Dietary protein levels are generally higher in Western populations than in the world average. We challenged three-stage continuous colonic model systems containing high protein levels and confirmed the production of potentially harmful metabolites from proteolysis, especially replicates of the transverse and distal colon. Fermentations of proteins with a prebiotic supplementation resulted in a change in the human gut microbiota and inhibited the production of some proteolytic metabolites. Moreover, we observed both bacterial and metabolic differences between fecal bacteria from omnivore donors and vegetarian donors. Proteins with prebiotic supplementation showed higher Bacteroides spp. and inhibited Clostridium cluster IX in omnivore models, while in vegetarian modes, Clostridium cluster IX was higher and Bacteroides spp. lower with high protein plus prebiotic supplementation. Synergy1 addition inhibited p-cresol production in vegetarian high p-cresol-producing models while the inhibitory effect was not seen in omnivore models.
KEYWORDS: gut microbiota, prebiotics, diet, vegetarian, protein fermentation
ABSTRACT
Dietary protein residue can result in microbial generation of various toxic metabolites in the gut, such as ammonia. A prebiotic is “a substrate that is selectively utilised by host microorganisms conferring a health benefit” (G. R. Gibson, R. Hutkins, M. E. Sanders, S. L. Prescott, et al., Nat Rev Gastroenterol Hepatol 14:491–502, 2017, https://doi.org/10.1038/nrgastro.2017.75). Prebiotics are carbohydrates that may have the potential to reverse the harmful effects of gut bacterial protein fermentation. Three-stage continuous colonic model systems were inoculated with fecal samples from omnivore and vegetarian volunteers. Casein (equivalent to 105 g protein consumption per day) was used within the systems as a protein source. Two different doses of inulin-type fructans (Synergy1) were later added (equivalent to 10 g per day in vivo and 15 g per day) to assess whether this influenced protein fermentation. Bacteria were enumerated by fluorescence in situ hybridization with flow cytometry. Metabolites from bacterial fermentation (short-chain fatty acid [SCFA], ammonia, phenol, indole, and p-cresol) were monitored to further analyze proteolysis and the prebiotic effect. A significantly higher number of bifidobacteria was observed with the addition of inulin together with reduction of Desulfovibrio spp. Furthermore, metabolites from protein fermentation, such as branched-chain fatty acids (BCFA) and ammonia, were significantly lowered with Synergy1. Production of p-cresol varied among donors, as we recognized four high producing models and two low producing models. Prebiotic addition reduced its production only in vegetarian high p-cresol producers.
IMPORTANCE Dietary protein levels are generally higher in Western populations than in the world average. We challenged three-stage continuous colonic model systems containing high protein levels and confirmed the production of potentially harmful metabolites from proteolysis, especially replicates of the transverse and distal colon. Fermentations of proteins with a prebiotic supplementation resulted in a change in the human gut microbiota and inhibited the production of some proteolytic metabolites. Moreover, we observed both bacterial and metabolic differences between fecal bacteria from omnivore donors and vegetarian donors. Proteins with prebiotic supplementation showed higher Bacteroides spp. and inhibited Clostridium cluster IX in omnivore models, while in vegetarian modes, Clostridium cluster IX was higher and Bacteroides spp. lower with high protein plus prebiotic supplementation. Synergy1 addition inhibited p-cresol production in vegetarian high p-cresol-producing models while the inhibitory effect was not seen in omnivore models.
INTRODUCTION
Protein consumption is increasing annually worldwide, and 2.5% of British adult males consume over 136 g protein per day according to a recent national dietary survey (1, 2). Some protein is metabolized or absorbed in the small intestine, but residual dietary protein may enter the large intestine where colonic bacteria can utilize it and produce various metabolites. Proteolysis gradually increases throughout the colon, being highest in distal regions where carbohydrate is depleted (3, 4). Bacterial degradation of amino acids produces ammonia, amines, CO2, short-chain fatty acids (SCFA), branched-chain fatty acids (BCFA), occasionally H2S, and aromatic molecules, such as indole, p-cresol, and skatole (5). These metabolites may interact with the host and potentially affect human health in many ways. In cell culture experiments, ammonia at relevant luminal concentrations was found to have a negative effect on barrier function resulting in elevated permeability (6). Two animal studies illustrated how ammonia can shorten mucosal cell life span and promote colon cancer in combination with other carcinogens in vivo (7, 8). Hydrogen sulfide can be generated from sulfur containing amino acids or reduction of dietary sulfates. Both rat and human colonocyte cell lines showed that H2S had a negative effect on butyrate and acetate oxidation by cells for energy uptake (9, 10). Bacterial H2S production rates are higher in ulcerative colitis patients compared to healthy adults, which may contribute to the pathogenesis of this inflammatory bowel disease (11). Aromatic amino acids, such as tyrosine, phenylalanine, and tryptophan, can be degraded by colonic bacteria producing phenol, indole, p-cresol, skatole, and other aromatic metabolites. Phenol and p-cresol have been shown to damage epithelial barrier function in vitro and can be potentially carcinogenic (6, 12, 13). Indole and p-cresol would be transformed to indoxyl sulfate and p-cresol sulfate after conjugation in the human body, and indoxyl sulfate and p-cresol sulfate levels in blood are correlated with renal disease progression and vascular dysfunction in chronic kidney disease (14, 15).
A prebiotic has been defined as “a substrate that is selectively utilised by host microorganisms conferring a health benefit” by Gibson et al. (16). One confirmed type of prebiotic is inulin-type fructans, which are carbohydrates comprised of fructose residues, sometimes with a glucose as the terminal residue. The d-fructose molecules are linked by β(2→1) linkages, and when there is a glucose, the chain is terminated by a d-glucose molecule bonded to fructose by an α(1↔2) linkage. A randomized, double-blind, placebo-controlled, crossover human study recently analyzed bacterial compositional changes after inulin-type fructan intervention by 16S rRNA gene sequencing and found the following: bifidobacteria and Anaerostipes increased while Bilophila decreased after prebiotic supplementation for 4 weeks, concomitant with an improvement in constipation (17). Other human intervention studies have investigated various health benefits of inulin-type fructans, such as mineral absorption, energy regulation, and lipid-lowering properties (18–20). Metabolism of inulin-type fructans not only promotes specific bacterial changes in the colon but also generates short-chain fatty acids (SCFA). Bifidobacteria can produce acetate and lactate from carbohydrates, and these two acids may be substrates for butyrate producers, such as Eubacterium rectale and Anaerostipes caccae, resulting in enhanced butyrate production in the gut (21, 22). SCFA, especially butyrate, can promote epithelial cell differentiation and apoptosis in vitro; therefore, they may possibly have an impact upon colorectal cancer risk (23).
A batch culture fermenter was previously used to evaluate the effect of prebiotic supplementation on the fecal microbiota of individuals consuming a high-protein diet (24). Prebiotic addition inhibited protein fermentation by modulating microbiota composition and bacterial metabolism within 48 h. Fecal samples obtained from omnivores and vegetarians showed different responses to protein/carbohydrate intervention and showed adaptation to various dietary protein sources. In this paper, we are interested in how changing the host diet would modify bacteria composition and metabolism in the whole colon using a more realistic and well-established model system. This system simulates the proximal, transverse, and distal colon using three interconnected fermentation vessels with temperature and pH control. Samples are taken after equilibrium is reached for each treatment, after at least 8 turnovers of the operating volume with a stable medium flow rate. This system is a well-established tool in gut microbial ecology research and allows study of the changes in the ecosystem along the colon (25). The model has been validated by comparing both microbial and chemical characteristics of the model over time with human sudden death victims (25).
Out of four protein sources (meat protein, casein, soy protein, and Quorn protein) that were studied in the batch culture experiment (24), casein was selected for the continuous culture because dairy products are consumed both in vegetarians and omnivores, and casein had the highest phenol and indole production in the batch culture experiment.
In the present paper, prebiotic effects on the possible negative consequence of a high-protein diet (change of bacterial composition and metabolism with higher production of ammonia, phenols, and indoles) in the gut were studied using a validated in vitro multiple-stage continuous culture system. How bacterial communities from different hosts respond to carbohydrates and protein was also studied.
RESULTS
Bacterial enumeration.
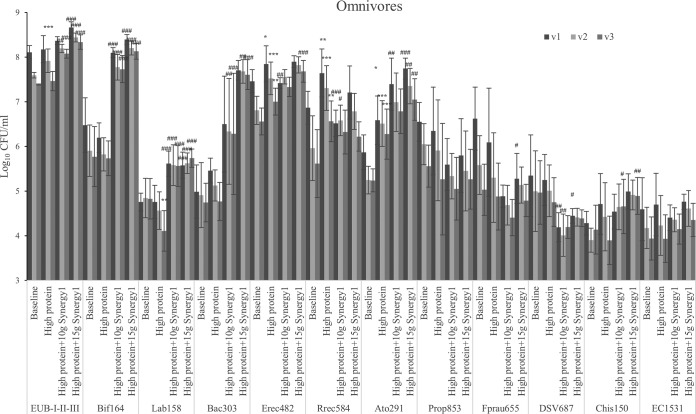
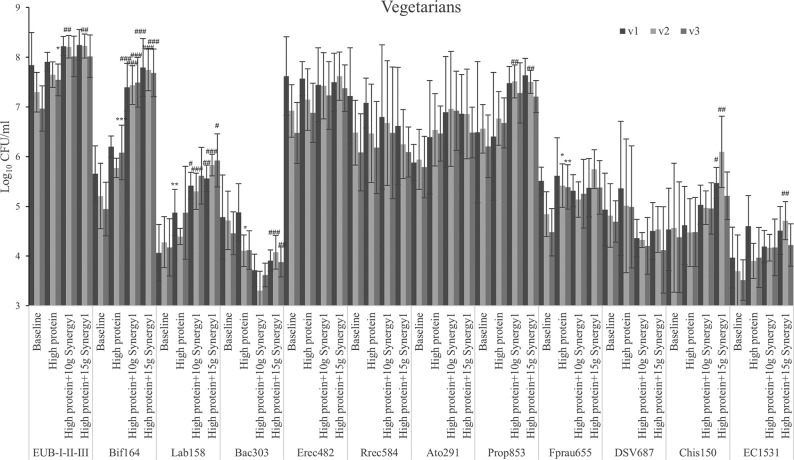
High-protein feeding did not affect bacterial numbers in the proximal colon simulation; however, the simulated transverse colon of omnivore models and the simulated distal colon of vegetarian models had significantly higher bacterial populations after protein feeding (Fig. 1 and 2). Following prebiotic addition, total bacterial populations in gut models from both omnivore and vegetarian donors were slightly higher, especially at the higher dose in the models inoculated with omnivore donors (P < 0.001 in all three regions). Meanwhile, populations of bifidobacteria and lactobacilli increased by growth on Synergy1 in all models (P < 0.001) (Fig. 1 to 3).
FIG 1.
Bacterial counts with different steady states as log10 CFU/ml in three different vessels of a human in vitro colonic model as analyzed by fluorescent in situ hybridization (FISH). Values are means over 3 consecutive days from 3 omnivores’ microbiota ± standard deviation. For each measurement, significant differences in each vessel among four steady states are labeled. *, mean values were significantly different from the baseline steady state (P < 0.05). **, mean values were significantly different from the baseline steady state (P < 0.01). ***, mean values were significantly different from the baseline steady state (P < 0.001). #, mean values were significantly different from the high-protein steady state (P < 0.05). ##, mean values were significantly different from the high-protein steady state (P < 0.01). ###, mean values were significantly different from the high-protein steady state (P < 0.001).
FIG 2.
Bacterial counts with different steady states as log10 CFU/ml in three different vessels of a human in vitro colonic model as analyzed by fluorescent in situ hybridization (FISH). Values are means over 3 consecutive days from 3 vegetarians’ microbiota ± standard deviation. For each measurement, significant differences in each vessel among four steady states are labeled. *, mean values were significantly different from the baseline steady state (P < 0.05). **, mean values were significantly different from the baseline steady state (P < 0.01). ***, mean values were significantly different from the baseline steady state (P < 0.001). #, mean values were significantly different from the high-protein steady state (P < 0.05). ##, mean values were significantly different from the high-protein steady state (P < 0.01). ###, mean values were significantly different from the high-protein steady state (P < 0.001).
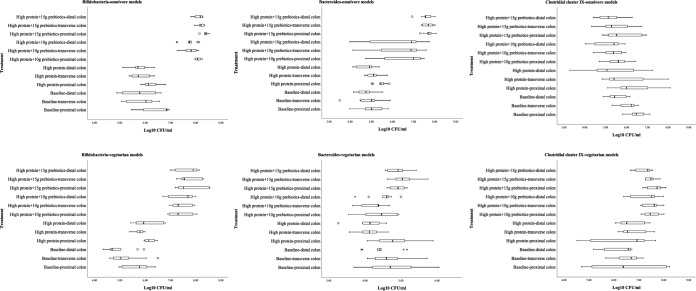
FIG 3.
Bacterial counts with different steady states as log10 CFU/ml in three different vessels of a human in vitro colonic model as analyzed by fluorescent in situ hybridization (FISH).
Bacteroides species populations in the omnivore models displayed a trend to increase when grown on the protein and kept growing on Synergy1, especially in the transverse and distal colon simulations (Fig. 3). Meanwhile in the vegetarian models, Bacteroides species populations decreased with protein addition and significantly so with Synergy1 in the transverse and distal colon. Bacteroides spp. are propionate producers, and the genus contains proteolytic species (26). Clostridium cluster IX, which is another propionate-producing group, decreased in the omnivore models with Synergy1 supplementation though not significantly, while in vegetarian models, the group increased significantly in the transverse and distal colon with Synergy1 treatment.
Within Clostridium subclusters XIVa and XIVb, there are proteolytic bacteria, and this group responded to a high-protein dose (Fig. 1 and 2) as follows: these bacteria from omnivore donors increased in all vessels while vegetarian microbiota only had an increasing trend. The Atopobium cluster from all donors grew through all steady states.
Desulfovibrio can reduce dietary sulfate to produce H2S (27). These organisms were not affected by the protein-containing medium; however, both doses of prebiotics decreased their numbers (Fig. 1 and 2). In omnivore-inoculated models, reduction was significant, while in vegetarian-inoculated models, the change was only a trend. Vegetarian models had lower Desulfovibrio counts at baseline (4.81 ± 0.59 log10 CFU/ml) compared to those of omnivore models (5.11 ± 0.84 log10 CFU/ml), which could contribute to the reduction being less significant (proximal colon, P = 0.074 after prebiotic addition) in vegetarian models; however, this difference was not significant (P = 0.07).
Organic acids.
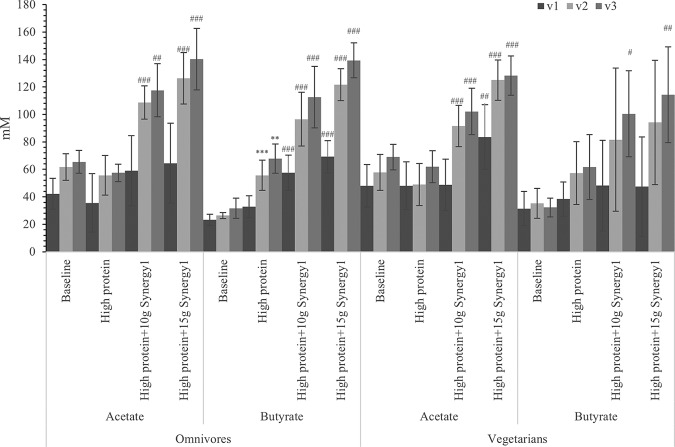
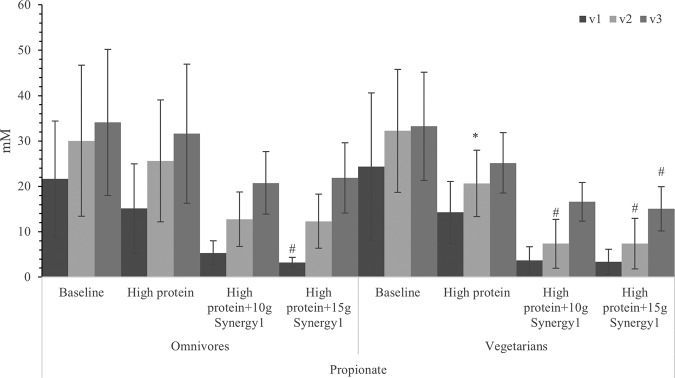
Acetate levels were maintained with protein treatment but increased significantly in all three regions after prebiotic steady state, which can be correlated with higher total bacterial numbers in both prebiotic treatments (Fig. 4). As discussed before, bifidobacterial numbers were significantly higher after adding the prebiotic. A higher number of bifidobacteria would be expected to promote acetate production. Butyric acid was significantly increased in the later steady states when grown on Synergy1, which could be explained by increases in the butyrate-producing Clostridium coccoides-Eubacterium rectale group (Fig. 4). Concentrations of propionic acid were reduced by protein supplementation and further reduced by prebiotic supplementation (Fig. 5).
FIG 4.
Acetate and butyrate concentrations in samples with different steady states in three different vessels of a human in vitro colonic model as analyzed by gas chromatography (GC). Values are means over 3 consecutive days from 3 omnivores’ microbiota or 3 vegetarians’ microbiota ± standard deviation. For each measurement, significant differences in each vessel among four steady states are labeled. *, mean values were significantly different from the baseline steady state (P < 0.05). **, mean values were significantly different from the baseline steady state (P < 0.01). ***, mean values were significantly different from the baseline steady state (P < 0.001). #, mean values were significantly different from the high-protein steady state (P < 0.05). ##, mean values were significantly different from the high-protein steady state (P < 0.01). ###, mean values were significantly different from the high-protein steady state (P < 0.001).
FIG 5.
Propionate concentration in samples with different steady states in three different vessels of a human in vitro colonic model as analyzed by gas chromatography (GC). Values are means over 3 consecutive days from 3 omnivores’ microbiota or 3 vegetarians’ microbiota ± standard deviation. For each measurement, significant differences in each vessel among four steady states are labeled. *, mean values were significantly different from the baseline steady state (P < 0.05). **, mean values were significantly different from the baseline steady state (P < 0.01). ***, mean values were significantly different from the baseline steady state (P < 0.001). #, mean values were significantly different from the high-protein steady state (P < 0.05). ##, mean values were significantly different from the high-protein steady state (P < 0.01). ###, mean values were significantly different from the high-protein steady state (P < 0.001).
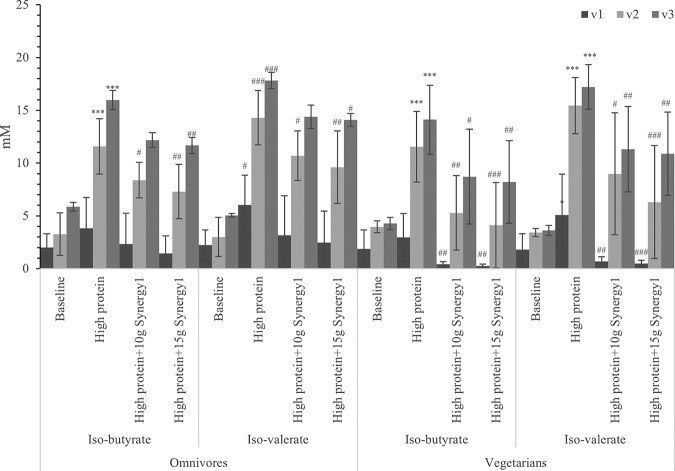
Bacterial metabolism of valine and leucine produces isobutyrate and isovalerate, respectively, and these metabolites indicate gut bacterial proteolysis (28). Both isovaleric acid and isobutyric acid concentrations increased after extra protein was added, especially in the transverse and distal colon simulations (Fig. 6). Increased production from protein fermentation was inhibited by prebiotic supplementation.
FIG 6.
BCFA concentrations in samples with different steady states in three different vessels of a human in vitro colonic model as analyzed by gas chromatography (GC). Values are means over 3 consecutive days from 3 omnivores’ microbiota or 3 vegetarians’ microbiota ± standard deviation. For each measurement, significant differences in each vessel among four steady states are labeled. *, mean values were significantly different from the baseline steady state (P < 0.05). **, mean values were significantly different from the baseline steady state (P < 0.01). ***, mean values were significantly different from the baseline steady state (P < 0.001). #, mean values were significantly different from the high-protein steady state (P < 0.05). ##, mean values were significantly different from the high-protein steady state (P < 0.01). ###, mean values were significantly different from the high-protein steady state (P < 0.001).
Volatile organic compounds.
Three potentially detrimental volatile organic compounds (indole, p-cresol, and skatole) were quantified. Concentrations of skatole were under the detection limit, and indole production was very low in all steady states (data not shown). Production of p-cresol displayed high individual variation, as omnivore donors 1 and 2 and vegetarian donors 1 and 3 displayed higher production than omnivore donor 3 and vegetarian donor 2 (Tables 1 and 2). At high-protein steady state, its concentration was significantly higher in distal vessels in these high-p-cresol-producing models (211.92 ± 125.55) compared to that of low producers (7.50 ± 4.77) (P < 0.001). Protein addition significantly promoted p-cresol production (from 45.72 ± 24.00 to 211.92 ± 125.55; P < 0.001) in vessel three (distal area) of the high producing models. Vegetarian models had reduced p-cresol production after adding a lower dose of prebiotics. Concentrations in omnivore models kept increasing with lower doses of prebiotics and only started to decrease in steady state 4; however, the level was still not lower than that of the high-protein steady state.
TABLE 1.
p-Cresol concentration in omnivore donor samples with different steady states of a human in vitro colonic model in three different vessels as analyzed by GC
| Steady state |
p-Cresol concentration (μg/ml)a
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Omnivore donor 1 |
Omnivore donor 2 |
Omnivore donor 3 |
|||||||
| v1 | v2 | v3 | v1 | v2 | v3 | v1 | v2 | v3 | |
| Baseline | 0 ± 0 | 45.59 ± 26.92 | 73.13 ± 28.65 | 0 ± 0 | 23.77 ± 3.12 | 46.29 ± 1.38 | 0 ± 0 | 0 ± 0 | 5.79 ± 1.24 |
| High protein | 0 ± 0 | 98.01 ± 36.86 | 153.56 ± 39.77 | 88.57 ± 36.76 | 162.26 ± 16.71 | 207.42 ± 27.61 | 0 ± 0 | 7.47 ± 7.45 | 7.85 ± 7.48 |
| High protein + 10 g Synergy1 | 0 ± 0 | 150.19 ± 16.65 | 244.35 ± 25.00 | 17.29 ± 14.39 | 284.24 ± 19.27 | 549.94 ± 141.86 | 0 ± 0 | 3.58 ± 3.15 | 15.26 ± 4.11 |
| High protein + 15 g Synergy1 | 1.67 ± 1.45 | 43.11 ± 16.44 | 113.77 ± 63.18 | 10.32 ± 1.95 | 227.45 ± 32.60 | 353.66 ± 17.33 | 4.60 ± 5.81 | 16.53 ± 0.52 | 36.41 ± 16.71 |
Values are means over 3 consecutive days from each donor ± standard deviation.
TABLE 2.
p-Cresol concentration in vegetarian donor samples with different steady states of a human in vitro colonic model in three different vessels as analyzed by GC
| Steady state |
p-Cresol concentration (μg/ml)a
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Vegetarian donor 1 |
Vegetarian donor 2 |
Vegetarian donor 3 |
|||||||
| v1 | v2 | v3 | v1 | v2 | v3 | v1 | v2 | v3 | |
| Baseline | 0 ± 0 | 27.22 ± 3.05 | 44.92 ± 7.64 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 10.83 ± 0.84 | 13.62 ± 2.64 | 18.55 ± 7.20 |
| High protein | 0 ± 0 | 119.52 ± 18.33 | 194.43 ± 10.09 | 0 ± 0 | 0 ± 0 | 7.15 ± 0.73 | 5.19 ± 0.77 | 16.66 ± 11.56 | 146.78 ± 52.17 |
| High protein + 10 g Synergy1 | 0 ± 0 | 131.04 ± 46.17 | 138.24 ± 14.74 | 0 ± 0 | 0 ± 0 | 27.49 ± 9.32 | 2.55 ± 0.28 | 6.20 ± 1.78 | 36.98 ± 5.97 |
| High protein + 15 g Synergy1 | 0 ± 0 | 87.75 ± 36.79 | 106.11 ± 48.15 | 0 ± 0 | 0 ± 0 | 24.66 ± 6.62 | 0 ± 0 | 6.59 ± 5.41 | 15.63 ± 14.94 |
Values are means over 3 consecutive days from each donor ± standard deviation.
Ammonia.
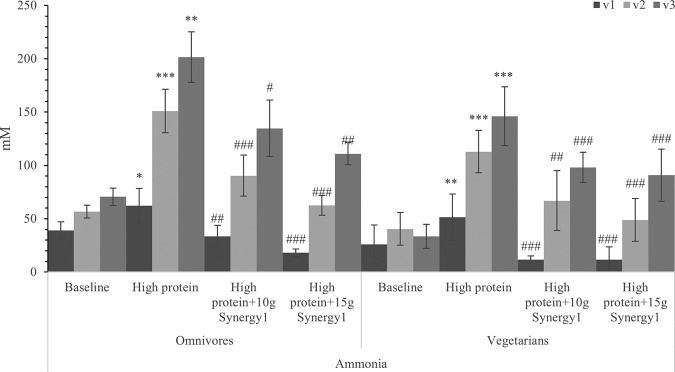
Ammonia is a major metabolite of the fermentation of protein by fecal bacteria as a result of deamination of amino acids. Addition of protein to the colonic model significantly increased the production of ammonia in all three regions, especially in the transverse and distal vessels (Fig. 7). The higher prebiotic supplementation dose suppressed ammonia production to a greater degree than the 10 g of prebiotic addition (further reduced ammonia from 67% less than high-protein treatment to 55% for omnivore models and 62% for vegetarian models after giving 15 g prebiotics per day), with significant differences in all three regions of all six models.
FIG 7.
Ammonia concentration in samples with different steady states of a human in vitro colonic model in three different vessels. Values are means over 3 consecutive days from 3 omnivores’ microbiota or 3 vegetarians’ microbiota ± standard deviation. Significant differences in each vessel among four steady states are labeled. *, mean values were significantly different from the baseline steady state (P < 0.05). **, mean values were significantly different from the baseline steady state (P < 0.01). ***, mean values were significantly different from the baseline steady state (P < 0.001). #, mean values were significantly different from the high-protein steady state (P < 0.05). ##, mean values were significantly different from the high-protein steady state (P < 0.01). ###, mean values were significantly different from the high-protein steady state (P < 0.001).
DISCUSSION
Three subjects following omnivore diets and three following vegetarian diets were recruited to donate their fecal samples for inoculation of the colonic models. Among these six donors, some different patterns of microbiota were observed. At the genus level, fecal bacterial composition differences between omnivores and vegetarians were not clearly seen; however, metabolic activities of the microbiota may differ. After feeding standardized medium until the fermentation equilibrated, total bacterial counts were lower in vegetarian models than those in omnivore models, and this difference persisted in all steady states. A possible explanation for in vitro culture differences is that this three-stage continuous culture system has been previously validated with six sudden death subjects who were probably not vegetarians (25). Therefore, carbohydrate and protein content in vegetarians’ digesta can be different from the validated medium used by Macfarlane et al. (25), resulting in differing bacterial growth in the gut model. In the present study, some bacterial groups from omnivore- and vegetarian-inoculated models displayed different responses on protein and prebiotic addition indicating that, at the genus level at least, bacteria have different metabolic activities. With various food sources entering the digestive tract, bacteria from different individuals adapt and may display different metabolic activities (29).
According to Gibson et al. (30), sulfate-reducing bacteria (SRB) are not always present in human populations because some individuals possess methanogens to dispose of hydrogen in the gut instead, which may explain the high variation of SRB count in this study. Hydrogen sulfide possibly contributes to the pathogenesis of ulcerative colitis, potentially offering a role for prebiotics in management of this disease (31). In this study, SRB counts were slightly higher in the proximal region, which is the opposite of that described in another three-stage continuous fermentation study looking at mucin metabolism, sulfate reduction, and methanogenesis (32). By analyzing colonic contents from three sudden death subjects, it was seen in subjects without methanogenic activity that sulfate reduction rates decreased gradually from the proximal to the distal colon (4).
Inulin-type fructans can promote bifidobacterial growth and be utilized by bifidobacteria to produce acetate and lactate. These organic acids can be further metabolized by other bacteria, such as butyrate-producing clostridia, which might be why Clostridium coccoides-Eubacterium rectale group and butyrate increased after the prebiotic intervention (22).
We saw interindividual differences in both batch culture and gut model work. Some study groups are in favor of mixing volunteers’ fecal samples to diminish the variance; however, these interindividual differences would not be seen in this case. Each individual has a distinct microbial community with distinct function. Mixing multiple ecosystems does not result in a natural ecosystem, and it does not represent any individual.
p-Cresol had a similar pattern to SRB as follows: concentrations were highly variable in different models. Out of six donors, four had gut bacteria producing high levels of p-cresol with the simulated high-protein diet (two vegetarians and two omnivores). In vegetarian models with high p-cresol levels, p-cresol concentration reduced with both doses of prebiotic supplementation. However, in omnivore models with high p-cresol content, its production was promoted by 10 g of prebiotic addition and only started to decrease with 15 g of prebiotics. Vegetarians are more likely to have a high-fiber diet compared to omnivores according to diet survey studies, and possibly their microbes are better adapted to complex carbohydrates (33). Therefore, after a high-protein diet challenge, it may be easier for bacteria from vegetarian donors to return to utilizing carbohydrates.
The current study confirmed that the main proteolytic regions in the colon are the transverse and distal regions (3, 4). Both BCFA and ammonia were detected at the highest concentration in the distal colon simulation. This can result from accumulation of metabolites due to a lack of absorption in this in vitro continuous culture system. When comparing concentration differences, the transverse colon had the highest production of BCFA and ammonia. The simulated transverse vessel has a pH of 6.2, which is close to the optimum pH of proteolytic enzymes (4).
One study on rats revealed that protein dose was correlated with negative consequences of colonic DNA damage, but DNA damage was not seen with whey protein (34). In a human intervention study with 3 g or 10 g of prebiotic galactooligosaccharides (GOS) fed to 5 healthy subjects with a sequential design, proteolysis inhibitory effects of GOS were seen; however, there was no difference between 3 g and 10 g of GOS per day (35). This may due to huge variation among the volunteers, small sample size (n = 5), and the short time of intervention (1 week). A better proteolysis inhibitory effect of higher dose prebiotics was seen in this in vitro study, which may result from elimination of variation by standardized medium flow.
Vegetarian models had lower production of ammonia and p-cresol than omnivore models. Potentially, a vegetarian diet has more complex carbohydrates, and vegetarians’ gut bacteria had adapted to a relatively low-protein diet. Therefore, it is likely that there was less potential for proteolytic metabolism within the microbiota. In the previous batch culture study, we also showed that nonanimal-based protein (soy protein and Quorn protein) produce less phenol and indole (24). In terms of gut health, this may indicate that a vegetarian diet is beneficial if consuming a high-protein diet. As a vegetarian diet is usually accompanied by a considerable amount of dietary fiber, this would further support that vegetarians are less likely to get high proteolysis product accumulation in the gut.
In the batch distal colon simulation setup, without continuous feeding, we also observed increased bifidobacterial growth, less BCFA, and ammonia production while adding prebiotics in a high-protein diet (24). We observed lower BCFA production in models with vegetarian donors’ samples in batch fermentation. In our continuous models, with addition of prebiotics, BCFA in the proximal colon in the vegetarian models (0.90 ± 0.62) were less than those in omnivore models (4.70 ± 5.63) (P = 0.01). This showed that even under 2 weeks high-protein selection pressure, communities from vegetarian donors did not evolve to express high proteolytic activities. Production of p-cresol by prebiotics was inhibited in high p-cresol producers from both omnivore and vegetarian donors in batch fermentation; however, in continuous fermentation, its production was inhibited only in vegetarian models. Under carbohydrate selection pressure after communities adapted to high-protein feeding, microbes from vegetarians are still able to evolve to lower p-cresol production. According to host-diet-driven BCFA and p-cresol differences, adaptation to protein source in the omnivore models is more likely to happen, whereas adaptation to carbohydrate source in the vegetarian models is more likely. These findings in different models might have practical implications as follows: different results could be obtained when translating in vitro work to in vivo experiment if there is a different time scale and human clinical trials often find responders and nonresponders when using diet to modulate the gut microbiome.
In conclusion, supplementation of prebiotic Synergy1 to three-stage, continuous, pH-controlled, colonic fermentation models shifted the microbiota to a more favorable profile by stimulating bifidobacteria and lactobacilli while repressing Desulfovibrio spp. The prebiotic addition also significantly decreased the concentration of protein metabolites (ammonia and BCFA); however, inhibitory effects of Synergy1 on p-cresol production was only seen in vegetarian high producing models. Our study also implies that host diet consumption plays an important role in response to prebiotic interventions.
MATERIALS AND METHODS
Proteins.
Protein substrates used were casein hydrolysates (Sigma-Aldrich, Poole, UK).
Prebiotic.
Inulin-type fructan was a mixture of oligofructose and inulin as follows: 50% ± 10% degree of polymerization (DP) of 3 to 9 and 50% ± 10% DP ≥ 10 (Orafti Synergy1; BENEO-Orafti, Tienen, Belgium).
Protein and prebiotic dose determination.
A miniature gut model system, 33% of the size of the version described by Macfarlane et al. (25), was used in this study. Prebiotic dosage was 33% of what would be used in human adults. Approximately 16 g protein will reach the colon following ingestion of 105 g protein/day of which 8 g are endogenous and 8 g are exogenous (36, 37). Steady state 1, which was the baseline, was just with standardized medium flow. The in vitro dose required to simulate prebiotic and protein effects in vitro was calculated as follows (Table 3).
TABLE 3.
Protein dosage calculation for continuous gut model fermentation
| Steady state | Protein dosage (g/day) |
|
|---|---|---|
| In vivo | In vitro fermentation | |
| 2 | 8 (dietary protein) | 2.67 |
| 3 | 8 (dietary protein) | 2.67 |
| 10 (prebiotic) | 3.33 | |
| 4 | 8 (dietary protein) | 2.67 |
| 15 (prebiotic) | 5 | |
In vitro gut model fermentation. (i) Fecal sample preparation.
Ethical approval of collecting fecal samples from healthy volunteers was obtained from the University of Reading Research Ethics Committee in 2014. Fecal samples were obtained from three healthy meat-eating individuals and three healthy vegetarian volunteers between the ages of 18 and 60 (vegetarians, 26.67 ± 5.69 years old; omnivores, 28.00 ± 4.36), who had not taken antibiotics for at least 6 months prior to the experiment and had no history of gastrointestinal disorders. All volunteers were following their diet for at least 5 years.
Fecal samples were diluted 1 in 20 (wt/vol) using 1 M phosphate-buffered saline (PBS) (Oxoid, Hampshire, UK), pH 7.4. This suspension was homogenized in a stomacher (Stomacher 80 Biomaster; Seward) for 120 s at normal speed.
(ii) Gut model medium.
Gut model medium was prepared with chemicals obtained from Sigma-Aldrich (Poole, UK) unless otherwise stated. In 1 liter, 5 g starch, 5 g peptone water, 5 g tryptone, 4.5 g yeast extract (Oxoid, Hampshire, UK), 4.5 g NaCl, 4.5 g KCl, 4 g mucin (porcine gastric type III), 3 g casein, 2 g pectin (citrus), 2 g xylan from beech wood pure (Serva, Heidelberg, Germany), 2 g arabinogalactan (larch wood), 1.5 g NaHCO3 (Fisher Scientific, Loughborough, UK), 1.25 g MgSO4·7H2O (Fisher Scientific, Loughborough, UK), 1 g guar gum, 1 g inulin (BENEO-Orafti, Tienen, Belgium), 0.8 g l-cystine HCl, 0.5 g KH2PO4, 0.5 g K2HPO4, 0.4 g bile salts number 3, 0.15 g CaCl2·6H2O, 0.005 g FeSO4·7H2O, 0.05 g hemin, 10 μl vitamin K, 1 ml Tween 80, and 4 ml resazurin (0.025 g/100 ml, pH 7).
(iii) Three-stage continuous pH-controlled, gut model fermentation.
A three-stage continuous fermentation culture system was used to simulate luminal conditions in each of the three distinct regions of the human colon, the proximal, transverse, and distal colon (V1, V2, and V3) (25). Vessels with operating volumes of 80 ml, 100 ml, and 120 ml were set up in sequence. Autoclaved culture medium (51.43 ml [V1], 66.67 ml [V2], 82.5 ml [V3]) was aseptically poured into sterile vessels. This system was left overnight with oxygen-free nitrogen pumping through the medium at a rate of 15 ml/min. Each vessel was temperature controlled at 37°C and stirred using a magnetic stirrer. Fecal slurry at 20% (wt/vol) was inoculated into the culture vessels (28.57 ml [V1], 33.33 ml [V2], 37.5 ml [V3]) and left to equilibrate for 24 h as a batch culture system prior to commencing the continuous medium flow. Control of pH was achieved by pH meters (FerMac 260 pH controller; Electrolab, Tewksbury, UK) connected to each vessel to regulate pH at 5.4 to 5.6 (V1), 6.1 to 6.3 (V2), and 6.7 to 6.9 (V3) with the aid of 0.5 M HCl and NaOH. Oxygen-free nitrogen flow and pH were maintained throughout the whole experiment.
After 8 turnovers (16 days) of the operating volume (300 ml in total) at a medium flow rate of 6.25 ml/h, SCFA were analyzed for three consecutive days to confirm the establishment of steady state. Samples were taken for three consecutive days after confirmation of the equilibrium for analysis of bacterial populations and metabolite concentrations.
Enumeration of fecal microbial populations by flow cytometry fluorescence in situ hybridization.
A 750-μl sample of batch culture fluid was centrifuged at 11,337 × g for 5 min and the supernatant discarded. The pellet was then suspended in 375 μl filtered 0.1 M PBS solution. Filtered cold (4°C) 4% paraformaldehyde (PFA) (1,125 μl) was added and samples stored at 4°C for 4 h. These were then washed thoroughly with PBS to remove PFA and resuspended in a mixture containing 300 μl PBS and 300 μl 99% ethanol. Samples were then stored at −20°C prior to fluorescence in situ hybridization (FISH) analysis by flow cytometry. Filtered cold (4°C) 0.1 M PBS (500 μl) was mixed with fixed samples (75 μl) before being centrifuged at 11,337 × g for 3 min. The pellets were then resuspended in 100 μl of TE-FISH (Tris-HCl 1 M, pH 8; EDTA 0.5 M, pH 8; and filtered distilled water with the ratio of 1:1:8) containing lysozyme solution (1 mg/ml of 50,000 U/mg protein). Samples were then incubated in the dark at room temperature for 10 min and then centrifuged at 11,337 × g for 3 min. Pellets were washed with 500 μl filtered cold PBS and then washed with 150 μl hybridization buffer (5 M NaCl, 1 M Tris-HCl [pH 8], formamide, double-distilled H2O, and 10% SDS in the ratio 180:20:300:499:1) and centrifuged at 11,337 × g for 3 min. Pellets were then resuspended in 1 ml of hybridization buffer. Aliquots (50 μl) with 4 μl of different probes (50 ng μl−1) were incubated at 35°C for at least 10 h. The probes used in this study are listed in Table 4. Non-EUB and EUB338-I-II-III (equal mix of EUB338 I, II, and III) are attached with fluorescence Alexa 488 at the 5′ end, and other specific probes are attached with Alexa 647. A set of non-EUB and EUB338-I-II-III (equal mix of EUB338 I, II, and III) is attached with fluorescence Alexa 647 at the 5′ end to be the controls. For samples to detect specific groups, 4 μl of EUB338-I-II-III was added together with 4 μl specific probes. Hybridization buffer (150 μl) was added to each aliquot after incubation, followed by 3 min centrifugation at 11,337 × g. Supernatants (150 μl) were carefully removed before samples were centrifuged at 11,337 × g for 3 min. Remaining supernatant was then removed, and pellets were resuspended in 200 μl washing buffer. Washing buffer was prepared as 12.8 μl of 5 M NaCl, 20 μl of 1 M Tris-HCl (pH 8), 10 μl of 0.5 M EDTA (pH 8), and 1 μl of 10% SDS in 956.2 μl of filtered cold distilled water. Samples were then incubated at 37°C for 20 min and centrifuged at 11,337 × g for 3 min. After supernatant removal, pellets were resuspended in different volume of filtered cold PBS based on flow cytometry load. Bacterial counts were then calculated through consideration of flow cytometry reading and PBS dilution.
TABLE 4.
Name, sequence, and target group of oligonucleotide probes used in this study for bacterial enumeration by fluorescent in situ hybridization
| Probe name | Sequence (5′ to 3′) | Target group(s) | Reference |
|---|---|---|---|
| Non Eub | ACTCCTACGGGAGGCAGC | Control probe complementary to EUB338 | 39 |
| Eub338I | GCTGCCTCCCGTAGGAGT | Most bacteria | 40 |
| Eub338II | GCAGCCACCCGTAGGTGT | Planctomycetales | 41 |
| Eub338III | GCTGCCACCCGTAGGTGT | Verrucomicrobiales | 41 |
| Bif164 | CATCCGGCATTACCACCC | Bifidobacterium spp. | 42 |
| Lab158 | GGTATTAGCAYCTGTTTCCA | Lactobacillus and Enterococcus | 43 |
| Bac303 | CCAATGTGGGGGACCTT | Most Bacteroidaceae and Prevotellaceae, some Porphyromonadaceae | 44 |
| Erec482 | GCTTCTTAGTCARGTACCG | Most of the Clostridium coccoides-Eubacterium rectale group (Clostridium subclusters XIVa and XIVb) | 45 |
| Rrec584 | TCAGACTTGCCGYACCGC | Roseburia genus | 46 |
| Ato291 | GGTCGGTCTCTCAACCC | Atopobium cluster | 47 |
| Prop853 | ATTGCGTTAACTCCGGCAC | Clostridium cluster IX | 46 |
| Fprau655 | CGCCTACCTCTGCACTAC | Faecalibacterium prausnitzii and relatives | 48 |
| DSV687 | TACGGATTTCACTCCT | Desulfovibrio genus | 49 |
| Chis150 | TTATGCGGTATTAATCTYCCTTT | Most of the Clostridium histolyticum group (Clostridium clusters I and II) | 45 |
| EC 1531 | CACCGTAGTGCCTCGTCATCA | Escherichia coli BJ4 | 50 |
Short chain fatty acid and volatile organic compounds analysis by gas chromatography.
Samples (1 ml) were collected and extractions performed with some modifications of a method from Richardson et al. (38). The sample (1 ml) was transferred into a 100-mm by 16-mm glass tube (International Scientific Supplies Ltd, Bradford, England) with 50 μl of 2-ethylbutyric acid (0.1 M internal standard) (Sigma, Poole, UK). Concentrated HCl (500 μl) and 1 ml diethyl ether were added to each glass tube and samples vortexed for 1 min. Samples were centrifuged at 2,000 × g for 10 min. The diethyl ether (upper) layer of each sample was transferred to a clean glass tube. Ether extract (400 μl) and 50 μl N-(tert-butyldimethylsilyl)-N-methyltrifluoroacetamide (MTBSTFA; Sigma-Aldrich, Poole, UK) were added into a gas chromatography (GC) screw-cap vial. Samples were left at room temperature for 72 h to allow lactic acid in the samples to completely derivatize.
An Agilent/HP 6890 gas chromatograph (Hewlett Packard, UK) using an HP-5MS 30-m by 0.25-mm column with a 0.25-μm coating of cross-linked (5%-phenyl)-methylpolysiloxane (Hewlett Packard, UK) was used for analysis of SCFA. Temperatures of injector and detector were 275°C, with the column temperature programmed from 63°C to 190°C at 15°C min−1 followed by 190°C for 3 min. Helium was the carrier gas (flow rate, 1.7 ml min−1; head pressure, 133 KPa). A split ratio of 100:1 was used. Quantification of samples was achieved by calibration with lactic, acetic, formic, propionic, butyric, and valeric acids and branched SCFA (isobutyric and isovaleric) in concentrations between 12.5 and 100 mM.
Ammonia analysis.
Samples were diluted 1 in 50 vol/vol prior to analysis. Ammonia concentrations in diluted fermentation samples were analyzed by using a Sigma-Aldrich (Poole, U.K.) FluoroSelect ammonia kit. Reagent was prepared by combining 100 μl assay buffer, 4 μl reagent A, and 4 μl reagent B provided in the kit. H2O (10 μl) served as a blank, and 10 μl of each sample was added to a glass vial. Reagent (100 μl) was then added to each tube. Samples were kept in the dark for 15 min at room temperature before they were read in a fluorimeter. Ammonia standards were prepared by diluting 20 mmol/liter NH4Cl into distilled water, and the concentration range was 0.25 to 1 mmol/liter.
Statistical analysis.
All statistical tests were performed with the use of IBM SPSS Statistics version 24 (IBM Corp., USA). Results are presented as means ± standard deviation (SD). Changes in specific bacterial groups, organic acids, volatile organic compounds, and ammonia were assessed among different treatments/steady states using one-way analysis of variance (ANOVA). Significant differences were assessed by post hoc Tukey honestly significant difference (HSD) test among the four steady states.
ACKNOWLEDGMENTS
We acknowledge partial financial support from BENEO.
We thank technicians from the Food and Nutritional Sciences department at the University of Reading for their role in supporting this study.
REFERENCES
- 1.FAO. 2017. Food balance/food balance sheets. FAO, Rome, Italy: http://faostat3.fao.org/browse/FB/FBS/E. Accessed 15 December 2017. [Google Scholar]
- 2.Food Standards Agency. 2016. National diet and nutrition survey results from years 5 and 6 (combined) of the rolling programme (2012/2013–2013/2014). Food Standards Agency, London, England. [Google Scholar]
- 3.Macfarlane GT, Allison C, Gibson GR. 1988. Effect of pH on protease activities in the large-intestine. Lett Appl Microbiol 7:161–164. doi: 10.1111/j.1472-765X.1988.tb01269.x. [DOI] [Google Scholar]
- 4.Macfarlane GT, Gibson GR, Cummings JH. 1992. Comparison of fermentation reactions in different regions of the human colon. J Appl Bacteriol 72:57–64. doi: 10.1111/j.1365-2672.1992.tb04882.x. [DOI] [PubMed] [Google Scholar]
- 5.Smith EA, Macfarlane GT. 1997. Dissimilatory amino acid metabolism in human colonic bacteria. Anaerobe 3:327–337. doi: 10.1006/anae.1997.0121. [DOI] [PubMed] [Google Scholar]
- 6.Hughes R, Kurth MJ, McGilligan V, McGlynn H, Rowland I. 2008. Effect of colonic bacterial metabolites on Caco-2 cell paracellular permeability in vitro. Nutr Cancer 60:259–266. doi: 10.1080/01635580701649644. [DOI] [PubMed] [Google Scholar]
- 7.Clinton SK, Bostwick DG, Olson LM, Mangian HJ, Visek WJ. 1988. Effects of ammonium acetate and sodium cholate on N-methyl-N’-nitro-N-nitrosoguanidine-induced colon carcinogenesis of rats. Cancer Res 48:3035–3039. [PubMed] [Google Scholar]
- 8.Lin HC, Visek WJ. 1991. Colon mucosal cell-damage by ammonia in rats. J Nutr 121:887–893. doi: 10.1093/jn/121.6.887. [DOI] [PubMed] [Google Scholar]
- 9.Roediger WEW, Duncan A, Kapaniris O, Millard S. 1993. Sulfide impairment of substrate oxidation in rat colonocytes: a biochemical basis for ulcerative-colitis. Clin Sci (Lond) 85:623–627. doi: 10.1042/cs0850623. [DOI] [PubMed] [Google Scholar]
- 10.Leschelle X, Goubern M, Andriamihaja M, Blottiere HM, Couplan E, Gonzalez-Barroso MD, Petit C, Pagniez A, Chaumontet C, Mignotte B, Bouillaud F, Blachier F. 2005. Adaptative metabolic response of human colonic epithelial cells to the adverse effects of the luminal compound sulfide. Biochim Biophys Acta 1725:201–212. doi: 10.1016/j.bbagen.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 11.Gibson GR, Cummings JH, Macfarlane GT. 1991. Growth and activities of sulfate-reducing bacteria in gut contents of healthy subjects and patients with ulcerative colitis. FEMS Microbiol Ecol 86:103–111. doi: 10.1111/j.1574-6968.1991.tb04799.x. [DOI] [Google Scholar]
- 12.Cerini C, Dou L, Anfosso F, Sabatier F, Moal S, Glorieux G, De Smet R, Vanholder R, Dignat-George F, Sampol J, Berland Y, Brunet P. 2004. p-Cresol, a uremic retention solute, alters the endothelial barrier function in vitro. Thromb Haemost 92:140–150. doi: 10.1160/TH03-07-0491. [DOI] [PubMed] [Google Scholar]
- 13.McCall IC, Betanzos A, Weber DA, Nava P, Miller GW, Parkos CA. 2009. Effects of phenol on barrier function of a human intestinal epithelial cell line correlate with altered tight junction protein localization. Toxicol Appl Pharmacol 241:61–70. doi: 10.1016/j.taap.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barreto FC, European Uremic Toxin Work Group (EUTox), Barreto DV, Liabeuf S, Meert N, Glorieux G, Temmar M, Choukroun G, Vanholder R, Massy ZA. 2009. Serum indoxyl sulfate is associated with vascular disease and mortality in chronic kidney disease patients. Clin J Am Soc Nephrol 4:1551–1558. doi: 10.2215/CJN.03980609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vanholder R, Schepers E, Pletinck A, Nagler EV, Glorieux G. 2014. The uremic toxicity of indoxyl sulfate and p-cresyl sulfate: a systematic review. J Am Soc Nephrol 25:1897–1907. doi: 10.1681/ASN.2013101062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gibson GR, Hutkins R, Sanders ME, Prescott SL, Reimer RA, Salminen SJ, Scott K, Stanton C, Swanson KS, Cani PD, Verbeke K, Reid G. 2017. Expert consensus document: the International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat Rev Gastroenterol Hepatol 14:491–502. doi: 10.1038/nrgastro.2017.75. [DOI] [PubMed] [Google Scholar]
- 17.Vandeputte D, Falony G, Vieira-Silva S, Wang J, Sailer M, Theis S, Verbeke K, Raes J. 2017. Prebiotic inulin-type fructans induce specific changes in the human gut microbiota. Gut 66:1968–1974. doi: 10.1136/gutjnl-2016-313271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holloway L, Moynihan S, Abrams SA, Kent K, Hsu AR, Friedlander AL. 2007. Effects of oligofructose-enriched inulin on intestinal absorption of calcium and magnesium and bone turnover markers in postmenopausal women. Br J Nutr 97:365–372. doi: 10.1017/S000711450733674X. [DOI] [PubMed] [Google Scholar]
- 19.Russo F, Clemente C, Linsalata M, Chiloiro M, Orlando A, Marconi E, Chimienti G, Riezzo G. 2011. Effects of a diet with inulin-enriched pasta on gut peptides and gastric emptying rates in healthy young volunteers. Eur J Nutr 50:271–277. doi: 10.1007/s00394-010-0135-6. [DOI] [PubMed] [Google Scholar]
- 20.Jackson KG, Taylor GR, Clohessy AM, Williams CM. 1999. The effect of the daily intake of inulin on fasting lipid, insulin and glucose concentrations in middle-aged men and women. Br J Nutr 82:23–30. doi: 10.1017/s0007114599001087. [DOI] [PubMed] [Google Scholar]
- 21.Riviere A, Gagnon M, Weckx S, Roy D, De Vuyst L. 2015. Mutual cross-feeding interactions between Bifidobacterium longum subsp. longum NCC2705 and Eubacterium rectale ATCC 33656 explain the bifidogenic and butyrogenic effects of arabinoxylan oligosaccharides. Appl Environ Microbiol 81:7767–7781. doi: 10.1128/AEM.02089-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moens F, Verce M, De Vuyst L. 2017. Lactate- and acetate-based cross-feeding interactions between selected strains of lactobacilli, bifidobacteria and colon bacteria in the presence of inulin-type fructans. Int J Food Microbiol 241:225–236. doi: 10.1016/j.ijfoodmicro.2016.10.019. [DOI] [PubMed] [Google Scholar]
- 23.Hague A, Elder DJE, Hicks DJ, Paraskeva C. 1995. Apoptosis in colorectal tumor-cells: induction by the short-chain fatty-acids butyrate, propionate and acetate and by the bile-salt deoxycholate. Int J Cancer 60:400–406. doi: 10.1002/ijc.2910600322. [DOI] [PubMed] [Google Scholar]
- 24.Wang X, Gibson GR, Costabile A, Sailer M, Theis S, Rastall RA. 2019. Prebiotic supplementation of in vitro fecal fermentations inhibits proteolysis by gut bacteria and host diet shapes gut bacterial metabolism and response to intervention. Appl Environ Microbiol 85:e02749-18. doi: 10.1128/AEM.02749-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macfarlane GT, Macfarlane S, Gibson GR. 1998. Validation of a three-stage compound continuous culture system for investigating the effect of retention time on the ecology and metabolism of bacteria in the human colon. Microb Ecol 35:180–187. doi: 10.1007/s002489900072. [DOI] [PubMed] [Google Scholar]
- 26.Macfarlane GT, Cummings JH, Allison C. 1986. Protein degradation by human intestinal bacteria. J Gen Microbiol 132:1647–1656. doi: 10.1099/00221287-132-6-1647. [DOI] [PubMed] [Google Scholar]
- 27.Gibson GR, Cummings JH, Macfarlane GT, Allison C, Segal I, Vorster HH, Walker A. 1990. Alternative pathways for hydrogen disposal during fermentation in the human colon. Gut 31:679–683. doi: 10.1136/gut.31.6.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macfarlane GT, Gibson GR, Beatty E, Cummings JH. 1992. Estimation of short-chain fatty acid production from protein by human intestinal bacteria based on branched-chain fatty acid measurements. FEMS Microbiol Lett 101:81–88. [Google Scholar]
- 29.Giraud A, Matic I, Tenaillon O, Clara A, Radman M, Fons M, Taddei F. 2001. Costs and benefits of high mutation rates: adaptive evolution of bacteria in the mouse gut. Science 291:2606–2608. doi: 10.1126/science.1056421. [DOI] [PubMed] [Google Scholar]
- 30.Gibson GR, Macfarlane GT, Cummings JH. 1988. Occurrence of sulphate-reducing bacteria in human faeces and the relationship of dissimilatory sulphate reduction to methanogenesis in the large gut. J Appl Bacteriol 65:103–111. doi: 10.1111/j.1365-2672.1988.tb01498.x. [DOI] [PubMed] [Google Scholar]
- 31.Roediger WEW, Moore J, Babidge W. 1997. Colonic sulfide in pathogenesis and treatment of ulcerative colitis. Dig Dis Sci 42:1571–1579. doi: 10.1023/A:1018851723920. [DOI] [PubMed] [Google Scholar]
- 32.Gibson GR, Cummings JH, Macfarlane GT. 1988. Use of a three-stage continuous culture system to study the effect of mucin on dissimilatory sulfate reduction and methanogenesis by mixed populations of human gut bacteria. Appl Environ Microbiol 54:2750–2755. doi: 10.1128/AEM.54.11.2750-2755.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clarys P, Deliens T, Huybrechts I, Deriemaeker P, Vanaelst B, De Keyzer W, Hebbelinck M, Mullie P. 2014. Comparison of nutritional quality of the vegan, vegetarian, semi-vegetarian, pesco-vegetarian and omnivorous diet. Nutrients 6:1318–1332. doi: 10.3390/nu6031318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toden S, Bird AR, Topping DL, Conlon MA. 2007. Differential effects of dietary whey, casein and soya on colonic DNA damage and large bowel SCFA in rats fed diets low and high in resistant starch. Br J Nutr 97:535–543. doi: 10.1017/S0007114507336817. [DOI] [PubMed] [Google Scholar]
- 35.Tanaka R, Takayama H, Morotomi M, Kuroshima T, Ueyama S, Matsumoto K, Kuroda A, Mutai M. 1983. Effects of administration of TOS and Bifidobacterium breve 4006 on the human fecal flora. Bifidobacteria Microflora 2:17–24. doi: 10.12938/bifidus1982.2.1_17. [DOI] [Google Scholar]
- 36.Gibson JA, Sladen GE, Dawson AM. 1976. Protein absorption and ammonia production: the effects of dietary protein and removal of the colon. Br J Nutr 35:61–65. doi: 10.1079/bjn19760009. [DOI] [PubMed] [Google Scholar]
- 37.Moughan PJ, Butts CA, Rowan AM, Deglaire A. 2005. Dietary peptides increase endogenous amino acid losses from the gut in adults. Am J Clin Nutr 81:1359–1365. doi: 10.1093/ajcn/81.6.1359. [DOI] [PubMed] [Google Scholar]
- 38.Richardson AJ, Calder AG, Stewart CS, Smith A. 1989. Simultaneous determination of volatile and non-volatile acidic fermentation products of anaerobes by capillary gas-chromatography. Lett Appl Microbiol 9:5–8. doi: 10.1111/j.1472-765X.1989.tb00278.x. [DOI] [Google Scholar]
- 39.Wallner G, Amann R, Beisker W. 1993. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 14:136–143. doi: 10.1002/cyto.990140205. [DOI] [PubMed] [Google Scholar]
- 40.Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA. 1990. Combination of 16S ribosomal-RNA-targeted oligonucleotide probes with flow-cytometry for analyzing mixed microbial-populations. Appl Environ Microbiol 56:1919–1925. doi: 10.1128/AEM.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Daims H, Bruhl A, Amann R, Schleifer KH, Wagner M. 1999. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol 22:434–444. doi: 10.1016/S0723-2020(99)80053-8. [DOI] [PubMed] [Google Scholar]
- 42.Langendijk PS, Schut F, Jansen GJ, Raangs GC, Kamphuis GR, Wilkinson MHF, Welling GW. 1995. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl Environ Microbiol 61:3069–3075. doi: 10.1128/AEM.61.8.3069-3075.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harmsen HJM, Elfferich P, Schut F, Welling GW. 1999. A 16S rRNA-targeted probe for detection of lactobacilli and enterococci in faecal samples by fluorescent in situ hybridization. Microb Ecol Health Dis 11:3–12. [Google Scholar]
- 44.Manz W, Amann R, Ludwig W, Vancanneyt M, Schleifer KH. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum cytophaga-flavobacter-bacteroides in the natural environment. Microbiology 142:1097–1106. doi: 10.1099/13500872-142-5-1097. [DOI] [PubMed] [Google Scholar]
- 45.Franks AH, Harmsen HJ, Raangs GC, Jansen GJ, Schut F, Welling GW. 1998. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl Environ Microbiol 64:3336–3345. doi: 10.1128/AEM.64.9.3336-3345.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walker AW, Duncan SH, Leitch ECM, Child MW, Flint HJ. 2005. pH and peptide supply can radically alter bacterial populations and short-chain fatty acid ratios within microbial communities from the human colon. Appl Environ Microbiol 71:3692–3700. doi: 10.1128/AEM.71.7.3692-3700.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harmsen HJM, Wildeboer-Veloo ACM, Grijpstra J, Knol J, Degener JE, Welling GW. 2000. Development of 16S rRNA-based probes for the Coriobacterium group and the Atopobium cluster and their application for enumeration of Coriobacteriaceae in human feces from volunteers of different age groups. Appl Environ Microbiol 66:4523–4527. doi: 10.1128/aem.66.10.4523-4527.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suau A, Rochet V, Sghir A, Gramet G, Brewaeys S, Sutren M, Rigottier-Gois L, Doré J. 2001. Fusobacterium prausnitzii and related species represent a dominant group within the human fecal flora. Syst Appl Microbiol 24:139–145. doi: 10.1078/0723-2020-00015. [DOI] [PubMed] [Google Scholar]
- 49.Ramsing NB, Fossing H, Ferdelman TG, Andersen F, Thamdrup B. 1996. Distribution of bacterial populations in a stratified fjord (Mariager fjord, Denmark) quantified by in situ hybridization and related to chemical gradients in the water column. Appl Environ Microbiol 62:1391–1404. doi: 10.1128/AEM.62.4.1391-1404.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Poulsen LK, Lan FS, Kristensen CS, Hobolth P, Molin S, Krogfelt KA. 1994. Spatial distribution of Escherichia coli in the mouse large intestine inferred from ribosomal RNA in situ hybridization. Infect Immun 62:5191–5194. doi: 10.1128/IAI.62.11.5191-5194.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]