We quantified the effects of environment (diet and water sources) and host early ontogenetic development on the diversity of and compositional changes in gut microbial communities based on massively parallel sequencing of the 16S rRNA genes from the GI tracts of larval lake sturgeon (Acipenser fulvescens). The gut microbial community diversity declined and the community composition differed significantly among ontogenetic stages; however, only modest differences associated with dietary or water source treatments were documented. Selectivity associated with microbe-host GI tract interactions through early ontogenetic stages was evident. The results have implications for lake sturgeon and early larval ecology and survival in their natural habitat and for conservation and aquaculture production specifically, as well as applications of microbe-based management in teleost fish generally.
KEYWORDS: community ecology, diet, freshwater fish, gut microbiota, ontogeny
ABSTRACT
Gastrointestinal (GI) or gut microbiotas play essential roles in host development and physiology. These roles are influenced partly by the microbial community composition. During early developmental stages, the ecological processes underlying the assembly and successional changes in host GI community composition are influenced by numerous factors, including dispersal from the surrounding environment, age-dependent changes in the gut environment, and changes in dietary regimes. However, the relative importance of these factors to the gut microbiota is not well understood. We examined the effects of environmental (diet and water sources) and host early ontogenetic development on the diversity of and the compositional changes in the gut microbiota of a primitive teleost fish, the lake sturgeon (Acipenser fulvescens), based on massively parallel sequencing of the 16S rRNA gene. Fish larvae were raised in environments that differed in water source (stream versus filtered groundwater) and diet (supplemented versus nonsupplemented Artemia fish). We quantified the gut microbial community structure at three stages (prefeeding and 1 and 2 weeks after exogenous feeding began). The diversity declined and the community composition differed significantly among stages; however, only modest differences associated with dietary or water source treatments were documented. Many taxa present in the gut were over- or underrepresented relative to neutral expectations in each sampling period. The findings indicate dynamic relationships between the gut microbiota composition and host gastrointestinal physiology, with comparatively smaller influences being associated with the rearing environments. Neutral models of community assembly could not be rejected, but selectivity associated with microbe-host GI tract interactions through early ontogenetic stages was evident. The results have implications for sturgeon conservation and aquaculture production specifically and applications of microbe-based management in teleost fish generally.
IMPORTANCE We quantified the effects of environment (diet and water sources) and host early ontogenetic development on the diversity of and compositional changes in gut microbial communities based on massively parallel sequencing of the 16S rRNA genes from the GI tracts of larval lake sturgeon (Acipenser fulvescens). The gut microbial community diversity declined and the community composition differed significantly among ontogenetic stages; however, only modest differences associated with dietary or water source treatments were documented. Selectivity associated with microbe-host GI tract interactions through early ontogenetic stages was evident. The results have implications for lake sturgeon and early larval ecology and survival in their natural habitat and for conservation and aquaculture production specifically, as well as applications of microbe-based management in teleost fish generally.
INTRODUCTION
One of the primary goals in community ecology is to understand community composition and diversity and the relative influences of the ecological forces underlying the patterns of distribution and abundance across space and time (1). In parallel with community ecology, the field of microbial ecology focuses on studying free-living and host-associated microbiota (2, 3). The microbial communities in host-microbial ecosystems have been extensively studied, and findings reveal that the microbiotas from different habitats are taxonomically diverse and that most have ecological importance. Yet, there are common ecological principles that govern microbial community assembly and composition (2–5).
The importance of the gut microbiota to fish has long been recognized, as microbial taxa perform important roles associated with nutritional provisioning, metabolic homeostasis, and immune defense (6–8). Compositional information pertaining to the fish gut microbiota (reviewed in references 9 and 10) has characterized microbial communities as being highly variable in composition across ontogenetic stages (11–14). Comparatively, the ecological processes governing microbial community formation, diversity, and dynamic compositional changes are typically less understood due to the challenges of detangling the complexities of many underlying factors (9, 15–19). Gut microbiota assembly usually occurs early in fish host development, involving factors such as fish host physiology, surrounding environmental conditions, and diet, providing opportunities to study microbial community formation and succession, including exchanges between the aquatic environment and the gut (11, 12, 20).
Examinations of the community composition and diversity of microbiota associated with fish gastrointestinal (GI) tracts would benefit from a statistical (5) exploration of the relative importance of four processes described by Vellend (1), specifically, selection, drift, dispersal, and speciation. These processes underlie the patterns in many other ecological communities and are also applicable to studies of microbial communities (5, 21). In our examination of the processes affecting the fish GI tract microbiota, we evaluated alternative predictions from neutrality theory, as introduced by Hubbell (22), with niche-based hypotheses (1). The ecological neutrality theory emphasizes the absence of differences in deterministic processes, such as ontogenetic shifts in the gut environment that place certain taxa at a selective advantage over others, including the per capita growth rate, death, and dispersal among species, and that assumes equal fitness across species (1, 22–24).
The neutral model in community ecology theory predicts that patterns in community composition and taxonomic diversity are likely the outcomes of stochastic processes of dispersal and drift (23, 25). By incorporating this conceptual framework together with advanced sequencing technologies, an improved understanding of the processes governing fish gut microbiota assembly during early life stages is possible. Understanding the principles associated with the bacterial colonization of GI tracts and with compositional changes across life stages will also help managers, particularly in aquaculture settings, to manipulate gut microbial communities to promote animal health, performance, and production (9, 26, 27).
Sturgeons belong to one of the oldest groups of the bony fish (Osteichthyes), and many are species of conservation concern. Of all 27 sturgeon species, lake sturgeon (Acipenser fulvescens) is the only sturgeon species endemic to the North American Great Lakes Basin and the only sturgeon in the genus Acipenser that spends its life solely in freshwater (28, 29). This species has experienced significant declines in abundance and distribution due to overfishing and the loss and degradation of its habitat (30). In recent years, sturgeon conservation aquaculture has increased greatly as part of restoration actions.
Successful lake sturgeon production can be limited due to high mortality, especially during early ontogenetic stages. The low survival of sturgeon larvae is tied to the nutritional regimes associated with diet formulation, feeding schedule, food presentation, and preference (31, 32). After hatch, yolk-sac larvae gradually develop gastrointestinal (GI) tracts that eventually resemble the adult structures by 10 to 11 days posthatch (dph). Once the yolk-sac stage has completed (i.e., the mouth fully develops and opens), the fish start their transition to exogenous feeding, often on brine shrimp nauplii (Artemia salina) (33, 34). During this stage, early exposure to and the interaction of the fish gut with microbial colonists from surrounding water are possible, and shifts in the intestinal microbiota can be documented as the fish continue to develop. Early ontogenetic changes in host diet and physiology can shape gut community dynamics. Previous studies have demonstrated that the gut microbiota exhibits variations in community composition during early life stages (5, 9, 21) and that these can have important consequences during later stages (i.e., ontogenetic contingency [35–37], including disease susceptibility [38]).
Our objectives in this study were to characterize the gut microbiota of lake sturgeon during the early larval stages, at the time before the fish begin feeding, until 14 days following the onset of exogenous feeding, using Illumina MiSeq high-throughput sequencing of a portion of the 16S rRNA gene. Here, we define microbiotas/microbiomes as the assemblages of microorganisms existing in or associated with a defined habitat (in this case, the gastrointestinal tract), including active and interacting members as well transient or inactive members. The gut microbiotas colonizing lake sturgeon larvae were experimentally manipulated to quantify the associations of host factors, water supply, and diet on the lake sturgeon gut microbiota. An understanding of the ecological principles that govern the stability or transiency of fish microbial community composition and diversity is essential to successfully altering microbial communities for therapeutic and agricultural benefits. We also quantified whether the gut microbiota assembly was consistent with neutral expectations. Our findings have implications for the management of nutrition, disease, and potential probiotic use in lake sturgeon culture and characterize the dynamic relationships between the host ontogeny and the environmental epibiota associated with the temporal variability of the microbiota residing in the gut.
(This research was conducted by Shairah Abdul Razak in partial fulfillment of the requirements for a doctoral degree from Michigan State University [39].)
RESULTS
Sequencing summary and fish morphometric data.
The four treatment groups evaluated in this study are as follows: fish raised in natal stream water and fed live Artemia fish (denoted group S), fish raised in natal stream water and fed live Artemia fish mixed with retentate (denoted group Sp), fish raised in groundwater and fed live Artemia fish (denoted group GW), and fish raised in groundwater and fed live Artemia fish mixed with retentate (denoted group GWp). These group designations are used throughout the paper.
Rarefaction analyses showed that sequencing efforts were consistent across replicate samples and treatment groups at a depth of 5,775 sequences per sample, as denoted by a total percentage of coverage of higher than 98%. We were able to sample a large portion of the operational taxonomic units (OTUs) and diversity present while still retaining a large number of samples within fish of each age. After quality filtering, our 16S rRNA amplicon data set produced 6,034,269 high-quality reads. In total, we observed 4,137 OTUs (2,894 OTUs when omitting singleton OTUs), defined at 97% sequence identity. From 118 samples submitted for 16S rRNA sequencing, rarefaction at 5,775 sequences per sample eliminated five samples with coverage below this sequencing coverage.
A total of 56,989 sequence reads were obtained across four retentate samples after quality filtering. Eukaryotic profiling based on the 18S rRNA amplicon data set detected 1,131 taxa overall when rarefaction was achieved at 6,000 sequences per sample, yet only 201 taxa were retained after singletons were omitted. Many of these taxa detected could not be classified beyond the class level, as 85% of the total sequences from nonsingleton taxa remained unclassified but are known dietary items described in other larval fish. Among the predominant classes detected were Podocopida (seed and muscle shrimps; 2.54%), Peridiniphycidae (dinoflagellates; 1.81%), and Bacillariophytina (diatoms), Spirotrichea (protozoan), Synurales (algae), and Peronosporomycetes (water molds) (each at 1.1%).
Morphometric data included mean ± standard deviation (SD) fish weight (in grams) and length (in millimeters) and revealed that fish growth was consistent across all four treatments throughout the duration of the study (Tables 1 and 2). By the end of the experiment, fish raised in stream water grew significantly larger than fish raised in UV-treated groundwater. No fish health issues were detected. The survival of fish was nearly 100% in all treatments and during all developmental stages.
TABLE 1.
Length and weight of sampled larval lake sturgeon from the four treatment groups across developmental stagesa
| Stage | Treatment group | Mean ± SE length (mm) | Mean wt (g) | Temp (oC) |
|---|---|---|---|---|
| Prefeeding | GW | 18.97 ± 0.18A | 0.034A | 13.8 |
| GWp | 18.95 ± 0.21A | 0.034A | ||
| S | 21.05 ± 0.19B | 0.038B | 15.7 | |
| Sp | 21.18 ± 0.13B | 0.038B | ||
| 1 wk after active feeding | GW | 28.63 ± 0.23C | 0.087C | 16.1 |
| GWp | 28.92 ± 0.26C,E | 0.084C | ||
| S | 30.03 ± 0.19D | 0.096D | 17.2 | |
| Sp | 29.81 ± 0.35D,E | 0.095D | ||
| 2 wk after active feeding | GW | 36.48 ± 0.41F | 0.178E | 15.6 |
| GWp | 35.71 ± 0.30F | 0.166E | ||
| S | 40.42 ± 0.22G | 0.225F | 19.5 | |
| Sp | 40.40 ± 0.40G | 0.217F |
The mortality rate during the experimental duration was too small and is thus not presented here. The capital letters indicate significant differences in the means based on a post hoc Tukey HSD test (P < 0.05).
TABLE 2.
Influence of temperature of rearing water on lengtha
| Parameter | Estimate | SE | t value | P value (>t) |
|---|---|---|---|---|
| Intercept | 7.141 | 3.587 | 1.991 | 0.05105 |
| GWp | −0.022 | 0.381 | −0.057 | 0.95469 |
| S | 0.451 | 0.738 | 0.611 | 0.54326 |
| Sp | 0.575 | 0.826 | 0.696 | 0.48883 |
| 1 wk after active feeding | 7.677 | 0.833 | 9.212 | 4.35E−13* |
| 2 wk after active feeding | 15.965 | 0.800 | 19.952 | <2.00E−16* |
| Temp | 0.857 | 0.269 | 3.186 | 0.00229* |
| GWp × 1 wk after active feeding | 0.315 | 0.538 | 0.585 | 0.56097 |
| S × 1 wk after active feeding | 0.008 | 0.628 | 0.012 | 0.99014 |
| Sp × 1 wk after active feeding | −0.336 | 0.672 | −0.5 | 0.61903 |
| GWp × 2 wk after active feeding | −0.752 | 0.538 | −1.396 | 0.16777 |
| S × 2 wk after active feeding | 0.146 | 0.538 | 0.271 | 0.78756 |
| Sp × 2 wk after active feeding | NA | NA | NA | NA |
Analysis of covariance indicated that the temperature of the rearing water significantly influenced the length. The residual standard error was 0.659 on 60 degrees of freedom, the multiple R2 value was 0.994; the adjusted R2 value was 0.993, the F statistic was 864.3 on 11 and 60 degrees of freedom; and the P value for temperature was 0.00229. NA, not applicable. *, parameter with a significant P value.
Ontogenetic changes of dominant bacterial taxa in lake sturgeon larva rearing.
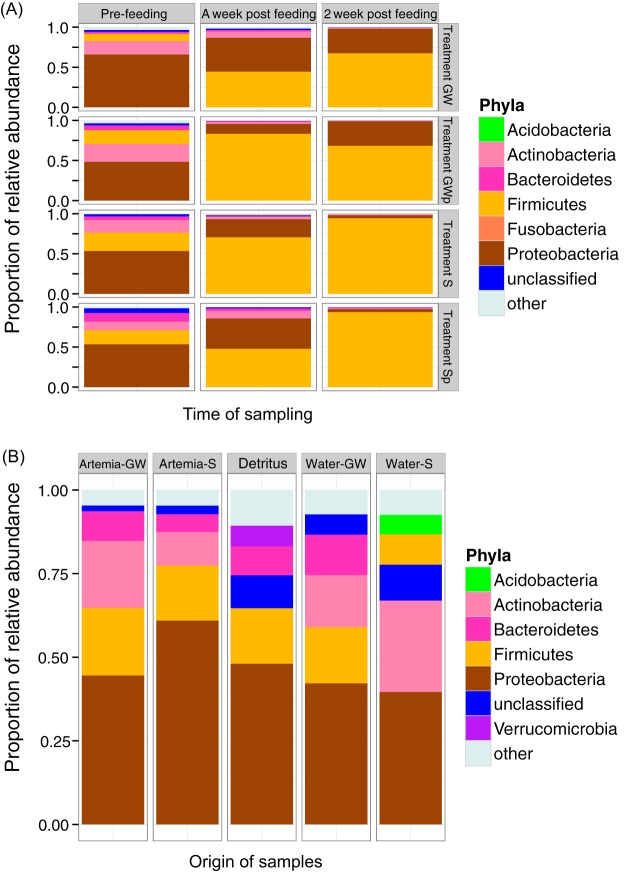
We quantified the number of sequences that were represented by each phylum from the GI tracts of all fish sampled in each developmental period. We found that the gut bacterial communities comprised 26 microbial phyla; however, the most abundant phyla, covering more than 95% of all sequences, included, in order of abundance, Proteobacteria, Firmicutes, Bacteroidetes, Actinobacteria, Acidobacteria, Planctomycetes, Chloroflexi, and Fusobacteria. General patterns of bacterial phylum-level contributions to the gut microbiota were shown with regard to treatment group and the corresponding developmental stages (Fig. 1). Across all treatment groups, the microbiota composition shifted from communities dominated by Proteobacteria and several other phyla prior to the initiation of exogenous feeding to communities dominated by Firmicutes after feeding began. The relative abundance of all other phyla was reduced after the fish began actively feeding.
FIG 1.
Bacterial composition of different communities identified from lake sturgeon larval gut (A) and environmental samples (B). (A) Relative abundance of dominant bacterial phyla found in lake sturgeon larval gut microbiota across treatments and during different developmental stages. Only the dominant phyla are shown in the bar chart (Acidobacteria, Actinobacteria, Bacteroidetes, Firmicutes, Fusobacteria, Proteobacteria). The remaining taxa were assigned to “other.” The four treatment groups are denoted S, Sp, GW, and GWp, as defined in the text. (B) Relative abundance of dominant bacterial phyla found in environmental microbiota. Only the dominant phyla are shown in the bar chart (Acidobacteria, Actinobacteria, Bacteroidetes, Firmicutes, Proteobacteria, Verrucomicrobia). The remaining taxa were assigned to “other.”
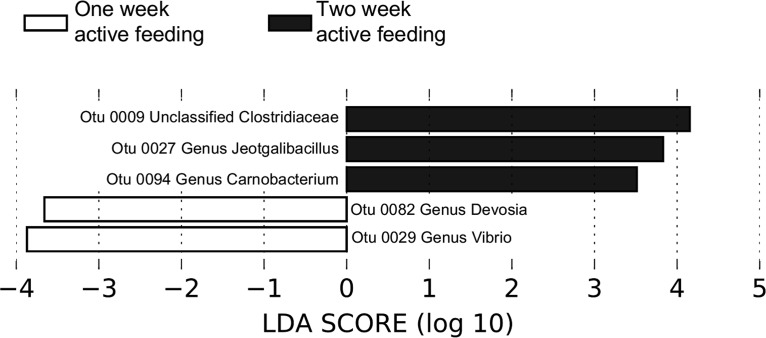
Linear discriminant analyses (LDA) effect size (LEfSe) scores were computed for taxa differentially abundant among the three ontogenetic stages (prefeeding and 1 and 2 weeks after active feeding began) (40). Among all 45 detected OTUs that were statistically and biologically different between lake sturgeon gut microbial communities compared across stages, 5 OTUs explained the greatest differences (Fig. 2). All other OTUs with significant LEfSe scores are listed in Table S1 in the supplemental material. The abundance of two OTUs (OTU82, Alphaproteobacteria genus Devosia; OTU29, Gammaproteobacteria genus Vibrio) at 1 week after feeding was different from that at the other two stages. Three OTUs (OTU9, unclassified Clostridiaceae from the phylum Firmicutes; OTU27, Firmicutes genus Jeotgalibacillus; OTU94, Firmicutes genus Carnobacterium) were present at a statistically significantly higher relative abundance in fish gut communities at 2 weeks after the initiation of feeding.
FIG 2.
Linear discriminant analysis (LDA) effect size (LEfSe) analyses identify OTUs in fish communities that respond significantly to feeding progression (from prefeeding and 1 week and 2 weeks after active feeding). Relative abundance was significant when P was <0.05 and the logarithmic LDA score was ≥2.0.
We tested whether changes in the larval gut microbiota occurred when fish were raised on the different experimental diets and in the different rearing water sources across life stages. Over the course of 3 weeks of development, we found that the microbiota diversity varied among age cohorts. Overall, the community diversity decreased as fish transitioned to active feeding (Fig. 3A and B). Multiple-factor analysis of variance (ANOVA) quantified the sources of the variability in the alpha diversity indices (inverse Simpson diversity index [1/D]) and indicated statistically significant differences in diversity as a function of sampling time (F value for inverse Simpson diversity index [F valueinv] = 11.31, number of degrees of freedom [df] = 2, P < 0.001), whereas OTU richness did not differ across sampling times (F value for OTU richness [F valuerich] = 0.91, df = 2, P = 0.407) or treatment (F value = 1.92, df = 3, P = 0.137). No significant interaction was found between water and food treatments for the inverse Simpson diversity index or for OTU richness (for the inverse Simpson diversity index, F valueinv = 1.13, df = 6, and P = 0.358; for OTU richness, F valuerich = 0.38, df = 6, and P = 0.892).
FIG 3.
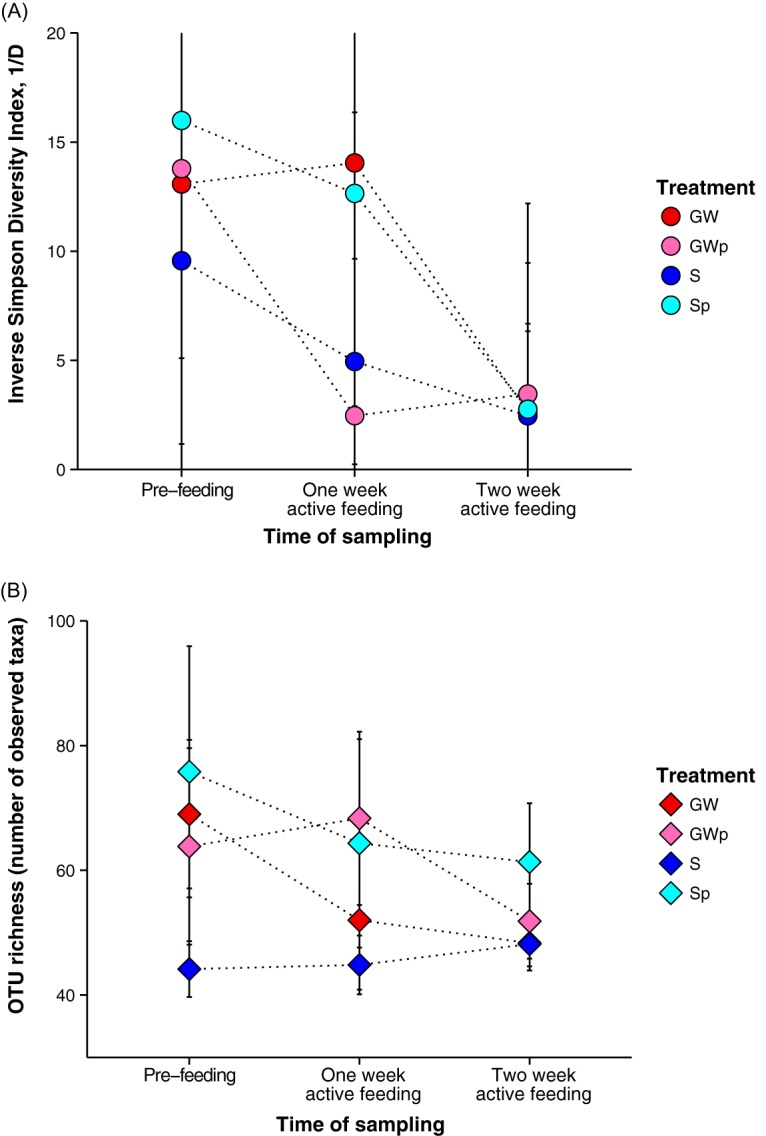
Estimates of alpha diversity for lake sturgeon gut microbial communities from all treatments across all developmental stages. The differences in the gut microbiota composition across the treatments at the different ages were evaluated using a two-way ANOVA. Each point indicates the mean value of the diversity index, colored by the different treatments. The four treatment groups are denoted S, Sp, GW, and GWp, as defined in the text. (A) Alpha diversity in the gut microbiota at each time point, as measured by the inverse Simpson diversity index. (B) OTU richness based on the number of taxa observed in the gut microbiota from all treatments and times.
Variation in gut bacterial community profiles in association with water, diet, and time.
To visualize the relationships between the gut microbial community composition and the composition of the environmental sources (water and diet), a principal-component analysis (PCoA) was performed to analyze the samples in a reduced-dimensional space using ordination plots. The variation in community membership among the gut microbiota from all treatment groups across time periods indicated age-dependent changes in the prevalent microbial taxa (Fig. 4A). The variation in community membership was less evident when comparisons were made across environmental (water and food) microbial communities (Fig. 4B) sampled concurrently during the same time period.
FIG 4.

Principal-coordinate analysis among microbial communities originating from the lake sturgeon larval gut across all four different treatments at three developmental stages (prefeeding and 1 week and 2 weeks after active feeding initiation) (A) and from environmental samples, including water, detritus, and Artemia fish (B). These environmental samples were collected at times corresponding to the times of fish sampling. Axes represent the first two principal coordinates maximizing the variance in the data (PCoA1 and PCoA2). Dissimilarity was calculated based on Bray-Curtis distances. The fish gut microbial communities from the four treatment groups are denoted S, Sp, GW, and GWp, as defined in the text. Water-GW and Water-S are water samples from groundwater and river water, respectively. Artemia-GW and Artemia-S are brine shrimp prepared using the respective water sources, and detritus was collected from a sock filter. Var, variation.
Statistical analyses of the beta diversity among sampling periods (fish developmental stages) revealed significant microbial community taxonomic compositional divergence (Table 3; permutational multivariate analysis of variance [PERMANOVA] test pseudo-F value = 2.077, R2 = 0.059, P < 0.001). We rejected the null hypothesis of no temporal (developmental) differences in the multivariate centroid location characterizing the microbial community composition. In addition, the significant results of a test for the homogeneity of multivariate dispersion (PERMDISP) (P < 0.001) indicated that the compositional dispersion within each group was heterogeneous (Tables 4 and 5).
TABLE 3.
Variability of fish gut microbial communities across treatment and ontogenetic stagesa
| Parameter | No. of df | Sum-of-square value | Mean square value | F model | R2 | P value (>F) |
|---|---|---|---|---|---|---|
| Stage | 2 | 1.907 | 0.954 | 2.077 | 0.059 | <0.001* |
| Treatment | 3 | 1.471 | 0.490 | 1.068 | 0.045 | 0.120 |
| Stage × treatment | 6 | 2.863 | 0.477 | 1.039 | 0.088 | 0.170 |
| Residuals | 57 | 26.173 | 0.459 | 0.807 | ||
| Total | 68 | 32.414 | 1.000 |
The variability of the fish gut microbial communities across treatment and ontogenetic stages was analyzed by PERMANOVA. The results revealed that the ontogenetic stages significantly influenced the gut microbial community composition for at least one sample (PERMANOVA pseudo-F value = 2.077, R2 = 0.059, P < 0.001, number of permutations = 1,000). *, parameter with a significant P value.
TABLE 4.
Permutation test for homogeneity of multivariate dispersionsa
| Parameter | No. of df | Sum-of-square value | Mean square value | F model | Permutation | P value (>F) |
|---|---|---|---|---|---|---|
| Stage group | 2 | 0.025 | 0.0124 | 12.518 | 999 | <0.001* |
| Residuals | 66 | 0.065 | 0.001 | 0.807 |
The multivariate dispersions (number of permutations = 999) indicate that the dispersion of the distances in microbial communities among three ontogenetic stages is significantly heterogeneous. *, parameter with a significant P value.
TABLE 5.
Pairwise comparisons
| Stage | Observed P value/permuted P valuea
|
||
|---|---|---|---|
| Prefeeding | 1 wk after active feeding | 2 wk after active feeding | |
| Prefeeding | 0.843 | 0.002* | |
| 1 wk after active feeding | 0.856 | 0.001* | |
| 2 wk after active feeding | <0.001* | <0.001* | |
The observed P values are given below the diagonal, and the permuted P values are given above the diagonal. *, pairwise comparisons with a significant P value.
Community composition data were further analyzed to decouple the treatment effects of water source and diet from the pervasive effects of sampling time. Under the null hypotheses, the water type and the food treatments administered were not expected to significantly affect the gut community taxonomic composition, and there would be no interaction between water and diet treatments within each time point. During the prefeeding period (sampling period 1), we found significant water treatment effects on PCoA axis 6 since the fish had not begun feeding (Table 6). As the fish transitioned to active feeding, both water and food treatments interacted significantly to affect the gut community composition after 1 week of feeding (sampling period 2; PCoA axis 1; Fig. 5A). The effect of water treatment on PCoA axis 5 was significant (Table 6). During the third sampling period, after the second week of active feeding, the gut community composition was statistically significantly different between water treatments. There was no effect of diet (PCoA axis 1; Fig. 5B). Least-squares (LS) mean values for all important axes across all times are shown in Table 6.
TABLE 6.
Results of analyses of significant PCoA axes for each sampling period
| Linear regression model |
Least-squares means |
||||||
|---|---|---|---|---|---|---|---|
| Stream water |
Groundwater |
||||||
| Stage, important axisa | Significant treatment | P value | R2 value | Food with supplement | Food without supplement | Food with supplement | Food without supplement |
| Prefeeding, PCoA axis 6 | Water type | 0.045 | 0.206 | 0.044 ± 0.06 | −0.137 ± 0.05 | 0.070 ± 0.05 | 0.030 ± 0.05 |
| 1 wk after active feeding, PCoA axis 1 | Interaction (water type and food type) | 0.004 | 0.444 | −0.079 ± 0.09 | −0.048 ± 0.09 | 0.327 ± 0.09 | −0.300 ± 0.12 |
| 1 wk after active feeding, PCoA axis 5 | Water type | 0.016 | 0.233 | 0.064 ± 0.05 | 0.079 ± 0.05 | −0.049 ± 0.05 | −0.142 ± 0.06 |
| 2 wk after active feeding, PCoA axis 1 | Water type | 0.020 | 0.267 | −0.189 ± 0.10 | −0.125 ± 0.10 | 0.086 ± 0.10 | 0.227 ± 0.10 |
The four axes across all three stages that showed a significant effect of either water, food, or an interaction of both water and food treatments on the microbial community composition were axis 6 for prefeeding, axis 1 and axis 5 for 1 week after active feeding, and axis 1 for 2 weeks after active feeding. Interaction plots are shown in Fig. 5.
FIG 5.
Interaction plots of marginal (least-squares [LS]) means for the first PCoA axes (axes that explained the largest variation in the data set) at different developmental stages. Additional information pertaining to the LS means is compiled in Table 6. (A) First PCoA axis at 1 week after active feeding; (B) first PCoA axis at 2 weeks after active feeding. The information in panel A indicates that significant interactions occurred between food and water treatments. Water and food treatments influenced the gut microbial community composition (represented by the first PCoA axis) during the sampling period at 1 week after active feeding. No significant difference in gut community composition between supplemented and nonsupplemented food treatments within the stream water environment was detected. However, a significant difference between the gut microbiota of fish raised in groundwater was observed on the basis of the food treatment (see the information in panel B). Significant effects of water treatment on gut composition (represented by the first PCoA axis) were observed in fish at 2 weeks after active feeding began. Diet effects were no longer observed. Food Suppl., food treatment in which live Artemia fish mixed with retentate was offered to the fish; Food No suppl., food treatment in which live Artemia fish only was offered to the fish.
Focusing on the first coordinate axis during sampling periods 2 and 3, following the initiation of feeding, we investigated whether there was evidence for the co-occurrence of taxa in the gut communities. The genus Clostridium (OTU001), associated with the family Clostridiaceae (phylum Firmicutes), was found to have a strong, positive correlation with the first axis during the first week of feeding (Pearson correlation [r] = 0.884). During the second week of feeding, two other bacterial taxa (Sarcina [OTU002] and an unclassified genus [OTU004], both of which are members of the Clostridiaceae family), showed strong correlations (for Sarcina, r = −0.791; for the unclassified Clostridiales genus, r = 0.743) with the first coordinate axis. Another taxon in the genus Deefgea, a member of the family Neisseriaceae (phylum Proteobacteria, order Gammaproteobacteria), was also present at a high abundance and was positively correlated on the same axis (r = 0.779). The strong correlations of these genera are reflected by the presence of taxa at high relative abundances, quantified in Tables S2 and S3.
Neutral processes are not the dominant mechanisms generating and maintaining microbial community composition during early gut microbiome assembly.
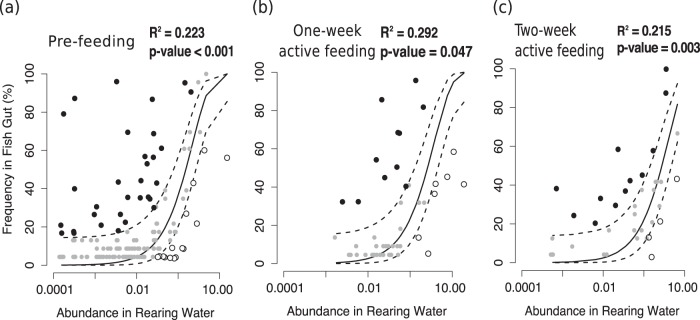
LS mean analyses of PCoA axis 1 at sampling point 3 (2 weeks after the initiation of feeding, 22 dph) indicated that the dispersal of bacteria from environmental (water) sources into the gut was important in shaping the community composition in the gut (Fig. 5B; Table 6). However, the analyses did not address the following question: is the fish gut community at each developmental stage a neutral subset of the source community? We wished to distinguish between species that were detected in the gut in association with neutral processes (e.g., dispersal associated with water source) and species that were detected in the gut in association with deterministic processes that may be associated with age-dependent gut conditions. To answer this question, we applied the neutral model theory, based on the neutral model for prokaryotes of Sloan et al. (24), to investigate the developmental processes underlying gut microbiome compositional and successional changes over time. The neutral model assumes that the community composition can be explained by the dispersal of microbial taxa from the environment (water) and ecological drift (stochastic change) within the source community (23). Based on LS mean analyses, we found that water was a more important source of gut community members than diet was.
Neutral processes were detected as important contributors to microbial community formation during each sampling period (developmental stage), as indicated by significant P values (Fig. 6a to c). These processes were significant, yet they were not strongly predictive. Using the R2 value as a measure of the goodness of fit, we determined that the gut community composition from across all treatments poorly fit a neutral model, based on the low R2 value of the model fit (R2 value range, 0.215 to 0.292).
FIG 6.
Results of neutral model testing with water as the source of gut microbial communities at the prefeeding stage (a), at 1 week after active feeding initiation (b), and at 2 weeks after active feeding initiation (c). The solid black line represents the best-fit neutral model, generated using a beta probability distribution. The model was developed based only on taxa found in both gut and water sources. The dashed lines represents the 95% confidence intervals around the best-fitting neutral model. Species within the confidence intervals (gray points) are classified as neutrally dispersed taxa that were likely present in the gut as a result of neutral processes (such as dispersal or ecological drift). Species deviating from neutral model and identified by black and white points were classified as underrepresented and overrepresented taxa, respectively. These taxa were likely affected by deterministic processes or may have had a dispersal ability different from that of the other taxa in the water. The coefficient of determination (R2) represents the goodness of fit of the relative abundance under the neutral model. The value ranges from ≤0 (no fit) to 1 (perfect fit). The P value indicates that the neutral processes that were detected are significant and did not occur by random chance. In general, neutrality could not be rejected during all three developmental stages, but the fit of the data to the neutrality expectations was poor, as shown by the relatively low R2 values.
Across sequential sampling periods, as the fish aged, the number of shared microbial OTUs between the gut and the water decreased (Tables 7 and 8). At the prefeeding stage, larval GI tracts contained the most neutrally dispersed taxa (number of taxa, 114), but this number declined dramatically as the fish began active feeding (number of taxa, 45). At 2 weeks after active feeding, only 38 taxa were shared between the gut and water communities, and about half of these taxa were underrepresented or overrepresented in the gut (Table 7). We conclude from these analyses that the OTUs of the gut microbiomes are likely under selection.
TABLE 7.
Results of neutral models applied to gut microbiotaa
| Parameter | No. (%) of OTUs for the following developmental stage: |
||
|---|---|---|---|
| Prefeeding | 1 wk after active feeding | 2 wk after active feeding | |
| All OTUs shared between source (rearing water) and target (fish gut) communities | 160 | 45 | 38 |
| Neutrally dispersed OTUs | 114 (71.25) | 27 (60.00) | 22 (57.89) |
| Overrepresented OTUs | 31 (19.38) | 11 (24.44) | 12 (31.58) |
| Underrepresented OTUs | 15 (9.37) | 7 (15.56) | 4 (10.53) |
The number and proportion of shared OTUs detected in both the gut microbial communities and the environmental microbial source (water) are shown. Overrepresented taxa are those that were presumed to be selected for (i.e., detection at a low abundance in water but detection at a higher abundance in the gut), whereas underrepresented taxa were present at a lower abundance in the gut than in water.
TABLE 8.
Taxa overrepresented, underrepresented, and neutrally represented in all three ontogenetic stages
| Representation | OTU (taxonomy) | Developmental stage in which OTU was detected | OTU detection frequencies in rearing water/fish gut |
||
|---|---|---|---|---|---|
| Prefeeding | At 1 wk after active feeding | At 2 wk after active feeding | |||
| Neutrally represented | OTU062 (genus Finegoldia) | All stages | 0.091/0.087 | 0.500/0.045 | 0.333/0.250 |
| Overrepresented | OTU016 (genus Pseudomonas) | All stages | 0.455/0.957 | 0.500/0.864 | 0.667/0.375 |
| OTU041 (genus Janibacter) | All stages | 0.455/0.609 | 0.500/0.318 | 0.333/0.458 | |
| Underrepresented | OTU015 (genus Turicella) | All stages | 0.818/0.609 | 1.000/0.409 | 0.6667/0.412 |
The taxa present in the gut are not a completely neutral subset of the taxa present in the source water communities (see Tables S4 to S6, which detail the taxa that were overrepresented, underrepresented, and neutrally represented during all three stages). Over the time course covering all ontogenetic stages, the abundance of several taxa consistently conformed to neutral expectations (OTU062, genus Finegoldia), whereas other taxa were overrepresented (OTU016, genus Pseudomonas; OTU041, genus Janibacter) or underrepresented (OTU015, genus Turicella) in the fish gut in relation to their frequencies in rearing water (Table 8).
DISCUSSION
Many basic microbial community ecology questions concerning the dynamics of a community composition at the onset of colonization and early ontogenetic changes are of importance to understand host-microbial relationships in the wild and under domestic (i.e., fish culture) conditions. Information pertaining to the source(s) of microbial communities that establish in the fish gut, the relative importance of neutral versus nonneutral factors during community assembly, and animal host age/growth effects on gut community establishment is incompletely known.
In this study, we were interested in characterizing the development of the fish gut microbiota during important early developmental stages. Our experimental system enhanced our understanding of the factors affecting the initial colonization and development of the lake sturgeon larval gut microbiota prior to and during the critical transition from endogenous to exogenous feeding. Our research contrasts with previous studies on other sturgeon species associated with diet and gut microbiota, which were performed on either larger or older fish. For example, two studies involved Siberian sturgeon (Acipenser barii) (41) and white sterlet sturgeon (Acipenser ruthenus) (42) used fish at the juvenile stage (average weight, 15 g to 30 g; age, more than 3 months posthatch). Other studies used fish that had been exposed to cultivation for extended periods of time (53, 79).
This research setting provided an experimental system amenable to the evaluation of environmental factors (focusing both on the rearing water type and on the diet administered) and host-associated factors that affect the compositional dynamics of the intestinal microbiota in lake sturgeon, an important aquaculture teleost fish species and a species of conservation concern.
Lake sturgeon larvae were raised in constant, controlled environments through manipulation of the water and diet and evaluated over three sampling periods from 11 to 29 dph. Our results suggest that major compositional shifts in the gut community composition corresponded to the different developmental stages. The taxonomic profiles changed across sequential larval stages, extending from before the onset of exogenous feeding (prefeeding) through 29 dph, when the fish were actively feeding and when the GI tract structures resembled the adult anatomical structures (33, 43). The findings indicate that during these early ontogenetic periods, host physiological development likely serves as a strong deterministic force directing the formation of gut communities, regardless of the food type or surrounding (water) environmental communities. The temporal shift in bacterial community composition has also been documented in others studies involving zebrafish (12, 20) and rainbow trout (Oncorhynchus mykiss) larvae (44) during periods of constant diet and environmental conditions. However, while the rainbow trout studies reported a strong influence of diet type and environmental factors on the gut microbiota composition, the findings from the zebrafish studies differed from those of the rainbow trout studies (12, 20, 44, 45).
Pronounced temporal changes in microbial community composition occurred from the prefeeding period to the exogenous feeding periods. Proteobacteria dominated the gut communities during the prefeeding stage, whereas Firmicutes dominated the communities after 2 weeks of feeding. Large decreases in community taxonomic diversity were also documented between the prefeeding and postfeeding periods. These results are concordant with the findings for other fish species, which documented Proteobacteria and/or Firmicutes to be among the most abundant phyla in fish gut communities. Studies on zebrafish indicated that Firmicutes and Proteobacteria are most common in larvae (12) but that the microbiota in adults was dominated by Fusobacteria (46). Studies on microbiota ontogenetic shifts in rainbow trout fry also indicated the presence of Proteobacteria or Firmicutes in association with either a marine-based or a freshwater plant-based diet offered during the first feeding (18). Meanwhile, the administration of prebiotics to Siberian sturgeon (Acipenser barii) shifted the composition of the gut microbiota primarily in the phylum Firmicutes (41). Comparative analyses of the gut microbiota from 8 freshwater fish species encompassing fish with different feeding habits revealed that Proteobacteria and Firmicutes were the dominant phyla in all fish species (47). Another cross-sectional gut microbiota study performed on channel catfish also indicated the prevalence of Proteobacteria in fish at the earlier stage and the appearance of Firmicutes along with Proteobacteria when the fish reached 65 dph (48) or when the fish were stocked into a nursery pond (11).
We also found that the predominant taxonomic assemblages tended to consist of closely related taxa from the family Clostridiaceae. These included the genera Carnobacterium, Jeotgalibacillus, Sarcina, and Clostridium and an unclassified genus from the family Clostridiaceae. This might indicate that traits underlying assemblage membership were often shared among related host organisms. Taxa of a type of lactic acid bacterium, Carnobacterium, are common commensals of fish, in which they have been detected in the intestinal content and in the mucosal layer (49). One of these taxa, the genus Clostridium, was enriched following the transition to active feeding. In humans, this genus is part of the important commensal microbiota that begins to colonize the intestines of breast-fed infants as early as the first month of life and has been shown to play roles in modulating gut homeostasis over the entire life span (for a review, see reference 50). Studies in mice indicated that commensal clostridia populate specific regions in the intestinal mucosa, thus establishing a close relationship with gut cells that perform critical physiological functions (50).
Another taxon, Sarcina, was also abundant by the time that the fish reached 22 dph and were actively feeding. Members of the genus Sarcina are fermenting bacteria that are frequently found in the gastric contents and feces of human patients with gastrointestinal disorders. All the strains were obligate anaerobes, fermented cellulose, and required a carbohydrate for growth. One species, Sarcina maxima, fermented carbohydrates mainly to butyrate, acetate, CO2, and H2. Another species, S. ventriculi, produces ethanol as a major product but not butyric acid from glucose and can also produce acid and gas from sugar, like glucose, fructose, sucrose, maltose, lactose, galactose, and raffinose (51). S. ventriculi is widespread in the soil and may be considered part of the intestinal flora of human, although its significance remains unknown (51, 52). Given the biology of both microbial taxa and the benthic feeding habits of sturgeon, it is possible that Sarcina and Clostridium are predominant taxa in the distal gut of sturgeon in natural habitats. Sturgeon possess a valvular hindgut (spiral valve) that serves as the primary region of digestion and nutrient absorption and thus might provide an abundance of nutrients for bacteria like Sarcina and Clostridium to flourish (33, 53) to maintain gut physiological functions. Previous studies (53) showed that anaerobic bacterial fermentation takes place in the spiral valve, producing volatile fatty acids (VFAs) and hydrogen gas as by-products, supporting the idea that these bacteria may enhance digestive efficiency.
Two important Proteobacteria taxa present during all stages were Pseudomonas and Deefgea. The presence of Pseudomonas at a relatively high abundance has also been reported as part of the lake sturgeon egg-associated community (54). Another study found Pseudomonas spp. to be present in abundance within the gastrointestinal tracts of juvenile coho salmon (Oncorhynchus kisutch) (55) as well as on eggs, but not in the water source or in food. This was likely attributed to the vertical transmission of a pioneering strain from the eggs to the fish GI tracts. Pseudomonas is also commonly observed in the gut microbiota of mature fish (9, 56, 57). A number of diet-related studies have reported variability in the relative abundance of Pseudomonas spp. that were affected by differential food treatment (for a review, see reference 6).
Little information pertaining to Deefgea spp. is available. Studies have so far found only two species in the genus Deefgea (family Neisseriaceae, order Betaproteobacteria) (58). Those species were described as Deefgea rivuli and Deefgea chitinilytica (58, 59). However, these two taxa originated from hard water and wetland samples, respectively. The association of Deefgea isolates with fish was first documented in another study (60). That study reported six isolates of D. chitinilytica that were cultured from swabs of skin and internal organs of two freshwater ornamental fish species raised on farms, gold tench (Tinca tinca) and goldfish (Carassius auratus auratus). Several other bacterial taxa from the same family were described to be chitin-hydrolyzing species, and D. chitinilytica was suspected to have a similar function, too. Due to this, Deefgea may have significant importance and may be listed among opportunistic taxa that may play roles in infections of aquatic organisms (60).
Throughout the assembly of early gut bacterial communities, stochastic and deterministic factors associated with water and food epibiota play roles in shaping these ecological communities (9, 55, 61). As we have shown by ANOVA and multivariate analyses of the gut microbiota at each time point, the water rearing environment initially had a strong influence on the community composition during the prefeeding stage, as the gut communities reflected the aquatic communities of both water treatments. Water appears to serve as a primary inoculant before and during the transition from endogenous to exogenous feeding. The communities in the early stages of gut microbiota colonization were temporally unstable in our study. Stochastic processes, such as the random recruitment of water epibiota into the gut, often occur at the beginning of exposure of the early upper GI tract and gill surface to ambient water (56). As the fish in our study developed further, the gut communities changed coincident with the initiation of active feeding and diverged in composition from the surrounding aquatic communities. Previous studies of the egg surface communities of lake sturgeon performed by our group also documented directional changes in bacterial community composition and diversity across sequential egg developmental stages (54). Another study on tilapia larvae (62) also documented changes in the gut community structure over time, with significant contributions of water bacterial communities.
As feeding continued, significant interactions were observed. We observed no significant differences in the gut communities between the supplemented and the nonsupplemented food treatment groups within the stream water environment. In contrast, significant differences in the gut microbiota of fish raised in groundwater were observed on the basis of the food treatment administered (a supplemented versus a nonsupplemented diet). The gut communities of fish raised in groundwater differed according to their diet, indicating that diet influences gut community membership. However, further analyses showed that the effect of diet on the gut community composition after the prefeeding stage was not evident. Only water treatment was significantly associated with the gut community composition at 2 weeks after active feeding. At this point, the gut communities differed between the stream water and the groundwater treatments.
We failed to detect significant differences in the gut microbial communities of fish raised using different feeding treatments. We were surprised to see that there was not much variation in the microbial taxonomic composition associated with water and food (Artemia), even though the water sources used for rearing the fish and preparing the live Artemia were different. A lack of community compositional heterogeneity may have been because the Artemia shrimp prepared in stream and groundwater had comparable nutrient properties. However, the ordination of detritus was segregated at the bottom right side of the PCoA plot, likely attributed to natural food items in the samples influencing their microbial community structure. We demonstrated the presence of natural food items in detritus/retentate based on eukaryotic profiling using 18S rRNA sequence data. Several groups of lower eukaryotes were detected in the retentate collected from sock filters, including protozoans, diatoms, and dinoflagellates.
The dynamics of the community compositional change during gut assembly processes can be attributed to many processes (2, 5). First, the composition of the gut community could be determined by environmental selection. In this study, the environments were associated with the rearing condition of the fish, as fish raised within similar water environments were exposed to similar pools of microbial taxa present in each water source. Thus, the local community may also be under the influence of neutral processes. The taxa present in the gut may be a random draw of the species present in water.
With respect to the rearing temperature, fish raised in stream water were exposed to ambient temperatures of 12 to 18°C, which are within the typical range of temperatures in the Upper Black River, Cheboygan, MI, during spawning season. During the experiment, the temperatures of both the surface water and the groundwater were standardized as much as possible to be different by about 2°C. Although stream temperature fluctuations are considered ecologically relevant, we are aware that we cannot completely overrule the effect that a higher temperature may have on the physiology and metabolism of larval sturgeon (34, 63). This was especially true at the final stage of 2 weeks after active feeding initiation, as the stream water had a warmer temperature than the groundwater (Tables 1 and 2). Changes in fish physiology may affect gut anatomical development and contribute to a gut selective environment (28). Subsequently, maturation of the fish gut may impose selective pressures that favor particular subsets of taxa or that inhibit the growth of certain subsets of taxa.
Evaluations of the relative importance of neutral versus deterministic processes were achieved through the implementation of a neutral model. Our findings indicated that neutral processes (e.g., a random dispersal of taxa from water) were ongoing but were not the pervasive ecological force shaping the gut community during all three developmental stages. The low model predictability (low R2 values) indicated that neutral processes are not dominant. The results suggest that deterministic processes are also responsible for changes in the gut community composition associated with development of the fish GI tract, as evidenced by reductions in the number of taxa between the gut and water (Fig. 6a to c). Meanwhile, the presence of overrepresented taxa, such as Pseudomonas and Janibacter, may play important roles for fish hosts. For example, Pseudomonas species are important decomposers of organic matter in soil, water, and food products, but several species are also known to be pathogens in plants, animals, and human (64). Pseudomonas is also commonly found as part of the fish intestinal and fish egg microbial communities (55). Janibacter, on the other hand, is less common, yet a study has reported the presence of this genus in the midgut of mosquito, indicating its possible role as part of the gut microbiota (65).
We recommend that future studies include experimental designs that sample over a longer duration, including through manipulations of the water community. Further studies focusing on taxa that increase in relative abundance over time would be useful to establish the ecological functions that may be used at a greater efficiency in applications of microbe-based fish management in aquaculture. Such data could also be used to identify suitable sites for the release of fish from conservation hatchery programs.
Overall, the significant changes in the diversity and the taxonomic composition of the lake sturgeon gut microbiota that occurred were principally associated with early developmental stages in connection with the initiation of the first feeding. Following the initiation of exogenous feeding, the microbial communities diverged from those in the surrounding water community and the epibiota of the food provided. Our understanding and ability to control (i.e., through the application of probiotics) underlying deterministic and stochastic factors associated with the source of microbial inocula appeared to be tied in part to the communities in the water when feeding began. Advancing the use of microbial manipulation (pre- and probiotics) is a goal for the aquaculture industry to promote fish growth and health. Accordingly, advancing understandings of the compositional dynamics that naturally occur in the gut microbiota of cultured fish species, like sturgeon, has relevance to commercial and conservation aquaculture. Future studies may profitably explore the effects of manipulations of communities in rearing water and food samples to understand the dynamics of microbial community assembly associated with these factors.
MATERIALS AND METHODS
Experimental design and feeding regime.
Lake sturgeon larvae were produced from a single mated pair collected during the sturgeon spawning season on the Upper Black River, Cheboygan, MI, in May 2013. Full-sib individuals were used to reduce the potential variability in the microbiota associated with host genetic background. All individuals were raised in a flowthrough water system under four different rearing conditions. We used a 2-by-2 treatment factor design associated with water type and feeding regime. The water types included river water from the natal stream (S) and UV-treated groundwater (GW), reflecting water sources commonly used in traditional hatchery operations. These water types were used throughout the culture process, including food (brine shrimp or Artemia fish) preparation. Fish were fed either live Artemia nauplii, which are commonly used in sturgeon hatcheries (66), or Artemia nauplii supplemented with organic retentate, including detritus and aquatic zooplankton obtained from serial filtration through 100-μm- and 50-μm-pore-size filters used to filter river water entering the hatchery (referred to here as retentate). The presence of digestible taxa in the retentate was confirmed using massively parallel sequencing of the V9 region of the 18S rRNA gene.
Each treatment included six 3.0-liter polycarbonate tanks (Aquatic Habitat, Inc.) that served as biological replicates. Each tank held 70 individuals to achieve an estimated statistical power of 0.8. The power analyses were performed based on our preliminary microbial studies (54). The fish were exposed to the same water type beginning at the time of egg fertilization and during the incubation stages. A newly hatched larval group of 70 fish was then distributed into each treatment replicate at 10 days posthatch (dph). The food was offered at 12 dph. However, only at 16 dph, we began to consistently offer food when at least half of the fish were feeding. To ensure that the fish received consistent amounts of food throughout the experiment, previously established dry weight feeding rates for sturgeon (67) were utilized, whereby the larvae in all tanks were fed at 26% of body weight daily (BWD). Prior to the first feeding each day, the retentate was added to the freshly prepared Artemia fish for treatment groups Sp and GWp. The amount of retentate (in grams) collected daily varied depending on each day of collection, yet the food amount was adjusted accordingly to ensure that the fish in each treatment group consumed the same amount of food. Fish were fed to satiation three times daily. Mortality was recorded daily, and the body weight (in grams) was recorded once every third day. All experiments were conducted at the Sturgeon Streamside Rearing Facility, managed by the Michigan Department of Natural Resources (MDNR) and Michigan State University (MSU) at Onaway, MI, using an approved MSU Institutional Animal Care and Use Committee (IACUC) protocol (animal use and care procedure numbers 03/04-058-00 and 05/07-086-00).
Sample collection.
We sampled 10 sturgeon larvae from each replicate for each treatment at three ontogenetic developmental stages: time 1, which was before active feeding (11 dph); time 2, which was after 1 week of fish active feeding (22 dph); and time 3, which was after 2 weeks of active feeding (29 dph). These time points were selected to capture the critical phases of GI tract development after the fish completely absorbed their yolk sac and the gastrointestinal tract anatomy was completed (which occurs at approximately 10 dph). Lake sturgeon started actively feeding at between 13 and 16 dph when food was first offered (33, 43). The fish were sampled for microbial community interrogation at each time period and were euthanized with an overdose of ethyl 3-aminobenzoate methanesulfonate (MS-222; Sigma-Aldrich, St. Louis, MO, USA) at the time of sampling. Each individual larva was photographed at the time of euthanization, and images were analyzed, using ImageJ software, to determine the total body length (in millimeters). The fish were then transferred to and preserved in 50-ml Corning centrifuge tubes containing 80% filter-sterilized ethanol until GI tract dissection and bacterial DNA extraction were performed.
Other environmental samples, including water and food, were also collected during each sampling period. Water samples (250 ml) from stream water and groundwater were collected from the hatchery reservoir tank and filtered through 0.45-μm-pore-size filters with a 47-mm-diameter filter membrane (Fisher Scientific), using a hand pump to obtain the aquatic microbial communities on the filter paper. These filters were then transferred to and preserved in 50-ml Corning centrifuge tubes containing 80% ethanol until bacterial DNA extraction was performed. For food samples, approximately 200 μl of food was pipetted into a 2-ml Eppendorf tube and was preserved in the same manner as the fish and water samples. All environmental samples from the different treatment groups are denoted by Artemia-GW (food treatment in which Artemia was prepared using groundwater), Artemia-S (food treatment in which live Artemia was prepared using stream water), detritus (food treatment in which retentates were collected from a sock filter prior to mixing with live Artemia), Water-GW (UV-treated groundwater), and Water-S (treatment with stream water).
DNA extraction and 16S rRNA amplicon sequencing.
The gut microbiota from lake sturgeon larvae was surveyed using high-throughput sequencing of the V4 region of the 16S rRNA gene. The distal gut (spiral valve) was recovered from each sturgeon larva following aseptic techniques. The distal gut was defined as the section extending from the beginning of the intestine to the spiral valve. Exterior surfaces were swabbed with 100% ethanol before dissections of the whole digestive tract using sterile instruments. Dissections were performed as previously described (68) with slight modification. The intact GI tracts were cut from the fish body cavities, and the excised GI tract was immediately transferred into filter-sterilized 80% ethanol for DNA isolation. Due to the small size of the gut, a composite of at least four GI tract samples from larvae were combined for each tank replicate within each treatment group at each time point. Each tube containing GI tract samples was first centrifuged for 15 min at 4°C to pellet the tissues and bacteria, before the DNA was extracted. A MoBio PowerSoil DNA isolation kit (Carlsbad, CA, USA), including a bead-beating step, was used following the protocols for low-biomass samples, as suggested by the manufacturer, with slight modification. The integrity of each DNA sample was assessed based on the amplification of approximately 1.4 kbp of the 16S rRNA gene (with forward primer 27F and reverse primer 1389R), followed by gel agarose electrophoresis, and the DNA concentration was determined using a microplate spectrophotometer (BioTek, Winooski, VT, USA).
A total of 20 filter membranes (consisting of two biological replicates from each of the water treatments, stream and UV-filtered water) containing trapped aquatic microbial communities and 20 food samples (two biological replicates of fresh brine shrimp prepared using the respective water sources from both water treatments) were randomly collected five times throughout the period of fish rearing. The membranes and food samples were first vortexed vigorously for 5 min to wash the bacteria from the membrane or food. The homogenates were then centrifuged (10,000 rpm, 4°C, 15 min) to pellet the tissues and bacteria before the DNA was extracted. The pellets were suspended with 500 μl of buffer solution from the power bead tube of the MoBio PowerSoil DNA isolation kit (Carlsbad, CA, USA) and then transferred back into the power bead tube. The isolation of DNA from the water and food bacterial communities proceeded using protocols for low-biomass samples, as suggested by the manufacturer.
One hundred nineteen DNA samples, consisting of 79 samples from fish gut, 36 samples from water and food (here referred as environmental microbial communities), and 4 positive-control samples (microbial DNA present in samples from activated sludge and a pure culture of Escherichia coli), were then validated to contain qualitatively sufficient bacterial DNA, as indicated by the presence of PCR amplicon bands following gel electrophoresis, and later were submitted for sequencing at the Michigan State University Research Technology Support Facility (RTSF; East Lansing, MI, USA). All of the sequencing procedures, including the construction of the Illumina sequencing library, emulsion PCR, and MiSeq paired-end sequencing (with the V2 platform) of the V4 region (∼250 bp; with primers 515F [GTGCCAGCMGCCGCGGTAA] and 806R [TAATCTWTGGGVHCATCAGGTGCAGG]) (69, 70), followed standard Illumina (San Diego, CA, USA) protocols. For reference, this primer pair amplifies the region from positions 533 to 786 in the E. coli strain 83972 sequence (70). Michigan State’s Genomics RTSF (https://rtsf.natsci.msu.edu/genomics/) provided standard Illumina quality control, including base calling by Illumina real-time analysis (v1.18.61), demultiplexing, adaptor and barcode removal, and real-time analysis (RTA) conversion to the FastQ format with the Illumina Bcl2Fastq (v1.8.4) program.
Sequence processing.
Sequence data were processed using the default sequencing data analysis pipeline and computing workflow. Briefly, paired-end sequence merging, quality filtering, denoising, singleton sequence removal, chimera checking, and taxonomic assignment of the operational taxonomic unit (OTU) selection were conducted using a protocol within an open-source workflow implemented by the mothur (v1.36.1) program (69).
Alignments were performed using the SILVA-based bacterial reference database. De novo OTU clustering was performed using hierarchical algorithms as the default option offered in mothur to cluster sequences defined with 97% identity (71), whereas the taxonomic assignment was achieved using the training file set provided by mothur, which was derived from the Ribosomal Database Project (RDP).
To minimize the effects of undersampling while maintaining as large a data set as possible, the final OTU table was rarefied to a depth of 5,775 sequences per sample. Five DNA samples with low sequence depths were discarded in downstream analyses. Rarefaction analyses were performed to evaluate the sampling coverage for each sample based on the selected sequence depth.
Retentate samples and eukaryotic 18S rRNA V9 metabarcoding for diet characterization.
To assess the biodiversity inventories and edible items in the detritus recovered from the sock filter, we performed massively parallel amplicon sequencing of the V9 region using a total of four retentate DNA samples (including both biological and technical replicates). Samples were submitted for sequencing to the Michigan State University Research Technology Support Facility (RTSF; East Lansing, MI, USA). All sequencing procedures, including the construction of the Illumina sequencing library, emulsion PCR, and MiSeq paired-end sequencing (with the V2 platform) of the V9 region (∼175 bp; with primers Euk 1391f [GTACACACCGCCCGTC] and Euk Br [TGATCCTTCTGCAGGTTCACCTAC]) (72), followed standard Illumina (San Diego, CA, USA) protocols. Michigan State’s Genomics RTSF (https://rtsf.natsci.msu.edu/genomics/) provided standard Illumina quality control, as mentioned above.
Sequence reads were analyzed based on the protocol provided elsewhere (69), as mentioned above. Lower eukaryotic profiling was achieved with slight modification during sequence alignment and taxonomic identification. mothur program-provided SILVA reference files were customized based on Saccharomyces cerevisiae V9 region sequences and later properly formatted for taxonomic classification to the lowest level possible for eukaryotic analysis in mothur. Due to the short length of the amplified region, the sequence data allowed identification only to higher taxonomic levels, such as the genus, family, and order levels.
Alpha diversity and temporal and differential abundance of OTUs.
All measures of community diversity and similarity, including the inverse Simpson diversity index (1/D) and the OTU richness of each sample, were calculated from the sequence data within mothur to quantify alpha (α) diversity. To test for significant differences in diversity indices among treatment groups (water and diet) and among time periods, a multiple-factor ANOVA was performed on the summary files provided by mothur using the programming and statistical software R (v3.0.2) base package. The test was followed by Tukey’s honest significant difference (HSD) post hoc tests. P values below 0.05 indicated significant differences in pairwise mean comparisons.
A custom R code was used to calculate the relative abundance and identify the dominant phyla and taxa (OTUs) in all communities across sampling times (gut microbiota, water, and Artemia fish-associated epibiota). Codes were written and implemented using the packages dplyr and reshape2. The relative abundance of all taxa within community samples was calculated, and taxa with a normalized abundance that exceeded 0.1% were considered predominant taxa. The 20 most abundant taxa were a subset of the total number of OTUs based on the 0.1% cutoff, and relative abundances were prepared in a tabular format to show the temporal variability in fish gut community composition. The remaining taxa were grouped as “others.”
Linear discriminant analysis (LDA) effect size (LEfSe) scores were determined using freely available online software (https://huttenhower.sph.harvard.edu/galaxy/) (40) to determine the microbial communities that were statistically significantly associated with community compositional differences across stages and across treatments. More specifically, the nonparametric factorial Kruskal-Wallis (KW) sum-rank test was first used to detect microbial communities with significant differences in taxon abundance among developmental stages (i.e., prefeeding and 1 week and 2 weeks following the initiation of active feeding) (P < 0.05) and across rearing (water) environments. The unpaired Wilcoxon rank-sum test was used to compare the significant differences in abundance among taxa under influences of developmental stages and treatments (P < 0.05). Linear discriminant analysis was applied to calculate the effective size of the differences in abundance. The LDA scores were normalized by log10.
Beta diversity.
We used several packages implemented in the program R to perform comparative community compositional analysis of beta (β) diversity and other community ecological statistics using the tabulated OTU data set of the predominant taxa. Briefly, we used the vegan function to generate estimates of Bray-Curtis (BC) distances among sample microbial communities (73). Subsequently, we used the cmdscale function to perform ordination (principal-coordinate analyses [PCoA]) based on BC distance (74). The ggplot and ggplots2 packages (75) were used to create ordination plots to visually compare the gut and environmental bacterial community compositions among samples collected from different treatments and among sampling periods based on the two largest eigenvalues. Two multivariate hypothesis tests were implemented using two functions. The adonis function was used to perform multivariate hypothesis testing on differences between the locations of the centroids of treatment group coordinate ordinations based on permutational multivariate analyses of variance (PERMANOVA). The betadisper function was used to perform a test for the homogeneity of multivariate dispersion (PERMDISP) test on community BC matrices (76, 77). These tests were employed because of the nonparametric and skewed nature of the microbial community compositional data. The OTUs that had the highest correlations with the PCoA x and y component axes were identified based on Pearson correlation coefficients using the corr function.
Influence of water and diet treatments of GI tract microbiota.
To analyze the treatment effects of water type and diets on the fish gut microbiota, PCoAs were performed separately on the fish gut communities for all four treatment groups within each developmental stage. Important PCoA axes denoted by eigenvalues comparatively larger than the average eigenvalues were selected. Linear regression models were fit, where each axis was a response variable given the predictor variables of water type and food type. Under the null hypothesis, we expected the gut community composition at each time point to be unaffected by treatment. The axes represent the linear correlations of the bacterial taxonomic compositions present in gut communities. The axes that showed significant effects of treatments or interactions between water and diet treatments were analyzed using the lsmean function to determine the effect of each factor (or combinations of factors in the interaction) on the bacterial taxonomic composition and relative abundance in larval fish GI tracts.
Tests evaluating whether GI tract communities were a neutral subset of the environmental (source) communities.
We used a neutral community ecological model adapted from previous work (24) to explore the relative importance of neutral processes (i.e., dispersal and ecological drift) and selection in the gut microbiota at a given time of sampling (prefeeding and 1 week and 2 weeks after active feeding began). This model also distinguished members of the gut microbiota whose presence was consistent with dispersal from surrounding environmental communities (water as a source) and those that deviated from the neutral model [i.e., those that were over- or underrepresented in the gut relative to the water source(s)]. In general, the model predicts that taxa that are abundant in the source will be widespread taxa which occur in a set of target communities (larval fish gut), since these taxa are more likely to disperse by chance and be randomly sampled by an individual host, while rare taxa are more likely to be lost from fish hosts due to ecological drift (23, 24).
To quantify the relative importance of stochastic processes (neutral drift and gene flow) versus deterministic processes (selection) on the lake sturgeon gut community composition during the three ontogenetic stages, we adapted the approach of Venkataraman and colleagues (25) using their custom R scripts, with slight modification, referring to our data set. Water sources that were interrogated at the time that each group of fish was sampled were considered sources for the fish gut microbiome. The water source microbial communities were created by pooling surveys of both stream water and groundwater taxa at each respective sampling time, whereas the target communities were those microbial taxa that were collectively detected in the gut of fish larvae across all treatments.
Under the neutral model, the probability of observing a microbial OTU in the gut (target) was based on the relative frequency of occurrence in the source (water) at the time of sampling. The relative abundance of a given OTU in the source community was calculated as the number of sequences with the OTU in the source community/total number of sequences in the source community. Similarly, the empirically observed frequency of detection for each OTU in the gut was calculated as the number of gut samples in which the OTU was detected/total number of gut samples across all treatments. Next, the expected frequency of detection was calculated based on a beta probability distribution for each OTU shared between the gut and the source community. Briefly, the lower limit of this probability density function was the relative abundance of the least-abundant OTU in the source community, while the upper limit was 1. The neutral model of Sloan et al. (24) was fit to an overall fitting parameter (Ntm) and the relative abundance of the OTU in the source community. Ntm is the minimized sum of squares of residuals between observed detection frequency and predicted detection frequency for total number of OTUs (25). The fitting parameter reflects the dispersal of microbes from the source community to the fish gut. The fitting of this parameter was performed using a least-squares approach (25). Finally, the variability around this expected detection frequency was calculated using 95% binomial proportion confidence intervals (the Wilson score interval method) with the HMisc package in R (80). Calculating this for all OTUs yields the best-fit neutral model curve and the 95% confidence intervals shown in Fig. 6. The goodness of fit of this curve was assessed using the coefficient of determination (R2) (77, 78). The level of fit of the neutral model served as an adequate representation of the gut community composition if the R2 value was high, whereas the significance of neutral processes was indicated by the P value.
Supplementary Material
ACKNOWLEDGMENTS
Funding for this study was provided by the Michigan Department of Natural Resources, the Great Lakes Fishery Trust, and Michigan State University Ag-Bio Research. S.A.R. was supported by the Universiti Kebangsaan Malaysia (UKM) and the Ministry of Higher Education (MOHE), Government of Malaysia, and a dissertation completion fellowship from the College of Agriculture and Natural Resources while attending MSU.
We thank Brian Maurer, Andrew Denhaart, and Nick Sard for helping with data analyses and R. We also thank John Bauman, Kari Dammerman, Nathan Barton, Sarah Watson, Troy Smith, Adam Unstead, Lindsay Adams, and James Garavaglia for assistance in construction of the experiment and collection of the data. Mohamed Faisal and Terence Marsh provided valuable advice on the experimental design, analyses, and interpretation of the data.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Vellend M. 2010. Conceptual synthesis in community ecology. Q Rev Biol 85:183–206. doi: 10.1086/652373. [DOI] [PubMed] [Google Scholar]
- 2.Miller ET, Svanbäck R, Bohannan B. 2018. Microbiomes as metacommunities: understanding host-associated microbes through metacommunity ecology. Trends Ecol Evol 33:926–935. doi: 10.1016/j.tree.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 3.Gilbert JA, Lynch SV. 2019. Community ecology as a framework for human microbiome research. Nat Med 25:884–889. doi: 10.1038/s41591-019-0464-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.García-Bayona L, Comstock LE. 2018. Bacterial antagonism in host-associated microbial communities. Science 361:eaat2456. doi: 10.1126/science.aat2456. [DOI] [PubMed] [Google Scholar]
- 5.Costello EK, Stagaman K, Dethlefsen L, Bohannan BJM, Relman DA. 2012. The application of ecological theory toward an understanding of the human microbiome. Science 336:1255–1262. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ringø E, Zhou Z, Vecino JLG, Wadsworth S, Romero J, Krogdahl Å, Olsen RE, Dimitroglou A, Foey A, Davies S, Owen M, Lauzon HL, Martinsen LL, De Schryver P, Bossier P, Sperstad S, Merrifield DL. 2016. Effect of dietary components on the gut microbiota of aquatic animals. A never-ending story? Aquacult Nutr 22:219–282. doi: 10.1111/anu.12346. [DOI] [Google Scholar]
- 7.Nayak SK. 2010. Probiotics and immunity: a fish perspective. Fish Shellfish Immunol 29:2–14. doi: 10.1016/j.fsi.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 8.Gómez GD, Balcázar JL. 2008. A review on the interactions between gut microbiota and innate immunity of fish. FEMS Immunol Med Microbiol 52:145–154. doi: 10.1111/j.1574-695X.2007.00343.x. [DOI] [PubMed] [Google Scholar]
- 9.Llewellyn MS, Boutin S, Hoseinifar SH, Derome N. 2014. Teleost microbiomes: the state of the art in their characterization, manipulation and importance in aquaculture and fisheries. Front Microbiol 5:207. doi: 10.3389/fmicb.2014.00207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egerton S, Culloty S, Whooley J, Stanton C, Ross RP. 2018. The gut microbiota of marine fish. Front Microbiol 9:873. doi: 10.3389/fmicb.2018.00873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abdul Razak S, Griffin MJ, Mischke CC, Bosworth BG, Waldbieser GC, Wise DJ, Marsh TL, Scribner KT. 2019. Biotic and abiotic factors influencing channel catfish egg and gut microbiome dynamics during early life stages. Aquaculture 498:556–567. doi: 10.1016/j.aquaculture.2018.08.073. [DOI] [Google Scholar]
- 12.Stephens ZW, Burns AR, Stagaman K, Wong S, Rawls JF, Guillemin K, Bohannan B. 2016. The composition of the zebrafish intestinal microbial community varies across development. ISME J 10:644–654. doi: 10.1038/ismej.2015.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohd Nosi MZ, Syed Jamil Fadaak SNE, Muhammad MDD, Iehata S. 2018. Assessment of gut microbiota in different developmental stages of Malaysian mahseer (Tor tambroides). Aquac Res 49:2977–2987. doi: 10.1111/are.13757. [DOI] [Google Scholar]
- 14.Wang W, Wu S, Zheng Y, Cheng Y, Li W, Zou H, Wang G. 2015. Characterization of the bacterial community associated with early-developmental stages of grass carp (Ctenopharyngodon idella). Aquac Res 46:2728–2735. doi: 10.1111/are.12428. [DOI] [Google Scholar]
- 15.Hata H, Tanabe AS, Yamamoto S, Toju H, Kohda M, Hori M. 2014. Diet disparity among sympatric herbivorous cichlids in the same ecomorphs in Lake Tanganyika: amplicon pyrosequences on algal farms and stomach contents. BMC Biol 12:90. doi: 10.1186/s12915-014-0090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sullam KE, Rubin BE, Dalton CM, Kilham SS, Flecker AS, Russell JA. 2015. Divergence across diet, time and populations rules out parallel evolution in the gut microbiomes of Trinidadian guppies. ISME J 9:1508–1522. doi: 10.1038/ismej.2014.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baldo L, Riera JL, Tooming-Klunderud A, Albà MM, Salzburger W. 2015. Gut microbiota dynamics during dietary shift in eastern African cichlid fishes. PLoS One 10:e0127462. doi: 10.1371/journal.pone.0127462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baldo L, Pretus JL, Riera JL, Musilova Z, Bitja Nyom AR, Salzburger W. 2017. Convergence of gut microbiotas in the adaptive radiations of African cichlid fishes. ISME J 11:1975–1987. doi: 10.1038/ismej.2017.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith P, Willemsen D, Popkes M, Metge F, Gandiwa E, Reichard M, Valenzano DR. 2017. Regulation of life span by the gut microbiota in the short-lived African turquoise killifish. Elife 6:e27014. doi: 10.7554/eLife.27014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong S, Stephens WZ, Burns AR, Stagaman K, David LA, Bohannan BJM, Guillemin K, Rawls JF. 2015. Ontogenetic differences in dietary fat influence microbiota assembly in the zebrafish gut. mBio 6:e00687-15. doi: 10.1128/mBio.00687-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. 2012. Human gut microbiome viewed across age and geography. Nature 486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hubbell SP. 2001. The unified neutral theory of biodiversity and biogeography (MPB-32). Princeton University Press, Princeton, NJ. [Google Scholar]
- 23.Burns AR, Stephens WZ, Stagaman K, Wong S, Rawls JF, Guillemin K, Bohannan B. 2016. Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. ISME J 10:655–664. doi: 10.1038/ismej.2015.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sloan WT, Lunn M, Woodcock S, Head IM, Nee S, Curtis TP. 2006. Quantifying the roles of immigration and chance in shaping prokaryote community structure. Environ Microbiol 8:732–740. doi: 10.1111/j.1462-2920.2005.00956.x. [DOI] [PubMed] [Google Scholar]
- 25.Venkataraman A, Bassis CM, Beck JM, Young VB, Curtis JL, Huffnagle GB, Schmidt TM. 2015. Application of a neutral community model to assess structuring of the human lung microbiome. mBio 6:e02284-14. doi: 10.1128/mBio.02284-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Schryver P, Vadstein O. 2014. Ecological theory as a foundation to control pathogenic invasion in aquaculture. ISME J 8:2360–2368. doi: 10.1038/ismej.2014.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vadstein O, Bergh Ø, Gatesoupe F-J, Galindo-Villegas J, Mulero V, Picchietti S, Scapigliati G, Makridis P, Olsen Y, Dierckens K, Defoirdt T, Boon N, De Schryver P, Bossier P. 2013. Microbiology and immunology of fish larvae. Rev Aquacult 5:S1–S25. doi: 10.1111/j.1753-5131.2012.01082.x. [DOI] [Google Scholar]
- 28.Wilson JM, Castro LFC. 2010. Morphological diversity of the gastrointestinal tract in fishes, p 1–55. In Fish physiology: the multifunctional gut of fish. Elsevier, Philadelphia, PA. [Google Scholar]
- 29.Detlaf TA, Ginsburg AS, Schmanlhausen OI. 1993. Sturgeon fishes developmental biology and aquaculture. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 30.Saffron I. 2004. The decline of the North American species, p 1–21. In Lebreton GTO, Beamish FWH, McKinley RS (ed), Sturgeon & paddlefish of North America. Kluwer Academic Publishers, Amsterdam, Netherlands. [Google Scholar]
- 31.Mim SD, Lazur A, Shelton WL, Gomelsky B, Chapman F. 2002. Production of sturgeon. South Regional Aquaculture Center, Stoneville. MS. [Google Scholar]
- 32.Vedrasco A, Lobchenko V, Pirtu I, Billard R. 2002. The culture of live food for sturgeon juveniles, a mini review of the Russian literature. Int Rev Hydrobiol 87:569–575. doi:. [DOI] [Google Scholar]
- 33.Buddington RK, Christofferson JP. 1985. Digestive and feeding characteristics of the chondrosteans. Environ Biol Fish 14:31–41. doi: 10.1007/BF00001574. [DOI] [Google Scholar]
- 34.Wang YL, Binkowski FP, Doroshov SI. 1985. Effect of temperature on early development of white and lake sturgeon. Environ Biol Fish 14:43–50. doi: 10.1007/BF00001575. [DOI] [Google Scholar]
- 35.Diggle PK. 1994. The expression of andromonoecy in Solanum hirtum (Solanaceae): phenotypic plasticity and ontogenetic contingency. Am J Bot 81:1354–1365. doi: 10.1002/j.1537-2197.1994.tb11457.x. [DOI] [Google Scholar]
- 36.Orizaola G, Dahl E, Laurila A. 2010. Compensating for delayed hatching across consecutive life-history stages in an amphibian. Oikos 119:980–987. doi: 10.1111/j.1600-0706.2009.17956.x. [DOI] [Google Scholar]
- 37.Huey RB, Kearney MR, Krockenberger A, Holtum JAM, Jess M, Williams SE. 2012. Predicting organismal vulnerability to climate warming: roles of behaviour, physiology and adaptation. Philos Trans R Soc Lond B Biol Sci 367:1665–1679. doi: 10.1098/rstb.2012.0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gensollen T, Iyer SS, Kasper DL, Blumberg RS. 2016. How colonization by microbiota in early life shapes the immune system. Science 352:539–544. doi: 10.1126/science.aad9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abdul Razak S. 2017. Biotic and abiotic factors influence formation and ontogenic dynamics of molecularly defined gastro-intestinal microbial communities in lake sturgeon. PhD dissertation. Michigan State University, East Lansing, MI. [Google Scholar]
- 40.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. 2011. Metagenomic biomarker discovery and explanation. Genome Biol 12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geraylou Z, Souffreau C, Rurangwa E, Maes GE, Spanier KI, Courtin CM, Delcour JA, Buyse J, Ollevier F. 2013. Prebiotic effects of arabinoxylan oligosaccharides on juvenile Siberian sturgeon (Acipenser baerii) with emphasis on the modulation of the gut microbiota using 454 pyrosequencing. FEMS Microbiol Ecol 86:357–371. doi: 10.1111/1574-6941.12169. [DOI] [PubMed] [Google Scholar]
- 42.Bacanu GM, Oprea L. 2013. Differences in the gut microbiota between wild and domestic Acipenser ruthenus evaluated by denaturing gradient gel electrophoresis. Rom Biotechnol Lett 18:8069–8076. [Google Scholar]
- 43.Buddington RK, Doroshov SI. 1986. Structural and functional relations of the white sturgeon alimentary canal (Acipenser transmontanus). J Morphol 190:201–213. doi: 10.1002/jmor.1051900205. [DOI] [PubMed] [Google Scholar]
- 44.Ingerslev HC, von Gersdorff Jorgensen L, Lenz Strube M, Larsen N, Dalsgaard I, Boye M, Madsen L. 2014. The development of the gut microbiota in rainbow trout (Oncorhynchus mykiss) is affected by first feeding and diet type. Aquaculture 424-425:24–34. doi: 10.1016/j.aquaculture.2013.12.032. [DOI] [Google Scholar]
- 45.Desai AR, Links MG, Collins SA, Mansfield GS, Drew MD, Van Kessel AG, Hill JE. 2012. Effects of plant-based diets on the distal gut microbiome of rainbow trout (Oncorhynchus mykiss). Aquaculture 350–353:134–142. doi: 10.1016/j.aquaculture.2012.04.005. [DOI] [Google Scholar]
- 46.Rawls JF, Mahowald MA, Ley RE, Gordon JI. 2006. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell 127:423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li J, Ni J, Li J, Wang C, Li X, Wu S, Zhang T, Yu Y, Yan Q. 2014. Comparative study on gastrointestinal microbiota of eight fish species with different feeding habits. J Appl Microbiol 117:1750–1760. doi: 10.1111/jam.12663. [DOI] [PubMed] [Google Scholar]
- 48.Bledsoe JW, Peterson BC, Swanson KS, Small BC. 2016. Ontogenetic characterization of the intestinal microbiota of channel catfish through 16S rRNA gene sequencing reveals insights on temporal shifts and the influence of environmental microbes. PLoS One 11:e0166379. doi: 10.1371/journal.pone.0166379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mansfield GS, Desai AR, Nilson SA, Van Kessel AG, Drew MD, Hill JE. 2010. Characterization of rainbow trout (Oncorhynchus mykiss) intestinal microbiota and inflammatory marker gene expression in a recirculating aquaculture system. Aquaculture 307:95–104. doi: 10.1016/j.aquaculture.2010.07.014. [DOI] [Google Scholar]
- 50.Lopetuso LR, Scaldaferri F, Petito V, Gasbarrini A. 2013. Commensal Clostridia: leading players in the maintenance of gut homeostasis. Gut Pathog 5:23. doi: 10.1186/1757-4749-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Crowther JS. 1971. Sarcina ventriculi in human feces. J Med Microbiol 4:343–350. doi: 10.1099/00222615-4-3-343. [DOI] [PubMed] [Google Scholar]
- 52.Smit J. 1911. The biology of the fermenting Sarcina. J Pathol 35:455–468. doi: 10.1002/path.1700360310. [DOI] [Google Scholar]
- 53.Callman JL, Macy JM. 1984. The predominant anaerobe from the spiral intestine of hatchery-raised sturgeon (Acipenser transmontanus), a new Bacteroides species. Arch Microbiol 140:57–65. doi: 10.1007/BF00409772. [DOI] [Google Scholar]
- 54.Fujimoto M, Crossman JA, Scribner KT, Marsh TL. 2013. Microbial community assembly and succession on lake sturgeon egg surfaces as a function of simulated spawning stream flow rate. Microb Ecol 66:500–511. doi: 10.1007/s00248-013-0256-6. [DOI] [PubMed] [Google Scholar]
- 55.Romero J, Navarrete P. 2006. 16S rDNA-based analysis of dominant bacterial populations associated with early life stages of coho salmon (Oncorhynchus kisutch). Microb Ecol 51:422–430. doi: 10.1007/s00248-006-9037-9. [DOI] [PubMed] [Google Scholar]
- 56.Hansen GH, Olafsen JA. 1999. Bacterial interactions in early life stages of marine cold water fish. Microb Ecol 38:1–26. doi: 10.1007/s002489900158. [DOI] [PubMed] [Google Scholar]
- 57.Navarrete P, Mardones P, Opazo R, Espejo R, Romero J. 2008. Oxytetracycline treatment reduces bacterial diversity of intestinal microbiota of Atlantic salmon. J Aquat Anim Health 20:177–183. doi: 10.1577/H07-043.1. [DOI] [PubMed] [Google Scholar]
- 58.Chen W-M, Chung Y-N, Chiu T-F, Cheng C-Y, Arun AB, Sheu S-Y. 2010. Deefgea chitinilytica sp. nov., isolated from a wetland. Int J Syst Evol Microbiol 60:1450–1453. doi: 10.1099/ijs.0.015263-0. [DOI] [PubMed] [Google Scholar]
- 59.Stackebrandt E, Lang E, Cousin S, Päuker O, Brambilla E, Kroppenstedt R, Lünsdorf H. 2007. Deefgea rivuli gen. nov., sp. nov., a member of the class Betaproteobacteria. Int J Syst Evol Microbiol 57:639–645. doi: 10.1099/ijs.0.64771-0. [DOI] [PubMed] [Google Scholar]
- 60.Jung A, Jung-Schroers V. 2011. Detection of Deefgea chitinilytica in freshwater ornamental fish. Lett Appl Microbiol 52:497–500. doi: 10.1111/j.1472-765X.2011.03030.x. [DOI] [PubMed] [Google Scholar]
- 61.Navarrete P, Espejo RT, Romero J. 2009. Molecular analysis of microbiota along the digestive tract of juvenile Atlantic salmon (Salmo salar L.). Microb Ecol 57:550–561. doi: 10.1007/s00248-008-9448-x. [DOI] [PubMed] [Google Scholar]
- 62.Giatsis C, Sipkema D, Smidt H, Heilig H, Benvenuti G, Verreth J, Verdegem M. 2015. The impact of rearing environment on the development of gut microbiota in tilapia larvae. Sci Rep 5:18206–18215. doi: 10.1038/srep18206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wassink L, Bussy U, Li W, Scribner KT. 2019. High-stress rearing temperature in Acipenser fulvescens affects physiology, behaviour and predation rates. Anim Behav 157:153–165. doi: 10.1016/j.anbehav.2019.09.005. [DOI] [Google Scholar]
- 64.Palleroni NJ. 1993. Pseudomonas classification. A new case history in the taxonomy of Gram-negative bacteria. Antonie Van Leeuwenhoek 64:231–251. doi: 10.1007/bf00873084. [DOI] [PubMed] [Google Scholar]
- 65.Kampfer P, Terenius O, Lindh JM, Faye I. 2006. Janibacter anophelis sp. nov., isolated from the midgut of Anopheles arabiensis. Int J Syst Evol Microbiol 56:389–392. doi: 10.1099/ijs.0.63905-0. [DOI] [PubMed] [Google Scholar]
- 66.Bauman JM. 2015. Investigations of aquaculture methodologies to enhance success of Great Lakes Lake Sturgeon streamside facilities. MS thesis. Michigan State University, East Lansing, MI. [Google Scholar]
- 67.Deng D-F, Koshio S, Yokoyama S, Bai SC, Shao Q, Cui Y, Hung SS. 2003. Effects of feeding rate on growth performance of white sturgeon (Acipenser transmontanus) larvae. Aquaculture 217:589–598. doi: 10.1016/S0044-8486(02)00461-1. [DOI] [Google Scholar]
- 68.Milligan-Myhre K, Charette JR, Phennicie RT, Stephens WZ, Rawls JF, Guillemin K, Kim CH. 2011. Study of host-microbe interactions in zebrafish. Methods Cell Biol 105:87–116. doi: 10.1016/B978-0-12-381320-6.00004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Caporaso JG, Lauber C, Walters W, Berg-Lyons D, Lozupone C, Turnbaugh P, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108(Suppl):4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schloss PD, Westcott SL. 2011. Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Appl Environ Microbiol 77:3219–3226. doi: 10.1128/AEM.02810-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Amaral-Zettler LA, McCliment EA, Ducklow HW, Huse SM. 2009. A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PLoS One 4:e6372. doi: 10.1371/journal.pone.0006372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H. 2015. Vegan: community ecology package. R package version 2.2-1 R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 74.Bray JR, Curtis JT. 1957. An ordination of the upland forest communities of southern Wisconsin. Ecol Monogr 27:325–349. doi: 10.2307/1942268. [DOI] [Google Scholar]
- 75.Wickham H. 2009. ggplot2: elegant graphics for data analysis with R. Springer-Verlag, New York, NY. [Google Scholar]
- 76.Anderson M. 2001. A new method for non parametric multivariate analysis of variance. Austral Ecol 26:32–46. doi: 10.1046/j.1442-9993.2001.01070.x. [DOI] [Google Scholar]
- 77.Anderson MJ. 2006. Distance-based tests for homogeneity of multivariate dispersions. Biometrics 62:245–253. doi: 10.1111/j.1541-0420.2005.00440.x. [DOI] [PubMed] [Google Scholar]
- 78.Wilson EB. 1927. Probable inference, the law of succession, and statistical inference. J Am Stat Assoc 22:209–212. doi: 10.1080/01621459.1927.10502953. [DOI] [Google Scholar]
- 79.Masouleh AS, Sharifpour I, Arani AS. 2006. The aerobic bacterial flora of hatchery reared Caspian Sea sturgeon fingerlings. J Appl Ichthyol 22:261–264. doi: 10.1111/j.1439-0426.2007.00964.x. [DOI] [Google Scholar]
- 80.Harrell FE. 2014. Hmisc: Harrell miscellaneous. R package version 3.14-5 http://CRAN.R-project.org/package=Hmisc.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.