Associated microorganisms (“microbiota”) are intimately connected to the behavior and physiology of their animal hosts, and defining the mechanisms of these interactions is an urgent imperative. This study focuses on how microorganisms influence the life span of a model host, the fruit fly Drosophila melanogaster. First, we performed a screen that suggested a strong influence of bacterial methionine metabolism on host life span. Follow-up analyses of gene expression and metabolite abundance identified stronger roles for vitamin B6 and glucose than methionine metabolism among the tested mutants, possibly suggesting a more limited role for bacterial methionine metabolism genes in host life span effects. In a parallel set of experiments, we created a distinct bacterial strain that expressed life span-extending methionine metabolism genes and showed that this strain can extend fly life span. Therefore, this work identifies specific bacterial genes that influence host life span, including in ways that are consistent with the expectations of methionine restriction.
KEYWORDS: Acetobacter, Lactobacillus, Drosophila melanogaster, microbiota, metagenome-wide association, MGWA, life span, vitamin B6, methionine restriction, glucose
ABSTRACT
To better understand how associated microorganisms (“microbiota”) influence organismal aging, we focused on the model organism Drosophila melanogaster. We conducted a metagenome-wide association (MGWA) as a screen to identify bacterial genes associated with variation in the D. melanogaster life span. The results of the MGWA predicted that bacterial cysteine and methionine metabolism genes influence fruit fly longevity. A mutant analysis, in which flies were inoculated with Escherichia coli strains bearing mutations in various methionine cycle genes, confirmed a role for some methionine cycle genes in extending or shortening fruit fly life span. Initially, we predicted these genes might influence longevity by mimicking or opposing methionine restriction, an established mechanism for life span extension in fruit flies. However, follow-up transcriptome sequencing (RNA-seq) and metabolomic experiments were generally inconsistent with this conclusion and instead implicated glucose and vitamin B6 metabolism in these influences. We then tested if bacteria could influence life span through methionine restriction using a different set of bacterial strains. Flies reared with a bacterial strain that ectopically expressed bacterial transsulfuration genes and lowered the methionine content of the fly diet also extended female D. melanogaster life span. Taken together, the microbial influences shown here overlap with established host genetic mechanisms for aging and therefore suggest overlapping roles for host and microbial metabolism genes in organismal aging.
IMPORTANCE Associated microorganisms (“microbiota”) are intimately connected to the behavior and physiology of their animal hosts, and defining the mechanisms of these interactions is an urgent imperative. This study focuses on how microorganisms influence the life span of a model host, the fruit fly Drosophila melanogaster. First, we performed a screen that suggested a strong influence of bacterial methionine metabolism on host life span. Follow-up analyses of gene expression and metabolite abundance identified stronger roles for vitamin B6 and glucose than methionine metabolism among the tested mutants, possibly suggesting a more limited role for bacterial methionine metabolism genes in host life span effects. In a parallel set of experiments, we created a distinct bacterial strain that expressed life span-extending methionine metabolism genes and showed that this strain can extend fly life span. Therefore, this work identifies specific bacterial genes that influence host life span, including in ways that are consistent with the expectations of methionine restriction.
INTRODUCTION
Model organisms, especially Caenorhabditis elegans and Drosophila melanogaster, have dramatically influenced our understanding of aging in mammals, including humans. For example, work on these models has helped reveal that lowering organismal caloric intake, oxidative stress, and inflammation can all promote a long and healthy life (1–9). In particular, calorie restriction extends longevity across the tree of life, underscoring its potential as a therapeutic target (10–13). In some cases, restriction of individual nutrients can fully or partially reproduce the longevity-promoting benefits of total calorie restriction, including methionine (8). Here, we focus on methionine metabolism genes in associated microorganisms (“microbiota”) and their relationship to life span in the model host D. melanogaster.
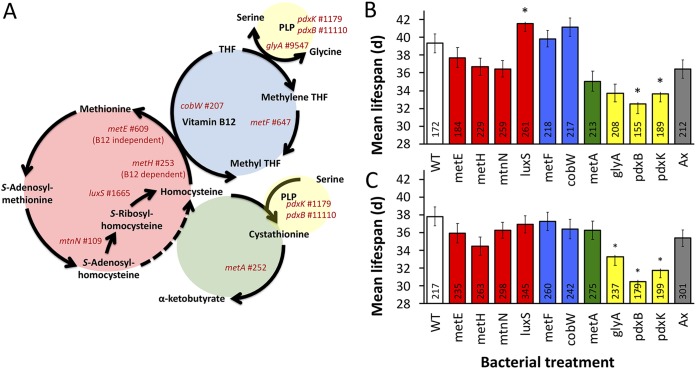
Methionine restriction is well documented to extend the life span of Drosophila and other model organisms. For example, fruit fly longevity can be promoted either by restricting the methionine content of the diet or by differentially expressing genes that decrease the accumulation of methionine cycle intermediates (8, 12, 14–17). Still, the mechanistic basis for Drosophila responses to dietary and methionine restriction is not fully understood and is complex. For example, the paradox that naturally long-lived flies display increased methionine contents in early life has been explained as an increased flux through the methionine cycle that decreases accumulation of S-adenosyl methionine (SAM) and S-adenosyl homocysteine (SAH) intermediates, rather than simply minimizing levels of methionine (see Fig. 2A) (17). Consistent with this expectation, constitutive expression of cystathionine beta synthase (CBS), which drives flux through transsulfuration, extends Drosophila melanogaster life span (15). Also, mimicking methionine restriction can slow organismal development (18). These varied findings underscore the complexity of interactions between methionine metabolism and aging and emphasize the need for continued study.
FIG 2.
Methionine metabolism mutants and D. melanogaster life span. (A) Diagram of central methionine metabolism (red), vitamin B12-dependent conversion of homocysteine to methionine (blue), transsulfuration (green), and vitamin B6, pyridoxal phosphate (PLP) (yellow). Gene names that were tested with E. coli mutants, together with their rank in the MGWA, are presented. Average life spans of male (B) and female (C) D. melanogaster monoassociated with E. coli mutant strains (WT, wild-type E. coli; Ax, bacterium-free). Colors of the bars correspond to colors shown in panel A. Numbers inside the bars report the number of flies tested per treatment. *, P < 0.05 versus WT bacteria, determined by a Cox mixed-effects survival model.
In addition to dietary and genetic interventions, the microbiota can substantially influence organismal longevity. In D. melanogaster, numerous studies have revealed a positive, neutral, or antagonistic role for the microbiota in animal life span (19–23, 25). A unifying explanation for the varied influence of bacteria on D. melanogaster life span is that the species-specific influences of the microbiota are interactive with diet (26). For example, when flies are reared on a nutrient-poor diet, microbes may provision nutrients that aid in healthy growth and development and promote longevity, but rearing on a nutrient-rich diet may spare the flies from microbial dependence (22). Other effects, such as age of the host (20, 21) or changes in nutrient allocation to, e.g., immunity, may also play a role. Finally, the functions harbored by the different microorganisms can also lead to distinct outcomes, although the genetic mechanisms for such functions are poorly described. The different members of the microbiota in wild and laboratory flies are of low diversity—usually 2 to 100 different species per fly—and typically dominated by acetic acid bacteria (AAB), lactic acid bacteria (LAB), and enterobacteria (21, 24, 27–29). Order-, species-, and strain-level effects on life span have been observed (20), but fly life span is usually shortest when flies bear high loads of AAB.
In this work, we investigate the relationship between life span and the microbiota in D. melanogaster reared on a nutrient-rich diet. Our main goal was to address two key questions: first, what are the specific bacterial genes that influence D. melanogaster life span, and second, do these genes influence the levels of methionine cycle metabolites in D. melanogaster? We address these questions using metagenome-wide association (MGWA) as a surrogate genetic screen, followed by analysis of life span and metabolomes in fruit flies reared with bacterial mutants in the MGWA-identified pathways. The results of these experiments reveal that, of the bacterial methionine metabolism genes that influence the life span of the flies, some but not all alter the levels of methionine and its related metabolites in the flies or the flies’ diets.
RESULTS
There are strain-specific influences of the microbiota on D. melanogaster life span.
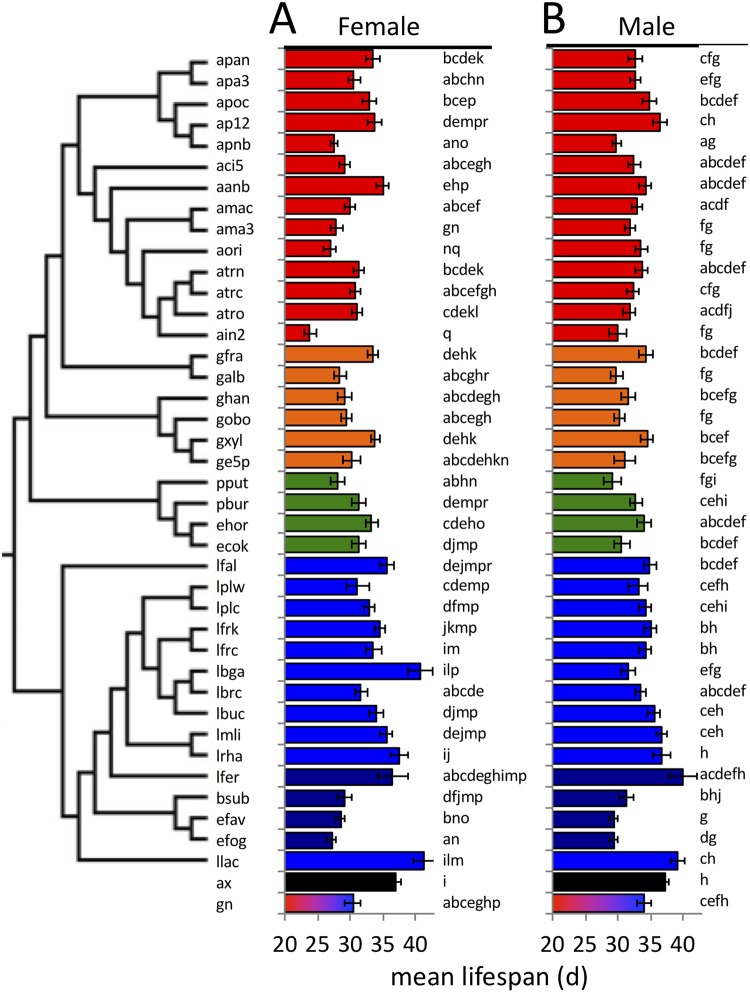
To study the influences of different bacterial species on the life span of Drosophila melanogaster, we monoassociated the flies with each of 39 bacterial strains and measured fly life span. For controls, we measured the life spans of bacterium-free flies and flies that were associated with a representative four-species bacterial community containing members from our previous work (30). The mean life span values we detected were generally low compared to those from other surveys of life span in D. melanogaster Canton-S (31, 32), a result that may be due, in part, to our use of a high-nutrient diet and mixed-sex populations (33, 34). Microbial presence generally decreased fruit fly life span relative to that of microorganism-free flies, with variation in life span effects within and between bacterial species (Fig. 1; see also Fig. S1 and raw data in Table S1 in the supplemental material) (bacterial species are listed in Table 1). For example, the Acetobacteraceae tended to confer a shorter life span on the flies than Lactobacillus species, but some species did not follow this trend. Altogether, these data are consistent with the expectation that rearing the flies on high-nutrient diets and under mixed-sex conditions leads to relatively short fly life spans. The data also show that the different strains significantly altered fly life span.
FIG 1.
Bacteria influence the mean life spans of female and male flies. The mean life spans of flies reared in monoassociation with a panel of bacterial isolates is presented (total n = 13,350; n for each treatment is shown in Table S1A in the supplemental material). Four-character strain designations are listed in Table 1, and the phylogenetic relationship between the strains, based on 16S sequences, is shown in the tree at left. Coloring represents Acetobacter (red), non-Acetobacter Acetobacteraceae (orange), Gammaproteobacteria (green), Lactobacillus (blue), non-Lactobacillus Firmicutes (purple), bacterium free (ax; black), and 4-species gnotobiotic (gn; multicolored, containing strains atrc, apoc, lbrc, and lplc). Different lowercase letters by the bars represent statistically significant differences between treatments. d, days.
TABLE 1.
Strains used in this study
| Identifier | Relevant characteristics | Medium | Oxygen conditions |
|---|---|---|---|
| 7636 | Escherichia coli BW25113, CGSC wild type; PMIDs: 10829079, 16738554 | LB | Oxic |
| 8422 | CGSC 7636 Δpfs-773::kan; Kmr; PMIDs: 10829079, 16738554 | LB | Oxic |
| 9859 | CGSC 7636 ΔpdxB729::kan; Kmr; PMIDs: 10829079, 16738554 | LB | Oxic |
| 9920 | CGSC 7636 ΔpdxK747::kan; Kmr; PMIDs: 10829079, 16738554 | LB | Oxic |
| 10018 | CGSC 7636 ΔglyA725::kan; Kmr; PMIDs: 10829079, 16738554 | LB | Oxic |
| 10100 | CGSC 7636 ΔluxS768::kan; Kmr; PMIDs: 10829079, 16738554 | LB | Oxic |
| 10758 | CGSC 7636 ΔmetE774::kan; Kmr; PMIDs: 10829079, 16738554 | LB | Oxic |
| 10826 | CGSC 7636 ΔmetF728::kan; Kmr; PMIDs: 10829079, 16738554 | LB | Oxic |
| 10856 | CGSC 7636 ΔmetA780::kan; Kmr; PMIDs: 10829079, 16738554 | LB | Oxic |
| 10862 | CGSC 7636 ΔmetH786::kan; Kmr; PMIDs: 10829079, 16738554 | LB | Oxic |
| 11994 | CGSC 7636 ΔyjiA750::kan; Kmr; PMIDs: 10829079, 16738554 | LB | Oxic |
| aanb | Acetobacter aceti NBRC 14818; accession BABW00000000 | mMRS | Oxic |
| aci5 | Acetobacter sp. strain DmW_043; accession JOMN00000000 | mMRS | Oxic |
| afab | Acetobacter fabarum DsW_054; PMID 29487183 | mMRS | Oxic |
| afab-pAH1 | A. fabarum DsW_054 expressing pAH1; Tcr (this study) | LB | Oxic |
| ain2 | Acetobacter indonesiensis DmW_046; accession JOMP00000000 | mMRS | Oxic |
| ama3 | Acetobacter malorum DsW_057; accession JOPG00000000 | mMRS | Oxic |
| amac | Acetobacter malorum DmCS_005; accession JOJU00000000 | mMRS | Oxic |
| aori | Acetobacter orientalis DmW_045; accession JOMO00000000 | mMRS | Oxic |
| apa3 | Acetobacter pasteurianus 3P3; accession CADQ00000000 | mMRS | Oxic |
| apan | Acetobacter pasteurianus NBRC 101655; accession BACF00000000 | mMRS | Oxic |
| apnb | Acetobacter pasteurianus NBRC 106471 or LMG1262; accession PRJDA65547 | mMRS | Oxic |
| apoc | Acetobacter pomorum DmCS_004; accession JOKL00000000 | mMRS | Oxic |
| atrc | Acetobacter tropicalis DmCS_006; accession JOKM00000000 | mMRS | Oxic |
| atrn | Acetobacter tropicalis NBRC 101654; accession BABS00000000 | mMRS | Oxic |
| atro | Acetobacter tropicalis DmW_042; accession JOMM00000000 | mMRS | Oxic |
| bsub | Bacillus subtilis subsp. subtilis strain 168; accession NC_000964.3 | LB | Oxic |
| ecok | Escherichia coli K-12 substrain MG1655; accession NC_000913.3 | LB | Oxic |
| efav | Enterococcus faecalis V583; accession NC_004668.1 | BHI | Oxic |
| efog | Enterococcus faecalis OG1RF; accession NC_017316.1 | BHI | Oxic |
| ehor | Enterobacter hormaechei ATCC 49162; accession AFHR00000000 | LB | Oxic |
| galb | Gluconobacter sp. strain DsW_056; accession JOPF00000000 | Potato | Oxic |
| ge5p | Gluconacetobacter europaeus 5p3; accession CADS00000000 | Potato | Oxic |
| gfra | Gluconobacter frateurii NBRC 101659; accession BADZ00000000 | Potato | Oxic |
| ghan | Gluconacetobacter hansenii ATCC 23769; accession ADTV01000000 | Potato | Oxic |
| gobo | Gluconacetobacter oboediens 174Bp2; accession CADT00000000 | Potato | Oxic |
| gxyl | Gluconacetobacter xylinus NBRC 3288; accession NC_016037.1 | Potato | Oxic |
| lbga | Lactobacillus brevis subsp. gravesensis ATCC 27305; accession NZ_ACGG01000000 | mMRS | Microoxic |
| lbrc | Lactobacillus brevis DmCS_003; accession JOKA00000000 | mMRS | Microoxic |
| lbuc | Lactobacillus buchneri NRRLB-30929; accession NC_015428.1 | mMRS | Microoxic |
| lfal | Leuconostoc fallax KCTC 3537; accession AEIZ00000000 | mMRS | Microoxic |
| lfer | Lactobacillus fermentum ATCC 14931; accession ACGI00000000 | mMRS | Microoxic |
| lfrc | Lactobacillus fructivorans DmCS_002; accession JOJZ00000000 | mMRS | Microoxic |
| lfrk | Lactobacillus fructivorans KCTC 3543; accession AEQY00000000 | mMRS | Microoxic |
| llac | Lactococcus lactis BPL1; accession JRFX00000000 | mMRS | Microoxic |
| lmli | Lactobacillus mali KCTC 3596 (DSM 20444); accession BACP00000000 | mMRS | Microoxic |
| lplc | Lactobacillus plantarum DmCS_001; accession JOJT00000000 | mMRS | Microoxic |
| lplw | Lactobacillus plantarum WCFS1; accession NC_004567.2 | mMRS | Microoxic |
| lrha | Lactobacillus rhamnosus GG; accession NC_013198.1 | mMRS | Microoxic |
| Klebsiella variicola ATCC BAA-830 | LB | Oxic | |
| pAH1 | Plasmid, pCM62 containing CBS::CGL from K. variicola inserted at the XbaI and EcoRI restriction sites; Tcr; this study | LB | Oxic |
| pbur | Providencia burhodogranariea DSM 19968; accession AKKL00000000 | LB | Oxic |
| pCM62 | Plasmid, hybrid of pUC19 and pCM51, improved bhr cloning vector, Tcr; 11495985 | LB | Oxic |
| pput | Pseudomonas putida F1; accession NC_009512.1 | LB | Oxic |
We tested if two factors that can vary with bacterial strain were associated with variation in fly life span: development rate of the flies and the duration of bacterial colonization in the flies. The average life span varied from about 23 to 41 days in female flies, but the mean developmental period, between egg deposition and eclosion, was 9 to 11 days long, indicating that variation in fly development rates alone did not explain the variation in life span. A second factor we considered was the period of bacterial colonization. The frequent transfer of flies to fresh diets is necessary in life span experiments and can cause loss or reduction in the continued detection of some, but not all, bacteria associated with Drosophila (35–38). We defined bacterial persistence as the number of days over which bacteria were able to be transferred from the parental (P) generation to the first-generation (F1) offspring during twice- to thrice-weekly transfers to sterile diet, and it varied significantly across the tested strains (Kruskal-Wallis [KW] χ2 = 187.91, P < 10−15) (see Fig. S2, raw data from Table S1 and mean values in Table S2) and was significantly negatively correlated with mean life spans of both male and female flies (see Fig. S3). A negative association between life span and bacterial persistence is consistent with the idea that the bacteria generally had negative, not positive, impacts on fly life span and that at least some of the life span influence depends on the continued presence of the bacteria (21). We could not test if the abundance of bacteria in the flies contributed to the species-specific effects because the microbial load assay is destructive. We anticipated that specific microbial functions contributed to the influence of the bacteria on life span and sought to define these by MGWA.
Bacterial cysteine and methionine metabolism are predicted to influence D. melanogaster life span.
We performed MGWA to identify bacterial functions that influence D. melanogaster life span. Using genome sequences from public repositories that correspond to the exact strains used in our analysis, we clustered into orthologous groups (OGs) all the genes from the strains tested for life span influence and defined the presence/absence pattern of each gene in each strain. Of 14,225 OGs, there were 5,855 distinct phylogenetic distribution groups (PDGs), defined as the exact set of bacterial strains in which an OG was present. Of these, 4,822 PDGs contained exactly one OG, whereas the remaining 1,022 PDGs collectively bore the remaining 9,403 OGs (see Fig. S4). From these, MGWA statistics were calculated for 3,539 PDGs (input data in Table S3), identifying 170 OGs that were significantly associated with changes in D. melanogaster longevity (false-discovery rate [FDR]-corrected P < 0.01) (top results are shown in Table 2; see Table S4 for the full results). Among the 170 most significant OGs, two KEGG categories were significantly enriched: glucagon signaling and cysteine and methionine metabolism (Table 3; see also Table S5). Since glucagon signaling is an animal pathway and bacteria only bear homology to scattered genes in the pathway, we focused our remaining efforts on testing the hypothesis that microbial cysteine and methionine metabolism influences D. melanogaster life span.
TABLE 2.
MGWA predictions
| OGa | P valueb | Mean life span (days)c |
No. of bacteria or identifier of species containing the OGd | Representative annotation (KEGG identifier) | |
|---|---|---|---|---|---|
| +OG | −OG | ||||
| stl00615 | 7.37E−06 | 29.4 | 34.7 | 12 Acetobacter, 5 Komagataeibacter, 2 Lactobacillus, 2 Gammaproteobacteria, efav, efog | Transport system permease (K02015) |
| stl00806 | 9.93E−05 | 34.1 | 29.2 | 9 Lactobacillus, 2 Gammaproteobacteria, bsub | Membrane protein |
| stl00883, stl00888, stl01007 | 1.32E−03 | 30.0 | 33.8 | 12 Acetobacter, efav, efog, 4 Gammaproteobacteria, 5 Komagataeibacter | Ankyrin (K06867); putative metal chaperone (Zn); deoxyribodipyrimidine photolyase (K01669) |
| stl02563, stl02568, stl02572, stl02573 | 1.32E−03 | 33.8 | 30.0 | 9 Lactobacillus, bsub | Pheromone precursor lipoprotein CamS; cell division FtsL; RibT (K02859); hypothetical |
| stl02493, stl02927, stl02930, stl03103, stl01237 | 2.44E−03 | 33.8 | 30.1 | 8 Lactobacillus, bsub | Membrane protein; hypothetical; septum site-determining MinC (K03610); trypsin-like serine protease; MFS family transporter |
| stl01019 | 5.11E−03 | 29.5 | 34.2 | 11 Acetobacter, 5 Gammaproteobacteria, 3 Gammaproteobacteria, bsub, efav, efog | Ribonuclease PH |
| stl02777 | 1.20E−02 | 34.2 | 29.6 | 7 Lactobacillus, bsub, 2 Gammaproteobacteria | Ribosomal-protein-L7/L12-serine acetyltransferase (K03817) |
| stl02172 | 1.48E−02 | 33.6 | 30.1 | 8 Lactobacillus, ghan, bsub | NAD-dependent epimerase/dehydratase |
| stl03062 | 1.99E−02 | 33.3 | 30.4 | 7 Lactobacillus, bsub, pbur | ABC transporter metal ion transporter periplasmic component/surface antigen (K02073) |
| stl02146, stl00713 | 2.05E−02 | 34.2 | 29.5 | 9 Lactobacillus, ecok, ehor | rRNA [cytosine-C(5)]-methyltransferase RsmF, hydrolase (HAD superfamily) (K07757) |
| stl02072, stl01315 | 3.06E−02 | 34.2 | 30.2 | 8 Lactobacillus | Hypothetical protein, l-2-hydroxyisocaproate dehydrogenase (K00016) |
| stl01428 | 3.17E−02 | 34.1 | 29.6 | 6 Lactobacillus, 3 Gammaproteobacteria, bsub | NAD(P)H dehydrogenase (quinone) |
| stl00675 | 3.65E−02 | 29.3 | 33.6 | 12 Acetobacter, 5 Komagataeibacter, pbur, efav, efog | Dipeptide ABC transporter substrate-binding protein (K02035) |
| stl00558 | 3.72E−02 | 33.2 | 29.5 | 9 Lactobacillus, 3 Gammaproteobacteria, bsub | S-Methylmethionine transport protein (K11733) |
| stl02793, stl03291 | 3.91E−02 | 33.8 | 30.3 | 7 Lactobacillus, bsub | Sodium/hydrogen exchanger (K03455); transport protein (K06994) |
OG, orthologous group.
Bonferroni corrected P value.
+OG, conferred by bacteria containing the OG; –OG, conferred by bacteria lacking the OG.
Four-character designations are listed in Table 1.
TABLE 3.
KEGG enrichment analysis of significant MGWA predictionsa
| KOb | Annotation | No. of genes |
P valuec | |
|---|---|---|---|---|
| All (n = 4,953) | Top predictions (n =149) | |||
| ko00270 | Cysteine and methionine metabolism | 72 | 7 | 6.25E−03 |
| ko04922 | Glucagon signaling pathway | 16 | 3 | 8.74E−03 |
| ko05230 | Central carbon metabolism in cancer | 10 | 2 | 0.05 |
| ko02010 | ABC transporters | 279 | 3 | 0.10 |
| ko01100 | Metabolic pathways | 1,138 | 24 | 0.13 |
| ko00790 | Folate biosynthesis | 32 | 3 | 0.14 |
| ko00230 | Purine metabolism | 106 | 6 | 0.22 |
| ko01120 | Microbial metabolism in diverse environments | 424 | 8 | 0.26 |
| ko04122 | Sulfur relay system | 23 | 2 | 0.37 |
| ko00240 | Pyrimidine metabolism | 75 | 4 | 0.43 |
| ko00561 | Glycerolipid metabolism | 28 | 2 | 0.50 |
| ko00620 | Pyruvate metabolism | 86 | 4 | 0.59 |
| ko02024 | Quorum sensing | 152 | 3 | 0.63 |
| ko00250 | Alanine, aspartate and glutamate metabolism | 34 | 2 | 0.66 |
| ko00480 | Glutathione metabolism | 35 | 2 | 0.69 |
| ko01200 | Carbon metabolism | 181 | 4 | 0.70 |
| ko01230 | Biosynthesis of amino acids | 196 | 7 | 0.82 |
| ko00564 | Glycerophospholipid metabolism | 43 | 2 | 0.87 |
| ko01130 | Biosynthesis of antibiotics | 327 | 9 | 0.93 |
| ko00920 | Sulfur metabolism | 48 | 2 | 0.98 |
| ko01110 | Biosynthesis of secondary metabolites | 443 | 14 | 0.98 |
| ko00010 | Glycolysis/gluconeogenesis | 83 | 3 | 1.00 |
| ko00260 | Glycine, serine and threonine metabolism | 75 | 2 | 1.00 |
| ko00330 | Arginine and proline metabolism | 57 | 2 | 1.00 |
Only pathways containing at least 2 genes among top predictions are included. Full results are shown in Table S5 in the supplemental material.
KO, KEGG orthology.
Chi-square test.
Bacterial methionine metabolism genes influence D. melanogaster life span.
We performed a mutant analysis to test the prediction that bacterial cysteine and methionine metabolism genes influence D. melanogaster life span. We measured life span in D. melanogaster monoassociated with Escherichia coli strains bearing mutations in methionine metabolism genes (Fig. 2). Because a different strain of E. coli bacteria persisted poorly during thrice-weekly fly transfers to fresh diets in our first set of experiments (Fig. S2), we inoculated one set of fresh diet vials per week with the corresponding bacteria to ensure continued exposure to the bacteria. The luxS mutant was the only mutant that significantly extended fruit fly life span, and the significant effect was only detectable in male flies (Fig. 2B, see also Fig. S5, raw data in Table S6, full statistics in Table S7). Conversely, two bacterial mutants related to vitamin B6 biosynthesis—pdxB and pdxK mutants—each significantly shortened fly life span relative to that with wild-type E. coli strains in male and female flies, and a mutation in the glyA gene, which participates in the vitamin B6-dependent conversion of glycine to serine, shortened female life span (Fig. 2B and C). Together, these findings reveal that disruption of methionine metabolism genes in associated bacteria can reduce or extend fruit fly life span.
Life span-influencing bacteria do not alter methionine levels in old or young D. melanogaster.
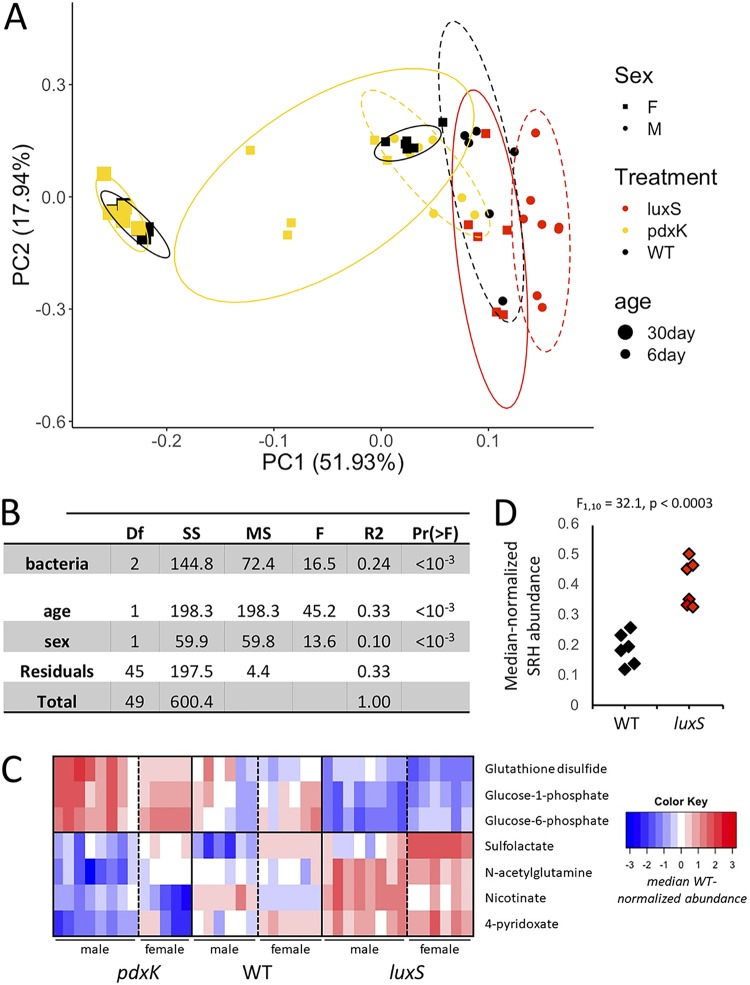
To begin to understand why the pdxBK mutants shortened Drosophila life span whereas the luxS mutant promoted longevity, we performed a global metabolomic analysis of flies or diets that had been monoassociated with the different mutants. When we compared the metabolomes of the flies bearing mutant E. coli strains versus those bearing a wild-type E. coli strain, there was a significant effect of fly age, fly sex, and the bacterial treatment on the adult fly metabolome, with fly age and bacterial treatment explaining the largest parts of the variance (Fig. 3A and B; raw data in Table S8). The differences were apparent only in 5- to 7-day-old adults, whereas by the time adults reached 30 days old, there were no differences between flies bearing wild-type bacteria and flies bearing mutant bacteria (see Table S9). Among the differentially abundant metabolites (not FDR corrected), there was no significant enrichment for metabolites from specific pathways, including methionine metabolism (see Table S10). The most striking pattern was that just 7 metabolites were differentially abundant between both male and female flies that bore three different E. coli strains: the wild type, a pdxK mutant, and a luxS mutant (Fig. 3C). The metabolites fell into two clusters relative to their abundance in flies bearing wild-type bacteria: (i) a group upregulated in flies bearing the pdxK mutant and downregulated in flies colonized with the luxS mutant, which included two glucose derivatives and (ii) a group displaying the opposite trend, including a vitamin B6 derivative, 4-pyridoxate. Together, these findings reveal that the bacterial mutants had a more striking influence on host glucose regulation than on host methionine metabolism.
FIG 3.
Metabolomics of aging flies and diets exposed to E. coli mutants. (A) Principal-component analysis (PCA) plot showing the clustering of samples from flies inoculated with mutant bacteria (luxS and pdxK) in an analysis of global metabolomes (∼100 metabolites). (B) PERMANOVA table showing significant variations in the metabolome with bacterial treatment and with age and sex of the flies. Df, degrees of freedom; SS, sum of squares; MS, mean squares; F, F value; R2, R2; Pr(>F), P value. (C) Heat map showing the differences in specific metabolite levels between flies bearing mutant (luxS and pdxK) and wild-type bacteria (WT). (D) Differences in the abundance of S-ribosyl homocysteine (SRH) in fly diets exposed to flies and either wild-type (WT) or luxS mutant bacteria.
We reasoned that methionine cycle metabolites in the flies might remain relatively constant when reared with bacterial mutants because of compensatory transcription of fly genes to maintain homeostasis. To test this idea, we performed a transcriptomic analysis of 5- to 7-day-old and 30-day-old flies bearing pdxK mutant or wild-type bacteria (see Table S11). More genes were differentially expressed with fly aging than with bacterial treatment (see Table S12), and there was no enrichment for methionine metabolism genes among differentially expressed genes (see Table S13). We note that the raw sequence data were inadvertently deleted after calculation of fragments per kilobase per million (FPKM) values, blocking independent reproduction of our analysis and tempering the logical extension of the presented data that differences in fly transcription did not normalize the different influences of the bacterial mutants on methionine cycle metabolites in the flies. Regardless, the expression data are consistent with the general conclusions in this section, that methionine metabolism was not dramatically altered in flies reared with bacterial mutants that significantly alter the fly life span.
In a final test of the relationship between bacterial mutants and methionine metabolism, we performed a metabolomic analysis of the diets of flies reared with wild-type bacteria versus those reared with pdxK or luxS mutant bacteria. Relative to that in diets of flies inoculated with wild-type E. coli, there were no differences in the metabolite contents of the diet from flies inoculated with the pdxK mutant and only one metabolite that differed from the diet of flies inoculated with the luxS mutant (see Table S14), i.e., S-ribosyl homocysteine, the product metabolized by the luxS gene (Fig. 3D; see also Table S15). luxS is the second gene in the 2-step conversion of SAH to homocysteine via the S-ribosyl homocysteine (SRH) intermediate, and the accumulation of SRH in the diet is consistent with the luxS-dependent conversion of SRH to homocysteine. Additionally, xanthosine levels were significantly higher in the diets of flies bearing either bacterial mutant than in those with the wild-type strain, although not after correction for multiple tests. Since only one metabolite differed between the diets and many differed between the flies, these findings show that mutant-specific variation in the diets was in one case consistent with the expected influence of the bacterial mutant but was not a direct predictor of changes in the flies’ metabolomes and bore no relationship to methionine metabolism in the flies.
Bacterial transsulfuration extends fruit fly life span.
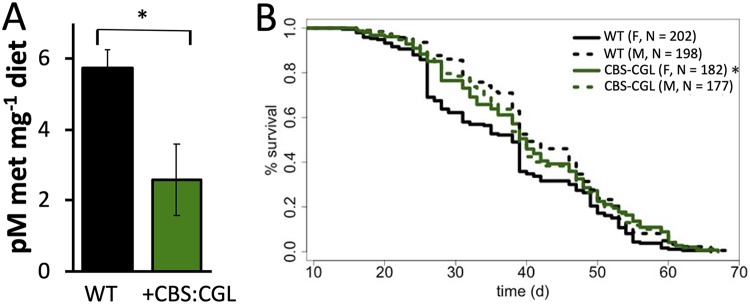
The results of the previous section provided poor support for the MGWA prediction that disruption of bacterial methionine metabolism can influence D. melanogaster life span. Therefore, we designed a second set of experiments to directly test if a microbiota-dependent reduction in dietary methionine could extend the life span of D. melanogaster. Because fruit fly life span is longer when the CBS gene is constitutively expressed in the D. melanogaster gut, we constructed an Acetobacter fabarum strain, using plasmid pAH1, that ectopically expressed a single genomic locus containing CBS and cystathionine gamma lyase (CGL) genes from Klebsiella variicola. Together, these genes are involved in transsulfuration (Fig. 2A) and are poorly conserved in bacteria. We expressed them from K. variicola because it was a readily accessible strain that expressed both genes from a single locus. The methionine content of diets of flies bearing the CBS::CGL expression mutant was lower than that of diets on which flies were inoculated with a wild-type A. fabarum strain (Fig. 4A; raw data in Table S16). Furthermore, female, but not male, flies lived significantly longer when inoculated with the CBS::CGL expression strain (Fig. 4B; raw data in Table S17). Therefore, even though the influence of the luxS and pdxK mutants on fly life span were not attributable to metabolic changes in methionine cycle metabolites, these data demonstrate a correlation between the methionine-restricting activity of the CBS::CGL expression strain and life span extension that is consistent with the known life span-extending influence of methionine restriction (12). Together, these findings identify a link between bacterial methionine metabolism and host life span, with the caveat that at least some bacterial genes influence D. melanogaster life span without any evidence of metabolic patterns consistent with the expectations of methionine restriction.
FIG 4.
The influence of transsulfuration gene-expressing Acetobacter on fly life span and methionine content. (A) The influence of CBS::CGL-expressing bacteria on dietary methionine content; n = 6 readings per treatment. *, P < 0.05. (B) Mean life spans of female (F) and male (M) flies bearing wild-type (WT) or CBS::CGL-expressing bacteria (+CGS-CGL). *, P < 0.05 versus female flies bearing wild-type (WT) bacteria.
DISCUSSION
In this work, we show that altered regulation of specific bacterial functions, including methionine metabolism, can significantly influence D. melanogaster life span. Initially, our discovery that vitamin B6 and SRH bacterial mutants shortened and extended the fly life span, respectively, appeared consistent with the idea that blocking flux through the methionine pathway shortens fly life span (15, 17). For example, we expected that bacterial mutants for vitamin B6, which serves as a cofactor in transsulfuration and vitamin B12-dependent conversion of homocysteine to methionine, shortened fly life span by blocking the escape of methionine cycle intermediates from the methionine cycle. Conversely, we expected that permitting storage by the microbiota of methionine cycle intermediates in SRH, which is metabolically inaccessible to the flies, would extend fly life span by lowering the abundances of methionine cycle intermediates in the flies or the diets. Instead, neither the flies nor (with the exception of SRH) the diets of flies inoculated with these mutants displayed significant variation in the abundance of methionine intermediates, suggesting either that we were looking for the wrong output (metabolite levels instead of measuring flux) or that these mutants influence life span independent of methionine restriction. A follow-up experiment replicating the effects of increasing transsulfuration by ectopic expression of fly genes in the fly gut (15) did show that bacterial transsulfuration can restrict the methionine content of the diet and extend the life span of female flies. Thus, our findings reveal methionine restriction-independent and -dependent mechanisms of bacterial influence on D. melanogaster life span.
Because of established links in the literature between aging and methionine restriction, we focused our follow-up work from the MGWA on bacterial genes involved in methionine metabolism (10–13). Also, our previous work showed that E. coli methionine mutants can influence D. melanogaster starvation resistance (30), a life history trait that is positively correlated with natural variation in fruit fly life span (39). Therefore, we expected that many of the E. coli methionine mutants already in our possession from that analysis would also influence life span, even though most of the mutants we tested were selected for their location in the methionine pathway and not for a high predicted MGWA score (Fig. 2A). For example, pdxB, which did influence life span, was ranked 11,110 of 12,980 results in the MGWA. We tested it because it was in our possession and directly linked to methionine metabolism. However, we did not test metC, ranked 81, because we did not have it in our possession and its mechanism was tested by CBS::CGL expression in A. fabarum. Therefore, the fact that few of the E. coli methionine cycle genes we tested influence D. melanogaster life span may suggest incorrect assumptions or experimental design flaws. For example, we expected that because methionine metabolism genes were enriched in the MGWA, many genes related to methionine metabolism would influence life span regardless of their MGWA-predicted impact. We also expected that E. coli mutants would be good models of fruit fly microbes. Instead, further tests on individual predictions from the MGWA, using E. coli or other microorganisms, may or may not reveal additional bacterial genes with life span influence.
Several MGWA-predicted genes we did not test are conspicuous candidates for further testing. A pyrroloquinoline quinone (PQQ) aldehyde dehydrogenase was previously shown to influence development rate in D. melanogaster (40), a life history trait that is negatively correlated with changes in host life span (39, 41), and the pqqC and pqqB genes from the same family were among the top 100 predictions. Three top MGWA predictions were genes encoding cell division proteins (ftsB, ftsL, and minC), possibly suggesting bacterial division in the flies or fly diet can shape fly life span. Finally, several significant MGWA results were related to molybdenum transport and cofactor biosynthesis. These include modE, moaA, and moaC. A deficiency in this cofactor causes severe disabilities early in life and death within 12 days of birth in mice, but death can be suppressed by bacterially provisioned “precursor Z,” identifying a link between these bacterial genes and a mammalian animal host (42). It would be interesting to test if this link is detectable in the interactions between D. melanogaster and its microbiota. Taken together, these considerations show that while our study focused on only a small number of genes in one area of bacterial metabolism, there are likely to be many other bacterial functions that influence fruit fly life span, and some may be among other top MGWA predictions.
We used E. coli mutants to validate bacterial genetic influences on host traits according to the model from previous works by us and others in dipteran hosts (30, 43). Although the use of E. coli mutants has some obvious strengths, it does come with caveats. For example, E. coli is not an abundant partner of wild D. melanogaster and does not persist in the flies during frequent transfers as well as many other strains naturally isolated from Drosophila. Some of the E. coli mutants we tested influence fly life span, providing support to previous findings that E. coli can be a powerful tool to understand host-microbe interactions in Drosophila and other animals that do not naturally bear this organism abundantly (30, 43). However, one area that has not been conclusively addressed is if E. coli mutants influence their hosts in the same ways as the host’s natural partners do. We hypothesize that such influences, especially on traits with polygenic influences, are likely to be bacterial strain specific whether they are a natural partner or otherwise. For example, epistasis or the presence of background effects could mask or expose the effects of certain genes. In this case, studying the same mutants in different bacterial strain backgrounds could potentially unveil additional genetic interactions between hosts and microbes. Future studies that explicitly compare the influence of the same mutations when made in natural or other partners of D. melanogaster or other hosts are necessary to address this issue.
Our findings are consistent with the idea that microbial influence on D. melanogaster life span is primarily antagonistic when the flies are reared on a nutrient-rich diet. Of the 39 strains we tested, none significantly extended host life span relative to that of axenic flies. We attributed some of these antagonistic influences to bacterial methionine metabolism. Other differences were associated with changes in the abundance of glucose in the flies. Flies reared with a vitamin B6 mutant displayed lower levels of 4-pyridoxate, the only version of vitamin B6 we were able to detect in our analysis, possibly suggesting that restriction of vitamin B6 has negative effects on fly life span. A commensurate increase in glucose levels mirrors the effects of fly Δpdx-1 mutants, which display higher glucose levels than wild-type flies and are linked to increased chromosome damage (44). Such damage could explain why the bacterial pdxBK mutants shortened fruit fly life span. The parallel host phenotypes observed from manipulating homologous host or bacterial genes suggests a similar mechanistic outcome and provides justification for further research in the vitamin B6-dependent bacterial mechanisms of life span influence.
We previously showed that bacterial methionine metabolism genes influence starvation resistance in D. melanogaster and speculated that bacterial methionine metabolism might also be related to fruit fly life span (30). This idea was supported by established connections between fruit fly longevity and both methionine restriction and starvation resistance. We have already discussed the methionine restriction connection here. Starvation resistance is related to life span as a life history trait that influences the survival and reproduction of flies in natural settings along with other critical traits such as reproduction, fat storage, and development rate (39). The influences of different bacterial strains on life history traits in fruit flies reared on our nutrient-rich diet led to correlated phenotypes, where bacteria that conferred shorter starvation resistance and life span also conferred higher early reproduction and development rates and lower fat storage (41). Since bacterial methionine metabolism influences both life span and starvation resistance, though this work shows such effects are not necessarily through the same genes, and because of established work showing that levels of methionine cycle metabolites in the diet influence fly reproduction and development rate (8, 12, 45), bacterial methionine metabolism may be a key trait underlying microbial influences on D. melanogaster life history traits. A counterargument is that tradeoffs between reproduction and fecundity can be decoupled by rearing flies on different diets and that these relationships may only be apparent under particular dietary or other scenarios (8, 12). Regardless, we propose that methionine metabolism may be linked to the bacterial influence on Drosophila life history, possibly through the host insulin-like signaling/target of rapamycin (IlS/Tor) pathway with its established connections to both methionine restriction and the microbiota (12, 40, 46).
We also show an Acetobacter strain that constitutively expresses nonnative transsulfuration genes CBS and CGL can extend the life span of D. melanogaster. This finding is fully consistent with a previous study that extended D. melanogaster life span by expressing Drosophila CBS in the fly gut, with males displaying a more modest response than females (in our study, there was no difference in male life span) (15). Our finding that this bacterial strain lowered the methionine content of the diet raises the question of whether the transgenic Drosophila CBS-expressing line’s influence is correlated with or independent of dietary changes. Although we did not measure the metabolite content of flies bearing the CBS::CGL expression strain, we suspect that measuring flux through the methionine cycle, rather than absolute metabolite content, may be a better indicator of the influence of such mutants on the methionine cycle. Our findings from flies reared with pdxK and luxS mutants suggest that it may be difficult to detect changes in fly metabolite abundances when rearing flies with different bacterial strains, and current working models are that flux through the pathway may be more important than a snapshot of absolute metabolite content (17).
Taken together, our work links the influence of the microbiota on fruit fly life span with vitamin B6, glucose, and methionine metabolism. Our finding that genetic manipulations in bacterial partners can recapitulate host phenotypes already known to be induced by host genetic manipulations suggests that associated microorganisms can adopt overlapping metabolic influences relative to those of host genes. Thus, we identify key bacterial functions and provide some underlying explanations for the influence of these functions on the life span of a model host.
MATERIALS AND METHODS
Fly and bacterial culture.
All experiments were conducted with Wolbachia-free D. melanogaster Canton-S obtained from Mariana Wolfner and cultured at 25°C on a 12-h light-dark cycle. Under standard culture (maintenance) conditions, Drosophila melanogaster was fed on a yeast-glucose diet (1 liter H2O, 100 g inactive brewer’s yeast, 100 g glucose, 12 g agar, 0.84% propionic acid, and 0.08% phosphoric acid) as in our previous work (47, 48).
The bacterial strains (Table 1) were cultured as in our previous work on clade-specific media: modified MRS medium (mMRS; 1.25% peptone, 0.75% yeast extract, 2% glucose, 0.5% sodium acetate, 0.2% dipotassium hydrogen phosphate, 0.2% triammonium citrate, 0.02% magnesium sulfate heptahydrate, 0.005% manganese sulfate tetrahydrate, 1.2% agar), potato medium (0.5% glucose, 1% yeast extract, 1% peptone, 0.8% potato extract), lysogeny broth (LB; 1% tryptone, 0.5% yeast extract, 0.5% sodium chloride), and brain heart infusion (BHI) broth. Growth of auxotrophs was confirmed on M9 medium supplemented with the auxotrophic nutrient. E. coli were grown at 37°C, and all other strains were grown at 30°C. Transposon insertion or ectopic expression mutants were cultured with antibiotics: 50 mg/ml kanamycin for E. coli transposon insertion mutants, and 20 μg/ml chlortetracycline for the Acetobacter fabarum DsW_54 ectopic expression strain. Strains grown under oxic conditions were grown in liquid culture with shaking or with no atmospheric treatment in solid culture. Strains grown under microoxic conditions were grown statically (liquid) or in a sealed CO2-flooded chamber (solid).
Preparation of axenic and gnotobiotic flies.
Flies were reared under axenic or gnotobiotic conditions as in our previous work (48). Briefly, <20-h-old eggs laid on grape juice agar plates were collected from D. melanogaster Canton-S and surface sterilized with a 0.6% sodium hypochlorite solution in two 2.5-min washes. Hypochlorite was washed away by three rinses with sterile water, and 30 to 60 eggs, qualitatively estimated, were aseptically transferred to sterile diet in a sterile biosafety cabinet. Then, 7.5 ml of sterile yeast-glucose diet (omitting the acid) was inoculated in 50-ml conical tubes, autoclaved, and allowed to cool before transferring sterile eggs. To rear under axenic conditions, the eggs were left undisturbed. To rear with defined bacterial species, the sterile eggs were inoculated with 50 μl of bacterial cultures normalized to an optical density at 600 nm (OD600) of 0.1. If more than one species was added, the multiple strains were normalized as above and pooled in equal ratios before inoculating the flies with a 50-μl volume of bacteria. For each analysis, we sought to collect data from triplicate vials of flies in each of three separate experiments; however, some treatments could not be collected at this level of replication, and after discarding contaminants, all data were used regardless of replication. For example, in the Fig. 1 analysis, one treatment, lfer (Table 1), had an n of just 1 vial. All other treatments had an n of at least 4 vials, and the total number of vials and flies used in the experiment are presented in Table S1A in the supplemental material.
Life span analysis.
Drosophila melanogaster life span was measured by recording the number and sex of dead flies and transferring surviving flies to fresh sterile diet every 2 to 3 days until all flies in a vial were dead. The spent vials were incubated at room temperature (∼22°C) until eggs laid during the 2- to 3-day interval grew to adulthood. Bacterial residence in the flies was also tracked on a weekly basis by measuring bacterial presence in F1 progeny of the P generation flies we examined. Each week, a mixed-sex pool of five flies from each vial was homogenized in 125 μl homogenization buffer (10 mM Tris [pH 8], 1 mM EDTA, 0.1% Triton X-100, as in reference 49) with 125 μl Lysing Matrix D ceramic beads (116540434; MP Biomedicals) by shaking for 30 to 60 s at 4.0 m/s in a FastPrep-24, dilution plated onto mMRS medium with a limit of detection of 20 CFU/fly, incubated under oxic and microoxic conditions, and visually inspected by colony morphology to confirm strain identity. If at least 200 CFU fly−1 of the expected bacterial strain was detected, the strain was deemed “present” (see below for incorporation into statistical models). If at least 200 CFU fly−1 of an unexpected bacterial species was detected in 2 consecutive weeks, the vial was deemed contaminated. Differences between Acetobacter strains could usually not be determined by colony morphology, and so Acetobacter contamination of other Acetobacter strains cannot be ruled out. These cutoffs are in line with those in our previous work (30, 50). Initially, three replicated experiments were performed, each with triplicate vials per treatment. Three additional blocks were subsequently performed to add more replication for vials that were discarded as contaminated from the first three experiments, together with controls and a few randomly selected (different for each block) treatments to provide a reference during the blocking.
The life span analysis for flies bearing E. coli mutants was conducted exactly as described above with one exception: because E. coli persisted poorly in flies during the first life span experiment (see Fig. S2), each P-generation fly vial was reinoculated with the corresponding E. coli mutant once per week after eclosion. Auxotrophs were distinguished by their ability to grow on M9 medium supplemented with the auxotrophic nutrient. Three replicated experiments were performed, each with triplicate vials per treatment.
Differences in fly life span with bacterial treatments as the main effect were determined by a left- and right-censored Cox mixed-effects survival model in R (51, 52) to account for differences in bacterial residence in the flies. All flies entered the experiment at the time of egg transfer to sterile diet, and bacterial presence was indicated as a fixed effect with the value of “1.” If no bacteria were detected in two consecutive weeks, all flies in the corresponding vial were marked to exit the experiment at the last day bacteria were detected in F1 vials (right censored). Then, we added a second row of data with identical metadata that indicated that a fly colonized with the same bacteria entered the experiment on that day with a bacterial presence value of “0.” Thus, the total number of live flies in each treatment remained the same, but the bacterial colonization status of the flies could vary. Finally, when a fly died, it was marked to exit the experiment regardless of its colonization status (which was still recorded and included in the model). Also, if flies were lost during transfer or deemed as contaminated in 2 consecutive weeks (see the method for detecting contamination above), they were marked to leave the experiment permanently at the time of loss or the last time point they were known to be uncontaminated. Flies contaminated from the first transfer onward were excluded from the analysis entirely. Data in Fig. 2 and 4 were only right censored because E. coli were reinoculated weekly (Fig. 2) and because A. fabarum persists during vial transfers (Fig. 4).
Correlations between bacterial persistence in the flies and life span were calculated in R using a Spearman rank correlation. The calculation was between the number of days flies were detected in each vial and the average life span of all dying flies in that vial (flies that were lost during bumping were excluded before calculating the mean). Any vial that was deemed contaminated at any time was completely excluded from the analysis.
Metagenome-wide association.
To predict bacterial genes that influenced life span, a meta-genome wide association (MGWA) approach was used, as in our previous work (49). Amino acid sequences from the exact bacterial strains used in the monoassociation experiments were obtained from GenBank and were clustered in orthologous groups (OGs) using a local installation of the OrthoMCL software with an inflation factor of 1.5. MGWA was performed using the MAGNAMWAR R package (53). Differences in life spans of the flies relative to the presence or absence of each OG as the main fixed effect were determined by the right- and left-censored Cox mixed-effects model described above. OGs were ranked according to P value, with a false-discovery rate (FDR)-corrected P value of ≤0.01 considered significant.
To identify functional categories that were enriched among the significantly associated OGs, a KEGG enrichment analysis was performed. KEGG categories were assigned to a representative sequence from each OG using BlastKOALA (54). The KEGG pathway assignments to each OG were retrieved in KEGG PATHWAY, and chi-square tests were performed to test for pathways that were enriched in the top OGs relative to all OGs. FDR correction was applied. Chi-square tests and FDR correction were performed in R (55).
The phylogenetic tree for the figures was built by manually extracting 16S rRNA gene sequences from the nucleotide sequences of each sequenced genome, aligning with the EMBL-EBI online MUSCLE tool (http://www.ebi.ac.uk/Tools/msa/muscle/), manually trimming to an aligned (with gaps) partial 16S rRNA sequence length of 1,348 bp, and rerunning the alignment with default ClustalW parameters. A neighbor-joining tree without distance correction was downloaded from the online module and manually formatted in FigTree v 1.4.3 (56). A 16S rRNA gene sequence for Halobacterium jilantaiense JCM 13558, accession NR_113425, was used as an outgroup (not shown in figures).
Metabolomics.
For experiments shown in Fig. 3, shotgun metabolomics data were collected for diet and fly samples at 5 to 7 or 30 days posteclosion between 4 and 7 h into the daily light cycle. From triplicate vials, flies were anesthetized on CO2 and immediately frozen at –80°C, and 0.2 mg of diet was collected at the surface of the fly vial and freeze-dried in a Thermo Savant ModulyoD freeze dryer. All samples were shipped on dry ice overnight to the biological and small molecule mass spectrometry facility at the University of Tennessee, Knoxville, TN, for shotgun metabolomic analysis. To determine significant differences in the abundances of individual metabolites, metabolite abundances were median centered, significant differences between metabolite abundances with microbial treatment or time were determined by a linear model, and P values were FDR corrected. Overall differences between samples were determined by principal-component analysis and permutational multivariate analysis of variance (PERMANOVA) of the median centered values, all conducted in R (55, 57). KEGG enrichments were calculated as for the MGWA.
For experiments shown in Fig. 4, we used high-performance liquid chromatography (HPLC)-UV to detect the abundance of methionine in diet samples. Twenty-five milligrams of diet was removed from vials containing 5- to 7-day-old D. melanogaster reared in monoassociation with either A. fabarum DsW_054 or A. fabarum DsW_054 containing pAH1, making sure to remove all D. melanogaster eggs or larvae from the sample. The sample was lyophilized for ∼24 h in a Thermo Savant ModulyoD freeze dryer until the food was completely visibly dry. Then, the dry sample was resuspended in 100 μl TET buffer (10% Tris, 1% EDTA, 0.1% Triton X-100), mixed with an equivalent volume of MP Biomedicals Lysing Matrix D beads, and shaken at 1,250 rpm for 120 s on a Geno/Grinder 2010. The particulate matter was removed by first centrifuging the samples at 4°C and 15,000 × g for 5 min in a microcentrifuge and then filtering through a 0.45-μm low-binding hydrophilic polytetrafluoroethylene (PTFE) Multiscreen Solvinert plate (MSRLN0410; Millipore) by centrifuging at 1,500 × g for 5 min. The plate was previously primed with 100 μl TET buffer by centrifuging at 4°C and 1,500 × g for 10 min. The filtrate was stored at −80°C until it was derivatized using the Waters AccQ Tag Ultra derivatization kit (186003836). Briefly, the AccQ Tag Ultra reagent powder was dissolved in AccQ Tag Ultra reagent diluent by heating at 55°C for less than 15 min. Then, 70 μl of AccQ Tag Ultra borate buffer was mixed sequentially with 10 μl of sample and 20 μl of AccQ Tag Ultra reagent. Samples were vortexed, heated to 55°C for 10 min, and placed in an Agilent model 1260 instrument for analysis at the BYU Chromatography Center. Samples were run on a Luna C18 (2) 5-μm 4.6 mm by 100 mm column. Buffer A was 0.14 M sodium acetate plus 0.05% triethylamine adjusted with phosphoric acid at pH 6.4 plus 60 ml acetonitrile (ACN) (buffer/ACN, 940:60). Buffer B was ACN/H2O (60:40). A flow rate of 1 ml/min was used. The methionine peak was defined by comparison with retention time and UV spectrum at 254 nm for an external standard (Sigma amino acid standard, SLBR6088V) that was diluted in water to 100 pmol μl−1 and derivatized exactly as described above. Significant differences with microbial treatment for dietary methionine abundance normalized to weight of the diet sample were determined by a Student’s t test. Protein contents were measured from 10 μl of the sample prior to derivatization by a Bio-Rad DC protein assay kit, according to the manufacturer’s instructions.
RNA-seq.
Transcriptome sequencing (RNA-seq) libraries were prepared using the NEBNext poly(A) mRNA magnetic isolation module (NEB E7490) and Ultra RNA Library Prep kit for Illumina (NEB E7530). Flies were reared from birth with either wild-type E. coli or an E. coli pdxK mutant. At 5 or 31 days posteclosion, 30 female flies were frozen on dry ice and homogenized with MP Biomedicals Lysing Matrix D beads in 490 μl buffer RLT (Qiagen) mixed with 10 μl beta-mercaptoethanol on a Geno/Grinder 2010 at 1,250 rpm for 120 s. For each time point and treatment, three biological replicates were performed (flies dechorionated and inoculated with bacteria on different days), and each biological replicate was derived from 1 or 2 replicate fly vials in the same experiment. The homogenate was centrifuged to pellet debris, and 166 μl of clarified supernatant was used for RNA extraction with the Zymo Direct-zol RNA Miniprep kit (11-330) according to the manufacturer’s instructions, including the on-column DNase I digestion. mRNA isolation, fragmentation, and priming from total RNA, first- and second-strand synthesis, purification, end preparation, and adapter ligation were performed according to the manufacturer’s protocol using a subset of the 8 by 12 TS HT dual index mixed adapter plate of 96 duplexed Illumina barcodes through IDT. The samples were sequenced on two lanes of rapid run mode 50-bp single-end chemistry on an Illumina HiSeq at the BYU DNA sequencing center. Output files from each lane were concatenated and run through TopHat (58) and CuffLinks (59) using the UCSC dm6 reference. Significant differences in FPKMs of each gene with treatment were calculated by a Student’s t test, and KEGG enrichments were calculated as described above.
Expression of Klebsiella variicola CBS::cystathionine gamma lyase in A. fabarum DsW_054.
Plasmid pAH1 was constructed by insertion of the 2.5-kb CBS::CGL Klebsiella variicola operon into expression vector pCM62. Genomic DNA was isolated from 1.5 ml of K. variicola culture using the DNeasy blood and tissue kit (Qiagen). The CBS::CGL fragment was amplified from the K. variicola genomic DNA with Pfx polymerase (Thermo Fisher Scientific) and primers CBSCGL-xbaI-for (5′-NNNNtctagaATGTCACTGTTTCATTCC-3′) and CGL-ecoRI-rev (5′-NNNNgaattcCGGAATAATCACTCCTCC-3′) (restriction enzymes in lowercase letters). The pCM62 plasmid was digested using XbaI and EcoRI (New England BioLabs) according to the manufacturer’s recommendations, and the CBS::CGL fragment was ligated into the plasmid using T4 DNA ligase (New England BioLabs). The assembled pAH1 plasmids were then electroporated into E. coli S17 λpir and selected on LB plates containing chlortetracycline at a concentration of 5 ng/ml. Successful cloning was initially screened by PCR across the polylinker, selecting for colonies with the 2.5-kb insert, and the sequence of the insert was verified in its entirety by Sanger sequencing at the BYU DNA sequencing center using primers cbscgl-500 (5′-ACCACTTATTCAGCGAACC-3′), CBS_CGL-1000 (5′-GGAGGAACGATAATCGAAG-3′), CBS_CGL_1500 (5′-GATCCTGGCTGGTCGAAG-3′), CBS_CGL-2000 (5′-CCAACGCTTCTCCCTGCC-3′), CBS_CGL_2500 (5′-TGGATAAGGACAGTCACC-3′), and CBS_CGL_3000 (5′-GAAGTGGAGCGAGTCTGG-3′). This created plasmid pAH1. pAH1 was conjugated into Acetobacter fabarum DsW_054 as described previously (60). Briefly, S17 λpir E. coli containing pAH1 was grown overnight at 37°C in LB-chlortetracycline, and A. fabarum DsW_054 was grown at 30°C in potato medium. Five hundred microliters of cells from each culture was centrifuged, the supernatants were discarded, and each pellet was resuspended in 50 μl of potato medium. Fifty microliters of each culture was mixed in a microcentrifuge tube and incubated at 30°C for 16 h. The 100-μl mixture was then plated on YPG medium (1.5% agar, 1% glycerol, 0.5% peptone, 0.5% yeast extract, 0.2% acetic acid, and 20 mg/liter chlortetracycline) (49). YPG plates containing transformed A. fabarum DsW_054 were incubated for 72 h, after which, colony PCR and plasmid isolation were performed to confirm the presence of the plasmid. The flies and fly diets were inoculated with this strain and the untransformed control strain to measure the life span of the flies and methionine content of the diet as described above.
Supplementary Material
ACKNOWLEDGMENTS
We thank three anonymous reviewers for feedback that helped us improve the manuscript.
This research was possible through funds received from BYU, including an Office of Research and Creative Activities award to Melinda K. Matthews, startup funds to John M. Chaston, and a grant from the BYU Gerontology Program.
We thank the Yale Coli Genetic Stock Center, Adam Dobson, Parvin Shahrestani, and Jiping Zhou for reagents or assistance with work. Mass spectrometric analyses were performed at the University of Tennessee, Knoxville, TN, biological and small molecule mass spectrometry core with the assistance of Shawn R. Campagna, Hector F. Castro, and Sara Howard.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Zhao L, Zhao Y, Liu R, Zheng X, Zhang M, Guo H, Zhang H, Ren F. 2017. The transcription factor DAF-16 is essential for increased longevity in C. elegans exposed to Bifidobacterium longum BB68. Sci Rep 7:7408. doi: 10.1038/s41598-017-07974-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smolentseva O, Gusarov I, Gautier L, Shamovsky I, DeFrancesco AS, Losick R, Nudler E. 2017. Mechanism of biofilm-mediated stress resistance and lifespan extension in C. elegans. Sci Rep 7:7137. doi: 10.1038/s41598-017-07222-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khanna A, Kumar J, Vargas MA, Barrett L, Katewa S, Li P, McCloskey T, Sharma A, Naude N, Nelson C, Brem R, Killilea DW, Mooney SD, Gill M, Kapahi P. 2016. A genome-wide screen of bacterial mutants that enhance dauer formation in C. elegans. Sci Rep 6:38764. doi: 10.1038/srep38764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwon G, Lee J, Lim YH. 2016. Dairy Propionibacterium extends the mean lifespan of Caenorhabditis elegans via activation of the innate immune system. Sci Rep 6:31713. doi: 10.1038/srep31713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gruber J, Kennedy BK. 2017. Microbiome and longevity: gut microbes send signals to host mitochondria. Cell 169:1168–1169. doi: 10.1016/j.cell.2017.05.048. [DOI] [PubMed] [Google Scholar]
- 6.Lee J, Kwon G, Lim YH. 2015. Elucidating the mechanism of Weissella-dependent lifespan extension in Caenorhabditis elegans. Sci Rep 5:17128. doi: 10.1038/srep17128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sohal RS, Forster MJ. 2014. Caloric restriction and the aging process: a critique. Free Radic Biol Med 73:366–382. doi: 10.1016/j.freeradbiomed.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grandison RC, Piper MD, Partridge L. 2009. Amino-acid imbalance explains extension of lifespan by dietary restriction in Drosophila. Nature 462:1061–1064. doi: 10.1038/nature08619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee KP, Simpson SJ, Clissold FJ, Brooks R, Ballard JW, Taylor PW, Soran N, Raubenheimer D. 2008. Lifespan and reproduction in Drosophila: new insights from nutritional geometry. Proc Natl Acad Sci U S A 105:2498–2503. doi: 10.1073/pnas.0710787105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orentreich N, Matias JR, DeFelice A, Zimmerman JA. 1993. Low methionine ingestion by rats extends life span. J Nutr 123:269–274. doi: 10.1093/jn/123.2.269. [DOI] [PubMed] [Google Scholar]
- 11.Kozieł R, Ruckenstuhl C, Albertini E, Neuhaus M, Netzberger C, Bust M, Madeo F, Wiesner RJ, Jansen-Dürr P. 2014. Methionine restriction slows down senescence in human diploid fibroblasts. Aging Cell 13:1038–1048. doi: 10.1111/acel.12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee BC, Kaya A, Ma S, Kim G, Gerashchenko MV, Yim SH, Hu Z, Harshman LG, Gladyshev VN. 2014. Methionine restriction extends lifespan of Drosophila melanogaster under conditions of low amino-acid status. Nat Commun 5:3592. doi: 10.1038/ncomms4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McIsaac RS, Lewis KN, Gibney PA, Buffenstein R. 2016. From yeast to human: exploring the comparative biology of methionine restriction in extending eukaryotic life span. Ann N Y Acad Sci 1363:155–170. doi: 10.1111/nyas.13032. [DOI] [PubMed] [Google Scholar]
- 14.Troen AM, French EE, Roberts JF, Selhub J, Ordovas JM, Parnell LD, Lai CQ. 2007. Lifespan modification by glucose and methionine in Drosophila melanogaster fed a chemically defined diet. Age (Dordr) 29:29–39. doi: 10.1007/s11357-006-9018-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kabil H, Kabil O, Banerjee R, Harshman LG, Pletcher SD. 2011. Increased transsulfuration mediates longevity and dietary restriction in Drosophila. Proc Natl Acad Sci U S A 108:16831–16836. doi: 10.1073/pnas.1102008108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Obata F, Miura M. 2015. Enhancing S-adenosyl-methionine catabolism extends Drosophila lifespan. Nat Commun 6:8332. doi: 10.1038/ncomms9332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parkhitko AA, Binari R, Zhang N, Asara JM, Demontis F, Perrimon N. 2016. Tissue-specific down-regulation of S-adenosyl-homocysteine via suppression of dAhcyL1/dAhcyL2 extends health span and life span in Drosophila. Genes Dev 30:1409–1422. doi: 10.1101/gad.282277.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang MY, Wei D, Li R, Jia HT, Liu YW, Taning CNT, Wang JJ, Smagghe G. 2018. Cytoplasmic glutamine synthetase gene expression regulates larval development in Bactrocera dorsalis (Hendel). Arch Insect Biochem Physiol 97:21447. doi: 10.1002/arch.21447. [DOI] [PubMed] [Google Scholar]
- 19.Fast D, Duggal A, Foley E. 2018. Monoassociation with Lactobacillus plantarum disrupts intestinal homeostasis in adult Drosophila melanogaster. mBio 9:e01114-18. doi: 10.1128/mBio.01114-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Douglas AE. 2018. Contradictory results in microbiome science exemplified by recent Drosophila research. mBio 9:e01758-18. doi: 10.1128/mBio.01758-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brummel T, Ching A, Seroude L, Simon AF, Benzer S. 2004. Drosophila lifespan enhancement by exogenous bacteria. Proc Natl Acad Sci U S A 101:12974–12979. doi: 10.1073/pnas.0405207101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamada R, Deshpande SA, Bruce KD, Mak EM, Ja WW. 2015. Microbes promote amino acid harvest to rescue undernutrition in Drosophila. Cell Rep 10:865–872. doi: 10.1016/j.celrep.2015.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deshpande SA, Yamada R, Mak CM, Hunter B, Soto Obando A, Hoxha S, Ja WW. 2015. Acidic food pH Increases palatability and consumption and extends Drosophila lifespan. J Nutr 145:2789–2796. doi: 10.3945/jn.115.222380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cox CR, Gilmore MS. 2007. Native microbial colonization of Drosophila melanogaster and its use as a model of Enterococcus faecalis pathogenesis. Infect Immun 75:1565–1576. doi: 10.1128/IAI.01496-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keebaugh ES, Yamada R, Obadia B, Ludington WB, Ja WW. 2018. Microbial quantity impacts Drosophila nutrition, development, and lifespan. iScience 4:247–259. doi: 10.1016/j.isci.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keebaugh ES, Yamada R, Ja WW. 2019. The nutritional environment influences the impact of microbes on Drosophila melanogaster life span. mBio 10:e00885-19. doi: 10.1128/mBio.00885-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bakula M. 1969. The persistence of a microbial flora during postembryogenesis of Drosophila melanogaster. J Invertebr Pathol 14:365–374. doi: 10.1016/0022-2011(69)90163-3. [DOI] [PubMed] [Google Scholar]
- 28.Chandler JA, Lang JM, Bhatnagar S, Eisen JA, Kopp A. 2011. Bacterial communities of diverse Drosophila species: ecological context of a host-microbe model system. PLoS Genet 7:e1002272. doi: 10.1371/journal.pgen.1002272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong CN, Ng P, Douglas AE. 2011. Low-diversity bacterial community in the gut of the fruitfly Drosophila melanogaster. Environ Microbiol 13:1889–1900. doi: 10.1111/j.1462-2920.2011.02511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Judd AM, Matthews MK, Hughes R, Veloz M, Sexton CE, Chaston JM. 2018. Bacterial methionine metabolism genes influence Drosophila melanogaster starvation resistance. Appl Environ Microbiol 84:e00662-18. doi: 10.1128/AEM.00662-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Linford NJ, Bilgir C, Ro J, Pletcher SD. 2013. Measurement of lifespan in Drosophila melanogaster. J Vis Exp 2013:50068. doi: 10.3791/50068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bauer JH, Goupil S, Garber GB, Helfand SL. 2004. An accelerated assay for the identification of lifespan-extending interventions in Drosophila melanogaster. Proc Natl Acad Sci U S A 101:12980–12985. doi: 10.1073/pnas.0403493101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gendron CM, Kuo TH, Harvanek ZM, Chung BY, Yew JY, Dierick HA, Pletcher SD. 2014. Drosophila life span and physiology are modulated by sexual perception and reward. Science 343:544–548. doi: 10.1126/science.1243339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skorupa DA, Dervisefendic A, Zwiener J, Pletcher SD. 2008. Dietary composition specifies consumption, obesity, and lifespan in Drosophila melanogaster. Aging Cell 7:478–490. doi: 10.1111/j.1474-9726.2008.00400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blum JE, Fischer CN, Miles J, Handelsman J. 2013. Frequent replenishment sustains the beneficial microbiome of Drosophila melanogaster. mBio 4:e00860-13. doi: 10.1128/mBio.00860-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Inamine H, Ellner SP, Newell PD, Luo Y, Buchon N, Douglas AE. 2018. Spatiotemporally heterogeneous population dynamics of gut bacteria inferred from fecal time series data. mBio 9:e01453-17. doi: 10.1128/mBio.01453-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Obadia B, Guvener ZT, Zhang V, Ceja-Navarro JA, Brodie EL, Ja WW, Ludington WB. 2017. Probabilistic invasion underlies natural gut microbiome stability. Curr Biol 27:1999–2006. doi: 10.1016/j.cub.2017.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pais IS, Valente RS, Sporniak M, Teixeira L. 2018. Drosophila melanogaster establishes a species-specific mutualistic interaction with stable gut-colonizing bacteria. PLoS Biol 16:e2005710. doi: 10.1371/journal.pbio.2005710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoffmann AA, Harshman LG. 1999. Desiccation and starvation resistance in Drosophila: patterns of variation at the species, population and intrapopulation levels. Heredity 83:637–643. doi: 10.1046/j.1365-2540.1999.00649.x. [DOI] [PubMed] [Google Scholar]
- 40.Shin SC, Kim SH, You H, Kim B, Kim AC, Lee KA, Yoon JH, Ryu JH, Lee WJ. 2011. Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science 334:670–674. doi: 10.1126/science.1212782. [DOI] [PubMed] [Google Scholar]
- 41.Walters AW, Matthews MK, Hughes RC, Malcolm J, Call TB, Walker CJ, Rudman S, Newell PD, Douglas AE, Schmidt PS, Chaston JM. 2018. The microbiota influences the Drosophila melanogaster life history strategy. BioRxiv doi: 10.1101/471540. [DOI] [PMC free article] [PubMed]
- 42.Schwarz G, Santamaria-Araujo JA, Wolf S, Lee HJ, Adham IM, Grone HJ, Schwegler H, Sass JO, Otte T, Hanzelmann P, Mendel RR, Engel W, Reiss J. 2004. Rescue of lethal molybdenum cofactor deficiency by a biosynthetic precursor from Escherichia coli. Hum Mol Genet 13:1249–1255. doi: 10.1093/hmg/ddh136. [DOI] [PubMed] [Google Scholar]
- 43.Coon KL, Vogel KJ, Brown MR, Strand MR. 2014. Mosquitoes rely on their gut microbiota for development. Mol Ecol 23:2727–2739. doi: 10.1111/mec.12771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marzio A, Merigliano C, Gatti M, Verni F. 2014. Sugar and chromosome stability: clastogenic effects of sugars in vitamin B6-deficient cells. PLoS Genet 10:e1004199. doi: 10.1371/journal.pgen.1004199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blatch SA, Stabler SP, Harrison JF. 2015. The effects of folate intake on DNA and single-carbon pathway metabolism in the fruit fly Drosophila melanogaster compared to mammals. Comp Biochem Physiol B Biochem Mol Biol 189:34–39. doi: 10.1016/j.cbpb.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 46.Storelli G, Defaye A, Erkosar B, Hols P, Royet J, Leulier F. 2011. Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab 14:403–414. doi: 10.1016/j.cmet.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 47.Newell PD, Douglas AE. 2014. Interspecies interactions determine the impact of the gut microbiota on nutrient allocation in Drosophila melanogaster. Appl Environ Microbiol 80:788–796. doi: 10.1128/AEM.02742-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koyle ML, Veloz M, Judd AM, Wong AC, Newell PD, Douglas AE, Chaston JM. 2016. Rearing the fruit fly Drosophila melanogaster under axenic and gnotobiotic conditions. J Vis Exp 2016:54219. doi: 10.3791/54219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chaston JM, Newell PD, Douglas AE. 2014. Metagenome-wide association of microbial determinants of host phenotype in Drosophila melanogaster. mBio 5:e01631-14. doi: 10.1128/mBio.01631-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dobson AJ, Chaston JM, Newell PD, Donahue L, Hermann SL, Sannino DR, Westmiller S, Wong AC, Clark AG, Lazzaro BP, Douglas AE. 2015. Host genetic determinants of microbiota-dependent nutrition revealed by genome-wide analysis of Drosophila melanogaster. Nat Commun 6:6312. doi: 10.1038/ncomms7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Therneau T. 2014. A package for survival analysis in S, v2.37-7. http://cran.r-project.org/package=survival.
- 52.Therneau T. 2012. Mixed effects cox models, v2.2–3. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 53.Sexton CE, Smith HZ, Newell PD, Douglas AE, Chaston JM. 2018. MAGNAMWAR: an R package for genome-wide association studies of bacterial orthologs. Bioinformatics 34:1951–1952. doi:10.1093/bioinformatics/bty001. doi: 10.1093/bioinformatics/bty001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kanehisa M, Sato Y, Morishima K. 2016. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol 428:726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 55.R Core Team. 2018. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org. [Google Scholar]
- 56.Rambaut A. 2016. FigTree version 1.4.3. http://tree.bio.ed.ac.uk.
- 57.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Henry M, Stevens H, Szoecs E, Wagner H. 2018. vegan: community ecology package. https://CRAN.R-project.org/package=vegan.
- 58.Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.White KM, Matthews MK, Hughes RC, Sommer AJ, Griffitts JS, Newell PD, Chaston JM. 2018. A metagenome-wide association study and arrayed mutant library confirm Acetobacter lipopolysaccharide genes are necessary for association with Drosophila melanogaster. G3 (Bethesda) 8:1119–1127. doi: 10.1534/g3.117.300530. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.