ANOVA followed by Bonferroni test was used for multiple comparisons. Data are presented as dot plot with the mean ± SEM. §P < 0.05 versus Sham; #P < 0.05 versus CLP + Vehicle. CLP, cecal ligation and puncture; RNase, ribonuclease 1; Bcl-2, B-cell lymphoma 2.
After detecting elevated serum levels of RNH1 and total eRNA in patients with sepsis, we investigated serum levels of RNase 1, RNH1, and total eRNA in murine polymicrobial sepsis. We also analyzed the TNF serum levels in CLP-sepsis to gain a better understanding of the underlying mechanisms and associations between the eRNA concentration and the proinflammatory/proapoptotic cardiac response. In sham-operated mice treated with vehicle or RNase 1, RNase 1 serum levels were 236.86 ± 8.11 ng/mL and 287.32 ± 19.37 ng/mL (Figure 7B). In contrast, untreated sepsis (24 hours after CLP challenge) resulted in significantly decreased RNase 1 serum levels (69.68 ± 27.16 ng/mL RNase 1; P < 0.05 when compared with sham-operated mice; Figure 7B), whereas septic mice treated with RNase 1 showed a mean RNase 1 concentration of 108.5 ± 22.70 ng/mL (Figure 7B). Furthermore, the treatment of CLP mice with RNase 1 resulted in an improved renal dysfunction and hepatocellular injury (Supplemental Figure 1). In sham-operated mice treated with vehicle, the TNF and RNH1 concentrations were below the detection limit of the ELISA used (31.3 pg/mL and 0.78 ng/mL; Figure 7, A and C). CLP animals showed TNF and RNH1 concentrations of 215.4 ± 57.86 pg/mL and 4.68 ± 0.73 ng/mL, respectively (both n = 8; Figure 7, A and C). However, when compared with vehicle-treated animals, the treatment with RNase 1 resulted in a decrease in the concentrations of TNF (41.1 ± 17.90 pg/mL; p < 0.05; Figure 7A) and RNH1 (3.83 ± 1.15 ng/mL; Figure 7C). In addition, we found a significant correlation between TNF and RNase 1 concentration in CLP mice treated with vehicle or RNase 1 (both p < 0.05; Figure 7, F and G). Figure 7D shows the corresponding eRNA levels of the individual groups (Figure 7D). To gain a better understanding of the RNase 1 expression in sepsis, we performed a time course of RNase 1 serum levels in CLP-sepsis (Figure 7E). We detected a significant decrease of RNase 1 serum levels in CLP-sepsis compared with sham-operated mice at all time points (all p < 0.05; Figure 7E). The highest RNase 1 serum levels in CLP-sepsis could be measured 1 hour after sepsis induction by CLP (177.21 ± 62.78). We detected the lowest RNase 1 serum levels at 3 hours after surgery (11.43 ± 1.20), with the levels increasing again (52.14 ± 18.59) at 24 hours after surgery.
Figure 7. RNase 1 treatment of mice with polymicrobial sepsis resulted in a decreased TNF expression.
(A) TNF, (B) RNase 1 and (C) RNH1 serum levels of sham-operated mice treated with vehicle (n = 11) or RNase 1 (n = 5) or mice with polymicrobial sepsis induced by CLP treated with vehicle (n = 8) or RNase 1 (n = 9) were analyzed by ELISA. An unpaired t test (2-tailed) was used for statistical analysis (A and C). In B, a 1-way ANOVA followed by Bonferroni test was used for multiple comparisons. Data are presented as dot plot with the mean ± SEM. (D) Total extracellular RNA in serum of mice are demonstrated. A 1-way ANOVA followed by Bonferroni test was used for multiple comparisons. Data are presented as dot plot with the mean ± SEM. (E) RNase 1 serum levels over the time in sham-operated mice (n = 3) and mice with polymicrobial sepsis induced by CLP treated with vehicle (n = 4). Correlation of TNF and RNase 1 of mice with polymicrobial sepsis induced by CLP (F) treated with vehicle and (G) treated with RNase 1. §P < 0.05 versus sham; #P < 0.05 versus CLP + vehicle. CLP, cecal ligation and puncture; RNase, ribonuclease 1; RNH1, ribonuclease-inhibitor 1.
Discussion
Septic cardiomyopathy is a life-threatening organ dysfunction associated with a complicated postoperative course and increased mortality. Despite intense efforts in basic and clinical research, therapy for this condition is supportive in nature, as no specific therapy is available (5). This urgently necessitates a better understanding of the underlying pathophysiology and most importantly, the identification of new therapeutic targets and strategies. This study provides the first evidence to our knowledge that RNase 1 reduces cardiac apoptosis and attenuates septic cardiomyopathy by reducing the levels of circulating eRNA in a murine model of polymicrobial sepsis.
Specifically, we report for the first time to our knowledge that the levels of RNH1, the endogenous inhibitor of RNase 1, are significantly elevated on the day of diagnosis and 3 days after diagnosis of sepsis (Figure 1A). We have also recently reported an increased serum expression of RNase 1 at the onset of sepsis (22). RNH1 specifically binds to RNase 1 and inhibits its activity (20). Based on these findings, we hypothesized that an increase in RNH1 expression results in an increase of eRNA levels, owing to inhibition of the enzymatic activity of RNase 1 by RNH1 (Figure 8). Indeed, we found significantly elevated levels of free circulating eRNA in serum of septic patients (Figure 1B). The tissue injury and cell death in sepsis and septic shock leads to a loss of tissue integrity and subsequently to the release of both cell-free DNA and eRNA into the circulation (26). Although there is good evidence that cell-free DNA plays a crucial role in the crosstalk between tissue injury, inflammation, and coagulation, very little is known about the physiological and pathophysiological roles of eRNA in sepsis (27). It has been suggested that the negative charge of eRNA builds the structural feature rendering eRNA capable of activating factor XII and, hence, coagulation (28, 29). Moreover, Chen and colleagues report that eRNA contributes to myocardial inflammation via TLR-3-Trif signaling (23). Whether eRNA plays a role in cardiac apoptosis, which plays a pivotal role in the pathophysiology of septic cardiomyopathy, however, was not known. To address this important question, we exposed cardiomyocytes to NC or serum of septic shock patients (Figure 2, Figure 3, and Figure 5). In line with the work from Chen and colleagues, we used NC (as our source of eRNA) to stimulate cardiomyocytes with eRNA (23). This approach enabled us to expose cardiomyocytes to the eRNA contained in cardiomyocytes, which contain several types of RNA (e.g., miRNA, tRNA, and siRNA) (10). In addition to the free circulating, extracellular RNA, in both stimulants (NC and serum of septic patients) a part of the RNA may be encapsulated in extracellular vesicles such as exosomes or microvesicles and, thus, be protected from the digestion of RNase 1 (30, 31). Nevertheless, we were able to show that RNase 1 significantly reduces both apoptosis and inflammation in murine cardiomyocytes (Figure 2, Figure 3, and Figure 4). To standardize our model, we first measured the total amount of eRNA in the cell supernatant of cardiomyocytes exposed to NC (Figure 2D). In fact, we found an increased release of eRNA into the cell supernatant of cardiomyocytes exposed to NC compared with the cell supernatant of untreated cardiomyocytes (Figure 2D). Additionally, we found a significant activation (measured as cleavage) of caspase-3 and consecutive induction of apoptosis by exposing murine cardiomyocytes to either NC or, notably, serum of septic patients (Figure 2 and Figure 4). Furthermore, to address the question as to whether the observed increase in apoptosis is caused by eRNA, we exposed cardiomyocytes to eRNA isolated from cardiomyocytes. Indeed, we found a significant increase in caspase-3 activation in cardiomyocytes challenged with eRNA compared with unstimulated cells (Figure 4D). In line with Chen and colleagues, we showed that eRNA contributes to induction of apoptosis (caspase-3 activation) via TLR3 signaling (Figure 4D and ref. 23). Having shown that sepsis is associated with elevated RNH1 levels, we next aimed to investigate the potential protective/therapeutic effects of RNase 1.
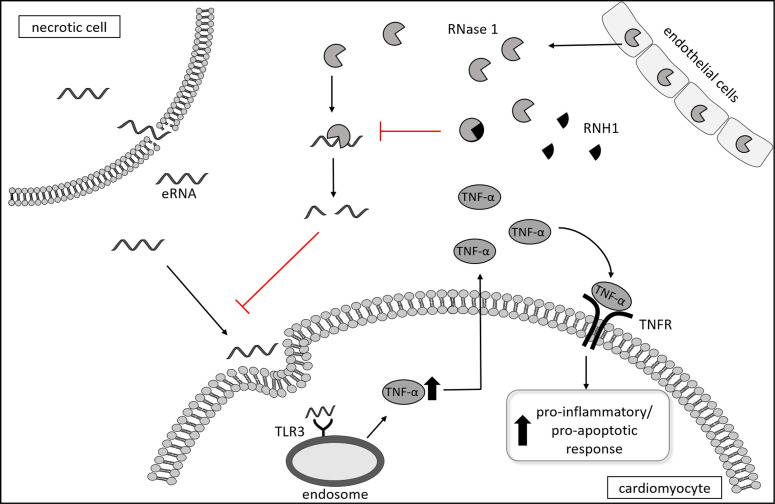
Figure 8. Proposed pathway and pathophysiologic role of eRNA, RNase 1, and RNH1 in septic cardiomyopathy.
eRNA belongs to the heterogeneous group of DAMPs and is released as a result of sepsis-associated tissue injury and necrotic cell death. eRNA binds to the pattern recognition receptors toll-like receptor 3 located on the endosome, which results in the formation of TNF in cardiomyocytes. TNF binds to TNFR and induces pro-apoptotic as well as pro-inflammatory pathways. RNases belong to a group of host-defense peptides secreted from endothelial cells and thereby cleave eRNA. RNH1 binds to RNase 1 and inactivates the host-defense function of RNases, which allows eRNA to bind to TLR. eRNA, extracellular RNA; RNase 1, ribonuclease 1; RNH1, ribonuclease-inhibitor 1; TNFR, TNF receptor.
Here, we report for the first time to our knowledge that RNase 1 attenuates caspase-3 activation and apoptosis in cardiomyocytes exposed to either NC or, as a reverse translational approach, to the serum of septic patients. Previous studies have investigated the therapeutic potential of a reduction/inhibition of apoptosis by direct/specific caspase-inhibition in animal models of cardiomyopathy induced by ischemia/reperfusion and rapid ventricular pacing (32, 33). Cabrera-Fuentes et al. reported that RNase 1 treatment reduces the infarct size caused by myocardial ischemia/reperfusion (34). Furthermore, they measured a reduction in necrotic cell death in ischemic hearts treated with RNase 1 compared vehicle treatment (34). In addition, there is also good evidence that eRNA released from necrotic cells triggers the release of proinflammatory cytokines, whereas RNase 1 reduces the formation of cytokines (23, 24), which play a key role in septic cardiomyopathy and cardiac apoptosis (35, 36). We report here that RNase 1 attenuates TNF mRNA expression in cardiomyocytes exposed to either NC or serum of septic patients. Thus, RNase 1 may attenuate septic cardiomyopathy by reducing both cardiac apoptosis and cytokine production.
To gain a better insight into the roles of RNH1, RNase 1 and eRNA in septic cardiomyopathy in vivo, we used a murine model of polymicrobial sepsis. Having shown increased serum levels of RNase 1 in patients at the early phase (onset) of sepsis (22), we detected significantly lower RNase 1 serum levels in CLP mice (at 24 hours after onset of sepsis). Owing to the very short half-life of RNase 1 as well as the inhibition of RNase 1 by RNH1 may all be reasons, which either individually or in combination, contributing to the differences observed. In line with our findings in human sepsis, we found that the levels of RNH1 are significantly elevated in mice with polymicrobial sepsis (Figure 7C). Based on these findings, we propose that an increase in RNH1 expression results in a decreased RNase 1 expression/enzymatic activity, which results in an increase in circulating eRNA, leading to a release of proinflammatory cytokines (Figure 8). To substantiate this hypothesis, we have investigated the effect of RNase 1 as a therapeutic target on the cardiac dysfunction associated with sepsis. We confirm that the model of polymicrobial sepsis used here, indeed, results in a substantial cardiac dysfunction defined by a significant decrease in EF, FS, and fraction area change (Figure 5). Stieger et al. reported an increase of FS in mice treated with RNase 1 (50 μg/kg) compared with mice treated with saline 24 hours after myocardial infraction (37).We also detected a significant reduction in left ventricular cardiac dysfunction in CLP mice treated with RNase 1, as determined by a substantial increase in EF, FS, fraction area change, and cardiac output (Figure 5). Moreover, we detected a significant reduction in the release of TNF in CLP mice treated with RNase 1 when compared with CLP mice treated with vehicle (Figure 7A). Additionally, we found a negative correlation between the RNase 1 and TNF concentration; higher RNase 1 serum levels were associated with lower TNF levels (Figure 7, E and F). These findings support the hypothesis that eRNA released from necrotic cells triggers the release of proinflammatory cytokines, whereas RNase 1 reduces the formation of cytokines by cleaving eRNA (Figure 8). Notably, treatment of CLP mice with RNase 1 reduced the cardiac dysfunction associated with sepsis.
We also investigated the sepsis-associated degree of cardiac apoptosis in our murine model of polymicrobial sepsis. The induction of apoptosis can be triggered by either the intrinsic or the extrinsic pathway. Several studies have shown that cytochrome C and Apaf-1 form an apoptosome, which activates caspase-9 (38), which, in turn, activates caspase-3. Activated caspase-3 then initiates and drives apoptosis. Indeed, we observed a relevant activation of the intrinsic apoptosis signaling pathway in murine hearts from mice with polymicrobial sepsis, indicated by a significant increase in Bax levels, activation of caspase-9 and caspase-3 and a decrease of Bcl-2 (Figure 6). Furthermore, we measured increased apoptosis induction in heart tissue by TUNEL labeling (Figure 6). In line with our findings, Hotchkiss et al. reported as early as 1999 that apoptosis in the spleen of septic patients also occurs through the intrinsic apoptosis pathway (39). Treatment of CLP mice with RNase 1 resulted in a reduction of the activation of apoptosis in the heart. Most notably, treatment of septic mice with RNase 1 attenuated the rise in cardiac Bax levels, activation of caspase-9 and caspase-3, the decrease in Bcl-2 and TUNEL+ cells caused by sepsis in the heart (Figure 6). In line with our findings, Stieger et al. reported a reduction of apoptosis 24 hours after ligation of the left anterior descending artery (LAD) in mice treated with RNase 1 (37). Treatment with RNase 1 in sham-operated mice did not show any apoptosis induction (Figure 6). These results indicate that excessive amounts of eRNA drive apoptosis in the heart by activating the intrinsic apoptosis pathway. However, in contrast to the eRNA serum levels of septic patients, we could not detect any difference in eRNA expression in CLP mice compared with sham-operated mice (Figure 7D). One possible reason for the different results may be the different time points of the measurements. Whereas eRNA concentration was measured in patients with sepsis 3 days after diagnosis, the concentration of eRNA in mice was measured 24 hours after CLP-sepsis induction. This hypothesis is also confirmed by Stieger and colleagues, who reported increased eRNA levels within 30 minutes and maximal eRNA levels at 2 hours after ligation of the LAD. However, 4 hours after ligation of LAD, the eRNA levels had returned to baseline (37).
In conclusion, our data show for the first time to our knowledge that sepsis results in a significant increase in eRNA and RNH1, the endogenous inhibitor of RNase 1. We found that RNase 1 reduces apoptosis as well as TNF expression in murine cardiomyocytes induced by NC or serum of septic patients. In addition, treatment with RNase 1 reduces the degree of (a) cardiac apoptosis induced by the intrinsic pathway, (b) TNF expression, and (c) cardiac dysfunction caused by polymicrobial sepsis in mice. Based on these findings, we propose that (a) eRNA plays a significant role in the pathophysiology of the organ (cardiac) dysfunction in sepsis and (b) RNase may be a new therapeutic strategy to reduce the cardiac injury and dysfunction caused by sepsis.
Methods
Study population.
The blood samples of septic ICU patients (n = 21) were collected during the first 18 hours and 3 days after diagnosis of sepsis defined by the 2012 Surviving Sepsis Campaign Guidelines using an intravascular catheter (40). Age- and sex-matched healthy subjects were used as controls (n = 10).
Serum sampling.
After 10 minutes of coagulation at room temperature, the serum samples were centrifuged at 2000 g for 10 minutes and stored at –80°C (22). The characteristics of the study population are shown in Supplemental Table 1 according to the groups: healthy subjects, patient with sepsis on the day of diagnosis (Sepsi DD), and patients with sepsis 3 days after diagnosis (Sepsis D3).
ELISAs.
The levels of the RNH1 and RNase 1 in human and mice serum were determined using a commercial ELISA kit (LifeSpan BioSciences). According to the manufacturer, no significant cross-reactivity or interference between Ribonuclease A/RNASE1 and analogs is known. Furthermore, the ELISAs do not distinguish between bound RNase 1 and unbound RNase 1 with RNH1. Mouse TNF-α DuoSet ELISA kit (R&D Systems) was used to measure the TNF serum levels in mice. The optical density was measured at 450 nm using a microplate reader (Tecan Group).
Total eRNA analyzation and screening.
Total eRNA in serum of healthy subjects (n = 10) and septic patients 3 days after diagnosis (n = 21) were analyzed using QuantiFlour RNA System and Quantus Fluorometer (Promega). For RNA size quantification cell-free RNA was isolated from serum of healthy subjects and patients with sepsis by the Quick-cfRNA Serum & Plasma Kit (Zymo Research). The 4200 TapStation system and the high sensitivity RNA ScreenTape Assay (Agilent) were used for analyzing the RNA molecules from 50 to 6000 nt.
Cell culture.
HL-1 cells, a mouse atrial cell line, were provided by William C. Claycomb (Departments of Biochemistry and Molecular Biology and Cell Biology and Anatomy, Louisiana State University Medical Center, New Orleans, Louisiana, USA) (41). Briefly, HL-1 cells were cultured on gelatin/fibronectin-coated plates and maintained in Claycomb-Medium (Sigma-Aldrich) supplemented with 10% fetal calf serum (Biochrom), 0.1 mM norepinephrine (Sigma-Aldrich), 2 mM L-glutamine (Sigma-Aldrich), and 1% penicillin/streptomycin (Invitrogen) under an atmosphere of 5% CO2 at 37°C (42–45). Cardiomyocytes were exposed to 1 × 105 NC per milliliter of medium or to 5% septic patient serum in the presence or absence of 2.8 U/mL RNase 1 (Invitrogen) for 16 hours. Unstimulated cells were used as negative control.
NC.
NC were generated by 3 circles of freezing at –80°C and thawing at 37°C (23). Necrosis was verified by trypan blue staining. Necrotic cells (1 × 106 cells/mL) were stored at –80°C.
RNA extraction of cardiomyocytes.
HL-1 cells were cultivated as described before and harvested by scraping (45). After 3 washing cycles with PBS the RNA was isolated using the NucleoSpin RNA Kit (Macherey-Nagel).
Cell stimulation.
Cardiomyocytes were exposed to 1 × 105 necrotic cardiomyocytes per milliliter medium or to 5% serum of septic patients in the presence or absence of 2.8 U/mL RNase 1 (Invitrogen) for 16 hours. Unstimulated cells were used as negative control. Furthermore, cardiomyocytes were exposed to 100 ng/mL RNA in presence or absence of 2 μM CU CPT 4a (TLR3 inhibitor, Tocris Bioscience).
Immunofluorescence of cleaved caspase-3.
The amount of cleaved caspase-3 was analyzed using immunofluorescence. As described previously (45), HL-1 cells were grown on coverslips and exposed to necrotic cardiomyocytes or serum of septic patients in presence or absence of 2.8 U/mL RNase 1 for 16 hours. Afterward, cells were fixed with 4% formaldehyde (Sigma-Aldrich) for 15 minutes at room temperature. After 3 washing cycles using PBS, cells were blocked with blocking buffer (5% BSA and 0.3% Triton X-100 in PBS) for 1 hour. The cardiomyocytes were, then, incubated with a primary antibody directed against cleaved caspase-3 (Cell Signaling), diluted 1:200 in antibody dilution buffer (1% BSA and 0.3% Triton X-100 in PBS) over night at 4°C. After 3 washing cycles, the cardiomyocytes were incubated with a secondary antibody (Invitrogen) diluted 1:1000 in antibody dilution buffer for 2 hours at room temperature in the dark. Coverslips were washed as described before and covered with ProLong Gold (Invitrogen) containing DAPI for nuclei staining. Immunofluorescence was detected using LSM 710 confocal microscope (Zeiss) and quantified by ImageJ software (NIH).
TUNEL labeling.
As described previously (45), HL-1 cells were grown in 12-well plates on coverslips coated with gelatin/fibronectin. HL-1 cells were fixed with 4% formaldehyde for 1 hour at room temperature after exposure with necrotic cells in the presence or absence of RNase 1 for 16 hours. After washing, cells were permeabilized with 0.1% Triton X-100 for 2 minutes on ice. Next, cells were labeled using the In Situ Cell Death Detection Kit TMR red (Roche) as described (45). Coverslips were covered with 15 μL of ProLong Gold. Unstimulated cells were used as negative control. The paraffin-embedded heart tissues were dewaxed by a series of alcohol and washed twice in PBS. The tissues were permeabilized in the microwave with 0.1 M citrate buffer (pH 6.0) for 5 minutes by 350 W. After cooling down the tissues were labeled also using the In Situ Cell Death Detection Kit TMR red (Roche) and covered with ProLong Gold. An LSM 710 confocal microscope (Zeiss) and ImageJ software were used to detect and quantify apoptotic cells.
Relative TNF mRNA expression.
RNA was isolated using Trizol reagent, as described previously (45). The following primers were used to analyze the relative mRNA expression of TNF in the quantitative real-time PCR (StepOnePlus Real-Time PCR System; Thermo Fisher Scientific, MA, USA): TNF 5′ TCCCCAAAGGGATGAGAAG 3′ (for) and 5′ GCACCACTAGTTGGTTGTC 3′ (rev), S7 5′ GGTGGTCGGAAAGCTATCA 3′ (for) and 5′ AAGTCCTCAAGGATGGCGT 3′ (rev). Relative quantification was performed by using Microsoft Excel.
Flow cytometry.
The amount of cell death was analyzed as described previously by using flow cytometry (45). Stimulated cardiomyocytes were harvested by scraping. After 3 washing cycles with PBS, the supernatant was discarded and cells were resuspended in binding buffer as described in the APC Annexin V Apoptosis Detection Kit (BD Pharmingen). Cells were stained with APC Annexin V Apoptosis Detection Kit and analyzed by FACSCanto-II (BD Biosciences). In addition, renal dysfunction and hepatocellular injury were analyzed in the serum of all mice, as shown in the Supplemental Methods
Caspase-3 activity.
To analyze caspase-3 activity 1.5 × 104 cardiomyocytes/well were seeded in a 96-well white plate. After 24 hours the cells were exposed with 100 μL of medium containing RNA in presence and absence of TLR3 inhibitor. Untreated cells were used as negative control. The caspase-3 activity was analyzed by adding 100 μL of Caspase-Glo 3/7 Assay reagent (Promega Corporation). The luminescence was detected after 30 minutes of incubation at room temperature using microplate reader (Tecan Group).
Murine polymicrobial sepsis model.
For this study, 10-week-old, male C57BL/6 mice were used. CLP was performed to induce polymicrobial sepsis as previously described (46, 47). Surgery was performed under anesthesia by isoflurane (3%) delivered in oxygen at a rate of 1 L/min in an anesthetic chamber and maintained with isoflurane (2%) and oxygen (1 L/min) via a face mask. Buprenorphine (0.05 mg/kg, i.p.) was administered just before starting the surgery to obtain adequate analgesia. Under anesthesia, the abdomen was opened, the cecum was totally ligated below the ileocecal valve and punctured at both opposite ends using an 18-G needle, where a small amount of feces was squeezed out. As control, sham- operated mice (without CLP) were used. For fluid resuscitation, normal saline (0.9% NaCl) was administered into the mouse’s abdomen before closure (5 mL/kg) and administrated directly after surgery (10 mL/kg s.c.). Antibiotics (Imipenem/Cilastatin; 0.25 mg dissolved in fluid resuscitation 7.5 mL/kg 0.9% NaCl, s.c.) and analgesia (Buprenorphine; 0.05 mg/kg, i.p.) were injected 6 and 18 hours after surgery. RNase 1 or its vehicle was administrated before sepsis induction (50 μg/kg RNase 1 or 0.9% NaCl, i.v.), directly after surgery (200 μg/100 μL RNase 1 or 100 μL 0.9% NaCl, s.c.), and 4 hours after surgery (500 μg/100 μL RNase 1 or 100 μL 0.9% NaCl, s.c.). The surgeon was blinded to vehicle or treatment group. After 24 hours, cardiac function was assessed by echocardiography in vivo.
Echocardiography.
Cardiac function was assessed 24 hours after CLP on the Vevo 3100 imaging system (VisualSonics). Briefly, mice were sedated in an anesthetic chamber with isoflurane (3%) delivered in oxygen at a rate of 1 L/min and then transferred onto a thermoregulatory platform in the supine position, where anesthetic was maintained at: isoflurane (0.5%–2.0%) and oxygen (1 L/min). The fur was removed from the chest by Veet hair removal cream and prewarmed echo gel was then placed onto the shaven chest, where the following parameters of cardiac function were measured via the MX550D imaging probe: percentage EF, percentage FS, and percentage FAC. After sacrifice of the animals, the organs and blood were collected for further experiments. All analysis was conducted offline and blinded.
Protein isolation and Western blot analysis.
Semiquantitative Western blot analysis of caspase-3, caspase-9, Bax, and Bcl-2 was performed in murine heart tissue as previously before (48, 49). For protein isolation, the tissue was homogenized in homogenization buffer (20 mM HEPES, pH 7.9, 1 mM MgCl2, 0.5 mM EDTA, 1% Nonidet P-40, 1 mM EGTA, 1 mM DTT, 0.5 mM PMSF, 1 μL/mL PIC) and centrifuged by 18,800 g for 40 minutes at 4°C. The protein concentration of the supernatant was determined using the bicinchoninic acid (BCA) protein assay (Thermo Fisher Scientific). Proteins were separated by size using sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membrane. Unspecific bonds were blocked by incubating the membrane for 1 hour in blocking buffer (10% BSA in TBS containing 0.1% Tween). Following, the membrane was incubated with the primary antibody over night at 4°C diluted in 5% BSA in TBS containing 0.1% Tween. After 3 washing cycles, the membrane was incubated for 30 minutes with the HRP-conjugated secondary antibody. Protein bands were detected with enhanced chemiluminescent (ECL) detection system. The immunoreactive bands were visualized by autoradiography and the densitometric analysis was performed using Gel Pro Analyzer 4.5, 2000 software (Media Cybernetics). Results were normalized by using Tubulin as an endogenous control. The following primary antibodies from Cell Signaling were used: caspase-9 (C9) mouse (catalog 9508), Bcl-2 (D17C4) rabbit (catalog 3498), caspase-3 rabbit (catalog 9662), and Bax (D3R2M) rabbit (catalog 14796).
Statistics.
The statistical analysis was performed with and the graphs were created with GraphPad Prism 7. A 1-way ANOVA followed by Bonferroni test or unpaired 2-tailed t test was used for multiple comparisons with a significance level of P < 0.05. The data represent the mean ± SEM of 3 independent experiments.
Study approval.
All serum samples were collected in a study carried out by the Department of Intensive Care and Intermediate Care at University Hospital RWTH Aachen and were stored in the centralized biomaterial bank (RWTH cBMB) after obtaining informed consent of all study participants and approval by the local ethics committee (University Hospital RWTH Aachen; EK 206/09).
The Animal Welfare Ethics Review Board of the Queen Mary University of London approved the animal experimental protocols used in this study (Procedure Project License: PC5F29685). Animal care was in accordance with the guidelines of the Home Office guidance on Operation of Animals (Scientific Procedures Act 1986) published by Her Majesty’s Stationery Office and the Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011).
Author contributions
EZ, LM, and CT conceived of the manuscript and design. EZ, NW, LM, TS, SE, and GMN conducted all in vitro experiments and analyzed the data. CEO, EZ, LS, LM, CT, SMC, and BW conducted all in vivo experiments and data analyses. BD, TPS, CS, EZ, and LM performed all of the eRNA measurements and analyzed the data. FC, DC, MC, EZ, CT, and LM performed all Western blots and analyzed the data. EZ wrote the manuscript. EZ, GM, LM, and CT revised the manuscript for important intellectual content. All of the authors reviewed and finally approved the manuscript.
Supplementary Material
Acknowledgments
This work was supported by an intramural grant to EZ (START 113/17), a grant from the German Research Foundation to LM (DFG, MA 7082/3-1), and a grant from the Federal Ministry of Education and Research, Germany to SMC (BMBF 03Z22JN12, Research Group Translational Septomics, Centre for Innovation Competence Septomics). CT is funded by the Centre for Diabetic Kidney Disease, which is funded by the Barts and London Charity, and the William Harvey Research Foundation (WHRF). This work was supported by the RWTH centralized Biomaterial Database (RWTH cBMB) of the University Hospital RWTH Aachen and by the Confocal Microscopy Unit, a core facility of the Interdisciplinary Center for Clinical Research (IZKF) Aachen within the Faculty of Medicine at RWTH Aachen University. The authors thank Nadine Frank, Daniel Hinkelmann, and Oliver Krenkel for excellent technical support.
Version 1. 03/26/2020
In-Press Preview
Version 2. 04/23/2020
Electronic publication
Version 3. 01/07/2021
The author contribution notes where updated.
Funding Statement
This work was supported by an intramural grant to EZ.
This work was supported by a grant of the Deutsche Forschungsgemeinschaft to LM.
This work was supported by a grant of the Federal Ministry of Education and Research, Germany to SMC.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(8):e131571.https://doi.org/10.1172/jci.insight.131571.
Contributor Information
Elisabeth Zechendorf, Email: ezechendorf@ukaachen.de.
Lara Stiehler, Email: lara.stiehler@rwth-aachen.de.
Natalie Wischmeyer, Email: n.wischmeyer@t-online.de.
Fausto Chiazza, Email: fausto.chiazza@unito.it.
Debora Collotta, Email: debora.collotta@unito.it.
Bernd Denecke, Email: bernd.denecke@rwth-aachen.de.
Sabrina Ernst, Email: sabrina.ernst@rwth-aachen.de.
Gerhard Müller-Newen, Email: mueller-newen@rwth-aachen.de.
Sina M. Coldewey, Email: S.m.coldewey@qmul.ac.uk.
Bianka Wissuwa, Email: Bianka.Wissuwa@med.uni-jena.de.
Massimo Collino, Email: massimo.collino@unito.it.
Tim-Philipp Simon, Email: tsimon@ukaachen.de.
Tobias Schuerholz, Email: tobias.schuerholz@med.uni-rostock.de.
Christian Stoppe, Email: christian.stoppe@gmail.com.
Gernot Marx, Email: gmarx@ukaachen.de.
Christoph Thiemermann, Email: c.thiemermann@qmul.ac.uk.
Lukas Martin, Email: lmartin@ukaachen.de.
References
- 1.Singer M, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3) JAMA. 2016;315(8):801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.SepNet Critical Care Trials Group. Incidence of severe sepsis and septic shock in German intensive care units: the prospective, multicentre INSEP study. Intensive Care Med. 2016;42(12):1980–1989. doi: 10.1007/s00134-016-4504-3. [DOI] [PubMed] [Google Scholar]
- 3.Sato R, Kuriyama A, Takada T, Nasu M, Luthe SK. Prevalence and risk factors of sepsis-induced cardiomyopathy: a retrospective cohort study. Medicine (Baltimore) 2016;95(39):e5031. doi: 10.1097/MD.0000000000005031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Narváez I, et al. Incidence and evolution of sepsis-induced cardiomyopathy in a cohort of patients with sepsis and septic shock. Med Intensiva. 2018;42(5):283–291. doi: 10.1016/j.medin.2017.08.008. [DOI] [PubMed] [Google Scholar]
- 5.Martin L, et al. The septic heart: current understanding of molecular mechanisms and clinical implications. Chest. 2019;155(2):427–437. doi: 10.1016/j.chest.2018.08.1037. [DOI] [PubMed] [Google Scholar]
- 6.Coldewey SM, et al. Elevation of serum sphingosine-1-phosphate attenuates impaired cardiac function in experimental sepsis. Sci Rep. 2016;6:27594. doi: 10.1038/srep27594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bianchi ME. DAMPs, PAMPs and alarmins: all we need to know about danger. J Leukoc Biol. 2007;81(1):1–5. doi: 10.1189/jlb.0306164. [DOI] [PubMed] [Google Scholar]
- 8.Chan JK, et al. Alarmins: awaiting a clinical response. J Clin Invest. 2012;122(8):2711–2719. doi: 10.1172/JCI62423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zernecke A, Preissner KT. Extracellular Ribonucleic Acids (RNA) enter the stage in cardiovascular disease. Circ Res. 2016;118(3):469–479. doi: 10.1161/CIRCRESAHA.115.307961. [DOI] [PubMed] [Google Scholar]
- 10.Dinger ME, Mercer TR, Mattick JS. RNAs as extracellular signaling molecules. J Mol Endocrinol. 2008;40(4):151–159. doi: 10.1677/JME-07-0160. [DOI] [PubMed] [Google Scholar]
- 11.Creemers EE, Tijsen AJ, Pinto YM. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ Res. 2012;110(3):483–495. doi: 10.1161/CIRCRESAHA.111.247452. [DOI] [PubMed] [Google Scholar]
- 12.Tijsen AJ, Pinto YM, Creemers EE. Circulating microRNAs as diagnostic biomarkers for cardiovascular diseases. Am J Physiol Heart Circ Physiol. 2012;303(9):H1085–H1095. doi: 10.1152/ajpheart.00191.2012. [DOI] [PubMed] [Google Scholar]
- 13.Roy S, Sen CK. MiRNA in innate immune responses: novel players in wound inflammation. Physiol Genomics. 2011;43(10):557–565. doi: 10.1152/physiolgenomics.00160.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fabbri M, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A. 2012;109(31):E2110–E2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim NH, Kang PM. Apoptosis in cardiovascular diseases: mechanism and clinical implications. Korean Circ J. 2010;40(7):299–305. doi: 10.4070/kcj.2010.40.7.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benner SA. Extracellular ‘communicator RNA’. FEBS Lett. 1988;233(2):225–228. doi: 10.1016/0014-5793(88)80431-9. [DOI] [PubMed] [Google Scholar]
- 17.Koczera P, Martin L, Marx G, Schuerholz T. The Ribonuclease A superfamily in humans: canonical rnases as the buttress of innate immunity. Int J Mol Sci. 2016;17(8):E1278. doi: 10.3390/ijms17081278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Futami J, et al. Tissue-specific expression of pancreatic-type RNases and RNase inhibitor in humans. DNA Cell Biol. 1997;16(4):413–419. doi: 10.1089/dna.1997.16.413. [DOI] [PubMed] [Google Scholar]
- 19.Dickson KA, Haigis MC, Raines RT. Ribonuclease inhibitor: structure and function. Prog Nucleic Acid Res Mol Biol. 2005;80:349–374. doi: 10.1016/S0079-6603(05)80009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lomax JE, Bianchetti CM, Chang A, Phillips GN, Fox BG, Raines RT. Functional evolution of ribonuclease inhibitor: insights from birds and reptiles. J Mol Biol. 2014;426(17):3041–3056. doi: 10.1016/j.jmb.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abtin A, et al. Degradation by stratum corneum proteases prevents endogenous RNase inhibitor from blocking antimicrobial activities of RNase 5 and RNase 7. J Invest Dermatol. 2009;129(9):2193–2201. doi: 10.1038/jid.2009.35. [DOI] [PubMed] [Google Scholar]
- 22.Martin L, et al. The Human Host Defense Ribonucleases 1, 3 and 7 are elevated in patients with sepsis after major surgery — A Pilot Study. Int J Mol Sci. 2016;17(3):294. doi: 10.3390/ijms17030294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen C, et al. Role of extracellular RNA and TLR3-Trif signaling in myocardial ischemia-reperfusion injury. J Am Heart Assoc. 2014;3(1):e000683. doi: 10.1161/JAHA.113.000683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng Y, et al. Cardiac RNA induces inflammatory responses in cardiomyocytes and immune cells via Toll-like receptor 7 signaling. J Biol Chem. 2015;290(44):26688–26698. doi: 10.1074/jbc.M115.661835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fischer S, Cabrera-Fuentes HA, Noll T, Preissner KT. Impact of extracellular RNA on endothelial barrier function. Cell Tissue Res. 2014;355(3):635–645. doi: 10.1007/s00441-014-1850-8. [DOI] [PubMed] [Google Scholar]
- 26.Schneck E, et al. Plasma DNA and RNA differentially impact coagulation during abdominal sepsis-an explorative study. J Surg Res. 2017;210:231–243. doi: 10.1016/j.jss.2016.11.044. [DOI] [PubMed] [Google Scholar]
- 27.Gould TJ, et al. Cell-free DNA modulates clot structure and impairs fibrinolysis in sepsis. Arterioscler Thromb Vasc Biol. 2015;35(12):2544–2553. doi: 10.1161/ATVBAHA.115.306035. [DOI] [PubMed] [Google Scholar]
- 28.Kannemeier C, et al. Extracellular RNA constitutes a natural procoagulant cofactor in blood coagulation. Proc Natl Acad Sci U S A. 2007;104(15):6388–6393. doi: 10.1073/pnas.0608647104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garnett ER, et al. Phenotype of ribonuclease 1 deficiency in mice. Rna. 2019;25(8):921–934. doi: 10.1261/rna.070433.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waldenström A, Gennebäck N, Hellman U, Ronquist G. Cardiomyocyte microvesicles contain DNA/RNA and convey biological messages to target cells. PLoS One. 2012;7(4):e34653. doi: 10.1371/journal.pone.0034653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chistiakov DA, Orekhov AN, Bobryshev YV. Cardiac extracellular vesicles in normal and infarcted heart. Int J Mol Sci. 2016;17(1):E63. doi: 10.3390/ijms17010063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laugwitz KL, et al. Blocking caspase-activated apoptosis improves contractility in failing myocardium. Hum Gene Ther. 2001;12(17):2051–2063. doi: 10.1089/10430340152677403. [DOI] [PubMed] [Google Scholar]
- 33.Yaoita H, Ogawa K, Maehara K, Maruyama Y. Attenuation of ischemia/reperfusion injury in rats by a caspase inhibitor. Circulation. 1998;97(3):276–281. doi: 10.1161/01.CIR.97.3.276. [DOI] [PubMed] [Google Scholar]
- 34.Cabrera-Fuentes HA, et al. RNase1 prevents the damaging interplay between extracellular RNA and tumour necrosis factor-α in cardiac ischaemia/reperfusion injury. Thromb Haemost. 2014;112(6):1110–1119. doi: 10.1160/TH14-08-0703. [DOI] [PubMed] [Google Scholar]
- 35.Müller-Werdan U, Engelmann H, Werdan K. Cardiodepression by tumor necrosis factor-α. Eur Cytokine Netw. 1998;9(4):689–691. [PubMed] [Google Scholar]
- 36.Krown KA, et al. Tumor necrosis factor α-induced apoptosis in cardiac myocytes. Involvement of the sphingolipid signaling cascade in cardiac cell death. J Clin Invest. 1996;98(12):2854–2865. doi: 10.1172/JCI119114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stieger P, et al. Targeting of extracellular RNA reduces edema formation and infarct size and improves survival after myocardial infarction in mice. J Am Heart Assoc. 2017;6(6):e004541. doi: 10.1161/JAHA.116.004541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li P, et al. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91(4):479–489. doi: 10.1016/S0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 39.Hotchkiss RS, et al. Apoptotic cell death in patients with sepsis, shock, and multiple organ dysfunction. Crit Care Med. 1999;27(7):1230–1251. doi: 10.1097/00003246-199907000-00002. [DOI] [PubMed] [Google Scholar]
- 40.Dellinger RP, et al. Surviving Sepsis Campaign: international guidelines for management of severe sepsis and septic shock, 2012. Intensive Care Med. 2013;39(2):165–228. doi: 10.1007/s00134-012-2769-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Claycomb WC, et al. HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc Natl Acad Sci U S A. 1998;95(6):2979–2984. doi: 10.1073/pnas.95.6.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin L, et al. Peptide 19-2.5 inhibits heparan sulfate-triggered inflammation in murine cardiomyocytes stimulated with human sepsis serum. PLoS One. 2015;10(5):e0127584. doi: 10.1371/journal.pone.0127584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin L, Koczera P, Zechendorf E, Schuerholz T. The endothelial glycocalyx: new diagnostic and therapeutic approaches in sepsis. Biomed Res Int. 2016;2016:3758278. doi: 10.1155/2016/3758278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin L, et al. Soluble heparan sulfate in serum of septic shock patients induces mitochondrial dysfunction in murine cardiomyocytes. Shock. 2015;44(6):569–577. doi: 10.1097/SHK.0000000000000462. [DOI] [PubMed] [Google Scholar]
- 45.Zechendorf E, et al. Heparan sulfate induces necroptosis in murine cardiomyocytes: a medical-in silico approach combining in vitro experiments and machine learning. Front Immunol. 2018;9:393. doi: 10.3389/fimmu.2018.00393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Al Zoubi S, et al. Linagliptin attenuates the cardiac dysfunction associated with experimental sepsis in mice with pre-existing type 2 diabetes by inhibiting NF-κB. Front Immunol. 2018;9:2996. doi: 10.3389/fimmu.2018.02996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martin L, et al. The synthetic antimicrobial peptide 19-2.5 attenuates septic cardiomyopathy and prevents down-regulation of SERCA2 in polymicrobial sepsis. Sci Rep. 2016;6:37277. doi: 10.1038/srep37277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamada N, et al. Novel synthetic, host-defense peptide protects against organ injury/dysfunction in a rat model of severe hemorrhagic shock. Ann Surg. 2018;268(2):348–356. doi: 10.1097/SLA.0000000000002186. [DOI] [PubMed] [Google Scholar]
- 49.Sordi R, et al. Inhibition of IκB kinase attenuates the organ injury and dysfunction associated with hemorrhagic shock. Mol Med. 2015;21:563–575. doi: 10.2119/molmed.2015.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.