An investigator may be interested in the effect of an exposure such as a single nucleotide polymorphism (SNP) on the outcome of interest through a mediator or intermediate variable (i.e. the mediated or indirect path) as seen in Figure 1. Mediation analysis can be used in this scenario to assess the direct effect of the SNP on the outcome (i.e. the direct path) and the effect of the SNP on the outcome through the mediator (i.e. the indirect or mediated path). (VanderWeele et al., 2016) For example, mediation analysis has been used to determine the effect of SNPs on chromosome 15 [CHRNA5/3B4] on pulmonary function through cigarette smoking. (Lutz & Hokanson, 2014; Siedlinski et al., 2013)
Figure 1:
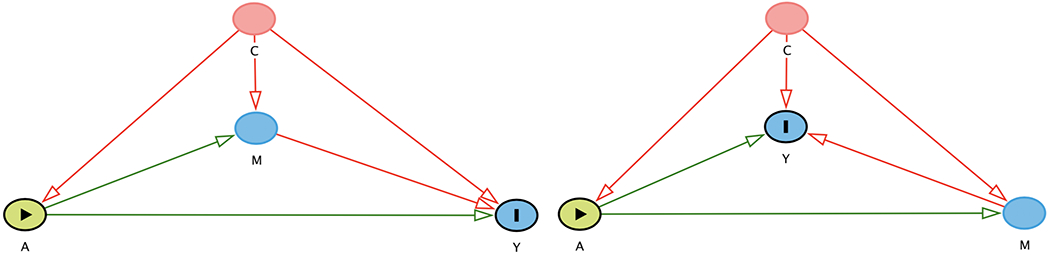
The Directed Acyclic Graph (DAG) on the left shows how the exposure A acts on the outcome Y both directly (i.e. A->Y) and indirectly through the mediator M (i.e. A->M->Y) given measured confounders C. When the outcome Y is treated as the mediator as seen in the figure on the right, then the outcome Y becomes a collider on the path from the exposure A to the mediator M. These figures were generated in DAGitty. (Textor et al., 2016)
However, in order to use standard mediation analysis methods, one needs to specify the directed acyclic graph (DAG) in Figure 1. Nevertheless, sometimes the investigator is not sure which variable is the outcome variable in this context of the analysis. For instance, if an investigator wants to use mediation analysis to determine the effect of a SNP on nicotine dependence and depression, it may not be clear which variable is the mediator and which is the outcome given the complex relationship between nicotine dependence and depression. (Boden et al., 2010; Manafo et al., 2010) The literature on the prospective association between smoking and depression is inconsistent in terms of the direction of association (i.e. whether smoking is the mediator or outcome). (Fluharty et al., 2016) In such scenarios, investigators may run the mediation analysis twice: 1) with nicotine dependence as the mediator and depression as the outcome, and 2) with nicotine dependence as the outcome and depression as the mediator.
It is not clear whether this analysis strategy is adequate to identify the mediator and the outcome variable. Through simulation studies, we examined the results of a counterfactual based mediation analysis when the role of the mediator and outcome were correctly specified and when they were reversed (i.e. reverse causality). (Imai et al., 2010a; Imai et al., 2010b; Tingley et al., 2014) For 1,000 subjects, we generated an exposure (i.e. SNP) from a binomial distribution with a minor allele frequency of 20%. We considered the following 3 scenarios.
In scenario 1, we generated an indirect effect of the exposure on the outcome through the mediator but no direct effect. We generated a normally distributed mediator with a genetic effect size of 0.2 and a variance of 1. The mediator had an effect size varying from 0.1, 0.2, to 0.3 on the normally distributed outcome, which also has a variance of 1. As seen in Figure 2a which displays the results of our simulation study based on 5,000 replicates, if the outcome and mediator are incorrectly specified, there is a significant direct effect but no significant indirect effect, which is the opposite of what was simulated. In practice, this would provide misleading analysis results in terms of the identification of the mediator and the outcome variable.
Figure 2:
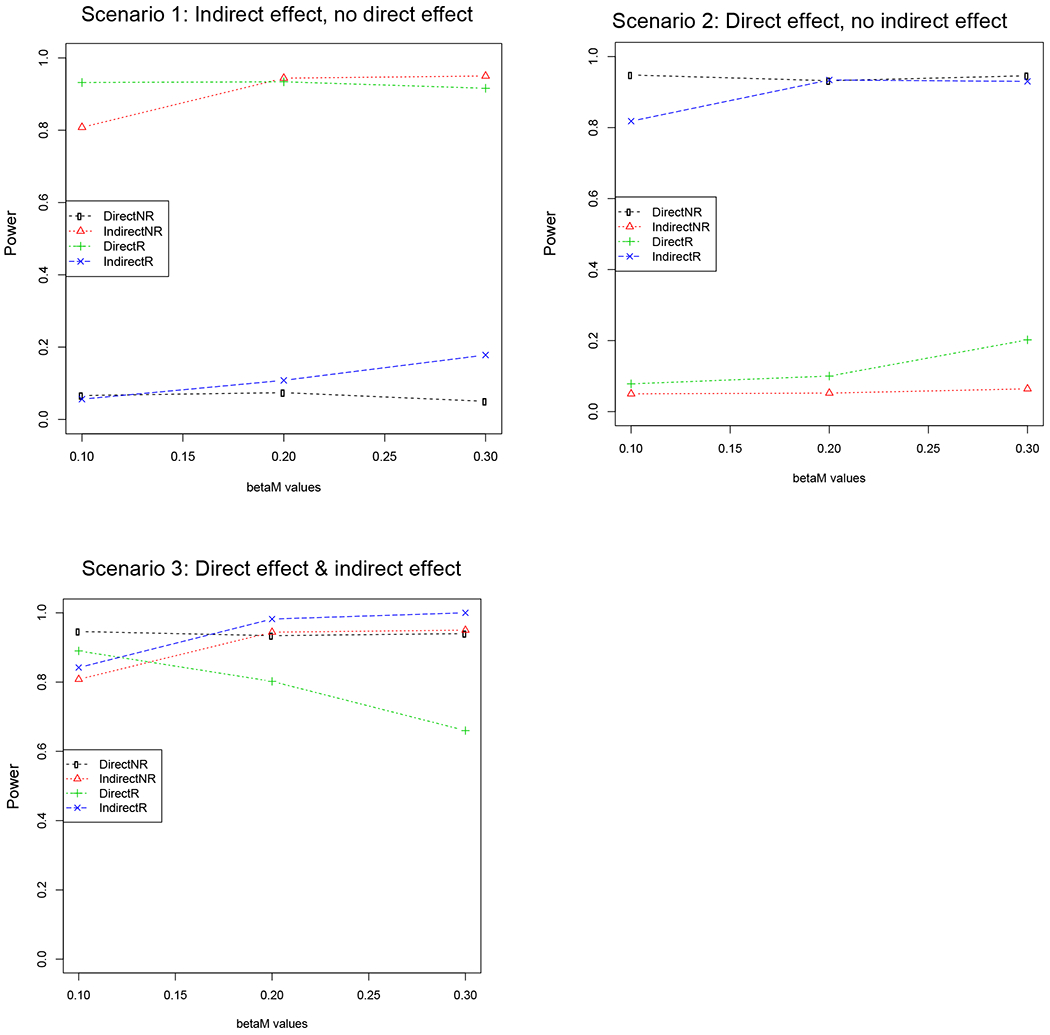
We considered 3 scenarios for 5,000 replicates. In the plots below, NR refers to the mediator and outcome being correctly specified and R refers to the mediator and outcome being incorrectly specified (i.e. reversed). The y axis shows the power and the x axis is the effect of the mediator on the outcome. In scenario 1, we generated an indirect effect of the exposure on the outcome through the mediator but no direct path. As seen below, if the outcome and mediator are incorrectly specified, there is a significant direct effect but no significant indirect effect. In scenario 2, we generated a direct effect of the exposure on the outcome but no indirect effect of the exposure on the outcome through the mediator. As seen below, if the outcome and mediator are incorrectly specified, there is a significant indirect effect but no significant direct effect. In scenario 3, we generated both a direct and indirect effect of the exposure on the outcome. As seen below, if the outcome and mediator are incorrectly specified, there is a significant indirect and direct effect and it is hard to distinguish the results when the mediator and outcome are correctly specified or not.
In scenario 2, we generated a direct effect of the exposure on the outcome but no indirect effect of the exposure on the outcome through the mediator. We generated a normally distributed mediator with a mean of 0 and a variance of 1 and the mediator had an effect size varying from 0.1, 0.2, to 0.3 on the normally distributed outcome. The normally distributed outcome is also a function of the exposure with a genetic effect size of 0.2 and a variance of 1. The results of our simulation study are shown Figure 2b. We observed that if the outcome and mediator are incorrectly specified, there is a significant indirect effect but no significant direct effect, which is the opposite of what was simulated. Again, as in the first simulation study, when the mediator and outcome are switched, the results of the mediation analysis are the opposite of when the mediator and outcome are correctly specified.
In scenario 3, we generated both a direct and indirect effect of the exposure on the outcome. We generated a normally distributed mediator with a genetic effect size of 0.2 and a variance of 1. The mediator had an effect size varying from 0.1, 0.2, to 0.3 on the normally distributed outcome. The normally distributed outcome is also a function of the exposure with a genetic effect size of 0.2 and a variance of 1. As seen in Figure 2c, if the outcome and mediator are incorrectly specified, there is both a significant indirect and direct effect of the exposure on the mediator, but it is hard to distinguish the results when the mediator and outcome are correctly specified or not.
To evaluate the approach based on real data, we applied this approach to the COPDGene, a study of current and former smokers. Among the 6,659 non-Hispanic White COPDGene subjects, we examined the effect of the SNP rs16969968 [CHRNA5] on chromosome 15q25 on cigarette smoking and forced expiratory volume in 1 second (FEV1) since this SNP and region have previously been associated with both lung function and smoking burden. (Lutz et al., 2015; Lutz et al., 2019) In this scenario, it is clear that the mediator is pack-years of cigarette smoking and the outcome is FEV1 given that smoking cigarettes decreases lung function. As seen in Table 1, when cigarette smoking is correctly set as the mediator, the estimate for the proportion mediated is 0.20. As seen in Table 1, when cigarette smoking is incorrectly set as the outcome, the estimate for the proportion mediated is 0.37. The estimate for the proportion mediated is higher when the mediator and outcome are incorrectly specified. This data analysis example illustrates that the results of the mediation analysis can be overestimated if the mediator and outcome are incorrectly specified.
Table 1:
Below are the estimates and p-values for the mediation analysis for the effect of rs16969968 on the outcome FEV1 through the mediator pack-years of cigarette smoking and then reversing the roles and specifying FEV1 as the mediator and pack-years of cigarette smoking as the outcome in the COPDGene study.
| Direct Effect | Indirect Effect | Proportion Mediated | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Mediator/Outcome | Estimate | p-value | 95% CI | Estimate | p-value | 95% CI | Estimate | p-value | 95% CI |
| Pack-Years/FEV1 | −0.08 | <2e-16 | (−0.11, −0.06) | −0.02 | <2e-16 | (−0.03, −0.01) | 0.20 | <2e-16 | (0.12, 0.28) |
| FEV1/Pack-Years | 1.47 | <2e-16 | (0.60, 2.33) | 0.86 | <2e-16 | (0.62, 1.13) | 0.37 | <2e-16 | (0.25, 0.59) |
While we only provide results for a limited set of scenarios, our results and conclusions seem generally representative, as we tested numerous other scenarios (data not shown). To facilitate examination of alternative scenarios, we have also created an R package on Github called reverseMA (https://github.com/SharonLutz/reverseMA) which runs simulations for user-specified scenarios in order to examine the results of this mediation analysis method if the role of the mediator and outcome are reversed.
In conclusion, we recommend against switching the role of the mediator and the outcome in mediation analysis if one is unsure of the DAG, as this can lead to incorrect conclusions. Standard mediation analysis approaches are not suitable to identify the directionality of the causal relationships, and collider bias may be introduced.
Acknowledgements:
We would like to thank Annie Thwing for her help with creating the reverseMA R package. The COPDGene study (NCT00608764) is also supported by the COPD Foundation through contributions made to an Industry Advisory Committee comprised of AstraZeneca, Boehringer-Ingelheim, GlaxoSmithKline, Novartis, and Sunovion. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Funding: Research reported in this publication was supported by the National Heart, Lung, And Blood Institute of the National Institutes of Health grants K01HL125858 (SML). This work was also supported by NHLBI U01HL089897 and U01HL089856 and NICHD R01HD085993 (AW) and the Cure Alzheimer’s Fund (CL).
Footnotes
Data Availability Statement: The COPDGene dataset used in this paper can be found on dbGaP http.www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000179.v1.p1
References:
- Boden JM, Fergusson DM and Horwood LJ (2010) Cigarette smoking and depression: tests of causal linkages using a longitudinal birth cohort. The British Journal of Psychiatry, 196, 440–446. 10.1192/bjp.bp.109.065912 [DOI] [PubMed] [Google Scholar]
- Fluharty M, Taylor AE Grabski M, Munafò MR.(2016) The Association of Cigarette Smoking With Depression and Anxiety: A Systematic Review. Nicotine & Tobacco Research, 3–13 10.1093/ntr/ntw140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai K, Keele L, & Tingley D (2010a). A general approach to causal mediation analysis. Psychological Methods, 15, 309–334. 10.1037/a0020761 [DOI] [PubMed] [Google Scholar]
- Imai K, Keele L, & Yamamoto T (2010b). Identification, Inference and Sensitivity Analysis for Causal Mediation Effects. Statistical Science, 25, 21 10.1214/10-STS321 [DOI] [Google Scholar]
- Lutz SM, & Hokanson JE (2014). Genetic influences on smoking and clinical disease. Understanding behavioral and biological pathways with mediation analysis. Annals of the American Thoracic Society, 11, 1082–1083. 10.1513/AnnalsATS.201407-315ED [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz SM, Cho MH, Young K, Hersh CP, Castaldi P, McDonald ML, …, ECLIPSE Investigators, and COPDGene Investigators. (2015) A Genome Wide Association Study Identified Risk Loci for Spirometric Measures among Smokers of European and African Ancestry. BMC Genet. 16:138 10.1186/s12863-015-0299-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz SM, Frederiksen B, Begum F, McDonald ML, Cho MH, Hobbs B, Parker MM, …, ECLIPSE and COPDGene Investigators. (2019) Common and Rare Variants Genetic Association Analysis of Cigarettes Per Day Among Ever Smokers in COPD Cases and Controls. NTR. 21(6):714–722. 10.1093/ntr/nty095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munafo MR and Araya R (2010) Editorial: Cigarette smoking and depression: a question of causation. The British Journal of Psychiatry, 196, 425–26. 10.1192/bjp.bp.109.074880 [DOI] [PubMed] [Google Scholar]
- Siedlinski M, Tingley D, Lipman PJ, Cho MH, Litonjua AA, Sparrow D, …, COPDGene and ECLIPSE Investigators. (2013). Dissecting direct and indirect genetic effects on chronic obstructive pulmonary disease (COPD) susceptibility. Human Genetics, 132, 431–441. 10.1007/s00439-012-1262-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Textor J, van der Zander B, Gilthorpe MS, Liskiewicz M, & Ellison GT (2016). Robust causal inference using directed acyclic graphs: The R package “dagitty.” International Journal of Epidemiology, 45, 1887–1894. 10.1093/ije/dyw341 [DOI] [PubMed] [Google Scholar]
- Tingley D, Yamamoto T, Hirose K, Keele L, & Imai K (2014). mediation: R Package for Causal Mediation Analysis. Journal of Statistical Software, 59, 38 10.18637/jss.v059.i05 [DOI] [Google Scholar]


