Abstract
Infections due to carbapenem-resistant Klebsiella pneumoniae have emerged as a global threat due to its widespread antimicrobial resistance. Transplant recipients and patients with hematologic malignancies have high mortality rate, suggesting host factors in susceptibility. We developed a model of pulmonary infection using ST258 strain C4, KPC-2 clone, which are predominant K. pneumoniae carbapenemase–producing (KPC-producing) bacteria, and demonstrated that Rag2–/– Il2rg–/– mice — but not WT C57BL/6 or Rag2–/– mice — were susceptible to this opportunistic infection. Using single cell RNA sequencing in infected Rag2–/– mice, we identified distinct clusters of Ifng+ NK cells and Il17a+, Il22+, and inducible T cell costimulatory molecule–positive (ICOS+) group 3 innate lymphoid cells (ILCs) that were critical for host resistance. As solid organ transplantation is a risk factor, we generated a more clinically relevant model using FK506 in WT C57BL/6 mice. We further demonstrated that immunotherapy with recombinant IL-22 treatment ameliorated the ST258 pulmonary infection in both FK506-treated WT mice and Rag2–/– Il2rg–/– mice via hepatic IL-22ra1 signaling. These data support the development of host-directed immunotherapy as an adjunct treatment to new antibiotics.
Keywords: Infectious disease, Pulmonology
Keywords: Bacterial infections, Cytokines, T cells
Introduction
Klebsiella pneumoniae (K. pneumoniae) is one of the leading pathogens in healthcare-associated infections worldwide. The emergence of carbapenem-resistant K. pneumoniae has been globally recognized as a threat due to its rapid increase and limited therapeutic options (1–3). K. pneumoniae consists of approximately 90% carbapenem-resistant Enterobacteriaceae (CRE), and 92% of carbapenem-resistant K. pneumoniae produce the carbapenemase in the United States, which is occupied almost entirely by sequence type 258 (ST258) (4, 5). Carbapenemases are the major cause of expanding resistance; their genes can easily be transmitted by plasmids, and they often confer the resistance to other classes of antimicrobials, including fluoroquinolones and aminoglycosides (6). ST258 is identified by multilocus sequencing of housekeeping genes and has obtained resistance to almost all classes of antibiotics (7). Clinically, this multidrug-resistant pathogen has represented a significant and continued threat to patients, especially in patients with high prevalence of prior hospitalizations and a discharge to long-term care setting (8, 9), and is often associated with a high mortality rate (10). In a multicenter study in New York/New Jersey hospitals, 50% of patients with CRE bacteremia had cancer or history of transplantation, implicating host factors as important risk factors for the infection (4). Interestingly, recent epidemiology has reported a close relationship between ST258 infection and solid organ or stem cell transplant recipients (1).
As further evidence of the opportunistic nature of ST258, this pathogen has been reported to be virtually avirulent for immunocompetent animals and highly susceptible to serum killing in vitro (11). Understanding of the immunological mechanisms of this opportunistic infection is indispensable in exploring counter-measures against this infection and would allow for the development of innovative treatments. Importantly, due to the limitations of small molecule antibiotics, alternative therapies should be considered. For example, various reports suggest that using antibodies for the enhancement of complement-mediated bactericidal activity (7, 12) may be effective against this pathogen. Interestingly, Xiong et al. reported that there are differing requirements for ST258 versus more virulent strains of K. pneumoniae; for ST258 infection, the role of CCR2+ monocytes was essential, whereas neutrophils were dispensable (13).
We initially took a genetic approach using Rag2–/– Il2rg–/– mice and Rag2–/– mice to understand critical host factors for ST258 infection. Single cell RNA sequencing (scRNAseq) revealed that Rag2–/– mice were able to recruit an IFN-γ+ NK cell population and ICOS+IL-17A+IL-22+ group 3 ILCs (ILC3), and both populations were required for resistance to the infection in the Rag2–/– Il2rg–/– background. We next developed a clinically relevant model using FK506, a drug used to manage transplant rejection, and found that this drug renders WT C57BL/6 mice susceptible to ST258 strain C4 infection and was associated with reduction in Il17a, Il22, and Ifng gene expression in the lung. Finally, we confirmed the capability of fusion protein IL-22:Fc to rescue both the genetic and pharmacological model through IL-22ra1 signaling in liver. Thus, these data show that lymphoid cell populations expressing type 1 and type 17 cytokines mediate host resistance to infection and that recombinant IL-22 can improve host defense against this opportunistic infection via hepatic IL-22ra1 signaling.
Results
Il2rg-dependent cells are required for host resistance to ST258 Infection.
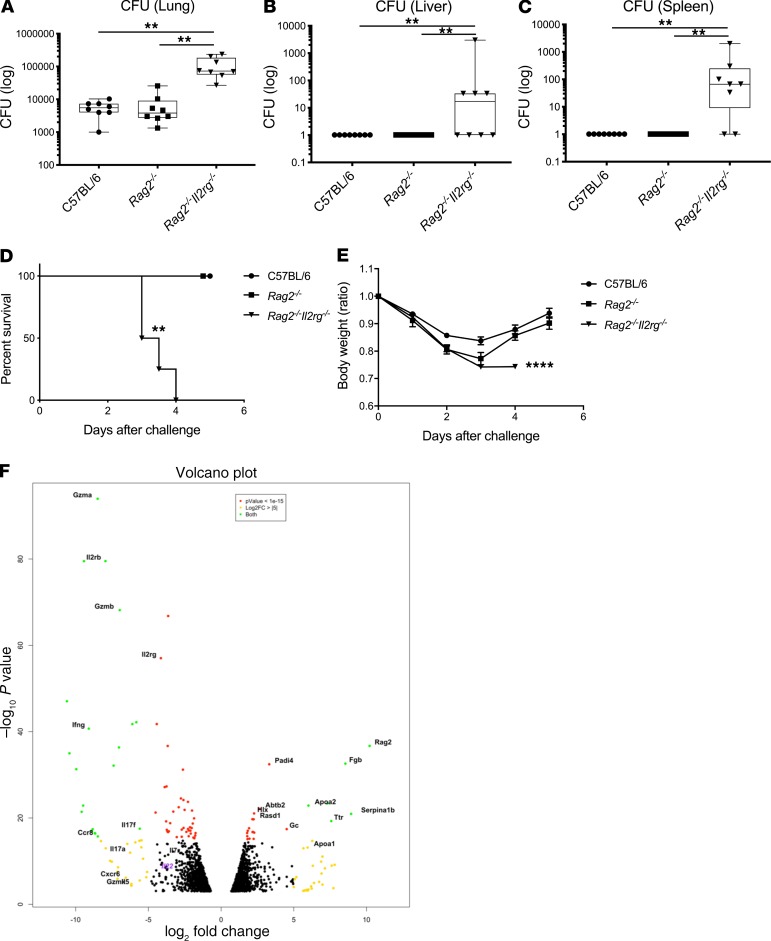
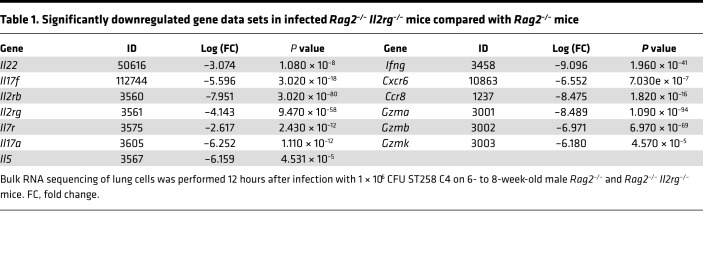
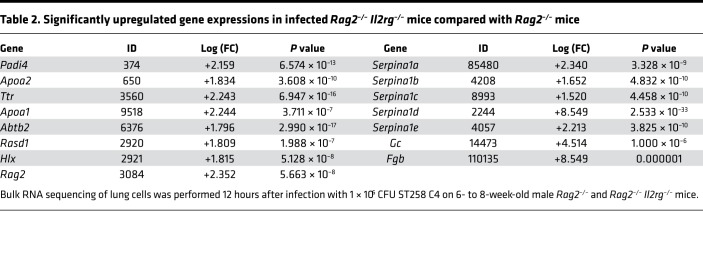
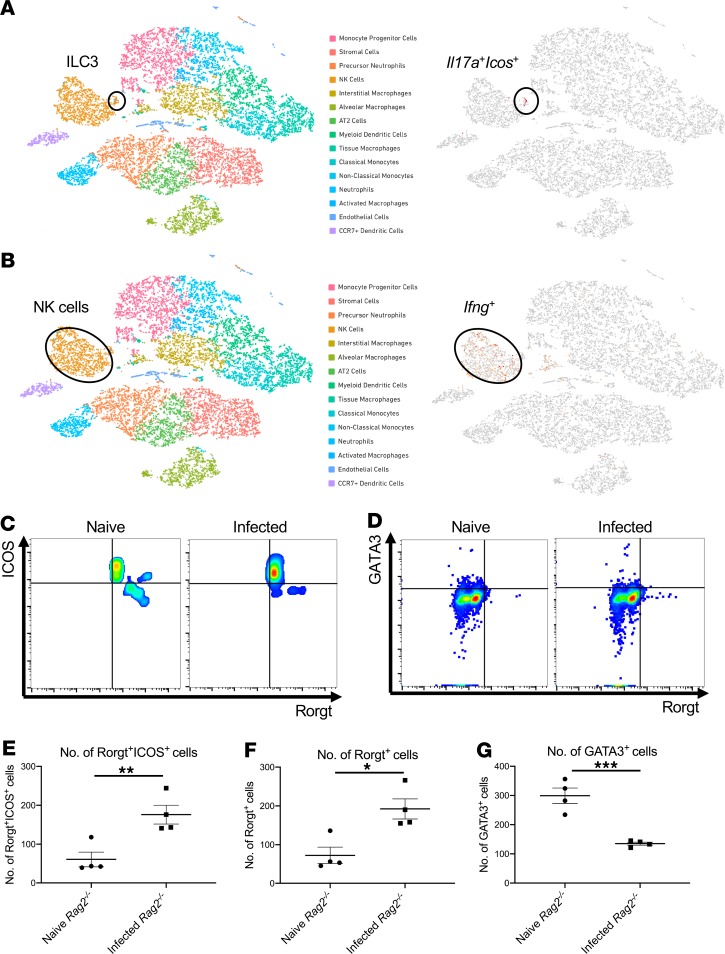
To determine host factors that are required for host resilience to this infection, we developed a model of pulmonary infection using ST258 strain C4, a K. pneumoniae carbapenemase 2–producing (KPC-2–producing) clone that was isolated from bronchoalveolar lavage fluid of a lung transplant patient in 2010 (BioSample, SAMN06445930; SRA, SRS2000639; BioProject PRJNA375812; ref. 14). Rag2–/– Il2rg–/– mice, which lack T, B, NK, and innate lymphoid cells, showed substantially greater bacterial burdens in the lung compared with WT C57BL/6 and Rag2–/– mice at a dose of 1 × 106 CFU ST258 C4 (Figure 1A). Moreover, these mice showed greater bacterial dissemination to the liver (Figure 1B) and spleen (Figure 1C), along with a significant trend toward increased mortality (Figure 1D) and weight loss (Figure 1E). There were no significant differences between C57BL/6 mice and Rag2–/– mice, which suggested that adaptive immunity was not required for initial ST258 C4 pulmonary infection within the observed period. Based on these data, we designed an experiment in which we infected resistant Rag2–/– and susceptible Rag2–/– Il2rg–/– mice with 1 × 106 CFU ST258 C4, euthanized them at 12 hours, and harvested lung tissue for bacterial burden and the other lobe for RNA extraction. At this time point, there were no differences in CFU (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.135591DS1), so any differences in gene expression would not be due to differences in lung bacterial burdens. Bulk RNAseq analysis showed that Rag2–/– Il2rg–/– mice had substantial reduction of genes associated with NK cells (Ifng, Gzma, Gzmb, Gzmk) and ILC3 (Il17a, Il17f, Il22) (Figure 1F and Table 1), as well as significantly downregulated KEGG pathways, including cytokine-cytokine receptor pathways (Supplemental Table 1). These mice also showed several overexpressed genes (Figure 1F and Table 2), and pathway analysis also elucidated upregulated KEGG pathways (Supplemental Table 1). To confirm the downregulated genes at the single cell level, we performed scRNAseq in infected Rag2–/– mice, which showed distinct clusters of Il17a+, Il22+, and Icos+ ILC3 (Figure 2A) and Ifng+ NK cells (Figure 2B), confirming that these cytokines are expressed by distinct cell populations in the lung. Because ICOS expression was largely restricted to the ILC3 cluster, we confirmed that ICOS surface expression was detectable in ILC3 in the ST258 infection model (Figure 2, C and E). Moreover, we observed an increase of Rorgt+ cells in the lung 12 hours after ST258 C4 infection in Rag2–/– mice by FACS (Figure 2, D and F). RNAseq data were also confirmed by quantitative PCR (qPCR) on ILCs and NK cells in naive Rag2–/– and infected Rag2–/– mice. These data demonstrate a significant increase in lung Il17a (Supplemental Figure 2A), Il17f (Supplemental Figure 2B), Il22 (Supplemental Figure 2C), Ifng (Supplemental Figure 2D), and Rorc (Supplemental Figure 2E) gene expression in infected Rag2–/– mice. Additional analysis of the scRNAseq data demonstrated that the Icos+Il17a+ (Supplemental Figure 3A) cell cluster also expressed Il22 (Supplemental Figure 3B), Ccr6 (Supplemental Figure 3C), Rora (Supplemental Figure 3D), and Rorc (Supplemental Figure 3E). We also detected Ncr1 (Supplemental Figure 3F) expression in a subgroup of Il17a+Rorc+ cells, but most of the Ncr1 expression was clustered in the NK cell cluster. The relative lack of Ncr1 expression might be due to a lack of transcriptome coverage in the ICL3 population (15). Also, we could detect an ILC2 population in infected Rag2–/– mice (Supplemental Figure 3G), while the increase of GATA3-producing cells in lung of infected Rag2–/– mice was not observed by FACS (Figure 2, D and G). Thus, Il2rg-dependent ILC3 and NK cells might be essential for the ST258 C4–infected Rag2–/– mice.
Figure 1. Rag2–/– Il2rg–/– mice are susceptible to the ST258 C4 pulmonary infection and lack type1 and type3 cytokine expression.
C57BL/6 mice, Rag2–/– mice and Rag2–/– Il2rg–/– mice were infected intratracheally with 1 × 106 CFU ST258 C4 and euthanized at 24 hours. (A–C) Bacterial CFU in lungs (A), liver (B), and spleen (C) are representative as box and whisker plots showing median, first and third quartiles, and maximum and minimum values (n = 8, two independent experiments). Significant differences are designated by using Kruskal-Wallis test followed by Dunn’s multiple comparisons test. **P < 0.01 (D) Kaplan–Meier survival curves of mice infected with 1 × 107 CFU ST258 C4. Significant differences were designated by log-rank test (n = 5, 2 independent experiments). **P < 0.01 versus C57BL/6 and Rag2–/– mice. (E) Weight change for comparing C57BL/6 mice, Rag2–/– mice, and Rag2–/– Il2rg–/– mice were also observed (n = 4–5, two independent experiments). Significant differences are designated by using 1-way ANOVA followed by Tukey’s multiple comparisons test. ****P < 0.0001 versus C57BL/6 and Rag2–/– mice. (F) Volcano plot of differentially expressed genes in lungs between infected Rag2–/– versus Rag2–/– Il2rg–/– mice 12 hours after infection (n = 3).
Table 1. Significantly downregulated gene data sets in infected Rag2–/– Il2rg–/– mice compared with Rag2–/– mice.
Table 2. Significantly upregulated gene expressions in infected Rag2–/– Il2rg–/– mice compared with Rag2–/– mice.
Figure 2. scRNAseq analysis reveals ICOS– NK cells and ICOS+ group 3 innate lymphoid cells in the lung after infection.
scRNAseq was performed 12 hours after infection with 1 × 106 CFU ST258 C4 on 6- to 8-week-old male Rag2–/– lung cells using CD45 selection after depletion of Ly6G+ cells (n = 2). (A and B) t-SNE plots of scRNAseq data revealed that Rag2–/– mice have a distinct group 3 ILC population with preferential expression of ICOS and Il17a (A), whereas Ifng was primarily confined to the NK cells cluster (B). (C and E) Representative FACS plots and the quantification showed surface ICOS expression in both naive and infected lung. (D, F, and G) Rorgt+ ILC3 (D and F) in the lung increased post infection whereas GATA3+ ILC2 (D and G) cells decreased. Flow gating strategies for Rorgt+ or GATA3+ cells are conducted as CD45+CD127+Lin–. Data are presented as mean ± SEM (n = 4, two independent experiments). Significant differences are designated using unpaired t test. *P < 0.05, **P < 0.01, ***P < 0.001.
Requirement of NK cells and ILC3 for host resistance to ST258 infection.
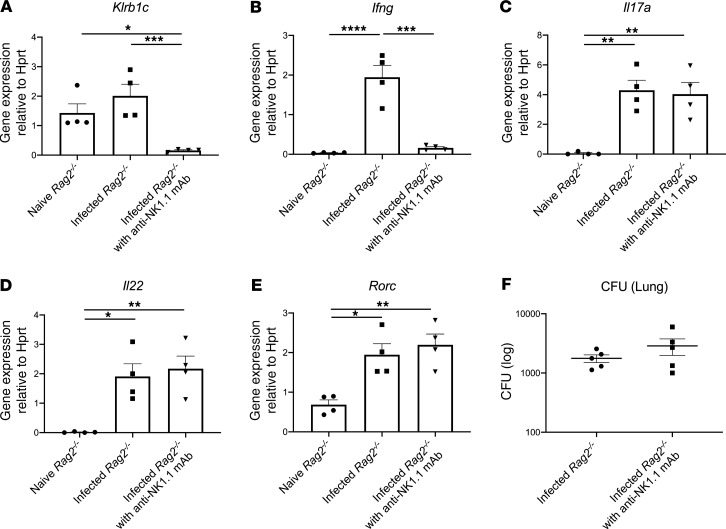
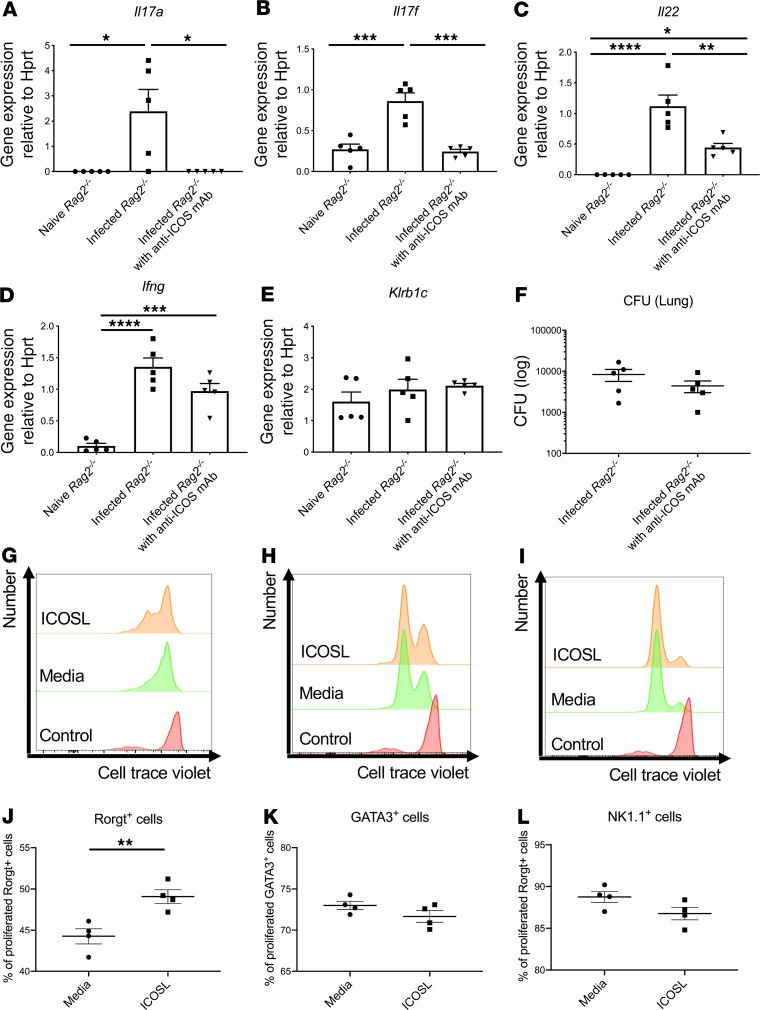
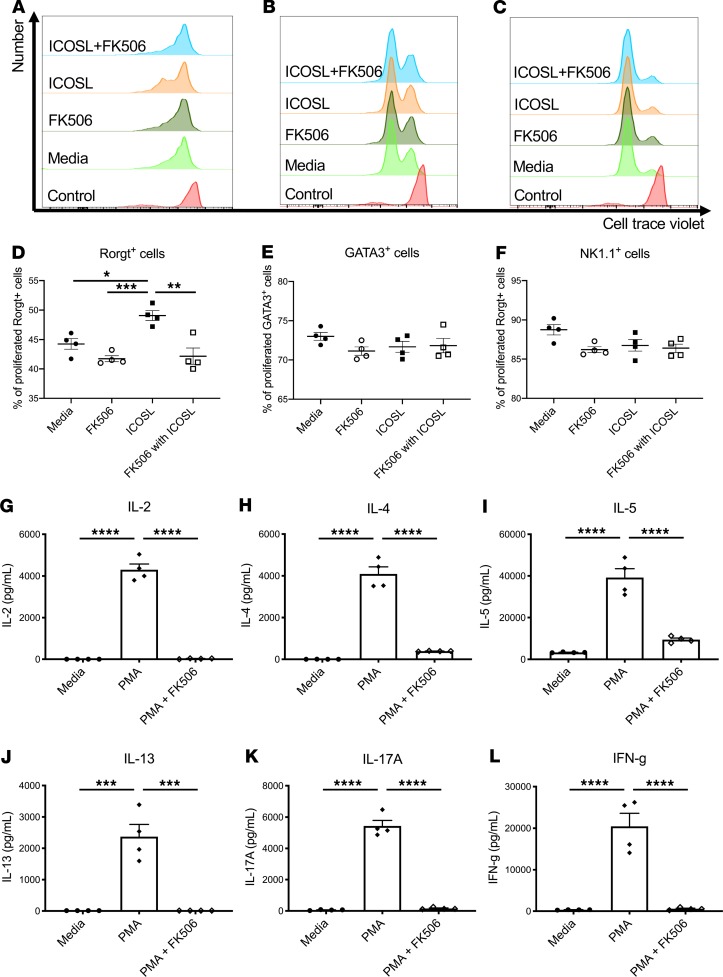
To determine the relative contributions of NK cells versus ILC3 to host resistance, we first depleted NK cells in Rag2–/– mice with anti-NK1.1 antibody. Anti-NK1.1 antibody administration 2 hours prior to infection completely depleted NK cells in the lung, as measured by Klrb1c gene expression (Figure 3A), and reduced Ifng gene expression (Figure 3B) to essentially undetectable levels. NK cell depletion had no effect on the expression of Il17a, Il22, or Rorc (Figure 3, C–E). Interestingly, NK cell depletion did not perturb bacterial clearance (Figure 3F). These data suggest that NK cells were dispensable for host defense against ST258 C4 K. pneumoniae pulmonary infection. As mentioned above, mining our scRNAseq data, we determined that ICOS expression was confined largely to the ILC3 cluster and absent in the NK cell cluster (Figure 2, A and B). To determine if ICOS was required for ILC3 activation, we administered 500 μg of a neutralizing anti-ICOS antibody i.p. 2 hours prior to ST258 C4 pulmonary infection. Neutralization of ICOS substantially reduced Il17a, Il17f, and Il22 expression (Figure 4, A–C) but resulted in retained expression of Klrb1c and Ifng gene expression (Figure 4, D and E), demonstrating an essential role of ICOS in the ILC3 response but not the NK cell response. However, like NK1.1 depletion, anti-ICOS antibody did not perturb bacterial clearance (Figure 4F). Anti-ICOS–treated animals showed a small decrease in Icos gene expression by qPCR (Supplemental Figure 4A); thus, in addition to neutralization, anti-ICOS mAb may also affect the number of ICOS+ cells. To determine if anti-ICOS affected ILC2 activation, we assessed Il5 and Prg2 gene expression in whole lung. In contrast to reduced Il17a and Il22 expression with in vivo anti-ICOS mAb, neutralization of ICOS did not affect Il5 gene expression (Supplemental Figure 4B) or Prg2 gene expression (a marker of eosinophils; Supplemental Figure 4C) (16). Moreover FACS analysis showed no decrement of IL-5+ICOS+ cells (Supplemental Figure 4, D and F) in infected lungs of anti-ICOS–treated Rag2–/– mice. In contrast, IL-17A+ICOS+ cells were decreased (Supplemental Figure 4, E and G), suggesting the anti-ICOS mAb selectively suppressed ILC3 at this time point in infection. Because we observed a reduction in Icos gene expression in the lungs of infected anti-ICOS–treated Rag2–/– mice, we hypothesized that ICOS may provide a proliferative signal to ILC3 in the model. Ex vivo cultures of lung ILC (obtained 12 hours after infection in Rag2–/– mice) with recombinant ICOSL showed enhanced proliferation of lung ILC3 (Rorgt+ cells; Figure 4, G and J), but no effect was observed in lung ILC2 (GATA3+ cells; Figure 4, H and K) or lung ILC1 (NK1.1+ cells; Figure 4, I and L).
Figure 3. Depletion of NK cells with antibody reveals these cells are dispensable for host resistance to ST258 infection.
Six- to 8-week-old male Rag2–/– mice were administered 250 μg anti-NK1.1 antibody or isotype control once 2 hours prior to intratracheal challenge with 1 × 106 CFU ST258 C4 strain and euthanized 24 hours after infection. (A–F) Klrb1c (A), Ifng (B), Il17a (C), Il22 (D), and Rorc (E) mRNA expression in lung, and then lung CFU (F) 24 hours after infection are shown. Data are presented as mean ± SEM (n = 4–5, data shown representative of 2 separate experiments). Significant differences are designated using 1-way ANOVA followed by Tukey’s multiple comparisons test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Figure 4. Effect of ICOS blockade on group 3 ILCs during lung ST258 C4 infection.
Six- to 8-week-old male Rag2–/– mice were administered 500 μg anti-ICOS or control once 2 hours prior to intratracheal challenge with 1 × 106 CFU ST258 C4 strain and euthanized 24 hours after infection. (A–E) Gene expression in lung of Il17a (A), Il17f (B), and Il22 (C) were reduced, but not Ifng (D) or Klrb1c (E). (F) CFU in the lung was not affected by anti-ICOS. Data are presented as mean ± SEM (n = 5, two independent experiments). Significant differences are designated using 1-way ANOVA followed by Tukey’s multiple comparisons test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. (G–I) Ex vivo effects of ICOSL (1.5 μg/mL) on innate lymphoid cells (G and H) and NK cell (I) proliferation (as assayed by dye dilution) obtained from Rag2–/– mice 12 hours after infection (n = 4, two independent experiments). Flow gating strategies are conducted as CD45+CD127+Lin–Rorgt+ for Rorgt+ cells, CD45+CD127+Lin– GATA3+ for GATA3+ cells, and CD45+CD127+Lin–NK1.1+ for NK1.1+ cells. Representative histograms are showing Rorgt+ cells (G), GATA3+ cell (H), and NK1.1+ cells (I) gated. (J–L) Percentage of dye-diluted Rorgt+ cells (J), GATA3+ cell (K), and NK1.1+ cells (L) are shown. Significant differences are designated by using unpaired t test. **P < 0.01.
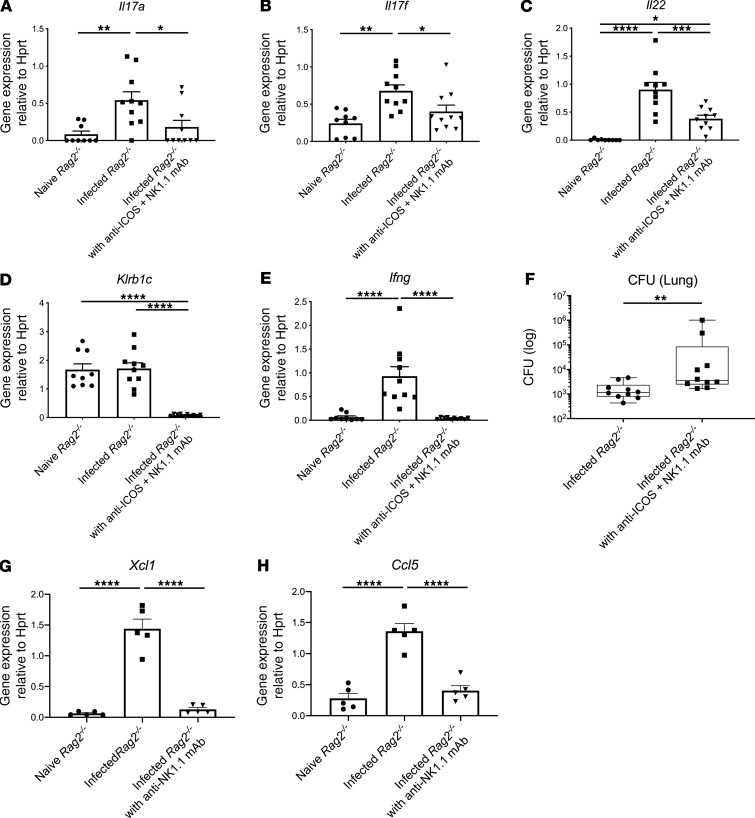
However, when both anti-NK1.1 and anti-ICOS antibody were administered in this infection model, we observed reduction of gene expression in both type 17 (Figure 5, A–C) and type 1 (Figure 5, D and E) cytokines, resulting in exacerbation in lung infection (Figure 5F). These data suggest a cooperativity of NK cells and ICOS+ ILC in ST258 clearance. In addition to Ifng, the scRNAseq data showed that NK cells also express Xcl1 and Ccl5, which was confirmed by qPCR (Figure 5, G and H). CCL5 can be chemotactic for DCs (17), and XCL1 has been also reported to have some activity on neutrophils (18), so these chemokines may have an additional role in host immunity in this model.
Figure 5. Dual blockade of ICOS and NK cells exacerbates lung infection.
Six- to 8-week-old male Rag2–/– mice were administered both 500 μg anti-ICOS and 250 μg anti-NK1.1 or isotype control once 2 hours prior to intratracheal challenge with 1 × 106 CFU ST258 C4 strain and euthanized 24 hours after infection. (A–F) Dual blockade reduced mRNA expression of Il17a (A), Il17f (B), Il22 (C), Klrb1c (D), and Ifng (E) and significantly increased CFU in the lung (F) (n = 9–10). Data are pooled from 3 independent experiments. (G and H) Additionally Xcl1 (G) and Ccl5 (H) mRNA expression in the naive Rag2–/–, infected Rag2–/–, and infected Rag2–/– pretreated with 250 μg anti-NK1.1 mAb i.p. once 2 hours prior to infection are shown (n = 5, data shown representative of 2 separate experiments). Data are presented as mean ± SEM. Significant differences are designated by using 1-way ANOVA followed by Tukey’s multiple comparisons test (A–E, G-H), while Mann-Whitney U test is used for the CFU comparison. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
FK506 increases host susceptibility to ST258 infection.
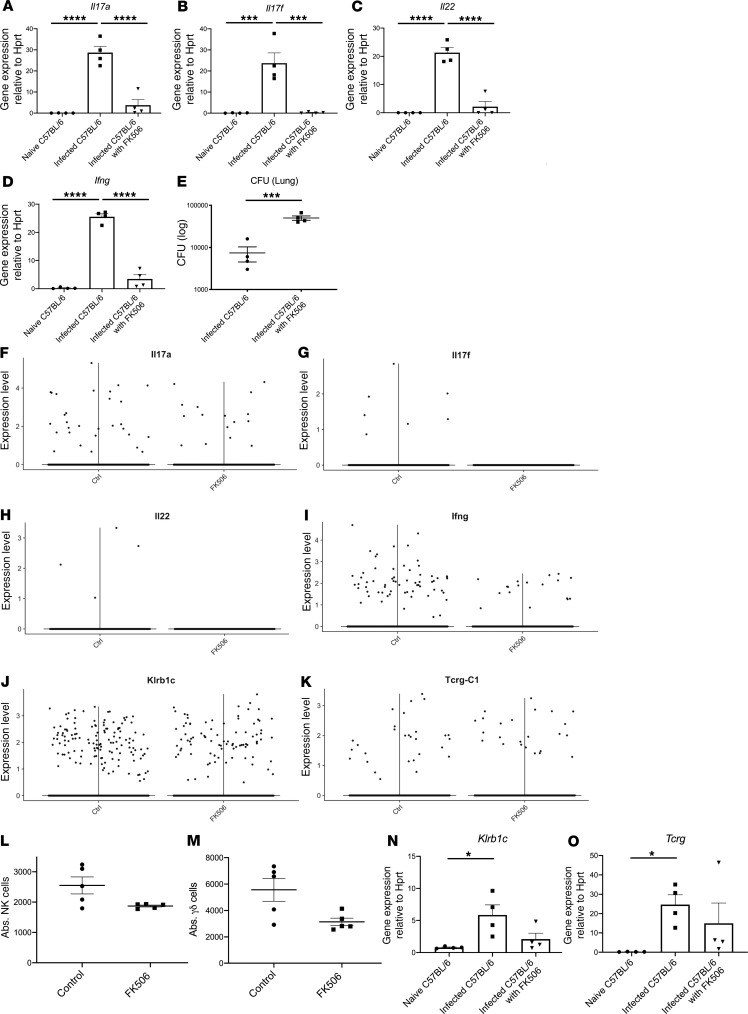
The genetic models have allowed us to identify key cellular events that are critical for host resistance against ST258 K. pneumoniae pulmonary infection; however, they do not mimic important clinical factors that may increase host susceptibility. To develop a more clinically relevant model, we investigated if FK506 — a clinically used calcineurin inhibitor — could be used to model susceptibility to infection in solid organ transplant recipients, like the patient from whom this strain C4 was identified. In these experiments, C57BL/6 mice were administered 10 mg/kg FK506 i.p. 24 hours prior to ST258 infection and every 24 hours until 24 hours before euthanasia. FK506, in contrast to vehicle control, substantially reduced the gene expression of Il17a, Il17f, Il22, and Ifng (Figure 6, A–D) and exacerbated bacterial burden in the lung 48 hours after infection (Figure 6E). To determine cellular sources of cytokines in this model, we performed scRNAseq comparing the FK506-treated C57BL/6 mice and WT mice at 24 hours after infection with 1 × 106 CFU ST258 C4. There was no significant difference of CFU at 24 hours, so we conducted scRNAseq at this time point to perform an unbiased comparison of gene expression. The violin plots from Seurat of scRNAseq data showed that FK506 treatment reduced both type 1 and type 17 cytokine genes, including Il17a, Il17f, Il22, and Ifng (Figure 6, F–I) — but not Klrb1c (the major expresser of Ifng) and Tcrg (the major expresser of Il17a, Il22) (Figure 6, J and K). Similarly, FACS analysis also showed that the absolute number of NK cells and γδ cells at 24 hours after 1 × 106 CFU ST258 C4 pulmonary infection was modestly decreased in FK506-treated C57BL/6 mice, but this was not statistically significant (Figure 6, L and M). Additionally, the Klrb1c and Tcrg gene expression were not so changed (Figure 6, N and O). T-distributed stochastic neighbor embedding (t-SNE) plots from the scRNAseq data in ST258 C4–infected C57BL/6 mice with FK506 treatment showed that Ifng expression was mostly observed in NK cells (Supplemental Figure 5A), whereas Il17a- and Il22-expressing cells were clustered in γδ T cells (Supplemental Figure 5B). Pathway analysis was also performed, and significant KEGG pathways are shown (Supplemental Table 2). To examine if FK506 also affected ILC responses, we performed an ex vivo analysis using lung ILCs from Rag2–/– mice (obtained 12 hours after infection with ST258 C4) and incubated these cells with vehicle or FK506. FK506 showed an inhibitory effect on proliferation in ILC3 (Rorgt+ cells; Figure 7, A and D) but not in lung ILC2 (GATA3+ cells; Figure 7, B and E) or ILC1 (NK1.1+ cells; Figure 7, C and F). Moreover, FK506 suppressed the phorbol 12-myristate 13-acetate (PMA) elicited type 1, type 2, and type 17 cytokine secretion in lung ILCs ex vivo in a cell-intrinsic manner (Figure 7, G–L). In addition, we examined the effect of FK506 on myeloid cells. FK506 treatment in Rag2–/– Il2rg–/– mice showed a slight exacerbation in lung CFU, but this was not statistically significant (Supplemental Figure 6A). Moreover bacterial dissemination to the liver and spleen of FK506-treated Rag2–/– Il2rg–/– mice was similar to untreated Rag2–/– Il2rg–/– mice (Supplemental Figure 6, B and C). These data suggest that the major effects of FK506 are on T cells and ILC3. In support of this, scRNAseq data in ST258 C4–infected Rag2–/– mice showed about 8.1% in Ly6g+ cells (Supplemental Figure 6D) and 60.7% in CD68+ cells (Supplemental Figure 6E) express Fkbp1a+, the target of FK506. In contrast, 98.7% in Rorc+ cells and 57.9% in Klrb1c+ cells expressed Fkbp1a, by scRNAseq.
Figure 6. Pharmacological model of host KPC susceptibility with FK506 mimics the susceptibility in Rag2–/– Il2rg–/– mice.
Six- to 8-week-old male C57BL/6 mice were treated with vehicle control or 10 mg/kg FK506 i.p. 24 hours prior to intratracheal challenge with 1 × 106 ST258 C4 strain. FK506 was administered every 24 hours until 24 hours before euthanasia. Lungs were harvested 48 hours after infection. (A–E, N, and O) Il17a (A), Il17f (B), Il22 (C), Ifng (D), Klrb1c (N), and Tcrg (O) mRNA expression, and CFU in the lungs (E) of naive C57BL/6, infected C57BL/6, and infected C57BL/6 mice treated with FK506 are shown (n=4, two independent experiments). Expression of mRNA was normalized to the constitutive expression of Hprt mRNA, and data are presented as mean ± SEM. (L and M) The absolute number of lung NK cells (L) and lung γδ cells (M) were also examined by FACS 24 hours after pulmonary challenge in 6- to 8-week-old male C57BL/6 mice treated with vehicle control or FK506. Data are presented as mean ± SEM (n=5, two independent experiments). Significant differences are designated by using 1-way ANOVA test followed by Tukey’s multiple comparisons test. *P < 0.05, ***P < 0.001, ****P < 0.0001. CFU was compared by using Mann-Whitney U test. ***P < 0.001 versus vehicle control. Six- to 8-week-old male FK506-treated WT C57BL/6 mice were infected with 1 × 106 CFU ST258 C4 strain intratracheally; 24 hours after inoculation, lungs were harvested to analyze single cell RNA sequencing. A total of 10 mg/kg FK506 was administered i.p. 24 hours prior to intratracheal challenge and every 24 hours until 24 hours before euthanasia. (F–K) The violin plots from Seurat of scRNAseq data showed FK506 treatment affected to reduce various cytokine gene expression such as Il17a (F), Il17f (G), Il22 (H), Ifng (I), but not Klrb1c (J) and Tcrg (K).
Figure 7. FK506 suppresses ILC function in vitro.
Six- to 8-week-old male Rag2–/– mice were infected with 1 × 106 CFU ST258 C4 strain intratracheally, as well as 12 hours after inoculated lungs were harvested and enriched for innate lymphoid cells for analyzing via flow cytometry. The lung ILCs were incubated for 24 hours with or without ICOSL (1.5 μg /mL) and FK506 (50 ng/mL) (n = 4). Afterward, flow cytometry was performed. Flow gating strategies were conducted as CD45+CD127+Lin– Rorgt+ for Rorgt+ cells, CD45+CD127+Lin– GATA3+ for GATA3+ cells, and CD45+CD127+Lin– NK1.1+ for NK1.1+ cells. (A–C) Representative histograms showing Rorgt+ cells (A), GATA3+ cell (B), and NK1.1+ cells (C) gated. (D–F) Percentage of dye diluted in Rorgt+ cells (D), GATA3+ cell (E), and NK1.1+ cells (F) are shown. Moreover, we examined to see if FK506 could suppress the production of cytokine in ILCs activated by phorbol 12-myristate 13-acetate (PMA; 40 ng/mL) with ionomycin (4 μg /mL) for 24 hours in vitro (n = 4). (G–L) The supernatant was assessed for IL-2 (G), IL-4 (H), IL-5 (I), IL-13 (J), IL-17A (K), and IFN-γ (L) by Luminex. Data are presented as mean ± SEM, and representative data are shown of 2 separate experiments. Significant differences are designated by using ANOVA followed by Tukey’s multiple comparisons test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
IL-22 but not IFN-γ rescues pulmonary immunity to ST258 infection.
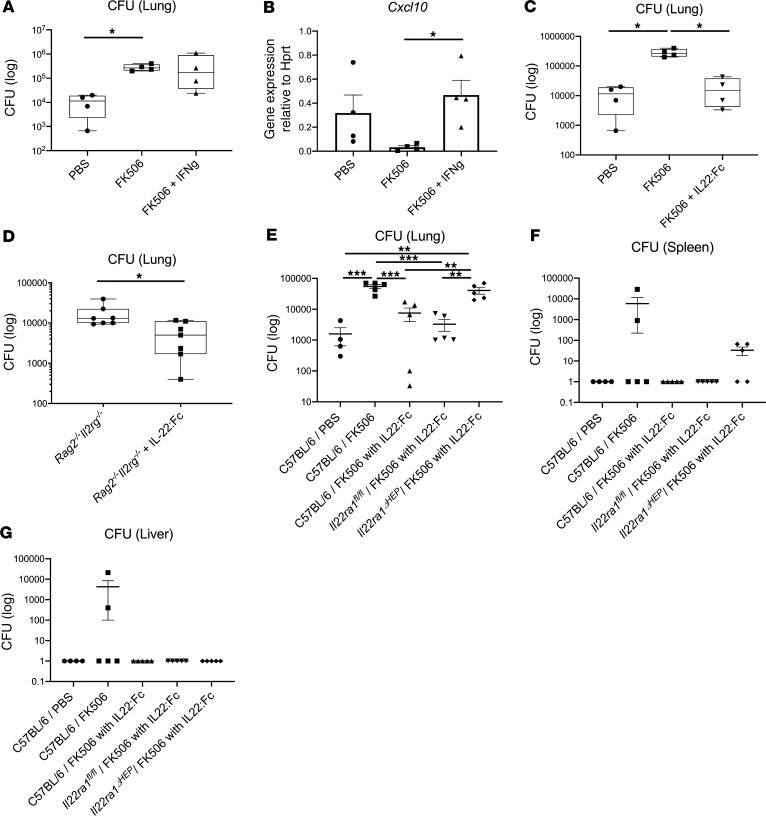
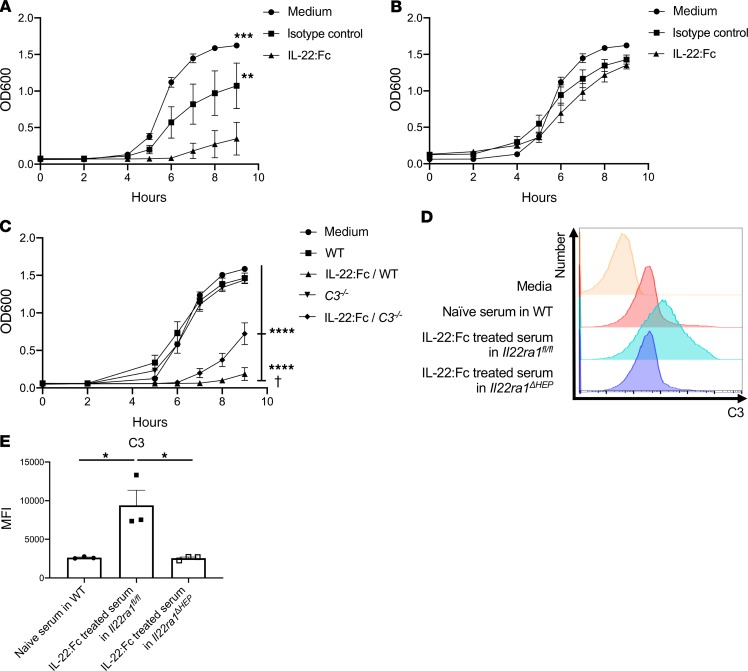
To determine the potential therapeutic benefit of cytokine immunotherapy, we examined if local IFN-γ or systemic IL-22 could be used as immunotherapy. Recombinant IFN-γ was ineffective in augmenting host resistance to infection (Figure 8A) despite evidence of its bioactivity, as measured by CXCL10 induction (Figure 8B) (19, 20). In contrast, recombinant IL-22:Fc administration 2 hours prior to infection in FK506-treated C57BL/6 mice substantially reduced bacterial burden in the lung (Figure 8C). IL-22:Fc was also effective in reducing bacterial burden in Rag2–/– Il2rg–/– mice (Figure 8D). IL22ra1 is expressed on airway epithelium (21) and on hepatocytes (22). To determine if hepatocyte signaling was required, we examined the effect of IL-22:Fc in mice with a conditional deletion of Il22ra1 in hepatocytes (23). In contrast to AlbCre–/– x Il22ra1fl/fl (Il22ra1fl/fl) mice, IL-22:Fc effects on bacterial control in the lung and spleen was substantially abrogated in AlbCre–/+ x Il22ra1fl/fl (Il22ra1ΔHEP) mice (Figure 8, E–G) (23, 24). Furthermore, we performed an experiment administering 1 μg IL-22:Fc by an oropharyngeal aspiration tongue-pull technique (hereafter referred to as intratracheally) once 30 minutes before infection in FK506-treated mice. Intratracheal administration of IL-22:Fc also ameliorated infection as measured by bacterial CFU in Il22ra1fl/fl mice, but the efficacy again was abrogated in Il22ra1ΔHEP mice, suggesting the efficacy might be attributed to the systemic circulation (Supplemental Figure 7). We next assessed serum killing or growth inhibition of ST258 C4 ex vivo. We observed that serum from parenteral IL-22:Fc–treated C57BL/6 mice could inhibit the growth of ST258 C4 (Figure 9A), and this effect was lost with heat inactivation consistent with complement (Figure 9B). The bacteriostatic effect by IL-22:Fc treatment was diminished in C3–/– mice compared with the WT mice (Figure 9C). With regard to these results, we could speculate that the efficacy was generated partially by C3 activity. In confirmation of this, IL-22:Fc serum in Il22ra1fl/fl mice showed greater C3 deposition by FACS on ST258 C4 after coincubation ex vivo, but the efficacy was abrogated in Il22ra1ΔHEP mice (Figure 9, D and E). These data suggest the IL-22–mediated augment of the immunity to ST258 pulmonary infection is largely dependent on hepatocytes, especially via acceleration of C3 deposition.
Figure 8. IL-22:Fc treatment rescued ST258 pulmonary infection in susceptible mice.
(A) A total of 1 μg IFN-γ was administered by intratracheal injection once 2 hours prior to infection in FK506-treated C57BL/6 mice, and lung CFU was measured 48 hours after ST258 infection (n = 4). (B) CXCL10 induction was analyzed in lung tissue after IFN-γ administration (n = 4). (C) A total of 4 μg IL-22:Fc was administered by i.p. injection once 2 hours prior to infection in FK506-treated C57BL/6 mice, and lung CFU was measured 48 hours after ST258 infection (n = 4). (A–C) Significant differences are designated by using Kruskal-Wallis test followed by Dunn’s multiple comparisons test. *P < 0.05. (D) A total of 4 μg IL-22:Fc was administered by i.p. injection once 2 hours prior to infection in Rag2–/– Il2rg–/– mice, and lung CFU was measured 24 hours after ST258 infection (n = 7). Significant differences are designated by using Mann-Whitney U test. Box and whisker plots show median, first and third quartiles, and maximum and minimum values. (E–G) A total of 4 μg IL-22:Fc was administered by i.p. injection once 2 hours prior to infection in FK506 treated C57BL/6, AlbCre–/– x Il22ra1fl/fl (Il22ra1fl/fl), and AlbCre–/+ x Il22ra1fl/fl (Il22ra1ΔHEP) mice for comparing to C57BL/6 mice treated with PBS or FK506 (n=5) and euthanized 48 hours after infection. Data are presented as mean ± SEM. Significant differences are designated by using ANOVA followed by Tukey’s multiple comparisons test. *P < 0.05, **P < 0.01, ***P < 0.001. All data are shown representative of 2 separate experiments.
Figure 9. IL-22:Fc–mediated bacteriostatic activity is partially dependent on complement produced by hepatocyte.
(A and B) A total of 4 μg IL-22:Fc–treated serum for 24 hours in WT C57BL/6 mice augmented bacteriostatic activity against ST258 C4 (A), and the bacteriostatic effect proved to be heat labile by using heat-inactivated serum (B). (C) The bacteriostatic effect by IL-22:Fc treatment was diminished in C3–/– mice more than WT mice. Lastly, 4- to 8-week-old male Il22ra1fl/fl mice and Il22ra1ΔHEP mice were treated with 4 μg IL-22:Fc i.p. for 24 hours, and blood were collected. (D and E) IL-22:Fc–treated serum in Il22ra1fl/fl mice results in more deposition of C3 binding to ST258 C4 after coincubation with 1 × 105 CFU ST258 C4 in vitro, but the effect was abrogated in Il22ra1ΔHEP mice. Data are presented as mean ± SEM. Representative data are shown of 2 separate experiments. Significant differences are designated by using ANOVA followed by Tukey’s multiple comparisons test. †P < 0.05 (vs. IL-22:Fc–treated C3–/– mice), ****P < 0.0001 (vs. medium, WT, C3–/– mice). *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
We show that Rag2–/– Il2rg–/– mice were susceptible to ST258 C4 pulmonary infection, while Rag2–/– and WT C57BL/6 mice were resistant to the infection. Both Rag2–/– and Rag2–/– Il2rg–/– mice have mutations in the rag gene, and they cannot form mature B or T cell receptors (25). Additionally, Rag2–/– Il2rg–/– mice also have deletions in the Il2rg gene, which causes this model to lack ILCs, NK cell, and functional receptors for many cytokines, such as IL-2, IL-4, IL-7, IL-9, IL-15, and IL-21 (26). Interestingly, bulk RNAseq of the ST258 C4–infected lungs shows that Rag2–/– Il2rg–/– mice lack both NK cells and ILCs, and scRNAseq attests the distinct clusters of Il17a+, Il22+, and Icos+ ILC3 (Figure 2A) and Ifng+ NK cells (Figure 2B) in ST258 C4–infected lungs in Rag2–/– mice. Considering that, early in infection, the majority of IL-17 and IL-22 are produced by γδ T cells and ILC3 (27, 28), there might be a substantial role for the ILC3 in protection in Rag2–/– mice. Similar data were reported in lung infection due to Streptococcus pneumoniae (29); thus, ILC3 could be identified as a major producer of IL-22, possibly contributing to early host defense against these pathogens. Concomitantly, several genes in the NK chemotaxis and cytotoxicity pathway were also deficient in the Rag2–/– Il2rg–/– mice; thus, these cells also contribute to host resilience to this infection in the Rag2–/– genetic background.
ICOS is one of the CD28 family of costimulatory molecules and is highly expressed on the activated T cell (30, 31), especially type 2 helper T cells (32) and ILC2 (33), contributing to the isotype switch and germinal center formation (34). However, in this infection model on the Rag2–/– background, ICOS is also a key regulator of ILC3. Our data are also consistent with data we recently reported that show ICOS was highly expressed in lung ILC3 during Mycobacteria tuberculosis infection (35); however, the functional role of ICOS was not studied in that paper. Further supporting our observation that ICOS is a regulator of ILC3, recombinant ICOSL enhanced proliferation of ILC3 ex vivo (Figure 4, G and J).
Considering the relationship between IL-17– or IL-22–producing ILC3 and IFN-γ–producing NK cells during innate immunity, which cell populations are responsible for the immunity against ST258 C4 pulmonary infection remains to be determined. Accumulating evidence has shown that IFN-γ, mainly produced by activated NK cells during K. pneumoniae infection, plays a critical role in models of infection with more virulent carbapenemase negative strains of K. pneumoniae (36, 37). Moreover, several reports have shown the additional requirements of IL-17 (38, 39) and IL-22 (29, 40, 41) in these models. In contrast, in the model developed with ST258, NK cells by themselves were dispensable. It was only in the setting of NK cell depletion and antagonism of ILC activation using anti-ICOS antibodies that Rag2–/– mice showed abnormal bacterial clearance. Our scRNAseq data in infected Rag2–/– mice also revealed that Icos expression was restricted mostly to the Il17a+Il22+ ILC3 cluster, which was well separated from Ifng+ NK cells, suggesting the cooperative role is resistant to ST258 infection.
Given the risk of solid organ transplantation and this infection, we investigated if calcineurin inhibitors would perturb bacterial clearance. To test this, we administered FK506 in WT C57BL/6 mice. FK506 is an immunosuppressant prevalent in the clinical setting, similar to cyclosporine A, especially for the prevention of allograft rejections in patients undergoing solid organ or stem cell transplantation. This drug inhibits the expression of IL-2 (42) and subsequent T cell proliferation (43) through blocking activation of the calcineurin pathway (44). Our data show that administration of FK506 renders WT C57BL/6 mice susceptible to ST258 infection, similar to infection in Rag2–/– Il2rg–/– mice, and qPCR elucidated the downregulation of Il17a, Il17f, Il22, and Ifng; however, the cellular source of these cytokines in WT C57BL/6 mice was unclear. scRNAseq showed that, although FK506 had little effect on the recruitment of NK cells or γδ T cells in the lungs, Ifng-, Il17a-, Il17f-, and Il22-expressing cells were all significantly decreased. Furthermore, Ifng expression was largely confined to NK cells, whereas most of Il17a- and Il22-expressing cells clustered in γδ T cells. Interestingly local IFN-γ was ineffective in augmenting bacterial clearance in FK506-treated C57BL/6 mice (Figure 8A), whereas IL-22:Fc could ameliorate the ST258 pulmonary infection in both Rag2–/– Il2rg–/– mice (Figure 8D) and FK506-treated C57BL/6 mice (Figure 8C). IL-22 primarily acts on epithelial cells to promote barrier function, such as enhancing the production of antimicrobial peptides (40). To date, there have been several reports demonstrating the effect of IL-22 in augmenting host resistance to infection, in part by inducing antimicrobial proteins such as lipocalin 2 (40, 45) and by augmenting hepatic C3 expression (46, 47). Meanwhile, ST258 isolates could be killed in healthy human serum, suggesting the prominent role for a complement system against ST258 bacteremia (7). Based on this, there have been some reports showing an antibody-based approach via complement-mediated bactericidal activity against this multidrug-resistant pathogen (48). We observed the effect of IL-22:Fc was abrogated in Il22raΔHEP mice (Figure 8, E–G), suggesting the essential role of hepatocytes for the effect of IL-22:Fc to ameliorate the ST258 infection. Moreover, intratracheal administration of IL-22:Fc also showed efficacy in ameliorating ST258 C4–infected Il22ra1fl/fl mice, but the efficacy was abrogated in Il22ra1ΔHEP mice (Supplemental Figure 7). Based on these data, hepatic IL-22ra1 signaling is essential for IL-22:Fc in this model. However, we cannot exclude a role of IL-22ra1 signaling in lung epithelium at this stage. The dominant role of hepatic IL-22ra1 in vivo is likely related to the pharmacological amount of IL-22:Fc provided and the large amount of cardiac output to the liver, as well as the fenestrated endothelium of the liver, which permits greater accessibility of IL-22 to hepatocytes. One mechanism underlying IL-22’s efficacy of an antibacterial effect in Il22ra1fl/fl mice is, at least partially, augmented in complement mediated protection (Figure 9, A–E).
While we show detailed data in ST258 pulmonary infection, there are several limitations to our investigation. We focused on NK cell and ILC genes in this study, since they were markedly different between Rag2–/– and Rag2–/– Il2rg–/– mice, but we also observed downregulation of Ccr2 and Nos2 in these data; this suggests that monocyte recruitment may also be perturbed in Rag2–/– Il2rg–/– mice, and these cells have previously been described as important in a higher-dose infection model (13). However, we did not observe differential expression of Ccr2 or Nos2 in the FK506 model (in our scRNAseq data). We observed a distinct population of NK cells and ILC3 by scRNAseq, and the antibodies to each population clearly downregulate the signature genes of each population. However, again, these data were generated using Rag2–/– mice, so the protection by NK and ILC3 cannot necessarily be applied to immunocompetent mice, where most Il17a and Il22 is expressed by γδ T cells. FK506 likely exacerbates infection by inhibiting these responses, but this would require additional experiments to define all of the cell populations that may be affected by this drug. Anti-ICOS antibodies showed that Il17a expression was substantially reduced (Figure 4A), but Il22 expression was only reduced by approximately 50% (Figure 4C). Considering that only IL-22:Fc could restore protective immunity in these immunocompromised hosts, this residual ICOS-independent IL-22–producing population may play a key role in immunity, and that may explain why anti-NK1.1– and anti-ICOS–treated mice exacerbate but not as severely as Rag2–/– Il2rg–/– mice that lack IL-22 completely. This concept might be inconsistent with the prior report by Ahn et al. showing that ST258 infection was similar in WT and Il22–/– mice (49). However, in this study, there were increases in Il1β and Il17 gene expressions in these mice, which may compensate for the lack of IL-22. Il22–/– and Reg3g–/– mice can have overgrowth of segmented filamentous bacteria (50, 51), and the latter mice have increased pulmonary IL-1 and IL-17 responses. Thus, it is possible that the increased IL-1 and IL-17 observed by Ahn et al. (49) may be due to gut dysbiosis and may explain the lack of a robust phenotype in Il22–/– mice in that study. Additionally, we observed anti–IL-22 treatment in WT C57BL/6 mice exacerbated ST258 C4 pulmonary infection (Supplemental Figure 8), supporting the concept that IL-22 is critical in this model. Additionally, in terms of rescuing with IL-22:Fc, we administered pharmacological doses that mediate robust STAT3 signaling in the liver; thus, any activator of hepatic STAT3 may be therapeutic in this model. Finally, we cannot currently exclude a role of airway epithelium as a critical target for the IL-22 response. However, in vivo, the data show that the efficacy of IL-22 was dependent on hepatic IL-22ra1 signaling using the liver (23). Although IL-22 can signal to the epithelium, we believe the liver may be dominant in vivo for the 3 reasons. First, we believe that the liver is a critical site for the systemic route of IL-22 administration, as the liver receives a much larger percentage of the cardiac output compared with the lung epithelium. Second, hepatocytes express abundant amounts of Il22ra1. Third, the liver has a fenestrated endothelium; thus, large amounts of the administered IL-22 are likely sequestered in the liver.
In conclusion, our findings show that both ILC3 and NK cells are associated with the resistance to ST258 C4 pulmonary infection. Considering the difficulty of treating this invasive infection due to multidrug resistance, immunomodulatory treatment or prevention — especially IL-22 treatment — might be considered to manage this infection.
Methods
Mice.
WT C57BL/6, Rag2–/– (B6.129S6-Rag2tm1Fwa N12), and Rag2–/– Il2rg–/– (B10; B6-Rag2tm1Fwa Il2rgtm1Wjl) mice were obtained from Taconic Farm. Il22ra1fl/fl mice, Il22ra1ΔHEP mice, and Il22ra1E2Acre mice, which are germline Il22ra1-KO mice, were inbred. All mice for experiments were 6- to 8-week-old male mice.
Experimental K.
pneumoniae infection. KPC-producing K. pneumoniae strain C4, provided by Yohei Doi at University of Pittsburgh (Pittsburgh, Pennsylvania, USA), were grown in 100 mL tryptic soy broth (Difco) containing the 5 μg/mL meropenem (Thermo Fisher Scientific) and 100 μg/mL carbenicillin (Thermo Fisher Scientific) for 18 hours at 37°C. The culture was then diluted at 1:100 and grown for an additional 2.5 hours to reach the early logarithmic phase. The concentration of K. pneumoniae was determined by measuring the absorbance at 600 nm. Bacteria were pelleted by centrifugation (3000 g, 4°C, 10 minutes) and washed twice in cold PBS; they were then resuspended to achieve the density of 1 × 107 CFU/mL. Mice were then infected with 1 × 106 CFU in 100 μL live K. pneumoniae intratracheally and sacrificed at 24 hours after infection. Bacterial burden in the left lung, liver, and spleen were analyzed by CFU on the homogenates. Simultaneously, the right middle lobe was resected in Trizol (Thermo Fisher Scientific) for RNA extraction.
qPCR.
RNA was isolated using Trizol reagent, and cDNA was prepared using iScript reverse transcriptase master mix (Bio-Rad). qPCR was carried out with Bio-Rad CFX96 system using TaqMan PCR Master Mix (Thermo Fisher Scientific) and premixed primers/probe sets from Thermo Fisher Scientific.
Flow cytometry and intracellular cytokine staining.
Fluorophore conjugated antibodies used mouse cell analysis are as follows: anti-RORγt (Q31-378, BD Bioscience, 562607), anti-GATA3 (16E10A23, BioLegend, 653813), anti-NK1.1 (PK136, BioLegend, 108715), anti–IL-5 (TRFK5, BioLegend, 504305), anti-CD278 (ICOS) (C398.4A, BioLegend, 313529), anti-CD45.2 (104, BioLegend, 109830), anti-CD127 (A7R34, BioLegend, 135021), anti-Lin (17A2/RB6-8C5/RA3-6B2/Ter-119/M1/70, BioLegend, 133313), anti–IL-17A (eBio17B7, Thermo Fisher Scientific, 12-7177-81; TC11-18H10.1, BioLegend, 506907), anti–IL-22 (1H8PWSR, Thermo Fisher Scientific, 46-7221-82; IL22JOP, Thermo Fisher Scientific, 17-7222-82), anti–TCR γδ (GL3, BioLegend, 118108), anti–TCRβ chain (H57-597, BD Bioscience, 553173), and anti–IFN-γ (XMG1.2, BioLegend, 505825). Nonspecific binding was blocked using a rat anti-mouse antibody directed against the FcγIII/II receptor (CD16/CD32) (2.4G2, BD Bioscience, 553141). All samples were resuspended in wash buffer and then subjected to flow cytometry analysis by using an Aurora (Cytek). For intracellular staining, each cell suspension was stimulated with PMA (Sigma-Aldrich) (40 ng/mL) with ionomycin (Sigma-Aldrich) (4 μg/mL) in 200 μL complete IMEM medium (Thermo Fisher Scientific), supplemented with 10% FBS (GE Healthcare) and 1% penicillin/streptomycin (Thermo Fisher Scientific), at 37°C and 5% CO2 for 6 hours; it was also stimulated with GolgiPlug (BD Bioscience) supplementation to the media for the last 5 hours. Intracellular staining was performed after cell permeabilization (BD Bioscience) according to the manufacturers’ instructions. All flow cytometry data were analyzed using Flow Jo Mac, version 9.6 (Tree Star Inc.).
Bulk RNAseq.
Total whole lung tissue RNA was used to perform RNAseq. RNA quantity and quality were assessed using NanoDrop and Agilent RNA 6000 Nano kit with Agilent 2100 Bioanalyzer instrument. Illumina TruSeq Stranded mRNA sample prep kit was used for library preparation, followed by Agilent DNA 1000 kit validation with Agilent 2100 Bioanalyzer and quantification by Qubit 2.0 fluorometer. The cDNA libraries were pooled at a final concentration 1.8 pM. Cluster generation and 75 bp paired read single-indexed sequencing was performed on Illumina NextSeq 550. Raw reads were processed and mapped; then, gene expression and nucleotide variation was evaluated by previously described methods (52). Data were deposited in the Gene Expression Omnibus (GEO accession no. GSE130045).
scRNAseq.
Mice were euthanized by exposure to CO2 and exsanguination. Lungs were isolated and minced with forceps and small scissors and digested in 2 mL serum-free medium with 2 mg/mL collagenase (MilliporeSigma) and 80 U/mL DNase I (MilliporeSigma) for 60 minutes at 37°C. Digested tissues were passed through a sterile 70-mm filter (BD Falcon) to get a single cell suspension. After washing, 2 mL ACK Lysing Buffer (Invitrogen) was added into the cell suspension and let sit for 2 minutes, washed 2 additional times. The whole lung single cell suspension obtained from this procedure was then tested for purity and quantity by Cellometer Auto 2000 (Nexcelom Bioscience) and found to be ready for scRNAseq. For scRNAseq analysis, 1 × 106 cells per condition were saved as whole lung single cell population. The remaining cells were then subjected to CD45 enrichment by applying Ly-6G negative selection (Miltenyi Biotec, 130-092-332) and followed by CD45 positive selection (Miltenyi Biotec, 130-052-301). Briefly, the cells were suspended in MACS separation buffer, incubated with the anti-Ly6G biotin followed by anti-biotin microbeads, and applied to a MACS LS separation column (Miltenyi Biotec, 130-042-401). The flow through was collected as the Ly6G– population. The Ly6G– cells were then incubated with anti-CD45 microbeads and applied to a MACS MS column. After washes, the CD45+ population was released from the column by flushing the column and collected into a 1.5-mL microtube. Both whole lung single cell and CD45-enriched populations were treated with 100 μL TrypLE for 1 minute to dissociate single cells from any small aggregates or clusters. Cell numbers and viabilities were validated by Cellometer prior to scRNAseq library preparation.
For 10× scRNAseq assay, 5000 live cells per sample were targeted by using 10× Single Cell RNAseq technology provided by 10× Genomics (10X Genomics). Briefly, viable single cell suspensions were partitioned into nanoliter-scale Gel Bead-In EMulsion (GEMs). Full-length barcoded cDNAs were then generated and amplified by PCR to obtain sufficient mass for library construction. Following enzymatic fragmentation, end-repair, A-tailing, and adaptor ligation, single cell 3′ libraries comprising standard Illumina P5 and P7 paired-end constructs were generated. Library quality controls were performed by using Agilent High Sensitive DNA kit with Agilent 2100 Bioanalyzer and quantified by Qubit 2.0 fluorometer. Pooled libraries at a final concentration of 1.8 pM were sequenced with paired end single index configuration by Illumina NextSeq 550. Cell Ranger version 2.1.1 (10X Genomics) was used to process raw sequencing data and Loupe Cell Browser (10X Genomics) to obtain differentially expressed genes between specified cell clusters. In addition, Seurat suite version 2.2.1 (52, 53) was used for further quality control and downstream analysis. Filtering was performed to remove multiplets and broken cells, and uninteresting sources of variation were regressed out. Variable genes were determined by iterative selection based on the dispersion versus average expression of the gene. For clustering, principal component (PC) analysis was performed for dimension reduction. Top 10 PCs were selected by using a permutation-based test implemented in Seurat and passed to t-SNE for clustering visualization.
Proliferation assay.
Single lung cell suspensions from 6- to 8-week-old male Rag2–/– mice 12 hours after infection with ST258 C4 were acquired following the digestion protocol as described above; then, lung ILCs were purified using Percoll (Sigma-Aldrich) gradient centrifugation. CellTrace Violet (CTV) labeling was performed as manufacturer described (Thermo Fisher Scientific). CTV-labeled lung ILCs (5 × 104/well) were coincubated with ICOSL (HK5.3, Bio X Cell, BE0028, 1.5 μg/mL) for 24 hours and analyzed for FACS. Finally, we examined the effect of FK506 (50 ng/mL) by coincubation on the proliferation of ICOSL-activated ILCs.
Antimicrobial assay.
To assay bacteriostatic activity, diluted serum from isotype control or IL-22:Fc–treated (Generon) WT C57BL/6 mice or conditioned medium was incubated with K. pneumoniae in a 96-well plate, and growth kinetics were analyzed over 9 hours in a heat-controlled shaking microplate reader and read at an OD of 600 nm (OD600) every hour (23). This procedure was repeated using heat-inactivated serum (56°C, 30 minutes). For C3 binding assay, a total of 1 × 105 CFU ST258 C4 was incubated for 20 minutes at 37°C (no CO2) with 5% diluted serum from isotype control or IL-22:Fc–treated WT C57BL/6 mice, and we then performed FACS analysis (24).
Antibody administration.
In some experiments, mice were injected i.p. with 500 μg anti-ICOS antibody (7E.17G9, Bio X Cell, BE0059), 250 μg anti-NK1.1 antibody (BE0036, Bio X Cell, 50562112), IgG2a isotype control (2A3, Bio X Cell, BE0089), or IgG2b isotype control (LTF-2, Bio X Cell, BE0090) once 2 hours before bacterial challenge and injected intratracheally with 50 μg anti–IL-22 mAb (R&D Systems, AF582) 30 minutes before bacterial challenge.
Cytokine immunotherapy.
For pharmacological model of Rag2–/– Il2rg–/– mice, 10 mg/kg FK506 was administered in C57BL/6, Il22ra1fl/fl, or Il22ra1ΔHEP mice i.p. 24 hours prior to intratracheal challenge with 1 × 106 ST258 C4 strain. FK506 was administered every 24 hours until 24 hours before euthanasia. These mice were euthanized at 48 hours after infection. To examine the efficacy of cytokine rescue, we administered 4 μg IL-22:Fc i.p. 2 hours before bacterial challenge, or we administered 1 μg recombinant murine IFN-γ (Peprotech) or 1 μg IL-22:Fc intratracheally 30 minutes before bacterial challenge.
Statistics.
Graphs were generated and statistical significance was analyzed using GraphPad Prism 8, and the used statistical tests are defined in the figure legends. P values of pairwise comparisons between 2 normally distributed groups were performed by simple 2-tailed unpaired Student’s t test, while other analyses for unequal variance were determined by Mann-Whitney U test or Welch’s 2-tailed t test. We used 1-way ANOVA with Tukey’s multiple comparison test or Kruskal-Wallis with Dunn’s multiple comparison test for multiple groups comparisons. Values are represented as means ± SEM. P values are annotated as follows *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001.
Study approval.
All mice were housed in accordance with the Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011) and in specific pathogen–free rooms within animal facilities at Tulane University. All mice experiments were approved by the Tulane IACUC.
Author contributions
Conceptualization was contributed by NI, IS, and JKK; methodology was contributed by NI, AW, KS, and JKK; validation was contributed by NI and JKK; formal analysis was contributed by NI; investigation was contributed by NI, IS, AW, KS, and JKK; all reagents and devices for analyzing data were contributed by JKK; writing the original draft was contributed by NI; review and editing of the manuscript was contributed by NI, IS, AW, JM, KS, and JKK; volcano plot and t-SNE visualization were contributed by NI and AW; and funding acquisition was contributed by JKK.
Supplementary Material
Acknowledgments
We thank Yohei Doi (University of Pittsburgh) for providing the carbapenemase-producing K. pneumoniae ST258 strain C4 for this project. This work was supported by the Louisiana Board of Regents Endowed Chairs for Eminent Scholars program, as well as by PHS grant R35HL139930.
Version 1. 03/26/2020
In-Press Preview
Version 2. 04/23/2020
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(8):e135591.https://doi.org/10.1172/jci.insight.135591.
Contributor Information
Naoki Iwanaga, Email: niwanaga@tulane.edu.
Ivy Sandquist, Email: isandqui@tulane.edu.
Alanna Wanek, Email: awanek@tulane.edu.
Kejing Song, Email: ksong1@tulane.edu.
References
- 1.Logan LK, Weinstein RA. The Epidemiology of Carbapenem-Resistant Enterobacteriaceae: The Impact and Evolution of a Global Menace. J Infect Dis. 2017;215(suppl_1):S28–S36. doi: 10.1093/infdis/jiw282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu L, Sun X, Ma X. Systematic review and meta-analysis of mortality of patients infected with carbapenem-resistant Klebsiella pneumoniae. Ann Clin Microbiol Antimicrob. 2017;16(1):18. doi: 10.1186/s12941-017-0191-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yigit H, et al. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45(4):1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Satlin MJ, et al. Multicenter Clinical and Molecular Epidemiological Analysis of Bacteremia Due to Carbapenem-Resistant Enterobacteriaceae (CRE) in the CRE Epicenter of the United States. Antimicrob Agents Chemother. 2017;61(4):e02349-16. doi: 10.1128/AAC.02349-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kitchel B, et al. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob Agents Chemother. 2009;53(8):3365–3370. doi: 10.1128/AAC.00126-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacoby GA, Munoz-Price LS. The new beta-lactamases. N Engl J Med. 2005;352(4):380–391. doi: 10.1056/NEJMra041359. [DOI] [PubMed] [Google Scholar]
- 7.Kobayashi SD, et al. Antibody-Mediated Killing of Carbapenem-Resistant ST258 Klebsiella pneumoniae by Human Neutrophils. mBio. 2018;9(2):e00297-18. doi: 10.1128/mBio.00297-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guh AY, et al. Epidemiology of Carbapenem-Resistant Enterobacteriaceae in 7 US Communities, 2012-2013. JAMA. 2015;314(14):1479–1487. doi: 10.1001/jama.2015.12480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han JH, Goldstein EJ, Wise J, Bilker WB, Tolomeo P, Lautenbach E. Epidemiology of Carbapenem-Resistant Klebsiella pneumoniae in a Network of Long-Term Acute Care Hospitals. Clin Infect Dis. 2017;64(7):839–844. doi: 10.1093/cid/ciw856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falagas ME, Tansarli GS, Karageorgopoulos DE, Vardakas KZ. Deaths attributable to carbapenem-resistant Enterobacteriaceae infections. Emerging Infect Dis. 2014;20(7):1170–1175. doi: 10.3201/eid2007.121004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tzouvelekis LS, et al. KPC-producing, multidrug-resistant Klebsiella pneumoniae sequence type 258 as a typical opportunistic pathogen. Antimicrob Agents Chemother. 2013;57(10):5144–5146. doi: 10.1128/AAC.01052-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeLeo FR, et al. Survival of Carbapenem-Resistant Klebsiella pneumoniae Sequence Type 258 in Human Blood. Antimicrob Agents Chemother. 2017;61(4):e02533-16. doi: 10.1128/AAC.02533-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiong H, Keith JW, Samilo DW, Carter RA, Leiner IM, Pamer EG. Innate Lymphocyte/Ly6C(hi) Monocyte Crosstalk Promotes Klebsiella Pneumoniae Clearance. Cell. 2016;165(3):679–689. doi: 10.1016/j.cell.2016.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leung LM, et al. Structural modification of LPS in colistin-resistant, KPC-producing Klebsiella pneumoniae. J Antimicrob Chemother. 2017;72(11):3035–3042. doi: 10.1093/jac/dkx234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sims D, Sudbery I, Ilott NE, Heger A, Ponting CP. Sequencing depth and coverage: key considerations in genomic analyses. Nat Rev Genet. 2014;15(2):121–132. doi: 10.1038/nrg3642. [DOI] [PubMed] [Google Scholar]
- 16.Eddens T, et al. Eosinophils Contribute to Early Clearance of Pneumocystis murina Infection. J Immunol. 2015;195(1):185–193. doi: 10.4049/jimmunol.1403162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Böttcher JP, et al. NK Cells Stimulate Recruitment of cDC1 into the Tumor Microenvironment Promoting Cancer Immune Control. Cell. 2018;172(5):1022–1037.e14. doi: 10.1016/j.cell.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang H, Li F, Cairns CM, Gordon JR, Xiang J. Neutrophils and B cells express XCR1 receptor and chemotactically respond to lymphotactin. Biochem Biophys Res Commun. 2001;281(2):378–382. doi: 10.1006/bbrc.2001.4363. [DOI] [PubMed] [Google Scholar]
- 19.Gauthier M, et al. Severe asthma in humans and mouse model suggests a CXCL10 signature underlies corticosteroid-resistant Th1 bias. JCI Insight. 2017;2(13):94580. doi: 10.1172/jci.insight.94580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raundhal M, et al. High IFN-γ and low SLPI mark severe asthma in mice and humans. J Clin Invest. 2015;125(8):3037–3050. doi: 10.1172/JCI80911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pociask DA, et al. IL-22 is essential for lung epithelial repair following influenza infection. Am J Pathol. 2013;182(4):1286–1296. doi: 10.1016/j.ajpath.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao B. Hepatoprotective and anti-inflammatory cytokines in alcoholic liver disease. J Gastroenterol Hepatol. 2012;27 Suppl 2:89–93. doi: 10.1111/j.1440-1746.2011.07003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng M, et al. Therapeutic Role of Interleukin 22 in Experimental Intra-abdominal Klebsiella pneumoniae Infection in Mice. Infect Immun. 2016;84(3):782–789. doi: 10.1128/IAI.01268-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trevejo-Nunez G, Elsegeiny W, Conboy P, Chen K, Kolls JK. Critical Role of IL-22/IL22-RA1 Signaling in Pneumococcal Pneumonia. J Immunol. 2016;197(5):1877–1883. doi: 10.4049/jimmunol.1600528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shinkai Y, et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68(5):855–867. doi: 10.1016/0092-8674(92)90029-C. [DOI] [PubMed] [Google Scholar]
- 26.Cao X, et al. Defective lymphoid development in mice lacking expression of the common cytokine receptor gamma chain. Immunity. 1995;2(3):223–238. doi: 10.1016/1074-7613(95)90047-0. [DOI] [PubMed] [Google Scholar]
- 27.Spits H, Cupedo T. Innate lymphoid cells: emerging insights in development, lineage relationships, and function. Annu Rev Immunol. 2012;30:647–675. doi: 10.1146/annurev-immunol-020711-075053. [DOI] [PubMed] [Google Scholar]
- 28.Chen K, et al. Th17 cells mediate clade-specific, serotype-independent mucosal immunity. Immunity. 2011;35(6):997–1009. doi: 10.1016/j.immuni.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Maele L, et al. Activation of Type 3 innate lymphoid cells and interleukin 22 secretion in the lungs during Streptococcus pneumoniae infection. J Infect Dis. 2014;210(3):493–503. doi: 10.1093/infdis/jiu106. [DOI] [PubMed] [Google Scholar]
- 30.Hutloff A, et al. ICOS is an inducible T-cell co-stimulator structurally and functionally related to CD28. Nature. 1999;397(6716):263–266. doi: 10.1038/16717. [DOI] [PubMed] [Google Scholar]
- 31.Rudd CE, Schneider H. Unifying concepts in CD28, ICOS and CTLA4 co-receptor signalling. Nat Rev Immunol. 2003;3(7):544–556. doi: 10.1038/nri1131. [DOI] [PubMed] [Google Scholar]
- 32.Simpson TR, Quezada SA, Allison JP. Regulation of CD4 T cell activation and effector function by inducible costimulator (ICOS) Curr Opin Immunol. 2010;22(3):326–332. doi: 10.1016/j.coi.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 33.Maazi H, et al. ICOS:ICOS-ligand interaction is required for type 2 innate lymphoid cell function, homeostasis, and induction of airway hyperreactivity. Immunity. 2015;42(3):538–551. doi: 10.1016/j.immuni.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bossaller L, et al. ICOS deficiency is associated with a severe reduction of CXCR5+CD4 germinal center Th cells. J Immunol. 2006;177(7):4927–4932. doi: 10.4049/jimmunol.177.7.4927. [DOI] [PubMed] [Google Scholar]
- 35.Ardain A, et al. Group 3 innate lymphoid cells mediate early protective immunity against tuberculosis. Nature. 2019;570(7762):528–532. doi: 10.1038/s41586-019-1276-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deng JC, et al. STAT4 is a critical mediator of early innate immune responses against pulmonary Klebsiella infection. J Immunol. 2004;173(6):4075–4083. doi: 10.4049/jimmunol.173.6.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ivin M, et al. Natural killer cell-intrinsic type I IFN signaling controls Klebsiella pneumoniae growth during lung infection. PLoS Pathog. 2017;13(11):e1006696. doi: 10.1371/journal.ppat.1006696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ye P, et al. Requirement of interleukin 17 receptor signaling for lung CXC chemokine and granulocyte colony-stimulating factor expression, neutrophil recruitment, and host defense. J Exp Med. 2001;194(4):519–527. doi: 10.1084/jem.194.4.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Happel KI, et al. Divergent roles of IL-23 and IL-12 in host defense against Klebsiella pneumoniae. J Exp Med. 2005;202(6):761–769. doi: 10.1084/jem.20050193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aujla SJ, et al. IL-22 mediates mucosal host defense against Gram-negative bacterial pneumonia. Nat Med. 2008;14(3):275–281. doi: 10.1038/nm1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu X, et al. Conventional NK cells can produce IL-22 and promote host defense in Klebsiella pneumoniae pneumonia. J Immunol. 2014;192(4):1778–1786. doi: 10.4049/jimmunol.1300039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bierer BE, et al. Two distinct signal transmission pathways in T lymphocytes are inhibited by complexes formed between an immunophilin and either FK506 or rapamycin. Proc Natl Acad Sci USA. 1990;87(23):9231–9235. doi: 10.1073/pnas.87.23.9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laskin BL, et al. The Effects of Tacrolimus on T-Cell Proliferation Are Short-Lived: A Pilot Analysis of Immune Function Testing. Transplant Direct. 2017;3(8):e199. doi: 10.1097/TXD.0000000000000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tonthat NK, et al. Structures of Pathogenic Fungal FKBP12s Reveal Possible Self-Catalysis Function. mBio. 2016;7(2):e00492–e00416. doi: 10.1128/mBio.00492-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coorens M, et al. Innate lymphoid cell type 3-derived interleukin-22 boosts lipocalin-2 production in intestinal epithelial cells via synergy between STAT3 and NF-κB. J Biol Chem. 2019;294(15):6027–6041. doi: 10.1074/jbc.RA118.007290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hasegawa M, et al. Interleukin-22 regulates the complement system to promote resistance against pathobionts after pathogen-induced intestinal damage. Immunity. 2014;41(4):620–632. doi: 10.1016/j.immuni.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trevejo-Nunez G, Elsegeiny W, Conboy P, Chen K, Kolls JK. Critical Role of IL-22/IL22-RA1 Signaling in Pneumococcal Pneumonia. J Immunol. 2016;197(5):1877–1883. doi: 10.4049/jimmunol.1600528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pennini ME, et al. Immune stealth-driven O2 serotype prevalence and potential for therapeutic antibodies against multidrug resistant Klebsiella pneumoniae. Nat Commun. 2017;8(1):1991. doi: 10.1038/s41467-017-02223-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ahn D, Wickersham M, Riquelme S, Prince A. The Effects of IFN-λ on Epithelial Barrier Function Contribute to Klebsiella pneumoniae ST258 Pneumonia. Am J Respir Cell Mol Biol. 2019;60(2):158–166. doi: 10.1165/rcmb.2018-0021OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shih VF, et al. Homeostatic IL-23 receptor signaling limits Th17 response through IL-22-mediated containment of commensal microbiota. Proc Natl Acad Sci USA. 2014;111(38):13942–13947. doi: 10.1073/pnas.1323852111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McAleer JP, et al. Pulmonary Th17 Antifungal Immunity Is Regulated by the Gut Microbiome. J Immunol. 2016;197(1):97–107. doi: 10.4049/jimmunol.1502566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Macosko EZ, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161(5):1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.