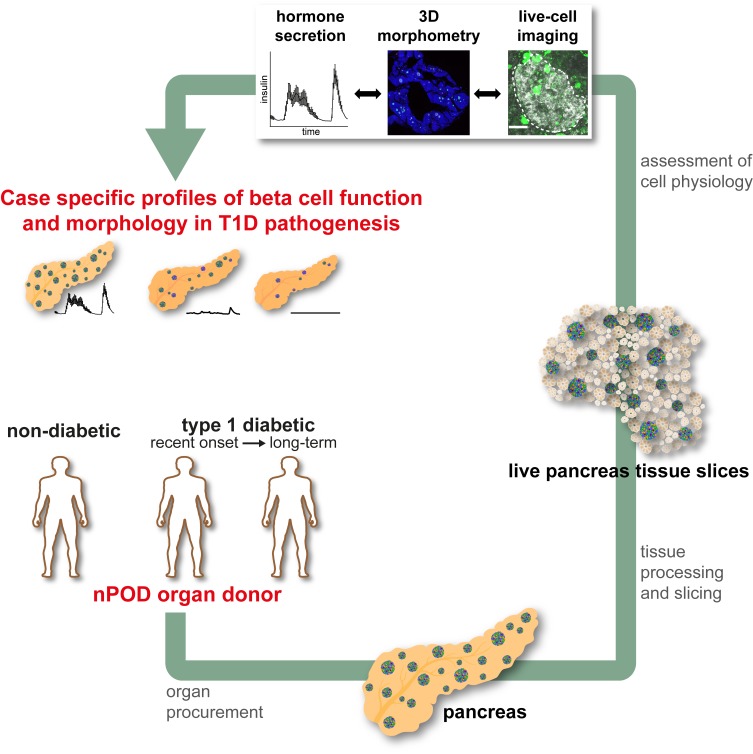
Abstract
In type 1 diabetes (T1D), autoimmune destruction of pancreatic β cells leads to insulin deficiency and loss of glycemic control. However, knowledge about human pancreas pathophysiology in T1D remains incomplete. To address this limitation, we established a pancreas tissue slice platform of donor organs with and without diabetes, facilitating the first live cell studies of human pancreas in T1D pathogenesis to our knowledge. We show that pancreas tissue slices from organ donors allow thorough assessment of processes critical for disease development, including insulin secretion, β cell physiology, endocrine cell morphology, and immune infiltration within the same donor organ. Using this approach, we compared detailed pathophysiological profiles for 4 pancreata from donors with T1D with 19 nondiabetic control donors. We demonstrate that β cell loss, β cell dysfunction, alterations of β cell physiology, and islet infiltration contributed differently to individual cases of T1D, allowing insight into pathophysiology and heterogeneity of T1D pathogenesis. Thus, our study demonstrates that organ donor pancreas tissue slices represent a promising and potentially novel approach in the search for successful prevention and reversal strategies of T1D.
Keywords: Autoimmunity, Endocrinology
Keywords: Autoimmune diseases, Beta cells, Diabetes
Live pancreas tissue slices from organ donors enable the study of islet of Langerhans pathophysiology during type 1 diabetes development.
Introduction
Type 1 diabetes (T1D) is a chronic autoimmune disease characterized by the selective destruction of insulin-producing β cells, resulting in massive reduction of β cell mass, insulin deficiency, and subsequent hyperglycemia (1–3). Although genetic susceptibility plays an evident role and environmental factors are suspected to contribute toward autoimmunity as well as loss of functional β cell mass, the precise mechanisms of T1D progression are not clearly defined (4–6). T1D is typically diagnosed when clinical symptoms occur, but the pathogenic changes affecting β cell and islet biology usually begin years before and continue after diagnosis. Yet, little is known regarding the sequential events occurring within the pancreas that lead to insulin insufficiency and hyperglycemia.
Much of our knowledge about the disease pathogenesis has been derived from studies performed in rodent models (7, 8). Although these models allow for insights into this complex disease, immune intervention strategies developed based on rodent data have not translated well to human T1D (9–11), likely due to species differences in disease processes (12–14). Developing successful therapeutic approaches requires a comprehensive understanding of how human islet biology is affected during T1D pathogenesis. To this end, the Network for Pancreatic Organ donors with Diabetes (nPOD) program has, since 2007, coordinated the procurement and study of pancreata from organ donors with T1D as well as from autoantibody+ and autoantibody– organ donors without diabetes (15). The availability of these tissues has significantly added to our understanding of T1D pathogenesis, including long-term resilience of β cells (16–18) and characteristics of immune infiltration (19). Most importantly, studies on human pancreas have further exemplified the evident heterogeneity of the disease.
However, a need exists to correlate anatomical and physiological knowledge of T1D pathogenesis along with its various subtypes (20). In particular, the physiological characterization of pancreatic islets in T1D is incomplete. Recent efforts using isolated islets from donors with T1D have provided valuable data on islet function in T1D (21, 22), but suffer from limitations related to the isolation procedure itself, which induces islet stress, eliminates potential cross-talk from the surrounding acinar tissues, and biases analyses toward the most resilient islets that survive the isolation procedure. Here, we implemented a platform that we believe to be novel for the study of pancreas tissue slices obtained from human organ donors. This approach, which was originally developed for mouse pancreas (23), was recently adapted for the study of human pancreas tissue obtained from surgical biopsy or resection (24, 25) and, in these settings, has been successfully used to study various aspects of rodent and human islet biology (23, 25–33). In this potentially novel study, we report that slices (a) can be readily prepared from pancreata from nondiabetic (ND) donors and donors with T1D; (b) have preserved endocrine, exocrine, and immune tissue compartments; and (c) allow correlation of anatomical, physiological, and clinical data. Most importantly, these findings provide unprecedented insight into the status of human β cell mass and function at distinct stages of T1D. Thus, our study demonstrates how the pancreas tissue slice technique from organ donors can help the research community gain new insight into human T1D pathogenesis and sources of disease heterogeneity.
Results
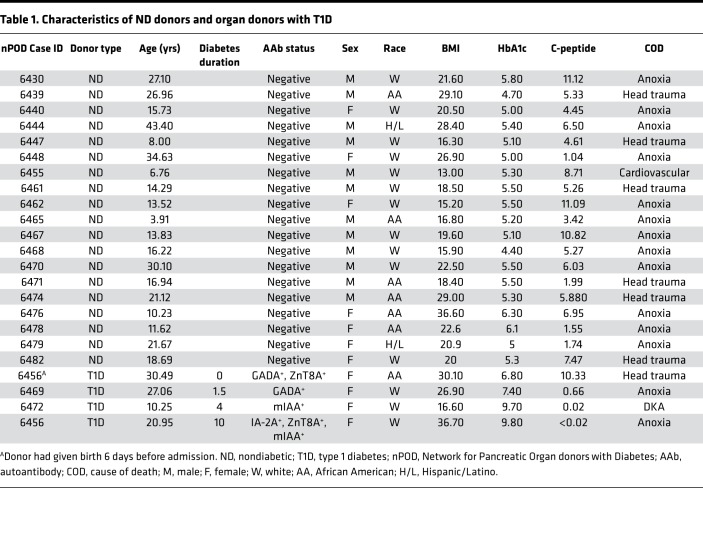
The living pancreas slice technique has allowed us and others to assess numerous parameters of islet cell function from tissues containing the intact endocrine and exocrine compartments (23, 25–33). We posited that this technological approach would facilitate investigation of structurally altered, infiltrated, or damaged islets during T1D disease pathogenesis, which are thought to be lost during classical islet isolation procedures. We therefore applied the slice technology to fresh tissue samples obtained from the nPOD program within a few hours after organ arrival to characterize islet function and morphology in ND organs and during T1D pathogenesis. We examined pancreas slices from 19 ND donors (age range, 3.91–43.40 years) and compared them to slices from 4 donors with T1D (age range, 10.25–30.49 years), with disease durations of 0, 1.5, 4, and 10 years. Detailed donor characteristics are shown in Table 1.
Table 1. Characteristics of ND donors and organ donors with T1D.
Pancreas tissue slices allow assessment of insulin secretion from donor organs and illustrate its rapid decline with increasing T1D disease duration.
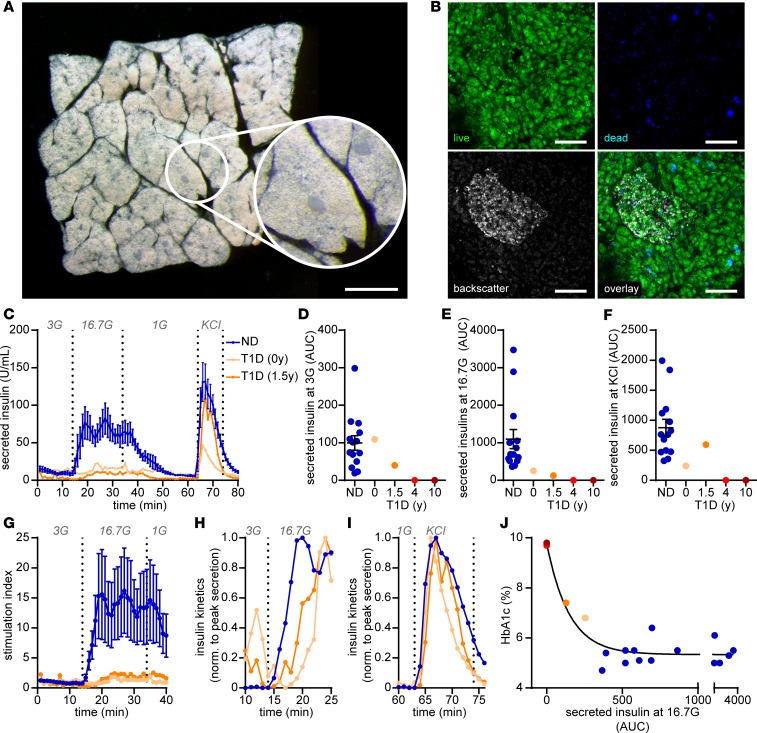
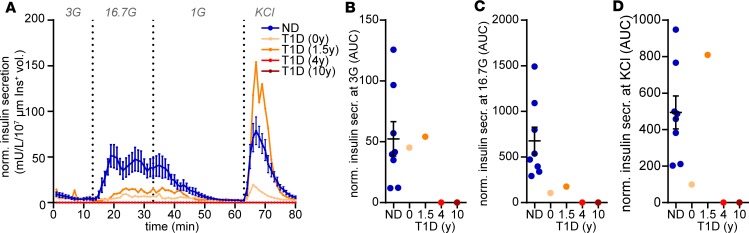
Preparation of tissue slices from donor organs procured by nPOD was straightforward and enabled the investigation of pancreatic tissue within a few hours after organ arrival. Despite the fact that pancreata from organ donors were flushed with preservative solutions and the tissue was processed after several hours of cold ischemia, slices showed preserved exocrine and endocrine morphology (Figure 1A). In addition, the vast majority of cells within pancreas tissue slices were viable and, except for the cutting surface, only a limited number of dead cells were detectable (Figure 1B). In dynamic perifusion assays, slices from ND organ donors demonstrated a substantial increase in insulin secretion in response to high glucose and KCl depolarization (Figure 1, C–F). By contrast, insulin responses to high glucose were severely diminished in tissue slices from organ donors with T1D, and impaired insulin secretion was more severe with longer disease duration. Specifically, in pancreata with recent-onset (nPOD 6456) and 1.5-year (nPOD 6469) T1D duration, glucose-stimulated insulin secretion only reached peak values of 22.8 and 11.4 U/mL, respectively, in contrast to 82.2 ± 20.6 U/mL from ND donor slices. Insulin release was not measurable under baseline or stimulated conditions for pancreas slices from the 2 donors with longer T1D durations. Further quantification confirmed the overall insulin secretory deficit in slices from donors with T1D (Figure 1, D–F) but also indicated noteworthy details for each case, including preserved basal insulin secretion (Figure 1D) with recent-onset T1D and maintained secretory capacity in response to KCl depolarization in slices from the donor with 1.5-year disease duration (Figure 1F). The diminished glucose responsiveness in both donors was further substantiated by a dramatically reduced stimulation index of 1.9 and 2.4 with recent onset and 1.5-year duration, respectively, in comparison to 16.2 ± 7.1 in slices from ND donors (Figure 1G). The kinetics of glucose-stimulated insulin secretion were also altered in T1D, showing an approximately 5-minute delay in comparison to ND slices (Figure 1H). In contrast, kinetics in response to KCl depolarization were unaffected (Figure 1I), indicating a functional deficit in the underlying mechanisms, leading specifically to glucose-induced insulin secretion. Strikingly, glucose-stimulated insulin secretion in situ was associated with HbA1c values of organ donors (Figure 1J), which increased with T1D disease duration (Table 1), demonstrating that the tissue slice technique can link functional measurements to systemic donor characteristics.
Figure 1. Assessment of β cell function in situ.
(A) Representative image of a human pancreas slice from a stereo microscope with dark-field illumination to demonstrate the morphology and anatomical location of islets in the slice (performed on every donor tissue). (B) Representative confocal microscope images of islets in human pancreas slice stained for viable (green) and dead (blue) cells and for backscattered light (gray) to locate islets (performed on every donor tissue). (C) Insulin secretory pattern of pancreas tissue slices from nondiabetic donors (ND, blue line) and 2 individual donors with type 1 diabetes (nPOD 6456, 6469, orange lines) during perifusion expressed as absolute insulin in mU/L. (D–F) Area under the curve (AUC) measurements of data presented in A for basal secretion at 3 mM glucose (3G) (D) and at 16.7 mM glucose (16.7G) (E) and perifusion with KCl (F) for nondiabetic donors (blue dots) and 4 donors with type 1 diabetes (orange dots), with duration as indicated. (G) Insulin secretory pattern shown in C (first 40 minutes) expressed as fold increase to the mean basal secretion at 3G (stimulation index). (H and I) Insulin secretory capacity normalized to the peak secretion at 16.7G (H) and KCl (I) for nondiabetic (blue line) and 2 individual type 1 diabetic (nPOD 6456, 6469, orange lines) donors. (J) Correlation of area under the curve at 16.7G with HbA1c all donors analyzed. Data for nondiabetic donors (n = 14) are expressed as mean ± SEM. Scale bar: 1 mm (A); 100 μm (B).
3D morphometry of tissue slices reveals distinct properties of β cell mass reduction in T1D pathogenesis.
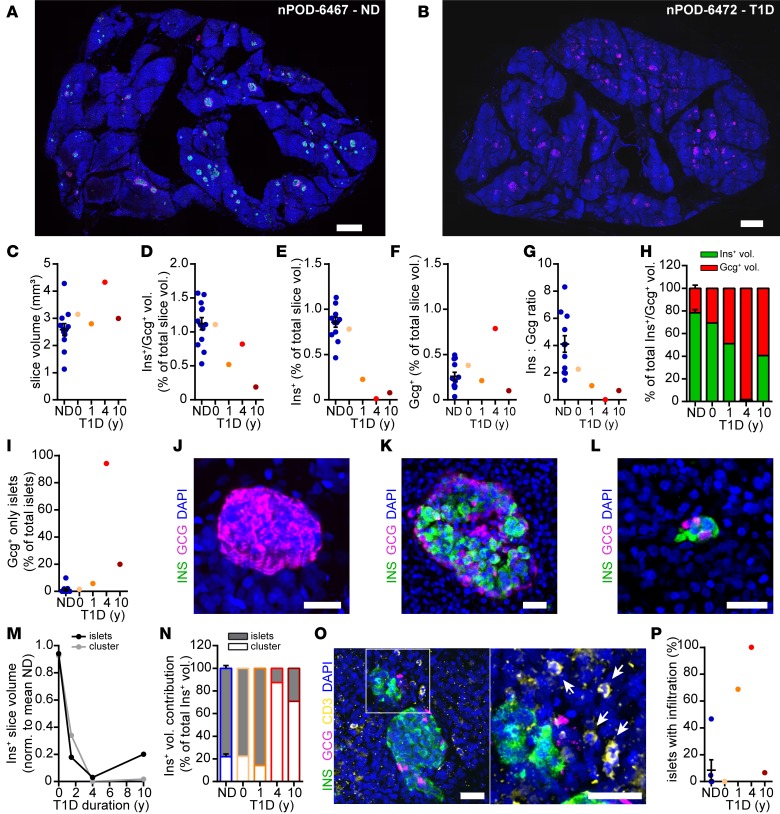
A major advantage of tissue slices includes the potential to assess detailed morphology of intact pancreas tissue after having studied physiological characteristics and function of the very same tissue. In addition, given the thickness of tissue slices (in our study, 120 μm), they enabled the assessment of tissue features within a 3D organ volume. Analysis in 3D adds morphological information on tissue structure. Thereby, it allows, e.g., the quantification of endocrine cell volumes and distributions from entire islets down to single-hormone+ cells within a preserved exocrine tissue environment. Following the completion of kinetic insulin secretion assays (Figure 1), we performed whole slice imaging and volumetric analyses to quantify endocrine mass in the perifused tissue slices of ND and T1D pancreata (Figure 2, A and B). In slices from ND donors (Figure 2A), hormone+ cells were observed to be dispersed throughout the pancreatic tissue as single cells, small-cell clusters (approximately 2–10 cells), or islets (>10 cells) with the typical islet cytoarchitecture. While more than 86% of all endocrine cells in a given pancreatic tissue volume were identified as single cells or in small-cell clusters, these only contributed approximately 25% to the total endocrine cell volume (Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.134525DS1). We did not observe any correlation of endocrine cell density or total endocrine cell volume with age, BMI, HbA1c, or C-peptide values in pancreata from ND organ donors (Supplemental Figure 1, C–J). This is in contrast to a recent study on pancreas slices from living tissue donors (25), where we found a positive correlation of endocrine cell volume with age, which is most likely due to the vastly different donor characteristics in the 2 studies. Volumetric analyses demonstrated that we reliably produced slices with similar total tissue volume from ND and T1D organs (Figure 2C), which allowed for quantitative comparisons between donors. As compared with ND, slices from T1D pancreas displayed decreased endocrine cell volume with increasing disease duration (Figure 2, B and D). This was primarily due to a diminished insulin+ volume after T1D onset, reflecting the progressive reduction in β cell mass (Figure 2E). Interestingly, β cell mass was comparable to that of ND organs in the recent-onset T1D case (nPOD 6456; 0-year duration, donor had given birth 6 days before) but was dramatically decreased in pancreata from donors with T1D with longer disease durations (1.5, 4, and 10 years, for nPOD 6469, 6472, and 6459, respectively). In contrast, the glucagon+ volume in T1D pancreas slices did not show the same dramatic reduction in comparison with that in ND pancreas (Figure 2F). As a consequence, the insulin-to-glucagon cell ratio decreased with T1D duration (Figure 2G), and the total endocrine population shifted from a β cell majority in ND subjects to an α cell majority in subjects with T1D (Figure 2H). Consequentially, we observed an increasing number of islets consisting only of glucagon+ cells in tissue slices from organ donors with T1D (Figure 2, I and J). Thus, pancreas tissue slices from organ donors provided an unprecedented detailed assessment of human pancreas morphology of functionally characterized tissue, enabling an exceedingly precise quantification of T1D-related pancreatic changes.
Figure 2. 3D histomorphometric analysis of tissue slices.
(A and B) Maximum intensity projections of human pancreas tissue slices from a nondiabetic donor (A) and a donor with a 4-year history of T1D (B) stained for insulin (green), glucagon (magenta), and DAPI (blue). (C) Total slice volumes analyzed for each donor. (D–F) Quantification of hormone+ volumes as a percentage of the total slice volumes for (D) Ins+/Gcg+ volume, (E) Ins+ volume, and (F) Gcg+ volume. (G) The Ins+/Gcg+ ratio within the slice volume of each individual donor. (H) The contribution of Ins+ and Gcg+ volumes to total endocrine volume of each individual donor. (I) The percentage of islets consisting of only Gcg+ cells. (J) Maximum intensity projection of an islet with only Gcg+ cells. (K and L) Maximum intensity projections of an example islet (K) and small-cell cluster (L). (M) Fractional loss of Ins+ volume in small-cell clusters (≤10 cells) and islets (>10 cells) normalized to ND. (N) Contribution of Ins+ volume in small-cell clusters (≤10 cells) and islets (>10 cells) to total residual Ins+ volume. (O) Maximum intensity projection of an islet in a pancreas tissue slice (donor nPOD-6469, 1.5y T1D) with surrounding CD3+ cells (the area in the white box is shown at high magnification to the right, with arrows indicating CD3+ cells). (P) Islets with infiltration for 3 nondiabetic donors and 4 donors with type 1 diabetes. Scale bars: 500 μm (A and B); 50 μm (J–L); 30 μm (O); data for nondiabetic donors (n = 13) are presented as mean ± SEM.
Previous studies have suggested a potential predominant destruction of β cells within islets and long-term survival of β cells within small-cell clusters or as single cells in T1D (34). If true, the underlying mechanisms for this selectivity could guide the development of intervention strategies to protect β cells from destruction. Using the tissue slice 3D approach, we examined whether β cell loss preferentially occurred within islets (>10 cells, Figure 2K) or at the level of single cells and small-cell clusters (2–10 cells, Figure 2L). Our results showed that β cell volume was reduced in both islets and small-cell clusters/single cells (Figure 2M). The fractional loss of β cell volume (normalized against mean insulin+ volume in ND slices) was comparable from islets and small-cell clusters/single cells (Figure 2M) but, as a result, after prolonged (i.e., 4- and 10-year) disease duration, the vast majority of remaining total β cell mass was composed of single cells and small-cell clusters (Figure 2N). Consequently, our data revealed β cell destruction to similarly affect islets as well as single cells and small-cell clusters in all 4 studied cases of T1D.
We next used 3D morphometry to assess immune infiltration of the pancreatic islets. Consistent with the previous consensus definition of insulitis (17), islets were considered infiltrated when 6 or more CD3+ cells were found within or in close proximity (Figure 2O). We found no infiltration in pancreas slices from 4 of 6 ND organ donors examined (Figure 2P). One ND donor (nPOD 6479) displayed 1 of 21 islets analyzed with infiltration while another (nPOD 6471) had 7 of 15 islets with infiltration. Of the donors with T1D, we observed no islets with infiltration in the recent-onset case (nPOD 6456; 0-year duration, 0 of 19 islets), whereas >68% (11 of 16) of islets in the donor with T1D with 1.5-year disease duration (nPOD 6469) showed infiltration. Interestingly, despite demonstrating an almost complete lack of β cells (Figure 2, E, G, and H), all investigated islets (15 of 15) of the 4-year duration T1D case (nPOD 6472) displayed infiltration. Finally, in the T1D case with 10-year disease duration (nPOD 6459), only 1 of 15 islets showed infiltration. Thus, 3D morphometry of tissue slices from organ donors enabled an exceptionally thorough depiction of pancreas morphology in T1D pathogenesis, providing details about alterations of the endocrine compartment and the extent of immune cell infiltration in functionally characterized tissue for each studied case.
Tissue slices uncover functional contributions to insulin insufficiency in T1D pathogenesis.
Because we measured hormone secretion and hormone+ cell volume from the exact same tissue slices (Figures 1 and 2), we were able to normalize insulin secretion to β cell mass. This allowed us to separate the contribution of cell mass to insulin secretion from that of cell function and to specifically compare functionality of the residual β cells across donors (Figure 3). β Cell mass adjusted insulin secretion in response to high glucose and KCl depolarization remained robust in slices from ND donors. In contrast, as β cell mass in the recent-onset (0 years) T1D pancreas was high (Figure 2E), mass-adjusted glucose and KCl-stimulated insulin secretion signified a functional defect of β cells in this organ. However, in slices from the short-duration (1.5 years) T1D case, β cell mass adjustment revealed strong insulin release, particularly under basal and KCl stimulatory conditions (Figure 3), revealing intact basal function and secretory capacity of residual β cells. As described above, insulin secretion from pancreas tissue slices of donors with longer disease duration was undetectable and therefore could not be adjusted to β cell mass.
Figure 3. β Cell function of residual β cell mass.
(A) Insulin secretory pattern of perifused pancreas tissue slices normalized to the assessed Ins+ volume from the same slices from nondiabetic (ND, blue line) and individual type 1 diabetic (orange and red lines) donors during perifusion. (B–D) Area under the curve (AUC) measurements of data presented in A for basal secretion at 3 mM glucose (3G) (B) and at 16.7 mM glucose (16.7G) (C) and perifusion with KCl (D) for nondiabetic donors (ND, blue bar) and 4 donors with type 1 diabetes (orange and red bars), with duration as indicated. Data from ND donors (n = 8) is presented as mean ± SEM.
Imaging of calcium responses in pancreas tissue slices facilitates functional characterization of islet cell signaling in T1D.
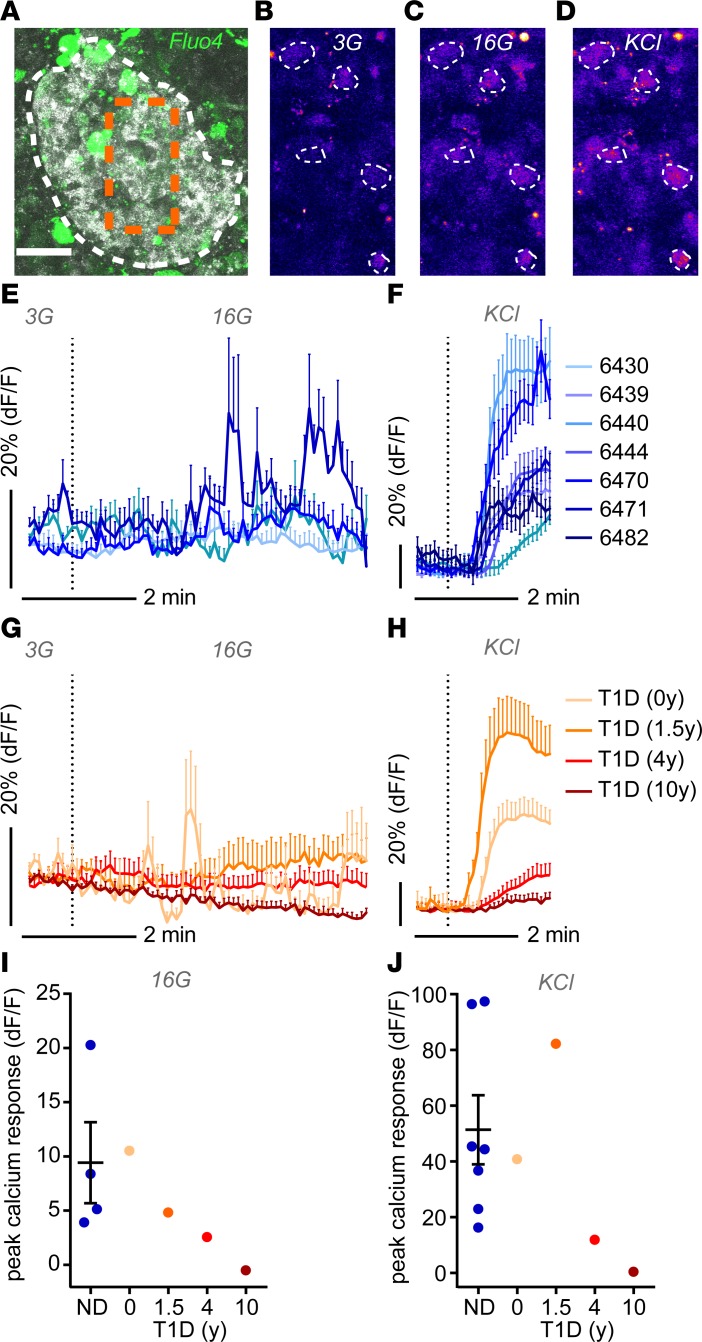
In addition to overall cell function of the studied piece of tissue, pancreas slices also facilitate a more detailed investigation of islet cell physiology. Intracellular Ca2+ kinetics provide an important indicator of islet cell function, as increases in Ca2+ concentration activate multiple physiological processes and trigger hormone release. Hence, we examined changes in intracellular Ca2+ concentration ([Ca2+]i) in response to elevated glucose levels and KCl depolarization (Figure 4). To this end, we loaded living human pancreas slices with the [Ca2+]i indicator Fluo-4 AM and recorded [Ca2+]i responses in single cells within islets by confocal time-lapse imaging (Figures 4, A–D). While the experimental approach used did not allow discrimination of endocrine cell types, we anticipated β cells to represent the majority of cells exhibiting increased [Ca2+]i during stimulation with elevated glucose and KCl levels. Accordingly, in slices from 7 ND donors, high glucose and KCl depolarization elicited [Ca2+]i responses in numerous islet cells (Figures 4, E and F). In pancreas slices from donors with recent-onset (0 years) and short-duration (1.5 years) T1D, [Ca2+]i responses to high glucose and KCl depolarization were comparable to those of ND slices (Figures 4, G–J). In contrast, in pancreata from donors with 4- and 10-year T1D duration, [Ca2+]i responses to either stimuli were strongly diminished (Figures 4, G–J).
Figure 4. Single-cell calcium dynamics in situ.
(A) Image of a human islet within a pancreas tissue slice stained with Fluo4 for live cell imaging of intracellular Ca2+ dynamics. Backscatter signal is shown in gray, Fluo4 signal is shown in green; the white dotted line delineates the islet, and the box represented by the orange dotted line is shown at higher magnification in B–D. (B–D) Fluo4 signal intensities of single cells (encircled with dotted lines) within the islet shown in A at basal glucose levels (3 mM glucose [3G]) (B), at high glucose (16 mM glucose [16G]) (C), and under depolarizing conditions (KCl) (D). (E–H) Ca2+ dynamics expressed as the fluorescent intensity of Fluo4 normalized to the basal signal intensity at 3 mM glucose (3G) at stimulation with high glucose (16G) and depolarizing conditions (KCl) from nondiabetic donors (E and F) and donors with type 1 diabetic (G and H). (I and J) Quantification of the peak calcium response at high glucose (16G) (I) and KCL (J) for all donors analyzed. Data in E and F are presented as mean ± SEM for 5–30 individual cells per donor; data for nondiabetic donors are presented as mean ± SEM, with n = 7 in (E, F, and J) and n = 4 in I. Scale bar: 50 μm (A).
Studies of tissue slices allow detailed characterization and facilitate comparison of pancreas physiology of donor organs.
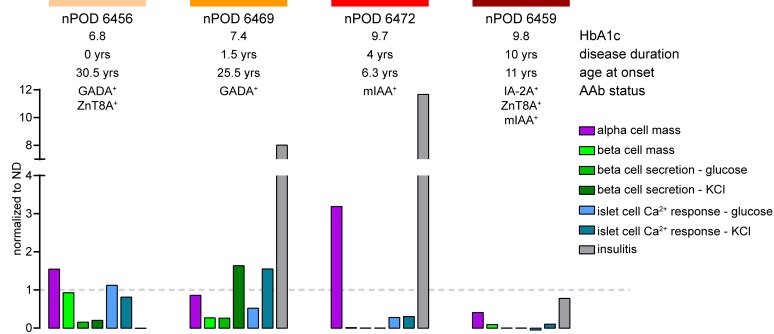
The combined assessment of several crucial parameters from the human pancreas in T1D pathogenesis, including β cell function, β cell mass, β cell Ca2+ activity, and immune cell infiltration, allowed us to establish specific profiles for all of the T1D cases assessed in this study (Figure 5). For each analyzed donor organ, these profiles provide detailed information about the relative contributions of β cell mass loss and β cell dysfunction to insulin insufficiency, possible underlying pathophysiological mechanisms affecting the insulin secretion deficit, and the potential role of ongoing immune infiltration (Figure 5). Our studies show that each studied T1D donor pancreas exhibits a unique profile of pancreas pathophysiology, which can then be correlated with clinical parameters, demonstrating the value of this technology to obtain potentially novel information about the pathogenesis and heterogeneity of human T1D.
Figure 5. Physiological profile of pancreas tissue slices for each T1D donor studied.
Summarized physiological parameters of tissue slices for all studied donors with T1D, including α cell mass, β cell mass, glucose- and KCl-induced β cell secretion, glucose- and KCl-induced islet cell Ca2+ activity, and infiltration, normalized to the mean for studied nondiabetic donors.
Discussion
More detailed knowledge of pathophysiological processes in the human pancreas during T1D pathogenesis is essential for the development of successful prevention and treatment protocols. Here, pancreas tissue slices from organ donors with and without diabetes represent a potentially novel approach for unprecedented in situ studies of T1D development, providing potentially insight into disease mechanisms and heterogeneity.
Concerted efforts to improve access to human pancreatic tissue (through nPOD and other tissue banks) and the establishment of human islet distribution networks (e.g., Integrated Islet Distribution Program) have enabled the study of T1D pathology and human islet biology primarily from fixed or frozen tissues and live isolated islets (14, 35, 36). While the anatomical characterization is advancing rapidly, physiological studies in T1D tissue are still rare. To date, we are aware of only 2 studies conducted on human islets isolated from donors with T1D (21, 22). However, the islet isolation procedure alters the tissue environment and islet biology, thus affecting downstream functional assessments. Moreover, the yield of islets from T1D pancreata is much lower than from ND donors, and there is concern that the islets that are most affected by T1D are unlikely to endure the procedure. The study of intact pancreas tissue slices helps mitigate these concerns, as islets remain undisturbed in their natural environment and are not exposed to the chemical and physical stressors associated with islet isolation.
The tissue slice technique was developed in the rodent model (23) and optimized for fresh human pancreatic tissue from surgical resections (24, 25). In rodents, tissue slices have been shown to be of major value for the study of not only endocrine cell types (e.g., α, β, and δ cells), but also immune, acinar, and vascular cells of the pancreas (24–28, 30, 37). Here, we successfully adapted the tissue slice approach to study islet anatomy and function/physiology in human pancreata from organ donors with and without T1D. We demonstrate that slices from donor organs show preserved tissue integrity and viability. Hence, we obtained detailed profiles of organ donor pancreas biology, including measurements of endocrine cell volume, insulin secretion, and intracellular Ca2+ activity in response to glucose and KCl depolarization, as well as islet infiltration by CD3+ cells. Comparing ND profiles with 4 T1D pancreata from different disease stages provided information about pancreas pathophysiology in each case. We observed a massive loss of insulin secretion and β cell mass with increasing disease duration. Importantly, in the 4 T1D cases studied here, we found the relative contribution of pancreatic β cell mass loss and β cell dysfunction to insulin insufficiency was not strictly related to disease duration. These findings demonstrate the ability of organ donor tissue slices to reveal potential mechanisms underlying disease heterogeneity by allowing a thorough characterization of pancreas physiology for each T1D case.
Interestingly, in the recent-onset T1D donor pancreas (0 years, nPOD 6456), diminished insulin secretion was not due to reduced β cell mass but rather β cell dysfunction. This, for the first time to our knowledge, provides secretion/functional evidence from intact pancreas tissue for the hypothesis that in some cases of T1D, significant β cell mass is preserved at disease onset and that insulin deficiency results from β cell dysfunction (38). Given that this donor was 30.5 years old at diagnosis and had no insulitis in the tissue slice examined, the relationship of mass loss versus dysfunction could be related to a less severe autoimmune attack, which has been suggested for late-onset diabetes (39). This supports the possibility that early and appropriate therapeutic intervention(s) could restore β cell function and glycemic control in a subset of patients with adult-onset T1D. Alternatively, impaired insulin secretion might be a sign of β cell exhaustion, supported by the observation that glucose and depolarization induced changes in islet cell cytosolic Ca2+ were comparable to ND donors. The β cell exhaustion in these slices from the pancreas tail could be due to loss of β cell function and mass in other parts of the organ. However, while histological sections of other parts of the organ showed signs of minor infiltration and β cell mass loss (see Supplemental Table 1 for histopathology report of the nPOD Organ Processing and Pathology Core [OPPC] Aperio database), the lack of substantial β cell loss, speaks against this scenario. A different explanation could be related to the fact that this donor had given birth only days prior to her demise, suggesting that β cell exhaustion might be related to pregnancy associated insulin demand in combination with T1D pathogenesis.
The donor with 1.5-year disease duration had also developed T1D in adulthood (25.5 years, nPOD 6469). The tissue examined showed infiltration in most islets, dramatically reduced β cell mass, and decreased glucose-stimulated β cell secretion, whereas β cell secretion in response to depolarization was intact. Both intracellular Ca2+ activity and insulin secretion showed the same pattern, indicating a selective dysfunction of the residual β cells in response to high glucose concentrations, despite having intact secretion capacity. The major contribution of β cell dysfunction to this case became even more apparent by the histopathology report (Supplemental Table 1) that confirmed the persistence of insulin+/glucagon+ islets in other parts of the pancreas. Thus, we were able to demonstrate β cell dysfunction in 2 donors who developed T1D as adults. The existence of β cell dysfunction, irrespective of β cell mass and infiltration, in the slices examined supports the concept that β cell dysfunction is a critical and possibly early event in T1D pathogenesis that could be targeted therapeutically. This finding contrasts a recent study that reported that the β cells in isolated islets of organ donors with T1D “maintained features of regulated insulin secretion” (22). This discrepancy could be due to several differences in the approaches used in both studies. Whereas Brissova and colleagues analyzed secretion from isolated islets within 72 hours after isolation and expressed insulin secretion as a percentage of insulin content (22), the present study assessed insulin secretion from pancreas tissue slices within 12 hours after organ processing and normalized insulin secretion to β cell mass quantified by 3D morphometry. Thus, both studies address a different status and functional component of β cells in T1D pathogenesis. Given the evidence of β cell dysfunction provided with the here presented approach, pancreas tissue slices from donors with T1D now provide the opportunity to study the underlying mechanisms of β cell dysfunction and may be used as a platform to test candidate treatments.
In contrast to these adult-onset cases, the pancreata with long-disease duration were from childhood-onset organ donors with T1D. Both showed no detectable insulin secretion, less than 10% β cell mass (compared with ND controls), and nearly complete loss of β cell function. Interestingly, case 6472 (4-year disease duration) displayed immune infiltration in or around every investigated islet in the tissue slices, despite the obvious lack of β cells. The presence of insulitis in insulin+ and insulin– islets was also observed in histological sections throughout the pancreas (see Supplemental Table 1 for histopathology report). Conversely, α cell mass was strongly elevated in comparison with slices from ND organ donors. Future studies of pancreas tissue slices will examine α cell mass and function as factors with the potential to promote hyperglycemia in T1D as well as the underlying mechanisms contributing toward ongoing autoimmune infiltration in insulin– tissues. Finally, despite having longer T1D duration (10 years) and the presence of multiple autoantibodies, we detected some residual insulin+ β cells in slices from nPOD 6459 (age at onset 11 years), which were also detectable in histological sections of other pancreatic areas (Supplemental Table 1), yet neither insulin secretion nor Ca2+ activity was observed following glucose or KCl stimulation. The very low frequency of islets with infiltration in this donor might suggest a fading off of the autoimmune attack, despite the presence of β cells in the tissue. We hypothesized that the residual β cells were in a dysfunctional state and perhaps as result, less visible to the immune system.
In summary, our study demonstrates that the pancreas tissue slice technique can be used to study transplant-quality organs from human donors and has the potential to deepen our understanding of the pathogenic processes related to T1D. Through the combination of this platform with additional methodologies, such as patch-clamp, imaging mass cytometry, or electron microscopy, future studies will be able to address important open questions of T1D pathogenesis, including the distinct roles of α, β, and δ cells; the cross-talk of endocrine and immune cells; or the functionality of single, persisting β cells. Thus, this potentially novel platform now allows the coupling of functional and anatomical parameters with genetic, histological, and other forms of data generated from the same organ/donor by investigators across the nPOD research network. Therefore, it is likely that the use of the pancreas tissue slice technology will provide insight into T1D pathogenesis and enable improved characterization of disease heterogeneity.
Limitations of the study.
In our study we only address a limited number of T1D donor organs, which not only differ in disease duration, but also in age at onset and AAb profile. Thus, application of our findings to general disease pathogenesis is limited. However, the cases included here demonstrate the feasibility of characterizing functional and morphological parameters of T1D pathogenesis, at different disease stages, and how utilization of the pancreas tissue slice platform can provide details about cellular processes and disease heterogeneity. To allow more general conclusions about disease mechanisms our studies need to be extended. Another limitation is that the study of tissue slices only assesses the biology of a given pancreatic region, which therefore can only allow extrapolation of processes occurring throughout the entire organ. We try to compensate for this limitation by evaluating considerable tissue volume via the simultaneous study of several tissue slices. In addition, histopathology reports from histological sections of numerous regions throughout the entire organ, which are prepared by the nPOD OPPC and are available in the nPOD database for each donor organ (see also Supplemental Table 1), represent an excellent resource and control to place the studied tissue slices in context of the histopathology of the entire organ. Finally, we are aware that using organs from brain-dead donors as tissue source might have an effect on organ physiology and disease pathogenesis. However, because it is not possible to obtain pancreas biopsies from living patients, the study of organ donors through the well-coordinated procurement of transplantation-grade organs, as implemented by nPOD, currently represents the most suitable, effective, and safe way to study the pancreas with T1D.
Methods
Human organ donors.
Pancreas tissue from human donors with or without T1D was procured from the nPOD program at the University of Florida (https://www.jdrfnpod.org). For each donor, a medical chart review was performed, and C-peptide was measured (40), with T1D diagnosed according to the guidelines established by the ADA. Demographic data, hospitalization duration, and organ transport time were obtained from hospital records. Donor pancreata were recovered, placed in transport media on ice, and shipped via organ courier to the University of Florida. Tissue was processed by a licensed pathology assistant (41). Detailed donor information is listed in Table 1.
Tissue processing and slicing.
Tissue pieces of approximately 1 g were obtained from the pancreas body-tail juncture and placed into ECS solution (125 mM NaCl, 2.5 mM KCl, 26 mM NaHCO3, 1.25 mM NaH2PO4, 1 mM MgCl2, 2 mM CaCl2, 10 mM HEPES, 3 mM glucose, pH 7.4) supplemented with aprotinin (25 KIU/mL, MilliporeSigma, catalog A6106). Tissue pieces were further processed: they were cut into smaller tissue blocks, and connective, adipose, and fibrotic tissue were removed. Small tissue blocks of about 0.5 cm3 were embedded in low-melting-point agarose (3.8%, MilliporeSigma, A0701) and mounted on a specimen holder using superglue (Superglue 90-120 CPS, World Precision Instruments Inc., catalog 7341). The holder was connected to the tray of a semiautomatic vibratome (Leica VT1200S, catalog 14048142066) and filled with cold ECS solution supplemented with aprotinin. Tissue slicing was performed at a step size of 120 μm, speed of 0.1 mm/s, amplitude of 1 mm, and an angle of 15 degrees. Slices were collected into a 60-mm Petri dish containing 3 mM glucose KRBH buffer (137 mM NaCl, 5.36 mM KCl, 0.34 mM Na2HPO4, 0.81 mM MgSO4, 4.17 mM NaHCO3, 1.26 mM CaCl2, 0.44 mM KH2PO4, 10 mM HEPES, 0.1% BSA, 3 mM glucose, pH 7.3) supplemented with aprotinin. After slices were generated, they were cultured for up to 12 hours in media (HyClone Dulbecco’s Low Glucose Modified Eagles Medium, Thermo Fisher, catalog SH3002101) containing FBS (10%, GE Healthcare HyClone Fetal Bovine Serum, Thermo Fisher, catalog SH3008803IR), antibiotic antimycotic solution (Corning, catalog 30-004-CL 1:100) and aprotinin in an incubator at 28°C and 5 % CO2 atmosphere. Prior to perifusion, slices were kept for 90–120 minutes in 3 mM KRBH buffer supplemented with aprotinin at room temperature while shaking. Additionally, a few slices were placed in cooled media (HyClone Dulbecco’s Low Glucose Modified Eagles Medium, Thermo Fisher, catalog SH3002101) supplemented with FBS (1%, GE Healthcare HyClone Fetal Bovine Serum, Thermo Fisher, catalog SH3008803IR), antibiotic antimycotic solution (1:1000 dilution, Corning, catalog 30-004-CL), and aprotinin 25 KIU/mL, immediately after slicing and shipped to the Division of Metabolism, Endocrinology and Diabetes at the University of Miami Miller School of Medicine, where tissue viability and cell function were assessed.
Tissue viability.
Cell viability within the tissue slices was evaluated by live/dead immunofluorescence staining (Live/Dead Cell Imaging Kit [488/570], Thermo Fisher, catalog R37601). Slices were incubated according to manufacturer’s protocol. Briefly, live green (Component A) and dead red (Component B) were mixed to create a 2× working solution. The solution was diluted with 1 mL ECS buffer, and Zinquin ethyl ester (1:200 dilution, Abcam, catalog 145478) and NucSpot Live 650 (2 μL, Invitrogen, catalog R37601) were added to the solution. Two to four tissue slices were incubated for 15–20 minutes at room temperature while shaking. Slices were transferred into PBS (MilliporeSigma, catalog D8537-500ml) and imaged using an inverted fluorescence phase contrast microscope (Keyence, BZ-X710).
Perifusion.
To assess glucose-stimulated insulin release, 3–4 viable tissue slices were placed into a closed perifusion chamber (Warner Instruments, catalog 64-0223 and catalog 64-0281 [P-5]) and connected to a perifusion system with automated tray handling (Biorep Technologies, catalog PERI4-02-230-FA). Tissue slices were perfused at a flow rate of 100 μL/minute, and samples were collected in 96-well plates with a 60-second interval. Slices were flushed for 60 minutes with 3 mM glucose KRBH containing aprotinin (25 KIU/mL) to wash out accumulated hormones and enzymes from the tissue. Then, slices were perfused with KRBH buffer with 3 mM glucose for 10 minutes, followed by 20 minutes in 16.7 mM glucose, 30 minutes with 1 mM glucose, 5 minutes with 3 mM and 60 mM KCL, and, after that, 10 minutes in 3 mM glucose. Perfusates were stored at –20°C until insulin content was measured using a commercially available ELISA kit (Mercodia, human insulin ELISA, catalog no. 10-1113-01 for ND organ donors and Mercodia, ultrasensitive human insulin ELISA, catalog no. 10-1132-01 for organ donors with T1D). After perifusion, tissue slices were gently removed from the chambers, fixed with 4% paraformaldehyde for 30 minutes at room temperature and kept in PBS at 4°C. Fixed slices were then shipped in PBS to the Paul Langerhans Institute Dresden to assess intact 3D islet morphology in the perfused slices by IHC.
Immunofluorescent staining.
All antibody solutions were prepared in 0.6% GSDB (30% goat serum, 900 mM NaCl, 40 mM sodium phosphate buffer [pH 7.4], 0.6% Triton X-100). Slices were incubated with primary antibodies against insulin (1:500, guinea pig, Dako, catalog A-0546), glucagon (1:2000, mouse, MilliporeSigma, catalog G2654), and CD3 (1:50, rabbit, Dako, catalog A-0452) overnight at 4°C while shaking. Slices were washed 3 times for at least 30 minutes with PBS and incubated with secondary antibodies: Alexa Fluor 488 goat anti–guinea pig (1:200, Invitrogen, catalog A-11073), Alexa Fluor 546 goat anti-rabbit (1:200, Invitrogen, catalog A-11035), Alexa Fluor 633 goat anti-mouse (1:200, Invitrogen, catalog A-21050), and DAPI (2.5 mg/ml, MilliporeSigma, catalog D9542) overnight at 4°C while shaking. Slices were washed 3 times for at least 30 minutes with PBS and kept at 4°C in the dark until imaging.
For single-islet imaging in order to quantify CD3+ cells, tissue slices were cleared using the recently described IsoScaleSQ solution (9.1 M urea, 22.5% D-sorbitol, 200 mM sodium chloride, 2% [v/w] Triton X-100) for 60 minutes (42).
Whole slice and single-islet imaging and analysis.
Stained slices were mounted under a glass coverslip submerged in PBS (pH 7.4) and imaged entirely using an upright laser scanning confocal microscope (LSM 780 NLO, Zeiss) with a Plan-Apochromat 20×/1.0 water-immersion objective (Zeiss) and an automated stage (Zeiss). Single tiles were image at a resolution of 0.75 μm × 0.75 μm, a tile overlap of 15%, and stack separation of 2.5 μm. DAPI and fluorescent secondary antibodies were excited at 405 nm, 488 nm, 561 nm, and 633 nm and detected at 405–465 nm (DAPI), 488–550 nm (insulin), 570–620 nm (CD3), and 640–720 nm (glucagon), respectively. Single islets were imaged in cleared mounted tissue slices submerged in IsoScaleSQ solution at a resolution of 0.19 μm × 0.19 μm and a Z interval of 1.5 μm.
Tile scan analysis.
Raw tile scans were stitched using the Fiji Stitching plugin (43, 44). Fusion was completed by maximum intensity, with a regression threshold of 0.3, maximum displacement threshold of 2.50, and absolute displacement threshold of 3.50. The overlap was computed with subpixel accuracy. Stitched tile scans were contoured manually for the total tissue area and for each of the individual clusters/islets on a maximum intensity projection, with the ROIs saved separately. For individual endocrine objects, the regions were individually cropped from the stitched tile scan. To account for bleed-through, signals were subtracted from each other (insulin subtracted from CD3, CD3 subtracted from glucagon). Images were then split by color, contrast enhanced (CLAHE), median filtered (3 × 3 × 1), and the background was subtracted (7.5 pixels, sliding paraboloid). Single planes with a pixel value standard deviation less than 1 were removed, a variance filter was applied to the images (3 × 3 × 1) and binarized (Li). Hole filling in 3D and size opening (limit, 500 μm3) was performed, followed by volume analysis (Euler connectivity: C26) using MorphoLibJ (45). For total slice volume, each channel of the original tile scan was added together and cropped to the total tissue ROI. The image was median filtered (3 × 3 × 1), and a mask of nearby points was generated within a distance of 10.0 μm and threshold of 12.75 using the built-in Fiji plugin Mask Of Nearby Points. Dark and bright outliers (respectively) with a radius of 20 pixels each were removed. Holes were filled, 3D volume closing was run (cube, 3 × 3 × 1), and volume was quantified using MorphoLibJ (45).
CD3 cell quantification.
Individual imaged islets were contoured and masked as described above. The mask was cropped to the main body of the islet by keeping the largest contiguous region (45). The convex hull (44) of each islet in 3D was generated and dilated by 50 μm to obtain a quantification volume, including the periphery of the islet. CD3+ cells within this volume were individually counted by 3 persons for each imaged islet.
Calcium imaging.
Two to three viable human slices were incubated with Fluo4-AM (6 μM, Invitrogen, catalog F1221) for 1 hour in 3 mM HEPES buffer (125 mmol/l NaCl, 5.9 mmol/l KCl, 2.56 mmol/l CaCl2, 1 mmol/l MgCl2, 25 mmol/l HEPES, 0.1% BSA [w/v], pH 7.4) supplemented with aprotinin (25 KIU, MilliporeSigma, catalog A6106) at room temperature in the dark. Slices were then placed on a coverslip in an imaging chamber (Warner instruments) for imaging on a Leica TCS SP5 upright laser-scanning confocal microscope (Leica Microsystems). Slices were continuously perfused with HEPES buffer solution with 3 mM glucose, 16 mM glucose, or 3 mM glucose with 30 mM KCl. Confocal images were acquired with LAS AF software (Leica Microsystems) using a ×40 water immersion objective (NA 0.8). A resonance scanner was used for fast image acquisition producing time-lapse recordings spanning 50–100 μm of the slice (Z step: 5 μm, stack of 10 confocal images with a size of 512 × 512 pixels) at 5-second resolution (XYZT imaging). Backscatter light was collected upon excitation with the 633 nm laser, and Fluo4-AM fluorescence was excited at 488 nm and emission detected at 510–550 nm.
Data were analyzed from maximum intensity projections over time using ImageJ (NIH). The average intensity of Fluo4-AM fluorescence was measured by drawing single ROIs for individual islet cells. Changes in cytosolic Ca2+ level response were calculated as increases relative to the mean baseline of each cell (ΔF/F) and are reported as percentage difference.
Statistics.
No specific statistical methods were used to predetermine sample size. All results are presented as mean ± SEM.
Study approval.
All procedures were performed according to the established standard operating procedures of the nPOD/OPPC and approved by the University of Florida Institutional Review Board (IRB201600029) and the United Network for Organ Sharing (UNOS) according to federal guidelines, with informed consent obtained from each donor’s legal representative.
Author contributions
Methods used in assigning the shared authorship were as follows: JKP established the slice technology from brain-dead organ donors, HH provided the perifusion data, CMC provided the morphometry data, and JA provided the live cell imaging data. MAA, AP, AC, IK, and SS conceived and designed the experiments. SJE, MB, SC, DMD, JRW, JS, MKH, MMFQ, EAP, and IK contributed to the conduction of the experiments. JKP, HH, CMC, JA, SJE, MB, SC, DMD, JRW, JS, MKH, MMFQ, RLP, JDB, EAP, AC, IK, and SS analyzed data. JKP, HH, CMC, JA, SJE, DMD, MAA, EAP, AP, AC, and SS prepared figures and/or contributed to the writing of the manuscript.
Supplementary Material
Acknowledgments
We acknowledge the support of the nPOD (RRID:SCR_014641), a collaborative T1D research project sponsored by JDRF (nPOD: 5-SRA-2018-557-Q-R), The Leona M. and Harry B. Helmsley Charitable Trust (grant 2018PG-T1D053, 2018PG-T1D060, and 2015PG-T1D052), and by the Diabetes Research Institute Foundation. The content and views expressed are the responsibility of the authors and do not necessarily reflect the official view of nPOD. Organ Procurement Organizations (OPO) partnering with nPOD to provide research resources are listed at http://www.jdrfnpod.org//for-partners/npod-partners Finally, we wholeheartedly thank the donors and their families for their invaluable contribution to science. Furthermore, this work was supported by NIH grant 1U01DK120456-01 and JRDF grant 2-SRA-2019-696-S-B as well as the Paul Langerhans Institute Dresden (PLID) of the Helmholtz Zentrum München at the University Clinic Carl Gustav Carus of Technische Universität Dresden, the German Ministry for Education and Research to the German Centre for Diabetes Research, the DFG Collaborative Research Centre/Transregio 127, the DFG — International Research Training Group 2251, and the DFG Center for Regenerative Therapies and Cluster of Excellence (CRTD).
Version 1. 04/23/2020
Electronic publication
Footnotes
JKP’s present address is: Department of Medicine, Division of Metabolism, Endocrinology and Diabetes, University of Miami Miller School of Medicine, Miami, Florida, USA.
JRW’s present address is: Department of Pediatrics, UCSD, San Diego, California, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(8):e134525.https://doi.org/10.1172/jci.insight.134525.
Contributor Information
Julia K. Panzer, Email: Julia.panzer@mailbox.tu-dresden.de.
Helmut Hiller, Email: hhiller@ufl.edu.
Christian M. Cohrs, Email: Christian.Cohrs@tu-dresden.de.
Joana Almaça, Email: jalmaca@med.miami.edu.
Stephen J. Enos, Email: stephen.enos@tu-dresden.de.
Sirlene Cechin, Email: scechin@med.miami.edu.
Denise M. Drotar, Email: denise-minerva.drotar@tu-dresden.de.
Mollie K. Huber, Email: mhuber@ufl.edu.
Mirza Muhammad Fahd Qadir, Email: fahd.qadir@med.miami.edu.
Juan Domínguez-Bendala, Email: jdominguez2@med.miami.edu.
Edward A. Phelps, Email: EPhelps@bme.ufl.edu.
Alberto Pugliese, Email: apuglies@med.miami.edu.
Alejandro Caicedo, Email: acaicedo@med.miami.edu.
Irina Kusmartseva, Email: inkusmartseva@ufl.edu.
Stephan Speier, Email: stephan.speier@tu-dresden.de.
References
- 1.Gepts W. Pathologic anatomy of the pancreas in juvenile diabetes mellitus. Diabetes. 1965;14(10):619–633. doi: 10.2337/diab.14.10.619. [DOI] [PubMed] [Google Scholar]
- 2.Gepts W, De Mey J. Islet cell survival determined by morphology. An immunocytochemical study of the islets of Langerhans in juvenile diabetes mellitus. Diabetes. 1978;27 Suppl 1:251–261. doi: 10.2337/diab.27.1.s251. [DOI] [PubMed] [Google Scholar]
- 3.Klöppel G, Drenck CR, Oberholzer M, Heitz PU. Morphometric evidence for a striking B-cell reduction at the clinical onset of type 1 diabetes. Virchows Arch A Pathol Anat Histopathol. 1984;403(4):441–452. doi: 10.1007/BF00737292. [DOI] [PubMed] [Google Scholar]
- 4.Skyler JS, Ricordi C. Stopping type 1 diabetes: attempts to prevent or cure type 1 diabetes in man. Diabetes. 2011;60(1):1–8. doi: 10.2337/db10-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rowe PA, Campbell-Thompson ML, Schatz DA, Atkinson MA. The pancreas in human type 1 diabetes. Semin Immunopathol. 2011;33(1):29–43. doi: 10.1007/s00281-010-0208-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jerram ST, Leslie RD. The genetic architecture of type 1 diabetes. Genes (Basel) 2017;8(8):E209. doi: 10.3390/genes8080209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Atkinson MA, Leiter EH. The NOD mouse model of type 1 diabetes: as good as it gets? Nat Med. 1999;5(6):601–604. doi: 10.1038/9442. [DOI] [PubMed] [Google Scholar]
- 8.Pearson JA, Wong FS, Wen L. The importance of the non obese diabetic (NOD) mouse model in autoimmune diabetes. J Autoimmun. 2016;66:76–88. doi: 10.1016/j.jaut.2015.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roep BO, Atkinson M, von Herrath M. Satisfaction (not) guaranteed: re-evaluating the use of animal models of type 1 diabetes. Nat Rev Immunol. 2004;4(12):989–997. doi: 10.1038/nri1502. [DOI] [PubMed] [Google Scholar]
- 10.Roep BO, Atkinson M. Animal models have little to teach us about type 1 diabetes: 1. In support of this proposal. Diabetologia. 2004;47(10):1650–1656. doi: 10.1007/s00125-004-1517-1. [DOI] [PubMed] [Google Scholar]
- 11.Roep BO, Buckner J, Sawcer S, Toes R, Zipp F. The problems and promises of research into human immunology and autoimmune disease. Nat Med. 2012;18(1):48–53. doi: 10.1038/nm.2626. [DOI] [PubMed] [Google Scholar]
- 12.Bosco D, et al. Unique arrangement of alpha- and beta-cells in human islets of Langerhans. Diabetes. 2010;59(5):1202–1210. doi: 10.2337/db09-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonner-Weir S, Sullivan BA, Weir GC. Human islet morphology revisited: human and rodent islets are not so different after all. J Histochem Cytochem. 2015;63(8):604–612. doi: 10.1369/0022155415570969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brissova M, et al. Assessment of human pancreatic islet architecture and composition by laser scanning confocal microscopy. J Histochem Cytochem. 2005;53(9):1087–1097. doi: 10.1369/jhc.5C6684.2005. [DOI] [PubMed] [Google Scholar]
- 15.Kaddis JS, Pugliese A, Atkinson MA. A run on the biobank: what have we learned about type 1 diabetes from the nPOD tissue repository? Curr Opin Endocrinol Diabetes Obes. 2015;22(4):290–295. doi: 10.1097/MED.0000000000000171. [DOI] [PubMed] [Google Scholar]
- 16.Keenan HA, et al. Residual insulin production and pancreatic ß-cell turnover after 50 years of diabetes: Joslin Medalist Study. Diabetes. 2010;59(11):2846–2853. doi: 10.2337/db10-0676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campbell-Thompson M, et al. Insulitis and β-cell mass in the natural history of type 1 diabetes. Diabetes. 2016;65(3):719–731. doi: 10.2337/db15-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coppieters KT, et al. Demonstration of islet-autoreactive CD8 T cells in insulitic lesions from recent onset and long-term type 1 diabetes patients. J Exp Med. 2012;209(1):51–60. doi: 10.1084/jem.20111187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pugliese A. Insulitis in the pathogenesis of type 1 diabetes. Pediatr Diabetes. 2016;17 Suppl 22:31–36. doi: 10.1111/pedi.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skyler JS. Characterizing subgroups of type 1 diabetes. Diabetes. 2014;63(11):3578–3580. doi: 10.2337/db14-1160. [DOI] [PubMed] [Google Scholar]
- 21.Krogvold L, et al. Function of isolated pancreatic islets from patients at onset of type 1 diabetes: insulin secretion can be restored after some days in a nondiabetogenic environment in vitro: results From the DiViD Study. Diabetes. 2015;64(7):2506–2512. doi: 10.2337/db14-1911. [DOI] [PubMed] [Google Scholar]
- 22.Brissova M, et al. α Cell function and gene expression are compromised in type 1 diabetes. Cell Rep. 2018;22(10):2667–2676. doi: 10.1016/j.celrep.2018.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Speier S, Rupnik M. A novel approach to in situ characterization of pancreatic beta-cells. Pflugers Arch. 2003;446(5):553–558. doi: 10.1007/s00424-003-1097-9. [DOI] [PubMed] [Google Scholar]
- 24.Marciniak A, et al. Using pancreas tissue slices for in situ studies of islet of Langerhans and acinar cell biology. Nat Protoc. 2014;9(12):2809–2822. doi: 10.1038/nprot.2014.195. [DOI] [PubMed] [Google Scholar]
- 25.Cohrs CM, et al. Dysfunction of persisting β cells is a key feature of early type 2 diabetes pathogenesis. Cell Rep. 2020;31(1):107469. doi: 10.1016/j.celrep.2020.03.033. [DOI] [PubMed] [Google Scholar]
- 26.Cohrs CM, et al. Vessel network architecture of adult human islets promotes distinct cell-cell interactions in situ and is altered after transplantation. Endocrinology. 2017;158(5):1373–1385. doi: 10.1210/en.2016-1184. [DOI] [PubMed] [Google Scholar]
- 27.Weitz JR, et al. Mouse pancreatic islet macrophages use locally released ATP to monitor beta cell activity. Diabetologia. 2018;61(1):182–192. doi: 10.1007/s00125-017-4416-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Almaça J, Weitz J, Rodriguez-Diaz R, Pereira E, Caicedo A. The pericyte of the pancreatic islet regulates capillary diameter and local blood flow. Cell Metab. 2018;27(3):630–644.e4. doi: 10.1016/j.cmet.2018.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meneghel-Rozzo T, Rozzo A, Poppi L, Rupnik M. In vivo and in vitro development of mouse pancreatic beta-cells in organotypic slices. Cell Tissue Res. 2004;316(3):295–303. doi: 10.1007/s00441-004-0886-6. [DOI] [PubMed] [Google Scholar]
- 30.Speier S, Gjinovci A, Charollais A, Meda P, Rupnik M. Cx36-mediated coupling reduces beta-cell heterogeneity, confines the stimulating glucose concentration range, and affects insulin release kinetics. Diabetes. 2007;56(4):1078–1086. doi: 10.2337/db06-0232. [DOI] [PubMed] [Google Scholar]
- 31.Huang YC, Rupnik M, Gaisano HY. Unperturbed islet α-cell function examined in mouse pancreas tissue slices. J Physiol (Lond) 2011;589(Pt 2):395–408. doi: 10.1113/jphysiol.2010.200345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dolenšek J, Stožer A, Skelin Klemen M, Miller EW, Slak Rupnik M. The relationship between membrane potential and calcium dynamics in glucose-stimulated beta cell syncytium in acute mouse pancreas tissue slices. PLoS One. 2013;8(12):e82374. doi: 10.1371/journal.pone.0082374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stožer A, et al. Functional connectivity in islets of Langerhans from mouse pancreas tissue slices. PLoS Comput Biol. 2013;9(2):e1002923. doi: 10.1371/journal.pcbi.1002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wasserfall C, et al. Persistence of pancreatic insulin mRNA expression and proinsulin protein in type 1 diabetes pancreata. Cell Metab. 2017;26(3):568–575.e3. doi: 10.1016/j.cmet.2017.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cabrera O, Berman DM, Kenyon NS, Ricordi C, Berggren PO, Caicedo A. The unique cytoarchitecture of human pancreatic islets has implications for islet cell function. Proc Natl Acad Sci USA. 2006;103(7):2334–2339. doi: 10.1073/pnas.0510790103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dorrell C, et al. Transcriptomes of the major human pancreatic cell types. Diabetologia. 2011;54(11):2832–2844. doi: 10.1007/s00125-011-2283-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marciniak A, Selck C, Friedrich B, Speier S. Mouse pancreas tissue slice culture facilitates long-term studies of exocrine and endocrine cell physiology in situ. PLoS One. 2013;8(11):e78706. doi: 10.1371/journal.pone.0078706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodriguez-Calvo T, et al. Increase in pancreatic proinsulin and preservation of β-cell mass in autoantibody-positive donors prior to type 1 diabetes onset. Diabetes. 2017;66(5):1334–1345. doi: 10.2337/db16-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poudel A, et al. Beta-cell destruction and preservation in childhood and adult onset type 1 diabetes. Endocrine. 2015;49(3):693–702. doi: 10.1007/s12020-015-0534-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Campbell-Thompson M, et al. Network for Pancreatic Organ Donors with Diabetes (nPOD): developing a tissue biobank for type 1 diabetes. Diabetes Metab Res Rev. 2012;28(7):608–617. doi: 10.1002/dmrr.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Campbell-Thompson ML, et al. Collection protocol for human pancreas. J Vis Exp. 2012;(63):e4039. doi: 10.3791/4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sato Y, Miyawaki T, Ouchi A, Noguchi A, Yamaguchi S, Ikegaya Y. Quick visualization of neurons in brain tissues using an optical clearing technique. Anat Sci Int. 2019;94(2):199–208. doi: 10.1007/s12565-018-00473-z. [DOI] [PubMed] [Google Scholar]
- 43.Preibisch S, Saalfeld S, Tomancak P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics. 2009;25(11):1463–1465. doi: 10.1093/bioinformatics/btp184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Legland D, Arganda-Carreras I, Andrey P. MorphoLibJ: integrated library and plugins for mathematical morphology with ImageJ. Bioinformatics. 2016;32(22):3532–3534. doi: 10.1093/bioinformatics/btw413. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.