Abstract
Background:
When β-blockers produce reverse-remodeling in idiopathic dilated cardiomyopathy (IDC) they partially reverse changes in fetal-adult/contractile protein, natriuretic peptide, SR-Ca2+-ATPase gene “program” constituents. The objective of the current study was to further test the hypothesis that reverse-remodeling is associated with favorable changes in myocardial gene expression by measuring additional contractile, signaling and metabolic genes that exhibit a fetal/adult expression predominance, are thyroid hormone-responsive, and/or are regulated by β1-adrenergic receptor signaling. A secondary objective was to identify which of these putative regulatory networks is most closely associated with observed changes.
Methods and Results:
Forty-seven IDC patients (LVEF 0.24±0.09) were randomized to the adrenergic-receptor blockers metoprolol (β1-selective), metoprolol+doxazosin (β1/α1), or carvedilol (β1/β2/α1). Serial radionuclide ventriculography and endomyocardial biopsies were performed at baseline, 3, and 12 months. Expression of 50 mRNA gene products was measured by quantitative PCR. Thirty-one patients achieved LVEF reverse-remodeling response defined as improvement by ≥0.08 at 12 months or by ≥0.05 at 3 months (ΔLVEF=0.21±0.10). Changes in gene expression in Responders vs. Nonresponders were decreases in NPPA and NPPB and increases in MYH6, ATP2A2, PLN, RYR2,ADRA1A, ADRB1, MYL3, PDFKM, PDHX, and CPT1B. All except PDHX involved increase in adult or decrease in fetal cardiac genes, but 100% were concordant with changes predicted by inhibition of β1-adrenergic signaling.
Conclusions:
In addition to known gene expression changes, additional calcium-handling, sarcomeric, adrenergic signaling, and metabolic genes were associated with reverse-remodeling. The pattern suggests a fetal-adult paradigm but may be due to reversal of gene expression controlled by a ‘β1-adrenergic receptor gene network.’
Trial Registration:
Keywords: myocardial gene expression, beta-blocker, dilated cardiomyopathy, heart failure
Subject Codes: [155]Physiological and pathological control of gene expression, [93]Receptor pharmacology, [110]Congestive heart failure, [115]Remodeling
INTRODUCTION
Idiopathic dilated cardiomyopathy (IDC) is a common type of nonischemic dilated cardiomyopathy (DCM) for which no cause is readily apparent.1 The main phenotypic feature of IDC and other types of Heart Failure with reduced Ejection Fraction (HFrEF) is ventricular chamber remodeling characterized by contractile dysfunction and “eccentric” pathologic hypertrophy. These processes are commonly detected and quantified by measurement of left ventricular (LV) volumes and the derived ejection fraction (EF), which relates a measure of systolic function (stroke volume) to the degree of eccentric hypertrophy (end diastolic volume, EDV). IDC in humans is associated with myocardial gene expression changes affecting contractile function and hypertrophy.2,3 Among these are up-regulation in mRNA and protein abundances of β-myosin heavy chain (MYH7) and atrial natriuretic peptide (NPPA) with down-regulation of α-myosin heavy chain (MYH6) and sarcoplasmic reticulum calcium-ATPase 2 (ATP2A2), a pattern also observed during fetal cardiac development.2,3 Reversal of these pathologic changes is associated with ventricular reverse-remodeling in response to both β1-selective and nonselective β-adrenergic receptor (AR) antagonists,3 suggesting that reversal of abnormalities in gene expression driven by chronic β1-AR stimulation may mediate salutary effects of β-blockers in IDC and potentially other forms of HFrEF.
Relationships between β1-adrenergic signaling, changes in contractile- and hypertrophy-related gene expression, progression of LV dysfunction and reversal of molecular and structural remodeling by blockade of β1-ARs are incompletely understood. Analysis of gene expression in septal endomyocardial biopsies obtained by right heart catheterization in the presence of β-blocker-associated reverse-remodeling constitutes a human model of gene function associated with changes in ventricular myocardial phenotype. We previously reported effects of β-blockers on human myocardial remodeling and gene expression using an early-generation method of reverse transcription-quantitative polymerase chain reaction (RT-qPCR) that could measure only a small number of mRNAs.3 That work was necessarily narrowly focused, and other gene categories potentially important in therapeutic response to β-blockers were not investigated. New tools using small amounts of RNA including improvements in qPCR methods and microarrays4 have greatly increased the potential yield of investigations where serial measurements of gene expression are coupled to therapeutic modulation of organ-specific phenotype.
We present here the β-blocker Effect On Remodeling and Gene Expression Trial (BORG, NCT01798992), which combined gene expression analysis of endomyocardial biopsy specimens with phenotypic measurements of LV structure and function. BORG is a next-generation longitudinal study of myocardial gene expression and reverse-remodeling in IDC patients treated with 3 different regimens of AR antagonists that have in common blockade of β1-AR. BORG investigated relationships between myocardial gene expression and cardiac phenotypic change, the contribution of α1- and β2-AR blockade, and changes in expression of gene families beyond the current cardiac fetal/adult gene program paradigm. The primary hypothesis was that ventricular reverse-remodeling associated with β1-AR blockade is driven by changes in contractile- or hypertrophy-modifying myocardial gene expression that either precedes or occurs contemporaneously with reverse-remodeling. Specifically, we hypothesized that candidate genes involved in AR signaling, renin-angiotensin and endothelin systems, cytokine signaling, muscle contraction, calcium handling, metabolism, and gene transcription would exhibit unique changes comprising a therapeutic molecular phenotype associated with ventricular reverse-remodeling and that observed changes would fit into a fetal/adult3, β1-AR driven5,6 or thyroid hormone-responsive pattern.7 To test this hypothesis, we quantified candidate gene mRNA expression using qPCR and microarray analysis in the context of changes in LV structure and function in response to β-blocker therapy.
METHODS
The primary outcome was a positive LVEF reverse-remodeling response defined as an improvement ≥8 EF units at 12 months or if not available, an improvement ≥5 units at 3 months (last-observation-carried-forward or LOCF). The LVEF definition of β-blocker response was based on previous observations of improved LVEF with β-blockers compared to placebo.3,8 The 3-month LVEF response cutoff was based on previously demonstrated favorable changes in fetal gene program components associated with an improvement ≥5 EF units after a longer treatment interval of 6 months.3 The 12-month cutoff was defined based on a blinded, prospective analysis of mean LVEF improvement at 12 months among those with improvement ≥5 EF units at 3 months. Nonresponse was defined as not meeting the positive response criteria or the occurrence of heart transplant, left ventricular assist device placement or death.
Expression of 50 candidate mRNA gene products and 2 reference genes (Table 1) was quantified by RT-qPCR. Gene expression changes associated with pathologic LV remodeling were identified by comparing differences in gene expression between IDC and nonfailing control patients. Changes associated with reverse-remodeling were identified by comparing gene expression at LOCF to baseline gene expression in patients with a positive LVEF response (“Responders”) to patients with LVEF nonresponse (“Nonresponders”).3 Analysis of differences in gene expression between the 3 AR-blocking groups was a secondary objective and will be reported separately.
Table 1 –
Genes measured by RT-qPCR
| Pathway | Symbol | Gene Name | Pathway | Symbol | Gene Name |
|---|---|---|---|---|---|
| Adrenergic signal transduction (N=11) | ADRB1 | Αdrenoceptor β1 | Contractile, cytoskeleton proteins (N=12) | MYH6 | Μyosin, heavy chain 6, cardiac, alpha |
| ADRB2 | Αdrenoceptor β2 | MYH7 | Μyosin, heavy chain 7, cardiac, beta | ||
| HNRNPD | Heterogeneous nuclear ribonucleoprotein D | ACTA1 | Actin, α1, skeletal | ||
| ADRBK1 | Adrenergic, beta, receptor kinase 1 | ACTC1 | Αctin, α1, cardiac | ||
| GNAS | GNAS complex locus | MYL2 | Myosin light chain 2, regulatory, cardiac, slow | ||
| GNAI2 | G protein, α inhibiting activity polypeptide 2 | MYL3 | Myosin light chain 3, alkali; vent., skeletal, slow | ||
| ADCY5 | Adenylate cyclase 5 | MYL4 | Myosin light chain 4, alkali; atrial, embryonic | ||
| ADRA1A | Adrenoceptor α 1A | MYL7 | Myosin light chain 7, regulatory | ||
| GNAQ | G protein, q polypeptide | TNNT2 | Troponin T type 2 | ||
| PRKCB | Protein kinase C, β | TNNI3 | Troponin I type 3 | ||
| SLC9A1 | Solute carrier family 9, (Na+/H+ exchanger), member 1 | TNNC1 | Troponin C type 1 | ||
| RAAS/endothelin (N=4) | ACE | Angiotensin I converting enzyme 1 | Counter-regulatory, transcription, other factors (N=6) | DMD | Dystrophin |
| AGT | Angiotensinogen | NPPA | Νatriuretic peptide A | ||
| AGTR1 | Angiotensin II receptor, type 1 | NPPB | Natriuretic peptide B | ||
| EDN1 | Endothelin 1 | TR-α1 | Thyroid hormone receptor-α, splice variant 1 | ||
| Metabolic pathways (N=5) | HK2 | Hexokinase 2 | TR-α2 | Thyroid hormone receptor-α, splice variant 2 | |
| PFKM | Phosphofructokinase, muscle | ERF | Ets2 repressor factor | ||
| PDK4 | Pyruvate dehydrogenase kinase, isozyme 4 | CSRP3 | Cysteine and glycine-rich protein 3 | ||
| PDHX | Pyruvate dehydrogenase complex, component X | NOS2 | Nitric oxide synthase 2, inducible | ||
| CPT1B | Carnitine palmitoyltransferase 1B | Cytokines (N=6) | TNF | Tumor necrosis factor | |
| Calcium handling or binding (N=6) | ATP2A2 | ATPase, Ca++ transporting, cardiac, slow twitch 2 | IL1B | Interleukin 1, β | |
| PLN | Phospholamban | CTF1 | Cardiotrophin 1 | ||
| RYR2 | Ryanodine receptor 2 | IL6 | Interleukin 6 | ||
| CASQ2 | Calsequestrin 2 | IL6ST | Interleukin 6 signal transducer | ||
| SLC8A1 | Solute carrier fam. 8 (Na+/Ca++ exchanger), member 1 | Normalization (N=2) | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | |
| CANX | Calnexin | RN18S1 | RNA, 18S ribosomal 1 |
RAAS=Renin-angiotensin-aldosterone system
Study design
This study was conducted between 2000 and 2008 at the Universities of Colorado and Utah according to the Declaration of Helsinki and included a Data Safety Monitoring Board. The study was approved by institutional review boards at both sites, and all subjects gave written informed consent. Patients were eligible for enrollment if they had IDC and New York Heart Association (NYHA) Class II-IV symptoms with an LVEF ≤40%, were ≥18 years old, had angiographically-confirmed unobstructed coronary arteries, and were receiving conventional medical HF therapy except β-blockers for ≥3 weeks prior to enrollment. Exclusion criteria included HF due to valvular disease; thyroid disease; obstructive or hypertrophic cardiomyopathy; pericardial disease; amyloidosis; myocarditis; heart transplant candidacy; decompensated HF; ongoing treatment with nondihydropyridine calcium channel blockers, theophylline, tricyclic antidepressants, monoamine oxidase inhibitors, β-agonists, β-blockers, or inotropes; life expectancy<2 years; active substance abuse; recently fired implantable cardiac defibrillator; bradycardia; uncontrolled insulin-dependent diabetes; high-degree atrioventricular block; or history of noncompliance.
Patients were randomized in an unblinded fashion to commercially available formulations of carvedilol (COREG®), metoprolol succinate (TOPROL-XL®), or metoprolol succinate+doxazosin mesylate (CARDURA®). The starting dose of carvedilol, a β1/β2/α1-AR blocking agent, was 3.125 mg twice daily with targets of 25 mg (patients<85 kg) or 50 mg (patients≥85 kg) twice daily.9 The starting dose of metoprolol succinate, a β1-AR selective blocking agent, was 12.5 mg daily with a target of 200 mg daily.10 Doxazosin mesylate, an α1-AR selective blocking agent, was started at 1 mg daily with a target of 8 mg daily.11 All medications were up-titrated weekly until target doses were met or limiting side effects developed.
Endomyocardial biopsy may be considered clinically in IDC patients to rule out causes of myocardial disease that might affect prognosis or therapy.1,12 Patients underwent right heart catheterization via right internal jugular vein with RV endomyocardial biopsy from the distal septum at baseline, 3, and 12 months where the 3 and 12 month biopsies were used for RNA extraction for research purposes only. Biopsies were performed on all IDC patients by personnel experienced in performing RV biopsies in non-transplant patients under echocardiographic guidance, and 15–25 mg of tissue was removed. Biopsies were also taken from 5 potential nonfailing control patients with LVEF≥45% who were studied for other indications to rule out myocardial disease. Histologic analysis of baseline biopsies reported hypertrophy in 71%, increased interstitial fibrosis in 39%, and evidence of inflammation/mononuclear infiltrate in 11%. One potential control was excluded due to giant cell myocarditis on histologic examination, whereas the 4 other potential control biopsies had no evidence of hypertrophy, fibrosis, or inflammation and were used in the study. LVEF was measured within days of each biopsy by radionuclide ventriculography (see Supplement). RNA was extracted from all IDC patient and nonfailing control biopsy samples, cDNA was synthesized, and expression of 50 candidate genes (Table 1) was quantified by RT-qPCR using threshold cycle (Ct) detection (see Supplement for methods and primer sequences). Gene expression was also measured by cDNA hybridization to the Affymetrix HG-U133 Plus 2.0 Gene Chip (see Supplement).
Statistical Analysis
Clinical and gene expression data were imported into the R statistical package (version 3.0.2, R Foundation for Statistical Computing, Vienna Austria). Patients were aggregated by LVEF response status, and clinical differences between responder groups were assessed using Fisher’s exact and Welch’s t-test for categorical and continuous variables. Marked skew was noted in some Ct distributions, and outliers were identified and removed using a median absolute deviation threshold>3.5, congruous with an alpha of 0.05 using parametric statistical analysis (276 of 13100 values, 2.1%).13 If a subject’s baseline sample was excluded or unavailable, follow-up samples were excluded. Changes in gene expression from baseline to LOCF (month 12 values or if not available, month 3 values) and with treatment were approximated using the ΔΔCt method with GAPDH as the reference gene.14 Changes in gene expression were compared between Responders and Nonresponders using the non-parametric Wilcoxon rank-sum test. Significance of changes in gene expression from baseline was determined using the non-parametric paired Wilcoxon signed-rank test in Responders and Nonresponders separately. Fold changes and aggregate statistics were calculated using normalized Ct differences (2-ΔΔCt). All tests were two-tailed, and a p<0.05 was considered significant.
Comparisons were repeated using the arithmetic mean of GAPDH and 18S rRNA expression to determine effect of normalization strategy on results. Microarray data were normalized by log-scale robust multi-array analysis.15 Cognate microarray data corresponding to the 50 RT-qPCR genes were also analyzed, and changes in expression from baseline were compared between Responders and Nonresponders using the Wilcoxon rank-sum test. A p<0.05 by microarray was considered confirmatory of a significant RT-qPCR finding.
RESULTS
Outcomes of Enrolled Patients
A total of 63 IDC patients met screening criteria and gave written informed consent (Figure 1). Of these, 6 subjects withdrew prior to baseline studies for personal or administrative reasons, 2 had normalization of a reduced LVEF obtained on a previous examination, and central venous access could not be obtained in 1 leaving 54 subjects who underwent baseline studies. In 2 subjects, radionuclide ventriculography revealed their LVEFs had normalized since screening, and their biopsy samples were analyzed as nonfailing controls. Of the randomized 52 subjects, 47 returned for 3 month studies and 40 returned for 12 month studies. Therefore, 47 patients had ≥1 follow-up biopsy and LVEF measurement after baseline and were included in all subsequent analyses. There were no complications from endomyocardial biopsy or right heart catheterization.
Figure 1 –
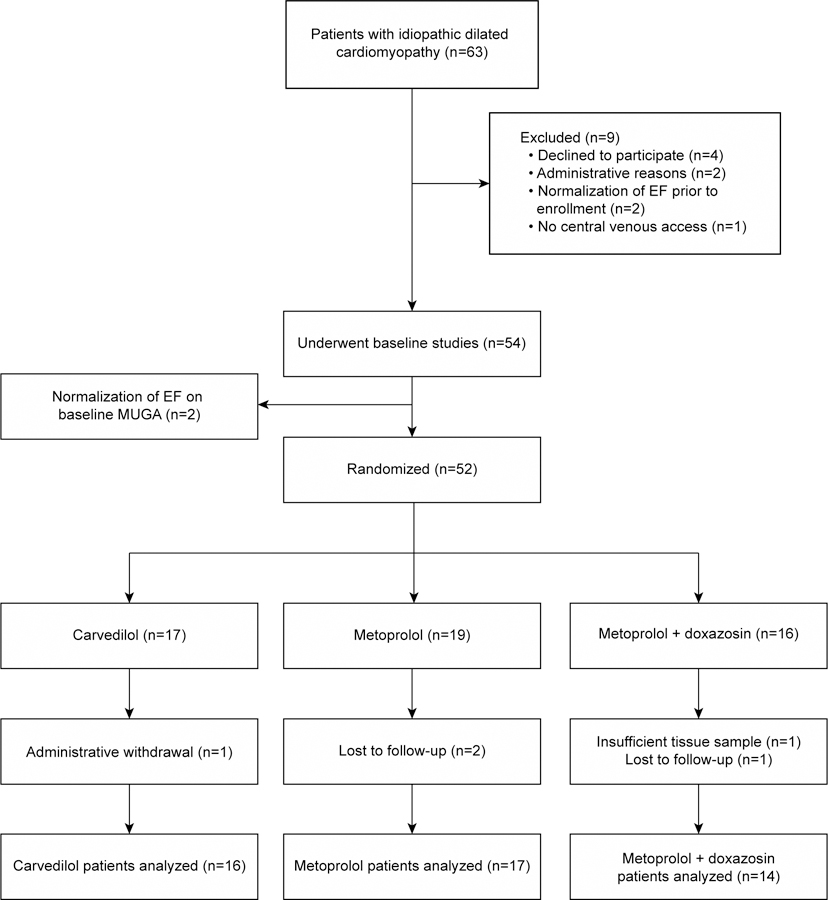
Screening, enrollment and randomization
In total 39 of 47 (83.0%) patients achieved target β-blocker dose including 16 of 16 carvedilol patients(mean daily dose 75±24 mg), 13 of 17 (76.5%) metoprolol patients (mean daily dose 163±60 mg), and 10 of 14 (71.4%) metoprolol+doxazosin patients(mean daily metoprolol dose 171±51 mg). Target doxazosin dose was reached in 10 of 14 patients (71.4%, mean daily dose 6±3 mg). Of the 5 patients not analyzed, 1 subject had inadequate tissue for RT-qPCR at the only follow-up visit, 2 relocated, and 2 were withdrawn for administrative reasons. One patient in the carvedilol arm died suddenly 10 months after randomization, and 1 patient underwent ventricular assist device placement after 16 months. There were no heart transplants during the study. A total of 8 patients (4 Responders, 4 Nonresponders) had 3 to 12 months LOCF imputation.
Baseline characteristics of IDC vs. nonfailing control subjects and LVEF Responders vs. Nonresponders are shown in Tables 2a–b. Significant differences between IDC and nonfailing subjects were limited to LVEF, RVEF, LV dimensions, NYHA class, and cardiac index. In total, 31 of 47 (66.0%) of patients met criteria for LVEF response. Baseline clinical features significantly associated with LVEF response included shorter duration of HF, higher estimated creatinine clearance, and narrower QRS duration. β-blocker doses were not significantly different between Responders and Nonresponders in any treatment arm (carvedilol: 78±23 vs. 71±27 mg, p=0.64; metoprolol: 179±50 vs 125±41 mg, p=0.1: metoprolol+doxazosin: 165±58 vs. 188±25 mg, p=0.29).
Table 2a –
Baseline clinical characteristics
| IDC (all BB) |
NF (Control) |
F vs. NF | R | NR | R vs. NR | |
|---|---|---|---|---|---|---|
| Clinical characteristic | 47 | 4 | p | 31 (66.0%) | 16 (34.0%) | p |
| Age, years | 45.8±13.1 | 41.0±17.1 | 0.62 | 43.8±13.2 | 49.7±12.3 | 0.20 |
| HF duration, months | 23.5±45.5 | - | - | 6.8±9.8 | 55.8±67.0 | 0.001 |
| Female | 13 (27.6%) | 1 (25.0%) | 1 | 9 (29.0%) | 4 (25.0%) | 1 |
| Race | 1 | 0.58 | ||||
| White | 31 (66.0%) | 4 (100.0%) | 22 (71.0%) | 9 (56.2%) | ||
| Black | 6 (12/8%) | 0 (0.0%) | 4 (12.9%) | 2 (12.5%) | ||
| Hispanic | 7 (14.9%) | 0 (0.0%) | 4 (12.9%) | 3 (18.8%) | ||
| Other | 3 (6.4%) | 0 (0.0%) | 1 (3.2%) | 2 (12.5%) | ||
| NYHA class | 0.002 | 0.55 | ||||
| I | 0 (0.0%) | 2 (50.0%) | 0 (0.0%) | 0 (0.0%) | ||
| II | 26 (55.3%) | 2 (50.0%) | 16 (51.6%) | 10 (62.5%) | ||
| III | 21 (44.7%) | 0 (0.0%) | 15 (48.4%) | 6 (37.5%) | ||
| Atrial fibrillation | 10 (21.3%) | 1 (25.0%) | 1 | 4 (12.9%) | 6 (37.5%) | 0.07 |
| Hypertension | 18 (38.3%) | 2 (50.0%) | 0.64 | 15 (48.4%) | 3 (18.8%) | 0.06 |
| Creatinine clearance, ml/min | 80.1±21.5 | 87.8±7.9 | 0.59 | 85.4±18.4 | 71.0±26.2 | 0.04 |
IDC=Idiopathic dilated cardiomyopathy; BB=β-blocker; F=Failing; NF=Non-failing; R=Responder; NR=Nonresponder
Table 2b –
Baseline cardiac parameters
| IDC (All BB) |
NF (Control) | F vs. NF | R | NR | R vs. NR | |
|---|---|---|---|---|---|---|
| Clinical characteristic | 47 | 4 | p | 31 (66.0%) | 16 (34.0%) | p |
| LVEF, % | 26.2±8.9 | 58.8±7.4 | 0.001 | 25.6±8.2 | 27.3±10.5 | 0.68 |
| RVEF, % | 27.1±8.9 | 38.3±4.7 | 0.03 | 27.0±8.7 | 27.2±9.7 | 0.95 |
| LV end diastolic vol., ml | 229±92 | - | - | 220±83 | 247±110 | 0.50 |
| Heart rate, bpm | 84.5±20.3 | 74.0±18.2 | 0.64 | 86.7±21.3 | 80.3±17.3 | 0.24 |
| QRS, ms | 115±31 | 101±9 | 0.51 | 107±24 | 133±39 | 0.03 |
| Norepinephrine, pg/ml | 489±327 | 372±152 | 0.69 | 450±261 | 574±438 | 0.74 |
| Systolic blood press., mm Hg | 107±14 | 118±29 | 0.56 | 106±13 | 109±17 | 0.46 |
| Mean PA pressure, mm Hg | 24.1±10.5 | 20.0±3.0 | 0.59 | 22.6±9.8 | 27.1±11.5 | 0.18 |
| Cardiac index, L/min/m2 | 2.2±0.6 | 3.0±0.5 | 0.05 | 2.3±0.7 | 2.2±0.6 | 0.91 |
IDC=Idiopathic dilated cardiomyopathy; BB=β-blocker; F=Failing; NF=Nonfailing; R=Responder; NR=Nonresponder; PA=Pulmonary artery
Changes in cardiac function, volume, and hemodynamic parameters are compared between Responders and Nonresponders in Table 3. Responders demonstrated significant improvement in LVEF (ΔLVEF=21.2±9.8 vs. 1.4±4.9 EF units in Nonresponders, p<0.001) and LV size (ΔLV EDV, −82±60 vs. 16±58 ml in Nonresponders, p<0.001). Heart rate decreased significantly more in Responders compared with Nonresponders (−18.2±20.6 vs. −4.7±13.3 bpm, p<0.001), as did pulmonary artery pressure (−4.6±8.4 vs. 1.7±8.8 mm Hg, p<0.05). Although RVEF improved significantly in Responders (27.7±8.7 to 37.0±7.6 EF units, p<0.001) and not in Nonresponders (27.2±9.7 to 32.0±11.6 EF units, p=0.14), the respective changes were not significantly different (p=0.27). Systolic blood pressure and cardiac index also increased significantly in Responders only compared to baseline, but differences between responder groups were not significant.
Table 3 –
Changes in cardiac structure and function according to responder status.
| Variable | Responder 31 (66.0%) |
Nonresponder 16 (34.0%) |
Responder vs. Nonresponder |
|---|---|---|---|
| LVEF, % | 21.2±9.8‡ | 1.4±4.9 | <0.0001 |
| RVEF, % | 9.8±11.4† | 4.9±10.4 | 0.27 |
| LV end diastolic vol., ml | −82.1±59.6‡ | 15.5±57.7 | <0.001 |
| Heart rate, bpm | −18.2±20.6† | −4.7±13.3 | <0.001 |
| Norepinephrine, pg/ml | −88.2±349.1 | −72.1±557.7 | 0.69 |
| Systolic blood pressure, mm Hg | 8.0±17.4* | 0.9±13.8 | 0.24 |
| Mean PA pressure, mm Hg | −4.6±8.4* | 1.7±8.8 | <0.05 |
| Cardiac index (L/min/m2) | 0.4±0.9* | 0.1±0.7 | 0.17 |
Compared to baseline:
p<0.05
p<0.001
p<0.0001
PA=pulmonary artery
Gene Expression Changes
Expression of the 50 candidate genes normalized to GAPDH at baseline in IDC patients is compared with control patients in Figure 2a, b, c. Compared to controls, 11 of 50 (22.0%) genes were differentially expressed in IDC patients. ADRB1, ATP2A2, MYH6 and ACTC1 were expressed at significantly lower levels in IDC patients, whereas MYL2, HK2, PDHX, CTF1, TNNI3, NPPA, and NPPB were expressed at significantly higher levels. The ACTC1/ACTA1 ratio was significantly lower in IDC patients compared to nonfailing controls (0.77±0.09 vs. 1.76±0.5, p=0.024), whereas MYH6/MYH7 (p=0.06) and ATP2A2/PLN (p=0.28) ratios were not significantly different.
Figure 2a – Baseline gene expression normalized to GAPDH: adrenergic signaling and contractile/cytoskeleton proteins in IDC (N=47) vs. controls (N=4).
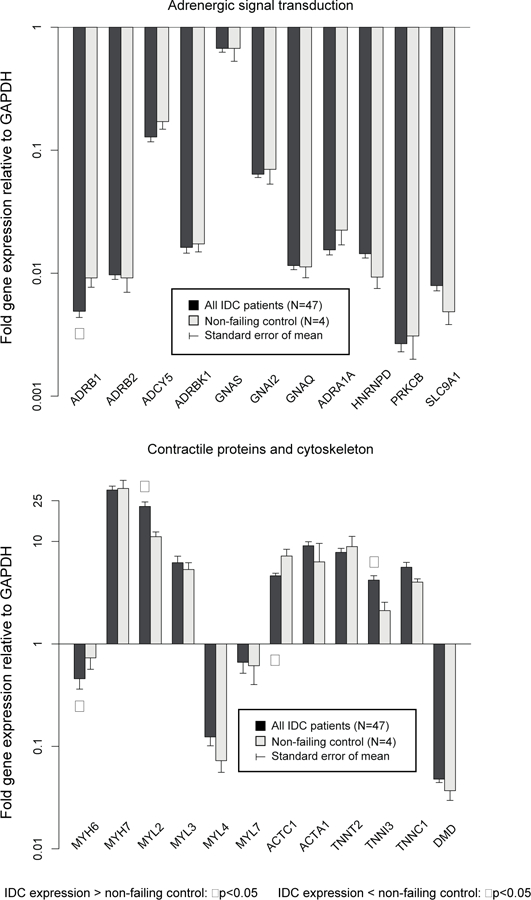
ADRB1, MYH6, and ACTC1 expressed at lower levels in IDC than controls, whereas MYH2, and TNNI3 were expressed at higher levels in IDC patients.
Figure 2b – Baseline gene expression normalized to GAPDH: calcium handling and cytokine pathways in IDC (N=47) vs. controls (N=4).
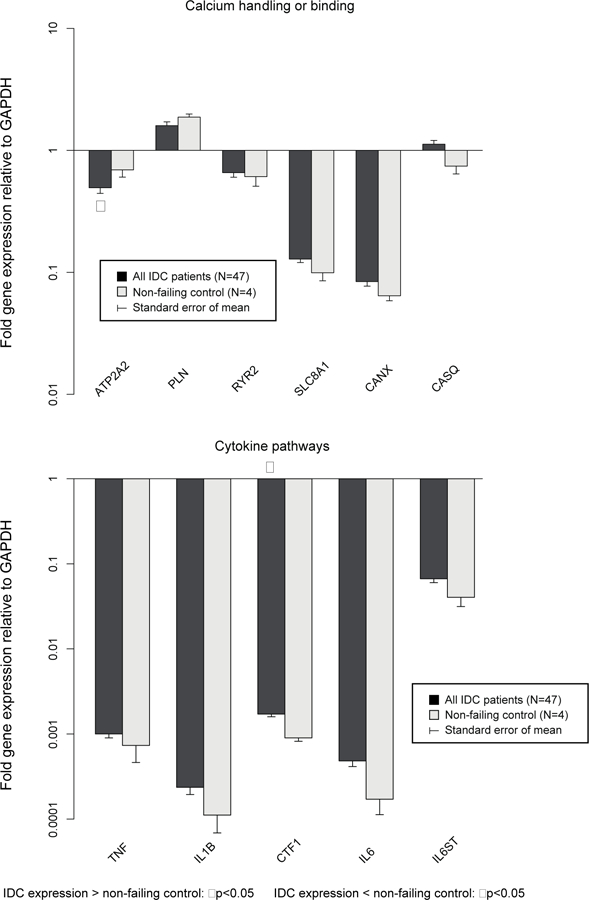
ATP2A2 was expressed at lower levels in IDC than controls, whereas CTF1 was expressed at higher levels in IDC patients.
Figure 2c – Baseline gene expression normalized to GAPDH: metabolic pathways and counter-regulatory, transcription factors, and other genes in IDC patients (N=47) vs. controls (N=4).
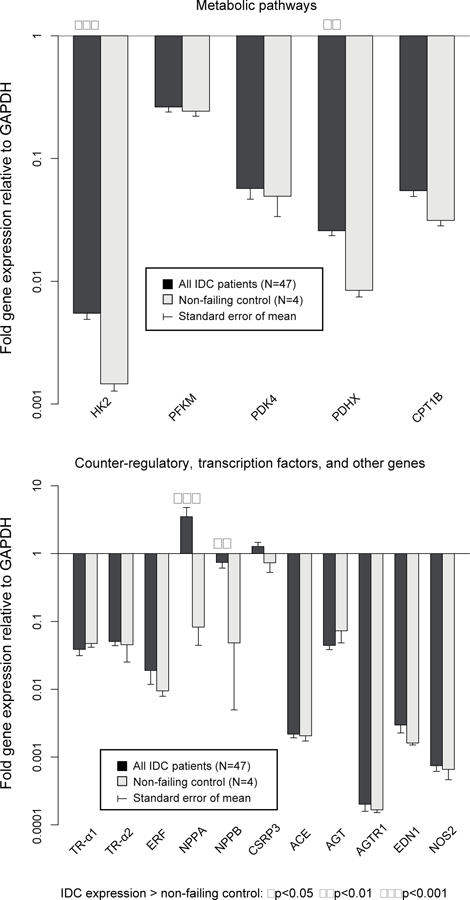
HK2, PDHX, NPPA and NPPB were expressed at higher levels in IDC patients.
Fold changes in myocardial gene expression from baseline by responder status are shown in Figure 3a, b, c. Expression of 13 of 50 (26.0%) genes (ACTA1, TNNC, TNNI3, IL6, GNAI2, GNAS, SLC8A1, SLC9A1, MYL2, ACTC1, HK2, CTF1, and CSRP3) decreased significantly AND 4 of 50 (8.0%) genes (TNNT2, CANX, PDK4, Tr-α1) increased significantly in Responders with no differences vs. changes in Nonresponders. Expression of 11 of 50 (22.0%) genes (ADRB1, ADRB2, ADRA1A, ATP2A2, PLN, RYR2, MYH6, MYL3, CPT1B, PDHX, and PFKM) increased significantly or decreased less in Responders vs. Nonresponders, whereas expression of 2 (4.0%) genes (NPPA and NPPB) decreased significantly in Responders vs. Nonresponders. MYH6/MYH7 (1.96±0.32 vs. 0.78±0.07, p<0.001) and ACTC1/ACTA1 (1.91±0.32 vs. 0.87±0.11,p<0.05) ratios also increased in Responders, whereas change in the ATP2A2/PLN ratio was not significantly different (p=0.37) between responder groups. When 3 (N=47) and 12 month (N=39) samples were considered separately, PFKM and RYR2 were significantly different in Responders vs. Nonresponders at 3 months only, ADRB1, ADRB2, ATP2A2, PLN, MYH6, and MYL2 were significantly different at 12 months only, and NPPA, NPPB, and MYL3 were significantly different at both time points. No genes were significant at either 3 or 12 months that were not significant in LOCF analysis. Neither GAPDH nor 18S rRNA changed significantly in analysis of nonnormalized Ct data (p>0.4, p>0.3, respectively). However expression of 18S rRNA decreased significantly relative to GAPDH on all 4 qPCR plates in Responders (fold change 0.47–0.59, p<0.01) but not in Nonresponders (p>0.14). All genes with significant differences between Responders and Nonresponders in serial gene expression normalized to GAPDH alone remained significant when normalized to the composite of GAPDH and 18s rRNA. The ratio of MYH6/MYH7 was also significantly higher in Responders vs. Nonresponders normalized to the composite of GAPDH and 18S rRNA (2.00±0.30 vs. 0.9±0.11, p<0.01), whereas the ACTC1/ACTA1 ratio was higher in Responders but not statistically significant (1.98±0.51 vs. 1.00±0.20, p=0.10).
Figure 3a – Fold change in gene expression: adrenergic signal transduction and contractile/cytoskeleton proteins in Responders (N=31) vs. Nonresponders (N=16).
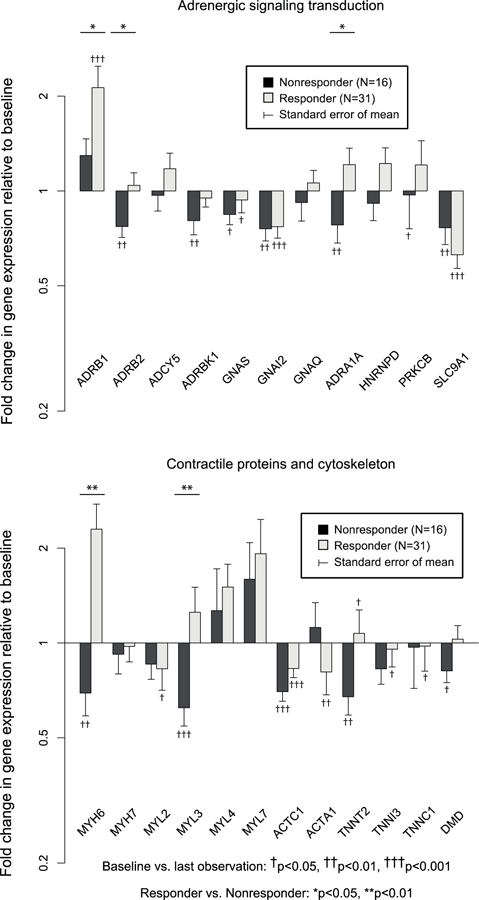
ADRB1, ADRB2, ADRA1A, MYH6, and MYL3 were upregulated in Responders compared with Nonresponders.
Figure 3b – Fold change in gene expression: calcium handling and cytokine pathways in Responders (N=31) vs. Nonresponders (N=16).
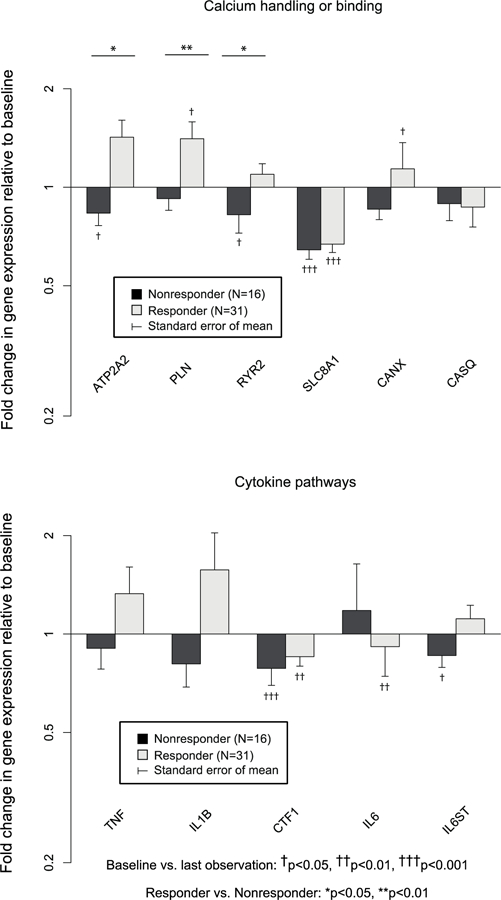
ATP2A2, PLN, and RYR2 were upregulated in Responders vs. Nonresponders.
Figure 3c – Fold change in gene expression: metabolic pathways and counter-regulatory, transcription factors, and other genes in Responders (N=31) vs. Nonresponders (N=16).
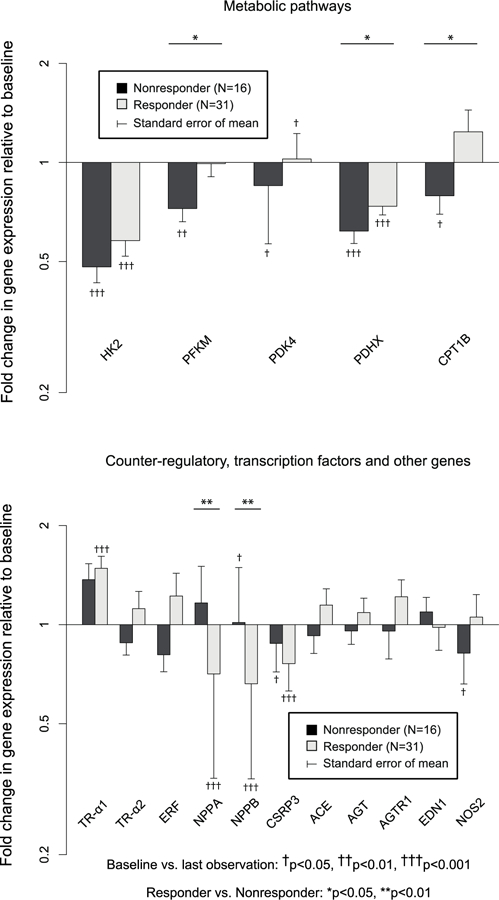
PFKM, PDHX, and CPT1B were upregulated in Responders vs. Nonresponders. NPPA and NPPB were downregulated in Responders vs. Nonresponders.
Genes that demonstrated differential expression changes between Responders and Nonresponders by RT-qPCR were also tested by microarray. Of these, 8 of 13 genes (MYH6, MYL3, PLN, RYR2, NPPA, NPPB, and PFKM) were also significantly differentially expressed on microarray with the same directionality (p<0.05). Increases in MYH6/MYH7 and ACTC1/ACTA1 ratios were also significant (p<0.01) by microarray in Responders compared to Nonresponders. Changes in ADRB1, ATP2A2, ADRA1A, CPT1B, and PDHX had the same directionality by microarray analysis and RT-qPCR, but differences between responder groups were not significant.
DISCUSSION
These data provide the most comprehensive longitudinal analysis reported to date of myocardial gene expression associated with reverse-remodeling in DCM patients. Because the intraventricular septum is shared between the ventricles and forms interdependent anatomic relationships with both the LV and RV free walls,16 molecular changes relative to LV structure and function will be detected by septal endomyocardial biopsy.3 In the current study Responders exhibited a robust improvement of 21±10 EF units, whereas changes in RVEF and hemodynamic parameters were not significantly different between Responders and Nonresponders. Therefore, it is likely that the observed changes in gene expression selectively present in Responders reflect intrinsic biologic changes in the LV chamber as opposed to changes in RV function, heart rate or LV loading conditions. In addition, the study design comparing Responders to Nonresponders who were given equivalent β1-AR blocking target doses3 at similar levels in Responders and Nonresponders minimizes the degree of β1-blockade as a mechanism for the observed differences. The present study provides insight into gene expression changes associated with LV reverse-remodeling with improvement in systolic function or regression of pathologic hypertrophy as well as myocardial response to β-blocker therapy irrespective of reverse-remodeling.
Gene Expression Changes in β-blocker Treatment Associated with Reverse-remodeling
The observed changes indicate increased expression in genes affecting contractile function, calcium handling, energy substrate utilization and adrenergic signaling plus decreases in natriuretic peptides, which are biomarkers of failing or hypertrophied myocardium. We reported previously that when β-blocking agents produce reverse-remodeling in IDC, they effect a partial reversal of HF-associated changes in expression of genes encoding β1-AR (ADRB1),17 contractile proteins (MYH6 and MYH6/MYH7 ratio),3 natriuretic peptides (NPPA),3 and sarcoplasmic reticulum calcium-ATPase 2 (ATP2A2).3 The results of BORG confirm those findings and extend the observations to increases in expression of additional contractile (ACTC1/ACTA1 ratio, MYL3) and calcium-handling genes (PLN, RYR2).
We also report changes in additional categories of genes associated with reverse-remodeling including the metabolic pathway genes CPT1B and PFKM and the α1-AR gene ADRA1A. Utilization of both fatty acids and glucose as energy sources is decreased in advanced HF18 limiting ATP synthesis. Coordinated up-regulation of both PFKM and CPT1B has been demonstrated in response to β-blockade suggesting a return to an adult metabolic gene expression pattern.19 If accompanied by cognate changes in protein abundance increased CPT1B should increase fatty acid metabolism,20 whereas increased PFKM should enhance glucose metabolism. Therefore, up-regulation of both free fatty acid and glucose metabolism occur to achieve higher energy production as LV function improves.
If mRNA abundance changes observed exclusively in Responders were translated to protein expression and activity, LV contractile function would be expected to increase due to the improved MYH6/MYH7 ratio and increased expression of MYH6,21 MYL3,22 ATP2A2,23 RYR2,24 ADRA1A,25 and ADRB1.26 In addition, decreases in MYH7/MYH627 and ACTA1/ACTC128 ratios and reduction in 18S rRNA29 should be anti-hypertrophic. Up-regulation in unphosphorylated PLN might be negatively inotropic,30 but increased ADRB1 expression would increase phosphorylated PLN potentially offsetting the impact of higher PLN expression.31 Decreases in NPPA and NPPB might be expected to be hypertrophic,32 but a previous study also observed that NPPA decreased in both Responders and Nonresponders, presumably as intracardiac filling pressures fell,3 and circulating B-type natriuretic peptide has been observed to decrease in association with reverse-remodeling.33 These data suggest that β1-AR blockade, common to all treatment arms, may effect reverse-remodeling by increasing expression of contractile, calcium-handling/-regulating, adrenergic receptor, and metabolic proteins that either improve LV contractile function or participate in its metabolic support.
Of 11 genes that exhibited altered baseline expression in IDC compared to nonfailing controls, 5 (ADRB1, MYH6, ATP2A2, NPPA, NPPB) were changed in the direction of control values (“normalized”) only in Responders, 2 (TNNI3 and MYL2) normalized in Responders without a significant difference vs. Nonresponders, and 3 (HK2, CTF1, PDHX) normalized in both Responders and Nonresponders. These data suggest that β-blocker therapy effects reverse-remodeling by partially normalizing some gene expression changes fundamentally associated with the DCM phenotype.
Many β-blocker associated gene expression changes in Responders vs. Nonresponders represent a partial reversal of the cardiac fetal/adult gene program,3,34,35 but they may also be consistent with alternative regulatory mechanisms including inhibition of β1-AR signaling5,6,36 and/or enhanced thyroid hormone receptor (TR-α) activity (Table 4).7 We previously reported changes in TR-α1 and TR-α2 expression in IDC LVs consistent with hypothyroidism at the TR-α level. Furthermore, myocardial expression of TR-α1 is positively correlated with MYH6 expression and negatively correlated with ANP, whereas TR-α2 expression is negatively correlated with MYH6 and positively with ANP expression, suggesting that TR-α expression may influence fetal/adult gene program regulation. In the current study changes in expression of either TR-α isoform were not different between responder groups, although TR-α1 expression increased significantly in Responders only, suggesting a TR-α effect was not completely ruled out.
Table 4 –
Significant differential changes in gene expression in Responders vs. Nonresponders compared with known gene regulatory pathways
| Concordance with regulatory pathways |
Verification |
|||||
|---|---|---|---|---|---|---|
| Gene name | Δ, R vs. NR |
*Adult/ Fetal |
†Inverse β1-AR stimulation |
‡THR | GAPDH+18S | Affymetrix |
| ADRB1 | ↑ | + | + | + | ✓ | NS |
| ADRB2 | ↑ | ? | + | + | ✓ | ✓ |
| ADRA1A | ↑ | + | + | + | ✓ | NS |
| MYH6 | ↑ | + | + | + | ✓ | ✓ |
| MYL3 | ↑ | + | + | X | ✓ | ✓ |
| NPPA | ↓ | + | + | + | ✓ | ✓ |
| NPPB | ↓ | + | + | + | ✓ | ✓ |
| ATP2A2 | ↑ | + | + | + | ✓ | NS |
| PLN | ↑ | + | + | − | ✓ | ✓ |
| RYR2 | ↑ | + | + | + | ✓ | ✓ |
| CPT1B | ↑ | + | + | + | ✓ | NS |
| PDHX | ↑ | ? | ? | ? | ✓ | NS |
| PFKM | ↑ | + | + | + | ✓ | ✓ |
| MYH6/MYH7 | ↑ | + | + | + | ✓ | ✓ |
| ACTC1/ACTA1 | ↑ | + | + | + | NS | ✓ |
Changes in gene expression concordant with thyroid hormone stimulation or TR-α1 expression.7
Directionally opposite regulation from that produced by β1-AR stimulation5,36 or transgenic cardiac overexpression;6
+=Concordant with pathway; −=Discordant with pathway; ?=Unknown if regulated by pathway; X=not regulated by pathway; ✓=Concordant, significant; NS=Concordant, not significant ; Δ=Directional change; R=Responder; NR=Nonresponder; THR=Thyroid hormone receptor. Decreased expression of genes expressed at higher levels in embryonic development (“fetal” genes) or increased expression of genes expressed at higher levels in the adult than in embryonic development (“adult” genes);
As shown in Table 4, changes in gene expression associated with LV reverse-remodeling were 100% concordant with changes expected from interruption of chronic β1-AR stimulation5,36 or cardiac β1-AR overexpression,6 both of which influence the fetal/adult gene program via the Ca2+/calmodulin-dependent kinase pathway.5 Thus the data are most consistent with a primary effect of a β1-blockade of a “β1-AR gene network” in Responders overlapping with effects expected for TR-α responsive and fetal/adult developmental gene expression patterns. The expansion of known genes whose expression changes in the setting of β-blocker therapy for IDC may provide additional insight into primary regulatory pathways involved in LV reverse-remodeling. Exploration of these findings may identify factors that determine whether β-blockade produces changes in myocardial gene expression that promote reverse-remodeling in a given individual, i.e. why the β1-AR network is affected by β1-blockade in reverse-remodeling patients only.
Gene expression changes associated with β-blocker therapy
Gene expression changes that occur in both Responders and Nonresponders are the likely result of pharmacologic effects of β1-AR blockade as opposed to being secondary to ventricular reverse-remodeling. Expression of genes encoding contractile (ACTC1), metabolic (HK2, PDHX), signaling (CSRP3, GNAI2, GNAS, SLC9A1), and calcium-handling proteins (SLC8A1) all decreased significantly in both Responders and Nonresponders. In some cases, these changes represent partial reversal of pathologic changes associated with IDC even when not associated with improvement in LVEF. For example, SLC8A1 has been shown to be up-regulated in the failing human heart, 37 and inhibition of SLC8A1 has been associated with reduced arrhythmia in experimental models of DCM, suggesting potential for improved survival even without reverse-remodeling.38
Comparison of Expression Changes in Responders to Previous Studies of Reverse-Remodeling in Human Dilated Cardiomyopathies
Previous studies using cardiac resynchronization therapy39–41 or β-blockers3,17,42–44 to effect reverse-remodeling have reported similar protein or mRNA gene expression changes in ADRB1+ADRB2,17,42 ADRB1,39 MYH6,3,40,41,43 MYH6/MYH7, 3,40,41,43 ATP2A2,3,41,44 PLN,40,41,44 NPPA3 and NPPB.41 Responder-specific gene expression changes in RYR2, ADRA1A, MYL3, PDKFM, PDHX, CPT1B, and ACTA1/ACTC1 have not been previously reported in reverse-remodeling of human dilated cardiomyopathies.
Limitations
Analysis was limited to gene expression changes, which may not be directly translated to changes in functional protein abundance. Because of the quantity of RNA required to perform the mRNA analyses reported in this study as well as additional microRNA and RNAseq measurements quantification of protein expression or post-translational modification was not feasible and was not a prespecified aim of the present study. Conclusions regarding significance of gene expression changes may vary according to assay and normalization standard. GAPDH was selected at the time of trial design based on its widespread use as a reference gene, its mid-range expression level and lack of common regulation, but use of GAPDH as a normalization standard has subsequently been questioned.45 Ribosomal 18S rRNA was quantified but is not generally used alone as a reference standard due to its high abundance and the unpredictable balance of rRNA and mRNA in different tissues.46 We repeated RT-qPCR analysis using a composite of GAPDH and 18S rRNA, and all genes associated with LV response remained significant. We used microarray analysis as an additional confirmatory assay although microarray analysis uses a distinct normalization strategy, and the importance of differences in statistical significance between RT-qPCR and microarray findings is unclear.
Carvedilol is a “biased ligand” for both β13,47–50 - and β2-ARs, meaning that it can G-protein independently activate MAP kinase ERK1,2 pathways via β-arrestin signaling as it produces receptor blockade. Metoprolol has also been shown to be a biased ligand for β1-ARs, in a pathway that appears to be different from carvedilol. In a previous study we found absolutely no differences between metoprolol vs. carvedilol for the 6 mRNAs measure by quantitative PCR. In the current study there were some differences between the carvedilol and the combined metoprolol groups, but the statistical results of the metoprolol groups remained unchanged when combined with the carvedilol group. We will be subsequently reporting a complete analysis of gene expression between groups, where the consequences of differences in biased ligand signaling in the carvedilol vs. metoprolol groups may become apparent.
CONCLUSIONS
LV reverse-remodeling in IDC patients receiving β-blocker therapy is associated with a distinct therapeutic molecular phenotype consisting of changes in expression of genes involved in AR signaling, contractile protein function, calcium handling, and metabolism. These changes are directionally opposite of previously described gene expression effects of chronic β1-AR stimulation,5,36 or transgenic myocardial overexpression,6 which collectively constitute a β1-AR gene network. The biologic effects of this gene network ultimately result in contractile dysfunction, hypertrophy and adverse effects on myocardial metabolism despite their positive inotropic acute effects.8,51 The explanation for this biologic dichotomy appears to be distinct differences in signaling with chronic gene expression changes mediated by protein kinase A (PKA)-independent pathways5 as opposed to PKA-dependent acute effects. Changes in expression of these gene families may provide opportunities for directed pharmacologic therapy, and further study is warranted to identify key regulatory nodes responsible for β-blocker therapy inhibiting the β1-AR gene network in some IDC patients (Responders) and not in others..
Supplementary Material
Acknowledgements:
The authors wish to thank Rachel Rosenberg and Katie Goodyear for their invaluable assistance.
Funding sources: National Heart, Lung, and Blood Institute (2R01 HL48013, 3R01 HL48013, R01 HL71118, PI:Bristow; P20 HL101435 PI:Lowes; T32 HL007822 PI:Buttrick) and research grants from GlaxoSmithKline and AstraZeneca.
Footnotes
Disclosures: The authors have no relevant disclosures.
References
- 1.Maron BJ, Towbin JA, Thiene G, Antzelevitch C, Corrado D, Arnett D, et al. Contemporary definitions and classification of the cardiomyopathies. Circulation. 2006;113(14):1807–16. [DOI] [PubMed] [Google Scholar]
- 2.Nakao K, Minobe W, Roden R, Bristow MR, and Leinwand LA. Myosin heavy chain gene expression in human heart failure. J Clin Invest. 1997;100(9):2362–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lowes BD, Gilbert EM, Abraham WT, Minobe WA, Larrabee P, Ferguson D, et al. Myocardial gene expression in dilated cardiomyopathy treated with beta-blocking agents. N Engl J Med. 2002;346(18):1357–65. [DOI] [PubMed] [Google Scholar]
- 4.Lowes BD, Zolty R, Minobe WA, Robertson AD, Leach S, Hunter L, et al. Serial gene expression profiling in the intact human heart. J Heart Lung Transplant. 2006;25(5):579–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sucharov CC, Mariner PD, Nunley KR, Long C, Leinwand L, and Bristow MR. A beta1-adrenergic receptor CaM kinase II-dependent pathway mediates cardiac myocyte fetal gene induction. Am J Physiol Heart Circ Physiol. 2006;291(3):H1299–308. [DOI] [PubMed] [Google Scholar]
- 6.Dockstader K, Nunley K, Karimpour-Fard A, Medway A, Nelson P, Port JD, et al. Temporal analysis of mRNA and miRNA expression in transgenic mice overexpressing Arg- and Gly389 polymorphic variants of the β1-adrenergic receptor. Physiol Genomics. 2011;43(23):1294–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kinugawa K, Minobe WA, Wood WM, Ridgway EC, Baxter JD, Ribeiro RC, et al. Signaling pathways responsible for fetal gene induction in the failing human heart: evidence for altered thyroid hormone receptor gene expression. Circulation. 2001;103(8):1089–94. [DOI] [PubMed] [Google Scholar]
- 8.Eichhorn EJ, and Bristow MR. Medical therapy can improve the biological properties of the chronically failing heart. A new era in the treatment of heart failure. Circulation. 1996;94(9):2285–96. [DOI] [PubMed] [Google Scholar]
- 9.Olsen SL, Gilbert EM, Renlund DG, Taylor DO, Yanowitz FD, and Bristow MR. Carvedilol improves left ventricular function and symptoms in chronic heart failure: a double-blind randomized study. J Am Coll Cardiol. 1995;25(6):1225–31. [DOI] [PubMed] [Google Scholar]
- 10.Hjalmarson A, Goldstein S, Fagerberg B, Wedel H, Waagstein F, Kjekshus J, et al. Effect of metoprolol CR/XL in chronic heart failure: Metoprolol CR XL Randomised Intervention Trial in Congestive Heart Failure (MERIT-HF). Lancet. 1999;353(9169):2001–2007. [PubMed] [Google Scholar]
- 11.Kukin ML, Kalman J, Mannino M, Freudenberger R, Buchholz C, and Ocampo O. Combined alpha-beta blockade (doxazosin plus metoprolol) compared with beta blockade alone in chronic congestive heart failure. Am J Cardiol. 1996;77(7):486–91. [DOI] [PubMed] [Google Scholar]
- 12.Felker GM, Hu W, Hare JM, Hruban RH, Baughman KL, and Kasper EK. The spectrum of dilated cardiomyopathy. The Johns Hopkins experience with 1,278 patients. Medicine (Baltimore). 1999;78(4):270–83. [DOI] [PubMed] [Google Scholar]
- 13.Iglewicz B, and Hoaglin DC. How to Detect and Handle Outliers. How to Detect and Handle Outliers. 1993. p. 11–13.
- 14.Schmittgen TD, and Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–8. [DOI] [PubMed] [Google Scholar]
- 15.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, and Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31(4):e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saleh S, Liakopoulos OJ, and Buckberg GD. The septal motor of biventricular function. Eur J Cardiothorac Surg. 2006;29 Suppl 1:S126–38. [DOI] [PubMed] [Google Scholar]
- 17.Gilbert EM, Abraham WT, Olsen S, Hattler B, White M, Mealy P, et al. Comparative hemodynamic, left ventricular functional, and antiadrenergic effects of chronic treatment with metoprolol versus carvedilol in the failing heart. Circulation. 1996;94(11):2817–25. [DOI] [PubMed] [Google Scholar]
- 18.Turer AT, Stevens RD, Bain JR, Muehlbauer MJ, van der Westhuizen J, Mathew JP, et al. Metabolomic profiling reveals distinct patterns of myocardial substrate use in humans with coronary artery disease or left ventricular dysfunction during surgical ischemia/reperfusion. Circulation. 2009;119(13):1736–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sack MN, Harrington LS, Jonassen AK, Mjøs OD, and Yellon DM. Coordinate Regulation of Metabolic Enzyme Encoding Genes During Cardiac Development and Following Carvedilol Therapy in Spontaneously Hypertensive Rats. Cardiovascular Drugs and Therapy. 2000;14(1):31–39. [DOI] [PubMed] [Google Scholar]
- 20.Cheng L, Ding G, Qin Q, Xiao Y, Woods D, Chen YE, et al. Peroxisome proliferator-activated receptor delta activates fatty acid oxidation in cultured neonatal and adult cardiomyocytes. Biochem Biophys Res Commun. 2004;313(2):277–86. [DOI] [PubMed] [Google Scholar]
- 21.Herron TJ, and McDonald KS. Small amounts of alpha-myosin heavy chain isoform expression significantly increase power output of rat cardiac myocyte fragments. Circ Res. 2002;90(11):1150–2. [DOI] [PubMed] [Google Scholar]
- 22.Schaub MC, Hefti MA, Zuellig RA, and Morano I. Modulation of contractility in human cardiac hypertrophy by myosin essential light chain isoforms. Cardiovasc Res. 1998;37(2):381–404. [DOI] [PubMed] [Google Scholar]
- 23.Müller OJ, Lange M, Rattunde H, Lorenzen HP, Müller M, Frey N, et al. Transgenic rat hearts overexpressing SERCA2a show improved contractility under baseline conditions and pressure overload. Cardiovasc Res. 2003;59(2):380–9. [DOI] [PubMed] [Google Scholar]
- 24.Jiang M, Xu A, Tokmakejian S, and Narayanan N. Thyroid hormone-induced overexpression of functional ryanodine receptors in the rabbit heart. Am J Physiol Heart Circ Physiol. 2000;278(5):H1429–38. [DOI] [PubMed] [Google Scholar]
- 25.Jensen BC, O’Connell TD, and Simpson PC. Alpha-1-adrenergic receptors: targets for agonist drugs to treat heart failure. J Mol Cell Cardiol. 2011;51(4):518–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mialet Perez J, Rathz DA, Petrashevskaya NN, Hahn HS, Wagoner LE, Schwartz A, et al. Beta 1-adrenergic receptor polymorphisms confer differential function and predisposition to heart failure. Nat Med. 2003;9(10):1300–5. [DOI] [PubMed] [Google Scholar]
- 27.Waspe LE, Ordahl CP, and Simpson PC. The cardiac beta-myosin heavy chain isogene is induced selectively in alpha 1-adrenergic receptor-stimulated hypertrophy of cultured rat heart myocytes. J Clin Invest. 1990;85(4):1206–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simpson PC, Long CS, Waspe LE, Henrich CJ, and Ordahl CP. Transcription of early developmental isogenes in cardiac myocyte hypertrophy. J Mol Cell Cardiol. 1989;21 Suppl 5:79–89. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Z, Liu R, Townsend PA, and Proud CG. p90(RSK)s mediate the activation of ribosomal RNA synthesis by the hypertrophic agonist phenylephrine in adult cardiomyocytes. J Mol Cell Cardiol. 2013;59:139–47. [DOI] [PubMed] [Google Scholar]
- 30.Kadambi VJ, Ponniah S, Harrer JM, Hoit BD, Dorn GW, Walsh RA, et al. Cardiac-specific overexpression of phospholamban alters calcium kinetics and resultant cardiomyocyte mechanics in transgenic mice. J Clin Invest. 1996;97(2):533–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindemann JP, Jones LR, Hathaway DR, Henry BG, and Watanabe AM. beta-Adrenergic stimulation of phospholamban phosphorylation and Ca2+-ATPase activity in guinea pig ventricles. J Biol Chem. 1983;258(1):464–71. [PubMed] [Google Scholar]
- 32.Knowles JW, Esposito G, Mao L, Hagaman JR, Fox JE, Smithies O, et al. Pressure-independent enhancement of cardiac hypertrophy in natriuretic peptide receptor A-deficient mice. J Clin Invest. 2001;107(8):975–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kubanek M, Sramko M, Maluskova J, Kautznerova D, Weichet J, Lupinek P, et al. Novel predictors of left ventricular reverse remodeling in individuals with recent-onset dilated cardiomyopathy. J Am Coll Cardiol. 2013;61(1):54–63. [DOI] [PubMed] [Google Scholar]
- 34.Taegtmeyer H, Sen S, and Vela D. Return to the fetal gene program: a suggested metabolic link to gene expression in the heart. Ann N Y Acad Sci. 2010;1188:191–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rajabi M, Kassiotis C, Razeghi P, and Taegtmeyer H. Return to the fetal gene program protects the stressed heart: a strong hypothesis. Heart Fail Rev. 2007;12(3–4):331–43. [DOI] [PubMed] [Google Scholar]
- 36.Stein B, Bartel S, Kirchhefer U, Kokott S, Krause EG, Neumann J, et al. Relation between contractile function and regulatory cardiac proteins in hypertrophied hearts. Am J Physiol. 1996;270(6 Pt 2):H2021–8. [DOI] [PubMed] [Google Scholar]
- 37.Reinecke H, Studer R, Vetter R, Holtz J, and Drexler H. Cardiac Na+/Ca2+ exchange activity in patients with end-stage heart failure. Cardiovasc Res. 1996;31(1):48–54. [PubMed] [Google Scholar]
- 38.Milberg P, Pott C, Frommeyer G, Fink M, Ruhe M, Matsuda T, et al. Acute inhibition of the Na(+)/Ca(2+) exchanger reduces proarrhythmia in an experimental model of chronic heart failure. Heart Rhythm. 2012;9(4):570–8. [DOI] [PubMed] [Google Scholar]
- 39.Vanderheyden M, Mullens W, Delrue L, Goethals M, Verstreken S, Wijns W, et al. Endomyocardial upregulation of beta1 adrenoreceptor gene expression and myocardial contractile reserve following cardiac resynchronization therapy. J Card Fail. 2008;14(2):172–8. [DOI] [PubMed] [Google Scholar]
- 40.Iyengar S, Haas G, Lamba S, Orsinelli DA, Babu GJ, Ferketich AK, et al. Effect of cardiac resynchronization therapy on myocardial gene expression in patients with nonischemic dilated cardiomyopathy. J Card Fail. 2007;13(4):304–11. [DOI] [PubMed] [Google Scholar]
- 41.Vanderheyden M, Mullens W, Delrue L, Goethals M, de Bruyne B, Wijns W, et al. Myocardial gene expression in heart failure patients treated with cardiac resynchronization therapy responders versus nonresponders. J Am Coll Cardiol. 2008;51(2):129–36. [DOI] [PubMed] [Google Scholar]
- 42.Heilbrunn SM, Shah P, Bristow MR, Valantine HA, Ginsburg R, and Fowler MB. Increased beta-receptor density and improved hemodynamic response to catecholamine stimulation during long-term metoprolol therapy in heart failure from dilated cardiomyopathy. Circulation. 1989;79(3):483–90. [DOI] [PubMed] [Google Scholar]
- 43.Abraham WT, Gilbert EM, Lowes BD, Minobe WA, Larrabee P, Roden RL, et al. Coordinate changes in Myosin heavy chain isoform gene expression are selectively associated with alterations in dilated cardiomyopathy phenotype. Mol Med. 2002;8(11):750–60. [PMC free article] [PubMed] [Google Scholar]
- 44.Yasumura Y, Takemura K, Sakamoto A, Kitakaze M, and Miyatake K. Changes in myocardial gene expression associated with beta-blocker therapy in patients with chronic heart failure. J Card Fail. 2003;9(6):469–74. [DOI] [PubMed] [Google Scholar]
- 45.Barber RD, Harmer DW, Coleman RA, and Clark BJ. GAPDH as a housekeeping gene: analysis of GAPDH mRNA expression in a panel of 72 human tissues. Physiol Genomics. 2005;21(3):389–95. [DOI] [PubMed] [Google Scholar]
- 46.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Galandrin S, and Bouvier M. Distinct signaling profiles of beta1 and beta2 adrenergic receptor ligands toward adenylyl cyclase and mitogen-activated protein kinase reveals the pluridimensionality of efficacy. Mol Pharmacol. 2006;70(5):1575–84. [DOI] [PubMed] [Google Scholar]
- 48.Wisler JW, DeWire SM, Whalen EJ, Violin JD, Drake MT, Ahn S, et al. A unique mechanism of beta-blocker action: carvedilol stimulates beta-arrestin signaling. Proc Natl Acad Sci U S A. 2007;104(42):16657–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakaya M, Chikura S, Watari K, Mizuno N, Mochinaga K, Mangmool S, et al. Induction of cardiac fibrosis by β-blocker in G protein-independent and G protein-coupled receptor kinase 5/β-arrestin2-dependent Signaling pathways. J Biol Chem. 2012;287(42):35669–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kao DP, Minobe W, Epperson LE, Meyer L, Ferguson D, Zolty R, et al. Comparison of changes in reverse remodeling associated myocardial gene expression between carvedilol and metoprolol in nonischemic dilated cardiomyopathy. Eur J Heart Fail. 2014;16(suppl2):362. [Google Scholar]
- 51.Bristow MR, and Gilbert EM. Improvement in cardiac myocyte function by biological effects of medical therapy: a new concept in the treatment of heart failure. Eur Heart J. 1995;16 Suppl F:20–31. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


